Abstract
The composition and fine structure of the vegetative cell wall peptidoglycan from Bacillus subtilis were determined by analysis of its constituent muropeptides. The structures of 39 muropeptides, representing 97% of the total peptidoglycan, were elucidated. About 99% analyzed muropeptides in B. subtilis vegetative cell peptidoglycan have the free carboxylic group of diaminopimelic acid amidated. Anhydromuropeptides and products missing a glucosamine at the nonreducing terminus account for 0.4 and 1.5%, respectively, of the total muropeptides. These two types of muropeptides are suggested to end glycan strands. An unexpected feature of B. subtilis muropeptides was the occurrence of a glycine residue in position 5 of the peptide side chain on monomers or oligomers, which account for 2.7% of the total muropeptides. This amount is, however, dependent on the composition of the growth media. Potential attachment sites for anionic polymers to peptidoglycan occur on dominant muropeptides and account for 2.1% of the total. B. subtilis peptidoglycan is incompletely digested by lysozyme due to de-N-acetylation of glucosamine, which occurs on 17.3% of muropeptides. The cross-linking index of the polymer changes with the growth phase. It is highest in late stationary phase, with a value of 33.2 or 44% per muramic acid residue, as determined by reverse-phase high-pressure liquid chromatography or gel filtration, respectively. Analysis of the muropeptide composition of a dacA (PBP 5) mutant shows a dramatic decrease of muropeptides with tripeptide side chains and an increase or appearance of muropeptides with pentapeptide side chains in monomers or oligomers. The total muropeptides with pentapeptide side chains accounts for almost 82% in the dacA mutant. This major low-molecular-weight PBP (dd-carboxypeptidase) is suggested to play a role in peptidoglycan maturation.
Cell wall peptidoglycan is present in most eubacteria and is essential for the maintenance of cellular viability and shape determination. B. subtilis is attractive as a model organism for peptidoglycan studies because of its life cycle, as nutrient deprivation results in a differentiation process leading to the production of a spore. This mechanism leads to a change in peptidoglycan structure and function between vegetative cells and spores (11). In vegetative cells, the peptidoglycan consists of glycan chains of alternating N-acetylglucosamine and N-acetylmuramic acid, which are frequently cross-linked to each other by short peptides. This rigid bag-shaped polymer surrounds the bacterial cell and therefore determines the shape of the bacterial cells. It also protects the cells against the turgor pressure exerted by the cytoplasm (1, 18). Although this highly cross-linked network forms a rigid, insoluble envelope in the shape of the bacterial cells, the peptidoglycan is nevertheless in a dynamic state throughout the life of the cell (9, 18).
Synthesis of peptidoglycan is a complex two-stage process. The first is the formation of a peptidoglycan subunit, disaccharide pentapeptide, in the cytoplasm. In many organisms, including Bacillus subtilis and Escherichia coli, the pentapeptide consists of l-Ala-d-Glu-l-meso-diaminopimelic acid-d-Ala-d-Ala. The second is the polymerization, which occurs on the outer surface of the cytoplasmic membrane (41). Polymerization of peptidoglycan requires two enzymatic activities, a transglycosylase that polymerizes the glycan strands and a transpeptidase that cross-links these strands by means of the peptide side chains (13, 34, 41). During the transpeptidation reaction the terminal d-alanine is removed from one of the peptide chains (donor), while its penultimate d-alanine becomes linked to the ɛ-amino group of diaminopimelic acid (A2pm) in another peptide chain (acceptor) (12, 17). The penicillin-binding proteins (PBPs) required for these activities are class A and B high-molecular-weight PBPs which are respectively bifunctional transpeptidases and transglycosylases or transpeptidases. The low-molecular-weight PBPs are generally involved in peptidoglycan maturation and can be carboxypeptidases or dd-endopeptidases (13, 18, 30). Although a number of genes encoding PBPs have been sequenced and inactivated, very little is known about the precise role of these genes in peptidoglycan biosynthesis (31–34, 39, 44). Analysis is further hampered by the functional redundancy in these enzymes (29, 30, 35).
Earlier studies of the peptidoglycan composition of B. subtilis revealed the presence of Ala, Glu, and A2pm in addition to N-acetylglucosamine and N-acetylmuramic acid. A dozen stem peptide structures have also been determined after hydrolysis of peptidoglycan by an amidase (43). Amidation of the free carboxylic group of A2pm was shown as a major feature of B. subtilis peptidoglycan (43). More recently, glycine was shown to occur in the peptidoglycan of outgrowing spores of this bacterium (29).
We have recently reported the fine structure of B. subtilis spore peptidoglycan and determined the structural dynamics which take place during the germination process (2, 3). In this paper, we report the structure of the vegetative cell wall peptidoglycan determined by a combination of reverse-phase high-pressure liquid chromatography (RP-HPLC) separation of muropeptides, amino acid analysis, mass spectrometry (MS), and nuclear magnetic resonance spectroscopy (NMR). Peptidoglycan analysis of dacA (PBP 5) and ponA (PBP 1) mutants has revealed the possible roles of these PBPs in peptidoglycan biosynthesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. subtilis 168 HR and the mutant strains were grown at 37°C in nutrient broth (Oxoid) with shaking at 250 rpm or on nutrient agar (1% [wt/vol]) plates at 37°C.
Preparation of cell wall peptidoglycan.
Cell cultures (100 ml) were boiled in a water bath for 7 min to avoid peptidoglycan autolysis and then collected by centrifugation (14,000 × g, 8 min, 4°C). Hot 5% (wt/vol) sodium dodecyl sulfate (SDS) was added to pelleted cells, which were then resuspended and boiled for 25 min. Insoluble material was recovered by centrifugation (14,000 × g, 8 min, 20°C) and boiled again in 4% (wt/vol) SDS (15 min) after resuspension. The resulting insoluble wall preparation was then washed with hot distilled water (60°C) at least five times until free of SDS. Covalently attached proteins are removed by treatment with 2 mg of pronase ml−1 for 1 h at 60°C (2). The walls were then recovered by centrifugation (14,000 × g, 8 min, 4°C), washed once in distilled water, and suspended in hydrofluoric acid (HF) (400 μl of a 48% [vol/vol] solution); the mixture was incubated at 2°C for 24 h. The insoluble material was collected by centrifugation (14,000 × g, 8 min, 4°C) and washed repeatedly by centrifugation and resuspension once with Tris-HCl buffer (50 mM, pH 7) and five times with cold distilled water until the pH was neutral. The material was then stored at −20°C.
Enzymatic hydrolysis of peptidoglycan and RP-HPLC separation of soluble muropeptides.
Samples containing muropeptides were digested with Cellosyl as previously reported (2). Soluble muropeptides were reduced by using sodium borohydride at a final concentration of 8 mg ml−1 as described previously (8). The reaction was stopped after 13 min by lowering the pH to 4 with phosphoric acid. Reduced muropeptides were separated with a Waters HPLC system and a Hypersil octadecylsilane column from Sigma (4.6 by 250 mm; particle size, 5 μm) as previously described (2). Elution buffers were as follows: A, 40 mM sodium phosphate (pH 4.5); B, 40 mM sodium phosphate (pH 4) containing 20% (vol/vol) methanol. A small amount of sodium azide (142 μl from a 1% [wt/vol] solution) was added to 1 liter of buffer A to equalize its A202 with that of buffer B. The column was equilibrated at 52°C with buffer A at a flow rate of 0.5 ml min−1 for 20 min. Soluble-reduced muropeptides (60 μl) were injected, and a linear gradient of 0 to 100% buffer B over a period of 270 min was started 5 min after sample injection. The flow rate was constant at 0.5 ml min−1 over the course of the gradient, and the eluted compounds were detected by monitoring A202. Muropeptides were collected individually at the detector outlet.
Desalting of HPLC-separated muropeptides, amino acid analysis, and MS.
Desalting of muropeptides, amino acid analysis, and MS were carried out as previously described (2).
Determination of cross-linking indices by gel filtration.
Cellosyl-digested muropeptides were loaded onto a TSK SW2000 (7.5 by 600 mm; Anachem, Luton, United Kingdom) gel filtration column. The muropeptides were eluted from the column at 0.3 ml min−1 with 40 mM phosphate buffer pH 6.5. A202 was monitored, and the cross-linking index was calculated as previously reported (27).
NMR analysis of muropeptides.
HPLC-purified muropeptides were lyophilized and dissolved in 90% H2O–10% D2O. NMR spectra were run at 298 K on a Bruker DRX-500 spectrometer. All spectra were acquired by using solvent presaturation and were processed by using low-frequency digital filtering to remove the water signal. Two-dimensional (2-D) double-quantum filtered correlated spectroscopy, total correlated spectroscopy (TOCSY), and rotating frame nuclear Overhauser effect spectroscopy (ROESY) spectra were acquired by using spectral widths of 12,500 Hz in F2 and 5,000 Hz in F1. Typically, spectra were acquired as 4,096 × 256 complex points, using the States-TPPI method. Spin-lock fields used were 10 kHz for TOCSY and 2.2 kHz for ROESY, and mixing times were 100 ms for both TOCSY and ROESY. All spectra were processed and plotted by using Felix 97.0 (Molecular Simulations, Inc.).
RESULTS
HPLC analysis of Cellosyl-digested peptidoglycan from B. subtilis 168.
Cultures used for muropeptide analysis were grown to stationary phase (8 h) prior to sampling. To determine the degree of peptidoglycan solubilization, the levels of A2pm (considered to be exclusive to peptidoglycan) were measured in total undigested peptidoglycan, Cellosyl-hydrolyzed soluble, and insoluble fractions. At least 98% of A2pm was solubilized by Cellosyl treatment with <3% contamination by nonpeptidoglycan amino acids.
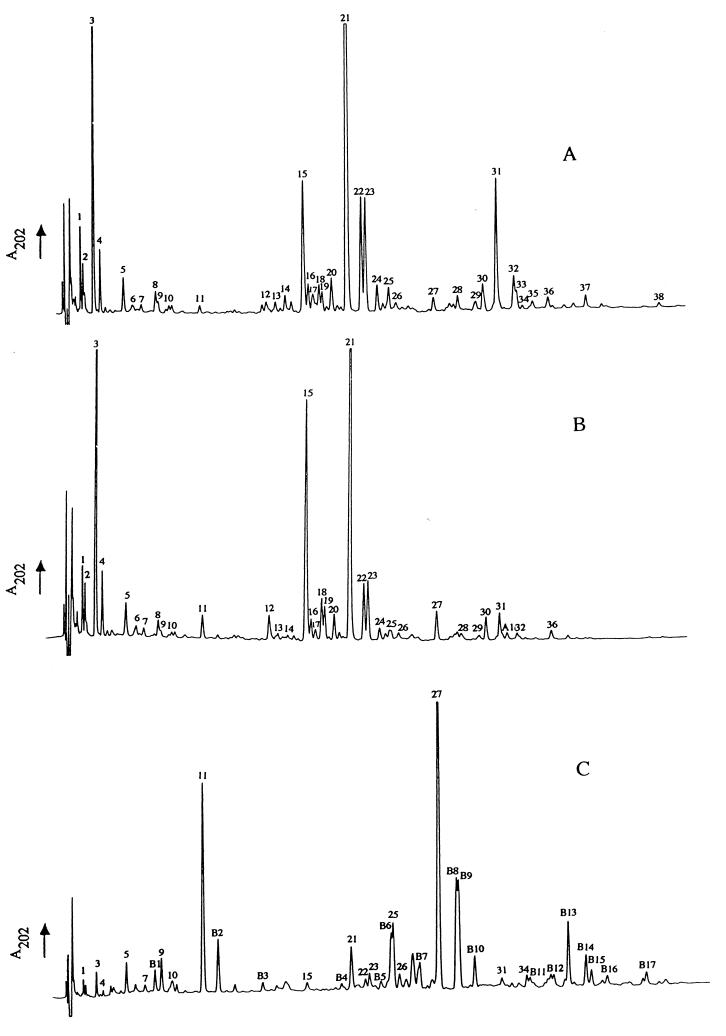
Separation of soluble reduced muropeptides was first carried out by using the conditions previously developed for muropeptide separation from B. subtilis spore peptidoglycan (2). However, after separation and the desalting process, some peaks appeared to contain more than one product. Optimization experiments comprised an increase in column temperature (from 40 to 52°C) and the use of pH 4.5 instead of 4.23 in buffer A. This optimization procedure allowed a better resolution of most muropeptides, as shown by a representative chromatogram in Fig. 1A.
FIG. 1.
RP-HPLC muropeptide elution patterns of peptidoglycan from B. subtilis 168 vegetative cells. (A) HR (wild type); (B) AA106 (ponA); (C) AA109 (dacA). Purified Cellosyl-digested peptidoglycan samples were separated on an octadecylsilane column, and the A202 of the eluate was monitored.
Effects of pronase and HF treatments on the muropeptide elution pattern of B. subtilis 168.
The omission of pronase treatment during peptidoglycan preparation did not alter the RP-HPLC muropeptide pattern. Quantification of muropeptides from pronase-treated and untreated samples also indicated no substantial loss of muropeptides in the pronase-untreated sample.
Peptidoglycan-bound anionic polymers are a common feature of gram-positive bacterial cell walls and those of B. subtilis in particular. The omission of HF treatment in the cell wall purification procedure leads to peptidoglycan with intact bound teichoic acids. Comparison of RP-HPLC profiles of HF-treated and untreated samples allowed the identification of the potential main teichoic acid-anchoring muropeptides. Indeed, muropeptides 2, 13, and 16 (Fig. 1A) were absent in the HF-untreated samples (results not shown). Also, a significant decrease of muropeptide 17 was noted.
Structural identification of the vegetative cell wall muropeptides.
Based on amino acid analysis, all numbered peaks in Fig. 1A are peptidoglycan-derived products (Table 1). The amino acids detected were Ala, Glu, A2pm, and Gly (although in small amounts). Determination of the precise relative molecular masses of the different muropeptides was crucial for identification of their structure. However, the potential occurrence of amidation (43), which causes a change of only one mass unit in some muropeptides, would be difficult to determine by matrix-assisted laser desorption ionization MS for dimers and oligomers. Therefore, in addition to amino acid and MS analyses, each muropeptide obtained in sufficient quantity was subjected to NMR for determination of the number of amide groups.
TABLE 1.
Calculated and observed m/z values for protonated, sodiated, and deprotonated molecular ions of vegetative cell wall muropeptides
| Muropeptidea | Ion |
m/z
|
Δm (Da)b | Muropeptide composition
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Observed | Calculated | Glcc | Murd | Ala | Glu | A2pm | Gly | |||
| 1 | [M + Na]+ | 892.7 | 893.9 | −1.1 | 1 | 1 | 1 | 1 | 1 | 0 |
| [M − H]− | 868.8 | 869.9 | −1.2 | |||||||
| 2 | [M + Na]+ | 972.6 | 893.9 | 78.5 | 1 | 1 | 1 | 1 | 1 | 0 |
| [M − H]− | 948.0 | 869.9 | 78.5 | |||||||
| 3 | [M + H]+ | 870.3 | 871.9 | −1.6 | 1 | 1 | 1 | 1 | 1 | 0 |
| [M − H]− | 868.1 | 869.9 | −1.8 | |||||||
| 4 | [M + H]+ | 829.1 | 871.9 | −42.8 | 1 | 1 | 1 | 1 | 1 | 0 |
| [M − H]− | 827.1 | 869.9 | −42.8 | |||||||
| 5 | [M + H]+ | 699.8 | 699.7 | 0.1 | 1 | 1 | 1 | 1 | 0 | 0 |
| [M − H]− | 698.2 | 697.7 | 0.5 | |||||||
| 6 | [M + H]+ | 941.7 | 942.9 | 1.2 | 1 | 1 | 2 | 1 | 2 | 0 |
| [M − H]− | 940.2 | 940.9 | 0.7 | |||||||
| 7 | [M + Na]+ | 1,136.9 | 1,137.0 | 0.1 | 1 | 1 | 2 | 1 | 1 | 0 |
| [M − H]− | 1,112.9 | 1,113.0 | 0.1 | |||||||
| 8 | [M + Na]+ | 1,136.4 | 1,137.0 | −0.6 | 1 | 1 | 2 | 1 | 2 | 0 |
| [M − H]− | 1,111.9 | 1,113.0 | −1.1 | |||||||
| 9 | [M + Na]+ | 1,022.3 | 1,021.9 | 0.4 | 1 | 1 | 2 | 1 | 1 | 1 |
| [M − H]− | 997.1 | 997.9 | −0.9 | |||||||
| 10 | [M + Na]+ | 1,334.5 | 1,337.2 | −2.7 | 1 | 1 | 3 | 2 | 2 | 0 |
| [M − H]− | 1,310.2 | 1,313.2 | −3 | |||||||
| 11 | [M + Na]+ | 1,034.7 | 1,036 | −1.2 | 1 | 1 | 3 | 1 | 1 | 0 |
| [M − H]− | 1,010.8 | 1,012 | −1.1 | |||||||
| 12 | [M + Na]+ | 1,612.8 | 1,817.8 | 205.0 | 1 | 2 | 3 | 2 | 2 | 0 |
| [M − H]− | 1,589.2 | 1,793.8 | 204.6 | |||||||
| 13 | [M + Na]+ | 1,896.5 | 1,817.8 | 78.7 | 2 | 2 | 3 | 2 | 2 | 0 |
| [M − H]− | 1,872.5 | 1,793.8 | 78.7 | |||||||
| 14 | [M + H]+ | 1,384.1 | 1,385.5 | 1.4 | 1 | 1 | 4 | 2 | 2 | 0 |
| [M − H]− | 1,382.6 | 1,383.5 | 0.9 | |||||||
| 15 | [M + Na]+ | 1,815.9 | 1,817.8 | −1.9 | 2 | 2 | 3 | 2 | 2 | 0 |
| [M − H]− | 1,792.3 | 1,793.8 | −1.5 | |||||||
| 16 | [M + H]+ | 1,872.6 | 1,795.8 | 76.8 | 2 | 2 | 3 | 2 | 2 | 0 |
| [M − H]− | 1,871.3 | 1,793.8 | 77.5 | |||||||
| 17a | [M + H]+ | 1,872.6 | 1,795.8 | 76.8 | 2 | 2 | 3 | 2 | 2 | 0 |
| [M − H]− | 1,871.5 | 1,793.8 | 77.3 | |||||||
| 17b | [M + H]+ | 1,590.7 | 1,795.8 | 205.1 | 1 | 2 | 3 | 2 | 2 | 0 |
| [M − H]− | 1,588.0 | 1,793.8 | 205.8 | |||||||
| 18 | [M + Na]+ | 1,773.4 | 1,817.8 | −44.4 | 2 | 2 | 3 | 2 | 2 | 0 |
| [M − H]− | 1,749.5 | 1,793.8 | −44.3 | |||||||
| 19 | [M + Na]+ | 1,773.4 | 1,817.8 | −44.4 | 2 | 2 | 3 | 2 | 2 | 0 |
| [M − H]− | 1,749.5 | 1,793.8 | −44.3 | |||||||
| 20 | [M + H]+ | 1,794.9 | 1,795.8 | 0.9 | 2 | 2 | 3 | 2 | 2 | 0 |
| [M − H]− | 1,792.8 | 1,793.8 | 1.0 | |||||||
| 21 | [M + Na]+ | 1,816.6 | 1,817.8 | 1.2 | 2 | 2 | 3 | 2 | 2 | 0 |
| [M − H]− | 1,792.8 | 1,793.8 | 1.0 | |||||||
| 22 | [M + H]+ | 1,753.0 | 1,795.8 | −42.8 | 2 | 2 | 3 | 2 | 2 | 0 |
| [M − H]− | 1,750.2 | 1,793.8 | −43.6 | |||||||
| 23 | [M + H]+ | 1,753.0 | 1,795.8 | −42.8 | 2 | 2 | 3 | 2 | 2 | 0 |
| [M − H]− | 1,750.2 | 1,793.8 | −43.6 | |||||||
| 24 | [M + Na]+ | 1,945.8 | 1,945.9 | −0.1 | 2e | 2e | 4e | 2e | 2e | 1e |
| [M − H]− | 1,920.5 | 1,921.9 | −1.4 | |||||||
| 25 | [M + Na]+ | 1,945.8 | 1,945.9 | −0.1 | 2 | 2 | 4 | 2 | 2 | 1 |
| [M − H]− | 1,921.5 | 1,921.9 | −0.4 | |||||||
| 26 | [M + Na]+ | 1,889.5 | 1,888.9 | 0.6 | 2 | 2 | 4 | 2 | 2 | 0 |
| [M − H]− | 1,865.1 | 1,864.9 | 0.2 | |||||||
| 27 | [M + Na]+ | 1,961.3 | 1,959.9 | 1.4 | 2 | 2 | 5 | 2 | 2 | 0 |
| [M − H]− | 1,936.8 | 1,935.9 | 0.9 | |||||||
| 28 | [M + Na]+ | 2,743.5 | 2,740.7 | −2.8 | 3 | 3 | 5 | 3 | 3 | 0 |
| [M − H]− | 2,718.3 | 2,716.7 | −1.6 | |||||||
| 29 | [M + Na]+ | 2,537.6 | 2,739.7 | 202.1 | 2 | 3 | 5 | 3 | 3 | 0 |
| [M − H]− | 2,513.4 | 2,716.7 | 203.3 | |||||||
| 30 | [M + H]+ | 2,719.9 | 2,718.7 | 1.2 | 3 | 3 | 5 | 3 | 3 | 0 |
| [M − H]− | 2,717.6 | 2,716.7 | 0.9 | |||||||
| 31 | [M + H]+ | 2,718.3 | 2,718.7 | −0.4 | 3 | 3 | 5 | 3 | 3 | 0 |
| [M − H]− | 2,716.1 | 2,716.7 | −0.1 | |||||||
| 32 | [M + Na]+ | 2,701.6 | 2,740.7 | −40.7 | 3 | 3 | 5 | 3 | 3 | 0 |
| [M − H]− | 2,674.7 | 2,716.7 | −42.0 | |||||||
| 33 | [M + Na]+ | 2,701.6 | 2,740.7 | 40.6 | 3 | 3 | 5 | 3 | 3 | 0 |
| [M − H]− | 2,674.7 | 2,716.7 | 42.0 | |||||||
| 34 | [M + Na]+ | 2,868.4 | 2,868.6 | 0.2 | 3 | 3 | 6 | 3 | 3 | 1 |
| [M − H]− | 2,843.0 | 2,844.6 | 1.1 | |||||||
| 35 | [M + H]+ | 3,642.8 | 3,641.6 | 1.2 | 4 | 4 | 7 | 4 | 4 | 0 |
| [M − H]− | 3,638.5 | 3,639.6 | 1.1 | |||||||
| 36 | [M + H]+ | 1,774.9 | 1,794.8 | −19.9 | 2 | 2 | 3 | 2 | 2 | 0 |
| [M − H]− | 1,772.7 | 1,793.8 | −21.1 | |||||||
| 37 | [M + Na]+ | 3,662.3 | 3,663.6 | −1.3 | 4 | 4 | 7 | 4 | 4 | 0 |
| [M − H]− | 3,635.5 | 3,639.6 | −4.1 | |||||||
| 38 | [M + H]+ | 2,700.1 | 2,718.7 | −18.6 | 3 | 3 | 5 | 3 | 3 | 0 |
| [M − H]− | 2,696.8 | 2,716.7 | −19.9 | |||||||
| A1 | [M + Na]+ | 1,800.5 | 1,817.8 | 17.3 | 2 | 2 | 3 | 2 | 2 | 0 |
| [M − H]− | 1,775.3 | 1,793.8 | 18.5 | |||||||
| B1 | [M + H]+ | 1,014.7 | 1,013.9 | 0.8 | 1 | 1 | 3 | 1 | 1 | 0 |
| [M − H]− | 1,012.8 | 1,011.9 | 0.9 | |||||||
| B2 | [M + Na]+ | 993.7 | 1,035.9 | −42.2 | 1 | 1 | 3 | 1 | 2 | 0 |
| [M − H]− | 969.3 | 1,011.9 | −42.6 | |||||||
| B3 | [M + H]+ | 1,255.5 | 1,257.2 | −1.7 | 1 | 1 | 4 | 1 | 1 | 0 |
| [M − H]− | 1,253.3 | 1,255.2 | −1.9 | |||||||
| B4 | [Na + H]+ | 1,946.0 | 1,945.9 | 0.1 | 2 | 2 | 4 | 2 | 2 | 1 |
| [M − H]− | 1,921.3 | 1,921.9 | −0.6 | |||||||
| B5 | [M + H]+ | 2,016.3 | 1,937.9 | 78.4 | 2 | 2 | 5 | 2 | 2 | 0 |
| [M − H]− | 2,013.7 | 1,935.9 | 77.8 | |||||||
| B6 | [M + H]+ | 1,937.1 | 1,937.9 | 0.8 | 2 | 2 | 5 | 2 | 2 | 0 |
| [M − H]− | 1,934.4 | 1,935.9 | 1.5 | |||||||
| B7 | [M + Na]+ | 1,959.8 | 1,959.9 | 0.1 | 2 | 2 | 5 | 2 | 2 | 0 |
| [M − H]− | 1,935.1 | 1,935.9 | 0.8 | |||||||
| B8 | [M + Na]+ | 1,916.3 | 1,959.9 | −43.6 | 2 | 2 | 5 | 2 | 2 | 0 |
| [M − H]− | 1,892.6 | 1,935.9 | −43.3 | |||||||
| B9 | [M + Na]+ | 1,916.3 | 1,959.9 | −43.6 | 2 | 2 | 5 | 2 | 2 | 0 |
| [M − H]− | 1,892.6 | 1,935.9 | −43.3 | |||||||
| B10 | [M + Na]+ | 1,875.3 | 1,959.9 | −84.2 | 2 | 2 | 5 | 2 | 2 | 0 |
| [M − H]− | 1,850.8 | 1,935.9 | −84.2 | |||||||
| B11 | [M + Na]+ | 2,882.1 | 2,882.8 | 0.7 | 3 | 3 | 7 | 3 | 3 | 0 |
| [M − H]− | 2,858.2 | 2,858.8 | 0.6 | |||||||
| B12 | [M + Na]+ | 2,881.5 | 2,882.8 | 1.3 | 3 | 3 | 7 | 3 | 3 | 0 |
| [M − H]− | 2,857.1 | 2,858.8 | 1.7 | |||||||
| B13 | [M + Na]+ | 2,879.6 | 2,882.8 | 3.2 | 3 | 3 | 7 | 3 | 3 | 0 |
| [M − H]− | 2,854.7 | 2,858.8 | 4.1 | |||||||
| B14 | [M + Na]+ | 2,837.8 | 2,882.8 | 45.0 | 3 | 3 | 7 | 3 | 3 | 0 |
| [M − H]− | 2,813.3 | 2,858.8 | 45.5 | |||||||
| B15 | [M + Na]+ | 2,837.3 | 2,882.8 | 45.5 | 3 | 3 | 7 | 3 | 3 | 0 |
| [M − H]− | 2,815.1 | 2,858.8 | 43.7 | |||||||
| B16 | [M + Na]+ | 3,804.0 | 3,805.7 | 1.7 | 4 | 4 | 9 | 4 | 4 | 0 |
| [M − H]− | 3,780.4 | 3,781.7 | 1.3 | |||||||
| B17 | [Na + H]+ | 1,916.1 | 1,937.9 | 21.8 | 2 | 2 | 5 | 2 | 2 | 0 |
| [M − H]− | 1,913.8 | 1,935.9 | 22.1 | |||||||
Muropeptides are numbered as indicated in Fig. 1.
Difference between observed and calculated protonated, sodiated, or deprotonated molecular mass values. Boldface characters denote m/z values calculated as the most likely combinations of the list components which nevertheless deviated largely from the observed values. For explanation, see text.
Glc, N-acetylglucosamine.
Mur, N-acetylmuramitol.
Not determined but estimated from mass value.
Vegetative cell wall peptidoglycan composition of B. subtilis.
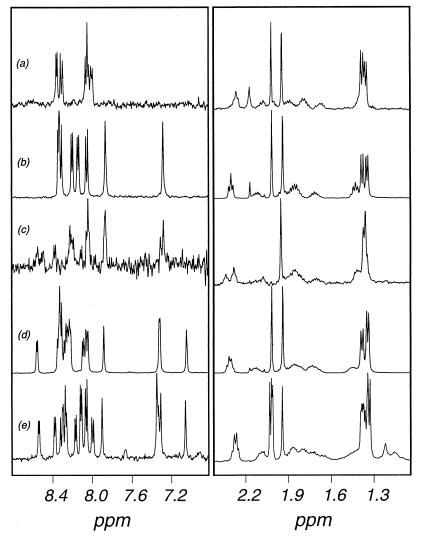
Amino acid analysis combined with MS and NMR allowed structural determination and quantification of all purified muropeptides (Fig. 1A and 2; Tables 1 and 2). Muropeptides 1 and 3 have the same MS and amino acid composition (Table 1) but different retention times (Fig. 1A). NMR analysis indicates the presence of an amide group on muropeptide 3, visible as a pair of singlet peaks at 7.29 and 7.86 ppm, which are clearly absent in muropeptide 1 (Table 2; Fig. 3). Detailed 2-D NMR investigations carried out on muropeptides 9 and 25 (Table 2) confirmed that the amide groups are in all cases incorporated as amidation of the A2pm carboxylate, by observation of rotating frame nuclear Overhauser enhancements in the ROESY spectrum between A2pm and amide protons (results not shown). Peak 2 can be seen (Fig. 1A) to be likely a mixture of two products. Desalting confirmed this, but the structure of only the major product could be determined (Tables 1 and 2). Muropeptides 15, 20, and 21 show the same amino acid composition and very similar masses (Table 1). The difference in the retention times of muropeptides 15 and 20, which show one amide group each (Table 2), is likely due to the position of the amide group on the carboxylic group of either A2pm in the dimer. Muropeptide 21 has two amide groups (Fig. 1A and 3; Table 2) and therefore has a retention time different from those of the former muropeptides. Under the conditions used for separation of muropeptides from B. subtilis, the retention times of otherwise identical muropeptides increase with the number of amide groups. This is the case for muropeptides 15, 20, and 21, muropeptides 30 and 31, and muropeptides 35 and 37 (Fig. 1A; Tables 1 and 2). Muropeptide 4 is a disaccharide tripeptide with an amide group but with a 42-Da mass defect (Tables 1 and 2). This mass defect corresponds to a lack of an N-acetyl group on an amino sugar, as demonstrated by the NMR spectrum (Fig. 3), in which the upfield N-acetyl signal (2.02 ppm) is absent. Fragmentation of the muropeptide by MS revealed that the de-N-acetylation occurs on the glucosamine (result not shown). Likewise, the 42-Da mass defect of muropeptides 18 and 19, 22 and 23, and 32 and 33 corresponds to de-N-acetylation of glucosamine (Table 2). The substitution of N-acetylmuramic acid with either a tripeptide (acceptor) or tetrapeptide (donor) in dimers or oligomers and the occurrence of de-N-acetylation of glucosamine explains the presence of more than one peak for each apparent product.
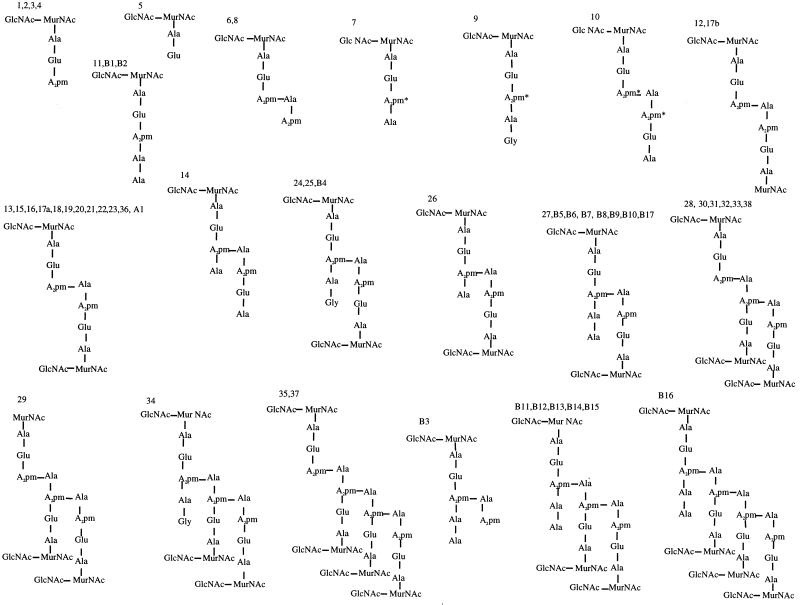
FIG. 2.
Proposed possible basic structures for muropeptides from B. subtilis 168 HR, AA106 (ponA) and AA109 (dacA). Numbers refer to peaks in Fig. 1. Amidation (∗) is shown only on unique monomer muropeptides where unequivocal positioning is known.
TABLE 2.
Muropeptide identities and quantification from peptidoglycan of B. subtilis HR, AA106 (ponA), and AA109 (dacA)
| Muro-peptidea | Identity | Mol%
|
||
|---|---|---|---|---|
| HR (wild type) | AA106 (ponA) | AA 109 (dacA) | ||
| 1 | Disaccharide tripeptide | 3.3 | 2.9 | 0.7 |
| 2 | Disaccharide tripeptide with 1 amidation and 1 phosphate | 0.9 | 1 | |
| 3 | Disaccharide tripeptide with 1 amidation | 20.5 | 28.9 | 1.3 |
| 4 | Disaccharide tripeptide 1 amidation missing an acetyl group | 3.4 | 4.3 | 0.3 |
| 5 | Disaccharide dipeptide | 3.8 | 4.4 | 3.7 |
| 6 | Disaccharide tripeptide dipeptide with 1 amidation | 0.3 | 0.7 | |
| 7 | Disaccharide tetrapeptide with 1 amidation | 0.5 | 0.9 | 0.5 |
| 8 | Disaccharide tripeptide dipeptide with 2 amidations | 1.6 | 0.3 | |
| 9 | Disaccharide pentapeptide (Ala-4–Gly-5) with 1 amidation | 0.5 | 1.6 | 1.9 |
| 10 | Disaccharide tripeptide tetrapeptide with 2 amidations | 0.5 | 0.2 | 0.4 |
| 11 | Disaccharide pentapeptide (Ala-4–Ala-5) with 1 amidation | 0.6 | 2.1 | 19.1 |
| 12b | Disaccharide tripeptide disaccharide tetrapeptide missing a glucosamine | 0.4 | 1.1 | |
| 13 | Disaccharide tripeptide disaccharide tetrapeptide with 1 phosphate and 1 amidation | 0.5 | 0.3 | |
| 14 | Disaccharide tetrapeptide tetrapeptide with 2 amidations | 0.3 | 0.1 | |
| 15 | Disaccharide tripeptide disaccharide tetrapeptide with 1 amidation | 6.3 | 11.7 | 0.4 |
| 16 | Disaccharide tripeptide disaccharide tetrapeptide with 1 phosphate and 2 amidations | 1.1 | 0.9 | |
| 17a | Disaccharide tripeptide disaccharide tetrapeptide with 1 phosphate and 2 amidations | 0.6 | 0.3 | |
| 17b | Disaccharide tripeptide disaccharide tetrapeptide with 2 amidations and missing a glucosamine | 0.6 | 0.2 | |
| 18 | Disaccharide tripeptide disaccharide tetrapeptide with 1 amidation and missing an acetyl group | 1.2 | 1.9 | |
| 19 | Disaccharide tripeptide disaccharide tetrapeptide with 1 amidation and missing an acetyl group | 0.8 | 1.5 | |
| 20 | Disaccharide tripeptide disaccharide tetrapeptide with 1 amidation | 1.7 | 1.3 | |
| 21 | Disaccharide tripeptide disaccharide tetrapeptide with 2 amidations | 27.7 | 20 | 2.2 |
| 22 | Disaccharide tripeptide disaccharide tetrapeptide with 2 amidations and missing an acetyl group | 5.2 | 2.7 | 0.5 |
| 23 | Disaccharide tripeptide disaccharide tetrapeptide with 2 amidations and missing an acetyl group | 5.0 | 2.9 | 0.8 |
| 24b | Disaccharide pentapeptide (Gly-5) disaccharide tetrapeptide | 1.3 | 0.6 | |
| 25 | Disaccharide pentapeptide (Gly-5) disaccharide tetrapeptide with 2 amidations | 0.8 | 0.4 | 4.1 |
| 26 | Disaccharide tetrapeptide disaccharide tetrapeptide with 2 amidations | 0.4 | 0.4 | 0.5 |
| 27 | Disaccharide pentapeptide disaccharide tetrapeptide with 2 amidations | 1.0 | 2.4 | 30.8 |
| 28 | Disaccharide tripeptide disaccharide tetrapeptide disaccharide tetrapeptide with 2 amidations | 0.5 | 0.4 | |
| 29b | Disaccharide tripeptide disaccharide tetrapeptide disaccharide tetrapeptide missing a glucosamine | 0.5 | 0.1 | |
| 30 | Disaccharide tripeptide disaccharide tetrapeptide disaccharide tetrapeptide with 2 amidations | 1.3 | 1.0 | |
| 31 | Disaccharide tripeptide disaccharide tetrapeptide disaccharide tetrapeptide with 3 amidations | 4.2 | 1.1 | 0.3 |
| 32 | Disaccharide tripeptide disaccharide tetrapeptide disaccharide tetrapeptide with 3 amidations and missing an acetyl group | 1.2 | 0.2 | |
| 33 | Disaccharide tripeptide disaccharide tetrapeptide disaccharide tetrapeptide with 3 amidations and missing an acetyl group | 0.5 | ||
| 34 | Disaccharide pentapeptide (Gly-5) disaccharide tetrapeptide disaccharide tetrapeptide with 2 or 3 amidations | 0.1 | ||
| 35 | Disaccharide tripeptide disaccharide tetrapeptide disaccharide tetrapeptide disaccharide tetrapeptide with 2 or 3 amidations | 0.1 | ||
| 36 | Anhydrodisaccharide tripeptide disaccharide tetrapeptide with 2 amidations | 0.3 | 0.2 | |
| 37 | Disaccharide tripeptide disaccharide tetrapeptide disaccharide tetrapeptide disaccharide tetrapeptide with 4 amidations | 0.2 | ||
| 38 | Anhydrodisaccharide tripeptide disaccharide tetrapeptide disaccharide tetrapeptide with 3 amidations | 0.1 | ||
| A1 | Anhydrodisaccharide tripeptide disaccharide tetrapeptide with 1 amidation | 0.6 | ||
| B1 | Disaccharide pentapeptide | 1.7 | ||
| B2 | Disaccharide pentapeptide missing an acetyl group | 5.0 | ||
| B3 | Disaccharide pentapeptide dipeptide | 1.1 | ||
| B4 | Disaccharide pentapeptide (Gly-5) disaccharide tetrapeptide | 0.3 | ||
| B5 | Disaccharide pentapeptide disaccharide tetrapeptide with 1 phosphate | 0.3 | ||
| B6 | Disaccharide pentapeptide disaccharide tetrapeptide | 1.9 | ||
| B7 | Disaccharide pentapeptide disaccharide tetrapeptide | 0.9 | ||
| B8 | Disaccharide pentapeptide disaccharide tetrapeptide missing an acetyl group | 7.0 | ||
| B9 | Disaccharide pentapeptide disaccharide tetrapeptide missing an acetyl group | 6.8 | ||
| B10 | Disaccharide pentapeptide disaccharide tetrapeptide missing 2 acetyl groups | 2.1 | ||
| B11 | Disaccharide pentapeptide disaccharide tetrapeptide disaccharide tetrapeptide | 0.3 | ||
| B12 | Disaccharide pentapeptide disaccharide tetrapeptide disaccharide tetrapeptide | 0.4 | ||
| B13 | Disaccharide pentapeptide disaccharide tetrapeptide disaccharide tetrapeptide | 1.8 | ||
| B14 | Disaccharide pentapeptide disaccharide tetrapeptide disaccharide tetrapeptide missing an acetyl group | 1.1 | ||
| B15 | Disaccharide pentapeptide disaccharide tetrapeptide disaccharide tetrapeptide missing an acetyl group | 0.6 | ||
| B16 | Disaccharide pentapeptide disaccharide tetrapeptide disaccharide tetrapeptide disaccharide tetrapeptide | 0.2 | ||
| B17 | Anhydrodisaccharide pentapeptide disaccharide tetrapeptide | 0.7 | ||
Numbered as indicated in Fig. 1.
Muropeptide from B. subtilis HR (wild type) not analyzed for amide levels due to lack of material.
FIG. 3.
NMR spectra of muropeptides 1 (a), 3 (b), 4 (c), 21 (d), and 36 (e). Low-field and high-field regions are shown with different horizontal and vertical expansions.
Muropeptides 36 (a dimer [disaccharide tripeptide disaccharide tetrapeptide with two amidations]) and 38 (a trimer [disaccharide tripeptide disaccharide tetrapeptide disaccharide tetrapeptide with three amidations]) (Fig. 1A and 2; Table 2) are eluted later than related muropeptides 21 and 31. MS analysis indicates that these muropeptides have an average 19.8-Da mass defect, suggesting a modification of muramic acid to an anhydro group (3). NMR analysis of muropeptide 36 confirms this structure, because one of the two muramic acid N-acetyl signals, which are normally coincident at 1.94 ppm, moves to 2.03 ppm (Fig. 3).
Muropeptides 2, 13, and 16 all show an extra 78 Da compared to respective related muropeptides 4, 15, and 21 (Fig. 1A; Tables 1 and 2). This extra 78 Da does not correspond to any known amino acid. Furthermore, these muropeptides disappear from the RP-HPLC profile of the HF-untreated sample and are eluted earlier than related ones. The 78-Da shift in the molecular mass fits our prediction of a phosphate bound to C6 of one N-acetylmuramic acid.
Peak 17 contains two different muropeptides, which are separated during the desalting process. Muropeptide 17a has the same MS, amino acid content, and amide levels as muropeptide 16 (Tables 1 and 2). The difference in the retention time between muropeptides 16 and 17a is likely therefore to be the position of the phosphate on either N-acetylmuramic acid of these cross-linked muropeptides. Muropeptides 9, 25, and 34 are respectively a monomer, dimer, and trimer, each containing a single glycine (Fig. 1A and 2; Tables 1 and 2). Analysis of pure muropeptides 9 and 25 (Fig. 1A and 2) by 2-D NMR showed sequential rotating frame nuclear Overhauser enhancements between A2pm and Ala-4 and between Ala-4 and Gly, indicating that Gly is in position 5 in both the monomer and the dimer (results not shown). Muropeptide 24 could not be recovered after desalting. Therefore, MS analysis was performed on a nondesalted sample, and a mass similar to that of muropeptide 25 was found. The difference in the retention times of these two muropeptides could be due to the presence of only one amide group on muropeptide 24.
The average 204-Da mass defect in muropeptides 12 and 17b (dimers) and 29 (trimer) corresponds to a loss of an N-acetylglucosamine. Analysis of these muropeptides by NMR clearly shows a loss of a sugar moiety (result not shown). The RP-HPLC profile of lysozyme-digested peptidoglycan resulted in the disappearance of all muropeptides with 42- and 78-Da defects in their masses (result not shown; Tables 1 and 2).
From the chemical structure assigned to most of the muropeptides of B. subtilis (Fig. 2; Table 2), it is clear that the structure of peptidoglycan is relatively simple. It is the amidation of the free carboxylic groups of A2pm and de-N-acetylation of glucosamine which has doubled the number of muropeptides shown in Fig. 1A.
Effects of different media and growth phases on peptidoglycan structure.
To determine whether B. subtilis cell wall peptidoglycan structure is altered as a function of medium composition, the cells were grown to early stationary phase in nutrient broth (Oxoid), Luria-Bertani (LB) medium or minimal medium (24). No new muropeptides were detected in cells grown in either LB or minimal medium. However, one striking difference between the peptidoglycan of cells grown in the last two media and nutrient broth is the virtual absence of muropeptides containing glycine (result not shown). Another minor difference is the substantial decrease in the level of muropeptides 2, 13, 16, and 17a, which contain phosphate.
Apart from the ratio of the different muropeptides, no other major differences characterize B. subtilis peptidoglycan structure during the different growth phases. The overall composition indicates a gradual decrease of monomers and an increase of dimers and trimers as the culture grows older (Table 3).
TABLE 3.
Distribution of muropeptides in B. subtilis HR (wild type) at different growth phases and in AA106 (ponA) and AA109 (dacA) in late stationary phase
| Strain | Growth phase (h) | % Muropeptidesa
|
% Cross-linking
|
||||
|---|---|---|---|---|---|---|---|
| Monomers | Dimers | Trimers | Tetramers | RP-HPLC | Gel filtration | ||
| HR (wild type) | Exponential (2) | 43.6 | 50.3 | 5.8 | 0.2 | 29.1 | 39.6 |
| Late exponential (4) | 39.5 | 53.4 | 6.8 | 0.3 | 31.4 | 41.4 | |
| Stationary (6) | 37.2 | 54.3 | 8.1 | 0.3 | 32.8 | 43.0 | |
| Late stationary (8) | 36.1 | 54.8 | 8.4 | 0.3 | 33.2 | 44.0 | |
| AA106 (ponA) | Late stationary (8) | 47.4 | 49.4 | 2.8 | 0 | 26.6 | NDb |
| AA109 (dacA) | Late stationary (8) | 36.0 | 58.8 | 4.5 | 0.2 | 32.5 | ND |
Determined from RP-HPLC-resolved muropeptides.
ND, not determined.
The cross-linking index calculated from RP-HPLC resolved muropeptides rises from 29.1% in the exponential phase to 33.2% in stationary phase (after 8 h of culture). The same trend was found by gel filtration analysis. However, the values are approximately 10% higher than those obtained by RP-HPLC (Table 3). This discrepancy in cross-linking calculation is due to errors in muropeptide quantification from the gel filtration profile. In terms of individual muropeptides, there is a relative increase of muropeptide 1 (disaccharide tripeptide) compared to all other monomers as cells enter the stationary phase (result not shown).
Role of PBP(s) in peptidoglycan biosynthesis.
PBPs are crucial enzymes for bacterial cells since they are involved in peptidoglycan biosynthesis. High-molecular-weight PBPs polymerize the peptidoglycan building block, disaccharide pentapeptide. However, low-molecular-weight PBPs are involved in peptidoglycan maturation. To determine the role of the high-molecular-weight PBP 1 (ponA) and low-molecular-weight PBP 5 (dacA), we compared the peptidoglycan structure and composition of the strains lacking these enzymes to those of the wild type.
The RP-HPLC muropeptide profile from strain AA106 (ponA) is shown in Fig. 1B. The obvious difference between the muropeptide profile from this strain and that of the wild type is the increase of muropeptides 11, 12, and 27, the decrease in muropeptide 31, and the absence of muropeptides 37 and 38 (Fig. 1B; Table 2). A minor new muropeptide termed A1 (between muropeptides 31 and 32) identified as anhydrodisaccharide tripeptide disaccharide tetrapeptide with one amidation is noted (Fig. 1B; Tables 1 and 2). This muropeptide is scarcely detectable in the wild-type strain (Fig. 1A). The overall composition of the peptidoglycan from ponA shows a decrease in dimers and trimers and an increase in monomers (Table 3). The cross-linking index is only 26.6%, compared to 33.2% of the wild type, in an 8-h culture (Table 3).
Comparison of the peptidoglycan profile of the wild-type strain to that of AA109 (dacA) (Fig. 1A and C) indicates that both the composition and relative amounts of a number of muropeptides change dramatically. Muropeptides 11 (disaccharide pentapeptide with one amidation) and 27 (disaccharide pentapeptide disaccharide tetrapeptide with two amidations), which represent respectively 0.6 and 1% of the total muropeptides in the wild-type strain, become the dominant muropeptides at 19.1 and 30.8% in this mutant (Fig. 1C and 2; Tables 1 and 2). Figure 1C also shows a dramatic decrease or disappearance of muropeptides with tripeptide side chains in monomers or cross-linked muropeptides (e.g., muropeptides 3, 15, and 21). A range of novel muropeptides (B1 to B17) were detected in AA109 (dacA) (Fig. 1C). The chemical structure determined by amino acid analysis and MS indicates the presence of a pentapeptide side chain in all of them (Tables 1 and 2; Fig. 2). Muropeptide B5 shows an extra 78 Da in comparison to equivalent ones (muropeptides B6 and B7). This shift in molecular mass corresponds to a phosphate as suggested for some of the wild-type muropeptides cited above. Likewise, the 42-Da defect in muropeptides B8 and B9 is attributed to de-N-acetylation of glucosamine. The structure of the products eluted between muropeptides 26 and B7 (Fig. 1C) could not be determined. The number of amidations in B1 to B17 was not verified by NMR but their level may account for the range of muropeptides (e.g., B6 to B7 and B11 to B13). The overall composition of peptidoglycan from AA109 (dacA) shows a slight increase of dimers and a decrease in trimers (Table 3). However, the cross-linking index does not differ significantly from that of the wild type (Table 3).
DISCUSSION
The composition and structure of peptidoglycan from vegetative cells of B. subtilis were investigated. The peptidoglycan purification procedure allowed solubilization of >97% muropeptides, and 39 muropeptides were identified.
A most distinctive feature of peptidoglycan in B. subtilis is amidation of the free carboxylic group of A2pm as reported earlier (43). In fact, of all muropeptides containing A2pm analyzed by NMR, only muropeptide 1 (disaccharide tripeptide) is not amidated. This muropeptide was previously reported to be part of the primordial cell wall of spore peptidoglycan (2, 3). Amidation of the free carboxylic group of A2pm has been found in peptidoglycan of a number of bacteria, including Corynebacterium diphtheriae (21) and B. stearothermophilus (16). The physiological significance of this peptidoglycan modification is unknown, although it contributes to a modification of peptidoglycan chemical properties. Amidation neutralizes the acidic carboxyl groups of A2pm and therefore reduces the charge density in the walls. It may be that in the native peptidoglycan, all free carboxy termini of A2pm are amidated and some are lost during muropeptides analysis.
De-N-acetylation of glucosamine is well known in B. subtilis vegetative cell peptidoglycan (1, 46). De-N-acetylation of glucosamine occurs on 17.3% of the total muropeptides. This value is half of that found in previous studies of B. subtilis 168 peptidoglycan using fluorodinitrobenzene (1, 46). De-N-acetylation of glucosamine is also known to cause resistance to lysozyme digestion (1, 46). Similarly, the presence of wall teichoic acid appears to protect against lysozyme digestion (28). Our results correlate with these observations, as the peptidoglycan in B. subtilis is incompletely digested with lysozyme. The muropeptides which disappear from the RP-HPLC profile of the lysozyme digested sample are indeed those with de-N-acetylated glucosamine and those containing phosphate. The role of glucosamine de-N-acetylation could be the regulation of peptidoglycan hydrolases involved in autolysis.
Anhydromuropeptides are a common feature in peptidoglycan of gram-negative bacteria (15, 40) and are proposed to end glycan strands (15). Recently these muropeptides have been shown to be associated with cortex hydrolysis during germination of B. subtilis and B. megaterium spores (3, 4). In E. coli, anhydromuropeptides represent almost 80% of cell wall degradation products released during autolysis induced by cephaloridine or trichloroacetic acid (22). Recent findings showed that anhydromuropeptides are recycled and play a role in gene regulation (19, 20). The lower amounts of these muropeptides in B. subtilis (0.4%) than in E. coli (3.71%) (15) could be explained by longer glycan strands in B. subtilis. Alternatively, other specific muropeptides may end the glycan strands. The absence of one N-acetylglucosamine moiety in some muropeptides indicates that N-acetylmuramic acid is at the nonreducing end of the native glycan strand. This modification would not be expected from the mechanism of peptidoglycan biosynthesis (43). Therefore, it is likely to have been generated by the activity of a glucosaminidase cleaving at specific locations. A 90-kDa glucosaminidase (LytD) from B. subtilis has already been characterized (37), and analysis of a mutant lacking this enzyme has shown its role in cell separation, cell wall turnover, and antibiotic-induced lysis as well as motility (5). Analysis of peptidoglycan structure of the mutant will determine whether this enzyme is involved in peptidoglycan structural determination.
Muropeptides 5 (disaccharide dipeptide) and 8 (disaccharide tripeptide dipeptide with two amidations) are both monomers likely to have been generated by an endopeptidase, cleaving the glutamyl-A2pm bond of the dominant muropeptide 21 (disaccharide tripeptide disaccharide tetrapeptide two amidations). These muropeptides may be generated by LytF, a glutamate-meso-diaminopimelate endopeptidase very recently identified in B. subtilis (26).
Muropeptides 10 and 14 are the products of an amidase. These muropeptides are unlikely to be due to cell autolysis since the cells were boiled just after harvesting. A 50-kDa amidase (LytC) from B. subtilis has already been characterized (37). This enzyme has recently been shown to be involved in cell autolysis induced by sodium azide (5).
An important and unexpected feature of the composition of B. subtilis peptidoglycan is the presence of muropeptides containing glycine (Fig. 2). The location of the glycine in position 5 of the peptide side chain as determined by 2-D NMR in the monomer (muropeptide 9) and the dimer (muropeptide 25) is similar to locations of some muropeptides of E. coli (15) and Thermus thermophilus (36). The mechanism which could explain their formation is the incorporation of Gly instead of d-Ala in the synthesis of d-Ala-d-Ala by d-Ala:d-Ala ligase. This mechanism has been suggested for both gram-positive and gram-negative bacteria (15, 41). Muropeptides containing glycine represent 2.7% of the total muropeptides (Table 2), and their levels depend on the composition of the growth medium. This is probably the reason why they have not been identified in earlier investigations of peptidoglycan chemical structure of B. subtilis using less sensitive techniques (43, 45).
Anionic polymers are covalently attached to peptidoglycan by a phosphodiester linkage to the hydroxyl group of an acetylmuramic residue in the glycan chain (1). Acid hydrolysis yields muramic acid 6-phosphate (25). Muropeptides 2, 13, 16, and 17a (Fig. 1A) are likely to be the muropeptides to which anionic polymers are attached in B. subtilis 168. The fact that these muropeptides have the same amide groups as the dominant muropeptides 3, 15, and 21 (Fig. 1A; Table 2) suggests that the attachment occurs on the dominant muropeptides.
The alteration in peptidoglycan cross-linking levels between cells in exponential and stationary phase (Table 3) could be explained by the change in peptidoglycan dynamics. In the stationary phase, the bacterial cells are unable to adapt quickly and could be more vulnerable to assaults; therefore, a higher cross-linking index is aimed to ensure their mechanical integrity. Similar results have also been reported for other bacteria (6, 10, 14).
Peptidoglycan structure determined by muropeptide analysis gives a snapshot of the wall composition. The interplay of a large number of components produces the final structure. These include enzymes involved in biosynthesis, maturation, and autolysis. By a study of the muropeptide profiles of specific mutants, their role in peptidoglycan structural determination can be elucidated.
PBP 1 is a class A high-molecular-weight PBP (34). A ponA mutation results in a significant decrease in growth rate and an abnormal colony phenotype (34). Our studies have shown a significant alteration in cross-linking which could explain the effect on other cellular processes.
A remarkable feature of peptidoglycan structure of strain AA109 (dacA) lacking PBP 5 is the number of muropeptides with pentapeptide side chains, which account for almost 82% of the total muropeptides. This result confirms that PBP 5 is a dd-carboxypeptidase and demonstrates its role in vegetative cell peptidoglycan maturation. This major modification of peptidoglycan does not affect the growth or the morphology of the cells (39). Earlier studies have shown that the loss of 95% of this enzyme activity by 6-aminopenicillanic acid treatment of exponentially growing cells did not affect cell growth or the extent of peptidoglycan cross-linking (7, 38). The cross-linking index of AA109 strain (dacA) determined by RP-HPLC is comparable to that of the wild type (Table 3). From the present results, it is not clear whether PBP 5 removes both d-alanine residues or only the final alanine and then another ld-carboxypeptidase gives the final mature muropeptides with tripeptide side chains. The presence of muropeptides with tripeptide side chains suggests that there is at least one other dd-carboxypeptidase involved in peptidoglycan maturation, although its action is not sufficient to give the mature structure. Membranes isolated from exponentially growing cells of the dacA mutant contain about 5% of the total d-alanine carboxypeptidase activity found in the wild type (39). Alternatively, disaccharide tripeptide could be incorporated directly into the peptidoglycan from lipid linked N-acetylmuramyl tripeptide, as has been demonstrated in E. coli (42).
This study has demonstrated that peptidoglycan structure is determined by many biosynthetic and autolytic reactions. The recent release of the B. subtilis genome has revealed a number of novel putative peptidoglycan hydrolases and PBPs homologous to known enzymes in B. subtilis or other organisms (23). Analysis of peptidoglycan structure in single and multiple mutants of these putative enzymes will reveal their combined roles in wall architecture. Muropeptide analysis has shown the presence of hitherto unknown features of peptidoglycan in B. subtilis. The spatial distribution of muropeptides within the wall and the dynamics of cell wall architecture during growth and division is the subject of our continuing research.
ACKNOWLEDGMENTS
This work was supported by the BBSRC (funding to A.A. and M.P.W. and for the NMR spectrometer), Royal Society (S.J.F.), the Fonds zur Förderung der wissenschaftlichen Forschung (MALDI MS, grant 11183 to G.A.) and the ARC Programme (UK/Austria travel fund).
REFERENCES
- 1.Archibald A R, Hancock I C, Harwood C R. Cell wall structure, synthesis, and turnover. In: Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria, biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 381–410. [Google Scholar]
- 2.Atrih A, Zöllner P, Allmaier G, Foster S J. Structural analysis of Bacillus subtilis 168 endospore peptidoglycan and its role during differentiation. J Bacteriol. 1996;178:6173–6183. doi: 10.1128/jb.178.21.6173-6183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atrih A, Zöllner P, Allmaier G, Williamson M P, Foster S J. Peptidoglycan dynamics during germination of Bacillus subtilis 168 endospores. J Bacteriol. 1998;180:4603–4612. doi: 10.1128/jb.180.17.4603-4612.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atrih A, Bacher G, Körner R, Allmaier G, Foster S J. Structural analysis of Bacillus megaterium KM spores peptidoglycan and its dynamics during germination. Microbiology. 1999;145:1033–1041. doi: 10.1099/13500872-145-5-1033. [DOI] [PubMed] [Google Scholar]
- 5.Blackman S A, Smith T J, Foster S J. The role of autolysins during vegetative growth of Bacillus subtilis 168. Microbiology. 1998;145:57–65. doi: 10.1099/00221287-144-1-73. [DOI] [PubMed] [Google Scholar]
- 6.Blasco B A, Pisabarro A G, de Pedro M A. Peptidoglycan biosynthesis in stationary-phase cells of Escherichia coli. J Bacteriol. 1988;170:5224–5228. doi: 10.1128/jb.170.11.5224-5228.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blumberg P M, Strominger J L. Inactivation of d-alanine carboxypeptidase by penicillins and cephalosporins is not lethal in Bacillus subtilis. Proc Natl Acad Sci USA. 1970;68:2814–2817. doi: 10.1073/pnas.68.11.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dougherty T J. Analysis of Neisseria gonorrhoeae peptidoglycan by reverse-phase high-pressure liquid chromatography. J Bacteriol. 1985;163:69–74. doi: 10.1128/jb.163.1.69-74.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiedler F, Glaser L. Assembly of bacterial cell walls. Biochem Biophys Acta. 1973;300:467–485. doi: 10.1016/0304-4157(73)90016-6. [DOI] [PubMed] [Google Scholar]
- 10.Fordham W D, Gilvarg C. Kinetics of cross-linking of peptidoglycan in Bacillus megaterium. J Biol Chem. 1974;249:2478–2482. [PubMed] [Google Scholar]
- 11.Foster S J. The role and regulation of cell wall structural dynamics during differentiation of endospore-forming bacteria. J Appl Bacteriol. 1994;76:25–39. doi: 10.1111/j.1365-2672.1994.tb04355.x. [DOI] [PubMed] [Google Scholar]
- 12.Gally D L, Hancock I C, Harwood C R, Archibald A R. Cell wall assembly in Bacillus megaterium: incorporation of new peptidoglycan by a monomer addition process. J Bacteriol. 1991;173:2548–2555. doi: 10.1128/jb.173.8.2548-2555.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghuysen J-M. Serine β-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 14.Glauner B, Höltje J-V. Growth pattern of the murein sacculus of Escherichia coli. J Biol Chem. 1990;265:18988–18996. [PubMed] [Google Scholar]
- 15.Glauner B, Höltje J-V, Schwarz U. The composition of the murein of Escherichia coli. J Biol Chem. 1988;263:10088–10095. [PubMed] [Google Scholar]
- 16.Grant W D, Wicken A J. Autolysis of cell walls of Bacillus stearothermophilus B65 and the chemical structure of the peptidoglycan. Biochem J. 1970;118:859–868. doi: 10.1042/bj1180859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Höltje J-V. A hypothetical holoenzyme involved in the replication of the murein sacculus of Escherichia coli. Microbiology. 1996;142:1911–1918. doi: 10.1099/13500872-142-8-1911. [DOI] [PubMed] [Google Scholar]
- 18.Höltje J-V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs C, Joris B, Jamin M, Klarsov K, van Heijenoort J, Parks J T, Normark S, Frere J M. AmpD, essential for both β-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetyl muramyl-l-alanine amidase. Mol Microbiol. 1995;15:553–559. doi: 10.1111/j.1365-2958.1995.tb02268.x. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs C, Frere J M, Normark S. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible β-lactamase resistance in Gram-negative bacteria. Cell. 1997;88:823–832. doi: 10.1016/s0092-8674(00)81928-5. [DOI] [PubMed] [Google Scholar]
- 21.Kato K, Strominger J L, Kotani S. Structure of the cell wall of Corynebacterium diphtheriae. I. Mechanism of hydrolysis by the L-3 enzyme and the structure of the peptide. Biochemistry. 1968;7:2762–2773. doi: 10.1021/bi00848a010. [DOI] [PubMed] [Google Scholar]
- 22.Kitano K, Tuomanen E, Tomasz A. Transglycosylase and endopeptidase participate in the degradation of murein during autolysis of Escherichia coli. J Bacteriol. 1986;167:759–765. doi: 10.1128/jb.167.3.759-765.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunst F, Ogasawara N, Moszer I, Albertini A M, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 24.Kunst F, Rapoport G. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J Bacteriol. 1995;177:2403–2407. doi: 10.1128/jb.177.9.2403-2407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu T Y, Gottschlick E C. Muramic acid phosphate as a component of the mucopeptide of Gram-positive bacteria. J Biol Chem. 1967;242:471–476. [PubMed] [Google Scholar]
- 26.Margot P, Pagni M, Karamata D. Bacillus subtilis 168 lytF encodes a γ-d-glutamate-meso-diaminopimelate muropeptidase expressed by the alternative vegetative sigma factor, ςD. Microbiology. 1999;145:57–65. doi: 10.1099/13500872-145-1-57. [DOI] [PubMed] [Google Scholar]
- 27.Martin H H, Gmeiner J. Modification of peptidoglycan structure by penicillin action in the cell walls of Proteus mirabilis. Eur J Biochem. 1979;95:487–495. doi: 10.1111/j.1432-1033.1979.tb12988.x. [DOI] [PubMed] [Google Scholar]
- 28.Mauck J, Glaser L. On the mode of in vivo assembly of the cell wall of Bacillus subtilis. J Biol Chem. 1972;247:1180–1187. [PubMed] [Google Scholar]
- 29.Murray T, Popham D L, Pearson C B, Hand A R, Setlow P. Analysis of outgrowth of Bacillus subtilis spores lacking penicillin-binding protein 2a. J Bacteriol. 1998;180:6493–6502. doi: 10.1128/jb.180.24.6493-6502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popham D L, Gilmore M E, Setlow P. Roles of low-molecular weight penicillin-binding proteins in Bacillus subtilis spore peptidoglycan synthesis and spore properties. J Bacteriol. 1999;181:126–132. doi: 10.1128/jb.181.1.126-132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popham D L, Setlow P. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis pbpE operon, which codes for penicillin-binding protein 4* and an apparent amino acid racemase. J Bacteriol. 1993;175:2917–2925. doi: 10.1128/jb.175.10.2917-2925.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Popham D L, Setlow P. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis pbpF gene, which codes for a putative class A high-molecular-weight penicillin-binding protein. J Bacteriol. 1993;175:4870–4876. doi: 10.1128/jb.175.15.4870-4876.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popham D L, Setlow P. Cloning, nucleotide sequence, mutagenesis, and mapping of the Bacillus subtilis pbpD gene, which codes for penicillin-binding protein 4. J Bacteriol. 1994;176:7197–7205. doi: 10.1128/jb.176.23.7197-7205.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Popham D L, Setlow P. Cloning, nucleotide sequence, and mutagenesis of the Bacillus subtilis ponA operon, which codes for penicillin-binding protein (PBP) 1 and a PBP-related factor. J Bacteriol. 1995;177:326–335. doi: 10.1128/jb.177.2.326-335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popham D L, Setlow P. Phenotypes of Bacillus subtilis mutants lacking multiple class A high-molecular-weight penicillin binding proteins. J Bacteriol. 1996;178:2079–2085. doi: 10.1128/jb.178.7.2079-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quintela J C, Pittennauer E, Allmaier G, Aran V, de Pedro M A. Structure of peptidoglycan from Thermus thermophilus HB8. J Bacteriol. 1995;177:4947–4962. doi: 10.1128/jb.177.17.4947-4962.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogers H J, Taylor C, Ryter S, Ward J B. Purification and properties of autolytic endo-β-N-acetylglucosaminidase and the N-acetyl muramoyl-l-alanine amidase from Bacillus subtilis strain 168. J Gen Microbiol. 1984;130:2395–2402. doi: 10.1099/00221287-130-9-2395. [DOI] [PubMed] [Google Scholar]
- 38.Sharpe A, Blumberg P M, Strominger J L. d-Alanine carboxypeptidase and cell wall cross-linking in Bacillus subtilis. J Bacteriol. 1974;117:926–927. doi: 10.1128/jb.117.2.926-927.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Todd J A, Roberts A N, Johnstone K, Piggot P J, Winter G, Ellar D J. Reduced heat resistance of mutant spores after cloning and mutagenesis of the Bacillus subtilis gene encoding penicillin-binding protein 5. J Bacteriol. 1986;167:257–264. doi: 10.1128/jb.167.1.257-264.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tuomanen E, Schwartz J, Sande S, Light K, Gage D. Unusual composition of peptidoglycan in Bordetella pertussis. J Biol Chem. 1989;264:11093–11098. [PubMed] [Google Scholar]
- 41.van Heijenoort J. Biosynthesis of the bacterial peptidoglycan unit. In: Ghuysen J M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier; 1994. pp. 39–54. [Google Scholar]
- 42.van Heijenoort Y, Gomez M, Derrien M, Ayala J, van Heijenoort J. Membrane intermediates in the peptidoglycan metabolism of Escherichia coli: possible roles of PBP 1b and PBP 3. J Bacteriol. 1992;174:3549–3559. doi: 10.1128/jb.174.11.3549-3557.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warth A D, Strominger J L. Structure of the peptidoglycan from vegetative cell wall of Bacillus subtilis. Biochemistry. 1971;10:4349–4358. doi: 10.1021/bi00800a001. [DOI] [PubMed] [Google Scholar]
- 44.Yanouri A, Richard R A, Errington J, Buchanan C E. Cloning and sequencing of the cell division gene pbpB, which encodes penicillin-binding protein 2B in Bacillus subtilis. J Bacteriol. 1993;175:7604–7616. doi: 10.1128/jb.175.23.7604-7616.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young F E. Variation in chemical composition of cell wall of Bacillus subtilis during growth in different media. Nature. 1965;207:104–105. doi: 10.1038/207104b0. [DOI] [PubMed] [Google Scholar]
- 46.Zipperle G F, Ezzell J W, Doyle R J. Glucosamine substitution and muramidase susceptibility in Bacillus anthracis. Can J Microbiol. 1984;30:553–559. doi: 10.1139/m84-083. [DOI] [PubMed] [Google Scholar]