Abstract
Background –
Spontaneous coronary artery dissection (SCAD) is a cause of acute coronary syndrome that predominantly affects women. Its pathophysiology remains unclear but connective tissue disorders (CTD) and other vasculopathies have been observed in a number of SCAD patients. A genetic component for SCAD is increasingly appreciated, although few genes have been robustly implicated. We sought to clarify the genetic etiology of SCAD using targeted and genome-wide methods in a cohort of sporadic cases to identify both common and rare disease-associated variants.
Methods –
A cohort of 91 unrelated sporadic SCAD cases was investigated for rare, deleterious variants in genes associated with either SCAD or CTD, while new candidate genes were sought using rare variant collapsing analysis and identification of novel loss-of-function variants in genes intolerant to such variation. Finally, two SCAD polygenic risk scores (PRS) were applied to assess the contribution of common variants.
Results –
We identified 10 cases with at least one rare, likely disease-causing variant in CTD-associated genes, although only one had a CTD phenotype. No genes were significantly associated with SCAD from genome-wide collapsing analysis, however, enrichment for TGF-β signaling pathway genes was found with analysis of 24 genes harboring novel loss-of-function variants. Both PRS demonstrated that sporadic SCAD cases have a significantly elevated genetic SCAD risk compared to controls.
Conclusions –
SCAD shares some genetic overlap with CTD, even in the absence of any major CTD phenotype. Consistent with a complex genetic architecture, SCAD patients also have a higher burden of common variants than controls.
Keywords: Spontaneous coronary artery dissection, genome sequencing, connective tissue disorders, rare variants
Introduction
Spontaneous coronary artery dissection (SCAD) is a potentially fatal disorder that predominantly affects women. It manifests as an acute coronary syndrome (myocardial infarction or unstable angina) or sudden cardiac death. SCAD arises due either to rupture of the vasa vasorum or to an intimal tear.1 The resulting intramural hematoma causes the wall of the culprit vessel to bulge into the lumen, thereby obstructing blood flow to the myocardium and culminating in myocardial injury. SCAD is the cause of up to 4% of acute coronary syndromes and is the primary cause of pregnancy-related myocardial infarction.2 Often those affected have few of the traditional risk factors associated with atherosclerotic coronary artery disease (e.g. dyslipidemia, diabetes, obesity).2
Factors known to precipitate or predispose to SCAD include pregnancy, the presence of extra-coronary vascular disorders (principally fibromuscular dysplasia [FMD]), hypertension, inflammatory diseases, migraine and major physical or emotional stress.1 Over the last decade SCAD cases have been reported in patients with connective tissue disorders (CTD)3 and previously undiagnosed CTD found in SCAD patients.4–6 CTD feature a vascular element, and a growing body of evidence now exists to link SCAD with other vasculopathies, such as FMD and migraine. Moreover, a high proportion of FMD-free SCAD cases show vascular abnormalities.7 Therefore, an overlap between SCAD and CTD, as well as other vasculopathies, merits further investigation.
A genetic component to SCAD etiology is suggested by the identification of several multigenerational SCAD pedigrees.8 A single nucleotide polymorphism (SNP) in the PHACTR1/EDN1 locus is associated not only with an increased risk of SCAD, but also an increased risk of migraine, FMD and cervical artery dissection.9, 10 A further six common independent loci are now associated with SCAD.9, 11, 12 Recently, rare variants in three genes with apparently unrelated function have been proposed to contribute specifically to SCAD pathophysiology, with TSR1 identified in a sporadic SCAD cohort;13 TLN1 found initially in a SCAD pedigree and then in sporadic cases;14 and a single rare F11R variant in an affected mother-daughter pair.15 Taken together, these findings suggest that the genetic architecture underlying SCAD is complex, involving both rare and common variants. Moreover, there appears to be an overlap between potential SCAD genes and those underlying CTD and vasculopathies.16 However, the prevalence of variants in CTD- or SCAD-related candidate genes, and the role of either common or rare variants across SCAD cohorts, has not yet been established. Using whole genome sequencing (WGS) of a cohort of 91 unrelated SCAD patients, we sought to: (i) identify potentially damaging coding and non-coding rare genetic variants in SCAD- and CTD-associated genes even in the absence of a CTD phenotype; (ii) further investigate the role of common variants; and (iii) identify candidate genes that might play a role in SCAD etiology.
Methods
Methods are available in the Supplementary Material. The study was approved by the St. Vincent’s Hospital Human Research Ethics Committee (HREC/16/SVH/338, protocol number SVH 16/245) and conducted in accordance with the Australian National Health and Medical Research Council’s National Statement on Ethical Conduct in Human Research and the CPMP/ICH Note for Guidance on Good Clinical Practice. All patients gave signed informed consent prior to enrolment into the study and SCAD diagnosis was confirmed by expert review of coronary angiogram images (blinded to the results of the genetic analysis).
Results
Clinical cohort characteristics
The cohort of 91 unrelated SCAD cases subjected to WGS (clinical characteristics summarized in Table 1) was 91.2% (83) female, predominantly European (88; 96.7%), and had an average age at the first SCAD event of 45 years (range: 24 – 69). The majority of patients had only one SCAD event, with 11 (12.1%) having suffered multiple episodes. Migraine was present in 42 (46.2%) patients and FMD in 12 of 43 patients screened (27.9%). Stressors at the time of SCAD were common, with 46 (50.5%) patients reporting chronic emotional stress, 14 (15.4%) reporting physical stress, and four (4.4%) reporting acute emotional stress. The most common cardiovascular risk factor was hypertension (18.7%, n = 17). Two (2.2%) cases had prior CTD diagnoses: vascular Ehlers Danlos syndrome (vEDS) and Alport syndrome (AS).
Table 1.
Cohort clinical characteristics in the total SCAD cohort and in subsets with/without CTD variants.
| Total SCAD cohort (n = 91) | SCAD cases carrying Tier 1 or Tier 2 CTD gene variants (n = 17) | SCAD cases not carrying Tier 1 or Tier 2 CTD variants (n = 74) | P-value† | |
|---|---|---|---|---|
| Mean age at first SCAD (y) | 45.4 | 40.8 | 46.5 | 0.028 |
| Female % | 91.2% | 94.1% | 90.5% | 1 |
| Mean Height (cm) | 167 | 167 (n = 16) | 167 (n = 72) | 0.92 |
| Mean BMI | 26.4 | 27.4 (n = 16) | 26.2 (n = 72) | 0.55 |
| Pregnancy-related SCAD* % (n/n female) | 12% (10/83) | 25% (4/16) | 9% (6/67) | 0.09 |
| Mean number of live births | 2.13 (n = 83) | 2.19 (n = 16) | 2.12 (n = 67) | 0.84 |
| >1 SCAD episode % (n) | 12.1% (11) | 23.5% (4) | 9.5% (7) | 0.21 |
| Comorbidities | ||||
| Connective tissue disorder % (n) | 2% (2) | |||
| Kidney disorder % (n) | 1% (1) | |||
| FMD % (n/n screened) | 27.9% (12/43) | 60% (3/5) | 23.7% (9/38) | 0.35 |
| Migraines % (n) | 46.2% (42) | 41.2% (7) | 47.3% (35) | 0.79 |
| T2 diabetes % (n) | 3.3% (3) | |||
| History of high cholesterol % (n) | 14.3% (13) | 17.6% (3) | 13.5% (10) | 0.70 |
| History of hypertension % (n) | 18.7% (17) | 11.8% (2) | 20.3% (15) | 0.51 |
| Physical stress % (n) | 15.4 (14) | 11.8% (2) | 16.2% (12) | 1 |
| History of anxiety/depression % (n) | 23.1% (21) | 35.3% (6) | 20.3% (15) | 0.21 |
| Chronic emotional stress % (n) | 50.5% (46) | 41.2% (7) | 52.7% (39) | 0.43 |
| Acute emotional stress % (n) | 4.4% (4) | |||
| Heart disease % (n) | 5.5% (5) | |||
| Dissection of the carotid or vertebral arteries % (n) | 6.6% (6) | |||
| Aneurysm % (n) | 4.4% (4) | |||
| Stroke % (n) | 2.2% (2) | |||
| Deep vein thrombosis % (n) | 3.3% (3) | |||
| Family history | ||||
| Heart disease % (n) | 72.5% (66) | 64.7% (11) | 74.3% (55) | 0.55 |
| Dissection of any artery % (n) | 4.4% (4) | |||
| Connective tissue disorder % (n) | 1.1% (1) | |||
| Aneurysm % (n) | 24.2% (22) | 35.3% (6) | 21.6% (16) | 0.34 |
| FMD % (n) | 2.2% (2) | |||
FMD, Fibromuscular dysplasia.
Gestation through to 5 months post-partum
P-values refer to comparisons between variant carriers and non-carriers, determined by t-tests for continuous variables and Fisher’s Exact for proportions, which were tested only when at least 10 cases meeting the criteria were observed.
Tier 1 and Tier 2 gene screen
We created a two-tiered list of 90 genes based on eight diagnostic gene panels for CTD and vasculopathies, and on previous publications that identified gene mutations in SCAD patients (accessed December 2019, Supplementary Table I). Tier 1 consisted of 75 genes that are associated with CTD with high confidence and Tier 2 contained 15 lower confidence CTD genes and genes found to be associated with SCAD (Supplementary Table II). The predicted pathogenicity of rare variants was assessed according to American College of Medical Genetics (ACMG) criteria. SCAD cases carrying clinically actionable variants in CTD-associated genes were contacted to ascertain phenotypic features relevant to the indicated CTD disorder (Supplementary Table III)
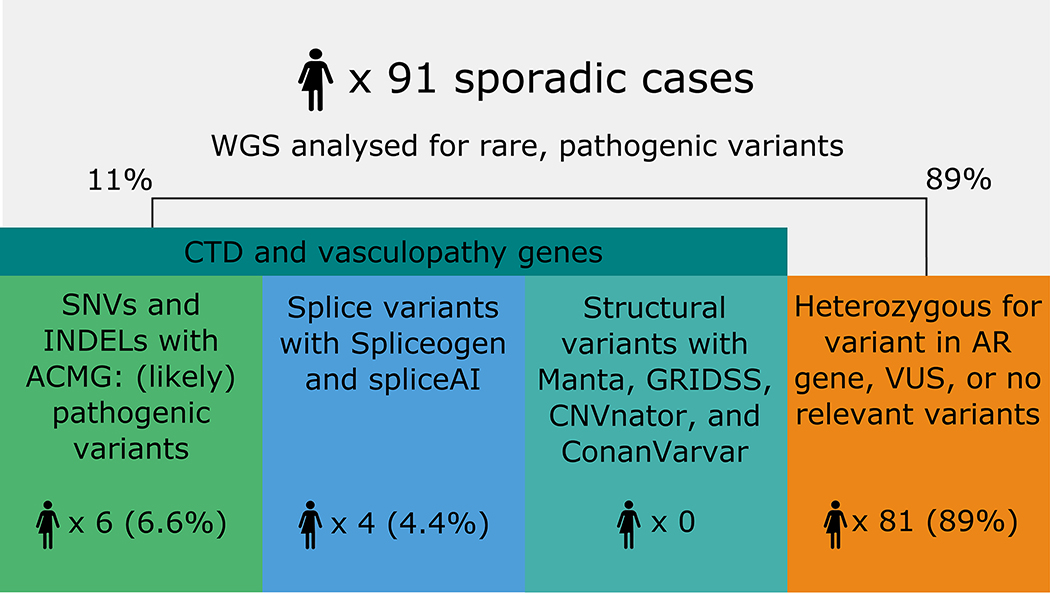
A total of six (6.6%) cases had a pathogenic or likely pathogenic variant in a Tier 1 gene (ALDH18A1, n=1; COL3A1, n=1; COL4A1, n=2; FBN1, n=2), implicating 4/75 genes in this Tier (Figure 1; Table 2). One additional case had a likely pathogenic variant in a Tier 2 gene, ACVR1, and one case, 090, harbored two likely pathogenic variants, one in ALDH18A1 (Tier 1) and one in ACVR1 (Tier 2, Table 2). All cases were heterozygous for their respective variants. A further two cases with heterozygous variants were identified in Tier 1 genes that cause disease in an autosomal recessive (AR) fashion (AEBP1, n=1; and SLC2A10, n=1) (Supplementary Table IV). Also of interest, a heterozygous variant in a Tier 2 gene, ABCC6 (NM_001171.5:c.2787+1G>T), was identified in two unrelated individuals (Supplementary Table IV), although ABCC6 predominantly causes AR disease. Only one of the 11 cases carrying potential disease-causing genes had any phenotypic evidence for the cognate CTD, although five of these cases had at least one second-degree relative with an aneurysm or dissection elsewhere (clinical details in Supplementary Table V). Variants of uncertain significance (VUS) were identified in 40/80 remaining cases, 35 of which had at least one VUS in a Tier 1 gene (Supplementary Table VI).
Figure 1.
Summary of pathogenic single nucleotide variants (SNVs) and insertion/deletions (INDELs) across CTD and vasculopathy genes identified in SCAD cases.
Table 2.
Likely pathogenic and pathogenic variants in both Tier 1 and Tier 2 genes
| Case | Gene | Nucleotide variant | Amino acid variant | ACMG classification* | gnomAD MAF^ | Varsome ACMG criteria† |
|---|---|---|---|---|---|---|
| Tier 1 | ||||||
| 090 | ALDH18A1 | NM_002860.4:c.1367G>A | p.Arg456His | Likely pathogenic | 0.00004600 | PM1, PM2, PP2, PP3 |
| 146 | COL3A1 | NM_000090.3:c.2798dupG | p.Ser934IlefsTer35 | Pathogenic | - | PVS1, PM2, PP3 |
| 171 | COL4A1 | NM_001845.6:c.3592G>A | p.Gly1198Arg | Likely pathogenic | - | PM1, PM2, PP2, PP3 |
| 170 | COL4A1 | NM_001845.6:c.4877C>G | p.Ala1626Gly | Likely pathogenic | 0.00000707 | PM1, PM2, PP2, PP3 |
| 115 | FBN1 | NM_000138.4:c.793A>T | p.Thr265Ser | Likely pathogenic | - | PM1, PM2, PP2, PP3 |
| 004 | FBN1 | NM_000138.4:c.256C>T | p. Arg86Trp | Likely pathogenic | 0.00001195 | PM1, PM2, PP2, PP3 |
| Tier 2 | ||||||
| 090 | ACVR1 | NM_001111067.4:c.1298A>G | p.Asp433Gly | Likely pathogenic | 0.00000398 | PM1, PM2, PP2, PP3 |
| 091 | ACVR1 | NM_001111067.4:c.1265G>A | p.Gly422Asp | Likely pathogenic | - | PM1, PM2, PP2, PP3 |
ACMG, American College of Medical Genetics; MAF, minor allele frequency.
ACMG classifications are automatically derived from Varsome and manually evaluated
gnomAD MAF refers to all populations.
ACMG interpretation criteria, detailed in Supplementary Table XV
Nine variants within Tier 1 genes were predicted to alter splicing. Four of these, each occurring in one of four cases (4.4%), are heterozygous variants in genes causing autosomal dominant (AD) disease (Table 3). The remaining five variants are heterozygous but in Tier 1 genes causing AR disease. These were identified in four additional cases and in one case with a splice-altering variant in an AD Tier 1 gene (Supplementary Table IV). One case, 091, had both a splice variant in PRKG1 and a likely pathogenic missense variant in ACVR1 (Tier 2; Table 2). Two cases showed some phenotypic evidence of the cognate CTD upon follow-up even though one of them only had a heterozygote variant in an AR gene (ADAMTS2) (Supplementary Table V). A third case had a second degree relative with an extra-coronary dissection, when again carrying a heterozygote variant in an AR gene (ADAMTS10) (Supplementary Table V). We also identified ultra-rare variants (with MAF <0.001) that are potentially splice-altering, although with a lower SpliceAI score (0.2 – 0.4, Supplementary Table VII). No abnormal splicing caused by the nine variants was expected to be able to be validated in whole blood as only two of the nine variants were predicted to escape nonsense-mediated decay, while all nine were lowly expressed in blood (Table 3).
Table 3.
Splice-altering variants identified within Tier 1 genes
| Case | Gene | Nucleotide variant | Predicted effect | gnomAD MAF* | GnomAD homozygotes | SpliceAI score | GTEx whole blood median TPM | Predicted to undergo NMD |
|---|---|---|---|---|---|---|---|---|
| 091 | PRKG1 | NM_006258.4:c.762+10598G>T | Donor gain | 0.00312 | 1 | 0.88 | 0.3744 | N |
| 074 | FBN2 | NM_001999.4:c.2095+1680T>C | Acceptor gain | 0.002548 | 0 | 0.48 | 0 | Y |
| 046 | MFAP5 | NM_003480.4:c.410-286T>C | Donor gain | 0.0003821 | 0 | 0.46 | 0.1031 | N |
| 108 | ABL1 | NM_007313.2:c.1571-130C>T | Acceptor gain | 0.001274 | 0 | 0.4 | 4.057 | Y |
MAF, minor allele frequency; TPM, transcripts per million; NMD, nonsense-mediated decay.
gnomAD MAF refers to overall frequencies across populations.
No structural variants (SV) were identified that directly impacted an exon within any of the 90 Tier 1 and Tier 2 genes investigated, nor caused a deletion of any of these genes. One VUS was identified in case 079, however: a heterozygous 60kb duplication of the entirety of the Tier 2 gene, MYLK2, together with FOXS1 and a partial duplication of DUPS15 and TPX2 (GRCh37: chr20:30387503–30447873).
In total, 17 cases were identified that carried potential disease-causing variants in Tier 1 and 2 genes that could underlie SCAD etiology, including cases heterozygous for variants in genes causing an AR CTD. Upon follow up, none of these patients had a prior CTD diagnosis, nor were any found to meet the criteria for the appropriate CTD diagnosis (Supplementary Table V). Clinical characteristics of these cases were compared to the other members of the SCAD cohort; variant carriers were significantly younger on average than non-carriers (mean age at first SCAD in carriers 40.8 years vs 46.5 years, p = 0.028, Table 1). No other differences were identified.
Rare Variant Collapsing Analysis
The general utility of our Tier 1 and Tier 2 gene lists in distinguishing SCAD from healthy controls was examined by rare variant collapsing analysis taking all 90 genes as the sole analysis unit. Rare variants (gnomAD MAF < 0.01) were included for analysis if the consequence was a frameshift or nonsense alteration, an alteration of a canonical splice site, or a missense mutation with phred-scaled CADD > 12 and PolyPhen HDIV “P” or “D”. Overall, a larger proportion of SCAD cases (n=88) carried at least one qualifying variant within this set of genes than did controls from the Medical Reference Genome Bank (MGRB) (n=1127) (Table 4, 86.4% vs 76.8%, p-value = 0.022), highlighting the general association of these genes with SCAD. When each tier was considered as a single unit for analysis, Tier 1 (75 genes) showed a significant enrichment of SCAD cases carrying at least one qualifying variant (85.2% vs 72% of controls, p-value = 0.0037), while Tier 2 (15 genes) did not (p-value = 0.15; Supplementary Table VIII). Thus, the higher confidence Tier 1 gene set is primarily driving the association of SCAD with CTD and vasculopathies.
Table 4.
Rare variant collapsing analysis results for the entire CTD and vasculopathy gene list or tiers therein
| Gene list | SCAD gene variant carriers n (%), n = 88 | Control gene variant carriers n (%), n = 1127 | OR | P-value* |
|---|---|---|---|---|
| All genes (n = 90) | 76 (86.4%) | 865 (76.8%) | 1.92 | 0.022 |
| Tier 1 genes (n = 75) | 75 (85.2%) | 812 (72%) | 2.24 | 0.0037 |
| Tier 2 genes (n = 15) | 15 (17%) | 142 (12.6%) | 1.42 | 0.15 |
OR, odds ratio: the odds of carrying variants in the SCAD group divided by the odds of carrying variants in the control group, calculated using a 2×2 contingency table.
P-values determined by one-sided Fisher’s exact test
Genome-wide Analysis
To broaden the search beyond CTD and vasculopathy genes, we used a rare variant collapsing analysis to investigate all genes, genome-wide, for a novel potential association with SCAD. No single gene had a significantly greater proportion of cases carrying qualifying variant(s) compared to the control cohort after multiple testing correction. However, 375 genes were nominally significantly enriched in SCAD cases compared to controls (Supplementary Table IX). Gene ontology analysis for biological processes did not identify any significant associations for these 375 genes.
We interrogated our dataset for novel loss of function (LoF) variants or likely damaging SVs within LoF intolerant genes. Twenty-four novel LoF variants were found across 24 different genes, in 22 cases (Supplementary Table X). For eight of the identified genes and variants, the carrier was found to also have one or more candidate variants within genes on our CTD list. Pathway analysis of these 24 genes showed a significant overrepresentation within the TGF-β signaling pathway (WP366, adjusted p-value = 0.0005). Seven cases each had one unique SV with exonic impact affecting a total of 10 LoF-intolerant genes, however, none of these genes were affected by SVs in more than one of the seven cases (Supplementary Table XI). No genes were found with both SVs and LoF variants. These 24 genes were assessed for LoF variants in a validation cohort of 384 UK SCAD samples16, which identified one further novel LoF variant in ACACA (Supplementary Table X).
Recently, reports have suggested a possible connection between the Fragile X premutation and a predisposition to SCAD.17 Hence, we applied three bioinformatics tools tailored to identify known pathogenic short tandem repeats (STRs) in WGS data. No SCAD cases carried a premutation length expansion of the Fragile X STR, nor any other known disease-associated STRs.
Polygenic Risk Score (PRS)
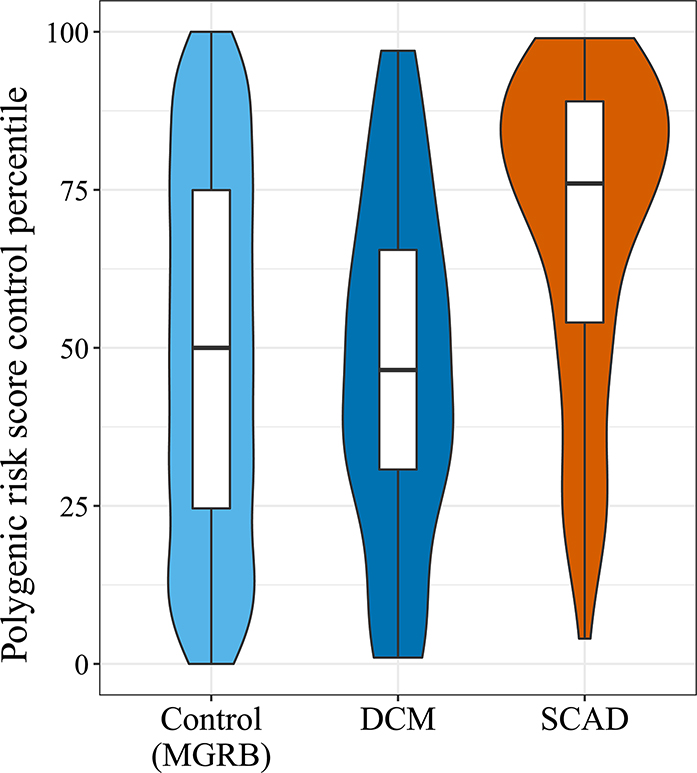
Using a recently constructed PRS for SCAD11 (Supplementary Table XII), we found significantly higher scores for our European SCAD cases (n = 88) versus MGRB controls (n = 1127), with the median SCAD score in the 76th percentile of control scores (p-value = 1.372e-09, Figure 2). To confirm that the SCAD PRS was specific to SCAD patients and not indicative of a general cardiovascular signal, the SCAD PRS was also calculated for a cohort of familial dilated cardiomyopathy (DCM) probands (n = 76). SCAD patients had significantly higher median scores compared to DCM patients (p-value = 2.08e-07), with the median score in DCM patients in the 46th control percentile, thereby not significantly different from control scores (p-value = 0.291). Similar results were found when using a PRS constructed from the five single nucleotide polymorphisms (SNPs) identified by Turley et al12 (Supplementary Figure I, Supplementary Table XII), with the two scores having three SNPs in common. Overall, this indicates that individual SCAD cases had a higher total burden of SCAD-associated common SNPs than controls.
Figure 2.
Polygenic Risk Score percentile amongst SCAD and dilated cardiomyopathy patients relative to MGRB control samples. Control scores were used to create a reference distribution of scores. Violin plots indicate the percentile score distribution relative to the control distribution, while boxplots indicate the score quartiles.
Discussion
The genetic architecture of SCAD is not well understood. In this study we performed WGS in a cohort of sporadic SCAD cases. Using a two-tiered candidate gene approach, we identified rare, likely pathogenic variants in 11% of our cohort (AD variants in Tier 1 genes), candidate disease-causing variants in another 9.9% of our cohort, as well as potential new disease genes. Our analysis included screening for single nucleotide variants, insertion/deletions, structural variants, splice-altering variants including deep intronic variants, and STR expansions, making this the first study to interrogate WGS data for all types of variation to identify potential causes of SCAD. Of interest, our yield for likely pathogenic or pathogenic variants in CTD-associated genes in SCAD patients is higher (11%) than reported previously (3.6–8.2%)4, 5,16
Epidemiologically, extra-coronary vascular disorders, such as CTD and vasculopathies, have often been reported in SCAD patients, indicating a potential common genetic etiology.3–6 Six of our SCAD patients carried pathogenic or likely pathogenic variants in four Tier 1 genes associated with CTD, however, none of these cases had a prior CTD diagnosis, nor did they meet the clinical criteria for CTD diagnosis upon follow-up. This suggests that SCAD is potentially part of the phenotypic spectrum of these disorders and may occur in people with subclinical CTD. A similar hypothesis has been advanced in relation to cervical artery dissection, where the majority of apparently sporadic cases show connective tissue anomalies, some of which appear severe yet occur in the absence of a clinically diagnosable CTD.18 In a few cases, where the CTD is inherited in an AR pattern, the heterozygote variants we identified may be dominant with respect to SCAD only, although more evidence of patients carrying these variants would be required. Variant carriers were younger than non-carriers at their first SCAD, as observed previously,5 which suggests that their variant(s) is contributing to their SCAD phenotype.
Likely pathogenic FBN1 variants were seen in two female cases, both lacking a previous Marfan syndrome (MFS) diagnosis or any Marfanoid features at follow-up. SCAD has previously been reported in MFS patients,4 however, there has only been one report of SCAD in a patient lacking a diagnosis of MFS prior to the discovery of an FBN1 mutation, albeit of unclear pathogenicity.19
Likely pathogenic variants in exons 42 and 51 of COL4A1 were found in two cases, both of whom were 35 years or younger at first SCAD, each suffering one pregnancy-associated SCAD and one additional SCAD event. While the case carrying c.3592G>A (exon 51) reported no further CTD features bar an abdominal aneurysm in her maternal grandfather, the case carrying the novel variant c.3592G>A (exon 42) reported a possible carotid dissection and strokes in multiple family members including an intracerebral hemorrhage in her son during the 3rd trimester of pregnancy, leading to cerebral palsy. Mutations in COL4A1 may cause brain small vessel disease 1 (BSVD1) or hereditary angiopathy with nephropathy, aneurysms, and muscle cramps and pathogenic COL4A1 variants have also been reported in cervical artery dissection families.20 Variants causing BSVD1 have almost exclusively been identified between exons 25 and 51.21 Confirmation of the novel exon 42 variant in both the SCAD-affected mother and her son suggests this variant may indeed be responsible for the vascular events in this family.
A heterozygous frameshift insertion leading to premature termination of the protein encoded by COL3A1 was identified in one male, as we have previously reported.16 Although variants in COL3A1 are the major genetic cause of vascular EDS, an autosomal dominant disorder, our patient did not exhibit any clinical characteristics of EDS upon follow-up. Different types of mutation in COL3A1 cause vEDS of varying severity: null variants, similar to the one described here, have been linked to a less typical, “subclinical” phenotype presenting with fewer diagnostic criteria than is typical of vEDS patients and showing variable penetrance.22, 23 Indeed, in one study of null COL3A1 variant carriers, 9/18 cases had arterial dissection as their initial complication, and generally no minor characteristics of disease.22 Reportable COL3A1 variants have been found previously in SCAD patients with only vascular characteristics.4, 6 These data suggest that null COL3A1 variants may have an important role in SCAD, which has possible treatment implications. β-blockers, for example, are often prescribed for both vEDS and SCAD patients, but differences in effectiveness of various β-blockers in vEDS/COL3A1 variant carriers and mouse models have been reported.24, 25 SCAD patients found to have a COL3A1 mutation may benefit from a tailored treatment plan.
In one case, a likely pathogenic missense variant was identified in ALDH18A1. To date, few such individuals have been identified and little is known about cardiovascular complications in the ALDH18A1-linked CTD, cutis laxa. However, dominant-negative mutation carriers with severe disease including arterial tortuosity are known26, as are recessive cutis laxa patients with cardiovascular abnormalities including aneurysms.27
Very few of our cohort had a pre-existing CTD diagnosis. Only one case was known to have vEDS and another, Alport syndrome (AS). The genes known to cause vEDS are included on most CTD panels and were thus on our gene lists. None of the three genes associated with AS - COL4A3, COL4A4, and COL4A5 - featured in any of our gene list sources, yet SCAD has been previously observed in two patients with AS3, 28 and other arteriopathies have been reported in AS patients.20 We thus report the third known case of SCAD in an AS patient, for whom we identified a pathogenic variant in COL4A4 after screening genes associated with AS. Screening of the remainder of the cohort identified two more carriers of rare heterozygote variants in COL4A4, a likely pathogenic variant and a splice-altering variant (Supplementary Table XIII). The inheritance of AS has been reported as both dominant and recessive with slightly different phenotypes.29 Neither of these two cases had a pre-existing diagnosis nor renal or hearing problems, which are commonly associated with AS. We are the first to systematically assess AS genes within a SCAD cohort and our high discovery rate compared to other CTD genes suggests these genes are worth considering in future studies.
We also looked for variants that would alter splicing, including deep intronic variants. We identified four variants in Tier 1 genes in four cases, of which three had no other variants in any gene. Validation of these variants in vivo has not been possible due to their very low expression in blood and the fact that the majority are predicted to undergo nonsense-mediated decay. Although splicing alterations outside of canonical sites have never before been considered in SCAD, our data suggest they are likely to have a role and that broadening the search space of variant types in SCAD will contribute to our understanding of disease mechanisms.
Using the Tier 2 gene list, we identified two likely pathogenic variants in ACVR1 in two SCAD cases. However, both cases also possessed variants in a Tier 1 gene – either ALDH18A1 or PRKG1. ACVR1 is a receptor for bone morphogenetic proteins; both receptor and ligands are thought to have a role in vascular homeostasis30 and mutations therein can cause fibrodysplasia ossificans progressiva. Although variants in Tier 2 genes potentially contributed to the genetic diagnosis of these patients, more evidence is required to consider these clinically actionable.
Across our Tier 1 and 2 ACMG and Tier 1 splice-altering variant analyses, we identified seven cases heterozygous for a variant of interest but only in a gene associated with AR disease. Interestingly, of these seven, one case has some phenotypic support for the associated CTD, another has an uncle with an aortic dissection, and a third has suffered three SCAD events. In one case, a variant in an essential splice site was identified in the Tier 1 gene, AEBP1. This alteration has been shown to cause loss of the final 22 nucleotides of exon 13, leading to a frameshift and has been previously reported as a biallelic variant causing EDS.31 AEBP1 heterozygote carriers have rarely been assessed clinically but generally are healthy.31 The variant identified in a 27 year old SCAD case in the Tier 1 gene, SLC2A10, has also been reported previously. This variant has been identified as one of two causative variants for arterial tortuosity syndrome in four compound heterozygous families32 with heterozygous carriers lacking obvious angiographic abnormalities.32 Mutations in ABCC6 (Tier 2), found in two cases including one who sustained three SCADs, causes pseudoxanthoma elasticum, typically with AR inheritance (MIM #264800), although an atypical form with AD inheritance (MIM #177850) is also recognized. Both cases carry the same pathogenic splice-altering variant. The role of these three variants and the variants in the other genes causing AR disease – ADAMTS2, ADAMTS10, LTBP4, and SLC39A13 – remains unclear, but the clinical findings of these patients suggest it may be worth considering heterozygous variants in such genes in SCAD patients.
Given our high rate of identification of CTD and vasculopathy gene variants, we sought to determine if these genes were collectively more likely to contain potentially damaging rare (“qualifying”) variants within SCAD cases compared to controls in a collapsing analysis. Such an analysis across both or either of our Tier genes indicated that SCAD cases were significantly more likely to have at least one qualifying variant than controls, supporting the general association of these genes with SCAD. However, a genome-wide rare variant collapsing analysis assessing individual genes, found no gene significantly enriched for cases with qualifying variants, although a total of 375 genes were nominally significantly enriched in SCAD cases compared to controls. A comparison of the seven genes with the lowest p-values in this study with our previously reported analysis of 384 UK patients16 identified PCDHA4 as nominally significant in both studies. Although both studies were limited by sample size, further larger studies may reveal a more conclusive link between SCAD and PCDHA4.
Twenty-four LoF-intolerant genes were found to have novel LoF variants, but no LoF-intolerant gene was identified in more than one case harboring structural variants. Additionally, one gene, ACACA, was independently validated in our UK SCAD cohort, with another LoF variant identified in one case. ACACA, Acetyl-CoA carboxylase alpha, has an important role in fatty acid biosynthesis, and changes in lipid metabolism and ACACA expression have been observed in mice overexpressing EDN133 – itself potentially associated with SCAD via a common intronic SNP9 – in the endothelium, with the suggestion it may contribute to vascular injury. Etiological links between the remaining LoF-intolerant genes and SCAD are plausible, e.g. AHR plays a role in cell adhesion and extracellular matrix remodeling through interactions with estrogen receptor, NF-κB, retinoic acid receptor and TGF-β signaling pathways, and influences EDN1 expression,34 while, NEDD4L, WWP1, TERT and SKIL interact with SMAD proteins in the TGF-β signaling pathway.35, 36 Additionally, NASP, with a LoF variant in one case, was also nominally enriched for potentially damaging non-synonymous single nucleotide variants in UK patients.16 Our identification of TGF-β signaling as a significantly overrepresented pathway is of interest, since gene variants causing disruption of the TGF-β signaling pathway are commonly associated with CTD and vasculopathies.37
Two individuals with fragile X FMR1 premutations and SCAD 17 and a third with fragile X syndrome and SCAD 38 have been reported to date. No case reported a family history of fragile X and we did not detect any expansions of the FMR1 repeat outside of the normal range across our 91 SCAD cases. However, as the prevalence of fragile X premutation carriers is estimated to be 1 in 150–300,39 a larger cohort may provide further insight to the possible involvement of this expansion in SCAD.
PRS calculated using common variants associated with SCAD9, 11, 12 showed that our SCAD cases carried a higher genetic risk than either controls or a DCM cohort, reinforcing that there is a common genetic element to SCAD. This is the first demonstration of a quantifiable polygenic burden of common variation in SCAD cases relative to a broad control population; a PRS having previously been applied only to distinguish SCAD risk within an FMD cohort.11 This also indicates the complex etiology of SCAD; the majority of cases potentially harboring multiple susceptibility alleles and only a minority having a monogenic cause.
Interestingly, we could not replicate significant associations between SCAD and genes identified in previous studies,13–15 despite our study being sufficiently powered to discover an association of equivalent magnitude. Indeed, in contrast to Sun et al13 and Fahey et al,15 we found more potentially pathogenic TSR1 and F11R variants in controls than in cases (Supplementary Table XIV). More potentially pathogenic TLN1 variants were observed in cases than controls, in agreement with Turley et al,14 however, this difference did not reach statistical significance in our cohort.
In summary, in-depth analysis of a sporadic SCAD cohort revealed a higher proportion, relative to previous studies, of cases with CTD-related genetic variants despite being otherwise asymptomatic; identified a new set of genes that may underlie SCAD pathophysiology, with possible functional overlap with pathways altered in CTD; and confirmed a role for common variants in SCAD risk. Further analysis of larger cohorts should increase our understanding of the complex genetic architecture of SCAD and the genetic etiology shared between SCAD and CTD.
Supplementary Material
Acknowledgments:
The authors acknowledge the MGRB,40 in particular, the study participants from the 45 and Up and ASPREE studies who were the controls for this study. The authors gratefully thank the Atlassian engineers Diego Berrueta Munoz, Luis Lainez, Peter Grasevski, and Alex Yakovlev for producing the jointsv software (used in structural variation analysis). The authors are also grateful to Profs Emmanuelle Plaisier and Pierre Ronco, INSERM, U1155, Paris F-75020 France for advice on genotype/phenotype correlations associated with COL4A1 mutations.
Sources of Funding:
This work was supported in part by grants from the Cardiac Society of Australia and New Zealand, the National Health and Medical Research Council (NHMRC), Australia (APP1161200), the St Vincent’s Clinic Foundation, the Catholic Archdiocese of Sydney, Perpetual Philanthropy, NSW Health and SCAD Research Inc. EG is supported by a NSW Health Early-Mid Career Fellowship, a NSW Health Early Mid-Career Cardiovascular Grant and a National Heart Foundation of Australia Future Leader Fellowship (101204). JK acknowledges research support from the NIH (R01HL130423, R01HL135093, R01HL148167–01A1) and NSW Health (RG194194). SLD is supported by an NHMRC Principal Research Fellowship (ID1135886) and a NSW Health Cardiovascular Senior Scientist Grant. DF is supported by NSW Health Cardiovascular Senior Scientist and Investigator Development Grants. The authors thank AstraZeneca’s Centre for Genomics Research, Discovery Sciences, BioPharmaceuticals R&D for funding the sequencing of and providing the bioinformatics support related to subjects recruited at Leicester University (UK).
Nonstandard Abbreviations and Acronyms
- ACMG
American College of Medical Genetics
- AD
Autosomal Dominant
- AR
Autosomal Recessive
- AS
Alport syndrome
- CTD
connective tissue disorders
- DCM
Dilated cardiomyopathy
- EDS
Ehlers Danlos syndrome
- FMD
fibromuscular dysplasia
- LoF
Loss of function
- MAF
Minor allele frequency
- MFS
Marfan syndrome
- MGRB
Medical Genome Reference Bank
- PRS
Polygenic risk score
- SCAD
Spontaneous coronary artery dissection
- SNP
Single nucleotide polymorphism
- SV
Structural variant
- WGS
Whole Genome Sequencing
Footnotes
Disclosures: KJC is an employee of AstraZeneca. The remaining authors have no conflicts of interest to declare.
Supplemental Materials:
References:
- 1.Iismaa SE, Hesselson S, McGrath-Cadell L, Muller DW, Fatkin D, Giannoulatou E, Kovacic J and Graham RM. Spontaneous Coronary Artery Dissection and Fibromuscular Dysplasia: Vasculopathies With a Predilection for Women. Heart Lung Circ. 2021;30:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayes SN, Kim ESH, Saw J, Adlam D, Arslanian-Engoren C, Economy KE, Ganesh SK, Gulati R, Lindsay ME, Mieres JH, et al. Spontaneous Coronary Artery Dissection: Current State of the Science: A Scientific Statement From the American Heart Association. Circulation. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diez-del Hoyo F, Sanz-Ruiz R, Diez-Villanueva P, Nunez-Garcia A, Casado-Plasencia A, Angulo-Llanos R, Clavero-Olmos M, Elizaga Corrales J and Fernandez-Aviles F. A novel cardiovascular presentation of Alport Syndrome: spontaneous coronary artery dissection. Int J Cardiol. 2014;177:e133–134. [DOI] [PubMed] [Google Scholar]
- 4.Henkin S, Negrotto SM, Tweet MS, Kirmani S, Deyle DR, Gulati R, Olson TM and Hayes SN. Spontaneous coronary artery dissection and its association with heritable connective tissue disorders. Heart. 2016;102:876–881. [DOI] [PubMed] [Google Scholar]
- 5.Kaadan MI, MacDonald C, Ponzini F, Duran J, Newell K, Pitler L, Lin A, Weinberg I, Wood MJ and Lindsay ME. Prospective Cardiovascular Genetics Evaluation in Spontaneous Coronary Artery Dissection. Circ Genom Precis Med. 2018;11:e001933. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura M, Yajima J, Oikawa Y, Ogasawara K, Uejima T, Abe K and Aizawa T. Vascular Ehlers-Danlos syndrome--all three coronary artery spontaneous dissections. J Cardiol. 2009;53:458–462. [DOI] [PubMed] [Google Scholar]
- 7.McNair PW, Parker A, Taylor A, Battle R, Norton P and Sharma AM. Spontaneous Coronary Artery Dissection and Its Association With Fibromuscular Dysplasia and Other Vascular Abnormalities. Am J Cardiol. 2020;125:34–39. [DOI] [PubMed] [Google Scholar]
- 8.Goel K, Tweet M, Olson TM, Maleszewski JJ, Gulati R and Hayes SN. Familial spontaneous coronary artery dissection: evidence for genetic susceptibility. JAMA Intern Med. 2015;175:821–826. [DOI] [PubMed] [Google Scholar]
- 9.Adlam D, Olson TM, Combaret N, Kovacic JC, Iismaa SE, Al-Hussaini A, O’Byrne MM, Bouajila S, Georges A, Mishra K, et al. Association of the PHACTR1/EDN1 Genetic Locus With Spontaneous Coronary Artery Dissection. J Am Coll Cardiol. 2019;73:58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debette S, Kamatani Y, Metso TM, Kloss M, Chauhan G, Engelter ST, Pezzini A, Thijs V, Markus HS, Dichgans M, et al. Common variation in PHACTR1 is associated with susceptibility to cervical artery dissection. Nat Genet. 2015;47:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saw J, Yang ML, Trinder M, Tcheandjieu C, Xu C, Starovoytov A, Birt I, Mathis MR, Hunker KL, Schmidt EM, et al. Chromosome 1q21.2 and additional loci influence risk of spontaneous coronary artery dissection and myocardial infarction. Nat Commun. 2020;11:4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turley TN, O’Byrne MM, Kosel ML, de Andrade M, Gulati R, Hayes SN, Tweet MS and Olson TM. Identification of Susceptibility Loci for Spontaneous Coronary Artery Dissection. JAMA Cardiol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Y, Chen Y, Li Y, Li Z, Li C, Yu T, Xiao L, Yu B, Zhao H, Tao M, et al. Association of TSR1 Variants and Spontaneous Coronary Artery Dissection. J Am Coll Cardiol. 2019;74:167–176. [DOI] [PubMed] [Google Scholar]
- 14.Turley TN, Theis JL, Sundsbak RS, Evans JM, O’Byrne MM, Gulati R, Tweet MS, Hayes SN and Olson TM. Rare Missense Variants in TLN1 Are Associated With Familial and Sporadic Spontaneous Coronary Artery Dissection. Circ Genom Precis Med. 2019;12:e002437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fahey JK, Williams SM, Tyagi S, Powell DR, Hallab JC, Chahal G, Ramialison MSM and White AJ. The Intercellular Tight Junction and Spontaneous Coronary Artery Dissection. J Am Coll Cardiol. 2018;72:1752–1753. [DOI] [PubMed] [Google Scholar]
- 16.Carss KJ, Baranowska AA, Armisen J, Webb TR, Hamby SE, Premawardhana D, Al-Hussaini A, Wood A, Wang Q, Deevi SVV, et al. Spontaneous Coronary Artery Dissection: Insights on Rare Genetic Variation from Genome Sequencing. Circ Genom Precis Med. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenzie FJ, Tassankijpanich N, Epps KC, March SK and Hagerman RJ. Spontaneous Coronary Artery Dissection in Females With the Fragile X FMR1 Premutation. JACC Case Rep. 2020;2:40–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandt T, Orberk E, Weber R, Werner I, Busse O, Muller BT, Wigger F, Grau A, Grond-Ginsbach C and Hausser I. Pathogenesis of cervical artery dissections: association with connective tissue abnormalities. Neurology. 2001;57:24–30. [DOI] [PubMed] [Google Scholar]
- 19.von Hundelshausen P, Oexle K, Bidzhekov K, Schmitt MM, Hristov M, Blanchet X, Kaemmerer H, Matyas G, Meitinger T and Weber C. Recurrent spontaneous coronary dissections in a patient with a de novo fibrillin-1 mutation without Marfan syndrome. Thromb Haemost. 2015;113:668–670. [DOI] [PubMed] [Google Scholar]
- 20.Traenka C, Kloss M, Strom T, Lyrer P, Brandt T, Bonati LH, Grond-Ginsbach C and Engelter S. Rare genetic variants in patients with cervical artery dissection. Eur Stroke J. 2019;4:355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plaisier E and Ronco P. COL4A1-Related Disorders. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Mirzaa G, et al. , eds. GeneReviews((R)). 1993–2021 ed. Seattle (WA); 2009. [PubMed] [Google Scholar]
- 22.Leistritz DF, Pepin MG, Schwarze U and Byers PH. COL3A1 haploinsufficiency results in a variety of Ehlers-Danlos syndrome type IV with delayed onset of complications and longer life expectancy. Genet Med. 2011;13:717–722. [DOI] [PubMed] [Google Scholar]
- 23.Frank M, Albuisson J, Ranque B, Golmard L, Mazzella JM, Bal-Theoleyre L, Fauret AL, Mirault T, Denarie N, Mousseaux E, et al. The type of variants at the COL3A1 gene associates with the phenotype and severity of vascular Ehlers-Danlos syndrome. Eur J Hum Genet. 2015;23:1657–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ong KT, Perdu J, De Backer J, Bozec E, Collignon P, Emmerich J, Fauret AL, Fiessinger JN, Germain DP, Georgesco G, et al. Effect of celiprolol on prevention of cardiovascular events in vascular Ehlers-Danlos syndrome: a prospective randomised, open, blinded-endpoints trial. Lancet. 2010;376:1476–1484. [DOI] [PubMed] [Google Scholar]
- 25.Gorosabel MC, Dubacher N, Meienberg J and Matyas G. Vascular Ehlers-Danlos syndrome: can the beneficial effect of celiprolol be extrapolated to bisoprolol? Eur Heart J Cardiovasc Pharmacother. 2020;6:199–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischer-Zirnsak B, Escande-Beillard N, Ganesh J, Tan YX, Al Bughaili M, Lin AE, Sahai I, Bahena P, Reichert SL, Loh A, et al. Recurrent De Novo Mutations Affecting Residue Arg138 of Pyrroline-5-Carboxylate Synthase Cause a Progeroid Form of Autosomal-Dominant Cutis Laxa. Am J Hum Genet. 2015;97:483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischer B, Callewaert B, Schroter P, Coucke PJ, Schlack C, Ott CE, Morroni M, Homann W, Mundlos S, Morava E, et al. Severe congenital cutis laxa with cardiovascular manifestations due to homozygous deletions in ALDH18A1. Mol Genet Metab. 2014;112:310–316. [DOI] [PubMed] [Google Scholar]
- 28.Anuwatworn A, Sethi P, Steffen K, Jonsson O and Petrasko M. Spontaneous Coronary Artery Dissection: A Rare Manifestation of Alport Syndrome. Case Rep Cardiol. 2017;2017:1705927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savige J Should We Diagnose Autosomal Dominant Alport Syndrome When There Is a Pathogenic Heterozygous COL4A3 or COL4A4 Variant? Kidney Int Rep. 2018;3:1239–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goumans MJ, Zwijsen A, Ten Dijke P and Bailly S. Bone Morphogenetic Proteins in Vascular Homeostasis and Disease. Cold Spring Harb Perspect Biol. 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alazami AM, Al-Qattan SM, Faqeih E, Alhashem A, Alshammari M, Alzahrani F, Al-Dosari MS, Patel N, Alsagheir A, Binabbas B, et al. Expanding the clinical and genetic heterogeneity of hereditary disorders of connective tissue. Hum Genet. 2016;135:525–540. [DOI] [PubMed] [Google Scholar]
- 32.Callewaert BL, Willaert A, Kerstjens-Frederikse WS, De Backer J, Devriendt K, Albrecht B, Ramos-Arroyo MA, Doco-Fenzy M, Hennekam RC, Pyeritz RE, et al. Arterial tortuosity syndrome: clinical and molecular findings in 12 newly identified families. Hum Mutat. 2008;29:150–158. [DOI] [PubMed] [Google Scholar]
- 33.Simeone SM, Li MW, Paradis P and Schiffrin EL. Vascular gene expression in mice overexpressing human endothelin-1 targeted to the endothelium. Physiol Genomics. 2011;43:148–160. [DOI] [PubMed] [Google Scholar]
- 34.Kung T, Murphy KA and White LA. The aryl hydrocarbon receptor (AhR) pathway as a regulatory pathway for cell adhesion and matrix metabolism. Biochem Pharmacol. 2009;77:536–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Xu D, Li J, Berndt MC and Liu JP. Transforming growth factor beta suppresses human telomerase reverse transcriptase (hTERT) by Smad3 interactions with c-Myc and the hTERT gene. J Biol Chem. 2006;281:25588–25600. [DOI] [PubMed] [Google Scholar]
- 36.Weiss A and Attisano L. The TGFbeta superfamily signaling pathway. Wiley Interdiscip Rev Dev Biol. 2013;2:47–63. [DOI] [PubMed] [Google Scholar]
- 37.Wheeler JB, Ikonomidis JS and Jones JA. Connective tissue disorders and cardiovascular complications: the indomitable role of transforming growth factor-beta signaling. Adv Exp Med Biol. 2014;802:107–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park HY, Cho HM, Kim DH, Park CB and Kim CJ. Spontaneous coronary artery dissection in a female patient with fragile X syndrome. Kosin Med J. 2017;32:240–243. [Google Scholar]
- 39.Hagerman RJ and Hagerman P. Fragile X-associated tremor/ataxia syndrome - features, mechanisms and management. Nat Rev Neurol. 2016;12:403–412. [DOI] [PubMed] [Google Scholar]
- 40.Pinese M, Lacaze P, Rath EM, Stone A, Brion MJ, Ameur A, Nagpal S, Puttick C, Husson S, Degrave D, et al. The Medical Genome Reference Bank contains whole genome and phenotype data of 2570 healthy elderly. Nat Commun. 2020;11:435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinese M, Lacaze P, Rath EM, Stone A, Brion MJ, Ameur A, Nagpal S, Puttick C, Husson S, Degrave D, et al. The Medical Genome Reference Bank contains whole genome and phenotype data of 2570 healthy elderly. Nat Commun. 2020;11:435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minoche AE, Horvat C, Johnson R, Gayevskiy V, Morton SU, Drew AP, Woo K, Statham AL, Lundie B, Bagnall RD, et al. Genome sequencing as a first-line genetic test in familial dilated cardiomyopathy. Genet Med. 2019;21:650–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carss KJ, Baranowska AA, Armisen J, Webb TR, Hamby SE, Premawardhana D, Al-Hussaini A, Wood A, Wang Q, Deevi SVV, et al. Spontaneous Coronary Artery Dissection: Insights on Rare Genetic Variation from Genome Sequencing. Circ Genom Precis Med. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ntusi NB and Mayosi BM. Aetiology and risk factors of peripartum cardiomyopathy: a systematic review. Int J Cardiol. 2009;131:168–179. [DOI] [PubMed] [Google Scholar]
- 45.Li H and Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang K, Li M and Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arthur R, Schulz-Trieglaff O, Cox AJ and O’Connell J. AKT: ancestry and kinship toolkit. Bioinformatics. 2017;33:142–144. [DOI] [PubMed] [Google Scholar]
- 49.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M and Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henkin S, Negrotto SM, Tweet MS, Kirmani S, Deyle DR, Gulati R, Olson TM and Hayes SN. Spontaneous coronary artery dissection and its association with heritable connective tissue disorders. Heart. 2016;102:876–881. [DOI] [PubMed] [Google Scholar]
- 51.Kaadan MI, MacDonald C, Ponzini F, Duran J, Newell K, Pitler L, Lin A, Weinberg I, Wood MJ and Lindsay ME. Prospective Cardiovascular Genetics Evaluation in Spontaneous Coronary Artery Dissection. Circ Genom Precis Med. 2018;11:e001933. [DOI] [PubMed] [Google Scholar]
- 52.Sun Y, Chen Y, Li Y, Li Z, Li C, Yu T, Xiao L, Yu B, Zhao H, Tao M, et al. Association of TSR1 Variants and Spontaneous Coronary Artery Dissection. J Am Coll Cardiol. 2019;74:167–176. [DOI] [PubMed] [Google Scholar]
- 53.Turley TN, Theis JL, Sundsbak RS, Evans JM, O’Byrne MM, Gulati R, Tweet MS, Hayes SN and Olson TM. Rare Missense Variants in TLN1 Are Associated With Familial and Sporadic Spontaneous Coronary Artery Dissection. Circ Genom Precis Med. 2019;12:e002437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fahey JK, Williams SM, Tyagi S, Powell DR, Hallab JC, Chahal G, Ramialison MSM and White AJ. The Intercellular Tight Junction and Spontaneous Coronary Artery Dissection. J Am Coll Cardiol. 2018;72:1752–1753. [DOI] [PubMed] [Google Scholar]
- 55.Ip E, Chapman G, Winlaw D, Dunwoodie SL and Giannoulatou E. VPOT: A Customizable Variant Prioritization Ordering Tool for Annotated Variants. Genomics Proteomics Bioinformatics. 2019;17:540–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kopanos C, Tsiolkas V, Kouris A, Chapple CE, Albarca Aguilera M, Meyer R and Massouras A. VarSome: the human genomic variant search engine. Bioinformatics. 2019;35:1978–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thorvaldsdottir H, Robinson JT and Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Monger S, Troup M, Ip E, Dunwoodie SL and Giannoulatou E. Spliceogen: an integrative, scalable tool for the discovery of splice-altering variants. Bioinformatics. 2019;35:4405–4407. [DOI] [PubMed] [Google Scholar]
- 60.Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, Darbandi SF, Knowles D, Li YI, Kosmicki JA, Arbelaez J, Cui W, Schwartz GB, et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176:535–548 e524. [DOI] [PubMed] [Google Scholar]
- 61.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coban-Akdemir Z, White JJ, Song X, Jhangiani SN, Fatih JM, Gambin T, Bayram Y, Chinn IK, Karaca E, Punetha J, et al. Identifying Genes Whose Mutant Transcripts Cause Dominant Disease Traits by Potential Gain-of-Function Alleles. Am J Hum Genet. 2018;103:171–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cameron DL, Schroder J, Penington JS, Do H, Molania R, Dobrovic A, Speed TP and Papenfuss AT. GRIDSS: sensitive and specific genomic rearrangement detection using positional de Bruijn graph assembly. Genome Res. 2017;27:2050–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Kallberg M, Cox AJ, Kruglyak S and Saunders CT. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. [DOI] [PubMed] [Google Scholar]
- 65.Abyzov A, Urban AE, Snyder M and Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Victor Chang Cardiac Research Institute. ConanVarvar: a versatile tool for the detection of large syndromic copy number variation from whole genome sequencing data. 2020. [DOI] [PMC free article] [PubMed]
- 67.Sifrim A, Hitz MP, Wilsdon A, Breckpot J, Turki SH, Thienpont B, McRae J, Fitzgerald TW, Singh T, Swaminathan GJ, et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat Genet. 2016;48:1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mi H, Muruganujan A, Ebert D, Huang X and Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47:D419–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tang H, Kirkness EF, Lippert C, Biggs WH, Fabani M, Guzman E, Ramakrishnan S, Lavrenko V, Kakaradov B, Hou C, et al. Profiling of Short-Tandem-Repeat Disease Alleles in 12,632 Human Whole Genomes. Am J Hum Genet. 2017;101:700–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dolzhenko E, Deshpande V, Schlesinger F, Krusche P, Petrovski R, Chen S, Emig-Agius D, Gross A, Narzisi G, Bowman B, et al. ExpansionHunter: a sequence-graph-based tool to analyze variation in short tandem repeat regions. Bioinformatics. 2019;35:4754–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tankard RM, Bennett MF, Degorski P, Delatycki MB, Lockhart PJ and Bahlo M. Detecting Expansions of Tandem Repeats in Cohorts Sequenced with Short-Read Sequencing Data. Am J Hum Genet. 2018;103:858–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Turley TN, O’Byrne MM, Kosel ML, de Andrade M, Gulati R, Hayes SN, Tweet MS and Olson TM. Identification of Susceptibility Loci for Spontaneous Coronary Artery Dissection. JAMA Cardiol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saw J, Yang ML, Trinder M, Tcheandjieu C, Xu C, Starovoytov A, Birt I, Mathis MR, Hunker KL, Schmidt EM, et al. Chromosome 1q21.2 and additional loci influence risk of spontaneous coronary artery dissection and myocardial infarction. Nat Commun. 2020;11:4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cummings BB, Karczewski KJ, Kosmicki JA, Seaby EG, Watts NA, Singer-Berk M, Mudge JM, Karjalainen J, Satterstrom FK, O’Donnell-Luria AH, et al. Transcript expression-aware annotation improves rare variant interpretation. Nature. 2020;581:452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.