Abstract
The polar organelle development gene, podJ, is expressed during the swarmer-to-stalked cell transition of the Caulobacter crescentus cell cycle. Mutants with insertions that inactivate the podJ gene are nonchemotactic, deficient in rosette formation, and resistant to polar bacteriophage, but they divide normally. In contrast, hyperexpression of podJ results in a lethal cell division defect. Nucleotide sequence analysis of the podJ promoter region revealed a binding site for the global response regulator, CtrA. Deletion of this site results in increased overall promoter activity, suggesting that CtrA is a negative regulator of the podJ promoter. Furthermore, synchronization studies have indicated that temporal regulation is not dependent on the presence of the CtrA binding site. Thus, although the level of podJ promoter activity is dependent on the CtrA binding site, the temporal control of podJ promoter expression is dependent on other factors.
Caulobacter crescentus is a gram-negative, dimorphic bacterium whose unique life cycle makes it an ideal organism for the study of prokaryotic differentiation and cell cycle control. The primary basis for its unusual life cycle is the asymmetric division that produces a motile swarmer cell and a sessile stalked cell (Fig. 1A) (40). After cell division, the stalked cell immediately initiates another round of cell division. By contrast, the swarmer cell must differentiate into a stalked cell before DNA replication and cell division can be initiated (8, 18).
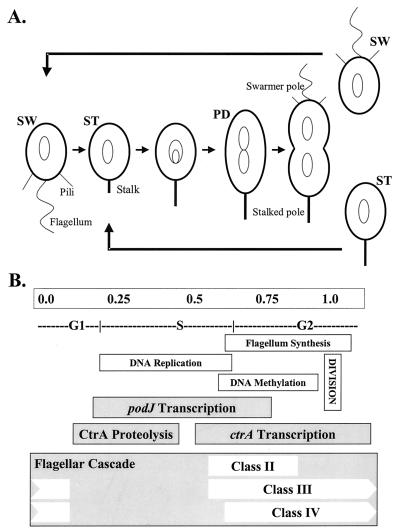
FIG. 1.
C. crescentus cell cycle. (A) Diagram of the cell cycle. During the swarmer-to-stalked cell transition, the swarmer cell (SW) differentiates into a stalked cell (ST); chromosome replication occurs in the stalked cell. Cytokinesis of the predivisional cell (PD) produces a motile swarmer and a sessile stalked cell. (B) Divisional units (1 U ∼ 90 min), phases of the cell cycle, and timing of various cell cycle regulated events in C. crescentus.
Much of the study of C. crescentus has focused on the analysis of genes whose transcription is temporally controlled. The genes of the flagellar hierarchy constitute the best-understood temporally controlled genes in C. crescentus. The majority of the estimated 50 genes necessary for motility (15) have been categorized into four classes that are expressed sequentially. Furthermore, the expression of each class of flagellar genes is required for the expression of the next class (Fig. 1B) (9, 11, 34, 38). This cascade of flagellar gene expression allows the transcription of genes just prior to the time they are needed for assembly (51).
The study of flagellar genes has led to the discovery of several factors that control temporal expression (7, 27, 28, 32). In addition to sigma factors, two types of proteins responsible for this regulation have been identified in C. crescentus: sensor and regulator proteins. The sensor proteins belong to a family of proteins called histidine protein kinases and are capable of autophosphorylation in response to unknown cues (20, 37, 39, 55). The regulator proteins belong to a family of proteins called response regulators which are activated by histidine protein kinase phosphorylation (3, 14, 20, 57) and regulate gene expression at the level of transcription. These factors constitute the proteins of a two-component signal transduction system and are thought to control the temporal expression of genes during the C. crescentus cell cycle (20, 37, 55, 56). Several response regulators have been identified in C. crescentus (3, 14, 20, 57). One of the best studied, CtrA, recently was shown to control both DNA replication and the initiation of the flagellar cascade in C. crescentus (41, 42). CtrA was found to be regulated by temporal expression, phosphorylation, and proteolysis (14, 42, 58).
In this study, we report that CtrA modulates the expression of podJ, a gene involved in the swarmer to stalked cell transition. Two podJ mutants were identified after Tn5 mutagenesis (16). They exhibited a normal cell morphology but were chemotaxis negative, resistant to polar bacteriophage (φCbK), and deficient in rosette formation (55). Also, 10 to 30% of the swarmer cells of this strain failed to release the flagellum, resulting in a flagellum on the end of the stalk. Our analysis demonstrates that the nucleotide sequence of the podJ promoter region contains a putative binding site for the response regulator, CtrA, and removal of the binding site results in elevated podJ expression. In addition, promoter fusion experiments demonstrated that podJ expression is temporally regulated with peak expression during the swarmer-to-stalked cell transition. Finally, hyperexpression of the podJ gene was shown to cause a lethal cell division defect.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Tables 1 and 2. C. crescentus strains were grown at 33°C in peptone-yeast extract (PYE) or minimal (M2) (24) medium. Escherichia coli strains were grown at 37°C in Luria-Bertani (LB) broth (48). When specified, PYE medium was supplemented with either 0.02 to 20 mM (0.0003 to 0.3%) xylose or 11 mM (0.2%) glucose. Antibiotics were used as required: in PYE medium, tetracycline (1 μg/ml [PYEtet]), chloramphenicol (1 μg/ml [PYEchl]), and kanamycin (50 μg/ml); in M2 medium, tetracycline (0.25 to 0.5 μg/ml); and in LB medium, ampicillin (100 μg/ml), tetracycline (5 μg/ml), and chloramphenicol (50 μg/ml). All chemicals were purchased from the Sigma Company (St. Louis, Mo.).
TABLE 1.
Strains used in this study
| Strains | Genotype | Reference |
|---|---|---|
| Caulobacter crescentus | ||
| CB15 | Wild type | 40 |
| LS107 | syn-1000 bla-6 | 17 |
| LS2195 | ctrA401 syn-1000 bla-6 | 41 |
| SC1119 | podJ152::Tn5 str-152 | 55 |
| Escherichia coli | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA lacF+ (proAB laclqΔM15 Tn10) | 54 |
| S17-1 | proA recA hsdR hsdM zzz::RP4 (tet::mu) (km::Tn7) | 52 |
TABLE 2.
Plasmids and oligonucleotides used in this study
| Plasmid or oligonucleotide | Descriptiona | Reference |
|---|---|---|
| General purpose vectors | ||
| pBKS+/− | General cloning vector (pBKS); Ampr | 54 |
| pBSK− | General cloning vector (pBSK); Ampr | 54 |
| pRKlac290 | lacZ transcriptional fusion; Tetr | 1 |
| pUJ142 | Xylose-inducible promoter; Chlr | 30 |
| Subclones of the podJ region | ||
| pBC1 | 151-bp PCR fragment (−128 to +24) generated with primers | This study |
| pDZ7 | 3-kb PstI subclone from K28 containing podJ in pBKS− | This study |
| pBC3 | 860-bp PstI/XhoI fragment of pDZ7 in pBSK+ | This study |
| pBC4 | 1,519-bp PstI/HindIII fragment of pDZ7 in pBSK+ | This study |
| pBC5 | 1,519-bp PstI/HindIII fragment of pDZ7 in pBKS+ | This study |
| pBC9 | 95-bp fragment (−128 to −33) in pBKS+ from the StyI/XbaI deletion of pBC1 | This study |
| pBC12 | 211-bp PCR fragment (−188 to +24) generated with primers BE364 and BE238, in HindIII site of pBKS+ | This study |
| pBC15 | 201-bp fragment (−188 to −148, −137 to +24) resulting from a HpaI deletion of pBC12 | This study |
| Expression fusion | ||
| pBC16 | 1,614-bp PCR fragment generated with primers BE300 and BE301, cloned into the SpeI/KpnI sites of pUJ142 (PxylX::podJ) | This study |
| Transcriptional fusions | ||
| pBC6 | pBC5 into the HindIII/KpnI sites of pRKlac290 | This study |
| pBC7 | pBC4 into the PstI/XbaI sites of pRKlac290 | This study |
| pBC10 | pBC1 into the HindIII/KpnI sites of pRKlac290 | This study |
| pBC11 | pBC9 into the HindIII/KpnI sites of pRKlac290 | This study |
| pBC13 | pBC12 into the KpnI site of pRKlac290 | This study |
| pBC17 | pBC15 into the KpnI site of pRKlac290 | This study |
| Oligonucleotides | ||
| BE176 | 5′-ACCAACCTCATCTGCCGGAT | This study |
| BE208 | 5′-AACTGCAGGCTTTTGAACCGCGCTACAGAG | This study |
| BE238 | 5′-CCCAAGCTTCGTCATGCGAATCGATCTCC | This study |
| BE364 | 5′-CCCAAGCTTGCGCGCTTCTAGGCCTGCGACA | This study |
| BE369 | 5′-CTAGGCGGCCAGATCTGATC | This study |
Antibiotic resistance markers: Ampr, ampicillin; Tetr, tetracycline; Chlr, chloramphenicol. Nucleotide positions given in parentheses are in relation to the podJ transcription start site. pBKS, pBluescript KS; pBSK, pBluescript SK.
DNA manipulations.
All cloning and general procedures were carried out as previously described (48). Plasmid DNA isolation from E. coli host strains was performed with a QIAprep Spin Miniprep kit (Qiagen Inc., Valencia, Calif.). Electroporation was performed on C. crescentus cells as previously described (26). All restriction and modifying enzymes were purchased from New England Biolabs Inc. (Beverly, Mass.) except for calf intestinal alkaline phosphatase, which was purchased from GIBCO (Gaithersburg, Md.). Radioisotopes ([γ-32P]ATP and [35S]methionine-cysteine) were purchased from NEN Life Sciences (Boston, Mass.). Anti-β-galactosidase antibody was purchased from Promega (Madison, Wis.). Nucleotide sequence analyses were performed with the dideoxy-chain termination method (49) on a LICOR (Lincoln, Neb.) automated sequencer. Nucleotide sequence data were analyzed with the Genetics Computer Group (University of Wisconsin, Madison) (13) Wisconsin Package (version 9.0) and the following programs from European Bioinformatics Institute’s web page (16a): PROPSEARCH (22, 46), MaxHom (50), and PHDsec (45, 47).
PCR amplifications were performed with approximately 20 ng of chromosomal DNA, cosmid (K28) or plasmid (pDZ7), as template. PCR mixtures contained 1× buffer [60 mM Tris-HCl, 15 mM (NH4)2SO4, 1 mM MgCl2 (pH 9.0)], 0.2 μM each oligonucleotide primer (Table 2), 0.06 μM each deoxynucleoside triphosphate, and 0.02 U of AmpliTaq DNA polymerase (Perkin-Elmer, Foster City, Calif.) per μl. Conditions used for PCR amplification of promoter fragments cloned in pBC1 and pBC12 were an initial denaturation for 6 min at 94°C, followed by 32 amplification cycles (denaturation, 45 s at 94°C; annealing, 30 s at 58°C; extension, 45 s at 72°C) and a final extension for 10 min at 72°C. Conditions used for PCR amplification of a 1,556-bp fragment containing the podJ coding region were an initial denaturation for 6 min at 94°C, followed by 32 amplification cycles (denaturation, 105 s at 94°C; annealing, 50 s at 58°C; extension, 60 s at 72°C) and a final extension for 10 min at 72°C.
S1 nuclease protection assay.
A 396-bp single-stranded DNA (ssDNA) fragment was synthesized by asymmetrical PCR (29) with primers BE208 and BE176 (Table 2). The ssDNA fragment was 5′ end labeled with [γ-32P]ATP by using T4 kinase and hybridized to total RNA (20 or 60 μg) overnight at 50°C as previously described (48). A nucleotide sequencing ladder was generated by using primer BE176 in dideoxy-chain termination reaction with Sequenase purchased from Amersham Life Sciences, Inc. (Cleveland, Ohio). The products were separated by electrophoresis on a 6% polyacrylamide DNA sequencing gel and visualized on Kodak BIOMAX MR autoradiograph film.
β-Galactosidase assay.
Mid-log-phase cultures (125 Klett units [optical density at 650 nm of ∼0.5]) grown in liquid PYEtet medium were centrifuged at 4°C, washed twice, and resuspended in 1/20 sample volume of cold 0.1× TE (1× TE is 10 mM Tris-HCl plus 1 mM EDTA [pH 8.0]). Samples were subjected to at least four rounds of sonication on ice (30 s on, 60 s off), and complete sonication was confirmed by microscopic evaluation. Cell debris was removed by centrifugation at 14,000 × g for 2 min at 4°C. A Bradford analysis was used to determine protein concentration (6), and 5 ug of total protein was used for the β-galactosidase assay (31).
Synchronization experiments.
Swarmer cells were isolated by differential centrifugation as previously described (2). The final sample was found to have ∼95% swarmer cells by microscopic evaluation. Synchronized cultures were allowed to proceed through the cell cycle, and samples were taken every 20 min and labeled with [35S]methionine-cysteine for 10 min at 33°C. The labeled samples were then subjected to immunoprecipitation with anti-β-galactosidase or antiflagellin antibody as previously described (19).
Nucleotide sequences.
The GenBank accession number for podJ is AF084609.
RESULTS
Cloning of the podJ gene.
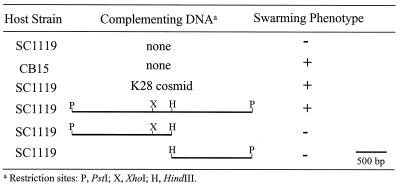
A cosmid clone (K28) was shown to complement the swarming defect of SC1119 (podJ::Tn5). A subclone, pDZ7, containing a ∼3.0-kb PstI fragment from K28 also was sufficient for complementation. This result was consistent with the fact that the Tn5 element in SC1119 had inserted into a 3.0-kb PstI fragment (data not shown). Subclones containing either the 1.6-kb or the 1.4-kb PstI-HindIII fragment could not complement the swarming defect (Fig. 2). These results suggested that the HindIII site was either in the podJ gene or in an operon containing the podJ gene.
FIG. 2.
Complementation analysis. SC1119 is a podJ insertional mutant. +, normal swarming compared to the wild-type strain (CB15) in soft agar PYE plates after overnight incubation.
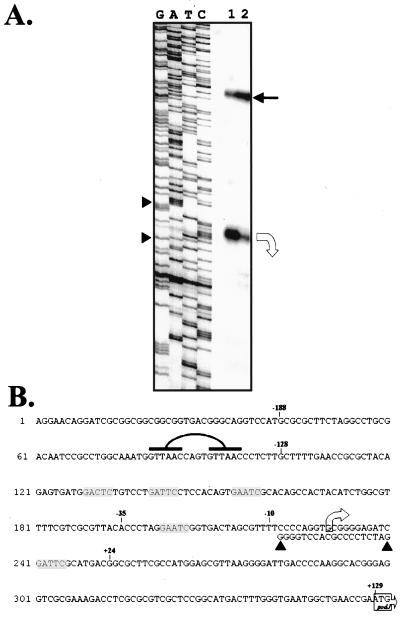
The nucleotide sequence of the 3-kb subclone revealed a 1,416-bp open reading frame (ORF) with an ATG start codon located at position +129 (Fig. 3). The ORF originating from this start codon had both a low frequency of rare codons and a high GC content at the third position of the codon. The gene was designated podJ because it included the HindIII site required for complementation, and no other candidate reading frames were present. PCR amplification using primers BE208 (podJ forward primer) and BE369 (Tn5 reverse primer) revealed the Tn5 position in SC1119 to be in the podJ ORF at position ca. +700 (Fig. 4), indicating that only half of the podJ protein would be synthesized in this mutant. The podJ gene is predicted to encode a peptide of 472 amino acids with a mass of 52 kDa and an isoelectric point of 10.47. A hydropathy plot (25) did not predict any transmembrane regions.
FIG. 3.
(A) S1 nuclease protection assay. The DNA sequence ladder was generated with primer BE176. Lane 1, ssDNA probe with 60 μg of C. crescentus total RNA; lane 2, ssDNA probe with 20 μg of total RNA. The bent open arrow indicates the 268-bp fragment of protected DNA indicating the transcription start site is 129 bp from the proposed translation start site; the solid arrow indicates the 396-bp free probe that is protected by contaminating complementary strand sequences. (B) Nucleotide sequence of the podJ promoter region. Linked bars indicate the putative CtrA binding site; shaded boxes indicate possible methylation sites (see Discussion). The transcription start site is labeled by the bent arrow, and the translation start site is labeled with a boxed arrow at the ATG start codon. The sequence, as read from the sequencing ladder, is indicated (▴ in panel B; ▸ in panel A).
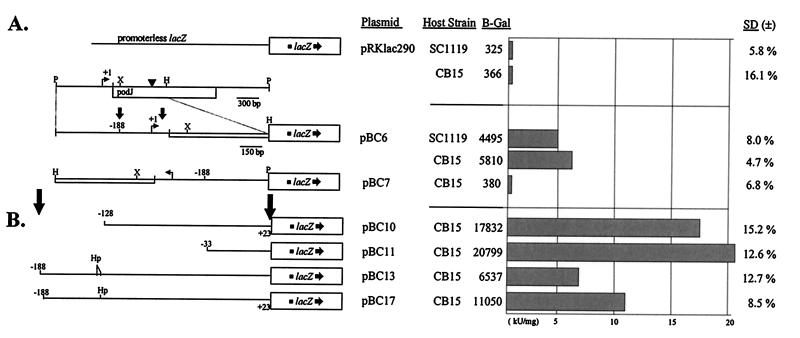
FIG. 4.
Characterization of podJ promoter regions and promoter activity. The promoter fusions are diagrammed along with the strains in which they were tested. The β-galactosidase (B-gal) activity is expressed numerically (in Miller units per milligram), along with a histogram and percent standard deviation. (A) Promoter fusions tested: the transcriptional fusion vector alone, as a negative control; a 3-kb subclone containing the podJ gene and an enlargement of a 1,519-bp PstI/HindIII fragment containing the upstream portion of the podJ gene in both orientations. ▾, insertion site of the Tn5 in SC1119; bent arrows, transcription start site. (B) Deletions of the podJ promoter. Filled arrows denote the boundaries of the expanded area. Restriction sites: P, PstI; X, XhoI; H, HindIII; Hp, HpaI.
To gain insight into the role of podJ in the C. crescentus cell cycle, we have examined the predicted PodJ peptide for homology to known proteins. Strict homology searches using National Center for Biotechnology Information BLAST and Genetics Computer Group BESTFIT programs have shown PodJ to have 28% identity over 342 amino acids to the sensory rhodopsin II transducer in Natronobacterium pharaonis and 26% identity over 283 amino acids to a methyl-accepting chemotaxis protein of E. coli. Another group of proteins which share homology to PodJ are specialized structural proteins. PodJ shares ∼30% homology over ∼100 amino acids to a group eukaryotic proteins in the β and 4.1 family (ezrin, moesin, and Band 4.1). Using the PROPSEARCH program and profiles generated by PHDsec and MaxHom (predicting secondary structural homology) (16a), we found an amino acid composition match to the DeaD helicase of Klebsiella pneumoniae. BESTFIT comparison of this sequence to PodJ shows 25% identity over 268 amino acids. PodJ also shares low levels of homology with eukaryotic myosin, tropomyosin, and troponin T chains. Because PodJ shares homology with a variety of proteins, any proposed function for PodJ would be speculative at best.
Localization and characterization of the podJ promoter.
We used an S1 nuclease protection assay to determine the probable transcription start site of the podJ gene. The protected fragment was 268 bp in length, indicating that the first nucleotide transcribed is 129 bp upstream of the proposed translation start site (Fig. 3A). To determine the extent of the promoter region, we generated transcriptional fusions using subclones of the podJ gene. Fusions containing the region spanning −654 to +876 of the podJ gene showed transcriptional activity in the direction of the podJ gene (pBC6) and not in the reverse orientation (pBC7) (Fig. 4B). These data rule out the possibility of bidirectional transcription from the podJ promoter region. A construct containing the −188 to +24 region (pBC13) showed levels of expression similar to those of pBC6 (Fig. 4B). By contrast, constructs containing regions −128 to +24 (pBC10) or −33 to +24 (pBC11) had three- to fivefold-higher levels of expression (Fig. 4B). These results suggest that the region between −128 and −188 negatively regulated transcription. Since this region contained a CtrA consensus binding site (Fig. 3B), half of this site was removed by deletion of 10 bp between the two HpaI sites that make up the site. Expression of the resulting construct (pBC17) was almost twofold higher than that of the undeleted control (Fig. 4B). Thus, CtrA appears to function as a negative regulator of podJ expression.
Temporal control of podJ expression.
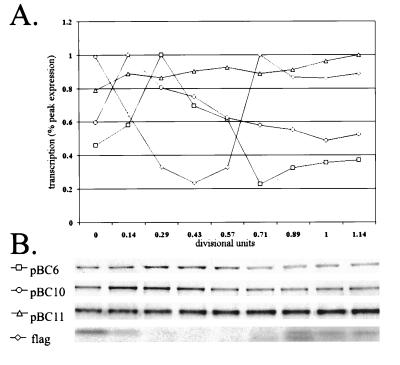
To identify the promoter regions responsible for the temporal expression of podJ, synchronization experiments were performed with cultures containing various plasmid-borne promoter fusions. Constructs containing the entire region, −654 to +876 (pBC6), and those lacking the CtrA binding site, −128 to +24 (pBC10), had similar patterns of temporal expression (Fig. 5). These data suggest that the CtrA site is not necessary for the cell cycle control of podJ expression. Constructs containing the region −33 to +24 (pBC11) exhibited a relatively constant level of expression throughout the cell cycle (Fig. 5). Thus, the region between −128 and −33 is necessary for the temporal regulation of podJ expression.
FIG. 5.
Cell cycle expression of various promoter regions. (A) Relative transcription levels throughout the cell cycle. (B) Key to symbols and autoradiographs of immunoprecipitations of the transcriptional fusions tested. Flagellin (flag) synthesis was monitored in each transcription experiment as a control.
Hyperexpression of podJ causes lethal cell division defect.
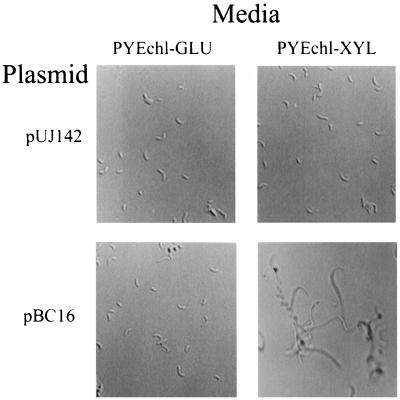
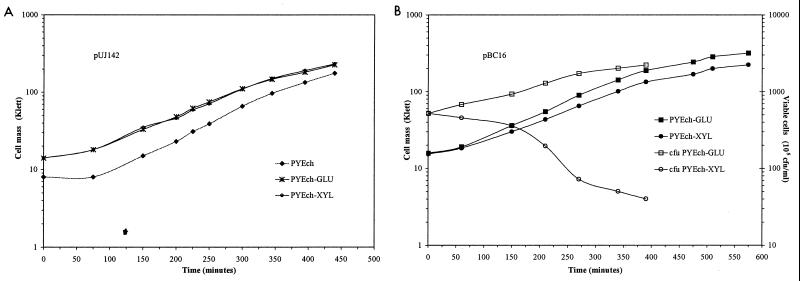
To test the effect of hyperexpression of podJ, the C. crescentus xylX promoter (PxylX) was used to control the expression of the podJ gene. The level of induction of PxylX is dependent on the time of exposure to a given xylose concentration; xylose levels of 20, 2, and 0.2 mM provide levels of expression roughly 10, 6, and 3 times that of uninduced expression after ∼150 min (30). Also, expression of PxylX is not under cell cycle control. When PxylX::podJ expression was induced in the presence of either 20 or 2 mM xylose, filamentous cells were observed after 180 min (Fig. 6). The inhibition of cell division was not observed when strains containing the vector alone pUJ142 (Fig. 6), PxylX::fliF, PxylX::lacZ (30), or PxylX::ctrA (41) were grown in xylose. Thus, growth in xylose is not responsible for filamentation. The filamentous cells continued to elongate, and cell mass doubled every 100 min (see below). These data indicate that cell division stopped soon after the induction of high levels of podJ expression. By contrast, when podJ expression was induced in 0.2 mM xylose, cell mass doubled at the same rate, but filamentous cells were not observed until cell mass had increased by a factor of 4 (data not shown). Thus, lower levels of podJ hyperexpression have a less immediate effect, but even an increase in expression of approximately threefold (30) results in a highly filamentous phenotype after overnight incubation. In contrast, concentrations of xylose found to induce PxylX twofold and lower (<0.02 mM) (30) had no effect on cell division after overnight growth. These data suggest that filamentation is caused by overexpression of PodJ at a critical time during the cell cycle since synchronization experiments using PpodJ::lacZ showed that the expression of PpodJ varies roughly threefold over the cell cycle (Fig. 5). Thus, filamentation appears to occur in response to a threefold increase in podJ expression at an inappropriate time in the cell cycle.
FIG. 6.
Hyperexpression of podJ causes cell division defect. Wild-type C. crescentus (CB15) was grown in medium containing either pUJ142 (PxylX) or pBC16 (PxylX::podJ) supplemented with 11 mM glucose (PYEchl-GLU) or 20 mM xylose (PYEchl-XYL) after 480 min; cells were photographed with Zeiss Photomicroscope III with a Plan-NEOFLUAR 40/0.9 Imm Ph3 lens (magnification, ×400).
To learn more about the effect of podJ hyperexpression on viability, colonies of CB15 containing either pBC16 (PxylX::podJ) or the control plasmid (pUJ142) were grown to stationary phase in PYEchl to select for the presence of the plasmid. The cells were then subcultured in PYEchl containing either 20 mM xylose to induce PxylX or 11 mM glucose to repress PxylX. As measured by CFU, the number of viable cells containing pBC16 (PxylX::podJ) decreased as a function of time in the presence of xylose even though cell mass continued to increase (Fig. 7B). These results indicate that cell growth initially continues as colony-forming ability decreases. The control growth curves and CFU data from cells containing pBC16 subcultured in glucose and pUJ142 subcultured in xylose or glucose were typical and unremarkable (Fig. 7). Thus, hyperexpression of podJ causes an immediate cell division arrest, followed by filamentation and finally cell death.
FIG. 7.
Hyperexpression of podJ causes a lethal cell division phenotype. (A) Growth curve of CB15 containing pUJ142 cultured in PYEchl with 11 mM glucose (PYEch-GLU) or 20 mM xylose (PYEch-XYL) or in PYEchl alone (PYEch). (B) Growth curve of CB15 containing pBC16 and CFU data showing a decrease in the number of viable cells per milliliter. Cell mass was measured by Klett readings. Data points for Klett readings represent the average of at least five samples for which the standard deviation was <8%.
The podJ hyperexpression phenotype is strikingly similar to that of the temperature-sensitive ctrA401 mutant in restrictive conditions (41). When the ctrA401 mutant is shifted to the restrictive temperature (37°C), the cells filament and die. We reasoned that the restrictive temperature could prohibit CtrA from binding to the podJ promoter and consequently allow the hyperexpression of podJ to cause the ctrA401 phenotype. To test this possibility, phage lysate (φCR30) prepared on SC1119 was used to transduce the podJ::Tn5 into the ctrA401 strain (LS2195). The double mutants were analyzed for both growth at 37°C and colony-forming ability under normal and restrictive conditions. During incubation at restrictive conditions, the double mutants exhibited mortality rates (>85% mortality after 400 min) similar to that of the ctrA401 controls (41). During growth in permissive conditions, the cell morphologies of the two strains appeared identical, but the double mutants had roughly fivefold more viable cells per unit of optical density at 650 nm than the ctrA401 mutant (data not shown), indicating that the presence of the podJ mutation enhanced viability. Furthermore, microscopic evaluation of wild-type strains of C. crescentus containing multiple copies of the podJ gene and promoter exhibit a very mild filamentous phenotype, though not as severe as that of the ctrA401 strain under restrictive conditions (data not shown). Taken together, these findings suggest that the interruption of the podJ gene (thus preventing hyperexpression) in the ctrA401 podJ::Tn5 double mutant may increase the viability of a ctrA mutant at permissive conditions, but it has no overall effect on growth rate and viability at restrictive conditions.
DISCUSSION
This study describes the isolation of the podJ gene and the characterization of the podJ promoter. Our analyses have identified regions of the promoter that are responsible for the repression and proper timing of podJ expression. In addition, hyperexpression of podJ resulted in a lethal cell division defect. These findings suggest that podJ expression is carefully controlled during the C. crescentus cell cycle.
In an attempt to further characterize the promoter, we looked for homology between the promoter region of podJ and known C. crescentus consensus promoter sequences. The closest homology was to the ς32-like promoters that are thought to be responsible for the heat shock response in C. crescentus (43, 44) and in E. coli (12). The −10 region of podJ matches six of seven bases of the consensus, but the −35 region matches only two out of five bases of the consensus. To test the possibility of heat shock regulation on the podJ promoter, we assayed PpodJ::lacZ at 33 and 42°C and found less than 5% variation in the level of expression. Thus, it is unlikely that ς32 is involved in podJ expression, and we conclude that the podJ promoter may represent a new class of cell cycle-regulated promoters. In addition, there is a unique nucleotide sequence (CTnGCnTTT) repeated three times in the podJ promoter: the first iteration located three bases from the CtrA binding site; a second located in the −50 region; and a third in the −10 region of the promoter. However, it is not known if these sites have a functional significance.
The podJ promoter region contains five CcrM methylation sites (Fig. 3B). DNA methylation was previously shown to affect DNA replication (4, 36), gene expression (5, 10, 23, 35), and DNA repair (33). Furthermore, the methyltransferase CcrM is essential for the progression of the cell cycle (53). Not surprisingly, the regulation of development in C. crescentus is affected by the methylation state of the chromosome by CcrM (59). In fact, Zweiger et al. (59) suggest that the temporal expression of promoters containing GAnTC sites could be regulated according to whether they were fully methylated or hemimethylated. The significance of the putative methylation sites in the podJ promoter remains unknown.
ACKNOWLEDGMENTS
We thank Guy Leclerc for the invigorating discussion and critical reading of the manuscript, and we thank Shui Ping Wang and Kim Quon for helpful suggestions.
This work was supported by NIH grant GM50547 to B. Ely.
REFERENCES
- 1.Alley M R, Gomes L, Alexander W, Shapiro L. Genetic analysis of a temporally transcribed chemotaxis gene cluster in Caulobacter crescentus. Genetics. 1991;129:333–341. doi: 10.1093/genetics/129.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender R A, Refson C M, O’Neill E A. Role of the flagellum in the cell-cycle-dependent expression of bacteriophage receptor activity in Caulobacter crescentus. J Bacteriol. 1989;171:1035–1046. doi: 10.1128/jb.171.2.1035-1040.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson A K, Ramakrishnun G, Ohta N, Feng J, Ninfa A, Newton A. The Caulobacter crescentus flbD protein acts at the ftr sequence elements both to activate and repress transcription of cell cycle regulated flagellar genes. Proc Natl Acad Sci USA. 1994;91:4989–4993. doi: 10.1073/pnas.91.11.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boye E, Lobner-Olesen A. The role of dam methyltransferase in the control of DNA replication of E. coli. Cell. 1990;62:981–989. doi: 10.1016/0092-8674(90)90272-g. [DOI] [PubMed] [Google Scholar]
- 5.Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. Rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Brun Y V, Shapiro L. A temporally controlled factor is required for polar morphogenesis and normal cell division in Caulobacter. Genes. 1992;6:2395–2408. doi: 10.1101/gad.6.12a.2395. [DOI] [PubMed] [Google Scholar]
- 8.Brun Y V, Marczynski G T, Shapiro L. The expression of asymmetry during Caulobacter cell differentiation. Annu Rev Biochem. 1994;63:419–450. doi: 10.1146/annurev.bi.63.070194.002223. [DOI] [PubMed] [Google Scholar]
- 9.Bryan R, Glaser D, Shapiro L. A genetic regulatory hierarchy in Caulobacter development. Adv Genet. 1990;27:1–31. doi: 10.1016/s0065-2660(08)60022-x. [DOI] [PubMed] [Google Scholar]
- 10.Busslinger M, Hurst J, Flavell R. DNA methylation and the regulation of globin gene expression. Cell. 1983;34:197–206. doi: 10.1016/0092-8674(83)90150-2. [DOI] [PubMed] [Google Scholar]
- 11.Champer R, Dingwall A, Shapiro L. Cascade regulation of Caulobacter flagellar and chemotaxis gene. J Mol Biol. 1987;194:71–80. doi: 10.1016/0022-2836(87)90716-9. [DOI] [PubMed] [Google Scholar]
- 12.Cowing D W, Bardwell J C, Craig E A, Woolford C, Hendrix R W, Gross C A. Consensus sequence for Escherichia coli heat shock gene promoters. Proc Natl Acad Sci USA. 1985;82:2679–2683. doi: 10.1073/pnas.82.9.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devereux D, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domian I J, Quon K Q, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 15.Ely B, Ely T W. Use of pulse field gel electrophoresis and transposon mutagenesis to estimate the minimal number of genes required for motility in Caulobacter crescentus. Genetics. 1989;114:719–730. doi: 10.1093/genetics/123.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ely B, Gerardot C J. Use of pulsed-field gradient gel electrophoresis to construct a physical map of the Caulobacter crescentus genome. Gene. 1988;68:323–333. doi: 10.1016/0378-1119(88)90035-2. [DOI] [PubMed] [Google Scholar]
- 16a.European Bioinformatics Institute. 15 June 1998, posting date. World-Wide Web (WWW) server. [Online.] http://www.ebi.ac.uk/ebi_home.html. EMBL Outstation, European Bioinformatics Institute, Cambridge, United Kingdom. [21 May 1999, last date accessed.]
- 17.Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gober J W, Marques M V. Regulation of cellular differentiation in Caulobacter crescentus. Microbiol Rev. 1995;59:31–47. doi: 10.1128/mr.59.1.31-47.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomes S L, Shapiro L. Differential expression and positioning of chemotaxis methylation proteins in Caulobacter. J Mol Biol. 1984;177:551–568. doi: 10.1016/0022-2836(84)90238-9. [DOI] [PubMed] [Google Scholar]
- 20.Hecht G, Lane T, Ohta N, Sommer J, Newton A. An essential domain response regulator required for normal cell division and differentiation in Caulobacter crescentus. EMBO J. 1995;14:3915–3924. doi: 10.1002/j.1460-2075.1995.tb00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hecht G B, Newton A. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J Bacteriol. 1995;177:6223–6229. doi: 10.1128/jb.177.21.6223-6229.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hobohm U, Sander C. A sequence property approach to searching protein databases. J Mol Biol. 1995;251:390–399. doi: 10.1006/jmbi.1995.0442. [DOI] [PubMed] [Google Scholar]
- 23.Iguchi-Ariga S, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGCCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 24.Johnson R C, Ely B. Isolation of the spontaneously-derived mutants from Caulobacter crescentus. Genetics. 1977;86:25–32. doi: 10.1093/genetics/86.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kyte J, Doolittle R F. A simple method for displaying the hydropathetic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 26.Malakooti J, Ely B. Identification and characterization of the ilvK gene encoding a LysR-type regulation of Caulobacter crescentus. J Bacteriol. 1994;176:1275–1281. doi: 10.1128/jb.176.5.1275-1281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marczynski G T, Lentine K, Shapiro L. A developmentally regulated chromosomal origin of replication uses essential transcription elements. Genes Dev. 1995;9:1543–1557. doi: 10.1101/gad.9.12.1543. [DOI] [PubMed] [Google Scholar]
- 28.Marczynski G T, Shapiro L. Cell-cycle control of a cloned chromosomal origin of replication from Caulobacter crescentus. J Mol Biol. 1992;229:959–977. doi: 10.1016/0022-2836(92)91045-q. [DOI] [PubMed] [Google Scholar]
- 29.McCabe P C. Production of single-stranded DNA by asymmetric PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. New York, N.Y: Academic Press; 1990. pp. 76–83. [Google Scholar]
- 30.Meisenzahl A C, Shapiro L, Jenal U. Isolation and characterization of the xylose-dependent promoter from Caulobacter crescentus. J Bacteriol. 1997;179:592–600. doi: 10.1128/jb.179.3.592-600.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 32.Minnich S A, Newton A. Promoter mapping and cell cycle regulation of transcription in Caulobacter crescentus. Proc Natl Acad Sci USA. 1987;84:1142–1146. doi: 10.1073/pnas.84.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Modrich P. Methyl-directed DNA mismatch correction. J Biol Chem. 1989;264:6597–6600. [PubMed] [Google Scholar]
- 34.Newton A, Ohta N, Ramakrishnan G, Mullin D, Raymon G. Genetic switching in the flagellar gene hierarchy of Caulobacter requires negative as well as positive regulation of transcription. Proc Natl Acad Sci USA. 1989;86:6651–6655. doi: 10.1073/pnas.86.17.6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nou X, Skinner B, Braaten B, Blynn L, Hirsch D, Low D. Regulation of the pyelonephritus-associated pili phase-variation in Escherichia coli: binding of the PapI and the Lrp regulatory protein is controlled by DNA methylation. Mol Microbiol. 1993;7:545–553. doi: 10.1111/j.1365-2958.1993.tb01145.x. [DOI] [PubMed] [Google Scholar]
- 36.Ogden G, Pratt M, Schaechter M. The replicative origin of the E. coli chromosome binds to cell membranes only when hemi-methylated. Cell. 1988;54:127–135. doi: 10.1016/0092-8674(88)90186-9. [DOI] [PubMed] [Google Scholar]
- 37.Ohta N, Lane T, Ninfa E G, Sommer J M, Newton A. A histidine protein kinase homologue required for regulation of bacterial cell division and differentiation. Proc Natl Acad Sci USA. 1992;89:10297–10301. doi: 10.1073/pnas.89.21.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohta N, Chen L S, Mullin D A, Newton A. Timing of flagellar gene expression in the Caulobacter cell cycle is determined by a transcriptional cascade of positive regulatory genes. J Bacteriol. 1991;173:1514–1522. doi: 10.1128/jb.173.4.1514-1522.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 40.Poindexter J S. Biological properties and classification of the Caulobacter group. Bacteriol Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quon K C, Marczynski G T, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 42.Quon K C, Yang B, Domian I J, Shapiro L, Marczynski G T. Negative control of DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci USA. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reisenauer A, Mohr C D, Shapiro L. Regulation of a heat shock ς32 homolog in Caulobacter crescentus. J Bacteriol. 1996;179:1919–1927. doi: 10.1128/jb.178.7.1919-1927.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts R C, Toochinda C, Avedissian M, Baldini R L, Gomes S L, Shapiro L S. Identification of Caulobacter crescentus operon encoding hrcA, involved in negatively regulating heat-inducible transcription, and the chaperone gene grpE. J Bacteriol. 1996;178:1829–1841. doi: 10.1128/jb.178.7.1829-1841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rost B. PHD: predicting one-dimensional protein structure by profile based neural networks. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 46.Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–77. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 47.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 49.Sanger F, Micklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sander C, Schneider R. Redefining the goals of protein secondary structure prediction. J Mol Biol. 1991;235:13–26. doi: 10.1016/s0022-2836(05)80007-5. [DOI] [PubMed] [Google Scholar]
- 51.Shapiro L. The bacterial flagellum: from genetic network to complex architecture. Cell. 1995;80:525–577. doi: 10.1016/0092-8674(95)90505-7. [DOI] [PubMed] [Google Scholar]
- 52.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 53.Stephens C, Reissennauer A, Wright R, Shapiro L. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc Natl Acad Sci USA. 1996;93:1210–1214. doi: 10.1073/pnas.93.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stratagene. Stratagene 1997/1998 catalog. La Jolla, Calif: Stratagene; 1997. [Google Scholar]
- 55.Wang S, Sharma P, Schoenlein P, Ely B. A histidine protein kinase is involved in polar organelle development in Caulobacter crescentus. Proc Natl Acad Sci USA. 1993;90:630–634. doi: 10.1073/pnas.90.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wingrove J A, Gober J W. Identification of an asymmetrically localized sensor histidine kinase responsible for temporally and spatially regulated transcription. Science. 1996;247:597–609. doi: 10.1126/science.274.5287.597. [DOI] [PubMed] [Google Scholar]
- 57.Wingrove J A, Mangan E K, Gober J W. Spatial and temporal phosphorylation of a transcriptional activator regulates pole-specific gene expression in Caulobacter. Genes Dev. 1993;7:1979–1992. doi: 10.1101/gad.7.10.1979. [DOI] [PubMed] [Google Scholar]
- 58.Wu J, Ohta N, Newton A. An essential multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter crescentus. Proc Natl Acad Sci USA. 1998;95:1443–1448. doi: 10.1073/pnas.95.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zweiger G, Marczynski G, Shapiro L. A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J Mol Biol. 1994;235:472–485. doi: 10.1006/jmbi.1994.1007. [DOI] [PubMed] [Google Scholar]