Abstract
Extracellular vesicles (EVs), is the umbrella term used for different types of vesicles produced by the cells, among which exosomes form the largest group. Exosomes perform intercellular communication by carrying several biologics from donor or parental cells and delivering them to recipient cells. Their unique cargo-carrying capacity has recently been explored for use as delivery vehicles of anticancer drugs and imaging agents. Being naturally produced, exosomes have many advantages over synthetic lipid-based nanoparticles currently being used clinically to treat cancer and other diseases. The finding of the role of exosomes in human diseases has led to numerous preclinical and clinical studies exploring their use as an amenable drug delivery vehicle and a theranostic in cancer diagnosis and treatment. However, there are certain limitations associated with exosomes, with the most important being the selection of the biological source for producing highly biocompatible exosomes on a large scale. This review article explores the various sources from which therapeutically viable exosomes can be isolated for use as drug carriers for cancer treatment. The methods of exosome isolation and the process of loading them with cancer therapeutics and imaging agents are also discussed in the follow-up sections. Finally, the article concludes with future directions for exosome-based applications in cancer diagnosis and treatment.
Keywords: Extracellular vesicles, exosomes, cancer, drug delivery, theranostics
Graphical Abstract
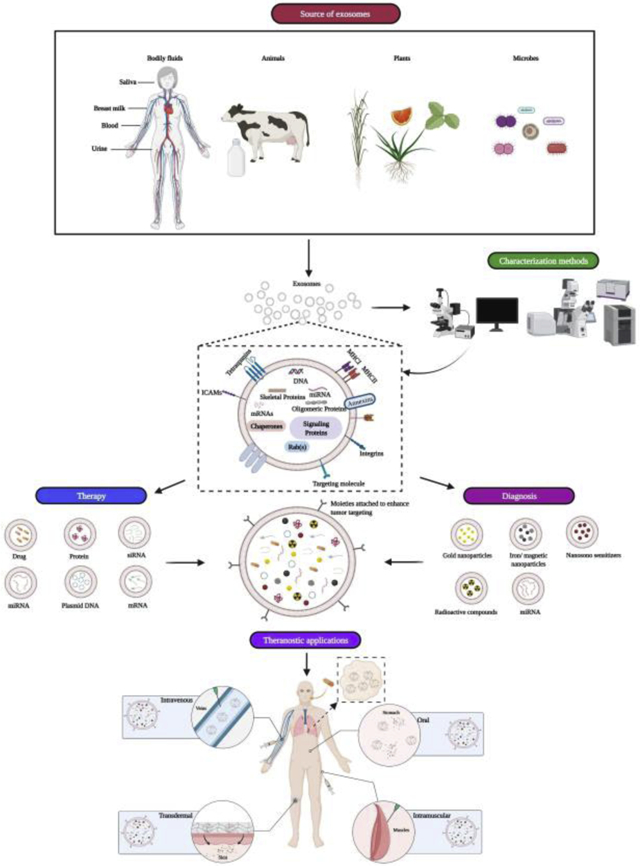
1. Introduction
The success of a chemotherapeutic treatment regimen largely depends on the efficient and precise delivery of therapeutic agents to cancer cells with minimal side effects or cytotoxicity to healthy cells [1]. An efficient and safe drug delivery system is desperately required to reduce the side effects of chemotherapy and improve patient outcomes. For many years, nanomedicine researchers have worked to develop multiple synthetic nanomaterials that can function as drug carriers and deliver anti-cancer therapeutics in a targeted fashion to cancer cells [2, 3]. Although significant progress has been made in this regard, few of the drug carriers have reached the clinic. Poor biocompatibility, inefficient bio-distribution, and unintended immunogenic responses are a few reasons for the failure of many synthetic drug delivery complexes. Liposomes, which are formulated using various compositions of phospholipids and cholesterol, have been the most successful drug delivery system developed and tested for cancer treatment to date. With a composition similar to that of the cell membrane, liposomes show better biocompatibility than do other synthetic drug delivery systems. Furthermore, liposomes are amenable to modifications to accommodate multiple therapeutic payloads (e.g., plasmid DNA, hydrophilic and hydrophobic drugs, siRNA, or miRNA) and tumor-targeting moieties (e.g., monoclonal antibodies, aptamers, or protein ligands) [4, 5]. Incorporation of tumor-targeted ligands facilitates tumor-targeted drug delivery, increased drug accumulation in the tumor, and enhanced antitumor activity with minimal off-target side effects. Active targeting in combination with the inherent enhanced permeability and retention (EPR) effect contributes to the enhanced antitumor activity [6]. These unique properties have made liposomes one of the most acceptable drug delivery method. Several such liposome formulations, including Doxil®, DaunoXome®, and Mariqibo®, and the more recently approved Onivyde®, Vyxeos®, and Onpattro®, are available for the treatment of cancer and other diseases [6, 7]. A search for clinical trials involving liposomes and cancer at “clinicaltrials.gov” reports a total of 1059 clinical trials that have used or will use liposomes in cancer (www.clinicaltrials.gov last accessed on May 7th 2021). However, despite their superior therapeutic carrying capability, liposomes have many drawbacks due to their synthetic nature. These include inferior targeting, generation of a cytokine storm and immune response, morphological instability, and their inability to pass through many stringent biological barriers, such as the blood brain barrier (BBB) [8–10].
As an alternative to synthetic liposomes, ‘natural nanoparticles,’ such as the extracellular vesicles (EV’s), produced by plants, microbes, or the body’s own cells, are under investigation as carriers of anti-cancer drugs. EV’s is a broad term used to define a spectrum of vesicle-like structures produced and released by cells into the extracellular milieu [11]. EVs are heterogeneous, and their classification is based on their mode of biogenesis, size, and morphology (Table 1A and B; Figure 1). Each type of EV has a functional role in the cell and contributes to different physiological processes [12]. Exosomes comprise the largest group of EVs and are classically defined as nano-sized (30–150 nm), lipid bilayer vesicles produced by the endosomal pathway by all types of cells [13, 14]. In 2018, The International Society of Extracellular Vesicles (ISEV) published the “Minimal Information for Studies of Extracellular Vesicles” (“MISEV”) which refrains from using the term exosomes in the absence of an exosome specific marker and rather suggests classifying EVs on the basis of size (small EVs < 100 nm or < 200 nm; medium/large EVs > 200 nm) [15]. Based on transmission electron microscopy (TEM), several studies report exosomes as cup-shaped organelles; however, the cryo TEM images of freshly frozen exosomes shows a near circular structure [16]. The biogenesis, and hence packaging of exosomes, occurs through a highly orchestrated endocytic pathway with the mature exosomes are released into the extracellular milieu by fusion of multivesicular bodies and the plasma membrane [17].
Table 1.
Popular modes of classifying extracellular vesicles (EVs)
| Type of EV | Size (Diameter in nm) | Morphology | References |
|---|---|---|---|
| Small EVs | < 100 nm < 200 nm | Spherical | [15] |
| Medium/large EVs | > 200 nm | Spherical | [15] |
| Type of EV | Production Mode | Morphology | References |
| Apoptotic bodies | Blebbing | Heterogeneous | [190] |
| Microvesicles | Outward budding and shedding | Spherical | [190] |
| Exosomes | MVBs fusion with plasma membrane | Spherical or cup shaped with bilayer membrane | [191] |
| Exomeres | MVBs fusion with plasma membrane | Non-membranous, spherical | [192] |
| Migrasomes | Formed during cell migration | Pomegranate-like structures-oval shaped | [193] |
| Oncosomes | vesicle budding and membrane scission | Spherical | [194] |
Figure 1.
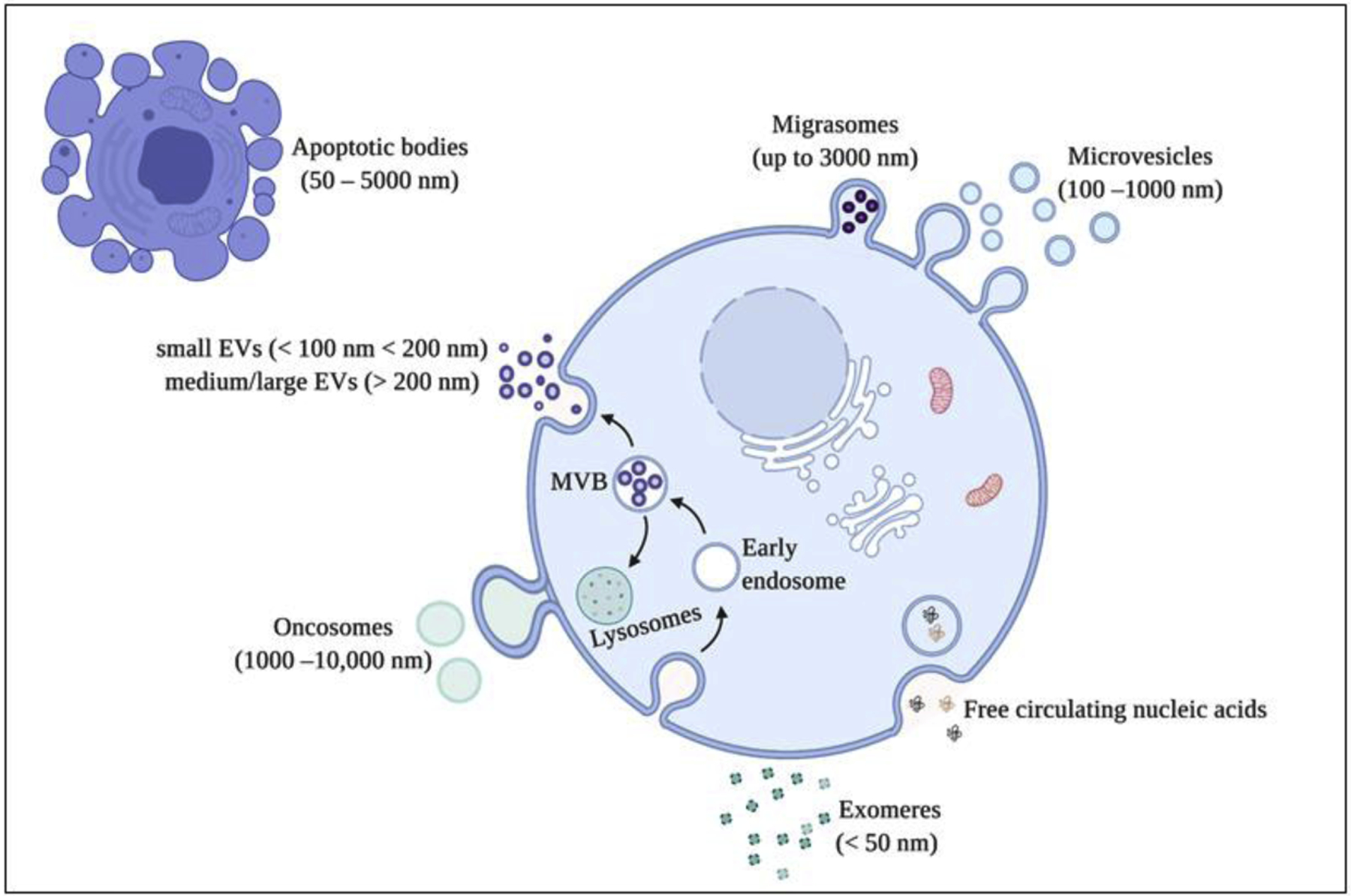
Different types of extracellular vesicles produced by the cell. These vesicles are categorized based on their mode of synthesis and size. Figure created with BioRender.com
During synthesis, exosomes are packaged with cargo consisting of different bioactive molecules, such as lipids, proteins, DNA, RNA, and several species of small non-coding (nc) RNAs and long non-coding (lnc) RNAs. The surfaces of the exosomes are embedded with several fusion proteins that are involved in navigation (targeting) and attachment to the surface of recipient cells (cellular adhesions). The most enriched proteins in exosomes are tetraspanins (CD63, CD81, and CD9) and targeting proteins (integrins) [18] [19]. Lipids also form a substantial part of exosome structure and cargo. Lipids that are enriched in the exosome membrane include cholesterol, sphingomyelins (SM), ceramide, lyso(bis)phosphatidic acid, lysophosphatidylcholines, phosphatidylcholines, phosphatidylethanolamines, phosphatidylserines (PS), phosphatidylinositols, phosphatidic acid, and phosphatidylglycerol. The composition of the lipids, akin to proteins and nucleic acids, present in the exosomes resembles that of the parental cells [20, 21].
The content of exosomes is significant as it reflects the composition and physiological status of its cell of origin. Since exosomes are present in all bodily fluids including urine, blood plasma/serum, and cerebrospinal fluid (CSF), and can be easily purified from such body fluids, they are used to study the pathobiology of disease and to develop novel liquid biopsy modalities for the diagnosis of diseases, including cancer [22, 23]. Exosomes were discovered in the early 1940s, and were primarily regarded as a mode of removing unwanted toxic materials from cells. However, in the late 2000s, several seminal discoveries highlighted the potential of exosomes in pathobiology and biomedical research [24]. This article aims to discuss the potential of exosomes isolated from several natural sources in the areas of cancer therapy and diagnosis.
2. Biogenesis of exosomes
The biogenesis of exosomes occurs when early endosomes mature to late endosomes. At this stage, the inner membrane folds inwards to form a small, vesicle-like structure. These vesicles are called intraluminal vesicles (ILVs), and the endosomes carrying them are known as multivesicular bodies (MVBs). There exists a cascade of signaling pathways that regulates the packaging of molecules into the lumen of ILVs [25]. The site of parental cells in the body and their physiological condition determines the biosynthesis and composition of exosomes [26]. In general, the endosomal sorting complex required for transport (ESCRT) proteins TSG101, STAM1, Alix, and CD9 are implicated in regulating the pathways involved in the packaging and biosynthesis process [27]. Following the packing and biosynthesis, the Rab-GTPase family of proteins, such as Rab2B, Rab5A, Rab7, Rab11, Rab27, and Rab35, are involved in the release of ILVs as exosomes into the extracellular cellular milieu as a result of fusion of MVBs with the plasma membrane [28, 29]. Finally, the SNARE family of proteins and other adhesion and targeting proteins guide and steer the exosomes to attach to recipient cells and release their contents [30]. Understanding the complex process involved during the biosynthesis of exosomes has facilitated development of strategies to regulate metastasis and tumor growth. Using mouse models, Ghoroghi et al. delineated the effect of GTPases of the Ral family on regulating exosome biogenesis by controlling the maturation of MVBs; they described how members of the Ral family, RalA and RalB or Ral A/B, are involved in the packaging of exosomes and dictate their organotropic capabilities, which enables the exosomes to reconfigure the tissue microenvironment to form a metastatic niche. The studies were performed on exosomes involved in lung metastasis of breast cancer cells [31].
Serum starvation also impedes exosome biosynthesis; THP-1 cells cultured in RPMI-1640 cell culture medium supplemented with 10% FCS secreted fewer exosomes than cells grown in cell culture medium supplemented with 1% FCS [32]. Similarly, hypoxia influences exosome biosynthesis, and multiple studies show that any kind of cellular stress induces the production of exosomes. This may be why many studies claim that cancer cells produce more exosomes than do their normal counterparts [33]. ESCRT-independent exosome biosynthesis has also been reported, indicating multiple mechanisms are operational in exosome biogenesis [34–36]. In addition to proteins, lipids and several other molecules have been associated with the biosynthesis pathway; these have been extensively described in review articles by Théry and Kowal [37, 38].
Understanding the mechanisms and constituents of exosome synthesis is essential for their application as diagnostic and therapeutic interventions, and thus attaining the goal of developing a potent theranostic for cancer treatment.
3. Characteristics of exosomes similar to liposomes
Exosomes can stably carry and transfer different biomolecules to recipient cells throughout the body, suggesting a new mode of intercellular communication. Building on this notion, scientists have also successfully engineered exosome contents in order to realize its potential as a natural drug delivery system [39]. Exosomes have many similarities with synthetic liposomes; the principal similarity is the composition of their membrane, which in both cases consists of lipoproteins. Additionally, liposomes and exosomes have similar size ranges [40], both have diameters of 20–200 nm, which allows them to move easily inside the body. Their size and composition also help in the packaging of cargo into their lumens.
4. Advantage of exosomes as a delivery vehicle
Exosomes are naturally produced by all cells in all living systems [41]. Despite having many similarities with synthetic drug delivery systems, including liposomes as discussed above, exosomes have certain unique characteristics that make them ideal for drug delivery. Exosomes are designed to transport cellular material from parental cell to recipient cells, and their membranes are composed of a constellation of proteins, including tetraspanins and other adhesion proteins. The presence of such proteins assists with assessing exosomes as drug delivery vehicles and reduces the off-target effects of any administered therapy [42].
Exosomes intrinsically possess many traits that are desirable for an ideal drug delivery vehicle. These include: (A) they do not elicit a measurable host immune response, i.e. they are non-immunogenic; (B) their uptake by recipient cells occurs through various mechanisms including receptor-mediated binding and internalization; (C) their surface can be functionalized for cell specific targeting; (D) they are stable and can be lyophilized without impacting their structural or biological properties; (E) they inherently carry small molecules and cargoes (e.g. mRNA, miRNA, and proteins among others,) and operate as biological carriers and cell signal inducers; and finally (F) the use of cross-species exosomes has shown no ill effect on recipient cells, suggesting the possibility of using several alternative sources for isolation of exosomes for use as drug carriers [43] [44]. Thus, exosomes have many properties that supersede liposomes (Table 2) and can be considered a potential replacement for liposomes as a drug delivery modality.
Table 2.
| Properties | Liposomes | Exosomes |
|---|---|---|
| Synthesis | Artificial | Synthesized naturally by cells by a well-regulated cellular mechanism |
| Scalability | Can be mass produced for clinical applications | Yet to establish a method to scale up the production of desired exosomes. |
| Polydispersity | Absent (monodisperse) | Present |
| Membrane potential | Variable | Negative |
| Immune stimulation | Present | Absent |
| Targeting | Targeting per se is absent, additional molecules can be added for targeting. | Several molecules (proteins) are present on the surface that de novo imparts targeting effect. |
| Organotropism | Absent | Present |
| Cellular uptake | Through undefined mechanism | Through well-defined mechanisms like micropinocytosis, receptor mediated absorption or diffusion |
| Multifunctionality (loading multiple therapeutics or signaling/targeting molecules) | Possible but involves multiple steps | Many molecules of therapeutic interest (like miRNAs) are naturally present in the exosomes in addition to several surface proteins that can help in targeting. Additionally, the molecules of interest can be consistently packaged into the exosomes by over expressing the parental cells. |
| Toxicity | Shows toxicity | Absent |
5. Contribution of Exosomes in imaging
In addition to effectively delivering therapeutics, exosomes as theranostics have also been exploited to develop imaging applications to monitor the physiological state of the body and observe the biodistribution of delivered exosomes carrying the therapeutics. Precise tracking of exosomes and their accumulation in the targeted site help investigators develop high-precision therapeutic modalities for efficient cancer treatment. Besides, the imaging of accumulated exosomes loaded with imaging agents enables physicians and scientists to diagnose and monitor the treatment response. Several methods have been developed to label exosomes for optical, fluorescence/bioluminescence, ionizing, and non-ionizing imaging modalities in the past few years.
5.1. Labeling exosomes for fluorescence and bioluminescence imaging
Exosomes can be labeled with different fluorescent molecules and studied for their role in disease pathophysiology. Lipophilic agents like PKH67, Di dyes (DiO, Dil, DiD, DiA, etc.), and CFSE are dyes routinely used for labeling exosomes. Among them, PKH67 is one of the most widely used lipophilic labeling dye for in vitro and in vivo tracking of exosomes [45]. Labeling of exosomes also aids in determining the loading efficiency of therapeutic molecules encapsulated as therapeutic cargo. Didiot et al. labeled the therapeutic siRNA (hsiRNA to silence Htt mRNA and HTT protein) with Cy3 and labeled exosomes with PKH67. The imaging result from this study strongly suggested that loading exosomes with hsiRNAHTT did not change the cellular uptake of exosomes, demonstrating that exosomes labeled with hsiRNAHTT were stable in the recipient primary neurons [46]. Tracking exosomes using lipophilic dyes is a popular and straightforward method, but it is certainly not the most favored one because of their non-specificity and high stability in the cells leading to false positive results [47].
Another approach to label exosomes is to transfect cells with green fluorescent protein (GFP)-expressing plasmid such as GFP-CD63 plasmid. Exosomes produced from such transfected cells will be de novo labeled with fluorescent molecules such as GFP. We have adopted this approach in our laboratory to produce and image GFP labeled exosomes. By using these labeled exosomes, the uptake of drug loaded exosomes (nanosomes) by the recipient lung cancer cells H1299 was demonstrated [48]. Satake et al., while working on metastatic pancreatic cancer nude-mouse models, developed color-coded imaging method to observe disease progression and metastasis. The study utilized the red fluorescent protein (RFP) labeled Mia-PaCa-2pancreatic cancer cell line. These cells were transduced by exosome-specific pCT-CD63-GFP to produce GFP labeled exosomes. The fluorescence imaging of cells and exosomes helped investigators determine the significance of exosomes in metastasis and their role in the pathophysiology of disease progression [49].
Along similar lines, Hikita et al. developed an imaging method that utilizes bioluminescence resonance energy transfer (BRET)-based reporter Antares2. Antares-2 is an Nluc-based luciferase conjugated with CyOFP1, a cyan-excitable red fluorescent protein. This imaging modality was intended for the long-term accumulation of exosomes in vivo. The investigators in this study conjugated CD63 with Antares2 to label exosomes. The emitted intense luminescence signal was used to determine the number of cancer-derived exosomes in a mouse model implanted with CD63-Antares2-expressing prostate cancer cells. The study further confirmed the homing property of exosomes towards their targeted organs and tissues. The investigators consider the CD63-Antares2 xenograft mouse model to be important for studying the role of exosomes in cancer pathophysiology and developing therapeutic and imaging modalities related to cancer treatment [50]. In a recent study, researchers have reported a more direct in situ one-step fluorescence labeling method for exosomes via bio-orthogonal click chemistry. In this strategy, exosome producing parental cancer cells were modified by treating with tetra-acetylated N-azidoacetyl-d-mannosamine (Ac4ManNAz) followed by labeling with near-infrared fluorescent dye-conjugated dibenzylcyclooctyne (DBCO-Cy5) via bioorthogonal click chemistry to the free azide group present on the surface of cells. This strategy resulted in the production of fluorescent exosomes. Investigators further tested the DBCO-Cy5-labeled exosomes for their biocompatibility and characteristics. They demonstrated that the labeled exosomes showed no difference in their uptake by the recipient cells and other functional characteristics. The study further evaluated the imaging capability of the Cy5-Exo after intravenously injecting into tumor-bearing mice and non-invasively tracking and imaging using near-infrared fluorescence (NIRF) imaging. The study thus provides a new tool for developing efficient theranostics using exosomes [51].
In addition to fluorescent labeling of exosomes, Chen et al. developed a single-molecule localization-based super-resolution imaging technique for imaging and tracking cancer-derived exosomes. In this strategy, the membrane receptors present on the surface of exosomes were labeled with photo-switchable probes that help perform super-resolution imaging by photo activated localization microscopy (PALM) or stochastic optical reconstruction microscopy (STORM). The researchers studied PALM/STORM imaging of breast cancer-derived exosomes and demonstrated the feasibility of imaging cancer exosomes in vivo when administered as a theranostic in a treatment regimen [52].
5.2. Exosomes loaded with metallic nanoparticles as a contrast agent for imaging.
Earlier, we discussed metallic nanoparticles loaded exosomes functioning as a therapeutic delivery vehicle. Besides functioning as a therapeutic vehicle, these metal nanoparticle-loaded exosomes can also be used in imaging as they (due to metal nanoparticles) serve as excellent contrasting agents. Cohen et al. used exosomes from two different sources, viz. squamous cell carcinoma A431 and mesenchymal stem cells (MSC-Exo), for adding gold nanoparticles (GNPs) and evaluated them for imaging through non-invasive computed tomography (CT) in in vivo mice model. This study demonstrated the homing ability of exosomes and the use of non-invasive CT strategy for determining the biodistribution of exosomes after they were injected into the body [53].
Developing exosomes as both, an imaging agent as well as a therapeutic cargo carrier simultaneously, has been an overarching goal of exosome-based theranostics. In the study by Bose et al., the authors demonstrated the theranostic potential of exosomes by coating them with gold–iron oxide nanoparticles (GIONs) and delivering anti-miR-21 in breast cancer cells. While the delivered anti-miR-21 imparted therapeutic effect by reducing doxorubicin resistance, the GIONs demonstrated excellent T2 contrast and hence magnetic resonance (MR) imaging [54]. Similar to loading of therapeutic cargo into exosomes, researchers interested in developing exosome-based theranostics have explored several approaches for efficient and stable loading of imaging agents and therapeutic molecules in exosomes without bringing any appreciable change into the physicochemical characteristics of exosomes. Silva et al. demonstrated one such strategy by adding magnetic nanoparticles and multiple therapeutic molecules such as doxorubicin, tissue-plasminogen activator (t-PA), and two photosensitizers (disulfonated tetraphenyl chlorin-TPCS2a and 5,10,15,20-tetra(m-hydroxyphenyl)chlorin-mTHPC) to macrophages. The exosomes derived from the macrophages showed the presence of all added therapeutics and magnetic nanoparticles. This study successfully demonstrated the MR imaging (MRI) and therapeutic delivery through this vehicle, suggesting that this approach could be an ideal strategy to develop exosome-based theranostic [55]. A few studies have been carried out to develop a multi-model or hybrid system where more than imaging modalities can be employed simultaneously, such as GNPs for PET imaging, SPIONs for MRI, or labeling with fluorescent proteins such as GFP, RFP, and other labeling molecules [56]. Shaikh et al. demonstrated the impact of a multimodal imaging system when they used multimodal imaging techniques such as CT, MRI and fluorescence, and demonstrated an enhanced sensitivity and specificity of imaging of tumor cells both in vitro and in vivo. The iridium and iron oxide nanoclusters (NCs) used to develop the multimodal imaging tool was also shown to be encapsulated into the tumor-derived exosomes thus serving as a potential biomarker for cancer diagnosis [57]. Tayaba et al. loaded exosomes with silver and iron oxide nanoclusters (NCs) produced in situ in HepG2 liver cancer cells. The authors of the study demonstrated the potential of Ag NCs as a fluorescent probe and Fe3O4 NCs as a contrast agent for CT and MRI. The research showed the significance of using NC-loaded exosomes for cancer early cancer diagnosis and detection [58].
5.3. Radiolabeled exosomes for imaging.
Non-invasive in vivo detection or imaging of exosomes can be performed by radiolabeling exosomes. In addition to being non-invasive, imaging of radiolabeled exosomes gives very precise information and shows a high degree of sensitivity compared to other available imaging tools. In a recent study, researchers demonstrated the improved imaging capabilities by developing a simple method for radiolabeling mouse RAW264.7 macrophage cell-derived exosome-mimetic nanovesicles (ENVs) with (99m)Tc-HMPAO under physiologic conditions. They evaluated the distribution of intravenously injected × labeled (99m)Tc-HMPAO-ENVs using SPECT/CT in living BALB/c mouse mice. The study results showed that the radiolabeled exosomes or ENVs were detectable in the serum even five hours after inoculation indicating their high stability in vivo with no marked changes in their morphology or size. The study concluded that non-invasive radiolabel-based imaging could be an ideal tool to enhance exosome-based research, especially while studying disease pathophysiology or therapeutics [59].
Faruqu et al. in their study explored a reliable and sustainable method to radiolabel exosomes. They used two different approaches to radiolabel melanoma (B16F10) cell-derived exosomes. The first approach of intraluminal labeling involved entrapment of 111Indium via tropolone shuttling, while the second approach involved membrane labeling which requires chelation of 111Indium via covalently attached bifunctional chelator, DTPA-anhydride. The study demonstrated that the membrane-labeled ExoB16 showed superior radiolabeling efficiency and radiochemical stability (19.2 ± 4.53 % and 80.4 ± 1.6 % respectively) compared to the intraluminal-labeled exosomes (4.73 ± 0.39 % and 14.21 ± 2.76 % respectively). Further, biodistribution study with the membrane-labeling approach carried out in both immunocompromised (NSG) and immunocompetent C57BL/6 mice showed similar outcomes with maximum accumulation of administered particles in liver and spleen followed by the kidney. On the one hand, this study establishes membrane radiolabeling as a preferred method of labeling exosomes with radioisotopes. At the same time, it also establishes a method to do live imaging of exosomes, thereby opening up a potential avenue to develop image-guided therapeutic approaches, which is one of the goals of developing a theranostic [60]. Molavipordanjani et al. exploited the radiolabeling of exosomes and the acquired imaging capacity to demonstrate the tumor-targeting effect of exosomes as a result of the addition of a tumor-targeting moiety. In this study, genetically engineered cells expressing HER2 receptor-ligand DARP in G3 were used to obtain exosomes that were further converted to 99mTc-radiolabeled HER2 targeted exosomes (99mTc-exosomes). The imaging of 99mTc-exosomes displayed a higher affinity toward high HER2 expressing SKOV-3 cells compared to low HER2 expressing MCF-7, HT29, U87-MG, and A549 cell lines. Further blockage of HER2 receptor using Trastuzumab inhibited the binding of 99mTc-exosomes to SKOV-3 cells by up to 40%. Besides, the biodistribution study in the SKOV-3 tumor mice model demonstrated the tumor targeting of 99mTc-exosomes by showing enhanced accumulation in the tumor. In addition to demonstrating the tumor-targeting effect of 99mTc-exosomes, this study also demonstrated how radiolabeled exosomes can be exploited for imaging in conjunction with therapeutic delivery, thereby furthering their use as potent theranostic tools for cancer treatment [61].
One of the challenges with the exosome-based delivery system is the systemic clearance of the injected exosomes by the liver, resulting in suboptimal tumor retention, thereby leading to a suboptimal treatment response. Shi et al. attempted this issue by designing radiolabeled PEGylated exosomes. The study showed that the radiolabeled PEGylated exosomes exhibited significantly enhanced tumor uptake, better in vivo PET imaging capability of tumor, and reduced hepatic clearance of exosomes [62].
The results from the studies described above establishes the use of exosomes for cancer imaging and diagnostic purposes and strategies for overcoming some of the limitations faced in exosome-based imaging.
The next set of pertinent issues that need to be addressed are: a). identification of a suitable source to isolate exosomes for developing theranostics, b). to formulate methodologies for efficient loading of desired therapeutic or diagnostic molecules, and finally, c). to isolate a homogenous population of exosomes in a large scale from the identified sources. Since the molecular formulation of exosomes is governed by its originating cells, it becomes critical to select the source of exosomes to be used for developing carriers for theranostics. Additionally, compatibility to functionalize with desired molecules and feasibility of isolation on a large scale are essential criteria that need to be considered to establish a ‘natural’ therapeutic and imaging modality with superior properties. These questions are discussed in the following sections of the article in an attempt to provide a snapshot into the development of novel exosome based theranostics, discuss potential bottlenecks as well as address the areas in need of improvement. This comprehensive information will enable the scientific community to design new ways to circumvent the current disadvantages and develop an amenable technology such that it can serve as a potential therapeutic intervention in cancer treatment.
6. Source of exosomes
6.1. Cell-derived exosomes
Cell-derived exosomes are the preferred resource for developing drug delivery vehicles. Cells can be cultured in a controlled and optimized environment to produce a near-homogenous population of exosomes in large quantities. Because exosomes have similar compositions to their cells of origin, researchers can explore different cell types in order to identify the most appropriate source of therapeutic-grade exosomes. Various parameters, such as production capacity, immunogenicity, ability to genetically manipulate to produce and load exosomes with the desired cargo, and the capacity to maintain long-term continuous exosomes production, are critical to examine prior to selecting the cell source for exosomes. Below we discuss some cell types that are being investigated as a source of exosomes for drug delivery.
6.1.1. Dendritic cells (DCs)
DCs are antigen-presenting cells that have been used in several studies to produce exosomes and are often referred to as Dexosomes or Dex. Alvarez–Erviti et al. were the first to show that small regulatory RNA, such as siRNA, can be successfully transferred to recipient cells and confer a therapeutic effect by engineering DC-derived exosomes [63]. DC-derived exosomes can mediate MHC-dependent immune responses and have been explored as cell-free vaccines for cancer treatment. Pitt et al. noted several advantages of Dex over DCs in design of potent immunotherapy interventions; these advantages included the ability to carry and maintain essential immunostimulatory properties of DCs, the greater stability of Dex compared to DCs, and an overall simpler approach lacking the complexities associated with using cells for treatment [64].
6.1.2. Natural killer (NK) cells
NK cells are a type of lymphoid cells that do not require any antigen priming and hence can vigorously execute an immune response against invading pathogens or aberrant cells such as cancer cells. NK cell-based immunotherapy in recent years has shown promising anti-tumor response. Similar to other cell types, NK cells also synthesize and release exosomes into their external environment. Several studies have recently reported that NK cell-exosomes can display a potent immune-modulatory and cytotoxic response against invading cancer cells [65] [66]. The content of NK cell-derived exosomes consists of several cytotoxic and tumor targeting proteins like FasL, perforin, Granzyme A and B, NKG2D, CD94, and CD40L that contribute to the observed toxic effects documented in the studies involving leukemia, neuroblastoma, and multiple other solid tumors. Additionally, NK-cell derived exosomes also carry several miRNAs that can stimulate other immune cells to cause immunomodulatory effects besides causing cytotoxicity to tumor cells [66–68] [66–68]. Exosomes isolated from resting and activated NK cells display cytotoxic property that selectively kills tumor cells and modulates activated immune cells. Besides, enhanced cellular uptake of NK cell-exosomes has also been observed while sparing the resting PBMCs [69].
Muller et al. in their study showed tumor-targeted killing by NK cell-derived exosomes. These exosomes carried miR-186 that subdued MYCN-amplified neuroblastoma cells and disrupted the TGFβ1-dependent inhibition of NK cells. Thus, this study presented convincingly an option of developing an NK cell-derived exosome-based therapeutic delivery system that simultaneously averts tumor growth and TGFβ1-dependent immune escape in high-risk neuroblastoma patients thereby augmenting NK cell-based immunotherapy [70]. As a result, augmentation of NK cell-derived exosomes has been seen as a potential modality to organically enhance cancer immunotherapy. To further adapt this premise of treatment into a pragmatic option, Jong et. al., proposed a methodology to enhance the isolation of NK-cell EV, while another group proposed the development of NK cell exosome mimetics to be used for the formulation of large-scale NK-cell exosome-based cancer therapy [71, 72]. In addition to this, Xu et al. also showed that NK-cell exosomes can be produced via autogenic and allogenic routes, increasing the scope of NK-cell exosome isolation for therapeutic purposes [67].
As another synthetic approach to enhance the NK cell-derived exosome production for theranostic application, Kang et. al. developed a microfluidic device that collects NK and CT cells from NSCLC patients for harvesting NK-cell derived exosomes on a chip. To demonstrate that the exosomes isolated from NK cells on-chip retain the theranostic capability, the study showed a difference in the number of NK cell derived exosomes isolated from NSCLC patients and normal individuals. These exosomes also exhibited a cytotoxic effect when administered to CTCs [73].
6.1.3. Macrophage cells
Macrophages are important components of the immune system involved in the removal of dead cells and found at the site of inflammation. Exosomes derived from macrophages are expected to retain the properties of immune cells and hence should show an enhanced anti-tumor effect when used as a carrier vehicle to deliver anticancer therapeutics. Li et. al. exploited this notion by designing a tumor targeted delivery vehicle based on exosomes isolated from macrophages and decorated them by poly(lactic-co-glycolic acid) and targeting peptide ligand to target the mesenchymal-epithelial transition factor (c-Met) of triple negative breast cancer cells (TNBC). The tumor targeted delivery vehicle that they developed enhanced cellular uptake and significant anti-tumor efficacy of doxorubicin in vitro and in vivo [74].
Photodynamic therapy is a novel cancer therapy that can treat cancerous lesions and tumors with minimal disease residue. The major disadvantage of photodynamic therapy is the resulting non-targeted cytotoxicity and inability of administration of drug/molecules in therapeutic dose range. The application of free photodynamic therapeutic yields unfavorable results. Combining exosomes and photodynamic therapeutic molecules has recently paved the way to evaluate a novel and effective modality for cancer treatment [75]. Iessi et al. used macrophage-derived exosomes to load Acridine Orange (AO), an acidophilic dye with fluorescence property that shows an anti-tumor effect when excited at a suitable wavelength. The AO loaded onto macrophage-derived exosomes (M ϕ Exo-AO) circumvented the off-target cytotoxicity caused by AO as a stand-alone therapeutic agent and improved the antitumor activity against sarcoma grown in 2D and 3D cell culture conditions. The study results demonstrated the potential utility for Exo-AO based theranostics in future [76].
One of the concerns of using nanomaterials in nanomedicine intervention is the accumulation of administered nanomaterials and their possible long-term toxic manifestations. Logozzi et al. in their study showed that exosomes isolated from macrophages support the cells by scavenging harmful molecules and clearing the body. The study found that 20nm gold nanoparticles (AuNPs) when administered to the body were effectively cleared via macrophage-derived exosomes [77]. This study substantiates the observations by Federici et al. that demonstrated through HPLC-Q-ICP-MS analysis the presence of native cisplatin in exosomes isolated from cisplatin treated tumor cells and hence were putatively responsible for causing resistance to the given cisplatin-based chemotherapy. An interesting observation of these two studies is that exosomes derived from cells can de novo carry metallic nano-molecules into their lumen [78].
6.1.4. Hybridoma cells
Monoclonal antibodies as therapeutic agents have been very successful treatment regimen for many cancers. Their high specificity and targeting ability are the vital properties that make them indispensable for cancer treatments. Extracellular vesicles or exosomes purified from hybridoma cells have the unique ability to package and express antibodies into their lumen and membrane, respectively. A study conducted on OKT3 hybridoma cells, responsible for producing murine monoclonal antibody IgG2a required for successful organ transplantation, showed that the OKT3 cells produce exosomes that can stimulate cytokine production in CD4 and CD8 T cells. This suggests that hybridoma-derived exosomes could be ideal for immunotherapy as they can stably and specifically carry and display antibodies (immunoglobins) and specific ligands useful for successful immunotherapeutic intervention [79]. Similarly, the chimeric antigen receptor engineered T (CAR-T) cells-derived exosomes also retain the tumor-targeting specificity thereby enhancing its efficacy [80]. These studies show that exosomes derived from hybridoma cells could become an attractive tool in the future to develop tumor-targeting vehicles carrying de novo antibodies to targeted tumor cells in the body.
6.1.5. Mesenchymal stem cells (MSCs)
MSCs are multipotent stem cells produced by many body tissues. In general, MSC-derived exosomes are involved in various processes related to the development and progression of cancer. They have been implicated in the initiation of the tumor niche, tumorigenesis, and cancer-promoting phenomena, such as angiogenesis. MSC-derived exosomes have received great interest as drug delivery vehicles in cancer treatment due to their immunomodulatory properties. MSC-derived exosomes direct towards tumors i.e. they exhibit tumor tropism, a property that has been exploited in several studies. Pascucci et al. packaged paclitaxel (PTX) into MSC-derived exosomes and successfully demonstrated anti-cancer activity in the human pancreatic cell line CFPAC-1 [81]. Kalimuthu et al. packaged PTX into exosomes, by loading MSC with PTX and then isolating exosomes from the treated MSCs, thus demonstrating a mechanism of isolating PTX-loaded exosomes that they termed PTX-loaded exosome mimetics (PTX-loaded Ems) [82]. MSC-derived exosomes have also been used to load small RNAs. Munoz et al. reported loading and transport of Cy5-tagged anti-miR-9 using temozolomide (TMZ)-resistant glioblastoma multiforme (GBM) cells [83] [83] and Kamerkar et al, showed a therapeutic effect of MSC-derived exosomes loaded with anti-KRAS siRNA against KRAS demonstrated a therapeutic effect in mouse pancreatic tumor models [43].
6.1.6. Tumor cells
Exosomes from tumor cells have been considered an ideal source of delivery vehicles for tumor-targeted delivery of anti-cancer therapeutics. In one of the earliest studies evaluating exosomes as carrier of molecules, Cossetti et al. made a remarkable discovery showing trans-generation delivery of RNA by exosomes from grafted melanoma (somatic) to germinal cells [84]. The observation from the study is pivotal not only because it reorients the conventional concept of epigenetics but also considers that exosomes from different somatic cells including tumor tissues can be used to develop multigenerational sustainable vaccines triggering passive immunity without bringing any germline modification in the genetic structure. Tumor cell-derived exosomes, or TEx, can be used to target specific tissues, since they possess many proteins or molecules that are responsible for their preferred and efficient uptake by tumor cells [85, 86]. The homing ability of TEx strengthens their conceptual use as an effective modality for cell vaccination. Efficient delivery of antigens and adjuvant to antigen-presenting cells is a challenge and is required to develop an effective immunotherapy modality for cancer treatment. Morishita et al. evaluated the use of murine melanoma (B16BL6) cell-derived exosomes genetically modified to express endogenous tumor antigens and immune stimulatory CpG DNA for cancer immunotherapy. Both in vitro and in vivo studies demonstrated that treatment with the modified exosomes led to activation of mouse dendritic (DC2.4) cells and efficient antigen presentation that resulted in greater antitumor response in B16BL6 tumor-bearing mice compared to treatment with a mixture of exosomes and CpG DNA. This data suggested that the use of engineered tumor-derived exosomes can be a useful tool for cancer immunotherapy [87]. In contrast, Rong et al showed that exosomes from human breast cancer cells suppressed T-cell proliferation and that this effect was mediated by transforming growth factor (TGF)-β present in the tumor-derived exosomes [88]. These results demonstrated the immunomodulatory properties of exosomes and underlined that the use of TEx for cancer therapy must be approached cautiously.
Qiao et al. explored using TEx to exploit their inherent tumor tropism for drug delivery. They used exosomes isolated from human colon cancer (HT1080) and cervical (Hela) cancer cell lines and tested their homing properties toward HT1080 tumor cells. Exosomes from both cell lines were loaded with Doxil (Doxorubicin) and exosome uptake and antitumor activity were assessed in vitro and in vivo using HT1080 cells. Both, the in vitro and in vivo studies demonstrated that drug loaded exosomes from HT1080 cells exhibited greater tumor tropism and enhanced antitumor activity towards HT1080 cells than did the drug loaded exosomes from Hela cells [86]. The findings show that in cancer, cell-derived exosomes fuse preferentially with their parent cancer cells and could potentially be exploited as drug carriers for cancer therapy. In another study, Sun et al used exosomes from mouse lymphoma (EL-4) cell lines to demonstrate the anti-inflammatory properties of curcumin-loaded exosomes in suppressing inflammation in a lipopolysaccharide (LPS)-induced septic shock mouse model [89, 90]. As a result exosomes from tumor cells can be useful for delivering anti-inflammatory agents.
Studies from our own laboratory show that TEx isolated from human lung cancer cell lines (H1299 and A549) when added back to parental tumor cells and to normal lung fibroblasts (MRC-9) result in increased cell proliferation of both cell types. Additionally, molecular studies revealed activation of oncogenic signaling pathways that favored cell survival and drug resistance (unpublished data).
The results from the studies described above provide strong evidence for the use of TEx in the treatment of cancer. However, advancing the use of TEx towards clinical translation requires careful consideration and appropriate caution.
6.1.7. Other cell-derived exosomes
In addition to the above cell types, several additional cell types have been explored as sources of exosomes. Stable (transformed) human embryonic kidney cells, HEK 293T cells, are most commonly used in studies related to exosome-mediated delivery of therapeutic molecules. Ohno et al. demonstrated the tumor-targeting property of exosomes isolated from HEK 293 cells by decorating them with the GE11 peptide or epidermal growth factor (EGF) on their surface, and then used them to deliver let-7a miRNA to EGFR-expressing breast cancer xenograft tissues leading to an enhanced therapeutic effect [91]. Similarly, HER-2 positive breast cancer cells were treated with therapeutic siRNA using engineered HEK298T-derived exosomes that displayed the synthetic peptides DARP in G3 on the surface to confer tumor-targeting ability [92]. HEK293-derived exosomes show a remarkable ability to carry and deliver chemotherapeutic drugs, such as doxorubicin, in such a manner that the drug causes minimal toxicity to non-targeted cells [93].
Our own studies have explored the use of normal lung fibroblast-derived exosomes as drug delivery carriers. We used normal human lung fibroblast (MRC-9)-derived exosomes and combined them with doxorubicin-conjugated gold nanoparticles (Dox-GNP) to develop a unique hybrid drug delivery vehicle that we termed ‘nanosomes.’ We reported enhanced tumor cell death and protection of cardiac cells (non-targeted cells) when nanosomes were used to deliver doxorubicin (Figure 2) [48]. Recently, we developed a multifunctional tumor-targeted drug delivery system by loading MRC-9 derived exosomes with cisplatin (CDDP) conjugated to iron-oxide nanoparticles (IONP) via a pH-sensitive linker. The rationale to incorporate a pH-sensitive linker was to exploit the intracellular pH difference between tumor cells (acidic) and normal cells (basic) and thus have selective drug release under acidic conditions i.e. in the tumor microenvironment. The CDDP-IONP loaded exosomes were further decorated on their surface with transferrin in order to achieve tumor-targeted drug delivery to transferrin-receptor (TfR) overexpressing lung cancer cells. These multifunctional tumor-targeted exosomes can act as theranostics by exerting CDDP-induced tumor cell cytotoxicity and IONP-mediated monitoring of tumor cell killing by magnetic resonance imaging (MRI). Additionally, we anticipate that these exosomes will have none to minimal toxicity towards normal cells. The theranostic potential of these exosomes is currently being evaluated and the data should be available within the next year.
Figure 2.
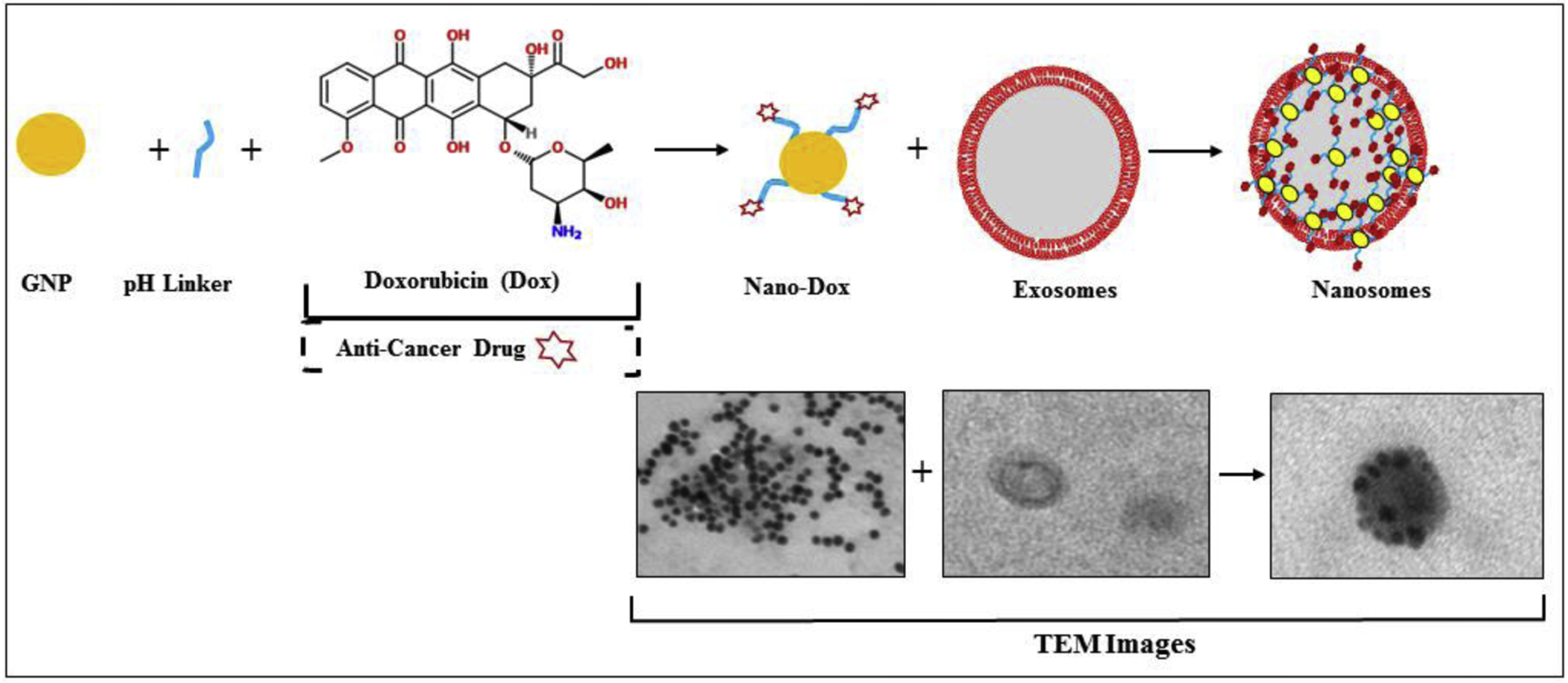
Schematic to show the synthesis of nanosomes. An exosome-based hybrid delivery system consisting of cell-derived exosomes and gold nanoparticles carrying doxorubicin was synthesized for lung cancer treatment. The bottom panel shows TEM images of each step of synthesis.
Figure modified from: Srivastava, A., Amreddy, N., Babu, A., Panneerselvam, J., Mehta, M., Muralidharan, R., et al. (2016). Nanosomes carrying doxorubicin exhibit potent anticancer activity against human lung cancer cells. Scientific Reports, 6, 38541.
https://doi.org/10.1038/srep38541.Copyright © 2016 Rights Managed by Nature Publishing Group.
6.1.8. 3D cell cultures for exosome production
Studies to develop exosome-based therapeutic modalities commonly use exosomes produced by cells growing in cell culture dishes or flasks in 2D. Although the cells may grow optimally in 2D space on a plastic-coated base, they seldom resemble the real growth conditions experienced in vivo. Cells grown in 3D, as organoids or spheroids, closely resemble cells grown in vivo. Recent studies have explored the isolation of exosomes from 3D-cultured cells and shown increased production of exosomes compared to 2D culture. They also compared the molecular composition of exosomes and showed that the small RNA profile of 3D- culture derived exosomes exhibited a higher similarity (~96%) to in vivo circulating exosomes derived from cervical cancer patient plasma [94, 95]. This finding demonstrates the feasibility of using 3D cell- derived exosomes as drug delivery vehicles for precise therapeutic interventions.
Although cell-derived exosomes appear to be a straightforward choice, changes in the physiology of cells as a consequence of growing them in plastic under artificial conditions may significantly impact both exosome production and composition. Thus, studies increasingly evaluate additional natural sources for exosomes.
6.2. Natural sources of exosomes
There are three major sources to isolate naturally produced exosomes: 1) plants, 2) microbes (e.g., bacteria, fungi), and 3) body fluids. The exosomes produced from these sources are more amenable and less toxic to use, while also having greater absorption potential in the recipient cells. Additionally, these sources have tremendous potential to produce exosomes on a large scale. In the following sections, these natural exosome reservoirs are described and some studies that have used natural exosomes as delivery vehicles are also discussed.
6.2.1. Plant-derived exosomes
Plant-derived extracellular vesicles are a group of nano-scaled vesicles that are produced by various plants and have been isolated from various edible vegetables and fruits [96] and shown to be involved in various physiological processes and in cell-to-cell communication [97, 98]. They are membrane-bound vesicles, like exosomes, and can be considered as a potential alternative to synthetic liposomes or nanoparticles. Plant-derived vesicles or exosomes thus have potential for developing green, sustainable, and biocompatible materials for the delivery of bioactive compounds.. The composition of plant-derived nanovesicles (exosomes) is similar to that of animal exosomes and is made up of lipids, proteins, and nucleic acids. Plant-derived exosomes have high degrees of biocompatibility with animal systems and can be used in cross-kingdom studies. Li et al. showed that plant-derived exosomes termed ‘botanosomes’, can carry RNAs and transfer them to humans. They showed the presence of exosome-like nanoparticles in plant-derived decoctions, referring to them as “decoctosomes” and showing they comprised of lipids, proteins, sRNAs, and chemical compounds. Oral administration of these decoctosomes containing sRNAs (HJT-sRNA-m7 and PGY-sRNA-6) offered therapeutic benefits in bleomycin-induced lung fibrosis and poly(I:C)-induced inflammation in mice [99]. Garaneva et al. used grapefruit juice-derived exosomes to successfully transport exogenous Alexa Fluor 647-labeled bovine serum albumin (BSA) and heat shock protein (HSP)70 to human peripheral blood mononuclear cells (PBMCs) and colon cancer cells; the presence of exosomes was confirmed using various methods, including Nanotracker Analysis (NTA), Dynamic Light Scattering (DLS), Atomic Force Microscopy (AFM), and cryo-electron microscopy (Cryo-EM) [100].
Various plant components have been used to isolate exosomes using conventional ultracentrifugation, filtration, and other methods (Table 3). Since plant-derived exosomes can be more easily scaled up for mass production and harvested in bulk than can synthetic nanoparticles, they hold great promise as a resource for exosomes production and use as drug carrier.
Table 3.
Plant derived exosomes and their isolation methods
| Source | Method | Study | Reference |
|---|---|---|---|
| Plant-derived Extracellular Vesicles-PDEVs (Lemon) | Ultracentrifugation and electrophoresis combined with Dialysis (named ELD) | Gastric cancer | [197] |
| Lemon | Differential centrifugation | Bone marrow and lung cancer | [102] |
| Ginger rhizome root juice | Sucrose gradient ultracentrifugation | Bio-distribution and uptake by mouse hepatocytes | [198] |
| Ginger juice | Ultracentrifugation | Colitis and colitis-associated colon cancer | [101] |
| Plant EVs: D. morbifera, P. densiflora, T. occidentalis, and C. obtusa |
Filtration and concentrator | Breast and skin cancer | [199] |
| Algae | Ultracentrifugation | Isolation and characterization | [104] |
| apoplastic fluids of Arabidopsis (Arabidopsis thaliana) leaves | Ultracentrifugation | PEN protein involved biotic and abiotic stress | [200] |
| Grapefruit juice | Differential centrifugation | Colon cancer | [100] |
| Grapefruit juice | Sucrose gradient centrifugation | Colitis in mice | [201] |
| Grapefruit juice | Differential centrifugation | Colon cancer mouse model | [202] |
| Arabidopsis thaliana and Brassicaceae vegetables | Sucrose gradient centrifugation | Characterization of exosomes | [203] |
| Herbal plants | Transfer of stable (s)RNA for treatment of lung fibrosis and inflammation | [99] | |
| Broccoli | Centrifugation and high pressure filtration | Mouse colitis | [204] |
Consumption of green vegetables and fruits and a vegetarian diet has been suggested as a way to reduce cancer incidence; some epidemiological studies have demonstrated that consumption of plant-based foods helps in reducing cancer incidence [101]. Other studies demonstrate the beneficial anti-cancer effect of various plant extracts and citrus fruit juice. Lemon-derived exosomes have anti-cancer activity against several solid tumors and hematological cancers [102]. Similarly, Zhang et al explored edible ginger-derived exosomes and showed an inhibitory effect in colitis-associated cancer [97]. In melanoma, ginseng-derived nanoparticles showed remarkable anti-cancer activity, effectively inhibiting tumor growth [103].
Plant-derived exosomes also show notable stability across pH and temperature ranges. Grapefruit-derived exosomes are stable across a wide pH ranges, suggesting their ability to work as an efficient delivery vehicle in the tumor microenvironment which is acidic in nature [96]. Algae produce particularly large numbers of exosomes, which supports the contention that scalability of exosomes needed for developing delivery vehicles can be easily achieved [104]. Thus, plant-derived exosomes not only have inbuilt anti-cancer activities, but also show remarkable stability, biocompatibility, and ability for large-scale production, strongly supporting that these exosomes represent a good resource for development of carrier vehicles.
6.2.2. Microbes and Bacteria
Small nano-sized vesicles that resemble EVs or exosomes are produced by several microbes, including bacteria, protozoans, and fungi. These vesicles are present in different groups or microbial kingdoms, and are often termed membrane vesicles, outer membrane vesicles (OMVs), exosomes, or shedding micro-vesicles. Regardless of the term used, the fundamental process of biogenesis that largely uses ESCRT proteins and their homologues to facilitate formation of vesicles/exosomes and their release is conserved [105]. The major function of these small vesicles is to transfer virulence factors from microbes to host cells, thereby aiding infection; these virulence factors are primarily proteins, lipids, nucleic acids, and other bioactive molecules. Thus, microbe-derived exosomes have the capability to deliver exogenic cargo to host cells, breaking the kingdom barrier. Physiologically, microbe-derived exosomes are also involved in promoting infections. Pathogen-associated molecular patterns (PAMPs) are key molecules involved in eliciting the host immune system by activation of pattern recognition receptors (PRRs) and Toll-like receptors (TLRs). It is now recognized that pathogens release PAMPS through exosomes or EVs, along with other immunomodulatory molecules to promote the infectious cycle [106].
The most extensively studied microbe-derived exosomes are those that are isolated from gram-negative bacteria. The first report of exosomes in gram-negative bacteria was from E.coli in the 1960s. In gram-negative bacteria, exosomes are called outer membrane vesicles (OMVs), as the bacteria have no membrane-bound organelles and the exosomes are usually generated from the outer membrane. The gram-negative OMVs are well characterized with respect to transferring biomolecules to host cells, including immune cells, to promote virulence and infection, develop resistance to antibiotics, and in some cases to degrade the host cells. OMVs have also been shown to be secreted in larger amounts when bacteria are under stress [107]. Gram-positive bacteria also release OMVs into the extracellular space to perform various biological activities. Species such as Staphylococcus aureus, Bacillus subtilis, Streptococcus pneumoniae, and Bacillus anthracis have been reported to produce OMVs [108].
For more than 20 years, protozoans have been known to produce extracellular vesicles. Leishmania spp. and Trypanosoma cruzi EVs (or exosomes) have recently been shown to have physiological roles. As for bacterial OMVs, protozoan-derived EVs carry and transfer various biomolecules that act as virulence factors and promote infection in the host cells. In a proteomic analysis of T. cruzi, a number of proteins containing nucleic acid-binding sites and ribosomal proteins were identified, suggesting that the exosomes from this protozoan can successfully carry nucleic acids to the host cells [109, 110].
Fungal cells are prolific producers of extracellular vesicles that are similar to exosomes. Unlike bacteria, fungi are eukaryotic cells, however, the exact mechanism of EV/exosome biogenesis is not fully understood. One reason is that, like gram positive bacteria, fungal cells are enclosed in a sturdy cell wall. Nevertheless, fungi-derived EVs have been reported to carry enzymes, proteins, and other biomolecules required to enhance fungal pathogenesis [111, 112].
In summary, microbe-derived EVs efficiently pass genetic and other biomolecules across kingdoms. Further, they can be easily isolated in high numbers from continuously growing microbial cultures and can be manipulated to transfer exogenous material, suggesting that they could be explored as ideal drug carriers and vaccines.
6.2.3. Body fluids
6.2.3.1. Milk-derived exosomes
Milk is an excellent source of exosomes; studies show that milk is enriched in exosomes. Milk-derived exosomes exhibit enhanced uptake by recipient cells, suggesting that they represent a potential efficient delivery system [113] [113]. In general, there are two sources of milk-derived exosomes: human breast milk and bovine milk. Since humans consume both types of milk, they have no immunogenic effect and retain bio-compatibility. Samuel et al. extensively compared the quality and quantity of exosomes isolated from bovine mature milk and colostrum samples at 24, 48, and 72 h postpartum. They showed the presence of many exosome-associated common and unique proteins, and also detected several proteins involved in vesicle docking and fusion with the cell membrane during release of exosomes. Rab family proteins and certain integrins responsible for anchoring and uptake of exosomes were enriched in all the tested milk samples [114]. Milk-derived exosomes are very robust; they can survive the harsh and degrading conditions present in the gut, and can cross biological barriers to reach peripheral tissues and deliver their content to those cells. Milk-derived exosomes exhibit inter-organismal and cross-species communication while being non-immunogenic, and milk is a natural source of abundant exosomes, is highly scalable and economically viable; these facts make milk-derived exosomes a significant candidate as drug carriers and vaccines for cancer therapy [115].
Anti-cancer drugs are often infused intravenously due to their poor oral bioavailability. This mode of administration, although necessary, involves drawbacks such as high cost, physical inconvenience, and issues associated with sterility. To circumvent these shortcomings, oral administration would be preferred and methods to achieve this are required. Milk-derived exosomes are an attractive option as they are non-toxic and available in abundance, they are also easily absorbed from the gastrointestinal tract in humans thereby increasing the bioavailability of any exosome packaged drugs. Built on these observation, multiple studies have explored the potential of milk-derived exosomes to develop an effective drug delivery vehicle; some of these studies are discussed below [116].
Munagala et al. were the first to report the utility of bovine milk as a potentially scalable source of exosomes that can be used as a drug delivery vehicle. They successfully demonstrated efficient loading and delivery of hydrophilic compounds, lipophilic agents, and chemotherapeutic drugs; several compounds, including curcumin (CUR), Withaferin A (WFA), anthocyanidins (Anthos), paclitaxel (PTX), and docetaxel (DOC), that vary in their lipophilicity, molecular weight, and functional groups were loaded onto the exosomes. Loaded exosomes were used to treat human lung (A549 and H1299) and breast (MDA-MB-231 and T47D) cancer cell lines and treatment with exosome-loaded vehicles led to improved treatment response compared to treatment with the comparable free compounds. The study further explored the efficacy of the milk exosome drug delivery vehicle in lung cancer A549 xenograft models, and therapeutic response was enhanced when exosomes were administered intraperitoneally (i.p.) or orally. The therapeutic effect was further increased by adding the tumor-targeting ligand folic acid (FA) to the milk-derived exosomes containing the drug [117]. Milk-derived exosomes membranes were conjugated to doxorubicin via a pH-responsive imine bond to leverage the acidic nature of the tumor microenvironment in aiding sustainable drug release. In addition to doxorubicin, administration of milk-derived exosomes loaded with anthracene endoperoxide derivative (EPT1) and chlorin e6 (Ce6) showed controlled drug release and biocompatibility, and was effective in treating oral squamous cell carcinoma (OSCC) [118].
Casein is an abundant protein in milk and causes reduced exosome release. Separation of casein by acid treatment using hydrochloric acid (HCl) or acetic acid facilitates exosome isolation and purification [119].
High quality bovine milk-derived exosomes isolated by casein chelation were engineered with modular surface tunability to enable loading of siRNA and administration via the oral route [120]. The study showed that polyethylene glycol (PEG) coating of exosomes can reduce their degradation in the acidic gastric environment and enhance their permeability through mucin. These findings show that milk-derived exosomes may be useful for the development of an effective, scalable platform technology for oral drug delivery of siRNA [120]; siRNA-based therapeutics and their delivery through exosomes is a promising treatment strategy. However, loading of negatively charged molecules like siRNA through standard methods such as electroporation is cumbersome and is accompanied by issues such as siRNA aggregation, toxicity, and other experimental complexities. To mitigate this, Shandilya, et al. exploited bovine lactoferrin, a natural ligand for its receptor GAPDH which is present in milk exosomes [121]. They used lactoferrin with poly-L-lysine to conjugate electrostatically with negatively charged siRNA in order to load the siRNA into milk exosomes. The lactoferrin-poly-L-lysine complexed with siRNA when added to milk exosomes, showed excellent loading into the exosomes resulting in transfection efficiency, colocalization percentage, and colocalization threshold equivalent to the electroporation method.
Aquil et al. used milk-derived exosomes to demonstrate successful loading of siRNAs targeting several genes (i.e. VEGF, EGFR, AKT, MAPK, and KRAS), albeit with varying loading efficiencies. Use of these siRNA-loaded exosomes resulted in 2- to 10-fold knockdown of the target protein expression levels in various cancers [122]. In the same study, the authors demonstrated the therapeutic effect of targeting KRASG12S in A549 lung tumors both in vitro and in vivo. Functionalization of the KRASG12S siRNA containing exosomes with folic acid further enhanced the therapeutic effect and resulted in significant suppression of A549 tumor growth in vivo.
miRNA is another class of regulatory RNA molecule that can used as anticancer drugs. Xhe et al. loaded raw bovine milk-derived exosomes with exogenous hsa-miR148a-3p and tested the delivery capability of the loaded system against hepatic (HepG2) and intestinal (Caco-2) cell lines. Results from this study strongly indicated that bovine milk-derived exosomes could be a cost-effective source for the production of nanocarriers of functional miRNAs and could be used in RNA-based therapy [123].
Important criteria of a successful drug delivery system are its bioavailability in the recipient system and the effective distribution of its cargo. Manca et al. used bovine, porcine and murine milk-derived exosomes labeled with fluorophores or fluorescent fusion proteins and studied their biodistribution within and across species boundaries. Their study revealed preferred accumulation of exosomes and their microRNA cargos in liver, spleen, and brain after suckling, oral gavage, and intravenous administration in mice and pigs. The study further showed unique distribution profiles and cross-species distribution when synthetic, fluorophore-labeled microRNA carrying bovine milk-derived exosomes were administered to mice [124].
The studies mentioned above, and many others, demonstrate that milk (human breast or bovine) is an excellent source of exosomes, and its scaling can easily be controlled. However, several issues prevent milk-derived exosomes from being explored at the commercial level as drug delivery modalities. One such issue regards the purity of exosomes [125], and a second question regards the status of exosomes in commercially available milk. Commercially available milk undergoes a number of processing steps, and these steps are speculated to change exosome properties including possibly their content. Kleinjan et al. compared exosomes (EVs) isolated from raw bovine milk with exosomes from commercially available processed milk, such as pasteurized and ultra-heat treated (UHT) milk. They concluded that the processing of milk affects the number of exosomes and their components. In UHT milk, no exosomes were detected. Although pasteurization had no effect on the number and morphology of exosomes, their protein and RNA profiles were different from those of raw milk, suggesting that industrial processing affects exosome characteristics [126].
6.2.3.2. Blood
Blood is a connective tissue that circulates throughout the body and plays an important role in communication between the various organs/structures of the body. Blood is also a central mechanism for transporting useful biological material and removing toxic substances from organs. These characteristics position blood as an enriched reservoir of exosomes. Indeed, blood is one of the most commonly used body fluid used for isolation of exosomes. The first report of isolation of exosomes from blood came from Marie-Pierre Caby and co-workers and they reported the detection of exosomes in blood plasma samples collected from 15 healthy donors. The results were substantiated by performing electron microscopy and identifying proteins characteristically present in exosomes, such as tetraspanins CD63, CD9, CD81, class I and class II MHC molecules, and Lamp-2 proteins [127]. Following this report, several others have focused on using blood-derived exosomes to identify novel biomarkers for cancer diagnosis and prognosis and as drug delivery vehicles. The benefits of using blood-derived exosomes for drug delivery is that they are biocompatible and exhibit reduced immunogenicity.
Wahlgren et al. investigated plasma-derived exosomes as a gene delivery vector (GDV). In their study, treatment of PBMC-derived monocytes and lymphocytes with mitogen-activated protein kinase (MAPK) 1 siRNA loaded exosomes resulted in effective suppression of MAPK1 protein expression [128]. Jakubec et al. reported human plasma-derived exosomes efficiently cross the blood brain barrier (BBB) and bind to endothelial cells [129], while also showing in comparisons with exosomes derived from PC3 cell line, that the human plasma-derived exosomes were enriched in lyso-phospholipids and lacked phosphatidylserine (PS). The presence of PS on cells, an indicator of cells undergoing apoptotic death, and on micro-particles, and micro-vesicles results in their rapid clearance by macrophages and removal from circulation. Thus, the lack of PS on plasma-derived exosomes explains their ability to evade macrophages and have a prolonged circulation time in vivo. This offers support for plasma-derived exosomes being preferable to liposomes for drug delivery. Plasma-derived exosomes carry functional biomolecules such as proteins and enzymes as cargo in their lumen. Logozzi et al. in their extensive studies concerning plasma exosomes in diagnostics and therapeutics, reported increased accumulation of enzymes such as carbonic anhydrase isoform IX (CA IX) and proteins like PSA in plasma exosomes from prostate cancer (PCa) patients, suggesting that plasma-derived exosomes can be used to deliver bioactive molecules of therapeutic value, de novo. The detection of these biomolecules in plasma exosomes can be exploited for use as biomarkers for detection and progression of cancer, thereby serving as a potent diagnostic tool. In addition to PCa, promising candidate biomarkers in plasma-derived exosomes have been detected, such as CD63 and caveolin-1 in melanoma [130–134]. In a clinical study, Osti et al. observed a change in exosome numbers in patients with GBM and a uniquely different protein profile in exosomes derived from the plasma of healthy individuals, providing an opportunity to develop diagnostic markers and target molecules for a favorable treatment response [135].
Red blood cells (RBCs) also serve as a rich source of exosomes that have been tested as drug carriers for cancer therapy. The benefit of RBC-derived exosomes is that they lack nuclear and mitochondrial DNA thus minimizing both potential toxicity and immune response. Additionally, concerns related to exosome scale-up is alleviated as exosomes can be easily isolated from blood samples, especially from group O-RBCs that can be sourced from established blood banks. Since blood, which contains billions of exosomes, when transfused to patients has not resulted in any exosome-related adverse events, it is accepted that blood-derived exosomes are safe and could be used for drug delivery [136]. Using RBC-derived exosomes, delivery of an antisense oligonucleotide against miR-125b resulted in suppression of leukemia and breast cell growth both in vitro and in vivo [137]. All of these studies demonstrate that exosomes from RBCs or plasma can be developed as drug carriers for cancer therapy.
6.2.3.4. Urine
In the previous section, we discussed the possible use of blood -derived exosomes as drug carriers. One current area of emphasis in cancer research is on precision medicine that heavily relies on innovations such as noninvasive “liquid biopsy” to reduce the dependency on tumor biopsies. Urine is a valuable resource for exosomes, and studies have explored the utility of urinary exosomes for the discovery of biomarkers to detect urologic cancers, including prostate and other urinary tract malignancies [138–140] [141]. In addition, urine, unlike blood, is relatively free of proteins and cells, and thus, the proteomic or molecular content of urinary exosomes usually lacks any other peripheral markers. Further, urinary exosomes are in more direct contact with tumor tissues than are blood-derived exosomes, which circulate throughout the body before they are harvested. These urinary exosomes should closely reflect the characteristics of (tumor) cell of origin and can be harnessed for a direct read of the tumor’s molecular profile and membrane proteins. In high-precision theranostics, this property can be useful for designing tumor-targeted drug delivery vehicles and identifying and characterizing tumor-specific biomarkers. Studies of urinary exosomes isolated from bladder cancer patients and healthy controls show that the concentration of urinary exosomes was significantly higher from patients with bladder cancer compared to healthy individuals [142].
miRNAs have been investigated as diagnostic biomarkers in bladder cancer; Armstrong et al. profiled miRNAs in the tumor tissues and body fluids, including urinary exosomes of bladder cancer patients. They identified miR-205, miR-2003c-3p, and miR-29b-3p as candidate biomarkers, since they had similar profiles in tissue and urinary exosomes, suggesting that urinary exosomes can be used as a source to discover diagnostic biomarkers in bladder cancer [143]. In a similar study, urinary exosomes showed SLC2A1, GPRC5A, and KRT17 were enriched in advanced-stage bladder cancer and could be used as biomarkers [144]. Our group has extensively explored the diagnostic potential of miRNAs and proteins contained in urinary exosomes for predicting treatment outcomes in non-small cell lung cancer patients treated with chemotherapy with or without radiation. We have identified a panel of miRNA signatures in urinary exosomes that are being validated for predicting treatment outcomes (unpublished data). In a separate ongoing study, we are investigating changes in the immune checkpoint (ICP) protein landscape in urinary exosomes isolated from lung cancer patients receiving anti-PD-1/anti-PD-L1 immunotherapy. The objective of this study is to identify changes in a set of ICP proteins during the course of immunotherapy treatment that can be used to predict resistance and treatment failure (unpublished data). Besides identifying genetic regulators, enrichment of specific metabolites also occur in the urinary exosomes from cancer patients compared to their normal healthy counterparts. In a study of patients with prostate cancer, the levels of glucuronate, D-ribose 5-phosphate, and isobutyryl-L-carnitine were lower in all pre-prostatectomy samples compared with healthy control and post-prostatectomy samples (p < 0.05) [145]. It is evident from these study results that urinary exosomes can be used for cancer prognosis and diagnosis.
6.2.3.5. Saliva
Saliva is a colorless biofluid that shares many characteristics with blood and has been extensively explored to discover biomarkers. Saliva is considered significant for developing diagnostic modalities because the salivary glands are densely surrounded by blood vessels, which leads to intense molecular exchange between circulating blood and salivary gland cells. Inturn, this exchange results in significant overlap between saliva and blood in the molecular characteristics and the biological information within their respective exosomes. Saliva offers many advantages over blood; it is easily collected in a non-invasive manner, there is essentially an unlimited supply of saliva, and, unlike blood, saliva does not coagulate and can be stored and transported easily [146]. Multiple studies show that saliva is an enriched source of exosomes containing DNA, several species of RNA, and proteins that can serve as prognostic and diagnostic biomarkers for several oral diseases, including oral cancer [147, 148]. Ogawa et al. reported presence of exosome-like vesicles in human saliva that contained dipeptidyl peptidase IV (DPP IV), galectin-3, and immunoglobulin A. The DPP IV present in the vesicles was metabolically active and able to catabolize proteins into peptides. DPP IV, galectin-3, and immunoglobulin A all reportedly participate in regulating the immune response, their presence in the vesicles of saliva and the role of saliva in preventing oral infection engender speculation that the exosome-like vesicles in saliva may promote local immunity [149].
Oral squamous cell carcinoma (OSCC) is one of the most prevalent oral cancers and generally has a poor prognosis as it is often diagnosed at an advanced stage. He et al. evaluated the miRNA content of salivary exosomes from OSCC patients compared to those from healthy controls. A total of 109 miRNAs were found to be enriched by at least 2-fold and in the saliva of patients with OSCC; significantly, miR-24-3p was one of top three miRNAs enriched in OSCC exosomes (121.54 fold over control) and is also reported to be upregulated and operate as an oncogene in head and neck squamous cell carcinoma (HNSCC). Elevated levels of miR-24-3p were also detected in OSCC neoplastic tissues, suggesting that circulating miR-24-3p may originate from the tumor cells [150]. Based on these findings, salivary exosomal miR-24-3p may serve as a biomarker in OSCC screening.
Zlotogorski-Hurvitz et al. took a unique approach towards diagnosing oral cancer. They used a Fourier-transform IR equipped (FTIR) and machine learning approach to study salivary exosomes and developed models to differentiate the absorbance levels of nucleic acids, proteins, and lipids to accurately differentiate oral cancer patients and healthy individuals [151]. Many other studies have also shown the utility of salivary exosomes in the diagnosis of oral cancers and are gaining attention as a promising non-invasive diagnostic modality for accurately detecting premalignant lesions and early-stage oral cancers. To describe the importance of salivary exosomes, Nonaka et al. coined the term “saliva-exosomics” as the next-generation salivaomics, i.e., detailed molecular studies of saliva exosomes and exploration of their molecular landscape and application of such, to develop diagnostics and studying the pathobiology of disease [152]. While salivary exosomes have demonstrated significance in the diagnostic space, their potential as therapeutic carriers is however yet to be investigated. Nevertheless, the ability to isolate exosomes from saliva in bulk and in a non-invasive manner, positions salivary exosomes as a strong candidate for developing theranostics for oral and other cancers.
7. Methods of loading therapeutics
The unique ability of exosomes to carry molecules has paved the way for their use as an alternate vehicle to carry anticancer drugs and molecular imaging agents. Exosomes have an inherent ability to carry many molecules in their lumen and deliver them to cells at distant sites in the body. Exploiting this feature for cancer therapy requires some modifications of the exosomes to ensure they specifically reach the tumor and deliver the therapeutic agent with minimal off-target effects to normal cells. Such reengineered exosomes, referred to as “designer’s exosomes,” are achieved using several different techniques for loading molecules into their lumen. This reengineering aspect of exosome research places exosomes at the cusp of their successful application as theranostics for cancer and other diseases. The various approaches undertaken to load biomolecules in exosomes can be broadly classified as either active/direct manipulation or passive/indirect loading. Both approaches have yielded interesting results and significant promise. Specific loading methods are discussed below.
7.1. Sonication
Sonication uses high-frequency ultrasound energy to agitate and partly disrupt the exosome membrane. This partial membrane disruption aids diffusion of therapeutic cargoes (drugs, proteins, or small RNAs). Kim et al. showed that drugs with low ater solubility, such as PTX, can be successfully packaged into exosomes by sonication [153], without any major structural changes in the exosomes that affect function. While sonication briefly disrupts the membrane, exosomes rapidly resumed their original state within one-hour post sonication when incubated at 37° C. Treatment of multi-drug resistant (MDR) murine lung cancer (3LL-M27) and Madin-Darby canine kidney (MDCKMDR1) cancer cells with exosomes loaded with PTX using this technique, resulted in enhanced antitumor activity. Similarly, Hanney et al. showed superior incorporation of both hydrophilic (doxorubicin) and hydrophobic (PTX) drugs into the lumens of macrophage-derived exosomes after sonication [154], Treatment of triple-negative breast cancer (TNBC) cells with these drug loaded exosomes resulted in antitumor activity. In another study, Li et al. demonstrated loading of gemcitabine (GEM) onto autologous exosomes by sonication resulted in higher drug loading efficiency and was superior to drug loading by simple incubation [155]. Treatment of pancreatic cancer with GEM loaded autologous exosomes (ExoGEM) resulted in antitumor activity and increased animal survival with no measurable non-specific toxicity.
While sonication is a straightforward method with favorable outcomes, its effects on membrane integrity and lack of a mechanism to precisely control the amount of drug loaded remain road blocks to developing this technique as a standard loading method.
7.2. Electroporation
Loading of drugs, and especially hydrophobic drugs and therapeutic nucleic acids such as siRNA and miRNAs, can be significantly enhanced by electroporation. The applied electric charge disrupts the phospholipid bilayer membrane of exosomes and creates transient, temporary micro-pores in the membrane. These micro-pores increase membrane permeability and develop an electrical potential, which allows efficient loading of cargo, and this method has been extensively used for loading siRNA or miRNA into exosomes. Alvarez-Elveti et al. used electroporation to load therapeutic siRNA into dendritic cell-derived exosomes [63]; the exosomal-membrane protein Lamp2b was fused to the neuron-specific RVG peptide for targeted delivery to mouse brain. They then showed that when BACE1 specific siRNA was loaded into the Lamp2b/RGV targeted exosomes by electroporation, and the exosomes were injected intravenously there was a significant reduction in BACE1 mRNA and protein expression in mice. Pomatto et al. demonstrated that loading of antitumor miRNAs (miR-31 and miR-451a) into plasma-derived EVs by electroporation was superior to conventional incubation; electroporation gave increased efficiency of miRNA loading and, better protection of miRNA from RNase degradation, and did not change the exosome characteristics in any way that would impact their uptake by the target cells [156]. Similarly, Liang et al. loaded exosomes with 5-Fluorouracil (5-FU) and anti-miR for miR-21 (miR-21i) by electroporation when administered to mice bearing 5-FU resistant colon tumors (HCT1165FR) the exosomes resulted in enhanced cytotoxicity and antitumor activity compared to treatment with either 5-FU or miR-21i [157].
The major drawbacks of electroporation are substantial cell death caused by high voltage pulses and only partly successful membrane repair. Thus electroporation, requires significantly greater quantities of cells than those chemical transfection method, the additional step of exosome purification after electroporation is also necessary. Aggregation of RNA is an additionally a reported drawback of electroporation [158].
7.3. Freeze-thaw
Freeze-thaw is a conventional method of cellular transfection and loading of drugs in liposomes [159]. This method involves incubation of exosomes and drugs at room temperature and at below freezing temperatures (−80°C) for multiple cycles. The freeze-thaw cycles permeabilize the membrane and allow the cargo to enter. This method has met with limited success using exosomes and conditions need to be carefully optimized to achieve loading. However, Sato et al. used the freeze-thaw method to fuse exosomes and liposomes and develop a novel exosome mimetic. The method can be useful once it is optimized for large-scale use [160].
7.4. Click chemistry or surface functionalization
The surface of exosomes can be functionalized with several molecules that are therapeutic or diagnostic. Small molecules and macromolecules of interest can be attached to the exosome surface via chemical bonds, such as covalent bonds. One form of such bioconjugation is copper-catalyzed azide-alkyne cycloaddition, or click chemistry [161]. This relatively rapid technique, is specific and suitable for reaction between an alkyne and an azide to form a triazole linkage. In their study involving bioconjugation of azide-fluor 545 to mouse breast cancer cell line 4T1-derived exosomes, Smyth et al. found that the conjugation was efficient and did not produce changes in the properties or structure of exosomes [162]. Wang et al. proposed a copper-free click chemistry to load mouse mesenchymal stem cell derived EVs with Alendronate, a bone-targeting ligand, and used this complex to treat osteoporosis [163]. Although studies using click chemistry as a means to load molecules of interest on exosomes are limited, the encouraging results suggest that this method can be used to load drugs and imaging agents and develop a powerful theranostic tool.
In addition to the above strategies, additional physical methods applied for drug loading into exosomes are to be included. For example, the extrusion method involves breaking the exosome membrane under physical pressure, mixing the exosome solution with drug of interest and allowing the membrane to recombine. Dialysis and use of surfactants to modify the exosomal membrane are additional strategies actively investigated for improving the loading of therapeutics into exosomes.
7.5. Physical loading
7.5.1. Incubation
Like cell membranes, the exosome membrane is dynamic in nature and allows passive movement of materials across it following a concentration gradient. This property exploited in many studies have demonstrated physical incubation is a simpler straightforward strategy for cargo loading into exosomes. For example, the hydrophobic drug curcumin has been efficiently loaded by incubating with mouse lymphoma-derived exosomes at 22°C in PBS [89]. In our laboratory, we have used simple incubation to load the hydrophilic drug doxorubicin (DOX) conjugated to gold nanoparticles (GNP) onto exosomes and test their in vitro therapeutic efficacy against lung cancer. The GNP-DOX loaded exosomes exhibited selective tumor cell killing with minimal toxicity towards normal cells [48]. We also assessed the imaging capability of exosomes by loading normal human lung fibroblast-derived exosomes with superparamagnetic iron oxide particles (SPIONS) and using them in MRI studies. In vitro phantom and cellular studies showed excellent T1 contrast images without losing the magnetic properties of the SPIONS (unpublished data). In addition to chemotherapeutic drugs, small nucleic acids like miRNAs have been loaded onto exosomes by direct incubation. Brynsaki et al. and Gong et al. showed successful fabrication of exosomes by loading them with miR150 and miR159, respectively [164, 165].
Incubation is a simple method that requires no specialized equipment and can be adapted to any laboratory setting. However, the major challenge with this method is ensuring consistency in drug loading which is essential to achieve reproducible results.
7.5.2. Transfection
In the transfection method, the cargo of interest is transfected into the parent cells and the cell packages the transfected molecules into the lumen of exosomes. This is a conventional way to load exosomes, especially with small proteins, siRNA, and miRNA molecules. Kooijmans et al. transfected the parental mouse neuroblastoma cell line, Neuro2A, with human glycosylphosphatidylinositol (GPI)-anchored protein decay-accelerating factor (GPI-DAF, or CD55) fused to targeting ligands for epidermal growth factor receptor (EGFR), and showed that the cells produced exosomes displaying EGFR-targeting ligands on their surface [166]. Along similar lines, Cho et al. expressed tumor-targeting human MUC1 (hMUC1) in the mouse cell lines CT26 and TA3HA; exosomes isolated from these cell lines had the target MUC1 on their surface, and both autologous and allogeneic exosomes were able to stimulate the activation of immune cells and suppress hMUC1-expressing tumor growth in a MUC1-specific and dose-dependent manner [167]. In addition to proteins and small peptides, several studies have reported packaging of siRNA or miRNA by transfecting the parental cells, followed by isolation of the engineered exosomes.
The CRISPR/Cas9 system is a powerful tool to change genome structure, resulting in changes in gene expression. Conventionally, the CRISPR/Cas9 system is engineered using large plasmids. More recent studies have explored using exosomes as carriers to deliver the CRISPR/Cas9 system into cells. Li et al. first engineered exosomes to express CD9-HuR by transfecting a protein fusion construct. HuR is an RNA-binding protein that was used to improve the loading efficiency of dCas9 mRNA into CD9-HuR exosomes. In this process, 3X AU rich elements (AREs) were cloned downstream of the dCas9 stop codon dCas9; the AREs were more efficiently encapsulated into CD9-HuR exosomes. Consistently, exosomes from the C/EBPα gRNA, CD9-HuR, and dCas9-ARE co-expressing cells significantly reduced C/EBPα expression in recipient adipogenic stem cells and liver cells [168]. Lin et al. used a different approach; they developed a hybrid exosome system by fusing HEK293FT cell- derived exosomes with liposomes to encapsulate large plasmids, including the CRISPR-Cas9 expression vectors [169]. These authors elegantly demonstrated successful loading of large nucleic acids such as the CRISPR-Cas9 by simple incubation of the plasmid with the hybrid exosomes. Addition of the CRISPR-Cas9 loaded hybrid exosomes to mesenchymal stem cells (MSCs) resulted in their expression and regulation of the target gene. This approach although exciting is in its infancy and additional studies are required prior to considering this gene delivery approach for clinical translation.
8. Isolation methods
Enrichment of exosomes for their application in the development of theranostic or drug delivery vehicles is a major bottleneck. To achieve clinically viable applications while following the requirements of precision and personalized medicine, exosomes should be isolated from body fluids. All the body fluids described above are complex and differ widely in their composition. The presence of proteins, lipids, or enzymes often necessitates a specific process to enrich exosomes. The development of an ideal therapeutic vehicle requires: 1) a uniformly sized, homogenous population of exosomes; 2) a continuous supply of large quantities of exosome particles; and 3) a methodology that is simple, economically viable, and that requires minimal input materials. To overcome the challenges and move the various methods described above into the clinic, exosome science is exploring novel ways to isolate exosomes from different resources efficiently. Table 4 shows some such methods currently under investigation.
Table 4.
Exosome isolation methods
| Isolation Techniques | Source | Advantages | Disadvantages | References |
|---|---|---|---|---|
| Differential centrifugation | Cell line | Isolation from large sample volumes, Simple method | Time-consuming. Requires expensive equipment | [205] |
| Density- gradient centrifugation | Cell line | High purity without the requirement of additional reagent | Time-consuming and complex procedure | [201] |
| Size-exclusion chromatography | Plasma | High yield and purity | Not suitable for large sample volumes. | [206] |
| Time-consuming | ||||
| Ultrafiltration | Cell line, plasma | No requirement of additional reagent | Risk of contamination | [207] |
| Simple procedure | Sample loss | |||
| Sequential filtration | Cell line | High purity | Time-consuming, requires skilled operator. | [208] |
| Homogeneous exosome size | Expensive | |||
| Hydrostatic Filtration Dialysis (HFD) | Urine | Minimum interference of soluble proteins | Time-consuming | [209] |
| High yield | ||||
| Tangential flow filtration (TFF) | Stem Cells | Higher yield of exosomes. | Time-consuming | [182] |
| Homogeneous exosome Size | Expensive | |||
| Fractionation | Stem cells | Homogeneous exosome size | Expensive | [210] |
| Flow Field-Flow Fractionation (FFFF) | Stem Cells | Homogeneous exosome size. | Expensive | [210] |
| Less starting material required | ||||
| Magneto-immunoprecipitation | Plasma, Cell line | Quick Method. | Less Yield | [201] |
| No requirement or expensive equipment | ||||
| Polymer-based precipitation | Serum | Easy to use | Risk of contamination. Interference of contaminants during detection | [211] |
| Biological activity of exosomes is retained | ||||
| Lectin-induced agglutination | Cell line, Urine | Less expensive | Co-precipitation of other soluble components may happen | [212] |
| Microfluidic-Based Isolation Techniques | Blood | Rapid, requires input | Expensive | [213] |
| Acoustic Nanofilter | Red blood cells | Simple and quick method | In developmental stage | [214] |
| ExoQuick Method | Serum | Less time-consuming | Expensive | [215] |
| Not suitable for large volume of sample | ||||
| Deterministic lateral displacement (DLD) pillar array | Urine | Inexpensive and sensitive method | Technology still in developmental stage | [216] |
| Integrated double-filtration microfluidic device | Urine | Less expensive | Not validated for clinical use | [217] |
| On-chip immunoelectrophoresis | Cell line | EVs and their subpopulations can be separated | Technology still in developmental stage | [218] |
| Sepharose and HiTrap Heparin HP column | Serum | Selective isolation of exosomes | [219] | |
| Cation- and anion-exchange chromatography | Cell line | Requires less time | Not suitable for all samples | [219] |
8.1. Conventional method
Exosome purification by conventional method includes ultracentrifugation and polymer-based precipitation. Both purification methods largely depend on physical properties like size, shape, and specific gravity of the exosomes.
8.1.1. Ultracentrifugation
Successive sets of high-speed centrifugation are used to separate cell debris, apoptotic bodies, microvesicles, and exosomes. High-speed centrifugation followed by ultracentrifugation is adopted standard method for isolating exosomes from less complex cell culture supernatants and from relatively complex biological fluids like blood, urine, cerebrospinal fluids, etc. [170–172]. Initially, the crude samples containing exosomes and other contaminants such as cell debris, larger vesicles etc., are separated at a low speed of 2000Xg and 10,000Xg in successive steps. Exosomes are sedimented through a final step of high-speed 100,000XG by ultracentrifugation. Generally, such isolations are performed in a fixed angle rotor, but often, the high ‘g’ force of ultracentrifugation causes damage to the structure of vesicles [173]. The addition of density gradient provides an alternative to circumvent this disadvantage. Through this method, the exosomes are separated based on their specific gravity on a carefully laid down density gradient medium such as sucrose and iodixanol that provide a cushion to the samples and aid in enhancing purity and absorbing the stress to maintain structural integrity [174, 175]. However, it has been noted that these methods considerably decrease exosome yield. Nevertheless, the ultracentrifugation process is the most popular method for the purification of exosomes, even though it is a labor-intensive and time consuming complex process requiring specific instruments.
8.1.2. Polymer based precipitation method
Polymer-based precipitation is another widely used method to purify exosomes. Traditionally this method was used for the purification of viral particles. In this method, an aqueous mixture of PolyEthylene Glycol (PEG) is used to entrap exosomes, which are then enriched by low-speed centrifugation [176, 177]. This process is straightforward and has isolation efficiency equivalent to the ultracentrifugation method with the advantage that it does not require any specific instrumentation and skilled personnel. However, the most significant disadvantage associated with this method is the inability to differentiate between various-sized particles present in the isolates such as microvesicles and co-precipitation of several non-exosomal proteins. Currently, several companies are selling precipitation-based exosome isolation kits based on their proprietary materials, although their efficiency needs to be evaluated for the production of a clinical-grade homogenous population of exosomes.
8.2. Size exclusion chromatography and ultrafiltration methods
One important point to consider to efficiently isolate exosomes for use as drug carriers is to purify a homogenous population of exosomes in consistently large numbers. The conventional methods specifically failed to achieve this. Hence, several novel exosome isolation techniques that rely on particle size rather than shape or specific gravity are being explored. Size exclusion chromatography (SEC), ultrafiltration, and sequential filtration are established methods that purify components based on size [178]. These methods are traditionally used to purify peptides, amino acids, antibodies, lipids, etc. [179]. These techniques have been repurposed to isolate a population of exosomes that are free of other vesicular structures and unwanted protein contaminants. SEC methods can help obtain an enriched population of exosomes from complex biological fluids such as blood, serum etc. SEC purifications are largely based on size, but the shape of eluting particles also influences the purification efficiency [180].
SEC is not suitable for the large-scale production of exosomes from a small volume of the initial sample like blood or saliva etc hence; it alone cannot be used as a stand-alone method for exosome isolation. On the other hand, ultrafiltration and sequential filtration are physical methods that purify based on size of the particles and membrane pore size. Though these methods are suitable for purificationthe biological materials obtained through this method are sometimes compromised because of unintended interactions of molecules, primarily proteins with filtration membranes often associated with solutions containing exosomes [181]. Tangential Flow Filtration (TFF) is a nano-scale filtration method that purifies by repetition of diafiltration and enrichment cycles. The complex biological fluid containing exosomes is sequentially passed through cellulose-based tangential cross filters in multiple cycles [182]. The repeated cycle of filtrations removes a wide array of contaminants like cell debris, proteins etc., enriching the population of exosomes. TFF method, due to its straightforward mechanism, relatively simple workflow and scope of automation, and the ability to purify a homogeneous population of exosomes, is a promising method that needs to be explored for producing clinical-grade exosomes [183].
8.3. Immunoaffinity and microfluidic based isolation methods
Isolation methods based on affinity towards a molecular structure or antigen-antibody interactions have been developed to enhance the quality and purity of exosomes for their use in critical clinical applications [184, 185]. The antibodies against the proteins present on the surface of exosomes are used to capture exosomes specifically expressing them. These interactions usually are very specific, and hence the expectation of using this method is to obtain a population of desired exosomes with very high purity. Antibodies against a particular set of proteins are coated on magnetic solid beads, or other solid surfaces, samples containing a heterogeneous population of EVs are incubated with such beads/ surfaces, following which the exosomes expressing the desired set of markers are eluted and purified [186] [187]. Combining this method with microfluidic devices can enhance the efficiency of exosome isolation. In such devices, antibodies are coated on fine capillaries through which samples containing exosomes are passed. This design enhances the surface area and increases incubation time, efficiently capturing desired exosomes [178] [188]. In a recent study, Zhao et al. purified exosomes from the blood of ovarian cancer patients using microfluidic chips coated with antibodies against EpCAM, Cancer Antigen-125 (CA-125) and CD24, proteins which are overexpressed in ovarian cancer [189]. This method, can efficiently purify exosomes from other EVs. Since the isolation depends on the expression of a set of proteins, it often induces a selective bias among different types of exosomes present in the sample. As a result, exosomes with low expression of capture protein are not isolated, which could reduce the overall population of exosomes. However, this approach could be a good strategy to develop diagnostics (biomarkers) but may not work well to develop delivery vehicles where large numbers of exosomes are required. Immunoaffinity methods have certain disadvantages as they can work with only a small volume of samples. They are expensive, and most importantly, in the present scenario where the scientific community has yet to identify a universal marker for exosomes, isolating a ubiquitous population of exosomes remains a challenge. Finally, from a drug delivery point of view, the essential requirement is to isolate exosomes of a homogenous size, and this cannot be achieved via this method as the immunocapture method does not discriminate based on size.
At present, there is no universally accepted method for isolation of exosomes that is widely available for unrestricted use. Exosome-based biotech companies such as Codiak Biosciences, have made claims for a proprietary platform technology for the enrichment of a homogenous population of exosomes for use and application in the treatment human diseases including cancer. Currently, several exosomes-based products are in clinical testing (Table 5) and results from these clinical trials are pending and necessary to determine their efficacy and treatment outcomes.
Table 5.
List of exosomes-based clinical trials completed
| NCT Number | Title | Status | Conditions | Interventions | Phases |
|---|---|---|---|---|---|
| NCT02702856 | Clinical Validation of a Urinary Exosome Gene Signature in Men Presenting for Suspicion of Prostate Cancer | Completed | Prostate Cancer | Exosome scoring | |
| NCT04529915 | Multicenter Clinical Research for Early Diagnosis of Lung Cancer Using Blood Plasma Derived Exosome | Active, not recruiting | Lung Cancer | Diagnostic Test: Exosome sampling | |
| NCT01294072 | Study Investigating the Ability of Plant Exosomes to Deliver Curcumin to Normal and Colon Cancer Tissue | Active, not recruiting | Colon Cancer | Curcumin conjugated with plant exosomes | Phase 1 |
| NCT01159288 | Trial of a Vaccination With Tumor Antigen-loaded Dendritic Cell-derived Exosomes | Completed | Non-Small Cell Lung Cancer | Biological: Dex2 | Phase 2 |
| NCT01668849 | Edible Plant Exosome Ability to Prevent Oral Mucositis Associated With Chemoradiation Treatment of Head and Neck Cancer | Active, not recruiting | Head and Neck Cancer and Oral Mucositis | Grape extract | Phase 1 |
| Drug: Lortab, Fentanyl patch, mouthwash | |||||
| NCT03109873 | Metformin Hydrochloride in Affecting Cytokines and Exosomes in Patients With Head and Neck Cancer | Completed | Larynx, Lip, Oral Cavity and Pharynx Cancer | Radiation: External Beam Radiation Therapy | Early Phase 1 |
| Drug: Metformin Hydrochloride | |||||
| NCT03032913 | Diagnostic Accuracy of Circulating Tumor Cells (CTCs) and Onco-exosome Quantification in the Diagnosis of Pancreatic Cancer - PANC-CTC | Completed | Pancreatic Ductal Adenocarcinoma (PDAC) | Blood samples for developing diagnostics | |
| NCT03488134 | Predicting Prognosis and Recurrence of Thyroid Cancer Via New Biomarkers, Urinary Exosomal Thyroglobulin and Galectin-3 | Active, not recruiting | Thyroid Cancer | ||
| NCT02662621 | Pilot Study With the Aim to Quantify a Stress Protein in the Blood and in the Urine for the Monitoring and Early Diagnosis of Malignant Solid Tumors | Completed | Cancer | blood samples | Not Applicable |
| Urine samples | |||||
| NCT03031418 | Clinical Evaluation of the ‘ExoDx Prostate IntelliScore’ (EPI) | Completed | Cancer of Prostate | ExoDx Prostate Intelliscore | |
| NCT03317080 | Dynamic Monitoring Circulating Tumor DNA in Surgical Patients With Lung Cancer | Active, not recruiting | Lung Cancer | ||
| NCT02862470 | Anaplastic Thyroid Cancer and Follicular Thyroid Cancer-derived Exosomal Analysis Via Treatment of Lovastatin and Vildagliptin and Pilot Prognostic Study Via Urine Exosomal Biological Markers in Thyroid Cancer Patients | Active, not recruiting | Thyroid Cancer | ||
| NCT03791073 | New Biomarkers in Pancreatic Cancer Using EXPEL Concept | Active, not recruiting | Oncology | ||
| NCT02063464 | Blood Collection From People With Ovarian Cancer | Completed | Ovarian Cancer, Cancer of the Ovary, Ovarian Neoplasms | ||
| NCT03235687 | Decision Impact Trial of the ExoDx Prostate (IntelliScore) | Active, not recruiting | Cancer of the Prostate | ExoDx Prostate (IntelliScore) | Not Applicable |
| NCT02921854 | Detection of Circulating Biomarkers of Immunogenic Cell Death | Completed | Non Small Cell Lung Cancer | Blood withdrawal | Not Applicable |
| NCT02507583 | Antisense102: Pilot Immunotherapy for Newly Diagnosed Malignant Glioma | Completed | Malignant Glioma, Neoplasms | Drug: IGF-1R/AS ODN; Simplanted for 48 hours. | Phase 1 |
| NCT01550523 | Pilot Immunotherapy Trial for Recurrent Malignant Gliomas | Completed | Malignant Glioma of Brain | Drug: IGF-1R/AS | Phase 1 |
| NCT02928432 | SWITCH: Study of the Prednisone to Dexamethasone Change in mCRPC Patients Treated With Abiraterone | Completed | Prostate Cancer | Steroids switch | Phase 2 |
| NCT03228277 | Olmutinib Trial in T790M (+) NSCLC Patients Detected by Liquid Biopsy Using BALF Extracellular Vesicular DNA | completed | Non Small Cell Lung Cancer | Olmutinib | Phase 2 |
| NCT02971761 | Pembrolizumab and Enobosarm in Treating Patients With Androgen Receptor Positive Metastatic Triple Negative Breast Cancer | Active, not recruiting | Androgen Receptor Positive, Estrogen Receptor Negative, HER2/Neu Negative|Metastatic Triple-Negative Breast Carcinoma, Progesterone Receptor Negative, Stage IV Breast Cancer | Enobosarm Laboratory Biomarker Analysis, Biological: Pembrolizumab |
Phase 2 |
| NCT00578240 | Molecular Studies and Clinical Correlations in Human Prostatic Disease | Active, not recruiting | Prostate Cancer | ||
| NCT02051101 | Pathogenic Mechanisms of Port Wine Stain and Repository of Port Wine Stain Biopsy Samples | Completed | Port-Wine Stain | Biopsy sample from Port Wine Stain Birthmark | Not Applicable |
| NCT02535247 | Study of MK-3475 Alone or in Combination With Copanlisib in Relapsed or Refractory NK and T-cell Non-Hodgkin Lymphoma | Active, not recruiting | Lymphoma, T-Cell, Peripheral | MK-3475 Drug: Copanlisib | Phase 1 |
| Phase 2 | |||||
9. Conclusion and future directions
In the present article, we discussed the development and use of exosomes for cancer diagnosis and treatment. Highlighting data from multiple studies, we present the premise of exosomes as a potential alternative to synthetic liposomes as drug carriers or delivery vehicles. Exosomes are naturally produced vesicles and thus have many advantages over synthetic delivery vehicles; this is the core idea behind using exosomes as drug delivery vehicles or carriers. Organotropism, non-immunogenicity, their efficient uptake by recipient cells, and the simultaneous packaging of multiple payloads in the same vesicle are some exemplary properties of exosomes that make them suitable as drug delivery vehicles. Nevertheless, important challenges must be overcome before exosome-based delivery systems can be translated into the clinic and replace the existing successful liposome-based modalities. Pertinent questions related to efficient isolation methods for purifying homogenous population of exosomes on a large scale and identification of a particular population of exosomes that shows maximum efficiency for therapeutic loading and transportation must be explored. Since exosome science is a relatively new area of study, there is a need to enhance our understanding of the biogenesis and basic physiology of exosome synthesis in order to develop processes for loading molecules of interest. Finally, the selection of the starting material for large-scale isolation of exosomes must be addressed. The properties of exosomes that make them ideal drug delivery vehicles also support their use as cancer theranostics, and we also address this aspect in the current article. The exosome science community has recently started exploring many of the described biological sources for exosome isolation and developing drug delivery vehicles/theranostics from them. The idea of an ‘organic exosome’ is in its infancy but is nevertheless an attractive area of study in order to develop products of clinical relevance. While there are several milestones yet to achieve, the tremendous progress and advances made in the field of EVs, emphasizes that successful application of exosomes for cancer therapy, diagnosis and as a theranostic in the clinic is not too distant.
Acknowledgments
This study was supported in part by grants received from the National Cancer Institute (NCI) of the National Institutes of Health (NIH; R01 CA167516, R01CA233201, R01 CA254192); from the National Institute of General Medical Sciences (P20 GM103639) of the National Institutes of Health; a Merit Grant (101 BX003420A1) from the Department of Veterans Affairs; a Pilot Grant and Seed Grant funded by the National Cancer Institute Cancer Center Support Grant P30 CA225520 awarded to the University of Oklahoma Stephenson Cancer Center; from the Department of Defense (DOD) through the Lung Cancer Research Program (LCRP) under award no.W81XWH-18-1-0637 & W81XWH-19-1-0647; a grant (HR18-088) from the Oklahoma Center for the Advancement of Science and Technology (OCAST); funds received from the Presbyterian Health Foundation Seed Grant, Presbyterian Health Foundation Bridge Grant, Oklahoma Tobacco Settlement Endowment Trust (TSET) awarded to the University of Oklahoma Stephenson Cancer Center, and the Jim and Christy Everest Endowed Chair in Cancer Developmental Therapeutics.
Rajagopal Ramesh is an Oklahoma TSET Research Scholar and holds the Jim and Christy Everest Endowed Chair in Cancer Developmental Therapeutics.
The content is solely the responsibility of the authors. The opinions, interpretations, conclusions and recommendations are those of the authors and are not necessarily endorsed by or representative of the official views by the NIH, DOD, Department of Veterans Affairs, OCAST, or TSET.
Funding Source
All sources of funding should also be acknowledged and you should declare any involvement of study sponsors in the study design; collection, analysis and interpretation of data; the writing of the manuscript; the decision to submit the manuscript for publication. If the study sponsors had no such involvement, this should be stated.
Abbreviations
- 2D
Two-Dimensional
- 3D
Three-Dimensional
- AFM
Atomic Force Microscopy
- BBB
Blood Brain Barrier
- BSA
Bovine Serum Albumin
- CpGDNA
5’-C-phosphate-G-3’DNA
- CRISPR/Cas9
Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR protein 9
- CSF
Cerebrospinal Fluid
- CUR
Curcumin
- DARP
Dopamine-Releasing Protein
- DARP
Designed Ankyrin Repeat Proteins
- DC
Dendritic Cells
- DLS
Dynamic Light Scattering
- DNA
Deoxyribonucleic acid
- DOC
Docetaxel
- EGFR
Epidermal Growth Factor Receptor
- EpCAM
Epithelial Cellular Adhesion Molecule
- EPR
Enhanced Permeability and Retention
- ESCRT
Endosomal Sorting Complex Required for Transport
- EV
Extracellular Vesicles
- FA
Folic Acid
- FCS
Fetal Calf Serum
- FTIR
Fourier-Transform Infrared Spectroscopy
- GBM
Glioblastoma Multiforme
- GDV
Gene Delivery Vector
- HCl
Hydrochloric Acid
- ILV
Intra Luminal Vesicles
- MAPK
Mitogen-Activated Protein Kinase
- MHC
Major Histocompatibility Complex
- miR
microRNA
- MRD
Multi-Drug Resistance
- MRI
Magnetic Resonance Imaging
- MS
Mass Spectroscopy
- MSC
Mesenchymal Stem Cells
- MVB
Multi Vesicular Bodies
- NMR
Nuclear Magnetic Resonance
- NTA
Nanotracker Analysis
- OMV
Outer Membrane Vesicles
- OSCC
Oral Squamous Cell Carcinoma
- PAMP
Pathogen-Associated Molecular Pattern
- PEG
Polyethylene Glycol
- PRR
Pattern Recognition Receptor
- PTX
Paclitaxel
- qRT-PCR
quantitative Real Time Polymerase Chain Reaction
- RBC
Red Blood Cells
- RNA
Ribonucleic Acid
- RPMI
Roswell Park Memorial Institute
- siRNA
Small interfering RNA
- SPIONs
Super Paramagnetic Iron Oxide Nanoparticles
- TEM
Transmission Electron Microscopy
- TEx
Tumor Exosomes
- TLR
Toll-Like Receptor
- TMZ
Temozolomide
- TNBC
Triple-Negative Breast Cancer
- UHT
Ultra Heat Treated
- VEGF
Vascular Endothelial Growth Factor
- WFA
Withaferin A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors report no conflicts of interest.
References
- [1].Senapati S, Mahanta AK, Kumar S, Maiti P, Controlled drug delivery vehicles for cancer treatment and their performance, Signal Transduct Target Ther 3 (2018) 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Li Z, Tan S, Li S, Shen Q, Wang K, Cancer drug delivery in the nano era: An overview and perspectives (Review), Oncol Rep 38(2) (2017) 611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen F, Shi Y, Zhang J, Liu Q, Nanoparticle-based Drug Delivery Systems for Targeted Epigenetics Cancer Therapy, Curr Drug Targets 21(11) (2020) 1084–1098. [DOI] [PubMed] [Google Scholar]
- [4].Akbarzadeh A, Rezaei-Sadabady R, Davaran S, Joo SW, Zarghami N, Hanifehpour Y, Samiei M, Kouhi M, Nejati-Koshki K, Liposome: classification, preparation, and applications, Nanoscale Res Lett 8(1) (2013) 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sercombe L, Veerati T, Moheimani F, Wu SY, Sood AK, Hua S, Advances and Challenges of Liposome Assisted Drug Delivery, Front Pharmacol 6 (2015) 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Almeida B, Nag OK, Rogers KE, Delehanty JB, Recent Progress in Bioconjugation Strategies for Liposome-Mediated Drug Delivery, Molecules 25(23) (2020) 5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pillai G, Nanomedicines for Cancer Therapy: An Update of FDA Approved and Those under Various Stages of Development, SOJ Pharmacy & Pharmaceutical Sciences 1 (2014). [Google Scholar]
- [8].Yue X, Dai Z, Recent advances in liposomal nanohybrid cerasomes as promising drug nanocarriers, Adv Colloid Interface Sci 207 (2014) 32–42. [DOI] [PubMed] [Google Scholar]
- [9].Raemdonck K, Braeckmans K, Demeester J, De Smedt SC, Merging the best of both worlds: hybrid lipid-enveloped matrix nanocomposites in drug delivery, Chem Soc Rev 43(1) (2014) 444–72. [DOI] [PubMed] [Google Scholar]
- [10].Shao J, Zaro J, Shen Y, Advances in Exosome-Based Drug Delivery and Tumor Targeting: From Tissue Distribution to Intracellular Fate, Int J Nanomedicine 15 (2020) 9355–9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bongiovanni L, Andriessen A, Wauben MHM, Hoen E, de Bruin A, Extracellular Vesicles: Novel Opportunities to Understand and Detect Neoplastic Diseases, Vet Pathol 58(3) (2021) 453–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Abels ER, Breakefield XO, Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake, Cell Mol Neurobiol 36(3) (2016) 301–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gyorgy B, Szabo TG, Pasztoi M, Pal Z, Misjak P, Aradi B, Laszlo V, Pallinger E, Pap E, Kittel A, Nagy G, Falus A, Buzas EI, Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles, Cell Mol Life Sci 68(16) (2011) 2667–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ailuno G, Baldassari S, Lai F, Florio T, Caviglioli G, Exosomes and Extracellular Vesicles as Emerging Theranostic Platforms in Cancer Research, Cells 9(12) (2020) 2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach JM, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borras FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan MA, Brigstock DR, Brisson A, Broekman ML, Bromberg JF, Bryl-Gorecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzas EI, Byrd JB, Camussi G, Carter DR, Caruso S, Chamley LW, Chang YT, Chen C, Chen S, Cheng L, Chin AR, Clayton A, Clerici SP, Cocks A, Cocucci E, Coffey RJ, Cordeiro-da-Silva A, Couch Y, Coumans FA, Coyle B, Crescitelli R, Criado MF, D’Souza-Schorey C, Das S, Datta Chaudhuri A, de Candia P, De Santana EF, De Wever O, Del Portillo HA, Demaret T, Deville S, Devitt A, Dhondt B, Di Vizio D, Dieterich LC, Dolo V, Dominguez Rubio AP, Dominici M, Dourado MR, Driedonks TA, Duarte FV, Duncan HM, Eichenberger RM, Ekstrom K, El Andaloussi S, Elie-Caille C, Erdbrugger U, Falcon-Perez JM, Fatima F, Fish JE, Flores-Bellver M, Forsonits A, Frelet-Barrand A, Fricke F, Fuhrmann G, Gabrielsson S, Gamez-Valero A, Gardiner C, Gartner K, Gaudin R, Gho YS, Giebel B, Gilbert C, Gimona M, Giusti I, Goberdhan DC, Gorgens A, Gorski SM, Greening DW, Gross JC, Gualerzi A, Gupta GN, Gustafson D, Handberg A, Haraszti RA, Harrison P, Hegyesi H, Hendrix A, Hill AF, Hochberg FH, Hoffmann KF, Holder B, Holthofer H, Hosseinkhani B, Hu G, Huang Y, Huber V, Hunt S, Ibrahim AG, Ikezu T, Inal JM, Isin M, Ivanova A, Jackson HK, Jacobsen S, Jay SM, Jayachandran M, Jenster G, Jiang L, Johnson SM, Jones JC, Jong A, Jovanovic-Talisman T, Jung S, Kalluri R, Kano SI, Kaur S, Kawamura Y, Keller ET, Khamari D, Khomyakova E, Khvorova A, Kierulf P, Kim KP, Kislinger T, Klingeborn M, Klinke DJ 2nd, Kornek M, Kosanovic MM, Kovacs AF, Kramer-Albers EM, Krasemann S, Krause M, Kurochkin IV, Kusuma GD, Kuypers S, Laitinen S, Langevin SM, Languino LR, Lannigan J, Lasser C, Laurent LC, Lavieu G, Lazaro-Ibanez E, Le Lay S, Lee MS, Lee YXF, Lemos DS, Lenassi M, Leszczynska A, Li IT, Liao K, Libregts SF, Ligeti E, Lim R, Lim SK, Line A, Linnemannstons K, Llorente A, Lombard CA, Lorenowicz MJ, Lorincz AM, Lotvall J, Lovett J, Lowry MC, Loyer X, Lu Q, Lukomska B, Lunavat TR, Maas SL, Malhi H, Marcilla A, Mariani J, Mariscal J, Martens-Uzunova ES, Martin-Jaular L, Martinez MC, Martins VR, Mathieu M, Mathivanan S, Maugeri M, McGinnis LK, McVey MJ, Meckes DG Jr., Meehan KL, Mertens I, Minciacchi VR, Moller A, Moller Jorgensen M, Morales-Kastresana A, Morhayim J, Mullier F, Muraca M, Musante L, Mussack V, Muth DC, Myburgh KH, Najrana T, Nawaz M, Nazarenko I, Nejsum P, Neri C, Neri T, Nieuwland R, Nimrichter L, Nolan JP, Nolte-’t Hoen EN, Noren Hooten N, O’Driscoll L, O’Grady T, O’Loghlen A, Ochiya T, Olivier M, Ortiz A, Ortiz LA, Osteikoetxea X, Ostergaard O, Ostrowski M, Park J, Pegtel DM, Peinado H, Perut F, Pfaffl MW, Phinney DG, Pieters BC, Pink RC, Pisetsky DS, Pogge von Strandmann E, Polakovicova I, Poon IK, Powell BH, Prada I, Pulliam L, Quesenberry P, Radeghieri A, Raffai RL, Raimondo S, Rak J, Ramirez MI, Raposo G, Rayyan MS, Regev-Rudzki N, Ricklefs FL, Robbins PD, Roberts DD, Rodrigues SC, Rohde E, Rome S, Rouschop KM, Rughetti A, Russell AE, Saa P, Sahoo S, Salas-Huenuleo E, Sanchez C, Saugstad JA, Saul MJ, Schiffelers RM, Schneider R, Schoyen TH, Scott A, Shahaj E, Sharma S, Shatnyeva O, Shekari F, Shelke GV, Shetty AK, Shiba K, Siljander PR, Silva AM, Skowronek A, Snyder OL 2nd, Soares RP, Sodar BW, Soekmadji C, Sotillo J, Stahl PD, Stoorvogel W, Stott SL, Strasser EF, Swift S, Tahara H, Tewari M, Timms K, Tiwari S, Tixeira R, Tkach M, Toh WS, Tomasini R, Torrecilhas AC, Tosar JP, Toxavidis V, Urbanelli L, Vader P, van Balkom BW, van der Grein SG, Van Deun J, van Herwijnen MJ, Van Keuren-Jensen K, van Niel G, van Royen ME, van Wijnen AJ, Vasconcelos MH, Vechetti IJ Jr., Veit TD, Vella LJ, Velot E, Verweij FJ, Vestad B, Vinas JL, Visnovitz T, Vukman KV, Wahlgren J, Watson DC, Wauben MH, Weaver A, Webber JP, Weber V, Wehman AM, Weiss DJ, Welsh JA, Wendt S, Wheelock AM, Wiener Z, Witte L, Wolfram J, Xagorari A, Xander P, Xu J, Yan X, Yanez-Mo M, Yin H, Yuana Y, Zappulli V, Zarubova J, Zekas V, Zhang JY, Zhao Z, Zheng L, Zheutlin AR, Zickler AM, Zimmermann P, Zivkovic AM, Zocco D, Zuba-Surma EK, Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines, J Extracell Vesicles 7(1) (2018) 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Emelyanov A, Shtam T, Kamyshinsky R, Garaeva L, Verlov N, Miliukhina I, Kudrevatykh A, Gavrilov G, Zabrodskaya Y, Pchelina S, Konevega A, Cryo-electron microscopy of extracellular vesicles from cerebrospinal fluid, PLoS One 15(1) (2020) e0227949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Srivastava A, Filant J, Moxley KM, Sood A, McMeekin S, Ramesh R, Exosomes: a role for naturally occurring nanovesicles in cancer growth, diagnosis and treatment, Curr Gene Ther 15(2) (2015) 182–92. [DOI] [PubMed] [Google Scholar]
- [18].Mathivanan S, Ji H, Simpson RJ, Exosomes: extracellular organelles important in intercellular communication, J Proteomics 73(10) (2010) 1907–20. [DOI] [PubMed] [Google Scholar]
- [19].Andreu Z, Yanez-Mo M, Tetraspanins in extracellular vesicle formation and function, Front Immunol 5 (2014) 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Makler A, Asghar W, Exosomal biomarkers for cancer diagnosis and patient monitoring, Expert Rev Mol Diagn 20(4) (2020) 387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Subra C, Laulagnier K, Perret B, Record M, Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies, Biochimie 89(2) (2007) 205–12. [DOI] [PubMed] [Google Scholar]
- [22].Soung YH, Ford S, Zhang V, Chung J, Exosomes in Cancer Diagnostics, Cancers (Basel) 9(1) (2017) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li W, Li C, Zhou T, Liu X, Liu X, Li X, Chen D, Role of exosomal proteins in cancer diagnosis, Mol Cancer 16(1) (2017) 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Srivastava A, Amreddy N, Pareek V, Chinnappan M, Ahmed R, Mehta M, Razaq M, Munshi A, Ramesh R, Progress in extracellular vesicle biology and their application in cancer medicine, Wiley Interdiscip Rev Nanomed Nanobiotechnol 12(4) (2020) e1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, Alahari SK, Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance, Mol Cancer 18(1) (2019) 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gurung S, Perocheau D, Touramanidou L, Baruteau J, The exosome journey: from biogenesis to uptake and intracellular signalling, Cell Commun Signal 19(1) (2021) 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Thery C, Raposo G, Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles, J Cell Sci 126(Pt 24) (2013) 5553–65. [DOI] [PubMed] [Google Scholar]
- [28].Blanc L, Vidal M, New insights into the function of Rab GTPases in the context of exosomal secretion, Small GTPases 9(1–2) (2018) 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hsu C, Morohashi Y, Yoshimura S, Manrique-Hoyos N, Jung S, Lauterbach MA, Bakhti M, Gronborg M, Mobius W, Rhee J, Barr FA, Simons M, Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C, J Cell Biol 189(2) (2010) 223–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bebelman MP, Smit MJ, Pegtel DM, Baglio SR, Biogenesis and function of extracellular vesicles in cancer, Pharmacol Ther 188 (2018) 1–11. [DOI] [PubMed] [Google Scholar]
- [31].Ghoroghi S, Mary B, Larnicol A, Asokan N, Klein A, Osmani N, Busnelli I, Delalande F, Paul N, Halary S, Gros F, Fouillen L, Haeberle AM, Royer C, Spiegelhalter C, Andre-Gregoire G, Mittelheisser V, Detappe A, Murphy K, Timpson P, Carapito R, Blot-Chabaud M, Gavard J, Carapito C, Vitale N, Lefebvre O, Goetz JG, Hyenne V, Ral GTPases promote breast cancer metastasis by controlling biogenesis and organ targeting of exosomes, Elife 10 (2021) e61539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gurunathan S, Kang MH, Kim JH, A Comprehensive Review on Factors Influences Biogenesis, Functions, Therapeutic and Clinical Implications of Exosomes, Int J Nanomedicine 16 (2021) 1281–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kumar A, Deep G, Hypoxia in tumor microenvironment regulates exosome biogenesis: Molecular mechanisms and translational opportunities, Cancer Lett 479 (2020) 23–30. [DOI] [PubMed] [Google Scholar]
- [34].Wei D, Zhan W, Gao Y, Huang L, Gong R, Wang W, Zhang R, Wu Y, Gao S, Kang T, RAB31 marks and controls an ESCRT-independent exosome pathway, Cell Res 31(2) (2021) 157–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wei H, Chen Q, Lin L, Sha C, Li T, Liu Y, Yin X, Xu Y, Chen L, Gao W, Li Y, Zhu X, Regulation of exosome production and cargo sorting, Int J Biol Sci 17(1) (2021) 163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Li SP, Lin ZX, Jiang XY, Yu XY, Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools, Acta Pharmacol Sin 39(4) (2018) 542–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Skryabin GO, Komelkov AV, Savelyeva EE, Tchevkina EM, Lipid Rafts in Exosome Biogenesis, Biochemistry (Mosc) 85(2) (2020) 177–191. [DOI] [PubMed] [Google Scholar]
- [38].Kowal J, Tkach M, Thery C, Biogenesis and secretion of exosomes, Curr Opin Cell Biol 29 (2014) 116–25. [DOI] [PubMed] [Google Scholar]
- [39].Elkhoury K, Kocak P, Kang A, Arab-Tehrany E, Ellis Ward J, Shin SR, Engineering Smart Targeting Nanovesicles and Their Combination with Hydrogels for Controlled Drug Delivery, Pharmaceutics 12(9) (2020) 849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Antimisiaris SG, Mourtas S, Marazioti A, Exosomes and Exosome-Inspired Vesicles for Targeted Drug Delivery, Pharmaceutics 10(4) (2018) 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhang Y, Liu Y, Liu H, Tang WH, Exosomes: biogenesis, biologic function and clinical potential, Cell Biosci (9) (2019) 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Jafari D, Shajari S, Jafari R, Mardi N, Gomari H, Ganji F, Forouzandeh Moghadam M, Samadikuchaksaraei A, Designer Exosomes: A New Platform for Biotechnology Therapeutics, BioDrugs 34(5) (2020) 567–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, Lee JJ, Kalluri R, Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer, Nature 546(7659) (2017) 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kibria G, Ramos EK, Wan Y, Gius DR, Liu H, Exosomes as a Drug Delivery System in Cancer Therapy: Potential and Challenges, Mol Pharm 15(9) (2018) 3625–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mondal A, Ashiq KA, Phulpagar P, Singh DK, Shiras A, Effective Visualization and Easy Tracking of Extracellular Vesicles in Glioma Cells, Biol Proced Online 21 (2019) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Didiot MC, Hall LM, Coles AH, Haraszti RA, Godinho BM, Chase K, Sapp E, Ly S, Alterman JF, Hassler MR, Echeverria D, Raj L, Morrissey DV, DiFiglia M, Aronin N, Khvorova A, Exosome-mediated Delivery of Hydrophobically Modified siRNA for Huntingtin mRNA Silencing, Mol Ther 24(10) (2016) 1836–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Takov K, Yellon DM, Davidson SM, Confounding factors in vesicle uptake studies using fluorescent lipophilic membrane dyes, J Extracell Vesicles 6(1) (2017) 1388731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Srivastava A, Amreddy N, Babu A, Panneerselvam J, Mehta M, Muralidharan R, Chen A, Zhao YD, Razaq M, Riedinger N, Kim H, Liu S, Wu S, Abdel-Mageed AB, Munshi A, Ramesh R, Nanosomes carrying doxorubicin exhibit potent anticancer activity against human lung cancer cells, Sci Rep 6 (2016) 38541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Satake T, Suetsugu A, Nakamura M, Kunisada T, Saji S, Moriwaki H, Shimizu M, Hoffman RM, Color-coded Imaging of the Fate of Cancer-cell-derived Exosomes During Pancreatic Cancer Metastases in a Nude-mouse Model, Anticancer Res 39(8) (2019) 4055–4060. [DOI] [PubMed] [Google Scholar]
- [50].Hikita T, Miyata M, Watanabe R, Oneyama C, In vivo imaging of long-term accumulation of cancer-derived exosomes using a BRET-based reporter, Sci Rep 10(1) (2020) 16616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Song S, Shim MK, Lim S, Moon Y, Yang S, Kim J, Hong Y, Yoon HY, Kim IS, Hwang KY, Kim K, In Situ One-Step Fluorescence Labeling Strategy of Exosomes via Bioorthogonal Click Chemistry for Real-Time Exosome Tracking In Vitro and In Vivo, Bioconjug Chem 31(5) (2020) 1562–1574. [DOI] [PubMed] [Google Scholar]
- [52].Chen C, Zong S, Wang Z, Lu J, Zhu D, Zhang Y, Cui Y, Imaging and Intracellular Tracking of Cancer-Derived Exosomes Using Single-Molecule Localization-Based Super-Resolution Microscope, ACS Appl Mater Interfaces 8(39) (2016) 25825–25833. [DOI] [PubMed] [Google Scholar]
- [53].Cohen O, Betzer O, Elmaliach-Pnini N, Motiei M, Sadan T, Cohen-Berkman M, Dagan O, Popovtzer A, Yosepovich A, Barhom H, Michaeli S, Popovtzer R, ‘Golden’ exosomes as delivery vehicles to target tumors and overcome intratumoral barriers: in vivo tracking in a model for head and neck cancer, Biomater Sci 9(6) (2021) 2103–2114. [DOI] [PubMed] [Google Scholar]
- [54].Bose RJC, Uday Kumar S, Zeng Y, Afjei R, Robinson E, Lau K, Bermudez A, Habte F, Pitteri SJ, Sinclair R, Willmann JK, Massoud TF, Gambhir SS, Paulmurugan R, Tumor Cell-Derived Extracellular Vesicle-Coated Nanocarriers: An Efficient Theranostic Platform for the Cancer-Specific Delivery of Anti-miR-21 and Imaging Agents, ACS Nano 12(11) (2018) 10817–10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Silva AK, Luciani N, Gazeau F, Aubertin K, Bonneau S, Chauvierre C, Letourneur D, Wilhelm C, Combining magnetic nanoparticles with cell derived microvesicles for drug loading and targeting, Nanomedicine 11(3) (2015) 645–55. [DOI] [PubMed] [Google Scholar]
- [56].Lorenc T, Chrzanowski J, Olejarz W, Current Perspectives on Clinical Use of Exosomes as a Personalized Contrast Media and Theranostics, Cancers (Basel) 12(11) (2020) 3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Shaikh S, Rehman FU, Du T, Jiang H, Yin L, Wang X, Chai R, Real-Time Multimodal Bioimaging of Cancer Cells and Exosomes through Biosynthesized Iridium and Iron Nanoclusters, ACS Appl Mater Interfaces 10(31) (2018) 26056–26063. [DOI] [PubMed] [Google Scholar]
- [58].Tayyaba, Rehman FU, Shaikh S, Tanziela, Semcheddine F, Du T, Jiang H, Wang X, In situ self-assembled Ag-Fe3O4 nanoclusters in exosomes for cancer diagnosis, J Mater Chem B 8(14) (2020) 2845–2855. [DOI] [PubMed] [Google Scholar]
- [59].Hwang DW, Choi H, Jang SC, Yoo MY, Park JY, Choi NE, Oh HJ, Ha S, Lee YS, Jeong JM, Gho YS, Lee DS, Noninvasive imaging of radiolabeled exosome-mimetic nanovesicle using (99m)Tc-HMPAO, Sci Rep 5 (2015) 15636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Faruqu FN, Wang JT, Xu L, McNickle L, Chong EM, Walters A, Gurney M, Clayton A, Smyth LA, Hider R, Sosabowski J, Al-Jamal KT, Membrane Radiolabelling of Exosomes for Comparative Biodistribution Analysis in Immunocompetent and Immunodeficient Mice - A Novel and Universal Approach, Theranostics 9(6) (2019) 1666–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Molavipordanjani S, Khodashenas S, Abedi SM, Moghadam MF, Mardanshahi A, Hosseinimehr SJ, (99m)Tc-radiolabeled HER2 targeted exosome for tumor imaging, Eur J Pharm Sci 148 (2020) 105312. [DOI] [PubMed] [Google Scholar]
- [62].Shi S, Li T, Wen X, Wu SY, Xiong C, Zhao J, Lincha VR, Chow DS, Liu Y, Sood AK, Li C, Copper-64 Labeled PEGylated Exosomes for In Vivo Positron Emission Tomography and Enhanced Tumor Retention, Bioconjug Chem 30(10) (2019) 2675–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ, Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes, Nat Biotechnol 29(4) (2011) 341–5. [DOI] [PubMed] [Google Scholar]
- [64].Pitt JM, Andre F, Amigorena S, Soria JC, Eggermont A, Kroemer G, Zitvogel L, Dendritic cell-derived exosomes for cancer therapy, J Clin Invest 126(4) (2016) 1224–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Fais S, NK cell-released exosomes: Natural nanobullets against tumors, Oncoimmunology 2(1) (2013) e22337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wu F, Xie M, Hun M, She Z, Li C, Luo S, Chen X, Wan W, Wen C, Tian J, Natural Killer Cell-Derived Extracellular Vesicles: Novel Players in Cancer Immunotherapy, Front Immunol 12 (2021) 658698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Xu Z, Zeng S, Gong Z, Yan Y, Exosome-based immunotherapy: a promising approach for cancer treatment, Mol Cancer 19(1) (2020) 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Enomoto Y, Li P, Jenkins LM, Anastasakis D, Lyons GC, Hafner M, Leonard WJ, Cytokine-enhanced cytolytic activity of exosomes from NK Cells, Cancer Gene Ther (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Lugini L, Cecchetti S, Huber V, Luciani F, Macchia G, Spadaro F, Paris L, Abalsamo L, Colone M, Molinari A, Podo F, Rivoltini L, Ramoni C, Fais S, Immune surveillance properties of human NK cell-derived exosomes, J Immunol 189(6) (2012) 2833–42. [DOI] [PubMed] [Google Scholar]
- [70].Neviani P, Wise PM, Murtadha M, Liu CW, Wu CH, Jong AY, Seeger RC, Fabbri M, Natural Killer-Derived Exosomal miR-186 Inhibits Neuroblastoma Growth and Immune Escape Mechanisms, Cancer Res 79(6) (2019) 1151–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Jong AY, Wu CH, Li J, Sun J, Fabbri M, Wayne AS, Seeger RC, Large-scale isolation and cytotoxicity of extracellular vesicles derived from activated human natural killer cells, J Extracell Vesicles 6(1) (2017) 1294368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Zhu L, Gangadaran P, Kalimuthu S, Oh JM, Baek SH, Jeong SY, Lee SW, Lee J, Ahn BC, Novel alternatives to extracellular vesicle-based immunotherapy - exosome mimetics derived from natural killer cells, Artif Cells Nanomed Biotechnol 46(sup3) (2018) S166–S179. [DOI] [PubMed] [Google Scholar]
- [73].Kang YT, Niu Z, Hadlock T, Purcell E, Lo TW, Zeinali M, Owen S, Keshamouni VG, Reddy R, Ramnath N, Nagrath S, On-Chip Biogenesis of Circulating NK Cell-Derived Exosomes in Non-Small Cell Lung Cancer Exhibits Antitumoral Activity, Adv Sci (Weinh) 8(6) (2021) 2003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Li S, Wu Y, Ding F, Yang J, Li J, Gao X, Zhang C, Feng J, Engineering macrophage-derived exosomes for targeted chemotherapy of triple-negative breast cancer, Nanoscale 12(19) (2020) 10854–10862. [DOI] [PubMed] [Google Scholar]
- [75].Kusuzaki K, Matsubara T, Murata H, Logozzi M, Iessi E, Di Raimo R, Carta F, Supuran CT, Fais S, Natural extracellular nanovesicles and photodynamic molecules: is there a future for drug delivery?, J Enzyme Inhib Med Chem 32(1) (2017) 908–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Iessi E, Logozzi M, Lugini L, Azzarito T, Federici C, Spugnini EP, Mizzoni D, Di Raimo R, Angelini DF, Battistini L, Cecchetti S, Fais S, Acridine Orange/exosomes increase the delivery and the effectiveness of Acridine Orange in human melanoma cells: A new prototype for theranostics of tumors, J Enzyme Inhib Med Chem 32(1) (2017) 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Logozzi M, Mizzoni D, Bocca B, Di Raimo R, Petrucci F, Caimi S, Alimonti A, Falchi M, Cappello F, Campanella C, Bavisotto CC, David S, Bucchieri F, Angelini DF, Battistini L, Fais S, Human primary macrophages scavenge AuNPs and eliminate it through exosomes. A natural shuttling for nanomaterials, Eur J Pharm Biopharm 137 (2019) 23–36. [DOI] [PubMed] [Google Scholar]
- [78].Federici C, Petrucci F, Caimi S, Cesolini A, Logozzi M, Borghi M, D’Ilio S, Lugini L, Violante N, Azzarito T, Majorani C, Brambilla D, Fais S, Exosome release and low pH belong to a framework of resistance of human melanoma cells to cisplatin, PLoS One 9(2) (2014) e88193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Logozzi M, Di Raimo R, Properzi F, Barca S, Angelini DF, Mizzoni D, Falchi M, Battistini L, Fais S, Nanovesicles released by OKT3 hybridoma express fully active antibodies, J Enzyme Inhib Med Chem 36(1) (2021) 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Gao D, Jiang L, Exosomes in cancer therapy: a novel experimental strategy, Am J Cancer Res 8(11) (2018) 2165–2175. [PMC free article] [PubMed] [Google Scholar]
- [81].Pascucci L, Cocce V, Bonomi A, Ami D, Ceccarelli P, Ciusani E, Vigano L, Locatelli A, Sisto F, Doglia SM, Parati E, Bernardo ME, Muraca M, Alessandri G, Bondiolotti G, Pessina A, Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: a new approach for drug delivery, J Control Release 192 (2014) 262–70. [DOI] [PubMed] [Google Scholar]
- [82].Kalimuthu S, Gangadaran P, Rajendran RL, Zhu L, Oh JM, Lee HW, Gopal A, Baek SH, Jeong SY, Lee SW, Lee J, Ahn BC, A New Approach for Loading Anticancer Drugs Into Mesenchymal Stem Cell-Derived Exosome Mimetics for Cancer Therapy, Front Pharmacol 9 (2018) 1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Munoz JL, Bliss SA, Greco SJ, Ramkissoon SH, Ligon KL, Rameshwar P, Delivery of Functional Anti-miR-9 by Mesenchymal Stem Cell-derived Exosomes to Glioblastoma Multiforme Cells Conferred Chemosensitivity, Mol Ther Nucleic Acids 2(10) (2013) e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Cossetti C, Lugini L, Astrologo L, Saggio I, Fais S, Spadafora C, Soma-to-germline transmission of RNA in mice xenografted with human tumour cells: possible transport by exosomes, PLoS One 9(7) (2014) e101629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Sun Y, Liu J, Potential of cancer cell-derived exosomes in clinical application: a review of recent research advances, Clin Ther 36(6) (2014) 863–72. [DOI] [PubMed] [Google Scholar]
- [86].Qiao L, Hu S, Huang K, Su T, Li Z, Vandergriff A, Cores J, Dinh PU, Allen T, Shen D, Liang H, Li Y, Cheng K, Tumor cell-derived exosomes home to their cells of origin and can be used as Trojan horses to deliver cancer drugs, Theranostics 10(8) (2020) 3474–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Morishita M, Takahashi Y, Matsumoto A, Nishikawa M, Takakura Y, Exosome-based tumor antigens-adjuvant co-delivery utilizing genetically engineered tumor cell-derived exosomes with immunostimulatory CpG DNA, Biomaterials 111 (2016) 55–65. [DOI] [PubMed] [Google Scholar]
- [88].Rong L, Li R, Li S, Luo R, Immunosuppression of breast cancer cells mediated by transforming growth factor-beta in exosomes from cancer cells, Oncol Lett 11(1) (2016) 500–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Ha D, Yang N, Nadithe V, Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges, Acta Pharm Sin B 6(4) (2016) 287–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Sun D, Zhuang X, Xiang X, Liu Y, Zhang S, Liu C, Barnes S, Grizzle W, Miller D, Zhang HG, A novel nanoparticle drug delivery system: the anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes, Mol Ther 18(9) (2010) 1606–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, Fujita K, Mizutani T, Ohgi T, Ochiya T, Gotoh N, Kuroda M, Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells, Mol Ther 21(1) (2013) 185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Limoni SK, Moghadam MF, Moazzeni SM, Gomari H, Salimi F, Engineered Exosomes for Targeted Transfer of siRNA to HER2 Positive Breast Cancer Cells, Appl Biochem Biotechnol 187(1) (2019) 352–364. [DOI] [PubMed] [Google Scholar]
- [93].Schindler C, Collinson A, Matthews C, Pointon A, Jenkinson L, Minter RR, Vaughan TJ, Tigue NJ, Exosomal delivery of doxorubicin enables rapid cell entry and enhanced in vitro potency, PLoS One 14(3) (2019) e0214545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Thippabhotla S, Zhong C, He M, 3D cell culture stimulates the secretion of in vivo like extracellular vesicles, Sci Rep 9(1) (2019) 13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Abdollahi S, Extracellular vesicles from organoids and 3D culture systems, Biotechnol Bioeng 118(3) (2021) 1029–1049. [DOI] [PubMed] [Google Scholar]
- [96].Yu L, Deng Z, Liu L, Zhang W, Wang C, Plant-Derived Nanovesicles: A Novel Form of Nanomedicine, Front Bioeng Biotechnol 8 (2020) 584391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Zhang M, Xiao B, Wang H, Han MK, Zhang Z, Viennois E, Xu C, Merlin D, Edible Ginger-derived Nano-lipids Loaded with Doxorubicin as a Novel Drug-delivery Approach for Colon Cancer Therapy, Mol Ther 24(10) (2016) 1783–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Yang C, Zhang M, Merlin D, Advances in Plant-derived Edible Nanoparticle-based lipid Nano-drug Delivery Systems as Therapeutic Nanomedicines, J Mater Chem B 6(9) (2018) 1312–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Li X, Liang Z, Du J, Wang Z, Mei S, Li Z, Zhao Y, Zhao D, Ma Y, Ye J, Xu J, Zhao Y, Chang J, Qin Y, Yu L, Wang C, Jiang C, Herbal decoctosome is a novel form of medicine, Sci China Life Sci 62(3) (2019) 333–348. [DOI] [PubMed] [Google Scholar]
- [100].Garaeva L, Kamyshinsky R, Kil Y, Varfolomeeva E, Verlov N, Komarova E, Garmay Y, Landa S, Burdakov V, Myasnikov A, Vinnikov IA, Margulis B, Guzhova I, Kagansky A, Konevega AL, Shtam T, Delivery of functional exogenous proteins by plant-derived vesicles to human cells in vitro, Sci Rep 11(1) (2021) 6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Zhang M, Viennois E, Xu C, Merlin D, Plant derived edible nanoparticles as a new therapeutic approach against diseases, Tissue Barriers 4(2) (2016) e1134415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Raimondo S, Naselli F, Fontana S, Monteleone F, Lo Dico A, Saieva L, Zito G, Flugy A, Manno M, Di Bella MA, De Leo G, Alessandro R, Citrus limon-derived nanovesicles inhibit cancer cell proliferation and suppress CML xenograft growth by inducing TRAIL-mediated cell death, Oncotarget 6(23) (2015) 19514–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Cao M, Yan H, Han X, Weng L, Wei Q, Sun X, Lu W, Wei Q, Ye J, Cai X, Hu C, Yin X, Cao P, Ginseng-derived nanoparticles alter macrophage polarization to inhibit melanoma growth, J Immunother Cancer 7(1) (2019) 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Kuruvinashetti K, Pakkiriswami S, Packirisamy M, Algal Extracellular Vesicles for Therapeutic Applications, 2020 IEEE 20th International Conference on Nanotechnology (IEEE-NANO), 2020, pp. 354–357. [Google Scholar]
- [105].Deatherage BL, Cookson BT, Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life, Infect Immun 80(6) (2012) 1948–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kuipers ME, Hokke CH, Smits HH, Nolte-’t Hoen ENM, Pathogen-Derived Extracellular Vesicle-Associated Molecules That Affect the Host Immune System: An Overview, Front Microbiol 9 (2018) 2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Brown L, Wolf JM, Prados-Rosales R, Casadevall A, Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi, Nat Rev Microbiol 13(10) (2015) 620–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Yu YJ, Wang XH, Fan GC, Versatile effects of bacterium-released membrane vesicles on mammalian cells and infectious/inflammatory diseases, Acta Pharmacol Sin 39(4) (2018) 514–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].de Souza W, Barrias ES, Membrane-bound extracellular vesicles secreted by parasitic protozoa: cellular structures involved in the communication between cells, Parasitol Res 119(7) (2020) 2005–2023. [DOI] [PubMed] [Google Scholar]
- [110].Szempruch AJ, Dennison L, Kieft R, Harrington JM, Hajduk SL, Sending a message: extracellular vesicles of pathogenic protozoan parasites, Nat Rev Microbiol 14(11) (2016) 669–675. [DOI] [PubMed] [Google Scholar]
- [111].Rizzo J, Rodrigues ML, Janbon G, Extracellular Vesicles in Fungi: Past, Present, and Future Perspectives, Front Cell Infect Microbiol 10 (2020) 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Samuel M, Bleackley M, Anderson M, Mathivanan S, Extracellular vesicles including exosomes in cross kingdom regulation: a viewpoint from plant-fungal interactions, Front Plant Sci 6 (2015) 766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Melnik BC, John SM, Schmitz G, Milk is not just food but most likely a genetic transfection system activating mTORC1 signaling for postnatal growth, Nutr J 12 (2013) 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Samuel M, Chisanga D, Liem M, Keerthikumar S, Anand S, Ang CS, Adda CG, Versteegen E, Jois M, Mathivanan S, Bovine milk-derived exosomes from colostrum are enriched with proteins implicated in immune response and growth, Sci Rep 7(1) (2017) 5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Sanwlani R, Fonseka P, Chitti SV, Mathivanan S, Milk-Derived Extracellular Vesicles in Inter-Organism, Cross-Species Communication and Drug Delivery, Proteomes 8(2) (2020) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Betker JL, Angle BM, Graner MW, Anchordoquy TJ, The Potential of Exosomes From Cow Milk for Oral Delivery, J Pharm Sci 108(4) (2019) 1496–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Munagala R, Aqil F, Jeyabalan J, Gupta RC, Bovine milk-derived exosomes for drug delivery, Cancer Lett 371(1) (2016) 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Zhang Q, Xiao Q, Yin H, Xia C, Pu Y, He Z, Hu Q, Wang J, Wang Y, Milk-exosome based pH/light sensitive drug system to enhance anticancer activity against oral squamous cell carcinoma, RSC Advances 10(47) (2020) 28314–28323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Rahman MM, Shimizu K, Yamauchi M, Takase H, Ugawa S, Okada A, Inoshima Y, Acidification effects on isolation of extracellular vesicles from bovine milk, PLoS One 14(9) (2019) e0222613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Warren MR, Zhang C, Vedadghavami A, Bokvist K, Dhal PK, Bajpayee AG, Milk exosomes with enhanced mucus penetrability for oral delivery of siRNA, Biomater Sci 9(12) (2021) 4260–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Shandilya S, Rani P, Onteru SK, Singh D, Natural ligand-receptor mediated loading of siRNA in milk derived exosomes, J Biotechnol 318 (2020) 1–9. [DOI] [PubMed] [Google Scholar]
- [122].Aqil F, Munagala R, Jeyabalan J, Agrawal AK, Kyakulaga AH, Wilcher SA, Gupta RC, Milk exosomes - Natural nanoparticles for siRNA delivery, Cancer Lett 449 (2019) 186–195. [DOI] [PubMed] [Google Scholar]
- [123].Del Pozo-Acebo L, Hazas MLL, Tome-Carneiro J, Gil-Cabrerizo P, San-Cristobal R, Busto R, Garcia-Ruiz A, Davalos A, Bovine Milk-Derived Exosomes as a Drug Delivery Vehicle for miRNA-Based Therapy, Int J Mol Sci 22(3) (2021) 1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Manca S, Upadhyaya B, Mutai E, Desaulniers AT, Cederberg RA, White BR, Zempleni J, Milk exosomes are bioavailable and distinct microRNA cargos have unique tissue distribution patterns, Sci Rep 8(1) (2018) 11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Somiya M, Yoshioka Y, Ochiya T, Biocompatibility of highly purified bovine milk-derived extracellular vesicles, J Extracell Vesicles 7(1) (2018) 1440132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Kleinjan M, van Herwijnen MJ, Libregts SF, van Neerven RJ, Feitsma AL, Wauben MH, Regular Industrial Processing of Bovine Milk Impacts the Integrity and Molecular Composition of Extracellular Vesicles, J Nutr 151(6) (2021) 1416–1425. [DOI] [PubMed] [Google Scholar]
- [127].Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C, Exosomal-like vesicles are present in human blood plasma, Int Immunol 17(7) (2005) 879–87. [DOI] [PubMed] [Google Scholar]
- [128].Wahlgren J, De LKT, Brisslert M, Vaziri Sani F, Telemo E, Sunnerhagen P, Valadi H, Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes, Nucleic Acids Res 40(17) (2012) e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Jakubec M, Maple-Grodem J, Akbari S, Nesse S, Halskau O, Mork-Jansson AE, Plasma-derived exosome-like vesicles are enriched in lyso-phospholipids and pass the blood-brain barrier, PLoS One 15(9) (2020) e0232442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Logozzi M, Angelini DF, Iessi E, Mizzoni D, Di Raimo R, Federici C, Lugini L, Borsellino G, Gentilucci A, Pierella F, Marzio V, Sciarra A, Battistini L, Fais S, Increased PSA expression on prostate cancer exosomes in in vitro condition and in cancer patients, Cancer Lett 403 (2017) 318–329. [DOI] [PubMed] [Google Scholar]
- [131].Logozzi M, Capasso C, Di Raimo R, Del Prete S, Mizzoni D, Falchi M, Supuran CT, Fais S, Prostate cancer cells and exosomes in acidic condition show increased carbonic anhydrase IX expression and activity, J Enzyme Inhib Med Chem 34(1) (2019) 272–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Logozzi M, Mizzoni D, Capasso C, Del Prete S, Di Raimo R, Falchi M, Angelini DF, Sciarra A, Maggi M, Supuran CT, Fais S, Plasmatic exosomes from prostate cancer patients show increased carbonic anhydrase IX expression and activity and low pH, J Enzyme Inhib Med Chem 35(1) (2020) 280–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Logozzi M, Angelini DF, Giuliani A, Mizzoni D, Di Raimo R, Maggi M, Gentilucci A, Marzio V, Salciccia S, Borsellino G, Battistini L, Sciarra A, Fais S, Increased Plasmatic Levels of PSA-Expressing Exosomes Distinguish Prostate Cancer Patients from Benign Prostatic Hyperplasia: A Prospective Study, Cancers (Basel) 11(10) (2019) 1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Logozzi M, De Milito A, Lugini L, Borghi M, Calabro L, Spada M, Perdicchio M, Marino ML, Federici C, Iessi E, Brambilla D, Venturi G, Lozupone F, Santinami M, Huber V, Maio M, Rivoltini L, Fais S, High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients, PLoS One 4(4) (2009) e5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Osti D, Del Bene M, Rappa G, Santos M, Matafora V, Richichi C, Faletti S, Beznoussenko GV, Mironov A, Bachi A, Fornasari L, Bongetta D, Gaetani P, DiMeco F, Lorico A, Pelicci G, Clinical Significance of Extracellular Vesicles in Plasma from Glioblastoma Patients, Clin Cancer Res 25(1) (2019) 266–276. [DOI] [PubMed] [Google Scholar]
- [136].Aslan C, Kiaie SH, Zolbanin NM, Lotfinejad P, Ramezani R, Kashanchi F, Jafari R, Exosomes for mRNA delivery: a novel biotherapeutic strategy with hurdles and hope, BMC Biotechnol 21(1) (2021) 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Usman WM, Pham TC, Kwok YY, Vu LT, Ma V, Peng B, Chan YS, Wei L, Chin SM, Azad A, He AB, Leung AYH, Yang M, Shyh-Chang N, Cho WC, Shi J, Le MTN, Efficient RNA drug delivery using red blood cell extracellular vesicles, Nat Commun 9(1) (2018) 2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Franzen CA, Blackwell RH, Foreman KE, Kuo PC, Flanigan RC, Gupta GN, Urinary Exosomes: The Potential for Biomarker Utility, Intercellular Signaling and Therapeutics in Urological Malignancy, J Urol 195(5) (2016) 1331–1339. [DOI] [PubMed] [Google Scholar]
- [139].Lourenco C, Constancio V, Henrique R, Carvalho A, Jeronimo C, Urinary Extracellular Vesicles as Potential Biomarkers for Urologic Cancers: An Overview of Current Methods and Advances, Cancers (Basel) 13(7) (2021) 1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Erozenci LA, Bottger F, Bijnsdorp IV, Jimenez CR, Urinary exosomal proteins as (pan-)cancer biomarkers: insights from the proteome, FEBS Lett 593(13) (2019) 1580–1597. [DOI] [PubMed] [Google Scholar]
- [141].Kang J-S, The potential of exosomes as theragnostics in various clinical situations, Exosomes (2020) 467–486. [Google Scholar]
- [142].Panfoli I, Cancer exosomes in urine: a promising biomarker source, Translational Cancer Research (2017) S1389–S1393. [Google Scholar]
- [143].Armstrong DA, Green BB, Seigne JD, Schned AR, Marsit CJ, MicroRNA molecular profiling from matched tumor and bio-fluids in bladder cancer, Mol Cancer 14 (2015) 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Chen CK, Liao J, Li MS, Khoo BL, Urine biopsy technologies: Cancer and beyond, Theranostics 10(17) (2020) 7872–7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Puhka M, Takatalo M, Nordberg ME, Valkonen S, Nandania J, Aatonen M, Yliperttula M, Laitinen S, Velagapudi V, Mirtti T, Kallioniemi O, Rannikko A, Siljander PR, Af Hallstrom TM, Metabolomic Profiling of Extracellular Vesicles and Alternative Normalization Methods Reveal Enriched Metabolites and Strategies to Study Prostate Cancer-Related Changes, Theranostics 7(16) (2017) 3824–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Cheng J, Nonaka T, Wong DTW, Salivary Exosomes as Nanocarriers for Cancer Biomarker Delivery, Materials (Basel) 12(4) (2019) 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Nair S, Tang KD, Kenny L, Punyadeera C, Salivary exosomes as potential biomarkers in cancer, Oral Oncol 84 (2018) 31–40. [DOI] [PubMed] [Google Scholar]
- [148].Zhan C, Yang X, Yin X, Hou J, Exosomes and other extracellular vesicles in oral and salivary gland cancers, Oral Dis 26(5) (2020) 865–875. [DOI] [PubMed] [Google Scholar]
- [149].Ogawa Y, Kanai-Azuma M, Akimoto Y, Kawakami H, Yanoshita R, Exosome-like vesicles with dipeptidyl peptidase IV in human saliva, Biol Pharm Bull 31(6) (2008) 1059–62. [DOI] [PubMed] [Google Scholar]
- [150].He L, Ping F, Fan Z, Zhang C, Deng M, Cheng B, Xia J, Salivary exosomal miR-24-3p serves as a potential detective biomarker for oral squamous cell carcinoma screening, Biomed Pharmacother 121 (2020) 109553. [DOI] [PubMed] [Google Scholar]
- [151].Zlotogorski-Hurvitz A, Dekel BZ, Malonek D, Yahalom R, Vered M, FTIR-based spectrum of salivary exosomes coupled with computational-aided discriminating analysis in the diagnosis of oral cancer, J Cancer Res Clin Oncol 145(3) (2019) 685–694. [DOI] [PubMed] [Google Scholar]
- [152].Nonaka T, Wong DTW, Saliva-Exosomics in Cancer: Molecular Characterization of Cancer-Derived Exosomes in Saliva, Enzymes 42 (2017) 125–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Kim MS, Haney MJ, Zhao Y, Mahajan V, Deygen I, Klyachko NL, Inskoe E, Piroyan A, Sokolsky M, Okolie O, Hingtgen SD, Kabanov AV, Batrakova EV, Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells, Nanomedicine 12(3) (2016) 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Haney MJ, Zhao Y, Jin YS, Li SM, Bago JR, Klyachko NL, Kabanov AV, Batrakova EV, Macrophage-Derived Extracellular Vesicles as Drug Delivery Systems for Triple Negative Breast Cancer (TNBC) Therapy, J Neuroimmune Pharmacol 15(3) (2020) 487–500. [DOI] [PubMed] [Google Scholar]
- [155].Li YJ, Wu JY, Wang JM, Hu XB, Cai JX, Xiang DX, Gemcitabine loaded autologous exosomes for effective and safe chemotherapy of pancreatic cancer, Acta Biomater 101 (2020) 519–530. [DOI] [PubMed] [Google Scholar]
- [156].Pomatto MAC, Bussolati B, D’Antico S, Ghiotto S, Tetta C, Brizzi MF, Camussi G, Improved Loading of Plasma-Derived Extracellular Vesicles to Encapsulate Antitumor miRNAs, Mol Ther Methods Clin Dev 13 (2019) 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Liang G, Zhu Y, Ali DJ, Tian T, Xu H, Si K, Sun B, Chen B, Xiao Z, Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer, J Nanobiotechnology 18(1) (2020) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Li X, Corbett AL, Taatizadeh E, Tasnim N, Little JP, Garnis C, Daugaard M, Guns E, Hoorfar M, Li ITS, Challenges and opportunities in exosome research-Perspectives from biology, engineering, and cancer therapy, APL Bioeng 3(1) (2019) 011503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Gangadaran P, Ahn BC, Extracellular Vesicle- and Extracellular Vesicle Mimetics-Based Drug Delivery Systems: New Perspectives, Challenges, and Clinical Developments, Pharmaceutics 12(5) (2020) 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Sato YT, Umezaki K, Sawada S, Mukai SA, Sasaki Y, Harada N, Shiku H, Akiyoshi K, Engineering hybrid exosomes by membrane fusion with liposomes, Sci Rep 6 (2016) 21933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Luan X, Sansanaphongpricha K, Myers I, Chen H, Yuan H, Sun D, Engineering exosomes as refined biological nanoplatforms for drug delivery, Acta Pharmacol Sin 38(6) (2017) 754–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Smyth T, Petrova K, Payton NM, Persaud I, Redzic JS, Graner MW, Smith-Jones P, Anchordoquy TJ, Surface functionalization of exosomes using click chemistry, Bioconjug Chem 25(10) (2014) 1777–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Wang Y, Yao J, Cai L, Liu T, Wang X, Zhang Y, Zhou Z, Li T, Liu M, Lai R, Liu X, Bone-Targeted Extracellular Vesicles from Mesenchymal Stem Cells for Osteoporosis Therapy, Int J Nanomedicine 15 (2020) 7967–7977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Bryniarski K, Ptak W, Martin E, Nazimek K, Szczepanik M, Sanak M, Askenase PW, Free Extracellular miRNA Functionally Targets Cells by Transfecting Exosomes from Their Companion Cells, PLoS One 10(4) (2015) e0122991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Gong C, Tian J, Wang Z, Gao Y, Wu X, Ding X, Qiang L, Li G, Han Z, Yuan Y, Gao S, Functional exosome-mediated co-delivery of doxorubicin and hydrophobically modified microRNA 159 for triple-negative breast cancer therapy, J Nanobiotechnology 17(1) (2019) 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Kooijmans SAA, Schiffelers RM, Zarovni N, Vago R, Modulation of tissue tropism and biological activity of exosomes and other extracellular vesicles: New nanotools for cancer treatment, Pharmacol Res 111 (2016) 487–500. [DOI] [PubMed] [Google Scholar]
- [167].Cho JA, Yeo DJ, Son HY, Kim HW, Jung DS, Ko JK, Koh JS, Kim YN, Kim CW, Exosomes: a new delivery system for tumor antigens in cancer immunotherapy, Int J Cancer 114(4) (2005) 613–22. [DOI] [PubMed] [Google Scholar]
- [168].Li Z, Zhou X, Wei M, Gao X, Zhao L, Shi R, Sun W, Duan Y, Yang G, Yuan L, In Vitro and in Vivo RNA Inhibition by CD9-HuR Functionalized Exosomes Encapsulated with miRNA or CRISPR/dCas9, Nano Lett 19(1) (2019) 19–28. [DOI] [PubMed] [Google Scholar]
- [169].Lin Y, Wu J, Gu W, Huang Y, Tong Z, Huang L, Tan J, Exosome-Liposome Hybrid Nanoparticles Deliver CRISPR/Cas9 System in MSCs, Adv Sci (Weinh) 5(4) (2018) 1700611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Livshits MA, Khomyakova E, Evtushenko EG, Lazarev VN, Kulemin NA, Semina SE, Generozov EV, Govorun VM, Isolation of exosomes by differential centrifugation: Theoretical analysis of a commonly used protocol, Sci Rep 5 (2015) 17319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Merchant ML, Rood IM, Deegens JKJ, Klein JB, Isolation and characterization of urinary extracellular vesicles: implications for biomarker discovery, Nat Rev Nephrol 13(12) (2017) 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Baranyai T, Herczeg K, Onodi Z, Voszka I, Modos K, Marton N, Nagy G, Mager I, Wood MJ, El Andaloussi S, Palinkas Z, Kumar V, Nagy P, Kittel A, Buzas EI, Ferdinandy P, Giricz Z, Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods, PLoS One 10(12) (2015) e0145686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Nordin JZ, Lee Y, Vader P, Mager I, Johansson HJ, Heusermann W, Wiklander OP, Hallbrink M, Seow Y, Bultema JJ, Gilthorpe J, Davies T, Fairchild PJ, Gabrielsson S, Meisner-Kober NC, Lehtio J, Smith CI, Wood MJ, El Andaloussi S, Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties, Nanomedicine 11(4) (2015) 879–83. [DOI] [PubMed] [Google Scholar]
- [174].Li K, Wong DK, Hong KY, Raffai RL, Cushioned-Density Gradient Ultracentrifugation (C-DGUC): A Refined and High Performance Method for the Isolation, Characterization, and Use of Exosomes, Methods Mol Biol 1740 (2018) 69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Gupta S, Rawat S, Arora V, Kottarath SK, Dinda AK, Vaishnav PK, Nayak B, Mohanty S, An improvised one-step sucrose cushion ultracentrifugation method for exosome isolation from culture supernatants of mesenchymal stem cells, Stem Cell Res Ther 9(1) (2018) 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Rider MA, Hurwitz SN, Meckes DG Jr., ExtraPEG: A Polyethylene Glycol-Based Method for Enrichment of Extracellular Vesicles, Sci Rep 6 (2016) 23978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Garcia-Romero N, Madurga R, Rackov G, Palacin-Aliana I, Nunez-Torres R, Asensi-Puig A, Carrion-Navarro J, Esteban-Rubio S, Peinado H, Gonzalez-Neira A, Gonzalez-Rumayor V, Belda-Iniesta C, Ayuso-Sacido A, Polyethylene glycol improves current methods for circulating extracellular vesicle-derived DNA isolation, J Transl Med 17(1) (2019) 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Akbar A, Malekian F, Baghban N, Kodam SP, Ullah M, Methodologies to Isolate and Purify Clinical Grade Extracellular Vesicles for Medical Applications, Cells 11(2) (2022) 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Hong P, Koza S, Bouvier ES, Size-Exclusion Chromatography for the Analysis of Protein Biotherapeutics and their Aggregates, J Liq Chromatogr Relat Technol 35(20) (2012) 2923–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Robertson JD, Rizzello L, Avila-Olias M, Gaitzsch J, Contini C, Magon MS, Renshaw SA, Battaglia G, Purification of Nanoparticles by Size and Shape, Sci Rep 6 (2016) 27494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Chen J, Li P, Zhang T, Xu Z, Huang X, Wang R, Du L, Review on Strategies and Technologies for Exosome Isolation and Purification, Front Bioeng Biotechnol 9 (2021) 811971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Haraszti RA, Miller R, Stoppato M, Sere YY, Coles A, Didiot MC, Wollacott R, Sapp E, Dubuke ML, Li X, Shaffer SA, DiFiglia M, Wang Y, Aronin N, Khvorova A, Exosomes Produced from 3D Cultures of MSCs by Tangential Flow Filtration Show Higher Yield and Improved Activity, Mol Ther 26(12) (2018) 2838–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Watson DC, Yung BC, Bergamaschi C, Chowdhury B, Bear J, Stellas D, Morales-Kastresana A, Jones JC, Felber BK, Chen X, Pavlakis GN, Scalable, cGMP-compatible purification of extracellular vesicles carrying bioactive human heterodimeric IL-15/lactadherin complexes, J Extracell Vesicles 7(1) (2018) 1442088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Nakai W, Yoshida T, Diez D, Miyatake Y, Nishibu T, Imawaka N, Naruse K, Sadamura Y, Hanayama R, A novel affinity-based method for the isolation of highly purified extracellular vesicles, Sci Rep 6 (2016) 33935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Burkova EE, Dmitrenok PS, Bulgakov DV, Vlassov VV, Ryabchikova EI, Nevinsky GA, Exosomes from human placenta purified by affinity chromatography on sepharose bearing immobilized antibodies against CD81 tetraspanin contain many peptides and small proteins, IUBMB Life 70(11) (2018) 1144–1155. [DOI] [PubMed] [Google Scholar]
- [186].Duijvesz D, Versluis CY, van der Fels CA, Vredenbregt-van den Berg MS, Leivo J, Peltola MT, Bangma CH, Pettersson KS, Jenster G, Immuno-based detection of extracellular vesicles in urine as diagnostic marker for prostate cancer, Int J Cancer 137(12) (2015) 2869–78. [DOI] [PubMed] [Google Scholar]
- [187].Jauregui R, Srinivasan S, Vojtech LN, Gammill HS, Chiu DT, Hladik F, Stayton PS, Lai JJ, Temperature-Responsive Magnetic Nanoparticles for Enabling Affinity Separation of Extracellular Vesicles, ACS Appl Mater Interfaces 10(40) (2018) 33847–33856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Contreras-Naranjo JC, Wu HJ, Ugaz VM, Microfluidics for exosome isolation and analysis: enabling liquid biopsy for personalized medicine, Lab Chip 17(21) (2017) 3558–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Zhao Z, Yang Y, Zeng Y, He M, A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis, Lab Chip 16(3) (2016) 489–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Jaiswal R, Sedger LM, Intercellular Vesicular Transfer by Exosomes, Microparticles and Oncosomes - Implications for Cancer Biology and Treatments, Front Oncol 9 (2019) 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Raposo G, Stoorvogel W, Extracellular vesicles: exosomes, microvesicles, and friends, J Cell Biol 200(4) (2013) 373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, Mark MT, Molina H, Martin AB, Bojmar L, Fang J, Rampersaud S, Hoshino A, Matei I, Kenific CM, Nakajima M, Mutvei AP, Sansone P, Buehring W, Wang H, Jimenez JP, Cohen-Gould L, Paknejad N, Brendel M, Manova-Todorova K, Magalhaes A, Ferreira JA, Osorio H, Silva AM, Massey A, Cubillos-Ruiz JR, Galletti G, Giannakakou P, Cuervo AM, Blenis J, Schwartz R, Brady MS, Peinado H, Bromberg J, Matsui H, Reis CA, Lyden D, Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation, Nat Cell Biol 20(3) (2018) 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Ma L, Li Y, Peng J, Wu D, Zhao X, Cui Y, Chen L, Yan X, Du Y, Yu L, Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration, Cell Res 25(1) (2015) 24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Li B, Antonyak MA, Zhang J, Cerione RA, RhoA triggers a specific signaling pathway that generates transforming microvesicles in cancer cells, Oncogene 31(45) (2012) 4740–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].van der Koog L, Gandek TB, Nagelkerke A, Liposomes and Extracellular Vesicles as Drug Delivery Systems: A Comparison of Composition, Pharmacokinetics, and Functionalization, Adv Healthc Mater (2021) e2100639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Kim MW, Kwon SH, Choi JH, Lee A, A Promising Biocompatible Platform: Lipid-Based and Bio-Inspired Smart Drug Delivery Systems for Cancer Therapy, Int J Mol Sci 19(12) (2018) 3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Yang M, Liu X, Luo Q, Xu L, Chen F, An efficient method to isolate lemon derived extracellular vesicles for gastric cancer therapy, J Nanobiotechnology 18(1) (2020) 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Zhuang X, Deng ZB, Mu J, Zhang L, Yan J, Miller D, Feng W, McClain CJ, Zhang HG, Ginger-derived nanoparticles protect against alcohol-induced liver damage, J Extracell Vesicles 4 (2015) 28713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Kim K, Yoo HJ, Jung JH, Lee R, Hyun JK, Park JH, Na D, Yeon JH, Cytotoxic Effects of Plant Sap-Derived Extracellular Vesicles on Various Tumor Cell Types, J Funct Biomater 11(2) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Rutter BD, Innes RW, Extracellular Vesicles Isolated from the Leaf Apoplast Carry Stress-Response Proteins, Plant Physiol 173(1) (2017) 728–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Wang B, Zhuang X, Deng ZB, Jiang H, Mu J, Wang Q, Xiang X, Guo H, Zhang L, Dryden G, Yan J, Miller D, Zhang HG, Targeted drug delivery to intestinal macrophages by bioactive nanovesicles released from grapefruit, Mol Ther 22(3) (2014) 522–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Wang Q, Zhuang X, Mu J, Deng ZB, Jiang H, Zhang L, Xiang X, Wang B, Yan J, Miller D, Zhang HG, Delivery of therapeutic agents by nanoparticles made of grapefruit-derived lipids, Nat Commun 4 (2013) 1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Liu Y, Wu S, Koo Y, Yang A, Dai Y, Khant H, Osman SR, Chowdhury M, Wei H, Li Y, Court K, Hwang E, Wen Y, Dasari SK, Nguyen M, Tang EC, Chehab EW, de Val N, Braam J, Sood AK, Characterization of and isolation methods for plant leaf nanovesicles and small extracellular vesicles, Nanomedicine 29 (2020) 102271. [DOI] [PubMed] [Google Scholar]
- [204].Deng Z, Rong Y, Teng Y, Mu J, Zhuang X, Tseng M, Samykutty A, Zhang L, Yan J, Miller D, Suttles J, Zhang HG, Broccoli-Derived Nanoparticle Inhibits Mouse Colitis by Activating Dendritic Cell AMP-Activated Protein Kinase, Mol Ther 25(7) (2017) 1641–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Jeppesen DK, Hvam ML, Primdahl-Bengtson B, Boysen AT, Whitehead B, Dyrskjot L, Orntoft TF, Howard KA, Ostenfeld MS, Comparative analysis of discrete exosome fractions obtained by differential centrifugation, J Extracell Vesicles 3 (2014) 25011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Boing AN, van der Pol E, Grootemaat AE, Coumans FA, Sturk A, Nieuwland R, Single-step isolation of extracellular vesicles by size-exclusion chromatography, J Extracell Vesicles 3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Lobb RJ, Becker M, Wen SW, Wong CS, Wiegmans AP, Leimgruber A, Moller A, Optimized exosome isolation protocol for cell culture supernatant and human plasma, J Extracell Vesicles 4 (2015) 27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Heinemann ML, Ilmer M, Silva LP, Hawke DH, Recio A, Vorontsova MA, Alt E, Vykoukal J, Benchtop isolation and characterization of functional exosomes by sequential filtration, J Chromatogr A 1371 (2014) 125–35. [DOI] [PubMed] [Google Scholar]
- [209].Musante L, Tataruch D, Gu D, Benito-Martin A, Calzaferri G, Aherne S, Holthofer H, A simplified method to recover urinary vesicles for clinical applications, and sample banking, Sci Rep 4 (2014) 7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Kang D, Oh S, Ahn SM, Lee BH, Moon MH, Proteomic analysis of exosomes from human neural stem cells by flow field-flow fractionation and nanoflow liquid chromatography-tandem mass spectrometry, J Proteome Res 7(8) (2008) 3475–80. [DOI] [PubMed] [Google Scholar]
- [211].Helwa I, Cai J, Drewry MD, Zimmerman A, Dinkins MB, Khaled ML, Seremwe M, Dismuke WM, Bieberich E, Stamer WD, Hamrick MW, Liu Y, A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents, PLoS One 12(1) (2017) e0170628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Samsonov R, Shtam T, Burdakov V, Glotov A, Tsyrlina E, Berstein L, Nosov A, Evtushenko V, Filatov M, Malek A, Lectin-induced agglutination method of urinary exosomes isolation followed by mi-RNA analysis: Application for prostate cancer diagnostic, Prostate 76(1) (2016) 68–79. [DOI] [PubMed] [Google Scholar]
- [213].Davies RT, Kim J, Jang SC, Choi EJ, Gho YS, Park J, Microfluidic filtration system to isolate extracellular vesicles from blood, Lab Chip 12(24) (2012) 5202–10. [DOI] [PubMed] [Google Scholar]
- [214].Lee K, Shao H, Weissleder R, Lee H, Acoustic purification of extracellular microvesicles, ACS Nano 9(3) (2015) 2321–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Cheng Y, Qu X, Dong Z, Zeng Q, Ma X, Jia Y, Li R, Jiang X, Williams C, Wang T, Xia W, Comparison of serum exosome isolation methods on co-precipitated free microRNAs, PeerJ 8 (2020) e9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Zeming KK, Salafi T, Shikha S, Zhang Y, Fluorescent label-free quantitative detection of nano-sized bioparticles using a pillar array, Nat Commun 9(1) (2018) 1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Liang LG, Sheng YF, Zhou S, Inci F, Li L, Demirci U, Wang S, An Integrated Double-Filtration Microfluidic Device for Detection of Extracellular Vesicles from Urine for Bladder Cancer Diagnosis, Methods Mol Biol 1660 (2017) 355–364. [DOI] [PubMed] [Google Scholar]
- [218].Akagi T, Kato K, Kobayashi M, Kosaka N, Ochiya T, Ichiki T, On-chip immunoelectrophoresis of extracellular vesicles released from human breast cancer cells, PLoS One 10(4) (2015) e0123603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Kim DK, Nishida H, An SY, Shetty AK, Bartosh TJ, Prockop DJ, Chromatographically isolated CD63+CD81+ extracellular vesicles from mesenchymal stromal cells rescue cognitive impairments after TBI, Proc Natl Acad Sci U S A 113(1) (2016) 170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]


