Abstract
The penicillin binding proteins (PBPs) synthesize and remodel peptidoglycan, the structural component of the bacterial cell wall. Much is known about the biochemistry of these proteins, but little is known about their biological roles. To better understand the contributions these proteins make to the physiology of Escherichia coli, we constructed 192 mutants from which eight PBP genes were deleted in every possible combination. The genes encoding PBPs 1a, 1b, 4, 5, 6, and 7, AmpC, and AmpH were cloned, and from each gene an internal coding sequence was removed and replaced with a kanamycin resistance cassette flanked by two res sites from plasmid RP4. Deletion of individual genes was accomplished by transferring each interrupted gene onto the chromosome of E. coli via λ phage transduction and selecting for kanamycin-resistant recombinants. Afterwards, the kanamycin resistance cassette was removed from each mutant strain by supplying ParA resolvase in trans, yielding a strain in which a long segment of the original PBP gene was deleted and replaced by an 8-bp res site. These kanamycin-sensitive mutants were used as recipients in further rounds of replacement mutagenesis, resulting in a set of strains lacking from one to seven PBPs. In addition, the dacD gene was deleted from two septuple mutants, creating strains lacking eight genes. The only deletion combinations not produced were those lacking both PBPs 1a and 1b because such a combination is lethal. Surprisingly, all other deletion mutants were viable even though, at the extreme, 8 of the 12 known PBPs had been eliminated. Furthermore, when both PBPs 2 and 3 were inactivated by the β-lactams mecillinam and aztreonam, respectively, several mutants did not lyse but continued to grow as enlarged spheres, so that one mutant synthesized osmotically resistant peptidoglycan when only 2 of 12 PBPs (PBPs 1b and 1c) remained active. These results have important implications for current models of peptidoglycan biosynthesis, for understanding the evolution of the bacterial sacculus, and for interpreting results derived by mutating unknown open reading frames in genome projects. In addition, members of the set of PBP mutants will provide excellent starting points for answering fundamental questions about other aspects of cell wall metabolism.
Peptidoglycan is a macromolecule of interlinked glycan chains and peptide bridges which forms the rigid structural component of the eubacterial cell wall, giving cells osmotic stability and imparting to them their shapes. Many of the final periplasmic steps in the synthesis and maturation of peptidoglycan are performed by the penicillin binding proteins (PBPs), enzymes to which β-lactam antibiotics bind covalently. The seven classic PBPs of Escherichia coli were first observed in polyacrylamide gels by Spratt (33, 34), who named the proteins in order of their decreasing molecular mass: PBPs 1a, 1b, 2, 3, 4, 5, and 6. Recently, five additional PBPs have been identified and authenticated by genetic and biochemical means, including PBP 7 and its proteolytic artifact PBP 8 (11, 12), DacD (1), AmpC and AmpH (13), and PBP 1c (29), thus bringing to 12 the number of PBPs in E. coli.
The physiological importance of four PBPs has been established by examining temperature-sensitive mutants or by inactivating individual proteins by specific β-lactam antibiotics. E. coli survives the loss of either PBP 1a or 1b, but inactivation of both proteins results in cell lysis (16, 37, 46). Because each of these proteins is a transglycosylase and transpeptidase, the implication is that these PBPs are required for initiating or continuing the elongation and cross-linking of glycan chains. PBP 2 helps govern cell shape; inactivating this protein causes cells to lose their rod shape and grow as enlarged spheres that eventually die unless compensatory mutations or conditions exist (26, 33, 35). Loss of PBP 3 inhibits cell septation, causing cells to grow as elongated filaments and establishing a central role for PBP 3 in this step of cell division (3, 33).
Seven low-molecular-weight PBPs are now known: PBPs 4, 5, 6, and 7, as well as DacD, AmpC, and AmpH. (Although AmpC and AmpH belong to the family of class C β-lactamases, we will also refer to these two proteins as PBPs because both enzymes bind covalently to at least one radioactively labeled β-lactam, which is the classical definition of a PBP [13].) Although the in vitro biochemical capabilities of these PBPs have been established and studied to various extents, the in vivo functions of these proteins remain mysterious. Any one of these PBPs can be deleted from the chromosome and E. coli will grow normally (1, 4, 9, 12, 13, 23, 24, 32), so that such PBPs are generally described as nonessential. However, many of these proteins have similar or identical enzymatic activities, making it possible that the loss of any one PBP could be masked by the presence of a surrogate PBP. For example, PBPs 4, 5, and 6 and DacD are carboxypeptidases, and PBPs 4 and 7 are endopeptidases (14, 27). Thus, within a particular enzymatic category, one PBP might substitute for the absence of another. If this were the case, then deletion of an individual PBP would not be lethal even though the biochemical activity that it represented might be essential for bacterial survival.
The question of whether several PBPs can substitute for one another can be addressed by creating E. coli strains in which multiple PBP genes have been mutated or deleted. However, only few such strains have been constructed. One of the earliest reports by Suzuki et al. (37) described five multiple mutants that contained combinations of temperature-sensitive (ts) and inactivating mutations (mutated PBP genes are listed in brackets): a double mutant [4 5]; three triple mutants [1a(ts) 4 5], [1b(ts) 4 5], and [3(ts) 4 5]; and one quadruple mutant [1a(ts) 1b(ts) 4 5]. Later, this same laboratory constructed three additional multiple mutants: [1a(ts) 3(ts) 4 5]; [1b(ts) 3(ts) 4 5]; and [1a(ts) 1b(ts) 4 5] (40). All the triple and quadruple mutants grew normally at 30°C but died, elongated, or lysed at 42°C, depending on which combination of the three high-molecular-weight PBPs was inactivated. In contrast, the mutant lacking PBPs 4 and 5 was viable at any temperature (37). Other laboratories have also constructed a few multiple mutants: a double mutant lacking PBPs 5 and 6 is viable (4), as is a triple mutant lacking PBPs 4, 5, and 6 (9) and a quadruple mutant lacking PBPs 4, 5, and 6 and DacD (1). In none of these cases was an obvious growth or morphological phenotype associated with the loss of the low-molecular-weight PBPs 4, 5, and 6 and DacD, either individually or in combination.
We have chosen to address the questions of PBP function and substitution in the most direct manner possible, by constructing a set of 192 E. coli strains containing every possible combination of deletions of the following eight PBPs: PBPs 1a, 1b, 4, 5, 6, and 7, AmpC, and AmpH. The results confirm that from among these proteins, PBPs 2 and 3 plus either PBP 1a or 1b are the only PBPs required for laboratory growth of regularly dividing rod-shaped cells. In addition, several mutants did not lyse when PBPs 2 and 3 were inhibited by specific β-lactam antibiotics, suggesting that the loss of some combination of low-molecular-weight PBPs produces this new phenotype. Finally, this set of E. coli strains supplies genetic backgrounds in which the physiological roles of nonessential PBPs and other peptidoglycan-reactive enzymes may be observed and investigated in greater detail.
MATERIALS AND METHODS
Bacteria, plasmids, phage, media, genetic techniques, and gel electrophoresis.
Individual PBP genes were deleted initially from E. coli JC9387 [thr-1 ara-14 leuB6 Δ(gpt-proA)62 lacY1 sbcC201 tsx-33 qsr galK2 rac sbcB15 hisG4(Oc) rfbD1 recB21 recC22 rpsL31(Sm) kdgK51 xylA5 mtl-1 argE3(Oc) thi-1) (strain 6613 from the E. coli Genetic Stock Center, Yale University). The parent from which individual and multiple PBP genes were deleted was E. coli CS109 (W1485 glnV rpoS rph) (from C. Schnaitman). E. coli S17-1λpir (recA thi pro hsdR [res− mod+][RP4::2-Tc::Mu-Km::Tn7] λpir phage lysogen) (20) was the donor for introducing parA-containing plasmids to mutated recipients. Plasmids pJMSB8 (containing a cloned parA gene) and pCK155 (the source of the res-npt-res kanamycin resistance cassette) were obtained from Claus Sternberg (20). The plasmid vector pBCSK− was from Stratagene (La Jolla, Calif.). Bacteriophages λ116, λ142, λ168, λ209, λ349, λ364, λ521, λ522, λ622, and λ650 were from the Kohara E. coli genomic library (18); all were supplied to us by Y. Kohara except for corrected versions of phages λ521 and λ522, provided by F. Blattner. Phage P1 was from laboratory stock, and P1 transductions were performed as described elsewhere (25).
Classical methods were followed for restriction enzyme digestion, ligation, and general cloning procedures (28), and restriction enzymes were from a variety of commercial sources. Southern blot hybridizations were performed as described elsewhere (31). Labeling, separation, and visualization of PBPs by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were performed as described previously (11). Bacteria were grown in Luria-Bertani (LB) medium or in M9-minimal glucose medium (25) containing the appropriate antibiotics: ampicillin (100 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (50 μg/ml). Ampicillin was never used in growth media when PBPs were to be assayed. Aztreonam was from E. R. Squibb & Sons (Princeton, N.J.), and mecillinam was from Leo Laboratories Ltd. (Dublin, Ireland). Each of the latter antibiotics was added to log-phase cultures of E. coli to a final concentration of 10 μg/ml. Yeast extract and tryptone were from Difco (Detroit, Mich.). Other chemicals were from Sigma Chemical Co. (St. Louis, Mo.).
Construction of PBP deletion mutants.
For each PBP, with the exception of PBPs 5 and 6, a DNA fragment containing the gene of interest was isolated from the appropriate λ phage in the Kohara genomic library and subcloned into the vector pBCSK− (Fig. 1). The plasmids were transformed into E. coli CS109, and overexpression of the PBP encoded by each subclone was confirmed by SDS-PAGE and autoradiography of 125I-penicillin X-labeled PBPs. A DNA fragment internal to each gene was removed by cutting the plasmid with selected restriction enzymes, the ends of the plasmid were blunted by incubation with DNA polymerase I Klenow fragment, HindIII linker oligonucleotides were ligated to each end, and the ends were cut with HindIII and ligated to one another to create a cloned sequence containing a deletion in the PBP gene (Fig. 1). In some cases, HindIII and other enzyme recognition sites were removed from plasmids prior to construction of the deletions (not shown). In all cases, enough flanking DNA was present for recombination to occur between the plasmid and chromosomal copies of the genes (described below). Loss of PBP expression from the derived plasmids was confirmed by SDS-PAGE of labeled PBPs.
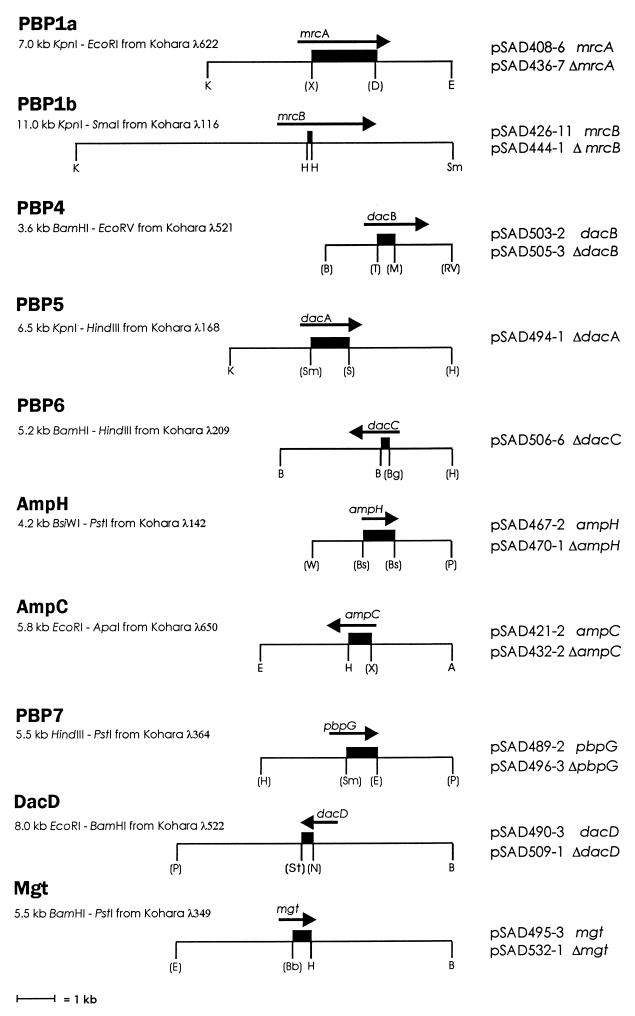
FIG. 1.
Cloned wild-type PBP genes and locations of internal deletions. Underneath the PBP name is listed the DNA fragment that was isolated and subcloned from the specified phage in the Kohara et al. genomic library (18). Each DNA fragment was subcloned into the vector pBCSK− (Stratagene) except for PBP 4, which was cloned into pBCKS−, and AmpH, which was cloned into pBCSK+. The light line represents the cloned DNA fragment, and the dark arrow above the line represents the position, length, and direction of the reading frame for an individual PBP gene. The heavy rectangle represents the internal DNA fragment that was deleted from the coding sequence and replaced with a HindIII linker. Names of the plasmids carrying these genes and deletions are listed at the right. Except for PBPs 5 and 6, each construct is represented by two plasmids: the first plasmid carries the entire DNA fragment and includes the cloned wild-type gene; the second plasmid carries the DNA fragment from which the internal segment was deleted and replaced by a HindIII linker (designated by the “Δ” prefix). For PBPs 5 and 6, only the deletion plasmids are represented because the wild-type genes were not cloned (Materials and Methods). Letters denote restriction enzyme sites (A, ApaI; B, BamHI; Bb, BstBI; Bg, BglI; Bs, BstEII; D, BspDI, E, EcoRI; H, HindIII; K, KpnI; M, MluI; N, NdeI; P, PstI; RV, EcoRV; S, SalI; Sm, SmaI; St, StuI; T, BstXI; W, BsiWI; X, XhoI). A letter enclosed in parentheses denotes a site destroyed during construction of the plasmid; letters in parentheses at the extreme ends of the entire DNA fragment were sites in the cloning vector that were destroyed, sometimes including neighboring sites in the multiple cloning site (data not shown). Sites that define the ends of the internal deletions were destroyed and replaced by a HindIII linker. To create marked genes for transfer to the E. coli chromosome, a res-npt-res (kanamycin-resistance) gene cassette from pCK155 (Materials and Methods) was inserted into the HindIII site of each deletion plasmid.
The cloning scheme described above was not used for creating deletions in the genes for PBP 5 (dacA) and PBP 6 (dacC) because overexpression of these proteins is lethal to E. coli. Therefore, an alternate method was used to construct plasmid-borne deletions of these two genes. For dacA, a 3.2-kb HindIII-SalI DNA fragment from Kohara phage λ168 was subcloned into pBCSK−. The HindIII site was deleted by opening, blunting, and religating, and a new HindIII site was inserted at the SalI position by opening, blunting, and inserting the appropriate linkers (pSAD488-2). This cloned fragment included the amino terminus of dacA and sequences immediately upstream of the gene. Next, from the same phage, a 2.5-kb KpnI-SmaI DNA fragment was cloned into pBCSK−, and HindIII linkers were inserted into the SmaI site (pSAD485-1). This cloned fragment included the carboxyl terminus of dacA and sequences immediately downstream of the gene. The 2.5-kb HindIII-KpnI carboxyl fragment from pSAD485-1 was moved into the same sites in pSAD488-2 to create, in effect, a dacA gene with an internal deletion of the SalI-SmaI DNA fragment (pSAD494-1) (Fig. 1). A deletion in the PBP 6 gene, dacC, was created in the same manner by combining two DNA fragments representing the amino and carboxyl termini of the gene into a single plasmid (pSAD506-6) (Fig. 1).
For marking the PBP gene deletions, a kanamycin resistance gene cassette flanked by two res sites was isolated as a 2.0-kb EcoRI DNA fragment from plasmid pCK155 (20). The ends of this DNA were blunt ended with E. coli DNA polymerase Klenow fragment, HindIII linker oligonucleotides were ligated to each end, and the ends were cut with HindIII. This cassette was ligated into the HindIII site of each PBP deletion plasmid, thereby replacing the internal gene sequences denoted by the heavy rectangles in Fig. 1.
PBP deletions were transferred from each plasmid to the chromosome of E. coli JC9387 by the λ transduction method of Kulakauskas et al. (21). Briefly, E. coli CS109 containing a plasmid with one of the kanamycin-marked gene deletions was infected with the Kohara λ phage which contained a wild-type copy of that particular PBP gene, and the plasmid-borne deletion and resistance marker were transferred by recombination to some of the phage in the resulting lysate. The lysate was used to transfer the PBP gene deletion to the chromosome of E. coli JC9387 by transduction and selection for kanamycin-resistant colonies. Deletion of the appropriate PBPs was confirmed by SDS-PAGE and autoradiography.
Nine mutants were constructed, each of which was missing the gene for one of PBPs 1a, 1b, 4, 5, 6, and 7, AmpC, AmpH, and DacD. In addition, the mgt gene, which encodes monofunctional glycosyltransferase, was deleted from a 10th strain. These individual JC9387-derived mutants were infected with phage P1, and the resulting lysates were used to move each of the kanamycin-marked PBP gene deletions into E. coli CS109 by P1 transduction (25), thus creating a set of 10 single-deletion mutants. Once again, the loss of PBPs from all strains was confirmed by SDS-PAGE and autoradiography or by Southern blotting to detect the loss of dacD and mgt. In addition, the strains were screened to certify that no auxotrophies were transduced from E. coli JC9387 to E. coli CS109.
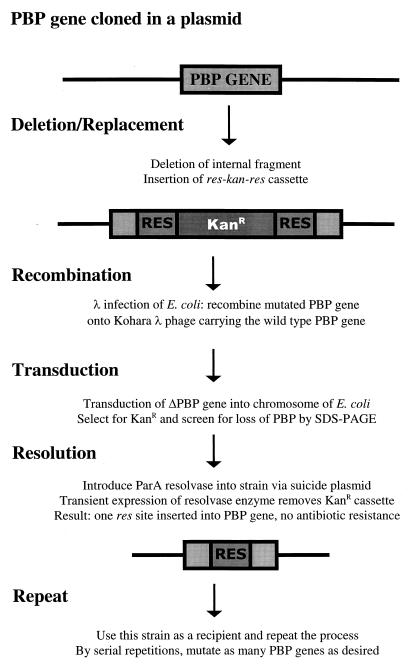
We wished to create strains that would eventually contain eight or more gene deletions, but it was not possible to mark each individual gene with a separate antibiotic resistance cassette to produce such combinations. This problem was avoided by combining the resolvase method described by Kristensen et al. (20) with the λ transduction method of Kulakauskas et al. (21). The generalized procedure is outlined schematically in Fig. 2. Briefly, a kanamycin-marked PBP gene deletion was moved into the chromosome of E. coli CS109 by P1 transduction, and the kanamycin resistance cassette was excised by transient expression of the RP4 ParA resolvase protein. Transient expression of resolvase was induced by conjugational mating of E. coli S17-1λpir(pJMSB8) to each kanamycin-marked strain. The transmissible plasmid pJMSB8 carries the parA gene under control of the lac promoter and is a suicide plasmid that replicates only in the presence of a λpir lysogen (20). Therefore, resolvase is expressed when the plasmid is transferred by conjugation into an E. coli recipient, but since the plasmid cannot replicate in this host, it is quickly lost as the recipient cell divides (20). In some of the recipients, resolvase excises the kanamycin marker from the site of the PBP deletion by acting on the two res sites that flank the gene cassette (20), leaving a single 8-bp res sequence at the site of the original deletion in the PBP gene. The mating mixture was plated onto M9-minimal glucose medium without kanamycin to select against the multiply auxotrophic donor, E. coli S17-1. E. coli CS109 colonies from these matings were replica plated onto minimal medium with and without kanamycin to identify those colonies that had become kanamycin sensitive, indicating that the resistance cassette had been removed from the chromosome. As before, the PBP profiles of kanamycin-sensitive deletion mutants were confirmed by SDS-PAGE and autoradiography.
FIG. 2.
Schematic illustration of the steps by which multiple genes were deleted from the chromosome of E. coli. KanR, kanamycin resistance.
Multiple PBP gene deletions were assembled into individual strains by repeated transduction and excision cycles: a kanamycin-marked PBP gene deletion was moved by P1 transduction into a previously constructed kanamycin-sensitive mutant, the newly added kanamycin resistance cassette was excised by resolvase, and the resulting strain was used as a kanamycin-sensitive recipient to which additional PBP gene deletions could be added. These two steps were repeated until the desired combinations of mutations were assembled in individual strains.
RESULTS
Construction of the set of PBP deletion mutants.
We constructed a total of 192 different strains that contained every possible combination of deletions involving the following eight PBPs: PBPs 1a, 1b, 4, 5, 6, and 7, AmpC, and AmpH. The only combinations that could not be produced were those that included deletions in both PBPs 1a and 1b, consistent with previous observations that strains lacking these two PBPs are inviable (16, 37, 46). In addition, the dacD gene was deleted from two strains lacking seven PBPs, and these constructs were confirmed by Southern blotting (data not shown).
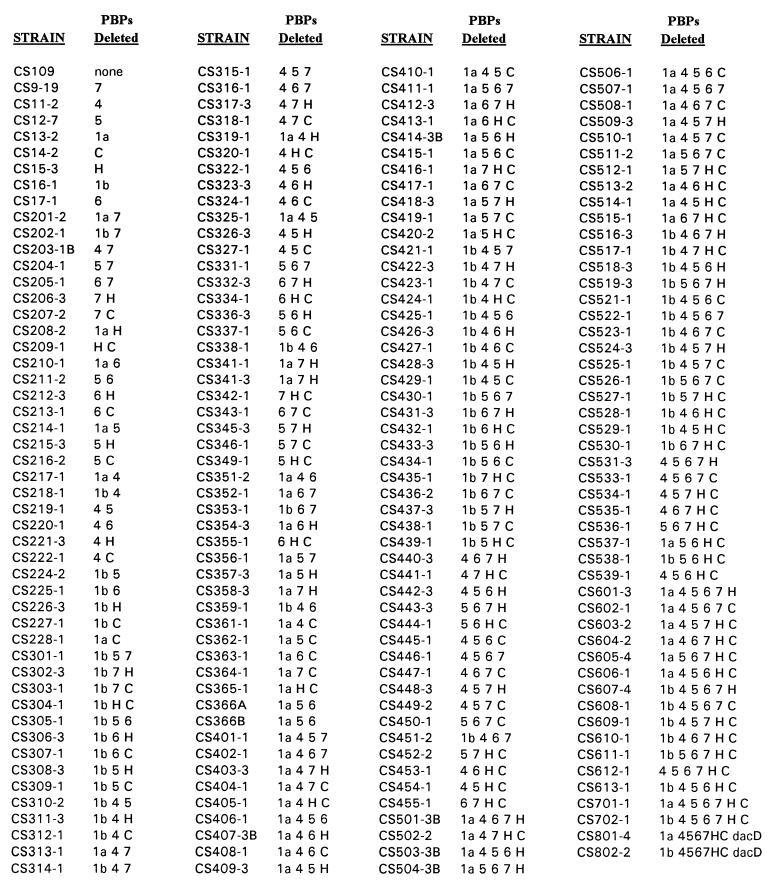
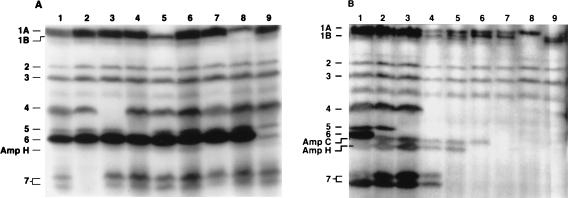
Table 1 lists the complete set of mutants and the PBPs that were deleted from each strain. The pathway by which each mutant was derived is displayed as a pedigree chart in Fig. 3. The PBP profile of every strain was observed at each step during construction of the mutant set, to confirm that those PBPs that should have been deleted were indeed absent. Figure 4 displays the PBP profiles of eight mutant strains from which a single PBP was deleted (Fig. 4A) and the profiles of a representative subset of mutants from which additional PBPs were deleted, culminating in the two mutants from which seven PBPs were removed (Fig. 4B).
TABLE 1.
Strains useda
The parental strain from which individual and multiple PBP genes were deleted was E. coli CS109 (W1485 glnV rpoS rph). PBPs deleted from individual E. coli strains are abbreviated as follows (PBP name and gene [in parentheses]): 1a, PBP 1a (mrcA); 1b, PBP 1b (mrcB); 4, PBP 4 (dacB); 5, PBP 5 (dacA); 6, PBP 6 (dacC); 7, PBP 7 (pbpG); C, AmpC (ampC); H, AmpH (ampH); dacD, DacD (dacD).
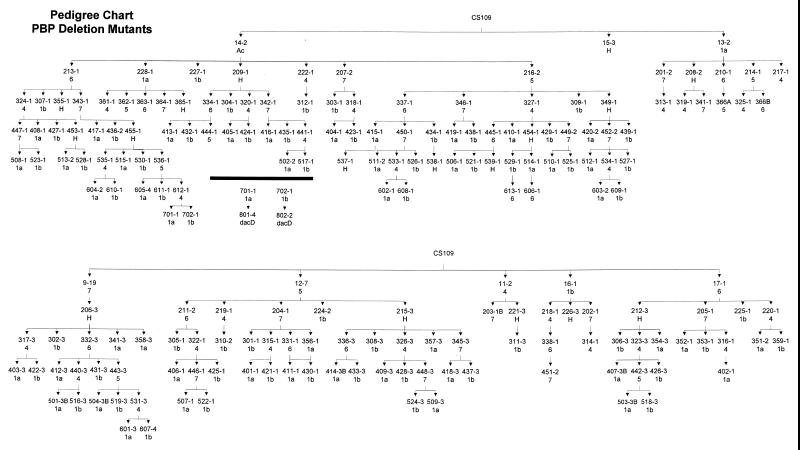
FIG. 3.
Pedigree of the PBP deletion mutants. The chart reflects the order in which genes were deleted from each strain. The original parental background was E. coli CS109. Mutants from which a single gene was deleted have names with numbers in the teens; mutants from which two genes were deleted have names with numbers in the 200’s; mutants with three genes deleted are numbered in the 300’s, etc. In the chart, the strain number is given but the “CS” prefix for each strain name was omitted for clarity. Underneath the number of each strain is listed the PBP gene that was deleted from the preceding parent to create that strain. Therefore, to determine the total complement of genes that were deleted from a particular strain, simply move “upward,” following the arrows backward to the ultimate parent, CS109, noting which genes were deleted in that lineage. More strains are shown than the minimum 192 required to make all possible mutants because some combinations were constructed by deleting genes in different orders. The mutants are listed in numerical order in Table 1, which includes a list of the genes deleted from each strain.
FIG. 4.
SDS-PAGE of 125I-penicillin-X-labeled PBPs from selected mutants. PBPs were labeled, separated by SDS-PAGE, and visualized by autoradiography as described elsewhere (11). (A) E. coli mutants from which a single PBP gene was deleted (deleted PBPs in parentheses): lane 1, CS109 (wild-type parent); lane 2, CS9-19 (PBP 7), lane 3, CS11-2 (PBP 4); lane 4, CS12-7 (PBP 5); lane 5, CS13-2 (PBP 1a); lane 6, CS14-2 (AmpC); lane 7, CS15-1 (AmpH); lane 8, CS16-1 (PBP 1b); lane 9, CS17-1 (PBP 6). (B) Representative subset of E. coli mutants from which a progressive number of PBPs were deleted: lane 1, CS109 (wild-type parent); lane 2, CS17-1 (PBP 6); lane 3, CS211-2 (PBPs 5 and 6); lane 4, CS322-1 (PBPs 4, 5, and 6); lane 5, CS446-1 (PBPs 4, 5, 6, and 7); lane 6, CS531-3 (PBPs 4, 5, 6, and 7 and AmpH); lane 7, CS612-1 (PBPs 4, 5, 6, and 7, AmpH, and AmpC); lane 8, CS702-1 (PBPs 1b, 4, 5, 6, and 7, AmpH, and AmpC); lane 9, CS701-1 (PBPs 1a, 4, 5, 6, and 7, AmpH, and AmpC). Note that in Fig. 4B, in lanes 8 and 9, some very faint bands equal to or smaller in size than PBP 4 are artifacts due to spillover from an adjacent lane (not shown); other gels of the samples in lanes 8 and 9 showed no such bands.
Complications associated with the strain construction procedure.
To our knowledge, this set of PBP mutants is the largest group of isogenic bacterial strains containing multiple mutations in a single enzyme family. Because the resolvase procedure of Kristensen et al. (20) promises to be extremely useful for creating multiply mutated strains, it is important to point out two problems that arose during application of the method. The first problem was the perpetuation of a phenotype unrelated to the genes being deleted. In our initial constructions, the ampH gene was deleted from CS109 to create the mutant CS15-1, and this strain served as the parent for over 50 multiply mutated strains. However, we later discovered that CS15-1 had become resistant to infection by phage λ and had passed this characteristic to all of its descendants (data not shown). Therefore, the entire branch of mutants was reconstructed by different routes: ampH was deleted from CS109 to create the new strain CS15-3, and the descendants of CS15-1 were reconstructed by deleting PBP genes from different parental mutants (the corrected lineage is reflected in Fig. 3). Resistance to phage λ disappeared in these new mutants (data not shown). However, there does exist a phage resistance phenotype that appears to be related to PBP genotype and which is spread among many branches of the pedigree tree (45). Thus, it is important that the results of any phenotypic test be compared to the pedigree to help determine if the phenomenon is real or possibly an artifact of the construction pathway.
The second problem that arose during this method of strain construction was that a few mutants acquired an unselected resistance to ampicillin. Although we avoided cloning into vectors containing β-lactamase genes (because expression of the enzyme prevents labeling PBPs with radioactive penicillin-X), six mutants were discovered to be resistant to ampicillin at 100 μg/ml. By using PCR and appropriate primers, we detected a chromosomal copy of the gene encoding TEM β-lactamase in these mutants (data not shown). The gene was most likely acquired during the removal of the res-npt-res cassette, when the ParA resolvase carried by the suicide plasmid pJMSB8 was introduced into the mutants by conjugation. Although the plasmid does not replicate in the absence of the λpir lysogen and is therefore usually lost from the recipients, portions of the plasmid evidently integrated into the chromosome of these six PBP mutants, thereby imparting ampicillin resistance. These strains were replaced by repeating the curing step to eliminate the res-npt-res cassette from the original mutants and by screening to avoid ampicillin-resistant colonies. Fortunately, none of the six mutants had served as the parent for other deletion mutations, and so only these strains needed to be reconstructed. Although ampicillin resistance arose at a low rate (less than 3% of the strains constructed), mutants produced by this procedure should be confirmed to be ampicillin sensitive.
Finally, there are 5,040 different possible pathways by which seven genes can be deleted from a single strain and, therefore, 10,080 pathways by which the two seven-deletion mutants could have been constructed. It is theoretically possible that the order in which genes are deleted can affect the phenotype of the descendent strains. As a preliminary test of this possibility, we constructed six sets of mutants lacking three or four PBPs, the strains in each set differing only in the order in which the same three or four PBPs were deleted (data not shown). We examined these strains for morphological differences, growth characteristics, temperature sensitivity, and antibiotic sensitivity to two β-lactams. There were no obvious phenotypic differences between the duplicated strains in each mutant subset. Nonetheless, it should be remembered that it is possible that deletion order effects exist for traits not yet tested.
Mutants lacking multiple PBPs are viable.
The most obvious result issuing from these mutants is that all were viable. Growth curves of mutants in LB medium at 37°C, including those of mutants from which seven or eight PBPs were deleted (representative data is shown in Fig. 5 and 6), were similar to the wild-type growth curves. Thus, the growth kinetics of E. coli were nearly normal if the following high-molecular-weight PBPs were present: either PBP 1a or 1b, and both of PBPs 2 and 3. Thus, for this particular set of proteins, the low-molecular-weight PBPs 4, 5, 6, and 7, AmpC, and AmpH, either individually or in any combination, were nonessential for growth of E. coli in a rich laboratory medium. In addition, the dacD gene was deleted from strains CS701-1 and CS702-1 to create two mutants that lacked eight different PBP genes (strains CS801-4 and CS802-2, respectively) (Table 1 and Fig. 3). These three octuple mutants were also viable, indicating that DacD was not essential even in the absence of all other low-molecular-weight PBPs.
FIG. 5.
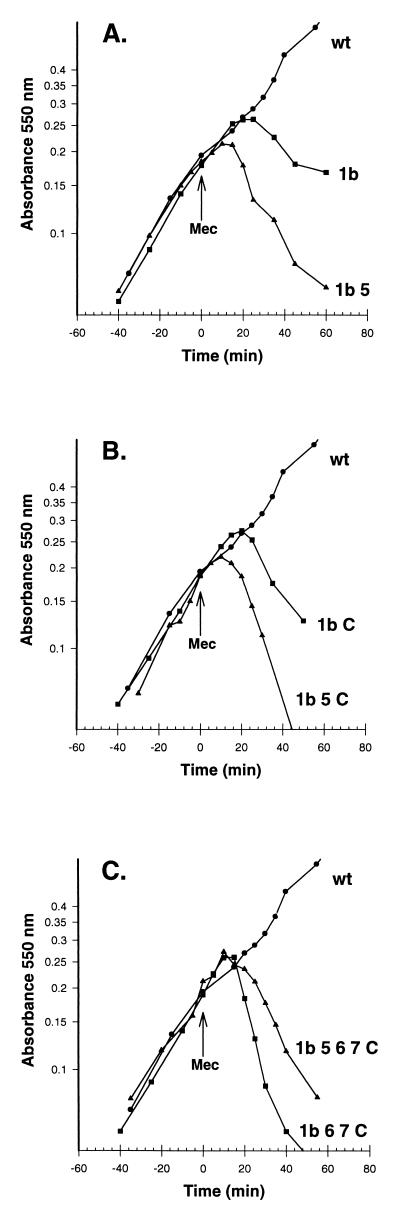
Lysis of PBP mutants by mecillinam. Cultures of E. coli mutants were grown in LB broth at 37°C, and the absorbance at 550 nm was measured. At time zero, mecillinam (10 μg/ml, final concentration) was added to each culture. Labels to the right of the curves list the PBPs that were deleted from the strain used to generate the data. (A) wt (wild type), CS109; 1b, CS16-1; 1b 5, CS224-2. (B) wt, CS109; 1b C (AmpC), CS227-1; 1b 5 C, CS309-1. (C) wt, CS109; 1b 6 7 C, CS436-2; 1b 5 6 7 C, CS526-1.
FIG. 6.
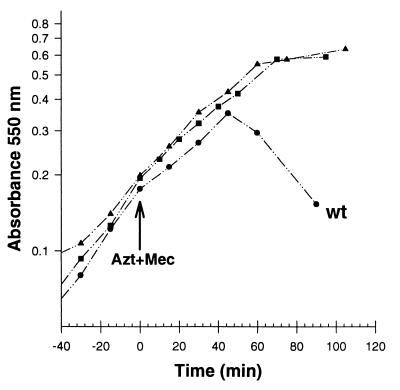
Growth of PBP mutants after simultaneous addition of mecillinam and aztreonam. Cultures of E. coli were grown in LB broth at 37°C, and the absorbance at 550 nm was measured. At time zero, aztreonam and mecillinam (Azt+Mec) were added to give a 10-μg/ml final concentration of each. Absorbance values for mutants CS403-3 and CS801-4 remained stable to at least 200 min (data not shown). Circles, CS109 (parental wild type [wt]); squares, CS801-4 (PBPs 1a, 4, 5, 6, and 7, AmpH, AmpC, and DacD deleted); triangles, CS403-3 (PBPs 1a, 4, and 7 and AmpH deleted).
Inactivation of PBP 2 induces lysis in mutants lacking PBP 1b.
The β-lactam mecillinam binds to and inactivates PBP 2, inhibiting cell wall elongation so that each E. coli cell grows as a continually enlarging sphere (10, 39). The absorbance of a mecillinam-treated culture continues to increase at a logarithmic rate, but the cells eventually stop growing unless there are compensatory mutations in aroK or cya (26, 43) or unless ppGpp or the FtsZ protein are overexpressed (42, 44). Thus, in the absence of PBP 2, peptidoglycan synthesis continues at approximately the wild-type rate but overall cell shape is altered.
We tested to see if any low-molecular-weight PBPs were required for peptidoglycan synthesis in the absence of PBP 2 by adding mecillinam to log-phase cultures of each of the members of the PBP deletion set. Wild-type E. coli and all mutants that retained active PBP 1b continued to enlarge normally, without lysis, in the presence of mecillinam (data not shown). The extreme example was that of E. coli CS801-4, which exhibited a normal response to mecillinam even though eight PBPs were deleted from this mutant (Table 1), leaving only four active PBPs (1b, 1c, 2, and 3). Thus, when PBP 2 was inactivated in these multiple mutants, the three remaining active PBPs (1b, 1c, and 3) could still synthesize osmotically stable peptidoglycan in the shape of a sphere.
In contrast, deletion of PBP 1b alone was sufficient to make E. coli sensitive to lysis by mecillinam (Fig. 5A), as del Portillo and de Pedro reported for a single mutant (6, 7). In addition, mecillinam induced lysis in each of the 64 mutants from which PBP 1b had been deleted (see Fig. 5 for examples of this effect). In most of these Δ1b mutants, lysis began ∼20 min after addition of the antibiotic and proceeded at a rate (having a negative slope) that was approximately equal to or slightly slower than the original growth rate. The results establish that the synthetic activity of PBP 1b does not require the aid of any of the low-molecular-weight PBPs and that in the absence of PBP 1b, PBP 2 becomes essential for synthesis of intact, osmotically protective peptidoglycan, regardless of the activity of any low-molecular-weight PBPs.
Deletion of PBP 5 sensitizes Δ1b mutants to inactivation of PBP 2.
Although all Δ1b mutants lysed when exposed to mecillinam, the time of lysis onset and its rate were accelerated in strains from which PBP 5 was also deleted. For example, the onset of mecillinam-induced lysis was decreased from 20 min in CS16-1 (Δ1b) to about 10 min in CS224-2 (Δ1b 5), and the ensuing lysis rate was more rapid in the double mutant (Fig. 5A). This trend was also exhibited by mutants from which multiple PBPs had been deleted. As a general rule, mutants lacking both PBPs 1b and 5 lysed more quickly and faster (e.g., Fig. 5B). However, some mutants lacking four or more PBPs lysed so quickly that removing PBP 5 reduced the time of lysis onset only slightly or not at all (e.g., Fig. 5C). Even so, in general, Δ[1b 5] mutant combinations lysed earlier and more rapidly than did their isogenic Δ1b relatives containing wild-type PBP 5. These observations indicate that active PBP 5 moderates the mecillinam sensitivity of Δ1b mutants.
Inactivation of PBP 3 induces lysis in mutants lacking PBP 1b.
Aztreonam is a β-lactam that inactivates PBP 3, thereby inhibiting septation so that each E. coli cell grows as a continually elongating, nonsegmented filament (38). The absorbance, but not cell number, of an aztreonam-treated culture continues to increase at a logarithmic rate. Schmidt et al. (30) and del Portillo and de Pedro (6) reported that inactivation of PBP 3 lyses an E. coli strain which contains a temperature-sensitive mutation in PBP 1b. We confirmed and extended these results by adding aztreonam to the entire set of PBP mutants. The only strains that lysed were those 64 from which PBP 1b had been deleted; all other strains in which PBP 1b remained active continued to grow and form filaments when PBP 3 was inactivated. Once again, the extreme example was that of E. coli CS801-4, which exhibited a normal response to aztreonam even though eight PBPs were missing, leaving PBPs 1b, 1c, 2, and 3 active. Thus, even when PBP 3 was inactivated, PBPs 1b, 1c, and 2 could synthesize osmotically stable peptidoglycan in the shape of an elongated filament. As with the experiments in which PBP 2 was inactivated, these results establish that the synthetic activity of PBP 1b does not require the aid of any of the low-molecular-weight PBPs. In addition, in the absence of PBP 1b, PBP 3 becomes essential for synthesis of intact, osmotically protective peptidoglycan, regardless of the presence or absence of any of the low-molecular-weight PBPs.
Synthesis of osmotically stable peptidoglycan when PBPs 2 and 3 are inactivated.
Simultaneous inactivation of PBPs 2 and 3 lyses E. coli (10, 30). This result has been interpreted as evidence that at least one or the other of PBPs 2 and 3 is required for continued peptidoglycan synthesis (10). To see if this observation was true for the set of PBP mutants, we grew individual strains in 2-ml aliquots of LB broth and tested for lysis by adding mecillinam and aztreonam to inactivate PBPs 2 and 3, respectively. All mutants lacking PBP 1b lysed (data not shown), consistent with previous observations that these strains were lysed by either antibiotic alone.
Fifteen strains that demonstrated resistance to lysis in 2-ml cultures were tested by growing them in a large volume of medium in well-aerated flasks, followed by addition of the two antibiotics. Several of these mutants did not lyse. Instead, one of three reactions occurred: growth of some strains leveled off without lysis; in other strains, lysis was delayed significantly and its rate was reduced; and the absorbance of a few strains continued to increase after addition of the two antibiotics (data not shown). When mecillinam or aztreonam was added individually to these strains, the cells became spherical or filamentous due to inactivation of PBP 2 or 3 (data not shown). This result proved that when administered individually, the antibiotics inactivated their respective targets. Nonetheless, some of the PBP mutants had become resistant to the lytic effect that normally accompanies exposure to both antibiotics.
Two examples of mutants that did not lyse after inactivation of PBPs 2 and 3 were strains CS403-3 (lacking PBPs 1a, 4, and 7, and AmpH) and CS801-4 (lacking PBPs 1a, 4, 5, 6, and 7, AmpC, AmpH, and DacD) (Fig. 6). These strains lacked PBP 1a and three or seven other low-molecular-weight PBPs, respectively. Thus, after PBPs 2 and 3 were inactivated in strain CS801-4, peptidoglycan was synthesized in the absence of 10 PBPs, and the structure of this peptidoglycan was sufficiently intact to prevent osmotic lysis in LB medium.
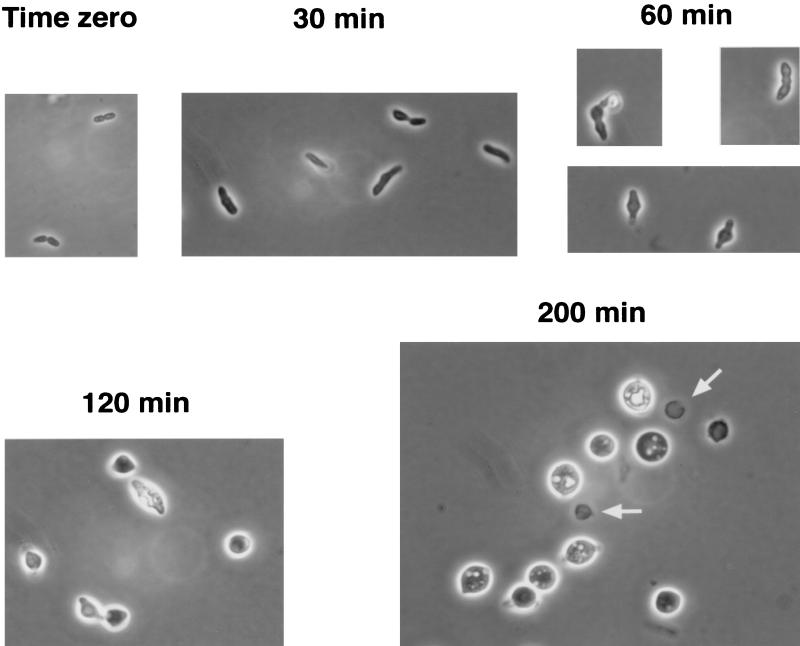
The microscopic appearance of strain CS403-3 was typical of the morphological responses of mutants that were resistant to the lytic effects of simultaneous addition of mecillinam and aztreonam (Fig. 7). Log-phase cells looked quite normal before exposure to the antibiotics (Fig. 7, time zero). However, as soon as 30 min after addition of both antibiotics, the mutants ceased dividing and began to expand (Fig. 7, 30 min). Cells at this early stage often had a ring or bulge around their midpoints, suggesting an inability to produce a normal septum (Fig. 7B, 30 and 60 min). After 120 min, almost all cells were spherical or nearly so, and these spheres continued to grow and enlarge until the diameter of individual cells reached 5 to 7 μm (Fig. 7, 120 and 200 min). Many of these huge cells contained phase-lucent vesicles of unknown origin and composition (Fig. 7, 200 min). In addition, the enlarging spherical cells often retained pairs of short protuberances, probably representing the original cell poles which are known to be inert to insertion of new peptidoglycan. Also observed in increasing numbers over time were ghost sacculi (peptidoglycan shells emptied of their cytoplasmic contents) (Fig. 7, 200 min). This latter observation is consistent with the fact that these mutants eventually stopped growing and did lyse, given enough time, even though the strains were resistant to the immediate lytic effect of losing PBPs 2 and 3.
FIG. 7.
Morphology of E. coli CS403-3 after simultaneous addition of mecillinam and aztreonam. E. coli CS403-3 was grown in LB broth at 37°C to an absorbance at 550 nm of 0.2, at which point both mecillinam and aztreonam were added to give a final concentration of 10 μg of each per ml. At the times indicated, samples were withdrawn for phase-contrast microscopy. All photographs were captured with a cold charge-coupled device camera at a magnification of ×1,000, and all images represent the same relative magnification so that direct comparisons of cell size can be made between time points. The size of an individual E. coli cell before addition of antibiotics was approximately 0.8 by 1.5 μm (see cells at time zero) and serves as a measure of the size of cells at other time points. Arrows indicate ghost cells that are emptied of cytoplasmic contents.
In summary, some mutants lacking multiple PBPs did not lyse in the absence of functional PBPs 2 and 3. Instead, individual cells continued to metabolize and enlarge, although they were unable to divide.
DISCUSSION
In 1978, Suzuki et al. suggested that it “might be the right time to construct a series of relevant mutants by combining these [PBP] mutations to dissect the roles of the binding proteins on cell growth, division, and penicillin-sensitive enzyme systems” (37). Their suggestion notwithstanding, only a handful of multiple mutants were constructed in the ensuing 20 years, and we believe that the absence of such a comprehensive set of mutants has delayed our understanding of the biological roles of individual PBPs. The mutants described in this work supply the genetic backgrounds in which the functions of this set of enzymes may be defined more exactly. Furthermore, preliminary examination of the phenotypes exhibited by these mutants places new constraints on current theories of peptidoglycan biosynthesis and suggests some novel ideas about how E. coli synthesizes its protective shell.
Preliminary caveats.
Before considering the ramifications of these results, we wish to emphasize some cautionary considerations. The foremost of these is that in the 192 strains lacking from one to seven PBPs, several peptidoglycan-specific enzymes remain active: DacD, a recently identified carboxypeptidase (1); PBP 1c, a newly discovered high-molecular-weight PBP (29); MepA, an endopeptidase similar to PBPs 4 and 7 (17); Mgt, a monofunctional transglycosylase with similarities to portions of PBPs 1a and 1b (8); all of the lytic transglycosylases (14); a penicillin-insensitive ld-carboxypeptidase (41); and a putative ld-transpeptidase (5). It is theoretically possible that one or more of these active enzymes can compensate for the loss of the inactived PBPs. However, the results reported here seem to decrease the likelihood that E. coli remains viable by a simple substitution of one PBP for another.
A second caveat is that during strain construction, some PBP mutants may compensate for the loss of one or more PBPs by accumulating cryptic adaptations. Such compensations could take the form of secondary mutations that relieve deleterious effects created by the absence of PBPs. We have no evidence of such mutations, and the high frequency of successful P1 transductions from one strain to another argues against the occurrence of such mutations, but the theoretical possibility should be kept in mind.
A third consideration is that during construction of the mutants, a strain might develop a phenotype unrelated to PBP loss. So far, we have observed only one case of this: one mutant became spontaneously resistant to phage λ and passed this resistance to all strains derived from it. We eliminated this particular problem by reconstructing the mutants from unaffected parental strains. However, other unknown mutations may have accumulated, affecting traits that we did not measure. Those who use these strains in the future should be aware that the best method for detecting such unrelated hereditary mutations is to compare newly measured phenotypes to the family tree and treating with some suspicion those characteristics exhibited by all descendants of a single strain.
Nonessential PBPs.
The most surprising result was that all the PBP mutants, including those mutants from which eight PBPs were deleted, survived and grew nearly as well as the parental E. coli strain. A long-standing argument is that inactivation or loss of one of the low-molecular-weight PBPs results in no phenotype because several proteins have similar functions and the remaining enzymes can substitute for one another. Based on this assumption, our original hypothesis was that sequential deletion of these PBPs would eventually yield a mutant in which one or more of the enzymes became essential for viability. The expectation was that at some point a multiply mutated strain would possess only one member of a particular substitution family. Instead, at least 8 of the 12 known PBPs could be eliminated with only slight morphological effects to untreated E. coli. Among these strains are mutants which grew in the absence of every known carboxypeptidase (PBPs 4, 5, and 6, and DacD), in the absence of two of the three known endopeptidases (PBPs 4 and 7), in the absence of both class C β-lactamases (AmpC and AmpH), and in the absence of all seven of these low-molecular-weight PBPs.
The fact that E. coli remains viable in the absence of so many PBPs raises the question of the normal physiological roles of these enzymes. There are at least three possible explanations for why no phenotype is associated with multiple mutations in the low-molecular-weight PBPs. First, these proteins may be completely dispensable for bacterial viability; second, another peptidoglycan-reactive enzyme may compensate for the loss of these PBPs; and third, the-low-molecular weight PBPs may not be essential for laboratory growth of E. coli but might, instead, affect bacterial viability or physiology under conditions not yet tested. We are currently screening the mutant set for phenotypes that might be expected to arise should PBP loss affect some aspect of the structure or function of peptidoglycan.
Interactions between high- and low-molecular-weight PBPs.
One of the unanswered questions about cell wall growth involves how the PBPs work together to synthesize functional peptidoglycan. We would like to know not only which PBPs are essential but which ones interact and how. In particular, one unknown is how the low-molecular-weight PBPs might influence interactions among the high-molecular-weight PBPs. To determine if there existed any such relationships, we inactivated PBPs 2 and/or 3 in the set of PBP mutants.
First, we reconfirmed the observations of del Portillo and de Pedro (6, 7) and of Schmidt et al. (30) that inactivation of either PBP 2 or PBP 3 lyses Δ1b mutants but not Δ1a mutants. Furthermore, we extended these observations to include 64 different Δ1a mutants and 64 different Δ1b mutants, in which up to six other PBPs were deleted in all possible combinations. The results establish that neither the lysis susceptibility of Δ1b mutants nor the lysis resistance of Δ1a mutants depends on the activity of the low-molecular-weight PBPs. Functional peptidoglycan is synthesized if PBPs 1b and 2 are active (in Δ1a mutants treated with aztreonam) or if PBPs 1b and 3 are active (in Δ1a mutants treated with mecillinam), regardless of the presence or absence of up to seven other PBPs. Thus, peptidoglycan synthesis by PBP 1b (possibly aided by PBP 1c) requires either one or the other of PBPs 2 and 3, and neither PBP 1a nor PBP 1b requires the low-molecular-weight PBPs.
Even though the low-molecular-weight PBPs did not affect the growth of cells with active high-molecular-weight PBPs, the smaller PBPs may play a role in cell lysis induced by inactivation of PBPs 2 and 3. A form of osmotically stable peptidoglycan was synthesized by some Δ1a PBP mutants lacking up to eight PBPs and in which PBPs 2 and 3 were inactivated by antibiotics. In one mutant, a form of peptidoglycan was synthesized in the absence of 10 of the 12 known PBPs. In making this statement, we draw a distinction between the synthesis of peptidoglycan that has the size and shape of a normal bacterium and the synthesis of peptidoglycan that has an abnormal size or shape but which serves to protect the cytoplasm from rupture by osmotic pressure in LB medium. Clearly, PBPs 2 and 3 are essential in the sense that without them E. coli grows incorrectly and fails to divide normally. Nonetheless, in some genetic backgrounds, PBPs 2 and 3 are not essential for synthesizing peptidoglycan that is at least partially osmotically resistant, in that cells continue to grow and enlarge in a rich medium. In the extreme case of E. coli CS801-4, from which eight PBPs are missing, the only known peptidoglycan synthetic enzymes that remain are PBP 1b, PBP 1c, and Mgt. The conclusion is that protective, presumably adequately cross-linked peptidoglycan can be synthesized either by PBP 1b alone or by PBP 1b in conjunction with PBP 1c and/or Mgt. Little is known about PBP 1c (29) or Mgt (8, 36), but because PBP 1b is the major peptidoglycan synthetic enzyme in E. coli (19), PBP 1b by itself may synthesize an osmotically stable sacculus.
Implications for the multienzyme complex hypothesis of peptidoglycan synthesis.
The results reported here place constraints on models of peptidoglycan synthesis. For example, the multienzyme hypothesis predicts that a complex of several different proteins executes a set of tightly orchestrated reactions to synthesize and insert new peptidoglycan strands into the older peptidoglycan of the intact sacculus (14, 15). In this scheme, an individual complex would include, at the very least, a synthetic transglycosylase (PBP 1a, 1b, or 1c), a synthetic transpeptidase (PBP 1a, 1b, 1c, 2, or 3), a lytic transglycosylase, and an endopeptidase (either PBP 4 or 7 or MepA) (14, 15). However, some of the mutants described here can synthesize a partially functional form of peptidoglycan in the absence of up to eight PBPs and when PBPs 2 and/or 3 are inactive. This means that some form of peptidoglycan, perhaps simplified in structure, can be synthesized by some combination of PBP 1b, PBP 1c, and Mgt (supplying transglycosylase and transpeptidase functions), MepA (the only known endopeptidase still remaining), any of the lytic transglycosylases, and, perhaps, a penicillin-insensitive ld-carboxypeptidase or ld-transpeptidase. A mutant lacking three of the five known lytic transglycosylases is viable (22), leaving only two other possible enzymes to fulfill this hypothesized role in the multienzyme complex. Thus, if a multienzyme complex is responsible for peptidoglycan synthesis, then in the mutants lacking eight PBPs there are only a limited number of potential combinations that remain. Perhaps a multienzyme complex provides specialized functions beyond the most basic requirements for peptidoglycan synthesis. In any case, the multiply mutated strains described in this report should make it easier to ask questions regarding these possibilities.
Implications for the dd-carboxypeptidase-dependent hypothesis.
Mutants lacking PBP 1b lyse when PBP 2 is inactivated by mecillinam, meaning that cells containing only PBPs 1a and 3 (of the classic PBPs) are unable to survive. We found that if PBP 5 is also deleted, then such Δ[1b 5] double mutants lyse even more rapidly. The conclusion is that in the absence of PBP 5, PBPs 1a and 3 cope even less well after inactivation of PBP 2. This result is consistent with the hypothesis that dd-carboxypeptidase activity may be required to supply PBP 3 with substrate (2). This “dd--carboxypeptidase-dependent” hypothesis would predict that when PBP 5 is absent trimers are less available as substrates for PBP 3, resulting in poorer survival (quicker lysis) when PBP 2 is inactivated. However, if a dd-carboxypeptidase is absolutely essential for the activity of PBP 2 or 3, it is not at all clear how E. coli can survive and septate in the absence of all the known dd-carboxypeptidases (as in CS801-4 and CS802-2, which lack PBPs 4, 5, 6, and 7 and DacD). Therefore, although some of the results presented here are consistent with the dd-carboxypeptidase-dependent hypothesis (2), other results call into question the necessity of having any dd-carboxypeptidases at all. (It should be noted that our results say nothing about the contribution of any ld-carboxypeptidases.)
Implications for evolution of the peptidoglycan sacculus.
It is difficult to envision a free-living bacterium without a cell wall because the peptidoglycan sacculus protects these cells from osmotic lysis in hypotonic environments. Therefore, protobacteria must have acquired, very rapidly, some means of osmotic protection. It is impossible to believe that the first cell walls required the organized activity of multiple enzymes. A more likely possibility is that a single enzyme evolved to fill this function and then, over time, additional enzymes were added to the wall-producing pathway. In this scenario, something as complex as a multienzyme peptidoglycan synthesis apparatus would have to be of relatively recent origin.
If peptidoglycan or some protopeptidoglycan structure were synthesized by a single enzyme, then there might be remnants of that mechanism in present-day bacteria. By removing or inactivating 10 of 12 PBPs from E. coli, we have approached a situation in which a single enzyme might be synthesizing osmotically resistant peptidoglycan. The best candidate for an enzyme with this capability is PBP 1b. As discussed earlier, we cannot rule out the possibility that PBP 1b is still participating in a multienzyme complex, and there is no evidence that E. coli itself should retain such a primordial enzyme. Nonetheless, we should seriously entertain the idea that the basic requirements for peptidoglycan synthesis are simpler than usually believed.
Implications for physiological studies based on completed genomic sequences.
We have entered an era in which we have more genetic information about many organisms than we have physiological information. For the organisms whose genomes are now completely sequenced, including even such a well-studied organism as E. coli, large numbers of sequences have no known genetic counterparts and many that do have counterparts have no established function. The most popular approach suggested for study of these myriad unidentified open reading frames is to mutate them individually and screen the resulting mutants for measurable phenotypes. It is likely that functions will be assigned to at least some unknown genes by using this approach. However, the results presented in this report suggest caution about what to expect from a strategy that uses mutation of individual genes.
We created a set of mutants in a group of eight genes that belong to a single related family. Despite knowing the general purpose and biochemical capabilities of members of the family, and therefore knowing what to look for, we observed no obvious phenotypes for most mutants. If this is the situation for a family of genes for which we have so much physiological and biochemical data, then it is likely to be true for other gene families for which we have much less information. Creation of strain sets lacking many genes, though tedious, may need to be the strategy of choice to yield clues about the functions of unknown genes.
Potential uses for the set of PBP mutants.
Finally, among many potential applications of the set of PBP mutants are three that should be particularly useful. First, selected mutants can be used as starting points for asking questions about the physiological functions of other enzymes, such as the lytic transglycosylases or non-PBP endopeptidases. In particular, questions about specific theories of peptidoglycan synthesis can be approached which would otherwise be difficult or impossible to address. Second, the physiological or biochemical functions of any two of the eight enzymes can be compared by examining individual strains from which competing PBPs have been deleted and which differ only in possession of the two PBPs of interest. This should be valuable in testing the roles of the various subfamilies of PBPs. Finally, some of the mutants may make good hosts from which to purify cloned PBPs. This will be especially useful for purifying PBPs that require an affinity chromatography step which might otherwise retain contaminating PBPs.
ACKNOWLEDGMENTS
The construction of some of the mutants described in this report was supported by a grant from SmithKline Beecham Pharmaceuticals.
We are deeply grateful to David Knowles and David Payne for their support and interest in the work. We especially thank Joachim-Volker Höltje for helpful discussions and comments on a previous version of the manuscript and for bringing to our attention the presence of TEM β-lactamase sequences in one of the original mutants. Finally, we thank Heather Skarhus and Hugh Nguyen for technical help, Bernadette Meberg for reformatting Fig. 1, and Victoria Swift for graphics assistance.
REFERENCES
- 1.Baquero M-R, Bouzon M, Quintela J C, Ayala J A, Moreno F. dacD, an Escherichia coli gene encoding a novel penicillin-binding protein (PBP6b) with dd-carboxypeptidase activity. J Bacteriol. 1996;178:7106–7111. doi: 10.1128/jb.178.24.7106-7111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Begg K J, Takasuga A, Edwards D H, Dewar S J, Spratt B G, Adachi H, Ohta T, Matsuzawa H, Donachie W D. The balance between different peptidoglycan precursors determines whether Escherichia coli cells will elongate or divide. J Bacteriol. 1990;172:6697–6703. doi: 10.1128/jb.172.12.6697-6703.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Botta G A, Park J T. Evidence for involvement of penicillin-binding protein 3 in murein synthesis during septation but not during cell elongation. J Bacteriol. 1981;145:333–340. doi: 10.1128/jb.145.1.333-340.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broome-Smith J K. Construction of a mutant of Escherichia coli that has deletions of both the penicillin-binding protein 5 and 6 genes. J Gen Microbiol. 1985;131:2115–2118. doi: 10.1099/00221287-131-8-2115. [DOI] [PubMed] [Google Scholar]
- 5.Caparrós M, Pisabarro A G, de Pedro M A. Effect of d-amino acids on structure and synthesis of peptidoglycan in Escherichia coli. J Bacteriol. 1992;174:5549–5559. doi: 10.1128/jb.174.17.5549-5559.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.del Portillo F G, de Pedro M A. Differential effect of mutational impairment of penicillin-binding proteins 1A and 1B on Escherichia coli strains harboring thermosensitive mutations in the cell division genes ftsA, ftsQ, ftsZ, and pbpB. J Bacteriol. 1990;172:5863–5870. doi: 10.1128/jb.172.10.5863-5870.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.del Portillo F G, de Pedro M A. Penicillin-binding protein 2 is essential for the integrity of growing cells of Escherichia coli ponB strains. J Bacteriol. 1991;173:4530–4532. doi: 10.1128/jb.173.14.4530-4532.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Berardino M, Dijkstra A, Stüber D, Keck W, Gubler M. The monofunctional glycosyltransferase of Escherichia coli is a member of a new class of peptidoglycan-synthesising enzymes. FEBS Lett. 1996;392:184–188. doi: 10.1016/0014-5793(96)00809-5. [DOI] [PubMed] [Google Scholar]
- 9.Edwards D H, Donachie W D. Construction of a triple deletion of penicillin-binding proteins 4, 5, and 6 in Escherichia coli. In: de Pedro M A, Höltje J-V, Löffelhardt W, editors. Bacterial growth and lysis. New York, N.Y: Plenum Press; 1993. pp. 369–374. [Google Scholar]
- 10.Gutmann L, Vincent S, Billot-Klein D, Acar J F, Mrèna E, Williamson R. Involvement of penicillin-binding protein 2 with other penicillin-binding proteins in lysis of Escherichia coli by some β-lactam antibiotics alone and in synergistic lytic effect of amdinocillin (mecillinam) Antimicrob Agents Chemother. 1986;30:906–912. doi: 10.1128/aac.30.6.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henderson T A, Dombrosky P M, Young K D. Artifactual processing of penicillin-binding proteins 7 and 1b by the OmpT protease of Escherichia coli. J Bacteriol. 1994;176:256–259. doi: 10.1128/jb.176.1.256-259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henderson T A, Templin M, Young K D. Identification and cloning of the gene encoding penicillin-binding protein 7 of Escherichia coli. J Bacteriol. 1995;177:2074–2079. doi: 10.1128/jb.177.8.2074-2079.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henderson T A, Young K D, Denome S A, Elf P K. AmpC and AmpH, proteins related to the class C β-lactamases, bind penicillin and contribute to the normal morphology of Escherichia coli. J Bacteriol. 1997;179:6112–6121. doi: 10.1128/jb.179.19.6112-6121.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Höltje J V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Höltje J V. A hypothetical holoenzyme involved in the replication of the murein sacculus of Escherichia coli. Microbiology. 1996;142:1911–1918. doi: 10.1099/13500872-142-8-1911. [DOI] [PubMed] [Google Scholar]
- 16.Kato J-I, Suzuki H, Hirota Y. Dispensability of either penicillin-binding protein-1a or -1b involved in the essential process for cell elongation in Escherichia coli. Mol Gen Genet. 1985;200:272–277. doi: 10.1007/BF00425435. [DOI] [PubMed] [Google Scholar]
- 17.Keck W, van Leeuwen A M, Huber M, Goodell E W. Cloning and characterization of mepA, the structural gene of the penicillin-insensitive murein endopeptidase from Escherichia coli. Mol Microbiol. 1990;4:209–219. doi: 10.1111/j.1365-2958.1990.tb00588.x. [DOI] [PubMed] [Google Scholar]
- 18.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 19.Kraus W, Höltje J-V. Two distinct transpeptidation reactions during murein synthesis in Escherichia coli. J Bacteriol. 1987;169:3099–3103. doi: 10.1128/jb.169.7.3099-3103.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kristensen C S, Eberl L, Sanchez-Romero J M, Givskov M, Molin S, de Lorenzo V. Site-specific deletions of chromosomally located DNA segments with the multimer resolution system of broad-host-range plasmid RP4. J Bacteriol. 1995;177:52–58. doi: 10.1128/jb.177.1.52-58.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulakauskas S, Wikström P M, Berg D E. Efficient introduction of cloned mutant alleles into the Escherichia coli chromosome. J Bacteriol. 1991;173:2633–2638. doi: 10.1128/jb.173.8.2633-2638.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lommatzsch J, Templin M F, Kraft A R, Vollmer W, Höltje J V. Outer membrane localization of murein hydrolases: MltA, a third lipoprotein lytic transglycosylase in Escherichia coli. J Bacteriol. 1997;179:5465–5470. doi: 10.1128/jb.179.17.5465-5470.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuhashi M, Maruyama I N, Takagaki Y, Tamaki S, Nishimura Y, Hirota Y. Isolation of a mutant of Escherichia coli lacking penicillin-sensitive d-alanine carboxypeptidase IA. Proc Natl Acad Sci USA. 1978;75:2631–2635. doi: 10.1073/pnas.75.6.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuhashi M, Takagaki Y, Maruyama I N, Tamaki S, Nishimura Y, Suzuki H, Ogino U, Hirota Y. Mutants of Escherichia coli lacking in highly penicillin-sensitive d-alanine carboxypeptidase activity. Proc Natl Acad Sci USA. 1977;74:2976–2979. doi: 10.1073/pnas.74.7.2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 26.Ogura T, Bouloc P, Niki H, D’Ari R, Hiraga S, Jaffé A. Penicillin-binding protein 2 is essential in wild-type Escherichia coli but not in lov or cya mutants. J Bacteriol. 1989;171:3025–3030. doi: 10.1128/jb.171.6.3025-3030.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romeis T, Höltje J-V. Penicillin-binding protein 7/8 of Escherichia coli is a DD-endopeptidase. Eur J Biochem. 1994;224:597–604. doi: 10.1111/j.1432-1033.1994.00597.x. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Schiffer G, Templin M F, Höltje J-V. Cloning and biochemical characterization of the bifunctional penicillin-binding protein 1C from Escherichia coli. GenBank accession no. U88571. 1997. [Google Scholar]
- 30.Schmidt L S, Botta G, Park J T. Effects of furazlocillin, a β-lactam antibiotic which binds selectively to penicillin-binding protein 3, on Escherichia coli mutants deficient in other penicillin-binding proteins. J Bacteriol. 1981;145:632–637. doi: 10.1128/jb.145.1.632-637.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 32.Spratt B G. Deletion of the penicillin-binding protein 5 gene of Escherichia coli. J Bacteriol. 1980;144:1190–1192. doi: 10.1128/jb.144.3.1190-1192.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spratt B G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci USA. 1975;72:2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spratt B G. Properties of the penicillin-binding proteins of Escherichia coli K12. Eur J Biochem. 1977;72:341–352. doi: 10.1111/j.1432-1033.1977.tb11258.x. [DOI] [PubMed] [Google Scholar]
- 35.Spratt B G, Pardee A B. Penicillin-binding proteins and cell shape in E. coli. Nature. 1975;254:516–517. doi: 10.1038/254516a0. [DOI] [PubMed] [Google Scholar]
- 36.Spratt B G, Zhou J, Taylor M, Merrick M J. Monofunctional biosynthetic peptidoglycan transglycosylases. Mol Microbiol. 1996;19:639–640. doi: 10.1046/j.1365-2958.1996.442924.x. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki H, Nishimura Y, Hirota Y. On the process of cellular division in Escherichia coli: a series of mutants of E. coli altered in the penicillin-binding proteins. Proc Natl Acad Sci USA. 1978;75:664–668. doi: 10.1073/pnas.75.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sykes R B, Bonner D P. Discovery and development of the monobactams. Rev Infect Dis. 1985;7:S579–S593. doi: 10.1093/clinids/7.supplement_4.s579. [DOI] [PubMed] [Google Scholar]
- 39.Tamaki S, Matsuzawa H, Matsuhashi M. Cluster of mrdA and mrdB genes responsible for the rod shape and mecillinam sensitivity of Escherichia coli. J Bacteriol. 1980;141:52–57. doi: 10.1128/jb.141.1.52-57.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamura T, Suzuki H, Nishimura Y, Mizoguchi J, Hirota Y. On the process of cellular division in Escherichia coli: isolation and characterization of penicillin-binding proteins 1a, 1b, and 3. Proc Natl Acad Sci USA. 1980;77:4499–4503. doi: 10.1073/pnas.77.8.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ursinus A, Heike S, Höltje J-V. Purification of a nocardicin A-sensitive ld-carboxypeptidase from Escherichia coli by affinity chromatography. J Bacteriol. 1992;174:441–446. doi: 10.1128/jb.174.2.441-446.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vinella D, D’Ari R, Bouloc P. Penicillin binding protein 2 is dispensable in Escherichia coli when ppGpp synthesis is induced. EMBO J. 1992;11:1493–1501. doi: 10.1002/j.1460-2075.1992.tb05194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vinella D, Gagny B, Joseleau-Petit D, D’Ari R, Cashel M. Mecillinam resistance in Escherichia coli is conferred by loss of a second activity of the AroK protein. J Bacteriol. 1996;178:3818–3828. doi: 10.1128/jb.178.13.3818-3828.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vinella D, Joseleau-Petit D, Thévenet D, Bouloc P, D’Ari R. Penicillin-binding protein 2 inactivation in Escherichia coli results in cell division inhibition, which is relieved by FtsZ overexpression. J Bacteriol. 1993;175:6704–6710. doi: 10.1128/jb.175.20.6704-6710.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young, K. D. Unpublished data.
- 46.Yousif S Y, Broome-Smith J K, Spratt B G. Lysis of Escherichia coli by β-lactam antibiotics: deletion analysis of the role of penicillin-binding proteins 1A and 1B. J Gen Microbiol. 1985;131:2839–2845. doi: 10.1099/00221287-131-10-2839. [DOI] [PubMed] [Google Scholar]