Abstract
Analysis of the genome of Synechocystis sp. strain PCC 6803 reveals three open reading frames (slr0851, slr1743, and sll1484) that may code for type 2 NAD(P)H dehydrogenases (NDH-2). The sequence similarity between the translated open reading frames and NDH-2s from other organisms is low, generally not exceeding 30% identity. However, NAD(P)H and flavin adenine dinucleotide binding motifs are conserved in all three putative NDH-2s in Synechocystis sp. strain PCC 6803. The three open reading frames were cloned, and deletion constructs were made for each. An expression construct containing one of the three open reading frames, slr1743, was able to functionally complement an Escherichia coli mutant lacking both NDH-1s and NDH-2s. Therefore, slr0851, slr1743, and sll1484 have been designated ndbA, ndbB, and ndbC, respectively. Strains that lacked one or more of the ndb genes were created in wild-type and photosystem (PS) I-less backgrounds. Deletion of ndb genes led to small changes in photoautotrophic growth rates and respiratory activities. Electron transfer rates into the plastoquinone pool in thylakoids in darkness were consistent with the presence of a small amount of NDH-2 activity in thylakoids. No difference was observed between wild-type and the Ndb-less strains in the banding patterns seen on native gels when stained for either NADH or NADPH dehydrogenase activity, indicating that the Ndb proteins do not accumulate to high levels. A striking phenotype of the PS I-less background strains lacking one or more of the NDH-2s is that they were able to grow at high light intensities that were lethal to the control strain but they retained normal PS II activity. We suggest that the Ndb proteins in Synechocystis sp. strain PCC 6803 are redox sensors and that they play a regulatory role responding to the redox state of the plastoquinone pool.
Cyanobacteria are photosynthetic prokaryotes that contain complete respiratory electron transport chains on both the thylakoid and cytoplasmic membranes. In addition, the thylakoid membrane is utilized for photosynthetic electron transport, involving some of the same redox-active electron transport intermediates (quinone pool, cytochrome b6f complex, and soluble electron carriers) that are used for respiratory electron transfer (see reference 24 for a review). The photosynthetic electron transport chain has been extensively studied. However, comparatively little is known about respiration in cyanobacteria and knowledge about the pyridine nucleotide dehydrogenases has been expanded only recently (24). Membrane-bound bacterial pyridine nucleotide dehydrogenases can be divided into two classes that are commonly called type 1 and type 2 NAD(P)H dehydrogenases (NDH-1 and NDH-2, respectively) (for extensive reviews, see references 34 to 36). NDH-1 is a multisubunit complex that has a minimal form of 14 subunits in Escherichia coli (31, 32) and that contains four to six iron-sulfur clusters, translocates protons across the membrane, has flavin mononucleotide as the prosthetic group, and is inhibited by rotenone (even though this inhibition is not always complete). The NDH-2 complex consists of a single subunit, does not contain iron-sulfur clusters, does not appear to translocate protons across the membrane even though the complex is membrane associated, has flavin adenine dinucleotide (FAD) as the prosthetic group, and may be inhibited by flavone but not by rotenone.
In Synechocystis sp. strain PCC 6803 NDH-1 has been shown to be present in both the thylakoid and cytoplasmic membranes (3, 7). Genes for 11 ndh genes are present in the genome of Synechocystis sp. strain PCC 6803 (12). In comparison with proteins in E. coli and Paracoccus denitrificans, proteins similar to subunits involved in pyridine dinucleotide binding (nuoE, nuoF, and nuoG in E. coli; nqoB, nqoA, and nqoC in P. denitrificans) are missing in Synechocystis sp. strain PCC 6803. Other proteins may have taken over this function in the cyanobacterial system. Inactivation of single-copy genes encoding subunits of NDH-1 in Synechocystis sp. strain PCC 6803 has shown that NDH-1 plays a role in both photosynthesis and respiration. These strains are unable to concentrate inorganic carbon and require an environment enriched in CO2 to grow (2% [vol/vol] CO2 in air). Respiratory studies of strains lacking ndhB or ndhL (ΔndhB or ΔndhL strains) have shown that oxygen consumption rates were severely impaired (reference 19 and references therein). By monitoring oxidation and reduction kinetics of P700, the primary electron donor in photosystem I (PS I), in the wild-type strain and ΔndhB or ΔndhK strains, it has been shown that thylakoid-bound NDH-1 mediates the cyclic electron transport around PS I that is dependent on both NADPH and ferredoxin (17). Recently, a functional NDH-1 complex has been isolated from Synechocystis sp. strain PCC 6803 (16).
The issue regarding the presence, identity, and role of NDH-2s in cyanobacteria has received very little attention. A soluble flavin-containing NAD(P)H dehydrogenase consisting of a noncovalently bound octamer consisting of identical subunits of approximately 5 kDa each was purified from Microcystis aeruginosa (30). Neither the size, oligomerization, nor solubility of this complex is reminiscent of traditional NDH-2. Alpes et al. (1) purified and characterized an NADH-oxidizing enzyme that utilized quinone as the electron acceptor and that was tightly bound to the thylakoid membrane of Anabaena variabilis. The purified enzyme consisted of a subunit of 17 kDa together with a substoichiometric subunit of 52 kDa. It was also insensitive to rotenone and contained FAD, both of which are characteristics of NDH-2s. A study involving Anabaena sp. strain PCC 7120 indicated that this organism also contained an NDH-2 on the thylakoid membrane (11). Activity staining of solubilized membrane complexes from Synechocystis sp. strain PCC 6803 on native gels has shown the presence of an NADH-specific enzyme that has been tentatively assigned as an NDH-2 (15).
Analysis of the genome sequence of Synechocystis sp. strain PCC 6803 (12) revealed the presence of three putative open reading frames for NDH-2s: slr0851, slr1743, and sll1484. As will be presented in Results, these three open reading frames code for polypeptides with low but significant similarity to known NDH-2s. In this study these three putative NDH-2 genes and their products were investigated through the use of functional complementation and deletion mutagenesis.
MATERIALS AND METHODS
Synechocystis sp. strain PCC 6803 was cultivated in air at 30°C in modified BG-11 medium buffered with 10 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid]–NaOH (pH 8.0) (23). The BG-11 modification consisted of partial replacement of NaNO3 with an equal concentration of NH4NO3 (the final concentration of ammonia was 4.5 mM). For photomixotrophic growth, the medium was supplemented with 5 mM glucose. For growth on plates, 1.5% (wt/vol) agar and 0.3% (wt/vol) sodium thiosulfate were added, and BG-11 was supplemented with antibiotics appropriate for the particular strain (15 μg of zeocin ml−1, 25 μg of erythromycin ml−1, 25 μg of spectinomycin ml−1 and/or 35 μg of chloramphenicol ml−1). Strains in the wild-type background were grown under normal illumination (50 microeinsteins m−2 s−1), while those in the PS I-less background were grown at low light intensity (5 microeinsteins m−2 s−1). Growth was monitored by measuring optical density of the cells at 730 nm with a Shimadzu UV-160 spectrophotometer.
slr0851 was amplified by PCR with the primers 5′ndbA (AAACCATCTTggatCCAAGGCAACCCC, nucleotides 1,336,921 to 1,336,947; numbering is according to the CyanoBase sequence [4a]; nucleotides in lowercase letters represent changes made to the genome sequence to facilitate cloning) and 3′ ndbA (CCTGACGCCAAGCTTCTACCCTCC, nucleotides 1,339,022 to 1,339,045) and cloned in pUC19 by using restriction sites in the primers. A 915-bp ScaI-HincII fragment from the ndbA coding region (nucleotides 1,337,507 to 1,338,423) was deleted and replaced by an erythromycin resistance cassette from pRL425 (9) (Fig. 1A). Wild-type Synechocystis sp. strain PCC 6803 and a strain in which part of the psaAB operon coding for the PS I reaction center had been deleted (26) were transformed with this construct, and transformants were selected for on plates containing erythromycin. Transformants were subcultured at increasing concentrations of erythromycin to allow segregation of wild-type and mutant genome copies to occur and thus to allow us to obtain homozygous strains (designated ΔndbA strains). Segregation was confirmed by PCR (Fig. 1D).
FIG. 1.
Construction of the ndb deletion plasmids and PCR analysis of segregation of the ΔndbA, ΔndbB, and ΔndbC strains. (A to C) Schematic representations of the ndbA (A), ndbB (B), and ndbC (C) clones and deletion constructs. Arrows indicate the direction of transcription of the genes. EmR, erythromycin resistance cassette; SmR, spectinomycin resistance cassette; ZeoR, zeocin resistance cassette. The scales for panels A, B, and C are identical. (D to G) PCR analysis of segregation of the ΔndbA (D), ΔndbB (E), and ΔndbC (F and G) mutations to homozygosity.
By similar methods, PCR fragments containing slr1743 and sll1484 (nucleotides 1,330,445 to 1,332,434 and 3,389,921 to 3,391,972, respectively) were cloned into pUC19, and regions of the genes from nucleotides 1,331,048 to 1,331,946 and 3,390,675 to 3,391,392 were replaced by spectinomycin (22) and zeocin (6) resistance cassettes, respectively (Fig. 1B and C). Wild-type and PS I-less strains of Synechocystis sp. strain PCC 6803 were transformed with these constructs, as were strains that had previously been transformed with either one or both of the other ndb deletion constructs, to give the appropriate double and triple mutants. Strains in which ndbB had been deleted were designated ΔndbB strains, while those in which ndbC had been deleted were designated ΔndbC strains. Segregation was confirmed by PCR (Fig. 1E to G).
DNA from Synechocystis sp. strain PCC 6803 was prepared essentially as described in reference 33. RNA was isolated according to the method described in reference 18. The RNA was separated by electrophoresis on formaldehyde gels containing 1.2% agarose and transferred to GeneScreen Plus according to the manufacturer’s instructions. Probes for Northern blots were prepared by hot PCR with [32P]dATP by using the cloned genes as the template. For Northern blot analysis the same membrane was probed with all four probes used. Between two hybridizations the membrane was stripped of radioactivity by incubation in a solution of boiling 1% sodium dodecyl sulfate and was then verified to be nonradioactive before hybridization with another probe.
The plasmid containing the zeocin resistance cassette (pZeo1) was constructed by cloning an MscI-EcoRI fragment from pSP109 (27) into the SmaI and EcoRI sites of pUC19. This resulted in insertion of the ble gene, conferring zeocin resistance (6), in frame with lacZ.
Oxygen consumption measurements were carried out in a manner similar to that described previously for oxygen uptake (29), with the cells being dark adapted for approximately 10 min prior to being assayed. Chlorophyll a concentrations were determined according to the method described in reference 21.
Fluorescence measurements were performed with a Walz fluorometer (model PAM101), with the intensity of the modulated measuring light minimized and with both gain and damping at their maximum settings. Cells from PS I-less cultures in mid-exponential growth phase (optical density at 730 nm, ∼0.6) were harvested and resuspended in 10 mM HEPES–NaOH (pH 7.4) to a final chlorophyll concentration of 2 μg ml−1. Cells were dark adapted for 1 min prior to the measurement of F0. Fluorescence intensity was measured for 10 s, after which the measuring light (<0.01 microeinsteins m−2 s−1) was turned off. The cells were dark adapted for 30 s, 5 mM KCN was added, the fluorescence level was measured immediately, and fluorescence was subsequently measured for 1 s at intervals of approximately 30 s until 6 min had elapsed. At this time the cells were illuminated for 3 s with strong actinic light (150 microeinsteins m−2 s−1) to determine the maximum fluorescence yield (Fmax). This light intensity was sufficient to obtain Fmax in the PS I-less strain (data not shown). After we turned off the strong actinic light, the decay of the fluorescence yield was monitored.
Membranes from Synechocystis sp. strain PCC 6803 were prepared according to the method of reference 37. Protein complexes were solubilized and run on native slab gels according to the method of reference 14. After electrophoresis the native gels were incubated in 100 mM MOPS–NaOH (pH 7.0) for 30 min. The incubation buffer was changed, and nitroblue tetrazolium was added to a final concentration of 0.5 μg ml−1. NADH or NADPH was then added to a final concentration of 0.5 mM, and staining was allowed to proceed until blue bands (formazan resulting from enzymatic reduction of the tetrazolium salt) appeared.
RESULTS
Search for NDH-2 open reading frames in the Synechocystis sp. strain PCC 6803 genome.
When searching the genome sequence of Synechocystis sp. strain PCC 6803 for sequences similar to those of known NDH-2s, we identified three open reading frames (slr0851, slr1743, and sll1484). The polypeptides predicted to be encoded by these open reading frames have modest sequence identity with known NDH-2s from other organisms and with each other (Table 1). However, known NDH-2s from different organisms have low percentages of overall sequence identity to each other (Table 1), and the three predicted Synechocystis proteins show no significant similarity to any other protein in the database. Moreover, all three Synechocystis proteins have characteristic FAD and NADH binding motifs (Fig. 2). The proteins are hydrophilic, and only the product of sll1484 contains a hydrophobic stretch that might be long enough to span a membrane. As a first step to test if the Synechocystis open reading frames encode functional NDH-2s, attempts were made to functionally complement a strain of E. coli that lacks both NDH-1 and NDH-2.
TABLE 1.
Percentages of amino acid sequence identity of the putative NDH-2s from Synechocystis sp. strain PCC 6803 to known NDH-2 proteins from other organismsa
| Species | Identity (%) to:
|
||||
|---|---|---|---|---|---|
| B. s. | E. c. | S. c. | Slr0851 | Slr1743 | |
| Sll1484 | 18 | 27 | 24 | 27 | 33 |
| Slr1743 | 17 | 30 | 27 | 32 | |
| Slr0851 | 16 | 29 | 28 | ||
| S. c. | 16 | 27 | |||
| E. c. | 15 | ||||
Sequence identities were determined with the FASTA program from the Genetics Computer Group package (5) at default settings. Abbreviations: B. s., Bacillus sp. strain YN-1; E. c., E. coli; S. c., Saccharomyces cerevisiae. Sll1484, Slr0851, and Slr1743 are the three putative NDH-2s from Synechocystis sp. strain PCC 6803.
FIG. 2.
Alignment of the FAD and NADH binding motifs from the putative NDH-2s from Synechocystis sp. strain PCC 6803 with the motifs from known NDH-2s. The consensus motif for FAD and NADH binding (see references 2 and 25 for reviews of NADH and FAD binding motifs, respectively) is indicated above the alignment. Alignments were made with the PileUp program from the Genetics Computer Group package (5). ○, conserved hydrophobic residue; −, conserved negatively charged residue; B. sp. YN-1, Bacillus sp. strain YN-1.
Functional complementation of E. coli.
Expression constructs for two of the putative NDH-2s (slr0851 and slr1743) were created by amplifying the open reading frames by PCR and cloning them into pUC120 by using NcoI such that the open reading frames were in frame with that of lacZ and the protein produced under the lacZ promoter was identical to that predicted to be produced by the Synechocystis sp. strain PCC 6803 open reading frames. The resulting plasmids were used to transform E. coli MCW232, which lacks both NDH-1 and NDH-2 and which is unable to grow on minimal media with mannitol as the sole carbon source (4). As a control the MCW232 strain was also transformed with pUC120. Ampicillin-resistant transformants were selected on Luria-Bertani medium, and plasmid miniprep experiments were performed to verify that the strains contained the correct plasmid. These E. coli transformants were grown to mid-exponential phase on Luria-Bertani medium and then plated on minimal medium that contained mannitol as the sole carbon source. The strain that contained the slr1743 expression construct was able to grow on this medium, whereas with the other expression construct no convincing restoration of growth was observed (data not shown). However, the lack of heterologous functional complementation does not necessarily imply that Slr0851 is not an NDH-2; it may not properly associate or function in the E. coli system, which has a different quinone composition. As Slr1743 has clear NDH-2 activity and as Slr0851 and Sll1484 are rather similar to Slr1743 but not to other proteins in the database, we designate the corresponding open reading frames ndbA (slr0851), ndbB (slr1743), and ndbC (sll1484). The “b” in the ndb designation signifies type 2.
Testing the deletion strains for homozygosity.
With the primers used to amplify ndbA and ndbB for cloning, PCR was performed to verify that the strains created were homozygous for the ndbA and ndbB loci (Fig. 1D and E). As the size of the inserted zeocin cassette was similar to that of the region of ndbC that was deleted, two PCRs were performed to test segregation for this locus. Both utilized the 5′ primer that was used to clone ndbC. However, in one reaction the 3′ primer was internal to the region of ndbC that was deleted (Fig. 1F). After this reaction, a PCR product was present in the wild-type sample but not in the ΔndbC strains, indicating that they were indeed homozygous for the deletion. To confirm that the DNA used was PCR competent, we performed a second PCR in which the 3′ primer hybridized to the zeocin cassette (Fig. 1G). As expected, no PCR product was seen in the wild type but one was present in all of the ΔndbC strains.
Analysis of the Ndb-less strains.
Total RNA was isolated from the Δndb strains in both the wild-type and PS I-less backgrounds. Northern blot analysis was performed with probes to ndbA, ndbB, and ndbC. No transcripts for any of the three genes could be detected in any of the strains (data not shown), suggestive of a very low transcript abundance under all of the conditions tested. These conditions were photoautotrophic and photomixotrophic growth for strains containing PS I and photoheterotrophic growth at low light intensity for strains in the PS I-less background. All cultures were harvested during the exponential growth phase. Note that photomixotrophic growth refers to growth when glucose is present and whole-chain photosynthetic electron transport is active but that photoheterotrophic growth refers to conditions under which no whole-chain photosynthetic electron transfer can occur due to the presence of an inhibitor or the lack of one of the photosystems.
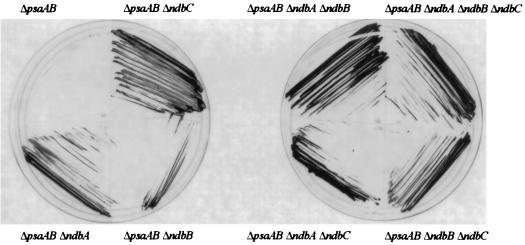
As shown in Table 2, the doubling times of the Δndb strains in the wild-type background were somewhat longer than that of the wild type when they were grown under photoautotrophic conditions, but there was no significant difference when they were grown photomixotrophically. In the PS I-less background at a light intensity of 5 microeinsteins m−2 s−1 the growth rates of the Δndb strains had the tendency to be a little higher than that of the parental strain (Table 2). Interestingly, these strains were able to survive and grow remarkably well at higher light intensities (50 microeinsteins m−2 s−1) that were lethal to the PS I-less strain. At higher light intensity, the plastoquinone (PQ) pool and cytochrome b6f complex are overreduced in the PS I-less strain, which is thought to lead to cell death (see references 26 and 29). To further illustrate the light tolerance of Ndb-less strains in a PS I-less background, in Fig. 3 vigorous growth of the PS I-less strains that lack one or more of the putative ndb genes is indicated at a light intensity of 30 microeinsteins m−2 s−1, at which intensity the parental strain dies.
TABLE 2.
Doubling times of Synechocystis sp. strain PCC 6803 mutants lacking one or more of the NDH-2s
| Genotype of strain | Doubling time (h)a
|
|||
|---|---|---|---|---|
| Wild type
|
PS I-less
|
|||
| PA | PM | PH at 5 microeinsteins m−2 s−1 | PH at 50 microeinsteins m−2 s−1 | |
| Parent | 10.6 ± 2.7 | 8.6 ± 2.6 | 21.9 ± 1.0 | ∞ |
| ΔndbA | 13.3 ± 3.6 | 10.9 ± 0.5 | 18.4 ± 2.2 | 27.5 ± 1.0 |
| ΔndbB | 13.3 ± 1.6 | 9.6 ± 1.9 | 19.3 ± 0.3 | 33.0 ± 3.1 |
| ΔndbC | 14.5 ± 1.5 | 9.4 ± 0.2 | 20.8 ± 0.2 | 19.5 ± 1.5 |
| ΔndbA ΔndbB | 14.9 ± 3.0 | 9.0 ± 0.7 | 18.5 ± 1.0 | 28.2 ± 3.5 |
| ΔndbA ΔndbC | 15.8 ± 3.2 | 9.1 ± 0.7 | 18.4 ± 1.0 | 21.0 ± 1.4 |
| ΔndbB ΔndbC | 12.9 ± 1.0 | 11.0 ± 0.9 | 20.2 ± 0.9 | 23.4 ± 1.5 |
| ΔndbA ΔndbB ΔndbC | 16.8 ± 2.0 | 9.9 ± 1.2 | 19.0 ± 0.7 | 26.5 ± 2.6 |
Values given are averages ± standard deviations from at least three determinations. Abbreviations for conditions: PA, photoautotrophic (with light but without glucose); PM, photomixotrophic (with light and glucose in the presence of linear photosynthetic electron transport); PH, photoheterotrophic (with light and glucose in the absence of linear photosynthetic electron transport).
FIG. 3.
Growth of the ΔpsaAB Δndb strains at a light intensity of 30 microeinsteins m−2 s−1. Cells were grown on plates at a light intensity of 5 microeinsteins m−2 s−1 and then streaked on fresh plates, which did not contain antibiotics, and incubated at a light intensity of 30 microeinsteins m−2 s−1 for 12 days. The ΔpsaAB control strain died at this intensity.
This increased light tolerance was not due to an impairment of PS II, as PS II activity was normal in all PS I-less, Ndb-less strains (data not shown). Table 3 shows the respiratory rates of Ndb-less strains and their parent strains. The increased respiratory rates in the PS I-less strains reflect the decreased amount of chlorophyll per cell; on a per cell basis the respiratory activities of wild-type and PS I-less control strains were similar. The respiratory rates in the Ndb-less strains were generally somewhat higher than in the parent strain. However, the slight increase in the respiratory rates of strains of the PS I-less background is insufficient to be able to account for the approximately 10-fold increase in light tolerance.
TABLE 3.
Respiratory rates of Synechocystis sp. strain PCC 6803 mutants lacking one or more NDH-2s
| Genotype of strain | Oxygen uptake (μmol of O2 mg of chl−1 h−1)a
|
|
|---|---|---|
| Wild type | PS I-less | |
| Parent | 50 ± 11 | 386 ± 49 |
| ΔndbA | 69 ± 15 | 470 ± 66 |
| ΔndbB | 77 ± 3 | 542 ± 74 |
| ΔndbC | 84 ± 14 | 377 ± 69 |
| ΔndbA ΔndbB | 81 ± 17 | 545 ± 110 |
| ΔndbA ΔndbC | 89 ± 10 | 524 ± 78 |
| ΔndbB ΔndbC | 93 ± 20 | 512 ± 142 |
| ΔndbA ΔndbB ΔndbC | 88 ± 17 | 509 ± 136 |
Values given are averages ± standard deviations from at least three determinations. Oxygen consumption of whole cells was measured in 10 mM HEPES–NaOH (pH 7.0). Strains in the wild-type background were grown photomixotrophically at 50 microeinsteins m−2 s−1, while those in the PS I-less background were grown photoheterotrophically at 5 microeinsteins m−2 s−1. chl, chlorophyll.
Monitoring PQ pool reduction with chlorophyll fluorescence.
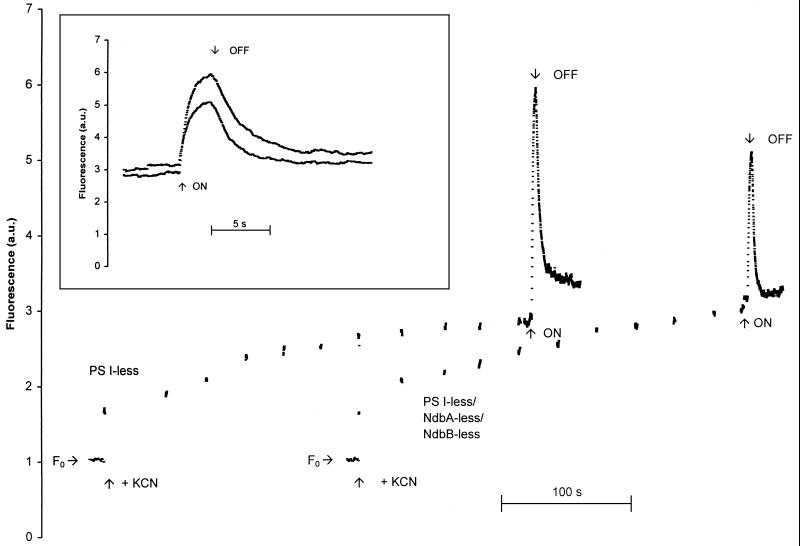
The results presented thus far do not provide evidence that NDH-2s contribute to respiratory electron flow. To test this further we wished to monitor the rate of electron flow into the PQ pool from sources other than PS II. A method of determining this electron flow is to monitor the reduction of the primary electron-accepting PQ in PS II (QA) in darkness in the presence of KCN, which blocks electron flow out of the PQ pool. When the PQ pool is sufficiently reduced, QA will become reduced as well, which can be monitored by the chlorophyll fluorescence yield. QA is a quencher whereas QA− is not. A typical fluorescence yield trace is shown in Fig. 4. A PS I-less strain was used for these studies because the variable fluorescence yield is much higher in such strains and because excitation of PS I by the measuring beam cannot oxidize the PQ pool. F0 was measured for 10 s prior to the addition of KCN, and no rise in variable fluorescence was seen, indicating that the measuring beam by itself did not lead to a significant accumulation of QA−. Upon addition of KCN, we detected an immediate (within several seconds) rise in variable fluorescence, reflecting reduction of the quinone pool and a partial reduction in QA due to inhibition of oxidases. This rise was independent of the concentration of KCN in the range of 1 to 5 mM and was not affected by the presence of an equal concentration of bicarbonate (data not shown). This rise in variable fluorescence upon the addition of KCN is therefore not due to phenomena such as the binding of cyanide to the nonheme iron of PS II (13), as the binding of cyanide to the nonheme iron of PS II is reversed by the addition of bicarbonate and requires concentrations of CN− that are at least an order of magnitude higher than those used in this study. Instead, we interpret the rise in variable fluorescence to be due to inhibition of the oxidase by KCN. After the initial nearly immediate QA reduction upon KCN addition, a slower increase in fluorescence yield was observed, with an initial nonlinear phase that possibly reflects additional QA reduction. This initial phase was followed by a linear, very slow increase in fluorescence yield. This slow increase continued for at least 15 min (data not shown) and may reflect further changes in the QA redox state. For this reason, this slow, linear increase is not considered further in this study. Illumination with strong actinic light showed that electron donation to the PQ pool in darkness, in the presence of KCN, did not reduce all QA (Fig. 4). After the actinic light was turned off, the level of fluorescence decreased slowly, indicating the presence of a pathway for electrons out of the PQ pool (presumably oxidation by oxygen), perhaps coupled to a reoxidation of QA− by components on the donor side of PS II. The changes seen in fluorescence yield cannot be attributed to state transitions, as dark-adapted PS I-less cells are predominantly in state 1 (26). Therefore, upon illumination no state transition can occur.
FIG. 4.
Monitoring of variable fluorescence in darkness in the PS I-less strain and the PS I-less-, NdbA-less-, and NdbB-less strain. Cells were harvested in mid-exponential growth phase and concentrated to a chlorophyll concentration of 2 μg ml−1 in 10 mM HEPES–NaOH (pH 7.4). KCN was added to a final concentration of 5 mM where indicated. The time points at which high-intensity actinic light (150 microeinsteins m−2s−1) was turned on and off have been indicated as well. The inset is an enlargement of the trace when Fmax and the subsequent decay of chlorophyll fluorescence were monitored during and after actinic illumination. The traces have been normalized to F0. The traces have been offset in the x direction in the main graph but not in the inset. a.u., arbitrary units.
The amplitudes of the immediate increases in fluorescence yields and the amplitudes and half-times (t1/2) of the noninstantaneous and nonlinear increases in variable fluorescence after the addition of KCN were similar in strains that lacked one or more of the ndb genes as compared to the control strain, even though they were found to vary significantly from culture to culture (Table 4). Therefore, the net flow of electrons into the PQ pool is essentially independent of the presence of NDH-2s. The level of the Fmax varied from culture to culture; however, the Fmax of any of the Ndb-less strains was not significantly different from that of the parental strain (Table 4). In the Δndb strains with multiple ndb genes deleted, the t1/2 of the decay of chlorophyll fluorescence after the actinic light was turned off was somewhat faster than in the control strain, which suggests a small role for the NDH-2s in electron transfer into the PQ pool, but this effect seems to be too small to be physiologically relevant (Table 4).
TABLE 4.
Characteristics of variable fluorescence (QA reduction and QA− reoxidation) of PS I-less strains measured in darkness in the presence of KCNa
| Genotype of strain | A (KCN) (AU) | A (increase) (AU) | t1/2 (increase) (s) | Fmax (AU) | t1/2 (decay) (s) |
|---|---|---|---|---|---|
| Parent | 0.39 ± 0.17 | 0.29 ± 0.12 | 35.3 ± 7.0 | 4.2 ± 1.0 | 2.5 ± 0.5 |
| ΔndbA | 0.44 ± 0.23 | 0.36 ± 0.20 | 44.3 ± 17.3 | 4.3 ± 1.0 | 2.3 ± 0.3 |
| ΔndbB | 0.38 ± 0.16 | 0.39 ± 0.17 | 44.8 ± 18.3 | 3.9 ± 0.7 | 1.8 ± 0.4 |
| ΔndbC | 0.30 ± 0.16 | 0.27 ± 0.06 | 33.7 ± 4.9 | 4.0 ± 1.1 | 2.1 ± 0.3 |
| ΔndbA ΔndbB | 0.29 ± 0.23 | 0.40 ± 0.28 | 41.4 ± 26.0 | 3.6 ± 1.0 | 1.4 ± 0.2 |
| ΔndbA ΔndbC | 0.28 ± 0.13 | 0.28 ± 0.23 | 39.8 ± 24.6 | 3.7 ± 1.2 | 1.8 ± 0.4 |
| ΔndbB ΔndbC | 0.20 ± 0.10 | 0.27 ± 0.12 | 41.6 ± 17.9 | 3.6 ± 1.1 | 1.7 ± 0.4 |
| ΔndbA ΔndbB ΔndbC | 0.31 ± 0.17 | 0.42 ± 0.37 | 42.0 ± 26.3 | 3.7 ± 1.2 | 1.7 ± 0.3 |
Values given are averages ± standard deviations from at least four determinations. Chlorophyll fluorescence of whole cells was measured in 10 mM HEPES–NaOH (pH 7.4) in the presence of 5 mM KCN. The amplitude of the immediate increase upon addition of KCN [A (KCN)], the amplitude and kinetics (t1/2) of the subsequent noninstantaneous and nonlinear increase in variable fluorescence [A (increase) and t1/2 (increase), respectively], the Fmax upon illumination with actinic light, and the t1/2 of decay after actinic illumination [t1/2 (decay)] are indicated. See Fig. 4 for an example of the experimental data from which these values were determined. AU, arbitrary units.
Native gel electrophoresis.
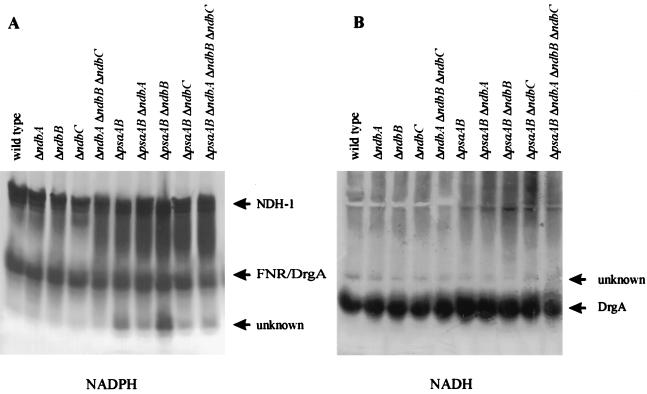
Membrane proteins were solubilized from total membranes of strains that lacked one or all three of the putative NDH-2s, separated on nondenaturing gels, and stained for NADH and NADPH activities. Each sample contained approximately the same amount of solubilized protein, as was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). No differences in NADPH- and NADH-oxidizing activities were seen between the control strains and strains that lacked one or more of the NDH-2s (Fig. 5). We presume that the upper NADPH-specific band (Fig. 5A) represents NDH-1, as an NADPH-specific band of similar size has been identified as NDH-1 by other workers (16); slight NADH-oxidizing activity is found at this position (Fig. 5B). This activity is more intense in strains that lack PS I. A smaller complex that oxidizes NADPH and NADH is also seen (Fig. 5). This complex has been identified as the product of drgA, and the purified protein has similar activity with both NADH and NADPH (15). In our system this band appears as a doublet upon NADPH staining, and we interpret the doublet to represent both DrgA and ferredoxin NADP+ oxidoreductase. Below this is an NADPH-specific band of unknown origin. A faint NADH-specific oxidizing activity is also present in all strains (Fig. 5B); therefore, this activity cannot be due to NDH-2 activity as was proposed by Matsuo et al. (15).
FIG. 5.
Native gels of membrane protein complexes from Synechocystis sp. strain PCC 6803. Protein complexes extracted from membranes of the wild-type strain, the PS I-less strain (ΔpsaAB) and Δndb mutants in these backgrounds were separated on a native gel and stained for NADPH (A)- and NADH (B)-oxidizing activities. Activity assignments have been indicated. Note that the two gels were run independently of each other; therefore, the relative positions of the activity bands cannot be compared directly between the two gels.
To address the possibility that in the cyanobacterial system NDH-2s are easily extracted from the membranes upon thylakoid preparation, the soluble (cytoplasmic, lumenal, and periplasmic) fraction obtained upon thylakoid preparation was also subjected to native gel electrophoresis. The results are shown in Fig. 6. The high-activity band that stains with both NADPH and NADH is probably DrgA, as it migrates to a position similar to that of the band presumed to be DrgA in the membrane samples (Fig. 5). The activity of this band upon NADPH staining is significantly higher than that upon NADH staining; we interpret this to be the result of comigration of ferredoxin NADP+ oxidoreductase with DrgA. The identities of the other bands are unknown. However, the upper NADPH-specific band that is present only in the PS I-less strains (Fig. 6A) may be the peripheral arm of NDH-1, which was removed from the membranes during sample preparation. The absence of this band from the strains in the wild-type background may simply be the result of insufficient amounts of the complex to stain, as in the membrane fractions the band presumed to be NDH-1 had higher activity in the strains which lack PS I (Fig. 5). We could distinguish no band in this gel that disappeared in the mutant lacking NDH-2s. For this reason, NDH-2s do not seem to accumulate in cyanobacteria in sufficient amounts to be visible after an activity staining. However, NDH-2s are expressed in Synechocystis sp. strain PCC 6803, as their deletion leads to a clear phenotype in the PS I-less strain. Moreover, the lowest NADH-specific band is present in somewhat reduced amounts in strains that lack the NDH-2s compared to the amount in the parental strain, indicating that the NDH-2s may be involved in the regulation of this protein.
FIG. 6.
Native gels of soluble protein complexes from Synechocystis sp. strain PCC 6803. Protein complexes from the soluble fraction of the wild-type strain, the PS I-less strain (ΔpsaAB), and the NDH-2-less mutants (ΔndbA ΔndbB ΔndbC) in these backgrounds were separated on a native gel and stained for NADPH (A)- and NADH (B)-oxidizing activities.
DISCUSSION
The ndb genes encode functional NDH-2s.
Structural analysis of the three putative NDH-2s revealed that they all contain NADH- and FAD-binding motifs (Fig. 2). Both motifs consist of a β sheet-α helix-β sheet structure which contains (i) a glycine-rich phosphate binding consensus sequence (GXGXXG); (ii) a conserved negatively charged residue (D or E) at the end of the second β sheet; and (iii) six positions typically occupied by small hydrophobic residues, four of which are marked in the NADH binding motif in Fig. 2 and the other two of which are generally positioned 2 and 4 residues before the conserved Asp or Glu. All six residues are marked in the FAD binding motif in Fig. 2 (see references 2 and 25 for reviews of NAD(P)H and FAD binding motifs, respectively). In NADPH binding proteins the third G in the GXGXXG motif associated with NADPH binding is generally replaced by S, A or P, and the negative charge at the end of the second β sheet is missing (2). As these features are absent in the NDH-2s, it is likely that all three NDH-2s in Synechocystis sp. strain PCC 6803 bind NADH and not NADPH.
All three NDH-2s contain the conserved nucleotide binding domain for FAD and also a second motif downstream from this that is characteristic of FAD binding motifs (Fig. 2). This motif contains a conserved aspartate that forms a hydrogen bond with the ribitol moiety of FAD (8). Thus, all three NDH-2s have the potential to bind both NADH and FAD, consistent with the expectation for functional NDH-2s.
No ndb homologs have been detected in chloroplast genomes. However, this does not necessarily imply that the corresponding proteins are missing in plant chloroplasts. As many genes coding for hydrophilic chloroplast proteins have been transferred to the nucleus, it is possible that chloroplast NDH-2 genes, if they exist, are in the nucleus.
The ability of ndbB to functionally complement a strain of E. coli that lacks NDHs shows that it encodes a functional NDH-2. This ability to complement also shows that NdhB is able to utilize other quinones besides PQ as the electron acceptor because E. coli does not contain PQ. Even though NdbA did not functionally complement MCW232, this does not imply that it does not encode a functional NDH-2. Instead, the protein may not have been functional in E. coli because of improper folding or because of an inability to utilize ubiquinone or menaquinone as a substrate.
As deletion of any one of the three NDH-2s resulted in greatly increased light tolerance of the PS I-less strain (Table 2 and Fig. 3), and as ndbB encodes a functional NDH-2, we presume that ndbA and ndbC also encode functional NDH-2s in Synechocystis sp. strain PCC 6803. This presumption is supported by the fact that they contain the structural motifs necessary to bind NADH and FAD. Moreover, in database searches with NdbA and NdbC as search sequences, known NDH-2s from other organisms come up with the highest level of identity.
The physiological role of NDH-2 in Synechocystis sp. strain PCC 6803.
The physiological role of the NDH-2s in Synechocystis sp. strain PCC 6803 does not appear to be a simple metabolic one to oxidize NADH. The increased light tolerance of the PS I-less strains that lack one or more of the ndb genes is not due to an impairment of PS II activity, as oxygen evolution rates and the kinetics of fluorescence induction in the presence of 3-(3,4-dichlorophenyl)-1,1-dimethyl urea are similar in the PS I-less strain and the strains lacking the NDH-2s (data not shown) and cannot be explained by the slight increase in respiratory rates (Table 3). It is unlikely that the high light tolerance is an artifact, as the single deletions represent three separate transformation events at different times. If the high light tolerance of the PS I-less and NDH-2-less strains were an artifact, it should also have been seen in the parental strain, which was maintained under the same conditions (Table 2 and Fig. 3).
The increase in respiratory rates is less than a doubling, and this is clearly not sufficient to explain the ability to tolerate a 10-fold increase in light intensity (Table 2) and thereby a 10-fold-higher flow of electrons into the PQ pool via the PS II pathway. One might argue that a small increase in respiratory rates does not necessarily equate to a similarly small increase in respiratory electron flow in thylakoids, as both cytoplasmic and thylakoid membranes participate in respiration. However, as thylakoids contribute very significantly to overall respiratory activity in cyanobacteria (20), and as respiratory electron flow into the thylakoid PQ pool in darkness is not significantly altered in strains lacking ndb genes (Fig. 4 and Table 4), a possible NDH-2 function in respiration cannot explain the drastic change in light tolerance in PS I-less strains upon deletion of ndb genes. Moreover, the ability of Δndb strains to grow at air levels of CO2 indicates that, unlike NDH-1 (19), the NDH-2s do not play an essential role in the CO2-concentrating mechanism that these organisms possess.
Even though NDH-2s do not appear to donate electrons to the PQ pool at a metabolically relevant rate, our data are suggestive of the presence of NDH-2s in the thylakoid membrane. The Δndb strains in the PS I-less background with multiple deletions had half-lives for the decay of chlorophyll fluorescence in the presence of KCN after illumination with strong actinic light that were somewhat faster than that of the parent strain (Table 4). This suggests a decreased rate of respiratory electron donation to the PQ pool in the absence of the NDH-2s, and therefore the three NDH-2s are expected to be present in thylakoids. As ΔndbA and ΔndbC single-deletion strains did not show this phenotype, individually, NDH-2s may be dispensable for a maximum rate of electron donation to the PQ pool; however, in the absence of two or three NDH-2s, reduction of the PQ pool in darkness is slowed down.
Activity staining of native gels for NADH and NADPH activities showed no changes in the banding patterns between strains that lacked one or more of the ndb genes and the control strain in either the wild-type or the PS I-less background (Fig. 5 and 6). The similarity in activity staining between wild-type and the Δndb strains on native gels confirms that the NDH-2s are minor components in cyanobacterial membranes and that they cannot be detected by activity measurements. These results, combined with the minor effects on electron flow into the PQ pool and the slight effect on respiration but the large change in light tolerance of the PS I-less strains, suggest that the NDH-2s do not have a significant catalytic role in respiration. Instead, we suggest that they play a regulatory role and that the most significant change seen in cell physiology in the absence of one or more of the NDH-2s (i.e., increased light tolerance of the PS I-less strains) may be due to an altered regulation of gene expression when the PQ pool is highly reduced.
One attractive possibility is that the NDH-2s serve as redox sensors of the membrane (PQ pool) and soluble fraction (NADH). It is known that the redox state of the membrane can modulate kinase activity (see references 10 and 28 for reviews), even though the details of signaling are unknown. In this scenario, in the absence of one or more of the NDH-2 sensors, regulatory processes may be altered, resulting in the increased light tolerance of the PS I-less strains that lack one or more of the NDH-2s.
ACKNOWLEDGMENTS
We thank Satoshi Tabata (Kazusa DNA Research Institute) for access to portions of the Synechocystis sp. strain PCC 6803 genome sequence prior to release. E. coli MCW232 was a gift from Bob Gennis (University of Illinois).
This work was supported by grant 97-35306-4881 from the U.S. Department of Agriculture National Research Initiative Competitive Grants Program. We also acknowledge support by the Human Frontiers Science Program Organization (RG0051/1997M). Pacer Udall was supported by a Howard Hughes undergraduate fellowship through the Biology Research Experience for Undergraduates Program at Arizona State University.
REFERENCES
- 1.Alpes I, Scherer S, Böger P. The respiratory NADH dehydrogenase of the cyanobacterium Anabaena variabilis: purification and characterisation. Biochim Biophys Acta. 1989;973:41–46. [Google Scholar]
- 2.Bellamacina C R. The nicotinamide dinucleotide binding motif: a comparison of nucleotide binding proteins. FASEB J. 1996;10:1257–1269. doi: 10.1096/fasebj.10.11.8836039. [DOI] [PubMed] [Google Scholar]
- 3.Berger S, Ellersiek U, Steinmüller K. Cyanobacteria contain a mitochondrial complex I-homologous NADH-dehydrogenase. FEBS Lett. 1991;286:129–132. doi: 10.1016/0014-5793(91)80957-5. [DOI] [PubMed] [Google Scholar]
- 4.Calhoun M W, Gennis R B. Demonstration of separate genetic loci encoding distinct membrane-bound respiratory NADH dehydrogenases in Escherichia coli. J Bacteriol. 1993;175:3013–3019. doi: 10.1128/jb.175.10.3013-3019.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.CyanoBase Website. 2 March 1999, revision date. Sequences. [Online.] Kazusa DNA Research Institute. http://www.kazusa.or.jp/cyano/cyano.html. [13 April 1999, last date accessed.]
- 5.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dracourt D, Calmels T P G, Reyens J P, Baron M, Tiraby G. Cassettes of the Streptoalloteichus hindustanus ble gene for transformation of lower and higher eukaryotes to phleomycin resistance. Nucleic Acids Res. 1990;18:4009. doi: 10.1093/nar/18.13.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dzelzkalns V A, Obinger C, Regelsberger G, Niederhauser H, Kamensek M, Peschek G A, Bogorad L. Deletion of the structural gene for the NADH-dehydrogenase subunit of Synechocystis 6803 alters respiratory properties. Plant Physiol. 1994;106:1435–1442. doi: 10.1104/pp.106.4.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eggink G, Engel H, Vriend G, Terpstra P, Witholt B. Rubredoxin reductase of Pseudomonas oleovorans. Structural relationship to other flavoprotein oxidoreductases based on one NAD and two FAD fingerprints. J Mol Biol. 1990;212:135–142. doi: 10.1016/0022-2836(90)90310-I. [DOI] [PubMed] [Google Scholar]
- 9.Elhai J, Wolk C P. A versatile class of positive selection vectors based on the non-viability of palindrome containing plasmids that allow cloning into long polylinkers. Gene. 1988;68:119–138. doi: 10.1016/0378-1119(88)90605-1. [DOI] [PubMed] [Google Scholar]
- 10.Gal A, Zer H, Ohad I. Redox-controlled thylakoid protein phosphorylation. News and views. Physiol Plant. 1997;100:869–885. [Google Scholar]
- 11.Howitt C A, Smith G D, Day D A. Cyanide-insensitive oxygen uptake and pyridine nucleotide dehydrogenases in the cyanobacterium Anabaena PCC7120. Biochim Biophys Acta. 1993;1141:313–320. [Google Scholar]
- 12.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosochi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 13.Koulougliotis D, Kostopoulos T, Petrouleas V, Diner B A. Evidence for CN− binding at the PS II non-heme Fe2+. Effects on the EPR signal for QA−Fe2+ and on QA/QB electron transfer. Biochim Biophys Acta. 1993;1141:275–282. [Google Scholar]
- 14.Kuonen D R, Roberts P J, Cottingham I R. Purification and analysis of mitochondrial proteins on non-denaturing gradient polyacrylamide gels. Anal Biochem. 1986;153:221–226. doi: 10.1016/0003-2697(86)90084-9. [DOI] [PubMed] [Google Scholar]
- 15.Matsuo M, Endo T, Asada K. Isolation of a novel NAD(P)H-quinone oxidoreductase from the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol. 1998;39:751–755. doi: 10.1093/oxfordjournals.pcp.a029430. [DOI] [PubMed] [Google Scholar]
- 16.Matsuo M, Endo T, Asada K. Properties of the respiratory NAD(P)H dehydrogenase isolated from the cyanobacterium Synechocystis PCC6803. Plant Cell Physiol. 1998;39:263–267. doi: 10.1093/oxfordjournals.pcp.a029366. [DOI] [PubMed] [Google Scholar]
- 17.Mi H, Endo T, Ogawa T, Asada K. Thylakoid membrane-bound, NADPH-specific pyridine nucleotide dehydrogenase complex mediates cyclic transport in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Cell Physiol. 1995;36:661–668. [Google Scholar]
- 18.Mohamed A, Jansson C. Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant Mol Biol. 1989;13:693–700. doi: 10.1007/BF00016024. [DOI] [PubMed] [Google Scholar]
- 19.Ogawa T. NAD(P)H dehydrogenase: a component of PS-I cyclic electron flow driving inorganic carbon transport in cyanobacteria. In: Murata N, editor. Research in photosynthesis. III. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1992. pp. 763–770. [Google Scholar]
- 20.Peschek G A. Cytochrome oxidases and the cta operon of cyanobacteria. Biochim Biophys Acta. 1996;1275:27–32. [Google Scholar]
- 21.Porra R J, Thompson W A, Kriedemann P E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of concentration of chlorophyll standards by atomic absorbtion spectroscopy. Biochim Biophys Acta. 1989;975:384–394. [Google Scholar]
- 22.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 23.Rippka R, Deruelles J, Waterbury J B, Herdmann M, Stanier R Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 24.Schmetterer G. Cyanobacterial respiration. In: Bryant D A, editor. The molecular biology of cyanobacteria. Vol. 1. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 409–435. [Google Scholar]
- 25.Schulz G E. Binding of nucleotides by proteins. Curr Opin Struct Biol. 1992;2:61–67. [Google Scholar]
- 26.Shen G, Boussiba S, Vermaas W F J. Synechocystis sp. PCC 6803 strains lacking photosystem I and phycobilisome function. Plant Cell. 1993;5:1853–1863. doi: 10.1105/tpc.5.12.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stevens D R, Rochaix J-R, Purton S. The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Mol Gen Genet. 1996;251:23–30. doi: 10.1007/BF02174340. [DOI] [PubMed] [Google Scholar]
- 28.Vener A V, Ohad I, Andersson B. Protein phosphorylation and redox sensing in chloroplast thylakoids. Curr Opin Plant Biol. 1998;1:217–223. doi: 10.1016/s1369-5266(98)80107-6. [DOI] [PubMed] [Google Scholar]
- 29.Vermaas W F J, Shen G, Styring S. Electrons generated by photosystem II are utilized by an oxidase in the absence of photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 1994;337:103–108. doi: 10.1016/0014-5793(94)80638-1. [DOI] [PubMed] [Google Scholar]
- 30.Viljoen C C, Cloete F, Scott W E. Isolation and characterisation of an NAD(P)H dehydrogenase from the cyanobacterium Microcystis aeruginosa. Biochim Biophys Acta. 1985;827:247–259. [Google Scholar]
- 31.Walker J E. The NADH:ubiquinone oxidoreductase (complex I) of respiratory chains. Q Rev Biophys. 1992;25:253–324. doi: 10.1017/s003358350000425x. [DOI] [PubMed] [Google Scholar]
- 32.Weidner U, Geire S, Ptock A, Freidrich T, Leif H, Weiss H. The gene locus of the proton-translocating NADH:ubiquinone oxidoreductase in Escherichia coli. Organisation of the 14 genes and relationship between the derived proteins and subunits of mitochondrial complex I. J Mol Biol. 1993;233:109–122. doi: 10.1006/jmbi.1993.1488. [DOI] [PubMed] [Google Scholar]
- 33.Williams J G K. Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 1988;167:766–778. [Google Scholar]
- 34.Yagi T. The bacterial energy-transducing NADH-quinone oxidoreductases. Biochim Biophys Acta. 1993;1141:1–17. doi: 10.1016/0005-2728(93)90182-f. [DOI] [PubMed] [Google Scholar]
- 35.Yagi T. Bacterial NADH-quinone oxidoreductases. J Bioenerg Biomembr. 1991;23:211–225. doi: 10.1007/BF00762218. [DOI] [PubMed] [Google Scholar]
- 36.Yagi T. Procaryotic complex I (NDH-1), an overview. Biochim Biophys Acta. 1998;1364:125–133. doi: 10.1016/s0005-2728(98)00023-1. [DOI] [PubMed] [Google Scholar]
- 37.Yu J, Vermaas W F J. Synthesis and turnover of photosystem II reaction center polypeptides in cyanobacterial D2 mutants. J Biol Chem. 1993;268:7407–7413. [PubMed] [Google Scholar]