Abstract
The PulC component of the Klebsiella oxytoca pullulanase secretion machinery (the secreton) was found by subcellular fractionation to be associated with both the cytoplasmic (inner) and outer membranes. Association with the outer membrane was independent of other secreton components, including the outer membrane protein PulD (secretin). The association of PulC with the inner membrane is mediated by the signal anchor sequence located close to its N terminus. These results suggest that PulC forms a bridge between the two membranes that is disrupted when bacteria are broken open for fractionation. Neither the signal anchor sequence nor the cytoplasmic N-terminal region that precedes it was found to be required for PulC function, indicating that PulC does not undergo sequence-specific interactions with other cytoplasmic membrane proteins. Cross-linking of whole cells resulted in the formation of a ca. 110-kDa band that reacted with PulC-specific serum and whose detection depended on the presence of PulD. However, antibodies against PulD failed to react with this band, suggesting that it could be a homo-PulC trimer whose formation requires PulD. The data are discussed in terms of the possible role of PulC in energy transduction for exoprotein secretion.
In gram-negative bacteria, the secretion of extracellular proteins (exoproteins) from the periplasm usually occurs via the secreton, the main terminal (type II) branch of the general secretory pathway (35). Studies of the secretons of several bacteria indicate similarities in the sequences and locations of their components, and in the arrangements of the genes that encode them, although there are several potentially important differences. For example, the number of proteins known or thought to be involved varies between 12 (Pseudomonas aeruginosa) and 15 (Aeromonas hydrophila) (35), and there are unexplained differences in energy requirements for secreton function in different bacteria (9, 23, 32). Nevertheless, conserved secreton components are often interchangeable (14, 24, 33). Despite these overall similarities, however, exoproteins secreted via the secreton in one species of bacteria are often not secreted when they are produced by a bacterium with a different secreton. This implies that secreton components recognize compatible exoproteins and fail to recognize exoproteins that are secreted by different secretons.
The most extensive study of the ability of individual secreton components to function in heterologous secretons (24) identified two proteins that might confer substrate specificity. These two proteins, OutC and OutD, are the only components of the Erwinia carotovora secreton that cannot function in the closely related Erwinia chrysanthemi. OutC and OutD are homologous to PulC and PulD of the Klebsiella oxytoca pullulanase secreton (11) and to XcpP (XcpC) and XcpQ (XcpD) of the P. aeruginosa secreton (1). PulD, OutD, and XcpQ are all members of the secretin family of multimeric outer membrane proteins (3, 10, 16, 17, 25, 42) that could form the channel through which exoproteins cross the outer membrane. One member of this family, OutD, is thought to function as the receptor for exoproteins that are secreted by the E. chrysanthemi Out secreton (42). PulC, OutC, and XcpP are all predicted to be anchored in the cytoplasmic membrane by a signal anchor sequence and to possess a short N-terminal cytoplasmic domain and a large C-terminal periplasmic domain (6, 11, 44) that includes a PDZ motif (PulC and OutC) or a predicted coiled-coil region (XcpP) (31). It has been proposed that XcpP might function in a manner analogous to that proposed for TonB (21, 22, 43) to transduce energy for the movement of exoproteins through the secretin channel (5).
This report describes studies of the PulC protein. In particular, we have determined its location in the cell envelope by subcellular fractionation and studied the effects of other secreton components on its location, its possible interaction with PulD, and the role of its signal anchor sequence.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The Escherichia coli K-12 strain used for most experiments was PAP105 [Δ(lac-pro) F′ lacIq pro+ Tn10]. E. coli K-12 PAP7232 (36) was used for transdominance assays. This strain carries the entire pullulanase secreton gene cluster from K. oxytoca inserted into the chromosome. pCHAP231 is a pBR322 derivative that carries the same gene cluster (12). Other plasmids are described below. Bacteria were grown in Luria-Bertani (LB) medium (28) at 30°C. When maltose (0.4%) was used, the medium was buffered with M63 salts solution (28) at 1/10 the normal concentration. Except where indicated otherwise, antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; and chloramphenicol, 25 μg/ml. Isopropyl β-d-thiogalactoside (IPTG) was used at 1 mM.
Cloning of pulC.
To facilitate genetic modification and analysis of pulC, we constructed four different plasmids. pCHAP5005 was constructed by subcloning a PstI-BglII fragment extending from the beginning of pulA through to close to the end of pulD from pCHAP231 into pUC19 in which the SmaI site has been destroyed by inserting a 12-bp XhoI linker. The insert in pCHAP5005 was subcloned into pSU18 (2) to give pCHAP5011. In pSU18 derivative pCHAP5001, PulC encoded by a PCR-amplified fragment carries eight substitutions as a result of mutations inadvertently introduced by PCR. Finally, a KpnI-ClaI fragment of pCHAP231 encompassing the whole of pulC and the beginning of pulD was cloned into pSU18 to give pCHAP2277. pCHAP2284 is a derivative of pCHAP2277 in which a StuI site was introduced by site-directed mutagenesis (20) at codon 25, thereby changing residue 25 (alanine) to arginine. These four plasmids were indistinguishable with respect to yields of PulC and complementation of ΔpulC.
Construction of pCHAP1229 (ΔpulC) and pCHAP1226 (ΔpulD).
The pulC mutation was constructed in pCHAP5005. The plasmid was linearized with NsiI (codons 7 to 8 of pulC) and then religated in the presence of a compatible linker that inserted a Bst1107I site to give pCHAP1203. This plasmid was then linearized with SmaI (codons 219 to 221 of pulC), and a second Bst1107I linker was inserted (to give pCHAP1208). This plasmid was then cleaved with Bst1107I and ligated in the presence of the nonpolar SmaI aphA-3 cassette (kan2) coding for resistance to kanamycin (26) to give pCHAP1222. The entire pulC::kan2 region was then subcloned from pCHAP1222 into pUC18Cm. The resulting plasmid was linearized with EcoRI and HindIII and electroporated into a recD mutant carrying pCHAP231. Plasmid DNA from kanamycin-resistant transformants was transferred into strain PAP105 with selection for resistance to ampicillin and kanamycin on medium containing maltose. DNA from one transformant (pCHAP1229) was shown by restriction analysis to carry the ΔpulC::kan2 mutation. This strain produced but failed to secrete pullulanase, but secretion was fully restored upon introduction of pCHAP5001, confirming the presence of a nonpolar pulC mutation.
To transfer the ΔpulD mutation in the chromosomal pullulanase secretion gene cluster in strain PAP7447 (16) onto pCHAP231, a kanamycin resistance cassette (kan1) was first inserted into the pulB gene, which is not needed for pullulanase secretion (11). This cassette was first inserted into the HincII site of the pulB gene of pCHAP590, a pUC19 derivative, pulB::kan1 was then subcloned into replicative DNA of the f1 phage derivative R704. f1 R704 carries the lacZp-lacZα and polylinker region of bacteriophage M13 mp18 inserted at nucleotide 5614 (constructed by K. Horiuchi), together with amber mutations in genes II and IV (constructed by M. Russel). This phage is similar to M13 mp10 (7) except that it accepts inserts that are considerably larger (up to 5 kb) than M13mp (1.5 to 2 kb). f1 R704 carrying the cloned pulB::kan1 DNA was used to lysogenize strain PAP7232. Cured lysogens (7) were screened for retention of kanamycin resistance. The presence of the pulB::kan1 insertion in the chromosome was confirmed by P1 transduction and by colony PCR. Pullulanase production and secretion in the resulting strain (PAP7452) were indistinguishable from those in strain PAP7232. The pulB::kan1 cassette was then transduced into strain PAP7447 with selection for resistance to kanamycin, and the transductants were screened for inability to secrete pullulanase (ΔpulD). The resulting strain (PAP7456) was then transformed with pCHAP231 and selection was carried out for resistance to kanamycin (250 μg/ml) on maltose-containing medium. Only clones in which the pulB::kan1 mutation had been transferred onto pCHAP231 were able to grow at this high level of kanamycin. Plasmid DNA from such clones was used to transform strain PAP105 with selection for resistance to ampicillin and kanamycin on medium containing maltose. Transformants were screened for production and secretion of pullulanase. DNA from a clone (pCHAP1226) that produced but failed to secrete pullulanase was shown by restriction analysis to carry ΔpulD. Pullulanase secretion was fully restored upon introduction of pCHAP3635, a pSU18 derivative carrying the cloned pulD gene under lacZp control (15).
PulC-specific antiserum.
PCR-amplified DNA coding for the periplasmic region of PulC (from codon 47) was cloned into pSU18 (2) (to give pCHAP5002). Sequencing revealed two substitutions in PulC, neither of which affected PulC function. The cloned fragment was subcloned into pMAL-p2 (New England Biolabs) so that MalE was fused in frame to ′PulC. Strains carrying the resulting plasmid (pCHAP5008) produced an IPTG-inducible 70-kDa polypeptide that reacted with MalE-specific antibodies. The soluble, signal peptide-processed hybrid was released from cells by converting them into spheroplasts, purified by amylose affinity chromatography (New England Biolabs), and injected into rabbits to obtain anti-MalE-′PulC serum (hereafter referred to as PulC antiserum).
Construction and analysis of pulC mutations.
Mutations in pCHAP5001 and pCHAP2277 were constructed by inserting linkers at or between unique restriction sites at codons 100 to 101 (EheI), 209 to 210 (HincII), 215 to 217 (EcoRV), and 219 to 221 (SmaI) (Table 1). The 5′ end of pulC in pCHAP1233 was created by inserting the kan2 cassette into the Bst1107I site created in pCHAP1203 (see above), thereby replacing the first six codons of pulC by 6 codons derived from the open reading frame that follows the aphA-3 gene (Table 2). pCHAP2303 carries a deletion of the region between the NsiI site and a StuI site in pCHAP2284 (see above). pCHAP2201 carries the first few codons of pSU18-derived lacZ fused to the same StuI site.
TABLE 1.
Mutations affecting PulC protein
| Plasmida | Original plasmid | Additional changesb | degP ompTc | Protein(s) detected (kDa) | Protein leveld | Complementation of ΔpulC (%)e |
|---|---|---|---|---|---|---|
| pCHAP1233 | pCHAP5001 | MTLQACM7 | NT | 31 | Normal | 100 |
| pCHAP2301 | pCHAP2284 | MTMITNSSSVPP27 | NT | 30 | Normal | 80 |
| pCHAP2303 | pCHAP2284 | K6RHSSTCP27 | NT | 30 | Normal | 95 |
| pCHAP2289 | pCHAP2277 | A100TSRWP101 | + | 31 | Normal | 90 |
| pCHAP2279 | pCHAP2277 | A100ΔG221 | + | 18 | Low | 0 |
| pCHAP1237 | pCHAP5001 | G215SPRGY216 | + | 31 | Normal | 100 |
| pCHAP2290 | pCHAP2277 | P220TSRWG221 | NT | 31 + 29 | Low | 100 |
| pCHAP1240 | pCHAP5001 | S214SGSWG221 | + | 30 + 24 | Low | 25 |
| pCHAP1250 | pCHAP5001 | V209HLEVN210 | + | 31 | Normal | 100 |
| pCHAP1251 | pCHAP5001 | V209QDPDR217 | + | 31 | Reduced | 0 |
| pCHAP1252 | pCHAP5001 | V209HGIPSG221 | + | 30 | Normal | 0 |
| pCHAP1262 | pCHAP5001 | V209GPN210 | + | 31 | Reduced | 0 |
| pCHAP1263 | pCHAP5001 | G215WAHR217 | + | 31 | Normal | 0 |
All are derived from pSU18 (2) with pulC under lacZp control.
Changes are indicated as amino acid insertions and/or deletions in PulC protein. Numbers indicate positions of amino acids in the PulC sequence; letters in italics indicate inserted amino acids. In pCHAP1233, the sequence MLTQAC replaces the sequence MSKGIK of normal (unmutated) PulC. Δ indicates deletion of intervening amino acids in PulC.
+, protein production and function tested in a strain carrying degP and ompT mutations; NT, not tested in this strain.
Normal, 60 to 100% of level normal PulC; reduced, 20 to 60% of normal level; low, 5 to 20% of normal level under conditions used for assay, as estimated by immunoblotting.
Complementation tested in strains carrying pCHAP1229 (ΔpulC). In the absence of complementing plasmid, <5% of total enzyme activity (lysed cells) is detected in whole cells. Complementation levels are the percentage of total enzyme activity that was detected with whole cells.
TABLE 2.
Effects of replacement of the signal anchor sequence of PulC
| Plasmida | Protein | Sequenceb | Complementation of ΔpulC
|
|
|---|---|---|---|---|
| Noninduced | Induced | |||
| pCHAP1305 | PulC | LAQLTWEFFKIIL | 80 | 100 |
| pCHAP1232 | MalE-′PulC | IEGRISEFFKIIL | 0 | 0 |
| pCHAP1292 | MalESP-′PulC | A*AKIDLEFFKIIL | 30 | 80 |
| pCHAP1315 | MalESA-′PulC | DAKIDLEFFKIIL | 50 | 80 |
| pCHAP1316 | PhoESP-′PulC | ASVQA*AEFFKIIL | 80 | 100 |
| pCHAP1298 | PulASPCD-′PulC | G*CDNSAEFFKIIL | 30 | 100 |
| pCHAP1299 | PulASPCS-′PulC | G*CSNSAEFFKIIL | 0 | 0 |
All are derivatives of pSU18 (2), with the gene fusion under lacZp control (induced with 1 mM IPTG).
Sequences indicated are at the junction between the sections of the protein near the beginning of the periplasmic domain of PulC. Residues in italics are derived from MalE, PhoE, or PulA and linker-encoded residues; other amino acids are from PulC. The last residue of the PulC sequence indicated is position 50. The sequence of unmutated PulC upstream of this position is LAQLTWKIIL. ∗, signal peptidase cleavage site.
The malE-′pulC gene fusion in pCHAP1232 was constructed by digesting pCHAP5008 with BglII (single site within malE of pMAL-p2) and HindIII (downstream from pulC) into the same sites in pCHAP3602, a pSU18 (2) derivative carrying the cloned malE gene with its own promoter and in the same orientation as lacZp (plasmid constructed by I. Guilvout). All other gene fusions (Table 2) were obtained by subcloning the entire sequence of ′pulC as an EcoRI-HindIII fragment from pCHAP5002 into pSU18 derivatives carrying DNA coding for the signal peptides and first few amino acids of MalE, PhoE (outer membrane protein PhoE), or PulA to give plasmids pCHAP1292 (MalESP-′PulC), pCHAP1316 (PhoESP-′PulC), and pCHAP1298 (PulASPCD-′PulC), respectively (superscript abbreviations in hybrid protein designations: SP, signal peptide; SA, uncleavable signal peptide; CD, cysteine-aspartate immediately after cleavage site; CS, cysteine-serine immediately after cleavage site). These signal peptide-coding fragments were constructed by PCR amplification such that its 3′ end was in the same reading frame as ′pulC (Table 2). pCHAP1315 (MalESA-′PulC) was constructed in the same way as pCHAP1292 except that the amplified malE DNA carried a mutation that removed the signal peptidase cleavage site (Table 2). pCHAP1299 (PulASPCS-′PulC) was the same as pCHAP1298 except that the aspartate +2 (D) was changed to a serine (S).
The strains used for complementation assays were PAP105(pCHAP1229) or SF110 (degP ompT [constructed by G. Georgiou])(pCHAP1229), which gave higher yields of some PulC variants. Assays were performed in maltose-induced cultures of fresh transformants grown in the absence or presence of IPTG to induce expression of the pulC gene in the pSU18 derivative. Transinhibition/transdominance, interference with pullulanase secretion when one component of the secretion (or a defective variant thereof) is produced at abnormally high levels, was tested in strain PAP7232, using fresh transformants grown in medium containing maltose and IPTG. Pullulanase assays were performed essentially as described previously (27, 36), using cells harvested from 5-ml cultures. Since pullulanase is anchored to the cell surface rather than being released into the medium (37), secretion levels were quantified as the percentage of total activity that was detectable in whole cells. Cell extracts were tested for the presence of PulC or PulD protein by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting.
Cloning of outC.
The outC gene was amplified from pCPP2236 (24) and cloned into pSU18. Transformants produced an IPTG-inducible protein that reacted with PulC antiserum. Four independent clones of outC gave identical results.
Membrane fractionation.
Cells were converted to spheroplasts, lysed by passage through a French cell, and then treated with RNase and DNase (10 μg/ml) and the serine protease inhibitor Pefabloc (100 μg/ml: Uptima-Interchim). Membranes were pelleted by centrifugation at 180,000 × g for 1 h, resuspended in HEPES buffer (25 mM, pH 7.4) containing 60% sucrose, loaded at the bottom of Beckman SW55 centrifuge tubes, and overlaid with steps of decreasing concentrations of sucrose solutions in HEPES buffer (from 53 to 35%). The tubes were then centrifuged for 24 h at 230,000 × g. Fractions collected from the top of the tubes were analyzed for proteins by SDS-PAGE and immunoblotting. Marker proteins were outer membrane porins (detected by staining the gels used for SDS-PAGE) and OmpA (detected by immunoblotting) and the cytoplasmic membrane protein DjlA (detected by immunoblotting with anti-DjlA serum; a gift from David Clarke).
Cross-linking.
Cells carrying pCHAP231 or its derivatives grown in L broth containing maltose to mid-exponential phase were washed three times in phosphate-buffered saline and resuspended in the same buffer to an optical density at 600 nm of 1.0. Dithiobis(succimidylproprionate) (DSP) was then added to 0.3 mM, and the mixture was incubated for 15 min at room temperature. Tris (100 mM, pH 7.4) was added to quench the cross-linker, and the cells were pelleted by centrifugation, resuspended in 0.2 ml of 100 mM Tris buffer (pH 7.4), and treated with 0.2 ml of phenol to dissociate the PulD complexes (16). Proteins were resuspended in SDS-PAGE sample buffer without or with 10 mM dithiothreitol (DTT; to disrupt the DSP-generated cross-links), heated to 100°C for 5 min, and examined by SDS-PAGE and immunoblotting with PulC antiserum or PulD antibodies (16).
Immunoblotting.
Procedures for immunoblotting were essentially as used previously (15). Proteins were separated by SDS-PAGE in gels containing 8, 9, or 10% acrylamide, then transferred onto nitrocellulose membranes and incubated first with specific antiserum (anti-PulC at 1:2,000, purified anti-PulD at 1:200, anti-OmpA at 1:10,000, or anti DjlA at 1:2,000) and then with horseradish peroxidase-coupled anti-rabbit immunoglobulin G. The membranes were developed by enhanced chemiluminescence.
RESULTS
Localization of PulC protein.
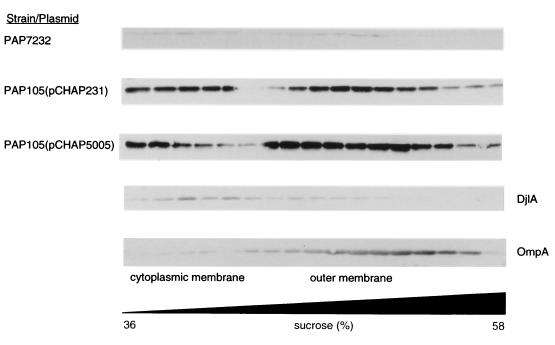
Ultracentrifugation of disrupted cells of PAP7232 (36) and PAP105 carrying pCHAP231 (11) or pCHAP5005 indicated that PulC was entirely in the pelleted (membrane fraction), irrespective of whether PulC was produced alone at moderate or high (noninduced/induced) levels (pCHAP5005) or at low or moderate levels together with all other components of the pullulanase secreton [maltose-induced PAP7232 or PAP105(pCHAP231), respectively] (data not shown). The pelleted fraction was therefore loaded at the bottom of a sucrose step gradient, and membrane subfractions were separated by flotation during centrifugation to determine the location of PulC. In all cases, PulC was found in two distinct peaks that contained, respectively, marker proteins of the cytoplasmic membrane (DjlA) and outer membrane (porins and OmpA) (Fig. 1). Thus, PulC seems to associate with both membrane fractions, as reported for PulG (40) and TonB (22). PulE, on the other hand, was located mainly in the cytoplasmic membrane (34), while PulD (16, 17, 34) and siderophore receptors (with which TonB interacts) were mainly in the outer membrane. A PulC-PhoA hybrid was previously shown to be located exclusively in the cytoplasmic membrane (11). Therefore, the N-terminal hydrophobic domain of PulC (present in the PulC-PhoA hybrid) presumably anchors it in the cytoplasmic membrane while association with the outer membrane is mediated by the C-terminal segment of PulC (amino acids 245 to 285) that is absent from the PulC-PhoA hybrid (11). The fact that PulC was found in both membrane fractions does not necessarily mean that the same PulC polypeptide is in simultaneous contact with both membranes.
FIG. 1.
Separation of membrane fractions by flotation through centrifuged sucrose gradients and detection of PulC by immunoblotting. The strains used were PAP7232 (pulC in chromosome together with all other pul genes), PAP105 (pCHAP231) (all pul genes cloned into pBR322), and PAP105 (pCHAP5005) (cloned pulC in pUC19). DjlA protein (cytoplasmic membrane maker) and OmpA were also detected by immunoblotting. The positions of cytoplasmic and outer membrane fractions are indicated.
Secretion-negative mutations in PulC are not transdominant.
The production of large amounts of a defective variant of certain secreton components (or even, in some cases, of large amounts of the active form) can prevent secreton function, presumably by titrating other secreton components or by stoichiometic imbalance (34, 36). To determine if this approach could be used to study the interaction between PulC and other secreton components, we created small deletion/insertion mutations in pulC and tested the mutated genes for the ability to complement the ΔpulC mutation in pCHAP1229 (pCHAP231 ΔpulC::kan2) and for transdominance in strain PAP7232.
Several of these protein constructs were unstable in E. coli K-12. Their stability was not improved by the other secreton components (encoded by pCHAP1229), but some were stabilized by a combination of degP and ompT mutations (Table 1). Stop codons introduced at positions 210, 217, and 221 of the 305 codon pulC gene caused failure to produce a protein that could be detected by immunoblotting with the PulC antiserum, even in the strain carrying the degP and ompT mutations (not shown), presumably due to their extreme instability or to lack of epitopes recognized by the antibodies. Other, less drastic mutations affecting the periplasmic region of PulC were without effect, whereas others abolished complementation of ΔpulC (Table 1). None of the latter were transdominant or transinhibitory. High-level production of wild-type PulC (encoded by pCHAP5005) did not affect pullulanase secretion in PAP7232. The cloned outC gene (the pulC homologue) from E. chrysanthemi failed to complement ΔpulC and was not transdominant. The outD gene is also unable to replace its homologue in the pullulanase secreton (15).
The signal anchor of PulC is not essential.
Some of the mutations that had no effect on PulC function truncated or altered the ca. 27-amino-acid N-terminal cytoplasmic region of the protein (Table 1). We were therefore able to test the effects of the complete removal of this region and of substitution of the signal anchor sequence.
The mainly periplasmic full-length MalE-′PulC hybrid protein used to raise the PulC antibodies (Materials and Methods) was nonfunctional in the complementation assay but was also not transdominant. Removal of almost all of the mature segment of MalE from this hybrid, to create MalESP-′PulC, restored complementation (Table 2). This result indicated that the precise sequence of the PulC signal anchor sequence (up to amino acid 46) is not essential for function, since it can be replaced by the MalE signal sequence. Analysis of cell lysates by immunoblotting revealed that the MalESP-′PulC protein was only partially processed by leader peptidase (Fig. 2), making it impossible to decide whether the processed or unprocessed form was functional. Therefore, two new variants were constructed. One was similar to MalESP-′PulC except that the leader peptidase recognition site was removed by an alanine-to-aspartate change at position −1 (13). This protein (MalESA-′PulC) retained ability to complement ΔpulC (Table 1) and, as expected (13), was very inefficiently processed (Fig. 2B). In the second construct, the signal peptide from the outer membrane protein PhoE was fused to the periplasmic domain of PulC (PhoESP-′PulC). This protein was also functional (Table 2), but although most of the protein present in the cell extracts was processed, unprocessed precursor was also detected (Fig. 2B). Since the level of unprocessed PhoESP-′PulC was greater than that of MalESA-′PulC, which was functional (Fig. 2B), we were unable to conclude that the presumably periplasmic processed PhoESP-′PulC was functional.
FIG. 2.
Immunodetection of PulC derivatives separated by SDS-PAGE and transferred onto a nitrocellulose membrane. Cells of strain PAP105(pCHAP1229) were grown in the presence of the inducer maltose and in the absence or presence of the inducer IPTG under conditions used to assay pullulanase secretion. The two dots indicate the positions of unprocessed (upper dot) and processed forms. MalE protein was also detected by the antiserum. (A) Extracts of cells with vector (pSU18) or plasmids encoding MalESP-′PulC (pCHAP5005), PulASPCD-′PulC (pCHAP1298), or PulASPCS-′PulC (pCHAP1299) in the absence or presence of IPTG (note that in pCHAP5005, the pulC gene is under its own, maltose-inducible promoter and is therefore fully induced in the absence of IPTG); (B) extracts of cells encoding PhoESP-MalE (pCHAP1316), MalESP-′PulC (pCHAP1292), or MalESA′PulC (pCHAP1315) grown in the presence of IPTG.
Finally, the signal peptide and the first four amino acids of pullulanase (PulA) were fused to the periplasmic region of PulC. After signal peptide processing, this PulC variant has the fatty acylated N-terminal cysteine residue of pullulanase (37) and the aspartate residue at position +2 that causes translocated lipoproteins to remain anchored to the cytoplasmic membrane (38, 39, 45). This PulC variant (PulASPCD-′PulC) was fully processed under the conditions used to assay pullulanase secretion (Fig. 2A), could be labeled with [3H]palmitate (indicating that it was fatty acylated; not shown), and retained functionality in the complementation assay (Table 2). However, replacement of the aspartate residue by a serine (PulASPCS-′PulC), which causes lipoproteins to be sorted to the periplasmic face of the outer membrane (38, 39, 45), resulted in complete loss of function without affecting processing, protein levels (Fig. 2A), or fatty acylation (not shown).
From these results, we concluded that neither the signal anchor nor the N-terminal cytoplasmic region of PulC is essential for its function. PulC that is predicted to be anchored to the periplasmic face of the cytoplasmic membrane is functional, but PulC whose N terminus is predicted to be anchored to the outer membrane is not.
Localization of PulC variants lacking the signal anchor sequence.
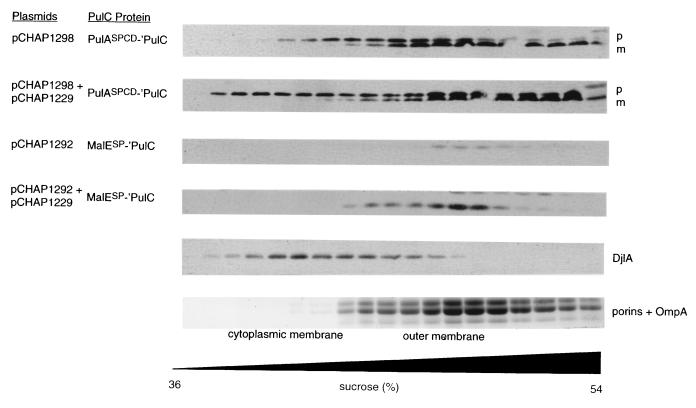
PulASPCD-′PulC is presumably anchored to the cytoplasmic membrane by fatty acids, whereas PulC is normally embedded in this membrane by a signal anchor sequence. To test whether this difference caused a change in the proportion of PulC that remained associated with the cytoplasmic and outer membranes, the two membranes were separated by flotation through a centrifuged sucrose gradient. In these particular experiments, the signal peptide was not fully processed, leading to the production of two forms of the protein that could be distinguished by immunoblotting (Fig. 3). The upper band, corresponding to unprocessed PulASPCD-′PulC, fractionated with both the cytoplasmic and outer membranes (Fig. 3) and is therefore similar to normal PulC (Fig. 1). However, the faster-migrating, presumably processed form of PulASPCD-′PulC fractionated mainly with the outer membrane (Fig. 3). Neither fractionation pattern was affected by the presence of other secreton components (encoded by pCHAP1229) (Fig. 3). The most plausible explanation for this observation is that the lipid anchor of mature PulASPCD-′PulC is unable to retain it in the cytoplasmic membrane when the cells are disrupted. Thus, the protein remains predominantly in the outer membrane fraction, whereas with the normal PulC and unprocessed PulASPCD-′PulC, the signal anchor and unprocessed signal peptide retain at least some of the protein in the cytoplasmic membrane. In accordance with this idea, the fractionation pattern of PulASPCD-′PulC was indistinguishable from that of PulASPCS-′PulC (not shown). This interpretation was further reenforced by analysis of MalESP-′PulC. A large proportion of the processed form of this protein was present in the soluble fraction, but some remained membrane associated and was found exclusively in the outer membrane fraction, irrespective of the presence or absence of the other secreton components (Fig. 3). Thus, PulC does not need a signal anchor sequence and does not need to be anchored in the cytoplasmic membrane in order to interact with the outer membrane.
FIG. 3.
Separation of membrane fractions of strain PAP105 carrying the plasmids indicated by flotation through centrifuged sucrose gradients. Proteins were detected by using SDS-PAGE and immunoblotting with antiserum against PulC protein or against DjlA protein. Porins and outer membrane protein OmpA were detected by Coomassie blue staining of an SDS-polyacrylamide gel. p, prePulASPCD-′PulC; m, mature PulASPCD-′PulC.
Probing possible interactions between PulC and PulD by chemical cross-linking.
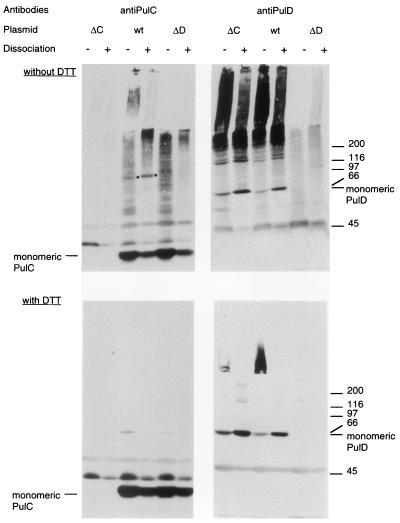
The fractionation pattern of PulC is similar to that of TonB (22). TonB has been shown to interact with at least two siderophore receptors (21, 29) and probably interacts with all of them and with the vitamin B12 receptor BtuB. The association of PulC with the outer membrane is independent of PulD, but this does not mean that they do not interact. This possibility was tested by cross-linking PulC in whole cells with the amine-specific cross-linker DSP. In cells carrying pCHAP231 (wild-type secreton), DSP treatment caused the appearance of a discrete band at approximately 110 kDa that reacted with the PulC antiserum (Fig. 4). The abundance of this band increased when cell extracts were treated with phenol, which dissociates PulD multimers (16), and it disappeared upon dissociation of the cross-link with DTT (Fig. 4). The 110-kDa band was absent when cells carrying pCHAP1229 (pCHAP231 ΔpulC), pCHAP5005 (pulC), or pCHAP1226 (ΔpulD) were tested in the same way (Fig. 4 and data not shown). The appearance of the 110-kDa cross-linked product was restored when pCHAP1298 coding for PulASPCD-′PulC was present in the strain carrying pCHAP1229 (not shown). Thus, the PulC signal anchor sequence is not required for formation of the 110-kDa cross-linked product. However, the 110-kDa band did not react with antibodies specific for PulD (Fig. 4). Thus, although the 110-kDa band is approximately the size of a PulC-PulD heterodimer (31 kDa + 68 kDa) and its appearance depended on the presence of PulD, it does not appear to contain PulD. It should also be noted the PulC antiserum reacted with material from DSP-treated bacteria that remained in the stacking gel when analyzed by SDS-PAGE. These large complexes were not detected in cells lacking either PulC or PulD (Fig. 4).
FIG. 4.
Formation of 110-kDa band upon cross-linking of cells from maltose-induced cultures of strains producing PulC protein. Whole cells of strains carrying pCHAP1229 (ΔpulC [ΔC]), pCHAP231 (wild type [wt]), or pCHAP1226 (ΔpulD [ΔD]) were incubated with 0.3 mM DSP for 15 min, extracted with phenol (dissociation), and dissolved in SDS-PAGE sample buffer without or with DTT to reduce the disulfide bond formed by the cross-linker. Proteins reacting with PulC or PulD antibodies were detected by SDS-PAGE and immunoblotting, using the same nitrocellulose sheet. The positions of PulC and PulD monomers are indicated, as are the positions of the ca. 110-kDa band that appears only after cross-linking of cells carrying pCHAP231 (dot) and molecular size markers (indicated at the right in kilodaltons).
DISCUSSION
Pullulanase secretion is apparently driven by the proton motive force (32), and it is therefore important to establish how energy is transduced to the outer membrane. Vitamin B12 and ferric siderophore uptake by gram-negative bacteria also appear to require the proton motive force (8, 41), and it has been proposed that TonB protein transduces energy to the vitamin and siderophore receptors in the outer membrane, thereby permitting the release of the bound ligand and its diffusion through the receptor’s β-barrel channel that is normally closed in the absence of ligand and TonB (30). Although PulC and TonB do not share any sequence similarity, they are associated with both membranes and are predicted to have long periplasmic domains (as expected for a protein that transduces energy between the two membranes). Are there further similarities between the two proteins that might support the idea that PulC has a TonB-like function?
TonB can be cross-linked to siderophore receptors (43), whereas PulC cannot be cross-linked to PulD. However, this might simply reflect the absence of suitably positioned amine groups on the two proteins. Although cross-linking of PulC failed to reveal any direct association with PulD, it did result in the appearance of a distinct 110-kDa product (as well as much larger complexes that remained in the stacking gel when analyzed by SDS-PAGE) whose appearance was PulD dependent (Fig. 4). The PulD-dependent formation of these cross-linked products, which presumably correspond to a PulC homo- or heteromultimers, constitutes the only evidence that PulC and PulD do interact, either directly or indirectly. Bleves (4) previously noted the formation of large complexes that reacted with XcpP antiserum when cells containing XcpQ were treated with the cross-linker formaldehyde. These large complexes are presumably the same as those reported here, but a cross-linked 110-kDa product was not observed by Bleves (4).
Another difference between TonB and PulC concerns the role of the signal anchor sequence. The specific sequence of the cytoplasmic membrane signal anchor of TonB protein is required for function, probably reflecting a specific interaction with other cytoplasmic membrane proteins such as ExbB and ExbD (18, 19). In contrast, the precise sequence of the PulC signal anchor is clearly not necessary because the protein remains functional irrespective of how its N terminus is anchored to the cytoplasmic membrane (and possibly even when it is periplasmic). These results indicate that PulC is fundamentally dissimilar to TonB, but they do not rule out a possible role for PulC in energy transduction.
Bleves et al. (5) have likewise considered the possibility that the P. aeruginosa PulC homologue XcpP transduces energy to the PulD homologue XcpQ to permit the opening of the XcpQ secretin channel and the release of specific exoproteins from the periplasm. This idea is again based on similarities between XcpP and TonB that are somewhat more extensive than those between TonB and PulC. For example, substitution of the XcpP signal anchor by another, similar sequence abolished function, and XcpP is stabilized by XcpQ (5). Furthermore, XcpP inhibits secretion when it is no longer retained in the cytoplasmic membrane (5), a phenomenon similar to the reported inhibition of siderophore uptake by periplasmic TonB (18) and suggestive of a specific interaction between XcpP and another secreton component. In contrast, we did not detect any difference in the yields of PulC in strains with or without PulD, indicating that PulD does not stabilize PulC (data not shown), and periplasmic forms of PulC (e.g., PhoESP-′PulC) did not inhibit secretion in strains with a chromosomally encoded, wild-type secreton. The latter results suggests that PulC is unable to titrate another secreton component in the same way as XcpP. Defective pulC alleles were also not transdominant, presumably because the mutations disrupted the ability of PulC to interact with the secreton or because this interaction is not saturable.
How can the apparent differences between XcpP and PulC be rationalized? One possibility is that the two proteins perform totally different functions. However, this seems unlikely since they share considerable sequence identity and have similar topologies, and since their structural genes are in the same position relative to their corresponding secretin genes. However, whereas pullulanase secretion is apparently proton motive force dependent (32), secretion in P. aeruginosa appears to be ATP dependent (9). Furthermore, while XcpP and PulC are closely related, they differ in that XcpP has a coiled-coil structure near the C terminus whereas PulC has a PDZ motif (31). Both PDZ and coiled-coil motifs are often involved in protein-protein interactions. These important differences might be sufficient to explain the discrepancies between our results and those of Bleves et al. (5) because they could alter the way the two proteins associate with the outer membrane.
The function of PulC protein remains unknown. Some linker/deletion mutations affecting the sequence of PulC near the beginning of the predicted PDZ domain (starting at or before position 215 and ending at or after position 273, both highly conserved glycines) caused loss of PulC function, whereas others did not (Table 1). More detailed investigation of the role of conserved residues in this domain is required to determine its role in PulC function. On the basis of data reported by Lindeberg et al. (24), it is conceivable that PulC interacts with pullulanase itself. However, data reported by Shevchik et al. (42) showing that the OutD secretin recognizes exoproteins secreted by E. chrysanthemi suggest that PulD could interact with pullulanase. Studies designed to test these ideas will form the focus of our future analysis of the pullulanase secreton.
ACKNOWLEDGMENTS
We are grateful to Sophie Bleves, Peter Braun, Alain Filloux, and Jan Tommassen for a frank and open exchange of information, to Tracy Letain for discussions regarding TonB, to Ken Horiuchi and Marjorie Russel for constructing and supplying the f1 phage derivative R704, and to Jean-Michel Betton, David Clarke, Alan Collmer, George Georgiou, Steve Lory, and Bénédicte Michel for strains, plasmids, or antibodies. We are also grateful to our collegues Nathalie Nadeau for technical assistance, Ingrid Guilvout for help with the cloning of outC and for pCHAP3635 and pCHAP3602, Olivera Francetic for the PhoESP fusion vector, Anke Seydel for helping to construct the PulASPCS fusion vector and for advice on sucrose gradient centrifugation, and to these and all other members of the secretion lab for their interest and encouragement.
The work was supported by the European Union (TMR grant FMRX-CT96-0004).
REFERENCES
- 1.Akrim M, Bally M, Ball G, Tommassen J, Teerink H, Filloux A, Lazdunski A. Xcp-mediated protein secretion in Pseudomonas aeruginosa: identification of two additional genes and evidence for regulation of xcp gene expression. Mol Microbiol. 1993;10:431–443. doi: 10.1111/j.1365-2958.1993.tb02674.x. [DOI] [PubMed] [Google Scholar]
- 2.Bartolomé B, Jubete Y, Martinez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 3.Bitter W, Koster M, Latijnhouwers M, de Cock H, Tommassen J. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol Microbiol. 1998;27:209–219. doi: 10.1046/j.1365-2958.1998.00677.x. [DOI] [PubMed] [Google Scholar]
- 4.Bleves S. Sécrétion de protéines chez Pseudomonas aeruginosa: étude des interactions protéine-protéine au sein de la machinerie Xcp. Ph.D. thesis. Marseille, France: Université de la Méditerranée Aix-Marseille II; 1998. [Google Scholar]
- 5.Bleves S, Gérard-Vincent M, Lazdunski A, Filloux A. Structure-function analysis of XcpP, a component involved in general secretory pathway-dependent protein secretion in Pseudomonas aeruginosa. J Bacteriol. 1999;181:4012–4019. doi: 10.1128/jb.181.13.4012-4019.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bleves S, Lazdunski A, Filloux A. Membrane topology of three Xcp proteins involved in exoprotein transport by Pseudomonas aeruginosa. J Bacteriol. 1996;178:4297–4300. doi: 10.1128/jb.178.14.4297-4300.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blum P, Holzschu D, Kwan H S, Riggs D, Artz S. Gene replacement and retrieval with recombinant M13 mp bacteriophages. J Bacteriol. 1989;171:538–546. doi: 10.1128/jb.171.1.538-546.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradbeer C. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J Bacteriol. 1993;175:146–150. doi: 10.1128/jb.175.10.3146-3150.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun, P., H. Adams, W. Bitter, and J. Tommassen. 1998. Personal communication.
- 10.Daefler S, Guilvout I, Hardie K R, Pugsley A P, Russel M. The C-terminal domain of the secretin PulD contains the binding site for its cognate chaperone, PulS, and confers PulS dependence on pIVf1 function. Mol Microbiol. 1997;24:465–475. doi: 10.1046/j.1365-2958.1997.3531727.x. [DOI] [PubMed] [Google Scholar]
- 11.d’Enfert C, Reyss I, Wandersman C, Pugsley A P. Protein secretion by Gram-negative bacteria: characterization of two membrane proteins required for pullulanase secretion by Escherichia coli K-12. J Biol Chem. 1989;264:17462–17468. [PubMed] [Google Scholar]
- 12.d’Enfert C, Ryter A, Pugsley A P. Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization and secretion of the lipoprotein pullulanase. EMBO J. 1987;6:3531–3538. doi: 10.1002/j.1460-2075.1987.tb02679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fikes J D, Bassford P J., Jr Export of unprocessed precursor maltose-binding protein to the periplasm of Escherichia coli cells. J Bacteriol. 1987;169:2352–2359. doi: 10.1128/jb.169.6.2352-2359.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francetic O, Pugsley A P. The cryptic general secretory pathway (gsp) operon of Escherichia coli K-12 encodes functional proteins. J Bacteriol. 1996;178:3544–3549. doi: 10.1128/jb.178.12.3544-3549.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Georgrou, G. Unpublished data.
- 15.Guilvout, I., N. Sauvonnet, K. R. Hardie, and A. P. Pugsley. 1999. Unpublished data. [DOI] [PMC free article] [PubMed]
- 16.Hardie K R, Lory S, Pugsley A P. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 1996;15:978–988. [PMC free article] [PubMed] [Google Scholar]
- 17.Hardie K R, Seydel A, Guilvout I, Pugsley A P. The secretin-specific, chaperone-like protein of the general secretory pathway: separation of proteolytic protection and piloting functions. Mol Microbiol. 1996;22:967–976. doi: 10.1046/j.1365-2958.1996.01539.x. [DOI] [PubMed] [Google Scholar]
- 18.Jaskula J C, Letain T E, Roof S K, Shale J T, Postle K. Role of the TonB amino terminus in energy transduction between membranes. J Bacteriol. 1994;176:2326–2338. doi: 10.1128/jb.176.8.2326-2338.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlsson M, Hannavy K, Higgins C F. A sequence-specific function for the N-terminal signal-like sequence of the TonB protein. Mol Microbiol. 1993;8:379–388. doi: 10.1111/j.1365-2958.1993.tb01581.x. [DOI] [PubMed] [Google Scholar]
- 20.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 21.Larsen R A, Foster-Hartnett D, McIntosh M A, Postle K. Regions of Escherichia coli TonB and FepA proteins essential for in vivo physical interactions. J Bacteriol. 1997;179:3213–3221. doi: 10.1128/jb.179.10.3213-3221.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Letain T E, Postle K. TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in Escherichia coli. Mol Microbiol. 1997;24:271–283. doi: 10.1046/j.1365-2958.1997.3331703.x. [DOI] [PubMed] [Google Scholar]
- 23.Letellier L, Howard S P, Buckley T J. Studies on the energetics of proaerolysin secretion across of the outer membrane of Aeromonas spp: evidence for requirement for both the protonmotive force and ATP. J Biol Chem. 1997;272:11109–11113. doi: 10.1074/jbc.272.17.11109. [DOI] [PubMed] [Google Scholar]
- 24.Lindeberg M, Salmond G P C, Collmer A. Complementation of deletion mutations in a cloned functional cluster of Erwinia chrysanthemi out genes with Erwinia carotovora out homologs reveals OutC and OutD as candidate gatekeepers of species-specific secretion of proteins via the type II pathway. Mol Microbiol. 1996;20:175–190. doi: 10.1111/j.1365-2958.1996.tb02499.x. [DOI] [PubMed] [Google Scholar]
- 25.Linderoth N A, Simon M N, Russel M. The filamentous phage pIV multimer visualized by scanning transmission electron microscopy. Science. 1997;278:1635–1638. doi: 10.1126/science.278.5343.1635. [DOI] [PubMed] [Google Scholar]
- 26.Menard R, Sansonetti P J, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michaelis S, Chapon C, d’Enfert C, Pugsley A P, Schwartz M. Characterization and expression of the structural gene for pullulanase, a maltose-inducible secreted protein of Klebsiella pneumoniae. J Bacteriol. 1985;164:633–638. doi: 10.1128/jb.164.2.633-638.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 29.Moeck G S, Coulton J W, Postle K. Cell envelope signaling in Escherichia coli. Ligand binding to the ferrichrome-iron receptor FhuA requires interaction with the energy-transducing protein TonB. J Biol Chem. 1997;272:28391–28397. doi: 10.1074/jbc.272.45.28391. [DOI] [PubMed] [Google Scholar]
- 30.Moeck G S, Coulton J W. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol Microbiol. 1998;28:675–681. doi: 10.1046/j.1365-2958.1998.00817.x. [DOI] [PubMed] [Google Scholar]
- 31.Pallen M J, Ponting C P. PDZ domains in bacterial proteins. Mol Microbiol. 1997;26:411–413. doi: 10.1046/j.1365-2958.1997.5591911.x. [DOI] [PubMed] [Google Scholar]
- 32.Possot O, Letellier L, Pugsley A P. Energy requirement for pullulanase secretion by the main terminal branch of the general secretory pathway. Mol Microbiol. 1997;24:457–464. doi: 10.1046/j.1365-2958.1997.3451726.x. [DOI] [PubMed] [Google Scholar]
- 33.Possot O, Pugsley A. The conserved tetracysteine motif in the general secretory pathway component PulE is required for efficient pullulanase secretion. Gene. 1997;192:45–50. doi: 10.1016/s0378-1119(97)00009-7. [DOI] [PubMed] [Google Scholar]
- 34.Possot O, Pugsley A P. Molecular characterization of PulE, a protein required for pullulanase secretion. Mol Microbiol. 1994;12:287–299. doi: 10.1111/j.1365-2958.1994.tb01017.x. [DOI] [PubMed] [Google Scholar]
- 35.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pugsley A P. Processing and methylation of PulG, a pilin-like component of the general secretory pathway of Klebsiella oxytoca. Mol Microbiol. 1993;9:295–308. doi: 10.1111/j.1365-2958.1993.tb01691.x. [DOI] [PubMed] [Google Scholar]
- 37.Pugsley A P, Chapon C, Schwartz M. Extracellular pullulanase of Klebsiella pneumoniae is a lipoprotein. J Bacteriol. 1986;166:1083–1088. doi: 10.1128/jb.166.3.1083-1088.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pugsley A P, Kornacker M G. Secretion of the cell surface lipoprotein pullulanase by Escherichia coli. Collaboration or competition between the specific secretion pathway and the lipoprotein sorting pathway. J Biol Chem. 1991;266:13640–13645. [PubMed] [Google Scholar]
- 39.Pugsley A P, Poquet I, Kornacker M G. Two distinct steps in pullulanase secretion by Escherichia coli K12. Mol Microbiol. 1991;5:865–873. doi: 10.1111/j.1365-2958.1991.tb00760.x. [DOI] [PubMed] [Google Scholar]
- 40.Pugsley A P, Possot O. The general secretory pathway of Klebsiella oxytoca: no evidence for relocalization or assembly of pilin-like PulG protein into a multiprotein complex. Mol Microbiol. 1993;10:665–674. doi: 10.1111/j.1365-2958.1993.tb00938.x. [DOI] [PubMed] [Google Scholar]
- 41.Pugsley A P, Reeves P. Uptake of ferrienterochelin by Escherichia coli: energy-dependent stage of uptake. J Bacteriol. 1977;130:26–36. doi: 10.1128/jb.130.1.26-36.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shevchik V E, Robert-Badouy J, Condemine G. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J. 1997;16:3007–3016. doi: 10.1093/emboj/16.11.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skare J T, Ahmer B M M, Seachord C L, Darveau R P, Postle K. Energy transduction between membranes. TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J Biol Chem. 1993;268:16302–16308. [PubMed] [Google Scholar]
- 44.Thomas J D, Reeves P J, Salmond G P C. The general secretion pathway of Erwinia carotovora subsp. carotovora: analysis of the membrane topology of OutC and OutF. Microbiology. 1997;143:713–720. doi: 10.1099/00221287-143-3-713. [DOI] [PubMed] [Google Scholar]
- 45.Yamaguchi K, Yu F, Inouye M. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell. 1988;53:423–432. doi: 10.1016/0092-8674(88)90162-6. [DOI] [PubMed] [Google Scholar]