Abstract
The general secretory pathway of Pseudomonas aeruginosa is required for the transport of signal peptide-containing exoproteins across the cell envelope. After completion of the Sec-dependent translocation of exoproteins across the inner membrane and cleavage of the signal peptide, the Xcp machinery mediates translocation across the outer membrane. This machinery consists of 12 components, of which XcpQ (GspD) is the sole outer membrane protein. XcpQ forms a multimeric ring-shaped structure, with a central opening through which exoproteins could pass to reach the medium. Surprisingly, all of the other Xcp proteins are located in or are associated with the cytoplasmic membrane. This study is focused on the characteristics of one such cytoplasmic membrane protein, XcpP. An xcpP mutant demonstrated that the product of this gene is indeed an essential element of the P. aeruginosa secretion machinery. Construction and analysis of truncated forms of XcpP made it possible to define essential domains for the function of the protein. Some of these domains, such as the N-terminal transmembrane domain and a coiled-coil structure identified at the C terminus of XcpP, may be involved in protein-protein interaction during the assembly of the secretory apparatus.
Protein secretion in Pseudomonas aeruginosa is driven by three separate secretion pathways that are widespread in gram-negative bacteria. Alkaline protease follows the type I pathway (22, 61), whereas exoenzymes S, T, and U follow the type III pathway (16). Most P. aeruginosa exoproteins, including elastase and exotoxin A, follow the type II pathway, or main terminal branch, of the general secretory pathway (GSP) (14, 15). GSP-dependent exoproteins are synthesized with an N-terminal signal sequence and are translocated in a Sec-dependent manner across the inner membrane. Transport across the outer membrane is mediated by specialized machinery, the type II secretory apparatus, comprising 12 proteins designated Xcp (15). The term Gsp is used to denote Xcp homologs from other bacteria (51). Although this machinery is involved in protein translocation across the outer membrane, most of its components are present in the inner membrane. Defects in any of the components lead to the periplasmic accumulation of exoproteins.
XcpA and XcpS are polytopic inner membrane proteins, as shown for their homologs of Erwinia carotovora, OutO and OutF, respectively (52, 60). XcpA functions as a prepilin leader peptidase (44) and is also responsible for the processing of five components of the Xcp machinery, the pseudopilins XcpT (GspG), -U (-H), -V (-I), -W (-J), and -X (-K) (4, 7, 45). The subcellular distribution of these pseudopilins is unclear. They have a hydrophobic segment typical of inner membrane proteins but have been found, in some conditions, to fractionate with both the inner and outer membrane fractions (4, 45). XcpR contains an ATP-binding motif (4) and, like several of its homologs (GspE proteins), is peripherally associated with the cytoplasmic membrane (3, 49, 55). Of the 12 Xcp components, only XcpQ (GspD), belonging to the newly defined secretin family, is located in the outer membrane. Secretins are involved in various secretion pathways in gram-negative bacteria (19), including filamentous phage secretion, and are large homomultimers of 10 to 12 subunits (5, 23, 29, 33, 56). The XcpQ secretin forms a ring-shaped structure with a central cavity 95 Å in diameter (5), which may be the outer membrane channel of the GSP through which exoproteins pass. The large diameter of the cavity is consistent with GSP-dependent exoproteins being secreted in a folded conformation (8, 9, 17, 24, 26). A pore of this kind in the outer membrane must be tightly gated to prevent the leakage of periplasmic components and cell death.
This gating may be a function of an Xcp component which directly interacts with XcpQ. Several Xcp proteins, including XcpP (GspC), -Y (-L), and -Z (-M), are bitopic inner membrane proteins, each with a large domain extending into the periplasm (6). These proteins could interact with the outer membrane XcpQ protein. We investigated in greater detail the role of XcpP for two main reasons: (i) the xcpP and xcpQ genes are organized into a single operon (2), which may reflect coordinated action of the corresponding proteins; and (ii) in Erwinia chrysanthemi, the XcpP and XcpQ homologs, OutC and OutD, respectively, have been proposed, on the basis of studies using the genetic approach, as gatekeepers of species-specific secretion via the GSP (37).
In this study, XcpP proteins with deletions and hybrid proteins were tested for complementation of an xcpP mutant. We identified several characteristic domains which appeared to be important for the function of the protein. In addition, we showed that XcpP was unstable in an xcpQ mutant, suggesting that there is an interaction between these two components. We finally propose that domains of XcpP may be involved in the controlled gating of the XcpQ pore in the outer membrane, thus regulating the functioning of the secretion machinery.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids are described in Table 1. Cells were grown at 37°C with aeration in Luria broth for Escherichia coli or tryptic soy broth (TSB) for P. aeruginosa. Plasmids were maintained by adding ampicillin, kanamycin, tetracycline, streptomycin, and HgCl2 50, 25, 15, 50, and 20 μg/ml, respectively) for E. coli and carbenicillin, tetracycline, and streptomycin (500, 200, and 1000 μg/ml, respectively) for P. aeruginosa. The conjugative properties of pRK2013 were used to transfer plasmids from E. coli to P. aeruginosa. Pseudomonas transconjugants were selected on Pseudomonas isolation agar containing antibiotics.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant genotype and characteristics | Reference or source |
|---|---|---|
| E. coli | ||
| BL21(DE3) | F−hsdS gal ompT, lysogen of DE3 carrying T7 polymerase gene | 58 |
| TG1 | supE Δ(lac-proAB) thi hsdRΔ5 (F′::traΔ36 proA+B+ lacIqZΔM15) | 39 |
| P. aeruginosa | ||
| PAO1 | Prototroph, chl-2 | B. Holloway |
| PAO1ΔP | PAO1 derivative, chromosomal deletion of xcpP | This work |
| PAG2 | PAO1 derivative, chromosomal deletion of xcpQ | A. de Groot |
| Plasmids | ||
| pT7.5 | E. coli vector, Apr, φ10 promoter | 59 |
| pMMB190 | Broad-host-range vector, Apr, lacUV5 promoter, lacZα | 43 |
| pMMB67HE | Broad-host-range vector, Apr, tac promoter | 18 |
| pACYC184 | E. coli vector, Tcr, Cmr | New England Biolabs |
| pJBS633 | E. coli vector, Tcr, Kmr | 10 |
| pGEX-2T | E. coli vector, Apr, GST gene fusion | AMRAD |
| pRK2013 | Kmr, ColE1 with Tra+ Mob+ | 13 |
| pKNG101 | Smr, oriR6K, oriTRK2, mobRK2, sacB R+ (suicide vector) | 32 |
| pHP45ΩHg | Hgr, Ω interposon | 11 |
| pMB4 | xcpQ gene in pMMB67HE | A. de Groot |
| pSB10, pSB58 | xcpP gene cloned in pMMB67HE or pUC19, Apr | This work |
| pSB95 | 510-bp xcpP internal deletion in pKNG101, Tcr, Smr | This work |
| pMG1, pMG2 | xcpP with an ATG→TGA or TTG→ATC change in pMMB67HE | This work |
| pSB60, pSB62 (Tc′XcpPp) | tc′-xcpP gene fusion in pJBS633 or pMMB190, Kmr or Apr | This work |
| pSB75 (XcpPΔX) | 3′ deletion in xcpP at the XhoI site in pMMB190 | This work |
| pSB94 (XcpPΔP) | 3′ deletion in xcpP at the PvuI site in pMMB190 | This work |
| pSB74 (XcpPΔM) | 3′ deletion in xcpP at the MscI site in pMMB190 | This work |
| pSB86 (XcpPΔ19) | xcpP with an internal deletion of 85 bp in pMMB67HE | This work |
| pSB82b (XcpPΔ20) | xcpP with an internal deletion of 273 bp in pMMB67HE | This work |
| pSB18 | PCR product encoding lasB signal sequence in pUC19, Apr | This work |
| pSB20 (lasB-′xcpP) | PCR product encoding ′XcpPp in pSB18 | This work |
| pSB25, pSB24 | lasB-xcpP fusion of pSB20 in pT7.5 or pMMB190 | This work |
| pSB46 (lasB-′xcpZ) | PCR product encoding ′XcpZp in pSB18 | This work |
| pSB51 | lasB-xcpZ fusion of pSB46 in pMMB190 | This work |
| pSB54 | PCR product encoding XcpP periplasmic domain in pGEX-2T | This work |
| pSB65 | xcpY gene in pGEX-2T | This work |
The xcpP mutant, PAO1ΔP, was produced as follows. An internal 510-bp PstI deletion was introduced into the xcpP gene cloned in pACYC184. This DNA fragment contains part of the xcpR gene at the 5′ end of the deleted xcpP gene; therefore, we added at the 3′ end a fragment encoding the downstream xcpQ gene to allow efficient recombination. The resulting plasmid was inserted into the suicide vector, pKNG101, to generate the xcpP mutator, pSB95. pSB95 was introduced into PAO1, and conjugants were selected for the first recombination event on Pseudomonas isolation agar containing streptomycin (pKNG101) and tetracycline (pACYC184). Several colonies were taken through several rounds of isolation on tryptic soy agar-skim milk plates containing 10% sucrose, which favors plasmid excision (loss of sacBR) and selection for the second recombination event (32). A secretion-defective clone was tested for its sensitivity to streptomycin and tetracycline and was complemented by the xcpP gene introduced in trans on pSB10. The clone was characterized by PCR as previously described (7), with the primers ORG4 (hybridizing at the 3′ end of xcpR) and AFO4 (hybridizing at the 3′ end of xcpP).
The lasB signal sequence was amplified by using the M13 reverse primer (−48) (New England Biolabs) and AFO5, which binds to a site behind the deduced signal peptide cleavage site of LasB (sslasB). The DNA fragment was cloned into pUC19, yielding pSB18. The part of the genes encoding the periplasmic domain of XcpP (residues 57 to 235) and XcpZ were amplified with AFO3 (5′-GGATCCCGCCTGCAACGCAGC-3′)-AFO4 and AFO13 (5′-CGCGGATCCCATCTGCAGT-3′)-AFO14 (5′-CCCAAGCTTACGGCCGCTC-3′), respectively. AFO5 introduced a BamHI site at the 3′ end of sslasB, in frame with the BamHI site created by the 5′ primer, AFO3 or AFO13. The 3′ regions of the xcpP and xcpZ genes were cloned into pSB18 to produce in-frame fusions with lasB, yielding pSB20 and pSB46, respectively. The chimeric genes were inserted into the broad-host-range vector pMMB190, under control of the tac promoter, yielding pSB24 and pSB51. The N-terminal sequence of the LasB′-′XcpP hybrid protein is MKKVSTLDLLFVAIMGVSPAAFA-ADLGSRLQRSP (LasB sequence underlined; position of the leader peptidase cleavage site indicated by a dash; XcpP sequence in italics). The gene encoding the lasB′-′xcpP fusion was also cloned into pT7.5 under control of the φ10 promoter, yielding pSB25.
A hybrid gene (tc′-xcpP) encoding amino acids 1 to 34 of TetA (including the first transmembrane domain) fused to the periplasmic domain of XcpP was constructed. The part of the xcpP gene encoding the periplasmic domain of XcpP was amplified by using oligonucleotides AFO3 and AFO4. The 450-bp PCR product was blunted with T4 DNA polymerase and cloned into the EcoRV site present in the tetracycline resistance region (tetA) of pJBS633 (10), yielding pSB60. The N-terminal amino acid sequence of the Tc′-XcpP hybrid protein is MKSNNALIVILGTVTLDAVGIGLVMPVLPGLLRDGSRLQ (TetA sequence in bold; transmembrane domain underlined; XcpP sequence in italics). The hybrid gene was recloned as a 1-kb EcoRI fragment into the broad-host-range vector pMMB190 under control of the tac promoter, yielding pSB62.
An internal deletion within xcpP, removing the region encoding the coiled-coil structure, was created by PCR with tail-to-tail primers (30). Primers AFO22 (5′-ATCGTTCTCGCTTGTAATG-3′) and AFO23 (5′-ATCGCCACGCCCATC-3′) were used, with pSB58. The amplified fragment was self-ligated, creating an EcoRV site, and reinserted into pMMB67HE, yielding pSB86. The fragment was sequenced and found to have the expected 81-bp deletion and the EcoRV site. We also produced another plasmid, pSB82b, which had a deletion of 300 bp, also in frame.
C-terminal deletions XcpPΔX, XcpPΔP, and XcpPΔM were obtained by cutting at XhoI, PvuI, and MscI restriction sites, respectively, within the xcpP gene. Stop codons were introduced at the 3′ end of the truncated xcpP genes by inserting an Ω-Hg interposon (pHP45ΩHg) (11).
The substitutions of Met for the UGA stop codon at position +1 and Leu for Ile at position +18 in XcpP were obtained by PCR using overlap extension site-directed mutagenesis (27). pSB58 was used as the template with primer AFO37A (5′-ACGAACTGCTTGAATCCCTCGGC-3′) and the M13 universal primer (−47) (New England Biolabs) or AFO37B (5′-GCCGAGGGATTCAAGCAGTTCGT-3′) and reverse primer for the substitution of Met1 in the stop codon and primer AFO38A (5′-AGTGATGTAATCCCTTTCTCC-3′) and the universal primer or AFO38B (5′-GGAGAAAGGGATTACATCACT-3′) and the reverse primer for the substitution of Leu18 in Ile. In each case, the two DNA fragments were mixed and joined by overlap extension PCR using the external universal and reverse primers. The fragments obtained were inserted into pMMB67HE, to give pMG1 and pMG2, respectively. The mutations were checked by sequencing.
Expression in vivo.
Pulse-chase experiments and subcellular fractionation were done with E. coli BL21(DE3), as described previously (14). Overexpression of genes cloned under the control of the tac promoter was induced by adding 1 or 2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (for E. coli or P. aeruginosa, respectively) to a culture grown to 0.5 A600 unit/ml. Samples were taken at various times. Extracellular medium was separated from the cells by centrifugation; proteins were precipitated with 10% trichloroacetic acid and washed with 90% acetone. For each sample, we analyzed the equivalent of 0.1 A600 unit of cells.
SDS-PAGE, immunoblotting, and autoradiography.
Samples were solubilized in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer and separated by electrophoresis in polyacrylamide gels containing SDS. Labeled proteins were detected by autoradiography. Immunoblotting was performed with antisera directed against XcpP, XcpQ, XcpY, and elastase, with peroxidase-conjugated goat anti-rabbit-immunoglobulin G used as the secondary antibody. Proteins were detected by chemiluminescence (Pierce). XcpP and XcpY antibodies were raised by using a purified glutathione S-transferase (GST)–XcpP fusion protein encoded by pSB54 or a GST-XcpY fusion protein encoded by pSB65 as previously described (7).
RESULTS
Sequence analysis and characteristic features of XcpP.
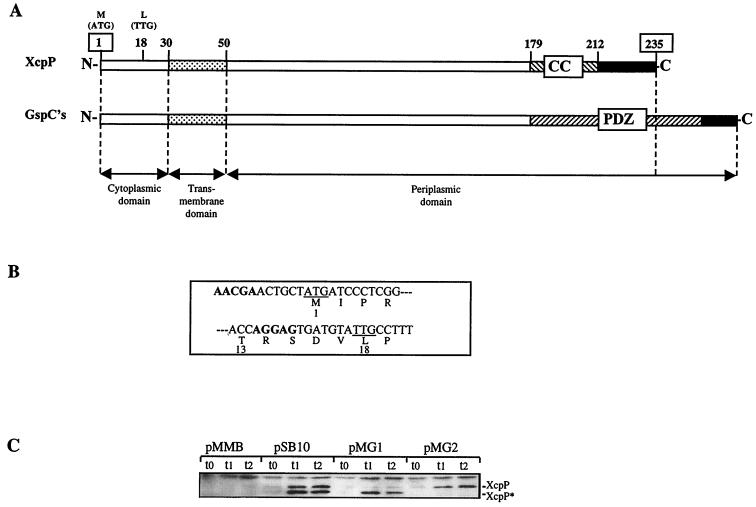
XcpP is a bitopic inner membrane protein (6) consisting of a short N-terminal cytoplasmic domain, a transmembrane domain, and a larger C-terminal periplasmic domain. A coiled-coil structure is predicted, according to the algorithm of Lupas et al. (38), near the C terminus of XcpP between residues 179 and 212. Surprisingly, such a coiled-coil structure is not found in homologs of the GspC family (Fig. 1A), even though those proteins should have similar functions. Instead, Pallen and Ponting (46) reported the presence of a PDZ domain near the C terminus of all GspC-related proteins except XcpP. PDZ domains, named after the three eukaryotic proteins in which they were first discovered (i.e., postsynaptic density, disc large, and zo-1), as well as coiled-coil domains, have been shown to be involved in protein-protein interactions (46). PDZ domains are 80 to 100 residues long (47), whereas the coiled-coil domain extends over 30 residues. The PDZ domain seems to substitute for the coiled-coil domain in the protein structure, since all GspC proteins are accordingly longer than the XcpP protein. P. aeruginosa XcpP is 235 residues long, whereas other GspCs range between 271 (E. coli) and 305 (Vibrio cholerae) residues. Interestingly, the recently sequenced xcpP gene of Pseudomonas alcaligenes (20) is predicted to have a coiled-coil and not a PDZ domain. The coiled-coil domain may thus be an original characteristic of GspC proteins in the Pseudomonaceae.
FIG. 1.
(A) Comparison of characteristic features of P. aeruginosa XcpP (235 amino acids) and other GspC members. Shown are the transmembrane domain ( ), coiled-coil region (CC) (▧), PDZ domain (▨), and extreme C terminus (■). The numbers indicated between XcpP domains correspond to residue positions in P. aeruginosa XcpP. The regions corresponding to the cytoplasmic, transmembrane, and periplasmic domains are delimited with double-headed arrows. The position of the alternative N-terminal residue is indicated (L18), and positions of the first and last XcpP residues are boxed. (B) 3′ region of the xcpP gene. The codons for Met1 and Leu18 are underlined. Boldface letters denote DNA stretches corresponding to the putative Shine-Dalgarno sequences. (C) Characterization of two xcpP gene products by immunodetection after separation by electrophoresis in an 11% acrylamide gel. Samples of TG1 producing both XcpP and XcpP* (pSB10), XcpP* only (pMG1), or XcpP only (pMG2) or containing the vector pMMB67HE were taken at various times after addition of IPTG (t0). t1 = 30 min; t2 = 1 h.
), coiled-coil region (CC) (▧), PDZ domain (▨), and extreme C terminus (■). The numbers indicated between XcpP domains correspond to residue positions in P. aeruginosa XcpP. The regions corresponding to the cytoplasmic, transmembrane, and periplasmic domains are delimited with double-headed arrows. The position of the alternative N-terminal residue is indicated (L18), and positions of the first and last XcpP residues are boxed. (B) 3′ region of the xcpP gene. The codons for Met1 and Leu18 are underlined. Boldface letters denote DNA stretches corresponding to the putative Shine-Dalgarno sequences. (C) Characterization of two xcpP gene products by immunodetection after separation by electrophoresis in an 11% acrylamide gel. Samples of TG1 producing both XcpP and XcpP* (pSB10), XcpP* only (pMG1), or XcpP only (pMG2) or containing the vector pMMB67HE were taken at various times after addition of IPTG (t0). t1 = 30 min; t2 = 1 h.
The xcpP gene encodes two different proteins.
XcpP antibodies were raised as described in Materials and Methods. The XcpP protein was detected as two distinct bands, XcpP and XcpP* (Fig. 1C). We identified a TTG codon that may act as an internal translation start site (Fig. 1B). It is preceded by an AGGAG sequence 6 bp upstream, which could function as a ribosome-binding site (2). The use of this TTG codon as an internal initiation codon would result in the production of a protein lacking the first 17 residues, XcpP*. We checked whether this was the case by performing site-directed mutagenesis to replace the predicted ATG start codon with a stop codon (TGA) and the TTG codon with an isoleucine codon (ATC). Western blot analysis (Fig. 1C) clearly showed that both codons were used as translation start sites. The construct lacking the ATG (pMG1) produced only the lower-molecular-weight form (XcpP*), whereas the construct lacking the TTG (pMG2) produced only the higher-molecular-weight form (XcpP).
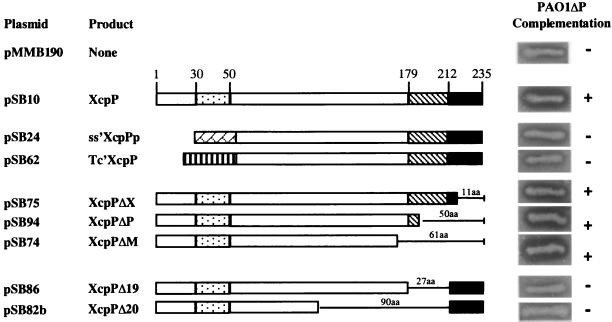
Construction and analysis of truncated XcpP proteins.
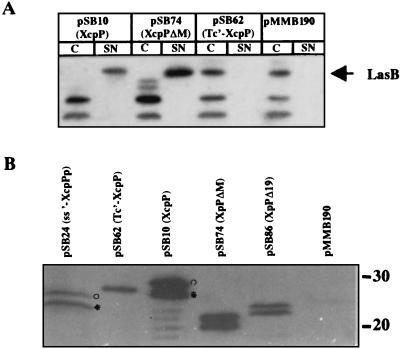
We further characterized the domains of XcpP that are important for function. Several constructs encoding truncated or hybrid XcpP proteins were produced as described in Materials and Methods and Table 1 and are schematically represented in Fig. 2: (i) constructs with replacement of the N terminus containing the hydrophobic segment with either the elastase (LasB) signal peptide (ss′-XcpPp) or a different transmembrane segment (Tc′-XcpP); (ii) C-terminally truncated products (XcpPΔX, XcpPΔP, and XcpPΔM); and (iii) a construct with precise deletion of the region corresponding to the coiled-coil structure (XcpPΔ19). We constructed an xcpP deletion mutant (PAO1ΔP) as described in Materials and Methods. All of the constructs were expressed in this strain, and all of the XcpP variants were detected by Western blotting using antibodies directed against XcpP (Fig. 3B and data not shown). Both forms of XcpP were detected with all variants, unless the N terminus was removed or replaced (Fig. 3B). We deduced that upon detection of two forms of the XcpP variants, the lower-molecular-weight form is truncated at the N terminus. We further tested whether the XcpP variants restored secretion of elastase in the xcpP mutant, by looking at halo formation on skim milk plates (Fig. 2) and by immunodetection of LasB in the cell and supernatant fractions (Fig. 3A). We found that replacement of the native N terminus (ss-′XcpPp and Tc′XcpP) containing the hydrophobic domain prevented the normal functioning of the protein. Moreover, deletion of the coiled-coil structure (XcpPΔ19) or a larger internal domain (XcpPΔ20) also resulted in loss of XcpP function. In contrast, protease plate assays showed that each of the C-terminally truncated XcpP variants, XcpPΔX, XcpPΔP, and XcpPΔM, was functional, producing a halo equal in size to or, especially in the case of XcpPΔM, even larger than that of a strain producing the wild-type form of XcpP (Fig. 2). This increased level of elastase secretion is also clearly seen in Western blot analysis using antibodies directed against LasB (Fig. 3A). Indeed, higher amounts of elastase could be detected in the supernatant of PAO1ΔP containing pSB74 (XcpPΔM) than in PAO1ΔP carrying pSB10 (XcpP). In the case of PAO1ΔP carrying the pMMB190 vector or pSB62 (Tc′XcpP), elastase is found only in the cell fraction. This secretion was specific and was not accompanied by a leakage of periplasmic enzymes such as β-lactamase (data not shown). These observations will be analyzed in Discussion.
FIG. 2.
Schematic representation and characterization of the various forms of XcpP. Sizes of the deletions and amino acid (aa) residue positions are indicated. XcpP domain motifs are as in Fig. 1. Also shown are the LasB signal peptide ( ) and TetA N terminus (
) and TetA N terminus ( ). For each construct, complementation of PAO1ΔP is shown by halo formation on skim milk plates containing 300 μg of carbenicillin per ml and 2 mM IPTG.
). For each construct, complementation of PAO1ΔP is shown by halo formation on skim milk plates containing 300 μg of carbenicillin per ml and 2 mM IPTG.
FIG. 3.
Complementation of the xcpP mutation in PAO1Δ with plasmids carrying recombinant genes encoding variants of XcpP. (A) Immunodetection of elastase in whole-cell extracts (C) or culture supernatants (SN) of PAO1ΔP containing pMMB190 or a plasmid encoding XcpP (pSB10), XcpPΔM (pSB74), or Tc-′XcpP (pSB62). Elastase is indicated by the arrow. (B) Immunodetection, using anti-XcpP antiserum, of variants of XcpP produced in PAO1ΔP containing pSB24 (ss-′XcpPp; precursor (°) and mature (*) forms), pSB62 (Tc′-XcpP), pSB10 (°XcpP and *XcpP*), pSB74 (XcpPΔM), pSB86 (XcpPΔ19), or pMMB190. In both cases, proteins were separated by electrophoresis in an 11% acrylamide gel.
Competitive inhibition of secretion by N-terminally truncated XcpP.
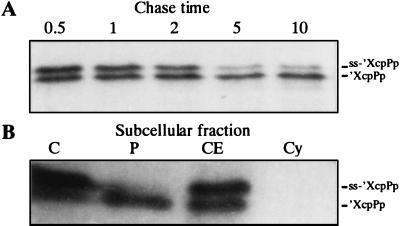
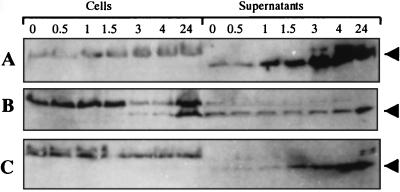
XcpP may be part of a macromolecular complex involving other Xcp proteins. Because of the presence of a large C-terminal periplasmic domain, one should expect that XcpP interacts with Xcp components accessible from this localization. The chimeric gene encoding the periplasmic domain of XcpP fused to the LasB signal peptide (ss′-XcpPp) facilitates transport of the truncated protein into the periplasm. The processing and periplasmic location of the fusion protein was checked by inserting the gene fusion behind the φ10 promoter (pSB25) and expressing it in E. coli BL21(DE3). A pulse-chase experiment showed that ss-′XcpPp was processed into its mature form, the N-terminally truncated XcpP protein (′XcpPp) (Fig. 4A). The cells were fractionated, and most of the mature form was present in the periplasm whereas the precursor form was exclusively associated with the membranes (Fig. 4B). Further, the chimeric gene was inserted into the broad-host-range vector pMMB190 (pSB24), and the resulting plasmid was introduced into PAO1. The cells were grown in TSB, IPTG was added, and samples were taken at various time points after IPTG addition. Western blot analysis was used to estimate the relative amounts of elastase in the cell-associated and extracellular fractions (Fig. 5). In P. aeruginosa carrying the vector (Fig. 5A), elastase was present exclusively in the supernatant; it was not detected in the cell-associated fraction. In contrast, elastase accumulated within bacteria producing ′XcpPp (Fig. 5B) as early as 3 h after IPTG induction. Thus, the C-terminal domain of XcpP competitively interfered with the function of its wild-type counterpart, probably by generating a nonfunctional interaction with its putative partner, thereby interfering with the functioning of the secretion machinery. The competitive inhibitory effect appears to be specific for XcpP, since similar LasB fusions with XcpZ do not have an inhibitory effect on secretion (Fig. 5C).
FIG. 4.
Processing (A) and subcellular location (B) of ss-′XcpPp. E. coli BL21(DE3) cells containing pSB25 encoding ss-′XcpPp were induced by adding IPTG. A 30-s pulse of [35S]methionine was followed by a chase with nonradioactive methionine (A; chase times are indicated in minutes) or subcellular fractionation of the cells (B). C, whole cells; P, periplasmic fraction; CE, cell envelope fraction; Cy, cytoplasmic fraction. Proteins were separated by electrophoresis in an 11% acrylamide gel and were detected by autoradiography. Precursor (ss-′XcpPp) and mature (′XcpPp) forms are indicated.
FIG. 5.
Competitive inhibition assay. Samples of PAO1 cells containing pMMB190 (A), pSB24 encoding ss-′XcpPp (B), or pSB51 encoding ss-′XcpZp (C) were taken at various times (indicated in hours) after addition of IPTG. Cells and supernatants were separated, and their protein contents were analyzed by electrophoresis in an 11% acrylamide gel and transfer onto nitrocellulose sheets. The blots were probed with an antielastase antiserum. The elastase band is indicated; the highest-molecular-weight band is a product of cross-reaction.
XcpP requires XcpQ for stability.
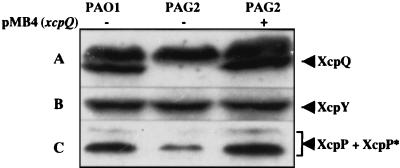
The gene encoding the XcpP protein is organized, together with xcpQ, in a divergent operon with respect to the other xcp genes. This may reflect a coordinate function of both proteins. XcpQ is a member of the secretin family (GspD), which are able to multimerize and to form pores in the outer membrane. The C terminus of the secretin appears to allow membrane insertion, whereas its N terminus extends into the periplasm. The observation that the stability of a protein is dependent on the presence of another protein strongly suggests that there is an interaction between the two components. We investigated whether the stability of XcpP depended on the presence of XcpQ. The wild-type strain PAO1, the isogenic xcpQ mutant PAG2, and PAG2 containing xcpQ on a plasmid were grown in TSB to 3 A600 units/ml. Samples were taken and subjected to electrophoresis, and their protein content was analyzed by Western blotting. There was clearly less XcpP in PAG2 (Fig. 6C, lane 2) than in PAO1 (Fig. 6C, lane 1). The introduction of the xcpQ gene into PAG2 resulted in levels of XcpP similar to those of PAO1 (Fig. 6C, lane 3). The low level of XcpP was specific to PAG2, because it was not observed for XcpY (Fig. 6B). XcpY is another protein of the type II secretory system with a large periplasmic domain. XcpP may be more susceptible to proteases in the absence of XcpQ because it is in a different conformation. We also analyzed the stability of XcpQ in an xcpP mutant strain (PAO1ΔP) but detected no decrease in XcpQ levels, either multimer or monomer (data not shown). Yet, these results are in favor of an interaction between XcpP and XcpQ.
FIG. 6.
XcpP stabilization by XcpQ. Total-cell extracts of PAO1 (lane 1), PAG2 (deletion of xcpQ; lane 2), and PAG2 containing XcpQ encoded by pMB4 (lane 3) were analyzed by SDS-PAGE (11% acrylamide) and immunoblotting with anti-XcpQ (A), anti-XcpY (B), or anti-XcpP (C) antiserum. The positions of XcpQ (the highest-molecular-weight band is a product of cross-reaction), XcpY, and XcpP (doublet, XcpP and -P*) are indicated.
DISCUSSION
In P. aeruginosa, most exoproteins use the type II secretory pathway, or main terminal branch, of the GSP, which consists of 12 Xcp proteins (15). Little is known about the mechanistic role of each Xcp protein in the secretion process. We focused this study on one such inner membrane protein, XcpP.
We initially tried to identify functionally important domains of XcpP. We showed that the protein needs to be anchored in the cytoplasmic membrane, because the periplasmic domain alone, ′XcpPp, is nonfunctional. Yet, the transmembrane segment is not a simple anchor, because no complementation of the xcpP mutation was observed if this domain was replaced with the first transmembrane segment of TetA. Therefore, the membrane anchor or the N-terminal cytoplasmic domain is probably involved in the function of XcpP. An internal translation product, XcpP* (pMG1), lacking the first 17 of the 30 N-terminal cytoplasmic residues is still functional (data not shown), suggesting the requirement of the transmembrane domain in XcpP function. Such a requirement may be due to specific interaction of this transmembrane domain with another membrane component of the Xcp macromolecular complex. This kind of interaction has been proposed in the case of the E. coli TonB-ExbB-ExbD and TolA-TolQ-TolR membrane complexes (1, 31, 35). Deletion of the putative coiled-coil domain, identified at the C terminus of the protein, also resulted in a loss of function, as seen by the noncomplementation of the xcpP mutant. Thus, the coiled-coil structure is also an important motif in XcpP function. Yet, this structure is predicted for XcpP but not for its homologs in the GspC family in which PDZ domains have been found at similar positions within the protein (46), possibly to fulfill the same function.
As part of a multiprotein complex, XcpP probably interacts with one or more components within the secretory apparatus, via the transmembrane domain, the coiled-coil structure, or other regions of the protein not yet characterized. The competitive inhibition assay of secretion, using overproduction of the C-terminal domain of the XcpP protein, shows that one such interaction may take place on the periplasmic side of the cytoplasmic membrane. There are several possible partners for XcpP. Several indications suggest that the XcpQ protein, which forms the ultimate channel of the secretory pathway, could be one of them. The xcpP and xcpQ genes are organized into a single operon at the 40-min locus on the chromosome (2), whereas the other xcp genes are organized into a divergently transcribed operon (4). This organization may indicate that XcpP and XcpQ act in coordination. The XcpP protein was not stable in the absence of XcpQ, suggesting an interaction between the two components. Indeed, in many instances, the absence of a member of a complex results in premature degradation of its partner(s). In the case of the type II secretory apparatus, complexes that have been proposed are PulS-PulD for Klebsiella oxytoca (25), XcpY-XcpZ for P. aeruginosa (40), and ExeA-ExeB for Aeromonas hydrophila (28); in the case of Agrobacterium tumefaciens, a network of stabilization in the T-complex transport apparatus has been proposed (12). Moreover, the interaction between a bitopic inner membrane protein and an outer membrane protein has been previously reported. Indeed, the E. coli TonB protein belongs to a cytoplasmic membrane macromolecular complex including ExbB and ExbD, but it also interacts with the outer membrane receptors for siderophores (34, 42, 57). This interaction induces a cascade of conformational changes, which allows the subsequent entry of the iron-siderophore complex into the cell (41). Such a transport across the outer membrane is supported energetically by the proton motive force (PMF) across the cytoplasmic membrane (50), and this energy source is coupled to the outer membrane siderophore receptor via the TonB protein. In such a system, the production of a truncated TonB protein (containing only the periplasmic domain) blocked the uptake of the substrates, by competing with the wild-type TonB and generating a nonfunctional interaction with the outer membrane receptor (31). Four features of the XcpP protein, (i) bitopic topology including a large periplasmic domain, (ii) specific requirement of the transmembrane domain, (iii) interaction with the outer membrane component, and (iv) competitive inhibition on production of the periplasmic domain, are also features of the TonB protein. The TonB-dependent uptake mechanism may thus provide the basis of a model for energization of the secretion process. Interestingly, PMF has been shown to be required for the GSP-dependent outer membrane translocation of periplasmic intermediates of aerolysin from Aeromonas species (36, 62) and of pullulanase from K. oxytoca (48). Despite the lack of sequence similarity to TonB, XcpP may be a candidate for energizing translocation of exoproteins across the outer membrane. In the A. hydrophila Gsp system, ExeB has been suggested to have a TonB-like function, based on the sequence similarity between ExeB and TonB (28). However, an ExeB-like protein has not been identified in the case of P. aeruginosa.
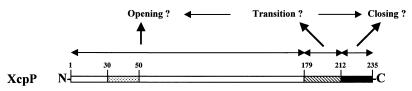
It is clear that the passage of large molecules across the outer membrane, avoiding disruption of its barrier function, requires a tight gating of the pore-forming proteins. The XcpQ channel has a large central cavity (95 Å) (5), and the pore must thus be open only transiently. Some of the results obtained with the C-terminally truncated forms of XcpP may provide evidence for such a statement. For example, it is very interesting that XcpPΔM complemented the xcpP mutation, even though it lacked the XcpP C terminus including the coiled-coil structure. Yet, we have shown that XcpPΔ19, which lacks only the coiled-coil structure, failed to complement the PAO1ΔP mutant, suggesting a key role of this particular domain in XcpP function. This result is original and puzzling, though it is still possible that the N-terminal domain of XcpP, present in XcpPΔM, is alone sufficient for the functioning of the Xcp machinery. Moreover, complementation with the XcpPΔM variant also resulted in an increased level of secretion. One possible explanation of these apparently contradictory results may be as follows. The N-terminal domain of XcpP, present in XcpPΔM, may, directly or via interaction with another Xcp protein, trigger opening of the XcpQ pores, which results in the efficient secretion observed. This large N-terminal domain is also present in the nonfunctional variant XcpPΔ19, which lacks only the coiled-coil structure. The only difference between XcpPΔ19 and XcpPΔM is the presence in XcpPΔ19 of the 22 C-terminal residues of the wild-type protein. This particular part of the C-terminal region may thus be inhibitory through the formation of inactive complexes. It may be involved in negatively controlling the secretion process, possibly by maintaining the XcpQ pores in a closed conformation, which results in the noncomplementation of the PAO1ΔP mutant strain by the XcpPΔ19 variant. This effect of the C terminus is thus dominant over the effect of the N terminus. However, these 22 C-terminal residues are normally present in XcpP and in this case do not prevent opening of the XcpQ pore and functioning of the Xcp machinery. Yet, the difference between XcpPΔ19, which does not complement PAO1ΔP, and XcpP, which does, is the presence in the latter of the coiled-coil domain. Therefore, XcpP may exist in two different states with the coiled-coil structure having a key function in shifting the protein from one conformation to the other, mediating the effect of the N or C terminus of the XcpP protein (Fig. 7). These two states may result from XcpP multimerization, or not, via the coiled-coil domain.
FIG. 7.
Hypothetical model for the role of XcpP domains (see text). XcpP domain motifs are as in Fig. 1.
A transport system more related to the type II secretion system than TonB-dependent uptake is the machinery involved in the assembly of filamentous bacteriophages (54). In this case, the ultimate channel in the outer membrane is the pIV protein, a homolog of XcpQ and member of the secretin family. A second accessory component in this system is the pI protein, a bitopic cytoplasmic membrane protein with no sequence homology with known Xcp proteins. Interestingly, and similar to XcpP, this protein is produced in two forms, pI and pI* (21). Moreover, pI interacts via its C-terminal periplasmic domain with pIV in order to energize the opening of this pore (53). Alternatively, the balance between the two XcpP products (XcpP and XcpP*) may also be involved in the fine tuning of XcpQ pore gating. The existence of an energizing mechanism controlling pore opening during protein secretion is likely, but whether it involves XcpP (GspC) could not be concluded from this study. Moreover, an energy source other than PMF could be transduced to the outer membrane, since pI has an ATP-binding site (21) as does ExeA, the inner membrane partner of ExeB (28).
ACKNOWLEDGMENTS
We thank A. de Groot for providing the PAG2 strain and pMB4 construct before publication, G. Michel for advice on protein purification, and W. Bitter for providing XcpQ antisera.
S. Bleves and M. Gérard-Vincent were supported by the Ministry of Research and Technology. This work was partly supported by the French Cystic Fibrosis Foundation and by Biotech Framework IV grant BIO4 CT960119 from the European Union as part of the Cell Factories Network.
REFERENCES
- 1.Ahmer B M, Thomas M G, Larsen R A, Postle K. Characterization of the exbBD operon of Escherichia coli and the role of ExbB and ExbD in TonB function and stability. J Bacteriol. 1995;177:4742–4747. doi: 10.1128/jb.177.16.4742-4747.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akrim M, Bally M, Ball G, Tommassen J, Teerink H, Filloux A, Lazdunski A. Xcp-mediated protein secretion in Pseudomonas aeruginosa: identification of two additional genes and evidence for regulation of xcp gene expression. Mol Microbiol. 1993;10:431–443. doi: 10.1111/j.1365-2958.1993.tb02674.x. [DOI] [PubMed] [Google Scholar]
- 3.Ball G, Chapon-Hervé V, Bleves S, Michel G, Bally M. Assembly of XcpR in the cytoplasmic membrane is required for extracellular protein secretion in Pseudomonas aeruginosa. J Bacteriol. 1999;181:382–388. doi: 10.1128/jb.181.2.382-388.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bally M, Filloux A, Akrim M, Ball G, Lazdunski A, Tommassen J. Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepilin peptidase. Mol Microbiol. 1992;6:1121–1131. doi: 10.1111/j.1365-2958.1992.tb01550.x. [DOI] [PubMed] [Google Scholar]
- 5.Bitter W, Koster M, Latijnhouwers M, de Cock H, Tommassen J. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol Microbiol. 1998;27:209–219. doi: 10.1046/j.1365-2958.1998.00677.x. [DOI] [PubMed] [Google Scholar]
- 6.Bleves S, Lazdunski A, Filloux A. Membrane topology of three Xcp proteins involved in exoprotein transport by Pseudomonas aeruginosa. J Bacteriol. 1996;178:4297–4300. doi: 10.1128/jb.178.14.4297-4300.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bleves S, Voulhoux R, Michel G, Lazdunski A, Tommassen J, Filloux A. The secretion apparatus of Pseudomonas aeruginosa: identification of a fifth pseudopilin, XcpX (GspK family) Mol Microbiol. 1998;27:31–40. doi: 10.1046/j.1365-2958.1998.00653.x. [DOI] [PubMed] [Google Scholar]
- 8.Bortoli-German I, Brun E, Py B, Chippaux M, Barras F. Periplasmic disulphide bond formation is essential for cellulase secretion by the plant pathogen Erwinia chrysanthemi. Mol Microbiol. 1994;11:545–553. doi: 10.1111/j.1365-2958.1994.tb00335.x. [DOI] [PubMed] [Google Scholar]
- 9.Braun P, Tommassen J, Filloux A. Role of the propeptide in folding and secretion of elastase of Pseudomonas aeruginosa. Mol Microbiol. 1996;19:297–306. doi: 10.1046/j.1365-2958.1996.381908.x. [DOI] [PubMed] [Google Scholar]
- 10.Broome-Smith J K, Spratt B G. A vector for the construction of translational fusions to TEM beta-lactamase and the analysis of protein export signals and membrane protein topology. Gene. 1986;49:341–349. doi: 10.1016/0378-1119(86)90370-7. [DOI] [PubMed] [Google Scholar]
- 11.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez D, Spudich G M, Zhou X R, Christie P J. The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J Bacteriol. 1996;178:3168–3176. doi: 10.1128/jb.178.11.3168-3176.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filloux A, Bally M, Ball G, Akrim M, Tommassen J, Lazdunski A. Protein secretion in gram-negative bacteria: transport across the outer membrane involves common mechanisms in different bacteria. EMBO J. 1990;9:4323–4329. doi: 10.1002/j.1460-2075.1990.tb07881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filloux A, Michel G, Bally M. GSP-dependent protein secretion in Gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol Rev. 1998;22:177–198. doi: 10.1111/j.1574-6976.1998.tb00366.x. [DOI] [PubMed] [Google Scholar]
- 16.Frank D W. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol Microbiol. 1997;26:621–629. doi: 10.1046/j.1365-2958.1997.6251991.x. [DOI] [PubMed] [Google Scholar]
- 17.Frenken L G, de Groot A, Tommassen J, Verrips C T. Role of the lipB gene product in the folding of the secreted lipase of Pseudomonas glumae. Mol Microbiol. 1993;9:591–599. doi: 10.1111/j.1365-2958.1993.tb01719.x. [DOI] [PubMed] [Google Scholar]
- 18.Furste J P, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 19.Genin S, Boucher C A. A superfamily of proteins involved in different secretion pathways in gram-negative bacteria: modular structure and specificity of the N-terminal domain. Mol Gen Genet. 1994;243:112–118. doi: 10.1007/BF00283883. [DOI] [PubMed] [Google Scholar]
- 20.Gerritse G, Ure R, Bizoullier F, Quax W J. The phenotype enhancement method identifies the Xcp outer membrane secretion machinery from Pseudomonas alcaligenes as a bottleneck for lipase production. J Biotechnol. 1998;64:23–38. doi: 10.1016/s0168-1656(98)00101-1. [DOI] [PubMed] [Google Scholar]
- 21.Guy-Caffey J K, Rapoza M P, Jolley K A, Webster R E. Membrane localization and topology of a viral assembly protein. J Bacteriol. 1992;174:2460–2465. doi: 10.1128/jb.174.8.2460-2465.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guzzo J, Duong F, Wandersman C, Murgier M, Lazdunski A. The secretion genes of Pseudomonas aeruginosa alkaline protease are functionally related to those of Erwinia chrysanthemi proteases and Escherichia coli alpha-haemolysin. Mol Microbiol. 1991;5:447–453. doi: 10.1111/j.1365-2958.1991.tb02128.x. [DOI] [PubMed] [Google Scholar]
- 23.Hardie K R, Lory S, Pugsley A P. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 1996;15:978–988. [PMC free article] [PubMed] [Google Scholar]
- 24.Hardie K R, Schulze A, Parker M W, Buckley J T. Vibrio spp. secrete proaerolysin as a folded dimer without the need for disulphide bond formation. Mol Microbiol. 1995;17:1035–1044. doi: 10.1111/j.1365-2958.1995.mmi_17061035.x. [DOI] [PubMed] [Google Scholar]
- 25.Hardie K R, Seydel A, Guilvout I, Pugsley A P. The secretin-specific, chaperone-like protein of the general secretory pathway: separation of proteolytic protection and piloting functions. Mol Microbiol. 1996;22:967–976. doi: 10.1046/j.1365-2958.1996.01539.x. [DOI] [PubMed] [Google Scholar]
- 26.Hirst T R, Holmgren J. Conformation of protein secreted across bacterial outer membranes: a study of enterotoxin translocation from Vibrio cholerae. Proc Natl Acad Sci USA. 1987;84:7418–7422. doi: 10.1073/pnas.84.21.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 28.Howard S P, Meiklejohn H G, Shivak D, Jahagirdar R. A TonB-like protein and a novel membrane protein containing an ATP-binding cassette function together in exotoxin secretion. Mol Microbiol. 1996;22:595–604. doi: 10.1046/j.1365-2958.1996.d01-1713.x. [DOI] [PubMed] [Google Scholar]
- 29.Hu N T, Hung M N, Chen D C, Tsai R T. Insertion mutagenesis of XpsD, an outer-membrane protein involved in extracellular protein secretion in Xanthomonas campestris pv. campestris. Microbiology. 1998;144:1479–1486. doi: 10.1099/00221287-144-6-1479. [DOI] [PubMed] [Google Scholar]
- 30.Imai Y, Matsushima Y, Sugimura T, Terada M. A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res. 1991;19:2785. doi: 10.1093/nar/19.10.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaskula J C, Letain T E, Roof S K, Skare J T, Postle K. Role of the TonB amino terminus in energy transduction between membranes. J Bacteriol. 1994;176:2326–2338. doi: 10.1128/jb.176.8.2326-2338.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaniga K, Delor I, Cornelis G R. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 33.Kazmierczak B I, Mielke D L, Russel M, Model P. pIV, a filamentous phage protein that mediates phage export across the bacterial cell envelope, forms a multimer. J Mol Biol. 1994;238:187–198. doi: 10.1006/jmbi.1994.1280. [DOI] [PubMed] [Google Scholar]
- 34.Larsen R A, Foster-Hartnett D, McIntosh M A, Postle K. Regions of Escherichia coli TonB and FepA proteins essential for in vivo physical interactions. J Bacteriol. 1997;179:3213–3221. doi: 10.1128/jb.179.10.3213-3221.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazzaroni J C, Vianney A, Popot J L, Benedetti H, Samatey F, Lazdunski C, Portalier R, Geli V. Transmembrane alpha-helix interactions are required for the functional assembly of the Escherichia coli Tol complex. J Mol Biol. 1995;246:1–7. doi: 10.1006/jmbi.1994.0058. [DOI] [PubMed] [Google Scholar]
- 36.Letellier L, Howard S P, Buckley J T. Studies on the energetics of proaerolysin secretion across the outer membrane of Aeromonas species. Evidence for a requirement for both the protonmotive force and ATP. J Biol Chem. 1997;272:11109–11113. doi: 10.1074/jbc.272.17.11109. [DOI] [PubMed] [Google Scholar]
- 37.Lindeberg M, Salmond G P, Collmer A. Complementation of deletion mutations in a cloned functional cluster of Erwinia chrysanthemi out genes with Erwinia carotovora out homologues reveals OutC and OutD as candidate gatekeepers of species-specific secretion of proteins via the type II pathway. Mol Microbiol. 1996;20:175–190. doi: 10.1111/j.1365-2958.1996.tb02499.x. [DOI] [PubMed] [Google Scholar]
- 38.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 39.Maniatis T E, Fritsch F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 40.Michel G, Bleves S, Ball G, Lazdunski A, Filloux A. Mutual stabilization between XcpZ and XcpY components of the secretory apparatus in Pseudomonas aeruginosa. Microbiology. 1998;144:3379–3386. doi: 10.1099/00221287-144-12-3379. [DOI] [PubMed] [Google Scholar]
- 41.Moeck G S, Coulton J W. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol Microbiol. 1998;28:675–681. doi: 10.1046/j.1365-2958.1998.00817.x. [DOI] [PubMed] [Google Scholar]
- 42.Moeck G S, Coulton J W, Postle K. Cell envelope signaling in Escherichia coli. Ligand binding to the ferrichrome-iron receptor fhua promotes interaction with the energy-transducing protein TonB. J Biol Chem. 1997;272:28391–28397. doi: 10.1074/jbc.272.45.28391. [DOI] [PubMed] [Google Scholar]
- 43.Morales V M, Backman A, Bagdasarian M. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene. 1991;97:39–47. doi: 10.1016/0378-1119(91)90007-x. [DOI] [PubMed] [Google Scholar]
- 44.Nunn D N, Lory S. Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc Natl Acad Sci USA. 1991;88:3281–3285. doi: 10.1073/pnas.88.8.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nunn D N, Lory S. Cleavage, methylation, and localization of the Pseudomonas aeruginosa export proteins XcpT, -U, -V, and -W. J Bacteriol. 1993;175:4375–4382. doi: 10.1128/jb.175.14.4375-4382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pallen M J, Ponting C P. PDZ domains in bacterial proteins. Mol Microbiol. 1997;26:411–413. doi: 10.1046/j.1365-2958.1997.5591911.x. [DOI] [PubMed] [Google Scholar]
- 47.Ponting C P, Phillips C, Davies K E, Blake D J. PDZ domains: targeting signalling molecules to sub-membranous sites. Bioessays. 1997;19:469–479. doi: 10.1002/bies.950190606. [DOI] [PubMed] [Google Scholar]
- 48.Possot O M, Letellier L, Pugsley A P. Energy requirement for pullulanase secretion by the main terminal branch of the general secretory pathway. Mol Microbiol. 1997;24:457–464. doi: 10.1046/j.1365-2958.1997.3451726.x. [DOI] [PubMed] [Google Scholar]
- 49.Possot O, Pugsley A P. Molecular characterization of PulE, a protein required for pullulanase secretion. Mol Microbiol. 1994;12:287–299. doi: 10.1111/j.1365-2958.1994.tb01017.x. [DOI] [PubMed] [Google Scholar]
- 50.Postle K. TonB protein and energy transduction between membranes. J Bioenerg Biomembr. 1993;25:591–601. doi: 10.1007/BF00770246. [DOI] [PubMed] [Google Scholar]
- 51.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reeves P J, Douglas P, Salmond G P. Beta-lactamase topology probe analysis of the OutO NMePhe peptidase, and six other Out protein components of the Erwinia carotovora general secretion pathway apparatus. Mol Microbiol. 1994;12:445–457. doi: 10.1111/j.1365-2958.1994.tb01033.x. [DOI] [PubMed] [Google Scholar]
- 53.Russel M. Protein-protein interactions during filamentous phage assembly. J Mol Biol. 1993;231:689–697. doi: 10.1006/jmbi.1993.1320. [DOI] [PubMed] [Google Scholar]
- 54.Russel M. Moving through the membrane with filamentous phages. Trends Microbiol. 1995;3:223–228. doi: 10.1016/s0966-842x(00)88929-5. [DOI] [PubMed] [Google Scholar]
- 55.Sandkvist M, Bagdasarian M, Howard S P, DiRita V J. Interaction between the autokinase EpsE and EpsL in the cytoplasmic membrane is required for extracellular secretion in Vibrio cholerae. EMBO J. 1995;14:1664–1673. doi: 10.1002/j.1460-2075.1995.tb07155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shevchik V E, Robert-Baudouy J, Condemine G. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J. 1997;16:3007–3016. doi: 10.1093/emboj/16.11.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skare J T, Ahmer B M, Seachord C L, Darveau R P, Postle K. Energy transduction between membranes. TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J Biol Chem. 1993;268:16302–16308. [PubMed] [Google Scholar]
- 58.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 59.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thomas J D, Reeves P J, Salmond G P. The general secretion pathway of Erwinia carotovora subsp. carotovora: analysis of the membrane topology of OutC and OutF. Microbiology. 1997;143:713–720. doi: 10.1099/00221287-143-3-713. [DOI] [PubMed] [Google Scholar]
- 61.Tommassen J, Filloux A, Bally M, Murgier M, Lazdunski A. Protein secretion in Pseudomonas aeruginosa. FEMS Microbiol Rev. 1992;9:73–90. doi: 10.1016/0378-1097(92)90336-m. [DOI] [PubMed] [Google Scholar]
- 62.Wong K R, Buckley J T. Proton motive force involved in protein transport across the outer membrane of Aeromonas salmonicida. Science. 1989;246:654–656. doi: 10.1126/science.2814486. [DOI] [PubMed] [Google Scholar]