Abstract
Introduction
Severe cutaneous adverse reactions (SCAR) are a group of T cell-mediated hypersensitivities associated with significant morbidity, mortality and hospital costs. Clinical phenotypes include Stevens-Johnson syndrome (SJS), toxic epidermal necrolysis (TEN), drug reaction with eosinophilia and systemic symptoms (DRESS) and acute generalised exanthematous pustulosis (AGEP). In this Australasian, multicentre, prospective registry, we plan to examine the clinical presentation, drug causality, genomic predictors, potential diagnostic approaches, treatments and long-term outcomes of SCAR in Australia and New Zealand.
Methods and analysis
Adult and adolescent patients with SCAR including SJS, TEN, DRESS, AGEP and another T cell-mediated hypersensitivity, generalised bullous fixed drug eruption, will be prospectively recruited. A waiver of consent has been granted for some sites to retrospectively include cases which result in early mortality. DNA will be collected for all prospective cases. Blood, blister fluid and skin biopsy sampling is optional and subject to patient consent and site capacity. To develop culprit drug identification and prevention, genomic testing will be performed to confirm human leukocyte antigen (HLA) type and ex vivo testing will be performed via interferon-γ release enzyme linked immunospot assay using collected peripheral blood mononuclear cells. The long-term outcomes of SCAR will be investigated with a 12-month quality of life survey and examination of prescribing and mortality data.
Ethics and dissemination
This study was reviewed and approved by the Austin Health Human Research Ethics Committee (HREC/50791/Austin-19). Results will be published in peer-reviewed journals and presented at relevant conferences.
Trial registration number
Australian New Zealand Clinical Trials Registry (ACTRN12619000241134).
Keywords: IMMUNOLOGY, Adverse events, EPIDEMIOLOGY, PREVENTIVE MEDICINE, CLINICAL PHARMACOLOGY, Dermatological epidemiology
Strengths and limitations of this study.
The strength of Australasian registry of Severe Cutaneous Adverse Reactions lies in its prospective design, allowing for the real-time collection of clinical data, standardised investigation and sampling for genomic evaluation and ex vivo diagnostics.
There will likely be selection bias favouring the recruitment of adults, less critically ill patients, patients living in metropolitan areas where the majority of study sites are located and patients whose first language is English.
A limitation of the sampling strategy is the large blood volume required due to the number of peripheral blood mononuclear cells (PBMCs) required for successful enzyme linked immunospot assay, complicated by the often poor viability of cells from acute bleeds due to critical illness. Many hospital-adjacent laboratories are also not set up to process PBMCs, blister fluid or skin specimens requiring same-day transport to specialised research labs.
Selection bias is also likely to affect the sampling strategy as treating teams may be less willing to overburden children and critically ill patients.
Finally, the registry is not designed to achieve complete case capture across the study period and recruiting the target sample size will be dependent on successful recognition of cases at each site.
Introduction
Severe cutaneous adverse reactions (SCAR) are a group of T cell-mediated hypersensitivities—including Stevens-Johnson syndrome (SJS), toxic epidermal necrolysis (TEN), drug reaction with eosinophilia and systemic symptoms (DRESS) and acute generalised exanthematous pustulosis (AGEP).1 Other severe T cell-mediated hypersensitivities include diseases such as generalised bullous fixed drug eruption (GBFDE), drug-induced liver injury (DILI) and acute interstitial nephritis (AIN).1 2 SCAR can be associated with significant morbidity, hospital costs, increased demand for specialist testing and high mortality.3–6 Despite this, an understanding of the clinical, genomic and pharmacological predictors of each phenotype remains absent from current practice. Further, there is currently a lack of consistency among the recommendations in treatment guidelines for these severe reactions.
Causality assessment in SCAR is often complex as multiple drugs are frequently involved.3 7–9 While the use of skin testing (in vivo) has been increasingly employed, it is hampered by low sensitivity and concerns around safety.10–12 Progress has been made in understanding the underlying immune mechanisms and genetic predisposition of SCAR,13 providing strong support for the role of human leukocyte antigen (HLA) typing in identifying specific drug-associated reactions (eg, HLA-B*57:01 for abacavir hypersensitivity, HLA-B*58:01 for allopurinol hypersensitivity).14 15 Further association studies are required, however, to capitalise on this knowledge and previously provided roadmap. The use of novel ex vivo T-cell diagnostics to aid drug causality assessment, including lymphocyte transformation testing (LTT) and interferon-γ enzyme linked immunospot (ELISpot) assay, has been used with some success in a research setting but their clinical utility is not yet established.12 16–22
The long-term outcomes of SCAR including quality of life, disease recurrence, inadvertent drug re-exposure and medication safety are not well-described. The development of a regional clinical and DNA registry of SCAR will allow investigators to (a) perform surveillance for new and emerging drug causality, (b) develop causality and phenotypic prediction rules, (c) understand best-practice treatment approaches, (d) discover genomic predictors that prevent SCAR onset and (e) improve long-term outcomes and medication safety. The additional biospecimen component of the study will allow investigators to assess the utility of T-cell diagnostics in aiding drug causality. While national SCAR registries have been set up successfully in Europe (RegiSCAR) and Korea (KoSCAR),23 24 Australasian Registry of Severe Cutaneous Adverse Reactions (AUS-SCAR) will be the first registry of severe drug allergy in the Australasian region.
Methods and analysis
AUS-SCAR is an Australasian, multicentre, prospective registry of severe cutaneous adverse reactions in adults and adolescents >12 years of age. Participating sites for recruitment include Austin Health, Alfred Health, Eastern Health, Peter MacCallum Cancer Centre, St Vincent’s Hospital Melbourne, Monash Health, Royal Melbourne Hospital, Epworth Healthcare, Campbelltown Hospital, Nepean Hospital, St Vincent’s Hospital Sydney, Fiona Stanley Hospital, Sir Charles Gairdner Hospital, Royal Brisbane and Women’s Hospital, Queensland Children’s Hospital, Royal Adelaide Hospital, Flinders Medical Centre, Royal Darwin Hospital, Launceston General Hospital and Auckland City Hospital. The Peter Doherty Institute for Infection and Immunity and Institute for Immunology and Infectious Diseases (IIID) are collaborating sites for sample analysis. Recruitment commenced in July 2019 and is planned to continue across all sites until accrual has reached n=500.
Study oversight
A 5–7 person Steering Committee will be formed following nominations from participating site principal investigators. Steering Committee members will meet regularly to discuss the direction of the project, assess requests from investigators and identify any potential risks or issues. The chair of the Steering Committee will be the chief investigator. A Steering Committee member can step down from this role at any time and nominations for a new member will be sought from participating site principal investigators. The Steering Committee should have at least one of the following at all times: infectious diseases physician, allergist/immunologist and dermatologist.
The clinical and DNA database and results of the study are the common property of AUS-SCAR investigators and cannot be used without the formal authorisation of the Steering Committee. The release of results at conference proceedings, meetings and/or in publication are decided on by the Steering Committee of AUS-SCAR.
Project development
Any principal investigator can propose a project using AUS-SCAR data. This proposal will be forwarded to all investigators for comment and the Steering Committee will provide (a) feedback to the proposing investigator and (b) approval/non-approval of the project.
Patient involvement
Although they have not been involved in the study design, patients will be invited to assist with identifying the best methods of disseminating results to patients and the public following publication. Patients or consumer representatives may also be considered for inclusion in the Steering Committee.
Participants
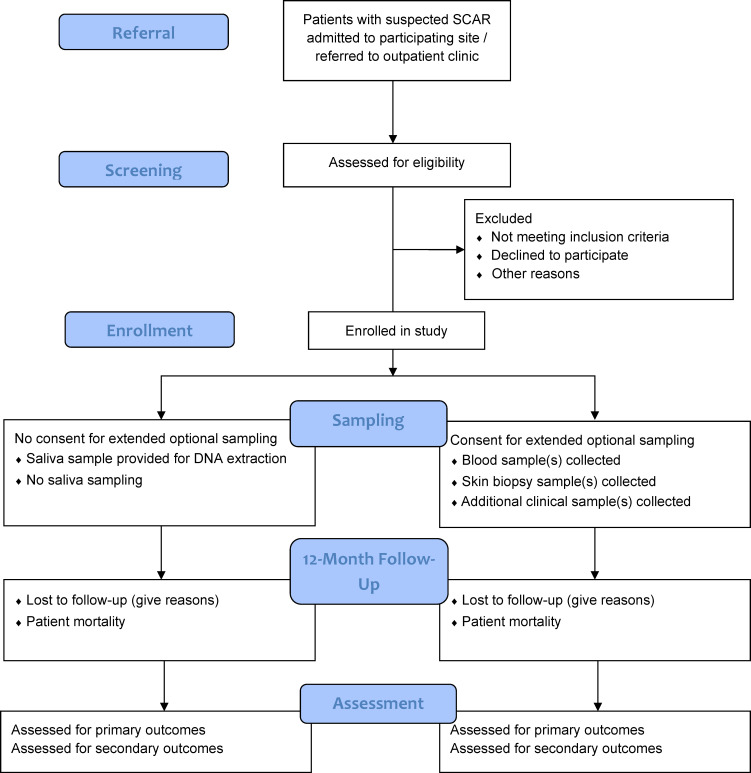
Patients will be invited to participate in the registry if (a) they are admitted to or referred to a participating site with a suspected SCAR, (b) two site investigators have agreed on inclusion, (c) SCAR is confirmed by a site dermatologist or immunologist or is biopsy proven and (d) the SCAR phenotype is consistent with disease-specific criteria and alternative diagnoses have been ruled out. Disease-specific criteria for SJS/TEN includes the presence of widespread erythema with skin detachment and more than one blister; patients with a suspected viral trigger will still be included if they meet eligibility criteria and case validation. For GBFDE, there must be multifocal, widespread, bullous-type fixed drug eruption characterised by sharply defined bullae at site of recurrent drug administration. Criteria for DRESS and AGEP are outlined in online supplemental table 1 and 2, respectively. The study design and biospecimen sampling schedule are outlined in figure 1.25
Figure 1.
Overview of Australasian Registry of Severe Cutaneous Adverse Reactions study design (adapted from Eldridge et al25). SCAR, severe cutaneous adverse reactions.
bmjopen-2021-055906supp001.pdf (276.2KB, pdf)
At a later stage of the project, extended recruitment may include the following phenotypes, following approval by >50% of principal investigators and the Project Steering Committee:
AIN: required hospitalisation and biopsy proven with acute kidney injury (increase in serum creatinine by ≥0.3 mg/dL within 48 hours or increase in serum creatinine to ≥1.5 times baseline within 7 days or urine output <0.5 mL/kg/hour for 6 hours)26 with other common causes excluded. Non-biopsy proven cases with a single implicated drug, urinary and peripheral eosinophilia, acute kidney injury and resolution of renal disease postdrug removal can be included.
DILI: required hospitalisation and biopsy proven with ≥5× upper limit of normal (ULN) for alanine aminotransferase (ALT) or ≥2 ULN for ALP, or ≥3 ULN for ALT with bilirubin ≥2 ULN.27 Non-biopsy proven cases with a single implicated drug, acute liver injury, autoimmune and other causes excluded and resolution of liver disease postdrug removal can be included.
FDE: dermatologist confirmed, well-defined, circular, hyper-pigmented single or a few plaques that recur in fixed locations on ingestion of drug (therefore requires two or more occurrences).
Biospecimen collection and analysis
Part 1: clinical data and DNA collection
Following informed consent, baseline patient demographics, medical history and clinical data including histopathology and clinical photography will be collected from the patient and medical record and entered into a secure REDCap database. Consent may be obtained from the patient, parent/guardian or medical treatment decision-maker, as appropriate and subject to local or national law. A waiver of consent has been obtained for some participating institutions to collect clinical data retrospectively for patients that die prior to obtaining informed consent.
DNA will be collected for all patients enrolled prospectively: this will be extracted from saliva for patients who do not consent to additional sample collection or from whole blood or peripheral blood mononuclear cells (PBMCs) for patients who agree to additional blood collection. The HLA testing process at the Institute for Immunology and Infectious Diseases (IIID) in Perth Western Australia is accredited by the American Society for Histocompatibility and Immunogenetics (ASHI) and the National Association of Testing Authorities (NATA). Specific HLA Loci will be PCR amplified using sample specific MID-tagged primers that amplify polymorphic exons from Class I (A, B, C Exons 2 and 3) and Class II (DQ, Exons 2 and 3; DRB and DPB1, Exon 2) MHC genes. MID tagged primers have been optimised to minimize allele dropouts and primer bias. Amplified DNA products from unique MID tagged products (up to 96 MIDs) will be pooled in equimolar ratios and subjected to library preparation using NEBNext Ultra II library prep kits (New England Biolabs). Libraries will be quantified using the Jetseq library quantitation kit (Meridian Bioscience) and High sensitivity D1000 screentape on an Agilent 2200 Tapestation (Agilent) for concentration and size distribution. Normalised libraries will then be sequenced on the Illumina MiSeq platform using the MiSeq V3 600-cycle kit (2X300bp reads). Sequences will be separated by MID tags and alleles called using an in house accredited HLA allele caller software pipeline that minimises the influence of sequencing errors. Alleles are called using the latest IMGT HLA allele database as the allele reference library. Sample to report integrity will be tracked and checked using proprietary and accredited Laboratory Information and Management System (LIMS) and HLA analyse reporting software that performs comprehensive allele balance and contamination checks on the final dataset. Further Genome Wide Association Studies, RNA-seq and virological assessment may be performed on stored DNA samples to determine if genetic and viral variants are associated with SCAR phenotype.
Part 2: additional biospecimen collection
This part of the study is optional for participating sites and for patients. Samples will need to be stored at the participating site or at a collaborating AUS-SCAR site if there is capacity. Informed consent must be obtained for all samples requested. The sampling schedule is outlined below and in online supplemental table 3.
Blood
For adults, an initial blood sample of 100–150 mL will be taken following informed consent. A reduced blood volume of 20–50 mL is taken for patients under 18 years of age. A convalescent follow-up blood draw of the same volume will be taken at 6 weeks to 12 months post-SCAR onset for consenting patients. Repeat blood draws may be requested at both time points if the full volume was unable to be collected or cell viability is poor, subject to ongoing patient consent.
Skin biopsy
A 3–4 mm punch biopsy may be taken from an affected area at the acute stage and 6 weeks to 12 months post-SCAR onset. For a subset of patients, repeat biopsies may be requested for comparison studies.
Blister fluid
For patients with blisters present, blister fluid of any volume may be collected into a heparinised tube for the extraction of PBMCs. For patients whose blisters reform, additional fluid collection may be requested.
Additional clinical samples
Additional clinical samples may be collected if considered to be of potential clinical relevance by the site investigator (eg, cerebrospinal fluid, lymph node aspirate).
Sample processing
PBMCs will be isolated from blood and blister fluid and stored at -80°C, to be utilised for DNA extraction and immunological studies such as IFN-γ release enzyme-linked immunosorbent spot (ELISpot) assay12, flow cytometry and alternative ex vivo diagnostics. Plasma will be separated from blood samples during the PBMC isolation process and stored at -80°C. Skin biopsy and clinical samples may be stored and processed for exploratory immunological studies. Samples are to be processed and stored at participating sites (or a collaborating AUS-SCAR site on request from participating site) and cannot be used in AUS-SCAR studies without written approval from the participating site. The participating site can choose to use these samples for any research project they have additional local ethics approval for, without consent required from AUS-SCAR investigators. The Doherty Institute for Infection and Immunity or IIID are available for assistance with sample processing where possible.
Part 3: patient follow-up
A 12-month follow-up will be performed to collect data regarding (a) drug utilisation, (b) health outcomes and (c) quality of life (online supplemental materials). Whether the patient received a referral for further allergy/immunology evaluation (left to the discretion of the treating team) and the results of any allergy testing performed will be captured. Mortality/morbidity and prescribing data will be extracted from the medical record. For available patients, a quality of life assessment will be performed using the previously validated and adapted Drug Hypersensitivity Quality of Life Questionnaire (DrHy-Q).28 A telephone script is provided with the DrHy-Q in online supplemental materials. The questionnaire may also be sent out via email. Prescribing and mortality data may be obtained from the Pharmaceutical Benefits Scheme (PBS) following approval or the participant’s general practitioner (GP) where not available from the medical record or patient.
Study aims
To develop a national registry of SCAR to ensure continuous surveillance of new and emerging causative drugs and improve pharmacovigilance.
To describe the causality and epidemiology of SCAR in Australasia.
To determine patient, drug and clinical factors that predict SCAR phenotypes and which treatments improve outcomes.
To determine the treatments employed for SCAR in Australasia and describe patient outcomes.
To determine pharmacogenomic and genomic associations in SCAR.
To examine the long-term impacts of SCAR including restricted medication use and quality of life (QOL) impact.
Primary outcome measures
-
Proportion (n, %) of SCAR secondary to antibiotics or non-antibiotics (addressed using the most likely implicated drug as determined through internal and external validation).
Proportion (n, %) for each drug class.
Secondary outcome measures
-
Proportion (n, %) of SCAR treated with a potentially disease-modifying therapy.
Proportion (n, %) for each treatment type, for example, topical or systemic corticosteroids, intravenous immunoglobulin, immunomodulators (eg, rituximab), antivirals (eg, ganciclovir).
For each treatment type: length-of-stay (days), inpatient mortality, all-cause mortality (90 days, 12 months), disease recurrence.
-
Proportion (n, %) of SCAR referred for in vivo/ex vivo allergy testing.
Proportion (n, %) of each testing modality: patch testing, intradermal testing, ex vivo (LTT, ELISpot, other).
Proportion (n, %) positive on skin testing.
For each SCAR phenotype: proportion (n, %) associated with inpatient and 12-month mortality.
For each SCAR phenotype: proportion (n, %) associated with relapse or drug re-exposure within 12 months.
-
Risk factors associated with development of each SCAR phenotype.
Patient (host factors), drug (pharmacological class), clinical (disease factors).
-
Risk factors associated with SCAR mortality.
Patient (host factors), drug (pharmacological class), disease (phenotype).
For each SCAR phenotype and implicated drug: Genomic associations within HLA class I and/or II.
Utility of in vitro/ex vivo diagnostics in assigning drug causality in SCAR.
The long-term sequelae of SCAR.
Sample size
Due to the registry study design, a sample size calculation is not required. The projected recruitment number over a 5-year period is 500 (80 cases are estimated over 5 years at a single institution3—allowing for 35% minimum case capture at 20 sites, the extrapolated numbers are 560 over this period).
Statistical analysis
Statistical analysis using Stata Statistical Software: Release 16 (College Station, TX: StataCorp LLC.) will be performed by study investigators with the assistance of a biostatistician as required. Categorical variables will be summarised using frequency and percentage and compared using a χ2 test or Fisher’s exact test. Continuous variables will first be assessed for significant skew using a Shapiro-Wilk test. They will then be summarised using mean and SD or median and IQR as appropriate and compared using a t-test or Wilcoxon signed-rank/Mann-Whitney U test. Multivariable logistic regression modelling will be used to examine secondary objectives related to variables/factors predicting specified outcomes.
Ethics and dissemination
This study was registered on 19 February 2019 and received ethical approval from the Austin Health Human Research Ethics Committee (HREC Number HREC/50791/Austin-19) in May 2019.
An initial publication of pilot data is planned after the recruitment of 100 patients. Patient demographics, SCAR phenotype, implicated drugs, ELISpot and HLA results will be included. Principal investigators from each participating site will be included as authors. Authorship will be guided by the Project Steering Committee with reference to the International Committee of Medical Journal Editors guidelines.
Further results of this research project are planned to be published and/or presented in a variety of scientific forums and journals. The data presented will explore SCAR epidemiology in Australasia, drug causality, genomic predictors, diagnostic tools and potential treatments.
Supplementary Material
Acknowledgments
EJP reports grants R01HG010863, R01AI152183, U01AI154659, R13AR078623, UAI109565) from the National Institutes of Health and additional funding from the National Health and Medical Research Council of Australia. She is co-director of IIID Pty Ltd that holds a patent for HLA-B*57:01 testing for abacavir hypersensitivity, and has a patent pending for Detection of Human Leukocyte Antigen-A*32:01 in connection with Diagnosing Drug Reaction with Eosinophilia and Systemic Symptoms without any financial remuneration and not directly related to the submitted work. EJP receives consulting fees and royalties from UpToDate and consulting fees from Janssen, AstraZeneca, Regeneron, Verve and Biocryst.
Footnotes
Twitter: @AnaCopaescu, @TrubianoJason
Contributors: FJ is the project manager and corresponding author, responsible for providing central operational oversight of the study. JT, EJP, JY, MSYG, WT, CZ, DG and CK contributed to the initial design of the study and drafting of the protocol. FJ, MSYG, EM, SV, KC, NEH, AA, A-MC, JFDL, CZ, DG, HC, AD, JK, CK, FT, SB, JY, WT, WS, AC, TA, AL, JB, LM, AKA, EJP and JT contributed to pilot data acquisition informing the study design and substantial revision of the manuscript. JT is the coordinating principal investigator, providing overall responsibility for the study. All authors have read and approved the final manuscript.
Funding: This research was funded by the Allergy and Immunology Foundation of Australasia (AIFA) and the Faculty of Medicine, Dentistry and Health Sciences, The University of Melbourne (Grant numbers not applicable).
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Ethics statements
Patient consent for publication
Not applicable.
References
- 1.Konvinse KC, Phillips EJ, White KD, et al. Old dog Begging for new tricks: current practices and future directions in the diagnosis of delayed antimicrobial hypersensitivity. Curr Opin Infect Dis 2016;29:561–76. 10.1097/QCO.0000000000000323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duong TA, Valeyrie-Allanore L, Wolkenstein P, et al. Severe cutaneous adverse reactions to drugs. Lancet 2017;390:1996–2011. 10.1016/S0140-6736(16)30378-6 [DOI] [PubMed] [Google Scholar]
- 3.Trubiano JA, Aung AK, Nguyen M, et al. A comparative analysis between Antibiotic- and Nonantibiotic-Associated delayed cutaneous adverse drug reactions. J Allergy Clin Immunol Pract 2016;4:1187–93. 10.1016/j.jaip.2016.04.026 [DOI] [PubMed] [Google Scholar]
- 4.Saka B, Barro-Traoré F, Atadokpédé FA, et al. Stevens-Johnson syndrome and toxic epidermal necrolysis in sub-Saharan Africa: a multicentric study in four countries. Int J Dermatol 2013;52:575–9. 10.1111/j.1365-4632.2012.05743.x [DOI] [PubMed] [Google Scholar]
- 5.Lin Y-F, Yang C-H, Sindy H, et al. Severe cutaneous adverse reactions related to systemic antibiotics. Clin Infect Dis 2014;58:1377–85. 10.1093/cid/ciu126 [DOI] [PubMed] [Google Scholar]
- 6.Su P, Aw CWD. Severe cutaneous adverse reactions in a local hospital setting: a 5-year retrospective study. Int J Dermatol 2014;53:1339–45. 10.1111/ijd.12118 [DOI] [PubMed] [Google Scholar]
- 7.Kardaun SH, Sekula P, Valeyrie-Allanore L, et al. Drug reaction with eosinophilia and systemic symptoms (dress): an original multisystem adverse drug reaction. results from the prospective RegiSCAR study. Br J Dermatol 2013;169:1071–80. 10.1111/bjd.12501 [DOI] [PubMed] [Google Scholar]
- 8.Rive CM, Bourke J, Phillips EJ. Testing for drug hypersensitivity syndromes. Clin Biochem Rev 2013;34:15–38. [PMC free article] [PubMed] [Google Scholar]
- 9.Blumenthal KG, Peter JG, Trubiano JA, et al. Antibiotic allergy. Lancet 2019;393:183–98. 10.1016/S0140-6736(18)32218-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phillips EJ, Bigliardi P, Bircher AJ, et al. Controversies in drug allergy: testing for delayed reactions. J Allergy Clin Immunol 2019;143:66–73. 10.1016/j.jaci.2018.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trubiano JA, Douglas AP, Goh M, et al. The safety of antibiotic skin testing in severe T-cell-mediated hypersensitivity of immunocompetent and immunocompromised hosts. J Allergy Clin Immunol Pract 2019;7:1341–3. 10.1016/j.jaip.2018.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trubiano JA, Strautins K, Redwood AJ, et al. The Combined Utility of Ex Vivo IFN-γ Release Enzyme-Linked ImmunoSpot Assay and In Vivo Skin Testing in Patients with Antibiotic-Associated Severe Cutaneous Adverse Reactions. J Allergy Clin Immunol Pract 2018;6:1287–96. 10.1016/j.jaip.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White KD, Abe R, Ardern-Jones M, et al. SJS/TEN 2017: building multidisciplinary networks to drive science and translation. J Allergy Clin Immunol Pract 2018;6:38–69. 10.1016/j.jaip.2017.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mallal S, Phillips E, Carosi G, et al. HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med 2008;358:568–79. 10.1056/NEJMoa0706135 [DOI] [PubMed] [Google Scholar]
- 15.Yun J, Marcaida MJ, Eriksson KK, et al. Oxypurinol directly and immediately activates the drug-specific T cells via the preferential use of HLA-B*58:01. J Immunol 2014;192:2984–93. 10.4049/jimmunol.1302306 [DOI] [PubMed] [Google Scholar]
- 16.Kano Y, Hirahara K, Mitsuyama Y, et al. Utility of the lymphocyte transformation test in the diagnosis of drug sensitivity: dependence on its timing and the type of drug eruption. Allergy 2007;62:1439–44. 10.1111/j.1398-9995.2007.01553.x [DOI] [PubMed] [Google Scholar]
- 17.Porebski G. In vitro assays in severe cutaneous adverse drug reactions: are they still research tools or diagnostic tests already? Int J Mol Sci 2017;18. 10.3390/ijms18081737. [Epub ahead of print: 10 Aug 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Redwood A, Trubiano J, Phillips EJ. Prevention and diagnosis of severe T-cell-mediated adverse drug reactions: are we there yet? J Allergy Clin Immunol Pract 2019;7:228–30. 10.1016/j.jaip.2018.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klaewsongkram J, Sukasem C, Thantiworasit P, et al. Analysis of HLA-B allelic variation and IFN-γ ELISpot responses in patients with severe cutaneous adverse reactions associated with drugs. J Allergy Clin Immunol Pract 2019;7:219–27. 10.1016/j.jaip.2018.05.004 [DOI] [PubMed] [Google Scholar]
- 20.Pichler WJ. Drug hypersensitivity: Karger publishers, 2007. [Google Scholar]
- 21.Copaescu A, Mouhtouris E, Vogrin S, et al. The Role of In Vivo and Ex Vivo Diagnostic Tools in Severe Delayed Immune-Mediated Adverse Antibiotic Drug Reactions. J Allergy Clin Immunol Pract 2021;9:2010–5. 10.1016/j.jaip.2020.12.052 [DOI] [PubMed] [Google Scholar]
- 22.Copaescu A, Gibson A, Li Y, et al. An updated review of the diagnostic methods in delayed drug hypersensitivity. Front Pharmacol 2020;11:573573. 10.3389/fphar.2020.573573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kardaun SH, Sekula P, Valeyrie-Allanore L, et al. Drug reaction with eosinophilia and systemic symptoms (DRESS): an original multisystem adverse drug reaction. Results from the prospective RegiSCAR study. British Journal of Dermatology 2013;169:1071–80. 10.1111/bjd.12501 [DOI] [PubMed] [Google Scholar]
- 24.Kang DY, Yun J, Lee SY. Korean severe cutaneous adverse reactions Consortium. A nationwide study of severe cutaneous adverse reactions based on the multicenter registry in Korea. J Allergy Clin Immunol Pract 2021;9:929–36. [DOI] [PubMed] [Google Scholar]
- 25.Eldridge SM, Chan CL, Campbell MJ, et al. Consort 2010 statement: extension to randomised pilot and feasibility trials. BMJ 2016;355:i5239. 10.1136/bmj.i5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kidney disease: improving global outcomes (KDIGO) acute kidney injury work group. KDIGO clinical practice guideline for acute kidney injury. Kidney Int 2012;2:1–138. [Google Scholar]
- 27.Aithal GP, Watkins PB, Andrade RJ, et al. Case definition and phenotype standardization in drug-induced liver injury. Clin Pharmacol Ther 2011;89:806–15. 10.1038/clpt.2011.58 [DOI] [PubMed] [Google Scholar]
- 28.Baiardini I, Braido F, Fassio O, et al. Development and validation of the drug hypersensitivity quality of life questionnaire. Ann Allergy Asthma Immunol 2011;106:330–5. 10.1016/j.anai.2010.12.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2021-055906supp001.pdf (276.2KB, pdf)