Summary
Borrelia burgdorferi (Bb), a vector-borne bacterial pathogen and the causative agent of Lyme disease, can spread to distant tissues in the human host by traveling in and through monolayers of endothelial cells (ECs) lining the vasculature. To examine whether Bb alters the physical forces of ECs to promote its dissemination, we exposed ECs to Bb and observed a sharp and transient increase in EC traction and intercellular forces, followed by a prolonged decrease in EC motility and physical forces. All variables returned to baseline at 24 h after exposure. RNA sequencing analysis revealed an upregulation of innate immune signaling pathways during early but not late Bb exposure. Exposure of ECs to heat-inactivated Bb recapitulated only the early weakening of EC mechanotransduction. The differential responses to live versus heat-inactivated Bb indicate a tight interplay between innate immune signaling and physical forces in host ECs and suggest their active modulation by Bb.
Subject areas: Immunology, Microbiology, Cell biology, Biophysics, Transcriptomics
Graphical abstract
Highlights
-
•
Early exposure to Borrelia decreases endothelial cell motility and physical forces
-
•
Early exposure to Borrelia also upregulates the host’s innate immune signaling pathways
-
•
Host cell mechanics and signaling return to steady state at late exposure times
-
•
Exposure to dead bacteria steadily reduces motility and physical forces of host cells
Immunology; Microbiology; Cell biology; Biophysics; Transcriptomics
Introduction
Lyme disease is a multisystem infectious disease and the most common and rapidly growing tick-borne infection in the northern hemisphere (Feria-Arroyo et al., 2014; Meriläinen et al., 2015). It is caused by a group of bacteria that belong to the Borrelia burgdorferi (sensu lato) complex, comprised more than 20 genospecies (Shan et al., 2021). In the majority of infected individuals, prompt treatment with antibiotics is sufficient to resolve the infection (CDC Lyme Disease). However, a subset of infected patients does not return to health and experiences symptoms including neurologic impairment, fatigue, cardiac and arthritic hallmarks as well as a progressive atrophy of the skin – which persist for over 6 months after antibiotic treatment. Although the cause of these continued symptoms is unknown, there is evidence of the continued presence of Bb antigens and ongoing inflammatory responses (Jutras et al., 2019; Rouse et al., 2021).
Infected ticks transmit Bb into the dermal interstitial fluid while feeding, and from there the pathogens can disseminate throughout the body (Coburn et al., 2021). A key step in bacterial dissemination is the ability of Bb to bypass the endothelial cell (EC) lining of blood vessels. To maintain their barrier function ECs form protein complexes at their surfaces that allow them to strongly anchor to the extracellular matrix (ECM) through focal adhesions and to each other through cell-cell junction complexes. The actin cytoskeleton connects these anchors to each other forming a network that allows the cells to transmit forces to their ECM (traction forces) and to each other (intercellular forces).
Various advances in biomechanical techniques have recently enabled the characterization of both the kinematics and dynamics of the motion of cells in monolayer in health and disease. For example, traction force microscopy (TFM) allows direct measurement of the traction forces generated by confluent ECs on their matrix through the active engagement of focal (cell-ECM) adhesions and actomyosin contractility (Rokhzan et al., 2019). An additional technique, monolayer stress microscopy (MSM) enables indirect calculation of the forces that cells in monolayer exert on one another, which is critical in revealing how cells regulate intercellular communication and barrier integrity (Bastounis et al., 2022; Bazellières et al., 2015). Calculation of cell-generated forces has also provided key insight into how certain intracellular bacterial pathogens modulate host cell forces to their own benefit, that is, to disseminate better through tissues (Faralla et al., 2018; Lamason et al., 2016). For example, intracellular Listeria monocytogenes hijacks host epithelial cell mechanotransduction weakening the traction stresses that host epithelial cells exert on their ECM, so that it can more efficiently transmigrate through the cell ventral surface and into the underlying basement membrane (Faralla et al., 2018). Although Bb is traditionally considered an extracellular pathogen, studies have shown that it can also get internalized within multiple cell types including ECs (Wu et al., 2011; Ma et al., 1991; Livengood and Gilmore, 2006; Larsen et al., 2003; Girschick et al., 1996), suggesting that Bb infection may be able to induce similar alterations in host EC biomechanics.
Of interest, gene expression analysis of host cells, including epithelial and endothelial cells, infected with Bb in vitro has revealed that many of the pathways that are related to the regulation of the actin cytoskeleton and focal adhesions show differential expression during the early and late stages of infection (LaFrance et al., 2011). Various integral components of host cell focal adhesions, like the integrin heterodimers α5β1 and αvβ3, facilitate adhesion of Bb to host cells (Coburn et al., 1998). They also mediate endothelial cell migration and mechanotransduction (Bastounis et al., 2019; Reinhart-King et al., 2005), suggesting that by interfering with integrin signaling Bb may alter host cell mechanics. In addition, exposure of different host cell types to Bb activates innate immune signaling pathways such as those dependent on the transcription factor NF-κB (Dev et al., 2011). Of interest, we recently showed that activation of innate immune signaling, particularly of NF-κB, by infection can lead to dramatic changes in host cell mechanics including a decrease in traction forces of infected as compared to uninfected cells (Bastounis et al., 2021). In the context of Bb infection, a weakening in intercellular force transduction could enhance paracellular permeability favoring bacterial transmigration through the endothelium.
Tick-transmitted Bb typically enters into the host bloodstream as single spirochetes whose diameter of 200 nm may be critical for transmigration through the endothelium and other tissues (Meriläinen et al., 2015; Burns et al., 1997). Bb can also localize intracellularly in many host cell types, including ECs, from which it can escape clearance and transmigrate transcellularly (Coleman et al., 1995; Tan et al., 2021; Kumar et al., 2015). Moreover, Bb can undergo different morphological transitions, forming for example spherical round bodies and aggregates (Anderson and Brissette, 2021; Sapi et al., 2012). Whether these alternative Bb forms emerge as a result of the harsh in vitro conditions, the multiplicity of infection, or represent a mechanism to evade in vivo host immune responses remains controversial (Murgia and Cinco, 2004; Anderson et al., 2003).
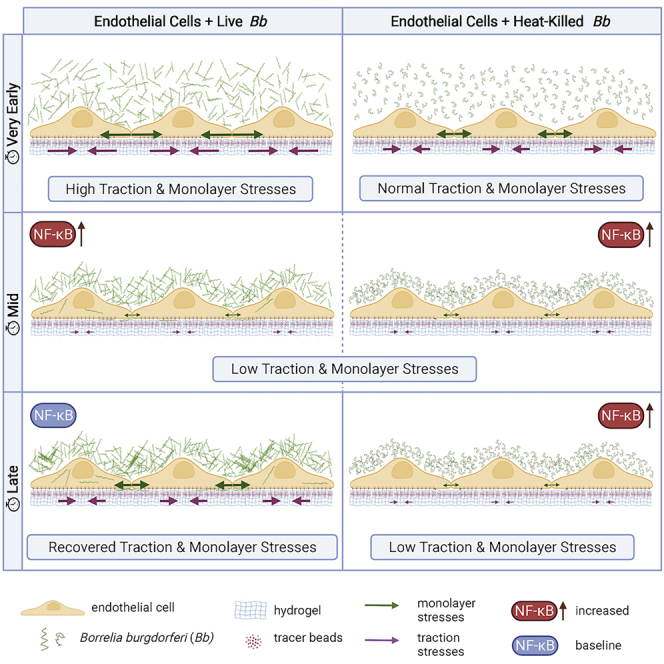
To determine whether exposure of ECs to Bb alters their biomechanics, we used video-microscopy to monitor EC monolayers for up to two days after exposure to Bb. We discovered a sharp and transient increase in EC traction forces and intercellular forces exerted on neighboring ECs, followed by a prolonged decrease in EC motility and physical forces up to 15 h after exposure (hpe). But all variables returned to levels similar to those of unexposed ECs at a later stage (24 hpe). Concomitantly, RNA sequencing analysis of Bb-exposed or unexposed ECs revealed that multiple innate immune signaling pathways (e.g., NF-κB, TNFα) were significantly upregulated at early but not late exposure. Of interest, we found that exposure of ECs to heat-inactivated Bb recapitulates the early weakening of EC mechanotransduction but not the reversal observed at late exposure. Altogether our findings suggest a tightly regulated interplay between innate immune signaling and physical forces in host ECs and a differential modulation of those processes at early versus late exposure to Bb.
Results
Bb are internally localized within ECs in a dosage-dependent manner at early exposure
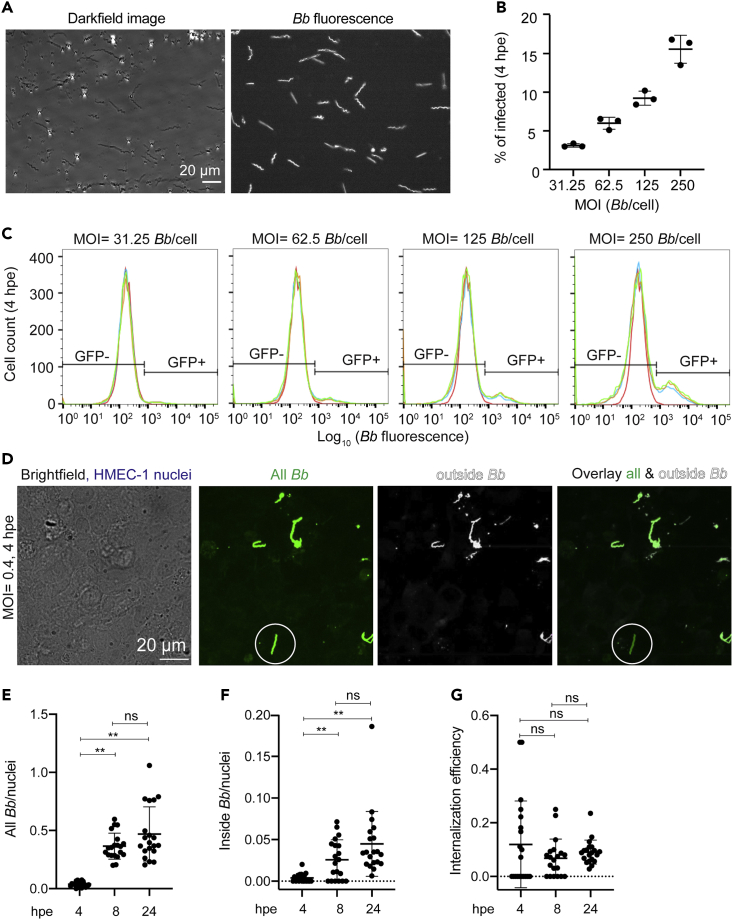
To infer whether exposure of ECs to Bb alters host cell mechanics, we first sought to evaluate how exactly Bb interacts with ECs and whether bacteria are localized extracellularly or also intracellularly. To this end, we grew Bb constitutively expressing GFP (Bb-GFP) in BSK-H media to mid-log phase (Dunham-Ems et al., 2009) and confirmed via microscopy that all Bb retained single spirochete morphology and expressed GFP (Figure 1A and Video S1). To quantify internalization efficiency of Bb into ECs, we exposed monolayers of human microvascular endothelial cells −1 (HMEC-1) to Bb-GFP for 4hat different multiplicities of infection (MOI). We used HMEC-1 as model host cells because they are dermal microvascular ECs and therefore probably the first type of ECs Bb interacts with after skin infection through a tick bite. In addition, this cell line has been previously used to interrogate interactions of Bb with ECs (LaFrance et al., 2011). Following extensive washing, we trypsinized the cell monolayers and determined the fraction of ECs that were GFP-positive in each well using flow cytometry (Figures 1B and 1C). At a constant host cell density, we found a monotonic increase in the number of Bb-infected ECs with increasing MOI. To confirm that Bb is internally localized within ECs, we also performed differential labeling of extracellular Bb-GFP cells (inside/outside labeling) followed by 3D confocal microscopy (Figures 1D and S1A–S1C). Samples were fixed shortly after exposure and specifically labeled with anti-Bb antibody under nonpermeabilizing conditions. This inside/outside labeling method allowed us to distinguish between extracellular Bb (GFP-positive and antibody-labeled Bb) and those that resided within ECs (only GFP-positive). We observed both internally-localized spirochetes as well as spirochetes adhering to the EC surface or that had transmigrated beneath the EC monolayer by 4 hpe. Although the number of bacteria adhering to host cells or being internally localized significantly increased from 4 to 8 hpe, we found no significant differences between 8 and 24 hpe (Figures 1E and 1F). In addition, the internalization efficiency remained constant irrespective of how long ECs were exposed to Bb (Figure 1G). These findings suggest that bacterial adhesion and internalization significantly increase during the first hpe. They also raise the question of whether the lack of changes observed between 8 hpe versus 24 hpe could be because of Bb persisting in a viable but not proliferative state onto or within ECs, thus resulting in a constant adhesion rate and internalization efficiency between 8 hpe and 24 hpe.
Figure 1.
ECs internalize Bb in a dose-dependent manner
(A) Darkfield (left) and fluorescence image (right) of Bb constitutively expressing GFP (Bb-GFP) grown to a concentration of 5 × 107 bacteria/mL in BSK-H media.
(B and C) Barplots of percentage of ECs infected with Bb versus MOI (mean ± SD) (B) and corresponding histograms of the logarithm of Bb-GFP fluorescence intensity per cell for ECs infected with different MOIs of Bb (N = 3 replicate wells shown in different colors) (C). The histogram of control unexposed cells is shown in red. Based on the autofluorescence of the control group, a gate is defined showing what is considered non-exposed (left, GFP-) and exposed (right, GFP+).
(D) Images of fixed samples of ECs exposed to Bb-GFP at a MOI = 11 at 4 hpe. Inside/outside staining was used to confirm internal localization of Bb. Left to right: brightfield image superimposed to maximum intensity projection of EC nuclei (blue), Bb-GFP fluorescence, antibody fluorescence of non-internalized adhering bacteria and overlay of the last two channels. White circle indicates an internalized spirochete.
(E–G) Barplots of all Bb-GFP spirochetes per host cell nucleus (E), inside Bb-GFP spirochetes per host cell nucleus (F), and invasion efficiency (inside/outside Bb) (mean ± SD, WRST: ∗∗p<0.01, ns: non signficant) at different time points after exposure. N = 20 fields of view were segmented and analyzed. See also Figure S1 and Video S1.
Time-lapse recording of Bb spirochetes constitutively expressing GFP (Bb1286, Bb-GFP) within a microscope slide. Phase contrast (left) and Bb-GFP fluorescence (right) are shown. Bb-GFP were grown to a concentration of 5x107 bacteria/mL in BSK-H medium and were imaged before EC exposure to determine exact Bb concentration. Scale bar and corresponding times are indicated.
Host EC motility transiently reduces during early Bb exposure
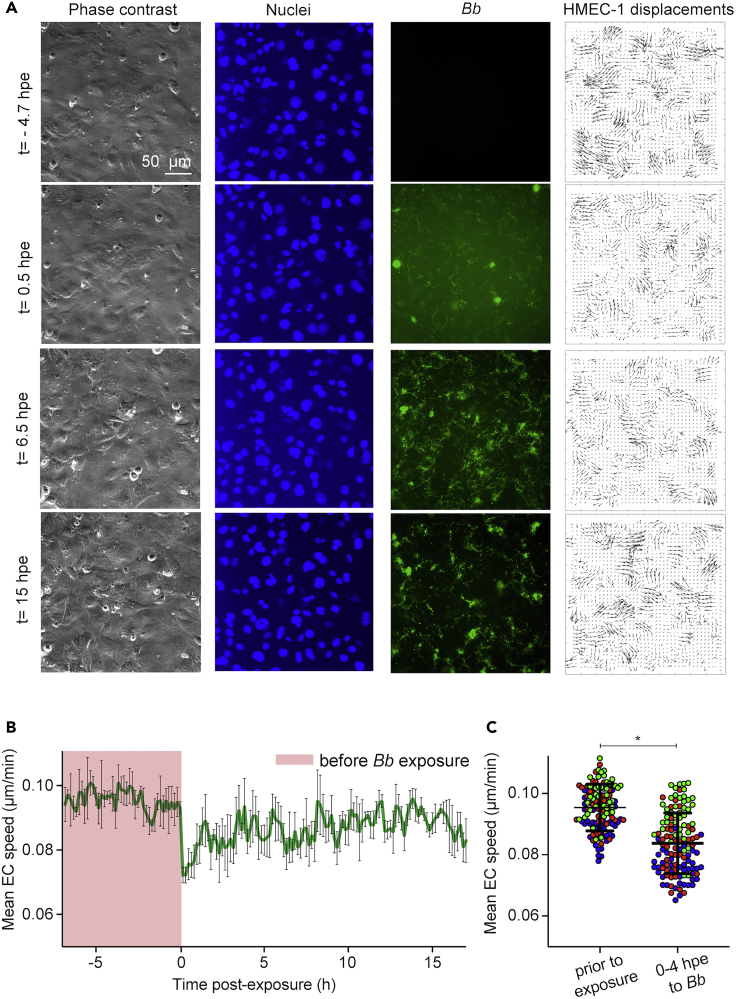
To examine in more detail how do Bb-GFP interact spatiotemporally with ECs in monolayer and what changes in EC kinematics those interactions could possibly induce, we monitored ECs exposed to Bb-GFP overtime using time-lapse epifluorescence microscopy (Figure 2A and Video S2). EC nuclei were stained with Hoechst so that the motion of ECs could be tracked 7 h before and up to 17 hpe to Bb-GFP by applying particle image velocimetry onto successive images of the EC nuclei (Gui and Wereley, 2002). We found that EC displacements and mean speed of motion decreased dramatically during the first few hours after exposure but at later time points ECs recovered their speed to levels just slightly lower than those before exposure (Figures 2A–2C). In our recordings we also observed that at later time points Bb formed aggregates. Motivated by these results, we used the Bb fluorescence images to calculate the integral Bb fluorescence across the whole field of view (FOV) to assess whether Bb continue to grow at later time points. We found that the integral of Bb fluorescence intensity increased progressively after bacteria had been added on top of the host ECs and reached a peak at a time point that depended on the MOI (the lower the MOI, the earlier this peak was observed, Figure S2A). Thereafter, Bb fluorescence decreased overtime until it reached a plateau and remained constant until the end of the recording (∼2 days after exposure). Irrespective of the MOI, no changes in mean Bb fluorescence or total area occupied by Bb occurred at 24 hpe and thereafter (Figures S2A and S2B). To characterize more precisely the changes in Bb morphology over the course of exposure to ECs and to identify whether single spirochetes are still present at late exposure, we used image segmentation followed by classification of the Bb into three distinct classes namely: single Bb (class A, blue); Bb networks (class B, magenta); and Bb aggregates (class C, green) (Figure S2C). We found that shortly after exposure to Bb were mostly in a single spirochete configuration, but overtime spirochetes formed networks and eventually bacterial aggregates although we could still observe motile single Bb spirochetes (Figure S2D and Video S2). We then performed propidium iodide (PI) staining of ECs exposed to Bb at 4 or 24 hpe to distinguish between live versus dead Bb (Figure 2E) (Krämer et al., 2016). Although we found an increase in the total PI fluorescence and in the area in whichBb-GFP co-localized with PI, at 24 hpe many of the bacterial aggregates present were PI-negative suggesting they were still viable (Figures S2E and S2F). To further corroborate this and rule out that the recovery of EC motility at late times after exposure was because of Bb death, we inspected supernatants of ECs exposed for 4 or 24 h to Bb-GFP at either MOI = 200 or MOI = 22 using higher resolution epifluorescence microscopy (Figures S3A and S3B). At 4 hpe at both MOIs most of the spirochetes were motile and no morphological aberrations could be observed either by dark-field or epifluorescence microscopy. At 24 hpe, some spirochetes lacked motility at both MOI = 22 and MOI = 200 and some developed blebs, but single motile spirochetes were still present. This finding was further confirmed when we incubated bacterial supernatants from EC-Bb co-cultures at 4 or 24 hpe into BSK-H medium and observed acidification of the medium, indicative of growth of the spirochetes under all conditions tested, albeit to a lesser extent compared to Bb never placed into the EC culture medium (data not shown). Thus, we can conclude that the recovery of EC motility at later times after Bb exposure is not because of bacterial death since viable spirochetes are still present, although their morphology and proliferation rate differ from those they would exhibit in BSK-H medium.
Figure 2.
ECs slow down during the early stages of Bb-exposure but their motility is recovered at later time points
(A) Representative time-lapse epifluorescence microscopy images of ECs in monolayer during exposure to Bb at a MOI =200. Columns show: phase contrast image; Hoechst-stained EC nuclei; Bb-GFP fluorescence; cellular displacements. Rows show different time points after exposure.
(B) Plot of mean host cell speed versus time (h) relative to the time point when host cells were exposed to Bb (mean ± SD, N = 3 recordings). Magenta dashed line corresponds to the time immediately after addition of Bb.
(C) Boxplots of mean EC speed for ECs before Bb-exposure (tracked for 4 h) and after exposure to Bb at MOI = 200 up to 4 hpe. Different colors correspond to different recordings and circles depict mean spread in the whole field of view (mean ± SD, WRST: ∗p<0.05). See also Figure S2 and Video S2.
Time-lapse epifluorescence video-microscopy of HMEC-1 cells in monolayer exposed to Bb-GFP at MOI = 200, followed by a second movie of cells that were exposed to Bb-GFP at MOI = 22. Recording started at 7 h before exposure and continued up to 33.2 hpe, with a time frame interval of 10 min. Upper left panel shows the phase contrast image, upper right panel shows the image of Hoechst-stained HMEC-1 nuclei (blue), bottom left panel shows the Bb fluorescence image, and bottom right panel the overlay of all channels. Scale bar and corresponding times are indicated. Note that although aggregates are formed at late exposure (∼24 hpe), viable motile spirochetes can still be observed at both MOIs.
Traction forces exerted by ECs onto their ECM are attenuated during early Bb exposure
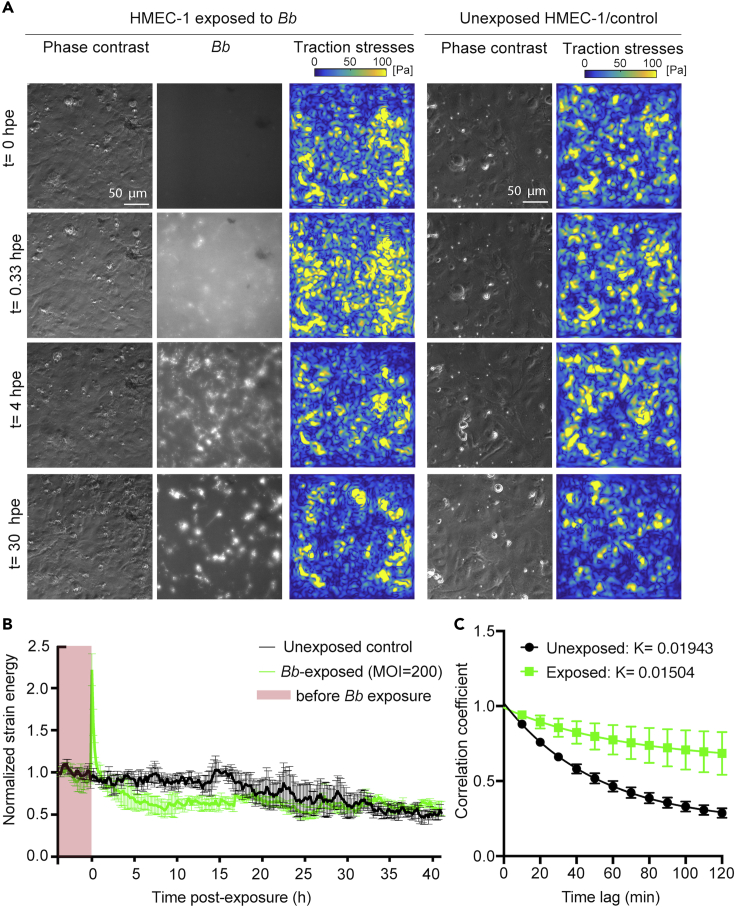
Adhesion of Bb onto ECs is mediated by the interaction between various bacterial adhesins and ECM components including different integrin subunits (Coburn et al., 1998, 2021; Ebady et al., 2016; Wu et al., 2011). We thus speculated that, during exposure, Bb could potentially interfere with focal adhesion organization and traction force generation of ECs in a stage-dependent manner through its interactions with integrin subunits, which could explain the attenuation we previously observed in EC motility. To test this hypothesis, we used TFM to monitor ECs 4 h before exposure and up to 2 days after exposure to a high dosage of Bb (MOI = 200) (Figure 3A, left). Although it might not accurately reflect a “normal in vivo infection”, a high MOI was chosen for this experiment to recognize a discernible effect in EC mechanotransduction and thereby gain new mechanistic information as it is often done in such assays. At the same time, we also recorded unexposed ECs to exclude the possibility of changes in traction forces because of an increase in cell confluence that can occur over the course of a day-long recording (Figure 3A, right). We found that upon addition of Bb-GFP to ECs, their strain energy (i.e., the mechanical work which ECs impact to deform their ECM) increased abruptly 2-fold within the first 0.5 hpe but decreased over time to 2-fold lower levels as compared to before exposure or to unexposed cells (Figure 3B). The decrease in traction stresses and strain energy as compared to unexposed cells was maintained up to approximately 15 hpe. After this point, the normalized strain energy appeared identical in Bb-exposed and unexposed cells and, in both cases, lower as compared to the beginning of the recording, likely as a result of increased cell confluence (Hur et al., 2012). Surprisingly, when we correlated the ECM deformation maps, we discovered that the turnover of traction adhesions (active EC adhesions that transduce force to the ECM) of Bb-exposed cells was significantly slower and less dynamic compared to that of unexposed cells (Figure 3C), a feature that can also be observed by inspection of the traction stress maps in Figure 3A.
Figure 3.
ECs weaken their traction stresses during the early but not late stages of Bb exposure
(A) Representative phase contrast image (first column), Bb-GFP fluorescence (second column) and cellular traction stress map (third column, Pa) for ECs in monolayer at different time points (rows) after exposure to Bb-GFP (MOI =200). TFM was performed for ECs residing on 3 kPa ECM. The fourth and fifth columns show the corresponding phase contrast image and cellular traction stress map for cells not exposed to Bb.
(B) Normalized strain energy (mechanical work) imparted by ECs during a TFM recording (mean ± SEM, N = 3 independent experiments). Strain energy has been normalized with respect to the first value at the beginning of each recording. Green: ECs exposed to Bb-GFP (MOI = 200); black: unexposed ECs. Time (h) is represented relative with respect to the time at whichBb was added. Time before exposure is shaded in red.
(C) Plot showing the cross-correlation coefficient versus time of the cellular deformation maps obtained via TFM for successive frames separated by different time delays for unexposed or Bb-exposed ECs (MOI = 200) tracked for 24 hpe. An exponential decay function was fitted into the data yielding a rate constant K= 0.01943 for unexposed ECs and K= 0.01504 for Bb-exposed ECs (see STAR Methods). See also Figure S3.
EC monolayer stresses are weakened only at early but not late Bb exposure
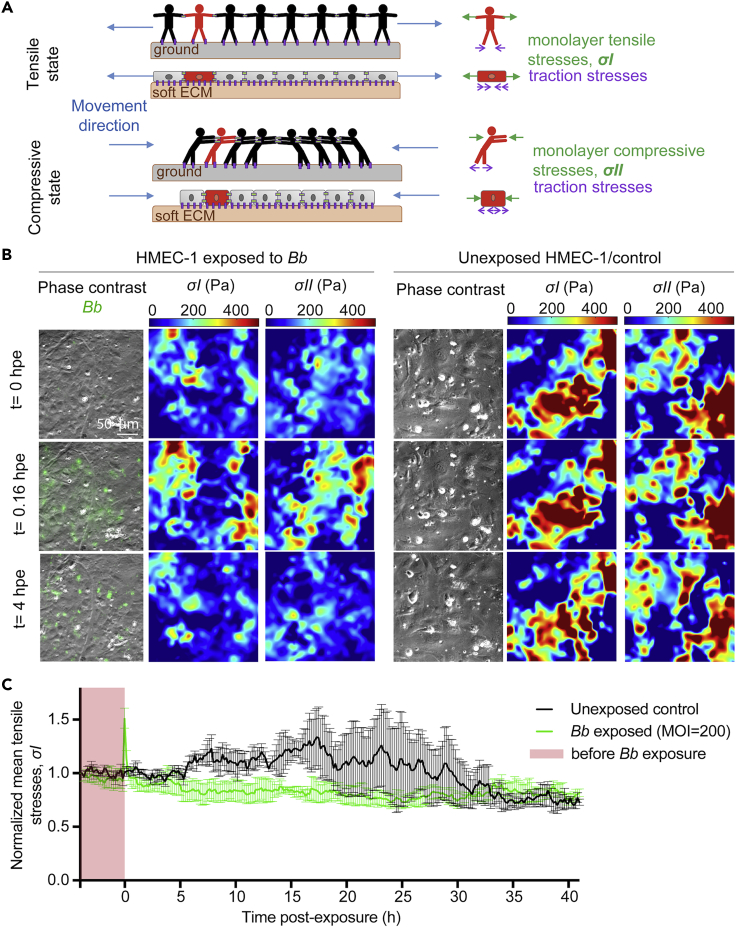
Our previous findings demonstrated that the traction stresses exerted by ECs on their ECM are weakened during early but not late exposure to Bb. However, cells in a monolayer are also able to transduce stresses to each other through their intercellular junctions which are key in regulating barrier function and tissue integrity (Tambe et al., 2011). Those stresses can be tensile or compressive, can result from cells contracting, expanding, or being at rest in different spatial locations and change overtime because of the reorganization of the cytoskeleton and adhesions (Figures 4 and S4A). To measure stresses everywhere within the EC monolayer and to examine how they might change during Bb exposure, we used MSM to indirectly estimate intra- and intercellular stresses within a cell monolayer (thereon referred to as monolayer stresses).
Figure 4.
EC monolayer stresses lower during early but not late Bbexposure
(A) Sketch of the physical forces present in collectives of cells, and analogue in humans, in 1D. Cells in the monolayer are subjected to tensional and compressive stresses which at any instance balance with the traction stresses on the substrate. Left sketches depict the whole monolayer while right sketches focus on a single cell/human (green). Top. Idealized situation in which all cells are pushed by adjacent cells toward the center of the layer. Cells are subjected to compressive stresses while there are no tensile stresses exerted between cells (). In the human analogue, humans are subjected to compressive stresses that balance with each other and with the friction exerted in the ground, resulting in a zero net force in each individual when there is no movement. Bottom. Idealized situation in which all the cells in the monolayer are pulled by adjacent cells away from the center of the layer and are thus subjected to tensile stresses only (). The analogue for humans is also shown.
(B) Representative phase contrast image with Bb-GFP fluorescence superimposed (first column), monolayer tensile stresses (, second column) and compressive stresses (, third column) for ECs in monolayer at different time points (rows) after exposure to Bb-GFP (MOI =200). Fourth-sixth columns show the corresponding phase contrast image, monolayer tensile stresses and absolute value of compressive stresses for ECs not exposed to Bb.
(C) Normalized mean monolayer tensile stresses () as a function of time after exposure (mean ± SEM, N = 3 independent experiments). Mean has been normalized with respect to the first value at the beginning of each recording. Green: ECs exposed to Bb-GFP (MOI = 200); black: unexposed ECs. Time (h) is represented relative to the time at whichBb was added. Time before exposure is shaded in red. See also Figure S4.
ECs not exposed to Bb, showed supracellular fluctuations and high heterogeneity in both monolayer tension and compression (, consistent with previous observations in epithelial cell monolayers (Figure 4B, right) (Bazellières et al., 2015). Interestingly, the mean normalized monolayer tension () in unexposed ECs remained nearly constant over the first 10 h of imaging and progressively increased over the following 24 h, potentially because of ECs becoming more confluent (Figure 4C). On the contrary, mean normalized monolayer tensionexhibited a sharp increase over the first 30 min after Bb exposure but remained markedly lower than that of unexposed cells thereafter and up to approximately 20 hpe (Figures 4B and 4C). After that time point, no significant differences were observed between ECs exposed or not to Bb (Figure 4C). Collectively, our findings suggest that both the EC traction stresses and monolayer stresses are significantly weakened in cells exposed to Bb at early stages (<15 hpe) but they display no differences at later points as compared to cells never exposed to Bb.
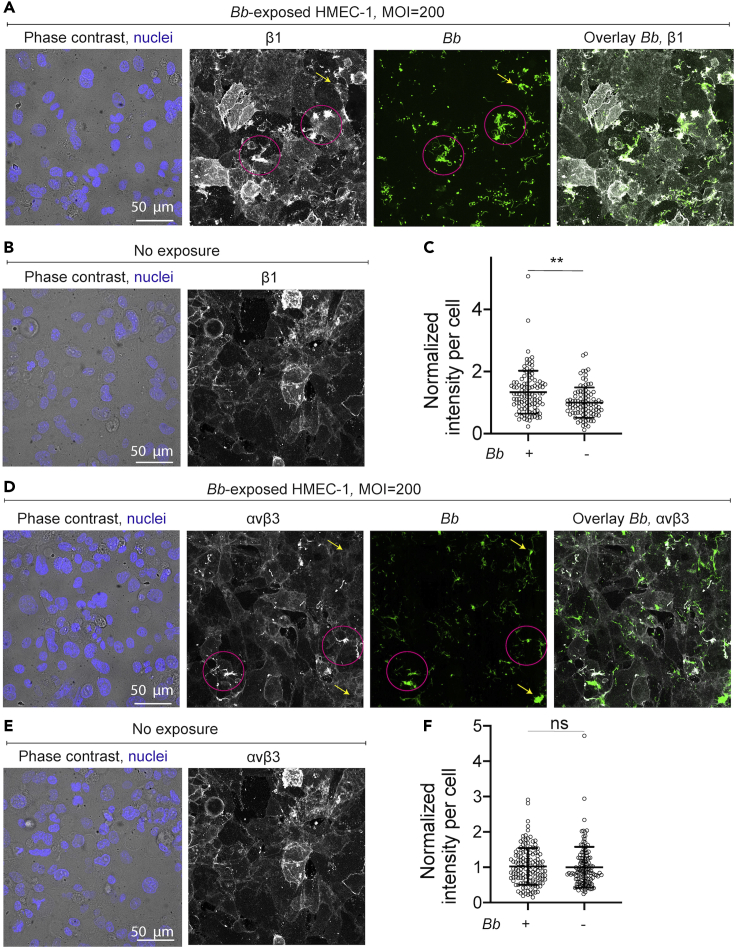
EC integrins β1 and αvβ3 colocalize with Bb but their ventral organization or that of F-actin is comparable to unexposed ECs
We next asked whether the changes in EC motility and traction force generation at early times after exposure were because of alterations in the organization of the cell cytoskeleton or of the focal adhesions. To address that, we grew ECs in monolayer and exposed them (or not) to Bb-GFP. Samples were fixed at 8 hpe, immunostained and inspected using 3D confocal microscopy. We first examined the F-actin cytoskeleton using fluorescently labeled phalloidin and observed no major differences when comparing non- or Bb-exposed cells (Figures S5A and S5B). Actin stress fibers were present in both cases, although slightly more abundant in unexposed as compared to Bb-exposed cells, however, the integral of phalloidin fluorescence intensity in a per cell basis was similar in both conditions (Figure S5C). Given the known involvement of integrins αvβ3 and α5β1 in the attachment of Bb to different host cell types (Coburn et al., 1998; Niddam et al., 2017), in the organization of focal adhesions and EC migration (Leavesley et al., 1993), we examined whether their localization or abundance differed between non- or Bb-exposed ECs by imaging fixed samples via 3D confocal microscopy (Figures 5A–5D). Inspection of the maximum projection images revealed that some but not all of the spirochetes co-localized with both anti-β1 and anti-αvβ3 antibodies, confirming that the observed signal did not originate from bleed-through fluorescence (Figures 5A and 5D). A more detailed analysis would be needed to conclude whether this co-localization is stochastic or not. The overall organization of those integrin subunits at the basal surface of ECs, from where traction forces are transduced to the extracellular matrix, did not differ dramatically as compared to unexposed ECs, although the integral of anti-β1 antibody fluorescence intensity per cell was increased in the Bb-exposed as compared to unexposed ECs (Figures S5D and S5E).
Figure 5.
EC integrins β1 and αvβ3 colocalize with Bb and β1 shows increased localization compared to unexposed ECs
(A) Representative brightfield image of cells superimposed with the Hoechst-stained nuclei image (first column), anti-β1 integrin antibody fluorescence (second column, maximum intensity projection), Bb-GFP fluorescence (third column, maximum intensity projection) and overlay of the last two channels (fourth column) for ECs exposed to Bb-GFP for 8 h.
(B) Representative brightfield image of cells superimposed with the Hoechst-stained nuclei image and anti-β1 integrin antibody fluorescence for HMEC-1 not exposed to Bb.
(C) Boxplots of normalized mean anti-β1 antibody fluorescence intensity per cell (mean ± SD, dots: individual cells) for ECs exposed to Bb-GFP for 8 h or unexposed ECs. Normalization is done with respect to the mean intensity of unexposed ECs. ∗∗: p<0.01, ns: not significant (Wilcoxon rank-sum test).
(D–F) Same as in (A–C) but showing anti-αvβ3 integrin antibody fluorescence. In panels (A) and (D) pink circles denote co-localization of Bb-GFP and the indicated integrins while arrows point to Bb-GFP cells that do not colocalize with integrins. See also Figure S5.
RNA sequencing reveals distinct transcriptional profiles in Βb-exposed versus unexposed ECs during early but not late exposure
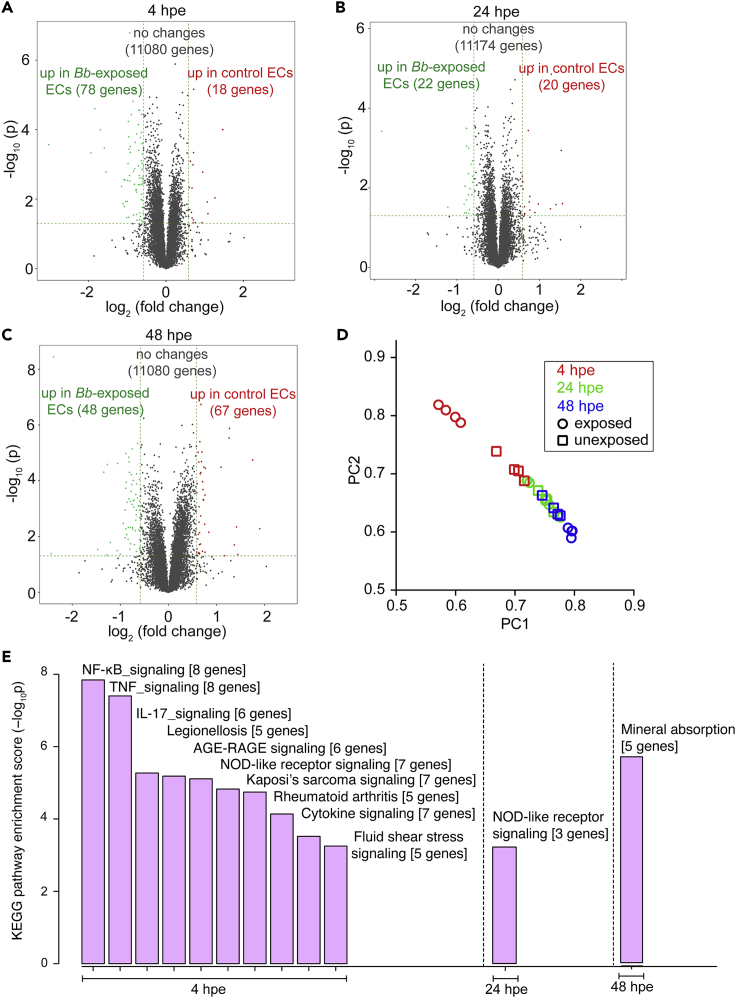
To better understand which signaling processes might regulate the changes in EC mechanotransduction in response to Bb-exposure and how they might differ between early versus late time points after exposure, we analyzed the EC transcriptome. To that end, we exposed ECs in monolayer to Bb and extracted their RNA at 4, 24 and 48 hpe. In parallel, at the same time points we also extracted the RNA from unexposed cells (control cells) that were seeded at the same density. We then performed RNA sequencing on these six different populations and used four replicates per condition to determine differentially expressed genes (DE-Gs) when comparing all six populations (Table S1, see Sheets 1–6). As shown by the volcano plot in Figure 6A, at 4 hpe a significant number of genes was upregulated in Bb-exposed ECs as compared to controls (78 genes) but only few were downregulated (18 genes). On the contrary, at 24 hpe only 22 and 20 genes were significantly upregulated and downregulated, respectively, in Bb-exposed ECs as compared to controls (Figure 6B). Interestingly and paradoxically, at 48 hpe 48 genes were significantly upregulated and 67 genes were significantly downregulated in Bb-exposed cells as compared to controls (Figure 6C). However, when we performed principal components analysis (PCA) on our samples (N = 4 replicates per condition), we observed two distinct clusters in the PCA space only for the Bb-exposed versus unexposed samples at 4 hpe, whereas the rest of the samples overlapped in the PCA space (Figure 6D). Moreover, as expected, the unexposed ECs at 4 hpe clustered closer to the rest of the samples in the PCA space whereas the Bb-exposed ECs at 4 hpe were the only group that clustered on its own as compared to the other samples.
Figure 6.
ECs upregulate innate immune signaling pathways at four but not 24 hpe to Bb
(A–C) Volcano plots of differentially expressed genes (DE-Gs). The-log10 pvalues are plotted against the average log2 fold changes in expression. For each pair of compared conditions the upregulated genes of each group are shown in the corresponding color. Each panel refers to a different time after exposure as indicated.
(D) PCA of top genes that have ANOVA p value ≤0.05 on FPKM abundance estimations. PC1 versus PC2.
(E) Pathway enrichment analysis. Bb-exposed ECs were compared to unexposed ECs based on their enrichment score (-log10p). Resulting barplots for the different times after exposure are shown only for pathways that had-log10p>3. See also Figure S6 and Table S1.
We then performed pathway enrichment analysis for the DE-Gs to revealwhich pathways were significantly perturbed when comparing the different groups (Figure 6E and Table S1, see Sheets 7–9). Compared to unexposed ECs, Bb-exposed ECs showed significant upregulation of 47 KEGG pathways and significant down regulation of only one pathway at 4 hpe (Table S1, see sheet 7). The pathways with an enrichment score larger than three and with at least three genes significantly upregulated in Bb-exposed ECs were in order of decreasing enrichment score: (1) NFκΒ signaling pathway; (2) TNF signaling pathway; (3) IL-17 signaling pathway; (4) Legionellosis; (5) AGE-RAGE signaling pathway in diabetic complications; (6) NOD-like receptor signaling pathway; (7) Kaposi’s sarcoma-associated herpesvirus infection; (8) Rheumatoid arthritis (refer to Table S1, sheet seven for a list of the specific DE-Gs pertaining to each pathway). Unlike Bb-exposed versus unexposed ECs at 4 hpe, the only KEGG pathway that complied to the criteria listed above at 24 hpe was the NOD-like receptor signaling pathway which contained only three genes differentially regulated as opposed to seven at 4 hpe. Finally, at 48 hpe the only KEGG pathway that complied to the criteria listed above was Mineral absorption, with the following five genes upregulated: MT1F, MT1G, MT1M, MT1X (genes encoding metallothioneins) and SLC30A1 (gene encoding zinc transporter 1), all involved in copper and zinc metabolism. Metallothioneins are cysteine-rich metal binding proteins with high binding affinity for several metals, such as copper and zinc (Calvo et al., 2017), both of which have previously been shown to regulate biofilm formation (Danilova et al., 2020). In addition, metallothioneins are upregulated by host cells in response to biofilm formation by other bacterial communities (Smolentseva et al., 2017).
Given the centrality of the NFκΒ and TNFα signaling pathways in many types of infection and their recent involvement in modulating host cell mechanics in response to infection with intracellular bacterial pathogens (Bastounis et al., 2021), we examined in more detailed which specific genes of these two pathways were differentially expressed during Bb-exposure (Figure S6). At 4 hpe, we found that genes encoding a number of cytokines were significantly upregulated in Bb-exposed ECs, namely: CXCL2 (gene encoding the chemokine CXCL2), CXCL1 (gene encoding the chemokine CXCL1), CXCL8 (gene encoding the chemokine IL-8) and CCL2 (gene encoding the chemokine MCP1). Moreover, upregulated were also the genes encoding the cell adhesion molecule ICAM1 and EDN1, a vasoconstrictor peptide that is often released by ECs in response to TNFα or NFκΒ activation (Quehenberger et al., 2000; Bourque et al., 2011). Out of all those genes only CCL2 and CXCL1 remained significantly upregulated at later times after exposure (i.e., 24 and 48 hpe) while the expression of the remaining genes returned to basal levels, i.e., to those of unexposed ECs. Altogether the upregulation of immunity pathways during the early but not late stages of exposure correlates with the early changes in biomechanics we observed in Bb-exposed ECs and is consistent with previous findings (LaFrance et al., 2011).
Exposure of ECs to heat-inactivated Bb recapitulates only the early weakening of EC mechanotransduction but not the reversal at late exposure
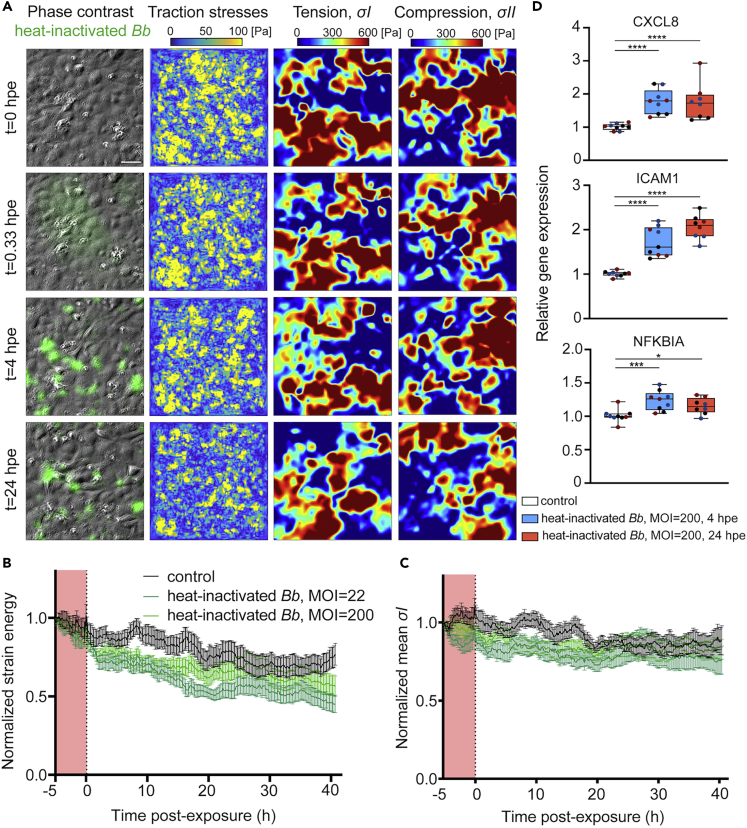
We speculated that the time-dependent changes in EC force transduction during exposure to Bb could have been actively triggered by the spirochetes. However, at later time points (24 hpe) we did observe a decrease in Bb viability as assessed by PI staining. To rule out the possibility that the observed changes in biomechanics were simply the response of ECs to dead Bb, we exposed ECs to heat-inactivated Bb-GFP at a high (MOI = 200) or low (MOI = 22) dosage and conducted TFM and MSM 5 h prior and over 24 hpe. We discovered that irrespective of the MOI, EC traction forces and strain energy weakened to a similar extent in ECs exposed to heat-inactivated Bb and in those exposed to live Bb (Figures 7A, 7B,3A and 3B). Of interest, in response to heat-inactivated Bb we observed neither the transient abrupt increase in traction forces and strain energy nor their recovery at late exposure (>15 hpe), which we observed upon exposure to live Bb. Monolayer maximum tensile and compressive stresses of ECs were also sustainably weakened upon exposure to heat-inactivated Bb-GFP at both MOIs, but we did not observe the transient and abrupt increase that occurred upon exposure to live Bb (Figures 7A–7C,4B and 4C). Given that the weakening of EC force transduction upon early exposure to live Bb was accompanied by an upregulation of innate immune signaling pathways such as the NFκΒ pathway, we wondered whether we were to observe such an effect also in response to heat-inactivated Bb. To that end, we performed RT-PCR analysis on ECs exposed to heat-inactivated Bb-GFP at 4 and 24 hpe, to test the expression of three NF-κB target genes (namely, CXCL8, ICAM and NFKBIA) which we had found upregulated at 4 but not 24 hpe to live Bb through RNA sequencing analysis. Of interest, we discovered an upregulation of all those genes at both 4 and 24 hpe and the extent of upregulation (approximately 2-fold) was similar to that observed just at 4 hpe for ECs exposed to live Bb.
Figure 7.
ECs sustainably weaken their force transduction and upregulate NF-κB target genes in response to heat-inactivated Bb
(A) Representative phase contrast image overlayed with heat-inactivated Bb-GFP fluorescence (first column), EC traction stress map (second column, Pa), monolayer tensile stresses , third column, Pa) and absolute value of compressive stresses, fourth column, Pa) for ECs in monolayer at different time points (rows) after exposure to heat-inactivated Bb-GFP (MOI =200). TFM was performed for ECs residing on 3 kPa ECM.
(B) Normalized strain energy imparted by ECs during a TFM recording (mean ± SEM, three independent experiments and N= 12 recordings in total). Strain energy has been normalized with respect to the first value at the beginning of each recording. Dark (light) green: ECs exposed to heat-inactivated Bb with an MOI = 200 (MOI= 22); black: unexposed cells. Time (h) is represented relative with respect to the time at whichBb was added. Time before exposure is shaded in red.
(C) Same as panel B but showing the normalized mean EC monolayer tensile stresses () as a function of time after exposure to heat-inactivated Bb-GFP.
(D) Relative with respect to GAPDH expression levels of the indicated NF-κB target genes obtained by RT-PCR. N = 3 independent experiments were performed. Three conditions were tested namely ECs exposed to nothing, to heat-inactivated Bb-GFP with an MOI = 200 for 4 h (blue) or for 24h (red). From top to bottom normalized expression of the following genes is shown: CXCL8, ICAM1, NFKBIA. Boxplots show the mean, 25th and 75th quartiles, different colors refer to replicates from independent experiments, ∗: p<0.05, ∗∗: p<0.01, ∗∗∗: p<0.001, ∗∗∗∗: p<0.0001 (Wilcoxon rank-sum test run for each condition’s distribution with respect to control distribution). See also Figure S7.
The intriguing correlation between weakening in EC mechanotransduction and upregulation of NFκΒ target genes led us wonder whether EC force transduction would change in response to a cytokine that is known to upregulate NFκΒ like tumor necrosis factor alpha (TNFα) or to muramyl dipeptide (MDP), which is found in the cell wall of many bacteria and is known to upregulate NOD signaling and ΝFκΒ in certain cell types (Stroka et al., 2012; Urbano et al., 2017; Lappas, 2013; Brandt et al., 2022). We discovered that addition of TNFα onto EC monolayers led to an immediate and sustained increase of traction and monolayer stresses while the biomechanical responses of MDP-exposed ECs appeared identical to those of non-exposed ECs (Figures S7A–S7C). When we assessed the expression of the three NF-κB target genes (CXCL8, ICAM and NFKBIA) via RT-PCR, we found a dramatic increase (10- to 100-fold compared to controls) in their expression both at 4 and 24 hpe to TNFα (Figure S7D) while there were no detectable changes in expression of those genes for cells exposed to MDP (data not shown). Thus, it appears that a dramatic upregulation of innate immune signaling as obtained in response to TNFα leads to a fast and sustained upregulation of EC force transduction. Whether TNFα is somehow related to the strong immediate upregulation of force transduction upon encounter of ECs with live Bb was not tested but could be the focus of future studies.
Discussion
Cell-cell and cell-matrix adhesions of ECs are continuously remodeled in response to biochemical and mechanical cues. This remodeling results in dynamic alterations of the forces that cells transmit to their matrix and to each other, which are regulated to a large extent through the action of the actomyosin contractile apparatus (Komarova et al., 2017). Pro-inflammatory mediators and secreted cytokines can reinforce (Brandt et al., 2022) or alternatively release the tensile stresses between ECs, thus enhancing vascular permeability (Lee et al., 2011). A decrease in cell-ECM forces can also lead to attenuation in EC monolayer stresses as a result of the crosstalk between adherens junctions and focal adhesions (Komarova et al., 2017). Such attenuation in traction and monolayer stresses, similar to what we observed during early exposure of ECs to Bb, could benefit the bacteria by enhancing their transmigration through the leaky EC monolayer. However, sustained, and prolonged impairment of cellular physical forces could lead to extensive leakiness of the vessels, a feature that could harm a pathogen like Bb in the first place by rapidly killing the host and making a persistent infection impossible.
The weakening in EC force transduction during early Bb exposure which we discovered, coincides with upregulation of a number of host cell genes involved in innate immune signaling. Previous studies have demonstrated an intricate link between innate immune signaling and alterations in either intercellular stresses or permeability (Bastounis et al., 2021; Kisseleva et al., 2006; Kempe et al., 2005). We recently showed that NFκΒ activation in response to infection with intracellular bacterial pathogens (L. monocytogenes and a Rickettsia parkerimutant), leads to a decrease in traction forces of epithelial cells in monolayer as compared to non-infected settings, and a decrease in monolayer stresses as measured indirectly through laser wounding (Bastounis et al., 2021). In vivo, NFκΒ activation has been explicitly linked to increased vascular EC permeability (Kisseleva et al., 2006). It is possible that Bb spirochetes might take advantage of the weakening in host cell mechanotransduction, which correlates with upregulation of innate immune signaling at early exposure, to facilitate their paracellular transmigration through ECs, or even their transcellular transmigration through ECs (i.e., by taking advantage of the EC intracellular tension weakening). Recently, intravital imaging of the microvasculature of mice revealed that infection with Bb triggers local recruitment of neutrophils which, instead of killing the pathogens, leads to production of various cytokines, particularly CCL2, thereby activating ECs and increasing barrier permeability (Tan et al., 2021). This in turn facilitates transcellular but not paracellular transmigration of Bb to reach distant tissues. Interestingly, we also found that ECs upregulated CCL2 expression throughout exposure to Bb, suggesting that some of the spirochetes we observed intracellularly may just be en route of transmigration through ECs. Production of additional cytokines, like TNFα and IL-10, was increased in the study of Tan et al., most likely because those cytokines are primarily produced by neutrophils. In vitro assays similar to the one we developed here but additionally involving co-culture with neutrophils could help shed more light into how ECs biomechanics in response to Bb exposure is altered in the presence of neutrophils. It should be noted that Bb might have additional ways of altering host cell mechanics, e.g., by inducing changes in the mechanical properties of the ECM to which host cells are anchored directly or indirectly. For example, the Bb surface exposed protease BbHtrA can directly degrade ECM proteins in various tissues including the skin, joints and brain, which is expected to induce concomitant changes in host cell mechanics (Russell et al., 2013). In addition, Bb infection leads to the upregulation of matrix metalloproteases (MMPs, ECM degrading enzymes) by chondrocytes (Behera et al., 2005), keratinocytes and macrophages (Gebbia et al., 2001). Although MMP upregulation, ECM stiffening and changes in cell contractility are tightly correlated processes in the context of cancer metastasis (Haage and Schneider, 2014), their interplay during Bb infection has yet to be uncovered.
The changes we found in EC kinematics, dynamics and gene expression in response to Bb exposure are transient and progressively vanish as time after exposure elapses. The absence of changes in EC gene expression at late times after exposure to Bb is consistent with a previous microarray analysis (LaFrance et al., 2011). However, in this study gene expression analysis of cells, including HMEC-1, at early Bb exposure (1 or 3 hpe) revealed upregulation of pathways related to the regulation of the actin cytoskeleton and focal adhesions which we do not find to be upregulated through RNA sequencing of HMEC-1 at 4 hpe. Rather, we found upregulation of a lot of pathways related to innate immune signaling. A possible reason for this discrepancy could be that in the LaFrance et al. study the noninfectious Bb strain HB19/KO1 was used while we used the infectious Bb strain B31 5A4 NP1. Different Bb strains present differences in genotype, clinical features and inflammatory potential, therefore the changes they can induce in EC gene expression and biomechanics could well be strain specific (Cerar et al., 2016).
At late EC exposure to Bb, we observed the formation of Bb aggregates, but single motile and viable spirochetes were still present in the culture medium. Past studies have suggested that multiple factors can lead to Bb aggregation (Anderson and Brissette, 2021; Sapi et al., 2012; Alban et al., 2000). However, in our assay we determined that this is the result of the high MOI used, since it also occurs when large amounts of spirochetes are cultured in their normal growth medium (data not shown). To determine herein, whether the lack of changes in EC gene expression and biomechanics at late exposure to live Bb could be related to the status of Bb, and to understand whether the time-dependent changes in EC force transduction represent and active or passive response to Bb, we exposed ECs to heat-inactivated Bb. We found that throughout the course of exposure EC mechanotransduction was weakened and NF-κB target genes were upregulated. It has been shown that Bb peptidoglycans (PG) and lipoproteins such as the outer surface protein OspC are resistant to heat and at least PG can induce NFκΒ activation in host cells (Jutras et al., 2016, 2019; Kumru et al., 2011) whereas other studies have shown that exposure of different cell types to dead Bb induce inflammatory mediators (Parthasarathy et al., 2013; Wang et al., 2008). Whether any of those components are responsible for the weakening in EC mechanotransduction could be tested in future studies experimentally by addition of recombinant proteins or PG components onto EC monolayers. However, when we added MDP onto ECs we did not observe any change in EC force transduction but also any upregulation of NFκΒ target genes in ECs (data not shown). On the contrary, when we exposed ECs to TNFα, NFκΒ target genes were dramatically upregulated (10- to 100-fold) and immediate and sustained upregulation of EC force transduction was observed, consistent with previous studies (Stroka et al., 2012; Urbano et al., 2017; Brandt et al., 2022). This result suggests that the relationship between EC physical forces and innate immune signaling is likely not monotonic. Finally, it is worth noting that the responses of ECs to live versus heat-inactivated Bb exposed ECs particularly differed at very early and late stages of exposure. First, upon exposure to heat-inactivated Bb there was no abrupt increase in EC force transduction, a feature that was observed upon exposure to live Bb. This result suggests that the initial signal conveyed to ECs may arise from adhesion of live Bb to ECs through some receptor that is possibly denatured upon heat inactivation. The late reversal of EC gene expression and biomechanics, a feature that was observed only in response to live but not heat-inactivated Bb, suggests that this is not a consequence of the loss of Bb viability because of prolonged exposure to EC culture media. Rather, it could be because of changes in the Bb state, including the metabolic or gene expression state. Whether this response reflects what would happen in an actual in vivo infection is unknown, but lack of host cell gene expression changes at one day after infection with Bb has previously been shown in vitro (LaFrance et al., 2011) while changes in gene expression of Bb, which have been shown to occur during actual in vivo infection, could well impact host cell responses in a time-dependent manner (Saputra et al., 2020).
To overcome a major limitation of in vivo approaches, namely, the fact that multiple parameters can change in an unpredictable manner at once limiting the strength of causal conclusions, herein we used an elaborate in vitro assay that enabled us to measure the spatiotemporal evolution of EC kinematics and dynamics during exposure to Bb in a controllable manner while tweaking one parameter at a time. This in turn allowed us to discover an intricate time-dependent link between EC innate immune signaling and biomechanics during Bb exposure. Our assay however is under static conditions and does not account for additional extracellular physical forces that ECs experience in vivo. For example, in the vasculature ECs are exposed to fluid shear stresses and gradients which can greatly impact EC behavior (e.g., proliferation, motility etc) and mechanotransduction (Barakat and Lieu, 2003; Brandt et al., 2022; Ostrowski et al., 2014; Tan et al., 2021). The fluid shear flow regime can also impact the way Bb adheres on ECs (Ebady et al., 2016). It is possible that apical fluid flow might impact both innate immune signaling and physical EC forces and thus the EC responses to Bb exposure might present distinct features as compared to static cultures. Examination of the impact of apically exposed fluid flow on EC biomechanics and the interactions with Bb can be the focus of future studies. Moreover, the high MOI used in our study might not reflect what would be observed in vivo, but we used it to be able to measure discernible effects in the EC traction-monolayer forces and signaling and thus gain new mechanistic information as it is often done in such assays. Future studies could focus on uncovering whether our conclusions are valid in in vivo systems.
Our discovery of the upregulation of innate immune signaling and weakening of host EC force transduction upon early but not late exposure to Bb underlines the importance of uncovering both the spatial and temporal changes that occur during the course of exposure. Studying how changes in host cell biomechanics and gene expression are temporally and spatially modulated will reveal how pathogens like Bb can subvert host physiology to their own benefit, that is, to disseminate or render infection chronic. Such studies can also lend key insights into how ECs regulate force transduction and barrier integrity in health and during bacterial infections.
Limitations of the study
Herein we used human skin microvascular ECs, HMEC-1, as our model ECs and pathogenic strains Bb1286 or GCB726, as our model Bb strains. Examination of additional EC types and Bb strains would allow assessing how generalizable or conversely EC type- or Bb strain-specific the mechanisms we discovered are. Moreover, in this study we assessed only the changes in the transcriptomics of host ECs. Performance of RNA sequencing to investigate the changes in gene expression that Bb undergoes during the course of exposure could reveal how changes in EC gene expression are linked to alterations in bacterial transcriptomics. Finally, our data suggest an active modulation of host EC mechanics and biochemistry by Bb. Experiments with specific Bbmutant strains could reveal specific Bb virulence factors that trigger the active responses of ECs and could facilitate the identification of the specific underlying mechanisms Bb employ to alter EC biomechanics. Finally, investigation of the physical and “dual” transcriptomic responses of host ECs and Bb in a more complex in vitro system which allows co-culture of ECs with professional immune cells, would have enabled examining the contribution of the host professional immune cells to Bb-ECs interactions, and would have thus been more physiological.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Borrelia burgdorferi antibody | Thermo Fisher Scientific | Cat# PA1-73004, RRID:AB_1016668 |
| Mouse monoclonal anti-integrin β1 | Millipore | Cat# MAB1987; RRID:AB_94493 |
| Mouse monoclonal anti-integrin αvβ3 | Sigma | Cat# MAB1976; RRID: AB_2296419 |
| Chemicals, peptides, and recombinant proteins | ||
| AlexaFluor546 phalloidin | Thermo Fisher Scientific | Cat# A22283 |
| SulfoSanpah | Thermo Fisher Scientific | Cat# 22589 |
| Collagen I rat tail | Fisher | Cat# A1048301 |
| Acrylamide 40% solution | Sigma | Cat# A4058 |
| Bisacrylamide solution 2% | Fisher | Cat# BP1404250 |
| Aminopropyltriethoxysilane | Sigma | Cat# 919302 |
| FluoSpheres™ Carboxylate-Modified Microspheres, 0.2 μm, yellow-green fluorescent (580/605), 2% solids | Invitrogen | Cat# F8810 |
| FluoSpheres™ Carboxylate-Modified Microspheres, 0.2 μm, yellow-green fluorescent (660/680), 2% solids | Invitrogen | Cat# F8807 |
| MCDB131 | Gibco | Cat# 10372019 |
| Leibovitz’s L-15 medium, no phenol red | Thermo Fisher Scientific | Cat# 21083027 |
| Fetal bovine serum | Gemini Bio-Prod | Cat# 900108 |
| Hydrocortisone | Sigma | Cat# H0888 |
| hEGF | Sigma | Cat# EG9644 |
| L-Glutamine | Fisher | Cat# SH3003401 |
| Hoechst 33342, trihydrochloride | Invitrogen | Cat# H3570 |
| BSK-H Medium | Sigma | Cat# B3528 |
| Rabbit serum | Sigma | Cat# R4505 |
| Gentamicin | Thermo Fisher Scientific | Cat# 15710064 |
| Propidium iodide | Sigma | Cat# P4170 |
| Recombinant human TNF-α | InvivoGen | Cat# rcyc-htnfa |
| Muramyldipeptide - MDP | NOD2 ligand | InvivoGen | Cat# tlrl-mdp |
| Critical commercial assays | ||
| QIAshredder Kit | Qiagen | Cat# 79656 |
| RNeasy Plus MicroKit | Qiagen | Cat# 74034 |
| PrimeScript™ RT Reagent Kit | Takara Bio | Cat# RR037A |
| KAPA SYBR® FAST | Kapa Biosystems | Cat# KK4601 |
| Deposited data | ||
| RNA-seq data | This paper | GSE174545 |
| Experimental models: Cell lines | ||
| HMEC-1 cells | M. Welch (Reed et al., 2014) | Previously obtained from Centers for Disease Control, Biological Products Branch |
| Experimental models: Organisms/strains | ||
| Borrelia burgdorferi (strain Bb1286) | J. Radolph (Dunham-Ems et al., 2009) | N/A |
| Borrelia burgdorferi (strain GCB726) | P. Kraiczy (Moriarty et al., 2008) | N/A |
| Oligonucleotides | ||
| Primers for ICAM1 (forward: 5′ GAGCTTCGTGTCCTGTATGGC -3′, reverse: 5′ ACAGAGGTAGGTGCCCTCAA -3′) | This paper | N/A |
| Primers for CXCL8 (forward: 5′ CAGAGACAGCAGAGCACACA -3′, reverse: 5′ GGCAAAACTGCACCTTCACA -3′) | This paper | N/A |
| Primers for NFKBIA (forward: 5′ ATGTCAATGCTCAGGAGCCC -3′, reverse: 5′ CAGGTGAGCTGGTAGGGAGA -3′) | This paper | N/A |
| Primers for GAPDH (forward: 5′ GGGAAACTGTGGCGTGAT -3′, reverse: 5′ GAGTGGGTGTCGCTGTTGA -3′) | This paper | N/A |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| MicroManager | Open Imaging | https://www.micro-manager.org/ |
| MATLAB | MathWorks | http://www.mathworks.com/products/matlab/?requestedDomain=www.mathworks.com |
| GraphPad Prism v6 | GraphPad | http://www.graphpad.com/scientific-software/prism/ |
| Hisat 2 | (Kim et al., 2015) | https://ccb.jhu.edu/software/hisat2/index.shtml) |
| Cutadapt | (Martin, 2011) | https://cutadapt.readthedocs.io/en/stable/ |
| R package GAGE | (Luo et al., 2009) | https://bioconductor.org/packages/release/bioc/html/gage.html |
| R package “Pathview” | (Luo et al., 2017) | https://www.bioconductor.org/packages/release/bioc/html/pathview.html |
| ABAQUS | Dassault systèmes | https://www.3ds.com/products-services/simulia/products/abaqus/ |
| QuantStudio™ Design and Analysis Software v2 | Applied Biosystems | https://www.thermofisher.com/de/en/home/global/forms/life-science/quantstudio-3-5-software.html |
| Imaris | Bitplane | https://imaris.oxinst.com/ |
Resource availability
Lead contact
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact Effie Bastounis effie.bastounis@uni-tuebingen.de (E.E.B.).
Materials availability
Materials developed in this study are available on request to the corresponding author.
Experimental model and subject details
Cell culture
Human dermal microvascular endothelial cells HMEC-1 (generous gift from the Welch lab, University of California, Berkeley previously obtained from Centers for Disease Control, Biological Products Branch) were maintained in MCDB131 medium (Fisher Scientific; 10372-019) supplemented with 10% fetal bovine serum (GemBio, 900108), 10 ng/mL epidermal growth factor (Sigma, E9644), 1 μg/mL hydrocortisone (Sigma, H0888), and 2 mM L-Glutamine (Sigma, 56-85-9) (Reed et al., 2014). Cells were passaged 1:6 at 90% confluence.
Bacterial strains used in this study
We used Borrelia burgdorferi (Bb) constitutively expressing GFP, strain Bb1286, indicated as Bb-GFP throughout the main text. This strain was a generous gift from Justin Radolf’s lab, University of Connecticut Health Center (Dunham-Ems et al., 2009). For the experiments involving exposure of human cells to heat-inactivated Bb-GFP we used strain GCB726. This strain, similar to Bb1286, was generated by electroporation of the infectious Bb strain B31 5A4 NP1 with the shuttle vector pTM61 that contains all plasmids except the circular plasmid cp9. The construction of this strain has been previously described (Moriarty et al., 2008). Bb1286 strain was generated also from B31 5A4 NP1 which was then electroporated with a suicide vector to produce a cp26 plasmid containing the GFP expressing gene (Caimano et al., 2015). Thus, the two strains we used are almost identical and just differ in only one plasmid.
Method details
Bacterial growth conditions and exposure of ECs to Borrelia burgdorferi
Borrelia burgdorferi (Bb) strain Bb1286 or GCB726 were grown to mid log phase (approximately 5x107 bacteria/mL) in BSK-H media (Sigma, B3528) supplemented with 50 μg/mL gentamicin (Dunham-Ems et al., 2009). For cultivation of strain GCB726, BSK-H medium (Bio&Sell, Feucht, Germany) supplemented with 5.4% rabbit serum (Sigma, R4505), 50 μg/mL gentamicin and 100 μg/mL kanamycin was used. Briefly, 10 μL of frozen glycerol stock was added into a tube containing 1.5 mL of BSK-H media supplemented with appropriate antibiotics. The tube was placed at 37°C and until the concentration of the bacteria reached 2-5x106 bacteria/mL. The bacterial solution was then diluted at 1x103 bacteria/mL and bacteria were grown in fresh BSK-H with antibiotics at 37°C until a density of 5x107 bacteria/mL growing as single spirochetes was reached (see Figure 1A). Bacterial concentrations were determined using a hemocytometer and a dark field microscope (generously provided by the Mougous lab, University of Washington).
The day prior toBb-GFP exposure, ECs were seeded at a density of 2 × 105 cells/well on glass bottom wells of 24-well plates coated with 0.25 mg/mL rat tail collagen I (Sigma-Aldrich, C3867) or on 3 kPa polyacrylamide hydrogels also coated with 0.25 mg/mL rat tail collagen I. For microscopy recordings, cell medium was removed and, instead of MCDB131, Leibovitz’s L-15 medium was used supplemented with 10% fetal bovine serum, 10 ng/mL epidermal growth factor, 1 μg/mL hydrocortisone, and 2 mM L-glutamine. For experiments involving tracking of host cell nuclei, 1 μg/mL Hoechst (Thermofisher, D1306) was added in each well to stain the cells’ nuclei for 10minat 37°C. Cells were then washed once in warm media to remove excess stain. Just prior to infection bacteria were spun down, washed once in PBS and then resuspended in cell media so that minimal volumes of bacteria (1-30 μL) were added in each well to achieve the target multiplicity of infection (MOI, bacteria/host cell).
For generation of heat-inactivated Bb-GFP, bacteria were incubated at 56°C for 30 min as previously reported (Replogle et al., 2021). Subsequently, heat-inactivated Bbwere stored at 4°C to avoid damage of the Bb membrane or morphology that can occur at lower temperatures.
Flow cytometry of ECs exposed to Bb-GFP
4 h post-exposure (hpe), ECs exposed to Bb-GFP at different MOIs were washed 4 times with sterile phosphate-buffered saline (PBS) to get rid of extracellular bacteria (Otte et al., 2021). Cells were then detached from the glass coverslip where they resided, by removing cell medium and adding 200 μL of 0.25% trypsin-EDTA in each well for 10 min at 37°C. Trypsin-EDTA cell-containing solutions in each well were then pipetted up and down 6 times to ensure single cell suspensions and 200 μL of complete medium were added to inactivate trypsin in each well. Suspensions were transferred into 35-μm cell strainers, (Falcon, 352235) and spun through at 500 x g followed by fixation in 1% paraformaldehyde for 10 min on ice. Samples were then washed once in PBS and stored in PBS with 1% BSA on ice or at 4°C. Flow cytometry analysis was performed on a BD FACS Canto RUO analyzer (University of Washington Cell Analysis Facility). 10,000-20,000 cells were analyzed per each replicate. To ensure analysis of single ECs, the bulk of the distribution of cell counts was gated using the forward versus side scatter plot and thus discarding debris or cell doublets or triplets from the analysis. In addition, a second gating step was applied after measuring the fluorescence of control, unexposed cells and gating the population of exposed cells accordingly to exclude cell autofluorescence.
Bb segmentation and morphology-dependent classification
To segment and classify bacteria depending on their morphology we used using IMARIS software (Bitplane) on the time-lapse epifluorescence images of bacteria. We opted for smoothing using surfaces detail parameter equal to 0.4 μm and thresholding was performed after background subtraction (local contrast) with the diameter of the largest sphere which fits into the object equal to 0.6 μm. An Imaris 9.6 built-in classifier was then used to classify bacteria to three distinct categories, namely: (1) single objects (spirochetes or spot-like structures); (2) bacterial networks; and (3) bacterial aggregates. For machine learning classification of surfaces Imaris 9.6 uses the existing statistics values and in addition computes more values that provide additional intensity and shape information. To provide additional shape descriptors the position and radius of the biggest sphere that fits entirely within the surface were computed and a number of features were derived from this. The original Surfaces statistics together with the machine learning statistics provide a lot of information about the shape of a surface and can be very useful for a machine learning classifier for surfaces as previously implemented elsewhere (Ranzato et al., 2007; Koenderink and van Doorn, 1987). For training, the user instigated 15 distinct objects for each class. For each instance of time the number of bacteria that fell in each category was extracted as well as average surface area (μm2) and mean object fluorescence intensity as illustrated in Figures 2D and S2A and S2B.
Fabrication of polyacrylamide hydrogels
Polyacrylamide hydrogel fabrication was done as previously described (Bastounis et al., 2018, 2021). Glass-bottom plates with 24 wells (MatTek, P24G-1.5-13-F) were incubated for 1 h with 500 μL of 1 M NaOH, then rinsed with distilled water, and incubated with 500 μL of 2% 3-aminopropyltriethoxysilane (Sigma, 919-30-2) in 95% ethanol for 5 min. After rinsing with water, 500 μL of 0.5% glutaraldehyde were added to each well for 30 min. Wells were then rinsed with water and dried at 60°C. To prepare polyacrylamide hydrogels of 3 kPa, mixtures containing 5% acrylamide (Sigma, A4058) and 0.1% bis-acrylamide (Fisher, BP1404-250) were prepared (Bastounis et al., 2018). Two mixtures were prepared, the second of which contained 0.2 μm fluorescent beads at 0.03% (Invitrogen, F8811) for traction force microscopy (TFM) experiments. 0.06% ammonium persulfate and 0.43% TEMED were then added to the first solution to initiate polymerization. First, 3.6 μL of the first mixture without beads were added at the center of each well, capped with 12-mm untreated circular glass coverslips, and allowed to polymerize for 20 min. After coverslip removal 2.4 μL of the mixture containing tracer beads were added and sandwiched again with a 12-mm untreated circular glass coverslip and allowed to polymerize for 20 min. Next, 50 mM HEPES at pH 7.5 was added to the wells, and coverslips were removed. Hydrogels were UV-sterilized for 1 h and then activated by adding 200 μL of 0.5% weight/volume heterobifunctional cross-linker Sulfo-SANPAH (Fisher, 22589) in 1% dimethyl sulfoxide (DMSO) and 50 mM HEPES, pH 7.5, on the upper surface of the hydrogels and exposing them to UV light for 10 min. Hydrogels were washed with 50 mM HEPES at pH 7.5 and were coated with 200 μL of 0.25 mg/mL rat tail collagen I (Fisher, A1048301) in 50 mM HEPES at pH 7.5 overnight at room temperature. Next morning, the collagen coated surfaces were washed with HEPES and gels were stored in HEPES.
Traction force microscopy (TFM)
TFM was performed as previously described (del Álamo et al., 2007; Lamason et al., 2016). Briefly, in TFM, cells actively pull on their ECM depending on how well their focal adhesions are organized and connected to the underlying cytoskeleton, and cellular force generation can be inferred from displacement of fluorescent tracer particles embedded in the deformable ECM (Bastounis et al., 2014; del Álamo et al., 2007). Prior to seeding ECs (as described above) hydrogels were equilibrated with MCDB131 medium for 30 min at 37°C. ECs were then seeded to a concentration of 2 x 105 cells/well directly onto the hydrogels 24 h prior to Bb-exposure. 1 h prior to initiation of the TFM recording, MCDB131 medium was replaced with L-15 medium supplemented appropriately. Multi-channel time-lapse sequences were acquired to image the tracer beads’ fluorescence, the bacterial fluorescence, and the phase contrast image of ECs. Images were acquired using an inverted Nikon Eclipse Ti2 with an EMCCD camera (Andor Technologies) using a 40X 0.60NA Plan Fluor air objective and the MicroManager software package (Edelstein et al., 2014). The microscope was surrounded by a box type incubator (Haison) maintained at 37°C. Images were acquired every 10 min for approximately 3-8 h before Bb-GFP bacteria were added (or not for control wells) on the wells at an MOI of ∼200Bb/cell. Subsequently, at each time interval we measured the 2D deformation of the substrate at each point using an image correlation technique similar to particle image velocimetry (Gui and Wereley, 2002).
We calculated the local deformation vector by performing image correlation between each image and a non-deformed reference image which we acquired by adding 10% SDS at the end of each recording to detach the cells from the hydrogels. We used interrogation windows of 32 x 16 pixels (window size x overlap). Calculations of the two-dimensional traction stresses that cell monolayers exert on the hydrogel are described elsewhere (Lamason et al., 2016; Bastounis et al., 2014). We calculated the strain energy () as the mechanical work imparted by cells to deform the hydrogel:
| (Equation 1) |
where u(z = h) is the measured displacement vector field on the free surface of the hydrogel, t is the calculated traction stress vector field, represents the surface integral, h is the height of the hydrogel and the vertical coordinate. For each field of view analyzed originating from different wells we normalized the strain energy value with that of time = 0 min, to avoid discrepancies due to slight differences in cell confluence among wells. To quantitate how dynamic the traction adhesions of ECs is we preformed 2D cross-correlations between successive cell deformation maps (obtained via TFM) separated by different time delays using the corr2 function in MATLAB (MathWorks) that returns the 2D correlation coefficient. The resulting data shown in Figure S3 were then fitted to an exponential decay function of the form:
| (Equation 2) |
where Y0 is the Y value at time equal 0, Plateau is the Y value at infinite times and K is the rate constant.
Monolayer stress microscopy (MSM)
We used MSM to measure the tensile and compressive stresses that ECs in monolayer experience. These stresses arise due to the cells contracting, expanding, or remaining in a resting state at different spatial locations within the monolayer and are due to reorganization of their cytoskeleton and adhesion complexes. These cytoskeletal rearrangements lead to pushing or pulling forces that affect adjacent cells, resulting in cell movements and changes in cell size and shape. Even if a given cell is also deformed due to its own remodelling and active behaviour, this cell deformation is a consequence of the stress distribution in the monolayer. This distribution is governed by the internal forces per unit of area, defining the monolayer configuration. Consequently, the stress distribution changes at each point (Figures 4A and S4). For measuring intercellular forces indirectly via MSM, we use the previously computed via TFM traction stresses that the cells in the monolayer exert on their substratum under the assumption that the traction stresses in the vertical direction are negligible, as considered previously (Lamason et al., 2016). We also assume that traction stresses in the hydrogel-monolayer interface occur only in the plane of the monolayer (i.e., there is no component of the traction stresses in the normal direction of the interface) as done previously (Trepat et al., 2009). Traction forces exerted by the cells on the hydrogel need to balance with the forces exerted by the cells to each other in the monolayer (third Newton’s law). To that end, we consider idealised perfect cell-substrate adhesions.
To determine the monolayer tensile and compressive stresses, we make several simplifications. First, we consider that the thickness of the monolayer is constant and uniform. Second, regarding material properties of the monolayer, we assume perfect cell-cell adhesions (the monolayer is assumed to be a continuum) and the properties of the monolayer are considered everywhere the same. That is all cells and adhesions in the monolayer have the same mechanical properties and the cell monolayer is considered as a linear elastic isotropic material (Tambe et al., 2011). Third, we work under the small strain assumption, so both the strains induced in the hydrogel and the configuration changes in the monolayer at each time increment (time distance between subsequent frames) are small. These assumptions dramatically simplify the formulation of the problem, since we can work in a two-dimensional framework assuming a plane stress formulation to solve the problem. Thus, the stress distribution is assumed constant through the monolayer thickness.
In this case, the equilibrium equations, compatibility, and constitutive equations yield the Beltrami differential formulation:
| (Equation 3) |
The advantage of using the Beltrami equation is that we can compute stresses at each point without knowing the elastic modulus of the monolayer. This equation, together with the Newton’s equilibrium equations, formulated at each point of the monolayer in a differential way, yield the distribution of the stresses in the monolayer:
| (Equation 4) |
where v is the Poisson’s coefficient, tx and tyare the traction forces in the monolayer in the and directions, respectively (components of traction stress vector field t); σx, σy are monolayer normal tension/compression stresses in the x and y directions, τyx and τxy are the monolayer shear stresses in x and y directions respectively. These shear stress components are equal (τyx=τxy) due to equilibrium. All these stresses will be different at each point of the monolayer (Figure S4A).
Although stresses in the directions are easy to retrieve, the physical meaning is difficult to interpret since the coordinate system is an arbitrary system, normally aligned with the image and not with the behaviour of the cell. To overcome this challenge, we transform our results into a new coordinate system (I, II), principal coordinate system, in which there is no shear component of stress (Figure S4B). By aligning with the axis of this coordinate system, we can compute the maximum monolayer tension (σI) and compressions (σII) in the plane of the monolayer, (σI>σII). In fact, the maximum tension occurs in a perpendicular direction to the maximum compression. Both values of maximum tension and compression and their directions vary along the different points of the monolayer.
We solve the equations of the problem through a finite element formulation (Oñate, 2013) and implement the final equations into a custom-made finite element code in MATLAB (R2020b). In the simulations, the monolayer is discretized with square elements of the same size as the pixels (3.648 × 3.648 μm2) resulting in 4225 nodes and 4096 elements. As boundary conditions, the displacements along the edge of the monolayer/image were set to zero. We choose arbitrary elastic modulus for the monolayer, since in this formulation the stresses are independent of this mechanical property of the monolayer. The Poisson’s coefficient was set to 0.48. The source code is provided at https://github.com/ebastoun/Monolayer-Stress-Microscopy.
Characterization of host cell kinematics and bacterial aggregation
ECs were seeded to a concentration of 2 x 105 cells/well directly onto the glass coverslips coated with 0.25 mg/mL collagen I rat tail, 24 h prior to Bb-exposure. 1 h prior to initiation of time-lapse imaging, cells were washed with PBS and incubated with 1 μg/mL Hoechst (Thermofisher, D1306) to stain the ECs’ nuclei for 10 min at 37°C. Wells were then washed with L-15 medium supplemented appropriately and 1 mL of this medium was added to each of the wells. Multi-channel time-lapse sequences were conducted to acquire at each instance of time the Hoechst-stained EC nuclear fluorescence, the bacterial fluorescence and the phase contrast image of ECs, using an inverted Nikon Eclipse Ti2 with an EMCCD camera (Andor Technologies) using a 40X 0.60NA Plan Fluor air objective and the MicroManager software package (Edelstein et al., 2014). The microscope was surrounded by a box type incubator (Haison) maintained at 37°C. Images were acquired every 10 min for approximately 7 h before Bb were added on the wells at an MOI of ∼200.
Subsequently, at each time interval we measured the 2D displacements based on the image of the host cell nuclei using an image correlation technique similar to particle image velocimetry (Gui and Wereley, 2002). We calculated the local displacement vectors by performing image correlation between subsequent images of the nuclei. We used interrogation windows of 48 x 24 pixels (window size x overlap). We calculated the mean displacement of cells before and after Bb-exposure by finding the average magnitude when considering all displacements in a particular field of view.
To quantify total bacterial fluorescence overtime, we used custom-built MATLAB code to integrate the Bb-GFP fluorescence intensity overtime. To characterize the aggregation of bacteria observed overtime we performed segmentation using IMARIS software (Bitplane) to identify all the objects (spirochetes or aggregates) at each instance of time. All identified objects and their associated attributes (i.e., area, mean Bb fluorescence intensity, ellipticity) were imported into MATLAB (MathWorks) for further analysis, e.g., for creating scatter plots of object ellipticity versus object area or object mean fluorescence versus object area.
RNA isolation and RNA sequencing
For sample preparation HMEC-1 cells were cultured in MCDB 131 medium (Fisher Scientific, 10372-019) supplemented with 10% fetal bovine serum (GemBio; 900-108), 10 ng/mL epidermal growth factor (Sigma; E9644), 1 μg/mL hydrocortisone (Sigma; H0888), and 2 mM L-glutamine (Sigma; 56-85-9). To generate confluent cell monolayers, 24-well plates glass-bottom for microscopy were coated with 50 μg/mL rat-tail collagen-I (diluted in 0.2 N acetic acid) for 1hat 37°C, air-dried for 15 min, and UV-sterilized for 30 min in a biosafety cabinet. ECs were seeded at a density of 2 × 105 cells/well. 24 h post-seeding ECs were exposed to Bb (MOI = 200) or not. At different times post-Bb-exposure, namely 4 h, 24 h or 48 hpe, four replicates per condition were lysed using the QIAshredder Kit by adding 200 μL of lysis buffer in each well (Qiagen, 79656). mRNA was harvested using the RNeasy Plus MicroKit (Qiagen, 74004) and eluted in 30 μL RNAase free water. A NanoDrop ND-1000 spectrophotometer was used to determine concentration (abs 260) and purity (abs260/abs230) of total RNA samples. Total RNA from each sample was 1-2 μg. Agarose gel electrophoresis was used to check the integrity of the RNA in all the samples (performed by Arraystar Inc.).
For sequencing library preparation, the subsequent steps were followed: 1. Total RNA was enriched by oligo (dT) magnetic beads (rRNA removed); 2. RNA-seq library preparation using KAPA Stranded RNA-Seq Library Prep Kit (Illumina), which incorporates dUTP into the second cDNA strand and renders the RNA-seq library strand-specific. The completed libraries were qualified with Agilent 2100 Bioanalyzer and quantified by absolute quantification qPCR method. To sequence the libraries on the Illumina NovaSeq 6000 instrument, the barcoded libraries were mixed, denatured to single stranded DNA in NaOH, captured on Illumina flow cell, amplified in situ, and subsequently sequenced for 150 cycles for both ends on Illumina NovaSeq 6000 instrument. Raw sequencing data that passed the Illumina chastity filter were used for further analysis. Trimmed reads (trimmed 5′, 3′-adaptor bases) were aligned to the reference genome (GRCh37). Based on alignment statistical analysis (mapping ratio, rRNA/mtRNA content, fragment sequence bias), we determined whether the results could be used for subsequent data analysis. To examine the sequencing quality, the quality score plot of each sample was plotted. The quality score Q is logarithmically related to the base calling error probability (P): Q = −10log10(P). For example, Q30 means the incorrect base calling probability to be 0.001 or 99.9% base calling accuracy. After quality control, the fragments were 5′, 3′-adaptor trimmed and filtered ≤20 bp reads with Cutadapt software (Martin, 2011). The trimmed reads were aligned to the reference genome with Hisat 2 software (Kim et al., 2015). In a typical experiment, it is possible to align 40 ∼ 90% of the fragments to the reference genome. However, this percentage depends on multiple factors, including sample quality, library quality and sequencing quality. Sequencing reads were classified into the following classes: (1) Mapped: reads aligned to the reference genome (including mRNA, pre-mRNA, poly-A tailed lncRNA and pri-miRNA); (2) mtRNA and rRNA: fragments aligned to rRNA, mtRNA; and (3) Unmapped: reads that are not aligned.
Differentially expressed genes and differentially expressed transcripts were calculated. The novel genes and transcripts were also predicted. The expression level (fragments per kilobase of transcript per million mapped reads (FPKM) value) of known genes and transcripts were calculated using R package ballgown by estimating the transcript abundances with StringTie. The number of identified genes and transcripts per group was calculated based on the mean of FPKM in group ≥0.5. The FPKM value was calculated with the formula: , where C is the number of fragments that map to a certain gene/transcript, L is the length of the gene/transcript in Kb and N is the fragments number that maps to all genes/transcripts. Differentially expressed gene and transcript analyses were performed with R package ballgown. Fold change (cutoff 1.5), p-value (≤0.05) and FPKM (≥0.5 mean in one group) were used for filtering differentially expressed genes and transcripts.
Principal component analysis (PCA) and mRNA function enrichment analysis
Principal Component Analysis (PCA) and Hierarchical Clustering scatter plots and volcano plots were calculated for the differentially expressed genes in R or Python environment for statistical computing and graphics. PCA was performed using the plotPCA function in R with genes that had the ANOVA p value ≤0.05 on FPKM abundance estimations (Not available for samples with no replicates). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of the whole data set of DEG were performed using the R package GAGE “Generally Acceptable Gene set Enrichment” (GAGE v.2.22.0) package implemented in R (Luo et al., 2009). The analysis allowed us to determine whether the differentially expressed mRNAs were enriched in certain biological pathways. The p-values calculated by Fisher’s exact test are used to estimate the statistical significance of the enrichment of the pathways between the two groups. The R package “Pathview” v.1.12.0 and KEGGraph v1.30.0 were used to visualize gene set expression data in the context of functional pathways (Luo et al., 2017).
Inside/outside bacterial labeling, immunofluorescence microscopy and image processing
Unexposed or Bb-exposed ECs in monolayer on glass coverslips coated with 0.25 mg/mL collagen I were washed with PBS and incubated with 1 μg/mL Hoechst (Thermofisher, D1306) to stain the cells’ nuclei for 10 min at 37°C. Wells were then washed twice in PBS and then fixed with 4% EM grade formaldehyde in PBS for 10 min. Samples were washed again with PBS and stored until further processing.
For staining only extracellular Bb cells inside/outside labeling was performed: samples were blocked for 30 min with 5% BSA in PBS and then incubated with primary rabbit polyclonal anti-Bb antibody (Thermo Fisher Scientific, PA1-73004) diluted 1:100 in PBS containing 2% BSA for 1 h, at room temperature. Samples were washed in PBS three times and then incubated with Alexa Fluor 546 goat anti-rabbit secondary antibody (Invitrogen, A-11035) diluted 1:250 in PBS containing 2% BSA for 1 h. Samples were washed three times in PBS and stored in 1 mL PBS for imaging. N>1000 cells were analyzed per condition. For imaging, we used an inverted Nikon Diaphot 200 with a charge-coupled device (CCD) camera (Andor Technologies) and a 40× air Plan Fluor NA 0.60 objective. The microscope was controlled by the MicroManager software package. For differential immunostaining, all “green” bacteria associated with individual cells were counted as adherent (because at the time of fixation they were adhering on the host cell surface or because prior to internalization they were adherent); bacteria that were both “green” and “red” (due to antibody binding) were counted as non-internalized. Nuclei number was identified by running a custom-made script in MATLAB (Mathworks).
For immunostaining of cytoskeletal elements, fixed cells were first permeabilized for 5 min in 0.2% Triton X-100 in PBS and washed again with PBS. Samples were then blocked for 30 min with 5% BSA in PBS and then incubated with primary antibodies (mouse monoclonal anti-integrin β1, Millipore, MAB1987; mouse monoclonal anti-integrin αvβ3, Sigma, MAB1976) diluted 1:100 in PBS containing 2% BSA for 1 h. Samples were washed in PBS three times and then incubated with the appropriate secondary fluorescent antibodies (Invitrogen) diluted 1:250 in PBS containing 2% BSA for 1 h. For actin staining we used 0.2 μM AlexaFluor546 phalloidin (Thermo Fisher Scientific, A22283). Samples were washed three times in PBS and stored in 1 mL PBS for imaging. For confocal imaging we used a Yokogawa W1 Spinning Disk Confocal with Borealis upgrade on a Leica DMi6 inverted microscope with a 50um Disk pattern, a 60x 1.4NA Plan Apo oil objective and MicroManager software. Z-stacks were taken with an interval of 0.2 μm.
For quantification of the relative abundance of F-actin or integrins β1 and αvβ3 on Bb-exposed versus not EC monolayers on a per cell basis the pipeline described was followed. Custom made codes were written in MATLAB (Mathowrks). Briefly, the images of the Hoechst-stained nuclei were used to segment the host cell nuclei and identify their centroids. The MATLAB function voronoi2mask was used to perform centroidal voronoi tessellation and thus approximate the boundaries of each cell in the monolayer. For the area occupied by each cell the integral of fluorescence intensity of each cytoskeletal component was then calculated and depicted as a single dot on the boxplots shown in Figures 5C, 5F and S5B.
Propidium iodide (PI) staining, imaging and quantitation of co-localization of Bb with PI
PI (Millipore Sigma, P4170-10MG) at a concentration of 5 μM was added in ECs in monolayer exposed to Bb (MOI = 200) at the time of exposure, as described previously to assess dynamically bacterial death (Krämer et al., 2016). ECs were residing in wells on 24-well plates were imaged for 24hat 37 °C using the IncuCyte S3 time lapse microscopy system (Sartorius) equipped with an IncuCyte S3 20 x PLAN FLUOR objective. Phase contrast images, images of bacterial fluorescence and of PI fluorescence were acquired every 90 min. For quantification of co-localization between Bb-GFP and PI stain the pipeline described below was followed. Images of bacterial fluorescence were segmented so that bacterial objects were identified. Next, images of PI fluorescence were also segmented. Then the percentage of pixels occupied by bacteria that are also PI positive was determined. Co-localization index was defined as the ratio of pixels that are both occupied by bacteria and are PI-positive to the total number of pixels occupied by bacteria. The source code is provided at https://github.com/ebastoun/borrelia_PI_colocalisation.
RT-PCR
To assess expression of NFκΒ target genes, ECs were seeded until monolayers were formed (∼4x105 cells/well of 24 well plate) as described above. At different time points, namely 4 h or 24 h post-treatment with TNFα or MDP, or post heat-killed Bb exposure, two to three replicates per condition per experiment were lysed using the QIAshredder Kit by adding 350 μL of lysis buffer in each well (Qiagen, 79656). mRNA was harvested using the RNeasy Plus MicroKit (Qiagen, 74034) and eluted in 30μL RNAase free water. RNA concentrations were measured spectrophotometrically (NanoDrop) and were comparable between conditions. cDNA was prepared using the PrimeScript™ RT Reagent Kit (Takara Bio, RR037A). RT-PCR was performed using the KAPA SYBR® FAST mix (Kapa Biosystems, KK4601) and the QuantStudio 3 Real-Time PCR system (Applied Biosystems). Genes of interest were amplified using the appropriate primers for ICAM1 (forward: 5′ GAGCTTCGTGTCCTGTATGGC -3′, reverse: 5′ ACAGAGGTAGGTGCCCTCAA-3′); for CXCL8 (forward: 5′ CAGAGACAGCAGAGCACACA -3′, reverse: 5′ GGCAAAACTGCACCTTCACA -3′); for NFKBIA (forward: 5′ ATGTCAATGCTCAGGAGCCC -3′, reverse: 5′ CAGGTGAGCTGGTAGGGAGA -3′); for GAPDH (forward:5′ GGGAAACTGTGGCGTGAT -3′, reverse: 5′ GAGTGGGTGTCGCTGTTGA -3′). Analysis was performed using the QuantStudio™ Design and Analysis Software v2 and the Comparative CT Method (2−ΔΔCt). GAPDH was used as the reference gene.
Quantification and statistical analysis
Statistical parameters and significance are reported in the Figures and the Figure Legends. Data are determined to be statistically significant when p < 0.05 or p <0.01 by an unpaired Student’s T-Test (USTT), or Wilcoxon Rank Sum Test (WRST), where indicated. As such, asterisk denotes statistical significance as compared to indicated controls. For inside/outside labeling 20 fields of view (FOV) were analyzed consisting of an average 158 ± 60 host cell nuclei per FOV. For statistical analysis of EC kinematics a large number of cells pertained in each field of view (FOV) and average of cells’ displacements were calculated in each FOV and overtime for three independent experiments. The same applies for the traction force microscopy recordings. In the boxplots and barplots midlines denote mean value and whiskers the standard deviation (vertical bars). Statistical analysis was performed in GraphPad PRISM 8.
Acknowledgments
We are grateful to Julie A. Theriot for her insight, scientific discussions and sharing her resources. We are grateful to Joseph D. Mougous for sharing his resources and to Ben Ross for discussions, experimental support and revising the manuscript. We thank Justin Radolf and Tara Moriarty for sharing the GFP-expressing borrelial strains with us. We thank Matthew Footer for discussions and experimental support. We thank Libera Lo Presti for discussions and for revising the manuscript and rest of the materials. We acknowledge that part of our MSM codes are based on the original code (plane stress problem) written by Siva Srinivas Kolukula and we hence used some functions from his code. This research was supported by the Cell Analysis Facility Flow Cytometry and Imaging Core in the Department of Immunology at the University of Washington. RNA Seq and RT-qPCR were performed by Arraystar Inc. The graphical abstract was created with BioRender.com. This work was supported in part by the American Heart Association, Award number: 18CDA34070047 (E.E.B.), the Lyme Disease Association (E.E.B.), the Stanford Lyme Disease Seed Grant (E.E.B.) and the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) underGermany's Excellence Strategy – EXC 2124–390838134 (E.E.B., K.A., M.M., A.R.), the Erasmus+ Programme of the European Union (K.A.), the European Research Council (ICoMICS Adg grant agreement: 101018587, (R.A.-Y. and M.J.G.-B.)), the Spanish Ministry of Universities (grant FPU 20/05274 (R.A.-Y.)), grant PID2021-124271OB-I00 founded by MCIN/AEI/10.13039/501100011033 (R.A-Y, M.J.G-B) and ERDF A way of making Europe (R.A.-Y, and M.J.G.-B.). Finally, we acknowledge support by Open Access Publishing Fund of the University of Tübingen.
Author contributions
Conceptualization, E.E.B.; Methodology, E.E.B., M.J.G.-B., M.M., K.A., P.K., R.A.-Y., M.C.T.; Software, E.E.B; M.J.G.-B., M.M., R.A.-Y., A.R.; Investigation, E.E.B., P.K., M.M., K.A., R.A.-Y., A.R., B.R.; Writing – Original Draft, E.E.B.; Writing – Review & Editing, E.E.B., K.A., P.K., M.C.T., M.J.G.-B.; Resources, E.E.B., P.K., M.C.T., M.J.G.-B.; Supervision, E.E.B.
Declaration of interests
The authors declare no competing interests.
Published: August 19, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104793.
Supplemental information
Sheets 1-2 of the table list the DEGs identified when comparing the transcriptome of unexposed to Bb-exposed ECs. Shown are genes upregulated (1st sheet, up_U4_vs_I4) or downregulated in unexposed cells (2nd sheet, down_U4_vs_I4) at 4 hpe. In an analogous manner, sheets 3-4 refer to similar samples examined at 24 hpe and sheets 5-6 refer to similar samples examined at 48 hpe. In each sheet rows correspond to the genes identified and columns A-K correspond to different parameters explained within the table. Columns L-S provide the FPMK (see STAR Methods) of genes for each sample from the corresponding groups compared. Sheets 7-9 of the table show KEGG pathways significantly perturbed when comparing the transcriptome of unexposed to Bb-exposed cells at 4 (sheet 7), 24 (sheet 8) or 48 (sheet 9) hpe. Pathways that are downregulated in unexposed cells (upregulated for Bb-exposed) are highlighted in pink, while those upregulated are highlighted in green. The gage package was used for pathway analysis which has precompiled databases for mapping genes to KEGG pathways so to perform pathway enrichment analysis of DEGs among all three populations. Rows show the KEGG pathways and columns the different parameters explained in the table, including the specific genes of each category that are differentially regulated.
Data and code availability
Data collected and computer codes are available on request to the corresponding authors and are also publicly available at: https://github.com/ebastoun/Monolayer-Stress Microscopy (codes for calculating monolayer stresses) and https://github.com/ebastoun/borrelia_PI_colocalisation (codes for calculating colocalization between bacteria and propidium iodide staining). The RNA sequencing data (FASTq files) generated during this study and subsequent analysis are available at the Gene Expression Omnibus (GEO) database (weblink: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE174545, series record number: GSE174545). All differential expression analysis results of this study are included as supplementary tables in this article. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- CDC Lyme Disease. https://www.cdc.gov/lyme/index.html.
- Alban P.S., Johnson P.W., Nelson D.R. Serum-starvation-induced changes in protein synthesis and morphology of Borrelia burgdorferi. Microbiology (Read.) 2000;146:119–127. doi: 10.1099/00221287-146-1-119. [DOI] [PubMed] [Google Scholar]
- Anderson C., Brissette C.A. The brilliance of Borrelia: mechanisms of host immune evasion by lyme disease-causing spirochetes. Pathogens. 2021;10:281. doi: 10.3390/pathogens10030281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson G.G., Palermo J.J., Schilling J.D., Roth R., Heuser J., Hultgren S.J. Intracellular bacterial biofilm-like pods in urinary tract infections. Science. 2003;301:105–107. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- Barakat A., Lieu D. Differential responsiveness of vascular endothelial cells to different types of fluid mechanical shear stress. Cell Biochem. Biophys. 2003;38:323–343. doi: 10.1385/cbb:38:3:323. [DOI] [PubMed] [Google Scholar]
- Bastounis E., Meili R., Álvarez-González B., Francois J., del Álamo J.C., Firtel R.A., Lasheras J.C. Both contractile axial and lateral traction force dynamics drive amoeboid cell motility. J. Cell Biol. 2014;204:1045–1061. doi: 10.1083/jcb.201307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastounis E.E., Ortega F.E., Serrano R., Theriot J.A. A multi-well format polyacrylamide-based assay for studying the effect of extracellular matrix stiffness on the bacterial infection of adherent cells. J. Vis. Exp. 2018:57361. doi: 10.3791/57361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastounis E.E., Radhakrishnan P., Prinz C.K., Theriot J.A. Mechanical forces govern interactions of host cells with intracellular bacterial pathogens. Microbiol. Mol. Biol. Rev. 2022:e0009420. doi: 10.1128/mmbr.00094-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastounis E.E., Serrano-Alcalde F., Radhakrishnan P., Engström P., Gómez-Benito M.J., Oswald M.S., Yeh Y.-T., Smith J.G., Welch M.D., García-Aznar J.M., Theriot J.A. Mechanical competition triggered by innate immune signaling drives the collective extrusion of bacterially infected epithelial cells. Dev. Cell. 2021;56:443–460.e11. doi: 10.1016/j.devcel.2021.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastounis E.E., Yeh Y.T., Theriot J.A. Subendothelial stiffness alters endothelial cell traction force generation while exerting a minimal effect on the transcriptome. Sci. Rep. 2019;9:18209. doi: 10.1038/s41598-019-54336-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazellières E., Conte V., Elosegui-Artola A., Serra-Picamal X., Bintanel-Morcillo M., Roca-Cusachs P., Muñoz J.J., Sales-Pardo M., Guimerà R., Trepat X. Control of cell–cell forces and collective cell dynamics by the intercellular adhesome. Nat. Cell Biol. 2015;17:409–420. doi: 10.1038/ncb3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera A.K., Hildebrand E., Scagliotti J., Steere A.C., Hu L.T. Induction of host matrix metalloproteinases by Borrelia burgdorferi differs in human and murine lyme arthritis. Infect. Immun. 2005;73:126–134. doi: 10.1128/IAI.73.1.126-134.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque S.L., Davidge S.T., Adams M.A. The interaction between endothelin-1 and nitric oxide in the vasculature: new perspectives. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011;300:R1288–R1295. doi: 10.1152/ajpregu.00397.2010. [DOI] [PubMed] [Google Scholar]
- Brandt M., Gerke V., Betz T. Human endothelial cells display a rapid and fluid flow dependent tensional stress increase in response to tumor necrosis factor-α. Preprint at bioRxiv. 2022 doi: 10.1101/2022.01.12.476017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns M.J., Sellati T.J., Teng E.I., Furie M.B. Production of interleukin-8 (IL-8) by cultured endothelial cells in response to Borrelia burgdorferi occurs independently of secreted [corrected] IL-1 and tumor necrosis factor alpha and is required for subsequent transendothelial migration of neutrophils. Infect. Immun. 1997;65:1217–1222. doi: 10.1128/iai.65.4.1217-1222.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano M.J., Dunham-Ems S., Allard A.M., Cassera M.B., Kenedy M., Radolf J.D. Cyclic di-GMP modulates gene expression in Lyme disease spirochetes at the tick-mammal interface to promote spirochete survival during the blood meal and tick-to-mammal transmission. Infect. Immun. 2015;83:3043–3060. doi: 10.1128/IAI.00315-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo J., Jung H., Meloni G. Copper metallothioneins. IUBMB Life. 2017;69:236–245. doi: 10.1002/iub.1618. [DOI] [PubMed] [Google Scholar]
- Cerar T., Strle F., Stupica D., Ruzic-Sabljic E., Mchugh G., Steere A.C., Strle K. Differences in genotype, clinical features, and inflammatory potential of Borrelia burgdorferi sensu stricto strains from Europe and the United States. Emerg. Infect. Dis. 2016;22:818–827. doi: 10.3201/eid2205.151806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn J., Garcia B., Hu L.T., Jewett M.W., Kraiczy P., Norris S.J., Skare J. Lyme disease pathogenesis. Curr. Issues Mol. Biol. 2021;42:473–518. doi: 10.21775/cimb.042.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn J., Magoun L., Bodary S.C., Leong J.M. Integrins alpha(v)beta3 and alpha5beta1 mediate attachment of lyme disease spirochetes to human cells. Infect. Immun. 1998;66:1946–1952. doi: 10.1128/iai.66.5.1946-1952.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J.L., Sellati T.J., Testa J.E., Kew R.R., Furie M.B., Benach J.L. Borrelia burgdorferi binds plasminogen, resulting in enhanced penetration of endothelial monolayers. Infect. Immun. 1995;63:2478–2484. doi: 10.1128/iai.63.7.2478-2484.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova T.A., Danilina G.A., Adzhieva A.A., Vostrova E.I., Zhukhovitskii V.G., Cheknev S.B. Inhibitory effect of copper and zinc ions on the growth of Streptococcus pyogenes and Escherichia coli biofilms. Bull. Exp. Biol. Med. 2020;169:648–652. doi: 10.1007/s10517-020-04946-y. [DOI] [PubMed] [Google Scholar]
- del Álamo J.C., Meili R., Alonso-Latorre B., Rodríguez-Rodríguez J., Aliseda A., Firtel R.A., Lasheras J.C. Spatio-temporal analysis of eukaryotic cell motility by improved force cytometry. Proc. Natl. Acad. Sci. USA. 2007;104:13343–13348. doi: 10.1073/pnas.0705815104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dev A., Iyer S., Razani B., Cheng G. NF-kappaB and innate immunity. Curr.Top.Microbiol. Immunol. 2011;349:115–143. doi: 10.1007/82_2010_102. [DOI] [PubMed] [Google Scholar]
- Dunham-Ems S.M., Caimano M.J., Pal U., Wolgemuth C.W., Eggers C.H., Balic A., Radolf J.D. Live imaging reveals a biphasic mode of dissemination of Borrelia burgdorferi within ticks. J. Clin. Invest. 2009;119:3652–3665. doi: 10.1172/JCI39401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebady R., Niddam A.F., Boczula A.E., Kim Y.R., Gupta N., Tang T.T., Odisho T., Zhi H., Simmons C.A., Skare J.T., Moriarty T.J. Biomechanics of Borrelia burgdorferi vascular interactions. Cell Rep. 2016;16:2593–2604. doi: 10.1016/j.celrep.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A.D., Tsuchida M.A., Amodaj N., Pinkard H., Vale R.D., Stuurman N. Advanced methods of microscope control using μManager software. J. Biol. Methods. 2014;1:e10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faralla C., Bastounis E.E., Ortega F.E., Light S.H., Rizzuto G., Gao L., Marciano D.K., Nocadello S., Anderson W.F., Robbins J.R., et al. Listeria monocytogenes InlP interacts with afadin and facilitates basement membrane crossing. PLoS Pathog. 2018;14:e1007094. doi: 10.1371/journal.ppat.1007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feria-Arroyo T.P., Castro-Arellano I., Gordillo-Perez G., Cavazos A.L., Vargas-Sandoval M., Grover A., Torres J., Medina R.F., de León A.A.P., Esteve-Gassent M.D. Implications of climate change on the distribution of the tick vector Ixodes scapularis and risk for Lyme disease in the Texas-Mexico transboundary region. Parasit. Vectors. 2014;7:199. doi: 10.1186/1756-3305-7-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebbia J.A., Coleman J.L., Benach J.L. Borrelia spirochetes upregulate release and activation of matrix metalloproteinase gelatinase B (MMP-9) and collagenase 1 (MMP-1) in human cells. Infect. Immun. 2001;69:456–462. doi: 10.1128/IAI.69.1.456-462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girschick H.J., Huppertz H.I., Rüssmann H., Krenn V., Karch H. Intracellular persistence ofBorrelia burgdorferi in human synovial cells. Rheumatol. Int. 1996;16:125–132. doi: 10.1007/BF01409985. [DOI] [PubMed] [Google Scholar]
- Gui L., Wereley S.T. A correlation-based continuous window-shift technique to reduce the peak-locking effect in digital PIV image evaluation. Exp. Fluids. 2002;32:506–517. [Google Scholar]
- Haage A., Schneider I.C. Cellular contractility and extracellular matrix stiffness regulate matrix metalloproteinase activity in pancreatic cancer cells. FASEB J. 2014;28:3589–3599. doi: 10.1096/fj.13-245613. [DOI] [PubMed] [Google Scholar]
- Hur S.S., del Álamo J.C., Park J.S., Li Y.S., Nguyen H.A., Teng D., Wang K.C., Flores L., Alonso-Latorre B., Lasheras J.C., Chien S. Roles of cell confluency and fluid shear in 3-dimensional intracellular forces in endothelial cells. Proc. Natl. Acad. Sci. USA. 2012;109:11110–11115. doi: 10.1073/pnas.1207326109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutras B.L., Lochhead R.B., Kloos Z.A., Biboy J., Strle K., Booth C.J., Govers S.K., Gray J., Schumann P., Vollmer W., et al. Borrelia burgdorferi peptidoglycan is a persistent antigen in patients with Lyme arthritis. Proc. Natl. Acad. Sci. USA. 2019;116:13498–13507. doi: 10.1073/pnas.1904170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutras B.L., Scott M., Parry B., Biboy J., Gray J., Vollmer W., Jacobs-Wagner C. Lyme disease and relapsing fever Borrelia elongate through zones of peptidoglycan synthesis that mark division sites of daughter cells. Proc. Natl. Acad. Sci. USA. 2016;113:9162–9170. doi: 10.1073/pnas.1610805113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempe S., Kestler H., Lasar A., Wirth T. NF0ºB controls the global pro-inflammatory response in endothelial cells: evidence for the regulation of a pro-atherogenic program. Nucleic Acids Res. 2005;33:5308–5319. doi: 10.1093/nar/gki836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisseleva T., Song L., Vorontchikhina M., Feirt N., Kitajewski J., Schindler C. NF-kappaB regulation of endothelial cell function during LPS-induced toxemia and cancer. J. Clin. Invest. 2006;116:2955–2963. doi: 10.1172/JCI27392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenderink J.J., van Doorn A.J. Representation of local geometry in the visual system. Biol. Cybern. 1987;55:367–375. doi: 10.1007/BF00318371. [DOI] [PubMed] [Google Scholar]
- Komarova Y.A., Kruse K., Mehta D., Malik A.B. Protein interactions at endothelial junctions and signaling mechanisms regulating endothelial permeability. Circ. Res. 2017;120:179–206. doi: 10.1161/CIRCRESAHA.116.306534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer C.E.M., Wiechert W., Kohlheyer D. Time-resolved, single-cell analysis of induced and programmed cell death via non-invasive propidium iodide and counterstain perfusion. Sci. Rep. 2016;6:32104. doi: 10.1038/srep32104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D., Ristow L.C., Shi M., Mukherjee P., Caine J.A., Lee W.-Y., Kubes P., Coburn J., Chaconas G. Intravital imaging of vascular transmigration by the lyme spirochete: requirement for the integrin binding residues of the B. Burgdorferi P66 protein. PLoS Pathog. 2015;11:e1005333. doi: 10.1371/journal.ppat.1005333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumru O.S., Schulze R.J., Rodnin M.V., Ladokhin A.S., Zückert W.R. Surface localization determinants of Borrelia OspC/Vsp family lipoproteins. J. Bacteriol. 2011;193:2814–2825. doi: 10.1128/JB.00015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFrance M.E., Pierce J.V., Antonara S., Coburn J. The Borrelia burgdorferi integrin ligand P66 affects gene expression by human cells in culture. Infect. Immun. 2011;79:3249–3261. doi: 10.1128/IAI.05122-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamason R.L., Bastounis E., Kafai N.M., Serrano R., Del Álamo J.C., Theriot J.A., Welch M.D. Rickettsia Sca4 reduces vinculin-mediated intercellular tension to promote spread. Cell. 2016;167:670–683.e10. doi: 10.1016/j.cell.2016.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappas M. NOD1 and NOD2 regulate proinflammatory and prolabor mediators in human fetal membranes and myometrium via nuclear factor-kappa B. Biol. Reprod. 2013;89:14. doi: 10.1095/biolreprod.113.110056. [DOI] [PubMed] [Google Scholar]
- Larsen M., Tremblay M.L., Yamada K.M. Phosphatases in cell-matrix adhesion and migration. Nat. Rev. Mol. Cell Biol. 2003;4:700–711. doi: 10.1038/nrm1199. [DOI] [PubMed] [Google Scholar]
- Leavesley D.I., Schwartz M.A., Rosenfeld M., Cheresh D.A. Integrin beta 1- and beta 3-mediated endothelial cell migration is triggered through distinct signaling mechanisms. J. Cell Biol. 1993;121:163–170. doi: 10.1083/jcb.121.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.Y., Zaske A.M., Novellino T., Danila D., Ferrari M., Conyers J., Decuzzi P. Probing the mechanical properties of TNF-α stimulated endothelial cell with atomic force microscopy. Int. J. Nanomed. 2011;6:179–195. doi: 10.2147/IJN.S12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livengood J.A., Gilmore R.D., Jr. Invasion of human neuronal and glial cells by an infectious strain of Borrelia burgdorferi. Microbes Infect. 2006;8:2832–2840. doi: 10.1016/j.micinf.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Luo W., Friedman M.S., Shedden K., Hankenson K.D., Woolf P.J. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinf. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Pant G., Bhavnasi Y.K., Blanchard S.G., Jr., Brouwer C. Pathview Web: user friendly pathway visualization and data integration. Nucleic Acids Res. 2017;45:W501–W508. doi: 10.1093/nar/gkx372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Sturrock A., Weis J.J. Intracellular localization of Borrelia burgdorferi within human endothelial cells. Infect. Immun. 1991;59:671–678. doi: 10.1128/iai.59.2.671-678.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 2011;17:10. [Google Scholar]
- Meriläinen L., Herranen A., Schwarzbach A., Gilbert L. Morphological and biochemical features of Borrelia burgdorferi pleomorphic forms. Microbiology. 2015;161:516–527. doi: 10.1099/mic.0.000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty T.J., Norman M.U., Colarusso P., Bankhead T., Kubes P., Chaconas G. Real-time high resolution 3D imaging of the lyme disease spirochete adhering to and escaping from the vasculature of a living host. PLoS Pathog. 2008;4:e1000090. doi: 10.1371/journal.ppat.1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgia R., Cinco M. Induction of cystic forms by different stress conditions in Borrelia burgdorferi. APMIS. 2004;112:57–62. doi: 10.1111/j.1600-0463.2004.apm1120110.x. [DOI] [PubMed] [Google Scholar]
- Niddam A.F., Ebady R., Bansal A., Koehler A., Hinz B., Moriarty T.J. Plasma fibronectin stabilizes Borrelia burgdorferi-endothelial interactions under vascular shear stress by a catch-bond mechanism. Proc. Natl. Acad. Sci. USA. 2017;114 doi: 10.1073/pnas.1615007114. E3490–e3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski M.A., Huang N.F., Walker T.W., VerwijleN T., Poplawski C., Khoo A.S., Cooke J.P., Fuller G.G., Dunn A.R. Microvascular endothelial cells migrate upstream and align against the shear stress field created by impinging flow. Biophys. J. 2014;106:366–374. doi: 10.1016/j.bpj.2013.11.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte S., Ipiña E.P., Pontier-Bres R., Czerucka D., Peruani F. Statistics of pathogenic bacteria in the search of host cells. Nat. Commun. 2021;12:1990. doi: 10.1038/s41467-021-22156-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate E. Vol. 2. Springer; 2013. (Structural Analysis with the Finite Element Method. Linear Statics). [Google Scholar]
- Parthasarathy G., Fevrier H.B., Philipp M.T. Non-viable Borrelia burgdorferi induce inflammatory mediators and apoptosis in human oligodendrocytes. Neurosci. Lett. 2013;556:200–203. doi: 10.1016/j.neulet.2013.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quehenberger P., Bierhaus A., Fasching P., Muellner C., Klevesath M., Hong M., Stier G., Sattler M., Schleicher E., Speiser W., Nawroth P.P. Endothelin 1 transcription is controlled by nuclear factor-kappaB in AGE-stimulated cultured endothelial cells. Diabetes. 2000;49:1561–1570. doi: 10.2337/diabetes.49.9.1561. [DOI] [PubMed] [Google Scholar]
- Ranzato M., Taylor P.E., House J.M., Flagan R.C., Lecun Y., Perona P. Automatic recognition of biological particles in microscopic images. Pattern Recogn. Lett. 2007;28:31–39. [Google Scholar]
- Reed S.C.O., Lamason R.L., Risca V.I., Abernathy E., Welch M.D. Rickettsia actin-based motility occurs in distinct phases mediated by different actin nucleators. Curr. Biol. 2014;24:98–103. doi: 10.1016/j.cub.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart-King C.A., Dembo M., Hammer D.A. The dynamics and mechanics of endothelial cell spreading. Biophys. J. 2005;89:676–689. doi: 10.1529/biophysj.104.054320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Replogle A.J., Sexton C., Young J., Kingry L.C., Schriefer M.E., Dolan M., Johnson T.L., Connally N.P., Padgett K.A., Petersen J.M. Isolation of Borrelia miyamotoi and other Borreliae using a modified BSK medium. Sci. Rep. 2021;11:1926. doi: 10.1038/s41598-021-81252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokhzan R., Ghosh C.C., Schaible N., Notbohm J., Yoshie H., Ehrlicher A.J., Higgins S.J., Zhang R., Haller H., Hardin C.C., et al. Multiplexed, high-throughput measurements of cell contraction and endothelial barrier function. Lab. Invest. 2019;99:138–145. doi: 10.1038/s41374-018-0136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J., Wahhab A., Danner R., Jutras B., Lochhead R. Borrelia burgdorferi peptidoglycan triggers inflammatory responses in Lyme arthritis. J. Immunol. 2021;206 52.17. [Google Scholar]
- Russell T.M., Delorey M.J., Johnson B.J.B. Borrelia burgdorferi BbHtrA degrades host ECM proteins and stimulates release of inflammatory cytokines in vitro. Mol. Microbiol. 2013;90:241–251. doi: 10.1111/mmi.12377. [DOI] [PubMed] [Google Scholar]
- Sapi E., Bastian S.L., Mpoy C.M., Scott S., Rattelle A., Pabbati N., Poruri A., Burugu D., Theophilus P.A.S., Pham T.V., et al. Characterization of biofilm formation by Borrelia burgdorferi in vitro. PLoS One. 2012;7:e48277. doi: 10.1371/journal.pone.0048277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saputra E.P., Trzeciakowski J.P., Hyde J.A. Borrelia burgdorferi spatiotemporal regulation of transcriptional regulator bosR and decorin binding protein during murine infection. Sci. Rep. 2020;10:12534. doi: 10.1038/s41598-020-69212-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J., Jia Y., Teulières L., Patel F., Clokie M.R.J. Targeting multicopy prophage genes for the increased detection of Borrelia burgdorferi sensu lato (s.l.), the causative agents of lyme disease, in blood. Front. Microbiol. 2021;12:464. doi: 10.3389/fmicb.2021.651217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolentseva O., Gusarov I., Gautier L., Shamovsky I., Defrancesco A.S., Losick R., Nudler E. Mechanism of biofilm-mediated stress resistance and lifespan extension in C. elegans. Sci. Rep. 2017;7:7137. doi: 10.1038/s41598-017-07222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroka K.M., Vaitkus J.A., Aranda-Espinoza H. Endothelial cells undergo morphological, biomechanical, and dynamic changes in response to tumor necrosis factor-α. Eur. Biophys. J. 2012;41:939–947. doi: 10.1007/s00249-012-0851-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambe D.T., Hardin C.C., Angelini T.E., Rajendran K., Park C.Y., Serra-Picamal X., Zhou E.H., Zaman M.H., Butler J.P., Weitz D.A., et al. Collective cell guidance by cooperative intercellular forces. Nat. Mater. 2011;10:469–475. doi: 10.1038/nmat3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Petri B., Devinney R., Jenne C.N., Chaconas G. The Lyme disease spirochete can hijack the host immune system for extravasation from the microvasculature. Mol. Microbiol. 2021;116:498–515. doi: 10.1111/mmi.14728. [DOI] [PubMed] [Google Scholar]
- Trepat X., Wasserman M.R., Angelini T.E., Millet E., Weitz D.A., Butler J.P., Fredberg J.J. Physical forces during collective cell migration. Nat. Phys. 2009;5:426–430. [Google Scholar]
- Urbano R.L., Furia C., Basehore S., Clyne A.M. Stiff substrates increase inflammation-induced endothelial monolayer tension and permeability. Biophys. J. 2017;113:645–655. doi: 10.1016/j.bpj.2017.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Petzke M.M., Iyer R., Wu H., Schwartz I. Pattern of proinflammatory cytokine induction in RAW264.7 mouse macrophages is identical for virulent and attenuated Borrelia burgdorferi. J. Immunol. 2008;180:8306–8315. doi: 10.4049/jimmunol.180.12.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Weening E.H., Faske J.B., Höök M., Skare J.T. Invasion of eukaryotic cells by Borrelia burgdorferi requires beta(1) integrins and Src kinase activity. Infect. Immun. 2011;79:1338–1348. doi: 10.1128/IAI.01188-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time-lapse recording of Bb spirochetes constitutively expressing GFP (Bb1286, Bb-GFP) within a microscope slide. Phase contrast (left) and Bb-GFP fluorescence (right) are shown. Bb-GFP were grown to a concentration of 5x107 bacteria/mL in BSK-H medium and were imaged before EC exposure to determine exact Bb concentration. Scale bar and corresponding times are indicated.
Time-lapse epifluorescence video-microscopy of HMEC-1 cells in monolayer exposed to Bb-GFP at MOI = 200, followed by a second movie of cells that were exposed to Bb-GFP at MOI = 22. Recording started at 7 h before exposure and continued up to 33.2 hpe, with a time frame interval of 10 min. Upper left panel shows the phase contrast image, upper right panel shows the image of Hoechst-stained HMEC-1 nuclei (blue), bottom left panel shows the Bb fluorescence image, and bottom right panel the overlay of all channels. Scale bar and corresponding times are indicated. Note that although aggregates are formed at late exposure (∼24 hpe), viable motile spirochetes can still be observed at both MOIs.
Sheets 1-2 of the table list the DEGs identified when comparing the transcriptome of unexposed to Bb-exposed ECs. Shown are genes upregulated (1st sheet, up_U4_vs_I4) or downregulated in unexposed cells (2nd sheet, down_U4_vs_I4) at 4 hpe. In an analogous manner, sheets 3-4 refer to similar samples examined at 24 hpe and sheets 5-6 refer to similar samples examined at 48 hpe. In each sheet rows correspond to the genes identified and columns A-K correspond to different parameters explained within the table. Columns L-S provide the FPMK (see STAR Methods) of genes for each sample from the corresponding groups compared. Sheets 7-9 of the table show KEGG pathways significantly perturbed when comparing the transcriptome of unexposed to Bb-exposed cells at 4 (sheet 7), 24 (sheet 8) or 48 (sheet 9) hpe. Pathways that are downregulated in unexposed cells (upregulated for Bb-exposed) are highlighted in pink, while those upregulated are highlighted in green. The gage package was used for pathway analysis which has precompiled databases for mapping genes to KEGG pathways so to perform pathway enrichment analysis of DEGs among all three populations. Rows show the KEGG pathways and columns the different parameters explained in the table, including the specific genes of each category that are differentially regulated.
Data Availability Statement
Data collected and computer codes are available on request to the corresponding authors and are also publicly available at: https://github.com/ebastoun/Monolayer-Stress Microscopy (codes for calculating monolayer stresses) and https://github.com/ebastoun/borrelia_PI_colocalisation (codes for calculating colocalization between bacteria and propidium iodide staining). The RNA sequencing data (FASTq files) generated during this study and subsequent analysis are available at the Gene Expression Omnibus (GEO) database (weblink: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE174545, series record number: GSE174545). All differential expression analysis results of this study are included as supplementary tables in this article. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.