Abstract
The uspA1 and uspA2 genes of M. catarrhalis O35E encode two different surface-exposed proteins which were previously shown to share a 140-amino-acid region with 93% identity (C. Aebi, I. Maciver, J. L. Latimer, L. D. Cope, M. K. Stevens, S. E. Thomas, G. H. McCracken, Jr., and E. J. Hansen, Infect. Immun. 65:4367–4377, 1997). The N-terminal amino acid sequences of the mature forms of both UspA1 and UspA2 from strain O35E were determined after enzymatic treatment to remove the N-terminal pyroglutamyl residue that had blocked Edman degradation. Mass spectrometric analysis indicated that the molecular mass of UspA1 from M. catarrhalis O35E was 83,500 ± 116 Da. Nucleotide sequence analysis of the uspA1 and uspA2 genes from three other M. catarrhalis strains (TTA24, ATCC 25238, and V1171) revealed that the encoded protein products were very similar to those from strain O35E. Western blot analysis was used to confirm that each of these three strains of M. catarrhalis expressed both UspA1 and UspA2 proteins. Several different and repetitive amino acid motifs were present in both UspA1 and UspA2 from these four strains, and some of these were predicted to form coiled coils. Linear DNA templates were used in an in vitro transcription-translation system to determine the sizes of the monomeric forms of the UspA1 and UspA2 proteins from strains O35E and TTA24.
Moraxella catarrhalis is recognized as a significant cause of disease in the respiratory tracts of children and adults (36), accounting for up to 20% of cases of acute bacterial otitis media in the former group (9). M. catarrhalis is associated with approximately one-third of infectious exacerbations of chronic obstructive pulmonary disease in adults (22, 39). Consequently, M. catarrhalis has become the focus of increased research effort, most of which involves elucidation of the interaction of this bacterium with its human host, with the ultimate goal of identifying appropriate vaccine candidates (36).
Among the surface antigens of M. catarrhalis, outer membrane proteins have received the most attention as possible vaccine candidates (10, 24, 25, 33, 37), although recent efforts have indicated that M. catarrhalis lipooligosaccharide may contain potential vaccine components (21). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of M. catarrhalis outer membrane proteins revealed little apparent strain-specific variability in these proteins among different M. catarrhalis strains (7), although fairly extensive genetic diversity among M. catarrhalis strains has been documented by PCR-based methods (49). A few of these outer membrane proteins, especially CopB (OMP B2) (3, 42), OMP CD (27), and UspA (ubiquitous surface protein A or HMW-OMP) (25, 28), which consists of two related proteins, UspA1 and UspA2 (1, 2), have been characterized in some detail. Furthermore, changes in expression of M. catarrhalis outer membrane proteins have been shown to affect the ability of this organism to resist clearance from the lungs of animals (30).
The UspA1 and UspA2 surface proteins are of interest for several reasons. First, these related proteins have different biological functions, with UspA1 having been shown to be essential for attachment of M. catarrhalis O35E to Chang conjunctival cells in vitro, whereas UspA2 is involved directly or indirectly in serum resistance of this strain (1). Second, after solubilization of M. catarrhalis cells at 37°C, these two proteins apparently are present as oligomers or aggregates, each of which migrates during SDS-PAGE with an apparent molecular weight greater than 250,000 (2). In addition, it was recently reported that purified forms of these two proteins in solution have molecular weights in excess of 800,000 (33). Nucleotide sequence analysis indicated that the deduced UspA1 and UspA2 proteins of strain O35E possessed molecular masses of 88 and 62 kDa, respectively (2). The amino acid sequences of UspA1 and UspA2 are only 43% identical, but each possesses an internal segment of 140 amino acids with 93% identity; this region contains an epitope which binds the monoclonal antibody (MAb) 17C7 and is present in all disease-associated isolates of M. catarrhalis tested to date (25).
There is still little known about the physical characteristics of the UspA1 and UspA2 proteins and whether these proteins are conserved among M. catarrhalis strains. In the present study, we identified the N-terminal amino acid sequence of the mature UspA1 and UspA2 proteins from strain O35E, determined the molecular mass of UspA1, and performed nucleotide sequence analysis of the uspA1 and uspA2 genes from three additional strains of M. catarrhalis. After establishing that the UspA1 and UspA2 proteins from these M. catarrhalis strains were remarkably similar to those from strain O35E, we used an in vitro transcription-translation system to investigate expression of the uspA1 and uspA2 genes.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. catarrhalis O35E and TTA24 have been described previously (24, 25, 47). M. catarrhalis ATCC 25238 was obtained from the American Type Culture Collection, Manassas, Va. M. catarrhalis V1171 was obtained from the nasopharynx of a healthy child in Chapel Hill, N.C. M. catarrhalis strains were routinely cultured at 37°C in brain heart infusion (BHI) broth (Difco Laboratories, Detroit, Mich.) or on BHI agar plates in an atmosphere of 95% air–5% CO2.
MAbs.
MAb 17C7 is reactive with a surface-exposed epitope present in both UspA1 and UspA2 from M. catarrhalis O35E and binds all M. catarrhalis disease isolates tested (2). The immunoglobulin G (IgG) MAb 24B5 is specific for UspA1 and binds the UspA1 protein of all M. catarrhalis disease isolates tested to date; this MAb was obtained from a hybridoma fusion that used splenocytes from mice immunized with purified UspA1 protein from M. catarrhalis O35E (1). The IgG MAb 17H4 is specific for the UspA2 protein of strain O35E and does not bind the UspA2 proteins of other M. catarrhalis strains (1).
Characterization of UspA1 and UspA2 proteins.
Whole-cell lysates of M. catarrhalis strains were prepared as previously described (38, 40) with the following exceptions. After addition of the concentrated sample buffer, these lysates were heated either at 100°C or at 37°C for 10 min. The lysates were then stored at −20°C until used in SDS-PAGE. Immediately prior to SDS-PAGE, the lysates were again heated as described above. Proteins present in these preparations were resolved by SDS-PAGE in the absence of reducing agents, transferred to nitrocellulose, and detected by Western blot analysis using MAbs (1, 2, 25).
PCR techniques.
Two different PCR systems were used in this study. Taq polymerase (Promega) was used to amplify DNA containing the uspA1 and uspA2 genes from strains TTA24, ATCC 25238, and V1171 for use in nucleotide sequence analysis. To prepare DNA templates for use in the in vitro expression system, rTth DNA polymerase was used as described in the instructions for the Gene Amp XL PCR kit (Perkin-Elmer, Foster City, Calif.) Chromosomal DNA purified from M. catarrhalis strains was used as the template for all PCR systems. The primers used to amplify individual uspA1 and uspA2 genes from the chromosome of M. catarrhalis strains were 5′-TGTGAGCAAATGACTGGC-3′ and 5′-TTTTGCTCAGCGTCATGG-3′ (for uspA1) and 5′-CGCTCTCTGCCAATCAGTACACTAC-3′ and 5′-GGATCCCGCTGTATGCCGCTACTCGCAGCT-3′ (for uspA2).
Expression of M. catarrhalis proteins in vitro.
The primers used to amplify uspA1 and uspA2 genes from M. catarrhalis O35E and TTA24 were those described immediately above. These PCR products were used in an Escherichia coli S30 extract system for linear DNA templates (Promega, Madison, Wis.). [3H]leucine was used to radiolabel proteins expressed in this coupled transcription-translation system. The radiolabeled proteins were precipitated with acetone, heated at 100°C in SDS sample buffer for 5 min, and then resolved by SDS-PAGE and detected by fluorography (23).
N-terminal amino acid sequence analysis.
Purified UspA1 and UspA2 from strain O35E (33) were heated at 60°C prior to SDS-PAGE. The UspA1 and UspA2 proteins were electroblotted to Immobilon PSQ (Millipore Corp., Bedford, Mass.), stained with amido black, and excised. The excised bands were treated with heat-stable Pfu pyroglutamate aminopeptidase (Panvera Corporation, Madison, Wis.). Digestion was performed essentially as previously described (31) except that the enzyme reaction was allowed to proceed for 3 h at 75°C. Automated Edman degradation was performed by using a PE Biosystems model 494 sequencer.
Determination of UspA1 subunit size by MALDI-TOF mass spectrometry.
A 13-μg quantity of purified UspA1 (33) was heated at 60°C for 10 min and then subjected to SDS-PAGE. The very-high-molecular-weight UspA1 antigen was located by copper staining and eluted in the presence of saturated α-cyano-4-hydroxycinnamic acid according to the method of Cohen and Chait (11). Approximately one-third of the eluate was spotted onto a 3-by-3-mm square of 3M octadecyl membrane (Fisher Scientific, Pittsburgh, Pa.) (51). The membrane was placed on a target for matrix-assisted laser desorption/ionization (MALDI), and spectra were acquired by time-of-flight (TOF) mass analysis on a Perseptive Biosystems Voyager DE mass spectrometer.
Nucleotide sequence analysis.
Nucleotide sequence analysis was performed with a model 373A automated DNA sequencer (Applied Biosystems, Foster City, Calif.). Both strands of the PCR products containing the uspA1 and uspA2 genes from strains TTA24, ATCC 25238, and V1171 were sequenced in their entirety. DNA sequence information was analyzed through the National Center for Biotechnology Information (NCBI) by using the BLAST network service to search GenBank (4) and with MacVector sequence analysis software (version 6; Oxford Molecular Group, Campbell, Calif.). The Multicoil program (50) was used to predict dimeric and trimeric coiled coils.
Nucleotide sequence accession numbers.
The complete nucleotide sequences of the uspA1 and uspA2 genes from M. catarrhalis TTA24, ATCC 25238, and V1171 were deposited in GenBank and given the following accession numbers: TTA24 uspA1, AF113608; TTA24 uspA2, AF113609; ATCC 25238 uspA1, AF113606; ATCC 25238 uspA2, AF113607; V1171 uspA1, AF113610; V1171 uspA2, AF113611.
RESULTS
N-terminal amino acid sequence analysis of native UspA1 and UspA2.
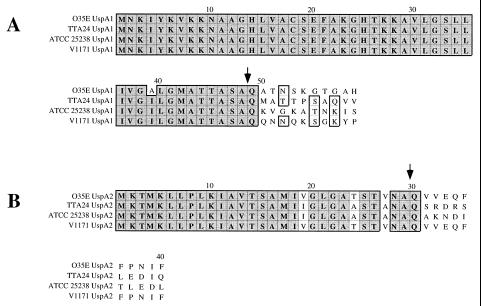
Investigation of the molecular basis for the unusual migration characteristics of both UspA1 and UspA2 during SDS-PAGE (1, 2) required determination of the amino acid sequences and molecular masses of both of these proteins. Previous efforts to determine the N-terminal amino acid sequences of the mature forms of the UspA1 and UspA2 proteins from strain O35E were unsuccessful, as both proteins appeared to be refractory to Edman degradation (2, 33). It was recently determined that the N terminus of the filamentous hemagglutinin (FHA) of Bordetella pertussis, which is also resistant to Edman degradation (14, 16), contained a pyroglutamyl residue that was removed by treatment with pyroglutamate aminopeptidase (31). Removal of this pyroglutamyl moiety, which was derived from a glutamine residue, allowed Edman degradation of the FHA protein. When purified UspA1 from strain O35E was treated with this enzyme and then subjected to Edman degradation, the N-terminal amino acid sequence ATNSKGTG was obtained (Fig. 1). When purified UspA2 was treated similarly, the N-terminal sequence VVEQFFP was obtained. In both UspA1 and UspA2, a glutamine (Q) was present in the deduced amino acid sequence immediately preceding both of these sequences (Fig. 1). It can be inferred from these results that both UspA1 and UspA2 contain signal peptides and that the signal peptide of the former macromolecule is unusually long (i.e., 48 amino acids). With a pyroglutamyl residue at the N terminus, the calculated molecular weight of the mature UspA1 protein was 83,364 and that of the mature UspA2 protein was 59,528.
FIG. 1.
Comparison of the deduced amino acid sequence of the N-terminal regions of the UspA1 and UspA2 proteins from four M. catarrhalis strains. The cleavage sites determined for the UspA1 and UspA2 proteins from strain O35E are indicated with arrows. The signal peptide for UspA1 was 48 residues in length whereas that for UspA2 contained 29 residues. The amino acid sequence comparison was made by using the Clustal-W Alignment program in MacVector, version 6. Dark shading indicates residues that are identical. Light shading indicates residues that are similar.
Determination of the molecular mass of UspA1.
Previous work from another laboratory (33) established that the molecular mass of UspA2 purified from M. catarrhalis O35E was 59,518 Da, which is in close agreement with the predicted size (i.e., 59,528 Da) of the mature UspA2 protein. To determine the molecular mass of UspA1, purified strain O35E UspA1 protein was subjected to SDS-PAGE and then the very-high-molecular-weight form of this antigen was subjected to MALDI-TOF mass spectrometric analysis. Singly and doubly charged ions from the protein were observed in two spectra. The molecular mass of UspA1 was estimated to be 83,500 ± 116 Da by averaging the values from the four available signals. The error of this measurement, estimated from the standard deviation of comparable measurements performed using standard proteins, was ±200 Da.
Detection of UspA1 and UspA2 protein expression.
Use of either UspA1 or UspA2 for vaccine purposes would require that these proteins be both expressed and well conserved among most if not all strains of M. catarrhalis. To address these issues directly, three M. catarrhalis strains with diverse origins were chosen for nucleotide sequence analysis of their uspA1 and uspA2 genes. Strain TTA24 was a disease isolate obtained from a transtracheal aspirate. Strain ATCC 25238 was a type strain whose genome has been physically mapped (18), whereas strain V1171 was isolated from the nasopharynx of a healthy child.
Western blot analysis was used to confirm that both UspA1 and UspA2 were expressed by these M. catarrhalis strains. A MAb (24B5) specific for the UspA1 protein and a second MAb (17H4) specific for the UspA2 protein of strain O35E were used to confirm protein solubilization conditions (33) which would allow differentiation of UspA1 and UspA2 by using just a single MAb (MAb 17C7) reactive with both of these proteins. (MAb 17C7 has been shown to bind an epitope common to both UspA1 and UspA2 [2].) This approach was necessitated by the fact that, whereas the UspA1-specific MAb 24B5 binds to all M. catarrhalis strains tested to date, the UspA2-specific MAb 17H4 binds only to strain O35E.
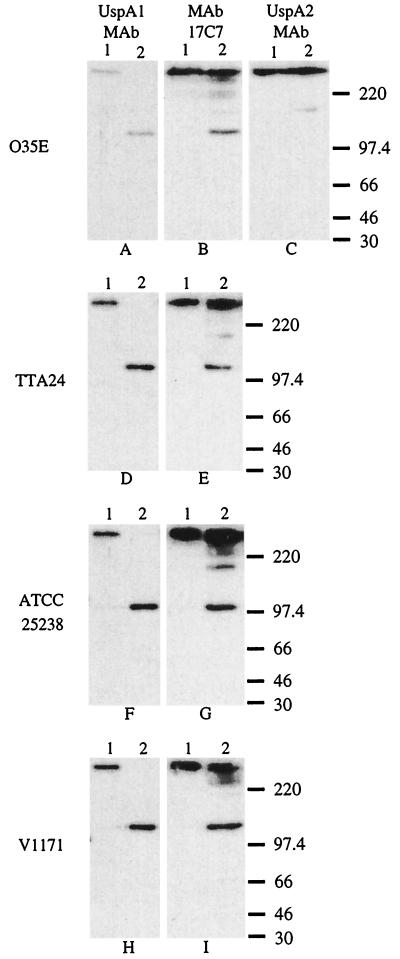
When whole-cell lysates of the four M. catarrhalis strains were solubilized at 37°C, the UspA1-specific MAb 24B5 bound to a very-high-molecular-mass (i.e., greater than 220 kDa) antigen from each strain (Fig. 2A, D, F, and H, lane 1). Therefore, all four of these strains expressed a UspA1 protein. However, when the whole-cell lysates were heated at 100°C for 10 min (33), this same UspA1-specific MAb bound to an antigen with an apparent molecular mass of approximately 120 to 140 kDa (Fig. 2A, D, F, and H, lane 2); no high-molecular-weight form of UspA1 was detectable in the latter samples probed with MAb 24B5.
FIG. 2.
Western blot-based detection of UspA1 and UspA2 proteins in selected M. catarrhalis strains. Whole-cell lysates of M. catarrhalis O35E (A, B, and C), TTA24 (D and E), ATCC 25238 (F and G), and V1171 (H and I) were heated in SDS sample buffer at 37°C (lane 1) or at 100°C (lane 2) for 10 min. Panels A, D, F, and H were probed with the UspA1-specific MAb 24B5. As seen in lane 1, this MAb bound the high-molecular-mass form of UspA1 in the 37°C sample. As seen in lane 2, this same MAb bound the putative monomeric 120 to 130 kDa form of UspA1 in the 100°C sample. Panels B, E, G, and I were probed with the UspA1- and UspA2-reactive MAb 17C7. Panel C was probed with the UspA2-specific MAb 17H4 which reacted with the high-molecular-weight aggregate of UspA2 from strain O35E in both the 37°C sample (lane 1) and the 100°C sample (lane 2); heating at this latter temperature did not reduce the size of the UspA2 antigen in this assay system. MAb 17C7 also recognizes aggregates or degradation products from UspA2 as confirmed by previous analysis of isogenic uspA1 and uspA2 mutants of strain O35E (1, 2). Molecular mass position markers (in kDa) are shown on the right side of the figure.
When probed with the O35E UspA2-specific MAb 17H4, whole-cell lysates of strain O35E heated at 37 or 100°C (Fig. 2C, lanes 1 and 2, respectively), both exhibited a very-high-molecular-weight antigen which bound this MAb, a finding which indicated that heating at 100°C for 10 min did not reduce the size of the UspA2 antigen (33). When probed with the UspA1- and UspA2-reactive MAb 17C7 (2), the whole-cell lysate of strain O35E heated at 100°C (Fig. 2B, lane 2) exhibited a very intense reaction in the very-high-molecular-weight range (i.e., UspA2) as well as a discrete 120 to 130 kDa band (i.e., UspA1). The sample solubilized at 37°C exhibited a very-high-molecular-weight antigen which contained both UspA1 and UspA2 (Fig. 2B, lane 1).
When whole-cell lysates of M. catarrhalis TTA24 (Fig. 2E, lane 1), ATCC 25238 (Fig. 2G, lane 1), and V1171 (Fig. 2I, lane 1) were heated at 37°C, each exhibited a very-high-molecular-weight band (containing both UspA1 and UspA2) reactive with MAb 17C7. When whole-cell lysates of these same M. catarrhalis strains were heated at 100°C and probed with MAb 17C7 (Fig. 2E, G, and I, lane 2), each exhibited the 120-to-140-kDa form of the UspA1 protein as well as the very-high-molecular-weight UspA2 antigen. Therefore, UspA2 was expressed by strains TTA24, ATCC 25238, and V1171.
Nucleotide sequence analysis of uspA1 and uspA2 genes from three M. catarrhalis strains.
PCR products containing the uspA1 and uspA2 genes from M. catarrhalis strains TTA24, ATCC 25238, and V1171 were obtained by using the oligonucleotide primers described in Materials and Methods together with purified chromosomal DNA from these three strains. These PCR products ranged in size from 3.2 to 3.5 kb for the uspA1 genes and from 2.6 to 2.8 kb for the uspA2 genes.
Features of the three uspA1 genes and their encoded protein products.
The uspA1 open reading frames (ORFs) from strains TTA24, ATCC 25238, and V1171 varied in size from 2,589 to 2,823 nucleotides (nt) (Table 1). It should be noted that the region 5′ from the uspA1 ORF in strains TTA24 (Fig. 3A), V1171, and O35E (2) contained a poly(G) tract (11 residues) located approximately 30 bp upstream from the translational start codon. Strain ATCC 25238 had six consecutive G residues in the same approximate position. An inverted repeat which might function as a transcriptional terminator was located approximately 24 nt from the translational stop codon of each uspA1 ORF (data not shown).
TABLE 1.
Characteristics of the uspA1 and uspA2 genes and their encoded products from four M. catarrhalis strains
| Gene | ORF size (nucleotides) | Predicted protein size (amino acids) | Calculated mol wt |
|---|---|---|---|
| O35E uspA1a | 2,496 | 832 | 88,257 |
| TTA24 uspA1 | 2,823 | 941 | 99,070 |
| ATCC 25238 uspA1 | 2,589 | 863 | 90,533 |
| V1171 uspA1 | 2,736 | 912 | 95,757 |
| O35E uspA2a | 1,728 | 572 | 62,483 |
| TTA24 uspA2 | 1,839 | 613 | 66,700 |
| ATCC 25238 uspA2 | 1,890 | 630 | 68,480 |
| V1171 uspA2 | 2,022 | 674 | 73,270 |
Data from Aebi et al. (2); included for purposes of comparison.
FIG. 3.
Location of nucleotide repeat motifs observed in the 5′ region preceding the uspA1 and uspA2 ORFs in M. catarrhalis TTA24. The poly(G) repeat preceding the uspA1 ORF (A) and the tetranucleotide repeat (AGAT) in front of the uspA2 ORF (B) are underlined.
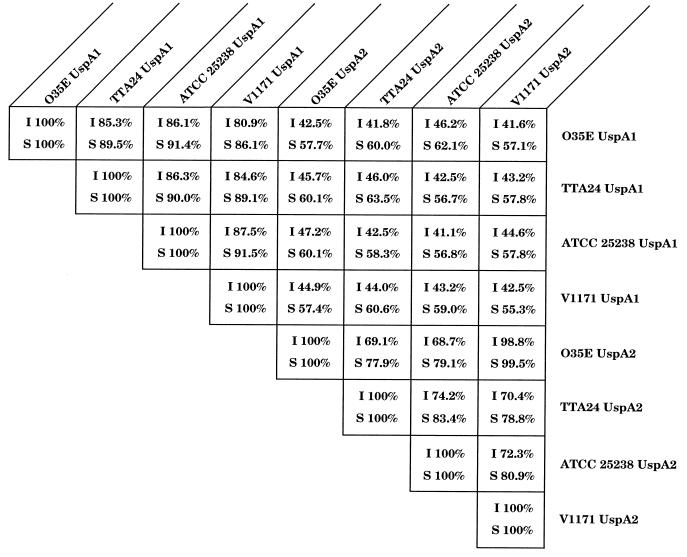
The deduced amino acid sequences of the encoded proteins ranged in length from 863 to 941 amino acids with calculated molecular weights of 90,533 to 99,070. The UspA1 proteins from these three strains were 81 to 86% identical with the UspA1 protein from strain O35E (Fig. 4). The UspA1 proteins from strains ATCC 25238 and V1171 had the highest degree of identity (88%). The putative leader peptides of the UspA1 proteins from these three strains were identical (Fig. 1A). In addition, the C-terminal 193 amino acids of the four UspA1 proteins were 98 to 100% identical (data not shown). When the amino acid sequences of these four UspA1 proteins were analyzed through NCBI by using the BLAST network service (4, 20), the most similar prokaryotic proteins were found to be adhesins from other bacterial pathogens, including Hsf (44, 45) and Hia (6, 46) from Haemophilus influenzae and YadA from Yersinia enterocolitica (43). It should be noted that the Hia and YadA proteins have also been shown to exhibit anomalous migration characteristics during SDS-PAGE (6, 43, 46). Interestingly, myosin heavy chains from a number of eukaryotes proved to be even more similar to some of these UspA1 proteins than did the prokaryotic adhesins (data not shown).
FIG. 4.
Comparison of the amino acid sequences of the UspA1 and UspA2 proteins from four M. catarrhalis strains. The amino acid sequences of the UspA1 and UspA2 proteins from strain O35E were obtained from Aebi et al. (2). The degrees of identity (I) and similarity (S) were determined by use of the GAP alignment program from the University of Wisconsin Genetics Computer Group software analysis package (version 8.1) (15).
Features of the three uspA2 genes and their encoded protein products.
The uspA2 ORFs from strains TTA24, ATCC 25238, and V1171 varied in size from 1,839 to 2,022 nt (Table 1). Similar to the situation with the uspA1 genes, the DNA immediately upstream from the translational start codon of each uspA2 ORF contained a distinctive nucleotide repeat motif. Fourteen repeats of the tetranucleotide AGAT were located approximately 130 nt upstream from the uspA1 ORF in both TTA24 (Fig. 3B) and ATCC 25238; 17 repeats of this motif were located similarly in V1171. The uspA2 gene from strain O35E had 15 repeats of the AGAT motif (2). Again, an inverted repeat that could comprise a transcriptional terminator was located approximately 16 nt 3′ from the translational stop codon of the uspA2 ORFs (data not shown).
The deduced amino acid sequences of the encoded proteins ranged in length from 613 to 674 amino acids, with calculated molecular weights of 66,700 to 73,270 (Table 1). These UspA2 proteins were 69 to 99% identical with UspA2 from strain O35E (Fig. 4). The UspA2 proteins from strains O35E and V1171 had the highest degree of identity (99%). The putative leader peptides of these three proteins were nearly identical (Fig. 1B). Similar to the UspA1 proteins, the C-terminal 149 amino acids of the four UspA2 proteins were 98 to 100% identical. Database searches indicated that YadA (43) was the prokaryotic protein whose amino acid sequence was most similar to those of the UspA2 proteins.
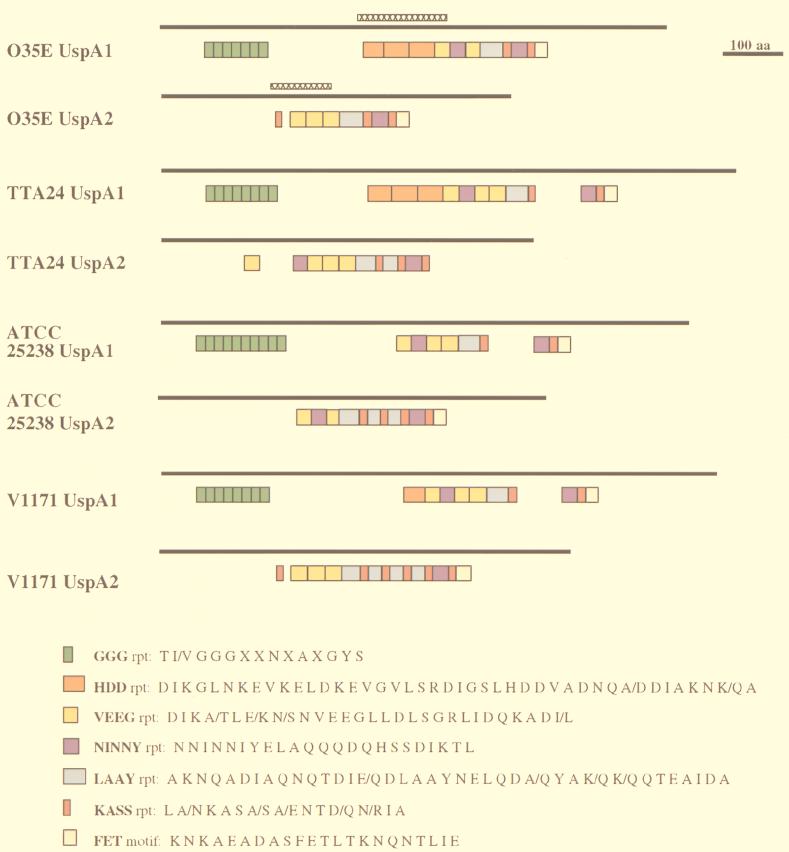
Amino acid repeat motifs common to UspA1 and UspA2.
It was previously shown that the UspA1 and UspA2 proteins of strain O35E had in common a 23-amino-acid motif that likely contained the epitope for the protective MAb 17C7 (2). This same 23-amino-acid motif (designated NINNY in Fig. 5) was present in one or two copies in all six of the new proteins included in this study. In addition, it became apparent that there were two amino acid repeats (designated GGG and HDD in Fig. 5) shared among the UspA1 proteins (GGG in all four strains and HDD in three strains). Furthermore, there were several amino acid repeats or motifs (designated VEEG, LAAY, KASS, and FET in Fig. 5) that were shared by both the UspA1 and UspA2 proteins from each strain.
FIG. 5.
Schematic diagram of the repetitive amino acid sequences and other motifs present in the UspA1 and UspA2 proteins of M. catarrhalis O35E, TTA24, ATCC 25238, and V1171. The solid bars represent the lengths of the entire proteins. The colored boxes designate the positions and lengths of the repeats and other motifs. The consensus amino acid sequence for each repeat or motif is listed beside each colored box at bottom. The regions of UspA1 and UspA2 from strain O35E that are predicted to be most likely to form coiled coils are indicated by the cross-hatched bars.
Prediction of coiled coil structure in UspA1 and UspA2.
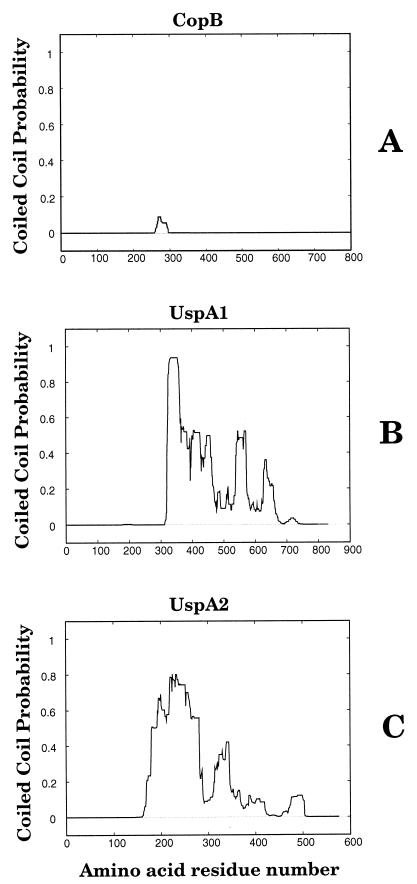
The similarity of UspA1 and UspA2 to other proteins (i.e., myosin) with proven coiled coil structures prompted us to analyze the amino acid sequences of these M. catarrhalis proteins for their potential to form coiled coils. A typical M. catarrhalis outer membrane protein (i.e., CopB) (24) was included in this analysis for the purpose of comparison. The use of the Multicoil program (50) revealed that regions of both UspA1 (Fig. 6B) and UspA2 (Fig. 6C) from strain O35E were predicted to form coiled coils. In contrast, no region of the CopB outer membrane protein was predicted to be likely to form a coiled coil (Fig. 6A). Amino acid residues 323 to 469 in UspA1 had the highest probability of forming a coiled coil (Fig. 6B). This region of the UspA1 protein includes three repeats of the HDD motif and a single VEEG motif (Fig. 5). Amino acids 180 to 281 of UspA2 had the highest probability of forming a coiled coil (Fig. 6C); this region of UspA2 contained a single KASS motif and three VEEG motifs (Fig. 5). The use of the Multicoil program to analyze the UspA1 and UspA2 proteins from the other three strains of M. catarrhalis showed that all six proteins contained areas with a very high probability (i.e., greater than 0.6) of forming coiled coils (data not shown).
FIG. 6.
Probability of coiled coil formation by the M. catarrhalis O35E CopB (A), UspA1 (B), and UspA2 (C) proteins as determined by Multicoil analysis (50). The total probability prediction is plotted on the y axis over the entire length of each protein.
Expression of UspA1 and UspA2 proteins in vitro.
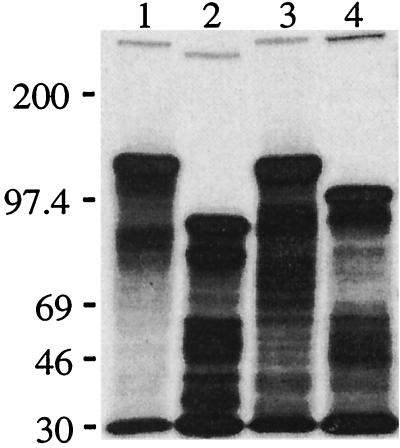
To determine whether the unusual SDS-PAGE migration characteristics of these proteins are dependent on their expression in M. catarrhalis, we used an E. coli-derived in vitro coupled transcription-translation system to express the UspA1 and UspA2 proteins from M. catarrhalis O35E and TTA24 (Fig. 7). Both UspA1 proteins synthesized in vitro had apparent molecular masses of 120 to 130 kDa (Fig. 7, lanes 1 and 3). The in vitro-synthesized UspA2 proteins of strains O35E and TTA24 had apparent molecular masses of 95 kDa (Fig. 7, lane 2) and 110 kDa (Fig. 7, lane 4), respectively. In all four instances, the apparent molecular weights of the in vitro-synthesized products derived from both the uspA1 and uspA2 genes were significantly larger than the calculated molecular weights for these proteins (Table 1). A control experiment using the hxuC gene from H. influenzae type b (12) showed that this typical outer membrane protein, with a calculated molecular weight of 78,000, migrated with an apparent molecular mass of approximately 79 kDa after being synthesized in this same in vitro system (data not shown). It should be noted that the in vitro-synthesized UspA1 and UspA2 proteins also formed small but detectable quantities of very-high-molecular-weight complexes with apparent molecular masses well in excess of 200 kDa (Fig. 7, lanes 1 to 4).
FIG. 7.
Fluorographic detection of M. catarrhalis UspA1 and UspA2 proteins expressed in vitro. The M. catarrhalis O35E uspA1 gene (lane 1), the O35E uspA2 gene (lane 2), the TTA24 uspA1 gene (lane 3), and the TTA24 uspA2 (lane 4) genes were each amplified from the chromosome of their respective M. catarrhalis strains and used in an in vitro coupled transcription-translation system for linear DNA templates to establish the size (as determined by SDS-PAGE) of the protein product encoded by each gene. Radiolabeled (14C-labeled) standards (not shown) were used to determine the position of the molecular mass markers (in kilodaltons) present on the left side of the figure. Degradation products are visible beneath each primary protein product.
DISCUSSION
Interest in the UspA1 and UspA2 proteins of M. catarrhalis as potential vaccine candidates has been stimulated by the finding that purified forms of both of these proteins can induce the synthesis of antibodies that are biologically active against this pathogen in both in vitro and in vivo systems (33). Both UspA1 and UspA2 were previously shown to be exposed on the surface of whole cells of M. catarrhalis O35E by virtue of their ability to bind MAbs specific for these proteins (1). The surface localization of these macromolecules suggested that both UspA1 and UspA2 likely were synthesized with signal peptides, and the hydrophobic nature of the N-terminal region of the deduced amino acid sequences of the complete UspA1 and UspA2 proteins (data not shown) was consistent with the presence of a possible signal peptide in both proteins. However, previous efforts at N-terminal sequence analysis of these proteins had been unsuccessful (2, 33). The use of pyroglutamyl aminopeptidase (31) removed from both UspA1 and UspA2 what was likely a pyroglutamyl residue and permitted determination of the N terminus of the mature forms of these two proteins.
Both UspA1 and UspA2 were found to be synthesized with signal peptides (Fig. 1), and that of UspA1 was unusually long (i.e., 48 amino acids) with some positively charged residues located in the middle of this amino acid sequence, similar to that of the E. coli AIDA-I adhesin (8). It must be noted, however, that we have not formally excluded the possibility that the translation initiation codon for the uspA1 ORF could appear later in this nucleotide sequence; there are two GTG codons that could serve in this capacity, located 16 and 43 nt, respectively, downstream from the proposed ATG start codon (Fig. 3). It was recently suggested that UspA1 may be a member of the autotransporter family of secreted proteins which includes numerous, relatively large bacterial macromolecules which often function in adherence to host cells (26).
Determination of the length of the signal peptides of both UspA1 and UspA2 allowed correlation of the size of the predicted mature form of each of these two proteins with the molecular mass of each protein as determined by mass spectrometric analysis. The calculated molecular weights of the mature forms of UspA1 and UspA2 (83,364 and 59,528, respectively) were very similar to the molecular weights obtained for the purified UspA1 and UspA2 proteins (83,504 and 59,518, respectively). Despite three lines of evidence (i.e., nucleotide sequence analysis, N-terminal amino acid sequence findings, and mass spectrometric data) now pointing to the existence of an 83-kDa UspA1 protein and a 60-kDa UspA2 protein in the M. catarrhalis outer membrane, Western blot analysis of 37°C-treated whole-cell lysates of M. catarrhalis strains detected only very-high-molecular-weight forms of these two antigens (Fig. 2A and C, lane 1).
In an attempt to begin to address the aberrant SDS-PAGE migration characteristics of both UspA1 and UspA2, both of these proteins were synthesized in an E. coli-derived in vitro coupled transcription-translation system. Interestingly, the in vitro-synthesized UspA1 protein from strain O35E exhibited an apparent molecular mass (120 to 130 kDa) (Fig. 7) that was approximately 50% greater than its actual molecular mass (83 kDa for the mature protein). Similarly, the O35E UspA2 protein produced in this cell-free system migrated during SDS-PAGE with an apparent molecular mass of approximately 95 kDa (Fig. 7), which is approximately 50% greater than its actual mass of 60 kDa. These differences between the calculated molecular weight of each monomeric protein and its apparent molecular weight in SDS-PAGE are much greater than those typically observed with well-studied heat-modifiable outer membrane proteins, including E. coli OmpA (19) and H. influenzae P1 (48). The molecular basis for the aberrant migration of the in vitro-synthesized, monomeric forms of UspA1 and UspA2 in SDS-PAGE remains to be determined.
In contrast, the unusual SDS-PAGE migration characteristics of native forms of both UspA1 (Fig. 2A, lane 1) and UspA2 (Fig. 2C, lane 1) may be explained, at least in part, by the finding that both of these proteins contain regions which are predicted to have a high probability of forming coiled coils (Fig. 6). Another bacterial surface protein predicted to form a coiled coil structure, the fibrinogen-binding protein of Streptococcus equi subsp. equi (34), has been reported to migrate anomalously in SDS-PAGE, with an apparent molecular weight much greater than that of the native monomeric protein. Similarly, the H. influenzae Hia protein, which has homology with UspA1 and exhibits aberrant migration in SDS-PAGE (6), contains a predicted coiled-coil motif (data not shown). If coiled coils are formed by UspA1 and UspA2 in M. catarrhalis, at least those formed by UspA2 are very resistant to heating at 100°C in SDS (Fig. 2C, lane 2). In this regard, the ability of very-high-molecular-weight UspA2 complexes to resist dissociation by heating in the presence of SDS is reminiscent of that of several other proteins, including the PilQ (OMP-MC) protein of Neisseria gonorrhoeae (17), the YscC protein of Yersinia pestis (29), and the pIV protein encoded by the E. coli filamentous phage f1 (32); all three of these outer membrane proteins form SDS-resistant, very-high-molecular-weight multimers. However, none of these latter three proteins have regions predicted to form coiled coils (data not shown).
Whether the very-high-molecular-weight forms of UspA1 and UspA2 detected in M. catarrhalis whole-cell lysates (Fig. 2A and C, lane 1) are homoaggregates of each individual protein or heteroaggregates involving both proteins is not known. The purified form of each protein gave rise to very-high-molecular-weight complexes (33). In addition, very-high-molecular-weight UspA1 and UspA2 proteins were also detected when these two proteins were synthesized individually in vitro (Fig. 7), suggesting that these very-high-molecular-weight aggregates or multimers can form spontaneously outside of the environment of the M. catarrhalis cell. It should also be noted that previous mutant analyses indicated that the very-high-molecular-weight form of UspA1 can be detected in a uspA2 mutant and, similarly, that a uspA1 mutant still expressed the very-high-molecular-weight form of UspA2 (1). Finally, because the mature forms of UspA1 and UspA2 both lack cysteine residues, disulfide bond formation cannot be involved in this phenomenon.
Nucleotide sequence analysis of the uspA1 and uspA2 genes from three other M. catarrhalis strains revealed that their encoded proteins were very similar to the UspA1 and UspA2 proteins of strain O35E. The UspA1 proteins were 81 to 86% identical and the UspA2 proteins had 69 to 99% identity. Interestingly, the signal peptides for the four UspA1 proteins were nearly identical to each other, as were the signal peptides for the UspA2 proteins (Fig. 1). More important from the standpoint of M. catarrhalis vaccine development is the finding that each of these proteins contained one or two copies of the NINNY motif (Fig. 5) which likely contains the epitope for a protective antibody (2). Whether other regions of these proteins can be targets for protective antibodies remains to be determined, but the conservation of different amino acid motifs and repeats in these two proteins among different M. catarrhalis strains (Fig. 5) indicates that there does exist significant potential for antigenic cross-reactivity among UspA1 proteins and among UspA2 proteins.
It is also interesting to note that certain of these amino acid motifs were actually present in both UspA1 and UspA2. In addition to the NINNY motif discussed above, the VEEG, LAAY, KASS, and FET motifs were present in both UspA1 and UspA2 (Fig. 5). This extensive sharing of sequences between UspA1 and UspA2 did not include the N- and C-terminal regions of these proteins. Whether one of these genes was originally derived from the other by some type of duplication event is not known, but mutant analysis indicates that these two proteins have different functions, at least in M. catarrhalis O35E (1). It is also known that the uspA1 and uspA2 genes are not physically linked because they have been mapped to sites approximately 600 kb apart in the M. catarrhalis ATCC 25238 genome (38a). The molecular basis for this extensive sharing of amino acid motifs between UspA1 and UspA2 proteins remains to be determined.
At this time, there are no data available which indicate whether expression of the uspA1 and uspA2 genes in M. catarrhalis is constitutive or regulated in some manner. The presence of a poly(G) tract in front of all four uspA1 ORFs (Fig. 3A) and a tetranucleotide repeat (AGAT) located 5′ of the four uspA2 ORFs (Fig. 3B) suggests that there could be regulation of expression of these two genes at the level of transcription. For example, in Neisseria meningitidis, a poly(G) tract located upstream of the porA ORF has been shown to be involved in transcription of this gene encoding the porin protein (5). Interestingly, there are long poly(T) tracts in the region 5′ of both the H. influenzae hia and hsf ORFs (6, 45), whose encoded proteins resemble UspA1. Tetra- and pentanucleotide repeats can be involved in expression of ORFs in gram-negative pathogens, although these repeats are usually contained within the ORF and function through a slipped-strand mispairing mechanism (35). However, a heptanucleotide repeat within the untranslated upstream region is involved in regulating expression of the Vibrio cholerae enterotoxin gene (41), and it has been reported recently that a heptanucleotide repeat located in a 5′ untranslated region is involved in control of expression of the HMW1 and HMW2 proteins of nontypeable H. influenzae (13).
In summary, we have established that the amino acid sequence of the UspA1 protein as well as that of the UspA2 protein of M. catarrhalis are well conserved among strains of this pathogen. Both of these proteins share the unusual characteristic of having a pyroglutamyl residue at the N termini of their mature forms. These two proteins also exhibit anomalous behavior during SDS-PAGE, whether they are derived directly from M. catarrhalis or synthesized in vitro. The exact composition of the very-high-molecular-weight forms of UspA1 and UspA2 expressed in vivo remains to be determined.
ACKNOWLEDGMENTS
This study was supported by U.S. Public Health Service grant AI36344 and by Texas Advanced Technology Program Award 003660-087 to E.J.H. C.A. was supported by a research grant for young investigators from Novartis AG, Basel, Switzerland.
Purified UspA1 and UspA2 from M. catarrhalis O35E were obtained from John McMichael. We thank John D. Nelson and Steven Berk for providing isolates of M. catarrhalis, and Steve Afendis and Carolyn Moomaw for technical assistance.
REFERENCES
- 1.Aebi C, Lafontaine E R, Cope L D, Latimer J L, Lumbley S R, McCracken G H, Jr, Hansen E J. Phenotypic effect of isogenic uapA1 and uspA2 mutations on Moraxella catarrhalis O35E. Infect Immun. 1998;66:3113–3119. doi: 10.1128/iai.66.7.3113-3119.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebi C, Maciver I, Latimer J L, Cope L D, Stevens M K, Thomas S E, McCracken G H, Jr, Hansen E J. A protective epitope of Moraxella catarrhalis is encoded by two different genes. Infect Immun. 1997;65:4367–4377. doi: 10.1128/iai.65.11.4367-4377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aebi C, Stone B, Beucher M, Cope L D, Maciver I, Thomas S E, McCracken G H, Jr, Sparling P F, Hansen E J. Expression of the CopB outer membrane protein by Moraxella catarrhalis is regulated by iron and affects iron acquisition from transferrin and lactoferrin. Infect Immun. 1996;64:2024–2030. doi: 10.1128/iai.64.6.2024-2030.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Arhin F F, Moreau F, Coulton J W, Mills E L. Sequencing of porA from clinical isolates of Neisseria meningitidis defines a subtyping scheme and its genetic regulation. Can J Microbiol. 1998;44:56–63. [PubMed] [Google Scholar]
- 6.Barenkamp S J, St. Geme J W., III Identification of a second family of high-molecular-weight adhesion proteins expressed by non-typable Haemophilus influenzae. Mol Microbiol. 1996;19:1215–1223. doi: 10.1111/j.1365-2958.1996.tb02467.x. [DOI] [PubMed] [Google Scholar]
- 7.Bartos L C, Murphy T F. Comparison of the outer membrane proteins of 50 strains of Branhamella catarrhalis. J Infect Dis. 1988;158:761–765. doi: 10.1093/infdis/158.4.761. [DOI] [PubMed] [Google Scholar]
- 8.Benz I, Schmidt M A. AIDA-I, the adhesin involved in diffuse adherence of the diarrhoeagenic Escherichia coli strain 2787 (O126:H27), is synthesized via a precursor molecule. Mol Microbiol. 1992;6:1539–1546. doi: 10.1111/j.1365-2958.1992.tb00875.x. [DOI] [PubMed] [Google Scholar]
- 9.Bluestone C D, Stephenson J S, Martin L M. Ten-year review of otitis media pathogens. Pediatr Infect Dis J. 1992;11:S7–S11. doi: 10.1097/00006454-199208001-00002. [DOI] [PubMed] [Google Scholar]
- 10.Chen D, McMichael J C, van der Meid K R, Hahn D, Mininni T, Cowell J, Eldridge J. Evaluation of purified UspA from Moraxella catarrhalis as a vaccine in a murine model after active immunization. Infect Immun. 1996;64:1900–1905. doi: 10.1128/iai.64.6.1900-1905.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen S L, Chait B T. Mass spectrometry of whole proteins eluted from sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. Anal Biochem. 1997;247:257–267. doi: 10.1006/abio.1997.2072. [DOI] [PubMed] [Google Scholar]
- 12.Cope L D, Yogev R, Muller-Eberhard U, Hansen E J. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J Bacteriol. 1995;177:2644–2653. doi: 10.1128/jb.177.10.2644-2653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawid S, Barenkamp S J, St. Geme J W., III Variation in expression of the Haemophilus influenzae HMW adhesins: a prokaryotic system reminiscent of eukaryotes. Proc Natl Acad Sci USA. 1999;96:1077–1082. doi: 10.1073/pnas.96.3.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delisse-Gathoye A-M, Locht C, Jacob F, Raaschou-Nielsen M, Heron I, Ruelle J-L, De Wilde M, Cabezon T. Cloning, partial sequence, expression, and antigenic analysis of the filamentous hemagglutinin gene of Bordetella pertussis. Infect Immun. 1990;58:2895–2905. doi: 10.1128/iai.58.9.2895-2905.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domenighini M, Relman D A, Capiau C, Falkow S, Prugnola A, Scarlato V, Rappuoli R. Genetic characterization of Bordetella pertussis filamentous haemagglutinin: a protein processed from an unusually large precursor. Mol Microbiol. 1990;4:787–800. doi: 10.1111/j.1365-2958.1990.tb00649.x. [DOI] [PubMed] [Google Scholar]
- 17.Drake S L, Koomey M. The product of the pilQ gene is essential for the biogenesis of type IV pili in Neisseria gonorrhoeae. Mol Microbiol. 1995;18:975–986. doi: 10.1111/j.1365-2958.1995.18050975.x. [DOI] [PubMed] [Google Scholar]
- 18.Furihata K, Sato K, Matsumoto H. Construction of a combined NotI/SmaI physical and genetic map of Moraxella (Branhamella) catarrhalis strain ATCC 25238. Microbiol Immunol. 1995;39:745–751. doi: 10.1111/j.1348-0421.1995.tb03266.x. [DOI] [PubMed] [Google Scholar]
- 19.Garten W, Hindennach I, Henning U. The major proteins of the Escherichia coli outer cell envelope membrane. Characterization of proteins II* and III, comparison of all proteins. Eur J Biochem. 1975;59:215–221. doi: 10.1111/j.1432-1033.1975.tb02444.x. [DOI] [PubMed] [Google Scholar]
- 20.Gish W, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 21.Gu X-X, Chen J, Barenkamp S J, Robbins J B, Tsai C-M, Lim D J, Battey J. Synthesis and characterization of lipooligosaccharide-based conjugates as vaccine candidates for Moraxella (Branhamella) catarrhalis. Infect Immun. 1998;66:1891–1897. doi: 10.1128/iai.66.5.1891-1897.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hager H, Verghese A, Alvarez S, Berk S L. Branhamella catarrhalis respiratory infections. Rev Infect Dis. 1987;9:1140–1149. doi: 10.1093/clinids/9.6.1140. [DOI] [PubMed] [Google Scholar]
- 23.Hanson M S, Slaughter C, Hansen E J. The hbpA gene of Haemophilus influenzae type b encodes a heme-binding lipoprotein conserved among heme-dependent Haemophilus species. Infect Immun. 1992;60:2257–2266. doi: 10.1128/iai.60.6.2257-2266.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helminen M E, Maciver I, Latimer J L, Cope L D, McCracken G H, Jr, Hansen E J. A major outer membrane protein of Moraxella catarrhalis is a target for antibodies that enhance pulmonary clearance of the pathogen in an animal model. Infect Immun. 1993;61:2003–2010. doi: 10.1128/iai.61.5.2003-2010.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helminen M E, Maciver I, Latimer J L, Klesney-Tait J, Cope L D, Paris M M, McCracken G H, Jr, Hansen E J. A large, antigenically conserved protein on the surface of Moraxella catarrhalis is a target for protective antibodies. J Infect Dis. 1994;170:867–872. doi: 10.1093/infdis/170.4.867. [DOI] [PubMed] [Google Scholar]
- 26.Henderson I R, Navarro-Garcia F, Nataro J P. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 1998;6:370–378. doi: 10.1016/s0966-842x(98)01318-3. [DOI] [PubMed] [Google Scholar]
- 27.Hsiao C B, Sethi S, Murphy T F. Outer membrane protein CD of Branhamella catarrhalis: sequence conservation in strains recovered from the human respiratory tract. Microb Pathog. 1995;19:215–225. doi: 10.1016/s0882-4010(95)90272-4. [DOI] [PubMed] [Google Scholar]
- 28.Klingman K L, Murphy T F. Purification and characterization of a high-molecular-weight outer membrane protein of Moraxella (Branhamella) catarrhalis. Infect Immun. 1994;62:1150–1155. doi: 10.1128/iai.62.4.1150-1155.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koster M, Bitter W, de Cock H, Allaoui A, Cornelis G R, Tommassen J. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia entrocolitica forms a ring-shaped multimeric complex. Mol Microbiol. 1997;26:789–797. doi: 10.1046/j.1365-2958.1997.6141981.x. [DOI] [PubMed] [Google Scholar]
- 30.Kyd J M, Cripps A W, Murphy T F. Outer-membrane antigen expression by Moraxella (Branhamella) catarrhalis influences pulmonary clearance. J Med Microbiol. 1998;47:159–168. doi: 10.1099/00222615-47-2-159. [DOI] [PubMed] [Google Scholar]
- 31.Lambert-Buisine C, Willery E, Locht C, Jacob-Dubuisson F. N-terminal characterization of the Bordetella pertussis filamentous haemagglutinin. Mol Microbiol. 1998;28:1283–1293. doi: 10.1046/j.1365-2958.1998.00892.x. [DOI] [PubMed] [Google Scholar]
- 32.Liunderoth N A, Model P, Russel M. Essential role of a sodium dodecyl sulfate-resistant protein IV multimer in assembly-export of filamentous phage. J Bacteriol. 1996;178:1962–1970. doi: 10.1128/jb.178.7.1962-1970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMichael J C, Fiske M J, Fredenburg R A, Chakravarti D N, VanDerMeid K R, Barniak V, Caplan J, Bortell E, Baker S, Arumugham R, Chen D. Isolation and characterization of two proteins from Moraxella catarrhalis that bear a common epitope. Infect Immun. 1998;66:4374–4381. doi: 10.1128/iai.66.9.4374-4381.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meehan M, Nowlan P, Owen P. Affinity purification and characterization of a fibrinogen-binding protein complex which protects mice against lethal challenge with Streptococcus equi subsp. equi. Microbiology. 1998;144:993–1003. doi: 10.1099/00221287-144-4-993. [DOI] [PubMed] [Google Scholar]
- 35.Murphy G L, Connell T D, Barritt D S, Koomey M, Cannon J G. Phase variation of gonococcal protein II: regulation of gene expression by slipped-strand mispairing of a repetitive DNA sequence. Cell. 1989;56:539–547. doi: 10.1016/0092-8674(89)90577-1. [DOI] [PubMed] [Google Scholar]
- 36.Murphy T F. Branhamella catarrhalis: epidemiology, surface antigenic structure, and immune response. Microbiol Rev. 1996;60:267–279. doi: 10.1128/mr.60.2.267-279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy T F, Kyd J M, John A, Kirkham C, Cripps A W. Enhancement of pulmonary clearance of Moraxella (Branhamella) catarrhalis following immunization with outer membrane protein CD in a mouse model. J Infect Dis. 1998;178:1667–1675. doi: 10.1086/314501. [DOI] [PubMed] [Google Scholar]
- 38.Murphy T F, Loeb M R. Isolation of the outer membrane of Branhamella catarrhalis. Microb Pathog. 1989;6:159–174. doi: 10.1016/0882-4010(89)90066-1. [DOI] [PubMed] [Google Scholar]
- 38a.Nguyen, K., E. J. Hansen, and M. A. Farinha. Construction of a physical and genetic map of Moraxella (Branhamella) catarrhalis ATCC 25238. Can. J. Microbiol., in press. [PubMed]
- 39.Nicotra B, Rivera M, Liman J I, Wallace R J. Branhamella catarrhalis as a lower respiratory tract pathogen in patients with chronic lung disease. Arch Intern Med. 1986;146:890–893. [PubMed] [Google Scholar]
- 40.Patrick C C, Kimura A, Jackson M A, Hermanstorfer L, Hood A, McCracken G H, Jr, Hansen E J. Antigenic characterization of the oligosaccharide portion of the lipooligosaccharide of nontypable Haemophilus influenzae. Infect Immun. 1987;55:2902–2911. doi: 10.1128/iai.55.12.2902-2911.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfau J D, Taylor R K. Genetic footprint of the ToxR-binding site in the promoter for cholera toxin. Mol Microbiol. 1996;21:213–222. doi: 10.1111/j.1365-2958.1996.tb02502.x. [DOI] [PubMed] [Google Scholar]
- 42.Sethi S, Surface J M, Murphy T F. Antigenic heterogeneity and molecular analysis of CopB of Moraxella (Branhamella) catarrhalis. Infect Immun. 1997;65:3666–3671. doi: 10.1128/iai.65.9.3666-3671.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skurnik M, Wolf-Watz H. Analysis of the yopA gene encoding the Yop1 virulence determinants of Yersinia spp. Mol Microbiol. 1989;3:517–529. doi: 10.1111/j.1365-2958.1989.tb00198.x. [DOI] [PubMed] [Google Scholar]
- 44.St. Geme J W, III, Cutter D. Evidence that surface fibrils expressed by Haemophilus influenzae type b promote attachment to human epithelial cells. Mol Microbiol. 1995;15:77–85. doi: 10.1111/j.1365-2958.1995.tb02222.x. [DOI] [PubMed] [Google Scholar]
- 45.St. Geme J W, III, Cutter D, Barenkamp S J. Characterization of the genetic locus encoding Haemophilus influenzae type b surface fibrils. J Bacteriol. 1996;178:6281–6287. doi: 10.1128/jb.178.21.6281-6287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.St. Geme J W, III, Kumar V V, Cutter D, Barenkamp S J. Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect Immun. 1998;66:364–368. doi: 10.1128/iai.66.1.364-368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Unhanand M, Maciver I, Ramilo O, Arencibia-Mireles O, Argyle J C, McCracken G H, Jr, Hansen E H. Pulmonary clearance of Moraxella catarrhalis in an animal model. J Infect Dis. 1992;165:644–650. doi: 10.1093/infdis/165.4.644. [DOI] [PubMed] [Google Scholar]
- 48.van Alphen L, Riemens T, Poolman J, Zanen H C. Characterization of major outer membrane proteins of Haemophilus influenzae. J Bacteriol. 1983;155:878–885. doi: 10.1128/jb.155.2.878-885.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker E S, Preston R A, Post J C, Ehrlich G D, Kalbfleisch J H, Klingman K L. Genetic diversity among strains of Moraxella catarrhalis: analysis using multiple DNA probes and a single-locus PCR-restriction fragment length polymorphism method. J Clin Microbiol. 1998;36:1977–1983. doi: 10.1128/jcm.36.7.1977-1983.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolf E, Kim P S, Berger B. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Worrall T A, Cotter R J, Woods A S. Purification of contaminated peptides and proteins on synthetic membrane surfaces for matrix-assisted laser desorption/ionization mass spectrometry. Anal Chem. 1998;70:750–756. doi: 10.1021/ac970969e. [DOI] [PubMed] [Google Scholar]