Abstract
In this study, we demonstrate that the cellulosome of Clostridium cellulolyticum grown on xylan is not associated with the bacterial cell. Indeed, the large majority of the activity (about 90%) is localized in the cell-free fraction when the bacterium is grown on xylan. Furthermore, about 70% of the detected xylanase activity is associated with cell-free high-molecular-weight complexes containing avicelase activity and the cellulosomal scaffolding protein CipC. The same repartition is observed with carboxymethyl cellulase activity. The cellulose adhesion of xylan-grown cells is sharply reduced in comparison with cellulose-grown cells. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis revealed that cellulosomes derived from xylan- and cellulose-grown cells have different compositions. In both cases, the scaffolding protein CipC is present, but the relative proportions of the other components is dramatically changed depending on the growth substrate. We propose that, depending on the growth substrate, C. cellulolyticum is able to regulate the cell association and cellulose adhesion of cellulosomes and regulate cellulosomal composition.
Plant cell walls, the major reservoir of fixed carbon in nature, have three major polymers: cellulose (comprised of insoluble fibers of β-1,4-glucan), hemicellulose (comprised of polysaccharides including glucans, mannans, and xylans), and lignin (comprised of a complex polyphenolic structure). In anaerobic environments rich in decaying plant material, complex communities of interacting microorganisms (5, 24) carry out the decomposition of lignocellulose. Because the substrate, lignocellulose, is insoluble, bacterial degradation occurs exocellularly, either in association with the outer envelope layer or extracellularly (5, 24, 34).
Among lignocellulolytic bacteria, cellulolytic clostridia play important roles in plant biomass turnover (24). Recent work in our laboratory has contributed to a better understanding of the physiology and cellulose-adhered life cycle of the bacterium Clostridium cellulolyticum (15, 18–20, 30). The primary event in the degradation of cellulose is the tight adhesion of the cellulolytic bacterium to its substrate. The growth phase of C. cellulolyticum occurs when bacteria are bound to insoluble cellulose. Like a majority of cellulolytic clostridia (e.g., Clostridium thermocellum [2, 4, 5], Clostridium cellulovorans [11], and Clostridium papyrosolvens [31, 32]), C. cellulolyticum produces a large cellulolytic complex (about 600 kDa) called cellulosome, in which several cellulases are tightly bound to a scaffolding protein called CipC (7, 14, 16, 26). Several cellulase-encoding genes from C. cellulolyticum have been sequenced, and the recombinant proteins have been characterized (1, 6, 7, 12, 13, 24). In C. thermocellum, the best-characterized cellulase system, the cellulosome has been shown to be responsible for cellular adherence to cellulose and for the degradation of cellulose (3, 22).
Evidence that xylan is utilized by C. cellulolyticum was recently published, and a β-xylosidase produced intracellularly by this organism was characterized (33). Little research has been done, however, on the extracellular xylanolytic activity of C. cellulolyticum (26). It was therefore of interest to characterize in general terms this extracellular xylanolytic activity. In addition, we were interested in understanding the interaction between C. cellulolyticum, which possesses a cellulose-adhered life cycle, and xylan, a major constituent of hemicellulose.
MATERIALS AND METHODS
Organism, substrates, and culture conditions.
Cultures of C. cellulolyticum were grown anaerobically at 34°C in serum bottles containing a previously described medium (19), which included a desired carbon source (cellulose [7.5 g/liter], xylan [5 g/liter], or cellobiose [5 g/liter]). Dialysis cultures were performed in 3-liter flasks, and fermentations were carried out in dialysis tubing (Visking) (molecular weight cutoff, 6,000 to 8,000) as described previously (17). No alteration of the tubing was observed during the course of our experiments.
Substrates.
Medium-viscosity carboxymethyl cellulose (CMC) and Avicel PH101 were obtained from Fluka. Oat-spelt xylan was purchased from Sigma. Unless otherwise specified, all other chemicals used in this study were reagent grade and purchased from Sigma.
Fractionation of xylan substrate.
To prepare soluble and insoluble fractions of xylan, a suspension of commercial xylan (0.5% in deionized water) was prepared and stirred for 1 h at room temperature. The mixture was centrifuged for 10 min at 17,300 × g, and the supernatant, comprising the soluble fraction, was removed and saved. The pellet, comprising the insoluble xylan, was collected and saved (28).
Preparation of extracellular material.
Extracellular (cell-free) material was obtained from the growth cultures by centrifuging the cells at 12,000 × g for 30 min. The supernatant fluids were collected. The pellets were washed and resuspended in the same volume of 25 mM phosphate buffer (pH 7.0). This attached fraction corresponded to both cell-associated and insoluble-xylan-associated material.
Bacterial biomass estimation.
The determination of biomass in the xylan-grown cultures was based on a bacterial-protein estimation as described by Bensadoun and Weinstein (9). A 2-ml aliquot was centrifuged. The pellet was washed twice with 2 ml of NaCl (0.9%) and incubated with 0.025 ml of sodium deoxycholate (2%) for 15 min. A 1-ml volume of 24% trichloroacetic acid was added to the medium, and the assay mixture was centrifuged at 6,000 × g for 15 min. Under these conditions, the cells burst, and intracellular proteins were found in the pellet. The protein concentration was determined by the method of Lowry et al. (25). The values obtained were converted to milligrams of cells per milliliter. Calibrations were performed with cells grown on cellobiose to estimate the relationship between dry weight and protein content of the cells (19).
Fractionation of extracellular xylanase.
Extracellular constituents of xylan-grown cells were fractionated either according to size or according to their interaction with cellulose or insoluble xylan. In the former case, the extracellular material was concentrated (40-fold) on Millipore PTGC (10-kDa cutoff) before being subjected to gel filtration on a Superose 12 HR column by using the fast protein liquid chromatography (FPLC) system (Pharmacia). The column was equilibrated and eluted with 100 mM Tris hydrochloride buffer (pH 7.0). Fractionation on either cellulose or xylan was performed by bringing the unprocessed, cell-free medium to a 2% dilution with the desired polymer. After an incubation of 30 min at room temperature, the stirred suspension was centrifuged (10,000 × g for 5 min), and the clarified supernatant was collected.
Cellulosome purification.
Cultures were grown either on xylan or cellulose in three pieces of dialysis tubing, each of which contained 100 ml of culture to allow a better biomass production (17).
On xylan, at the end of the exponential growth phase, cells and residual insoluble xylan were removed by centrifugation. The supernatant was applied to an Avicel cellulose-packed column (1.5 by 30 cm) equilibrated with 25 mM phosphate buffer (pH 7.0). The column was washed with the same buffer to elute the unattached fraction. The adhered fraction was eluted with deionized water and concentrated on Millipore PTGC (10-kDa cutoff), before being subjected to gel filtration on a superose 12 HR column as described above. The void volume fraction was used as cellulosome fraction.
On cellulose, after 5 days of growth, the culture was harvested by centrifugation. The pellet was washed in 50 ml of 25 mM Tris-HCl (pH 7.0) in order to eliminate nonspecifically adsorbed proteins. The cellulosome was then eluted with deionized water (150 ml) and concentrated on Millipore PTGC (10-kDa cutoff), before being subjected to gel filtration on a Superose 12 HR column as described above.
Cellulose binding experiments.
Binding experiments were performed with cellulose Avicel or insoluble xylan in 50 mM phosphate buffer (pH 7.5), giving a final volume of 1 ml. The cellulosome (70 μg) was mixed with the cellulose or insoluble xylan for 30 min at room temperature with slow shaking. After centrifugation (10,000 × g, 5 min), the supernatant (free fraction) was collected and then tested to determine its xylanase activity. The bound fraction was calculated by subtracting the xylanase activity of the free fraction from the initial xylanase activity.
Enzyme assays.
The protein concentrations were determined by using the method of Bradford (10) with bovine serum albumin as the standard. Levels of Avicelase, carboxymethyl cellulase (CMCase), and xylanase were determined as described previously by using the method of Miller et al. (27) with a solution of 1% CMC or xylan in a 25 mM citrate-phosphate buffer (pH 7.0). One international unit of activity corresponds to 1 μmol of d-glucose or d-xylose released per min.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on 10%-acrylamide gels by the procedure developed by Laemmli (21). The samples used for the SDS-PAGE were boiled in sample buffer and stored at −20°C. The profile analyses were performed by using the Gel Doc 1000 System (Bio-Rad). When gels were used to prepare zymograms, samples were incubated in sample buffer for 1 h at room temperature (mild denaturation) before SDS-PAGE (32).
Zymograms.
CMCase or xylanase activity in gels subjected to electrophoresis was detected by use of polyacrylamide gel zymograms that contained CMC or xylan, respectively, as a substrate. CMCase and xylanase zymograms were prepared as described previously (32).
Sequencing of the amino-terminal peptide of the 135-kDa protein.
For amino-terminal sequencing, cellulosomes were subjected to SDS-PAGE and transferred onto a hydrophobic polyvinylidene difluoride membrane (Bio-Rad) by using the conditions recommended by the manufacturer. The 135-kDa band was stained with 0.1% Ponceau red and cut out, and the amino-terminal sequence of the polypeptide was determined by the Edman method by using a model 476 A sequencer (Applied Biosystems).
Bacterial adhesion measurement.
Both xylan- and cellulose-grown cells were harvested in mid-exponential phase and separated from the insoluble substrates by using the decantation method described previously (19, 20). The adhesion experiments were performed by using the method developed by Gelhaye et al. (19, 20), with adhesion brought about by adding 5 ml of bacterial suspension (0.15 mg [dry weight]/ml of Tris-HCl buffer, pH 7.5) with 0.1 g of Whatman filter paper (1 by 10 cm).
RESULTS
Xylanase activity in C. cellulolyticum.
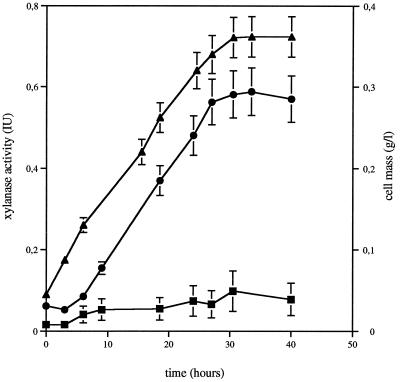
C. cellulolyticum was grown on washed insoluble-xylan medium. The respective fractions were examined for degradative activity on xylan (Fig. 1). During growth, the pellet-associated activity remained very low (reaching about 0.08 μmol of xylose per ml of broth per min). At the same time, nearly 90% of the total xylanase activity was detected in the cell-free fraction (0.58 μmol of xylose per ml of broth per min). Xylanase production correlated with cell growth. A similar partition of CMCase activity was observed. Maximal CMCase activities detected in the cell-free fraction and in the pellet-associated fraction were 0.22 and 0.04 μmol of glucose per ml of broth per min, respectively.
FIG. 1.
Growth and production of attached (in pellet fraction after centrifugation) and cell-free xylanase activities by C. cellulolyticum cultivated on insoluble xylan. Each point is the mean ± the standard deviation (SD) (indicated by error bar) of four determinations for two different cultures. Symbols: ■, attached xylanase activities; ●, cell-free xylanase activities; ▴, cell mass.
Cells were cultured with soluble or untreated (i.e., containing both soluble and insoluble xylan) xylan. The same partition of the xylanase and CMCase activity was observed in both cases. Indeed, about 90% of both activities were found in the cell-free fraction of these cultures. Previously published data have shown that when C. cellulolyticum was cultured with cellulose as growth substrate, both CMCase and xylanase activities were found largely in the pellet (about 90 and 70% of the total activity, respectively) (8).
Fractionation of extracellular xylanase activity.
To determine the distribution of the observed xylanase activity, C. cellulolyticum was grown on xylan-containing medium in dialysis culture conditions, as this culturing procedure allows better biomass production (17). The cultures were harvested in the late-exponential phase of growth. Ninety percent of the total xylanase activity was detected in the cell-free fraction supernatant (0.9 μmol of xylose per ml of broth per min). The extracellular constituents were fractionated either according to size or according to their interaction with cellulose or xylan.
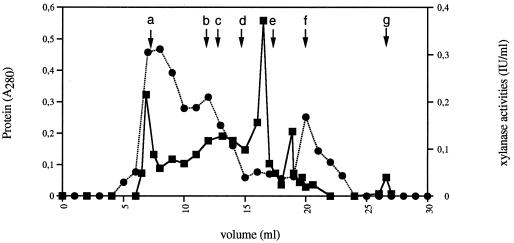
The cell-free growth medium was therefore subjected to gel filtration on a FPLC Superose column (Fig. 2). Three major fractions containing xylanase activity were isolated. The first one eluted with the void volume and the second corresponded to an apparent molecular mass of 600 kDa. As shown by Gal et al. (14), who studied cellulose-grown cultures, the first fraction could result from the assembly of several (20 to 25) 600-kDa cellulosome units. The results showed that about 70% of the xylanase activity was associated with these high-molecular-weight fractions. A third fraction containing xylanase activity was detected, the maximal activity corresponding to proteins with an apparent molecular mass of about 30 kDa. In the same way, about 70% of the total CMCase activity was detected in the high-molecular-weight fractions. Furthermore, avicelase activity was only detected with the high-molecular-weight fractions containing xylanase activity (i.e., fractions eluted with the void volume and fractions corresponding to an apparent molecular mass of 600 kDa), but no avicelase activity was detected in the third fraction containing xylanase activity.
FIG. 2.
FPLC gel filtration chromatography on Superose 12 HR column of the cell-free fraction of C. cellulolyticum grown on xylan. The fractions were analyzed for protein content (■) and xylanase activity (●) under the following conditions: protein load, 0.38 mg; flow rate, 0.5 ml/min; and buffer, 100 mM Tris-HCl (pH 7.0). Arrows indicate the retention times of some standards: a, blue dextran (indicating void volume); b, thyroglobulin (669 kDa); c, apoferritin (443 kDa); d, β-amylase (200 kDa); e, albumin (66 kDa); f, carbonic anhydrase (29 kDa); g, total volume.
In parallel, cellulose and xylan (2%) were introduced to cell-free growth medium, and after a 30-min incubation, the amount of xylanase activity adsorbed onto the corresponding matrix (after its removal by centrifugation) was determined. With cellulose, about 75% of the xylanase activity was bound to the insoluble polymer. In contrast, with xylan, all the activity remained in the unbound state.
The association of xylanase activity with high-molecular-weight cellulose-bound fraction containing avicelase activity suggested that a major portion of this activity in C. cellulolyticum is contained in the cellulosome-like complex.
Purification of the cellulosome.
A xylan, cell-free growth medium was applied to a cellulose-packed column (see Materials and Methods). In these conditions, between 80 and 90% of the xylanase activity was bound to the cellulose. Extensive buffer washing eliminated nonspecifically bound proteins. The cellulosomal proteins were then released by extensive washing with distilled water and concentrated, this fraction constituting the H2O fraction. This purification procedure led to the recovery of 75 mg of proteins per liter of culture with a specific CMCase activity of 2 IU/mg and a specific xylanase activity of 3.2 IU/mg in the H2O fraction.
When this fraction was analyzed by gel filtration on a FPLC Superose HR 12 column, we observed a major absorbance peak which was eluted in the same volume as the void volume (2 MDa). Furthermore, several minor fractions associated with xylanase activity were eluted in significantly larger volumes than the void volume, corresponding to proteins with apparent molecular masses of between about 50 and 15 kDa. Zymogram analysis showed that the same pattern of xylanases is present in both high- and low-molecular-weight fractions (data not shown). These findings suggested that at least a part of the xylanases detected in low-molecular-weight fractions could be provided from cellulosomes fragmented during the growth or during purification steps.
The cellulosomal fraction was obtained after a two-step process of affinity chromatography on cellulose and gel filtration on a FPLC Superose 12 HR column. Therefore, the experiments described here involved freshly prepared cellulosome.
SDS-PAGE analysis and zymograms.
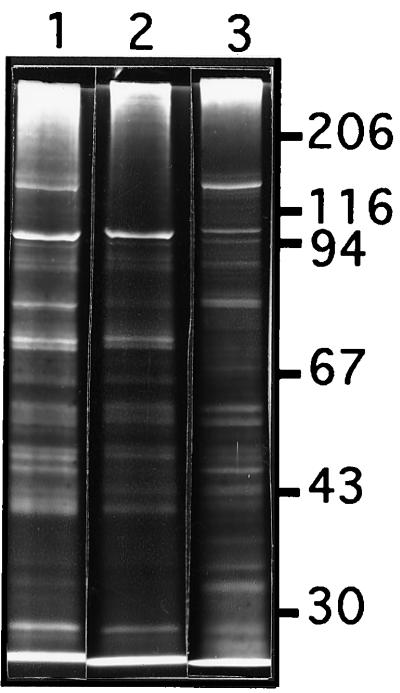
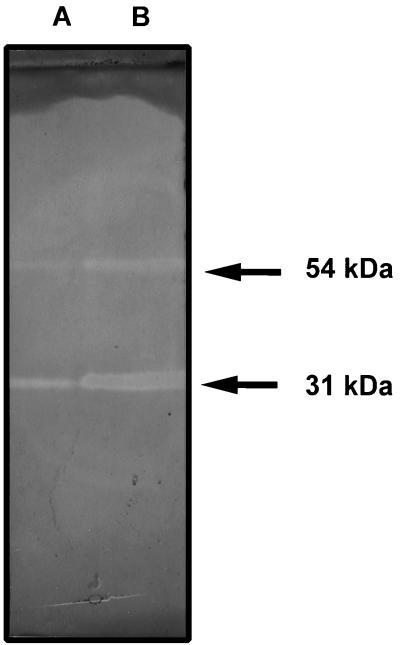
When subjected to SDS-PAGE, the cellulosome was found to consist of at least 17 proteins with molecular masses ranging from 25 to 160 kDa (Fig. 3). Among these proteins, the 135-kDa fraction was abundant. To identify this protein, its 135-kDa amino terminus was sequenced. The obtained sequence (AGTGVVSVQ) is identical to the amino acid 28 to 36 segment of the cellulosomal scaffolding protein CipC (GenBank accession no. U40345), a cellulose binding protein which might facilitate the anchoring of the cellulosome to cellulose (14, 29). Zymograms were performed on xylan with pure cellulosome and total extracellular material derived from xylan-grown cells (Fig. 4). In both tested materials the results showed the existence of two (or two groups of) proteins harboring xylanase activities with apparent molecular masses of 54 and 31 kDa.
FIG. 3.
Electrophoretic characterization of components of the cell-free fraction of C. cellulolyticum grown on xylan. The different samples were analyzed by SDS-PAGE under the conditions described in Materials and Methods. Lanes: 1, whole cell-free fraction (9 μg); 2, unattached fraction (9 μg); 3, cellulosomal fraction (fraction eluted from cellulose affinity chromatography and subjected to gel filtration chromatography) (9 μg). Molecular mass in kilodaltons is indicated on the right.
FIG. 4.
Xylanase zymograms of total extracellular material (lane A) and pure cellulosome (lane B) derived from xylan-grown cultures of C. cellulolyticum. Samples were mildly denatured prior to SDS-PAGE, and proteins were analyzed for xylanase activity with a xylan-polyacrylamide gel overlay. All samples contained 9 μg of protein.
Interaction of cellulosome with xylan.
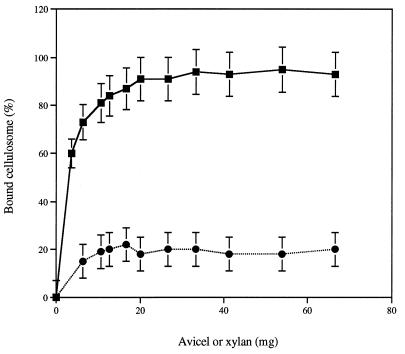
C. cellulolyticum is able to grow on xylan. Since the cellulosome is an exocellular complex responsible for the adhesion of the bacterium to the insoluble cellulose substrate, the binding of cellulosome derived from xylan-grown cells to xylan was determined. Furthermore, a positive-control experiment in which cellulose was used as the adsorbent was performed. With cellulose, about 90% of the cellulosome was adsorbed to relatively low amounts of the insoluble polymer (2.6%) (Fig. 5). In contrast, with xylan, the adsorption of the cellulosome remained very weak, since about 80% of the activity remained in the unbound state, even when high amounts of xylan (7%) were used.
FIG. 5.
Adsorption of the cellulosome to Avicel or insoluble xylan. The capacity of a constant amount (70 μg) of the purified cellulosome to bind to various amounts of crystalline cellulose or insoluble xylan (1 to 70 mg per tube) was determined as described in Materials and Methods. The results are expressed as percentages of bound xylanase activity as a function of various amounts of Avicel or xylan. Each point is the mean ± SD (indicated by error bar) of four determinations for two different assays. Symbols: ■, Avicel; ●, xylan.
Bacterial adhesion.
On xylan, cell-associated xylanase and CMCase activities remained very weak, suggesting a lack of cell-associated cellulosomes. Since the cellulosome has been shown to be responsible for cellular adherence to cellulose, xylan-grown cells were tested for binding to cellulose. As a control, the adhesion to filter paper of cells obtained from cellobiose and cellulose cultures was measured. For the same initial biomass concentration (0.15 mg of adhered biomass [dry weight]/ml), the adhesion of xylan-grown cells remained weak (23% ± 2% of adhered biomass) in comparison with cellobiose-grown cells (32% ± 3% of adhered biomass) and cellulose-grown cells (50% ± 5% of adhered biomass).
Cellulosomes derived from xylan- and cellulose-grown cells.
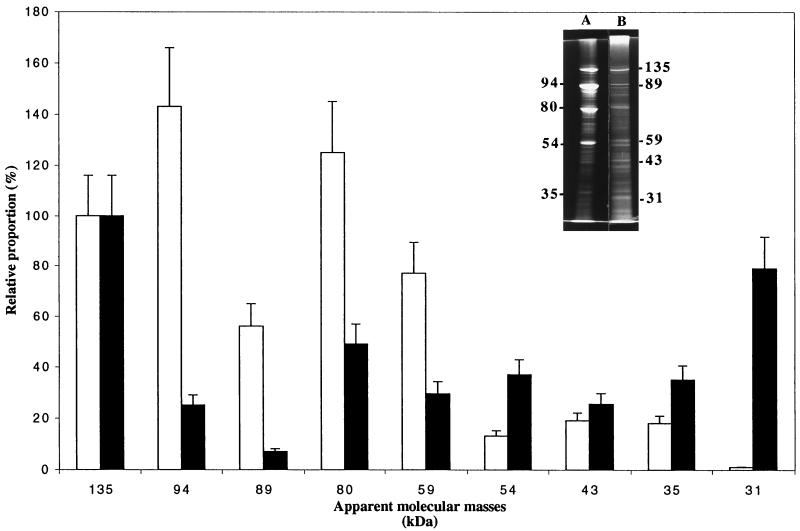
Cellulosomes derived from cellulose-grown cells were purified as described in Materials and Methods. As suggested previously, the cellulase complex found extracellularly at the end of growth may be very similar or even identical to the cell-associated enzyme system (26). When subjected to SDS-PAGE, the cellulosomes derived from xylan- and cellulose-grown cells exhibited great profile differences. In both cases, the 135-kDa protein was abundant (about 18% of total detected fractions). The cellulosome derived from cellulose-grown cells was found to consist of at least 14 proteins with molecular masses ranging from 160 to 20 kDa. The relative proportions of several components are given Fig. 6, the intensity of the 135-kDa fraction being arbitrarily fixed at 100%. Among these constituents, the 94- and 80-kDa fractions were the most abundant (125 and 140%, respectively), these data being consistent with the results obtained by Gal et al. (14). The relative proportions of the same constituents are dramatically changed in cellulosomes derived from xylan-grown cells. Particularly, the relative proportion of the 94-kDa fraction decreased from about 125% to about 25%, and the relative proportion of the 31-kDa fraction simultaneously increased from 1% to about 80%, this fraction being associated with xylanase activity (Fig. 4). Zymograms (data not shown) were performed on xylan and CMC with various quantities of the pure cellulosome derived from xylan-grown and cellulose-grown cells. The results showed the existence of five (or five groups of) proteins harboring CMCase activities with apparent molecular masses ranging from 90 to 30 kDa. Identical patterns have been previously reported during the analysis of cellulosomes derived from cellulose-grown cells (14, 16, 26). On xylan, the results for both tested materials showed the existence of two (or two groups of) proteins harboring xylanase activities with apparent molecular masses of 54 and 31 kDa.
FIG. 6.
Relative proportions of several components of cellulosomes derived from cellulose-grown (white columns) and xylan-grown cells (black columns). The amount of the 135-kDa fraction has been arbitrarily fixed to 100%. Each set of data is the mean ± SD (indicated by error bars) of separate analyses of four separate SDS-PAGE experiments. An example gel from an SDS-PAGE experiment, performed as described in Materials and Methods, is shown. Lanes: A, cellulosome derived from cellulose-grown cells (9 μg); B, cellulosome derived from xylan-grown cells (9 μg). Molecular mass in kilodaltons is indicated.
DISCUSSION
In this study, we have shown that when C. cellulolyticum is cultured with xylan as growth substrate, extracellular xylanase and CMCase activities were largely found in the cell-free fraction associated with high-molecular-weight complexes containing cellulolytic activity and the cellulosomal scaffolding protein CipC. These data clearly suggest that on xylan the cellulosome of C. cellulolyticum is not associated with the cells. However, when cells are cultured in a cellulose-containing medium, C. cellulolyticum has been shown to grow on cellulose when bacteria are bound to their insoluble substrate (19), this adhesion probably being mediated by cell-associated cellulosomes (7). Under these conditions, the xylanase and CMCase activities were found largely associated with the fraction containing the cells and the substrate (8). This difference suggests that C. cellulolyticum is able to regulate the cell association of its cellulosomal complexes, depending on the insoluble substrate used.
The cellulose adhesion of the xylan-grown cells remains very feeble in comparison with cellulose-grown cells. These data seem to indirectly confirm that the cellulosome is responsible for the adherence of bacteria to cellulose in C. cellulolyticum. In this context, the role of cellulosome cell-association regulation in this bacterium, which possesses cellulose-adhered life cycle, remains unclear. The cellulosome, which exhibits high affinity to cellulose, appears to be a targeting tool to bring xylanase enzymes to the surface of the insoluble hemicellulose- and cellulose-containing substrate. Nevertheless, as previously shown in C. thermocellum, interaction of purified cellulosome with insoluble xylans remained very weak (28). The degradation of hemicellulose by free cellulosomes would enhance the ability of the organism to reach the cellulose-containing component(s) of the plant cell surface, as suggested for the noncellulosomal xylanases of C. thermocellum (28).
It was recently demonstrated that, in C. thermocellum, the cellulosome was integrated into the cell wall by the scaffolding protein (CipA) C-terminal dockerin domain, which binds a cohesin domain of a cell wall integrated protein (SdbA) (23). In C. cellulolyticum, the scaffolding protein (CipC) does not contain a dockerin domain at its C terminus, suggesting that C. cellulolyticum cellulosomes are embedded in a very different manner than those existing in C. thermocellum (7). Further research is needed to elucidate the mechanism of cellulosome-cell association and its regulation.
In this study, we have shown that C. cellulolyticum is not only able to regulate cellulosome-cell association, but that the organism might also alter the production of various cellulosomal components in response to various growth conditions (i.e., the presence of xylan or cellulose). In both studies, growth substrates (cellulose and xylan) and the scaffolding protein (CipC) were always present, but the relative proportions of the various components were different. In particular, we observed a large increase of the relative proportion of the 31-kDa protein in cellulosomes derived from xylan cultures, this fraction being associated with xylanase activity. Furthermore, in both cases we observed a high proportion of CipC with an apparent molecular mass of 135 kDa, even when fresh preparations were used. The apparent molecular mass of CipC was previously reported as being either around 135 kDa (16) or between 160 and 170 kDa (14, 26). This difference may be due to differences between the bacterial strains studied and/or differences between the growth conditions and the methods of cellulosome preparation and characterization used.
Indeed, C. cellulolyticum is able to adapt the enzyme system responsible for the hydrolysis of crystalline cellulose and xylan, regulating both composition and cell-association of the cellulosomes. These regulations are very interesting and will be dealt with in further studies in our laboratory.
ACKNOWLEDGMENTS
We thank F. Saulnier at the Service Commun de Séquences des Protéines (Université H. Poincaré, France) for performing the amino-terminal Edman sequencing.
This research was supported by grants from the Programme intergouvernemental Algero-Français (number 96/10171), the commission of the European communities (Fair number CT95-0191), and from the Programme Agrice (number 97.01.041).
REFERENCES
- 1.Bagnara-Tardif C, Gaudin C, Belaich A, Hoest P, Citard T, Belaich J P. Sequence analysis of a gene cluster encoding cellulases from Clostridium cellulolyticum. Gene. 1992;119:17–28. doi: 10.1016/0378-1119(92)90062-t. [DOI] [PubMed] [Google Scholar]
- 2.Bayer E A, Setter E, Lamed R. Organization and distribution of the cellulosome in Clostridium thermocellum. J Bacteriol. 1985;163:552–559. doi: 10.1128/jb.163.2.552-559.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer E A, Lamed R. Ultrastructure of the cell surface cellulosome of C. thermocellum and its interaction with cellulose. J Bacteriol. 1986;167:828–836. doi: 10.1128/jb.167.3.828-836.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayer E A, Morag E, Lamed R. The cellulosome—a treasure trove for biotechnology. Trends Biotechnol. 1994;12:379–386. doi: 10.1016/0167-7799(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 5.Béguin P, Aubert J P. The biological degradation of cellulose. FEMS Microbiol Rev. 1994;13:25–58. doi: 10.1111/j.1574-6976.1994.tb00033.x. [DOI] [PubMed] [Google Scholar]
- 6.Belaich J P, Gaudin A, Belaich A, Bagnara C. Genes and proteins involved in cellulose degradation by mesophilic Clostridia. In: Sebald, editor. Genetics and molecular biology of anaerobic bacteria. New York, N.Y: Springer-Verlag; 1993. pp. 407–411. [Google Scholar]
- 7.Belaich J P, Tardif C, Belaich A, Gaudin A. The cellulolytic system of Clostridium cellulolyticum. J Biotechnol. 1997;57:3–14. doi: 10.1016/s0168-1656(97)00085-0. [DOI] [PubMed] [Google Scholar]
- 8.Benoit L, Cailliez C, Petitdemange E, Gitton J. Isolation of cellulolytic mesophilic clostridia from a municipal solid waste digestor. Microb Ecol. 1992;23:117–125. doi: 10.1007/BF00172634. [DOI] [PubMed] [Google Scholar]
- 9.Bensadoun A, Weinstein D. Assay of proteins in the presence of interfering materials. Anal Biochem. 1976;70:241–250. doi: 10.1016/s0003-2697(76)80064-4. [DOI] [PubMed] [Google Scholar]
- 10.Bradford M M. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principles of protein dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 11.Doi R H, Goldstein M, Hashiba S, Park J S, Takagi M. The Clostridium cellulovorans cellulosome. Crit Rev Microbiol. 1994;20:87–93. doi: 10.3109/10408419409113548. [DOI] [PubMed] [Google Scholar]
- 12.Faure E, Belaich A, Bagnara C, Gaudin C, Belaich J P. Sequence analysis of the Clostridium cellulolyticum endoglucanase-A-encoding gene, celCCA. Gene. 1989;84:39–46. doi: 10.1016/0378-1119(89)90137-6. [DOI] [PubMed] [Google Scholar]
- 13.Gal L, Gaudin C, Belaich A, Pages S, Tardif C, Belaich J P. CelG from Clostridium cellulolyticum: a multidomain endoglucanase acting efficiently on crystalline cellulose. J Bacteriol. 1997;179:6595–6601. doi: 10.1128/jb.179.21.6595-6601.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gal L, Pages S, Gaudin C, Belaich A, Reverbel-Leroy C, Tardif C, Belaich J P. Characterization of the cellulolytic complex (cellulosome) produced by Clostridium cellulolyticum. Appl Environ Microbiol. 1997;63:903–909. doi: 10.1128/aem.63.3.903-909.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gehin A, Gelhaye E, Raval G, Petitdemange H. Clostridium cellulolyticum viability and sporulation under cellobiose starvation conditions. Appl Environ Microbiol. 1995;61:868–871. doi: 10.1128/aem.61.3.868-871.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gehin A, Petitdemange H. The effects of tunicamycin on secretion, adhesion and activities of the cellulase complex of Clostridium cellulolyticum ATCC 35319. Res Microbiol. 1995;146:251–262. doi: 10.1016/0923-2508(96)80281-6. [DOI] [PubMed] [Google Scholar]
- 17.Gehin A, Cailliez C, Petitdemange E, Benoit L. Studies of Clostridium cellulolyticum ATCC 35319 under dialysis and co-culture conditions. Lett Appl Microbiol. 1996;23:208–212. doi: 10.1111/j.1472-765x.1996.tb00067.x. [DOI] [PubMed] [Google Scholar]
- 18.Gelhaye E, Gehin A, Petitdemange H. Colonization of crystalline cellulose by Clostridium cellulolyticum ATCC 35319. Appl Environ Microbiol. 1993;59:3154–3156. doi: 10.1128/aem.59.9.3154-3156.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gelhaye E, Petitdemange H, Gay R. Adhesion and growth rate of Clostridium cellulolyticum ATCC 35319 on crystalline cellulose. J Bacteriol. 1993;175:3452–3458. doi: 10.1128/jb.175.11.3452-3458.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gelhaye E, Claude B, Cailliez C, Burle C, Petitdemange H. Multilayer adhesion to filter paper of two mesophilic cellulolytic clostridia. Curr Microbiol. 1992;25:307–311. [Google Scholar]
- 21.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Lamed R, Setter E, Kenig R, Bayer E A. The cellulosome: a discrete cell surface organelle of Clostridium thermocellum which exhibits separate antigenic, cellulose-binding and various cellulolytic activities. Biotechnol Bioeng Symp. 1983;13:163–181. [Google Scholar]
- 23.Leibovitz E, Béguin P. A new type of cohesin domain of the Clostridium thermocellum cellulosome-integrating protein CipA. J Bacteriol. 1996;178:3077–3084. doi: 10.1128/jb.178.11.3077-3084.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leschine S B. Cellulose degradation in anaerobic environments. Annu Rev Microbiol. 1995;49:399–426. doi: 10.1146/annurev.mi.49.100195.002151. [DOI] [PubMed] [Google Scholar]
- 25.Lowry O H, Rosebrough N, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 26.Madarro A, Perra J L, Lequerica J L, Valles S, Gay R, Flors A. Partial purification and characterization of the cellulases from Clostridium cellulolyticum H10. J Chem Technol Biotechnol. 1991;52:393–406. [Google Scholar]
- 27.Miller G L R, Blum W E, Burton A L. Measurements of carboxymethylcellulase activity. Anal Biochem. 1960;2:127–132. [Google Scholar]
- 28.Morag E, Bayer E A, Lamed R. Relationship of cellulosomal and noncellulosomal xylanases of Clostridium thermocellum to cellulose-degrading enzymes. J Bacteriol. 1990;172:6098–6105. doi: 10.1128/jb.172.10.6098-6105.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pages S, Gal L, Belaich A, Gaudin C, Tardif C, Belaich J P. Role of scaffolding protein CipC of Clostridium cellulolyticum in cellulose degradation. J Bacteriol. 1997;179:2810–2816. doi: 10.1128/jb.179.9.2810-2816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Payot S, Guedon E, Cailliez C, Gelhaye E, Petitdemange H. Metabolism of cellobiose by Clostridium cellulolyticum growing in continuous culture: evidence for decreased NADH reoxidation as a factor limiting growth. Microbiology. 1998;144:375–384. doi: 10.1099/00221287-144-2-375. [DOI] [PubMed] [Google Scholar]
- 31.Pohlschröder M, Leschine S B, Canale-Parola E. Ultrastructural diversity of the cellulose complexes of Clostridium papyrosolvens C7. J Bacteriol. 1995;177:6625–6629. doi: 10.1128/jb.177.22.6625-6629.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pohlschröder M, Leschine S B, Canale-Parola E. Multicomplex cellulase-xylanase system of Clostridium papyrosolvens C7. J Bacteriol. 1994;176:70–76. doi: 10.1128/jb.176.1.70-76.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saxena S, Fierobe H P, Gaudin C, Guerlesquin F, Belaich J P. Biochemical properties of a β-xylosidase from Clostridium cellulolyticum. Appl Environ Microbiol. 1995;61:3509–3512. doi: 10.1128/aem.61.9.3509-3512.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomson J A. Molecular biology of xylan degradation. FEMS Microbiol Rev. 1993;104:65–82. doi: 10.1111/j.1574-6968.1993.tb05864.x. [DOI] [PubMed] [Google Scholar]