Abstract
Chronic sleep deprivation (SD) is a common problem for humans and can lead to many deleterious effects, including depression, anxiety, stroke, permanent cognitive deficits, stress, and other physiological diseases. It is vital to acquire information about the relevant neural activities at the whole-brain level to systematically explore the mechanisms of brain dysfunction related to SD. Expression of the immediate-early gene (IEG) Fos in the mouse brain has been widely used as a functional marker of brain activity in the field of neuroscience. However, most previous studies only analyzed the change of c-Fos in several specific brain regions using traditional research methods or in short-term SD model. Here, we applied c-Fos mapping through the fluorescence micro-optical sectioning tomography (fMOST) technique and AAV-PHP.eB to comprehensive analysis the state of cumulative activation across the whole brain in a mouse model of chronic SD. The chronic rapid eyes movement (REM) SD model was induced by moving mice to a separate holding area filled with water. The experimental period lasted for 6 h per day. The results showed that after 14 days of SD, the mice displayed anxiety-like behaviors in open field test and elevated plus maze test, and displayed depression-like behaviors in tail suspension test and the sucrose preference test. The c-Fos + cells were detected in a maximum of 230 brain regions. SD-induced stress model evoked c-Fos expression in several brain regions compared to the control group. In particular, the isocortex-cerebral cortex plate area, including the retrosplenial, anterior cingulate, agranular insular, gustatory, and parasubiculum, appear to be the most sensitive regions after chronic REM SD.
Keywords: Sleep deprivation, Immediate-early genes, c-Fos, Fluorescence micro-optical sectioning tomography
1. Introduction
Sleep deprivation (SD), which includes decreased quantity or impaired quality of sleep, is a common problem worldwide (Bandyopadhyay and Sigua, 2019), and its prevalence increases every year (Abrams, 2015; Knutson et al., 2010). Currently, it is a consensus that insufficient sleep contributes to many deleterious effects, including a general slowing of response speed, increased risks of cognitive deficits, cancer, stroke, obesity and diabetes (Abrams, 2015; Killgore, 2010; Liu and Chen, 2019). Moreover, depression, as the third leading cause of global non-fatal burden, severely affects quality of life and limits psychosocial functioning (Collaborators, 2018). Repeated exposure to stressors, such as sleep disruption, has been considered to be one of the factors that may induce depression. Previous studies have shown that after sleep disruption, adolescent mice display significantly greater depression-like behavior (Murack et al., 2021). In addition, sleep deprivation will also lead to increased anxiety levels (Goldstein-Piekarski et al., 2018; Pires et al., 2016). These findings suggest that chronic SD can lead to various emotional disorders and physical diseases (Okun et al., 2018; Yu et al., 2016). Nevertheless, the underlying mechanisms of these effects still remain unclear.
The expression of Fos, an immediate early gene (IEG), has been widely used as a functional marker of brain activity in the field of neuroscience, including sleep research (Terao et al., 2003; Thompson et al., 2010). Fos encodes the transcription factor c-Fos, which can act as a “third messenger” to regulate the expression of target genes. When induced by stimulating factors, c-Fos forms the heterodimer activator protein-1 (AP-1), which binds to the promoter region of numerous “later response” or target genes, through dimerizing with other DNA-binding proteins (Morgan and Curran, 1991). Generally, the basal level of c-Fos expression is relatively low without any stimuli. Whereas, the gene can be transiently and rapidly induced by a virous degree of extracellular stimuli, including seizure activity, peripheral sensory stimulation, kindling, and photic-induced phase shifts of the circadian system. Thus, the protein product of Fos gene expression has been used as an important marker of neuronal activation and the immunohistochemical method is widely used to identify specific brain regions whose activities are altered by stimuli. It has been shown that the expression level of Fos and c-Fos protein in the brain significantly changes from sleep to wakefulness patterns (Pompeiano et al., 1995; Sagar and Sharp, 1993). Such differences in expression implicate alterations in transcriptional regulation across behavioral states. However, most previous studies only analyzed several specific brain regions and reported various results regarding the significant differences in brain regions using traditional research methods (Cirelli et al., 1995; Jha et al., 2017; Ledoux et al., 1996; Montes-Rodríguez et al., 2019; O'Hara et al., 1993; Semba et al., 2001; Terao et al., 2003). Understanding the changes in neurons under stimuli that induces SD is far from sufficient.
It is known that acquiring information on neural activities at the whole-brain level is essential for exploring the mechanisms of brain function and dysfunction. Under these circumstances, fluorescence micro-optical sectioning tomography (fMOST), composed of a microtome, light microscope, and image recorder, has emerged. This technique allows rapid visualization of neural structures based on the number of neurons which are labeled by fluorescence (Li et al., 2010; Yang et al., 2018). Recently, AAV-PHP.eB, a highly blood brain barrier-permeable capsid variant of AAV serotype 9, has been applied systemically in the whole brain transduction (Konno and Hirai, 2020). Combined application of AAV-PHP.eB and fMOST will greatly facilitate observational studies of the whole brain. In the current study, we applied c-Fos mapping through the fMOST technique to build a comprehensive mapping of cumulatively activated brain regions in chronic SD mice.
2. Materials and methods
2.1. Animals
Male C57BL/6 mice (8–10 weeks of age) were obtained from the Fourth Military Medical University. The animals were kept in a temperature-controlled room (22–26 °C) under a 12-h light/dark cycle with free access to food and water. All experimental processes complied with the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health (NIH) and were approved by the Institutional Animal Care and Use Committee (IACUC) at the Fourth Military Medical University.
2.2. Experimental protocol and groups
2.2.1. Part I
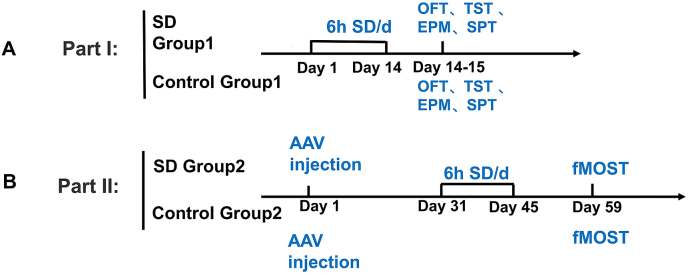
Mice (n = 6 per group) were randomized into a chronic rapid eye movement (REM) sleep deprivation group and a control group. The chronic REM sleep deprivation was induced by moving mice to a separate holding area filled with water (23 ± 2 °C) to a height of 1.5 cm. The water tank contained nine narrow platforms on which the mice were placed. Under this condition, they would try to avoid water and could not sleep or keep a resting position soundly (Suchecki and Tufik, 2000). The experimental period lasted for 6 h per day and continued for 14 days. At the end of the 6-h sleep disruption session, the mice were returned to the housing facility. In the 6-h experimental period, control mice were not manipulated. After sleep deprivation, all the mice were subjected to open-field test (OFT), elevated plus maze experiment (EPM), tail suspension tests (TST) and sucrose preference test (SPT) (Fig. 1A).
Fig. 1.
Experimental protocol and groups. (A) Mice were randomized into chronic sleep deprivation group and control groups. The experimental period lasted 6 h per day and continued for 14 days. Control mice were not manipulated during the 6-h experimental period. After sleep deprivation, the mice were subjected to open field test, elevated plus-maze, tail suspension teat, and sucrose preference tests. (B) Mice were injected with AAV virus one month before the sleep deprivation experiment. After 14 days of sleep deprivation, the mice were fed normally for 2 weeks and then were killed during the light cycle for whole brain imaging with fluorescence micro-optical sectioning tomography (fMOST).
2.2.2. Part II
Mice (n = 6 per group) were randomized into a chronic REM sleep deprivation group and a control group. Two groups of mice were injected with AAV virus one month before the sleep deprivation experiment. The sleep deprivation experimental period lasted for 6 h per day and continued for 14 days. In the 6-h experimental period control mice were not manipulated. After the 14 days REM sleep deprivation period, the mice were fed normally for 2 weeks and then subjected to the whole brain imaging with fMOST (Fig. 1B).
2.3. Open field test and tail suspension test
2.3.1. Open field test
The experimental process was as described in a previous study (Scott et al., 2017). Briefly, mice were placed in a white plastic box with a length, width and height of 40 cm × 40 cm × 40 cm under full-light conditions (1000 lx), and their movements were recorded. At the beginning, the mouse was placed in the center of the white plastic box and their activities was filmed. Each video recording lasted 5 min. After this the mice were placed back to their home cages. The automated analysis system (SMART 3.0, Panlab S.L.U.) was used to analyze the time spent in the center of the box, which reflected the level of anxiety.
2.3.2. Elevated plus maze experiment
The experimental process was as described in the previous study (Cai et al., 2020; Scott et al., 2017). Briefly, mice were tested in equipment with four arms measuring 50 cm × 5 cm each, which were rested on a platform 1 m from the ground. There are two closed arms with 15-cm-high walls and two open arms without walls. At the beginning of the experiment, the mouse was placed in the center of a platform facing one of the open arms. The duration of each video recording was 5 min, and we used an automated analysis system (SMART 3.0, Panlab S.L.U.) to analyze the time spent in the open arms, which was used to evaluate anxiety levels.
2.3.3. Tail suspension test
The experimental process was as described in a previous study (Kim et al., 2017). Briefly, acoustically and visually isolated mice were suspended 50 cm above the floor using adhesive tape. The tape was placed at one-third of the way from the tip of the tail. The hang time lasted for 6 min and the last 4 min period was used to analyze immobility time, which reflected the level of depression.
2.3.4. Sucrose preference test (SPT)
The experimental process was as described in a previous study (Yin et al., 2021). Mice need to be deprived of water and food for 15 h before the preference test. Remove all the food and water bottles from 6 p.m. on day 1–9 a.m. on day 2. The electronic SPT device and the software MDA were used to conduct preference experiments on day 2. Immediately after deprivation, transfer mice from home cages into chambers of the apparatus randomly. The duration of the experimental test was 10 h, and the software-MDA would pause automatically after the preset detection duration. Return all mice to their home cages for group housing with ad libitum access to food and water.
2.4. AAV viruses and viral injections
The AAV vector rAAV-PHP.eB-cFos-EYFP was purchased from Brain Case Biotechnology Co., Ltd. The viral stocks had titers of 3.89 × 1012 viral genomes/mL and were stored at −80 °C. rAAV-PHP.eB-cFos-EYFP was administered to six chronic sleep deprived mice and six control mice one month before the sleep deprivation experiment (section 2.2.2). Mice were administered 1 × 1011 viral genomes/mouse [in 100 μL sterile phosphate buffered saline (PBS)] via the lateral caudal vein.
2.5. fMOST
2.5.1. Tissue preparation
The mice were anesthetized with 1.5% isoflurane in oxygen (flow rate of 1 L/min) and perfused with 4% paraformaldehyde and 0.01 M PBS. The brains were removed and post-fixed in 4% paraformaldehyde at 4 °C for 24 h. An solution was prepared with a mixture of agarose powder (Sigma-Aldrich) and distilled water at a concentration of 3%–5%. The post-fixed mouse brains were embedded in the agarose solution, and the position was adjusted to ensure that the main axis of the mouse brain was parallel to the edge of the sample container. The sample was rested for 8–10 min until it solidified. The solid blocks were removed and fixed on the sample holder with glue.
2.5.2. Whole-brain imaging
The procedures of fMOST samples in the present study were similar to those used in a previous study (Yang et al., 2018). Before imaging, the mouse whole-brain sample was immobilized in the anterior-posterior direction on a 3D translation stage in a water bath, which was filled with 0.01 M PBS. Then the sample was immersed into the bath in order to provide a matched refractive index for the objective lens during imaging. The PBS solution enhanced EGFP fluorescence. The light sheet imaging system adopts a tomographic imaging strategy, and the thick of each image is 40 μm. The procedure was divided into four steps: (1) vibrating section with a customized vibratome of vibrating frequency to 77 Hz and 0.2 mm/s sectioning speed along the x-axis; (2) the movement of the platform to the starting point of imaging; (3) light sheet scanning imaging; (4) the return of the platform to the cutting tool setting point ready for the next layer of imaging. The images from fMOST were detected using the BioMapping 9000 system (Oebio Biological Co., Ltd, Wuhan, China), consisting of a 20X objective in an Olympus microscope (LUMPLFLN NA 0.5, Tokyo, Japan), two lasers with wavelengths of 473 nm and 561 nm (Cobolt 06-MLD, Cobolt), and sCMOS (ORCA-Flash 4.0, Hamamatsu).
2.5.3. Image processing
All image preprocessing for the acquired data was performed for one detection channel. First, lateral illumination correction for each stripe was performed. We used a polynomial curve to fit the mean intensity perpendicular to the stripe and determined the correction coefficient. Then the stripes of each imaging plane were stitched using their spatial orientation and overlap between adjacent stripes. Finally, the stripes were stitched to form one image at the imaging plane for image storage in a 16-bit depth TIFF format using LZW compression. Image preprocessing was implemented using Matlab and C++. Image preprocesses for mouse brain data set at the voxel resolution of 1.30 × 1.30 × 0.92 μm were executed on a computing server (32 cores, 2.9 GHz per core).
Then, the data were downsampled to align with the Common Coordinate Framework version 3 (CCFv3) (https://download.alleninstitute.org/informatics-archive/current-release/mouse_ccf/) average template to perform high-precision registration via the Brain Registration, Identification, and Encoding Framework (BRIEF) (unpublished method) interface (Wang X, 2020). The down-sampled data were deformed using the registration parameters for the visualization of co-localization. The CCF annotation was further used to obtain an inverse transformation for the purpose of mapping the preprocessed data. The number of cells in each region could be counted by the full pipeline of BRIEF, including interactive 3D cell detection, accurate classification of cells through a convolutional neural network (CNN), and the computation of the cell location in the annotation.
2.5.4. Visualization and reconstruction
We visualized the data set using Amira software (v 2020.1, FEI, Me'rignac Cedex, France) and Imaris software (v.9.7.2, Bitplane, Switzerland) to generate the figures and movies. The preprocessed dataset was imported into Amira software using a desktop graphical workstation (7920 with one Intel Xeon Gold 6226R CPU, 512 GB memory, and an Nvidia GeForce RTX 3090 graphics card, Dell Inc., Round Rock, Texas, USA). The data format was transformed from TIFF to the native LDA type using Amira. The visualization process was as follows: extracting the data in the range of interest, sampling or interpolation, re-slicing the images, identifying the maximum intensity projection, volume, and surface rendering, and generating the movies using the main module of Amira.
2.6. Brain-wide c-Fos expression analysis
Brain c-Fos expression in the mouse brain was mapped at single-cell resolution using the brain-wide c-Fos fMOST procedure, which evaluates more than 250 anatomical regions from the mouse brain. The data are reported as bar graph representations of anatomical visualizations.
2.7. Statistical data analysis
The Student's t-test is presented with statistical significance indicated at the P < 0.01–0.001 level. All statistical analyses were performed using SPSS (17.0).
3. Results
3.1. The SD model induces anxiety-like and depression-like behaviors
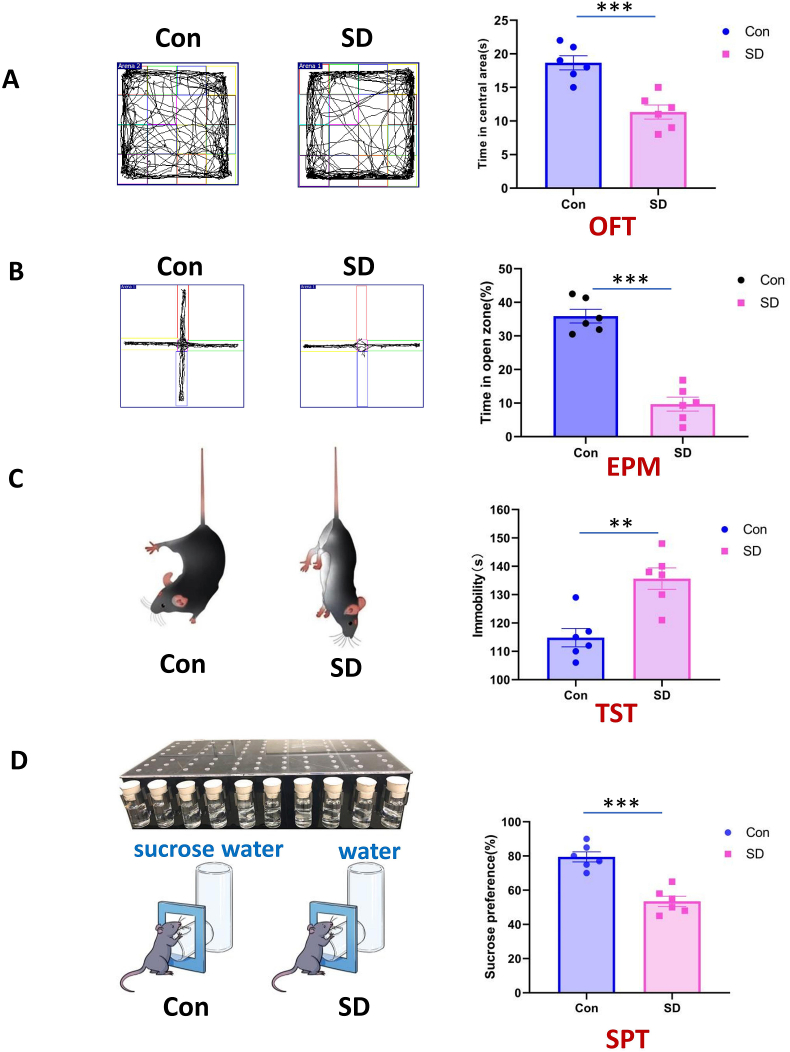
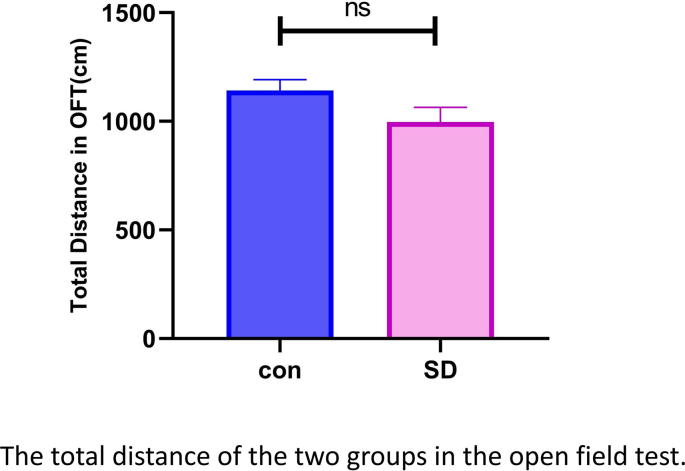
The anxiety-related exploratory behavior of mice was evaluated by the OFT and EPM after 14 days of SD. Compared with the control group mice, the SD group mice spent significantly less time in the center area of the box of OFT (Fig. 2A) and in the open arms of EPM (Fig. 2B). These data indicated that chronic SD mice exhibited increased anxiety-like behaviors. The total distance of OFT results showed no significant difference between the two groups (Supplementary Fig. 1), indicated that the motor function of SD mice remains normal.
Fig. 2.
Behavioral tests. (A) Compared with the control group, sleep deprived mice spent significantly less time in the center area of the open field test. (B) Compared with the control group, sleep deprived mice spent significantly less time in the open arms in the elevated plus-maze test. (C) Compared with the control group, sleep deprived mice spent more time immobile in tail suspension test. (D) Compared with the control group, sleep deprived mice showed significantly decreased consumption of sucrose in the sucrose preference tests. N = 6,Student's t-test. **P < 0.01, ***P < 0.001. Error bars indicate s.e.m.
The depression-like behavior of mice was evaluated by the TST and SPT after 14 days of SD. Compared with the control group, the SD group spent more time immobile in TST (Fig. 2C) and decreased consumption of sucrose in SPT (Fig. 2D). These results suggested that chronic SD mice exhibited depression-like symptoms.
3.2. Whole brain expression distribution of c-Fos positive cells after chronic sleep deprivation
Transfection efficiency of rAAV-PHP.eB-cFos-EYFP in the whole brain of mice was very good, and a representative GFP fluorescent image is shown in Supplementary Figs. 2 and 4. We tested the accuracy of rAAV-PHP.eB-cFos-EYFP by local injection and compared with antibodies expression. Our results showed that most of the c-fos antibody labeled cells were co-labeled with c-fos virus-labeled cells in medial prefrontal cortex (mPFC) after electric foot shocks (EFS)(see Supplementary Fig. 3). The c-Fos + cells were detected in a maximum of 230 brain regions (see Supplementary Table 1 and Videos 1–2 for the original data).
Supplementary video related to this article can be found at https://doi.org/10.1016/j.ynstr.2022.100478
The following is the supplementary data related to this article:
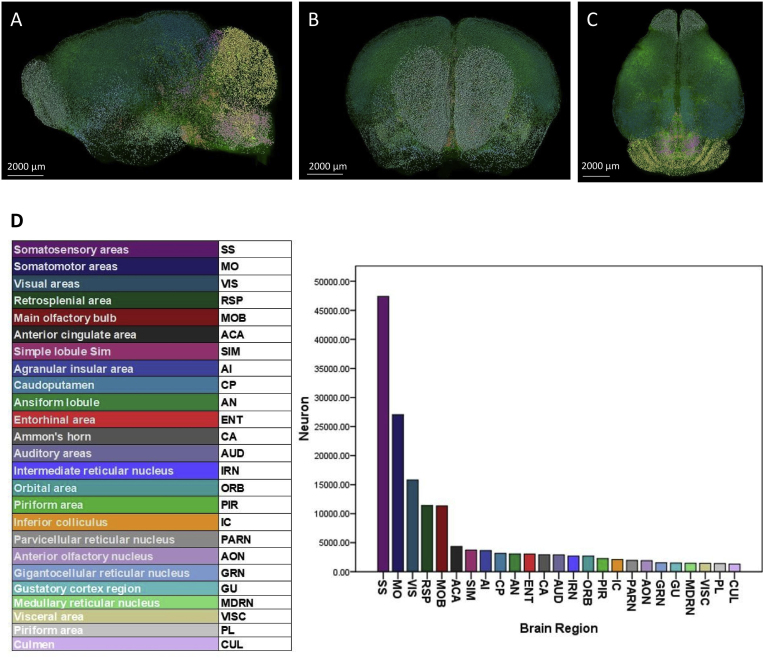
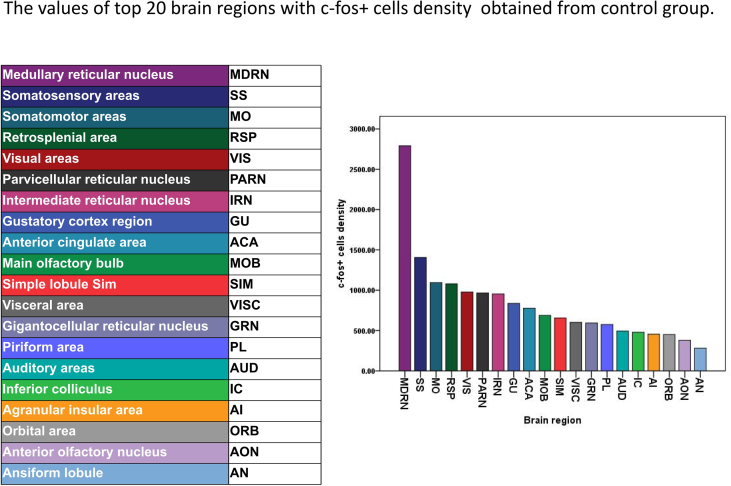
In the control group, the top 25 brain regions with more than 1000 c-Fos + cells included somatosensory areas (SS), somatomotor areas (MO), visual areas (VIS), retrosplenial area (RSP), main olfactory bulb (MOB), anterior cingulate area (ACA), simple lobule (SIM), agranular insular area (AI), caudoputamen (CP), ansiform lobule (AN), entorhinal area (ENT), Ammon's horn (CA), auditory areas (AUD), intermediate reticular nucleus (IRN), orbital area (ORB), piriform area (PIR), inferior colliculus (IC), parvicellular reticular nucleus (PARN), anterior olfactory nucleus (AON), gigantocellular reticular nucleus (GRN), gustatory areas (GU), medullary reticular nucleus (MDRN), visceral area (VISC), piriform area (PL) and culmen (CUL) (Fig. 3). In addition, the top 20 brain regions of c-Fos + density were MDRN, SS, MO, RSP, VIS, PARN, IRN, GU, ACA, MOB, SIM, VISC, GRN, PL, AUD, IC, AI, ORB, AON and AN, which was consistent with the top 25 brain regions with more than 1000 c-Fos + cells (Supplementary Fig. 5).
Fig. 3.
A-C were the images of c-Fos mapping from a representative animal in control group. (A) Sagittal plane; (B) Coronal plane; (C) Horizontal plane. Scale bar, 2000 μm. (D) The top 25 brain regions with more than 1000 c-Fos + expressing cells in the control group. N = 3,Student's t-test. Error bars indicate s.e.m.
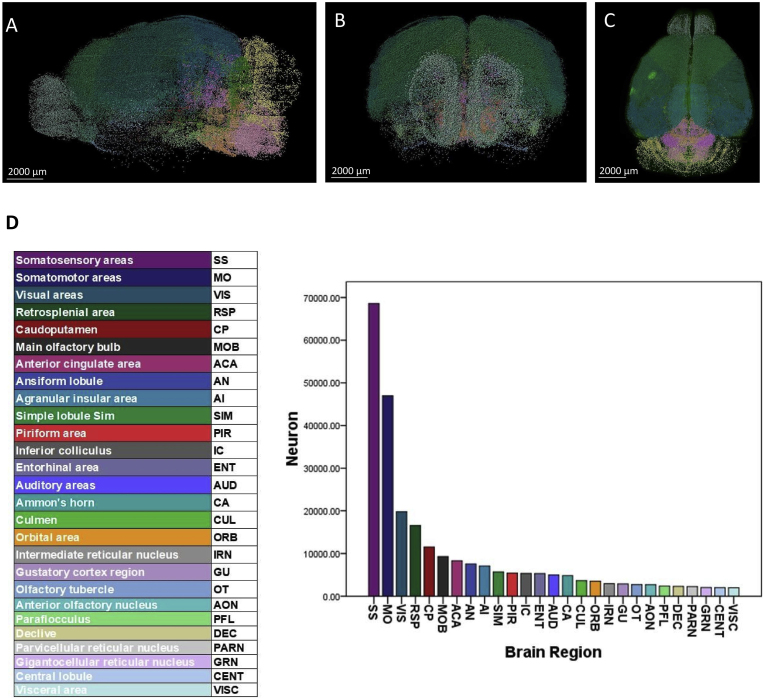
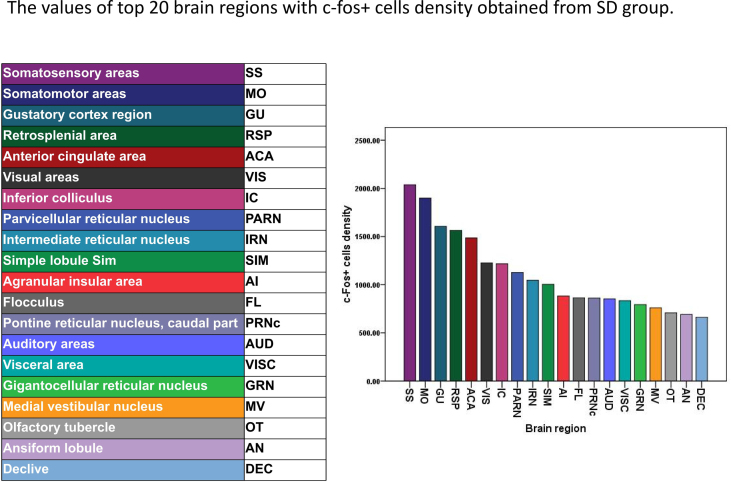
In the chronic SD group, there were 44 brain regions with more than 1000 c-Fos + cells. We show the top 27 brain regions with more than 2000 c-Fos + cells, including SS, MO, VIS, RSP, CP, MOB, ACA, AN, AI, SIM, PIR, IC, ENT, AUD, CA, CUL, ORB, IRN, GU, olfactory tubercle (OT), AON, paraflocculus (PFL), declive (DEC), PRAN, GRN, central lobule (CENT) and VISC (Fig. 4). The top 20 brain regions of c-Fos + density were SS, MO, GU, RSP, ACA, VIS, IC, PARN, IRN, SIM, AI, flocculus (FL), pontine reticular nucleus, caudal part (PRNc), AUD, VISC, GRN, medial vestibular nucleus (MV), OT, AN and DEC (Supplementary Fig. 6).
Fig. 4.
A-C were the images of c-Fos mapping from a representative animal in SD group. (A) Sagittal plane; (B) Coronal plane; (C) Horizontal plane. Scale bar, 2000 μm. (D) The top 27 brain regions with more than 2000 c-Fos + expressing cells in the chronic sleep deprivation group. N = 3,Student's t-test. Error bars indicate s.e.m.
3.3. The key brain regions displaying c-Fos expression difference after chronic sleep deprivation
The comparative analysis of c-Fos expression in the brains of the SD and control groups showed significant differences. There were 140 brain regions with c-Fos + cells. The most pronounced activation patterns (increased numbers of c-Fos + cells) were observed in the isocortex-cerebral cortex plate (CTXpl) area. Compared with the control group, there was a statistically significant increase in c-Fos + cells in certain brain areas in the chronic SD group, including RSP (P = 0.049), ACA (P = 0.030), AI (P = 0.005), GU (P = 0.013), parasubiculum (PAR) (P = 0.002), medial superior olivary complex (SOCm) (P = 0.029), retrorubral area in the midbrain reticular nucleus, (RR) (P = 0.034), and triangular nucleus of septum (TRS) (P = 0.039). Among them, RSP, ACA, AI, GU, and PAR were all located at CTXpl. TRS was located at the pallidum (PAL). The RR was located at the midbrain, motor related (MBmot). The SOCm was located at P (pons).
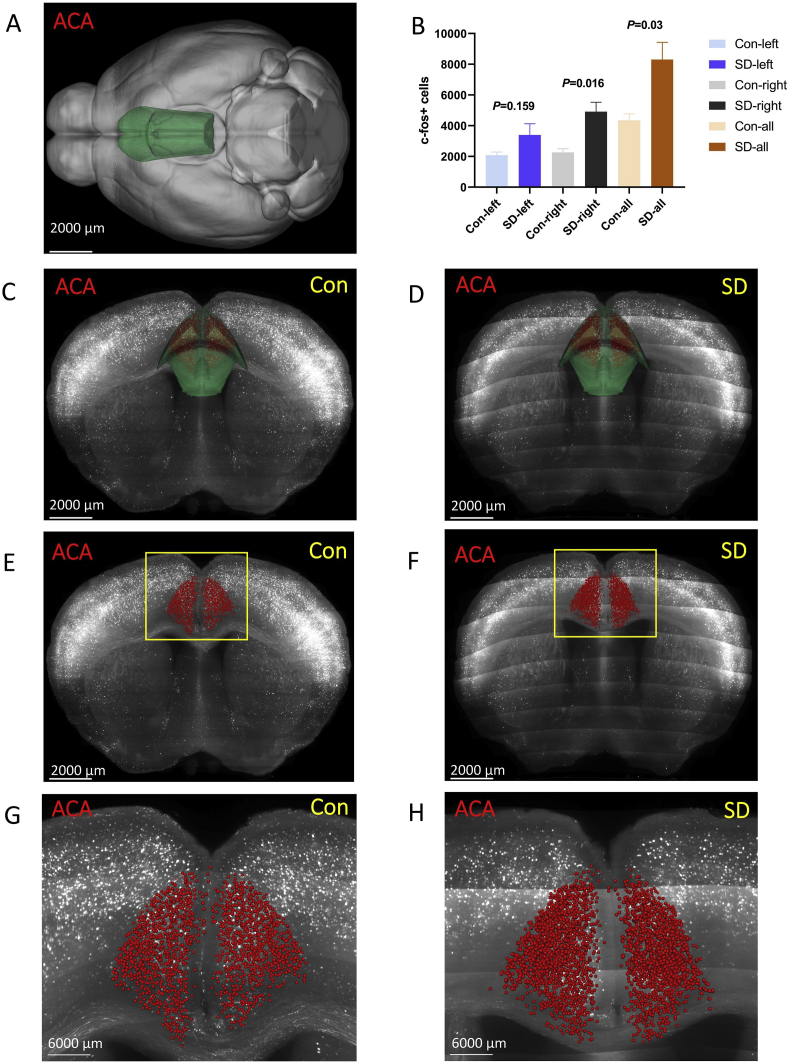
3.3.1. Anterior cingulate area (ACA)
The ACA was located at the isocortex (Fig. 5 A, C, and D). In general, the number of c-Fos + cells in the ACA of the SD group was higher than that in the control group (P = 0.03). Further analysis showed that there was no statistical difference between the two groups in the left ACA (P = 0.159), while the right ACA of the SD group increased significantly (P = 0.016) (Fig. 5 B, E-H).
Fig. 5.
The number of c-Fos + expressing cells in the anterior cingulate area (ACA).
(A, C, D) ACA was located at the isocortex. Green marks the ACA according to Allen Mouse Brain Common Coordinate Framework V3. (B, E, F) The number of c-Fos + cells in the ACA of the sleep deprivation group was more than that of the control group. Red marks the c-Fos + cells. (G, H) The enlarged view from (E, F). Scale bar: (A–F) 2000 μm, (G, H) 6000 μm. N = 3,Student's t-test. Error bars indicate s.e.m. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
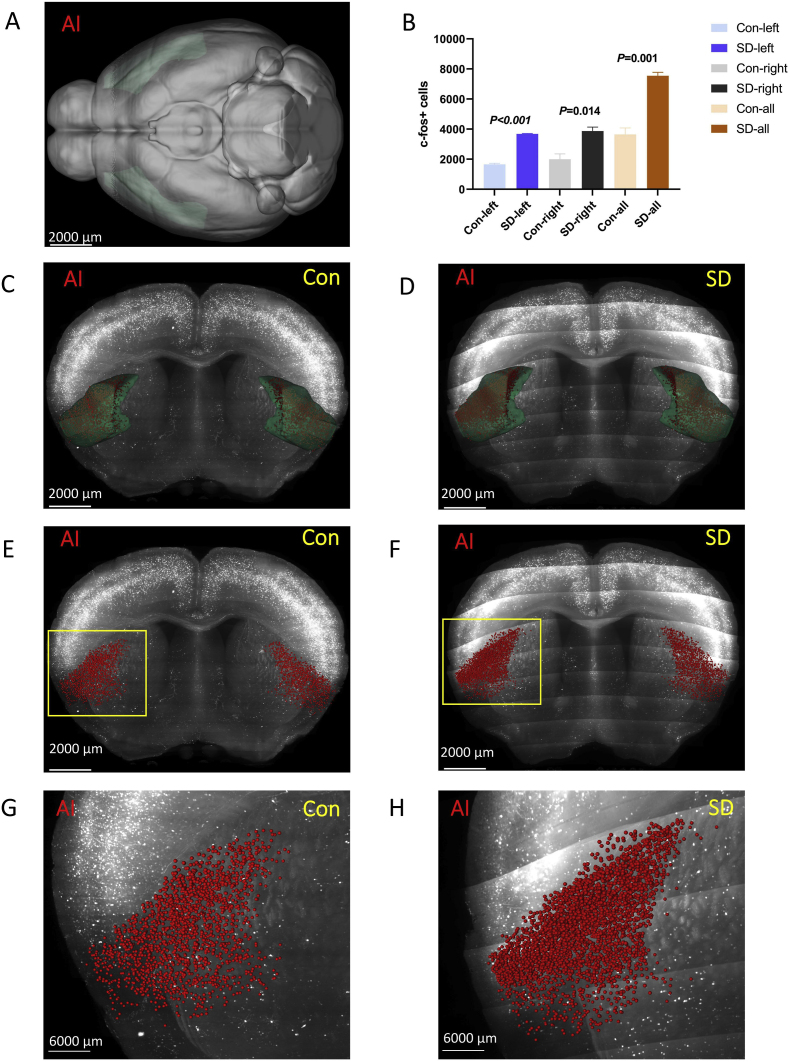
3.3.2. Agranular insular area (AI)
AI was located at the isocortex (Fig. 6 A, C, and D). In general, the number of c-Fos + cells in the AI of the SD group was higher than that in the control group (P = 0.001). Further analysis showed that there was a statistically significant difference between the two groups of left AI (P < 0.001) and right AI (P = 0.014). (Fig. 6 B, E-H).
Fig. 6.
The number of c-Fos + expressing cells in the agranular insular area (AI).
(A,C,D) AI was located at the isocortex (green). Green marks the AI according to Allen Mouse Brain Common Coordinate Framework V3. (B, E, F) The number of c-Fos + cells in the AI of the sleep deprivation group was more than that of the control group. Red denotes c-Fos + cells. (G, H) The enlarged view from (E, F). Scale bar: (A–F) 2000 μm, (G, H) 6000 μm. N = 3,Student's t-test. Error bars indicate s.e.m. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
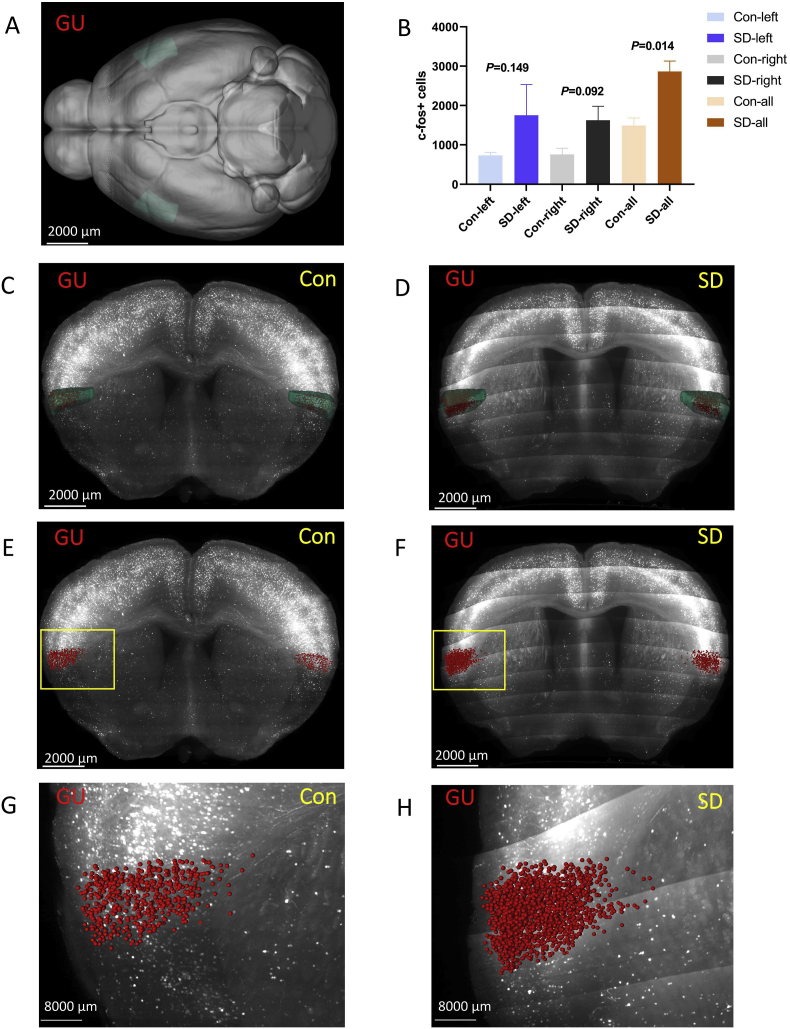
3.3.3. Gustatory areas (GU)
GU was located at the isocortex (Fig. 7 A, C, and D). In general, the number of c-Fos + cells in the GU of the SD group was higher than that of the control group (P = 0.014). Further analysis found that there was a statistically significant difference between the two groups of left GU(P = 0.149) and right GU (P = 0.092) (Fig. 7 B, E-H).
Fig. 7.
The number of c-Fos + expressing cells in gustatory areas (GU).
(A,C,D) GU was located at the isocortex. Green marks the GU according to Allen Mouse Brain Common Coordinate Framework V3. (B,E,F) The number of c-Fos + expressing cells in the GU of the sleep deprivation group was more than that of the control group. Red marks c-Fos + cells. (G, H) The enlarged view from (E, F). Scale bar: (A–F) 2000 μm, (G, H) 8000 μm. N = 3,Student's t-test. Error bars indicate s.e.m. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
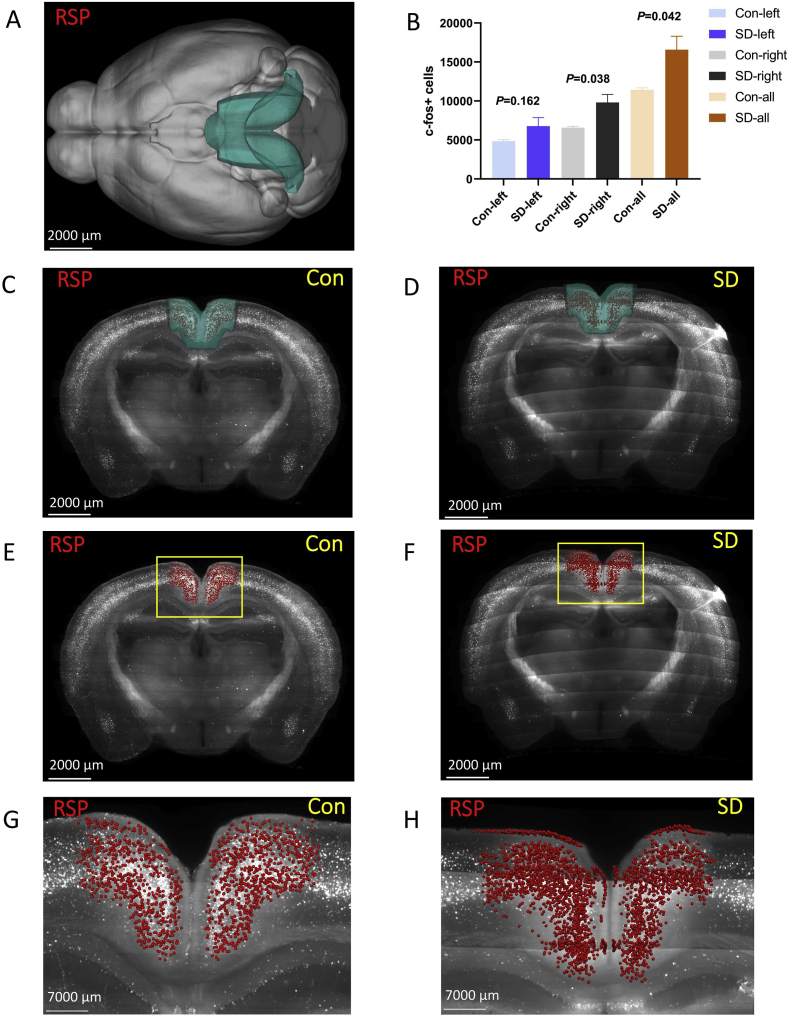
3.3.4. Retrosplenial area (RSP)
The RSP was located at the isocortex (Fig. 8 A, C, and D). In general, the number of c-Fos + cells in the RSP of the SD group was higher than that of the control group (P = 0.042). Further analysis revealed that there was no statistical difference between the two groups of left RSP (P = 0.162), while the right RSP of the SD group increased significantly (P = 0.038) (Fig. 8 B, E-H).
Fig. 8.
The number of c-Fos + expressing cells in the retrosplenial area (RSP).
(A,C,D). RSP was located at the isocortex. Green marks the RSP according to Allen Mouse Brain Common Coordinate Framework V3. (B,E,F) The number of c-Fos + expressing cells in the RSP of the sleep deprivation group was more than that of the control group. c-Fos + cells are red. (G, H) The enlarged view from (E, F). Scale bar: (A–F) 2000 μm, (G, H) 7000 μm. N = 3,Student's t-test. Error bars indicate s.e.m. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
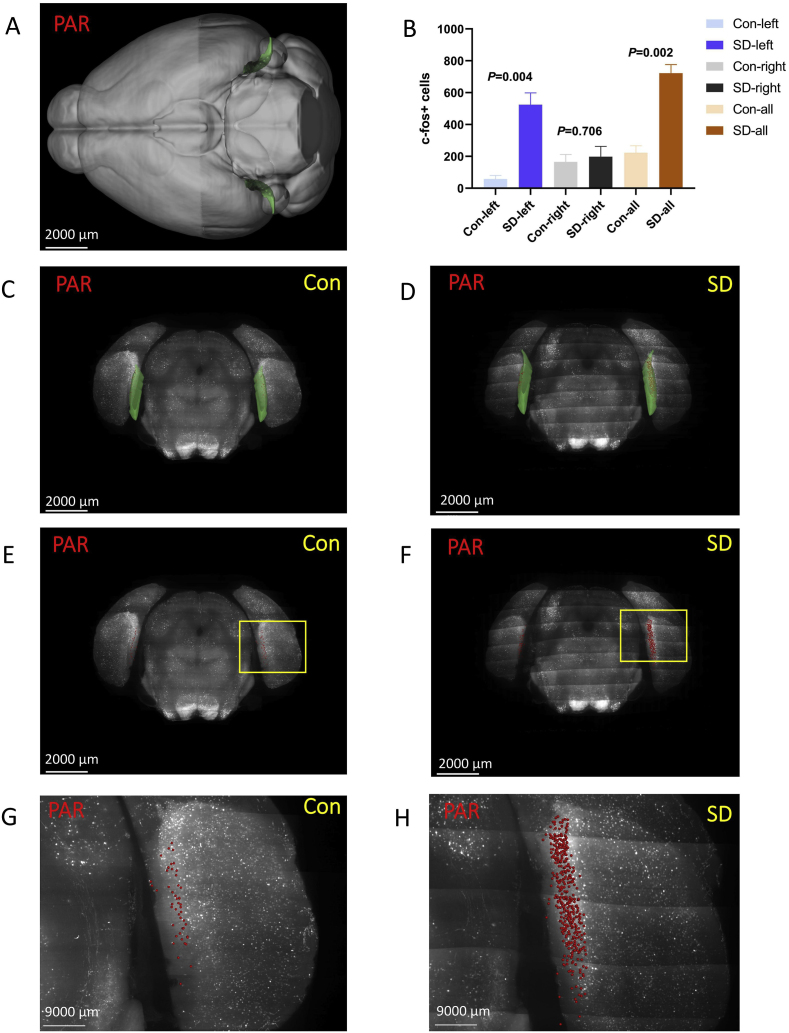
3.3.5. Parasubiculum (PAR)
PAR was located at the hippocampal formation (Fig. 9 A, C, and D). In general, the number of c-Fos + cells in the PAR of the SD group was higher than that of the control group (P = 0.002). Further analysis showed that there was no statistical difference between the two groups of right PAR (P = 0.706), while the left PAR of the SD group increased significantly (P = 0.004). (Fig. 9 B, E-H).
Fig. 9.
The number of c-Fos + expressing cells in the parasubiculum (PAR).
(A,C,D) PAR was located at the hippocampal formation. Green marks the PAR according to Allen Mouse Brain Common Coordinate Framework V3. (B,E,F) The number of c-Fos + cells in the PAR of the sleep deprivation group was more than that of the control group. The c-Fos + cells are red. (G, H) The enlarged view from (E, F). Scale bar: (A–F) 2000 μm, (G, H) 9000 μm. N = 3,Student's t-test. Error bars indicate s.e.m. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
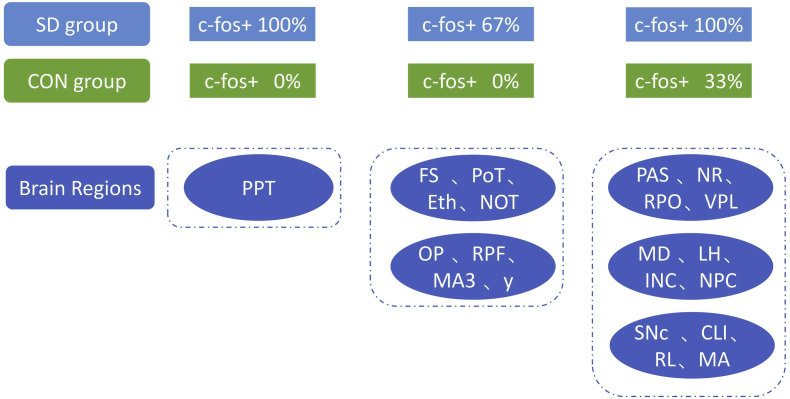
In addition, most of the brain areas showed a trend of increased numbers of c-Fos + cells in the chronic SD group, although the difference failed to reach statistical significance. We further compared the ratios of mouse models with c-Fos + expression in different brain regions of the two groups. The brain area with 100% c-Fos + expression in the mouse models in the SD group but 0% in the control group was PPT (posterior pretectal nucleus). The brain areas with 67% positive expression of c-Fos+ in the SD model mice but 0% positive in the control group included FS (fundus of striatum), PoT (posterior thalamic nuclear group, triangular part), Eth (ethmoid thalamic nucleus), NOT (nucleus of the optic tract), OP (ovary pretectal nucleus), RPF (retroparafascicular nucleus), MA3 (medial accessory oculomotor nucleus), and nucleus y. The brain regions with 100% c-Fos + expression in SD model mice but 33% in the control group included parasolitary nucleus (PAS), nucleus of Roller (NR), nucleus raphe pontis (RPO), ventral posterolateral nucleus of the thalamus (VPL), mediodorsal nucleus of the thalamus (MD), lateral habenula (LH), interstitial nucleus of Cajal (INC), nucleus of the posterior commissure (NPC), compact part of the substantia nigra (SNc), central linear nucleus raphe (CLI), rostral linear nucleus raphe (RL), and magnocellular nucleus (MA). (Fig. 10).
Fig. 10.
The comparison of the positive ratios of c-Fos expression in different brain regions between the sleep deprivation and control groups. (A) The brain areas with 100% c-Fos + expression in the sleep deprivation group but 0% in the control group. (B) The brain areas with 67% c-Fos + expression in the sleep deprivation group but 0% in the control group. (C) The brain regions with 100% c-Fos + expression in the sleep deprived mouse models but 33% in the controls. Abbreviations: PPT, posterior pretectal nucleus; FS, fundus of striatum; PoT, posterior thalamic nuclear group, triangular part; Eth, ethmoid thalamic nucleus; NOT, nucleus of the optic tract; OP, ovary pretectal nucleus; RPF, retroparafascicular nucleus; MA3, medial accessory oculomotor nucleus; PAS, parasolitary nucleus; NR, nucleus of Roller; RPO, nucleus raphe pontis; VPL, ventral posterolateral nucleus of the thalamus; MD, mediodorsal nucleus of the thalamus; LH, lateral habenula; INC, interstitial nucleus of Cajal; NPC, nucleus of the posterior commissure; SNc, compact part of the substantia nigra; CLI, central linear nucleus raphe; RL, rostral linear nucleus raphe; MA, magnocellular nucleus. Scale bar, 2000 μm.
4. Discussion
The current study investigated the accumulative effects of chronic REM sleep deprivation on the brain and behavior, and provided a comprehensive mapping of stress-activated brain regions in mice. Using the fMOST technology and AAV-PHP.eB, we described for the first time a clear map of the cumulative expression of c-Fos in the whole brain of a chronic REM SD model, which provides a wealth of knowledge for future studies.
Most previous studies have used a gentle SD model and analyzed the changes in c-Fos expression after SD in parts of brain regions. However, they failed to provide an accurate comprehensive view of c-Fos mapping in the whole brain. The brains were usually imaged one slice at a time, which is laborious and carries the risk of information loss (Cui et al., 2013). It is one of the most challenging research goals to elucidate information at the whole-brain level. By using a neurotropic virus, a gene containing a fluorescent protein can be transported into the nervous system. Fluorescent labeling of specific types of neurons can be achieved by combining a virus and recombinase system. Additionally, fMOST can achieve real-time imaging, which can feasibly image whole-brain distributions of specific cell types (Zhang et al., 2017). In this study, we used the fMOST system to map the brain-wide distribution of fluorescently labeled c-Fos positive cells and the quantitative cell data were obtained in 3D at the whole-brain level (Quan et al., 2013).
IEG are induced by various electrical or chemical signals to neural cells. And then their protein products act as transcription factors to regulate the expression of other genes. A remarkable finding in recent years is that the transition from sleep to wakefulness is accompanied by widespread activation of c-Fos (Cirelli and Tononi, 2000). Sleep-disrupted mice displayed significantly richer c-Fos expression compared to their non-sleep-disrupted counterparts. In the study by O'Hara et al., the cerebellum generally displayed the greatest increase in IEG mRNA levels in the sleep-deprived animals. The results also supported the hypothesis that brain regions may respond differentially to sleep deprivation (O'Hara et al., 1993). The data from published studies that focused mainly on the expression of c-Fos after sleep deprivation are summarized in Table 1. Montes-Rodríguez et al. found a significant increase in c-Fos in CTXpl in a total sleep deprivation model (Montes-Rodríguez et al., 2019). Consistent with previous studies, in our study, the most pronounced activation patterns (displaying increased numbers of c-Fos + cells) were also observed in the CTXpl area, including RSP, ACA, AI, GU, and PAR. The RSP is located behind the splenium, the most caudal part of the corpus callosum. The RSP supports spatial memory in conjunction with at least two other sites, the hippocampus and anterior thalamic nuclei (Vann et al., 2009). In addition, RSP function in rodents is not limited to spatial memory. A recent study showed the importance of RSP for the simultaneous processing of multiple stimuli. RSP dysfunction is likely to be a major contributor to cognitive deficits associated with mild cognitive impairment (MCI) and Alzheimer's disease (Villain et al., 2008). In addition, the ACA is a critical area in the processing of affective components of nociception in morphological, electrophysiological, neuroimaging, and behavioral studies (Zhuo, 2008). It is well known that a balance between excitation and inhibition of the cortical area, including the ACA, contributes to sleep-wake states (Brown et al., 2012). Zhang et al. found that 30 h SD could decrease the effective connection strength from the right insular lobe to the left anterior cingulate cortex (Zhang et al., 2021). Yamashita et al. observed that astrocytic activation in the ACA was critical for sleep disorder (Yamashita et al., 2014). These results all demonstrated the important role of ACA and RSP in sleep.
Table 1.
The summarization of previous studies that focused on the expression of c-Fos after sleep deprivation.
| Year | Journal | Animals | Stimuli methods | Test methods | Test level | Testing brain resions | Brain regions of significant increasing of c-fos |
|---|---|---|---|---|---|---|---|
| 1992 | Brain research | rats | auditory stimulation or sleep deprivation | Immunohistochemistry | Proteins | Brain stem | PPT,PB,SubC,NTS |
| 1993 | Sleep | rats | Cage rotation and cage tapping and gentle handling | Northern blots | mRNA | Thalamus, hypothalamus, cortex, cerebellum, pons | Pons, cerebellum |
| 1995 | Brain research | rats | auditory stimulation or sleep deprivation | Immunohistochemistry | Proteins | Brain stem, Diencephalon, Forebrain | SOL,PCR,PB,PPT,LDTG, SubCD, RD,SCH, LH, BLA |
| 1995 | J Sleep Res | rats | Gentle manipulation SD | in situ hybridization and immunocytochemistry | Proteins/mRNA | Cerebral Cortex, Basal Ganglia, Hypothlamus,Thalamus, Brainstem | cortical regions, MPO, PH, thalamic nulei and nuclei of the dorsal protine tegmentum |
| 1996 | Brain research | cats | Gentle manipulation SD and water tank technique | Immunohistochemistry | Proteins | Preoptic area; lateral hypothalamus; cingulate cortex, the colliculi, pontine grey | MPO and LPO |
| 2001 | Behavioural Brain Research | rats | Forced, slow locomotion by a wheel driven by electric motor | Immunoreactivity | Proteins | PVH | MPA,cortex,PVA,PVP,amygdala,cPu and LDTg |
| 2003 | Neuroscience | mouse | Gentle manipulation SD | Immunohistochemistry/RT-PCR | mRNA | cerebral cortex, basal forebrain, thalamus and cerebellum | cerebellum, cortex and thalamus |
| 2017 | journal of Neuroscience | rats | Early night SD | Immunohistochemistry | Proteins | SCN | dorsal and the ventral SCH |
| 2019 | Brain Research | rats | Gentle manipulation SD(24hTSD) | Immunohistochemistry | Proteins | hippocampus and amygdala | hippocampal CA1 and CA3 regions and the basolateral amygdala (BLA) |
SD, sleep deprivation; SCN,suprachiasmatic nuclei; LPO, lateral preoptic; MPO, medial preoptic; PH, posterior hypothalamic; PPT,pedunculo pontine tegmental nucleus; PB, parabrachial nucleus; SubC, subcoeruleus nucleus; SubCD, subcoeruleus (dorsal); NTS/SOL, solitary tract nucleus; PCR,parvocellular nucleus; LDTG, lateral dorsalis tegmental nucleus; RD, raphe dorsalis; SCH,suprachiasmatic nucleus; LH,lateral hypothalamic area; BLA, basolateral amygdaloid nucleus.
In addition, our study also found that c-Fos in GU and AI were significantly increased after chronic SD. The insula is known to be the primary gustatory cortex region. It plays a comprehensive role in processing sensory information and is the primary reception area in the cerebral cortex for interoceptive sensory information from the internal milieu. It has been suggested that insular abnormalities contributed to mental disorders that involve emotional dysregulation. Functional neuroimaging studies have found the abnormal grey matter volumes or hyperactivity of the insula in various disorders including schizophrenia, addiction, eating disorders, autism, and chronic pain (Gasquoine, 2014). In particular, insular hyper-reactivity has been considered as evidence in patients with anxiety and depressive disorders (Paulus and Stein, 2006). Previous study has established the relationship between neuroimaged activity levels in the insula and patient response to treatment. A better response to behavioral therapy was observed in regions in the anterior insula with lower levels of neuroimaging activity (McGrath et al., 2013).
In our results, the isocortex (ACA, AI, GU, RSP) and hippocampal formation showed an increase of cFos expression after REM sleep deprivation, which may be related to the activation of microglia and astrocyte. Bellesi et al. reported that the astrocytic phagocytosis in mouse frontal cortex increased after both acute and chronic sleep loss relative to sleep and wake (Bellesi et al., 2017). Chronic sleep restriction mice showed morphological signs of microglial activation and enhanced microglial phagocytosis of synaptic elements in mouse frontal cortex. In addition, Xue, et al. found the activation of microglia and astrocyte accompanied by down-regulation of α7 nicotinic acetylcholine receptor (α7-nAChR) and reduced activation of downstream PI3K/AKT/GSK-3β in the hippocampus of mice which were subjected to sleep deprivation for 7 days (Xue et al., 2019). The neuroinflammation in the brain may related to the increase in HPA axis activation and gut dysbiosis in SD mice. Alterations of the gut microbiome cause inflammatory response through impairments in the intestinal barrier. Because of an increase in blood–brain barrier permeability, peripheral inflammation and short-chain fatty acids further affect central immunity, contributing to an increase of inflammation in central nervous system (Wang et al., 2021).
Moreover, previous studies have shown that these activated brain areas play important roles in mental disorders including depression and anxiety as well. There is considerable evidence that ACA is involved in emotion. Stimulation in the subcallosal ACA has been widely used to treat depression (Rolls et al., 2019). In addition, it was suggested that ACA had features predictive of treatment outcome in depression (Pantazatos et al., 2020). And insula was related to the key symptoms in patients with depression by previous neuroimaging studies (Wu et al., 2021). The most robust findings were that psychological therapies for depression and anxiety resulted in decreased activation in the left ACC and left insula (Marwood et al., 2018). The study by Hogeveen J et al. Found the significant network of anterior insula (aINS) and RSP. And the aINA-RSP connectivity was associated with higher anxious-depressed symptoms in autism (Hogeveen et al., 2018). There results further supported that the activated brain regions in the current study was involved in chronic REM SD related depression and anxiety.
In total, we found a significant increase in c-Fos expression in eight brain regions. Although the difference failed to reach statistical significance in the rest of brain regions, an increased trend of c-Fos + cells was observed. The potential reasons for this phenomenon are as follows: First, we used fMOST as a measurement technique for detecting c-Fos + cells. In previous studies, immunohistochemistry and RT-PCR were used to detect the expression levels of c-Fos. However, fMOST allows the 3D imaging of neurons and neuronal processes and can simultaneously achieve imaging and sectioning. One of the important advantages of fMOST is that it can reflect the cumulative effect of chronic SD on the expression of c-Fos when combined with anterograde adeno-associated virus labeling, which is totally different from other research methods. This technical principle used a neurotropic virus to achieve nerve cell transfection, in which a gene containing a fluorescent protein can be carried into the nervous system (Beier et al., 2011). This characteristic could provide a cumulative expression level of c-Fos during the whole process of chronic SD, which is considered closer to reality. Second, as shown in Table 1, gentle manipulation SD or short time (24 h) total SD was used in most of the previous studies. It was considered that 24 h total SD could dramatically impair memory acquisition and consolidation. Currently, there is no consensus regarding the specific pattern of the SD model. The goal of the stimulus pattern is to maintain wakefulness in animals. It has been suggested that gentle, continuous handling is less stressful than other approaches to SD (Rechtschaffen et al., 1999). Forced, slow locomotion with short intervals has been considered a useful approach (Semba et al., 2001). Zhang et al. included 64 subjects and found that compared to 24 h SD, 30 h SD significantly decreased accuracy hit rates and induced larger waveform differences. In addition, Thompson et al. analyzed the signatures of SD on neuronal activity in the mouse brain using a short-term SD model, which could not reflect the cumulative effect of activation of brain regions (Thompson et al., 2010). In our study, we used a chronic SD model and the SD period lasted for 6 h per day and continued for 14 days, which induced the anxiety and depression-like behaviors in the OFT, EPM, SPT and TST. We considered that the long-term and chronic SD was closer to the situation of humans in real life.
Interestingly, most of the brain areas showed a trend of increased numbers of c-Fos + cells in the chronic SD group, although the difference failed to reach statistical significance. The brain area with 100% c-Fos + expression in the mouse models in the SD group but 0% in the control group was PPT. In two studies by Merchant-Nancy H et al., PPT was also found with significant increases in c-Fos + after SD (Merchant-Nancy et al., 1992). The brain areas with 67% positive expression of c-Fos+ in the SD model mice but 0% positive in the control group included FS, PoT, Eth, NOT, OP, RPF, MA3, and nucleus y. The brain regions with 100% c-Fos + expression in SD model mice but 33% in the control group included PAS, NR, RPO, VPL, MD, LH, INC, NPC, SNc, CLI, RL, and MA. Most of them were located at MBmot and TH. MBmot is an important area that regulates sleep/wakefulness through dopaminergic systems (Takata et al., 2018). And the previous studies have shown that TH had an important role in sleep onset, stabilization and termination (Gent et al., 2018).
In conclusion, the present study in mice revealed that the chronic sleep deprivation approach activated c-Fos expression in several brain regions by fMOST. Among the brain regions examined, CTXpl area including RSP, ACA, AI, GU, and PAR appeared to be the most sensitive regions after chronic SD. This atlas provides a wealth of knowledge for future studies. Understanding the functional differences of different brain regions in chronic REM sleep deprivation could shed light on the treatment of mental disorders associated with chronic SD.
Funding
This work was supported by Natural Science Foundation of China (Program No. 81900489 to YZ, No. 82071373 to JC, No. 82101294 to GC, No. 81730035 to SW), Natural Science Foundation of Shaanxi Province (Program No. 2022JM-456 to YZ); Shaanxi Provincial Key Research and Development Program (No. 2022SF-011 to GC).
CRediT authorship contribution statement
Guohong Cai: Investigation, Formal analysis, Writing – original draft. Yifan Lu: Investigation, FMOST, Formal analysis. Jing Chen: Writing – review & editing. Dingding Yang: Methodology. Ruixuan Yan: Formal analysis. Mudan Ren: Investigation. Shuixiang He: Writing – review & editing. Shengxi Wu: Conceptualization, Methodology, Writing – review & editing, Supervision. Yan Zhao: Conceptualization, Writing – review & editing, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank the members of our laboratories for helpful discussions and technical assistance with the experiments.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ynstr.2022.100478.
Contributor Information
Shengxi Wu, Email: shengxi@fmmu.edu.cn.
Yan Zhao, Email: yanzhao211@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. S1.
The total distance of the two group mice in the open field test. NS indicates no significant difference. N=6, Error bars indicate s.e.m.
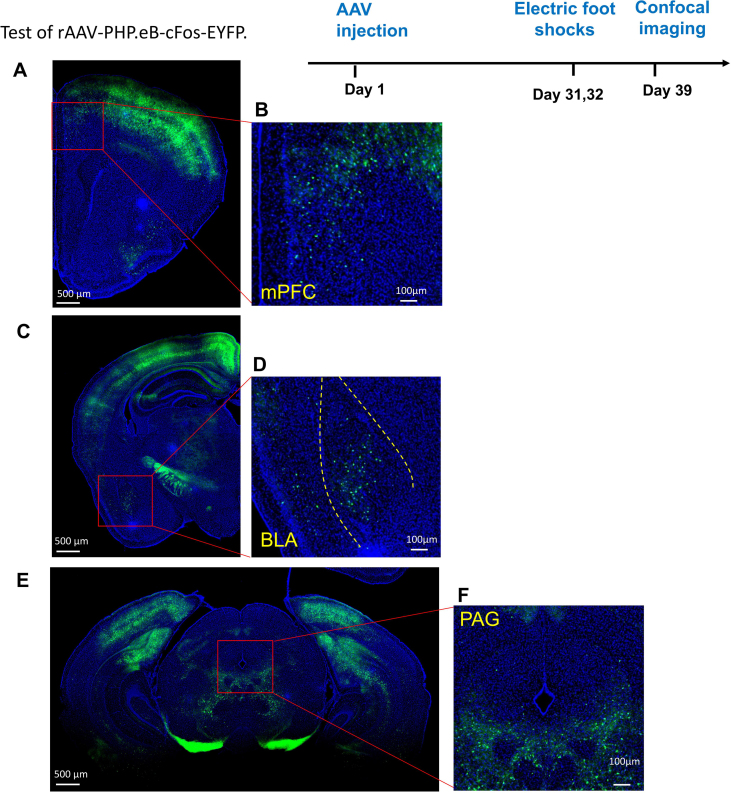
Fig. S2.
Test of rAAV-PHP.eB-cFos-EYFP (1).The test of rAAV-PHP.eB-cFos-EYFP was performed according to the electric foot shocks (EFS) experimental protocol which was provided in the study by Lin et al. In 2018. Briefly, four weeks after virus injection, mice were subjected to electrical foot shocks. Ten foot shocks of 0.8 mA intensity of 2s duration with random inter-shock intervals (30–60s) were delivered over 30 min to produce EFS stress. EFS experiments were performed once a day for 2 days. After the EFS experiment was completed, the mice were reared normally for a week, and then the mice were sacrificed, and the brain slices were cut for observation by Laser Scanning Confocal Microscope (OLYMPUS, FV3000). Consistent with their experimental results, our results showed that the c-Fos activation brain regions in response to EFS included (A,B) mPFC, (C,D) BLA and (E,F) PAG. (B, D, F) The enlarged view from (A, C, E). Scale bar: (A, C, E) 500μm, (B, D, F) 100μm.
Fig. S3.
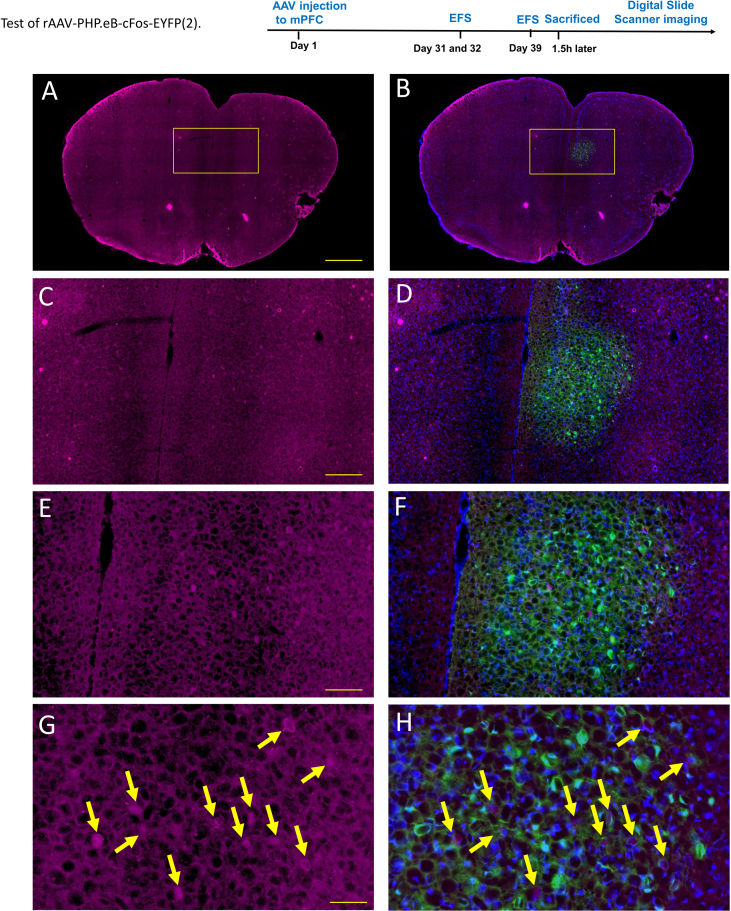
Test of rAAV-PHP.eB-cFos-EYFP (2).The test of rAAV-PHP.eB-cFos-EYFP was performed according to the electric foot shocks (EFS) experimental protocol which was provided in the study by Lin et al. In 2018. Briefly, four weeks after virus injection in mPFC, mice were subjected to electrical foot shocks. Ten foot shocks of 0.8 mA intensity of 2s duration with random inter-shock intervals (30–60s) were delivered over 30 min to produce EFS stress. EFS experiments were performed once a day for 2 days. After the EFS experiment was completed, the mice were reared normally for a week. Then the mice were subjected to EFS experiments again and the mice were sacrificed in 1.5 hours after EFS. The brain slices were cut for Immunofluorescence staining. Specifically, the brains were cut into 30μm-thick frontal sections on a freezing microtome. The sections were incubated overnight with Rabbit anti-c-Fos antibodies (1:2000, Cell Signaling Technology, #2250). After the sections were washed, they were incubated for 4h with Alexa Fluor 647-conjugated anti- rabbit IgG (1:500, Jackson, 711-605-152) to visualize c-Fos, which were observation by Digital Slide Scanner (OLYMPUS, VS200). The rAAV-PHP.eB-cFos-EYFP was successfully expressed in mPFC (B, D, F. Green). Consistent with previous experimental results, the results of the immunofluorescence experiments in our study showed that the c-Fos in the mPFC was activated in response to EFS (A, C, E. Purple). In addition, most of the c-fos antibody labeled cells were co-labeled with c-fos virus-labeled cells (G, H. Yellow arrow). Scale bar: (A, B) 1mm, (C, D)200μm, (E, F) 100μm, (G, H) 50μm. Blue: DAPI(1:1000, SIGMA, D9564).
Fig. S4.
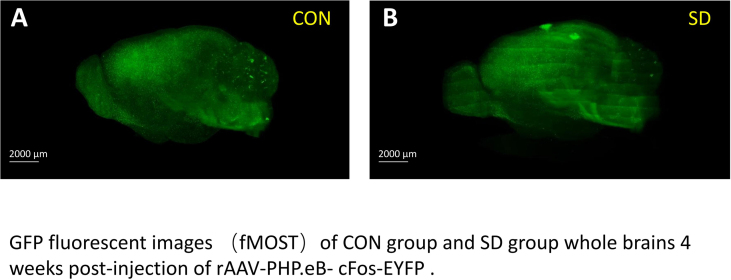
The GFP fluorescent image indicates the good transfection efficiency of rAAV-PHP.eB-cFos-EYFP in whole brains of mice. (A) GFP fluorescent images of whole brains 4 weeks after injection of rAAV-PHP.eB- cFos-EYFP in the control group. (B) GFP fluorescent images of whole brains 4 weeks after injection of rAAV-PHP.eB- cFos-EYFP in the chronic sleep deprivation group. Scale bar, 2000μm.
Fig. S5.
The top 20 brain regions of c-Fos+ density in the control group. The size of each brain region was based on Allen Mouse Brain Common Coordinate Framework V3.
Fig. S6.
The top 20 brain regions of c-Fos+ density in the chronic sleep deprivation group. The size of each brain region was based on Allen Mouse Brain Common Coordinate Framework V3.
Data availability
Data will be made available on request.
References
- Abrams R.M. Sleep deprivation. Obstet. Gynecol. Clin. N. Am. 2015;42:493–506. doi: 10.1016/j.ogc.2015.05.013. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A., Sigua N.L. What is sleep deprivation? Am. J. Respir. Crit. Care Med. 2019;199:P11–p12. doi: 10.1164/rccm.1996P11. [DOI] [PubMed] [Google Scholar]
- Beier K.T., Saunders A., Oldenburg I.A., Miyamichi K., Akhtar N., Luo L., Whelan S.P., Sabatini B., Cepko C.L. Anterograde or retrograde transsynaptic labeling of CNS neurons with vesicular stomatitis virus vectors. Proc. Natl. Acad. Sci. U.S.A. 2011;108:15414–15419. doi: 10.1073/pnas.1110854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellesi M., de Vivo L., Chini M., Gilli F., Tononi G., Cirelli C. Sleep loss promotes astrocytic phagocytosis and microglial activation in mouse cerebral cortex. J. Neurosci. : off. j. Soc. Neurosci. 2017;37:5263–5273. doi: 10.1523/JNEUROSCI.3981-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R.E., Basheer R., McKenna J.T., Strecker R.E., McCarley R.W. Control of sleep and wakefulness. Physiol. Rev. 2012;92:1087–1187. doi: 10.1152/physrev.00032.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G., Zhu Y., Chen J., Zhao S., Wang L., Wang M., Huang J., Wu S. Analysis of the gut microbiota and inflammatory factors in mGluR5-knockout mice. Front. Psychiatr. 2020;11:335. doi: 10.3389/fpsyt.2020.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C., Pompeiano M., Tononi G. Sleep deprivation and c-fos expression in the rat brain. J. Sleep Res. 1995;4:92–106. doi: 10.1111/j.1365-2869.1995.tb00157.x. [DOI] [PubMed] [Google Scholar]
- Cirelli C., Tononi G. On the functional significance of c-fos induction during the sleep-waking cycle. Sleep. 2000;23:453–469. [PubMed] [Google Scholar]
- Collaborators, Disease and Injury Incidence and Prevalence Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet (London, England) 2018;392:1789–1858. doi: 10.1016/S0140-6736(18)32279-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G., Jun S.B., Jin X., Pham M.D., Vogel S.S., Lovinger D.M., Costa R.M. Concurrent activation of striatal direct and indirect pathways during action initiation. Nature. 2013;494:238–242. doi: 10.1038/nature11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasquoine P.G. Contributions of the insula to cognition and emotion. Neuropsychol. Rev. 2014;24:77–87. doi: 10.1007/s11065-014-9246-9. [DOI] [PubMed] [Google Scholar]
- Gent T.C., Bassetti C., Adamantidis A.R. Sleep-wake control and the thalamus. Curr. Opin. Neurobiol. 2018;52:188–197. doi: 10.1016/j.conb.2018.08.002. [DOI] [PubMed] [Google Scholar]
- Goldstein-Piekarski A.N., Greer S.M., Saletin J.M., Harvey A.G., Williams L.M., Walker M.P. Sex, sleep deprivation, and the anxious brain. J. Cognit. Neurosci. 2018;30:565–578. doi: 10.1162/jocn_a_01225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogeveen J., Krug M.K., Elliott M.V., Solomon M. Insula-retrosplenial cortex overconnectivity increases internalizing via reduced insight in autism. Biol. Psychiatr. 2018;84:287–294. doi: 10.1016/j.biopsych.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha P.K., Bouâouda H., Gourmelen S., Dumont S., Fuchs F., Goumon Y., Bourgin P., Kalsbeek A., Challet E. Sleep deprivation and caffeine treatment potentiate photic resetting of the master circadian clock in a diurnal rodent. J. Neurosci. : off. j. Soc. Neurosci. 2017;37:4343–4358. doi: 10.1523/JNEUROSCI.3241-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killgore W.D. Effects of sleep deprivation on cognition. Prog. Brain Res. 2010;185:105–129. doi: 10.1016/B978-0-444-53702-7.00007-5. [DOI] [PubMed] [Google Scholar]
- Kim Y., Lee H.Y., Cho S.H. Antidepressant effects of pharmacopuncture on behavior and brain-derived neurotrophic factor (BDNF) expression in chronic stress model of mice. J. acupuncture and meridian stud. 2017;10:402–408. doi: 10.1016/j.jams.2017.08.007. [DOI] [PubMed] [Google Scholar]
- Knutson K.L., Van Cauter E., Rathouz P.J., DeLeire T., Lauderdale D.S. Trends in the prevalence of short sleepers in the USA: 1975-2006. Sleep. 2010;33:37–45. doi: 10.1093/sleep/33.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno A., Hirai H. Efficient whole brain transduction by systemic infusion of minimally purified AAV-PHP. eB. J. neurosci. methods. 2020;346 doi: 10.1016/j.jneumeth.2020.108914. [DOI] [PubMed] [Google Scholar]
- Ledoux L., Sastre J.P., Buda C., Luppi P.H., Jouvet M. Alterations in c-fos expression after different experimental procedures of sleep deprivation in the cat. Brain Res. 1996;735:108–118. doi: 10.1016/0006-8993(96)00599-9. [DOI] [PubMed] [Google Scholar]
- Li A., Gong H., Zhang B., Wang Q., Yan C., Wu J., Liu Q., Zeng S., Luo Q. Micro-optical sectioning tomography to obtain a high-resolution atlas of the mouse brain. Science. 2010;330:1404–1408. doi: 10.1126/science.1191776. New York, NY. [DOI] [PubMed] [Google Scholar]
- Liu H., Chen A. Roles of sleep deprivation in cardiovascular dysfunctions. Life Sci. 2019;219:231–237. doi: 10.1016/j.lfs.2019.01.006. [DOI] [PubMed] [Google Scholar]
- Marwood L., Wise T., Perkins A.M., Cleare A.J. Meta-analyses of the neural mechanisms and predictors of response to psychotherapy in depression and anxiety. Neurosci. Biobehav. Rev. 2018;95:61–72. doi: 10.1016/j.neubiorev.2018.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath C.L., Kelley M.E., Holtzheimer P.E., Dunlop B.W., Craighead W.E., Franco A.R., Craddock R.C., Mayberg H.S. Toward a neuroimaging treatment selection biomarker for major depressive disorder. JAMA Psychiatr. 2013;70:821–829. doi: 10.1001/jamapsychiatry.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant-Nancy H., Vázquez J., Aguilar-Roblero R., Drucker-Colín R. c-fos proto-oncogene changes in relation to REM sleep duration. Brain Res. 1992;579:342–346. doi: 10.1016/0006-8993(92)90072-h. [DOI] [PubMed] [Google Scholar]
- Montes-Rodríguez C.J., Rueda-Orozco P.E., Prospéro-García O. Total sleep deprivation impairs fear memory retrieval by decreasing the basolateral amygdala activity. Brain Res. 2019;1719:17–23. doi: 10.1016/j.brainres.2019.05.030. [DOI] [PubMed] [Google Scholar]
- Morgan J.I., Curran T. Stimulus-transcription coupling in the nervous system: involvement of the inducible proto-oncogenes fos and jun. Annu. Rev. Neurosci. 1991;14:421–451. doi: 10.1146/annurev.ne.14.030191.002225. [DOI] [PubMed] [Google Scholar]
- Murack M., Chandrasegaram R., Smith K.B., Ah-Yen E.G., Rheaume É., Malette-Guyon É., Nanji Z., Semchishen S.N., Latus O., Messier C., et al. Chronic sleep disruption induces depression-like behavior in adolescent male and female mice and sensitization of the hypothalamic-pituitary-adrenal axis in adolescent female mice. Behav. Brain Res. 2021;399 doi: 10.1016/j.bbr.2020.113001. [DOI] [PubMed] [Google Scholar]
- O'Hara B.F., Young K.A., Watson F.L., Heller H.C., Kilduff T.S. Immediate early gene expression in brain during sleep deprivation: preliminary observations. Sleep. 1993;16:1–7. doi: 10.1093/sleep/16.1.1. [DOI] [PubMed] [Google Scholar]
- Okun M.L., Mancuso R.A., Hobel C.J., Schetter C.D., Coussons-Read M. Poor sleep quality increases symptoms of depression and anxiety in postpartum women. J. Behav. Med. 2018;41:703–710. doi: 10.1007/s10865-018-9950-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazatos S.P., Yttredahl A., Rubin-Falcone H., Kishon R., Oquendo M.A., John Mann J., Miller J.M. Depression-related anterior cingulate prefrontal resting state connectivity normalizes following cognitive behavioral therapy. Eur. Psychiatr. : j. Assoc. Eur. Psychiatr. 2020;63:e37. doi: 10.1192/j.eurpsy.2020.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulus M.P., Stein M.B. An insular view of anxiety. Biol. Psychiatr. 2006;60:383–387. doi: 10.1016/j.biopsych.2006.03.042. [DOI] [PubMed] [Google Scholar]
- Pires G.N., Bezerra A.G., Tufik S., Andersen M.L. Effects of acute sleep deprivation on state anxiety levels: a systematic review and meta-analysis. Sleep Med. 2016;24:109–118. doi: 10.1016/j.sleep.2016.07.019. [DOI] [PubMed] [Google Scholar]
- Pompeiano M., Cirelli C., Arrighi P., Tononi G. c-Fos expression during wakefulness and sleep. Neurophysiol. clinique = Clin. neurophysiol. 1995;25:329–341. doi: 10.1016/0987-7053(96)84906-9. [DOI] [PubMed] [Google Scholar]
- Quan T., Zheng T., Yang Z., Ding W., Li S., Li J., Zhou H., Luo Q., Gong H., Zeng S. NeuroGPS: automated localization of neurons for brain circuits using L1 minimization model. Sci. Rep. 2013;3:1414. doi: 10.1038/srep01414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechtschaffen A., Bergmann B.M., Gilliland M.A., Bauer K. Effects of method, duration, and sleep stage on rebounds from sleep deprivation in the rat. Sleep. 1999;22:11–31. doi: 10.1093/sleep/22.1.11. [DOI] [PubMed] [Google Scholar]
- Rolls E.T., Cheng W., Gong W., Qiu J., Zhou C., Zhang J., Lv W., Ruan H., Wei D., Cheng K., et al. Functional connectivity of the anterior cingulate cortex in depression and in Health. Cerebr. Cortex. 2019;29:3617–3630. doi: 10.1093/cercor/bhy236. New York, NY : 1991. [DOI] [PubMed] [Google Scholar]
- Sagar S.M., Sharp F.R. Early response genes as markers of neuronal activity and growth factor action. Adv. Neurol. 1993;59:273–284. [PubMed] [Google Scholar]
- Scott K.A., Ida M., Peterson V.L., Prenderville J.A., Moloney G.M., Izumo T., Murphy K., Murphy A., Ross R.P., Stanton C., et al. Revisiting Metchnikoff: age-related alterations in microbiota-gut-brain axis in the mouse. Brain Behav. Immun. 2017;65:20–32. doi: 10.1016/j.bbi.2017.02.004. [DOI] [PubMed] [Google Scholar]
- Semba K., Pastorius J., Wilkinson M., Rusak B. Sleep deprivation-induced c-fos and junB expression in the rat brain: effects of duration and timing. Behav. Brain Res. 2001;120:75–86. doi: 10.1016/s0166-4328(00)00362-4. [DOI] [PubMed] [Google Scholar]
- Suchecki D., Tufik S. Social stability attenuates the stress in the modified multiple platform method for paradoxical sleep deprivation in the rat. Physiol. Behav. 2000;68:309–316. doi: 10.1016/s0031-9384(99)00181-x. [DOI] [PubMed] [Google Scholar]
- Takata Y., Oishi Y., Zhou X.Z., Hasegawa E., Takahashi K., Cherasse Y., Sakurai T., Lazarus M. Sleep and wakefulness are controlled by ventral medial midbrain/pons GABAergic neurons in mice. J. Neurosci. : off. j. Soc. Neurosci. 2018;38:10080–10092. doi: 10.1523/JNEUROSCI.0598-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terao A., Greco M.A., Davis R.W., Heller H.C., Kilduff T.S. Region-specific changes in immediate early gene expression in response to sleep deprivation and recovery sleep in the mouse brain. Neuroscience. 2003;120:1115–1124. doi: 10.1016/s0306-4522(03)00395-6. [DOI] [PubMed] [Google Scholar]
- Thompson C.L., Wisor J.P., Lee C.K., Pathak S.D., Gerashchenko D., Smith K.A., Fischer S.R., Kuan C.L., Sunkin S.M., Ng L.L., et al. Molecular and anatomical signatures of sleep deprivation in the mouse brain. Front. Neurosci. 2010;4:165. doi: 10.3389/fnins.2010.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vann S.D., Aggleton J.P., Maguire E.A. What does the retrosplenial cortex do? Nat. Rev. Neurosci. 2009;10:792–802. doi: 10.1038/nrn2733. [DOI] [PubMed] [Google Scholar]
- Villain N., Desgranges B., Viader F., de la Sayette V., Mézenge F., Landeau B., Baron J.C., Eustache F., Chételat G. Relationships between hippocampal atrophy, white matter disruption, and gray matter hypometabolism in Alzheimer's disease. J. Neurosci. : off. j. Soc. Neurosci. 2008;28:6174–6181. doi: 10.1523/JNEUROSCI.1392-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Z Y., Wang X., Dai J., Hua R., Zeng S., Li H. Anxiety-related cell-type-specific neural circuits in the anterior-dorsal bed nucleus of the stria terminalis. Sci. Bull. 2020;65:1203–1216. doi: 10.1016/j.scib.2020.03.028. [DOI] [PubMed] [Google Scholar]
- Wang Z., Chen W.H., Li S.X., He Z.M., Zhu W.L., Ji Y.B., Wang Z., Zhu X.M., Yuan K., Bao Y.P., et al. Gut microbiota modulates the inflammatory response and cognitive impairment induced by sleep deprivation. Mol. Psychiatr. 2021;26:6277–6292. doi: 10.1038/s41380-021-01113-1. [DOI] [PubMed] [Google Scholar]
- Wu H., Zheng Y., Zhan Q., Dong J., Peng H., Zhai J., Zhao J., She S., Wu C. Covariation between spontaneous neural activity in the insula and affective temperaments is related to sleep disturbance in individuals with major depressive disorder. Psychol. Med. 2021;51:731–740. doi: 10.1017/S0033291719003647. [DOI] [PubMed] [Google Scholar]
- Xue R., Wan Y., Sun X., Zhang X., Gao W., Wu W. Nicotinic mitigation of neuroinflammation and oxidative stress after chronic sleep deprivation. Front. Immunol. 2019;10:2546. doi: 10.3389/fimmu.2019.02546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., Hamada A., Suhara Y., Kawabe R., Yanase M., Kuzumaki N., Narita M., Matsui R., Okano H., Narita M. Astrocytic activation in the anterior cingulate cortex is critical for sleep disorder under neuropathic pain. Synapse. 2014;68:235–247. doi: 10.1002/syn.21733. New York, NY. [DOI] [PubMed] [Google Scholar]
- Yang X., Zhang Q., Huang F., Bai K., Guo Y., Zhang Y., Li N., Cui Y., Sun P., Zeng S., et al. High-throughput light sheet tomography platform for automated fast imaging of whole mouse brain. J. Biophot. 2018;11 doi: 10.1002/jbio.201800047. [DOI] [PubMed] [Google Scholar]
- Yin C.Y., Li L.D., Xu C., Du Z.W., Wu J.M., Chen X., Xia T., Huang S.Y., Meng F., Zhang J., et al. A novel method for automatic pharmacological evaluation of sucrose preference change in depression mice. Pharmacol. Res. 2021;168 doi: 10.1016/j.phrs.2021.105601. [DOI] [PubMed] [Google Scholar]
- Yu J., Rawtaer I., Fam J., Jiang M.J., Feng L., Kua E.H., Mahendran R. Sleep correlates of depression and anxiety in an elderly Asian population. Psychogeriatrics : Off. j. Japan. Psychogeriatr. Soc. 2016;16:191–195. doi: 10.1111/psyg.12138. [DOI] [PubMed] [Google Scholar]
- Zhang C., Yan C., Ren M., Li A., Quan T., Gong H., Yuan J. A platform for stereological quantitative analysis of the brain-wide distribution of type-specific neurons. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-14699-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Shao Y., Jin X., Cai X., Du F. Decreased effective connectivity between insula and anterior cingulate cortex during a working memory task after prolonged sleep deprivation. Behav. Brain Res. 2021;409 doi: 10.1016/j.bbr.2021.113263. [DOI] [PubMed] [Google Scholar]
- Zhuo M. Cortical excitation and chronic pain. Trends Neurosci. 2008;31:199–207. doi: 10.1016/j.tins.2008.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.