Abstract
Background
Evaluation of tumor-tissue images stained with hematoxylin and eosin (H&E) is pivotal in diagnosis, yet only a fraction of the rich phenotypic information is considered for clinical care. Here, we propose a survival deep learning (SDL) framework to extract this information to predict glioma survival.
Methods
Digitized whole slide images were downloaded from The Cancer Genome Atlas (TCGA) for 766 diffuse glioma patients, including isocitrate dehydrogenase (IDH)-mutant/1p19q-codeleted oligodendroglioma, IDH-mutant/1p19q-intact astrocytoma, and IDH-wildtype astrocytoma/glioblastoma. Our SDL framework employs a residual convolutional neural network with a survival model to predict patient risk from H&E-stained whole-slide images. We used statistical sampling techniques and randomized the transformation of images to address challenges in learning from histology images. The SDL risk score was evaluated in traditional and recursive partitioning (RPA) survival models.
Results
The SDL risk score demonstrated substantial univariate prognostic power (median concordance index of 0.79 [se: 0.01]). After adjusting for age and World Health Organization 2016 subtype, the SDL risk score was significantly associated with overall survival (OS; hazard ratio = 2.45; 95% CI: 2.01 to 3.00). Four distinct survival risk groups were characterized by RPA based on SDL risk score, IDH status, and age with markedly different median OS ranging from 1.03 years to 14.14 years.
Conclusions
The present study highlights the independent prognostic power of the SDL risk score for objective and accurate prediction of glioma outcomes. Further, we show that the RPA delineation of patient-specific risk scores and clinical prognostic factors can successfully demarcate the OS of glioma patients.
Keywords: digital pathology, glioma, H&E images, risk stratification, survival deep learning
Key Points.
The survival deep learning (SDL) risk score can predict patient-specific survival from whole slide images and the prediction accuracy exceeds other approaches.
An interaction between IDH status, SDL risk score, and age can delineate significantly different survival risk groups within glioma subtypes.
Importance of the Study.
Current pathologic evaluation of hematoxylin and eosin tumor tissue images focuses on only a small amount of the rich phenotypic information available. Here, we developed a deep learning approach to extract all additional information from the images to predict overall survival across distinct molecular subtypes of glioma patients. Our integrated survival deep learning framework has substantial prognostic power and combined with isocitrate dehydrogenase (IDH)-status and age can delineate significantly different survival risk groups. Interestingly, these groups identify higher-risk IDH-wildtype astrocytomas as well as lower risk IDH-wildtype glioblastomas and separate IDH-mutant subgroups with varying survival. The ability of a computational approach to histologic images to capture diverse, clinically relevant information may facilitate a more personalized patient evaluation in the neuro-oncology clinic.
A pathologist’s examination of tumor tissue stained with hematoxylin and eosin (H&E) is an important component of the decision-making process in oncology. The phenotypic information present in histology slides contains data on tumor aggressiveness and markers of disease progression that are crucial for prognostication.1 Historically, histologic grading of diffuse glioma was the clinical gold standard to determine the course of treatment or the need for additional testing, such as molecular profiling.2 More recently, molecular alterations identified in specific subsets of diffuse glioma, including 1p/19q-codeletion, EGFR amplifications, and isocitrate dehydrogenase 1/2 (IDH) mutations, informed major revisions, and the emergence of molecular subtyping in glioma.3–5 In 2016, the World Health Organization (WHO) identified several new entities of diffuse glioma based on genetic and epigenetic alterations in addition to the histologic phenotypes of tumors,6,7 and even greater integration of molecular information for diagnosis is incorporated in the fifth edition of the WHO Classification of Tumors of the Central Nervous System.8
Although research on the molecular determinants of glioma is ongoing, microscopic analysis of H&E-stained tumor tissue can reveal many characteristics of the disease and plays a critical role in the diagnosis and treatment of diffuse glioma.9 Such features include proliferation, nuclear and cellular atypia, vascular features, tumor cell infiltration, and extent of necrosis. However, diagnostic interpretation of histopathology images depends on the manual assessment of stained slides, which can be time-consuming and subject to inter-pathologist variability.1,10,11 The emergence of computational analysis of histological imaging has received significant attention. With the recent boost in artificial intelligence, an increasing number of methods have been developed to leverage the state-of-the-art deep learning techniques for the automatic classification of tumor subtypes, identification of metastases, and nuclei segmentation.12–18 Specifically, deep convolutional neural networks (CNNs) have become the de-facto standard in histopathological image analysis with performance on par with human experts for diagnostic tasks such as tumor detection and histologic grading.14,19,20
Several prior studies have implemented deep learning to address survival prediction. For instance, Faraggi and Simon17 introduced the first approach to combined Cox proportional hazards (CoxPH) with neural networks in 1995. And in 2016, Yousefi et al.18 built upon Faraggi and Simon’s work to combined the CoxPH model with more modern artificial learning techniques. More recently, Katzman et al.21 introduced, DeepSurv, a CoxPH-based deep neural network to predict the survival rate based solely on structured clinical data without leveraging histopathology images. DeepConvSurv, a similar approach by Zhu et al.,22 uses a modified deep CNN on whole slide images (WSIs) to predict survival outcomes and achieved marginally better performance (concordance index [c-index] of 0.62 for lung cancer) than DeepSurv (c-index of 0.60). Mobadersany et al.23 developed Survival CNNsto predict patient survival outcomes using high power fields extracted from different regions of interest (ROI) that showed superior performance in predicting survival compared to the conventional CoxPH model; however, this study was limited by requiring subjective interpretation to define risk group thresholds, limiting its application in the clinical setting. Chen et al.16 recently introduced Pathomic Fusion to allow the combination of histology and genomic features for survival prediction.
Despite the recent success in the application of deep learning in predicting survival outcomes from histopathology images, these techniques have not yet made a clinical impact by providing the necessary prognostic interpretation for cancer patients. One clinically relevant goal of prognostic models is risk stratification. Risk stratification for glioma patients is critical as it can help tailor treatments to reduce aggressive therapeutic regimens for low-risk patients while increasing the likelihood of those regimens in high-risk patients. While prior studies have emphasized determining complex interactions between histologic characteristics, clinical data, and molecular biomarkers,16,23–25 here we present a more practical and rigorous approach to understanding these interactions. We hypothesize that integrating deep learning-based patient survival outcomes with prognostic molecular and clinical covariates delineates patients into more homogenous risk groups and improves predictive accuracy necessary for the clinical management of gliomas.
In this study, we extend on the previously published work22,23 by exploring deep learning with a transfer learning technique26 for survival outcome prediction using images from H&E-stained tumor tissue and propose a clinically significant risk stratification model for diffuse gliomas. While previously published work has focused on identifying risk groups in the distribution of patient-specific survival outcomes,23,27–30 our study takes us one step closer to mapping out the relationship between deep learning-based patient outcomes and prognostic clinical and molecular parameters.
Materials and Methods
Data Cohort
Digitized WSIs from diagnostic formalin-fixed paraffin-embedded specimens stained with H&E were obtained from The Cancer Genome Atlas (TCGA) along with clinical information accessed via the Genomic Data Commons Data Portal (https://gdc.cancer.gov). The dataset contained a total of 1061 whole-slide images from 769 unique patients from TCGA-Glioblastoma Multiforme (GBM) and TCGA-Low Grade Glioma (LGG) cohorts. These images were classified based on the 2016 WHO paradigm that stratifies diffuse gliomas based on phenotypic and molecular genetic features such as IDH1/ IDH2 gene mutation status and 1p19q chromosome co-deletion status. Tumor subtypes include IDH-mutant and 1p/19q-codeleted oligodendroglioma, IDH-mutant/-wildtype astrocytoma, IDH-mutant/-wildtype glioblastoma. Additional information on overall survival (OS), clinical, and molecular biomarkers for the patient cohort were obtained from the cBioPortal for Cancer Genomics website (https://www.cbioportal.org/). Data were ascertained in accordance with the World Medical Association Declaration of Helsinki. Three patients were excluded from the initial set of 769 patients as they had missing survival information and conflicting IDH status. A summary of the dataset is provided in Table 1.
Table 1.
Summary of Patient Characteristics
| TCGA Cohort (N = 766) | Hazard Ratio | 95% CI | P | |
|---|---|---|---|---|
| Clinical and Demographics Variables | ||||
| Sex | ||||
| Female | 307 (41.8%) | — | ||
| Male | 427 (58.2%) | 1.12 | (0.92,1.38) | .262 |
| Age at diagnosis | ||||
| Mean (SD) | 49.7 (15.4) | 1.06 | (1.05, 1.07) | < .001 |
| Median | 51.0 | |||
| Q1, Q3 | 37.0, 61.0 | |||
| Range | 10.0–88.0 | |||
| Grade | ||||
| II | 181 (24.7%) | — | ||
| III | 205 (27.9%) | 2.98 | (1.84, 4.82) | < .001 |
| IV | 348 (47.4%) | 14.92 | (9.72, 22.91) | < .001 |
| WHO classification | ||||
| IDH-mutant astrocytoma | 203 (29.0%) | — | ||
| IDH-mutant GBM | 20 (2.9%) | 4.58 | (2.52, 8.34) | < .001 |
| IDH-mutant oligodendroglioma | 141 (20.1%) | 0.65 | (0.35,1.22) | .183 |
| IDH-wildtype astrocytoma | 72 (10.3%) | 5.66 | (3.52, 9.10) | < .001 |
| IDH-wildtype gbm | 264 (37.7%) | 11.90 | (8.22, 17.23) | < .001 |
| IDH status | ||||
| Mutant | 364 (52.0%) | — | ||
| Wildtype | 336 (48.0%) | 9.45 | (7.14, 12.50) | < .001 |
| ATRX status | ||||
| Mutant | 162 (21.1%) | — | ||
| Wildtype | 409 (53.4%) | 2.74 | (1.91, 3.93) | < .001 |
| 1p19q status | ||||
| Codeleted | 142 (18.5%) | — | ||
| Non-codeleted | 620 (80.9%) | 7.75 | (4.62, 13.00) | < .001 |
Abbreviations: IDH, isocitrate dehydrogenase 1 or 2 gene; ATRX, α-thalassemia, mental retardation, X-linked protein; 1p19q, deletion status of short arm of chromosome 1 and long arm of chromosome 19; GBM, glioblastoma multiforme.
Data Preparation
Due to the high dimensionality and gigapixel resolution of WSIs, the proposed model was trained on multiple ROI extracted from H&E-stained slides.23 These ROIs (1024 × 1024) were extracted at 20× magnification using Openslide software and accounted for all artifacts such as air bubbles, blurry regions, and folds.23 Data augmentation techniques such as morphological rotation along the center (90°, 180°, 270°), vertical and horizontal mirroring, and image scaling were applied to each ROI to accommodate limited cohort data, color variation, and image artifact. Furthermore, we also employed color augmentation using the transformation of brightness, hue, and contrast to adjust pixel-level image values.
Workflow
Our proposed integrated survival deep learning framework uses a pretrained CNN)model to extract visual features from ROIs. These high-level image-derived features are aggregated by a fully connected layer and global pooling strategy and then introduced to a final CoxPH layer. The output is a single risk value indicating patient cancer-specific survival. The learning process is guided by a precise loss function that accommodates time-to-event and censoring information. We further illustrate the pre-trained CNN model and integrated survival training in the following subsections.
Neural network architecture.
—A pretrained CNN, together with fine-tuning and transfer learning, leads to faster convergence and often outperforms training from scratch 26. We used a ResNet-50 architecture pre-trained on an ImageNet dataset with input resized from (1024 × 1024) to (256 × 256). We chose this family of architecture as it is designed to simplify training deep neural networks by adding residual connections to avoid information loss during deep network training. Our integrated deep learning system’s fundamental constituents comprised multiple convolution layers with weights initialized using a pre-trained model and a global average pooling layer. These sequential layers were followed by a fully connected layer and a final linear output layer modeled as the Cox layer that produced risk for each sample. A dropout layer was added to the fully connected layer before the Cox layer to control for overfitting. We trained the model using the Adam optimizer for gradient descent optimization with a total number of epochs set to 100 and a mini-batch size of 32. Parameters to the Adam optimizer include an initial learning rate of 1e-04, the momentum of 0.9, and inverse time decay factor of 0.1. To prevent overfitting during the training phase, we applied the Leaky-Relu activation function and dropout with a ratio of 0.35. Due to histological structure differences in H&E-stained images fine-tuning of the last layers was adopted to accommodate the difference in the glioma cancer dataset from the ImageNet dataset. Prediction models were trained using TensorFlow (v1.15.0) on NVIDIA:TESLA-V100 GPU. An overview of the model workflow is presented in Figure 1.
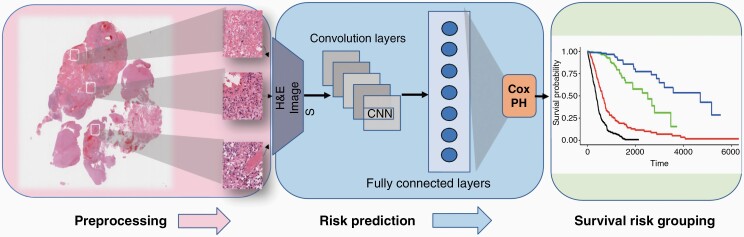
Figure 1.
Overview of proposed integrated survival deep learning model. (1) Multiple regions of interest are extracted from the whole slide image of H&E stained tumor tissue containing viable tumor. (2) These regions are then sent through a network of convolutional, pooling and fully connected layers that extract survival discriminative features. A Cox proportional hazards model was integrated with the fully connected layer which outputs patient specific risk scores. (3) Survival risk grouping: Recursive partitioning analysis was employed for risk stratification of the patient cohort based on predicted risk scores and prognostic molecular variables. H&E, hematoxylin and eosin.
Deep learning training and validation
The integrated survival deep learning model was trained with Monte Carlo cross-validation (MCCV).31 In MCCV, the sample is randomly split into a learning and test set numerous times. For each split, the patient cohort was randomly split into training (80%) and testing (20%) sets. Two advantages of MCCV are to decrease the bias associated with the split sample approach and decrease the variance over v-fold cross-validation.32 Each time, the training set trained the model, and the testing set assessed the corresponding model’s performance. This procedure was repeated 20 times (as previously seen there is a minimal advantage in increasing iterations to 50 or 1000 while the computational burden escalates)32 by changing random states while maintaining the same train-test split ratio. Z-score normalization was applied to each training/test image ROI before feeding into the model. The final model used for evaluation was aggregated by taking the exponential moving average of model weights across training steps with a decay constant of 0.99 to ensure stability across training epochs. The training process was guided by the negative log partial-likelihood loss function appropriate for CoxPH models and censored data. During training optimization, the loss function was evaluated over a small batch size of 32 samples instead of the entire dataset to improve generalization and allow a small memory footprint. The predicted SDL risk score at the patient level was aggregated by taking the median risk values for all samples across the patient.
Statistical analysis
Survival analysis was performed with univariate and multivariate CoxPH models to estimate hazard ratios (HRs) and 95% CIs for the association of predicted SDL risk score and other baseline clinical variables with OS, aggregated over training/testing sets. For multivariable analysis, we examined the additional prognostic value of predicted patient risk scores with and without controlling for known prognostic factors (ie, IDH-status, age at diagnosis, histologic grade). Prognostic prediction performance was evaluated using the c-index, defined as the ratio of all pairs of samples whose predicted survival times are correctly ordered among all uncensored patients.33
Internal validity of Cox regression models was determined using a bootstrapping technique.34 One-thousand, random bootstrap samples were drawn with replacement from the development data set. Then, the bootstrap sample estimated model was evaluated in the entire development dataset. The difference between the performance in the bootstrap sample and that in the development dataset was used to obtain the estimates of optimism in the development dataset.35
We employed recursive partitioning analysis (RPA), via the partDSA algorithm,36 to model OS. RPA enables the stratification of the patient into more homogenous survival groups based on multiple input variables. The variables included for the model building were age at diagnosis (as a continuous variable), SDL risk score, WHO-subtype, IDH1/2 mutation status, α-thalassemia, mental retardation, X-linked (ATRX) mutation status, and 1p19q co-deletion status. The partDSA tree that minimized the 5-fold cross-validated integrated Brier error was selected, and terminal nodes of the resulting tree defined the final risk groups from which the corresponding Kaplan-Meier curves were generated. HRs and 95% CIs for the risk groups were calculated via the CoxPH model. All statistical analyses were done in the R software, version 4.0.2. The significance level for statistical tests was 0.05.
Results
Characteristics of the Study Cohort
This study derived a risk score using an integrated survival deep learning framework on H&E-stained WSIs. The score was built and evaluated on the TCGA cohort consisting of both low-grade and high-grade diffuse gliomas. A summary of TCGA patient cohort characteristics is presented in Table 1.
Among the 766 unique patients from the TCGA cohort included in the analysis, the median age of diagnosis was 51 (interquartile range [IQR]: 37–61) years, and the median OS (mOS) for the combined LGG/GBM cohort was 2.5 years (95% CI: 2.16 to 3.13). Based on 2016 WHO classification of tumors of the central nervous systems,6 we classified the diffuse gliomas into 5 subtypes based on IDH mutations and co-deletion of chromosome 1p and 19q. Forty-eight percent (336 out of 700 with known IDH status) were IDH-wildtype, including 264 GBM (mOS of 1.16 years, 95% CI: 1.01 to 1.25) and 72 astrocytoma (mOS of 1.75 years, 95% CI: 1.53 to 2.24). Of the 52% (364 out of 700) that were IDH-mutant: 141 were oligodendroglioma with 1p/19q-codeletion (mOS of 14.14 years, 95% CI: 12.85 to not applicable [NA]), 203 were astrocytoma (mOS of 8.18 years, 95% CI: 6.26 to NA), and 20 were GBM (mOS of 2.95 years, 95% CI: 1.89 to 7.64). In the cohort, 47% were grade IV and 52% grade II/III gliomas. A detailed description of the patient characteristics, based on WHO subtype, is presented in Supplementary Table 1.
In univariate survival models, age (HR: 1.06; 95% CI: 1.05 to 1.07; P < .001), IDH-mutation status (mutant vs wildtype, HR: 9.45, 95% CI: 7.14 to 12.50, P < .001), histologic WHO grade (grade III vs grade II, HR: 2.98, 95% CI: 1.84 to 4.82, P < 0.001; grade IV vs grade II, HR: 14.92, 95% CI: 9.72 to 22.91, P < .001), WHO 2016 diffuse glioma subtype (IDH-mutant GBM vs IDH-mutant astrocytoma, HR: 4.58, 95% CI: 2.52 to 8.34, P < .001; IDH-mutant oligodendroglioma vs IDH-mutant astrocytoma, HR: 0.65, 95% CI: 0.35 to 1.22, P = .183; IDH-wildtype astrocytoma vs IDH-mutant astrocytoma, HR: 5.66, 95% CI: 3.52 to 9.10, P < .001; IDH-wildtype GBM vs IDH-mutant astrocytoma, HR: 11.90, 95% CI: 8.22 to 17.23, P < 0.001), along with ATRX-status (wildtype vs mutant, HR: 2.74, 95% CI: 1.91 to 3.93, P < .001) and codeletion of 1p19q (non-codeleted vs codeleted, HR: 7.75, 95% CI: 4.62 to 13.0, P < .001) were associated with OS while sex was not (male vs female, HR: 1.12, 95% CI: 0.92 to 1.38, P = 0.262) (Table 1).
Characteristics of the Risk Score From the Integrated Survival Deep Learning Framework
The survival deep learning model’s output produced a continuous patient-specific risk score calculated by taking the median risk score across all the patient samples. The mean SDL risk score across all patients was 0.1 (±1.3) and ranged from (−4.9 to 2.8). Performance of the SDL risk score was evaluated over 20 bootstrap iterations which showed substantial prognostic ability, achieving a median c-index of 0.79 (0.782, 0.794).
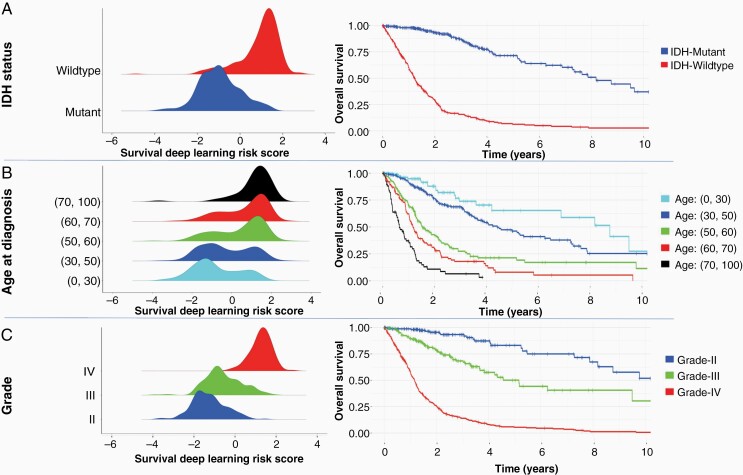
Next, we explored the association between the SDL risk score and clinical and molecular variables (Figure 2). An increase in SDL risk score was observed in IDH-wildtype versus IDH-mutant patients, as well as with an increase in age at diagnosis and histologic grade (Figure 2). Within the IDH subgroups the SDL risk score was higher for the IDH-wildtype subgroup with a median of 1.19 (0.62, 1.55) compared to the IDH-mutant subgroup with a median of −1.02 (−1.64, −0.35). Examining OS within the IDH subgroups, the patients with IDH-wildtype tumors had an mOS of 1.23 years (95% CI: 1.11 to 1.35) compared to those with IDH-mutant tumors which had an mOS of 8.18 years (95% CI: 7.28 to NA) (Figure 2A).
Figure 2.
Distribution of SDL risk score with prognostic molecular variables along with Kaplan-Meier survival curves. (A) Predicted SDL risk score strongly correlates IDH-status, showing strong association within genetic subtypes. (B) Correlation of SDL risk with age at discrete intervals, shows gradually changing peak toward higher risk values for older age group patients. (C) SDL risk score association with histologic grade. IDH, isocitrate dehydrogenase; SDL, survival deep learning.
Higher SDL risk scores were correlated with older age groups. Figure 2B shows the Kaplan-Meier analysis by age. Earlier empirical studies revealed an association of age with molecular characteristics of diffuse glioma patients.37–40 On average, patients with IDH-wildtype GBM have the highest age at diagnosis (median 59 years) and worst prognosis (mOS 1.16 years, 95% CI: 1.01 to 1.25). Patients belonging to IDH-mutant oligodendroglioma are relatively younger (median age 45 years) and have the longest mOS (14.1 years, 95% CI: 12.85 to NA).
For histologic grade, the SDL risk score increases from a median of −1.32 (−1.80, −0.82) at grade II, to −0.69 (−1.12, 0.04) at grade III, to 1.31 (1.00, 1.61) at grade IV (Supplementary Table 2). Histologic grade is associated with worse outcomes for grade IV (mOS of 1.16 years [95% CI: 1.03 to 1.25]) as compared to grade II with an mOS of 12.85 years (95% CI: 8.18 to NA) and grade III with an mOS 5.16 years (95% CI: 3.84 to NA) (Figure 2C).
To explore the histologic features associated with the SDL risk score, histologic features were compared for 68 ROIs of which 23 were designated as higher risk and 45 were designated as lower risk. A total of 12 histologic features were scored for each image by a neuropathologist (J.J.P) who was blinded to both the risk score and overall histologic diagnosis. A clear pattern emerged where images from higher-risk ROIs contained histologic features associated with tumor aggressiveness, including mitoses (16/23 [70%]), simple or complex microvascular hyperplasia (11/23 [48%]), increased cellular density (8/23 [35%]), or necrosis (5/23 [22%]). In contrast, images from lower-risk ROIs contained cells with uniform nuclei (32/45 [71%]), abundant eosinophilic cytoplasm (18/45 [40%]), and perinuclear halos (13/45 [29%]).
SDL Risk as a Prognostic Factor in Univariate and Multivariate Models
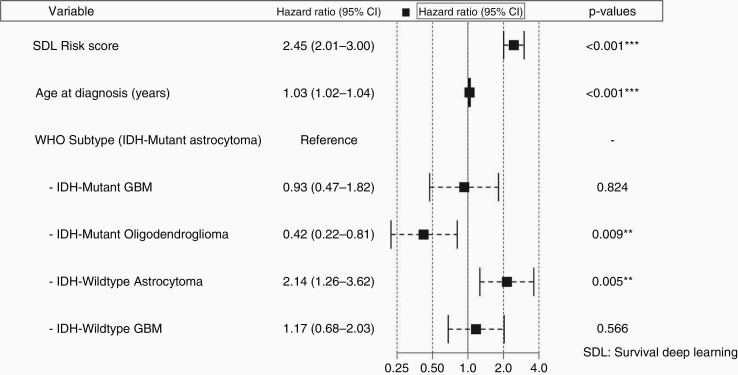
Cox-regression analysis was performed to assess the association of SDL risk score with OS. In a univariate model, the SDL risk score was associated with poor outcomes (HR: 3.29, 95% CI: 2.88 to 3.76; P < .001). That is, for every one-point increase in SDL risk score, the risk of dying increased more than 3-fold. In multivariate models, we controlled for prognostic clinical and molecular variables. We included age at diagnosis, sex, histologic grade, IDH status, and WHO 2016 diffuse glioma subtype. After forward and backward feature selection, the significant variables remaining were SDL risk score, age at diagnosis, and WHO 2016 diffuse subtype (Figure 3). The forest plot shows the substantial prognostic power of SDL risk scores in the presence of clinical and molecular variables with a hazard ratio of 2.45 (95% CI: 2.01 to 3.0).
Figure 3.
Forest plot of the HRs for multivariate survival model. The figure illustrates the HR and 95% CI of the SDL risk score in the presence of other clinical variables, including age at diagnosis and WHO 2016 subtype. HR = 1: No effect; HR < 1: Reduction in risk; HR > 1: Increase in risk. HR, hazard ratio; SDL, survival deep learning.
Performance of the SDL risk score model was compared by assessing the predictive accuracy of a baseline Cox model generated using clinical variables: WHO 2016 diffuse glioma subtype and age at diagnosis. This model performed slightly better (c-index: 0.82 [95% CI: 0.81 to 0.82]) than the Cox model with SDL risk score alone (c-index: 0.81 [95% CI: 0.813 to 0.813). Overall, the multivariate Cox model that included clinical variables, molecular variables, and the SDL risk score achieved a higher c-index of 0.84 (95% CI: 0.83 to 0.84)
Integrated SDL Framework Improves Patient Stratification
The RPA to classify the patients for OS is depicted in Figure 4A. The optimal tree elucidated interactions between significant clinical variables: IDH status, age at diagnosis, and SDL risk scores that separated the patients into 4 mutually exclusive risk groups. Group 1 patients had the worst outcome and were comprised of 2 IDH-wildtype subgroups: those with an SDL risk score greater than 1.08; and those with an SDL risk score less than 1.08 and over 54 years of age (n = 327; mOS of 1.03 years [95% CI: 0.97 to 1.16]). Group 2 patients had better survival than Group 1 and included patients who had an IDH-wildtype tumor, an SDL risk score less than 1.08, and were under 54 years of age (n = 75; mOS of 2.14 years [95% CI: 2.04 to 3.92]). Group 3 patients had better survival than those in Group 2 and included those with IDH-mutant tumors and an SDL risk score over −0.98 (n = 176; mOS of 5.29 years [95% CI: 4.21 to 7.64]). Group 4 patients experienced the best survival and were those with an IDH-mutant tumor and an SDL risk score less than −0.98 (n = 188; mOS of 14.14 years [95% CI: 9.50 to NA]). Clinical characteristics, HRs, and Kaplan-Meier curves for these four risk groups are shown in Table 2 and Figure 4B.
Figure 4.
RPA for TCGA cohort (n = 766). (A) RPA model defines 4 risk groups based on IDH mutation status, age at diagnosis, and SDL risk score. (B) Kaplan-Meier curves, number at risk, median OS, and HRs for the 4 risk groups as determined in (A). Group 1 has the worst OS, Group 2 and 3 have intermediate OS, and Group 4 has the best OS. (C) Kaplan-Meier curves, number at risk, and median OS of IDH-Wildtype split by Group 1 and Group 2. The solid two lines represent IDH-wildtype astrocytoma within Groups 1 and 2 resepectively whereas dashed represent IDH-wildtype GBM within Groups 1 and 2. HR, hazard ratio; OS, overall survival; SDL, survival deep learning; RPA, recursive partitioning analysis; IDH, isocitrate dehydrogenase; GBM, Glioblastoma multiforme.
Table 2.
Demographics Table for RPA risk Groups for TCGA Cohort
| Group 1 (N = 327) | Group2 (N = 75) | Group 3 (N = 176) | Group 4 (N = 188) | Total (N = 766) | |
|---|---|---|---|---|---|
| Clinical and Demographics Variables | |||||
| Sex | |||||
| Female | 128 (39.1%) | 38 (51.4%) | 68 (41.7%) | 73 (42.9%) | 307 (41.8%) |
| Male | 199 (60.9%) | 36 (48.6%) | 95 (58.3%) | 97 (57.1%) | 427 (58.2%) |
| Age at diagnosis | |||||
| Mean (SD) | 60.5 (11.9) | 41.1 (10.6) | 42.3 (12.5) | 40.0 (12.4) | 49.7 (15.4) |
| Median | 61.0 | 44.0 | 40.0 | 38.5 | 51.0 |
| Q1, Q3 | 54.5, 69.0 | 34.2, 50.0 | 33.0, 51.0 | 30.0, 49.8 | 37.0, 61.0 |
| Range | 14.0–88.0 | 10.0–54.0 | 20.0–75.0 | 14.0–74.0 | 10.0–88.0 |
| Grade | |||||
| II | 6 (1.8%) | 10 (13.5%) | 52 (31.9%) | 113 (66.5%) | 181 (24.7%) |
| III | 38 (11.6%) | 19 (25.7%) | 91 (55.8%) | 57 (33.5%) | 205 (27.9%) |
| IV | 283 (86.5%) | 45 (60.8%) | 20 (12.3%) | 0 (0.0%) | 348 (47.4%) |
| WHO grouping | |||||
| IDH-mutant Astrocytoma | 0 (0.0%) | 0 (0.0%) | 86 (48.9%) | 117 (62.2%) | 203 (29.0%) |
| IDH-mutant GBM | 0 (0.0%) | 0 (0.0%) | 20 (11.4%) | 0 (0.0%) | 20 (2.9%) |
| IDH-mutant oligodendroglioma | 0(0.0%) | 0 (0.0%) | 70 (39.8%) | 71 (37.8%) | 141 (20.1%) |
| IDH-wildtype astrocytoma | 43 (15.8%) | 29 (46.0%) | 0 (0.0%) | 0 (0.0%) | 72 (10.3%) |
| IDH-wildtype GBM | 230 (84.2%) | 34 (54.0%) | 0 (0.0%) | 0 (0.0%) | 264 (37.7%) |
| IDH status | |||||
| Wildtype | 273 (100.0%) | 63(100.0%) | 0 (0.0%) | 0 (0.0%) | 336 (48.0%) |
| Mutant | 0 (0.0%) | 0 (0.0%) | 176 (100.0%) | 188 (100.0%) | 364 (52.0%) |
| ATRX status | |||||
| Wildtype | 170 (52.0%) | 36 (48.0%) | 98 (55.7%) | 105 (55.9%) | 409 (53.4%) |
| Mutant | 9 (2.8%) | 3 (4.0%) | 67 (38.1%) | 83 (44.1%) | 162 (21.1%) |
| Vital status | |||||
| Alive | 53 (16.2%) | 26 (34.7%) | 126 (71.6%) | 175 (93.1%) | 380 (49.6%) |
| Deceased | 274 (83.8%) | 49 (65.3%) | 50 (28.4%) | 13 (6.9%) | 386 (50.4%) |
| Survival time (years) | |||||
| Median | 1.03 | 2.14 | 5.29 | 14.14 | 2.50 |
| 95% CI | (0.97 to 1.16) | (2.04 to 3.92) | (4.21 to 7.64) | (9.50 to NA) | (2.16 to 3.13) |
| SDL risk | |||||
| Mean (SD) | 1.2 (0.7) | 0.2 (1.1) | −0.2 (0.7) | −1.7 (0.6) | 0.1 (1.3) |
| Median | 1.3 | 0.6 | −0.3 | −1.6 | 0.2 |
| Q1, Q3 | 1.0, 1.6 | 0.1, 0.9 | −0.7, 0.2 | −1.9, −1.3 | −1.0, 1.3 |
| Range | −1.4 to 2.8 | −4.9 to 1.1 | −1.0 to 1.5 | −3.9 to -1.0 | −4.9 to 2.8 |
Abbreviations: IDH, isocitrate dehydrogenase 1 or 2 gene; ATRX, α-thalassemia, mental retardation, X-linked protein; 1p19q, deletion status of short arm of chromosome 1 and long arm of chromosome 19; GBM, glioblastoma multiforme; SDL risk, survival deep learning risk; RPA, recursive partitioning analysis.
Figure 4C shows the Kaplan-Meier plot for the IDH-wildtype tumor patients (Groups 1 and 2) split by Group and GBM/astrocytoma status. Interestingly, the combination of SDL risk score and age accurately delineated higher risk IDH-wildtype astrocytomas as well as lower-risk IDH-wildtype GBMs. For example, in Group 1 (defined by a high SDL risk score or a lower SDL risk score and higher age at diagnosis) the majority were IDH-wildtype GBM tumors; however, 16% of Group 1 (ie, 43 out of 273) were IDH-wildtype astrocytoma (solid black line in Figure 4C) and exhibited survival characteristics similar to IDH-wildtype GBM. In Group 2 (defined by a lower SDL risk score and younger age at diagnosis), approximately 45% (34 out of 75) of the patients were diagnosed with a GBM tumor (dotted red line in Figure 4C). A lower SDL risk score and younger age identified those patients as having a better prognosis than might be expected based on histologic grade alone.
Discussion
This study presents a clinically significant deep learning-based survival model to predict patient outcomes directly from images of H&E-stained tumor tissue. The proposed SDL model uses a residual deep learning framework and traditional CoxPH model to predict time-to-event outcomes. In this study, we showed that employing residual networks and utilizing randomized transformation of images addresses challenges in model overfitting when dealing with a small sample size. Furthermore, using a pre-trained model from the published literature and fine-tuning the model on glioma pathology images increases the network’s performance.26
We demonstrated that our SDL risk, which is derived from a modified ResNet model, is associated with histologic features of tumor aggressiveness in higher risk ROIs, has the ability to predict patient-specific survival from WSIs, and that the prediction accuracy exceeds other H&E-stained tissue imaging based deep learning approaches.23,29 In a multivariable regression model with age at diagnosis, WHO subtype, and SDL risk score, the SDL risk score remained a significant predictor associated with OS. Further, we introduced a novel recursive partitioning model, leveraging the SDL risk score and clinical variables to predict OS. These results demonstrate that the SDL model captures complex patterns non-redundant with known prognostic variables. Thus, this study takes us one step closer to systematically mapping out the relationship between histology-derived survival outcomes and prognostic molecular variables to strengthen significant risk group separation and overall prognostic performance.
A significant conclusion from the study is that the integrated SDL model, together with RPA, improved the prediction accuracy and accurate stratification of the patient cohort. Additionally, it highlights the relative importance of utilizing histologic features from H&E-stained tumor tissue to predict survival outcomes. The RPA indicated that patients with an IDH-mutant tumor and lower SDL risk score had a better prognosis than patients with an IDH-mutant tumor and higher SDL risk score. Furthermore, Kaplan-Meier analysis showed remarkable similarity in the discriminative power of SDL risk score and current WHO paradigm consistent with expected patient outcomes.
This work represents a proof-of-concept study to integrate deep learning in the analysis of H&E image and has some limitations. Foremost, the findings presented here require additional validation in a large, independent cohort. The TCGA cohort was classified with the now outdated WHO 2016 classification. Highlighting the importance of our findings, 47% (20/43) of lower grade IDH-wildtype tumors with higher SDL risk scores had gain of chromosome 7 and loss of chromosome 10 and would be considered WHO grade 4. Our use of Monte-Carlo cross-validation may include a bias, similar to using a split-sample approach, if each sample was not represented at least once in the training set and at least once in the test set. Although we attempted multiple steps to avoid additional biases in tuning parameter selection, we acknowledge it is best to separate tuning parameter selection from model building.41 The retrospective dataset used for training suffers from a previously documented selection bias.42 Furthermore, the proposed method relies on a small portion of regions from WSIs. In contrast, automated region extraction may lead to a better understanding of heterogeneity across the entire slide. Nevertheless, our study shows that the SDL framework can identify clinically relevant features associated with increased risk, and combining it with molecular and clinical data may lead to more homogenous patient cohorts and may have the potential to serve as noninvasive tool guiding patient management for clinical trials.
Supplementary Material
Contributor Information
Pranathi Chunduru, Department of Neurological Surgery, University of California San Francisco, San Francisco, California, USA.
Joanna J Phillips, Department of Neurological Surgery, University of California San Francisco, San Francisco, California, USA; Department of Pathology, University of California San Francisco, San Francisco, California, USA.
Annette M Molinaro, Department of Neurological Surgery, University of California San Francisco, San Francisco, California, USA; Department of Epidemiology and Biostatistics, University of California San Francisco, San Francisco, California, USA.
Funding
This work was supported by NIH NCI Brain Tumor SPORE Developmental Research Project and Biostatistics and Clinical CORE grant no. 5P50CA097257-18.
Conflict of Interest
The authors have no conflicts of interest to report with respect to the content of this manuscript.
Author Contributions
Experimental design: P.C., A.M.M.
Implementation: P.C., J.J.P., A.M.M.
Analysis and interpretation of the data: P.C., J.J.P., A.M.M.
All authors were involved in the writing of the manuscript and have read and approved the final version.
Prior Presentation
This research was presented as an on-demand prerecorded oral presentation at the Society for Neuro-Oncology (SNO) Annual Meeting (November 19–22, 2020).
References
- 1. Srinidhi CL, Ciga O, Martel AL. Deep neural network models for computational histopathology: a survey. Med Image Anal. 2021;67:101813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kros JM. Grading of gliomas: the road from eminence to evidence. J Neuropathol Exp Neurol. 2011;70(2):101–109. [DOI] [PubMed] [Google Scholar]
- 3. Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brat DJ, Aldape K, Colman H, et al. cIMPACT-NOW update 3: recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol. 2018;136(5):805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Molinaro AM, Taylor JW, Wiencke JK, Wrensch MR. Genetic and molecular epidemiology of adult diffuse glioma. Nat Rev Neurol. 2019;15(7):405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 7. Olar A, Aldape KD. Using the molecular classification of glioblastoma to inform personalized treatment. J Pathol. 2014;232(2):165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. WHO Classification of Tumours Editorial Board. World Health Organization Classification of Tumours of the Central Nervous System. Vol 6. 5th ed. Lyon, France: International Agency for Research on Cancer; 2021. [Google Scholar]
- 9. Arie Perry PW. Histologic classification of gliomas. In: Berger MS, Weller M, eds. Handbook of Clinical Neurology. Amsterdam, The Netherlands: Elsevier; 2016:71–95. [DOI] [PubMed] [Google Scholar]
- 10. Acs B, Rantalainen M, Hartman J. Artificial intelligence as the next step towards precision pathology. J Intern Med. 2020;288(1):62–81. [DOI] [PubMed] [Google Scholar]
- 11. van den Bent MJ. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: a clinician’s perspective. Acta Neuropathol. 2010;120(3):297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rathore S, Niazi T, Iftikhar MA, Chaddad A. Glioma grading via analysis of digital pathology images using machine learning. Cancers (Basel) 2020;12(3):578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu S, Shah Z, Sav A, et al. Isocitrate dehydrogenase (IDH) status prediction in histopathology images of gliomas using deep learning. Sci Rep. 2020;10(1):7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ertosun MG, Rubin DL. Automated grading of gliomas using deep learning in digital pathology images: A modular approach with ensemble of convolutional neural networks. AMIA Annu Symp Proc. 2015;2015:1899–1908. [PMC free article] [PubMed] [Google Scholar]
- 15. Kurc T, Bakas S, Ren X, et al. Segmentation and classification in digital pathology for glioma research: challenges and deep learning approaches. Front Neurosci. 2020;14:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen RJ, Lu MY, Wang J, et al. Pathomic fusion: an integrated framework for fusing histopathology and genomic features for cancer diagnosis and prognosis. IEEE Trans Med Imaging. 2022;41(4):757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Faraggi D, Simon R. A neural network model for survival data. Stat Med. 1995;14(1):73–82. [DOI] [PubMed] [Google Scholar]
- 18. Yousefi S, Song C, Nauata N, Cooper L. Learning genomic representations to predict clinical outcomes in cancer. arXiv:180105512. 2016. [Google Scholar]
- 19. Zhuge Y, Ning H, Mathen P, et al. Automated glioma grading on conventional MRI images using deep convolutional neural networks. Med Phys. 2020;47(7):3044–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matsui Y, Maruyama T, Nitta M, et al. Prediction of lower-grade glioma molecular subtypes using deep learning. J Neurooncol. 2020;146(2):321–327. [DOI] [PubMed] [Google Scholar]
- 21. Katzman JL, Shaham U, Cloninger A, et al. DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med Res Methodol. 2018;18(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu X, Yao J. Deep convolutional neural network for survival analysis with pathological images. Paper presented at: IEEE International Conference on Bioinformatics and Biomedicine (BIBM); December 15–18; 2016; Shenzhen, China, 2016. [Google Scholar]
- 23. Mobadersany P, Yousefi S, Amgad M, et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc Natl Acad Sci USA. 2018;115(13):E2970–E2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bice N, Kirby N, Bahr T, et al. Deep learning-based survival analysis for brain metastasis patients with the national cancer database. J Appl Clin Med Phys. 2020;21(9):187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bychkov D, Linder N, Turkki R, et al. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci Rep. 2018;8(1):3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Talo M. Automated classification of histopathology images using transfer learning. Artif Intell Med. 2019;101:101743. [DOI] [PubMed] [Google Scholar]
- 27. Cui L, Li H, Hui W, et al. A deep learning-based framework for lung cancer survival analysis with biomarker interpretation. BMC Bioinf. 2020;21(1):112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wulczyn E, Steiner DF, Xu Z, et al. Deep learning-based survival prediction for multiple cancer types using histopathology images. PLoS One. 2020;15(6):e0233678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu X, Yao J, Zhu F, Huang J. WSISA: making survival prediction from whole slide histopathological images. Paper presented at: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); July 21–26; 2017; Honolulu, HI, 2017. [Google Scholar]
- 30. Hao J, Kosaraju SC, Tsaku NZ, Song DH, Kang M. PAGE-Net: interpretable and integrative deep learning for survival analysis using histopathological images and genomic data. Pac Symp Biocomput. 2020;25:355–366. [PubMed] [Google Scholar]
- 31. Molinaro AM, Lostritto K. Statistical resampling techniques for large biological data analysis. In: Lee JK, ed. Statistical Bioinformatics: A Guide for Life and Biomedical Science Researchers. Hoboken, NJ; John Wiley & Sons, Inc.; 2010. [Google Scholar]
- 32. Molinaro AM, Simon R, Pfeiffer RM. Prediction error estimation: a comparison of resampling methods. Bioinformatics 2005;21(15):3301–3307. [DOI] [PubMed] [Google Scholar]
- 33. Harrell FE, Jr., Califf RM, Pryor DB, Lee KL, Rosati RA. Evaluating the yield of medical tests. JAMA 1982;247(18):2543–2546. [PubMed] [Google Scholar]
- 34. Iba K, Shinozaki T, Maruo K, Noma H. Re-evaluation of the comparative effectiveness of bootstrap-based optimism correction methods in the development of multivariable clinical prediction models. BMC Med Res Methodol. 2021;21(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moons KG, Kengne AP, Woodward M, et al. Risk prediction models: I. Development, internal validation, and assessing the incremental value of a new (bio)marker. Heart. 2012;98(9):683–690. [DOI] [PubMed] [Google Scholar]
- 36. Molinaro AM, Lostritto K, van der Laan M. partDSA: deletion/substitution/addition algorithm for partitioning the covariate space in prediction. Bioinformatics 2010;26(10):1357–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Curran WJ, Jr., Scott CB, Horton J, et al. Recursive partitioning analysis of prognostic factors in three Radiation Therapy Oncology Group malignant glioma trials. J Natl Cancer Inst. 1993;85(9):704–710. [DOI] [PubMed] [Google Scholar]
- 38. Delgado-López PD, Corrales-García EM. Survival in glioblastoma: a review on the impact of treatment modalities. Clin Transl Oncol. 2016;18(11):1062–1071. [DOI] [PubMed] [Google Scholar]
- 39. Lu J, Cowperthwaite MC, Burnett MG, Shpak M. Molecular predictors of long-term survival in glioblastoma multiforme patients. PLoS One. 2016;11(4):e0154313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Di Cristofori A, Zarino B, Fanizzi C, et al. Analysis of factors influencing the access to concomitant chemo-radiotherapy in elderly patients with high grade gliomas: role of MMSE, age and tumor volume. J Neurooncol. 2017;134(2):377–385. [DOI] [PubMed] [Google Scholar]
- 41. Molinaro AM, Wrensch MR, Jenkins RB, Eckel-Passow JE. Statistical considerations on prognostic models for glioma. Neuro Oncol 2016;18(5):609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Solomon DA, Kim JS, Ressom HW, et al. Sample type bias in the analysis of cancer genomes. Cancer Res. 2009;69(14):5630–5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.