Abstract
Recent work has proposed a new mechanism of bacterial iron regulation: riboswitches that undergo a conformational change in response to FeII. The czcD (NiCo) riboswitch was initially proposed to be specific for NiII and CoII, but we recently showed via a czcD-based fluorescent sensor that FeII is also a plausible physiological ligand for this riboswitch class. Here, we provide direct evidence that this riboswitch class responds to FeII. Isothermal titration calorimetry studies of the native czcD riboswitches from three organisms show no response to MnII, a weak response to ZnII, and similar dissociation constants (∼1 μM) and conformational responses for FeII, CoII, and NiII. Only the iron response is in the physiological concentration regime; the riboswitches’ responses to CoII, NiII, and ZnII require 103-, 105-, and 106-fold higher “free” metal ion concentrations, respectively, than the typical availability of those metal ions in cells. By contrast, the “Sensei” RNA, recently claimed to be an iron-specific riboswitch, exhibits no response to FeII. Our results demonstrate that iron responsiveness is a conserved property of czcD riboswitches and clarify that this is the only family of iron-responsive riboswitch identified to date, setting the stage for characterization of their physiological function.
Keywords: Irving-Williams series, metalloregulation, RNA, iron overload, metal selectivity, metal sensor
Introduction
Life has long wrestled with the beneficial and deleterious catalytic potential of iron. The ubiquity of FeII in the early earth has led to proposals that it played important roles in prebiotic chemistry1−4 and in hydrolysis and electron transfer chemistry in the hypothesized RNA world.5,6 Through the utility of its Lewis acidity, the FeII,III redox couple, its assembly into more complex cofactors such as iron–sulfur clusters7,8 and heme,9 and its facile reactivity with O2,10 the chemistry of iron has found its way into many of the core metabolic reactions of nearly every known extant organism.11 At the same time, however, iron also catalyzes destructive radical chemistry under oxic conditions, which may result in oxidative stress if improperly regulated;12−14 the possibility that high iron levels also may be toxic under anoxic conditions is tantalizing but has been less explored.4,15
Bacterial metalloregulatory systems for iron and other metal ions selectively sense the bioavailable (also called labile or “free”) concentrations of metal ions and control the expression of proteins involved in metal ion uptake, efflux, storage, and utilization, to maintain cellular free metal ion concentrations within a narrow range.16−18 On the basis of numerous studies and across several organisms,19−25 this range for iron appears to be high nanomolar to low micromolar. Iron is maintained at a relatively high labile concentration because its affinity for biological ligands tends to be relatively low, according to the Irving-Williams (IW) series, which ranks the affinities of metal–ligand complexes of divalent metal ions: MgII < MnII < FeII < CoII < NiII < CuII > ZnII.26 This trend dictates the relative buffered concentration ranges and consequently the affinities of the metalloregulators for each metal ion, spanning tens of micromolar for MnII, low micromolar for FeII, to picomolar for ZnII and attomolar for copper (CuI).18 Importantly, the fidelity of the regulators for metals low in the IW series, such as the ferric uptake regulator (Fur) for FeII, is frequently not the result of that regulator being highly specific for that metal;27 in fact, Fur binds CoII and ZnII with higher affinity than FeII in vitro.20 Instead, the cell maintains low labile concentrations of the metals higher in the IW series, thereby ensuring that the bioavailable concentrations of the high-IW metals are too low to aberrantly activate the low-IW regulator under most conditions.24,25
Within this framework, bacteria often possess different regulators to sense deficiency and excess of a particular metal ion. For example, ZnII deficiency and excess are sensed by two different transcription factors (Zur and ZntR) that regulate ZnII importers and exporters/storage, respectively.16 MnII levels are controlled by a transcription factor (MntR), regulating uptake and export, and riboswitches—RNA elements, located in the 5′-untranslated regions (5′-UTRs) of genes, which bind ligands to cause a conformational change to alter transcription or translation of the downstream gene28−30—as sensors of manganese excess, regulating the expression of MnII efflux proteins.31−33 In the case of iron, characterized regulators such as Fur20 are primarily charged with sensing deficiency or sufficiency, either directly as a repressor or as activator via the small antisense RNA, ryhB.34 Only recently have studies identifying iron-exporting ATPases pointed to the relevance of iron excess in bacteria under aerobic conditions35−37 and its intriguing links to pathogenesis.38−40 However, there is little understanding of mechanisms of toxicity and sensing of iron excess, and particularly whether they extend to anaerobic conditions, where oxidative stress is unlikely.41
Two recent publications have proposed riboswitches as an additional mechanism of sensing iron in bacteria. [Like the bacterial riboswitches, eukaryotic iron-responsive elements (IREs) may also sense iron by direct FeII binding.42] In 2020,43 we reported evidence that the czcD or “NiCo” riboswitch could respond to iron. This riboswitch is found in a number of obligate and facultative anaerobes, many of which are associated with the human gut as commensals and/or are human pathogens, such as Erysipelotrichaceae bacterium (Eba; this riboswitch was crystallographically characterized with CoII bound44), Listeria monocytogenes (Lmo), Enterococcus faecium, Clostridium botulinum, and other nonpathogenic clostridioides such as C. cellulolyticum (Cce). In 2015, the czcD riboswitch had been initially characterized as responsive to NiII and CoII by Breaker, Winkler, and co-workers and was proposed to sense cobalt and nickel excess and regulate putative efflux proteins.44 We hypothesized that FeII binding to this riboswitch had been overlooked, because those studies had been performed under aerobic conditions, in which FeII rapidly oxidizes to insoluble FeIII.43 Indeed, we showed that fluorescent sensors constructed from the Eba riboswitch respond to FeII (as well as to CoII, NiII, and other divalent first-row transition metals), and one of these sensors also responded to iron when expressed in Escherichia coli. Furthermore, the genes regulated by the Eba and Lmo riboswitches rescued iron toxicity when expressed in Bacillus subtilis, presumably by mediating FeII efflux. Therefore, although our in vitro studies were conducted with the czcD-based sensor, we proposed that the native czcD riboswitch itself might be iron-responsive in vivo.
A subsequent report from Ramesh and co-workers,45 premised on the nonresponsiveness of czcD to FeII, used bioinformatic methods to identify putative riboswitches that would be similar to czcD but would bind FeII instead of CoII and NiII. The RNAs identified, dubbed “Sensei,” were proposed to be structurally similar to czcD riboswitches. Isothermal titration calorimetry (ITC) studies suggested that the Sensei RNA from Haemophilus ducreyi (Hdu) was conformationally responsive to FeII but not CoII. Their claim that both czcD and Sensei were specific for their proposed ligands—CoII/NiII and FeII, respectively—was surprising, because it conflicted with the understanding of metalloregulation presented above, implying that metal selectivity rules differ for RNA and protein regulators.
Here, using two independent approaches, riboswitch-based fluorescent sensors and ITC, we demonstrate that czcD riboswitches from three organisms undergo a similar conformational change to FeII as to CoII and NiII, all with similar apparent dissociation constants (0.5–2 μM). These results demonstrate that iron responsiveness is a general property of the czcD family, and we propose that iron is the most likely cellular ligand for these riboswitches. By contrast, the Sensei RNA does not respond to FeII or CoII. These results establish the czcD riboswitch as the only known family of iron-responsive riboswitches, support a unified framework for protein- and RNA-based metalloregulation, and suggest a new player in the physiology of iron in numerous bacteria relevant to human health and disease.
Experimental Section
General Considerations
Chemical reagents were obtained from Thermo Fisher Scientific or Millipore Sigma, unless noted otherwise. Primers and gBlocks were ordered from Integrated DNA Technologies (IDT). Reagents used for PCR amplification and RNA transcription (NTPs, dNTPs, Q5 DNA polymerase, OneTaq DNA polymerase, T7 RNA polymerase) were obtained from New England Biolabs. The Sequagel reagents used for urea polyacrylamide gel electrophoresis were purchased from National Diagnostics. DFHBI-1T was purchased from Tocris. PCR cleanup kits were from Omega Bio-Tek. All riboswitch and sensor constructs used in this study are listed in Table S1. All primers used to generate the riboswitches and riboswitch-based sensors are listed in Table S2.
Nucleic acid UV–visible absorption spectra were obtained on a Mettler Toledo UV5Nano. Well plate analyses (for the Lmo sensors) were carried out using a BioTek Synergy H1 microplate reader. Experiments utilizing FeII were conducted within a vinyl anaerobic chamber (Coy Lab Products, used for ITC studies) or an MBraun Unilab anaerobic box (used for sensor studies). All glassware was acid-washed with TraceMetal-grade nitric acid (Fisher) followed by extensive rinsing with filtered ddH2O prior to use. All buffers were treated with Bio-Rad Chelex 100 resin (prior to metal addition) and sterile filtered. Aerobic and anaerobic isothermal calorimetry titration experiments were conducted using a TA Instruments Affinity ITC system.
Lmo Sensor Construction and Characterization
RNA sensor template library generation, screening, and citrate-buffered metal titrations were conducted as described (Table S3).43 Buffer conditions for titrations were 30 mM MOPS, 100 mM KCl, 3 mM MgCl2, 1 mM citrate, pH 7.2, and experiments were carried out at 20 °C.
Isothermal Titration Calorimetry
RNA samples were transcribed and gel-purified as described.43 In a typical experiment, the transcription reaction was carried out on a 1.5 mL scale to afford enough RNA for three titrations at a ∼10 μM final RNA concentration. After the overnight crush-soak, the buffer containing RNA was decanted and concentrated using a 10 000 MWCO Amicon Ultra centrifugal filtration device, followed by two cycles of 15× dilution and concentration with Buffer A (30 mM MOPS, 100 mM KCl, 3 mM MgCl2, pH 7.2) using the same centrifugal filter, to exchange out of the crush-soak buffer. The resulting RNA sample (1.5 mL) was transferred into a Thermo-Scientific Slide-A-Lyzer gamma-irradiated dialysis cassette (10 000 MWCO) and dialyzed in a 275–300× volume of Buffer A for 3–4 h. After dialysis, the RNA concentration was measured, and the RNA sample was refolded as described.43 This procedure yielded RNA concentrations of approximately 10 μM. The buffer following dialysis was used for blank runs of the ITC experiment, and all metal stocks for the ITC syringe were made using this buffer. Titrations of the metal stock solutions into the blank buffer generated minimal heats (<0.03 μJ/s per injection), and these heats were subtracted from the riboswitch titration data. Titrations with MnII (manganese(II) chloride tetrahydrate, >99%), CoII (cobalt(II) chloride hexahydrate, >99%), NiII (nickel(II) chloride hexahydrate, >99%), and ZnII (zinc(II) sulfate heptahydrate, >99%) were carried out aerobically. Titrations with FeII (iron(II) ammonium sulfate hexahydrate, >99%) were carried out anaerobically, in which case both the RNA sample and the dialysis buffer were deoxygenated by three cycles of degassing and refilling with nitrogen on a Schlenk line, and they were brought into the anaerobic chamber for RNA concentration measurement, metal stock preparation, and the ITC experiment. The concentrations of the metal ion solutions loaded into the ITC syringe were 600 μM for Eba and Cce titrations and 400 μM for Lmo titrations. The first injection was 0.1 μL (disregarded in data analysis), followed by 30 × 1.2 μL injections. Equilibration times were 180 s between injections, except for injections #2–6, which were 240 s. The sample cell was stirred at 125 rpm. The temperature was 20 °C. Samples in the autosampler were kept at 4 °C.
ITC data were analyzed using TA Instrument’s NanoAnalyze software. In order to remove the contribution from nonspecific metal–RNA interactions, the baseline value at saturating metal concentrations was first subtracted from the integrated heats for each titration, before fitting the corrected data using the “Independent” (one set of equivalent binding sites) ITC model to calculate binding parameters. As discussed below, the validity of the assumption that the baseline represents nonspecific metal binding is supported by the similarity between the magnitude of these heats and those observed for the nonresponsive M3 Eba riboswitch variant.
Results
Characterization of Fluorescent Sensors Based on the Lmo czcD Riboswitch
Determining the cognate metal ion of a regulator like the czcD riboswitch is challenging, because the tightest-binding metal ion is frequently not the physiologically relevant one. With iron, there is the added practical complication that FeII is readily oxidized under aerobic conditions. Furthermore, many of the typical methods to study conformational changes of RNAs are not easily adapted to anaerobic conditions. Therefore, to assess czcD riboswitch–metal ion interactions in our previous study,43 we constructed Eba czcD riboswitch-based fluorescent sensors by fusing the Spinach2 aptamer with the Eba riboswitch via its P1 stem (Figure S1C). The approach of fusion to Spinach2 has been used to generate fluorescent sensors from other riboswitches with minimal perturbation of function.46−48 The sensors bind metal and undergo a conformational change to bind (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylene]-3,5-dihydro-2-methyl-3-(2,2,2-trifluoroethyl)-4H-imidazol-4-one (DFHBI-1T),49 an analogue of the green fluorescent protein chromophore, leading to fluorescence activation. Our three sensors, czcD-1, czcD-2, and czcD-3, exhibited apparent binding affinities for divalent first-row transition metals, including FeII in an anaerobic-plate-reader-based assay, in accordance with the IW series.
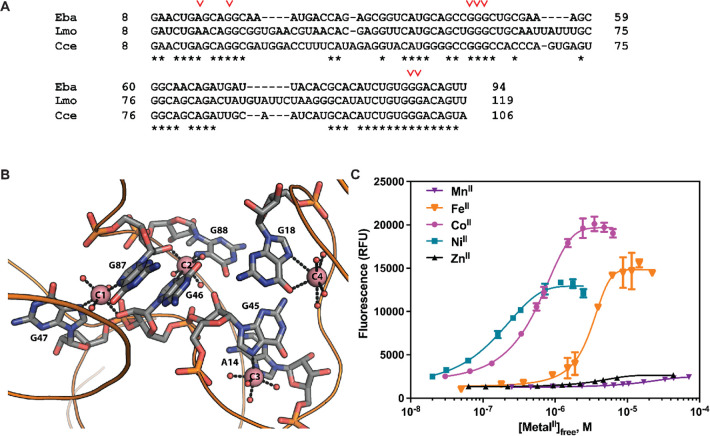
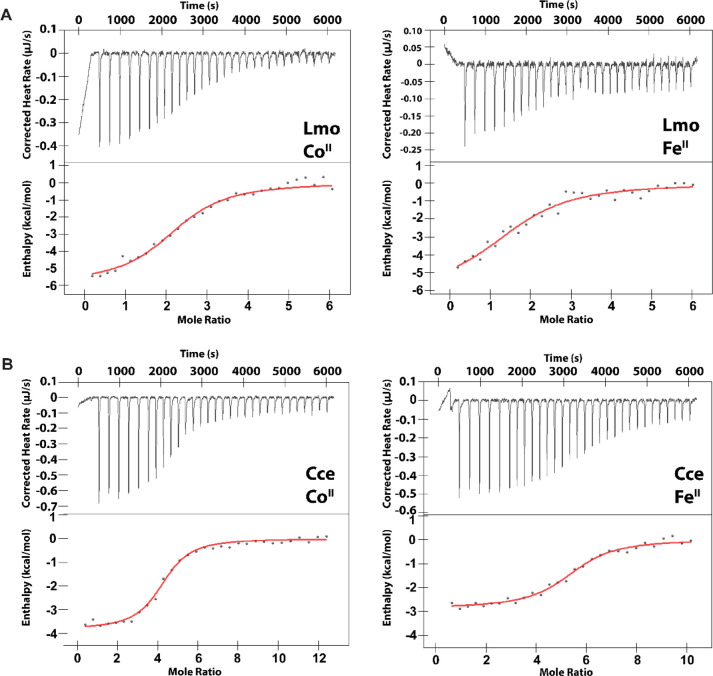
The czcD riboswitch from the human pathogen, Listeria monocytogenes (Lmo), retains all of the metal-binding nucleotides of the Eba riboswitch (Figure 1A,B). To probe the potential iron responsiveness of this riboswitch and to establish whether attachment at P1 is a general strategy to create sensors from this family of riboswitches, here we used the same approach of Spinach2 fusion to generate three Lmo riboswitch-based sensors, Lmo-1, Lmo-2, and Lmo-3. The numerical designation refers to the number of base pairs of the P1 stem in the original riboswitch retained in the sensor. Because the Spinach2 aptamer is expected to contribute four base pairs to the stem, Lmo-1 is anticipated to have the most similar P1 stem to the native riboswitch itself—predicted to have four base pairs by analogy to the Eba riboswitch (Figure S1C).
Figure 1.
(A) Sequence alignment of the Eba, Lmo, and Cce riboswitch constructs used in this study. Metal ligands are indicated with a red inverse caret (∨). (B) X-ray crystal structure of the Eba riboswitch with CoII ions bound (PDB code 4RUM). Metal-binding nucleotides are numbered according to Figure 1A, and CoII ions are shown as salmon spheres. The occupancy of C4 (anomalous signal 5σ) was significantly lower than that of C1, C2, and C3 (anomalous signals ∼12σ).44 Reprinted with permission from Xu and Cotruvo,43 copyright 2020 American Chemical Society. (C) Fluorescence response of Lmo-1 to first-row transition metal ions, with free metal concentrations buffered with 1 mM citrate. Full details and parameters from fits to the Hill equation (one set of sites) are given in Table 1.
Because RNA contains many potential nonspecific sites for metal binding, and because the predicted apparent Kd (Kd,app) values are on the same order as the sensor concentrations,43,50,51 we used 1 mM citrate to buffer the free concentration of metal ions in affinity titrations with MnII, FeII, CoII, NiII, and ZnII to determine Kd,app values. Interestingly, the Lmo-based sensors displayed a somewhat different selectivity pattern relative to the Eba-based czcD sensors. The Lmo-1 (Figure 1C) and Lmo-3 (Figure S1B) constructs exhibited responses to FeII, CoII, and NiII in accordance with the IW series, but no significant response to MnII and ZnII. The Lmo-2 sensor (Figure S1A) responded preferentially to FeII, CoII, and NiII, but it also responded to MnII and ZnII, albeit with higher Kd,app values, as we had observed with czcD-1.43 The Hill coefficients of all the sensors were 2 to 3, demonstrating similar cooperativity as the Eba-based sensors and suggesting that the attachment of the aptamer does not greatly perturb metal binding to the riboswitch. The results of the titrations are summarized in Table 1 (Lmo-1) and Tables S4 and S5 (Lmo-2 and Lmo-3). The improved in vitro selectivity and fluorescence response of Lmo-1 in particular suggests that it may perform more robustly in future bacterial imaging experiments than czcD-2 did in our prior work. These studies suggest that the Lmo riboswitch may also be iron-responsive and show the generality of using the P1 stem to create fluorescent sensors based on czcD riboswitches, an observation that will be exploited below to probe the Sensei RNA.
Table 1. Fluorescence Response of Lmo-1 to First-Row Transition Metal Ionsa.
Native Eba czcD Riboswitch Responds to FeII but Sensei Does Not
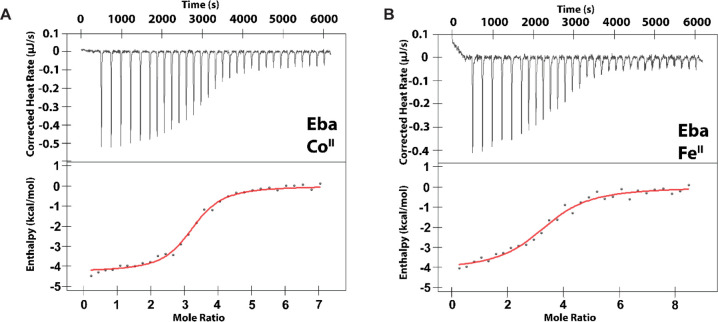
Whereas the riboswitch-based fluorescent sensors suggest that czcD riboswitches may respond to FeII, the specific Kd,app values may be perturbed by fusion with the Spinach2 aptamer. Therefore, we used ITC to further characterize metal binding; this method allows extrapolation of thermodynamic parameters including Kd, stoichiometry (n), and enthalpy (ΔH) for metal binding and the accompanying conformational change.52 We designed the ITC constructs by truncating the riboswitch at the P1 stem (Figure 1A, Table S1), similar to previous work on MnII-responsive riboswitches.33,53 We first carried out titrations of the Eba czcD riboswitch with MnII, FeII, CoII, NiII, and ZnII (Figures 2 and S2); the results are summarized in Table 2. The enthalpies associated with metal–Eba interactions are exothermic and similar in magnitude to those for MnII-responsive riboswitches (1–6 kcal/mol).33,53 We assign the small residual heats observed at the end of all titrations (0.05–0.1 μJ/s) to nonspecific binding of the metal ions to the RNA.33,53 In support of this assignment, the M3 variant of the Eba riboswitch, previously shown to ablate the RNA’s conformational response,44 exhibits background heats of the same magnitude but without the larger heat associated with a conformational response (Figure S3). Eba showed the highest-affinity interaction with CoII, NiII, and FeII, with Kd,app values of 0.5–1 μM. The Kd,app values for CoII and NiII binding are in good agreement with our fluorescent sensor studies43 as well as values determined by Nesbitt and co-workers54 and Unrau, Trachman, and co-workers55 by independent methods, although those investigators did not study FeII binding. We note especially the quantitative agreement between our Kd,app values and those of Nesbitt determined using single-molecule FRET, therefore supporting our interpretation that the observed enthalpy changes reflect the metal-induced conformational change. We were unable to detect any response with MnII (Figure S2), perhaps owing to a Kd,app for MnII–Eba being significantly greater than the concentration of riboswitch used for this experiment. Simulation of ITC traces assuming n = 3, ΔH = −4.5 kcal/mol, and [RNA] = 10 μM allows us to estimate that this Kd,app value is 0.1–1 mM (Figure S4). Together, these results strongly suggest that FeII elicits a similar conformational response in the riboswitch as CoII and NiII.
Figure 2.
Representative thermograms from ITC studies of the Eba czcD riboswitch (8.4–9.2 μM RNA) with CoII (A) or FeII (B). The data are fitted to a model with one set of equivalent binding sites. Fitted parameters are provided in Table 2. Conditions: 30 mM MOPS, 100 mM KCl, 3 mM MgCl2, pH 7.2, 20 °C.
Table 2. Thermodynamic Parameters for Metal Ion Binding to Eba, Eba Variants, Lmo, and Cce Riboswitches, Assessed by ITC.
| riboswitch | metal ion | Kd,app (μM) | n | ΔH (kcal/mol) | ΔS (cal/mol·K) |
|---|---|---|---|---|---|
| Eba | MnII | N.R.a | N.D.b | N.D. | N.D. |
| FeII | 0.99 ± 0.11 | 3.2 ± 0.3 | –4.2 ± 0.3 | 12.3 ± 2.4 | |
| CoII | 0.57 ± 0.15 | 3.2 ± 0.2 | –4.1 ± 0.3 | 14.5 ± 1.6 | |
| NiII | 0.45 ± 0.14 | 3.3 ± 0.1 | –4.1 ± 0.3 | 14.6 ± 1.2 | |
| ZnII | 2.70 ± 0.71 | 3.5 ± 0.8 | –3.2 ± 0.3 | 16.1 ± 1.3 | |
| Eba-CG | CoII | 0.88 ± 0.14 | 3.2 ± 0.4 | –5.0 ± 0.2 | 10.7 ± 0.8 |
| Eba-UA | CoII | 0.70 ± 0.20 | 3.2 ± 0.3 | –4.9 ± 0.3 | 11.3 ± 2.0 |
| Lmo | MnII | N.R. | N.D. | N.D. | N.D. |
| FeII | 2.21 ± 0.29 | 1.8 ± 0.1 | –6.2 ± 0.7 | 4.8 ± 2.4 | |
| CoII | 1.97 ± 0.32 | 2.3 ± 0.1 | –6.1 ± 0.4 | 5.3 ± 1.5 | |
| NiII | 1.20 ± 0.50 | 2.9 ± 0.1 | –4.5 ± 0.6 | 12.0 ± 1.4 | |
| ZnII | 1.96 ± 0.13 | 3.0 ± 0.5 | –2.5 ± 0.6 | 17.4 ± 2.0 | |
| Cce | FeII | 1.24 ± 0.12 | 4.8 ± 0.5 | –2.7 ± 0.2 | 18.0 ± 0.6 |
| CoII | 1.22 ± 0.18 | 4.5 ± 0.4 | –4.2 ± 0.4 | 12.8 ± 1.0 |
N.R.: No response detected.
N.D.: Not determined. Conditions: 30 mM MOPS, 100 mM KCl, 3 mM MgCl2, pH 7.2, 20 °C; 8–15 μM riboswitch, titrated with 400 μM (Lmo) or 600 μM (Eba, variants, and Cce) of each metal ion.
The metal-binding stoichiometry we observed for the Eba riboswitch contrasts with the four bound cobalt ions in the X-ray crystal structure of Eba determined by Furukawa et al. (Figure 1B).44 As the fourth CoII ion, C4 in Figure 1B, is only bound to one riboswitch nucleotide (G18) and exhibited lower occupancy than the others in the crystal structure, we hypothesized that its presence was an artifact of the high CoII concentration (2 mM) in the crystallization condition. Therefore, we designed two new constructs of the riboswitch with the base pair G18–C44 on the P2 stem mutated to remove the G18 metal ligand, in order to investigate whether this change would perturb metal-binding stoichiometry. As the nonfunctional M3 variant is made by mutating two base pairs into mismatching guanines in the P2 stem,44 the stability of this stem is important to the function of the riboswitch; therefore, we made mutations that will still retain the base pair at the G18–C44 positions: either CG (G18C/C44G, “Eba–CG”) or UA (G18U/C44A, “Eba–UA”). In titrations with CoII, we observed similar Kd,app and n values compared to the wild-type riboswitch (Table 2, Figure S5). These results suggest that the C4 site, in Eba at least, is not important for the riboswitch’s conformational response.
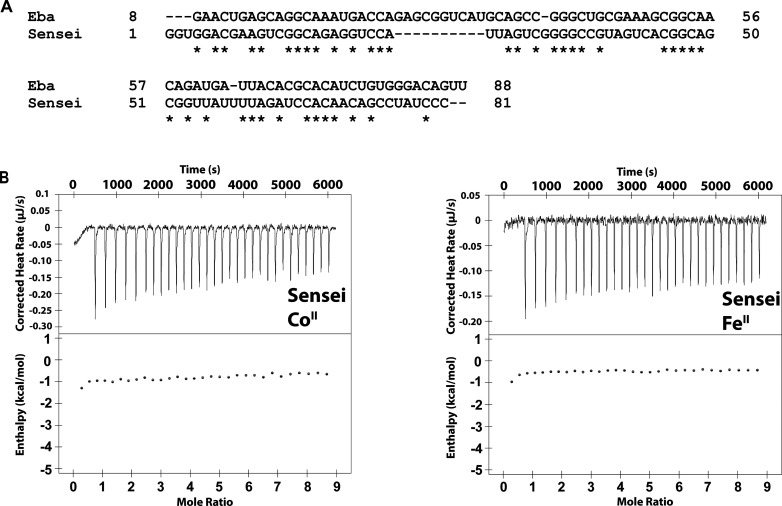
Our ITC data contrast with the results of Ramesh and co-workers,45 who reported little to no binding of FeII to Eba in equilibrium dialysis experiments. Conversely, their ITC analysis of the Hdu Sensei RNA indicated a Kd of 1.7 μM and 1:1 binding for FeII–Sensei, but no significant response to CoII. Because Sensei and czcD were putatively structurally similar, we reasoned that a Sensei-based fluorescent sensor analogous to the several czcD-based sensors we have designed would allow us to investigate this apparent discrepancy. Therefore, we inserted the Spinach2 aptamer at the P1 stem of the Hdu Sensei RNA; the sequences of the resulting three constructs are provided in Table S1. We did not observe any fluorescence activation during citrate-buffered metal titrations, with FeII or CoII, for any of the constructs (Figure S6). We also carried out ITC experiments with FeII and CoII using the same Hdu Sensei RNA construct that was reported by Ramesh and co-workers; we did not observe a significant change in enthalpy, indicating that neither FeII nor CoII induces a significant conformational change (Figure 3). After our work was completed, Ramesh et al. retracted their paper, but the retraction notice left open the question of whether the Sensei RNA was an FeII-specific riboswitch. Our fluorescence and ITC results clearly demonstrate that it is not an FeII-responsive riboswitch.
Figure 3.
Sensei RNA does not respond to iron. (A) Sequence alignment of the Eba czcD riboswitch and the Hdu Sensei RNA constructs used for ITC. The Sensei construct is the same one described by Ramesh and co-workers.45 (B) ITC thermograms of Hdu Sensei (15 μM) titrated aerobically with 225 μM CoII (left) and anaerobically with 225 μM FeII (right). The thermograms do not show evidence of significant metal binding and conformational change, beyond nonspecific interactions; compare Figures 2 and 4 for czcD riboswitches. Conditions: 30 mM MOPS, 100 mM KCl, 3 mM MgCl2, pH 7.2, 20 °C.
Iron Responsiveness Is a General Property of czcD Riboswitches
Next, we assessed the czcD riboswitches from two other organisms, Lmo and Cce, in order to determine whether the iron responsiveness and overall thermodynamic parameters of the Eba riboswitch were representative for this riboswitch family. The Lmo riboswitch was selected for study because of our Lmo sensors (Figure 1C) and because we have shown the riboswitch-regulated ATPase, LMO3884, to rescue iron toxicity in B. subtilis when overexpressed.43Cce was selected because RT-qPCR studies of the riboswitch-regulated gene had been carried out in the initial report of the czcD riboswitches.44 All three riboswitches have the metal-binding residues in the Eba X-ray structure fully conserved (Figure 1A). Characterization of the Lmo riboswitch (Figure 4A, Table 2, Figure S7) revealed a similar trend as with the Eba riboswitch, albeit with slightly higher Kd,app values and a metal-binding stoichiometry of only ∼2 for FeII and CoII. The ITC data were also in good agreement with the Lmo fluorescent sensor results (Figure 1C, Table 1). By titrating CoII and FeII into Cce (Figure 4B), we observed very similar metal–riboswitch affinities as with Eba and Lmo (Table 2). These results confirm that Cce is also a metal-responsive riboswitch. Interestingly, the stoichiometry for Cce is approximately 5, higher than the values observed for both Eba and Lmo. In all three cases, the response to FeII occurs with similar thermodynamic parameters, Kd,app, n, and ΔH as to CoII and NiII. Therefore, these results emphasize the generality of iron responsiveness in the czcD riboswitch class.
Figure 4.
Representative thermograms from ITC studies of (A) Lmo and (B) Cce czcD riboswitches (8.2–9.9 μM RNA) with CoII (left) or FeII (right). The data are fitted to a model with one set of equivalent binding sites. Fitted parameters are provided in Table 2. Conditions: 30 mM MOPS, 100 mM KCl, 3 mM MgCl2, pH 7.2, 20 °C.
Our ITC results establish that the czcD riboswitches bind and conformationally respond to FeII, just as they do to CoII and NiII (as well as ZnII, though with lower affinity). Despite conservation of the metal-binding nucleotides initially identified through the studies of Furukawa et al., these three riboswitches exhibit apparent differences in metal-binding stoichiometry. These differences may reflect the challenges of studying weak (micromolar affinity) metal-binding sites; nonspecific binding sites that are not linked to the conformational change may be of sufficiently similar affinity to the specific binding sites to affect the ITC stoichiometries. Nevertheless, all three czcD riboswitches bind at least two metal ions—suggesting that the two interconnected metal sites, C1 and C2 at the four-way junction (Figure 1B), may be the most critical for metal responsiveness and accounting for the cooperative binding observed in previous studies.43,44,54 These results highlight how the rules for iron ligation by RNA are not yet understood, motivating future structural biology studies of iron-bound riboswitches.
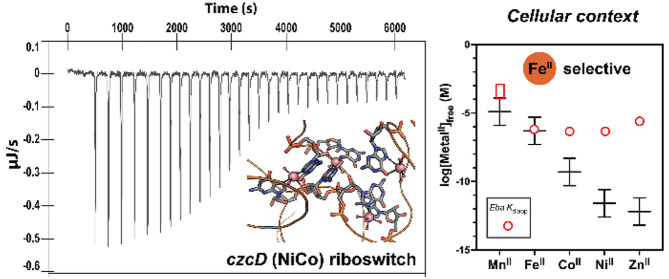
The riboswitches’ similar responsiveness to most of the divalent first-row transition metal ions motivates the question of which metal ion(s) is (are) physiologically relevant. The Kd,app values for FeII–czcD complexes fall squarely within the range of characterized metalloregulatory proteins for FeII, such as E. coli and S. enterica Fur (1.2 μM20 and 0.53 μM,24 respectively) and even an FeII-chaperoning complex from H. sapiens (0.8 μM).23 The Kd,app values for CoII–czcD complexes, however, are 1000-fold greater than for CoII–RcnR (0.51 nM),24 a model cobalt regulatory protein. The mismatches for NiII (200 000-fold; 0.45 μM for NiII–Eba versus 2.5 pM for NiII–NikR) and ZnII (4 000 000-fold; 2.7 μM for ZnII–Eba versus 0.64 pM for ZnII–Zur) are even more profound (Figure 5).24 The differences in riboswitch/transcription factor Kd,app values for CoII, NiII, and ZnII are even greater than was evident from the czcD fluorescent sensors.43 Therefore, the riboswitches’ Kd,app values are well matched to those of iron regulatory proteins but not to the corresponding cobalt and nickel regulators that sense labile metal concentrations in cells. This observation argues that the most likely physiological function for the czcD riboswitches is to sense iron.
Figure 5.
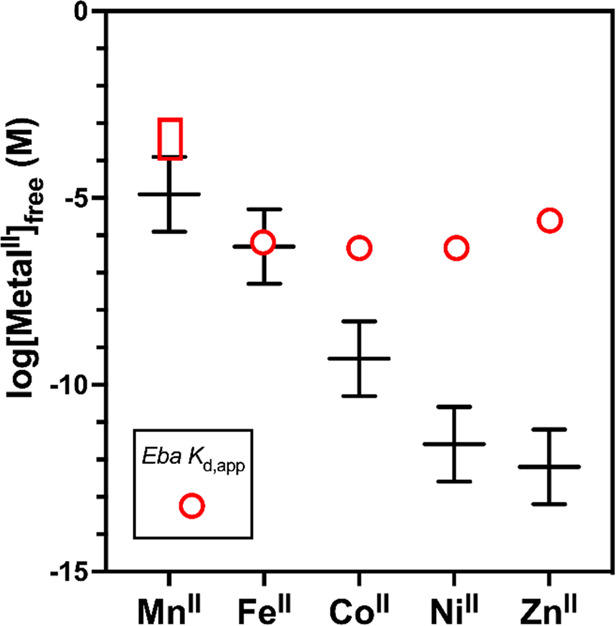
Comparison of Kd,app values for response of the Eba czcD riboswitch to transition metal ions (open red box for MnII, open red circles for FeII, CoII, NiII, and ZnII) with the calculated ranges of intracellular labile metal ion concentrations (black lines) determined for the Salmonella model system.24 The latter ranges consist of the free metal concentrations at which the relevant metal-sensing transcription factor gives 10, 50 (center), or 90% of its transcriptional response. Concept adapted from Young, Robinson, and co-workers,25 with our data added. See http://creativecommons.org/licenses/by/4.0/ for license information. In the case of MnII, the red box indicates that the Kd,app for the riboswitch is estimated to be 100–1000 μM (Figure S4). Because our experiments were carried out at 20 °C, but the prior ones were at 25 °C, our Kd values are slight underestimates relative to the intracellular labile metal ion concentrations. We also note that labile metal concentrations may differ somewhat from the Salmonella model, depending on the organism and aerobic versus anaerobic conditions.
Discussion
Our results demonstrate that the czcD riboswitches from three different organisms respond to FeII, CoII, and NiII with similar dissociation constants in the 1 μM range. Interestingly, this conclusion differs somewhat from that based on analysis of riboswitch-derived fluorescent sensors,43 which exhibited responses with greater metal dependence, in accordance with the Irving-Williams series. The native czcD riboswitches have flattened the typical affinity trend, with the exception of MnII (Figure 5). Viewing this trend in the context of buffered labile metal ion concentrations (see Introduction), with FeII and MnII being regulated in similar ranges, we hypothesize that the riboswitch is primarily tuned to suppress responsiveness to physiological concentrations of MnII and, as a result, is less selective for FeII against CoII and NiII (and ZnII). Presumably, however, the potential response to CoII and NiII, not just to FeII, is mitigated in vivo, because the labile concentrations of CoII and NiII are buffered by intracellular protein and small-molecule ligands and regulated at labile concentrations below that which is necessary to induce a response from the riboswitch, except when confronted with the highest levels of metal stress.24,25
Of course, it is necessary to obtain explicit in vivo information about czcD’s metal selectivity, which is a focus of our current efforts. Nevertheless, indirect in vivo information is currently available to support our argument. Wang et al. recently reported that an mCherry transcriptional reporter regulated by the czcD riboswitch requires 50 μM CoII or at least 1 mM NiII added to rich media (LB) in order to observe a significant response in E. coli.56 This result is in line with the increasing discrepancy between intracellular buffered metal concentration and the metal–czcD Kd value (Figure 5) from FeII to CoII to NiII, suggesting that response to NiII would only be relevant under the most extreme conditions. Furthermore, although Wang et al. did not assess whether their reporter was responding to basal iron levels, the czcD-2 sensor, with a Kd (0.4 μM) for FeII similar to that of the native riboswitch (1.0 μM), did show a basal response to FeII even in minimal media with no added iron.43 Therefore, whereas we cannot rule out the possibility that in some organisms or under certain conditions other metals (primarily CoII)44 might be able to transiently induce an aberrant response,27 our results herein clarify that the czcD riboswitch’s selectivity seems to be well tuned for iron when examined in the cellular context, and we hypothesize that iron is the primary ligand for the czcD riboswitches.
Having established czcD as the only known family of iron-responsive riboswitches, next we consider potential roles of these riboswitches and the genes that they regulate. Many of the predicted czcD riboswitches are present upstream of putative metal-effluxing P1B4-type ATPases.44 The identification of the primary substrate of this family of ATPases has long been contentious; while many can transport CoII under certain conditions,57,58 recent work has revealed that the physiological function of at least some of these ATPases is iron detoxification.38−40 It remains to be determined whether this function is shared by the riboswitch-regulated P1B4-type ATPases, although results in a heterologous system suggest that the riboswitch-regulated Lmo P1B4 ATPase LMO3448 exports iron efficiently.43 This gene also has been recently shown to be induced by lactic acid stress, by an unknown mechanism.59 In other organisms, the association between metal excess and the riboswitch-regulated genes is less clear. The Ccel_1038 gene positively regulated by the riboswitch in Cce is annotated as an MgtA-like P3-type ATPase. In E. coli and Salmonella enterica serovar Typhimurium, MgtA is an importer of MgII, and the activity of the purified enzyme is inhibited by CoII, NiII, and ZnII (FeII was not tested).60 This connection suggests that high levels of iron may interfere with magnesium homeostasis in Cce, leading to increased expression of a riboswitch-regulated MgII importer as a compensatory mechanism. We note that prediction of direction of transport and metal selectivity in P-type ATPases is not trivial and sometimes controversial;61,62 therefore, determination of whether these annotated mgtA genes indeed encode MgII importers will require additional investigation. In still other organisms, the genes putatively regulated by this class of riboswitch do not seem to be ATPases at all, suggesting an even broader array of cellular physiology that might be impacted by iron levels. Detailed examination of these systems promises to shed significant light on the mechanisms and consequences of iron overload in a multitude of bacteria.
Lmo strains possessing the riboswitch might yield particular insight into its function. Listeriaceae encode a chromosomal P1B4 ATPase, FrvA (LMO0641), that is regulated by Fur and the iron-dependent hydrogen peroxide sensor PerR and is implicated in iron efflux in response to iron overload, at least under aerobic conditions.39,41 Many clinical and environmental isolates, but not wild-type Listeria strains like EGD-e, also possess an accessory plasmid that contains the riboswitch-regulated P1B4 ATPase mentioned above, LMO3448 (42% sequence identity with FrvA).44,59,63 Why two seemingly redundant ATPases with different regulatory mechanisms exist in these strains remains to be explored, but the redundancy might suggest the particular importance of defending against iron excess. For example, the riboswitch-mediated mechanism might enable a more rapid response to iron excess; alternatively, the two regulatory mechanisms might be tuned to respond at slightly different labile iron levels. The Kd of the Lmo riboswitch (2.2 μM) is slightly higher than the Kds of the Eba and Cce riboswitches (1 μM), which are more similar to the Kd of E. coli FeII–Fur (1.2 μM).20 Perhaps most intriguingly, the riboswitch and its downstream P1B4 ATPase may provide a mechanism for sensing and responding to iron excess specifically under anaerobic conditions, when PerR may not be active. This proposal is supported by the observation that all bacteria containing czcD are either obligate or facultative anaerobes; furthermore, labile FeII levels would likely be higher in anaerobiosis than in aerobiosis,64 which might account for the higher FeII–riboswitch Kd value. We are currently interrogating the riboswitch’s place in Listeriaceae metallophysiology.
Conclusions
We have demonstrated that three native czcD riboswitches respond to FeII with Kd ∼ 1 μM, with similar affinities for CoII, NiII, and ZnII. Consideration of these results in the context of cellular metal regulation suggests that the riboswitch is tuned to preferentially respond to FeII in vivo. By contrast, our studies of the Sensei RNA indicate that it is not an iron-responsive riboswitch. We propose that the primary physiological function of czcD riboswitches is to respond to high iron levels, and in vivo investigations are in progress to test this hypothesis.
Acknowledgments
This work was supported by grants from the National Institutes of Health (R35GM138308) and the Charles E. Kaufman Foundation of the Pittsburgh Foundation as well as Penn State University (a Louis Martarano Career Development Professorship), to J.A.C. ITC studies were supported by NIH grant S10 OD025145 for the TA Instruments Low Volume AutoAffinity ITC, to N.H. Yennawar, Automated Biological Calorimetry core facility, Penn State Huck Institutes of the Life Sciences. We thank J.A. Fecko for training on the ITC instrument, S.J. Booker for use of his laboratory’s anaerobic chamber, and J.J. DeVos for experimental assistance. The authors are also grateful to J. Stubbe for helpful comments on this manuscript.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsbiomedchemau.1c00069.
Supplemental tables (including RNA constructs, primers, and titration data) and figures (additional fluorescence and ITC data) (PDF)
Accession Codes
Eba3544, Uniprot entry E2SQM5; LMO3448, Uniprot entry T1YRD8; Ccel_1038, Uniprot entry B8I9E0; FrvA, Uniprot entry Q8Y992.
The authors declare no competing financial interest.
Supplementary Material
References
- Barge L. M.; Flores E.; Baum M. M.; VanderVelde D. G.; Russell M. J. Redox and pH gradients drive amino acid synthesis in iron oxyhydroxide mineral systems. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 4828–4833. 10.1073/pnas.1812098116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.-Y.; Wang Y.-C.; Hsiao C. Prebiotic iron originates the peptidyl transfer origin. Mol. Biol. Evol. 2019, 36, 999–1007. 10.1093/molbev/msz034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowska K. B.; Varma S. J.; Moran J. Synthesis and breakdown of universal metabolic precursors promoted by iron. Nature 2019, 569, 104–107. 10.1038/s41586-019-1151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guth-Metzler R.; Bray M. S; Frenkel-Pinter M.; Suttapitugsakul S.; Montllor-Albalate C.; Bowman J. C; Wu R.; Reddi A. R; Okafor C D.; Glass J. B; Williams L. D. Cutting in-line with iron: ribosomal function and non-oxidative RNA cleavage. Nucleic Acids Res. 2020, 48, 8663–8674. 10.1093/nar/gkaa586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao C.; Chou I. C.; Okafor C. D.; Bowman J. C.; O’Neill E. B.; Athavale S. S.; Petrov A. S.; Hud N. V.; Wartell R. M.; Harvey S. C.; Williams L. D. RNA with iron(II) as a cofactor catalyses electron transfer. Nat. Chem. 2013, 5, 525–528. 10.1038/nchem.1649. [DOI] [PubMed] [Google Scholar]
- Okafor C. D.; Lanier K. A.; Petrov A. S.; Athavale S. S.; Bowman J. C.; Hud N. V.; Williams L. D. Iron mediates catalysis of nucleic acid processing enzymes: support for Fe(II) as a cofactor before the great oxidation event. Nucleic Acids Res. 2017, 45, 3634–3642. 10.1093/nar/gkx171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R.; Freibert S.-A. Mechanisms of mitochondrial iron-sulfur protein biogenesis. Annu. Rev. Biochem. 2020, 89, 471–499. 10.1146/annurev-biochem-013118-111540. [DOI] [PubMed] [Google Scholar]
- Talib E. A.; Outten C. E. Iron-sulfur cluster biogenesis, trafficking, and signaling: Roles for CGFS glutaredoxins and BolA proteins. Biochim. Biophys. Acta - Mol. Cell Res. 2021, 1868, 118847. 10.1016/j.bbamcr.2020.118847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I. G.; Willoughby M. M.; Hamza I.; Reddi A. R. One ring to bring them all and in the darkness bind them: The trafficking of heme without deliverers. Biochim. Biophys. Acta - Mol. Cell Res. 2021, 1868, 118881. 10.1016/j.bbamcr.2020.118881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs C.; Galonić Fujimori D.; Walsh C. T.; Bollinger J. M. Non-heme Fe(IV)–oxo intermediates. Acc. Chem. Res. 2007, 40, 484–492. 10.1021/ar700066p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posey J. E.; Gherardini F. C. Lack of a role for iron in the Lyme disease pathogen. Science 2000, 288, 1651–1653. 10.1126/science.288.5471.1651. [DOI] [PubMed] [Google Scholar]
- Lee J. W.; Helmann J. D. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 2006, 440, 363–367. 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- Sobota J. M.; Imlay J. A. Iron enzyme ribulose-5-phosphate 3-epimerase in Escherichia coli is rapidly damaged by hydrogen peroxide but can be protected by manganese. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 5402–5407. 10.1073/pnas.1100410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay J. A. The mismetallation of enzymes during oxidative stress. J. Biol. Chem. 2014, 289, 28121–28128. 10.1074/jbc.R114.588814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrangsu P.; Rensing C.; Helmann J. D. Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 2017, 15, 338–350. 10.1038/nrmicro.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z.; Jacobsen F. E.; Giedroc D. P. Coordination chemistry of bacterial metal transport and sensing. Chem. Rev. 2009, 109, 4644–4681. 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D. Specificity of metal sensing: iron and manganese homeostasis in Bacillus subtilis. J. Biol. Chem. 2014, 289, 28112–28120. 10.1074/jbc.R114.587071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster A. W.; Young T. R.; Chivers P. T.; Robinson N. J. Protein metalation in biology. Curr. Opin. Chem. Biol. 2022, 66, 102095. 10.1016/j.cbpa.2021.102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyer K.; Imlay J. A. Superoxide accelerates DNA damage by elevating free-ironlevels. Proc. Natl. Acad. Sci. U.S.A. 1996, 93, 13635–13640. 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills S. A.; Marletta M. A. Metal binding characteristics and role of iron oxidation in the ferric uptake regulator from Escherichia coli. Biochemistry 2005, 44, 13553–13559. 10.1021/bi0507579. [DOI] [PubMed] [Google Scholar]
- Guedon E.; Helmann J. D. Origins of metal ion selectivity in the DtxR/MntR family of metalloregulators. Mol. Microbiol. 2003, 48, 495–506. 10.1046/j.1365-2958.2003.03445.x. [DOI] [PubMed] [Google Scholar]
- Cassat J. E.; Skaar E. P. Iron in infection and immunity. Cell Host Microbe 2013, 13, 509–519. 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. J.; Frey A. G.; Palenchar D. J.; Achar S.; Bullough K. Z.; Vashisht A.; Wohlschlegel J. A.; Philpott C. C. A PCBP1-BolA2 chaperone complex delivers iron for cytosolic [2Fe-2S] cluster assembly. Nat. Chem. Biol. 2019, 15, 872–881. 10.1038/s41589-019-0330-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman D.; Martini M. A.; Foster A. W.; Chen J.; Scott A. J. P.; Morton R. J.; Steed J. W.; Lurie-Luke E.; Huggins T. G.; Lawrence A. D.; Deery E.; Warren M. J.; Chivers P. T.; Robinson N. J. Bacterial sensors define intracellular free energies for correct enzyme metalation. Nat. Chem. Biol. 2019, 15, 241–249. 10.1038/s41589-018-0211-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young T. R.; Martini M. A.; Foster A. W.; Glasfeld A.; Osman D.; Morton R. J.; Deery E.; Warren M. J.; Robinson N. J. Calculating metalation in cells reveals CobW acquires CoII for vitamin B12 biosynthesis while related proteins prefer ZnII. Nat. Commun. 2021, 12, 1195. 10.1038/s41467-021-21479-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving H.; Williams R. J. P. Order of stability of metal complexes. Nature 1948, 162, 746–747. 10.1038/162746a0. [DOI] [Google Scholar]
- Osman D.; Foster A. W.; Chen J.; Svedaite K.; Steed J. W.; Lurie-Luke E.; Huggins T. G.; Robinson N. J. Fine control of metal concentrations is necessary for cells to discern zinc from cobalt. Nat. Commun. 2017, 8, 1884. 10.1038/s41467-017-02085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M.; Breaker R. R. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004, 5, 451–463. 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- McCown P. J.; Corbino K. A.; Stav S.; Sherlock M. E.; Breaker R. R. Riboswitch diversity and distribution. RNA 2017, 23, 995–1011. 10.1261/rna.061234.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann C. E.; Wakeman C. A.; Sieling C. L.; Baker S. C.; Irnov I.; Winkler W. C. Structure and mechanism of a metal-sensing regulatory RNA. Cell 2007, 130, 878–892. 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- Price I. R.; Gaballa A.; Ding F.; Helmann J. D.; Ke A. Mn2+-sensing mechanisms of yybP-ykoY orphan riboswitches. Mol. Cell 2015, 57, 1110–1123. 10.1016/j.molcel.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambach M.; Sandoval M.; Updegrove T. B.; Anantharaman V.; Aravind L.; Waters L. S.; Storz G. The ubiquitous yyb-ykoY riboswitch is a manganese-responsive regulatory element. Mol. Cell 2015, 57, 1099–1109. 10.1016/j.molcel.2015.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J. E.; Le M. T.; Bhattarai N.; Capdevila D. A.; Shen J.; Winkler M. E.; Giedroc D. P. A Mn-sensing riboswitch activates expression of a Mn2+/Ca2+ ATPase transporter in Streptococcus. Nucleic Acids Res. 2019, 47, 6885–6899. 10.1093/nar/gkz494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E.; Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 4620–4625. 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frawley E. R.; Fang F. C. The ins and outs of bacterial iron metabolism. Mol. Microbiol. 2014, 93, 609–616. 10.1111/mmi.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi H.; Helmann J. D. Ferrous iron efflux systems in bacteria. Metallomics 2017, 9, 840–851. 10.1039/C7MT00112F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. B.; Lee M. A.; Smith A. T. Ins and outs: Recent advancements in membrane protein-mediated prokaryotic ferrous iron transport. Biochemistry 2021, 60, 3277–3291. 10.1021/acs.biochem.1c00586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan G.; Pinochet-Barros A.; Gaballa A.; Patel S. J.; Argüello J. M.; Helmann J. D. PfeT, a P1B4-type ATPase, effluxes ferrous iron and protects Bacillus subtilis against iron intoxication. Mol. Microbiol. 2015, 98, 787–803. 10.1111/mmi.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi H.; Patel S. J.; Argüello J. M.; Helmann J. D. The Listeria monocytogenes Fur-regulated virulence protein FrvA is an Fe(II) efflux P1B4-type ATPase. Mol. Microbiol. 2016, 100, 1066–1079. 10.1111/mmi.13368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. J.; Lewis B. E.; Long J. E.; Nambi S.; Sassetti C. M.; Stemmler T. L.; Argüello J. M. Fine-tuning of substrate affinity leads to alternative roles of Mycobacterium tuberculosis Fe2+-ATPases. J. Biol. Chem. 2016, 291, 11529–11539. 10.1074/jbc.M116.718239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinochet-Barros A.; Helmann J. D. Bacillus subtilis Fur is a transcriptional activator for the PerR-repressed pfeT gene, encoding an iron efflux pump. J. Bacteriol. 2020, 202, e00697-19 10.1128/JB.00697-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.; Haldar S.; Khan M. A.; Sharma S. D.; Merrick W. C.; Theil E. C.; Goss D. J. Fe2+ binds iron responsive element-RNA, selectively changing protein-binding affinities and regulating mRNA repression and activation. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 8417–8422. 10.1073/pnas.1120045109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.; Cotruvo J. A. Jr. The czcD (NiCo) riboswitch responds to iron(II). Biochemistry 2020, 59, 1508–1516. 10.1021/acs.biochem.0c00074. [DOI] [PubMed] [Google Scholar]
- Furukawa K.; Ramesh A.; Zhou Z.; Weinberg Z.; Vallery T.; Winkler W. C.; Breaker R. R. Bacterial riboswitches cooperatively bind Ni2+ or Co2+ ions and control expression of heavy metal transporters. Mol. Cell 2015, 57, 1088–1098. 10.1016/j.molcel.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S.; Chaudhury S.; Mehta D.; Ramesh A. RETRACTED ARTICLE: Discovery of iron-sensing bacterial riboswitches. Nat. Chem. Biol. 2021, 17, 924. 10.1038/s41589-020-00665-7. [DOI] [PubMed] [Google Scholar]
- Kellenberger C. A.; Wilson S. C.; Sales-Lee J.; Hammond M. C. RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP. J. Am. Chem. Soc. 2013, 135, 4906–4909. 10.1021/ja311960g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You M.; Litke J. L.; Jaffrey S. R. Imaging metabolite dynamics in living cells using a Spinach-based riboswitch. Proc. Natl. Acad. Sci. U.S.A. 2015, 112, E2756–E2765. 10.1073/pnas.1504354112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. C.; Wilson S. C.; Hammond M. C. Next-generation RNA-based fluorescent biosensors enable anaerobic detection of cyclic di-GMP. Nucleic Acids Res. 2016, 44, e139 10.1093/nar/gkw580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W.; Strack R. L.; Svensen N.; Jaffrey S. R. Plug-and-play fluorophores extend the spectral properties of Spinach. J. Am. Chem. Soc. 2014, 136, 1198–1201. 10.1021/ja410819x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z.; Wedd A. G. The challenges of determining metal-protein affinities. Nat. Prod. Rep. 2010, 27, 768–789. 10.1039/b906690j. [DOI] [PubMed] [Google Scholar]
- Young T. R.; Xiao Z. Principles and practice of determining metal–protein affinities. Biochem. J. 2021, 478, 1085–1116. 10.1042/BCJ20200838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. P.; Piszczek G.; Ferré-D’Amaré A. R. Isothermal titration calorimetry measurement of riboswitch-ligand interactions. Microcalorimetry of Biological Molecules: Methods and Protocols 2019, 1964, 75–87. 10.1007/978-1-4939-9179-2_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachas S. T.; Ferré-D’Amaré A. R. Convergent use of heptacoordination for cation selectivity by RNA and protein metalloregulators. Cell Chem. Biol. 2018, 25, 962–973. e5 10.1016/j.chembiol.2018.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H.-L.; Nesbitt D. J. Sequential folding of the nickel/cobalt riboswitch is facilitated by a conformational intermediate: Insights from single-molecule kinetics and thermodynamics. J. Phys. Chem. B 2020, 124, 7348–7360. 10.1021/acs.jpcb.0c05625. [DOI] [PubMed] [Google Scholar]
- Jeng S. C. Y.; Trachman R. J.; Weissenboeck F.; Truong L.; Link K. A.; Jepsen M. D. E.; Knutson J. R.; Andersen E. S.; Ferré-D’Amaré A. R.; Unrau P. J. Fluorogenic aptamers resolve the flexibility of RNA junctions using orientation-dependent FRET. RNA 2021, 27, 433–444. 10.1261/rna.078220.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Wei W.; Zhao J. Using a riboswitch sensor to detect Co2+/Ni2+ transport in E. coli. Front. Chem. 2021, 9, 631909. 10.3389/fchem.2021.631909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielazinski E. L.; Cutsail G. E. III; Hoffman B. M.; Stemmler T. L.; Rosenzweig A. C. Characterization of a cobalt-specific P1B-ATPase. Biochemistry 2012, 51, 7891–900. 10.1021/bi3006708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimunda D.; Long J. E.; Padilla-Benavides T.; Sassetti C. M.; Argüello J. M. Differential roles for the Co2+/Ni2+ transporting ATPases, CtpD and CtpJ, in Mycobacterium tuberculosis virulence. Mol. Microbiol. 2014, 91, 185–197. 10.1111/mmi.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes B. W.; Naditz A. L.; Anast J. M.; Schmitz-Esser S. Transcriptome sequencing of Listeria monocytogenes reveals major gene expression changes in response to lactic acid stress exposure but a less pronounced response to oxidative stress. Front. Microbiol. 2020, 10, 3110. 10.3389/fmicb.2019.03110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S.; Perdreau-Dahl H.; Morth J. P. The magnesium transporter A is activated by cardiolipin and is highly sensitive to free magnesium in vitro. eLife 2016, 5, e11407 10.7554/eLife.11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. T.; Smith K. P.; Rosenzweig A. C. Diversity of the metal-transporting P1B-type ATPases. J. Biol. Inorg. Chem. 2014, 19, 947–960. 10.1007/s00775-014-1129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinson O.; Lee A. T.; Rees D. C. A P-type ATPase importer that discriminates between essential and toxic transition metals. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 4677–4682. 10.1073/pnas.0900666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBrun M.; Loulergue J.; Chaslus-Dancla E.; Audurier A. Plasmids in Listeria monocytogenes in relation to cadmium resistance. Appl. Environ. Microbiol. 1992, 58, 3183–3186. 10.1128/aem.58.9.3183-3186.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchene N. A.; Mettert E. L.; Moore L. J.; Keleş S.; Willey E. R.; Kiley P. J. O2 availability impacts iron homeostasis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, 12261–12266. 10.1073/pnas.1707189114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.