Abstract
Like most temperate bacteriophages, phage Mx8 integrates into a preferred locus on the genome of its host, Myxococcus xanthus, by a mechanism of site-specific recombination. The Mx8 int-attP genes required for integration map within a 2.2-kilobase-pair (kb) fragment of the phage genome. When this fragment is subcloned into a plasmid vector, it facilitates the site-specific integration of the plasmid into the 3′ ends of either of two tandem tRNAAsp genes, trnD1 and trnD2, located within the attB locus of the M. xanthus genome. Although Int-mediated site-specific recombination occurs between attP and either attB1 (within trnD1) or attB2 (within trnD2), the attP × attB1 reaction is highly favored and often is accompanied by a deletion between attB1 and attB2. The int gene is the only Mx8 gene required in trans for attP × attB recombination. The int promoter lies within the 106-bp region immediately upstream of one of two alternate GTG start codons, GTG-5208 (GTG at bp 5208) and GTG-5085, for integrase and likely is repressed in the prophage state. All but the C-terminal 30 amino acid residues of the Int protein are required for its ability to mediate attP × attB recombination efficiently. The attP core lies within the int coding sequence, and the product of integration is a prophage in which the 3′ end of int is replaced by host sequences. The prophage intX gene is predicted to encode an integrase with a different C terminus.
Temperate phage Mx8 infects Myxococcus xanthus, a social gram-negative bacterium that lives in soil. The myxobacteria undergo a complex, multicellular developmental cycle in response to starvation. Development involves the morphogenesis of fruiting structures comprised of hundreds of thousands of cells that support the differentiation of a minority of cells into spores. Mature spores are resistant to UV light, desiccation, and heat and can germinate to develop into vegetative cells when nutrients become available. Mx8 has coadapted with its host to respond to this complex developmental cycle. The Mx8 prophage is stable upon the passage of an M. xanthus lysogen through cycles of development and germination (22). How the prophage maintains lysogeny throughout the dramatic changes in host gene expression during both sporulation and germination remains a mystery.
In a previous report, we described the initial genetic analysis of the 9.5-kilobase-pair (kb) immunity (imm) region of the Mx8 genome that includes the genes necessary for prophage integration and superinfection immunity (28). These two functions are required for the stability of the Mx8 prophage throughout development. The genes in the Mx8 imm region are densely packed and transcribed in a single direction. Three genes in the Mx8 imm region, mox, uoi, and int, encode products with known or suspected functions. The mox gene encodes a nonessential DNA adenine methylase (18), whereas uoi encodes a putative Mx8 excisionase and int encodes the Mx8 integrase (28, 37).
Plasmid vectors carrying the Mx8 int and attP genes integrate at the attB locus (16, 28, 34, 37). Such vectors have been used extensively to introduce second copies of M. xanthus genes at this ectopic locus, allowing for the construction of merodiploids that are more stable than those formed by homologous recombination (35). The ability of int-attP+ plasmids to integrate is our starting point for the dissection of the mechanism of Mx8 site-specific recombination in M. xanthus.
An independent study of the mechanism of Mx8 integration has shown that, unlike those of most other temperate phages and viruses, the Mx8 attP phage attachment site lies within the int coding sequence (37). In this paper, we show that the int gene is the only gene required in trans for integration and define the int promoter and the extent of the int coding sequence required for integration. We also describe the primary structure of the M. xanthus attB locus, confirm that the Mx8 attP site lies within the int gene, and characterize the changes in the structure of a bipartite attB locus that occur upon integration. In the accompanying paper (19), we show that, because the attP site lies within the int coding sequence, the integration event alters the primary sequence of the int gene to regulate the specific activity of its product.
MATERIALS AND METHODS
Bacterial strains.
M. xanthus DK1622 (11) is the wild type. The multiple-mutant strain DZ1 (4) is the preferred host for the growth of phage Mx8 and was used to assay plasmid integration dependent on subcloned Mx8 site-specific recombination functions. Escherichia coli JM107 (39) was used for the construction of plasmids and the preparation of plasmid DNA. Electroporation (36) was used to introduce plasmids into JM107. Liquid CTPM medium (38) was used for the routine growth of M. xanthus. Derivatives of DZ1 with integrated plasmids were grown in CTPM medium with kanamycin (40 μg/ml) and/or the combination of spectinomycin (800 μg/ml) and streptomycin sulfate (1 mg/ml). Derivatives of JM107 with plasmids were grown in Luria-Bertani medium supplemented with ampicillin (100 μg/ml), kanamycin (40 μg/ml), or spectinomycin and streptomycin (50 μg/ml each). Antibiotics were from Sigma Chemical Co. Oligonucleotides used for plasmid construction and mutagenesis were made by Biosource Inc.
Integration-proficient parental plasmids.
Plasmids with portions of the Mx8 genome were derived in one or more steps from DNA isolated from the wild-type strain of Mx8 (20) and are listed in Table 1. These include the kanamycin-resistant (Kmr) plasmid pAY50, a derivative of pACYC177 (6) which carries the Mx8 functions both necessary and sufficient for integration and superinfection immunity on an 8.1-kb Sau3AI-PvuII fragment of Mx8 DNA (18). The sequence of a larger region of the Mx8 genome including this 8.1-kb fragment has been assigned GenBank accession no. U64984 (28); coordinates (base pairs) used throughout this paper are from this entry. The Kmr int-attP plasmids pAY60, pAY62, and pAY721 (28) are subclones of Mx8 DNA in plasmid pBGS18 (33). Plasmid pAY721 has a 2.2-kb insert of Mx8 DNA in pBGS18 with the int termination codon at its 3′ end (28) and is used as a positive control in all electroporation experiments measuring the function of int and/or attP. Plasmid pAY952 is an Spr Smr, integration-proficient derivative of plasmid pGB2 (17). pAY952 has no sequence homology with pAY721, except within their common EcoRI-HindIII inserts of Mx8 DNA. None of the parental plasmids (pACYC177, pBGS18, or pGB2) used for subcloning of the Mx8 int-attP genes can integrate into the M. xanthus genome, as evidenced by their failure to give rise to antibiotic-resistant recombinants when electroporated into M. xanthus. Other plasmids were derived in one or more steps from these integration-proficient parental plasmids by standard cloning methods (29). Derivatives of pAY721 with the intVA1 mutation (pAY979), the intVA42 mutation (pAY754), and a combination of the intVA1 and intVA42 mutations (pAY990) have been described elsewhere (28).
TABLE 1.
Plasmids
| Plasmid | bpa | Vectorb | Relevant genotypec | Source or reference |
|---|---|---|---|---|
| pACYC177 | Apr Kmr | 6 | ||
| pBGS18 | Kmr | 33 | ||
| pGB2 | Spr Smr | 7 | ||
| pLITMUS28 | Apr | New England Biolabs | ||
| pLITMUS29 | Apr | New England Biolabs | ||
| pRS552 | Apr Kmr | ′lacZ | 32 | |
| pAY50 | 1–8072 | pBGS18/Kmr | 18 | |
| pAY60 | 4585–8072 | pBGS18/Kmr | 28 | |
| pAY62 | 4585–8072 | pBGS18/Kmr | This study | |
| pAY721 | 4585–6809 | pBGS18/Kmr | uoi+ int+ attP+ | This study |
| pAY952 | 4585–6809 | pGB2/Spr Smr | uoi+ int+ attP+ | This study |
| pAY979 | 4585–6809 | pBGS18/Kmr | intVA42 | 28 |
| pAY754 | 4585–6809 | pBGS18/Kmr | intVA1 | 28 |
| pAY990 | 4585–6809 | pBGS18/Kmr | intVA1 intVA42 | 28 |
| pAY722 | 4808–6809 | pBGS18/Kmr | uoi+ int+ attP+ | This study |
| pAY759 | 4979–6809 | pBGS18/Kmr | uoi+ int+ attP+ | This study |
| pAY743 | 4585–5403 | pLITMUS29/Apr | uoi-14(Am) Pint-14 | This study |
| pAY951 | 4585–6809 | pBGS18/Kmr | uoi-14(Am) Pint-14 | This study |
| pAY995 | 4585–6809 | pBGS18/Kmr | uoi-14(Am) Pint-14 | This study |
| pAY950 | 4585–6809 | pBGS18/Kmr | uoi+ int+ attP+ | This study |
| pAY735 | 4585–6809 | pBGS18/Kmr | uoi+ int-nd attP+ | This study |
| pAY980 | 4585–6809 | pBGS18/Kmr | uoi+ Δint-5403/5851 attP+ | This study |
| pAY725 | 4585–6809 | pBGS18/Kmr | int-1(Am) | This study |
| pAY982 | 4585–6809 | pBGS18/Kmr | int-1(Am) | This study |
| pAY703 | pBGS18/Kmr | mglBA+ | 18 | |
| pAY971 | 5073–6809 | pBGS18/Kmr | mglBA+int+ attP+ | This study |
| pAY972 | 5208–6809 | pBGS18/Kmr | mglBA+int+ attP+ | This study |
| pAY987 | 5238–6809 | pBGS18/Kmr | mglBA+ Δint-5238 | This study |
| pAY992 | 5298–6809 | pBGS18/Kmr | mglBA+ Δint-5298 | This study |
| pAY993 | 5337–6809 | pBGS18/Kmr | mglBA+ Δint-5337 | This study |
| pAY733 | 4585–6504 | pBGS18/Kmr | Δint-6504 | This study |
| pAY734 | 4585–6711 | pBGS18/Kmr | Δint-6711 | This study |
| pAY146 | 4585–6716 | pBGS18/Kmr | Δint-6716 | This study |
| pAY997 | 1–8072 | pACYC177/Kmr | uoi+ int-NotI′ attP+ | This study |
| pAY738 | 4585–5210 | pRS552/Apr Kmr | uoi+ int-lacZ (fusion to GTG-5085) | This study |
| pAY739 | 4585–5087 | pRS552/Apr Kmr | Δuoi int-lacZ (fusion to GTG-5208) | This study |
Coordinates of Mx8 DNA are those of GenBank accession no. U64984.
Antibiotic resistance determinants retained from the vector are indicated.
The uoi-14 mutation inactivates the int promoter (Pint); int-nd is an uncharacterized PCR-induced mutation within bp 4585 to 6089 that inactivates the int gene. The int-NotI′ mutation is a 4-bp insertion within int made by filling in the NotI site and ligation.
Plasmids with deletions extending rightward into uoi and int.
To map the int promoter, two derivatives of pAY721 were made with deletions extending rightward from a unique MfeI site at bp 4585 in the 8.1-kb sequence of the Mx8 immunity (imm) region to different end points upstream of int, bp 4808 and bp 4979. To construct pAY722, the PCR was used with 5′ primers having the sequences CCCAAGCTTCCTAGGTAGCGGAAGGGCTCTC and AAAAGATCTACTGCGTGTGCATCG and template plasmid pAY60. The BglII-HindIII fragment from the product (bp 4808 to 6809) was ligated to the BamHI and HindIII sites of pBGS18. To make pAY759, the Acc65I-StuI fragment of pAY973 (bp 4979 to 6809) was ligated to the larger fragment of pAY721.
Plasmids with small substitutions or insertions in uoi or int.
The open reading frame immediately upstream of int, uoi, is predicted to encode a putative excisionase and overlaps the 5′ end of int (28). We replaced codon 14 (Ser) of uoi, AGC, with a TAG (amber) codon. Primer pairs with the sequences GGGGGAAAAGATCTCGTGGACTGCAATTG and GGTGGATGTCTAGACACTTGCGAGGGCTGCCGC and the sequences CACCAGTGCGAGCACGTGCCGACGCGTCAGCAC and GCAAGTGTCTAGACATCCACCATTCGCAAC were used to amplify Mx8 DNA. The products were annealed and then amplified in a second step with primers having the sequences GGGGGAAAAGATCTCGTGGACTGCAATTG and CACCAGTGCGAGCACGTGCCGACGCGTCAGCAC. The product resulting from the second step was cleaved with BglII and StuI and ligated to pLITMUS29 to make pAY743. The 819-bp MfeI-StuI fragment with the uoi-14(Am) allele and the 5′ end of int was joined together with the 1.4-kb StuI-HindIII fragment of pAY721 with the 3′ of int to make pAY951. Cleavage of plasmid pAY743 with XbaI confirmed the presence of the uoi-14(Am) mutation. To confirm that the integration-defective phenotype of plasmid pAY951 is due solely to the uoi-14(Am) mutation, the largest BglI fragment of pAY951 was ligated to the smaller BglI fragments of pAY721 to make the isogenic integration-defective plasmid pAY995. The sequence of the region upstream of the BglI site was determined to show that the only change in this region is the desired one to amber. Plasmid pAY950, the integration-proficient, otherwise isogenic partner of pAY995 without the uoi(Am) mutation, was made by use of primers with the sequences GGGGGAAAAGATCTCGTGGACTGCAATTG and CCCAAGCTTCCTAGGTAGCGGAAGGGCTCTC to amplify Mx8 DNA. The 2.2-kb product was cleaved with BglII and HindIII and then ligated to the BamHI and HindIII sites of pBGS18 to make pAY735. pAY735 and pAY721 were cleaved with StuI and HindIII, and the 1.4-kb fragment of pAY721 was ligated to the larger fragment of pAY735 to make pAY950. pAY950 differs from control plasmid pAY721 only in the extent of polylinker sequence that it retains from parental vector pBGS18. pAY980 is a derivative of pAY721 with a substitution of the 86-bp StuI-PstI fragment of pLITMUS28 for the StuI-PstI fragment internal to int (bp 5403 to 5851). A derivative of pAY721, pAY982, with the int-1(Am) mutation was constructed by two-step amplification of plasmid template pAY721 with primer pairs having the sequences CAGCACTGACCCTTTTG and CTGGGCACCTAGGTCGCGTCAAGAAGTCG and the sequences GCGGACCTAGGTGCCCAGCCGTCAGGAGT and AGCGGATAACAATTTCACACAGGA. The amplified 1.7- and 0.5-kb products were annealed and amplified with primers having the sequences CAGCACTGACCCTTTTG and AGCGGATAACAATTTCACACAGGA. The 2.2-bp product was cleaved with EcoRI and HindIII and ligated to pBGS18 to make pAY725. Plasmids pAY721 and pAY725 were cleaved with BglI, and the largest BglI fragment of plasmid pAY725 was ligated to the three smaller BglI fragments of plasmid pAY721 to make plasmid pAY982. Plasmids pAY725 and pAY982 were cleaved with AvrII to confirm the presence of the int-1(Am) mutation.
Plasmids that express int from the mgl promoter and their deletion derivatives.
To express int from the constitutive mglBA promoter, several DNA fragments with different 5′ ends were amplified and cloned downstream of the mglBA operon (9) in plasmid pAY703 (18). Primers with the sequences AAAGGTACCCTGACGGCTGGGCATCGTG and CCCGAATTCTCAGGTAGCGGAAGGGCTCT were used to amplify a 1.7-kb fragment from template pAY721 with the int coding sequence from bp 5073 to 6809. The product was cleaved with EcoRI and Acc65I and ligated to pAY703 to make plasmid pAY971. Primers with the sequences AAAGGTACCCTGACGGCTGGGCATCGTGGGTAACGTCTATCGCAAGAAGGCCACT and CCCGAATTCTCAGGTAGCGGAAGGGCTCT were used to amplify bp 5208 to 6809 from template pAY721. The 1.6-kb product was cleaved with EcoRI and Acc65I and ligated to pAY703 to make pAY972. Smaller fragments within int, bp 5238 to 5433, 5298 to 5433, and 5337 to 5433, were amplified with 5′ primers having the sequences AAAGGTACCCTGACGCTACTTGTGGATGTGGTGG, AAAGGTACCCTGAGAAGAAGACGAAGTGAAGGCG, and AAAGGTACCCTGAACCGAGAAGCGCGTGGCCGCG, respectively, the common 3′ primer having the sequence ACGCGCCCCTCCATCCACTTG, and template pAY721. DNA fragments were cleaved with Acc65I and StuI to yield products of 167, 106, and 56 bp, which were ligated to pAY973 (19) to make plasmids pAY987, pAY992, and pAY993, respectively.
Plasmids with deletions extending leftward into int.
To construct defined 3′ deletions of int, we used the PCR to introduce premature termination codons within int in place of codons at bp 6504 (Leu474) and bp 6711 (Ala543). The common 5′ primer with the sequence GGGGGAAAAGATCTCGTGGACTGCAATTG was used with a primer having the sequence CCCCCAAGCTTGGCTACTGGCGCGGATTGGCGTAGAA or CCCCCAAGCTTCCTAAAGCCGCTCCAGGGCCATCAG to amplify Mx8 DNA. BglII-HindIII fragments made from the products (bp 4585 to 6504 and 4585-6711) were ligated to the BamHI and HindIII sites of pBGS18 to make plasmids pAY733 and pAY734, respectively. Plasmid pAY146 is a derivative of pAY62 made by cleavage with NotI and HindIII. Cleaved plasmid DNA was treated with the E. coli DNA polymerase I large fragment in the presence of deoxyribonucleoside triphosphates to fill in single-stranded ends and ligated. In plasmid pAY146, codon 543 of int is fused in frame to the 5′ end of lacZ carried on pBGS18. The hybrid protein initiated at the GTG-5085 (GTG at bp 5085) start codon of int is predicted to have the first 543 amino acid residues of integrase and 56 residues of LacZ. Plasmid pAY997 is a derivative of pAY50 made by cleavage with NotI, filling in, and ligation. The expected sequence of the junction made by filling in was confirmed by cleavage of pAY997 with EagI, NgoMI, and NotI.
Plasmids that express int-lacZ translational fusions from the int promoter and β-galactosidase assays.
Apr Kmr plasmid pRS552, used for constructing int-lacZ fusions, carries unique EcoRI and BamHI sites immediately upstream of the majority of the lacZ coding sequence (32). The 5′ primer with the sequence AGCGGATAACAATTTCACACAGGA and the 3′ primers with the sequences GGGGGGGATCCCCACGGCGCCGCCCTGCCTT and GGGGGGGATCCACGATGCCCAGCCGTCAGGA were used to amplify template pAY721. The products were cleaved with EcoRI and BamHI and then ligated to the same sites of pRS552 to make pAY738 and pAY739, which have in-frame translational fusions of the GTG-5085 and GTG-5208 start codons of int, respectively, to the seventh codon of lacZ.
For β-galactosidase assays, Kmr derivatives of DZ1(Mx8) carrying pAY738 or pAY739 integrated into the prophage genome by homologous recombination were constructed by electroporation of these plasmids into the Kms lysogen and selection for Kmr recombinants. These recombinants and their parental strain were grown to a density of 4 × 108/ml in CTPM medium at 32°C. Eight milliliters of each culture was pelleted by low-speed centrifugation, resuspended in 0.10 volume of TPM buffer (10 mM Tris, 8 mM MgSO4, 1 mM potassium phosphate [pH 7.6]), and sonicated for three 15-s 5-W pulses with a Microson cell disrupter. After sonication, 0.25 volume of 0.1% sodium dodecyl sulfate was added, and assays for β-galactosidase activities and protein concentrations were performed as described previously (14). Activities are given in nanomoles of o-nitrophenol per minute per milligram produced by hydrolysis of the substrate o-nitrophenol-β-galactoside (Sigma).
PCR amplification of M. xanthus genomic DNA.
Colonies of Kmr electroporants of host DZ1 were grown for 5 to 7 days on CTPM plates, patched on CTPM agar, and grown for an additional 2 to 3 days at 32°C. Approximately 0.1 g of cell paste was scraped from each plate and suspended in 200 μl of TE buffer (10 mM Tris-HCl [pH 7.6], 0.1 mM EDTA) supplemented with 100 μg of RNase A. Cells were incubated at 85°C for 5 min, extracted with an equal volume of distilled, buffered phenol, and centrifuged at 15,000 × g for 15 min. The aqueous phase was extracted twice with an equal volume of chloroform, and 0.5 to 5.0 μl of supernatant was used as a template in 50-μl reaction mixtures for PCR. Conditions for PCR amplification were as follows: 0.2 μM each oligonucleotide primer, 1.5 mM MgCl2, 0.25 mM each nucleotide triphosphate, and 0.5 to 2.0 U of Taq DNA polymerase (Gibco-BRL).
Amplification of attP, attB, attL, and attR sites with specific primer pairs.
To determine where phages or attP-containing plasmids integrate within the attB locus (at site attB1 or attB2), we performed two separate PCRs to amplify the attL and attR sites generated by phage or plasmid integration. These reactions yield products of different sizes, depending on whether integration occurs at the attB1 or the attB2 site. As controls for these PCR assays, we often tested whether strains carrying a prophage or integrated plasmid retained the attB sites or also had an attP site. attB primers with the sequences GGGGGAATTCGTCGACTGCGCAGGTCCGCGGAGGA and AAAAAAGCTTCCGGGCGGCCTTGCGGAATGAT yield a 591-bp product when used to amplify template DK1622 or DZ1 genomic DNA (see below). attP primers with the sequences TCAGCGCTTCAGGTCCGGGACTGGGAC and CCCAAGCTTCCTAGGTAGCGGAAGGGCTCTC yield a 969-bp product when used to amplify phage Mx8 DNA. attL primers with the sequences GGGGGAATTCGTCGACTGCGCAGGTCCGCGGAGGA and CCCAAGCTTCCTAGGTAGCGGAAGGGCTCTC yield a product of 526 bp if integration occurs at attB2 (or integration is accompanied by a deletion between attB1 and attB2) or a product of 692 bp if integration occurs at attB1. attR primers with the sequences AAAAAAGCTTCCGGGCGGCCTTGCGGAATGAT and TCAGCGCTTCAGGTCCGGGACTGGGAC yield a 1,006-bp product if integration occurs at attB2 and an 840-bp product if integration occurs at attB1 (or integration is accompanied by a deletion between attB1 and attB2).
To distinguish whether derivatives of plasmid pAY703 which contain the mgl operon have integrated at either the mgl or the attB loci, genomic DNAs isolated from Kmr electroporants were amplified with the attR primers. We confirmed that templates that yielded no product in these reactions had a plasmid integrated at the mgl locus by showing that they yielded a product when amplified with the attP primer pair.
Electroporation assays for plasmid integration.
To quantify the efficiencies with which plasmids can integrate into the M. xanthus genome, 1 ml of DZ1 cells at an exponential density of 5 × 108/ml was concentrated 25-fold and electroporated with plasmid DNA (100 to 200 ng) under conditions standard for the electroporation of M. xanthus (12). After electroporation, cells were suspended in 3 ml of CTPM medium to a density of 1.7 × 108/ml and grown at 32°C for 16 h before being plated on CTPM agar plates with antibiotics. Coelectroporations were done in a similar way, with a mixture of 100 to 200 ng of each plasmid. Colonies formed by antibiotic-resistant electroporants were counted after 7 days of incubation at 32°C.
Nucleotide sequences of the int and uoi genes and the attB locus and their accession numbers.
The sequence of a region of the Mx8 genome including uoi and int has been assigned GenBank accession no. U64984 (28). To determine the sequence of the attB locus, we amplified a 591-bp fragment of the attB locus (37) from both mutant strain DZ1 and wild-type strain DK1622 by using the attB primers. The products were sequenced by the dideoxy method (30). Sequencing runs were performed by Commonwealth Biotechnologies, Inc., Richlands, Va., and resolved on an ABI Prism model 377 automated sequencing apparatus. The sequence of the attB locus, determined to be identical for both DK1622 and DZ1, was the same as that determined by Tojo et al. (37), which has been assigned GenBank accession no. D26557.
RESULTS
The int promoter is located in a 106-bp region upstream of the int structural gene.
The model for the site-specific integration of temperate phage genomes initially proposed by Campbell (3) has proven particularly robust. Almost all temperate phages integrate into their host chromosome by a mechanism of site-specific recombination, in which an integrase protein catalyzes a four-strand exchange event between an attP site on the circular phage chromosome and an attB site on the host chromosome. Integration generates a linear prophage chromosome with termini representing the attL and attR products of the integration reaction.
Like most temperate phages, bacteriophage Mx8 of M. xanthus integrates into the chromosome of its host by a mechanism similar to that for coliphage λ, as predicted by Campbell (3) over 25 years ago (Fig. 1). As for most temperate phages, the genetic elements necessary for the site-specific recombination of Mx8 are clustered on the phage genome. When subcloned onto a circular plasmid, these genes confer upon the plasmid the ability to integrate into the host chromosome.
FIG. 1.
Model for Mx8 prophage integration and excision. The integration of phage λ into its preferred site on the E. coli chromosome, attB (B), is catalyzed by the phage-encoded integrase (Int) enzyme, the product of the int gene. The attB × attP (P) recombination event generates the attL (L) and attR (R) prophage junctions (3). Excision of the λ prophage involves the reciprocal attL × attR event, in which the phage-encoded Xis (excisionase) protein controls the directionality of Int-catalyzed recombination. The integration of phage Mx8 into the attB locus on the M. xanthus chromosome is a more complex event. Integration can occur at either of two tandem sites (attB1 and attB2) within the attB locus, catalyzed by the phage-encoded integrase (Int). The preferred product of integration, formed by recombination between the attP and attB1 sites, is shown. Unlike the case for phage λ, the Mx8 attP site lies within the int gene, and the integration event changes the 3′ end of int and the C terminus of its predicted product. We presume that excision of the Mx8 prophage is catalyzed by the modified, prophage-encoded integrase (IntX) and may be stimulated by the Mx8 Uoi protein, the predicted sequence of which is similar to those of other excisionases (28).
Previously, we have shown that the addition of a 2.2-kb fragment of Mx8 DNA to Kmr plasmid pBGS18 enables the recombinant plasmid, pAY721, to integrate into the attB locus of M. xanthus (28). The Mx8 DNA fragment subcloned in pAY721 begins about 500 bp upstream of the first of two int start codons and ends at the int stop codon (Fig. 2). The int gene has two alternate translation start codons, GTG-5085 and GTG-5208. Derivatives of plasmid pAY721 in which only one of these GTG codons has been changed to GCG can form Kmr recombinants after electroporation into a sensitive M. xanthus host. In contrast, an otherwise isogenic plasmid in which both GTG codons have been changed to GCG cannot form Kmr recombinants (28).
FIG. 2.
Genetic elements required for the integration of plasmids with Mx8 DNA inserts. Coordinates of Mx8 DNA inserts (base pairs), based on GenBank accession no. U64984 (28), are shown above the open boxes representing the uoi and int coding sequences (top) which, like the coliphage λ xis and int genes (10), overlap. The hatched regions in the int gene correspond to conserved domains I and II of integrases (1, 15); conserved domain II includes the active-site tyrosine residue (23). The filled region within int, O, is the attP common core. The extents of Mx8 DNA inserts present in each plasmid are shown as horizontal thin lines (bottom left). Mutant plasmids are derived from pAY721, which contains a 2.2-kb region of the Mx8 genome with the functional int-attP genes. pAY754, pAY979, and pAY990 have missense GTG-to-GCG changes (open circles) of one or both of the two alternate int start codons (28). pAY722 and pAY759 have deletions extending into the 3′ end of the 2.2-kb region. pAY980 is a derivative of pAY721 with a deletion, Δint-5403/5851 (open box), that removes a portion of domain I. pAY995 and pAY982 have amber codons (filled circles) in place of codon 14 of uoi and the first start codon (GTG-5085) of int, respectively. pAY734 and pAY733 are deletion derivatives with int coding sequences that terminate prematurely at engineered stop codons. pAY146 is a deletion derivative of pAY721 that makes a mutant integrase with an altered C terminus (small black box) encoded by adjacent vector sequences. The new C terminus of the active integrase made by pAY997 is produced from a shifted reading frame within int, translated due to the presence of a 4-bp insertion introduced into the 3′ end of int by filling in of its internal, unique NotI site. The efficiencies of electroporation (EOE) of the plasmids are the numbers of Kmr recombinants arising per microgram of DNA electroporated into host DZ1 and are the averages of at least three independent determinations (see Materials and Methods).
To map the int promoter, we determined the extent of sequences within this 500-bp upstream region required for Int-mediated site-specific recombination and made two derivatives of pAY721 with shorter upstream regions. These derivatives, pAY722 and pAY759, contain 277 and 106 bp, respectively, upstream of GTG-5085. Both of these deletion derivatives give rise to Kmr recombinants when electroporated into host DZ1 and retain the ability to integrate (see Fig. 2). These results show that the natural Mx8 promoter for int must lie within 106 bp upstream of its translation start codon GTG-5085.
A critical element of the int promoter lies within the upstream uoi gene.
Both of these deletion derivatives, pAY722 and pAY759, also retain the uoi gene, which is predicted to encode a product of 74 amino acids. The sequence of the Uoi protein is similar to those of other phage and plasmid excisionases (28), site-specific DNA-binding proteins that control the directionality of the Int-mediated site-specific recombination reaction. pAY759 has only 12 bp of Mx8 DNA upstream of the start codon of uoi and is likely missing the uoi promoter. However, we considered the possibility that uoi might be required for integration and fortuitously expressed from this plasmid, because some temperate phages require the products of several genes for site-specific integration. To exclude this possibility, we made a derivative of pAY721, pAY995, with the change of an ACG codon (Ser14) of uoi to TAG (amber). To our surprise, we found that this plasmid lost the ability to integrate (Fig. 2).
The uoi-14(Am) mutation could result in a loss of function for one of two possible reasons. Either both uoi and int are required in trans for integration or the uoi-14 mutation inactivates the int promoter. To distinguish between these possibilities, we examined whether a plasmid with the uoi-14 mutation and a plasmid with an int mutation could complement one another for integration. In this complementation test, as for its controls, we assayed plasmid integration after the coelectroporation of two different plasmids.
As shown in Table 2, Spr Smr plasmid pAY952, with the same subcloned region of Mx8 DNA as that in Kmr integration-proficient plasmid pAY721, gives rise to Spr Smr electroporants efficiently after electroporation into sensitive host DZ1. Kmr plasmid pAY980 is a derivative of pAY721 with a substitution of a short polylinker sequence for a large deletion (bp 5403 to 5851) internal to int (intΔ5403/5851) that interrupts conserved domain I of integrase (Fig. 2). As expected, pAY980 (intΔ5403/5851) cannot integrate (Table 2). However, when plasmids pAY952 (int+ Spr Smr) and pAY980 (intΔ5403/5851 Kmr) are coelectroporated into DZ1, Kmr recombinants arise, and a significant fraction of these are Sps Sms. These results show that pAY980 (intΔ5403/5851) has a functional attP site and that int function supplied by pAY952 can act in trans to mediate the integration of pAY980. Although pAY995 (uoi-14 Kmr) cannot integrate, it too can be complemented for integration by pAY952 upon coelectroporation. However, when pAY980 (intΔ5403/5851 Kmr) is coelectroporated with pAY995 (uoi-14 Kmr), Kmr electroporants arise only at a low efficiency. This result suggests that the uoi-14 mutation acts in cis to impair integration and inactivates the int promoter. It is likely that the defect caused by the uoi-14 mutation is not due to a polar effect of this amber mutation. pAY982, an otherwise isogenic derivative of pAY721 with the int-1(Am) mutation, which replaces the first alternate start codon of int, GTG-5085, with TAG, can integrate as efficiently as its pAY721 parent (Fig. 2).
TABLE 2.
The uoi-14 mutation reduces the activity of the int promotera
| Plasmid(s) [relevant genotype(s)] | EOE (103 μg−1) for the following recombinants:
|
% Sms Sps electroporants | |
|---|---|---|---|
| Smr Spr | Kmr | ||
| pAY952 (int+ attP+) | 4.3 × 105 | 100 | |
| pAY980 (Δint-5403/5851) | <0.005 | ||
| pAY995 (uoi-14) | <0.005 | ||
| pAY952 (int+ attP+) + pAY980 (Δint-5403/5851) | 7.0 × 105 | 0.94 | 50 |
| pAY952 (int+ attP+) + pAY995 (uoi-14) | 5.0 × 105 | 0.81 | 58 |
| pAY980 (Δint-5403/5851) + pAY995 (uoi-14) | 0.008 | ||
The efficiency of electroporation (EOE) of each plasmid alone or in coelectroporations was measured as the numbers of either Spr Smr or Kmr recombinants of host DZ1 as described in Materials and Methods. To determine the percentage of Sps Sms electroporants, 100 independent Kmr electroporants were plated on CTPM medium with spectinomycin and streptomycin, and growth was scored after 72 h of incubation at 32°C.
The int gene is the only Mx8 gene required in trans for integration.
If the uoi-14 mutation inactivates the int promoter, then the expression of int alone (in the absence of uoi) from a heterologous promoter should be sufficient to promote plasmid integration. We have shown that when the phage Mx8 mox gene (18), the M. xanthus sglK gene (38), or the E. coli glk gene (unpublished results) is subcloned into pAY703 and the subclones are integrated by homologous recombination into the M. xanthus mgl locus, the genes are expressed as part of the mglBA operon. Therefore, we made a series of plasmids in which the int gene or versions of the int gene truncated at the 5′ end are expressed from the constitutive mglBA promoter. Portions of the int region in plasmid pAY721 were amplified and subcloned into plasmid pAY703 immediately downstream of mglA, the more distal gene in the mgl operon.
Because pAY703 has the mglBA genes, it can integrate into the mgl locus of DZ1 by homologous recombination and can give rise to Kmr recombinants after electroporation of this host (18). When we add the int-attP genes to pAY703, we give this plasmid the option of integrating either at the mgl locus by homologous recombination or at the attB locus by site-specific recombination. Site-specific recombination prevails over homologous recombination. Stephens and Kaiser (35) have shown that a plasmid with both the mglBA and the int-attP genes integrates preferentially at the attB locus.
Figure 3 shows that when int alone is expressed from the mgl promoter on pAY703, starting with either GTG-5085 (pAY971) or GTG-5208 (pAY972), Kmr electroporants are obtained at high efficiencies. These recombinants are formed by site-specific recombination. When chromosomal DNA is isolated from these recombinants and amplified with the attL and attR primer pairs, products of the expected sizes are observed, indicating the presence of recombinant attL and attR sites. This result confirms that uoi is not required for integration. When otherwise isogenic derivatives of pAY703 that express 5′ truncated versions of int beginning at potential start codons ATG-5250 (pAY987), GTG-5310 (pAY992), and GTG-5349 (pAY993) are electroporated into host DZ1, a different result is obtained. All of these Kmr recombinants carry plasmids integrated at the mgl locus. When chromosomal DNA is isolated from these recombinants and amplified, a primer pair specific for attL or attR yields no amplification products, whereas an attP-specific primer pair yields a product of the expected size (data not shown). These results indicate that the 5′ coding sequence between GTG-5208 and ATG-5250 is essential for int function and are consistent with our finding that int has two alternate start codons, GTG-5085 and GTG-5208 (28).
FIG. 3.
The int gene is the only Mx8 gene required in trans for site-specific integration. Amplified regions of the int gene beginning with its first five potential translation start codons and ending with its stop codon were subcloned downstream of the mglBA genes on plasmid pAY703 (18). The 5′ ends of the portions of Mx8 DNA subcloned into these derivatives of pAY703 are shown as horizontal lines below the map of uoi and the 5′ end of int. The relative positions of these genes with respect to the Mx8 imm sequence are shown (see Fig. 5). Each of the plasmids expresses int or a 5′ truncated version of int as the third gene in the constitutive mgl operon (arrows). All five plasmids give rise to Kmr recombinants when electroporated into host DZ1. These can arise due to either Int-mediated site-specific recombination into the attB locus or homologous recombination into the mgl locus (35). When DNAs isolated from eight independent recombinants carrying integrated plasmids pAY971 and pAY972 are amplified with the attL primer pair, all of these recombinants yield an attL junction site fragment, indicating that they have integrated at attB (see Fig. 6). In contrast, none of eight independent recombinants carrying plasmids pAY987, pAY992, and pAY993 yield an attL junction fragment in the same assay. EOE, efficiency of electroporation.
The C-terminal 30 amino acids of Int are dispensable for integrase activity.
To define the extent of the int coding sequence distal to the attP core required in trans for the attP × attB reaction, we made additional derivatives of plasmid pAY721. Deletion derivatives pAY733 and pAY734, in which the int coding sequence is terminated prematurely by amber stop codons in place of codons Leu474 (bp 6504) and Ala543 (bp 6711), respectively, do not integrate after electroporation into host DZ1 (Fig. 2). Neither does plasmid pAY146, in which the coding sequence for 56 amino acids in the N-terminal region of LacZ is added to the 3′ end of int, terminating with Ala543.
In contrast, plasmid pAY997, made by filling in the unique NotI site within int, does integrate. This plasmid is predicted to encode a version of the Int protein that diverges in primary sequence from the wild type after Ala544 and has 10 amino acids added after this residue before translation termination (Fig. 4). A derivative of pAY50 with a large insert of DNA present at the filled-in NotI site also is able to integrate (data not shown). Taken together, these results suggest that the most C-terminal 30 amino acid residues of Int are not essential for attP × attB recombination but that C-terminal alterations of Int that are predicted to remove 31 or more amino acids from the C terminus of Int abolish integration (Fig. 4).
FIG. 4.
The last 30 amino acid residues of the Int C terminus are not required for integration. The predicted amino acid sequences of the C termini of the mutant integrases made by plasmids with changes in the 3′ end of int (see Fig. 2) are shown; 60 refers to a stretch of 60 residues in the wild-type sequence that is not shown. Residues corresponding to the common core are shown in bold, and those encoded by vector sequences are underlined. Residues conserved between new C termini and the wild-type C terminus are indicated by asterisks. Integration-proficient plasmids pAY721 and pAY997 diverge in sequence after residue Ala544. pAY146, pAY734, and pAY733, which either change or remove Ala544, cannot integrate. In the accompanying paper (19), we show that the IntX integrase encoded by plasmid pAY999 is active.
Tojo et al. (37) reported that a similar plasmid with the filled-in NotI site lost the ability to integrate. However, they based their conclusion on their failure to find recombinants and on the sequence determination of the mutational change. To draw our conclusions, we have quantified the average efficiency of electroporation from at least three independent determinations for each plasmid and have measured the efficiency of electroporation of pAY721 as an internal, positive control for each preparation of electrocompetent host cells. In addition, we confirmed the genotype of the mutation carried by this plasmid by a method that does not rely solely on DNA sequence (see Materials and Methods). The dideoxy method (30) yields a highly compressed sequence ladder across this region of int with this mutation and is difficult to interpret.
Structure of the attB locus: two M. xanthus attB sites for phage Mx8 integration lie within a cluster of three tandem tRNA genes.
Tojo et al. (37) determined the sequences of the prophage junction sites, attL and attR, formed upon the integration of phage Mx8 into its preferred locus on the M. xanthus genome, attB. Their analysis revealed the surprising result that the Mx8 attP site lies within the int coding sequence. This result predicts that, upon integration of the Mx8 prophage, the sequence of the int gene will be modified by the integration reaction catalyzed by its product. If this is the case, the integration will result in the formation of a lysogen in which the recombinant derivative of the int gene, intX, encodes a product with a different C terminus. The C terminus of the prophage-encoded IntX protein is predicted to have 13 amino acid residues in place of the 112 residues that terminate the phage-encoded Int protein. However, our 3′ deletion analysis of the phage int gene shows that substitution mutations that are predicted to alter more than 30 C-terminal residues of the Int protein result in a loss of function. From these results, we might predict that the product of the prophage intX gene should be inactive.
On the other hand, lysogens of M. xanthus carrying a single Mx8 prophage release infectious particles by spontaneous induction. When DNA is isolated from these phage particles and analyzed, the int gene is found to be identical in structure to that of the parental phage used to construct the lysogens (data not shown). For Mx8, as for other temperate phages, we expect that prophage excision as well as integration will depend on integrase. If the excision of Mx8 depends on the prophage intX gene, then the product of intX should be active.
Therefore, we examined whether the attP site lies within the int gene and whether the prophage intX gene encodes an active product. In this study, we confirm the findings of Tojo et al. (37) and show that, indeed, the attP site is within int. We also find that the integration of a plasmid with the int-attP genes into the M. xanthus genome can occur at two different attB sites, attB1 and attB2. The integration of Mx8 causes changes in the primary structures of two transcription units, the phage int gene and either of two M. xanthus tRNA genes, within which the attB sites lie. In the accompanying paper (19), we show that, indeed, the intX gene encodes an active product, and we measure the relative abilities of the Int and IntX integrases to catalyze a variety of site-specific recombination reactions.
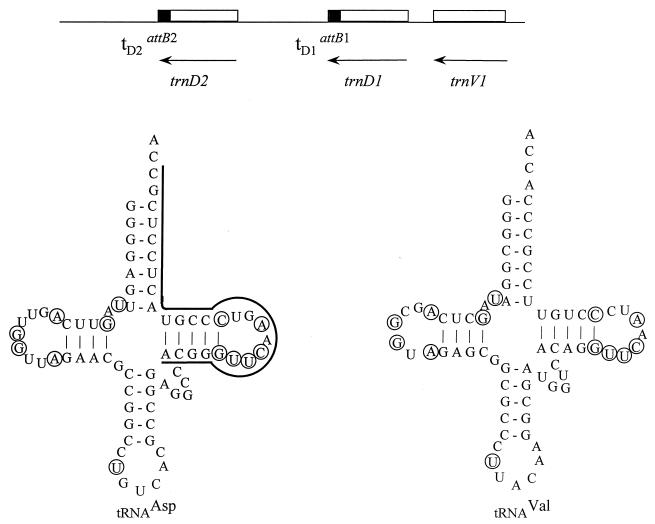
To confirm that attP lies within int and to characterize the attP and attB sites required in cis for integrative recombination, we first confirmed the sequence of the attB locus. From the published sequences of the attL and attR prophage junction sites, we designed oligonucleotide primers and used PCR to amplify and clone the attB locus from both wild-type M. xanthus DK1622 and its mutant derivative DZ1. The sequence of the attB locus was found to be identical for both strains. Analysis of this sequence shows that Mx8 integrates into the second gene of a cluster of three tRNA genes, trnD2, trnD1, and trnV1 (Fig. 5). Each of these genes encodes a class I or a class II tRNA (13, 40) with 4 bp in the stem of the dihydrouracil arm, 5 bases in the variable loop, and the 13 invariant bases shared by all tRNA molecules. Whereas trnD1 has a CCA acceptor stem within its coding sequence, the coding sequence for trnD2 terminates with CTA, and that for trnV1 terminates with CTG. We presume that, as for other class II tRNAs, the CCA acceptor stem is added enzymatically to the products of the class II trnD2 and trnV1 genes during their maturation.
FIG. 5.
Structure of the M. xanthus attB locus and its tRNA products. (Top) The attB locus contains three tandem tRNA genes, trnD2, trnD1, and trnV1, represented as open boxes. All three genes are transcribed from right to left, as indicated by the arrows. The two filled portions at the 3′ ends of the trnD1 and trnD2 genes indicate the positions of the 26-bp core sequence shared by the attB2, attB1, and phage Mx8 attP sites, the region of homology in which attP × attB recombination occurs. Two terminators, tD1 and tD2, follow the tRNAAsp genes that overlap the two attB sites within the attB locus. The trnV1 gene, which encodes tRNAVal, is located immediately upstream of trnD1, and these two genes may be cotranscribed. (Bottom) Predicted cloverleaf secondary structures of the tRNAAsp and tRNAVal products of these two genes. Circled nucleotides are the invariant tRNA bases (13, 40).
The attB1 and attB2 sites, located at the 3′ ends of tandem tRNAAsp genes, are both functional, but integration into the attB1 site is highly favored.
As diagrammed in Fig. 6, when pAY721 is electroporated into M. xanthus, Kmr recombinants can arise from site-specific recombination events involving the attP site on the plasmid and one or both of the attB sites. Alignment of the regions of homology between attP and the 3′ ends of trnD1 and trnD2 shows that all three sites have a common core of 26 bp within which site-specific recombination occurs (Fig. 7). Overlapping this area of homology, the attP site has a 29-bp sequence with striking dyad symmetry consisting of 7 bp flanked by inverted repeats of 11 bp. A shorter, dyad symmetry element consisting of this 7-bp sequence flanked by 5-bp repeats defines the common core, designated O. In the 5′ direction, the homology between attP and each of the tRNA genes extends beyond these 5-bp inverted repeats by 6 bp for attB1 (trnD1) but by only 3 bp for attB2 (trnD2). In the 3′ direction, both trnD genes share the same 48-bp (bottom-strand) coding sequence but diverge in sequence immediately before the first nucleotide of the coding sequence.
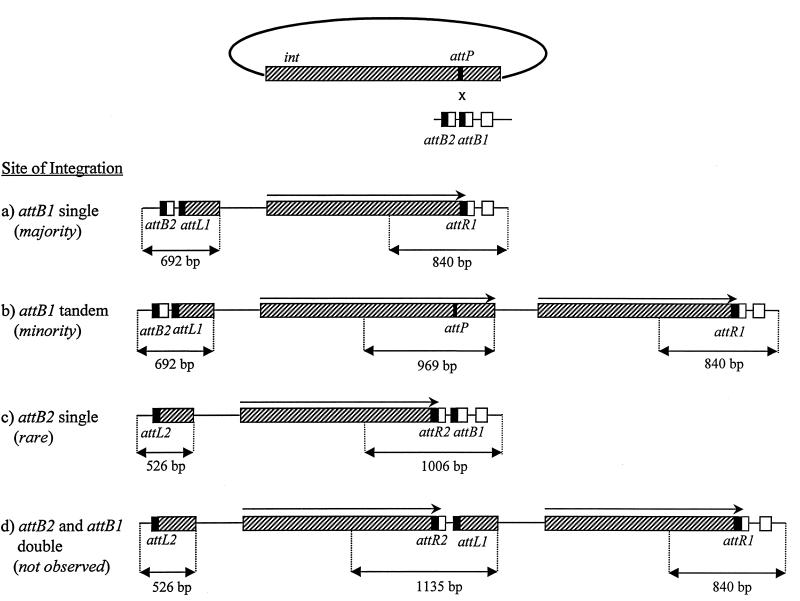
FIG. 6.
PCR can be used to distinguish the possible products of plasmid integration. Integration of a plasmid at the attB1 site generates prophage junctions that, when amplified with the attL and attR primer pairs, yield fragments of 692 and 840 bp, respectively. If integration at attB1 (or attB2) is accompanied by a deletion between the tandem attB sites, then we observe attL and attR products of 562 and 840 bp. Integration at attB2 generates junctions that yield amplified fragments of 526 and 1,006 bp. If two copies of a plasmid were to integrate, one at attB1 and the other at attB2, then amplification of the attP junction site would yield a product of 1,135 bp.
FIG. 7.
The integration of Mx8 or plasmids carrying the Mx8 int-attP genes results in changes in two transcription units. (a) The top-strand sequences of portions of the attP, attB1, and attB2 sites, located within the int, trnD1, and trnD2 genes, respectively, are aligned to show the 26-bp core sequence shared by these sites. Dyad sequences (arrows) flank the likely 7-bp target of cleavage and joining (bold). Bases identical to the corresponding bases in the preferred attB1 bacterial attachment site are shaded. (b) Embedded in the int coding sequence lies a terminator sequence, tMx8 (shaded), and the common core of attP (bold). A portion of the amino acid sequence of integrase is shown above the int coding sequence. Arrows indicate the directions of int and trnD1 transcription. The 3′ end of trnD1 contains the common core of attB1 (bold) and a terminator sequence, tD1 (shaded), distal to attB. The bottom sequence shows the attR product of attP × attB recombination in which the int coding sequence has been changed to intX and tMx8 replaces the tD1 terminator. The intX coding sequence, which terminates at a TAA stop codon, encodes a less active integrase with a new C terminus. (c) Predicted stem-loop secondary structures of the trnD gene terminators. The terminator sequences distal to both the trnD2 and the trnD1 genes, tD2 and tD1, respectively, are replaced by the weaker Mx8 terminator, tMx8, upon integration. Free energies for hairpin formation were calculated with the RNAFOLD program (5).
The integration of a plasmid or a phage into either tRNAAsp gene, trnD1 or trnD2, results in the simultaneous alteration of two different transcription units, one on the plasmid (or phage) and one on the host genome (Fig. 7). This is because attP lies within int and an attB core lies at the 3′ end of each trnD gene, before its transcription terminator. The change in the int gene results in a gene, intX, encoding an integrase with a new C terminus. This new C terminus of IntX is the same whether integration occurs at attB1 or at attB2. The change in the trnD1 and trnD2 transcription units involves the replacement of the natural terminators for these tRNA genes with an Mx8-specified terminator, tMx8. The tMx8 sequence is embedded within the antisense strand of the int gene, immediately distal to the attP core.
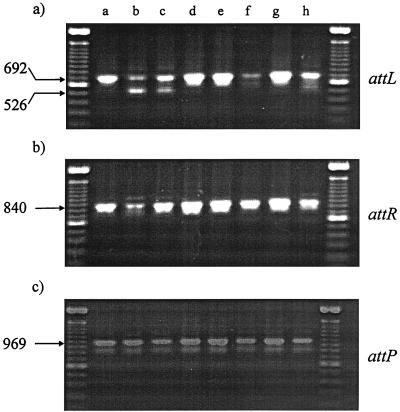
The majority of Kmr recombinants are formed by the simple integration of pAY721 into the attB1 site within the 3′ end of trnD1. A minority of Kmr recombinants are formed by the integration of pAY721 into either the attB1 or the attB2 site accompanied by a deletion between attB1 and attB2. Extremely rarely, Kmr recombinants are formed by the simple integration of pAY721 (or related int-attP plasmids) into attB2. As shown in Fig. 8, when DNA is isolated from eight independent Kmr electroporants of pAY721 and amplified with primer pairs for the attL and attR sites, five of eight templates (lanes a, d, e, f, and g) give rise to a 692-bp attL product and an 840-bp attR product. This is the result expected for strains carrying plasmids that have integrated into the (preferred) attB1 site. All of the DNA templates from these five strains give rise to an attP product of 969 bp, the size of the attP product obtained with circular plasmid pAY721 DNA as a template. This observation shows either that multiple copies of pAY721 can integrate into the same attB1 site or that, in some cells, pAY721 has excised from the M. xanthus chromosome.
FIG. 8.
Plasmid pAY721 prefers to integrate into the attB1 site. Chromosomal DNA was isolated from eight independent Kmr electroporants of host strain DZ1 (lanes a to h) with plasmid pAY721 and amplified with the attL (a), attR (b), and attP (c) primer pairs as described in Materials and Methods. Amplified products were resolved by electrophoresis through a 1.5% agarose gel and visualized after staining with ethidium bromide. Size standards flanking the amplified products are the Gibco-BRL 100-bp ladder.
The latter argument is sufficient to explain this observation, because it is known that integrated plasmid pAY721 can undergo excision in these recombinants to generate free, circular forms of plasmid DNA. Furthermore, when total chromosomal DNA is prepared from these Kmr recombinants and electroporated into E. coli, Kmr electroporants with circular plasmid pAY721 are recovered at low, but reproducible, efficiencies (ca. 10 to 100 μg−1). Thus, Kmr M. xanthus strains with pAY721 carry the plasmid in a dynamic equilibrium between integrated and excised forms.
Integration of a plasmid with the functional int-attP genes often is accompanied by a deletion within the attB locus.
Three of the electroporants analyzed in Fig. 8 (lanes b, c, and h) give rise to a mixture of amplified attL products but yield a single attR product of 840 bp, indicating that the right arm of the right prophage attachment site is derived from attB1. None of these recombinants gives rise to an amplified attP product of 1,135 bp, indicating that they do not have two copies of pAY721, one integrated at attB1 and another integrated at attB2 (Fig. 6). When segregants of these colonies are analyzed, many of them yield an attL fragment of the predicted size for a plasmid that has integrated into the attB2 site and an attR fragment of the predicted size for a plasmid that has integrated into the attB1 site. These segregants yield an attP product of 969 bp, not 1,135 bp. Together, these data show that a deletion between the attB2 and attB1 sites has occurred in these strains upon integration. Among 100 independent Kmr strains arising from the integration of pAY721 (and other, related int-attP plasmids) that we have analyzed by using the PCR with the attL and attR primers, only 3 have been found to have plasmids integrated only at attB2 (data not shown).
The int promoter is repressed in the prophage state.
When plasmids with the Mx8 attP-int genes are integrated at the attB locus, the recombinant strains maintain the integrated plasmids stably. This result suggests that, like the expression of other phage int genes, such as the phage λ int gene (31), the expression of the Mx8 intX gene may be repressed during the maintenance of the prophage state. Furthermore, we and others have found that when fusions of M. xanthus genes to the lacZ gene are subcloned onto a plasmid with the Mx8 int-attP genes and then integrated at the attB locus, these fusions are expressed at lower levels from the attB locus than from merodiploids with these fusions integrated at their normal chromosomal loci (8, 16). These results suggest that when M. xanthus genes are integrated ectopically at the attB locus, their expression is silenced.
Surprisingly, Tojo et al. (37) have reported that β-galactosidase activity is expressed from int-lacZ translational fusions on integrated plasmids at relatively high levels. To determine whether this is the case in our study as well, we constructed plasmids pAY738 and pAY739 with the int promoter and in-frame translational fusions of each of the two GTG start codons of int to the lacZ structural gene on Kmr plasmid pRS552 (32). These recombinant plasmids were integrated by homologous recombination into a lysogen of host DZ1 carrying wild-type Mx8 as a prophage. The β-galactosidase activity produced from the expression of the int-lacZ fusion genes in the recombinants was assayed. Each of the strains was found to produce <1 nmol of β-galactosidase activity per min per mg, a specific activity indistinguishable from the background level of activity made by the lysogen alone or present in control assays with no added source of enzyme. This result shows that the int promoter is expressed at an extremely low level from prophage Mx8.
We can account for the discrepancy between our results and those reported by Tojo et al. (37) because they did not control for the possibility that their int-lacZ fusions were expressed from a fortuitous upstream promoter within their plasmid vector sequence. Because we integrated our plasmids carrying int-lacZ fusion constructs by homologous recombination into an Mx8 prophage, our fusions could be expressed only from an upstream Mx8 promoter. Therefore, we can conclude that the expression of the int gene and likely of the int promoter is repressed in the prophage state.
DISCUSSION
Mx8 integrates into a preferred target locus, attB, on the M. xanthus genome. The only phage-encoded function required for the integration reaction in trans is that of integrase, the product of the Mx8 int gene, which has two alternate translation start codons. The int promoter is located immediately upstream of the int structural gene in a small, 106-bp region, and at least one sequence element required for promoter function lies within the overlapping uoi gene, which may encode an excisionase function.
Deletion analysis of the 3′ end of the int gene shows that, although the last 30 amino acid residues are not critical for Int function, deletions that are predicted to truncate the Int protein and alter the last 31 or more residues abolish function. Although this result predicts that the prophage-encoded IntX integrase should be inactive, the Mx8 prophage, as well as plasmids carrying functional int-attP genes that have integrated into the M. xanthus chromosome, can excise from the chromosome. The excision is site specific, because it regenerates a functional int gene with its internal attP site and occurs spontaneously at a measurable frequency. In the accompanying paper (19), we show that the prophage intX gene encodes an active integrase that can promote site-specific recombination events with a reduced efficiency relative to that of the phage-encoded Int protein.
Like many other temperate phages, as well as archaebacterium virus SSV1 of Sulfobolus shibatae (27), the preferred site for the integration of Mx8 lies within the 3′ end of a tRNA gene. The sequences of the three tRNA genes within the attB locus are the first reported sequences of tRNA genes for M. xanthus and offer few surprises. These three genes are predicted to encode tRNAs with all 13 invariant bases and the additional semivariant bases that contribute to tertiary tRNA structure (13, 40). In addition, these tRNAs are predicted to have a common secondary structure that includes a 7-bp amino acid acceptor stem, a 3- to 4-bp D stem with 7 to 10 bases in the D loop, a 5-bp anticodon stem, and a 7-base anticodon loop.
The common core within which site-specific recombination occurs between the Mx8 genome and its host genome is duplicated in close proximity on its host genome. Plasmids carrying the Mx8 int and attP functions required for site-specific recombination integrate frequently into the preferred attB1 site within trnD1 but only rarely into the attB2 site within trnD2. For several temperate phages, including λ, the sequences within the attB locus necessary and sufficient for efficient site-specific recombination do not extend far beyond the core sequence. For λ, the core sequence is comprised of 7-bp imperfect inverted repeats flanking a unique 7-bp sequence, and only an additional flanking 2 bp is required for the full activity of attB (21); even less of the core homology is required for attP function (2).
If the same is true for Mx8, because there are two different attB core sequences within the Mx8 attB locus (Fig. 3), we might expect Mx8 to integrate into either site at the 3′ end of either tRNAAsp gene (into attB1 or attB2) with approximately equal efficiencies. This is the case for temperate corynephages β, ω, and γ, which can integrate into either of two different attachment sites on the Corynebacterium diphtheriae C7 chromosome with equal efficiencies (24, 25). Like the M. xanthus attB1 and attB2 sites, the C. diphtheriae attB1 and attB2 sites lie within the 3′ ends of repeated tRNA genes close to one another on the host chromosome (26). When tox+ corynephages integrate at both sites, the lysogens are stable and produce higher levels of diphtheria toxin, perhaps conveying a selective advantage on their lysogenic hosts. In contrast, lysogens resulting from two corynephage genomes integrating at the same attachment site are unstable (24, 25).
The integration of Mx8 into the M. xanthus attB1 and attB2 sites follows different rules. Plasmids with the Mx8 attP-int genes prefer to integrate into the attB1 site. Integration into the attB1 site often results in a deletion between the attB1 and attB2 sites. Plasmids integrate only rarely into the attB2 site. Differences in the sequences flanking the attB1 and attB2 core sequences likely contribute to this site preference for integration. The integration of Mx8 into the M. xanthus attB locus is unusual in another respect. Unlike the situation for most other integrative elements, Mx8 site-specific recombination involves an attP site located within the int gene, and the integration reaction changes the structure of the phage int gene by altering its 3′ end to generate the prophage intX gene. In the accompanying paper (19), we show that and explain why Mx8 uses a mechanism of site-specific recombination in which the integration reaction serves to reduce the specific activity of the enzyme that catalyzes this reaction.
ACKNOWLEDGMENTS
We thank Trish Hartzell for critical reading of the manuscript.
This work was supported by a grant from the National Institute of General Medical Sciences (GM53392) to P.Y.
Footnotes
This work is dedicated to the memory of Hatch Echols, teacher and friend.
REFERENCES
- 1.Argos P, Landy A, Abremski K, Egan J B, Haggard-Ljungquist E, Hoess R H, Kahn M L, Kalionis B, Narayana S V, Pierson L S. The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986;5:433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer C E, Gardner J F, Gumport R I. Extent of sequence homology required for bacteriophage lambda site-specific recombination. J Mol Biol. 1985;181:187–197. doi: 10.1016/0022-2836(85)90084-1. [DOI] [PubMed] [Google Scholar]
- 3.Campbell A. The episomes. Adv Genet. 1962;11:101–116. [Google Scholar]
- 4.Campos J M, Geisselsoder J, Zusman D R. Isolation of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978;119:167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- 5.Cech T R, Tanner N K, Tinoco I J, Weir B R, Zuker M, Perlman P S. Secondary structure of the Tetrahymena ribosomal RNA intervening sequence: structural homology with fungal mitochondrial intervening sequences. Proc Natl Acad Sci USA. 1983;80:3903–3907. doi: 10.1073/pnas.80.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Churchward G, Belin D, Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984;31:165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- 8.Fisseha M, Gloudemans M, Gill R E, Kroos L. Characterization of the regulatory region of a cell interaction-dependent gene in Myxococcus xanthus. J Bacteriol. 1996;178:2539–2550. doi: 10.1128/jb.178.9.2539-2550.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartzell P, Kaiser D. Upstream gene of the mgl operon controls the level of MglA protein in Myxococcus xanthus. J Bacteriol. 1991;173:7625–7635. doi: 10.1128/jb.173.23.7625-7635.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoess R H, Foeller C, Bidwell K, Landy A. Site-specific recombination functions of bacteriophage lambda: DNA sequence of regulatory regions and overlapping structural genes for Int and Xis. Proc Natl Acad Sci USA. 1980;77:2482–2486. doi: 10.1073/pnas.77.5.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kashefi K, Hartzell P L. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF− defect. Mol Microbiol. 1995;15:483–494. doi: 10.1111/j.1365-2958.1995.tb02262.x. [DOI] [PubMed] [Google Scholar]
- 13.Kim S H. Three-dimensional structure of transfer RNA and its functional implications. Adv Enzymol Relat Areas Mol Biol. 1978;46:279–315. doi: 10.1002/9780470122914.ch4. [DOI] [PubMed] [Google Scholar]
- 14.Kroos L, Kuspa A, Kaiser D. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev Biol. 1986;117:252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- 15.Leong J M, Nunes-Duby S E, Oser A B, Lesser C F, Youderian P, Susskind M M, Landy A. Structural and regulatory divergence among site-specific recombination genes of lambdoid phage. J Mol Biol. 1986;189:603–616. doi: 10.1016/0022-2836(86)90491-2. [DOI] [PubMed] [Google Scholar]
- 16.Li S F, Shimkets L J. Site-specific integration and expression of a developmental promoter in Myxococcus xanthus. J Bacteriol. 1988;170:5552–5556. doi: 10.1128/jb.170.12.5552-5556.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magrini V, Creighton C, White D, Hartzell P L, Youderian P. The aadA gene of plasmid R100 confers resistance to spectinomycin and streptomycin in Myxococcus xanthus. J Bacteriol. 1998;180:6757–6760. doi: 10.1128/jb.180.24.6757-6760.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magrini V, Salmi D, Thomas D, Herbert S K, Hartzell P L, Youderian P. Temperate Myxococcus xanthus phage Mx8 encodes a DNA adenine methylase, Mox. J Bacteriol. 1997;179:4254–4263. doi: 10.1128/jb.179.13.4254-4263.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magrini V, Storms M L, Youderian P. Site-specific recombination of temperate Myxococcus xanthus phage Mx8: regulation of integrase activity by reversible, covalent modification. J Bacteriol. 1999;181:4062–4070. doi: 10.1128/jb.181.13.4062-4070.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin S, Sodergren E, Masuda T, Kaiser A D. Systematic isolation of transducing phages for Myxococcus xanthus. Virology. 1978;88:44–53. doi: 10.1016/0042-6822(78)90108-3. [DOI] [PubMed] [Google Scholar]
- 21.Mizuuchi M, Mizuuchi K. The extent of DNA sequence required for a functional bacterial attachment site of phage lambda. Nucleic Acids Res. 1985;13:1193–1208. doi: 10.1093/nar/13.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orndorff P, Stellwag E, Starich T, Dworkin M, Zissler J. Genetic and physical characterization of lysogeny by bacteriophage Mx8 in Myxococcus xanthus. J Bacteriol. 1983;154:772–779. doi: 10.1128/jb.154.2.772-779.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pargellis C A, Nunes-Duby S E, de Vargas L M, Landy A. Suicide recombination substrates yield covalent lambda integrase-DNA complexes and lead to identification of the active site tyrosine. J Biol Chem. 1988;263:7678–7685. [PubMed] [Google Scholar]
- 24.Rappuoli R, Ratti G. Physical map of the chromosomal region of Corynebacterium diphtheriae containing corynephage attachment sites attB1 and attB2. J Bacteriol. 1984;158:325–330. doi: 10.1128/jb.158.1.325-330.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rappuoli R, Ratti G, Perugini M, Murphy J R. Detection and physical map of a omega tox+-related defective prophage in Corynebacterium diphtheriae Belfanti 1030(−)tox−. J Virol. 1985;54:194–198. doi: 10.1128/jvi.54.1.194-198.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ratti G, Covacci A, Rappuoli R. A tRNA(2Arg) gene of Corynebacterium diphtheriae is the chromosomal integration site for toxinogenic bacteriophages. Mol Microbiol. 1997;25:1179–1181. doi: 10.1046/j.1365-2958.1997.5191887.x. [DOI] [PubMed] [Google Scholar]
- 27.Reiter W D, Palm P. Identification and characterization of a defective SSV1 genome integrated into a tRNA gene in the archaebacterium Sulfolobus sp. B12. Mol Gen Genet. 1990;221:65–71. doi: 10.1007/BF00280369. [DOI] [PubMed] [Google Scholar]
- 28.Salmi D, Magrini V, Hartzell P L, Youderian P. Genetic determinants of immunity and integration of temperate Myxococcus xanthus phage Mx8. J Bacteriol. 1998;180:614–621. doi: 10.1128/jb.180.3.614-621.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1998;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shulman M, Gottesman M. Lambda att2: a transducing phage capable of intramolecular int-xis promoted recombination. In: Hershey A D, editor. The bacteriophage lambda. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1971. pp. 477–487. [Google Scholar]
- 32.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 33.Spratt B G, Hedge P J, Te H S, Edelman A, Broome-Smith J K. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 34.Stellwag E, Fink J M, Zissler J. Physical characterization of the genome of the Myxococcus xanthus bacteriophage Mx8. Mol Gen Genet. 1998;199:123–132. doi: 10.1007/BF00327521. [DOI] [PubMed] [Google Scholar]
- 35.Stephens K, Kaiser A D. Genetics of gliding motility in Myxococcus xanthus: molecular cloning of the mgl locus. Mol Gen Genet. 1987;207:256–266. [Google Scholar]
- 36.Taketo A. DNA transfection of Escherichia coli by electroporation. Biochim Biophys Acta. 1988;949:318–324. doi: 10.1016/0167-4781(88)90158-3. [DOI] [PubMed] [Google Scholar]
- 37.Tojo N, Sanmiya K, Sugawara H, Inouye S, Komano T. Integration of bacteriophage Mx8 into the Myxococcus xanthus chromosome causes a structural alteration at the C-terminal region of the IntP protein. J Bacteriol. 1996;178:4004–4011. doi: 10.1128/jb.178.14.4004-4011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weimer R M, Creighton C, Stassinopoulos A, Youderian P, Hartzell P L. A chaperone in the HSP70 family controls production of extracellular fibrils in Myxococcus xanthus. J Bacteriol. 1998;180:5357–5368. doi: 10.1128/jb.180.20.5357-5368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 40.Yarus M. tRNA identity: a hair of the dogma that bit us. Cell. 1988;55:739–741. doi: 10.1016/0092-8674(88)90127-4. [DOI] [PubMed] [Google Scholar]