Abstract
In this Perspective we propose our current point of view and a suggestive roadmap on the field of high-resolution optical microscopy dedicated to bioimaging. Motivated by biological applications, researchers have indeed devised an impressive amount of strategies to address the diverse constraints of imaging and studying biological matter down to the molecular scale, making this interdisciplinary research field a vibrant forum for creativity. Throughout the discussion, we highlight several striking recent successes in this quest. We also identify some next challenges still ahead to apprehend biological questions in increasingly complex living organisms for integrative studies in a minimally invasive manner.
Keywords: optical bioimaging, biophotonics, super-resolution microscopy, optical microscopy, label-free imaging, fluorescence microscopy
Introduction
Biophotonics is a broad discipline involving gathering research and applications of light science to the study, understanding, and manipulation of biological matter. For several decades it has been a mature field encompassing a variety of topics not only driven by applications, but also often by technology. A dynamic subfield of biophotonics concerns optical bioimaging, the potential of which has yet to be fully exploited.
Vision is most likely our most prominent human sense and benefits from diverse light–matter interactions, especially in the visible wavelength range. This provides a rich representation of our remote environment. When it comes to vizualizing living organisms, the nature of these interactions is based on strong scattering and absorptions that make our perception of matter remain surface. In other words, light has a poor competency to penetrate the biological environment, as opposed to other radiations that could be commonly used in medical imaging for instance. So why is optical bioimaging the subject of so many new studies and developments?
A basic answer could be summarized by the maxim “Seeing is believing”, which is certainly the driving force behind optical bioimaging. For centuries (!), the whole field has pushed this concept to enlarge and resolve novel details of the specimens in order to go beyond our naked eye perception. This engendered the advent of optical microscopy, a simple instrument which conception has not fundamentally evolved since the first compound microscopes. The early motivation has been directed to the observation of biological specimens in order to reveal substructures that were too small or unbedewed into complex features. The term of cell has been coined immediately after their observation through an optical microscope back in the XVIIth century.1 Improvements of optical microscopes next aimed at augmenting the content of information obtained, which encompasses for instance our ability to gain specificity within images. In this context, several notions that are often intricated can be distinguished: resolutions, sensitivity and selectivity/contrast, and finally biological relevancy.
Different types of microscopies (not related to light) can reach ultimate resolution, down to the molecular or even the atomic scale. This is the case of electron microscopy, which has made fascinating progress over the last decades.2 So why would it still be useful to advance the field optical microscopy? The answer might seem trivial: no current approach allows imaging (resolving) molecular features in live complex organisms, and optical bioimaging is a promising route for combining ultrahigh resolution with live specimen imaging.
With our point of view, we will focus on what remains to be done to push the limit of the sample knowledge, in particular, to allow molecular scale studies with optics. Indeed, the molecular scale directly triggers structure, function and organization in cells and it is one of the key scales to analyze and understand biological matter. To attain such scales, the pioneer studies have been performed on isolated and fixed cell samples. Yet there is a growing need to perform experiments in more physiological conditions with living and 3D samples (including tissues, in vivo, ...), although there is still a long way to go to study intact samples or organisms.
Nonexhaustive Short Review of the Bioimaging Field: Pushing Fundamental Limits
As any investigation modality, optical bioimaging comes with fundamental and practical limitations. The last decades have seen an impressive amount of achievements that pushed away these limits. We will discuss here three main directions taken by difference communities in optical biological imaging: the quests for molecular scale resolution in fluorescence microscopy, the design of minimalist biological systems, and the advances in label-free imaging.
Super-Resolution Microscopy
The case of resolution is emblematic to the considerable progress in the field of fluorescence microscopy. Since the 19th century, it is known that the resolution of optical microscopes in the far-field imaging is ultimately limited by the diffraction of light which reaches the common limit of ∼λ/2 with high-numerical aperture objectives.3 In order to circumvent this limit in fluorescence microscopy, several strategies have been proposed, and proof of concepts started to emerge in the 1980–90s.4,5 Two general approaches can be distinguished. On the one hand, some methods rely on the concept of aperture synthesis, as commonly used in radars. With this concept in optical microcopy, a structuration of the excitation light is used to introduce some a priori knowledge and encodes high frequencies around a known (or deducible) frequency in the band-pass of the microscope.6 On the other hand, a large family of methods have in common to introduce some a priori knowledge that the number of emitters or the volume of emission can be precisely controlled akin multiphoton/nonlinear microscopy. This is realized by driving locally emitter fluorescent state populations5 or by reducing the densities of fluorescing probes at any given time. The concepts behind these methods are coined under the general term “super-resolution microscopy”.
Localization methods, which are based on the detection of isolated single molecules7 and their subwavelength localization, rely on a concept that is well-known in interferometry and spectroscopy. Namely, the precision reached in determining the central position of fringe (or line) is only given by the signal-to-noise ratio at which this fringe can be determined8 and not its intrinsic width (often given by λ/2). For instance, the precision of atomic clocks relies on this principle.9 The first propositions to achieve localization super-resolution microscopy were directly rooted in these considerations, and the initial demonstration can be attributed to Van Oijen et al. in 1998,10 where the authors were able to 3D resolve several fluorophores within a confocal volume at cryogenic temperatures. Later, the real advent of super-resolution microscopy at room temperature11−13 benefited from a combination of progress in laser beam engineering, spectroscopy, and photocontrol at the single molecule level or on fluorescent probe chemistry,14,15 to name a few.
Alternatively, to reduce the volume of emission beyond confocal microscopy, super-resolution methods like Stimulated Emission Depletion (STED) or REversible Saturable OpticaL Fluorescence Transitions (RESOLFT) microscopies (and related techniques) use nonlinear manipulations of emitters fluorescence properties in the confocal volume.16 These approaches are now routine in many applications, including 3D imaging of complex structures. An elegant evolution of these methods, Minflux, allowed minimizing the excitation levels to reduce emitter fluorescence fatigue down to minimum levels. By achieving single molecule localization by triangulation, Minflux bridges these methods with SMLM.17
Many articles and reviews provide an excellent overview of super-resolution approaches, including methodological descriptions, comparison of their performances and applications (e.g., refs (18−21)). As of today, these methods reached exquisite resolutions, even in 3D, but with one major constraints in the context of bioimaging: the samples need to be “optically simple”, that is, inducing minimal light scattering, absorption, or spatiotemporal optical aberrations. In practice, super-resolution imaging has been extremely performant to study minimalist biological systems (Figure 1).
Figure 1.
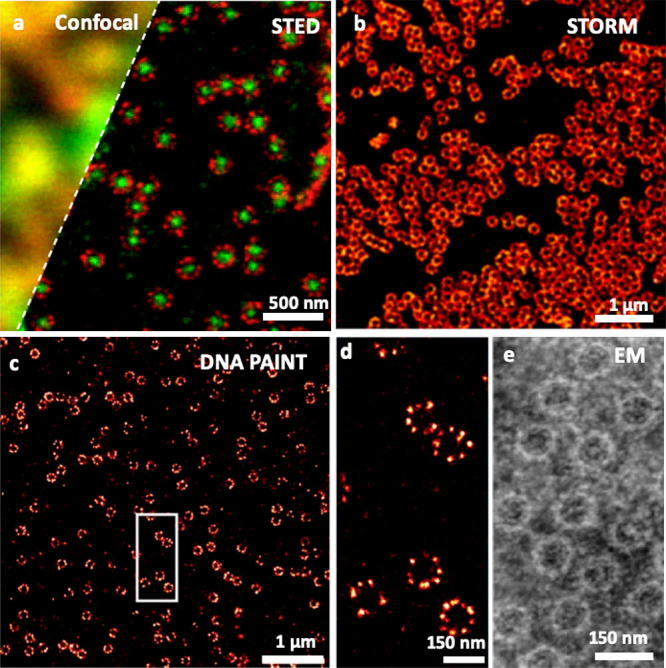
Super-resolution fluorescence microscopy of nuclear pore complexes with different modalities: STED microscopy (a), compared with confocal microscopy; STORM microscopy (b); DNA PAINT microscopy (c, d). Comparison to electron microscopy using negative staining. Republished with permission of Company of Biologist Ltd. from ref (22). Copyright 2012 The Company of Biologists, permission conveyed through Copyright Clearance Center, Inc.; Adapted with permission from ref (23). Copyright 2019 John Wiley and Sons, https://creativecommons.org/licenses/by/4.0/; Reprinted with permission from ref (24). Copyright 2013 Elsevier.
Bioimaging in and of Minimal(ist) or Optically Simplified Samples
Many developments of bioimaging strategies aiming to decipher basic biological processes have been strongly accelerated by the physicist methodology consisting in using minimalist systems to gain full control of the observable under study. To this end, biological samples have been purified and extracted from their complex environment to isolate basic components (proteins, DNA, phospholipids, etc.).
This has allowed the mechanistic study of, for example, molecular motors,25 DNA–protein interactions26 in reconstructed systems. In such pure systems, single molecule fluorescence microscopy resolutions were first developed and have become the working horse for many applications.27 On the methodological point of view, researchers pushed the performance of the optical microscopes to their paroxysms in terms of resolution and sensitivity reaching for instance subnanometer resolutions at high temporal resolutions as discussed above. In order to study basic molecular functions and interactions, advanced fluorescent microscopy methods where developed like fluorescence lifetime imaging, fluorescent energy transfer microscopy, or fluorescence correlation spectroscopy, which all rely or necessitate on a perfect understanding of fluorescent photophysics at the single-molecule level. In purified environments, single protein movements could also be detected label-free with interferometric scattering microscopy28 (see also below).
A more complex situation already deals with the study of molecular mechanisms in isolated cells. Yet, recent developments in imaging methods applied on fixed cells have been among the most fascinating in recent years. This is because plated cells, although more complex in terms of biochemistry/number of components than purified systems, remain optically inert in terms of optical aberrations, light scattering, and absorption. Single-cell analysis is the natural playground for todays most refined approaches in high resolution (i.e., molecular scale) microscopy, including 3D super resolution microscopy and lattice light sheet microscopy, for instance. This field is now reaching maturity.29
Importantly, all super-resolution methods require a long imaging time, and the study of live cell processes are thus challenging at molecular resolution. A common compromise consists of accepting sparsity by, for example, restraining the imaging zones or tracking individual molecules at low densities (eventually using photoactivation strategies) to ensure high localization precisions (typically 10–50 nm in live cells). When resolutions around 100 nm are sufficient, the approaches based on structured illuminations allow studying larger and denser zones.
The next degree of sample complexity arises when assemblies of cells are imaged (including tissues extracts, organoids, or whole organs). As will be discussed below, one main challenge arises from the difficulty to form a correct image through thick biological samples. At super-resolution scales, the problem arises already with few microns thick sample at visible wavelength. Additionally, imaging thick samples demands for 3D which is even more complex in super-resolution imaging regimes.
Interestingly, refractive index matching between the sample and the medium has been used for decades to improve the image formation in complex systems at depth. When applied to bioimaging, the ultimate approach has been developed when whole organs or even organisms (e.g., rodents) could be chemically cleared. Such transparisation methods (in fixed samples) allowed volumetric imaging of fluorescently labeled samples.30 Along the idea of a chemical manipulation of the sample’s properties prior to imaging, expansion microscopy has recently made it possible to recover molecular scale structural information (akin super-resolution imaging) but with standard diffraction-limited imaging methods.31 This is obtained by physically swelling the sample under study (inherently fixed in a matrix) and rescaling the image obtained with the a priori knowledge of the physical expansion applied to the sample and taking into account distortions that might have been introduced during the expansion process.
Label-Free Bioimaging: Back to the Origin of Microscopy
At the origin of optical microscopy label-free or colored samples were studied. Zernike phase-contrast microscopy and Nomarski differential interference microscopy were major milestones in the study of living matter. Fluorescence and nonlinear light–matter interaction only appears in the 1970s, but rapidly became the standard for biological sample imaging, chiefly thanks to the molecular specificity obtained in the images. However, fluorescence imaging has intrinsic limitations. Indeed, the inevitable alteration of the sample when imaged via fluorescence, due to both labeling and phototoxicity, remains a major issue, and the photoinstability of fluorescent probes limits quantification and long duration imaging.
Interferometry and in particular Michelson/Mach-Zender or Mirau interferometer have been implemented in microscopy in the 1950s.32 Yet, the democratization of quantitative measurements from interferences occurs in the 1990s with the accessibility to a matrix sensor and desktop computational capabilities. Moreover, the impressive improvements in sensor capability (including sensitivity, low noise, speed, and linearity) in the last 20 years have unlocked new horizons for label-free imaging. Coupled with real-time numerical signal processing, proper quantification allowed useful biophysical information to be extacted. This principle has been extensively applicable while keeping a very limited photon budget on the sample (the photons detected are the photons used to illuminate), especially when compared to fluorescence imaging. Initiated with digital holography,33 quantitative phase imaging is now a field of research in life sciences, but also in nanotechnologies when the sample cannot be labeled.
The core idea of quantitative phase imaging is to measure the distortion of the wavefront and the attenuation of light due the interaction with the sample itself. This measurement, usually in a single photon interaction regime, can be achieved in various manner. A reference arm (without the sample) can be used to interfere with the light that had interacted with the sample: this is the way holography is working. Self-reference methods can also be implemented: in this case the probed light is analyzed with respect to itself–either numerically or in an interferometric manner. In this case, usually, only gradients of the phase signal rather than the signal itself are measured, but it grants the capability to be performed without precise knowledge and control of the light source. The easiest way to obtain such a measurement without a priori knowledge about the sample is to record intensity images with various defocus and to invert the light propagation equation to retrieve the wavefront of the light.34 Modification of differential interference contrast, Zernike phase-contrast or other phase-related contrast-only methods (e.g., Hoffman contrast) have been proposed to reach quantification and not only contrast.35 Finally, wavefront sensor-based detection can also be used to perform quantitative phase imaging36 with the interest of being a very compact and stable method, adapted to any microscope and objectives.
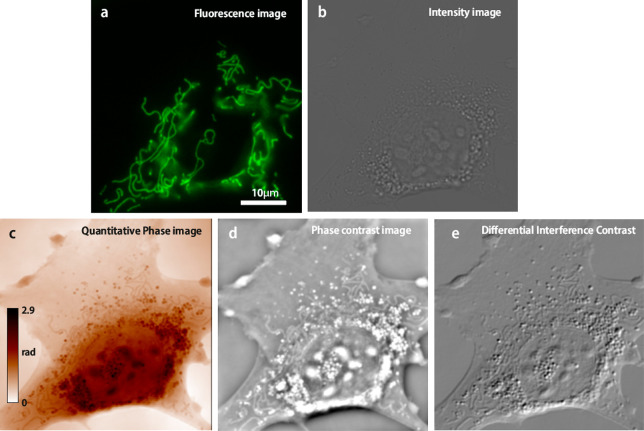
Quantitative phase imaging has many biological applications, including dry mass measurements, temperature variation mapping, ion flux in living cells, cellular identification, and noninvasive characterization. It is of great interest when coupled with fluorescence imaging to unravel the biological context of the fluorescence measurement (Figure 2). It also has applications in material sciences to characterize surface and nanoparticles and chemistry to follow the reaction via the refractive index changes.
Figure 2.
Living cell (mouse embryonic fibroblast) observed with (a) fluorescence imaging (mitochondrial labeling), (b) transmission intensity, (c) quantitative phase, (d) phase contrast (simulated from (c)), and (e) differential interference contrast (simulated from (c)). The setup used is described in ref (37).
Some Next Challenges: Placing the Complex Sample at the Core of the Image and Ensuring Quantitative Measurements
We believe that one big challenge in current bioimaging will lie in our capacity to discern what biological reality applies to the enormous amount of knowledge accumulated at ultrahigh spatiotemporal resolution in the minimalist systems mentioned above. In other words, one now needs to make the inverse routes where starting from minimalist systems, we need to reintroduce the native environment up to that of intact specimens. This means that we now need to put the complexity of the sample back into the heart of both the investigation and the development of optical instrumentation at high resolution; A corollary to this is that the native environment of the biological system under study must not only be present (even if not detected), but also ideally be imaged and taken into account to finally apprehend its impact on the system under study. The task becomes vertiginous and might even seem counterintuitive related to the quest for imaging molecular processes with high specificity, which triggered our habit to isolate particular processes from their environment in order to be deciphered.
Physicist can use (and are already using) a rigorous methodology in this quest, injecting a controlled amount of environmental complexity, starting from molecular processes, studied in cells to organs. Several optical challenges come with the complexification of the samples under study. The first one being to attain high resolution imaging in thick (poorly optically transparent) samples.
As mentioned above, some molecular resolution optical studies made on whole organs appeared in the 2010s thanks to the development of chemical processes inducing tissue clarification or allowing expansion microscopy or even their combination. However, super-resolution microscopy is difficult to implement at depths in tissue samples while maintaining compatibility with living organisms. Indeed, the complexity of the biological tissue prevents good light transmission, generating light absorption and scattering responsible for signal loss as well as optical aberrations, inducing distorted images.
Resolution and Specificity in Label-Free Imaging; toward Functional Imaging
Label-free imaging techniques and, in particular, phase-contrast and quantitative phase imaging techniques have two major drawbacks when compared to fluorescence imaging: they lack resolution and (molecular) specificity. Although not talking about super-resolution imaging techniques, phase imaging is generally performed with wide-field coherent illumination, which leads to a lateral resolution of about twice less than fluorescence lateral resolution (even worse in the axial direction). Many approaches have been developed to reach the incoherent imaging resolution, and the most used is diffraction tomography via multiple angle sequential acquisition.38 Other methods, including direct incoherent illumination, have also been proposed to match the fluorescence wide field. It is currently an important field of research, since this incoherent resolution is a threshold to reach in order to consider developments of actual label-free super-resolution methods. Of course, major adaptation from fluorescence super-resolution methods is then required in label-free but, currently, single particle tracking has already been demonstrated with holography on plasmonic or high-refractive index particles,39 and interferometric scattering microscopy has been used to track single nonfluorescent single proteins.40 Generalized methods to perform 3D super-resolution imaging without labeling will be a major milestone to unlock versatile application in living and preserved samples. Tissue imaging is also an important current field of research in the field.
The lack of molecular specificity, sensitivity and selectivity are also an intrinsic current limitation of label-free imaging. Indeed, by essence, the recorded signal rises from refractive index mismatches in the sample which are at the first order only depending on the local mass concentration of molecules and not the type of molecules, especially for biological compounds.41 However, it has been demonstrated that when coupled with image information, the identification of certain organelles, even cytoskeleton37 is possible thanks to structural a priori information. Recently, the development of machine learning has paved the way to a generalized cell compound identification.42 Although still being far from molecular specificity, it is a major milestone in this quest. To directly extract molecular specificity, label-free nonlinear spectroscopy has also triggered intense and prolific research focused on understanding the light/matter interaction of vibrational modes of molecules. Although penalized by weak signals, low readout speed, and thorough signal processing, this fascinating field of research, from spontaneous Raman scattering to stimulated Raman spectroscopy, is attracting highly deserved attention for bioimaging with application in biomedicine, for example, for cancer and drug characterization.43 Interestingly, machine-learning approaches are also becoming increasingly important to help analyze Raman spectral data in such methods.44
Functional imaging is also a current important field of research for label-free imaging. In 2010, a pioneer work has been performed showing that ion fluxes can be monitored with digital holography, the signal being carried by the change in refractive index due to osmotic equilibrium.45 By improving the acquisition framerate (1 kHz) and the sensitivity, Ling et al. have demonstrated that even action potential firing can be label-free monitored with interferometric imaging.46 Using the temporal fluctuation of the signal, dynamic optical coherence tomography can be applied to reveal metabolic information in living tissues.47 The intrinsic noninvasive capabilities of label-free methods and the high-temporal sensitivity will make them essential tools for living sample characterization.
Imaging Thick Samples: Off-Axis Microscopy and NIR Super-Resolution Microscopy
Bioimaging at high resolution within live organisms inherently implies that one can image through multicellular and tissue specimens. Unfortunately, this quest has been primarily hampered by the poor light penetration depth in most mammalian specimens, which are basically opaque at visible wavelengths (typ. 400–700 nm) where most fluorophores and detectors are effective. Contrast and resolutions are then degraded when imaging depth increases as a result of photon scattering and absorption by the tissue.
A well-known approach to ensure reasonable contrast in thick samples consists of restricting the excitation beam to well-defined and controllable regions. This is first the case of confocal microscopy and its nonlinear variants (see dedicated reviews, e.g., refs (48−50)), which have produced impressive achievements, for example, in neuroscience, often driven by technology developments (lasers, adaptive optics, etc.). Another class of approaches which also rely on volume excitation control, holds great promises for the future of the field: they are coined under the initial demonstration of single plane illumination microscopy (SPIM).51 The main concept behind SPIM is to get free from the common idea that illumination and detection should share the same optical axis. By using orthogonal or multiorientational excitations schemes, SPIM has opened additional degrees of freedom for handling complex, thick biological samples. The concept is now being generalized (and coupled) to multiview detection schemes aided by smart computational reconstructions, which allows to generate unmatched imaging data of live organisms. However, these approaches are intrinsically limited in resolution to the diffraction limit in the best case and to micrometer scales in the presence of strong light scattering and aberrations.
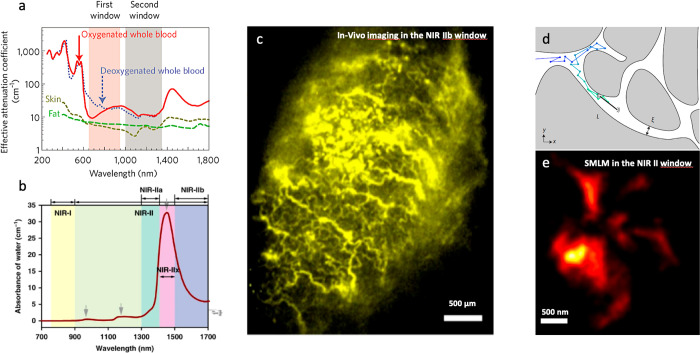
The limitations induced by photon scattering and absorption by the tissue at visible wavelengths have long been known as the optimal wavelength ranges for improved light tissue penetration were identified in the near-infrared NIR region typically between 800 and 1800 nm. In fact, several subregions have different properties52,53 (see Figure 3) when considering the combination of scattering by the tissue components (which decreases with increasing wavelengths) and light absorption, including by water (which increases with increasing wavelengths and displays some specific absorption bands).
Figure 3.
Near-infrared bioimaging with nanoparticles. (a, b) The different near-infrared (NIR) region biological transparency windows. (c) NIR in vivo imaging of colon cancer mouse model tumors injected with rare-earth nanoparticles. (c, d) NIRII single particle tracking of single wall carbon nanotubes (SWCNTs) in complex environments (d: schematics) allow the measurement of super-resolved maps of the extracellular space in live brain slices. Reprinted with permission from (52). Copyright 2009 Springer Customer Service Center GmbH: Springer Nature; Adapted with permission from ref (53). Copyright 2021 Springer Nature, https://creativecommons.org/licenses/by/4.0/; Reprinted with permission from ref (56). Copyright 2019 Springer Customer Service Center GmbH: Springer Nature; Reprinted with permission from ref (60). Copyright 2017 Springer Customer Service Center GmbH: Springer Nature.
A recognized strategy thus emerged over the last decades to adapt the imaging modalities to the different NIR windows. It relies on nonlinear microscopy techniques. The basic idea being to benefit from nonlinear light–matter interaction to use NIR excitation (multiphoton fluorescence, harmonic generation) while using “regular” detectors and fluorophores operating in the visible range. As mentioned above, the approach is highly effective, with impressive developments obtained over the last years, notably in the case of three-photon microscopy50 owing to the development of pulsed laser sources in the NIR IIa (∼1300 nm) and NIR IIb (∼1600 nm) regions. Several limiting factors accompany such approaches, in particular, the need to use energetic pulsed sources and to perform point-scanning microscopy, which impede high imaging rates.
Encouraged by the development of sensitive and affordable detectors in the NIR (especially based on InGaAs sensors), a current challenge consists of designing contrast agents that can absorb and luminesce in the near-infrared. In this case, both (one photon) excitation and emission would benefit from the NIR windows. This blooming and promising field of research gathers chemists and spectroscopist for the design and synthesis of novel infrared fluorophores having bright emitting properties or nanotechnologists to generate and shape the optical properties of NIR infrared emitting nanoparticles.54,55 One can cite the design of NIR quantum dots54 (e.g., based on PbS or CdTe) and rare-earth nanoparticles,56 as well as the recent development of gold nanoclusters with bright emission at 1000–1500 nm.57 Another type of NIR-emitting nanoparticle, namely, single wall carbon nanotubes (SWCNTs) deserve particular attention for their exquisite potentialities in the context of high-resolution deep tissue imaging.58,59 Indeed, SWCNTs combine high photoluminescence brightness and perfect photostability, NIR spectroscopic tunability in the different NIR windows, nanometer diameter, and tunable lengths, allowing morphological adaptability and could thus demonstrate promising applications in advanced bioimaging.60
All aforementioned NIR-based imaging modalities come with one inherent limitation, namely, that the resolution is linearly linked to the wavelength. Imaging at longer wavelength is thus accompanied by a degraded resolution which can be detrimental when molecular resolutions are targeted in live biological specimens. A next opportunity in the field will thus to marry super-resolution microscopy with NIR imaging.61 Of note, most organic NIR fluorophores do not compete with visible fluorophores in terms of brightness, which is however a decisive asset for super-resolution microscopy methods. Along the same line, it will be interesting to develop photo switchable NIR emitters for SMLM strategies. In this context, the design of such emitters based on (truly) NIR organic fluorophore or autofluorescent proteins would be decisive. Interestingly, our group showed that SWCNTs can offer a promising route for this quest. We developed two distinct strategies gathering SMLM approaches and SWCNT imaging. In the first one, we relied on single particle tracking concepts, where the superlocalizations of a single emitter are gathered along its diffusion in a complex structure.62 The approach can reveal the nanometer scale architectures of a living tissue when the emitter is bright enough to be detected at the single particle level inside the specimen (in the biological window), perfectly photostable and able to explore extensively its environment. SWCNTs fulfill all these requirements due to their NIR photophysical properties and 1D morphology, and they were applied to image the extracellular space of living brain tissue at unprecedented depth at nanoscale resolutions.60,63 The second strategy is based on the design of photoswitchable hybrid nanomaterial emitting at 1065 nm. The design consists of SWCNTs covalently functionalized with spiropyran-merocyanine, which opens the route for photoactivated localization microscopy in the biological windows.64
A lot of improvements are expected to expand the capabilities of NIR super-resolution microcopy, whether from nanostructure engineering like, for example, carbon nanotubes and quantum dots or from organic dye and protein developments. We can also envision that multiphoton approaches with NIR emitters become instrumental in this quest.
Probes for Function
Apart from the development of brighter and more photostable biological probes for fluorescence microscopy, we would like to briefly highlight the immense range of opportunities that will be available in bioimaging when efficient probes that are sensitive to their environment will be generated. We foresee that a large variety of physiological parameters will be attainable soon, such as pH, ionic concentrations, calcium, and analytes, but also different fields, such as electric, magnetic, or temperature, for instance. Many are already being developed, and when progress will be made to allow immersing them in a live specimen and imaging them at high resolution therein, the range of application and impact should be colossal.
In this expedition, fluorescent SWCNT represent again an archetypical platform owing to their high chemical sensitivity65,66 coupled with NIR imaging capabilities that can be engineered for analyte sensing.67,68 Several groups disseminated on the globe are working in this direction with demonstrations to sense, for example, proteins,69 neurotransmitters,70 arsenic,71 and plant pathogens.72
Another class of nanomaterials provide promising routes namely NV centers in diamonds73,74 as, for example, local temperature or magnetic optical sensors, owing to the presence of permanent spins that can be manipulated by light. Other probe developments with sensing capabilities will include fluorescent protein engineering or organic molecules.
Correlative Imaging
The quest for a unique imaging technique that can solve all problems is a chimera: advantages always come with drawbacks. We consider that multimodal imaging between complementary techniques is a key pathway for a better understanding of biological processes. A very basic example of that is the combination of fluorescence and label-free imaging. Indeed, fluorescence imaging can provide molecular specificity, but it is very challenging to perform long duration imaging without perturbing the sample and with a stable signal/noise ratio. Moreover, complementary staining and spectral multiplexing can be used to detect the cells and see the environment of the molecules interest. On the other hand, label-free imaging has an intrinsic poor specificity, but gives an overview of the whole sample structures. It is a highly stealthy method, to the sample point of view, and it provides stable-in-time signals essential for long duration imaging even at high framerate. Merging these two modalities greatly enhances the information retrieved from the sample. This is just one example of two complementary methods.
It is very efficient to perform not only multimodality, but also correlative imaging. To our opinion, we define the frontier between multimodal and correlative imaging when the complementary information from each modality are merged into one unique method with observables impossible to obtain otherwise. As an example, photoacoustics gives subcellular resolution at unprecedented depth (mm) by merging the signature of the acoustic wave into the photonic signal. We are convinced that novel correlative imaging approaches will be created in the future thank notably to the progress in computational imaging (including machine learning and high-resolution light shaping).
Conclusion
This Perspective was conceived as our view of a dynamic research field, bringing together a wide range of researchers motivated by fundamental, engineering, or application questions in topics where optics and photonics are the core discipline. We thus deliberately constructed it as less of a review and more as a stimulating call for ideas to face some challenges that we identified through our own practice of physicists evolving in multidisciplinary environments. In a nutshell, we shared our conviction that after having pushed the fundamental limits of optical microscopy to exquisite levels of resolution and sensitivity, an important challenge of this research field will now be to apprehend the complexity of biological samples. This will allow the placement of these samples at the core of the imaging processes and to ensure quantitative measurements.
Acknowledgments
P.B. acknowledges financial support from the European Research Council Starting Grant (848645) and L.C. from the European Research Council Synergy grant (951294).
The authors declare no competing financial interest.
References
- Hooke R.Micrographia: or Some Physiological Descriptions of Minute Bodies Made by Magnifying Glasses. with Observations and Inquiries Thereupon, 1st ed.; Royal Society: London, 1665. [Google Scholar]
- Cressey D.; Callaway E. Cryo-Electron Microscopy Wins Chemistry Nobel. Nature 2017, 550 (7675), 167–167. 10.1038/nature.2017.22738. [DOI] [PubMed] [Google Scholar]
- Born M.; Wolf E.. Principles of Optics; Cambridge University Press, 1959. [Google Scholar]
- Burns D. H.; Callis J. B.; Christian G. D.; Davidson E. R. Strategies for Attaining Superresolution Using Spectroscopic Data as Constraints. Appl. Opt. 1985, 24 (2), 154–161. 10.1364/AO.24.000154. [DOI] [PubMed] [Google Scholar]
- Hell S. W.; Wichmann J. Breaking the Diffraction Resolution Limit by Stimulated Emission: Stimulated-Emission-Depletion Fluorescence Microscopy. Opt. Lett. 1994, 19 (11), 780–782. 10.1364/OL.19.000780. [DOI] [PubMed] [Google Scholar]
- Gustafsson M. G. Surpassing the Lateral Resolution Limit by a Factor of Two Using Structured Illumination Microscopy. J. Microsc. 2000, 198 (2), 82–87. 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- Moerner W. E.; Orrit M. Illuminating Single Molecules in Condensed Matter. Science 1999, 283, 1670–1676. 10.1126/science.283.5408.1670. [DOI] [PubMed] [Google Scholar]
- Bobroff N. Position Measurement with a Resolution and Noise-Limited Instrument. Rev. Sci. Instrum. 1986, 57 (6), 1152. 10.1063/1.1138619. [DOI] [Google Scholar]
- Clairon A.; Salomon C.; Guellati S.; Phillips W. D. Ramsey Resonance in a Zacharias Fountain. EPL 1991, 16 (2), 165–170. 10.1209/0295-5075/16/2/008. [DOI] [Google Scholar]
- van Oijen A. M.; Köhler J.; Schmidt J.; Müller M.; Brakenhoff G. J. 3-Dimensional Super-Resolution by Spectrally Selective Imaging. Chem. Phys. Lett. 1998, 292 (1), 183–187. 10.1016/S0009-2614(98)00673-3. [DOI] [Google Scholar]
- Betzig E.; Patterson G. H.; Sougrat R.; Lindwasser O. W.; Olenych S.; Bonifacino J. S.; Davidson M. W.; Lippincott-Schwartz J.; Hess H. F. Imaging Intracellular Fluorescent Proteins at Nanometer Resolution. Science 2006, 313 (5793), 1642–1645. 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Hess S. T.; Girirajan T. P. K.; Mason M. D. Ultra-High Resolution Imaging by Fluorescence Photoactivation Localization Microscopy. Biophys. J. 2006, 91 (11), 4258–4272. 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust M. J.; Bates M.; Zhuang X. Sub-Diffraction-Limit Imaging by Stochastic Optical Reconstruction Microscopy (STORM). Nat. Methods 2006, 3 (10), 793–796. 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. M.; Cubitt A. B.; Tsien R. Y.; Moerner W. E. On/Off Blinking and Switching Behaviour of Single Molecules of Green Fluorescent Protein. Nature 1997, 388 (6640), 355–358. 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- Heilemann M.; van de Linde S.; Schüttpelz M.; Kasper R.; Seefeldt B.; Mukherjee A.; Tinnefeld P.; Sauer M. Subdiffraction-Resolution Fluorescence Imaging with Conventional Fluorescent Probes. Angew. Chem., Int. Ed. 2008, 47 (33), 6172–6176. 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- Sahl S. J.; Hell S. W.; Jakobs S. Fluorescence Nanoscopy in Cell Biology. Nat. Rev. Mol. Cell Biol. 2017, 18 (11), 685–701. 10.1038/nrm.2017.71. [DOI] [PubMed] [Google Scholar]
- Balzarotti F.; Eilers Y.; Gwosch K. C.; Gynnå A. H.; Westphal V.; Stefani F. D.; Elf J.; Hell S. W. Nanometer Resolution Imaging and Tracking of Fluorescent Molecules with Minimal Photon Fluxes. Science 2017, 355 (6325), 606–612. 10.1126/science.aak9913. [DOI] [PubMed] [Google Scholar]
- Huang B.; Bates M.; Zhuang X. Super-Resolution Fluorescence Microscopy. Annu. Rev. Biochem. 2009, 78 (1), 993–1016. 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin A. G.; Lounis B.; Cognet L. Super-Resolution Microscopy Approaches for Live Cell Imaging. Biophys. J. 2014, 107 (8), 1777–1784. 10.1016/j.bpj.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell S. W.; Sahl S. J.; Bates M.; Zhuang X.; Heintzmann R.; Booth M. J.; Bewersdorf J.; Shtengel G.; Hess H.; Tinnefeld P.; Honigmann A.; Jakobs S.; Testa I.; Cognet L.; Lounis B.; Ewers H.; Davis S. J.; Eggeling C.; Klenerman D.; Willig K. I.; Vicidomini G.; Castello M.; Diaspro A.; Cordes T. The 2015 Super-Resolution Microscopy Roadmap. J. Phys. D: Appl. Phys. 2015, 48 (44), 443001. 10.1088/0022-3727/48/44/443001. [DOI] [Google Scholar]
- Schermelleh L.; Ferrand A.; Huser T.; Eggeling C.; Sauer M.; Biehlmaier O.; Drummen G. P. C. Super-Resolution Microscopy Demystified. Nat. Cell Biol. 2019, 21, 72–84. 10.1038/s41556-018-0251-8. [DOI] [PubMed] [Google Scholar]
- Löschberger A.; van de Linde S.; Dabauvalle M.-C.; Rieger B.; Heilemann M.; Krohne G.; Sauer M. Super-Resolution Imaging Visualizes the Eightfold Symmetry of Gp210 Proteins Around the Nuclear Pore Complex and Resolves the Central Channel with Nanometer Resolution. Journal of Cell Science 2012, 125 (3), 570–575. 10.1242/jcs.098822. [DOI] [PubMed] [Google Scholar]
- Schlichthaerle T.; Strauss M. T.; Schueder F.; Auer A.; Nijmeijer B.; Kueblbeck M.; Jimenez Sabinina V.; Thevathasan J. V.; Ries J.; Ellenberg J.; Jungmann R. Direct Visualization of Single Nuclear Pore Complex Proteins Using Genetically-Encoded Probes for DNA-PAINT. Angew. Chem., Int. Ed. 2019, 58 (37), 13004–13008. 10.1002/anie.201905685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttfert F.; Wurm C. A.; Mueller V.; Berning S.; Cordes V. C.; Honigmann A.; Hell S. W. Coaligned Dual-Channel STED Nanoscopy and Molecular Diffusion Analysis at 20 nm Resolution. Biophys. J. 2013, 105 (1), L01–L03. 10.1016/j.bpj.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A.; Forkey J. N.; McKinney S. A.; Ha T.; Goldman Y. E.; Selvin P. R. Myosin v Walks Hand-Over-Hand: Single Fluorophore Imaging with 1.5-Nm Localization. Science 2003, 300 (5), 2061–2065. 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- Georgescu R. E.; Kurth I.; O’Donnell M. E. Single-Molecule Studies Reveal the Function of a Third Polymerase in the Replisome. Nat. Struct Mol. Biol. 2012, 19 (1), 113–116. 10.1038/nsmb.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C. J.; Chemla Y. R.; Liu S.; Wang M. D. Optical Tweezers in Single-Molecule Biophysics. Nat. Rev. Methods Primers 2021, 1, 25. 10.1038/s43586-021-00021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega Arroyo J.; Andrecka J.; Spillane K. M.; Billington N.; Takagi Y.; Sellers J. R.; Kukura P. Label-Free, All-Optical Detection, Imaging, and Tracking of a Single Protein. Nano Lett. 2014, 14 (4), 2065–2070. 10.1021/nl500234t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R.; Asano S. M.; Upadhyayula S.; Pisarev I.; Milkie D. E.; Liu T.-L.; Singh V.; Graves A.; Huynh G. H.; Zhao Y.; Bogovic J.; Colonell J.; Ott C. M.; Zugates C.; Tappan S.; Rodriguez A.; Mosaliganti K. R.; Sheu S.-H.; Pasolli H. A.; Pang S.; Xu C. S.; Megason S. G.; Hess H.; Lippincott-Schwartz J.; Hantman A.; Rubin G. M.; Kirchhausen T.; Saalfeld S.; Aso Y.; Boyden E. S.; Betzig E. Cortical Column and Whole-Brain Imaging with Molecular Contrast and Nanoscale Resolution. Science 2019, 363 (6424), eaau8302. 10.1126/science.aau8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson D. S.; Lichtman J. W. Clarifying Tissue Clearing. Cell 2015, 162 (2), 246–257. 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F.; Tillberg P. W.; Boyden E. S. Expansion Microscopy. Science 2015, 347 (6221), 543–548. 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn W.Interference Microscope with Transmitted Illumination. U.S. Patent US2950649A, 1960.
- Cuche E.; Bevilacqua F.; Depeursinge C. Digital Holography for Quantitative Phase-Contrast Imaging. Opt. Lett. 1999, 24 (5), 291–293. 10.1364/OL.24.000291. [DOI] [PubMed] [Google Scholar]
- Teague M. R. Image Formation in Terms of the Transport Equation. J. Opt Soc. Am. A 1985, 2 (11), 2019–2026. 10.1364/JOSAA.2.002019. [DOI] [Google Scholar]
- Liang R.; Erwin J. K.; Mansuripur M. Variation on Zernike’s Phase-Contrast Microscope. Appl. Opt. 2000, 39 (13), 2152–2158. 10.1364/AO.39.002152. [DOI] [PubMed] [Google Scholar]
- Bon P.; Maucort G.; Wattellier B.; Monneret S. Quadriwave Lateral Shearing Interferometry for Quantitative Phase Microscopy of Living Cells. Opt Express 2009, 17 (15), 13080–13094. 10.1364/OE.17.013080. [DOI] [PubMed] [Google Scholar]
- Bon P.; Lécart S.; Fort E.; Lévêque-Fort S. Fast Label-Free Cytoskeletal Network Imaging in Living Mammalian Cells. Biophysj 2014, 106 (8), 1588–1595. 10.1016/j.bpj.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debailleul M.; Georges V.; Simon B.; Morin R.; Haeberlé O. High-Resolution Three-Dimensional Tomographic Diffractive Microscopy of Transparent Inorganic and Biological Samples. Opt. Lett. 2009, 34 (1), 79–81. 10.1364/OL.34.000079. [DOI] [PubMed] [Google Scholar]
- Martinez-Marrades A.; Rupprecht J.-F.; Gross M.; Tessier G. Stochastic 3D Optical Mapping by Holographic Localization of Brownian Scatterers. Opt Express 2014, 22 (23), 29191–29203. 10.1364/OE.22.029191. [DOI] [PubMed] [Google Scholar]
- Ortega Arroyo J.; Kukura P. Interferometric Scattering Microscopy (iSCAT): New Frontiers in Ultrafast and Ultrasensitive Optical Microscopy. Phys. Chem. Chem. Phys. 2012, 14 (45), 15625–15636. 10.1039/c2cp41013c. [DOI] [PubMed] [Google Scholar]
- Barer R. Interference Microscopy and Mass Determination. Nature 1952, 169 (4296), 366–367. 10.1038/169366b0. [DOI] [PubMed] [Google Scholar]
- Ounkomol C.; Seshamani S.; Maleckar M. M.; Collman F.; Johnson G. R. Label-Free Prediction of Three-Dimensional Fluorescence Images From Transmitted-Light Microscopy. Nat. Methods 2018, 15, 917–920. 10.1038/s41592-018-0111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafft C.; Schmitt M.; Schie I. W.; Cialla-May D.; Matthäus C.; Bocklitz T.; Popp J. Label-Free Molecular Imaging of Biological Cells and Tissues by Linear and Nonlinear Raman Spectroscopic Approaches. Angew. Chem., Int. Ed. 2017, 56 (16), 4392–4430. 10.1002/anie.201607604. [DOI] [PubMed] [Google Scholar]
- El-Mashtoly S. F.; Gerwert K. Diagnostics and Therapy Assessment Using Label-Free Raman Imaging. Anal. Chem. 2022, 94 (1), 120–142. 10.1021/acs.analchem.1c04483. [DOI] [PubMed] [Google Scholar]
- Cotte Y.; Toy M. F.; Pavillon N.; Depeursinge C. Microscopy Image Resolution Improvement by Deconvolution of Complex Fields. Opt Express 2010, 18 (19), 19462–19478. 10.1364/OE.18.019462. [DOI] [PubMed] [Google Scholar]
- Ling T.; Boyle K. C.; Goetz G.; Zhou P.; Quan Y.; Alfonso F. S.; Huang T. W.; Palanker D. Full-Field Interferometric Imaging of Propagating Action Potentials. Light Sci. Appl. 2018, 7 (1), 107. 10.1038/s41377-018-0107-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apelian C.; Harms F.; Thouvenin O.; Boccara A. C. Dynamic Full Field Optical Coherence Tomography: Subcellular Metabolic Contrast Revealed in Tissues by Interferometric Signals Temporal Analysis. Biomed. Opt. Express 2016, 7 (4), 1511–1524. 10.1364/BOE.7.001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover E. E.; Squier J. A. Advances in Multiphoton Microscopy Technology. Nat. Photonics 2013, 7 (2), 93–101. 10.1038/nphoton.2012.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard C. J. R. Multiphoton Microscopy: a Personal Historical Review, with Some Future Predictions. J. Biomed. Opt. 2020, 25 (1), 1–11. 10.1117/1.JBO.25.1.014511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.; Xu C. Three-Photon Neuronal Imaging in Deep Mouse Brain. Optica 2020, 7 (8), 947. 10.1364/OPTICA.395825. [DOI] [Google Scholar]
- Huisken J.; Swoger J.; Del Bene F.; Wittbrodt J.; Stelzer E. H. K. Optical Sectioning Deep Inside Live Embryos by Selective Plane Illumination Microscopy. Science 2004, 305 (5686), 1007–1009. 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- Smith A. M.; Mancini M. C.; Nie S. Bioimaging: Second Window for in Vivo Imaging. Nat. Nanotechnol. 2009, 4 (11), 710–711. 10.1038/nnano.2009.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z.; Tang T.; Wu T.; Yu X.; Zhang Y.; Wang M.; Zheng J.; Ying Y.; Chen S.; Zhou J.; Fan X.; Zhang D.; Li S.; Zhang M.; Qian J. Perfecting and Extending the Near-Infrared imaging Window. Light Sci. Appl. 2021, 10, 197. 10.1038/s41377-021-00628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong G.; Antaris A. L.; Dai H. Near-Infrared Fluorophores for Biomedical Imaging. Nat. biomed. eng. 2017, 1 (1), 0010. 10.1038/s41551-016-0010. [DOI] [Google Scholar]
- Jackson C. T.; Jeong S.; Dorlhiac G. F.; Landry M. P. Advances in Engineering Near-Infrared Luminescent Materials. ISCIENCE 2021, 24 (3), 102156. 10.1016/j.isci.2021.102156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y.; Ma Z.; Wang F.; Wang X.; Yang Y.; Liu Y.; Zhao X.; Li J.; Du H.; Zhang M.; Cui Q.; Zhu S.; Sun Q.; Wan H.; Tian Y.; Liu Q.; Wang W.; Garcia K. C.; Dai H. In Vivo Molecular Imaging for Immunotherapy Using Ultra-Bright Near-Infrared-IIb Rare-Earth Nanoparticles. Nat. Biotechnol. 2019, 37, 1322–1331. 10.1038/s41587-019-0262-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z.; Musnier B.; Wegner K. D.; Henry M.; Chovelon B.; Desroches-Castan A.; Fertin A.; Resch-Genger U.; Bailly S.; Coll J.-L.; Usson Y.; Josserand V.; Le Guével X. High-Resolution Shortwave Infrared Imaging of Vascular Disorders Using Gold Nanoclusters. ACS Nano 2020, 14 (4), 4973–4981. 10.1021/acsnano.0c01174. [DOI] [PubMed] [Google Scholar]
- Cherukuri P.; Gannon C. J.; Leeuw T. K.; Schmidt H. K.; Smalley R. E.; Curley S. A.; Weisman R. B. Mammalian Pharmacokinetics of Carbon Nanotubes Using Intrinsic Near-Infrared Fluorescence. Proc. Natl. Acad. Sci. U.S.A. 2006, 103 (50), 18882–18886. 10.1073/pnas.0609265103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsher K.; Liu Z.; Sherlock S. P.; Robinson J. T.; Chen Z.; Daranciang D.; Dai H. A Route to Brightly Fluorescent Carbon Nanotubes for Near-Infrared Imaging in Mice. Nat. Nanotechnol. 2009, 4 (11), 773–780. 10.1038/nnano.2009.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin A. G.; Varela J. A.; Gao Z.; Danné N.; Dupuis J. P.; Lounis B.; Groc L.; Cognet L. Single-Nanotube Tracking Reveals the Nanoscale Organization of the Extracellular Space in the Live Brain. Nat. Nanotechnol. 2017, 12 (3), 238–243. 10.1038/nnano.2016.248. [DOI] [PubMed] [Google Scholar]
- Nandi S.; Caicedo K.; Cognet L. When Super-Resolution Localization Microscopy Meets Carbon Nanotubes. Nanomaterials 2022, 12 (9), 1433. 10.3390/nano12091433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paviolo C.; Soria F. N.; Ferreira J. S.; Lee A.; Groc L.; Bezard E.; Cognet L. Nanoscale Exploration of the Extracellular Space in the Live Brain by Combining Single Carbon Nanotube Tracking and Super-Resolution Imaging Analysis. Methods 2020, 174, 91–99. 10.1016/j.ymeth.2019.03.005. [DOI] [PubMed] [Google Scholar]
- Soria F. N.; Paviolo C.; Doudnikoff E.; Arotcarena M.-L.; Lee A.; Danné N.; Mandal A. K.; Gosset P.; Dehay B.; Groc L.; Cognet L.; Bezard E. Synucleinopathy Alters Nanoscale Organization and Diffusion in the Brain Extracellular Space Through Hyaluronan Remodeling. Nat. Commun. 2020, 11, 3440. 10.1038/s41467-020-17328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin A. G.; Setaro A.; Gandil M.; Haag R.; Adeli M.; Reich S.; Cognet L. Photoswitchable Single-Walled Carbon Nanotubes for Super-Resolution Microscopy in the Near-Infrared. Sci. Adv. 2019, 5 (9), eaax1166. 10.1126/sciadv.aax1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cognet L.; Tsyboulski D. A.; Rocha J.-D. R.; Doyle C. D.; Tour J. M.; Weisman R. B. Stepwise Quenching of Exciton Fluorescence in Carbon Nanotubes by Single-Molecule Reactions. Science 2007, 316 (5830), 1465–1468. 10.1126/science.1141316. [DOI] [PubMed] [Google Scholar]
- Cognet L.; Tsyboulski D. A.; Weisman R. B. Subdiffraction Far-Field Imaging of Luminescent Single-Walled Carbon Nanotubes. Nano Lett. 2008, 8 (2), 749–753. 10.1021/nl0725300. [DOI] [PubMed] [Google Scholar]
- Barone P. W.; Baik S.; Heller D. A.; Strano M. S. Near-Infrared Optical Sensors Based on Single-Walled Carbon Nanotubes. Nat. Mater. 2004, 4 (1), 86–92. 10.1038/nmat1276. [DOI] [PubMed] [Google Scholar]
- Heller D. A.; Jin H.; Martinez B. M.; Patel D.; Miller B. M.; Yeung T.-K.; Jena P. V.; Höbartner C.; Ha T.; Silverman S. K.; Strano M. S. Multimodal Optical Sensing and Analyte Specificity Using Single-Walled Carbon Nanotubes. Nat. Nanotechnol. 2009, 4 (2), 114–120. 10.1038/nnano.2008.369. [DOI] [PubMed] [Google Scholar]
- Budhathoki-Uprety J.; Langenbacher R. E.; Jena P. V.; Roxbury D.; Heller D. A. A Carbon Nanotube Optical Sensor Reports Nuclear Entry Viaa Noncanonical Pathway. ACS Nano 2017, 11 (4), 3875–3882. 10.1021/acsnano.7b00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyene A. G.; Delevich K.; Del Bonis-O’Donnell J. T.; Piekarski D. J.; Lin W. C.; Thomas A. W.; Yang S. J.; Kosillo P.; Yang D.; Prounis G. S.; Wilbrecht L.; Landry M. P. Imaging Striatal Dopamine Release Using a Nongenetically Encoded Near Infrared Fluorescent Catecholamine Nanosensor. Sci. Adv. 2019, 5 (7), eaaw3108. 10.1126/sciadv.aaw3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew T. T. S.; Park M.; Cui J.; Strano M. S. Plant Nanobionic Sensors for Arsenic Detection. Adv. Mater. 2021, 33, 2005683. 10.1002/adma.202005683. [DOI] [PubMed] [Google Scholar]
- Nißler R.; Müller A. T.; Dohrman F.; Kurth L.; Li H.; Cosio E. G.; Flavel B. S.; Giraldo J. P.; Mithöfer A.; Kruss S. Detection and Imaging of the Plant Pathogen Response by Near-Infrared Fluorescent Polyphenol Sensors. Angew. Chem., Int. Ed. 2022, 61 (2), e202108373 10.1002/anie.202108373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochalin V. N.; Shenderova O.; Ho D.; Gogotsi Y. The Properties and Applications of Nanodiamonds. Nat. Nanotechnol. 2012, 7 (1), 11–23. 10.1038/nnano.2011.209. [DOI] [PubMed] [Google Scholar]
- McGuinness L. P.; Yan Y.; Stacey A.; Simpson D. A.; Hall L. T.; Maclaurin D.; Prawer S.; Mulvaney P.; Wrachtrup J.; Caruso F.; Scholten R. E.; Hollenberg L. C. L. Quantum Measurement and Orientation Tracking Offluorescent Nanodiamonds Inside Living Cells. Nat. Nanotechnol. 2011, 6 (6), 358–363. 10.1038/nnano.2011.64. [DOI] [PubMed] [Google Scholar]