Abstract
A cadmium resistance gene, designated cadD, has been identified in and cloned from the Staphylococcus aureus plasmid pRW001. The gene is part of a two-component operon which contains the resistance gene cadD and an inactive regulatory gene, cadX*. A high degree of sequence similarity was observed between cadD and the cadB-like gene from S. lugdunensis, but no significant similarity was found with either cadA or cadB from the S. aureus plasmids pI258 and pII147. The positive regulatory gene cadX* is identical to cadX from pLUG10 over a stretch of 78 codons beginning at the N terminus, but it is truncated at this point and inactive. Sequence analysis showed that the cadmium resistance operon resides on a 3,972-bp element that is flanked by direct repeats of IS257. The expression of cadD in S. aureus and Bacillus subtilis resulted in low-level resistance to cadmium; in contrast, cadA and cadB from S. aureus induced higher level resistance. However, when the truncated version of cadX contained in pRW001 is complemented in trans with cadX from plasmid pLUG10, resistance increased approximately 10-fold suggesting that the cadmium resistance operons from pRW001 and pLUG10 are evolutionarily related. Moreover, the truncated version of cadX contained in pRW001 is nonfunctional and may have been generated by deletion during recombination to acquire the cadmium resistance element.
The cadA and cadB operons represent the two known mechanisms of plasmid-mediated cadmium resistance in Staphylococcus aureus (20, 22, 23, 26). The former is better characterized and is associated with plasmid pI258 (13). A 3.5-kb operon located on this plasmid contains two genes, cadA and cadC. cadA codes for a 727-amino acid (aa) protein that shows sequence similarity to the P class of ATPases (13, 18, 19). Cadmium enters S. aureus through an Mn2+-specific active transport system (27, 28) and accumulates to toxic levels. The cadmium resistance determinant (CadA) affords protection by functioning as an energy-dependent cadmium efflux ATPase (21, 23). The CadC protein is smaller, consisting of 122 aa, and serves as a transcription regulator of the cadmium operon (5). Both CadA and CadC gene products are required for resistance to cadmium and CadC can be provided either in cis or in trans (5, 29).
cadB is a less-defined mechanism of cadmium resistance and resides on an incompatibility group II plasmid, pII147 (14, 24). The mechanism of resistance afforded by cadB differs significantly from that for cadA. The operon contains two genes, designated cadB and cadX, the latter showing strong sequence similarity to the cadC protein (23). Even though the Mn2+-specific transport system is active, S. aureus cells containing pII147 do not accumulate cadmium intracellularly. It has been suggested that CadB does not promote cation efflux but may afford protection to the cell by binding cadmium in the membrane (14).
Recently Chaouni et al. (4) reported a third cadmium resistance element that was contained on plasmid pLUG10 from S. lugdunensis. It encodes two genes, a cadB-like cadmium resistance gene and a regulatory locus, cadX, that in concert result in high-level resistance to cadmium. CadX, like CadC, is a positive regulator of resistance, shares 40% of sequence of CadC, and increases resistance fourfold.
The S. aureus phage group II plasmid pRW001 (15), which contains the genes for exfoliative toxin B (16) and the bacteriocin BacR1 (17), also encodes resistance to cadmium (8). In this report, we show that the cadmium resistance determinant from pRW001 (cadD) is similar to the cadB-like operon from pLUG10 reported by Chaouni et al. (4). The determinant is localized to a 3.5-kb DNA fragment that has transposon-like characteristics. The element is flanked by direct repeats of IS257 and also encodes a transposase gene immediately 3′ of the leftward copy of IS257. The cadD gene is located at the other (3′) end of the element and is immediately upstream of a truncated version of cadX from pLUG10.
MATERIALS AND METHODS
Bacterial strains and media.
S. aureus RN4220 and Bacillus subtilis 168 were propagated at 37°C in tryptic soy broth (TSB). S. aureus RN4220 harboring plasmids pI258, pII147, pRW001, and/or pLUG314 was grown at 37°C in TSB containing cadmium sulfate (10 μg/ml). Escherichia coli DH5α was grown on LB agar and broth (8).
pLUG314, the generous gift from Francois Vandenesch (Lyon, France), contains the cadB-cadX operon from S. lugdenensis (4). The cadB gene has been inactivated by an internal deletion, but cadX is still expressed. The plasmid confers chloramphenicol resistance in S. aureus and was propagated in TSB containing 10 μg of the antibiotic per ml. It was transformed by electroporation (9) into S. aureus RN4220 and RN4220(pRW001). The presence of the plasmid in both strains was verified by agarose gel electrophoresis.
Cloning of cadmium resistance from pRW001.
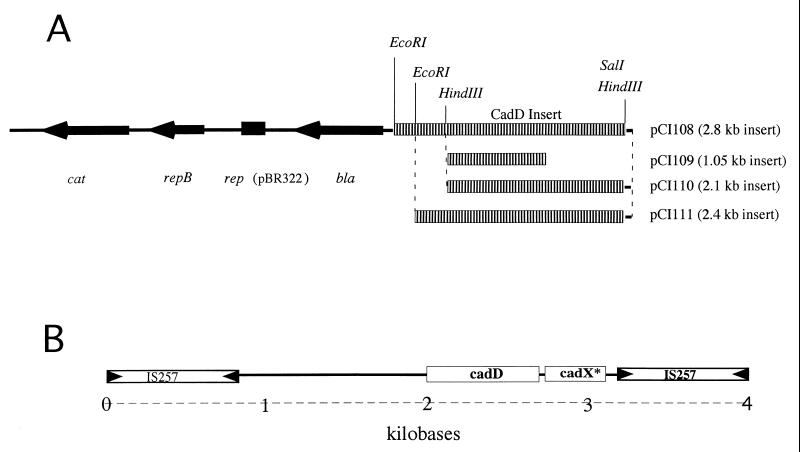
DNA fragments obtained from a partial Sau3A digest of pRW001 were cloned into the BamHI site of the shuttle plasmid pLI50 (11). The preparations were electroporated (9) into S. aureus RN4220, and clones that conferred cadmium resistance were identified (data not shown). A representative that contained a 2.8-kb plasmid insert, designated pCI108 (Fig. 1A), was chosen for further study. A 2.1-kb HindIII fragment from pCI108 containing the DNA sequence encoding cadD and an adjacent smaller open reading frame (ORF) was subcloned into the shuttle plasmid pLI50 to generate pCI110. In addition, a 2.4-kb EcoRI-SalI fragment containing the same genes was cloned in pLI50 to make pCI111 (Fig. 1A). All of these constructs were electrotransformed into S. aureus RN4220.
FIG. 1.
(A) Cloning strategy used to generate plasmids used in this work. Each subclone is shown on the pLI50 backbone; their construction is described in the text. pCI109 contains the 1.05-kb PCR fragment containing the cadD gene cloned into the HindIII site of pLI50. (B) Diagram of the transposon-like element that contains the cadmium resistance determinants. IS257 inverted repeats are indicated by arrowheads; genes locations are indicated in boxed areas.
Subcloning of cadD.
The primers 5′GAAGATAATAAAAAATAGACGACGC3′ (247 bp upstream of the putative translation start site) and 5′CTTCTTTAATCAAAGATAATATGA3′ (154 bp downstream of the CadD ORF) were used to amplify the cadD gene from pCI108 by PCR. The amplification was accomplished with 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min. The resulting 1,046-bp DNA fragment was cloned into the T/A cloning vector pT7Blue (Novagen). This fragment was subsequently cloned into the HindIII site of pLI50 to yield pCI109 (Fig. 1A). DNA sequencing of both strands confirmed the presence and correct sequence of the 205-aa ORF that we designated cadD.
DNA sequencing and analysis.
A Sau3A partial digest of pRW001 was size fractionated, and DNA in the 2-kb size range was recovered from the gel and cloned into the BamHI site of pBluescript. Random clones were sequenced in each direction, using the universal priming sites flanking the inserts. DNA was sequenced with a dideoxy termination kit (Applied Biosystems, Foster City, Calif.), using an Applied Biosystems model 373A automated DNA sequencer. Oligonucleotide primers used in DNA sequencing and PCR were purchased from Integrated DNA Technologies, Coralville, Iowa.
Approximately 400 bp of sequence was obtained from each end of 360 clones containing inserts. Overlapping regions of DNA were assembled by using Sequencher 3.1 (Gene Codes Inc., Ann Arbor, Mich.). These sequences were used to provide additional sequence surrounding the cadD operon contained on pCI110. Hydropathy analysis of putative protein products was carried out by the method of Kyte and Doolittle (10), using a window of seven residues. Molecular modeling was carried out with routines from the BCM Search Launcher (25) at the Baylor College of Medicine.
Determination of inhibitory dose.
Plasmids pC110 and pC111 were electroporated into S. aureus RN4220 and transformed into B. subtilis 168 trpC2 (2, 9). Cadmium resistance levels were determined after overnight growth at 37°C in TSB containing increasing amounts of cadmium sulfate. The maximum inhibitory concentration (MIC) was considered to be the concentration of cadmium sulfate which prevented the appearance of turbidity in the culture after overnight incubation at 37°C. Since CadD from pRW001 represents a third plasmid-encoded cadmium resistance mechanism found in S. aureus, a comparison of the resistance levels encoded by the three systems was carried out in S. aureus RN4220. Constructs containing pI258 (CadA), pII147 (CadB), or pRW001 were generated, and growth was assayed at increasing cadmium concentrations. Similar experiments were also performed to determine the MIC of S. aureus 4220(pRW001)(pLUG314).
Nucleotide sequence accession number.
The element (3,972 bases) containing the cadD operon was deposited in GenBank and assigned accession no. AF134905.
RESULTS AND DISCUSSION
Cloning and analysis of the cadmium resistance genes.
The cadmium resistance element was cloned on a 2.8-kb Sau3A fragment of plasmid pRW001 (Fig. 1, pCI108). A 2.1-kb HindIII fragment subcloned from the original insert (Fig. 1, pCI110) was sequenced at the Kansas State University sequencing facility. Analysis of the sequence revealed the presence of two adjacent ORFs, the first consisting of 209 codons and the second consisting of 78 codons.
BLAST search analysis (1, 6) showed that the cadmium resistance determinants of pRW001 are related to the cadB-like determinants of pLUG10 (4). Both plasmids contain operons composed of a cadmium resistance element and a regulatory gene required for full resistance. The predicted CadD protein and the CadB-like protein (4) share 84% of sequence, but only if the first methionine codon (Fig. 2, upper sequence) at base 236 is chosen as the translation start site. Using this start codon, we find that the first three residues of CadD and the CadB-like protein from pLUG10 match, but a significant divergence occurs between residues 4 and 14. The sequence divergence does not appear to represent a frameshift mutation. This DNA fragment was sequenced from eight independently isolated clones, and in each case the data were identical and unambiguous.
FIG. 2.
(A) Alignment of the CadD protein from pRW001 with the CadB-like protein from pLUG10 (top); (B) alignment of CadX with the putative CadX*. Identical residues are boxed.
There does not appear to be a consensus ribosome binding site upstream of this start site of either cadD or cadB of Chaouni et al. (4). Novick (12) has shown that staphylococcal ribosome binding sites vary from the canonical conserved sequence, but all contain purine-rich sites. With this in mind, translation may actually begin at the AUG codon 12 bases further downstream (CadD amino acid residue 5) which is preceded by a candidate ribosome binding site at bases 237 to 241 (TGAGG). Therefore, the probable translation start site of cadD is uncertain.
The CadB-like homologs are identical in size and share 84% of sequence, while the CadX homologs share 86% of sequence beginning over a 78-aa run at the N terminus (Fig. 2). However, the cadX gene from pRW001 (cadX*) is truncated and contains only 78 codons, compared to 115 for cadX from pLUG10. DNA sequencing of the cadX gene that was PCR amplified directly from pRW001 yielded the same nucleotide sequence as the cadX gene cloned from pRW001 (data not shown). Thus, this smaller ORF is also not the result of a cloning or sequencing artifact.
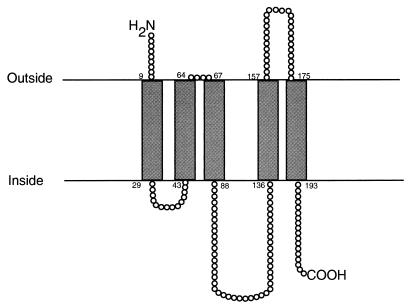
Hydropathy analysis of CadD revealed a substantial hydrophobic character, suggesting that it may function as an integral membrane protein. It is typical of prokaryotic membrane lipoproteins and contains a membrane lipoprotein lipid attachment site at residues 112 to 124. There are five prominent hydrophobic regions following the signal peptide region. Molecular modeling (25) has predicted these five transmembrane domains inserted in the membrane with the N terminus of the protein oriented toward the outside and the C terminus internal (Fig. 3). This arrangement would place cysteine residues located at positions 94 and 124 that could presumably interact with Cd2+ (13) oriented toward the cytosolic domain. Cadmium could then be bound at the membrane as described by Perry and Silver (14).
FIG. 3.
CadD membrane insertion model predicted from hydropathy analysis of transmembrane domains. The amino and carboxy termini are marked.
Cadmium resistance levels.
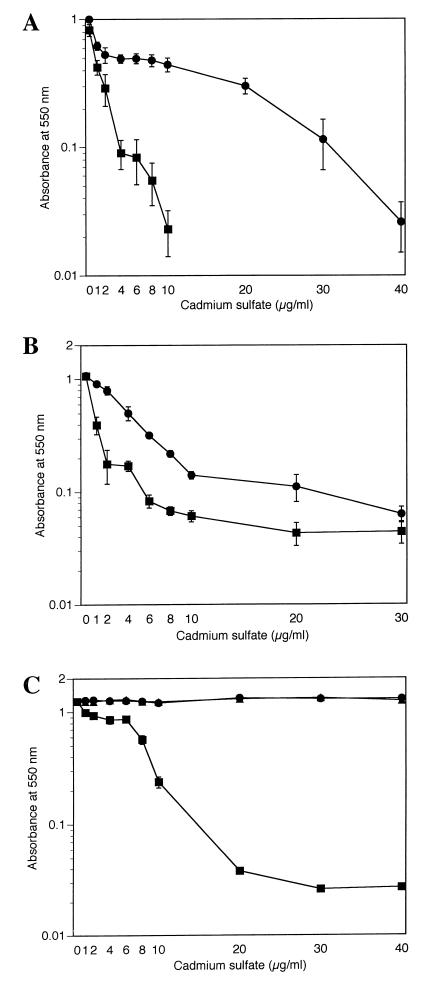
The cadD determinant confers a modest level of cadmium resistance. Cadmium sulfate at greater than 10 μg/ml completely inhibited the growth of S. aureus(pLI50), whereas measurable growth was detected at up to 40 μg/ml for S. aureus(pCI110) (Fig. 4A). The determinant also conferred cadmium resistance when present in B. subtilis, and although B. subtilis(pCI110) (Fig. 4B) was more resistant to cadmium than B. subtilis(pLI50), the overall level was markedly lower than in S. aureus. Low-level resistance similar to that of pCI110 was also obtained (data not shown) when pCI109 (PCR-amplified fragment containing only cadD) was tested for its ability to confer cadmium resistance in S. aureus and B. subtilis.
FIG. 4.
Cadmium resistance in S. aureus and B. subtilis containing pCI110. Overnight cultures were used to inoculate fresh in TSB containing variable amounts of cadmium sulfate to an initial A550 of 0.005. After growth of the cultures for 16 h at 37°C with shaking, the A550 of each culture was determined. Each data point is the mean of three experiments from duplicate cultures. (A) S. aureus(pCI110) (●) and S. aureus (pLI50) (■); (B) B. subtilis(pCI110) (●) and B. subtilis(pLI50) (■); (C) cadmium resistance in S. aureus containing either pI258 (●), pII147 (▴), or pRW001 (■). Overnight cultures were used to inoculate TSB containing variable amounts of cadmium sulfate to an initial A550 of 0.005. After growth for 16 h at 37°C with shaking, the A550 of the culture was determined. Each data point is the mean of three experiments measured in duplicate.
The degree of cadmium resistance conferred by cadD differs markedly from that resulting from the other characterized cadmium resistance determinants. As shown in Fig. 4C, the growth of S. aureus pRW001 steadily decreased at CdSO4 levels over 6 μg/ml, finally plateauing at about 20 μg/ml. However, 40 μg of CdSO4 per ml had no effect on the growth of S. aureus containing either pI258 or pII147. These strains also grew at cadmium sulfate concentrations as high as 500 μg/ml (data not shown).
The MIC of cadmium sulfate for S. aureus(pCI110) was 40 μg/ml, compared to approximately 10 μg/ml for B. subtilis(pCI110). The increased tolerance to cadmium of cells containing either the cadA or cadB determinant cannot be attributed to plasmid copy differences since all three resistance elements are contained on plasmids present in only a few copies per cell (12). Further support for this conclusion is provided by the observation that pCI110 and pCI111, which are present in higher copy numbers than pRW001, were unable to confer resistance to cadmium at concentrations similar to those of pI258 and pII147. Alternatively, since each organism possesses unique membrane properties, membrane insertion and conformation or an accessory protein present only in S. aureus membranes may be required for maximal resistance.
Another possible factor contributing to the notable difference in cadmium resistance conferred by the cadB-like and the cadD operons is the presence in cadD of a truncated version of the positive regulatory gene cadX. The gene from pRW001 contains only the first 78 of 115 codons of the authentic gene. We hypothesized that because it is truncated at the C-terminal end of a predicted helix-turn-helix DNA binding motif, it is probably nonfunctional. To test this hypothesis, we electrotransformed S. aureus(pRW001) with plasmid pLUG314, which contains a functional cadX and assessed levels of resistance of the resulting construct. High-level resistance was afforded by transcomplementation of cadX* by the cadX gene from pLUG10. This resulted in an approximately 10-fold increase in resistance level. The MIC of the transformant clones containing both plasmids was >150 μg/ml, or 195 μM, roughly equivalent to that conferred by pI258 or pII147. On the other hand, with pRW001 alone, which contains a defective copy of CadX, only low-level resistance (MIC of ca. 20 μg/ml [26 μM]) was observed. Neither the recipient strain alone nor the recipient containing pLUG314 showed any resistance to CdSO4. Thus, CadX appears to be necessary for expression of full cadmium resistance from pRW001.
Sequence analysis of cadX shows that it is a member of a regulatory family characterized by cadC, arsR, and smtB. The ArsR and SmtB repressor proteins bind DNA via a helix-turn-helix motif and dissociate from it in the presence of metal ions. They have been hypothesized to interact with cations, resulting in a conformational change that prohibits binding to DNA (3). This in turn results in increased transcription of the associated resistance elements.
This explanation, however, is inappropriate for CadC and CadX, which exert positive regulatory effects (4, 5). Therefore, a conformational change that prevents interaction with DNA would not upregulate transcription. In fact, in the absence of CadC or CadX, only low-level resistance is present. However, activation of cadD and cadB by other mechanisms such as a metal-dependent interaction of CadX with the genes cannot be ruled out.
Last, we also considered the possibility that cadD encodes another heavy metal resistance and functions in a limited fashion for cadmium resistance. However, cadD on pRW001 alone or upregulated by CadX on pLUG314 was unable to confer resistance to sodium arsenate, sodium arsenite, lead nitrate, mercuric nitrate, and zinc chloride when tested at several concentrations (data not shown).
Origin of the cadmium resistance determinants.
In some strains of S. aureus containing plasmids encoding the gene for exfoliative toxin B, cadmium resistance is conferred by a small plasmid of about 4 kb (15). However, pRW001 contains the determinants for cadmium resistance in the absence of small plasmids. Considering the similarity of CadD and CadX to counterparts in pLUG10 and the complementation of cadX* in trans by cadX, we suggest that the two operons are closely related. Acquisition of the cadmium element in pRW001 probably occurred by recombination. If this event was imprecise, it could be responsible for truncation of CadX* to an inactive form. To examine this further, we sequenced regions of pRW001 upstream and downstream of the 2.1-kb HindIII clone. Examination of the sequence showed that the CadD operon appears to reside on a 3,972-bp DNA fragment that is bounded by direct repeats of IS257 (Fig. 1B). The 3′-terminal IS257 element abuts the end of the cadX* gene, and it is likely that the truncated portion of cadX was lost during assembly of this element. The question of transposability is still open. We have not yet been able to demonstrate that it is capable of movement and illegitimate recombination. These experiments are under way and will be reported in a later communication.
ACKNOWLEDGMENTS
This work was supported by grants AI-17474 and AI43568 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bairoch A. A possible mechanism for metal-ion induced DNA-protein dissociation in a family of prokaryotic transcriptional regulators. Nucleic Acids Res. 1993;21:2515–2515. doi: 10.1093/nar/21.10.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaouni L B-A, Etienne J, Greenland T, Vandenesch F. Nucleic acid sequence and affiliation of pLUG10 of a novel cadmium resistance plasmid from Staphylococcus lugdenensis. Plasmid. 1996;36:1–8. doi: 10.1006/plas.1996.0025. [DOI] [PubMed] [Google Scholar]
- 5.Endo G, Silver S. CadC, the transcriptional regulatory protein of the cadmium resistance system of Staphylococcus aureus plasmid pI258. J Bacteriol. 1995;177:4437–4441. doi: 10.1128/jb.177.15.4437-4441.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gish W, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 7.Iandolo J J. Genetic analysis of extracellular toxins of Staphylococcus aureus. Annu Rev Microbiol. 1989;43:375–402. doi: 10.1146/annurev.mi.43.100189.002111. [DOI] [PubMed] [Google Scholar]
- 8.Jackson M P, Iandolo J J. Cloning and expression of the exfoliative toxin B gene from Staphylococcus aureus. J Bacteriol. 1985;166:574–580. doi: 10.1128/jb.166.2.574-580.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kraemer G H, Iandolo J J. High-frequency transformation of Staphylococcus aureus by electroporation. Curr Microbiol. 1990;21:373–376. [Google Scholar]
- 10.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 11.Lee C Y, Iandolo J J. Integration of staphylococcal phage L54a occurs by site-specific recombination: structural analysis of the attachment sites. Proc Natl Acad Sci USA. 1986;83:5474–5478. doi: 10.1073/pnas.83.15.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novick R P. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 13.Nucifora G, Chu L, Mirsa T K, Silver S. Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium efflux ATPase. Proc Natl Acad Sci USA. 1989;86:3544–3548. doi: 10.1073/pnas.86.10.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perry R D, Silver S. Cadmium and manganese transport in Staphylococcus aureus membrane vesicles. J Bacteriol. 1982;150:973–976. doi: 10.1128/jb.150.2.973-976.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogolsky M, Wiley B B, Glasgow L A. Phage group 2 staphylococcal strains with chromosomal and extrachromosomal genes for exfoliative toxin production. Infect Immun. 1976;13:44–52. doi: 10.1128/iai.13.1.44-52.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogolsky M. Nonenteric toxins of Staphylococcus aureus. Microbiol Rev. 1979;43:320–360. doi: 10.1128/mr.43.3.320-360.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rogolsky M, Wiley B B. Production and properties of a staphylococcin genetically controlled by the staphylococcal plasmid for exfoliative toxin synthesis. Infect Immun. 1977;15:726–732. doi: 10.1128/iai.15.3.726-732.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serrano R. Structure and function of proton translocating ATPase in plasma membranes of plants and fungi. Biochim Biophys Acta. 1988;947:1–28. doi: 10.1016/0304-4157(88)90017-2. [DOI] [PubMed] [Google Scholar]
- 19.Serrano R. Structure and function of plasma membrane ATPase. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:61–94. [Google Scholar]
- 20.Silver S, Laddaga R A. Molecular genetics of heavy metal resistance systems in Staphylococcus aureus plasmids. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH Publications; 1990. pp. 531–549. [Google Scholar]
- 21.Silver S, Nucifora G, Chu L, Misra T K. Bacterial resistance ATPases: primary pumps for exporting toxic cations and anions. Trends Biochem Sci. 1989;14:76–80. doi: 10.1016/0968-0004(89)90048-0. [DOI] [PubMed] [Google Scholar]
- 22.Silver S, Misra T K. Plasmid-mediated heavy metal resistances. Annu Rev Microbiol. 1988;42:717–743. doi: 10.1146/annurev.mi.42.100188.003441. [DOI] [PubMed] [Google Scholar]
- 23.Silver S, Walderhaug M. Gene regulation of plasmid- and chromosome-determined inorganic ion transport in bacteria. Microbiol Rev. 1992;56:195–228. doi: 10.1128/mr.56.1.195-228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith K, Novick R P. Genetic studies on plasmid-linked cadmium resistance in Staphylococcus aureus. J Bacteriol. 1972;112:761–772. doi: 10.1128/jb.112.2.761-772.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith R F, Wiese B A, Wojzynski M K, Davison D B, Worley K C. BCM Search Launcher—an integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res. 1996;6:454–462. doi: 10.1101/gr.6.5.454. [DOI] [PubMed] [Google Scholar]
- 26.Tisa L S, Rosen B P. Transport systems encoded by bacterial plasmids. J Bioenerg Biomembr. 1990;22:493–507. doi: 10.1007/BF00762959. [DOI] [PubMed] [Google Scholar]
- 27.Tynecka Z, Gos Z, Zajac J. Energy-dependent efflux of cadmium coded by a plasmid resistance determinant in Staphylococcus aureus. J Bacteriol. 1981;147:313–319. doi: 10.1128/jb.147.2.313-319.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiss A A, Silver S, Kinscherf T G. Cation transport alteration associated with plasmid-determined resistance to cadmium in Staphylococcus aureus. Antimicrob Agents Chemother. 1978;14:856–865. doi: 10.1128/aac.14.6.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon K P, Silver S. A second gene in the Staphylococcus aureus cadA cadmium resistance determinant of plasmid pI258. J Bacteriol. 1991;173:7636–7642. doi: 10.1128/jb.173.23.7636-7642.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]