Abstract
Lipid nanoparticles (LNPs) have shown great success as drug delivery systems, especially for mRNA vaccines, as those developed during the Covid-19 pandemics. Lipid analysis is critical to monitor the formulation process and control the quality of LNPs. The present study is focused on the development and validation of a high-performance liquid chromatography – diode array detector –evaporative light scattering detector (HPLC-DAD/ELSD) based method for the simultaneous quantification of 7 lipids, illustrating the main components of LNPs: ionizable lipids, the neutral co-lipid cholesterol, phospholipids, hydrophilic polymer-lipids for colloidal stability (e.g., a PEGylated lipid). In particular, this study focuses on two innovative synthetic lipids: a switchable cationic lipid (CSL3) which has demonstrated in vitro and in vivo siRNA transfection abilities, and the palmitic acid-grafted-poly(ethyloxazoline)5000 (PolyEtOx), used as an alternative polymer to address allergic reactions attributed to PEGylated lipids. The HPLC separation was achieved on a Poroshell C18 column at 50 °C using a step gradient of a mobile phase composed of water/methanol mixtures with 0.1% (v/v) trifluoroacetic acid (TFA). This method was validated following ICH Q2(R1) & (R2) guidelines in terms of linearity (R² ≥ 0.997), precision (relative standard deviation on peak areas < 5% for intermediate repeatability), accuracy (recoveries between 92.9% and 108.5%), and sensitivity. Indeed, low detection and quantitation limits were determined (between 0.02 and 0.04 µg and between 0.04 and 0.10 µg, respectively). Due to its high selectivity, this method allowed the analysis of lipid degradation products produced through degradation studies in basic, acidic, and oxidative conditions. Moreover, the method was successfully applied to the analysis of several liposome formulations at two key steps of the development process. Consequently, the reported HPLC method offers fast, versatile, selective and quantitative analysis of lipids, essential for development optimization, chemical characterization, and stability testing of LNP formulations.
Keywords: Reversed-phase HPLC, Evaporative light scattering detection, Lipid analysis, Lipid nanoparticles, Validation, Stability study
Graphical Abstract
1. Introduction
Liposomal formulations have been one of the most studied drug delivery systems. They have moved from the simple goal to improve bioavailability of poorly water-soluble drugs to multifunctional drug delivery platforms to enhance therapeutic efficacy by specific targeting or controlled and sustained drug release. Indeed, liposomes offer tunability of composition, flexibility in physico-chemical properties, ease of surface functionalization, biocompatibility, and biodegradability. Today, 18 pharmaceutical products based on liposomes are approved for clinical use, which makes liposomes the most advanced nanoparticle technology [1]. Their range of potential applications is constantly being expanded from the delivery of cancer agents, antifungal, pain, vaccines [2], and recently nucleic acid therapeutics, such as small interferent RNA or mRNA-based vaccines [3], [4].
Such complex nanomedicines require specific quality control and analytical tools to comply with the regulatory guidances. In particular, the identification and the quantification of all lipid species have been identified as critical quality attributes (CQAs) by the Food and Drug Administration (FDA) [5] and the European Medicines Agency (EMA) [6]. In addition, a general effort has been made in the past years to homogenize and standardize the characterization methods of nanomedicines, to improve their translation rate [7]. To contribute to this endeavor, it is therefore of great importance to develop efficient methods for lipid quantification in nanoparticles to support formulation of new products or to control the quality and the safety of final products.
High-performance liquid chromatography (HPLC) using reversed-phase (RP) mode has been used for the separation and the quantification of lipid components in nanoparticle formulations [8], such as cationic or ionizable lipids [9], [10], [11], zwitterionic phospholipids [9], [11], [12], [13], [14], [15], [16], neutral co-lipids, mainly cholesterol [11], [12], [13], [14], and PEGylated lipids [13], [16], [17], [18]. Importantly, most lipids do not possess chromophore in their structures, such as phospholipids, and require alternative UV detection modes. Although mass spectrometry (MS) offers high selectivity and sensitivity [19], [20], [21], its high cost, its specific and expensive maintenance, and the need for experienced operators make this detection technique incompatible with routine quality control activities [22]. Alternatively, evaporative light scattering detector (ELSD) [10], [11], [13], [14], [16] and charged aerosol detector (CAD) [9], [15], [17], [18], [23] can detect compounds less volatile than the mobile phase, which is first nebulized and evaporated to form analyte particles. As opposed to refractive index (RI) detection [24], ELSD and CAD detectors are compatible with the gradient modes often required for lipid separation. Even if both detectors show similar principle operation, ELSD benefits from a lower purchase cost, a simpler use, an easier and cheaper maintenance and a better robustness compared to CAD, making ELSD the apparent detector of choice in the quality control of liposome formulations. However, it is worth noting that CAD signal intensity appears to be more sensitive than ELSD for the quantification of low amounts of lipids [25].
In this study, a fast, selective and quantitative method was developed and validated in RP-HPLC-DAD/ELSD for the simultaneous analysis of 7 lipids involved in various LNP compositions: two ionizable ones, one neutral, two phospholipids, one PEG-derivative and one lipid-grafted hydrophilic polymer used as an alternative to PEG. The new synthetic cationic switchable lipid (CSL3) previously developed was investigated in this study [26], [27], [28]. This ionizable lipid has shown in vitro and in vivo transfection ability of microRNA (miRNA) and siRNA. A validated HPLC method is required to use it as an excipient in LNP formulations, as well as monitor its behavior during the formulation process. Since allergic reactions are attributed to PEG in LNP formulations, alternative to this hydrophilic polymer are being pursued [29], such as poly(ethyloxazoline) (PolyEtOx), a hydrophilic polymer reported to improve circulation times and prevent protein adsorption similarly to PEG [30]. In this study, a palmitic-anchored-poly(ethyloxazoline)5000 was synthesized as an alternative to 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)− 2000] (DSPE-PEG2000), usually reported in LNP composition. PolyEtOx and the ionizable one: 1,2-dioleyloxy-3-dimethylaminopropane (DODMA) have never been quantified in LNPs yet, to our knowledge. Commonly used lipids were also included, i.e. the neutral co-lipid cholesterol (Chol), the phospholipids 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), and the PEGylated phospholipid, DSPE-PEG2000. During method development, the influence of several parameters on lipid separation and detection was assessed, i.e. the nature of the stationary phase and the organic solvent, the type of acid modifier, and the ELSD critical detection parameters. Such in-depth method development improves the understanding of lipid chromatographic behavior, to reduce the analysis time and achieve a better selectivity of the resulting method. The developed method was finally validated and successfully applied to lipid quantitative analysis in LNPs of diverse compositions, proving the reliability of the developed HPLC-ELSD method for quality control.
2. Materials and methods
2.1. Materials
Ethanol (EtOH) 96%, methanol (MeOH), and trifluoroacetic acid (≥ 99.0%) were HPLC grade and purchased from VWR Chemicals (VWR International, Fontenay-sous-Bois, France). A water purification system (Millipore, Belford, MA, USA 18.2 MΩ.cm) was used to provide ultrapure water (18.2 MΩ.cm).
The cationic switchable lipid CSL3 was custom synthesized by Richman Chemicals (Lower Gwynedd, PA, USA) according to the previously described procedure [28]. 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dioleyloxy-3-dimethylaminopropane (DODMA), and 1,2-distearoyl-sn-glycero-3- phosphoethanolamine-N-[methoxy(polyethylene glycol)− 2000] (DSPE-PEG2000) were purchased from Avanti Polar Lipids (Alabaster, AL, USA). Cholesterol (Chol), stearic acid (≥ 98.5%), oleic acid (≥ 99.0%), 1-stearoyl-2-hydroxy-sn-glycero-3-phosphocholine (18:0 Lyso PC), and 1-oleoyl-2-hydroxy-sn-glycero-3-phosphoethanolamine (18:1 Lyso PE) and all chemicals used for Palmitic acid-grafted-poly(ethyloxazoline)5000 synthesis were supplied by Sigma Aldrich (Saint-Louis, MO, USA). Palmitic acid-grafted-poly(ethyloxazoline)5000 (PolyEtOx) was synthesized as described below.
2.2. Synthesis of PolyEtOx
In a round flask, poly(ethyloxazoline) (303 mg, 1 eq . Mn ≈ 5000 g/mol, PdI ≤ 1.3), palmitic acid (47 mg, 3 eq.), N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC-HCl) (53 mg, 4.5 eq.) and 4-(Dimethylamino)pyridine (DMAP) (24 mg, 3 eq.) were introduced and flushed with argon before the addition of 10 mL of dry chloroform. The solution was stirred 24 h at room temperature under argon atmosphere and evaporated under reduced pressure to yield a white solid. The crude was diluted in 3 mL ethanol and dialyzed against ethanol for 24 h (Spectra/Por Membranes, cutoff 2 kDa, Spectrum Laboratories). After evaporation, the white solid (264 mg, 87% yield) was analyzed by 1H NMR (300 MHz, Bruker) (Fig. S1). δ (ppm, CDCl3): 0.81 (t, J = 9 Hz, 3 H, CH3 palmitic); 1.03–1.08 (m, 42 H, CH3 EtOx chain); 1.18 (m, 26 H, CH2 palmitic); 1.52 (m, 2 H, CH2-CO palmitic); 2.19–2.39 (m, 28 H, CH2 EtOx), 2.96 (m, 3 H, CH3 terminal EtOx), 3.38–3.40 (m, 56 H, CH2CH2N EtOx).
2.3. Instrumentation
Chromatographic studies were performed on an HPLC Ultimate 3000 system from Dionex-Thermo Scientific (USA), composed of a quaternary pump (LPG-3400 SD), a thermostated autosampler (WPS-3000), and a column compartment using an outer heater CROCO-CIL. Diode Array Detector (DAD-3000) and Evaporative Light Scattering Detector (ELSD, Alltech Varex MKIII, Conquer Scientific, Poway, CA) dual detection was used. Due to its destructive nature, the ELSD detector was connected in series and after the DAD.
Three RP-HPLC columns were tested: Poroshell C18 30 * 3 mm, 2.7 µm (Agilent, Santa Clara, California); Accucore C18 50 * 4.6 mm, 2.6 µm (ThermoScientific, Waltham, Massachusetts); Zorbax SB-C18 50 * 2.1 mm, 1.8 µm (Agilent, Santa Clara, California).
Under optimal conditions, separation of lipids was carried out on the Poroshell C18 column at a temperature of 50 °C. Two eluents were used, eluent A: 100% water + 0.1% (v/v) TFA (pH = 2.01) and eluent B: 100% MeOH + 0.1% (v/v) TFA. After an equilibration step of 15 min, the initial mobile phase was composed of 13.5%/86.5% (v/v) A/B during 1 min and changed immediately to 100% B for 10 min. The flow rate was set at 0.3 mL.min−1. During method optimization, a volume of 2 µL of lipid mixture was injected. For optimized signal to noise ratios of lipids using the ELSD detector, the nebulizer gas flow rate was set at 1.5 standard liters per minute and the drift tube temperature was 40 °C.
Chromeleon 7.2 software allowed HPLC instrument control and data acquisition and processing.
2.4. Preparation of lipid standards
All lipid masses were weighed with an accuracy of 0.1 mg. Lipid standard stock solutions were prepared in EtOH 96% HPLC grade at a concentration of 10 mg.mL−1.
For HPLC method development, standard stock solutions were mixed and 100-fold diluted with EtOH 96%, to achieve a final concentration of 100 mg.L−1 of each lipid in EtOH.
Calibration was performed using concentration ranges between 20 and 200 mg.L−1 for Chol, DODMA, DSPE-PEG2000, and DSPC; 30–200 mg.L−1 for CSL3, PolyEtOx, and DOPE. Lipid standard solutions were prepared in EtOH 96% by diluting appropriately stock solutions.
2.5. HPLC method validation
Validation of the developed HPLC method was done in accordance to the International Conference of Harmonization (ICH) Q2A and Q2B (Q2(R1) & (R2)) guidelines.
Limits of detection (LOD) and quantitation (LOQ) were determined based on signal to noise ratios (S/N ratios) of 3 and 10, respectively. A series of progressively diluted standard lipid solutions were injected and signal-to-noise ratios were determined for each lipid compound.
Linearity was assessed with a linear least squares regression between log (peak area) and log (lipid mass concentration in mg.L−1). In fact, ELSD response of peak area (A) as function of the injected lipid mass (m) follows a non-linear empirical exponential relationship described by the equation: A = amb, where a and b are constants.
Repeatability and intermediate repeatability were studied by injecting 6 times standard mixtures of the 7 lipids, as the lipid mixtures used in the preparation of the solutions for the rapid mixing, each at 100 mg.L−1 in EtOH 96%, on a same day or on 3 different days (3 independent preparations of the standard mixture), respectively.
The accuracy of the method was checked by analyzing mixtures of the 7 lipids at known concentrations (each lipid at 100 or 50 mg.L−1, accounting for 100% and 50% of cationic lipid recovery after a 4-fold dilution in ethanol 96%, expected before and after the dialysis step, respectively) and expressed as the deviation in percentage (recovery) between the concentration calculated from the standard calibration curve of the lipid and the known concentration. For accuracy study, HPLC analyses were performed in triplicates at each concentration.
Method robustness was assessed through small variations in column temperature ( ± 2 °C), flow rate ( ± 0.03 mL.min−1), and ELSD gaz evaporation temperature ( ± 2 °C), using one factor at a time approach, since no interaction between these parameters was expected.
2.6. LNP preparation
Ionizable switchable LNPs were prepared by rapid mixing as previously described [26]. Briefly, an ionizable lipid (CSL3 or DODMA), Chol, a phospholipid (DSPC or DOPE), and DSPE-PEG2000 or PolyEtOx were mixed in molar ratio of 50.0:37.5:10.0:2.5 (LNP01, LNP02) or 60.0:30.0:8.0:2.0 (LNP03, LNP04), respectively. Then, 0.5 mL of this solution in EtOH (7 mM total lipid concentration) was mixed with 1.5 mL of 1x PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.76 mM KH2PO4, pH 7.4), using a rapid mixing home-made setup, using two syringe pumps (KDS-200, KdScientific, Holliston, MA, USA) connected by a T-junction (Chromtec, Apple Valley, MN, USA) and PEEK tubing (1/16″, 0.010″ between the pump and the T, 1/16″ 0.020″ after the T junction). The total flow rate was 12 mL.min−1 with an aqueous/lipid solution ratio of 3:1 (9 mL.min−1 for aqueous phase and 3 mL.min−1 for ethanolic phase). The obtained LNP suspension (2 mL) was dialyzed against 1 L of a 1x PBS buffer overnight at room temperature and under gentle stirring using Pur-A-Lyzer TM Maxi dialysis tubes MWCO 12–14 kDa (Sigma-Aldrich, Oakville, ON, Canada). Formulations were stored at 4 °C until further use.
2.7. Physico-chemical characterization of LNPs
For quality control of LNP formulations, mean hydrodynamic diameter, polydispersity index (PdI) and zêta potential were measured by Dynamic Light Scattering (DLS) using a Zetasizer Nano ZS (Malvern Instruments Ltd., Malvern, UK). For size and PdI measurements, LNPs were diluted 100-fold and three independent measurements were performed at 20 °C. For zêta potential, LNPs were diluted 20-fold in water, and loaded into a capillary cell (DT1060, Malvern Instruments). Three independent measurements were performed at 20 °C. Lipid amount before and after dialysis step was quantified by the developed HPLC-ELSD method. Before HPLC injection, a single 4-fold dilution step in EtOH 96% was performed to disrupt nanoparticles, and bring lipid concentrations in the linear working ranges. For quantitative analysis of LNPs, each formulation before and after dialysis was diluted and analyzed in HPLC in triplicates. LNPs were analyzed by the developed HPLC-DAD/ELSD method, under optimal chromatographic and detector conditions, as described above (part 2.3). LNP formulations were injected at 5 µL to quantify DSPC and DSPE-PEG2000 lipids and at 2 µL for others lipids.
3. Results and discussion
3.1. Method development and optimization
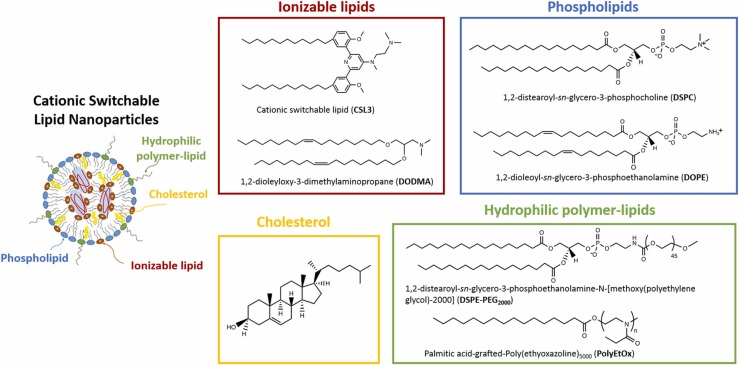
Lipid nanoparticles (LNPs) for nucleic acid delivery are mainly composed of four components ( Fig. 1): (i) an ionizable lipid, ideally ionizable with a pKa less than 7.0, which plays a critical role in nucleic acid complexation and endosomal escape; (ii) a phospholipid, that helps the formation of a stable lipid layer surrounding the nanoparticle; (iii) cholesterol, that provides membrane fluidity and biomimetic properties, and (iv) a lipid-anchored hydrophilic polymer, usually Poly(ethylene glycol) (PEG), that improves colloidal stability and circulation time in the blood, by preventing from undesired protein interactions [31].
Fig. 1.
Structures of the seven studied lipids.
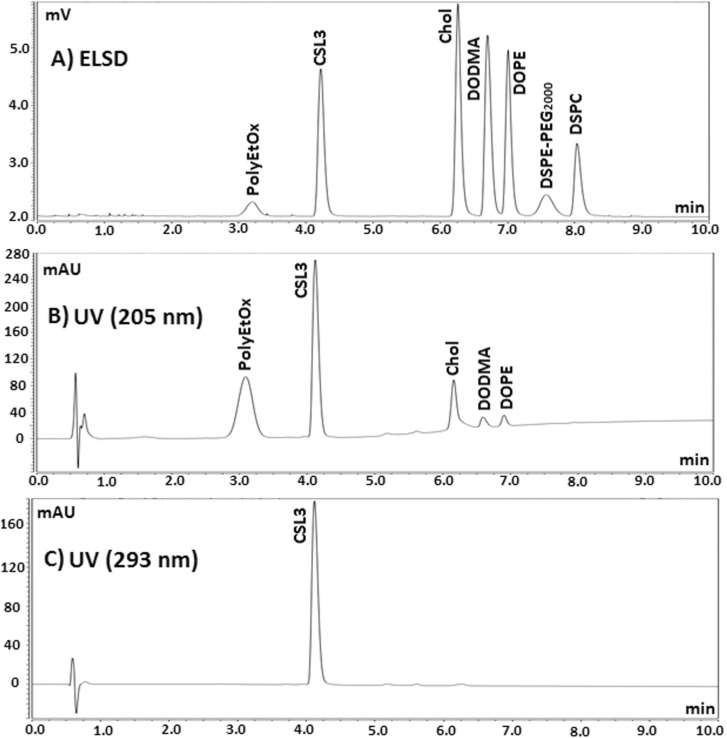
Given their lipophilic character, mainly due to fatty acid chains present in their structures, the different lipids involved in LNPs are classically separated on hydrophobic C18 stationary phases [8], [9], [15]. The objective of this work was to develop a HPLC method to separate 7 lipids (CSL3, DODMA, Chol, DOPE, DSPC, DSPE-PEG2000, PolyEtOx) within a short analysis time, for their quantitative determination in various ionizable LNP formulations. PolyEtOx was synthesized from palmitic acid and hydroxyl-terminated poly(ethyloxazoline) and characterized by 1H NMR (Fig. S1). Fig. 1 presents the chemical structures of the 7 studied lipids. Due to the absence of double bond in the fatty acid chains of DSPC and DSPE-PEG2000, these two compounds could only be detected by the ELSD detector. PolyEtOx, Chol, DODMA, and DOPE could be detected at a low UV wavelength of 205 nm ( Fig. 2). On the contrary, aromatic chromophores of CSL3 allowed its detection in UV with a maximum absorption wavelength of 293 nm (Fig. 2).
Fig. 2.
Chromatograms of a standard mixture of the 7 lipids of interest detected by A) ELSD, B) UV at 205 nm, C) UV at 293 nm. Column: Poroshell C18 30 * 3 mm, 2.7 µm. Column temperature: 50 °C. Mobile phase: 13.5%/86.5% (v/v) Water/MeOH + 0.1% (v/v) TFA during 1 min and 100% MeOH + 0.1% (v/v) TFA until 10 min. Flow rate: 0.3 mL.min−1. Each lipid in mixture at 100 mg.L−1 in EtOH. Injected volume: 2 µL.
During method development, the type of the column and the organic solvent, the nature and the concentration of the mobile phase acidic modifier, as well as ELSD detection parameters were investigated.
Three C18 columns were tested to compare selectivity and peak shape: two core-shell columns, Poroshell C18 30 * 3 mm, 2.7 µm and Accucore C18 50 * 4.6 mm, 2.6 µm, and one column filled with sub-2 µm porous particles: Zorbax SB-C18 50 * 2.1 mm, 1.8 µm. Main advantages of the Zorbax SB-C18 column are its high stability at low pH values and high temperatures (up to 90 °C) due to monofunctional silane, featuring two nonreactive and bulky diisopropyl groups, sterically protecting the siloxane bonds. Based on literature, the temperature of the column was set at 50 °C [10], [15].
A step gradient elution starting with 13.5% water and 86.5% MeOH (v/v) with 0.1% (v/v) TFA during 1 min and directly increasing to 100% MeOH with 0.1% (v/v) TFA was used. Such mobile phase composition allowed a good solubilization of lipids and has physicochemical properties compatible with ELSD detection due to its volatility. A 0.1% (v/v) TFA aqueous solution has a pH of 2.01. At such low pH, residual silanols at the surface of the stationary phase are mostly neutral, while CSL3 and DODMA are positively charged. Indeed, both ionizable lipids, CSL3 and DODMA, exhibit higher pKa values (predicted CSL3 pKa: 5.39 [26] and DODMA pKa: 8.65, measured DODMA pKa: 6.59 (TNS) and 5.83 (zêta potential) [32]. At pH 2.01, electrostatic interactions between lipids and silanols are highly reduced, improving peak shape of ionizable lipids, as already described in the literature [10].
The optimum chromatographic performance in terms of separation and peak shape of the 7 lipids was achieved on the Poroshell C18 column, as shown in Fig. S2.
Using this column, MeOH was then replaced by ACN to study the impact of the organic solvent on the chromatographic profile. In ACN conditions, only 4 peaks could be eluted, probably due to a problem of lipid solubility in ACN-rich mobile phases. It seems consistent with the bad peak shape observed for peaks that are eluted, in terms of asymmetry factors (As > 1.8 for 3 out of 4 peaks: As = 1.87 (RT = 2.40 min); 1.82 (RT = 6.01 min); 1.24 (RT = 6.65 min); 3.09 (RT = 7.47 min)) (Fig. S3).
The effects of the nature (acetic acid (AA) versus TFA) and the concentration (0.05% (v/v) TFA instead of 0.1% (v/v)) of the acidic modifier added to the mobile phase were then investigated. Poor peak asymmetry factors were observed using AA (As (DSPE-PEG2000) = 2.46 and As (DSPC) = 2.73) and 0.05% (v/v) TFA (As (DSPC) = 1.73), as compared to 0.1% (v/v) TFA (Table S2). As optimal conditions, 0.1% (v/v) TFA was therefore kept for further study.
Finally, to optimize the sensitivity of the detection, the two main ELSD detection parameters, i.e. the drift tube temperature and the nebulizer gas flow rate, were varied between 40 and 100 °C, and between 1.50 and 2.40 standard liter per minute (slpm), respectively. A global trend common to all lipids was a decrease of S/N ratios when increasing the evaporation temperature and the gas flow rate (Table S1). When increasing the gas flow rate from 1.50 to 2.40 slpm, a loss of the S/N ratio reaching 85% was calculated for DSPE-PEG2000. Similarly, increasing the drift tube temperature from 40° to 100°C, led to a decrease of S/N ratios up to 95% for Chol. Consequently, the drift tube temperature and the gas flow rate were fixed at 40 °C and 1.50 slpm, respectively.
Fig. 2 presents chromatograms of a standard mixture of the 7 lipids achieved in optimal HPLC conditions. To characterize the chromatographic performance of lipids, figures of merit (retention times, asymmetry factors, peak apparent efficiency, and resolutions) are gathered in Table S2. All lipids were separated in 8 min with resolutions superior to 1.5 (resolution of the critical DSPE-PEG2000/DSPC peak pair: 1.7). Satisfactory asymmetry factors As < 1.5 were achieved for all peaks. Lower peak apparent efficiency was noticed for PolyEtOx and DSPE-PEG2000 because of a heterogeneity in the length of polymer chains. As evidenced in Fig. 2, lipids featuring saturated aliphatic chains, DSPE-PEG2000 and DSPC, could not be detected in UV at 205 nm. Nevertheless, these lipids could be detected using ELSD detector since its response is a function of the injected mass of analyte [25; 33–35].
3.2. Method validation
Several criteria of the developed HPLC-DAD/ELSD method, i.e. sensitivity (detection and quantitation limits), linearity, precision (repeatability, intermediate repeatability), accuracy, and specificity were then validated to fullfill ICH Q2(R1) & (R2) guidelines. Validation results are all gathered in Table 1.
Table 1.
Validation results of the HPLC method for the analysis of the 7 lipids with their retention times, linearity ranges, coefficients of determination (R²), p-values for ANOVA test, limits of detection (LOD) and quantitation (LOQ), relative standard deviation values (RSDs) of retention times and peak areas using ELSD and UV detection for intermediate precision assessment (6 repeated injections per day over 3 days), and average recoveries (n = 3) for accuracy study. For calibration curves, all lipids were injected at 2 µL, except DSPC and DSPE-PEG2000 which were injected at 5 µL. nd: not detected.
| Lipid |
Retention times (min) (ELSD) |
Linearity ranges (mg.L−1) |
R² (ELSD) |
R² (UV 205 nm) |
ANOVA test p-values (ELSD) |
ANOVA test p-values (UV) |
LOD (µg) (ELSD) |
LOQ (µg) (ELSD) |
RSD (tR) (%) n = 18 (ELSD) |
RSD (peak areas) (%) n = 18 |
Recoveries (%) n = 3 (ELSD) |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| min | max | ELSD | UV (205 nm) | 50 mg.L−1 | 100 mg.L−1 | |||||||||
| PolyEtOx | 3.19 | 30 | 200 | 0.9970 | 0.9996 | 1.33.10−7 | 2.45.10−9 | 0.02 | 0.06 | 0.50 | 4.95 | 0.88 | 95.1 | 94.5 |
| CSL3 | 4.21 | 30 | 200 | 0.9993 | 0.9991 (205 nm) 0.9992 (293 nm) |
1.69.10−7 | 6.46.10−9 (205 nm) 4.41.10−9 (293 nm) |
0.02 | 0.06 | 0.80 | 4.47 | 0.74 (205 nm) 0.86 (293 nm) |
92.9 | 94.4 |
| Chol | 6.27 | 20 | 200 | 0.9993 | 0.9995 | 1.88.10−7 | 8.48.10−8 | 0.02 | 0.04 | 0.24 | 1.55 | 1.91 | 94.2 | 94.6 |
| DODMA | 6.70 | 20 | 200 | 0.9992 | 0.9980 | 3.83.10−7 | 9.78.10−7 | 0.02 | 0.04 | 0.50 | 2.15 | 2.09 | 98.6 | 102.9 |
| DOPE | 7.01 | 30 | 200 | 0.9990 | 0.9999 | 3.59.10−7 | 1.09.10−8 | 0.02 | 0.06 | 0.25 | 2.42 | 1.42 | 106.2 | 108.5 |
| DSPE-PEG2000 | 7.58 | 20 | 200 | 0.9990 | nd | 1.52.10−8 | nd | 0.04 | 0.10 | 0.30 | 1.87 | nd | 100.6 | 102.8 |
| DSPC | 8.04 | 20 | 200 | 0.9980 | nd | 1.09.10−8 | nd | 0.04 | 0.10 | 0.23 | 3.00 | nd | 100.0 | 102.8 |
Sensitivity of ELSD detection was studied by injecting gradually diluted standard lipid mixtures and calculating signal-to-noise ratios at each lipid concentration (Table 1). Limits of quantitation (LOQ) corresponding to a S/N ratio of 10 were found to be 0.04 µg for Chol and DODMA, 0.06 µg for PolyEtOx, CLS3, and DOPE, and 0.1 µg for DSPE-PEG2000 and DSPC. Limits of detection (LOD) displaying a S/N ratio of 3 were 0.04 µg for DSPE-PEG2000 and DSPC and 0.02 µg for other lipids. Compared to other reported LOQ and LOD values for lipid detection using ELSD detector, these values are in the same order of magnitude or even below [10], [14], reaching, in the case of DSPE-PEG2000 for example, the LOQ value obtained using CAD detection [15], [18].
The linearity of the method was assessed at 6 concentration levels (from LOQ value to 200 mg.L−1) for each of the 7 lipids. ELSD signal does not vary linearly as function of injected analyte mass but follows a power model like A = amb, with A, the peak area measured by ELSD; m, the injected analyte mass, and a and b, constants depending on the method [25], [33], [34], [35]. Consequently, to obtain linearity for quantitative analysis, calibration curves were established on a double logarithmic scale by plotting log(peak area) versus log (lipid mass concentration in mg.L−1) since injected sample volume remained constant [33], [35]. For all lipids, coefficients of determination (R²) superior or equal to 0.997 (Table 1) highlighted a strong linearity on studied concentration ranges. Dealing with UV detection, R² superior or equal to 0.998 were obtained for CSL3 at 293 nm and 205 nm and for PolyEtOx, Chol, DODMA, and DOPE at 205 nm. Model linearity was also assessed using ANOVA tests. Probability values (p-values) much lower than 0.05 (5% significant threshold) were obtained (Table 1), meaning that there was small probabilities that models were only due to the effect of the mean. The variations of peak area were therefore due to variations in lipid concentration, proving model significance. Moreover, residuals plotted as a function of the predicted responses, appeared to be randomly distributed.
The precision of the method was tested by analyzing standard mixtures of the 7 lipids at 100 mg.L−1 each in EtOH. Relative standard deviation (RSD) values of the retention times and peak areas measured by ELSD from 6 consecutive injections of a same standard lipid mixture, on one day were less than 0.5% and 2%, respectively for all lipids, proving the repeatability of the method (Table 1). Intermediate precision was also assessed by injecting 6 times a standard lipid mixture per day over 3 different days. Each day, a new standard mixture was prepared (3 independent preparations). RSDs calculated on retention times and ELSD peak areas were less than 1% and 5%, respectively. Small variations in retention times of CSL3 (RSD = 0.80%, n = 18) were observed from inter-day runs, probably due to small changes in TFA concentration in the mobile phase. Although satisfactory, the inter-day precision of the ELSD detector (RSDs on peak areas < 5.0%), was found to be lower compared with that of UV detection (RSDs on peak areas ≤ 2.1%) (Table 1). This was expected due to the processes of nebulization and evaporation of the mobile phase involved in ELSD before light scattering detection of lipids.
For accuracy assessment, mixtures of the 7 lipids were prepared at known concentrations of 50 mg.L−1 and 100 mg.L−1 in EtOH to cover the concentrations used in liposome formulations. Each mixture was analyzed in triplicates. The recoveries (%) between the known concentrations and the calculated concentrations of lipids, based on the calibration curves were determined (Table 1). All lipids showed a deviation from the nominal concentration lower than 10% with recoveries between 92.9% and 108.5% for all lipids, indicating satisfactory accuracy of the method.
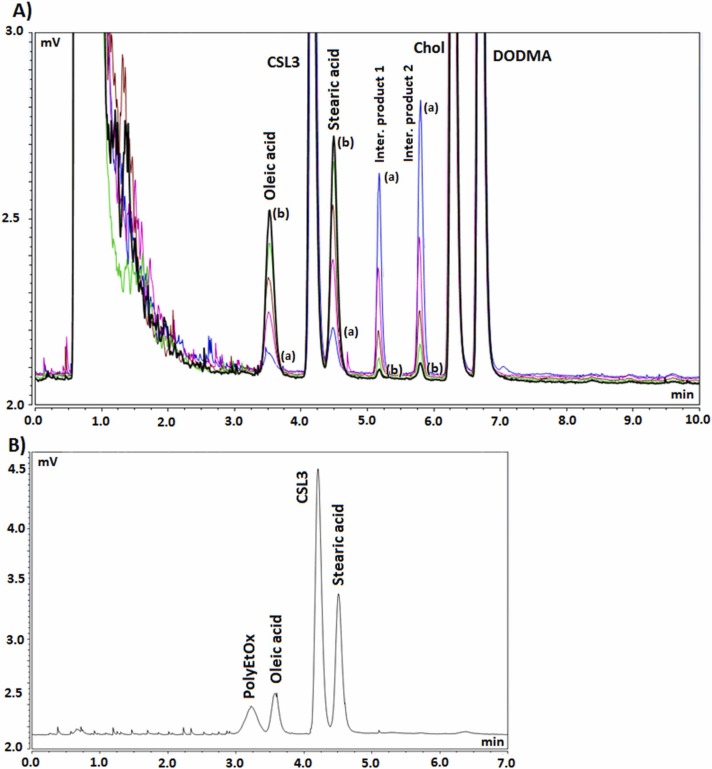
Finally, the specificity of the method was tested through forced degradation studies of standard lipids in mixture or individually under different stress conditions: basic medium (0.1 M NaOH), acidic medium (0.1 M HCl), and oxidizing medium (3% H2O2). Whatever the degradation conditions, several degradation products were separated from the lipids and could be quantified ( Fig. 3, S4-S7). Fig. 3 presents typical chromatograms of a standard mixture composed of the 7 lipids each at 100 mg.L−1 in EtOH undergoing stress condition of 0.1 M NaOH during 15 min and 2 h.
Fig. 3.
A) chromatograms of forced degradation of a standard mixture of 7 lipids using 0.1 M NaOH registered between (a) 15 min (blue) and (b) 2 h (black) after NaOH addition. Intermediate chromatograms were registered at 40 min (pink), 1 h (brown), and 1 h 30 (green). PolyEtOx (RT = 3.2 min), DOPE (RT =7.0 min), DSPE-PEG2000 (RT = 7.6 min), and DSPC (RT = 8.0 min) were totally degraded in these conditions. B) chromatogram showing the separation of a standard mixture composed of PolyEtOx, oleic acid, CSL3, and stearic acid using the developed HPLC method.
Under stress conditions of 0.1 M NaOH, phospholipids (DSPC, DOPE) and PEG-derivatives (DSPE-PEG2000 and PolyEtOx) were totally degraded in less than 15 min, whereas Chol, and ionizable lipids (CSL3 and DODMA) appeared stable (Fig. 3). By conducting similar forced degradation studies of individual lipids, main degradation products for the two phospholipids and DSPE-PEG2000 were identified as their corresponding fatty acids, i.e. oleic acid for DOPE and stearic acid for DSPC and DSPE-PEG2000. Peak identification of degradation products was achieved by injecting individual fatty acid standard samples. Two additional degradation products were detected; the first eluting compound (RT = 5.2 min) was produced by DOPE while the second eluting compound (RT = 5.8 min) was produced by DSPC and DSPE-PEG2000. They appeared to be intermediate degradation products as their peak area decreased over the studied degradation time (Fig. 3).
Chemical stability of lipids was also assessed under acidic conditions by stressing each individual lipid solution at 1 g.L−1 with 0.1 M HCl. Degradation occurred significantly slower than under basic conditions. A higher concentration of lipids was needed to detect all degradation products with a sufficient sensitivity. This prevented from degradation analysis of the lipid mixture, since the resolutions of lipid peaks were not optimal at such concentrations. Therefore, degradation studies were conducted on individual lipids. After 3 days, hydrolysis related byproducts could be detected for phospholipids (DSPC, DSPE-PEG2000 and DOPE) and PolyEtOx, evidencing degradation phenomenon (Figs. S4-S7). As previously reported, phospholipid acidic hydrolysis generated mainly the two 1- and 2-acyl lysophospholipid forms and free fatty acids (Figs. S4-S6) [8], [10], [14]. Peak identification of degradation products was achieved by matching retention times by injecting individual components. Similarly to NaOH conditions, non-identified peaks were detected for phospholipids, but in HCl conditions, their areas increased over the time of the study (between 3 and 5 days). All detected degradation products were satisfactory separated from each other with 2-acyl lysoforms eluting slightly ahead of 1-acyl lysoforms, as previously described [10], [14]. PolyEtOx also degraded in acidic medium into one main degradation product detected at dead time (Fig. S7). This suggests the release of the hydrophilic polymer from the lipid chain by saponification of the ester bond (Fig. 1). On the contrary, positively charged lipids (CSL3 and DODMA) and cholesterol remained stable under 0.1 M HCl up to 6 days (Table S3).
Finally, only CSL3 evidenced a degradation via oxidation in the presence of 3% H202. Losses of 32%, 78%, and 100% occurred in 1, 5, and 22 days, respectively (Table S4). Oxidation product of CSL3 eluted just before CSL3 peak but with a sufficient resolution to be able to quantify both peaks individually (data not shown).
To conclude on degradation studies, it is worth mentioning that whatever the stress conditions, basic, acidic, or oxidative, all observed peaks of degradation products were well separated from the lipids. The specificity of the method was checked for UV active impurities using peak purity tool, but cannot be ensured for non UV-active compounds using such universal ELSD detection.
As requested by ICH Q2(R1) & (R2) guidelines, the robustness of the developed HPLC-DAD/ELSD method was demonstrated by applying minor changes of method parameters, using one factor at a time approach, since the selected parameters were not expected to interact with each other. The flow rate was tested at 0.27 mL.min−1 and 0.33 mL.min−1 instead of 0.30 mL.min−1, column and ELSD evaporation tube temperatures were varied on ± 2 °C of the optimal temperatures, i.e. 48 °C and 52 °C for column temperature, and 38 °C and 42 °C for ELSD evaporation tube temperature. Whatever the conditions, resolutions between peaks, for the 7 lipids injected in mixture at 100 mg.L−1 were all ≥ 1.50, indicating a good robustness of the method within these variation ranges (Table S5).
3.3. Application to quality control of LNP formulations
The applicability of the validated HPLC-DAD/ELSD method for lipid quantification was assessed by analyzing 4 different formulations of ionizable LNPs of various compositions and lipid ratios. As stated previously, LNPs were composed of an ionizable lipid, cholesterol, a phospholipid and a lipid-anchored hydrophilic polymer. The nature and proportion of lipids need to be adapted to each application [31], therefore the quantitative analysis of each lipid is essential. In this study, the type of the ionizable lipid (CSL3 vs. DODMA), phospholipid (DSPC vs. DOPE) and the hydrophilic polymer (PEG vs. PolyEtOx) was varied ( Table 2). Liposomes were produced by rapid mixing between a lipid ethanolic solution and an aqueous PBS buffer, then dialyzed against 1x PBS buffer overnight at room temperature. Obtained LNP formulations were homogenously dispersed as evidenced by polydispersity indexes (PdI) ≤ 0.305 with measured hydrodynamic diameters between 74 nm and 114 nm, and measured zêta potentials between − 7.0 mV and 15.3 mV (Table 2).
Table 2.
Quantitative analysis of lipids in several ionizable LNPs using the validated HPLC-ELSD method and physico-chemical characterization of LNPs.
| Nanoparticule composition | Calculated concentrationa (mg.L−1) | Calculated lipid molar ratiob (%) | Targeted lipid molar ratio (%) | Hydrodynamic diameter (nm) | Polydispersity (PdI) | ζ-potential (mV) |
|---|---|---|---|---|---|---|
| LNP01: CSL3/Chol/DSPE-PEG2000/DSPC | 434.5 ± 8.4/150.1 ± 10.1/68.4 ± 7.4/60.7 ± 6.5 | 54.9/35.8/2.2/7.1 | 50.0/37.5/2.5/10.0 | 85 ± 1 | 0.192 ± 0.008 | -7.0 ± 0.6 |
| LNP02: DODMA/Chol/DSPE-PEG2000/DSPC | 307.8 ± 13.5/151.5 ± 11.1/61.2 ± 6.4/62.8 ± 4.8 | 50.2/39.6/2.2/8.0 | 50.0/37.5/2.5/10.0 | 74 ± 1 | 0.189 ± 0.006 | -4.7 ± 0.3 |
| LNP03: CSL3/Chol/PolyEtOx/DSPC | 625.0 ± 6.1/166.5 ± 11.2/154.1 ± 8.8/101.6 ± 8.6 | 59.3/29.8/2.0/8.9 | 60.0/30.0/2.0/8.0 | 81 ± 0 | 0.290 ± 0.019 | 14.3 ± 1.2 |
| LNP04: CSL3/Chol/DSPE-PEG2000/DOPE | 514.2 ± 17.7/149.8 ± 8.4/81.0 ± 9.1/69.5 ± 7.2 | 58.1/31.9/2.3/7.7 | 60.0/30.0/2.0/8.0 | 114 ± 2 | 0.305 ± 0.036 | 15.3 ± 3.7 |
Mean of three determinations (HPLC analyses performed in triplicate); confidence intervals (95%)
Lipid molar ratios calculated from mean lipid concentrations
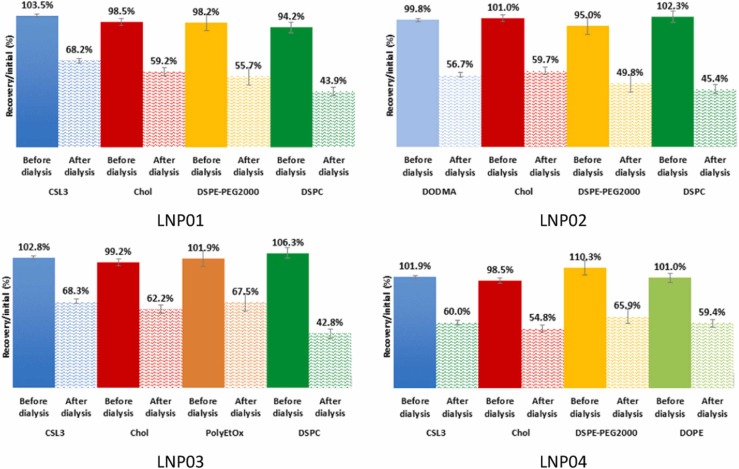
The HPLC method was then used as a quality control tool to monitor nanoparticles during manufacturing process and quantify lipids in final formulations. Chromatograms of the 4 ionizable LNP samples are shown in Figs. S8-S11. All lipids were satisfactory resolved with the HPLC method. 1x PBS buffer used during dialysis eluted at dead time. The lipid content was analyzed just after the mixing step (before dialysis) and after dialysis ( Fig. 4). Before dialysis, good recoveries (percentages of each lipid compared to initial quantities) were calculated for all lipids (between 94.0% and 110.3%), proving the disruption of LNPs by the dilution step and the reliability of the developed HPLC-ELSD method (Fig. 4). The dialysis of LNPs led to a loss between 32% and 55% in the total lipid concentration (Fig. 4), without inducing significant changes in lipid molar ratios (Table 2). This loss of lipids was probably due to elimination of lipids not involved into lipid nanoparticles that could pass through dialysis membrane. DSPC and DSPE-PEG2000 were faced with the lowest recoveries (42–45% and 49–66%, respectively), probably because of their higher water solubility. Table 2 shows quantitative analysis performed by the validated HPLC-ELSD method for the 4 studied ionizable LNPs of various lipid compositions. The calculated lipid molar ratios were in good agreement with the targeted lipid molar ratios for all LNPs, proving a similar loss of all lipids during dialysis. Interestingly, this loss of lipid has not been reported for LNP-based formulations, probably because the quantification of lipids after purification is often overlooked.
Fig. 4.
Lipid content of LNP formulations before and after dialysis step. Each bar represents the quantity of lipid compared to the quantity initially introduced in the formulation. LNP01: CSL3/Chol/DSPE-PEG2000/DSPC (50.0/37.5/2.5/10.0, mol%); LNP02: DODMA/Chol/DSPE-PEG2000/DSPC (50.0/37.5/2.5/10.0, mol%); LNP03: CSL3/Chol/PolyEtOx/DSPC (60.0/30.0/2.0/8.0, mol%); LNP04: CSL3/Chol/DSPE-PEG2000/DOPE (60.0/30.0/2.0/8.0, mol%). Each LNP formulation was diluted and analyzed 3 times.
This study highlights the ability of the developed and validated HPLC-ELSD method to quantify lipids in LNPs with accuracy, which is essential for quality control of developed LNP formulations (lipid composition, positive/negative charge ratio, loading ratios).
4. Conclusion
In this study, a simple and fast HPLC-DAD/ELSD method was developed for the quantitative analysis of 7 lipids involved in ionizable lipid nanoparticles. Prior to injection, a simple sample preparation step was applied, consisting in a single dilution of LNP formulations in ethanol. All validation criteria required by the guidelines ICH Q2(R1) & (R2) were fulfilled, proving the linearity, the accuracy, the repeatability, the intermediate repeatability, and the specificity of the HPLC method. Moreover, sufficient detection and quantification limits for all lipids were achieved and the method proved to be robust.
Due to its high selectivity, this method also offers the opportunity to test the stability of lipid excipients through the simultaneous determination of lipids of interest as well as their degradation products. Indeed, a baseline separation of main lipid hydrolysis products i.e. lysoforms and free fatty acids, from lipidic components was achieved.
Finally, the validated HPLC-DAD/ELSD method was successfully applied to the quantitative analysis of ionizable LNPs of various composition. Each lipid was individually quantified at the two main steps of the formulation process; after rapid mixing and after dialysis. A loss of lipids of about 40% was evidenced during the dialysis stage, without inducing changes in relative lipid molar ratios. This method therefore pointed out the importance of quantification of lipids after the purification step, which is often overlooked. Such method would also allow fine tuning of the process parameters to improve the formulation process and reduce the lipid loss in the final formulation.
The structural variety of analyzed lipids including new lipids never studied in the literature, such as the ionizable CSL3 and the PEG alternative PolyEtOx, proves the flexibility of the proposed method which could be easily transposed to a wide variety of lipids upon minor optimization. To conclude, the described method allows a broad range of applications in various industrial and research laboratories to optimize the formulation process of liposomes under development and for final quality controls.
CRediT authorship contribution statement
Yannick Mousli: Investigation, Writing – review & editing. Mathilde Brachet: Investigation, Validation. Jeanne Leblond Chain: Conceptualization, Resources, Writing – review & editing, Supervision, Project administration, Funding acquisition. Ludivine Ferey: Conceptualization, Methodology, Formal analysis, Resources, Writing – original draft, Writing – review & editing, Supervision, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by Vaincre La Mucoviscidose [grant number RF20200502713/1/1/57]. The authors thank Hong-Van PHAM for her help in the analytical development and Farrah CHRIGUI and Tuan-Nghia DINH for their help in nanoparticle formulation.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jpba.2022.115011.
Appendix A. Supplementary material
Supplementary material.
.
Data availability
No data was used for the research described in the article.
References
- 1.Large D.E., Abdelmessih R.G., Fink E.A., Auguste D.T. Liposome composition in drug delivery design, synthesis, characterization, and clinical application. Adv. Drug Deliv. Rev. 2021;176 doi: 10.1016/j.addr.2021.113851. [DOI] [PubMed] [Google Scholar]
- 2.Filipczak N., Pan J., Yalamarty S.S.K., Torchilin V.P. Recent advancements in liposome technology. Adv. Drug Deliv. Rev. 2020;156:4–22. doi: 10.1016/j.addr.2020.06.022. [DOI] [PubMed] [Google Scholar]
- 3.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kulkarni J.A., Witzigmann D., Thomson S.B., Chen S., Leavitt B.R., Cullis P.R., van der Meel R. The current landscape of nucleic acid therapeutics. Nat. Nanotechnol. 2021;16:630–643. doi: 10.1038/s41565-021-00898-0. [DOI] [PubMed] [Google Scholar]
- 5.U.S. Food, Drug Administration, Liposome Drug Products: Chemistry, Manufacturing, and Controls; Human Pharmacokinetics and Bioavailability; and Labeling Documentation, April 2018, 〈https://www.fda.gov/regulatory-information/search-fda-guidance-documents/liposome-drug-products-chemistry-manufacturing-and-controls-human-pharmacokinetics-and〉 (accessed on March 2022).
- 6.European Medicines Agency, Reflection paper on the data requirements for intravenous liposomal products developed with reference to an innovator liposomal product, February 2013, 〈https://www.ema.europa.eu/en/documents/scientific-guideline/reflection-paper-data-requirements-intravenous-liposomal-products-developed-reference-innovator_en.pdf〉 (accessed on March 2022).
- 7.Faria M., Björnmalm M., Thurecht K.J., Kent S.J., Parton R.G., Kavallaris M., Johnston A.P.R., Gooding J.J., Corrie S.R., Boyd B.J., Thordarson P., Whittaker A.K., Stevens M.M., Prestidge C.A., Porter C.J.H., Parak W.J., Davies T.P., Crampin E.J., Caruso F. Minimum information reporting in bio–nano experimental literature. Nat. Nanotechnol. 2018;13:777–785. doi: 10.1038/s41565-018-0246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan Y., Marioli M., Zhang K. Analytical characterization of liposomes and other lipid nanoparticles for drug delivery. J. Pharm. Biomed. Anal. 2021;192 doi: 10.1016/j.jpba.2020.113642. [DOI] [PubMed] [Google Scholar]
- 9.Li L., Foley J.P., Helmy R. Simultaneous separation of small interfering RNA and lipids using ion-pair reversed-phase liquid chromatography. J. Chromatogr. A. 2019;1601:145–154. doi: 10.1016/j.chroma.2019.04.061. [DOI] [PubMed] [Google Scholar]
- 10.Zhong Z., Ji Q., Zhang J.A. Analysis of cationic liposomes by reversed-phase HPLC with evaporative light-scattering detection. J. Pharm. Biomed. Anal. 2010;51:947–951. doi: 10.1016/j.jpba.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Roces C.B., Kastner E., Stone P., Lowry D., Perrie Y. Rapid quantification and validation of lipid concentrations within liposomes. Pharmaceutics. 2016;8(3):29. doi: 10.3390/pharmaceutics8030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh R., Ajagbe M., Bhamidipati S., Ahmad Z., Ahmad I. A rapid isocratic high-performance liquid chromatography method for determination of cholesterol and 1,2-dioleoyl-sn-glycero-3-phosphocholine in liposome-based drug formulations. J. Chromatogr. A. 2005;1073:347–353. doi: 10.1016/j.chroma.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 13.Oswald M., Platscher M., Geissler S., Goepferich A. HPLC analysis as a tool for assessing targeted liposome composition. Int. J. Pharm. 2016;497:293–300. doi: 10.1016/j.ijpharm.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 14.Jeschek D., Lhota G., Wallner J., Vorauer-Uhl K. A versatile, quantitative analytical method for pharmaceutical relevant lipids in drug delivery systems. J. Pharm. Biomed. Anal. 2016;119:37–44. doi: 10.1016/j.jpba.2015.11.020. [DOI] [PubMed] [Google Scholar]
- 15.Weber F., Rahnfel L., Luciani P. Analytical profiling and stability evaluation of liposomal drug delivery systems: a rapid UHPLC-CAD-based approach for phospholipids in research and quality control. Talanta. 2020;220 doi: 10.1016/j.talanta.2020.121320. [DOI] [PubMed] [Google Scholar]
- 16.Varache M., Ciancone M., Couffin A.-C. Development and validation of a novel UPLC-ELSD method for the assessment of lipid composition of nanomedicine formulation. Int. J. Pharm. 2019;566:11–23. doi: 10.1016/j.ijpharm.2019.05.038. [DOI] [PubMed] [Google Scholar]
- 17.Kurmi M., Suryavanshi V., Panduranga N.S., Jayaraman K., Bajpai L., Fish W., Hu Y., Bhutani H. Development of HPLC-CAD stability indicating assay method for polyethylene glycol-conjugated phospholipid (DMPE-PEG2000) and identification of its degradation products. J. Pharm. Biomed. Anal. 2021;198 doi: 10.1016/j.jpba.2021.113967. [DOI] [PubMed] [Google Scholar]
- 18.Díaz-López R., Libong D., Tsapis N., Fattal E., Chaminade P. Quantification of pegylated phospholipids decorating polymeric microcapsules of perfluorooctyl bromide by reverse phase HPLC with a charged aerosol detector. J. Pharm. Biomed. Anal. 2008;48:702–707. doi: 10.1016/j.jpba.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 19.Knittelfelder O.L., Weberhofer B.P., Eichmann T.O., Kohlwein S.D., Rechberger G.N. A versatile ultra-high performance LC-MS method for lipid profiling. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014;951–952:119–128. doi: 10.1016/j.jchromb.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harkewicz R., Dennis E.A. Applications of mass spectrometry to lipids and membranes. Annu. Rev. Biochem. 2011;80:301–325. doi: 10.1146/annurev-biochem-060409-092612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawthorne G., Harbach S., Wilson A. Cationic lipid bioanalysis: understanding distribution of lipid nanoparticles for delivery of RNA therapeutics. Bioanalysis. 2018;10(5):275–277. doi: 10.4155/bio-2017-0279. [DOI] [PubMed] [Google Scholar]
- 22.Rustam Y.H., Reid G.E. Analytical challenges and recent advances in mass spectrometry based lipidomics. Anal. Chem. 2018;90:374–397. doi: 10.1021/acs.analchem.7b04836. [DOI] [PubMed] [Google Scholar]
- 23.Ferey L., Albe Slabi S., Roy C.-E., Barthélémy P., Gaudin K. Chromatographic study of nucleoside-lipids by RP-UHPLC-DAD/CAD. Anal. Bioanal. Chem. 2018;410(29):7711–7721. doi: 10.1007/s00216-018-1388-9. [DOI] [PubMed] [Google Scholar]
- 24.Grit M., Crommelin D.J.A., Lang J. Determination of phosphatidylcholine, phosphatidylglycerol and their lyso forms from liposome dispersions by high-performance liquid-chromatography using high-sensitivity refractive-index detection. J. Chromatogr. 1991;585:239–246. doi: 10.1016/0021-9673(91)85083-R. [DOI] [Google Scholar]
- 25.Hazotte A., Libong D., Matoga M., Chaminade P. Comparison of universal detectors for high-temperature micro liquid chromatography. J. Chromatogr. A. 2007;1170:52–61. doi: 10.1016/j.chroma.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 26.Gibson V.P., Derbali R.M., Phan H.T., Tahiri H., Allen C., Hardy P., Leblond-Chain J. Survivin silencing improved the cytotoxicity of carboplatin and melphalan in Y79 and primary retinoblastoma cells. Int. J. Pharm. 2020;589 doi: 10.1016/j.ijpharm.2020.119824. [DOI] [PubMed] [Google Scholar]
- 27.Tabatabaei S.N., Derbali R.M., Yang C., Superstein R., Hamel P., Leblond-Chain J., Hardy P. Co-delivery of miR-181a and melphalan by lipid nanoparticles for treatment of seeded retinoblastoma. J. Control. Release. 2019;298:177–185. doi: 10.1016/j.jconrel.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 28.Viricel W., Poirier S., Mbarek A., Derbali R.M., Mayer G., Leblond J. Cationic switchable lipids: pH-triggered molecular switch for siRNA delivery. Nanoscale. 2017;9:31–36. doi: 10.1039/C6NR06701H. [DOI] [PubMed] [Google Scholar]
- 29.Khutoryanskiy V.V. Beyond PEGylation: alternative surface-modification of nanoparticles with mucus-inert biomaterials. Adv. Drug Deliv. Rev. 2018;124:140–149. doi: 10.1016/j.addr.2017.07.015. [DOI] [PubMed] [Google Scholar]
- 30.Simon L., Marcotte N., Devoisselle J.M., Begu S., Lapinte V. Recent advances and prospects in nano drug delivery systems using lipopolyoxazolines. Int. J. Pharm. 2020;585 doi: 10.1016/j.ijpharm.2020.119536. [DOI] [PubMed] [Google Scholar]
- 31.Eygeris Y., Gupta M., Kim J., Sahay G. Chemistry of lipid nanoparticles for RNA delivery. Acc. Chem. Res. 2022;55(1):2–12. doi: 10.1021/acs.accounts.1c00544. [DOI] [PubMed] [Google Scholar]
- 32.Carrasco M.J., Alishetty S., Alameh M.G., Said H., Wright L., Paige M., Soliman O., Weissman D., Cleveland T.E., IV, Grishaev A., Buschmann M.D. Ionization and structural properties of mRNA lipid nanoparticles influence expression in intramuscular and intravascular administration. Commun. Biol. 2021;4:956. doi: 10.1038/s42003-021-02441-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Héron S., Tchapla A. Comparison of the responses of triacylglycerols with an evaporative visible light scattering detector used in conventional, micro and capillary liquid chromatography. J. Chromatogr. A. 1999;848:95–104. doi: 10.1016/j.chroma.2011.01.062. [DOI] [Google Scholar]
- 34.Stolyhwo A., Colin H., Martin M., Guiochon G. Study of the qualitative and quantitative properties of the light-scattering detector. J. Chromatogr. A. 1984;288:253–275. doi: 10.1016/S0021-9673(01)93706-9. [DOI] [Google Scholar]
- 35.Kohler M., Haerdi W., Christen P., Veuthey J.-L. The evaporative light scattering detector: some applications in pharmaceutical analysis. Trends Anal. Chem. 1997;16(8):475–484. doi: 10.1016/S0165-9936(97)00072-1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Data Availability Statement
No data was used for the research described in the article.