Abstract
Temporal and spatial gene regulation during Bacillus subtilis sporulation involves the activation and inactivation of multiple sigma subunits of RNA polymerase in a cascade. In the mother cell compartment of sporulating cells, expression of the sigE gene, encoding the earlier-acting sigma factor, ςE, is negatively regulated by the later-acting sigma factor, ςK. Here, it is shown that the negative feedback loop does not require SinR, an inhibitor of sigE transcription. Production of ςK about 1 h earlier than normal does affect Spo0A, which when phosphorylated is an activator of sigE transcription. A mutation in the spo0A gene, which bypasses the phosphorelay leading to the phosphorylation of Spo0A, diminished the negative effect of early ςK production on sigE expression early in sporulation. Also, early production of ςK reduced expression of other Spo0A-dependent genes but not expression of the Spo0A-independent ald gene. In contrast, both sigE and ald were overexpressed late in development of cells that fail to make ςK. The ald promoter, like the sigE promoter, is believed to be recognized by ςA RNA polymerase, suggesting that ςK may inhibit ςA activity late in sporulation. To exert this negative effect, ςK must be transcriptionally active. A mutant form of ςK that associates with core RNA polymerase, but does not direct transcription of a ςK-dependent gene, failed to negatively regulate expression of sigE or ald late in development. On the other hand, the negative effect of early ςK production on sigE expression early in sporulation did not require transcriptional activity of ςK RNA polymerase. These results demonstrate that ςK can negatively regulate sigE expression by two different mechanisms, one observed when ςK is produced earlier than normal, which does not require ςK to be transcriptionally active and affects Spo0A, and the other observed when ςK is produced at the normal time, which requires ςK RNA polymerase transcriptional activity. The latter mechanism facilitates the switch from ςE to ςK in the cascade controlling mother cell gene expression.
In response to nutrient depletion, Bacillus subtilis undergoes a developmental process that culminates with the formation of a dormant spore (62). Two compartments, the mother cell and the forespore, are formed early during the sporulation process due to the synthesis of an asymmetric septum. The forespore is later engulfed within the mother cell, being completely surrounded by the two membranes of the septum. The mother cell contributes to the synthesis of many components necessary for forespore maturation, including a thick layer of peptidoglycan called cortex and a tough proteinaceous spore coat, and is discarded by lysis at the end of sporulation, releasing the mature spore.
Sporulation involves highly ordered programs of gene expression in the two compartments that are regulated primarily by the ordered appearance of two series of alternate sigma factors (33, 62). Upon starvation, multiple signals impinge on a phosphorelay system composed of protein kinases and phosphatases, a phosphotransferase, and at least one kinase inhibitor (5, 13, 22, 53, 65). The result is an elevated level of phosphorylated Spo0A (Spo0A∼P), a transcription factor that activates ςA RNA polymerase (RNAP) and ςH RNAP to transcribe the genes encoding ςE and ςF, respectively (2, 4, 5, 67). After formation of the asymmetric septum, ςF becomes active in the forespore and directs transcription of the gene encoding ςG (18, 41, 48, 52, 64). Similarly, ςE becomes active in the mother cell and directs transcription of the gene encoding ςK (10, 18, 36).
Communication between the mother cell and the forespore regulates sigma factor activity (33, 43). All the compartment-specific sigma factors are initially inactive. In the forespore, ςF and ςG are held inactive by an anti-sigma factor, SpoIIAB (11, 28, 31, 50). In the mother cell, ςE and ςK are first synthesized as inactive precursor proteins, pro-ςE and pro-ςK (8, 38, 44). Compartmentalized activation of these sigma factors, except for ςF, depends on intercompartmental signal transduction (33, 43). In this way, the programs of gene expression in the two compartments are coupled. In addition to controlling the synthesis and activation of subsequent sigma factors in the cascade, each sigma factor directs core RNAP to transcribe different genes whose products drive morphogenesis (62).
Although the synthesis and activation of sigma factors during B. subtilis sporulation have been relatively well studied, little is known about how later sigma factors replace the earlier ones. We showed previously that in the mother cell compartment, the appearance of ςK accelerates the disappearance of ςE (73). In mutants that fail to produce ςK, the ςE level at 5 to 8 h into development was two- to fivefold higher than in wild-type cells. In a mutant that produces ςK earlier than normal, twofold less ςE accumulated than in wild-type cells. ςK seems to affect the synthesis of ςE, because β-galactosidase activity from a lacZ transcriptional fusion to the promoter of the spoIIG operon (referred to as sigE-lacZ since spoIIGB, also called sigE, encodes pro-ςE) mirrored the ςE level during sporulation of wild-type cells or sigK mutant cells that either fail to make ςK or make ςK earlier than normal. Also, ςK did not detectably alter the stability of ςE. Taken together, these results suggest that ςK initiates a negative feedback loop that regulates transcription of sigE in the mother cell compartment of developing cells.
We have further investigated how ςK negatively regulates sigE expression during sporulation. Transcription of sigE is carried out by ςA RNAP and is activated by Spo0A∼P (2, 4, 30, 57) and repressed by SinR (46, 47). Here, we show that SinR is not required for the negative effect of ςK on sigE expression. The negative effect of early ςK production appears to involve Spo0A, but the negative effect of ςK late in sporulation was also observed for the Spo0A-independent ald gene. Since sigE is known to be transcribed by ςA RNAP, and ald is believed to be, we tested the idea that ςK inhibits ςA RNAP activity late in sporulation by competing with ςA for binding to core RNAP. We found instead that ςK must be transcriptionally active to exert its negative effect late in development. These results give further insight into why ςK activity is temporally regulated and how ςK switches the mother cell pattern of gene expression.
MATERIALS AND METHODS
Bacterial strains.
The B. subtilis strains used in this study are listed in Table 1. BZ48 was constructed by replacing the chloramphenicol resistance gene (cat) of VO48 with a spectinomycin resistance gene (spc) as described previously (60). To introduce gene fusions and mutations into the wild-type strain PY79 and its derivatives BK556 and BZ48, chromosomal DNA was prepared from a strain containing the desired fusion or mutation and used to transform competent cells of the recipient strain (19). Specialized transduction was used to move lacZ fusions carried on SPβ phages into various strains (19). Transformants or transductants were selected on LB plates containing appropriate antibiotics. Chloramphenicol was used at 5 μg/ml, and spectinomycin was used at 100 μg/ml. Resistance to macrolide-lincosamide-streptogramin B (MLS) was selected by using a combination of erythromycin (1 μg/ml) and lincomycin (25 μg/ml). Colonies of cells containing the sinR null mutation displayed a characteristic rough phenotype (12). The rvtA11 mutation in AG919 is 80 to 90% linked by cotransformation to a downstream chloramphenicol resistance gene marker (14). To verify the presence of the rvtA11 mutation in a chloramphenicol-resistant transformant, chromosomal DNA was used to transform competent AG1431 cells. DNA from isolates containing the rvtA11 mutation rescued the Spo− and Pig− AG1431 cells to Spo+ and Pig+ at a frequency of 80 to 90%. A derivative of PY79 containing ald::Tn917lac sporulated poorly in DS medium, consistent with a previous report (59). However, the sporulation efficiency of this strain was comparable to that of the wild-type strain in SM resuspension medium (data not shown). Apparently, the ald locus is dispensable for sporulation in SM medium.
TABLE 1.
B. subtilis strains used
| Strain | Relevant characteristics | Reference or source |
|---|---|---|
| PY79 | Spo+ prototroph | 70 |
| BK556 | spoIVCB23 | 34 |
| VO48 | spoIVCBΔ19 cat | 8 |
| BZ48 | spoIVCBΔ19 spc | This study |
| IS432 | sinR::cat | 12 |
| EU8743 | SPβ::spoIIG-lacZ | 29 |
| AG919 | rvtA11 cat | 14 |
| AG1431 | spo0FΔspo0BΔ | 14 |
| KI1202 | SPβ::spoIIA-lacZ | A. Grossman (66) |
| KY9 | SPβ::spoIIE-lacZ | 15 |
| KI220 | ald::Tn917lac MLS | 59 |
| BK410 | spoIIIC94 | 34 |
| BZ410 | spoIIIC94 sigKC109R spc | This study |
| SC284 | SPβ::gerE-lacZ | 9 |
| BSL50 | spoIIID83 bofA::Tn917Δlac::pTV21Δ2 cat | 45 |
| PS1 | bofA::Tn917Δlac::pTV21Δ2 cat | This study |
| PS2 | spoIIIC94 sigKC109R spc | This study |
| bofA::Tn917Δlac::pTV21Δ2 cat | ||
| BSL1 | sspB-lacZ cat | 49 |
| KI1261 | spo0A-lacZ cat | 24 |
| KH566 | spo0H-lacZ cat | 21 |
| KH586 | spo0K-lacZ cat | 21 |
| ZB456 | SPβ::spoVG42-lacZ | 74 |
Construction of a strain that makes transcriptionally inactive ςK during sporulation.
A 1.4-kb PstI-HindIII fragment containing the sigK gene (44) was cloned into phage M13mp19. A single base pair mutation was made in sigK by site-directed mutagenesis (37). The mutation resulted in a cysteine-to-arginine change at position 109 (C109R) in the region of ςK thought to interact with promoter −10 regions. The sequence of the oligonucleotide used to make the C109R mutation was 5′-CAGCGAGGCGTATTGAA-3′. The entire sigK gene was sequenced to confirm the desired mutation and to ensure that no other mutations had been introduced. A 1.2-kb SacI-HindIII DNA fragment containing the mutation was used to replace the corresponding fragment in pSL1 (44) to generate pBZ1, in which the mutated sigK gene was fused to the Pspac promoter. A 1.5-kb EcoRI-HindIII fragment from pBZ1 was then cloned into the integrational vector pUS19 (3). The resulting plasmid was transformed into BK410, where it integrated via homologous recombination. Recombination upstream of the mutation in sigK resulted in a copy of the mutated gene fused to the spoIVCB promoter followed by a wild-type copy of spoIVCB fused to Pspac. spoIVCB encodes the N-terminal part of ςK and is joined to spoIIIC, encoding the C-terminal part of ςK, by a DNA rearrangement that forms the composite sigK gene during B. subtilis sporulation (61). Since the spoIIIC94 mutation in BK410 is a deletion of spoIIIC (35), the wild-type copy of spoIVCB cannot recombine with spoIIIC during sporulation and no wild-type ςK is made. Therefore, transformants in which recombination occurred upstream of the mutation were expected to produce only mutant ςK and exhibit a Spo− phenotype. Spo− transformants were further screened by Western blot analysis (73), and one isolate (BZ410) that produced mutant ςK during development at a level similar to that observed in wild-type cells was used further. Mutations and lacZ fusions were introduced into BZ410 as described above.
Cell growth and sporulation.
LB medium (19) was used for growth of Escherichia coli and B. subtilis. Sporulation was induced by growing cells in the absence of antibiotic and resuspending cells in SM medium as described previously (19). The onset of sporulation (T0) is defined as the time of resuspension. The sporulation efficiency was measured as described previously (19).
Analysis of lacZ fusion expression.
Strains containing a lacZ fusion were constructed by transformation or transduction as described above. In each case, at least 10 isolates were screened by placing each isolate on DSM agar (19) containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (20 μg/ml). This qualitative assay was used to eliminate occasional isolates with abnormally high or low β-galactosidase activity. Two or more isolates that displayed average blue colony color were induced to sporulate by the resuspension method, and samples collected at hourly intervals were subjected to quantitative β-galactosidase assays, using toluene to permeabilize cells and o-nitrophenol-β-d-galactopyranoside as the substrate (19). One unit of enzyme hydrolyzes 1 μmol of substrate per min per A600 of initial cell density. We found that the maximum β-galactosidase specific activity of a given isolate often varied between experiments, but that the relative maximum β-galactosidase activity of different strains in the same experiment was reproducible. Therefore, strains to be compared were induced to sporulate in parallel, and β-galactosidase specific activities were normalized within each experiment. Minimally, two isolates of each strain were induced to sporulate in each of two separate experiments. The normalized data from separate experiments was averaged to obtain the points shown in the figures. Statistical analysis of the data was performed using the program GraphPad InStat (GraphPad Software).
RESULTS
SinR is not required for the negative effect of ςK on sigE expression.
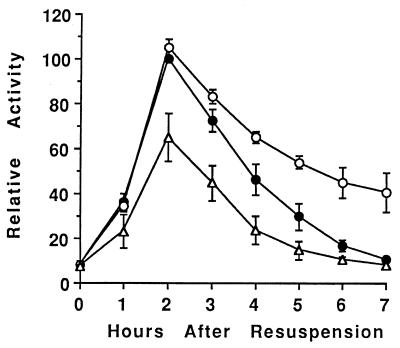
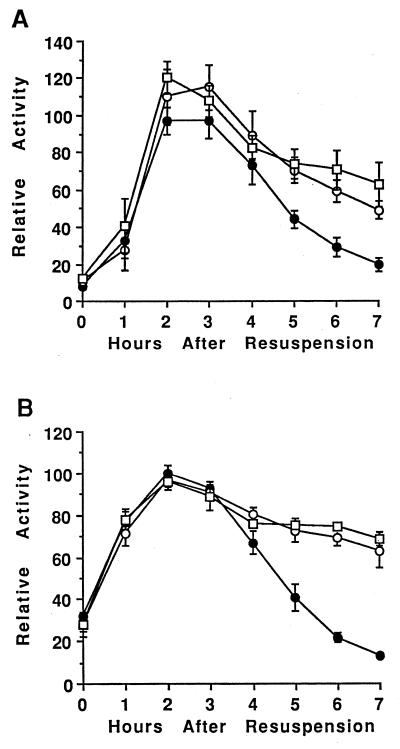
Using a transcriptional fusion between the spoIIG promoter and lacZ (29), which we referred to as sigE-lacZ since it provided an indirect measure of sigE transcription, we showed previously that in spoIVCB23 (spoIVCB encodes the N-terminal part of ςK) mutant cells that fail to produce ςK, sigE-lacZ was overexpressed late in development (73). In spoIVCBΔ19 cells that make active ςK 1 h earlier than normal due to a deletion in the prosequence of pro-ςK, sigE-lacZ expression was reduced (73). To further explore the mechanism by which ςK inhibits expression of sigE, we introduced a sinR null mutation into wild-type cells and mutants with altered ςK production. SinR is a transcription factor that inhibits the transcription of some early sporulation genes, including sigE (46, 47). In cells containing only the sinR mutation, sigE-lacZ expression increased and decreased with similar timing as in wild-type cells but reached a twofold-higher maximum level (130 U versus 70 U) (73), consistent with the finding reported previously that SinR inhibits sigE expression (46, 47). In sinR spoIVCB23 mutant cells that fail to make ςK, sigE-lacZ expression late in development was higher than in cells containing only the sinR mutation (Fig. 1). Statistical analysis of the data (i.e., a nonparametic test of the two-sided P value) indicated that at T4 to T7, sigE-lacZ expression was significantly higher in the strain that fails to make ςK. In sinR spoIVCBΔ19 cells that make ςK earlier than normal, the level of sigE-lacZ expression at T2 to T6 was significantly lower than that in sinR mutant cells (Fig. 1). Since the sinR null mutation did not change the effect of the spoIVCB23 or spoIVCBΔ19 mutation on sigE-lacZ expression, we conclude that ςK does not affect sigE expression by increasing the level or activity of SinR.
FIG. 1.
Effect of a sinR mutation on sigE-lacZ expression. The sinR null mutation in IS432 was transformed into wild-type cells (PY79 [●]), sigK (spoIVCB23) mutant cells (BK556 [○]), and spoIVCBΔ19 cells that produce ςK earlier than normal (BZ48 [▵]). The resulting strains were lysogenized with phage SPβ::sigE-lacZ, and expression of lacZ during development was analyzed as described in Materials and Methods. In each of two separate experiments, the β-galactosidase specific activities were normalized to the average maximum specific activity in two isolates containing the sinR mutation in the wild-type background (typically 130 U). Points on the graph are averages of the normalized values (four determinations), and error bars show 1 standard deviation of the data.
The negative effect of early ςK production depends on Spo0A, but the negative effect of ςK late in sporulation involves another mechanism.
A second mutation that we tested for an effect on the ςK-dependent inhibition of sigE expression was a missense mutation in spo0A called rvtA11 (58). At the onset of sporulation, multiple signals activate a multicomponent phosphorelay system to phosphorylate Spo0A (5, 13, 22, 53). Spo0A∼P then activates transcription of sigE and other early sporulation genes (2, 4). The rvtA11 mutation bypasses the need for the phosphorelay and renders Spo0A able to be phosphorylated by an alternate kinase (39). If ςK inhibits sigE transcription by affecting a component of the phosphorelay so as to lower the level of Spo0A∼P, then the rvtA11 mutation might bypass this effect and relieve the inhibition of sigE transcription by ςK.
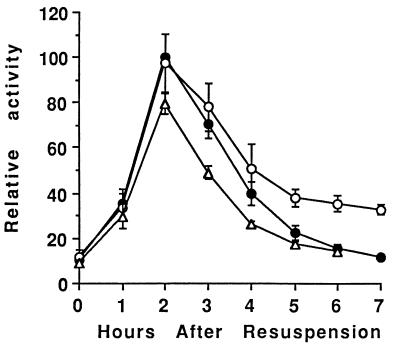
We introduced the rvtA11 mutation into wild-type cells and into spoIVCB23 and spoIVCBΔ19 mutant cells. The sigE-lacZ transcriptional fusion, carried on phage SPβ, was then integrated into the chromosomes of these strains, and developmental β-galactosidase activity was measured. Figure 2 shows that in general, the pattern of sigE-lacZ expression was the same in these strains as in the parental strains without the rvtA11 mutation (73). In cells containing only the rvtA11 mutation, sigE-lacZ expression increased and decreased with similar timing, and reached a similar maximum level, as in wild-type cells (73). In rvtA11 spoIVCB23 mutant cells that fail to make ςK, sigE-lacZ expression late in development was higher than in cells containing only the rvtA11 mutation (Fig. 2). The only difference was that the rvtA11 mutation diminished the negative effect of early ςK production in spoIVCBΔ19 cells. As shown in Fig. 2, sigE-lacZ expression in rvtA11 spoIVCBΔ19 cells reached, on average (five determinations), 80% of the maximum level observed (at T2) in cells containing only the rvtA11 mutation. Statistical analysis of the data collected for these two strains at T2 of sporulation yielded a two-sided P value of 0.095 in a nonparametric test, which is considered not quite a significant difference. In contrast, sigE-lacZ expression in spoIVCBΔ19 cells without the rvtA11 mutation reached only 55% of the maximum level observed in wild-type cells (73), a difference that is significant (P = 0.016) when the same statistical test is applied. Likewise, sigE-lacZ expression in sinR spoIVCBΔ19 cells reached only 65% of the maximum observed in cells containing only the sinR mutation (Fig. 1), which is a significant difference (P = 0.029). While the rvtA11 mutation clearly diminished the negative effect of early ςK production on sigE-lacZ expression, rvtA11 spoIVCBΔ19 cells did exhibit significantly (P ≤ 0.05) less sigE-directed β-galactosidase activity at T3 and T4 than cells containing only the rvtA11 mutation (Fig. 2). Taken together, these results suggest that early ςK production inhibits sigE-lacZ expression, in part, by reducing Spo0A∼P formation by the phosphorelay, because bypassing the phosphorelay with the rvtA11 mutation partially restored sigE-lacZ expression in spoIVCBΔ19 cells; however, early ςK production also inhibits sigE expression by another mechanism that is not bypassed by the rvtA11 mutation, because sigE-lacZ expression in rvtA11 spoIVCBΔ19 cells was not completely restored to the level observed in cells containing only the rvtA11 mutation.
FIG. 2.
Effect of bypassing the phosphorelay on sigE-lacZ expression. The rvtA11 mutation was introduced into wild-type cells (PY79 [●]), sigK (spoIVCB23) mutant cells (BK556 [○]), and spoIVCBΔ19 cells that produce ςK earlier than normal (BZ48 [▵]). The resulting strains were lysogenized with phage SPβ::sigE-lacZ, and expression of lacZ during development was analyzed as described in Materials and Methods. In each of two separate experiments, the β-galactosidase specific activities were normalized to the average maximum specific activity in two or three isolates containing the rvtA11 mutation in the wild-type background (typically 70 U). Points on the graph are averages of the normalized values (five determinations), and error bars show 1 standard deviation of the data.
If early ςK production reduces Spo0A∼P formation, then expression of other genes that depend on Spo0A∼P for activation may be reduced in spoIVCBΔ19 cells. Therefore, we examined expression of transcriptional lacZ fusions to the spoIIE and spoIIA promoters. The spoIIE promoter, like the spoIIG promoter (which drives sigE expression), is recognized by ςA RNAP (1, 69), and the spoIIA promoter is recognized by ςH RNAP (1, 66). All three promoters are activated by Spo0A∼P (2, 4, 5, 67, 69). Expression of the spoIIE-lacZ and spoIIA-lacZ fusions in spoIVCBΔ19 cells reached, on average (two or three determinations), 79 and 65%, respectively, of the maximum level observed in wild-type cells (data not shown), consistent with the idea that early ςK production reduces the Spo0A∼P level early in sporulation.
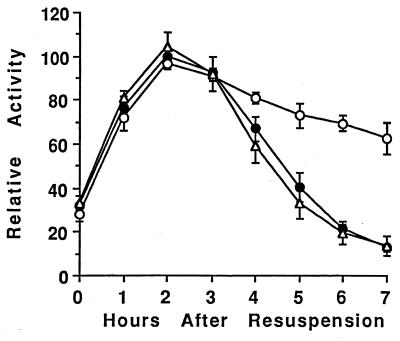
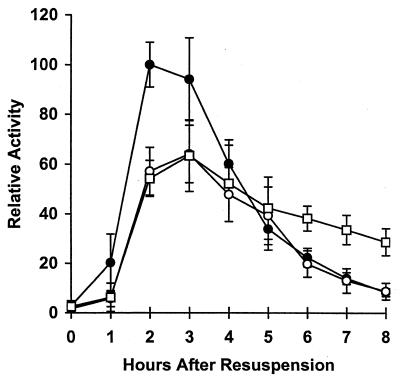
The incomplete restoration of sigE-lacZ expression in rvtA11 spoIVCBΔ19 cells (Fig. 2) indicated that early ςK production also inhibits sigE expression by another mechanism that is not bypassed by the rvtA11 mutation. To determine whether this is due to a general effect on transcription of genes induced early in sporulation, we examined expression of a Spo0A-independent gene in mutants with altered ςK production. The ald gene (encoding alanine dehydrogenase) is induced at the onset of sporulation by an unknown mechanism that does not depend on Spo0A (59). Figure 3 shows that expression of an ald-lacZ transcriptional fusion was unaffected by the spoIVCBΔ19 mutation. Thus, early ςK production does not inhibit expression of all genes induced early in sporulation. On the other hand, ald-lacZ, like sigE-lacZ (73) (Fig. 1 and 2), was overexpressed late in development of spoIVCB23 cells that fail to make ςK (Fig. 3). Taken together, these results suggest that the negative effect of early ςK production is exerted, at least in part, through Spo0A, but the negative effect of ςK produced at the normal time in sporulation involves another mechanism.
FIG. 3.
Effect of altered ςK production on ald-lacZ expression. Wild-type cells (PY79 [●]), sigK (spoIVCB23) mutant cells (BK556 [○]), and spoIVCBΔ19 cells that produce ςK earlier than normal (VO48 [▵]) were transformed with DNA from KI220 to introduce ald::Tn917lac. Expression of ald-lacZ was analyzed as described in Materials and Methods. In each of two separate experiments, the β-galactosidase specific activities were normalized to the average maximum specific activity in two isolates containing ald-lacZ in the wild-type background (typically 300 U). Points on the graph are averages of the normalized values (four determinations), and error bars show 1 standard deviation of the data.
Transcriptionally active ςK RNAP is required for the negative effect of ςK late in sporulation, but not for the negative effect of early ςK production.
One means by which ςK might inhibit expression of early genes is by competing with other sigma factors for binding to core RNAP. Alternatively, inhibition might require that ςK not only bind to core RNAP but also direct transcription. To distinguish between these possibilities, we mutated the sigK gene to produce a single amino acid substitution in ςK that was predicted to abolish transcriptional activity but not core RNAP binding ability. The mutation in ςK was C109R in subregion 2.4, which is thought to be involved in interaction with the −10 region of cognate promoters (20, 42). A similar mutation in ςE (C117R) did not prevent binding of the sigma to core RNAP or binding of the holoenzyme to a cognate promoter but did prevent initiation of transcription (25).
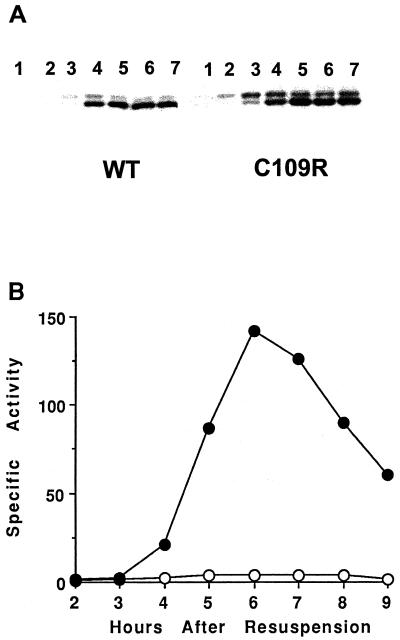
We mutated the sigK gene and integrated it into the chromosome of a sigK mutant, creating BZ410, in which only the sigKC109R allele is expressed (see Materials and Methods). Figure 4A shows that in this mutant pro-ςKC109R was processed to ςKC109R, which accumulated abundantly at T4 and persisted at least until T7, as did ςK in wild-type cells. The ςKC109R was transcriptionally inactive because it failed to direct expression of a gerE-lacZ fusion (Fig. 4B), and the cells failed to form heat-resistant spores (data not shown). When an extract of cells producing ςKC109R was fractionated by gel filtration chromatography as described previously (72), ςKC109R coeluted with the subunits of core RNAP (data not shown), demonstrating that ςKC109R binds to core RNAP.
FIG. 4.
Production of transcriptionally inactive ςK during sporulation. Wild-type PY79 cells (WT) and BZ410 cells engineered to produce transcriptionally inactive ςK (C109R) were lysogenized with phage SPβ::sigE-lacZ and induced to sporulate in SM medium. Samples were collected at the indicated hours after the onset of sporulation. (A) Whole-cell extracts (5 μg) were subjected to Western blot analysis using anti-pro-ςK antibodies as described previously (73). (B) β-Galactosidase specific activity from gerE-lacZ in wild-type (●) and sigKC109R mutant (○) cells.
Expression of sigE-lacZ (Fig. 5A) and ald-lacZ (Fig. 5B) was higher late in development of sigKC109R cells than wild-type cells. The levels of expression in sigKC109R cells making ςKC109R were not significantly different from the levels observed in spoIVCB23 cells which fail to make ςK (Fig. 5). These results suggest that ςK must be transcriptionally active to exert its negative effect on sigE-lacZ and ald-lacZ expression late in sporulation.
FIG. 5.
Effect of making transcriptionally inactive ςK during sporulation on sigE-lacZ and ald-lacZ expression. Wild-type cells (PY79 [●]), sigK (spoIVCB23) mutant cells (BK556 [○]), and sigKC109R cells that produce transcriptionally inactive ςK (BZ410 [□]) were lysogenized with phage SPβ::sigE-lacZ (A) or transformed with DNA from KI220 to introduce ald::Tn917lac (B). Expression of lacZ during development was analyzed as described in Materials and Methods. In each of two separate experiments, the β-galactosidase specific activities were normalized to the average maximum specific activity in two isolates containing the lacZ fusion in the wild-type background (typically 70 U for sigE-lacZ and 300 U for ald-lacZ). Points on each graph are averages of the normalized values (four determinations), and error bars show 1 standard deviation of the data.
To determine whether ςK RNAP transcriptional activity is also necessary for the negative effect of early ςK production on sigE expression, we introduced a bofA mutation into sigKC109R cells. The bofA mutation uncouples processing of pro-ςK from its normal dependence on a signal from the forespore, causing ςK to be produced about 1 h earlier than normal (8, 23, 56). Figure 6 shows that in cells containing only the bofA mutation, the level of sigE-lacZ expression at T2 and T3 was lower than that in wild-type cells. This effect is similar to that observed previously for bofB8 or spoIVCBΔ19 mutations (73), which also cause early ςK production (8, 16). In bofA sigKC109R cells, sigE-lacZ expression early in sporulation was indistinguishable from that in cells containing only the bofA mutation (Fig. 6). Thus, early production of transcriptionally inactive ςKC109R inhibited sigE expression early in sporulation as effectively as early production of wild-type ςK. On the other hand, the level of sigE-lacZ expression in bofA sigKC109R cells was significantly higher than that in bofA or wild-type cells at T6 to T8 (Fig. 6), consistent with the idea that transcriptionally active ςK RNAP is required for the negative effect on sigE expression late in development.
FIG. 6.
Effect on sigE-lacZ expression of making transcriptionally inactive ςK earlier than normal during sporulation. The bofA::cat mutation in BSL50 was transformed into wild-type PY79 cells and BZ410 cells engineered to produce transcriptionally inactive ςK, resulting in PS1 (○) and PS2 (□), respectively. These strains, and wild-type PY79 (●), were lysogenized with phage SPβ::sigE-lacZ, and expression of lacZ during development was analyzed as described in Materials and Methods. In each of two separate experiments, the β-galactosidase specific activities were normalized to the average maximum specific activity in two isolates containing sigE-lacZ in the wild-type background (typically 70 U). Points on the graph are averages of the normalized values (four determinations), and error bars show 1 standard deviation of the data.
DISCUSSION
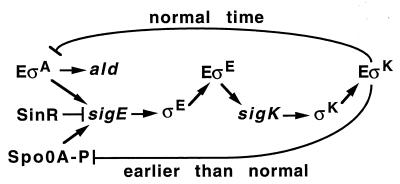
We have found that the appearance of ςK during B. subtilis sporulation can negatively regulate expression of the sigE gene by two different mechanisms, depending on whether ςK is produced at the normal time or 1 h earlier (Fig. 7). The mechanism operative when ςK is produced earlier than normal does not require ςK to be transcriptionally active (Fig. 6) and appears to affect Spo0A∼P, as evidenced by the partial restoration of sigE-lacZ expression in rvtA11 spoIVCBΔ19 cells (Fig. 2) and the reduced expression in spoIVCBΔ19 cells of both sigE (73) and other Spo0A-dependent genes (spoIIA and spoIIE [data not shown]) but not a Spo0A-independent gene (ald [Fig. 3]). As depicted in Fig. 7, a second mechanism, observed when ςK is produced at the normal time, inhibits sigE expression late in development. This mechanism requires ςK RNAP transcriptional activity (Fig. 5 and 6) and inhibits expression of the Spo0A-independent ald gene (Fig. 3 and 5B). Since ald is believed to be transcribed by ςA RNAP, we speculate that transcription of one or more genes by ςK RNAP creates a feedback loop that normally lowers transcription of early genes by ςA RNAP (Fig. 7). The resulting inhibition of sigE expression, together with turnover of ςE, would help switch the mother cell from ςE-directed transcription to the ςK-directed pattern.
FIG. 7.
Model showing the mother cell sigma factor cascade and two different mechanisms by which ςK can negatively regulate sigE expression. ςK produced earlier than normal may compete with other sigma factors for binding to core RNAP (E) and inhibit formation of Spo0A∼P, an activator of sigE transcription. Transcriptional activity of EςK produced at the normal time during sporulation may inhibit EςA activity, reducing transcription of sigE, ald, and other early genes late in development.
Previous work has shown that it is important not to make ςK too early during sporulation (8). Proteolytic processing of pro-ςK to ςK in the mother cell is governed by a signal transduction pathway that emanates from the forespore (7, 8, 33, 44). Bypassing this step by deleting the prosequence (i.e., the spoIVCBΔ19 mutation) or mutating components of the pathway (i.e., the bof mutations) causes a 10-fold decrease in sporulation efficiency, and the spores that are produced germinate poorly (8). Our results provide a plausible explanation for these defects. ςK produced earlier than normal inhibits expression of Spo0A-dependent sporulation genes, including sigE. This lowers the level of ςE (73), and SpoIIID (16) produced. SpoIIID is a transcription factor that activates or represses many genes in the ςE and ςK regulons (17, 71). The cumulative effects of aberrant early gene regulation presumably cause the observed sporulation and germination defects.
How might early production of ςK inhibit the expression of Spo0A-dependent genes? The finding that early production of transcriptionally inactive ςKC109R has the same effect (Fig. 6) suggests that competition of ς factors for binding to a limiting amount of core RNAP may be responsible. Evidence for such competition between ςA and ςH at the onset of sporulation has been presented previously (22). Recently, Ju et al. (27) have used velocity centrifugation and Western blot analysis to monitor the association of ςA, ςE, and ςK with core RNAP during sporulation. Their results suggest that ςE partially displaces ςA from core and that ςK further displaces ςA and also displaces ςE. Transcriptionally inactive ςEC117R and ςKC109R were as effective as their wild-type counterparts at displacing other ς factors from core RNAP. These results suggest that later-acting ς factors in the mother cell cascade have successively higher affinity for core RNAP. If this is the case, then early ςK production would prematurely displace ςA, ςH, and ςE from core RNAP. The resulting changes in the pattern of mother cell gene expression could reduce Spo0A-dependent gene expression in more than one way. For example, the complex phosphorelay system that governs the level of Spo0A∼P provides many potential regulatory targets (5, 13, 22, 53, 65). Reduced expression of a phosphorelay component that leads to the formation of Spo0A∼P may explain the portion of reduced sigE-lacZ expression in spoIVCBΔ19 cells (73) that was restored by the rvtA11 mutation which bypasses the phosphorelay (Fig. 2). Reduced expression of the spo0A gene, which is transcribed by ςA RNAP (6) and ςH RNAP (55, 63) from different promoters, might account for the reduced sigE-lacZ expression that could not be restored by the rvtA11 mutation (Fig. 2). Alternatively, early ςK production may act more directly to inhibit sigE expression, displacing enough ςA from core RNAP to reduce transcription of sigE by ςA RNAP.
Early ςK production did not lower ald expression (Fig. 3). If ald is transcribed by ςA RNAP, as has been proposed (59), then apparently the ςK produced earlier than normal in spoIVCBΔ19 cells does not displace enough ςA from core RNAP to inhibit ald transcription. It has been postulated that an unidentified regulatory factor is involved in ald induction early in sporulation (59). Perhaps this putative factor can stimulate ald transcription even when the ςA RNAP level is low.
Pro-ςK does not inhibit expression of Spo0A-dependent genes, even if it is produced earlier than normal (45), because it does not associate with core RNAP (72). The prosequence targets pro-ςK to membranes (72) and prevents premature ς competition in the mother cell.
Sigma competition does not seem to account for the negative effect of ςK produced at the normal time during sporulation, because ςKC109R was completely ineffective at inhibiting sigE-lacZ (Fig. 5A and 6) or ald-lacZ (Fig. 5B) expression late in development. How, then, does ςK RNAP transcriptional activity inhibit expression of early genes late in development? We considered the possibility that expression of genes in the ςK regulon causes morphological changes that make some of the products of early gene expression inaccessible, due to sequestering in the forespore. In our previous study (73) and in the experiments shown here, we used toluene to permeabilize developing cells to a substrate for β-galactosidase. Lysozyme treatment is much more effective than toluene for the detection of β-galactosidase activity in the forespore (7), a finding that we verified in assays using cells containing sspB-lacZ (data not shown), which is expressed specifically in the forespore (49). However, there was no significant difference between the two methods for detection of sigE-lacZ expression during sporulation (data not shown), indicating that β-galactosidase produced from this fusion is predominantly in the larger mother cell compartment. Perhaps β-galactosidase partitioned to the forespore is degraded, as appears to be the case for ςE produced in the forespore (26, 40, 54). Thus, it is very unlikely that ςK-dependent sequestering of early gene products in the forespore accounts for the lower level of sigE-directed β-galactosidase activity (73) (Fig. 1 and 2), ald-directed β-galactosidase activity (Fig. 3), or ςE protein (73) observed late in development of wild-type cells than spoIVCB23 mutant cells that fail to make ςK, nor do we think that ςK RNAP activity leads to increased turnover of ςE and β-galactosidase in the mother cell. The spoIVCB23 mutation did not affect turnover of pro-ςE or ςE at T3 to T5 in a pulse-chase experiment (73). Also, this mutation did not alter the decay of β-galactosidase activity after T1 in cells containing lacZ fused to the spo0A, spo0H, or spo0K promoter (data not shown). Unlike these promoters, which probably are no longer transcribed after T1, the spoIIA, spoIIE, and spoVG42 promoters continue to be transcribed, judging from the continued increase of β-galactosidase activity after T1 in cells containing lacZ fusions to these promoters. Interestingly, β-galactosidase activity from these fusions is reproducibly higher at T5 to T7 in spoIVCB23 cells than in wild-type cells (data not shown), although the difference is smaller than observed for sigE-lacZ (73) or ald-lacZ (Fig. 3). It is possible that transcription by ςK RNAP drains mother cell nucleotide pools late in development, inhibiting expression of all early genes. Alternatively, the product(s) of a specific gene(s) under ςK control may be involved in the feedback loop. In either case, it appears that the feedback loop inhibits expression of many early genes, including ones transcribed by ςA RNAP (sigE, ald, and spoIIE) and ones transcribed by ςH RNAP (spoIIA and spoVG42).
We do not know how important the feedback loop created by ςK RNAP transcriptional activity is for sporulation. The lowering of the ςE level (73) is expected to reduce transcription of all genes for which ςE RNAP is the limiting factor. In addition, ςK RNAP activity may inhibit ςE RNAP activity by draining nucleotide pools or by a more specific mechanism, as proposed above. At least one key gene in the ςE regulon, spoIIID, is overexpressed in mutants that fail to make ςK (34, 73). We have proposed previously that the feedback loop ensures the timely disappearance of SpoIIID from the mother cell, relieving SpoIIID repression of several cot genes that encode spore coat proteins (16, 17, 73). Both ςE RNAP and ςK RNAP transcribe genes whose products are involved in formation of the spore cortex and coat (62). In order for these structures to form properly, it may be critical to regulate the transition from ςE- to ςK-directed transcription.
Negative feedback loops control the transcription of genes involved in flagellar biosynthesis in E. coli (32) and Caulobacter crescentus (51, 68). In some cases, negative control turns off expression of genes encoding structural proteins once those proteins assemble properly. Determination of whether the feedback loop initiated by ςK RNAP activity monitors morphogenesis or whether it simply acts as a timer governing the switch from early to late gene expression must await further elucidation of the molecular mechanism.
ACKNOWLEDGMENTS
We are very grateful to R. Losick, A. Grossman, C. Moran, P. Setlow, I. Smith, P. Zuber, and P. Youngman for providing B. subtilis strains and to W. Haldenwang for aiding in the construction of BZ410 and communicating results prior to publication. We also thank P. Setlow and A. Grossman for helpful discussions.
This research was supported by the Michigan Agricultural Experiment Station and by grant GM43585 from the National Institutes of Health.
REFERENCES
- 1.Baldus J M, Buckner C M, Moran C P. Evidence that the transcriptional activator Spo0A interacts with two sigma factors in Bacillus subtilis. Mol Microbiol. 1995;17:281–290. doi: 10.1111/j.1365-2958.1995.mmi_17020281.x. [DOI] [PubMed] [Google Scholar]
- 2.Baldus J M, Green B D, Youngman P, Moran C P. Phosphorylation of Bacillus subtilis transcription factor Spo0A stimulates transcription from the spoIIG promoter by enhancing binding to weak 0A boxes. J Bacteriol. 1994;176:296–306. doi: 10.1128/jb.176.2.296-306.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson A K, Haldenwang W G. Bacillus subtilis ςB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc Natl Acad Sci USA. 1993;90:2330–2334. doi: 10.1073/pnas.90.6.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bird T H, Grimsley J K, Hoch J A, Spiegelman G B. The Bacillus subtilis response regulator Spo0A stimulates transcription of the spoIIG operon through modification of RNA polymerase promoter complex. J Mol Biol. 1996;256:436–448. doi: 10.1006/jmbi.1996.0099. [DOI] [PubMed] [Google Scholar]
- 5.Burbulys D, Trach K A, Hoch J A. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 6.Chibazakura T, Kawamura F, Takahashi H. Differential regulation of spo0A transcription in Bacillus subtilis: glucose represses promoter switching at the initiation of sporulation. J Bacteriol. 1991;173:2625–2632. doi: 10.1128/jb.173.8.2625-2632.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cutting S, Driks A, Schmidt R, Kunkel B, Losick R. Forespore-specific transcription of a gene in the signal transduction pathway that governs pro-ςK processing in Bacillus subtilis. Genes Dev. 1991;5:456–466. doi: 10.1101/gad.5.3.456. [DOI] [PubMed] [Google Scholar]
- 8.Cutting S, Oke V, Driks A, Losick R, Lu S, Kroos L. A forespore checkpoint for mother-cell gene expression during development in Bacillus subtilis. Cell. 1990;62:239–250. doi: 10.1016/0092-8674(90)90362-i. [DOI] [PubMed] [Google Scholar]
- 9.Cutting S, Panzer S, Losick R. Regulatory studies on the promoter for a gene governing synthesis and assembly of the spore coat in Bacillus subtilis. J Mol Biol. 1989;207:393–404. doi: 10.1016/0022-2836(89)90262-3. [DOI] [PubMed] [Google Scholar]
- 10.Driks A, Losick R. Compartmentalized expression of a gene under the control of sporulation transcription factor ςE in Bacillus subtilis. Proc Natl Acad Sci USA. 1991;88:9934–9938. doi: 10.1073/pnas.88.22.9934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan L, Losick R. SpoIIAB is an anti-ς factor that binds to and inhibits transcription by regulatory protein ςF from Bacillus subtilis. Proc Natl Acad Sci USA. 1993;90:2325–2329. doi: 10.1073/pnas.90.6.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaur N K, Oppenheim J, Smith I. The Bacillus subtilis sin gene, a regulator of alternate developmental processes, codes for a DNA-binding protein. J Bacteriol. 1991;173:678–686. doi: 10.1128/jb.173.2.678-686.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grossman A. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu Rev Genet. 1995;29:477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]
- 14.Grossman A D, Lewis T, Levin N, DeVivo R. Suppressors of a spo0A missense mutation and their effects on sporulation in Bacillus subtilis. Biochimie. 1992;74:679–688. doi: 10.1016/0300-9084(92)90140-a. [DOI] [PubMed] [Google Scholar]
- 15.Guzman P, Westpheling J, Youngman P. Characterization of the promoter region of the Bacillus subtilis spoIIE operon. J Bacteriol. 1988;170:1598–1609. doi: 10.1128/jb.170.4.1598-1609.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halberg R, Kroos L. Fate of the SpoIIID switch protein during Bacillus subtilis sporulation depends on the mother-cell sigma factor, ςK. J Mol Biol. 1992;228:840–849. doi: 10.1016/0022-2836(92)90868-k. [DOI] [PubMed] [Google Scholar]
- 17.Halberg R, Kroos L. Sporulation regulatory protein SpoIIID from Bacillus subtilis activates and represses transcription by both mother-cell-specific forms of RNA polymerase. J Mol Biol. 1994;243:425–436. doi: 10.1006/jmbi.1994.1670. [DOI] [PubMed] [Google Scholar]
- 18.Harry E J, Pogliano K, Losick R. Use of immunofluorescence to visualize cell-specific gene expression during sporulation in Bacillus subtilis. J Bacteriol. 1995;177:3386–3393. doi: 10.1128/jb.177.12.3386-3393.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harwood C R, Cutting S M. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons; 1990. [Google Scholar]
- 20.Helmann J D, Chamberlin M J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 21.Hicks K A, Grossman A D. Altering the level and regulation of the major sigma subunit of RNA polymerase affects gene expression and development in Bacillus subtilis. Mol Microbiol. 1996;20:201–212. doi: 10.1111/j.1365-2958.1996.tb02501.x. [DOI] [PubMed] [Google Scholar]
- 22.Hoch J. The phosphorelay signal transduction pathway in the initiation of sporulation. In: Piggot P J, Moran C P, Youngman P, editors. Regulation of bacterial differentiation. Washington, D.C: American Society for Microbiology; 1994. pp. 41–60. [Google Scholar]
- 23.Ireton K, Grossman A. Interaction among mutations that cause altered timing of gene expression during sporulation in Bacillus subtilis. J Bacteriol. 1992;174:3185–3195. doi: 10.1128/jb.174.10.3185-3195.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ireton K, Grossman A D. Coupling between gene expression and DNA synthesis early during development in Bacillus subtilis. Proc Natl Acad Sci USA. 1992;89:8808–8812. doi: 10.1073/pnas.89.18.8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones C H, Moran C P. Mutant ς factor blocks transition between promoter binding and initiation of transcription. Proc Natl Acad Sci USA. 1992;89:1958–1962. doi: 10.1073/pnas.89.5.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ju J, Luo T, Haldenwang W. Forespore expression and processing of the SigE transcription factor in wild-type and mutant Bacillus subtilis. J Bacteriol. 1998;180:1673–1681. doi: 10.1128/jb.180.7.1673-1681.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ju, J., T. Mitchell, H. Peters, and W. G. Haldenwang. Sigma factor displacement from RNA polymerase during Bacillus subtilis sporulation. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 28.Kellner E M, Decatur A, Moran C P. Two-stage regulation of an anti-sigma factor determines developmental fate during bacterial endospore formation. Mol Microbiol. 1996;21:913–924. doi: 10.1046/j.1365-2958.1996.461408.x. [DOI] [PubMed] [Google Scholar]
- 29.Kenney T J, Moran C P. Organization and regulation of an operon that encodes a sporulation-essential sigma factor in Bacillus subtilis. J Bacteriol. 1987;169:3329–3339. doi: 10.1128/jb.169.7.3329-3339.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kenney T J, York K, Youngman P, Moran C P. Genetic evidence that RNA polymerase associated with ςA uses a sporulation-specific promoter in Bacillus subtilis. Proc Natl Acad Sci USA. 1989;86:9109–9113. doi: 10.1073/pnas.86.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirchman P A, DeGrazia H, Kellner E M, Moran C P. Forespore-specific disappearance of the sigma-factor antagonist SpoIIAB: implications for its role in determination of cell fate in Bacillus subtilis. Mol Microbiol. 1993;8:663–671. doi: 10.1111/j.1365-2958.1993.tb01610.x. [DOI] [PubMed] [Google Scholar]
- 32.Komeda Y. Transcriptional control of flagellar genes in Escherichia coli K-12. J Bacteriol. 1986;168:1315–1318. doi: 10.1128/jb.168.3.1315-1318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kroos L, Zhang B, Ichikawa H, Yu Y-T N. Control of ς factor activity during Bacillus subtilis sporulation. Mol Microbiol. 1999;31:1285–1294. doi: 10.1046/j.1365-2958.1999.01214.x. [DOI] [PubMed] [Google Scholar]
- 34.Kunkel B, Kroos L, Poth H, Youngman P, Losick R. Temporal and spatial control of the mother-cell regulatory gene spoIIID of Bacillus subtilis. Genes Dev. 1989;3:1735–1744. doi: 10.1101/gad.3.11.1735. [DOI] [PubMed] [Google Scholar]
- 35.Kunkel B, Losick R, Stragier P. The Bacillus subtilis gene for the developmental transcription factor ςK is generated by excision of a dispensable DNA element containing a sporulation recombinase gene. Genes Dev. 1990;4:525–535. doi: 10.1101/gad.4.4.525. [DOI] [PubMed] [Google Scholar]
- 36.Kunkel B, Sandman K, Panzer S, Youngman P, Losick R. The promoter for a sporulation gene in the spoIVC locus of Bacillus subtilis and its use in studies of temporal and spatial control of gene expression. J Bacteriol. 1988;170:3513–3522. doi: 10.1128/jb.170.8.3513-3522.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 38.LaBell T L, Trempy J E, Haldenwang W G. Sporulation-specific sigma factor, ς29, of Bacillus subtilis is synthesized from a precursor protein, P31. Proc Natl Acad Sci USA. 1987;84:1784–1788. doi: 10.1073/pnas.84.7.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LeDeaux J R, Grossman A D. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J Bacteriol. 1995;177:166–175. doi: 10.1128/jb.177.1.166-175.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis P, Wu L, Errington J. Establishment of prespore-specific gene expression in Bacillus subtilis: localization of SpoIIE phosphatase and initiation of compartment-specific proteolysis. J Bacteriol. 1998;180:3276–3284. doi: 10.1128/jb.180.13.3276-3284.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis P J, Magnin T, Errington J. Compartmentalized distribution of the proteins controlling the prespore-specific transcription factor ςF of Bacillus subtilis. Genes Cells. 1996;1:881–894. doi: 10.1046/j.1365-2443.1996.750275.x. [DOI] [PubMed] [Google Scholar]
- 42.Lonetto M, Gribskov M, Gross C A. The ς70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Losick R, Stragier P. Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature. 1992;355:601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- 44.Lu S, Halberg R, Kroos L. Processing of the mother-cell ς factor, ςK, may depend on events occurring in the forespore during Bacillus subtilis development. Proc Natl Acad Sci USA. 1990;87:9722–9726. doi: 10.1073/pnas.87.24.9722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu S, Kroos L. Overproducing the Bacillus subtilis mother-cell sigma factor precursor, pro-ςK, uncouples ςK-dependent gene expression from dependence on intercompartmental communication. J Bacteriol. 1994;176:3936–3943. doi: 10.1128/jb.176.13.3936-3943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mandic-Mulec I, Doukhan L, Smith I. The Bacillus subtilis SinR protein is a repressor of the key sporulation gene spo0A. J Bacteriol. 1995;177:4619–4627. doi: 10.1128/jb.177.16.4619-4627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mandic-Mulec I, Gaur N, Bai U, Smith I. Sin, a stage-specific repressor of cellular differentiation. J Bacteriol. 1992;174:3561–3569. doi: 10.1128/jb.174.11.3561-3569.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Margolis P, Driks A, Losick R. Establishment of cell type by compartmentalized activation of a transcription factor. Science. 1991;254:562–565. doi: 10.1126/science.1948031. [DOI] [PubMed] [Google Scholar]
- 49.Mason J M, Hackett R H, Setlow P. Regulation of expression of genes coding for small, acid-soluble proteins of Bacillus subtilis spores: studies using lacZ gene fusions. J Bacteriol. 1988;170:239–244. doi: 10.1128/jb.170.1.239-244.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Min K, Hilditch C M, Diederich B, Errington J, Yudkin M D. ςF, the first compartment-specific transcription factor of Bacillus subtilis, is regulated by an anti-ς factor that is also a protein kinase. Cell. 1993;74:735–742. doi: 10.1016/0092-8674(93)90520-z. [DOI] [PubMed] [Google Scholar]
- 51.Newton A, Ohta N, Ramakrishnan G, Mullin D, Raymond G. Genetic switching in the flagellar gene hierarchy of Caulobacter requires negative as well as positive regulation of transcription. Proc Natl Acad Sci USA. 1989;86:6651–6655. doi: 10.1073/pnas.86.17.6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Partridge S R, Foulger D, Errington J. The role of ςF in prespore-specific transcription in Bacillus subtilis. Mol Microbiol. 1991;5:757–767. doi: 10.1111/j.1365-2958.1991.tb00746.x. [DOI] [PubMed] [Google Scholar]
- 53.Perego M, Glaser P, Hoch J A. Aspartyl-phosphate phosphatases deactivate the response regulator components of the sporulation signal transduction system in Bacillus subtilis. Mol Microbiol. 1996;19:1151–1157. doi: 10.1111/j.1365-2958.1996.tb02460.x. [DOI] [PubMed] [Google Scholar]
- 54.Pogliano K, Hofmeister A, Losick R. Disappearance of the ςE transcription factor from the forespore and the SpoIIE phosphatase from the mother cell contributes to establishment of cell-specific gene expression during sporulation in Bacillus subtilis. J Bacteriol. 1997;179:3331–3341. doi: 10.1128/jb.179.10.3331-3341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Predich M, Nair G, Smith I. Bacillus subtilis early sporulation genes kinA, spo0F, and spo0A are transcribed by RNA polymerase containing ςH. J Bacteriol. 1992;174:2771–2778. doi: 10.1128/jb.174.9.2771-2778.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ricca E, Cutting S, Losick R. Characterization of bofA, a gene involved in intercompartmental regulation of pro-ςK processing during sporulation in Bacillus subtilis. J Bacteriol. 1992;174:3177–3184. doi: 10.1128/jb.174.10.3177-3184.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Satola S W, Baldus J M, Moran C P. Binding of Spo0A stimulates spoIIG promoter activity in Bacillus subtilis. J Bacteriol. 1992;174:1448–1453. doi: 10.1128/jb.174.5.1448-1453.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharrock R A, Rubenstein S, Chan M, Leighton T. Intergenic suppression of spo0 phenotypes by the Bacillus subtilis mutation rvtA. Mol Gen Genet. 1984;194:260–264. doi: 10.1007/BF00383525. [DOI] [PubMed] [Google Scholar]
- 59.Siranosian K J, Ireton K, Grossman A D. Alanine dehydrogenase (ald) is required for normal sporulation in Bacillus subtilis. J Bacteriol. 1993;175:6789–6796. doi: 10.1128/jb.175.21.6789-6796.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steinmetz M, Richter R. Plasmids designed to alter the antibiotic resistance expression by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene. 1994;142:79–83. doi: 10.1016/0378-1119(94)90358-1. [DOI] [PubMed] [Google Scholar]
- 61.Stragier P, Kunkel B, Kroos L, Losick R. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science. 1989;243:507–512. doi: 10.1126/science.2536191. [DOI] [PubMed] [Google Scholar]
- 62.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 63.Strauch M A, Trach K A, Day J, Hoch J A. Spo0A activates and represses its own synthesis by binding at its dual promoters. Biochimie. 1992;74:619–626. doi: 10.1016/0300-9084(92)90133-y. [DOI] [PubMed] [Google Scholar]
- 64.Sun D, Cabrera-Martinez R M, Setlow P. Control of transcription of the Bacillus subtilis spoIIIG gene, which codes for the forespore- specific transcription factor ςG. J Bacteriol. 1991;173:2977–2984. doi: 10.1128/jb.173.9.2977-2984.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang L, Grau R, Perego M, Hoch J A. A novel histidine kinase inhibitor regulating development in Bacillus subtilis. Genes Dev. 1997;11:2569–2579. doi: 10.1101/gad.11.19.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu J-J, Piggot P J, Tatti K M, Moran C P. Transcription of the Bacillus subtilis spoIIA operon. Gene. 1991;101:113–116. doi: 10.1016/0378-1119(91)90231-y. [DOI] [PubMed] [Google Scholar]
- 67.Wu J-J, Schuch R, Piggot P. Characterization of a Bacillus subtilis sporulation operon that includes genes for an RNA polymerase ς factor and for a putative dd-carboxypeptidase. J Bacteriol. 1992;174:4885–4892. doi: 10.1128/jb.174.15.4885-4892.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu H, Dingwall A, Shapiro L. Negative transcriptional regulation in the Caulobacter flagellar hierarchy. Proc Natl Acad Sci USA. 1989;86:6656–6660. doi: 10.1073/pnas.86.17.6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.York K, Kenny T J, Satola S, Moran C P. Spo0A controls the ςA-dependent activation of Bacillus subtilis sporulation-specific transcription unit spoIIE. J Bacteriol. 1992;174:2648–2658. doi: 10.1128/jb.174.8.2648-2658.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Youngman P, Perkins J B, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- 71.Zhang B, Daniel R, Errington J, Kroos L. Bacillus subtilis SpoIIID protein binds to two sites in the spoVD promoter and represses transcription by ςE RNA polymerase. J Bacteriol. 1997;179:972–975. doi: 10.1128/jb.179.3.972-975.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang B, Hofmeister A, Kroos L. The prosequence of pro-ςK promotes membrane association and inhibits RNA polymerase core binding. J Bacteriol. 1998;180:2434–2441. doi: 10.1128/jb.180.9.2434-2441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang B, Kroos L. A feedback loop regulates the switch from one sigma factor to the next in the cascade controlling Bacillus subtilis mother cell gene expression. J Bacteriol. 1997;179:6138–6144. doi: 10.1128/jb.179.19.6138-6144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zuber P, Losick R. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J Bacteriol. 1987;169:2223–2230. doi: 10.1128/jb.169.5.2223-2230.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]