Abstract
Amyloid beta (Aβ) peptides are a major contributor to Alzheimers disease. They occur in differing lengths, each of which forms a multitude of assembly types. The most toxic soluble oligomers are formed by Aβ42; some of which have antiparallel β-sheets. Previously, our group proposed molecular models of Aβ42 hexamers in which the C-terminus third of the peptide (S3) forms an antiparallel 6-stranded β-barrel that is surrounded by an antiparallel barrel formed by the more polar N-terminus (S1) and middle (S2) portions. These hexamers were proposed to act as seeds from which dodecamers, octadecamers, both smooth and beaded annular protofibrils, and transmembrane channels form. Since then, numerous aspects of our models have been supported by experimental findings. Recently, NMR-based structures have been proposed for Aβ42 tetramers and octamers, and NMR studies have been reported for oligomers composed of ~ 32 monomers. Here we propose a range of concentric β-barrel models and compare their dimensions to image-averaged electron micrographs of both beaded annular protofibrils (bAPFs) and smooth annular protofibrils (sAPFs) of Aβ42. The smaller oligomers have 6, 8, 12, 16, and 18 monomers. These beads string together to form necklace-like bAPFs. These bAPRs gradually morph into sAPFs in which a S3 β-barrel is shielded on one or both sides by β-barrels formed from S1 and S2 segments.
Keywords: Amyloids, Concentric β-barrels, Amyloid beta 42, oligomers, annular protofibrils, structural models
Introduction
The quest to determine precise molecular mechanisms by which amyloid beta (Aβ) peptides wreak havoc on the human brain can appear hopeless. These peptides are shapeshifters; they assume countless forms that are often present simultaneously and likely have partially occupied secondary structures (see Deleanu et al.1 and Urban et al.2 for recent reviews). Numerous factors affect Aβ assemblies; e.g., ions and heavy cations, lipids, concentration, time, method of isolation and preparation, location in the body, initial seed conformation, length of the peptide, and oxidation. Also, they appear to interact with up to twenty receptors3, and with several other types of amyloid-forming peptides.
It remains unclear which of their many guises and interactions are the culprits. Much attention has focused on the most visible and stable forms: fibrils within the amyloid plaques that are the hallmark of Alzheimer’s Disease (AD). Their stability has allowed the molecular structure of some forms to be determined experimentally. However, fibrils come in multiple forms: some have U-shaped monomers4–7; others have S-shaped monomers5,8, some have twofold symmetry along their long axis and others have three-fold symmetry5,8; and most have inregister parallel β-sheets, but one has an elongated plus a β-hairpin conformation9, and at least one highly toxic mutant forms antiparallel β-sheets4–7,10. However, evidence is increasing that much smaller assemblies, called oligomers, are more detrimental (reviewed in11–13) and that the longer of the two major forms, Aβ42, is the most toxic12. A subset of the oligomer school contends that Aβ oligomers perturb neuronal signaling and eventually kill neurons by forming transmembrane ion channels13–20.
Previously we attempted to address these issues by developing atomically explicit models of the structures of Aβ42 hexamers, dodecamers, annular protofibrils, and an ion channel21,22. The starting point for our models was the hypotheses that Aβ42 hexamers can adopt a concentric antiparallel β-barrel structure with a hydrophobic core β-barrel, formed by the last third of the sequence (S3), surrounded by a more hydrophilic β-barrel formed by the first third (S1) and middle third (S2) of the sequence. In these hexamer models all monomers have well defined identical conformations and interactions with other monomers.
Aβ homologs comprise a large superfamily present in most vertebrates. Multiple studies indicate that they have a functional role23–25 including antimicrobial activity26. We aligned 250 homologous sequences from all types of vertebrates, mammals through bony fish (Supplement Fig. S1), to determine which residues are most essential.
S1, S2, and S3 segments have different properties. S1 (residues 1–14 DAEFRHDSGYEVHH) is the most hydrophilic and least well conserved segment. All hypervariable positions where five or more residue types and deletions occur are in S1 or in a putative loop region toward the end of S2. Only S1 positions Y10 and V12 are relatively conserved among distantly related homologs. In human Aβ S1 contains only four residues with hydrophobic side-chains (A2, F4, Y8 and V10); excluding the histidines, all charged side-chains (D1, E3, R5, and E9) occur at odd numbered positions. When modeled as a β-strand the hydrophobic side chains are on one side of the strand whereas charged side chains are on the other side. However, residues in its mid region (D7-S8-G9)27 have a high propensity for turn and coil conformations. S1 adopts a β conformation in some fibrils with a bend in this mid region. In some of our models S1 forms a β-hairpin with two β-strands, S1a and S1b. Its charged side-chains are exposed to water in all our models. Its importance in Aβ assemblies is uncertain. N-truncation of Aβ has been associated with some forms of AD (reviewed in Wirths et al, 201928) and some mutations within S1 are pathogenic while at least one (Icelandic mutant A2T) is protective (see 8 and 29 for reviews).
S2 is more hydrophobic and more highly conserved than S1. Its sequence resembles those of two homologous repeats that are highly conserved among α- and β-Synucleins.
QKLVFFAEDVGSN Aβ 17–27
QGVVAAAEKTKQG α-Syn 13–25
EGVLYVGSKTKEG α-Syn 35–47
These segments have been shown to adopt an amphipathic α-helix conformation in membranes and a β-strand conformation in fibrils; when helical hydrophobic or ambivalent residues are on the hydrophobic face of the helix, when β the charged K18 and E24 side-chains of Aβ are on the side of the strand that is exposed to water. Only one substitution occurs at K16, L17, V18, F19, and N27 in S2 in our alignment. Only three or four residue types occur at Q15, F20, A21, D23, and S26.The last three S2 residues have a high propensity for coil or turn structure and connect S2 to S3.
S3 is the most hydrophobic and most highly conserved segment. Its sequence is similar to that of the final hydrophobic segment of the α-Syn,
KGAIIGLMVGGVVIA Aβ 28–42
TVEGAGSIAAATGFV α-Syn 81–95
These segments form portions of hydrophobic β-barrels that are buried in the core of our soluble models and are in contact with lipid alkyl chains in our transmembrane models. Residues 29–42 of Aβ have no polar side-chain atoms. S3 residues are incredibly conserved among all vertebrates; the sequences are identical in all but three positions and even those substitutions are limited to large alkyl side chains: i.e., I32-V or L, V39-I, and V40-I (Fig. S1). The remarkable degree of conservation in S3 suggests that these proteins adopt a precise functional conformation or conformations in which at least some S3 segments are buried within the protein complex. Mutagenesis studies indicate that S3 residues I31, I32, L34, V39, V40 and I42 are key to Aβ oligomerization30.
Several aspects of our original hexamer model were unprecedented: (1) six-stranded antiparallel beta-barrels had never been reported. However; Laganowsky et al.31 have since found that a segment from an amyloid-forming protein, alpha B crystalline, indeed has the six-stranded antiparallel β-barrel motif (which they call Cylindrin), and Do et al.32 found that several eleven-residue peptides with the sequences of portions of S3 that includes methionine also form this motif. Their calculations confirm our findings that the presence of glycine facilitates packing of aliphatic side chains (especially methionine) in the interior of the barrel. The importance of these residues is supported by findings that mutation of Gly33 to Ala33 or oxidation of Met3534 reduces toxicity and alters oligomerization of Aβ42. (2) Our β structures were antiparallel whereas all known Aβ fibril β-structures were parallel. However, since then an Iowa mutant responsible for some forms of early onset AD has been shown to form fibrils with antiparallel β-sheets10. More important, recent experiments indicate that some Aβ42 oligomers do have an antiparallel β secondary structure that is similar to that of OMPA (an antiparallel β-barrel channel)35–37, and NMR studies of tetramers, octamers38, and 150 kDa oligomers39 indicate that adjacent S3 β-strands are antiparallel. Also, antiparallel oligomers are more toxic than those with parallel structures35,37. (3) Concentric β-barrels had never been observed when we proposed the structures. But recent studies have found that the channel-forming toxins Areolysin40 and Lysenin41 do, in fact, contain concentric β-barrels. (4) There was no experimental evidence supporting our proposal that the S1 segments form a β-strand or possibly a β-hairpin. However, subsequently two fibril structures with S-shaped monomers that include S1 have been solved (one with 3-fold symmetry5,8 and one with two-fold symmetry4). In both, the S1 and S2 segments comprise a parallel β-sheet with a bend near the center of S1. Banchelli et al.42 found that Cu++ causes formation of dimers by binding between His6 and His13 or His14 of two Aβ monomers. They concluded that at least portions of adjacent S1 segments are antiparallel (consistent with some of our models) rather than in-register parallel. Also, Tyr10 side chains can cross-link under oxidizing conditions to form dimers25, indicating that they are proximal in some oligomers as they are in some of our models. (5) There was no experimental evidence that Aβ42 forms β-barrels. However, Serra-Batiste et al.19 have recently discovered membrane-mimicking conditions under which Aβ42, but not Aβ40 peptides, form a well-defined β-barrel composed of only two monomeric conformations and Osterlund et al. 201943 found that in a similar environment some Aβ oligomers form β-barrel shaped hexamers. These results support our proposal that Aβ42 channels contain β-barrels resembling those we proposed for oligomers and annular protofibrils. (6) The hypothesis that numerous amyloid oligomers have β-barrel structures and evidence that various β-barrel proteins can form amyloid structures has gained popularity (see Sulatskaya et al 202144 for a recent review).
As pointed out in our paper on models of Synuclein assemblies45, our hypothetical models are likely over-structured. They do not adequately capture the dynamic nature of amyloid assemblies in which the number of monomers, how they interact, and even secondary structures appear to transition among numerous alternatives. The phrase ‘intrinsically disordered’ is probably appropriate for small Aβ oligomers in the aqueous phase where polar backbone atoms can hydrogen bond to water during initial stages of development. Numerous groups have performed molecular dynamic (MD) simulations of small Aβ assemblies in water with varying results (see Nguyen et al, 201546 for review). For example, Voelker et al. (2017)47 found that trimers through pentamers displayed coil and turn content (40–50%) with minor β-strand and α-helical content (<10%). These assemblies had water-filled pores, but they are much too small to correspond to the APFs analyzed here or with microscopy images of Aβ ion channels in membranes (Quist et al., 200548, Shafrir et al., 201021). In contrast, MD studies of Nguyen et al, (2019)49 found that Aβ42 in water tends to form β-barrel tetramers. Major limitations of MD simulations are their duration; typically lasting less than a millisecond, and difficulties in simulating large assemblies. Thus, they should not be expected to accurately simulate processes that can take days to develop and that often involve large assemblies.
With time the size of Aβ oligomers gradually increases and they become more ordered as a higher fraction of residues become buried. Their structural order likely increases even more when they are placed in less aqueous environments; e.g., in the presence of hexane, detergents, lipids, and/or air. The APFs analyzed here were developed from prefibril oligomers (PFOs) with the addition of 5% hexane followed by drying. The more symmetric APFs with consistent sizes and shapes are likely to be relatively ordered.
Methods
Experimental constraints used in developing the models presented are based primarily on results of the 2009 study of annular protofibrils by Kayed et al.50,51 Two types of Annular ProtoFibrils were reported: beaded APFs (bAPFs) that resemble necklaces formed by a string of beads, and smooth APFs (sAPF) that resemble smooth rings50. Portions of negatively-stained electron micrographs used in this study are shown in Fig. 1. Methods used to develop and analyze these assemblies were presented in their papers52. In short, the APFs of the EM images were formed form spherical Pre-Fibril Oligomers (PFOs) in the presence of 5% hexane with drying at an air interface. bAPFs formed initially from oligomers, then over a period of about seven days morphed into sAPFs. Circular dichroism studies indicated that the APFs have a high content of beta secondary structure. The precise beta content of any specific assembly is impossible to determine because the samples are highly heterogenous. APFs were less toxic and affected membrane conductances less than the PFOs. However, antibodies that bind to APFs, but not to PFOs or fibrils, did interact with lipid vesicles exposed to PFOs and at water-lipid and water-hexane interfaces, suggesting that APF-like structures do form in membranes. These antibodies also recognized synuclein and amylin APFs as well as α-hemolysin, a heptapeptide toxin that forms antiparallel β-barrel channels in membranes. These results support the hypothesis that these assemblies share a common structural feature and inspired our efforts to develop more precise models of these assemblies. Similarities among different amyloids have been reviewed recently (Nguyen et al 2021)53.
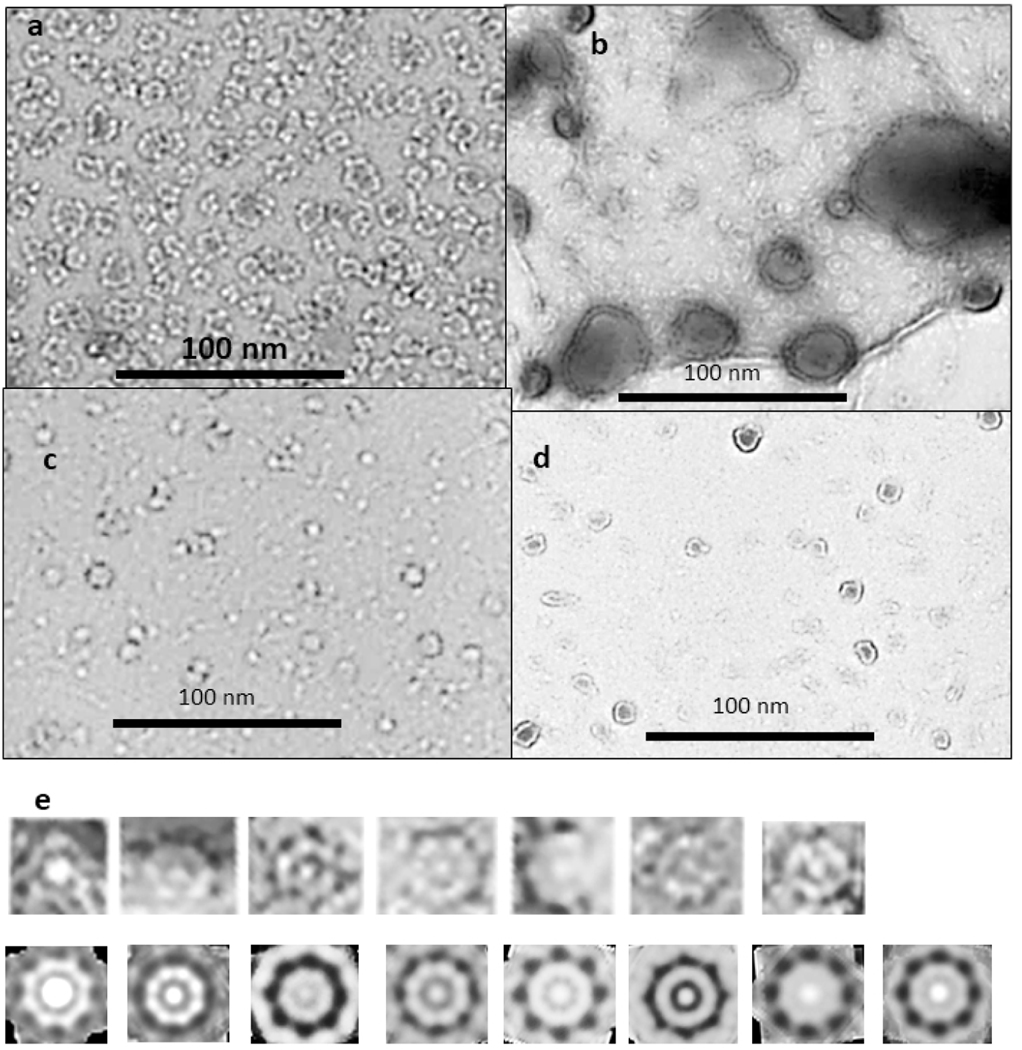
Figure 1.
Negative stained EM images of beaded (a) and smooth (b, c, & d) APFs (originals provided by Rakez Kayed). See50 for methods. (e) Example of how averaged images were developed. These images are from the micrograph shown in (b) for one size of WCsAPF that has 8-fold radial symmetry. Images of individual APFs were copied into Photoshop and intensity and contrast were adjusted to make their characteristics more visible (top row). Radial image averaging should be used cautiously because it can produce the appearance of radial symmetry even when none exists. Unaveraged images are shown to illustrate that some WCsAPFs, especially the larger ones, exhibit radial symmetry without radial averaging. Each of these was averaged with radial symmetry (bottom row). All radially averaged images of similar size and symmetry were aligned and averaged to obtain the final image for each specific sAPF (last image of the bottom row).
A major challenge in analyzing the EM images was to identify relatively ordered assemblies within the highly heterologous populations that included relatively disordered assemblies, assemblies composed of beads with differing sizes, and assemblies in transition between bAPFs and sAPFs. Photoshop54 was used to analyze APF images. Isolated bAPFs (Fig. 1a) were copied and grouped according to the number and size of beads. Radial image averaging was applied to relatively symmetric bAPFs in which all beads had the same approximate size. Multiple radially averaged images of the same size and shape were superimposed, aligned, and averaged to obtain the final image. A similar process was used to analyze sAPFs but with the additional constraint of placing the sAPFs into three categories: those with white centers and periodic dots on the perimeters (WCsAPF), those with dark centers and periodic dots (DCsAPF), and those with dark centers but with super smooth outer rings without periodic dots (SsAPF). The first two categories of images were grouped according to their diameters and then averaged radially with the symmetry based primarily on the number and spacing of the perimeter dots. Radially averaged images that were similar in size, dots, and shapes were then aligned to obtain images averaged over multiple sAPFs (Fig. 1e). When the symmetry was ambiguous numerous symmetries were tried. If a specific symmetry and diameter resembled those obtained from less ambiguous sAPFs the image was included in the multiple sAPF averaging process; otherwise, it was excluded.
In addition, we have incorporated results of recent solid-state NMR studies of Aβ oligomers38,39 into some of our latest models as described later.
Diameters of β-barrels comprising our models were calculated from the number of strands (N) and sheer number (S) (related to strand tilt) of the barrels (see Murzin et al.55 for early theory and Hayward and Milner-White56 for an extension and modification of this theory to radially symmetric β-barrels and β-helices). The latter analysis does not include the P2 2-fold perpendicular symmetry present in our models. These structures have axes of 2-fold symmetry perpendicular to the radial axis of symmetry and intersecting it at the assembly’s center of mass. This feature constrains symmetric models with concentric β-barrels because all barrels must have the same axes of symmetry. The gap distance between the walls of the adjacent barrels were constrained to be between 0.6 and 1.2 nm45, depending primarily on the side chain sizes. We favor models in which interacting pleats of adjacent β-barrels fit between each other in a manner that reduces clashes among side chains, and in which all pleats that intersect the axes of 2-fold symmetry have the same inward or outward side-chain orientation in all concentric β-barrels. We call this type of interaction “Intermeshing Pleats45.” We selected models that maximize hydrogen bonds, salt bridges, interactions among aromatic side chains, burial and tight packing of hydrophobic side chains that are highly conserved among Aβ homologs, and aqueous solvent exposure of hydrophilic side chains, especially for positions that are hypervariable among homologs. Residues with polar side chains and/or that have a high propensity for turn or coil secondary structure27 and that contain proline in some homologs were favored for connecting loops. The final constraints were experimental: the sizes, shapes, molecular weights, and secondary structures of assemblies as determined by EM images of annular protofibrils and other studies, and distance constraints based on NMR studies of tetramers, octamers, and 32mers.
Atomic-scale structures were generated with an in-house program and illustrated using the Chimera program57,58.
Sequences of 250 Aβ homologs were collected and aligned using the Blast program59.
Results and Discussion
Oligomers and beaded APFs
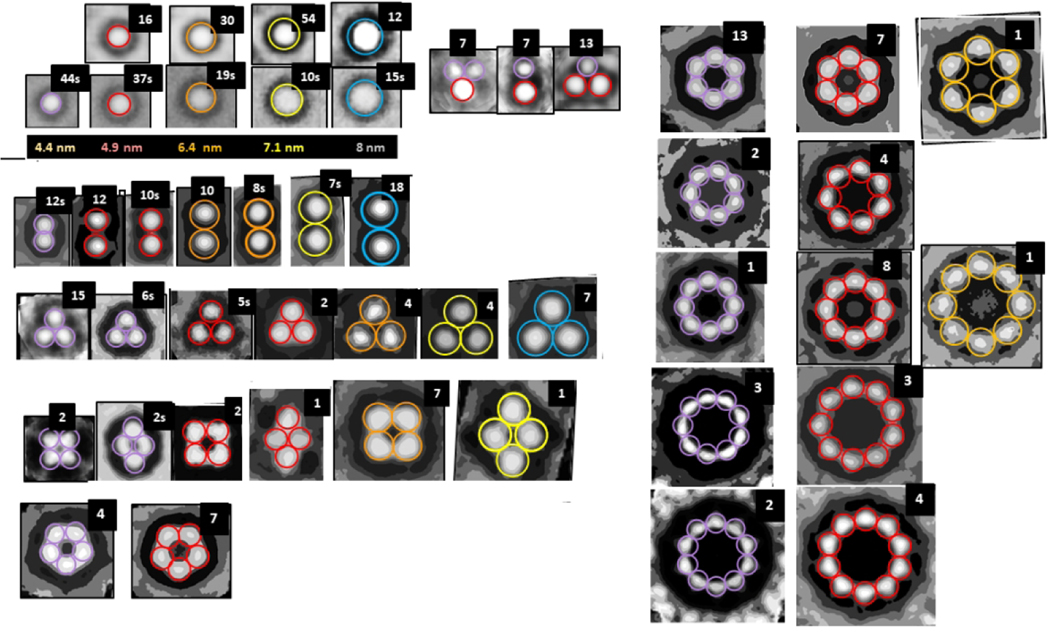
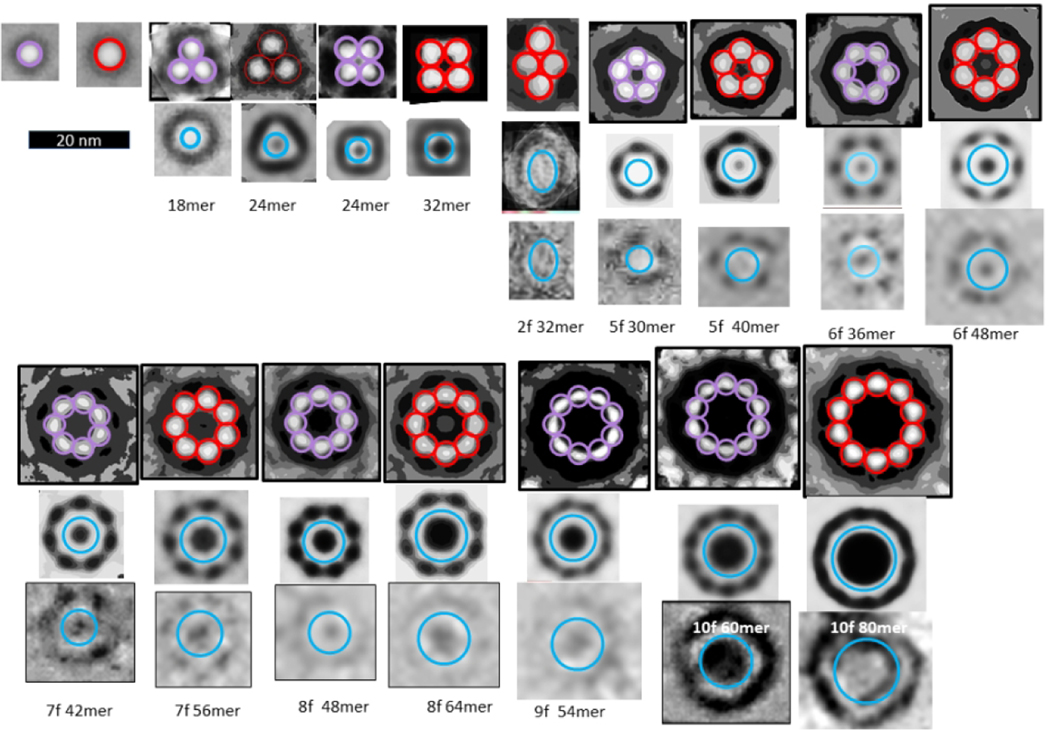
The first method of evaluating the sizes of the beads was to copy individual images of isolated beads in both bAPF and sAPF micrographs, classify them according to their sizes, superimpose the images in each category, and then use image averaging to obtain a single image for each size (first two rows of Fig. 2). We identified five sizes of beads. The major difficulty with this method is that the outer perimeter is not clear cut.
Figure 2.
Averaged images of isolated and symmetric assemblies of beads (oligomers). The colored circles have the diameters indicated below the second row. The number of images used to develop the averaged images is in the upper right corner; when followed by a “S” the images were made from one of the sAPF micrographs. There appears to be five sizes of beads with diameters ranging from about 4 to 8 nm. The first two rows on the left-half show isolated beads from both types of micrographs followed by three examples of small clusters composed of the two smallest-size beads. The remaining rows shows symmetric assemblies composed of 2 to 10 beads. Beads within each illustrated bAPF all have the same size, but the bead size varies among bAPFs that have the same number of beads. The two smallest sizes occur most often for bAPFs with five or more beads. The “cutout” facility of Photoshop was applied to most images to help identify centers of the beads.
The second method was to approximate distances between beads of bAPFs. The advantage of this approach is that the diameter of the beads can be approximated from the distance between the centers of adjacent beads, which can be measured more precisely than the location of the outer edges of isolated beads. The remaining rows in Fig. 2 are averaged images of assemblies of 2–10 beads.
Models of oligomers that correspond to “beads”.
The next challenge was to generate structural models consistent with these bead sizes. The following features are common to small non-APF oligomer (bead) models presented below.
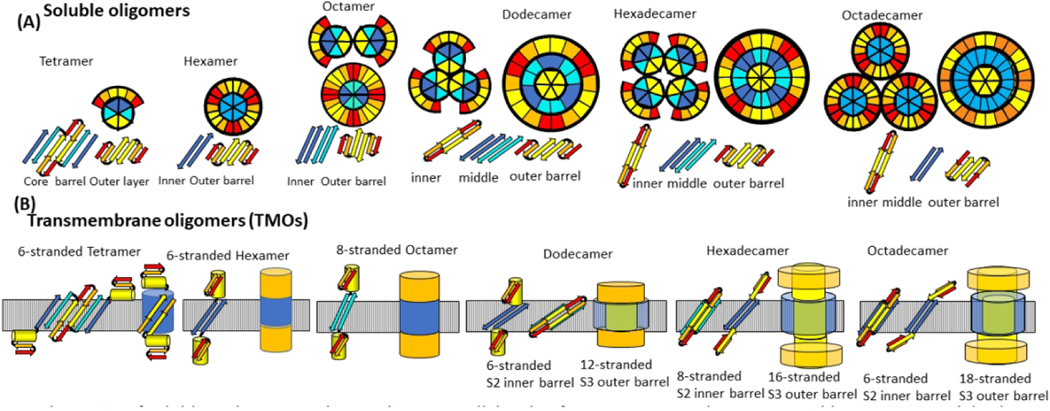
The number of monomers, M, of an oligomer is an integer multiple of six or eight. Models presented here are only for assemblies of six or more monomers. The selections of hexamers, dodecamers, and octadecamers are based on results of cross-linking studies60. Octamers are based on solid-state NMR studies38.
All models have two or three concentric β-barrels. Six or eight antiparallel S3 β-strands comprise the innermost β-barrel.
All have 2-fold perpendicular symmetry and 3-fold or 4-fold radial symmetry.
All adjacent β strands are antiparallel.
Except for the octadecamer, all putative β-strands are components of β-barrels. S1 can be either a S1a-S1b β-hairpin or a single β-strand.
S1a-S1b β-hairpins and S1 β-strands are on the surface; hydrophilic sides of the strands are exposed to water while hydrophobic side chains on the opposite side interact with S3 or S2.
The vast majority of side chains buried in the interior of the assembly are hydrophobic, the few buried polar side chain atoms from the outer strands can form H-bonds with other groups.
Electrostatic interactions are also favorable; almost all charged groups form salt-bridges and almost all polar backbone atoms form H-bonds.
Our concentric β-barrel models described below of dodecamer, hexadecamer, and octadecamer are not unique; some alternatives with differing details are described in the supplement and in the conclusions. The resulting structure of larger oligomers may depend on the process by which they develop. We considered two possibilities: In the first pathway positions of the hexamer S2 and S3 segments are unaltered as additional monomers bind to its outer surface. In the second pathway tetramers merge to form larger assemblies in which S3 strands form a single β-barrel that is sandwiched between an inner barrel formed by S2 strands and an outer barrel formed by S1a-S1b-S2 β-sheets (Fig. S2 of the supplement). We emphasize the first pathway in most of this text because soluble proteins usually have well-conserved hydrophobic cores. However, oligomers formed by the second pathway may be better suited for inserting through membranes (see conclusions). Resolutions of EM images and molecular modeling methods are not sufficient to determine which alternatives are better. However, all are consistent with the hypothesis of concentric β-barrel assemblies.
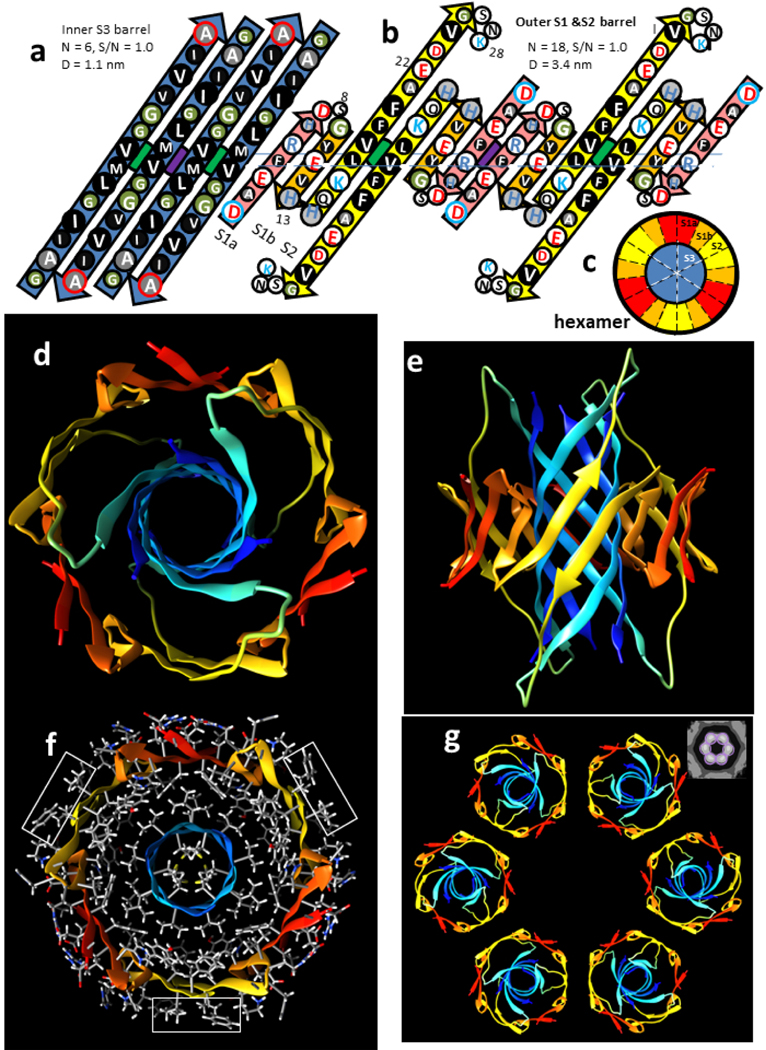
The smallest β-barrel oligomer modeled here is a hexamer (Fig. 3). S3 strands form a hydrophobic core as a six-stranded antiparallel β-barrel. Although six stranded β-barrels are rare, this one is made possible by the absence of side chains at the inward-pleated G33 and G37 residue backbones, and the flexibility of the M35 side chains that fit next to these glycines. The S3 core of the hexamer is surrounded by an 18-stranded antiparallel β-barrel formed by S1a, S1b, and S2 strands. The illustrated hexamer has an outer diameter of ~4.4 nm (Fig. 3a) assuming an outer diameter 1.0 nm greater than the diameter of the outer β-barrel’s wall. Side chains at the axes of 2-fold symmetry intermesh; i.e., V18 of S2 and V36 of S3 (green circle) are both oriented outwardly, and F4 of S1a and M35 of S3 (purple circle) are both oriented inwardly.
Figure 3.
Hexamer concentric β-barrel model. (a & b) Flattened representation of four subunits viewed from the exterior as if the barrel were split open on the back side and spread flat. The axes of 2-fold perpendicular symmetry are indicated by green and purple circles behind the β-sheets, and the plane containing these axes by the horizontal line. Parameters of the barrels are listed. The arrows represent β-strands colored by segment. The circles represent side chains; those oriented outwardly are larger than those oriented inwardly. They are colored by residue type: white = positively charged (blue letter), negatively charged (red letter), or uncharged polar (black letter), green = ambivalent Gly, light gray = ambivalent His (blue letter) or Thr (black letter), dark gray - white letter = slightly hydrophobic Ala, black - white letter = hydrophobic. The N- and C-termini residues have blue and red outlines to indicate their positive and negative charges. (a) Inner S3 β-barrel. (b) Outer β-barrel; S1a (pink), S1b (orange), and S2 (yellow). (c) Wedge representation cross-section on the plane containing axes of 2-fold symmetry illustrating relative positions of the β-strands. (d-g) Atomic-scale model. The backbone is illustrated as a rainbow-colored ribbon from red (N-termini) to blue (C-termini). (d) View down the radial axis of the β-barrels. (e) Side view along the 2-fold axis between the yellow S2 strands. (f) Radial cross-section through the central portion with side chains colored by element; hydrogens and carbons are white and gray, polar oxygen and nitrogen atoms are red and blue. Rectangles enclose exposed V18 and F20 side chains. (g) Ribbon representations of six hexamers forming a beaded APF; the insert in the upper right corner is an averaged EM image from Fig. 2.
The exterior surface of the hexamer model has three hydrophobic patches formed by V18 and F20 (rectangles of Fig. 3f) flanked by positively charged K18 and negatively charged E22. These are the most probable interaction regions between APFs in an aqueous solution due to a combination of hydrophobic and electrostatic interactions. Such interactions should be geometrically optimal when the number of beads is six due to the 2-fold symmetry of the interaction sites (Fig. 3g). In the beaded APF micrograph analyzed here, six-membered APFs dominated the smallest bead APFs; 13 six-membered rings were observed whereas the average number of other small bead bAPFs was only about two (Fig. 2).
Experimental studies indicate that Aβ42 can form dodecamers61,62 and octadecamers63,64, and that these sizes of oligomers can be isolated from brains of Alzheimer’s patients. Some of these may form when additional monomers bind to hexamers. If so, the S3 strands of the additional Aβ monomers will likely bind in a hydrophobic environment. Site-Directed spin-labeling studies indicate that mobility decreases and rigidity increases within Aβ oligomers from the N-terminus to the C-terminus, and that S3 segments are tightly packed65. We hypothesize that relatively dynamic S1 segments can be displaced as S2 and/or S3 segments of six additional monomers bind next to S3s and between S2s of the original hexamer (Figs. 4 & 5). The displaced S1a-S1b β-hairpins of the original hexamer may combine with S1a-S1b β-hairpins of the additional monomers to form an outer 24-stranded antiparallel β-barrel with approximate 6-fold radial symmetry (Fig 4), but the outermost barrel or layer is likely to be highly dynamic and difficult to predict precisely.
Figure 4.

Dodecamer models. Parameters of the barrel are listed below the octadecamer schematics. The color code is the same as in the previous figures. The structures of the core S2 and S3 strands of the hexamer are maintained; the core S1a-S1b hairpins are displaced outwardly to the third barrel. S2 and S3 strands of six additional subunits are added to the middle barrel between the core S2 strands. (a) Wedge representations of the central cross-section; the six additional peripheral subunits are stippled. (b-d) Flattened schematics of four subunits of (b) the inner 6-stranded S3 β-barrel (same as hexamer), (c) the middle 18-stranded S2 and S3 β-barrel, and (d) the outer 24-stranded S1a-S1b β-barrel. Strands of the six additional subunits have shaded backgrounds. (e-h) Atomic-scale models of the dodecamer. (e & f) Rainbow colored ribbon illustration of the backbone as viewed from the top and side. (g & h) Cross-sections with side chains colored by element viewed along the barrels’ axis and from the side. Note that most buried side chains are tightly packed and hydrophobic (gray carbons, white hydrogens, and yellow sulfurs), and that most surface side chains have polar red oxygen or blue nitrogen atoms with white hydrogens.
Figure 5.
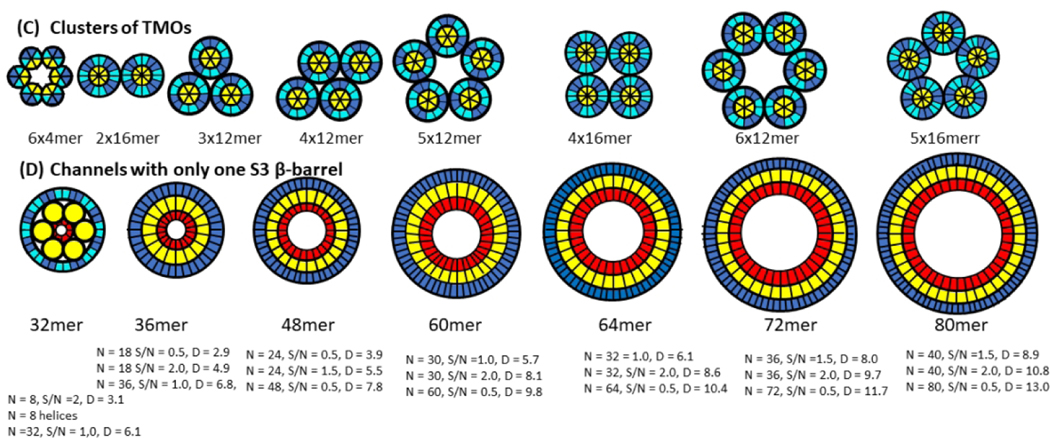
Octadecamer model. (a) Wedge representation of strand positions in the central cross-section; the twelve peripheral subunits are stippled. (b-e) Flattened representation of (b) the core 6-stranded S3 β-barrel, (c) the middle 18-stranded S2-S3 β-barrel; the poorly conserved S1a interacts with the poorly conserved portion near the end of S2 and its structure is likely more disordered than depicted, (d) the outer 24-stranded S1-S2 β-barrel, and (e) two peripheral S1a-S1b β-hairpins. Residues are colored according to the number of residue types at each position of a multi-sequence alignment. (f-i) Atomic-scale models. (f) Top view and (g) side view cross-section of backbone ribbon representation colored by rainbow. (h & i) Model with side chains colored by degree of conservation as in b-e; (h) middle cross-section, (i) side view cross-section. Schematics of alternative octadecamer models are in Supplement Fig. S3.
The octadecamer model of Fig. 5 was developed in part based on the hypothesis that all highly conserved hydrophobic residues form a core structure that is buried in this model. S1a is peripheral as are outwardly oriented side chains of S1b. Side chain packing between barrels is quite tight with few large cavities; all conserved hydrophobic side chains (blue and cyan) and most semi-conserved side chains (yellow and green) are buried, the hypervariable side chain positions (red) are on or near the outer surfaces. Backbone H-bonding is extensive, and almost all charged groups form salt bridges.
We suspect that the second largest oligomer (bead) is an octamer. We used atomic-scale modeling to explore the feasibility of octamer structures with concentric β-barrels. After developing numerous models, we concluded that the structure illustrated in Fig. 6 is the soundest that is also consistent with the size of the next to smallest beads, those circled in red in Fig. 2. Schematics of some alternative models are given in supplement Fig. S4.
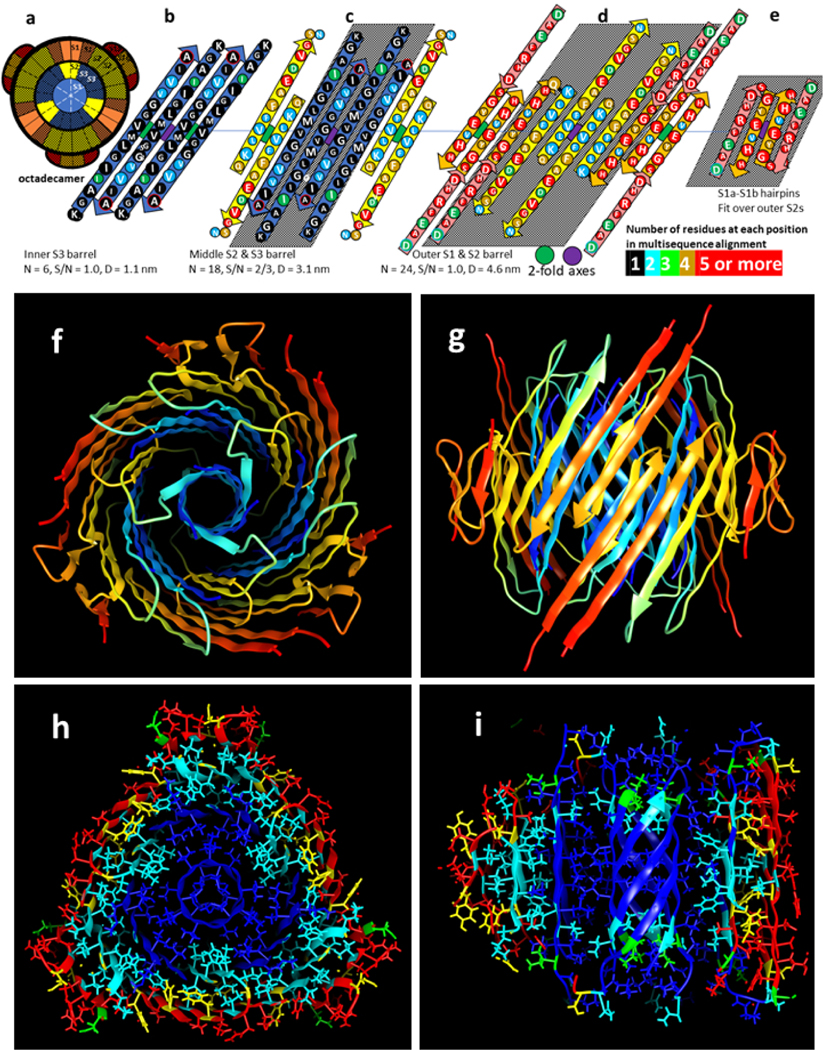
Figure 6.
Model of an Aβ42 octamer. Features and colors are like those of Fig. 3. (a) Flattened schematic of four subunits of the outer and inner β-barrels and parameters of the β-barrels. (b & c) Ribbon representation of the octamer backbone as viewed from the side through a perpendicular 2-fold axis between yellow S2 strands and from the top. (d-f) Cross-sections with side chains colored by element of the top region, the next region down, and the middle region.
Smooth Annular Protofibrils.
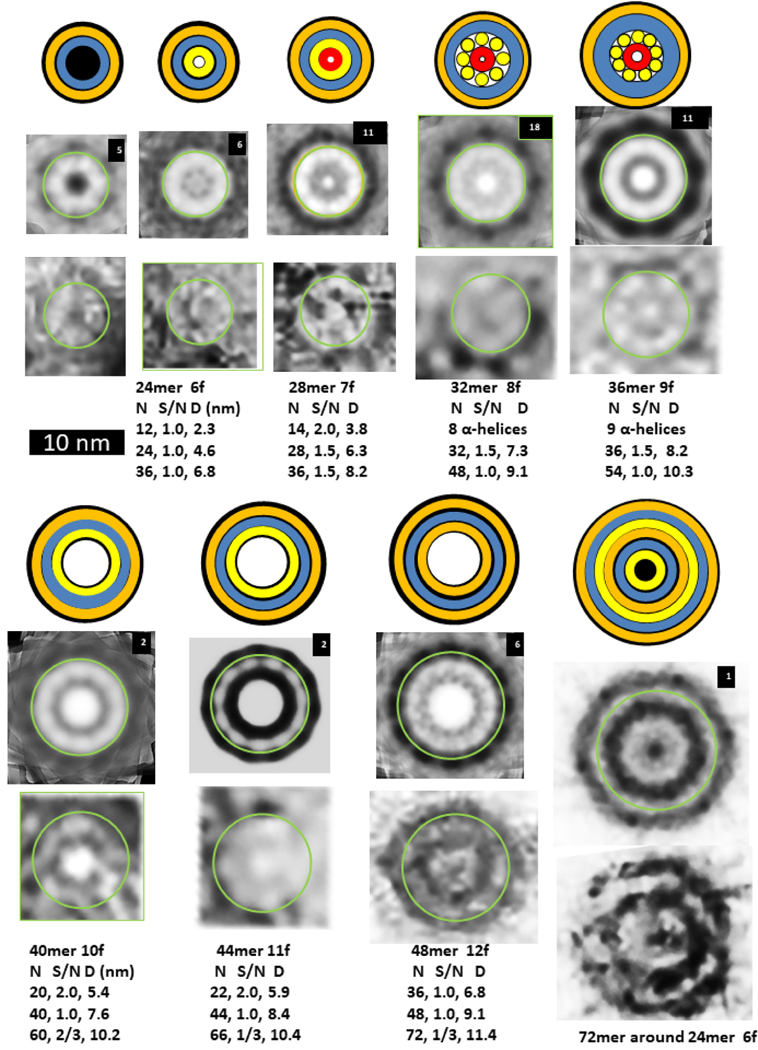
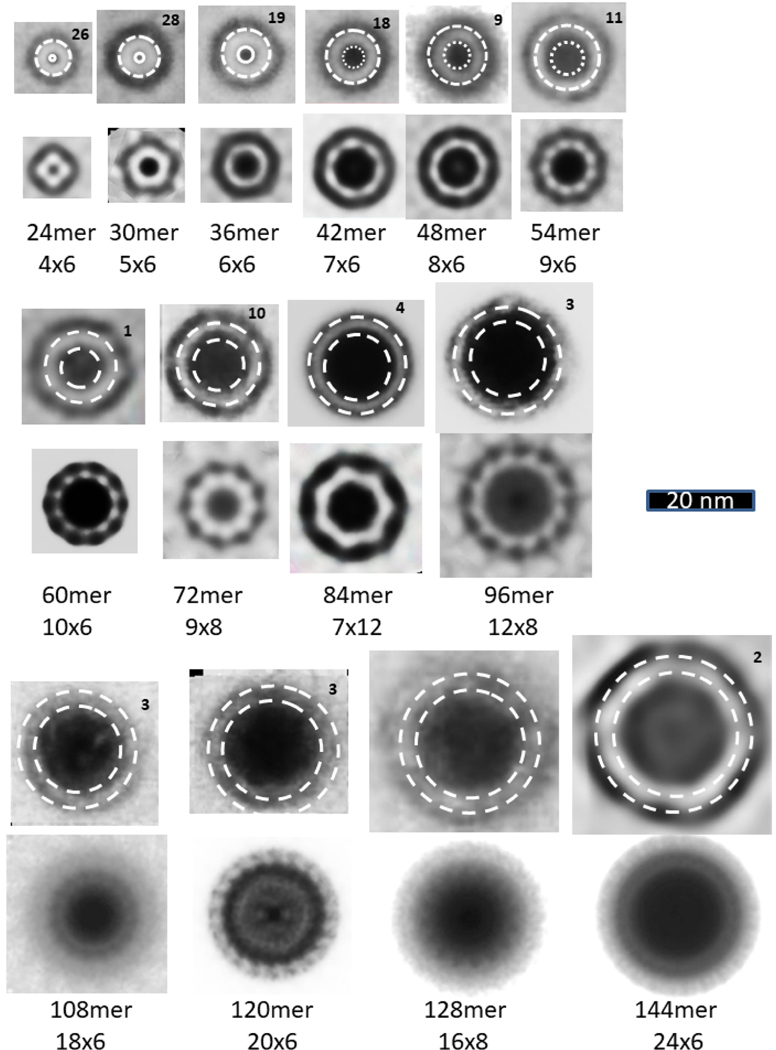
Beaded APFs gradually morph into smooth APFs. Most sAPFs in the upper portion of Fig. 1b exhibit concentric rings: an outer dark ring, followed by a light ring, followed by a second dark ring which surrounds a circular light center. These sAPFs resemble cog gears or cog wheels due to evenly spaced dark dots on the perimeter of the outer ring; thus, we call these dots cogs. White-centered cog APFs (WCsAPFs) are densely packed in the top region of the micrograph of Fig. 1b, which appears to be enclosed or covered by a film. The concentric rings of averaged WCsAPFs EM images provide the strongest pictorial evidence that Aβ42 peptides can assemble into concentric β-barrel structures (Fig. 7). WCsAPFs can be classified into seven sizes, We hypothesize that the outer white ring corresponds to a middle S3 β-barrel, that the number of strands in this barrel is the same as the number of monomers in the assembly, and that the S/N value is either 1.0 or 1.5. These assumptions permit the number of monomers in the assembly to be calculated from the diameter of the white ring (see parameters below the images). Our calculations suggest that the number of monomers increase by four for each size increase. Also, the number of cogs in the outer dark ring increases by one each time the number of monomers increases by four. Thus, these sAPFs may develop and grow from mergers of tetramers. If the tetramer structure of Aβ42 proposed by Cuidad et al.38 for S3 strands is approximated in the S3 barrel of the white-centered sAPFs, then the radial unit-cell will be composed of four monomers with two subunit conformations.
Figure 7.
Images of WCsAPFs and schematics of Aβ42 models with three concentric β-barrels. The outer orange rings of the schematics represent the outer β-barrel formed by S1 and S2, the blue ring represents a S3 β-barrel, the inner orange ring represents an inner β-barrel formed by S1 and S2, the inner yellow ring represents a β-barrel formed only by S2 strands, and the tiny red ring represents a highly tentative β-barrel formed by S1a segments. The yellow cylinders of the 32mer and 36mer models represent α-helices formed by S1b-S2 segments. Parameters of the models are listed below the images. The upper rows of images were averaged radially for the number of WCsAPF’s indicated in the upper right corner. The lower row of images are individual sAPFs. The green circles on the images indicate the proposed size and location of boundary between the outer and middle β-barrels. A small DCsAPF (the first image) was included because it also occurs frequently within the same region of the micrograph. The final schematic/EM image is larger and has more concentric rings than the others. The schematic illustrates that the assembly may have six concentric β-barrels with a three-barrel 72mer sAPF surrounding a three-barrel 24mer sAPF (also see Supplement Fig. S5).
The final image and schematic of Fig. 7 illustrates the most extreme example of multiple concentric β-barrels. The unaveraged image has a relatively hexagonal shape. Averaging with 6-fold symmetry reveals multiple concentric rings. Fig. S5 of the supplement compares images of putative 24mers and a 72mer to this image and illustrates details of a model in which a 72mer sAPF surrounds a 24mer sAPF. The central rings of the image resemble the first of the WCsAPF images, the putative 24mers. The outer diameter of the image is consistent with a 72mer sAPF with S/N = 0.5 for the S3 barrel. The model has six concentric β-barrels, but this type of assembly is rare; this is the only image we observed with these properties.
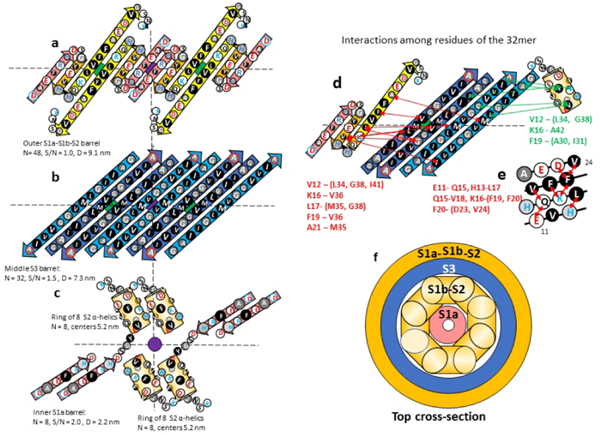
Like the tetramers proposed by Cuidad et al.38, our WCsAPFs models have two monomeric conformations and 2-fold perpendicular symmetry. An antiparallel S3 β-barrel is sandwiched between two cylinders formed by S1-S2 segments. In our models S1a-S1b-S2 β-strands of half the monomers form an outer β-barrel with a structural motif resembling that proposed above for hexamers (Fig. 8). S2 of these monomers connects to the S3 strands for which centrally located V36 side chains are oriented outwardly. We call the conformation of these monomers Cout and the conformation of the other half of the monomers Cin.
Figure 8.
Model of the 32mer WCsAPF. (a-c) Flattened representation of β-strands for four subunits. The green and purple circles behind the strands indicate the axes of 2-fold perpendicular symmetry and the horizontal dashed line indicates the plane containing those axes. Parameters of the β-barrels are indicated beside or below the schematics. (a) The 3-stranded S1a-S1b-S2 β-sheets that comprise the outer β-barrel. (b) The S3 β-strands of the middle β-barrel. Cout S3 strands are darker blue. (c) The S1b-S2 α-helices and S1b β-strands of two TIM barrel-like structures of Cin subunits. (d) Interactions indicated by NMR studies of Gao et al.39 between residues of S1b-S2 and S3. Red lines and residue list on the left side indicate possible interactions between S1b-S2 residues and outwardly-oriented S3 side chains; green lines and residue list on the right indicate possible interactions between the Cin S1b-S2 α-helix residues and inwardly-oriented S3 side chains. (e) A helical net representation of the S1b-S2 α-helix showing observed interactions among residues that are sequentially separated by three or four positions. (f) Schematic cross-section of the upper portion of the 32-mer model.
Gao et al.39 developed a method to stabilize and isolate 150-kDa Aβ42 oligomers, which they conclude are composed of 32 subunits. These oligomers may correspond to the 32mer WCsAPF of Fig. 7. Their solid-state NMR studies indicate that S3 forms antiparallel β strands centered at V36, consistent with our model and the structure of the central S3 strands of the tetramer. Their results for S2 strands are more complicated. Based on their analysis of chemical-shifts for backbone C sites, they concluded that residues 11–24 (S1b-S2) have a β secondary structure, but that these residues occupy multiple magnetically inequivalent sites. Additional NMR results indicated that proximal residues are sequentially either three or four positions apart on S1b-S2. They interpreted these data as indicative of out-of-register parallel β-sheets in which the strands are shifted three positions relative to one another. This arrangement is not possible if S2 segments form antiparallel β-barrels.
An alternative possibility is that the assembly contains two distinct S2 conformations, one that has β-secondary structure and another that has α-helical secondary structure in which residues three or four positions apart are proximal (Fig. 8e). If so, then some results may come from the β-structure of Cout while other results come from an α-conformation of Cin. We propose that S1b-S2 α-helices of Cin monomers comprise an inner ring of the 32mer sAPFs. These helices are substantially shorter than the β-strands and stack end-on inside the S3 barrel. S1a segments of Cin may form an inner parallel β-barrel. If so, the portion of the assembly inside the S3 β-barrel would resemble a TIM αβ-barrel in which eight parallel α-helices surround an eight stranded parallel β-barrel. This motif has been observed in numerous soluble proteins66 and could contribute to the stability of 150-kDa oligomers. A high stability of this structure could also explain why the putative 32mers are the most frequently observed WCsAPFs (Fig. 7).
Additional NMR results of Gao et al.39 regarding interactions between S1b-S2 residues and S3 residues are consistent with our model if one assumes some data are due to interactions with Cout and others with interactions with Cin (Fig. 8). Note that the three residues (V12, K16, and F19) proposed to interact with S3 residues near the beginning and ends of S3 strands are on the same face of the putative α-helix.
An atomic-scale model of a 32mer sAPF is shown in Fig. 9. Side chains of the S1b-S2 helix make direct contact with side chains of S3 β-strands indicated in green in Fig. 8. Some side chains of the S1b and S2 β-strands predicted by NRM to interact did not make direct contact, but distances between the closest side chain atoms were relatively small. One interaction proposed by the NMR study, between K15 and M35, was not satisfied. The EM images of the putative 32mer in Fig. 7 closely corresponds to the size and shape of the cross-section pictures of Fig. 9e and f (also see Supplement Fig. S6). The atomic-scale model is also consistent with our modeling criteria: almost all conserved hydrophobic side chains are buried and pack tightly in at least one of the two conformations, almost all charged groups form salt-bridges, and almost all backbone polar atoms form H-bonds due to the high content of β and α-secondary structure.
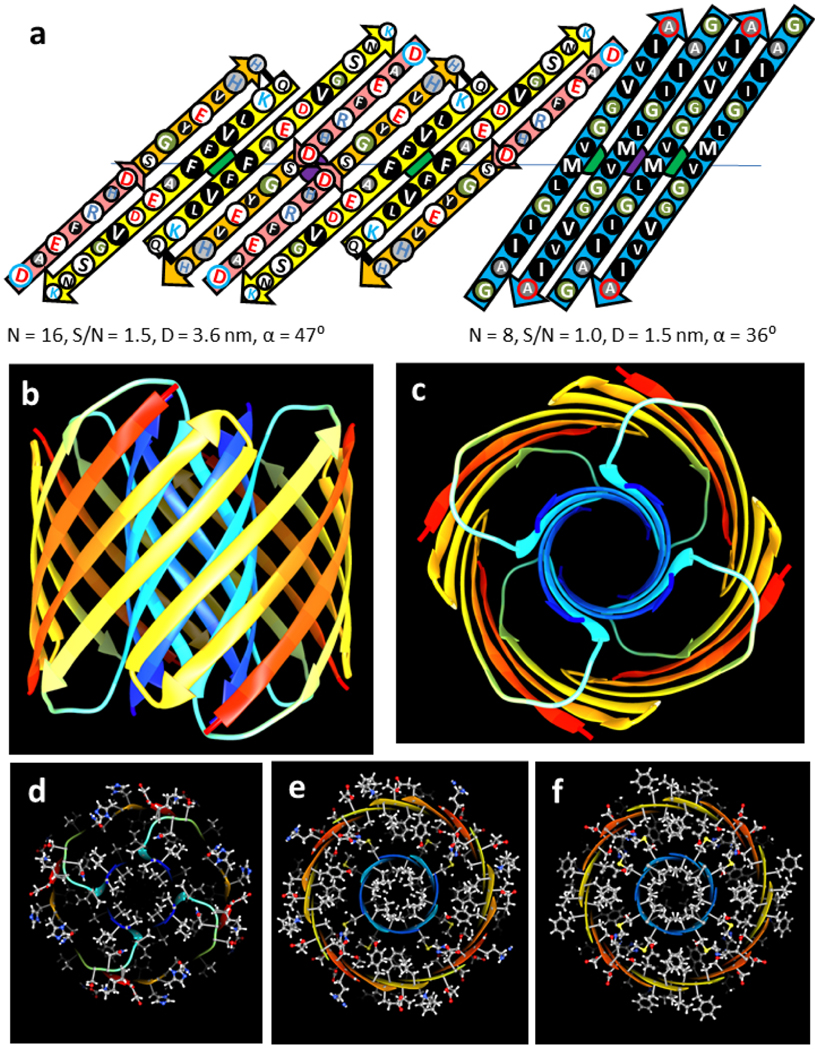
Figure 9.
Atomic-scale model of the 32mer WCsAPF. (a-c) Rainbow-colored ribbon representation of the backbone. (a) View from the side. (b) Same as (a) except the outer and S3 strands have been clipped away to show the inner S1b-S2 helices surrounding the S1a 8-stranded β-barrel. (c) View down the central axis of the assembly. (d) Sphere representation of side chains colored by element for a central cross-section illustrating tight side chain packing. (e-g) Stick and ball side chains colored by element and backbone ribbon for the (e) top, (f) slightly lower, and (g) middle cross-sections.
DCsAPFs and their relationship to bAPFs
The second category of sAPFs, DCsAPFs, have a dark outer ring with cogs surrounding a light ring, which surrounds a dark circular center. Unlike the white-centered sAPFs, DCsAPFs with the same number of cogs can have different diameters (Fig. 10). Also, WCsAPFs and DCsAPFs of the same approximate diameters often have differing number of cogs and thus different symmetries; e.g., the putative 32mer WCsAPF has eight cogs and 8-fold radial symmetry, whereas the putative 32mer DCsAPF has four cogs and 4-fold radial symmetry. Comparison of sizes and shapes of DCsAPFs to those of bAPFs suggest that the number and distribution of DCsAPF cogs are like those of the beads of bAPFs. Most DCsAPF appear to have morphed from bAPFs composed of the two smallest sizes of beads; i.e., the ones we propose to be hexamers and octamers (outlined in purple and red in Figs. 2 and 10). If so, then the radial unit-cells should have six and eight monomers for the two principal groups of DCsAPFs.
Figure 10.
Comparison of DCsAPFs images to bAPF images of Fig. 2. The bAPF images are those proposed to be formed by hexamers (purple circles) or octamers (red circles). Averaged and individual BCsAPF images are illustrated in the second and third rows below the bAPF images. The blue circles near the middle of the light-shaded rings of the BCsAPFs have diameters predicted for walls of S3 β-barrels (if N is the number of monomers proposed for the APF and S/N = 1.0 for 18–32mers, 2/3 for APFs developed from hexamers, and 3/4 for APFs developed from octamers).
The blue circles superimposed on the white rings of the DCsAPFs in Fig. 10 have the diameter predicted for a S3 β-barrel with the number of strands suggested by the number of monomers proposed for the associated bAPF; e.g., the bAPF composed of six small beads (hexamers) should have 36 monomers and the S3 β-barrel of the associated DCsAPF should have 36 β-strands. S/N values for these putative S3 β-barrels are 1.0 for the 18–32mers, and 2/3 or 3/4 for larger multiples of hexamers or octamers. Thus, these DCsAPFs are consistent with the hypothesis that an antiparallel S3 β-barrel is surrounded by a S1a-S1b-S2 β-barrel in which the S/N values change in a manner that maintains a gap distance between the walls of the barrels near 1.0 nm. It is difficult to predict based on these images whether there is a S1-S2 β-barrel inside the S3 barrel because the interior is completely dark. If the dark center is due to the presence of hexane, then hexane may shield the inner surface of the S3 β-barrel from water and any interior S1 and S2 segments may interact with the surface of a hexane layer.
An interesting exception to the general rule of relatively circular sAPFs are the putative oval-shaped 32mers proposed to have developed from diamond-shaped clusters of four octamers (7th image of Fig. 10). Seven such images were identified in the portion of the micrograph of Fig. 1b that contains the WCsAPFs. These were the only oval sAPFs observed in this region of the EM and they all have similar sizes and shapes. In this instance the sAPFs may have retained the general shape of its ancesteral bAPF rather than adopting β-barrel structures with circular crosssections. The other possible exception are the DCsAPFs we propose to have 10-fold symmetry. Some of the isolated images appear to have 5-fold symmetry, which when radially averaged with 10-fold symmetry, would appear to have ten cogs. 5-fold symmetry could result if each radial unit-cell has 12 or 16 monomers instead of six or eight, which could occur if they developed from bAPFs composed of five dodecamers or hexadecamers.
SsAPFs
Many sAPF images have no discernable cogs. These super smooth APFs (SsAPFs) simply have light-shaded rings with darker shading on their exterior and interiors. However, some do appear to have radial symmetry, as indicated by the radially-averaged images. Our working hypothesis is that cogs appear when triple-stranded S1a-S1b-S2 comprise the outer β-barrel because the S1a and S1b strands are much shorter than the S2 strands (Fig. 6), and that cogs do not appear when S1-S2 hairpins comprise the outer barrel because the continuous S1 strand is about the same length as the S2 strand (Fig. 11). There are two mechanisms by which the outer and inner S1-S2 β-barrels can have larger and smaller diameters than those of the central S3 β-barrels. In most cases the numbers of strands in the outer and inner barrels are likely greater and less than those of the S3 barrels. This situation could develop from bAPF precursors if S1-S2 strands inside the bAPF ring become inner barrel strands and those outside the ring become outer barrel strands. As the SsAPF’s increase in size, the ratio of outer to inner barrel strands should decrease, and for very-large SsAPF’s it likely becomes 1.0. In these large SsAPFs the S/N values of the outer and inner barrels likely become greater or less than those of the S3 barrels. Sizes of the SsAPF vary greatly, and many larger ones are less circular than those illustrated in Fig. 11.
Figure 11.
Images of SsAPFs averaged without and with radial symmetry. The top rows of circular SsAPFs images were generated by averaging multiple images of the same size but without radial averaging; the number of images that were averaged is indicated in the upper right corner. The second row of images were radially averaged. Dashed circles represent approximated edges of the SsAPFs walls; the gap distance between the two circles is 3.0 nm. The proposed numbers of monomers, of radial unit-cells, and of monomers/unit-cell are indicated below the images.
The thickness of the light rings of SsAPF’s appears to be between 3 and 4 nm regardless of the size or shape of the SsAPF. This thickness is consistent with three or four β-sheet layers that are each about 1 nm apart.
SUMMARY AND CONCLUSIONS
This is the second paper of a series on the concentric β-barrel hypothesis for non-fibril amyloid assemblies. The first was on Synucleins, especially α-synuclein associated with Parkinson’s Disease45. Others in progress involve Aβ42 channels, Amylin (or islet amyloid polypeptide, IAPP) assemblies, and interactions of various amyloids (synucleins, amylin, humanin, and human cystatin C) with Aβ42 hexamers. These papers are follow-ups to and an expansion of our earlier publications on concentric β-barrel models of Aβ42 oligomers, APFs, and ion channels21,22, utilizing recent experimental results and image averaging. Models presented in these papers are works in progress. Much more needs to be done both experimentally and theoretically to evaluate these models.
This manuscript focuses only on soluble oligomers and annular protofibrils. Some may consider Aβ42 APFs of secondary interest because they are not toxic and do not form transmembrane channels50. However, APF specific antibodies do interact with channels formed when PFO’s interact with membranes and with α-hemolysin antiparallel β-barrel channels. We have chosen to emphasize APFs because they provide the best pictorial evidence we have for concentric structural architectures. They also provide clues to the structure of smaller oligomers, which are thought to be important in Alzheimer’s disease and may be precursors to transmembrane channels.
The next paper in our series will present models of Aβ42 channel structures and both atomic force microscopy (Quist et al, 2005)48 and freeze fracture (Shafrir et al)21 images supporting them. Although we are still developing and evaluating these models, a schematic of our current working hypothesis is illustrated in Fig. 12. Atomic force microscopy (AFM) images of membrane-embedded Aβ assemblies resemble the bAPFs; i.e. three to six peaks of ~ 2 – 4 nm in diameter form radially symmetric clusters. We suspect that each peak corresponds to a transmembrane oligomer (TMO) and that their membrane spanning portions have the same structures as the hydrophobic cores of some bAPF oligomers. The major difference is that the S1 and S2 segments that shield the cores from water peel away and form the aqueous domains that extend beyond the membrane surfaces. The TMOs with diameters of ~ 2 nm are likely tetramers or hexamers and have only one 6-stranded membrane-spanning β-barrel formed by S3 strands in hexamers and four S3 plus two S2 strands in tetramers. Most putative TMOs have ~ 4nm diameters and may correspond to dodecamers, hexadecamers, and octadecamers. These TMOs have two concentric β-barrels; an inner barrel formed by six or eight S2 strands and an outer barrel formed by S3 strands. Some S2 strands become trapped in the centers of the larger TMOs when they form by mergers of multiple tetramer or hexamer TMOs (see Fig.12). It is unclear whether ions will pass through individual TMOs due to the small sizes of the core S2 barrels or between the TMOs due to possible blockage by lipids.
Figure 12.
Hypothesis for how Aβ42 oligomers transition from the aqueous phase to span a membrane, form clusters, and morph into channels with larger pores. (A) Wedge representations and side views of some β-strands colored by segment (red S1a, orange S1b, yellow S2, cyan S3 odd side-chains outward, blue S3 odd side-chains inward). Interactions of tetramers or hexamers are proposed to merge to form larger oligomers. S3 β barrels are sandwiched between an inner S2 barrel and an outer S1-S2 barrel for dodecamers, hexadecamers, and octadecamers in the aqueous phase. (B) Transmembrane octamers (TMOs). Subunit transmembrane topology (small yellow cylinders represent S2 α-helices) and schematic of transmembrane and soluble domains. Transmembrane strands are most tilted for dodecamers and least tilted for octadecamers. Only S3 barrels are in the transmembrane region for hexamers and octamers, S3 transmembrane barrels surround S2 barrels for dodecamers, hexadecamers, and octadecamers. S1 and S2 segments comprise the soluble domains (orange). (C) Wedge schematics of TMO aggregates. (D) Wedge representations of the transmembrane region of channels. An outer S3 barrel surrounds an inner S2 barrel, which may surround S1 barrels. S1 and S2 segments that are not in the transmembrane region form soluble domains (not illustrated). Dimensions and shapes of AFM and freeze-fracture images agree well with dimensions and shapes predicted by these models.
Some freeze-fracture images resemble the AFM images, but others are more circular and have smoother perimeters. Like our sAPF models, these may develop from the mergers of the TMOs to form channels in which a single S3 barrel surrounds pore-lining β-barrels formed by some S2 and possibly S1 segments.
Like all known β-barrel channels, our model transmembrane β-barrel are antiparallel, and, like all know concentric β-barrel structures, the inner pore-lining β-barrel has fewer strands than the surrounding outer barrel. Our models are consistent with β-barrel theory. The TMO assemblies have relatively large aqueous domains that should extend beyond the membrane surfaces. Alternative models proposed by some other groups (Jang et al, 2010 a & b)67,68 have parallel barrels, are inconsistent with β-barrel theory, have the same number of strands in the inner and outer β-barrels, and have smaller aqueous domains. In models of Strodel et al (2010)69 S2 and S3 form a triple-stranded antiparallel β-sheet that spans the membrane. (S3 forms a β-hairpin). The resulting structures do not have strand tilts typical of β-barrels and predicted by β-barrel theory. Only one β-sheet forms the channel and most side-chains that line the pore are hydrophobic. The dimensions of the structures do not match EM images of Aβ-channels.
With some reservation, we have deposited coordinates of the atomic scale models illustrated here in the Protein Data Bank, but readers are warned that these preliminary models are hypothetical and should be expected to contain errors.
Hypothetical models and concepts are vital for research because they may suggest experiments that otherwise would not be performed or funded. Possible approaches include:
Disulfide bridge formation: The same residue of adjacent monomers interacts at axes of 2-fold perpendicular symmetry, but the predicted interactions differ among models. Aβ has no cysteine residues. Introduction of a cysteine residue at these positions could create disulphide-linked dimers that stabilize some assemblies while preventing formation of others. Doing so could reduce the polymorphism and disorder that complicates structural studies of amyloids and help identify which models are better. Stabilization of Aβ analog hexamers by linking S2 to S3 with a disulfide bridge led to an NMR-based model that differs from ours (Lendel et al, 2014)70. The disulfide bridge forced S2-S3 to form a β-hairpin instead of a U-shaped pair as in our models and some fibril studies. It is difficult to say whether this modification stabilized the native structure or forced a nonnative conformation. Another group developed hexamer Aβ models, but considered only models in which S2 and S3 form a β-hairpin (Osterlund et al, 2019)43. Some of our models have two conformations; while a disulfide bridge may stabilize one conformation it could prevent the other.
Single molecule studies: The analysis presented here suggests that some relatively stable assemblies can be identified within a heterogeneous population of APFs. Single-particle Cryo-EM methods have been used to determine near atomic resolution structures of numerous antiparallel β-barrel toxin channels (see 71 for review). To our knowledge such methods have not succeeded in analyzing Aβ42 assemblies, but results of the APF analysis suggest that they could work.
Amyloid homologs: Studies of non-human ion channels, such as bacterial potassium and sodium channels, have proved invaluable in understanding their structures and functional mechanisms. Most amyloids also have long evolutionary histories, as indicated in Supplement Fig. S1 and Fig. 5 H & I. Experimental and computational analyses of distantly related amyloids may help determine what physiological functions and structural properties they share and how they differ. Some may be better suited for crystallography or cryo-EM studies.
Atomic force microscopy-infrared spectroscopy has been used analyze the structure of αSyn oligomers at different stages of aggregation72. The nanometer resolution of this approach has allowed the secondary structures of different aggregate species of α-Syn to be monitored as they develop with time. Transitions from primarily coil, α-helical, and antiparallel β-sheet assemblies to primarily parallel β-sheet fibrils were observed. Perhaps a similar approach could be used to analyze secondary structure transitions of PFO’s, bAPFs, and sAPFs.
Antibodies and drugs: Models of oligomers can be used to identify possible target segments to which binding of antibodies or drugs could preclude further growth of the assembly. Having a basic understanding of how amyloids develop and morph among many experimentally observed configurations may help identify which assemblies are toxic and which are essential for vital physiological processes. They may also provide new insights for development of molecular preventions, treatments, and cures of the devastating diseases associated with amyloid misfolding.
Supplementary Material
Acknowledgments
This work was supported in part by the Intramural Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
The authors have no competing interests.
Original EM images will be supplied by HRG at hrguy46@yahoo.com upon request. Coordinates of atomic scale model of Aβ42 hexamer, octamer, dodecamer, octadecamer, and 32mer oligomers have been deposited in the Protein Data Bank.
References
- 1.Deleanu M, Hernandez JF, Cipelletti L, et al. Unraveling the Speciation of beta-Amyloid Peptides during the Aggregation Process by Taylor Dispersion Analysis. Anal Chem. 2021;93(16):6523–6533. [DOI] [PubMed] [Google Scholar]
- 2.Urban AS, Pavlov KV, Kamynina AV, Okhrimenko IS, Arseniev AS, Bocharov EV. Structural Studies Providing Insights into Production and Conformational Behavior of Amyloid-beta Peptide Associated with Alzheimer’s Disease Development. Molecules. 2021;26(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mroczko B, Groblewska M, Litman-Zawadzka A, Kornhuber J, Lewczuk P. Cellular Receptors of Amyloid beta Oligomers (AbetaOs) in Alzheimer’s Disease. Int J Mol Sci. 2018;19(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gremer L, Schölzel D, Schenk C, et al. Fibril structure of amyloid-β(1–42) by cryo–electron microscopy. Science. 2017;358(6359):116–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Molecular structure of beta-amyloid fibrils in Alzheimer’s disease brain tissue. Cell. 2013;154(6):1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lührs T, Ritter C, Adrian M, et al. 3D structure of Alzheimer’s amyloid-β(1–42) fibrils. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(48):17342–17347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s beta-amyloid fibrils. Biochemistry. 2006;45(2):498–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soldner CA, Sticht H, Horn AHC. Role of the N-terminus for the stability of an amyloid-beta fibril with three-fold symmetry. PLoS One. 2017;12(10):e0186347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh U, Thurber KR, Yau WM, Tycko R. Molecular structure of a prevalent amyloid-beta fibril polymorph from Alzheimer’s disease brain tissue. Proc Natl Acad Sci U S A. 2021;118(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiang W, Yau WM, Luo Y, Mattson MP, Tycko R. Antiparallel beta-sheet architecture in Iowa-mutant beta-amyloid fibrils. Proc Natl Acad Sci U S A. 2012;109(12):4443–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mroczko B, Groblewska M, Litman-Zawadzka A, Kornhuber J, Lewczuk P. Amyloid β oligomers (AβOs) in Alzheimer’s disease. Journal of Neural Transmission. 2018;125(2):177–191. [DOI] [PubMed] [Google Scholar]
- 12.Cline EN, Bicca MA, Viola KL, Klein WL. The Amyloid-beta Oligomer Hypothesis: Beginning of the Third Decade. J Alzheimers Dis. 2018;64(s1):S567–S610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y-r Liu R-t. The Toxicity and Polymorphism of β-Amyloid Oligomers. International Journal of Molecular Sciences. 2020;21(12):4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu L, Sun Y, Guo Y, et al. Comparison of neurotoxicity of different aggregated forms of Aβ40, Aβ42 and Aβ43 in cell cultures. Journal of Peptide Science. 2017;23(3):245–251. [DOI] [PubMed] [Google Scholar]
- 15.Arispe N, Pollard HB, Rojas E. Zn2+ interaction with Alzheimer amyloid beta protein calcium channels. Proc Natl Acad Sci U S A. 1996;93(4):1710–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arispe N, Rojas E, Pollard HB. Alzheimer disease amyloid beta protein forms calcium channels in bilayer membranes: blockade by tromethamine and aluminum. Proc Natl Acad Sci U S A. 1993;90(2):567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirakura Y, Lin MC, Kagan BL. Alzheimer amyloid abeta1–42 channels: effects of solvent, pH, and Congo Red. J Neurosci Res. 1999;57(4):458–466. [PubMed] [Google Scholar]
- 18.Lin H, Bhatia R, Lal R. Amyloid beta protein forms ion channels: implications for Alzheimer’s disease pathophysiology. FASEB J. 2001;15(13):2433–2444. [DOI] [PubMed] [Google Scholar]
- 19.Serra-Batiste M, Ninot-Pedrosa M, Bayoumi M, Gairí M, Maglia G, Carulla N. Aβ42 assembles into specific β-barrel pore-forming oligomers in membrane-mimicking environments. Proceedings of the National Academy of Sciences. 2016;113(39):10866–10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bode DC, Baker MD, Viles JH. Ion Channel Formation by Amyloid-β42 Oligomers but Not Amyloid-β40 in Cellular Membranes. Journal of Biological Chemistry. 2017;292(4):1404–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shafrir Y, Durell S, Arispe N, Guy HR. Models of membrane-bound Alzheimer’s Abeta peptide assemblies. Proteins. 2010;78(16):3473–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shafrir Y, Durell SR, Anishkin A, Guy HR. Beta-barrel models of soluble amyloid beta oligomers and annular protofibrils. Proteins. 2010;78(16):3458–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pham CL, Kwan AH, Sunde M. Functional amyloid: widespread in Nature, diverse in purpose. Essays Biochem. 2014;56:207–219. [DOI] [PubMed] [Google Scholar]
- 24.Otzen D, Riek R. Functional Amyloids. Cold Spring Harb Perspect Biol. 2019;11(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urbanc B. Cross-Linked Amyloid beta-Protein Oligomers: A Missing Link in Alzheimer’s Disease Pathology? J Phys Chem B. 2021;125(5):1307–1316. [DOI] [PubMed] [Google Scholar]
- 26.Kumar DKV, Choi SH, Washicosky KJ, et al. Amyloid-beta peptide protects against microbial infection in mouse and worm models of Alzheimer’s disease. Sci Transl Med. 2016;8(340). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou PY, Fasman GD. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. [DOI] [PubMed] [Google Scholar]
- 28.Wirths O, Zampar S, Weggen S. N-Terminally Truncated Aβ Peptide Variants in Alzheimer’s Disease. In: Wisniewski T, ed. Alzheimer’s Disease. Brisbane (AU): Codon Publications; [PubMed] [Google Scholar]
- 29.Aggarwal L, Biswas P. Hydration Thermodynamics of the N-Terminal FAD Mutants of Amyloid-beta. J Chem Inf Model. 2021;61(1):298–310. [DOI] [PubMed] [Google Scholar]
- 30.Ngo S, Guo Z. Key residues for the oligomerization of Abeta42 protein in Alzheimer’s disease. Biochem Biophys Res Commun. 2011;414(3):512–516. [DOI] [PubMed] [Google Scholar]
- 31.Laganowsky A, Liu C, Sawaya MR, et al. Atomic view of a toxic amyloid small oligomer. Science. 2012;335(6073):1228–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Do TD, LaPointe NE, Nelson R, et al. Amyloid β-Protein C-Terminal Fragments: Formation of Cylindrins and β-Barrels. Journal of the American Chemical Society. 2016;138(2):549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harmeier A, Wozny C, Rost BR, et al. Role of amyloid-beta glycine 33 in oligomerization, toxicity, and neuronal plasticity. J Neurosci. 2009;29(23):7582–7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bitan G, Tarus B, Vollers SS, et al. A molecular switch in amyloid assembly: Met(35) and amyloid beta-protein oligomerization. Journal of the American Chemical Society. 2003;125(50):15359–15365. [DOI] [PubMed] [Google Scholar]
- 35.Cerf E, Sarroukh R, Tamamizu-Kato S, et al. Antiparallel beta-sheet: a signature structure of the oligomeric amyloid beta-peptide. Biochem J. 2009;421(3):415–423. [DOI] [PubMed] [Google Scholar]
- 36.Yoshiike Y, Kayed R, Milton SC, Takashima A, Glabe CG. Pore-forming proteins share structural and functional homology with amyloid oligomers. Neuromolecular medicine. 2007;9(3):270–275. [DOI] [PubMed] [Google Scholar]
- 37.Vignaud H, Bobo C, Lascu I, et al. A Structure-Toxicity Study of Aß42 Reveals a New Anti-Parallel Aggregation Pathway. PLOS ONE. 2013;8(11):e80262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ciudad S, Puig E, Botzanowski T, et al. Abeta(1–42) tetramer and octamer structures reveal edge conductivity pores as a mechanism for membrane damage. Nat Commun. 2020;11(1):3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao Y, Guo C, Watzlawik JO, et al. Out-of-Register Parallel beta-Sheets and Antiparallel beta-Sheets Coexist in 150-kDa Oligomers Formed by Amyloid-beta(1–42). J Mol Biol. 2020;432(16):4388–4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iacovache I, De Carlo S, Cirauqui N, Dal Peraro M, van der Goot FG, Zuber B. Cryo-EM structure of aerolysin variants reveals a novel protein fold and the pore-formation process. Nat Commun. 2016;7:12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bokori-Brown M, Martin TG, Naylor CE, Basak AK, Titball RW, Savva CG. Cryo-EM structure of lysenin pore elucidates membrane insertion by an aerolysin family protein. Nat Commun. 2016;7:11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banchelli M, Cascella R, D’Andrea C, et al. Probing the Structure of Toxic Amyloid-beta Oligomers with Electron Spin Resonance and Molecular Modeling. ACS Chem Neurosci. 2021;12(7):1150–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osterlund N, Moons R, Ilag LL, Sobott F, Graslund A. Native Ion Mobility-Mass Spectrometry Reveals the Formation of beta-Barrel Shaped Amyloid-beta Hexamers in a Membrane-Mimicking Environment. Journal of the American Chemical Society. 2019;141(26):10440–10450. [DOI] [PubMed] [Google Scholar]
- 44.Sulatskaya AI, Kosolapova AO, Bobylev AG, et al. beta-Barrels and Amyloids: Structural Transitions, Biological Functions, and Pathogenesis. International Journal of Molecular Sciences. 2021;22(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Durell SR, Guy HR. The amyloid concentric β-barrel hypothesis: models of Synuclein oligomers, annular protofibrils, lipoproteins, and transmembrane channels. Proteins: Structure, Function, and Bioinformatics. 2021:1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nasica-Labouze J, Nguyen PH, Sterpone F, et al. Amyloid beta Protein and Alzheimer’s Disease: When Computer Simulations Complement Experimental Studies. Chem Rev. 2015;115(9):3518–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voelker MJ, Barz B, Urbanc B. Fully Atomistic A beta 40 and A beta 42 Oligomers in Water: Observation of Porelike Conformations. J Chem Theory Comput. 2017;13(9):4567–4583. [DOI] [PubMed] [Google Scholar]
- 48.Quist A, Doudevski I, Lin H, et al. Amyloid ion channels: a common structural link for protein-misfolding disease. Proc Natl Acad Sci U S A. 2005;102(30):10427–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen PH, Campanera JM, Ngo ST, Loquet A, Derreumaux P. Tetrameric A beta 40 and A beta 42 beta-Barrel Structures by Extensive Atomistic Simulations. II. In Aqueous Solution. Journal of Physical Chemistry B. 2019;123(31):6750–6756. [DOI] [PubMed] [Google Scholar]
- 50.Kayed R, Pensalfini A, Margol L, et al. Annular Protofibrils Are a Structurally and Functionally Distinct Type of Amyloid Oligomer. Journal of Biological Chemistry. 2009;284(7):4230–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lasagna-Reeves CA, Glabe CG, Kayed R. Amyloid-beta Annular Protofibrils Evade Fibrillar Fate in Alzheimer Disease Brain. Journal of Biological Chemistry. 2011;286(25):22122–22130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kayed R, Glabe CG. Conformation-dependent anti-amyloid oligomer antibodies. Method Enzymol. 2006;413:326–344. [DOI] [PubMed] [Google Scholar]
- 53.Nguyen PH, Ramamoorthy A, Sahoo BR, et al. Amyloid Oligomers: A Joint Experimental/Computational Perspective on Alzheimer’s Disease, Parkinson’s Disease, Type II Diabetes, and Amyotrophic Lateral Sclerosis. Chem Rev. 2021;121(4):2545–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Photoshop. Adobe Photoshop, RRID:SCR_014199. Available at: https://www.adobe.com/products/photoshop.html.
- 55.Murzin AG, Lesk AM, Chothia C. Principles determining the structure of beta-sheet barrels in proteins. I. A theoretical analysis. J Mol Biol. 1994;236(5):1369–1381. [DOI] [PubMed] [Google Scholar]
- 56.Hayward S, Milner-White EJ. Geometrical principles of homomeric beta-barrels and beta-helices: Application to modeling amyloid protofilaments. Proteins. 2017;85(10):1866–1881. [DOI] [PubMed] [Google Scholar]
- 57.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. [DOI] [PubMed] [Google Scholar]
- 58.UCSF Chimera. http://www.rbvi.ucsf.edu/chimera.
- 59.Blastp. https://blast.ncbi.nlm.nih.gov/Blast.cgi.
- 60.Bernstein SL, Dupuis NF, Lazo ND, et al. Amyloid-beta protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer’s disease. Nat Chem. 2009;1(4):326–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lacor PN, Buniel MC, Chang L, et al. Synaptic targeting by Alzheimer’s-related amyloid beta oligomers. J Neurosci. 2004;24(45):10191–10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lesne S, Koh MT, Kotilinek L, et al. A specific amyloid-beta protein assembly in the brain impairs memory. Nature. 2006;440(7082):352–357. [DOI] [PubMed] [Google Scholar]
- 63.Stroud JC, Liu C, Teng PK, Eisenberg D. Toxic fibrillar oligomers of amyloid-beta have cross-beta structure. Proc Natl Acad Sci U S A. 2012;109(20):7717–7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sebollela A, Mustata GM, Luo K, et al. Elucidating molecular mass and shape of a neurotoxic Abeta oligomer. ACS Chem Neurosci. 2014;5(12):1238–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gu L, Liu C, Guo Z. Structural insights into Abeta42 oligomers using site-directed spin labeling. J Biol Chem. 2013;288(26):18673–18683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nagano N, Orengo CA, Thornton JM. One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J Mol Biol. 2002;321(5):741–765. [DOI] [PubMed] [Google Scholar]
- 67.Jang H, Arce FT, Ramachandran S, et al. Truncated beta-amyloid peptide channels provide an alternative mechanism for Alzheimer’s Disease and Down syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(14):6538–6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jang H, Arce FT, Ramachandran S, Capone R, Lal R, Nussinov R. beta-Barrel Topology of Alzheimer’s beta-Amyloid Ion Channels. Journal of Molecular Biology. 2010;404(5):917–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Strodel B, Lee JWL, Whittleston CS, Wales DJ. Transmembrane Structures for Alzheimer’s A beta(1–42) Oligomers. Journal of the American Chemical Society. 2010;132(38):13300–13312. [DOI] [PubMed] [Google Scholar]
- 70.Lendel C, Bjerring M, Dubnovitsky A, et al. A Hexameric Peptide Barrel as Building Block of Amyloid-beta Protofibrils. Angew Chem Int Edit. 2014;53(47):12756–12760. [DOI] [PubMed] [Google Scholar]
- 71.Gilbert RJC. Chapter Three - Electron microscopy as a critical tool in the determination of pore forming mechanisms in proteins. In: Heuck AP, ed. Methods in Enzymology. Vol 649. Academic Press; 2021:71–102. [DOI] [PubMed] [Google Scholar]
- 72.Zhou L, Kurouski D. Structural Characterization of Individual alpha-Synuclein Oligomers Formed at Different Stages of Protein Aggregation by Atomic Force Microscopy-Infrared Spectroscopy. Anal Chem. 2020;92(10):6806–6810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.