Abstract
This article reports the identification of a putative P-transporter operon in the genome of a Burkholderia sp. living in the cytoplasm of the arbuscular mycorrhizal fungus Gigaspora margarita. Its presence suggests that Burkholderia sp. has the potential for P uptake from this environment. This finding raises new questions concerning the importance of intracellular bacteria for mycorrhizal symbiosis.
Arbuscular mycorrhizal fungi (AMF) are obligate biotrophic organisms which establish symbiotic associations with root tissues of more than 80% of land plants. The level of complexity of arbuscular mycorrhizal symbiosis is especially increased by the presence of intracellular bacteria inside AMF. The initial observation by Mosse (15) that AMF harbor structures called bacterium-like organisms (BLOs) has been amply confirmed by other authors (for a review, see reference 22). However, as these BLOs cannot be grown in culture media (11, 22), their bacterial nature has been the subject of debate. Only recently, Bianciotto et al. (2) have demonstrated that true viable bacteria live in the spores of Gigaspora margarita (BEG 34) and move via the mycelium to its structures in the root of the host plant. These bacteria belong to the genus Burkholderia and constitute a homogeneous population present throughout the fungal life cycle. Four of five AMF species of the Gigasporaceae have been shown to possess intracellular Burkholderia organisms, confirming that they are widespread (3).
Morphological observations show that Burkholderia multiplies both inside the fungal spore and in the mycelium during differentiation of the colonization structures (4). This suggests that they possess all the biosynthetic machinery for DNA replication and energy production. Hence, it would be expected that they also possess a P-uptake system to accomplish their basal metabolism. It is well known that AMF take up P from the soil and transfer it to plants (23). We therefore wondered whether intracellular bacteria interact with the P uptake and transport by the fungus, i.e., by incorporating Pi from the fungal cytoplasm where they live. Two phosphate transport systems have been described in bacteria: a low-affinity phosphate inorganic transport system and a high-affinity phosphate-specific transport (Pst) system (5, 24, 28). These systems are multisubunit permeases composed of a soluble substrate-binding protein and a membrane-bound complex containing two to four proteins (5). To investigate the role of Burkholderia sp. in fungal P metabolism and its possible shunting off in P transfer from the fungus to the plant, we looked for bacterial genes involved in P transport. We have cloned and characterized an operon for a Pst-like system. This would seem to be the first operon described in a bacterium living in the cytoplasm of an AMF.
G. margarita Becker and Hall (isolate BEG 34) and Gigaspora rosea Nicolson and Schenck (isolate BEG 9) spores were recovered from pot cultures of Trifolium repens L. by wet sieving (7). Spores of a Scutellospora sp. were collected from sand dunes in Migliarino (Pisa, Italy). All spores were rinsed five times with sterile, filtered, and distilled water, surface sterilized with 4% chloramine-T and 300-ppm streptomycin for 30 min, and then rinsed seven times for 1 h (total) with sterile, filtered, and distilled water (2). Approximately 100 surface-sterilized spores were crushed with a plastic pestle in 300 μl of lysis buffer (50 mM Tris-HCl [pH 8], 25 mM Na-EDTA, 100 mM NaCl, 1% [wt/vol] sodium dodecyl sulfate, 0.1% [vol/vol] Triton X-100 and 0.1% [vol/vol] β-mercaptoethanol) and then treated with proteinase K (final concentration, 50 μg/ml) at 60°C for 1 h and with DNase-free RNase for 30 min at 37°C. Proteins were precipitated with 0.1 volume of 5 M potassium acetate and the supernatant was treated with 1 volume of phenol-chloroform (1/1). The genomic DNA was then precipitated under standard conditions (21).
After compilation of sequences for the PstC protein (Escherichia coli, Enterobacter cloacae, Pseudomonas aeruginosa, Aquifex aeolicus, and Synechocystis sp.), degenerate oligonucleotide primers were designed as described by Numberg et al. (18) (forward, 5′-TCIAT[T or C]GT[A or C]TA[T or C]GG[T or G]ATGTGG-3′; reverse, 5′-[T or A]AT[T or C]A[A or G][A or G]AA[T or G]A[A or G]CAT[T or C]A[A or G]ICC-3′). A 522-bp DNA fragment of the pstC gene was amplified with these primers by PCR. Conditions for PCR and thermal parameters were as described by McPherson et al. (12). The amplified DNA was purified from agarose gel with the QIAEX II Gel Extraction Kit (Qiagen) and cloned into pGME plasmid (Promega, Madison, Wis.). Plasmid DNA was isolated as described by Sambrook et al. (21). Sequencing was performed with an Applied Biosystems model 370A DNA sequencer (Genome Express Society, Grenoble, France).
A genomic library constructed for G. margarita and shown to be also representative of the bacterial genome (27) was screened for identification and sequencing of the five components of the Pst system in Burkholderia sp. For the screening, the probe consisted of the 522-bp DNA fragment of the pstC gene obtained with the degenerated primers defined above. The ECL Direct DNA Labelling and Detection System (Amersham, Little Chalfont, England) was used as recommended by the manufacturer. Sequencing was performed as described above. Sequences were analyzed with PC/GENE software (IntelliGenetics, Inc., Mountain View, Calif.), and the similarity searches of the EMBL data bank were carried out by using the FASTA program from the Wisconsin Package 8 (Genetics Computer Group, Madison, Wis.) or the BLAST software available through the National Center for Biotechnology Information. After sequence alignment, the phylogenetic tree was constructed by the neighbor-joining method from the software package Clustal X.
Specific primers were designed on two regions of the Pst system. These were primer pair 1 (forward 1 [5′-TTTCTTGACTGAACTCTCGCCTGC-3′] and reverse 1 [5′-ATGTTTAGGGCCTGTGCTCATCG-3′]) and primer pair 2 (forward 2 [5′-TTCATCCGTCAAAGCAAAGCTGC-3′] and reverse 2 [5′-AGAGGCGTTCAAACAATTTCATGC-3′]).
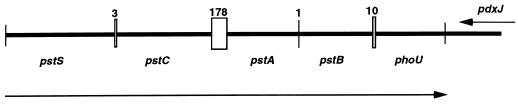
The complete nucleotide sequence of the genes coding for a high-affinity P transporter was determined in the intracellular Burkholderia of G. margarita. Six open reading frames (ORFs) were detected. Five were identified on the basis of the similarity of their amino acid sequences to those of known E. coli gene products. ORF1 codes for a protein similar to PstS (50 and 61% identity in amino acid and nucleotide sequences, respectively). ORF2 codes for a protein similar to PstC (65% identity in both sequences). ORF3 contains a protein similar to PstA (68% identity in both sequences). The fourth ORF codifies for a protein similar to PstB (72 and 70% identity in amino acid and nucleotide sequences, respectively). ORF5 contains a protein similar to PhoU (40 and 59% identity in amino acid and nucleotide sequences, respectively). Their predicted molecular masses range from 36.26 kDa for the biggest (PstS) to 26.17 kDa for the smallest (PhoU). The last ORF had no significant homologies. Figure 1 represents the gene order in the sequenced fragment, as well as the lengths of the intergenic regions in the Pst operon, which range from 1 to 178 nt. The organization of the P-transport system in Burkholderia sp. is similar that of enteric bacteria. The gene order in the E. coli chromosome is pstS, pstC, pstA, pstB, and phoU, and they are transcribed in a counterclockwise direction (17, 24). Our results on the hydropathy properties of the five proteins in Burkholderia sp. also agree with those for E. coli and other bacteria (5, 17, 24). In E. coli, PstS is a phosphate-binding protein, located in the periplasmic space. PstA and PstC are hydrophobic and constitute the transmembrane channel of the Pst system. PstB is also periplasmic and constitutes the ATP-binding subunit, and PhoU is a transcriptional regulatory protein. All these facts suggest that the genes of the Pst system in Burkholderia sp. are part of a single regulatory unit, which probably constitutes an operon, as proposed for the other bacteria (9, 24).
FIG. 1.
Gene order on the genome of Burkholderia sp. The arrows indicate the directions of transcription. Vertical lines delineate the extent of the genes, and numbers and boxes indicate the size of the intergenic regions in base pairs.
Part of the C-terminal end of a putative protein similar to a pyridoxal phosphate synthase of E. coli has been found in the complementary DNA strand of the Pst operon. The DNA from this putative gene is transcribed in the opposite direction to the previous ones (Fig. 1).
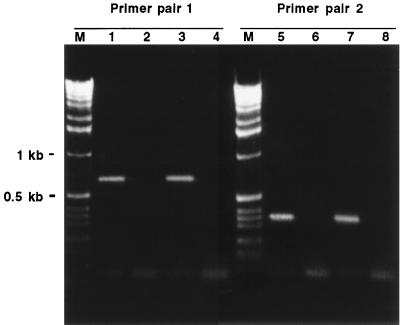
The P-transport system reported here was first cloned after PCR with degenerated primers on DNA from the complex of fungus-bacteria and then identified from a genomic library also containing representatives of the two genomes (27). Therefore, it could be derived from the genome of either of the partners. The lack of introns, characteristic of eukaryotic genes, which give discontinuous ORFs, itself suggests a bacterial origin for these genes. As a first step, the five genes were aligned with P transporters from the mycorrhizal fungus Glomus versiforme (8), from Medicago truncatula (10), or from Saccharomyces cerevisiae (6). Notable differences in the sequences have been found (data not shown), suggesting that this Pst operon is not related to that of eukaryotic cells. To unequivocally demonstrate that it represents genes from the intracellular Burkholderia, specific primers were designed in two of its regions. When these primers were used in PCR (Fig. 2), they successfully amplified the expected fragment from DNA extracted from G. margarita spores containing the intracellular Burkholderia and from Scutellospora sp. spores. Scutellospora is a member of the Gigasporaceae, and the isolate used possesses the same intracellular Burkholderia as G. margarita (3). In contrast, no amplification occurred on DNA from the related species Gigaspora rosea (1), which is devoid of intracellular bacteria.
FIG. 2.
Electrophoresis on 1.2% agarose gel for PCR products obtained with specific primers designed in two regions of the Pst operon. M, 1-kb molecular size marker. Lanes: 1 and 5, Scutellospora sp.; 2 and 6, G. rosea; 3 and 7, G. margarita; 4 and 8, controls containing no DNA.
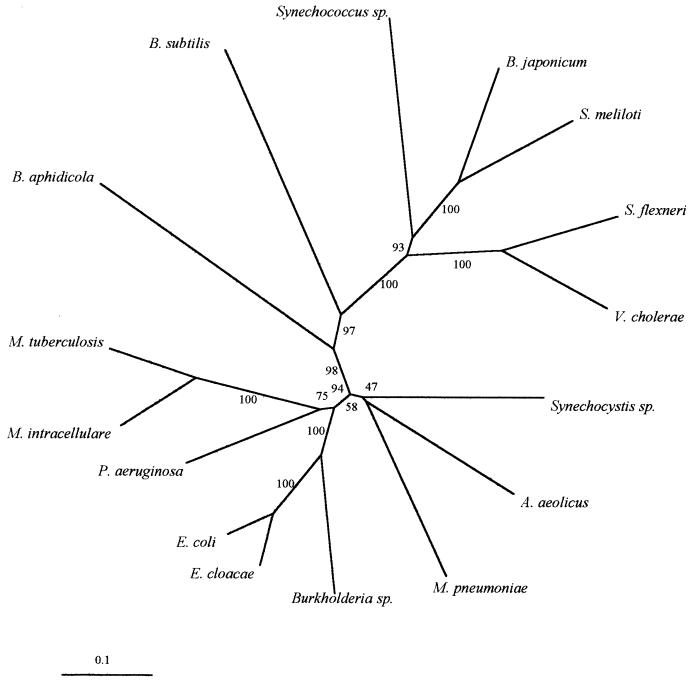
The nucleotide sequence of the Burkholderia pstB gene (the most conserved in the Pst operon) was compared with those of a variety of bacteria. As shown in Fig. 3, E. coli and Enterobacter cloacae were the closest relatives to Burkholderia. The Pst system seems to be a widespread operon in bacteria, since it has been found in free-living, intracellular, and symbiotic bacteria, such as Bradyrhizobium japonicum (13). Curiously, Buchnera aphidicola, another intracellular bacterium (16), is located far from Burkholderia sp., suggesting that their origins and evolutive pathways are different. It has been proposed that some Burkholderia isolates are versatile, can behave as opportunistic pathogens, and easily invade eukaryotic cells (26). From a phylogenetic point of view, it is of interest to determine when this bacterial population was acquired by the fungus and how it has evolved. Perotto and Bonfante (19) postulated that an AMF ancestor acquired bacteria through a single endocytotic event followed by their vertical transmission in the derived phylogenetic branches or that their acquisition is still in progress and their transfer is horizontal. Many fungal isolates will need to be analyzed to determine the evolution of this Burkholderia strain.
FIG. 3.
Phylogenetic tree of pstB sequences from Burkholderia sp. and other bacteria. Scale represents the estimated number of nucleotide substitutions per sequence position. Branch lengths in the phylogram are directly proportional to the number of base replacements. EMBL accession numbers of sequences used to construct the tree are as follows: Aquifex aeolicus, AE000720; Bacillus subtilis, D88802; Bradyrhizobium japonicum, AJ223073; Buchnera aphidicola, U11045; Burkholderia sp., AJ132617; Enterobacter cloacae, D89963; Escherichia coli, K01992; Mycobacterium intracellulare, X95538; Mycobacterium tuberculosis, Z95209; Mycoplasma pneumoniae, AE000023; Pseudomonas aeruginosa, D45195; Sinorhizobium meliloti, M96261; Shigella flexneri, X81000; Synechocystis sp., D64001; Synechococcus sp., U38917; and Vibrio cholerae, AF043352.
In conclusion, the present study shows that Burkholderia sp. contains a genomic region similar to the Pst operon of E. coli in sequences as well as in the order and number of genes and hence has the potential to take up P from its environment. It has been proposed that the rate of P uptake by AMF is regulated by their internal P concentrations (25). Thomson et al. (25), for example, found that P uptake by germ tubes of G. margarita was highest when the fungi had been P starved and suggested that the P uptake may be controlled by the ability of the fungus to translocate and transfer P to the host. Since the main effect of AMF is the uptake and transfer of phosphate to the host plant, the potential of the intracellular Burkholderia to take up P from the hyphae could be seen as a “cost” for the mycorrhizal symbiosis. However, as there is no in vivo evidence of a negative effect of Burkholderia sp. on the symbiotic efficiency of G. margarita in comparison to those of other fungal species without bacteria (20), if this cost really exists, it must be outweighed by a benefit, leading to a positive overall interaction. The fact that the intracellular Burkholderia possesses nif genes (14) and, hence, the potential to fix N2, may be the answer to this question.
Nucleotide sequence accession number.
The nucleotide sequence determined in this study has been deposited in the EMBL database under accession no. AJ132617. Alignments of the different protein sequences from Burkholderia sp. and other bacteria are also available in the EMBL under accession no. ds 38191 (PstC), ds 38258 (PstS), ds 38259 (PstB), ds 38260 (PhoU), and ds 38261 (PstA).
Acknowledgments
This work was supported by an EU research training grant to J. M. Ruiz-Lozano (contract BIO4-CT97-5118) and by an EU Biotechnology Project (IMPACT; contract BIO4-CT96-0027).
We thank S. Perotto and V. Bianciotto for helpful discussions concerning the manuscript, and L. Lanfranco for technical advice.
REFERENCES
- 1.Bentivenga S P, Morton J B. A monograph of the genus Gigaspora, incorporating developmental patterns of morphological characters. Mycologia. 1995;87:719–731. [Google Scholar]
- 2.Bianciotto V, Bandi C, Minerdi D, Sironi M, Ticky H V, Bonfante P. An obligately endosymbiotic mycorrhizal fungus itself harbors obligately intracellular bacteria. Appl Environ Microbiol. 1996;62:3005–3010. doi: 10.1128/aem.62.8.3005-3010.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bianciotto, V., D. Minerdi, L. Lanfranco, C. Bandi, and P. Bonfante. Unpublished data.
- 4.Bonfante P, Balestrini R, Mendgen K. Storage and secretion processes in the spores of Gigaspora margarita (Becker & Hall) as revealed by high-pressure freezing and freeze substitution. New Phytol. 1994;128:93–101. doi: 10.1111/j.1469-8137.1994.tb03991.x. [DOI] [PubMed] [Google Scholar]
- 5.Braibant M, Lefèvre P, Wit L, Ooms J, Peirs P, Huygen K, Wattiez R, Content J. Identification of a second Mycobacterium tuberculosis gene cluster encoding proteins of an ABC phosphate transporter. FEBS Lett. 1996;394:206–212. doi: 10.1016/0014-5793(96)00953-2. [DOI] [PubMed] [Google Scholar]
- 6.Bun-ya M, Nishimura M, Harashima S, Oshima Y. The PH084 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol Cell Biol. 1991;11:3229–3238. doi: 10.1128/mcb.11.6.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerdemann J W. Spores of mycorrhizal Endogone species extracted from soil by wet-sieving and decanting. Trans Br Mycol Soc. 1963;46:235–244. [Google Scholar]
- 8.Harrison M J, van Buuren M L. A phosphate transporter from the mycorrhizal fungus Glomus versiforme. Nature. 1995;378:626–629. doi: 10.1038/378626a0. [DOI] [PubMed] [Google Scholar]
- 9.Kato J, Sakai Y, Nikata T, Ohtake H. Cloning and characterization of a Pseudomonas aeruginosa gene involved in the negative regulation of phosphate taxis. J Bacteriol. 1994;176:5874–5877. doi: 10.1128/jb.176.18.5874-5877.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H, Trieu A T, Blaylock L A, Harrison M J. Cloning and characterization of two phosphate transporters from Medicago truncatula roots: regulation in response to phosphate and colonization by arbuscular mycorrhizal (AM) fungi. Mol Plant-Microbe Interact. 1997;11:14–22. doi: 10.1094/MPMI.1998.11.1.14. [DOI] [PubMed] [Google Scholar]
- 11.Macdonald R M, Chandler M R, Mosse B. The occurrence of bacterium-like organelles in vesicular-arbuscular mycorrhizal fungi. New Phytol. 1982;90:659–663. [Google Scholar]
- 12.McPherson M J, Jones K M, Gurr S J. PCR with highly degenerate primers. In: McPherson M J, Quirke P, Taylor G R, editors. PCR: a practical approach. Oxford, United Kingdom: Oxford University Press; 1991. pp. 171–186. [Google Scholar]
- 13.Minder A C, Narberhaus F, Fischer H M, Hennecke H. The Bradyrhizobium japonicum phoB gene is required for phosphate-limited growth but not for symbiotic nitrogen fixation. FEMS Microbiol Lett. 1998;161:47–52. doi: 10.1111/j.1574-6968.1998.tb12927.x. [DOI] [PubMed] [Google Scholar]
- 14.Minerdi D, Fani R, Gallo R, Bonfante P. Abstracts of the Second International Conference on Mycorrhiza, 1998. 1998. Identification of nitrogen fixation genes in Burkholderia endosymbionts of arbuscular mycorrhizal fungi; pp. 120–121. Uppsala, Sweden. [Google Scholar]
- 15.Mosse B. Honey-coloured sessile Endogone spores. II. Changes in the fine structure during spore development. Arch Mikrobiol. 1970;74:129–145. [Google Scholar]
- 16.Munson M A, Baumann P. Molecular cloning and nucleotide sequence of putative trpDC(F)BA operon in Buchnera aphidicola (endosymbiont of the aphid Schizaphis graminum) J Bacteriol. 1993;175:6426–6432. doi: 10.1128/jb.175.20.6426-6432.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novak R, Cauwels A, Charpentier E, Tuomanen E. Identification of a Streptococcus pneumoniae gene locus encoding proteins of an ABC phosphate transporter and a two-component regulatory system. J Bacteriol. 1999;181:1126–1133. doi: 10.1128/jb.181.4.1126-1133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Numberg J H, Wright D K, Cole G E, Petrovskis E A, Post L E, Compton T, Gilbert J H. Identification of the thymidine kinase gene of feline herpesvirus: use of degenerate oligonucleotides in the polymerase chain reaction to isolate herpesvirus gene homologs. J Virol. 1989;63:3240–3249. doi: 10.1128/jvi.63.8.3240-3249.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perotto S, Bonfante P. Bacterial associations with mycorrhizal fungi: close and distant friends in the rhizosphere. Trends Microbiol. 1997;5:496–501. doi: 10.1016/S0966-842X(97)01154-2. [DOI] [PubMed] [Google Scholar]
- 20.Ruiz-Lozano, J. M., and P. Bonfante. Unpublished data.
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Scannerini S, Bonfante P. Bacteria and bacteria-like objects in endomycorrhizal fungi (Glomaceae) In: Margulis L, Fester R, editors. Symbiosis as a source of evolutionary innovation: speciation and morphogenesis. Cambridge, Mass: MIT Press; 1991. pp. 273–287. [Google Scholar]
- 23.Smith S E, Read D J. Mycorrhizal symbiosis. London, United Kingdom: Academic Press; 1997. [Google Scholar]
- 24.Surin B P, Rosenberg H, Cox G B. Phosphate-specific transport system of Escherichia coli: nucleotide sequence and gene-polypeptide relationships. J Bacteriol. 1985;161:189–198. doi: 10.1128/jb.161.1.189-198.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomson B D, Clarkson D T, Brain P. Kinetics of phosphorus uptake by the germ-tubes of the vesicular-arbuscular mycorrhizal fungus Gigaspora margarita. New Phytol. 1990;116:647–653. [Google Scholar]
- 26.Tipper J, Ingham E, Cove J H, Todd N, Kerr K G. Survival and multiplication of Burkholderia cepacia within respiratory epithelial cells. Clin Microbiol Infect. 1998;4:450–459. [Google Scholar]
- 27.van Buuren M L, Lanfranco L, Longato S, Minerdi D, Harrison M J, Bonfante P. Construction and characterization of genomic libraries of two endomycorrhizal fungi: Glomus versiforme and Gigaspora margarita. Mycol Res. 1999;103:955–960. [Google Scholar]
- 28.Wilsky G R, Malamy M H. Characterization of two genetically separable inorganic phosphate transport systems in Escherichia coli. J Bacteriol. 1980;144:356–365. doi: 10.1128/jb.144.1.356-365.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]