Abstract
Mutations of RAS are commonly seen in human cancers, especially in lung, colorectal, and pancreatic adenocarcinoma. Despite huge effort for decades, targeting RAS mutations has been “undruggable” because of the molecular instability of RAS protein inhibition. However, the recent discovery of the KRAS G12C inhibitor paved the way to expand therapeutic options for patients with cancer harboring the KRAS G12C mutation. At the same time, the successful development of immune checkpoint inhibitors (ICIs) drastically changed the paradigm of cancer treatment and resulted in a better understanding of the tumor immune microenvironment in patients with KRAS-mutant cancer. This review describes the following: the clinical characteristics of cancer with KRAS mutation; successful development of the KRAS G12C inhibitor and its impact on the tumor immune microenvironment; and potential new avenues such as the combination strategy using KRAS inhibitor and ICI, with preclinical and clinical rationales for overcoming resistance to inhibition of KRAS to improve therapeutic efficacy for patients with cancer harboring KRAS mutations.
Keywords: KRAS mutation, KRAS G12C inhibitor, immune checkpoint inhibitor, tumor immune microenvironment
INTRODUCTION: RAS MUTATIONS IN CANCER
The RAS family of oncogenes, including Kirsten rat sarcoma viral oncogene homolog (KRAS), neuroblastoma rat sarcoma viral oncogene homolog (NRAS), and Harvey rat sarcoma viral oncogene homolog (HRAS), is the most frequently mutated gene family and accounts for approximately 30% of mutations in cancer cells.[1,2] RAS genes encode GTPases, which act as a gatekeeper to switch on and off RAS proteins, and thus, controls the downstream signaling pathways.[3] Therefore, RAS proteins are well known to have an important role in cell differentiation, division, proliferation, and survival by regulating its downstream pathways such as RAF-MEK-ERK (MAPK) and PI3K-PTEN-AKT pathways.[4] Hence, enormous effort was made to develop therapeutic options targeting these pathways, resulting in the successful development of clinically approved inhibitors of several proteins in these pathways such as MEK, BRAF, and epidermal growth factor receptor (EGFR) inhibitors in various types of cancer (Fig. 1).[5–8] Notably, KRAS is the most commonly mutated RAS isoform, making up approximately 85% of oncogenic RAS mutations in all cancer types.[1] KRAS mutation is seen in 61–86% of pancreatic ductal adenocarcinoma, 33–41% of colorectal adenocarcinoma, and 32% of lung adenocarcinoma.[2,9,10] Mutations in KRAS lead to a single amino acid substitution at a codon, and the substitution usually occurs in G12, G13, or Q61.[1] G12 mutations are known to account for more than 80% of all KRAS mutations.[1] In addition, the pattern of mutations of codons and substitutions of amino acid varies among tumor types. Among patients with cancer harboring KRAS mutation, KRAS G12C is seen in 46% of lung adenocarcinoma, and in contrast, KRAS G12D mutation is observed in approximately 45% of colorectal adenocarcinoma and pancreatic ductal adenocarcinoma.[9,11,12] Beyond these cancer types, analysis of the COSMIC database (version 95) demonstrated that KRAS G12D mutation is commonly seen in biliary tract cancer (45%), small intestine adenocarcinoma (41%), ovarian carcinoma (36%), and endometrial carcinoma (34%), whereas KRAS G12C mutation is seen in fewer than 10% of these cancer types.[13] The number of each KRAS point mutation across common cancer types and their histology is illustrated in Table 1. Therefore, targeting mutated KRAS rather than their downstream molecules, such as RAF, MEK, and mTOR, sounds reasonable to improve the survival outcome in a variety of tumors (Fig. 1). However, despite the effort for the past several decades, the strategy of targeting RAS proteins had not achieved a feasible therapeutic response. This is because of limited drug-binding pockets outside of the nucleotide-binding pocket in RAS proteins and of the high affinity of guanosine diphosphate (GTP) for RAS proteins resulting in difficulties in the development of GTP-competitive inhibitors.[14,15] Given RAS proteins are active when they are associated with the plasma membrane, another strategy inhibiting farnesyltransferase, which is involved in the process of connection of RAS proteins to the plasma membrane, was attempted with promising preclinical results but unfortunately demonstrated minimal clinical activity.[16–18] However, a recent study revealed that tipifarnib, another farnesyltransferase inhibitor, demonstrated antitumor activities against HRAS mutated head and neck squamous cell carcinoma both in patient-derived xenograft (PDX) models and in a phase II clinical trial.[19,20] The breakthrough of the strategy to inhibit KRAS mutations was driven by the discovery of small molecules that can bind to the acquired cysteine residue in KRAS G12C covalently leading to the clinical development of KRAS G12C inhibitors.[21]
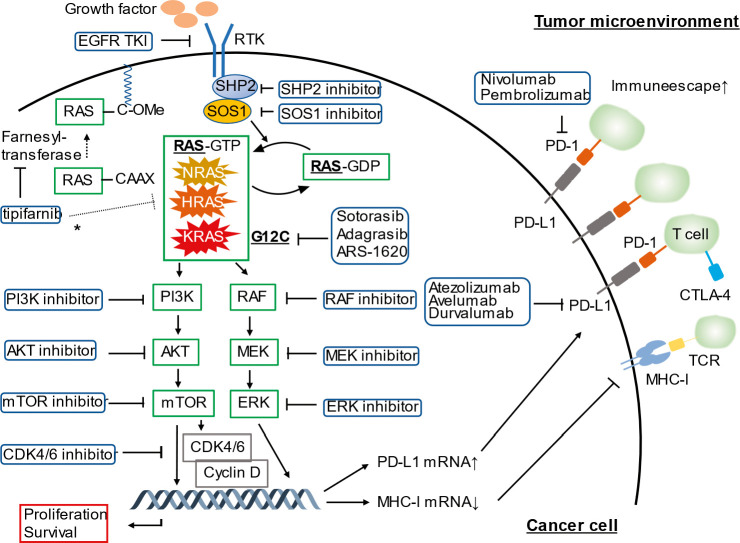
Figure 1.
A schema of the RAS-RAF-MEK-ERK pathway, the immune microenvironment in RAS-mutant cancer, and potential therapeutic strategies targeting RAS-mutant cancer. Oncogenic RAS signaling promotes PD-L1 expression through stabilization of PD-L1 mRNA, leading to immune escape in the tumor microenvironment. The inhibitors of the RAS-RAF-MEK-ERK pathway and the RAS-PI3K-AKT-mTOR pathway are potential agents to improve survival outcomes in patients with RAS mutations. *Tipifarnib is a farnesyltransferase inhibitor and demonstrated encouraging efficacy (objective response rate: 55%) in patients with head and neck squamous cell carcinoma harboring HRAS mutations.
AKT: protein kinase B; CDK: cyclin-dependent kinase; CTLA-4: cytotoxic T-lymphocyte–associated antigen 4; EGFR: epithelial growth factor receptor; ERK: extracellular signal regulated kinase; GDP: guanosine diphosphate; GTP: guanosine triphosphate; HRAS: Harvey rat sarcoma virus oncogene; KRAS: Kirsten rat sarcoma viral oncogene homologue; MEK: mitogen-activated protein kinase; MHC-1: major histocompatibility class I; mRNA: messenger RNA; mTOR: mammalian target of rapamycin; NRAS: neuroblastoma rat sarcoma virus oncogene; PD-1: programmed cell death 1; PD-L1: programmed death-ligand 1; PI3K: phosphatidylinositol 3-kinase; RAF: rapidly accelerated fibrosarcoma; RAS: rat sarcoma virus oncogene; RNA: ribonucleic acid; RTK: receptor tyrosine kinase; SHP2: Src homology 2 domain-containing protein tyrosine phosphatase-2; SOS1: Son of sevenless 1; TCR: T-cell receptor; TKI: tyrosine kinase inhibitor.
Table 1.
The overview of each point KRAS mutation across cancer types
|
Location
|
KRAS
Mutation and Cancer Type |
Others
|
Total
|
|||||||||||||||||||
|
G12
|
G13
|
Q61
|
||||||||||||||||||||
|
12A
|
12C
|
12D
|
12R
|
12S
|
12V
|
Others
|
13A
|
13C
|
13D
|
13R
|
13S
|
13V
|
Others
|
61H
|
61K
|
61L
|
61P
|
61R
|
Others
|
|||
| Anus | 2 | 1 | 6 | 4 | 1 | 14 | ||||||||||||||||
| Anal squamous cell carcinoma | 2 | 1 | 6 | 4 | 1 | 14 | ||||||||||||||||
| Biliary tract | 41 | 60 | 428 | 34 | 72 | 181 | 10 | 7 | 47 | 4 | 3 | 3 | 3 | 20 | 5 | 1 | 6 | 29 | 954 | |||
| Bile duct carcinoma | 38 | 55 | 331 | 24 | 55 | 164 | 10 | 5 | 37 | 2 | 3 | 3 | 2 | 17 | 5 | 1 | 6 | 25 | 783 | |||
| Bile duct carcinoma intraductal papillary neoplasm | 2 | 16 | 2 | 1 | 1 | 2 | 3 | 27 | ||||||||||||||
| Bile duct intraepithelial neoplasia | 13 | 1 | 14 | |||||||||||||||||||
| Gallbladder carcinoma | 1 | 5 | 68 | 10 | 17 | 14 | 1 | 9 | 2 | 1 | 1 | 1 | 130 | |||||||||
| Breast | 9 | 13 | 25 | 10 | 6 | 22 | 1 | 1 | 17 | 2 | 1 | 4 | 41 | 152 | ||||||||
| Carcinoma | 9 | 13 | 25 | 10 | 6 | 22 | 1 | 1 | 17 | 2 | 1 | 4 | 41 | 152 | ||||||||
| Central nervous system | 8 | 1 | 10 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 10 | 40 | |||||||||
| Glioma | 8 | 1 | 10 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 10 | 40 | |||||||||
| Cervix | 5 | 13 | 55 | 3 | 21 | 26 | 18 | 1 | 4 | 2 | 1 | 16 | 165 | |||||||||
| Carcinoma | 5 | 13 | 55 | 3 | 21 | 26 | 18 | 1 | 4 | 2 | 1 | 16 | 165 | |||||||||
| Colorectal | 271 | 420 | 1781 | 54 | 293 | 1116 | 9 | 33 | 1031 | 14 | 2 | 9 | 10 | 59 | 16 | 15 | 1 | 14 | 278 | 5426 | ||
| Anorectal adenocarcinoma | 5 | 1 | 1 | 1 | 2 | 2 | 1 | 13 | ||||||||||||||
| Colon adenocarcinoma | 207 | 326 | 1328 | 38 | 207 | 838 | 3 | 19 | 762 | 7 | 1 | 5 | 5 | 41 | 13 | 6 | 1 | 7 | 201 | 4015 | ||
| Rectal adenocarcinoma | 64 | 94 | 448 | 15 | 85 | 277 | 6 | 14 | 267 | 7 | 1 | 4 | 5 | 16 | 3 | 8 | 7 | 77 | 1398 | |||
| Endometrium | 74 | 59 | 241 | 3 | 20 | 157 | 2 | 12 | 78 | 1 | 2 | 3 | 6 | 4 | 49 | 711 | ||||||
| Carcinoma | 74 | 59 | 241 | 3 | 20 | 157 | 2 | 12 | 78 | 1 | 2 | 3 | 6 | 4 | 49 | 711 | ||||||
| Esophagus | 1 | 3 | 16 | 1 | 3 | 7 | 1 | 10 | 1 | 30 | 73 | |||||||||||
| Adenocarcinoma | 1 | 2 | 8 | 3 | 5 | 1 | 8 | 18 | 46 | |||||||||||||
| Squamous cell carcinoma | 1 | 8 | 1 | 2 | 2 | 1 | 12 | 27 | ||||||||||||||
| Germ cell tumor | 7 | 5 | 10 | 7 | 4 | 31 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 5 | 19 | 96 | ||||||
| Extragonadal | 2 | 1 | 3 | 2 | 8 | 2 | 1 | 5 | 13 | 37 | ||||||||||||
| Testicular | 5 | 4 | 7 | 7 | 2 | 23 | 1 | 1 | 1 | 1 | 1 | 6 | 59 | |||||||||
| Hematopoietic and lymphoid | 77 | 34 | 284 | 33 | 60 | 102 | 4 | 2 | 15 | 288 | 1 | 1 | 9 | 72 | 3 | 6 | 13 | 13 | 1 | 277 | 1295 | |
| Hematopoietic neoplasm | 36 | 15 | 144 | 17 | 32 | 49 | 2 | 1 | 9 | 129 | 7 | 24 | 1 | 5 | 5 | 2 | 107 | 585 | ||||
| Lymphoid neoplasm | 41 | 19 | 140 | 16 | 28 | 53 | 2 | 1 | 6 | 159 | 1 | 1 | 2 | 48 | 2 | 1 | 8 | 11 | 1 | 170 | 710 | |
| Kidney | 5 | 8 | 1 | 7 | 5 | 1 | 1 | 1 | 1 | 2 | 32 | |||||||||||
| Carcinoma | 5 | 8 | 1 | 7 | 5 | 1 | 1 | 1 | 1 | 2 | 32 | |||||||||||
| Liver | 4 | 19 | 4 | 2 | 6 | 1 | 6 | 43 | 85 | |||||||||||||
| Carcinoma | 4 | 19 | 4 | 2 | 6 | 1 | 6 | 43 | 85 | |||||||||||||
| Lung | 550 | 2578 | 1342 | 106 | 206 | 1543 | 42 | 4 | 206 | 212 | 7 | 6 | 7 | 6 | 64 | 13 | 20 | 2 | 11 | 4 | 58 | 6987 |
| Adenocarcinoma | 355 | 1425 | 715 | 48 | 107 | 879 | 28 | 3 | 108 | 129 | 2 | 4 | 4 | 2 | 38 | 5 | 13 | 4 | 38 | 3907 | ||
| Bronchioloalveolar adenocarcinoma | 10 | 42 | 69 | 2 | 3 | 47 | 1 | 3 | 177 | |||||||||||||
| Invasive mucinous adenocarcinoma | 1 | 11 | 22 | 1 | 3 | 26 | 1 | 1 | 66 | |||||||||||||
| Large cell carcinoma | 9 | 52 | 17 | 2 | 5 | 38 | 1 | 5 | 3 | 1 | 8 | 1 | 3 | 1 | 146 | |||||||
| Mixed adenosquamous carcinoma | 1 | 18 | 8 | 3 | 5 | 1 | 7 | 2 | 45 | |||||||||||||
| Neuroendocrine tumor | 2 | 3 | 1 | 2 | 8 | 1 | 4 | 21 | ||||||||||||||
| Non–small cell carcinoma | 149 | 915 | 434 | 44 | 72 | 480 | 10 | 87 | 64 | 4 | 1 | 3 | 4 | 12 | 5 | 3 | 1 | 5 | 1 | 5 | 2299 | |
| Pleomorphic carcinoma | 9 | 16 | 5 | 10 | 1 | 41 | ||||||||||||||||
| Sarcomatoid carcinoma | 3 | 37 | 14 | 1 | 17 | 1 | 2 | 1 | 76 | |||||||||||||
| Squamous cell carcinoma | 11 | 54 | 55 | 7 | 10 | 31 | 1 | 2 | 5 | 1 | 3 | 1 | 1 | 1 | 2 | 3 | 9 | 197 | ||||
| Undifferentiated carcinoma | 2 | 6 | 1 | 2 | 1 | 12 | ||||||||||||||||
| Melanoma | 3 | 4 | 12 | 4 | 8 | 11 | 2 | 1 | 14 | 1 | 1 | 2 | 7 | 1 | 5 | 35 | 111 | |||||
| Malignant melanoma | 3 | 4 | 12 | 4 | 8 | 11 | 2 | 1 | 14 | 1 | 1 | 2 | 7 | 1 | 5 | 35 | 111 | |||||
| Ovary | 40 | 36 | 278 | 35 | 14 | 264 | 4 | 9 | 48 | 1 | 2 | 1 | 9 | 3 | 23 | 767 | ||||||
| Carcinoma | 34 | 25 | 243 | 30 | 14 | 231 | 4 | 7 | 43 | 1 | 2 | 1 | 9 | 3 | 21 | 668 | ||||||
| Others | 6 | 11 | 35 | 5 | 33 | 2 | 5 | 2 | 99 | |||||||||||||
| Pancreas | 82 | 160 | 3097 | 835 | 99 | 2169 | 21 | 3 | 5 | 73 | 5 | 7 | 2 | 4 | 102 | 9 | 15 | 1 | 40 | 2 | 51 | 6782 |
| Ductal carcinoma | 79 | 133 | 2760 | 738 | 85 | 1867 | 16 | 3 | 5 | 56 | 5 | 5 | 2 | 4 | 87 | 9 | 8 | 1 | 26 | 2 | 42 | 5933 |
| Dysplasia-in situ neoplasm | 2 | 17 | 281 | 77 | 12 | 244 | 1 | 17 | 2 | 11 | 6 | 14 | 4 | 688 | ||||||||
| Neuroendocrine tumor | 1 | 3 | 3 | 2 | 4 | 1 | 5 | 19 | ||||||||||||||
| Osteoclast-like giant cell carcinoma | 2 | 8 | 3 | 2 | 7 | 22 | ||||||||||||||||
| Pancreatic intraepithelial neoplasia (PanIN) | 8 | 45 | 14 | 49 | 3 | 1 | 120 | |||||||||||||||
| Prostate | 2 | 10 | 23 | 3 | 3 | 41 | 23 | 3 | 1 | 7 | 4 | 1 | 19 | 140 | ||||||||
| Adenocarcinoma | 2 | 10 | 23 | 3 | 3 | 41 | 23 | 3 | 1 | 7 | 4 | 1 | 19 | 140 | ||||||||
| Salivary gland | 1 | 1 | 1 | 1 | 6 | 10 | ||||||||||||||||
| Carcinoma | 1 | 1 | 1 | 1 | 6 | 10 | ||||||||||||||||
| Skin | 7 | 13 | 1 | 1 | 6 | 1 | 1 | 24 | 54 | |||||||||||||
| Carcinoma | 7 | 13 | 1 | 1 | 6 | 1 | 1 | 24 | 54 | |||||||||||||
| Small intestine | 15 | 13 | 94 | 4 | 11 | 37 | 40 | 1 | 1 | 2 | 1 | 10 | 229 | |||||||||
| Adenocarcinoma | 15 | 13 | 94 | 4 | 11 | 37 | 40 | 1 | 1 | 2 | 1 | 10 | 229 | |||||||||
| Soft tissue | 3 | 15 | 21 | 4 | 1 | 1 | 33 | 3 | 1 | 1 | 2 | 85 | ||||||||||
| Angiosarcoma | 6 | 11 | 1 | 1 | 11 | 2 | 32 | |||||||||||||||
| Leiomyosarcoma | 4 | 2 | 1 | 1 | 1 | 1 | 10 | |||||||||||||||
| Pleomorphic sarcoma | 1 | 2 | 18 | 21 | ||||||||||||||||||
| Rhabdomyosarcoma | 3 | 5 | 7 | 3 | 1 | 1 | 2 | 22 | ||||||||||||||
| Stomach | 13 | 12 | 97 | 8 | 29 | 2 | 61 | 11 | 1 | 1 | 4 | 3 | 2 | 3 | 30 | 277 | ||||||
| Adenocarcinoma | 13 | 12 | 97 | 8 | 29 | 2 | 61 | 11 | 1 | 1 | 4 | 3 | 2 | 3 | 30 | 277 | ||||||
| Thyroid | 4 | 20 | 38 | 30 | 20 | 22 | 1 | 1 | 24 | 2 | 11 | 2 | 8 | 3 | 3 | 40 | 10 | 239 | ||||
| Anaplastic carcinoma | 2 | 12 | 15 | 3 | 9 | 6 | 11 | 4 | 1 | 11 | 1 | 75 | ||||||||||
| Follicular carcinoma | 1 | 3 | 3 | 1 | 3 | 5 | 7 | 1 | 1 | 2 | 10 | 37 | ||||||||||
| Medullary carcinoma | 24 | 2 | 4 | 1 | 1 | 2 | 2 | 2 | 3 | 5 | 46 | |||||||||||
| Mixed papillary and follicular carcinoma | 8 | 4 | 12 | |||||||||||||||||||
| Papillary carcinoma | 1 | 5 | 12 | 2 | 6 | 7 | 1 | 1 | 5 | 2 | 1 | 4 | 1 | 1 | 16 | 4 | 69 | |||||
| Unknown primary | 4 | 14 | 9 | 2 | 12 | 2 | 1 | 5 | 1 | 1 | 3 | 54 | ||||||||||
| Carcinoma | 4 | 14 | 9 | 2 | 12 | 2 | 1 | 5 | 1 | 1 | 3 | 54 | ||||||||||
| Upper aerodigestive tract | 3 | 7 | 31 | 1 | 7 | 2 | 2 | 8 | 1 | 1 | 1 | 1 | 1 | 22 | 88 | |||||||
| Adenocarcinoma | 1 | 19 | 1 | 1 | 1 | 5 | 1 | 29 | ||||||||||||||
| Squamous cell carcinoma | 3 | 6 | 12 | 1 | 6 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 22 | 59 | ||||||||
| Urinary tract | 10 | 20 | 44 | 11 | 7 | 32 | 13 | 1 | 3 | 2 | 3 | 21 | 167 | |||||||||
| Adenocarcinoma | 1 | 7 | 2 | 1 | 8 | 7 | 1 | 5 | 32 | |||||||||||||
| Carcinoma, unclassified | 2 | 7 | 11 | 3 | 6 | 2 | 1 | 1 | 1 | 5 | 39 | |||||||||||
| Transitional cell carcinoma | 7 | 13 | 26 | 6 | 6 | 18 | 4 | 1 | 2 | 1 | 1 | 11 | 96 | |||||||||
| Total | 1224 | 3514 | 7983 | 1179 | 865 | 5830 | 92 | 16 | 299 | 2058 | 35 | 52 | 30 | 42 | 362 | 66 | 98 | 26 | 137 | 16 | 1109 | 25,033 |
Data were extracted from the COSMIC database (version 95 released Mar 31, 2022). The number of tumors with a point mutation on the KRAS isoform is indicated in the table. Subtypes of tumors with the number of fewer than 10, benign tumors, and tumors with undetermined primary sites and unclear histology were removed from this analysis.
Bold values are the totals by location, mutation, and cancer type.
KRAS: Kirsten rat sarcoma viral oncogene homologue.
UNIQUE ROLE OF KRAS G12C MUTATION IN CANCER
Among patients with KRAS-mutant cancer, the prevalence of KRAS G12C mutation is higher in patients with non–small-cell lung cell carcinoma (NSCLC), accounts for approximately 25–46% of all KRAS mutations in NSCLC.[9,12,22,23] Therefore, clinical and molecular characteristics of KRAS G12C mutations were mainly derived from NSCLC. Among KRAS G12 mutations, G12C is frequently seen in former or current smokers than never smokers and is relatively more frequent in women than in men. In contrast, G12D mutation is the most common mutation in never smokers among patients with NSCLC with the KRAS mutation.[24] A recent analysis of a large series of patients with metastatic KRAS-mutant NSCLC revealed that G12C had a higher tumor mutation burden (TMB) and programmed cell death-ligand 1(PD-L1) expression, although overall survival from diagnosis was similar for G12C and non-G12C mutations.[23] In addition, in early-stage lung cancer, G12C mutation is also associated with an increase in higher TMB and recurrence of early-stage lung adenocarcinoma after resection.[25] On the other hand, lower TMB is accompanied by G12D mutation, suggesting different immunogenicity in each KRAS G12 mutation.[26] In NSCLC, TMB could be a potential biomarker to predict clinical benefit with anti-PD-L1 inhibitor, or atezolizumab, and therefore, the KRAS mutational status might become a predictor for immune checkpoint blockade therapy.[27]
Another cancer type that commonly accompanies KRAS mutation is colorectal cancer, and KRAS G12C mutation occurs in approximately 8% of metastatic colorectal cancers with KRAS mutation.[28] A recent comprehensive study[29] analyzed patients with colorectal cancer molecularly and clinically elucidated that the progression-free survival of patients with KRAS G12C mutation was poorer than those with KRAS non-G12C mutation, suggesting innate resistance to chemotherapy for this subpopulation in colorectal cancer. However, a robust description of clinical features of G12C mutation not only in colorectal cancer but also in other cancer types remains scarce, and further understanding of pathophysiological features, clinical characteristics such as responsiveness to systemic therapy, and prognosis of KRAS G12C mutations across a variety of cancer types, is needed.
CLINICAL ACTIVITY OF KRAS G12C INHIBITION IN CANCER
After the discovery of the small molecules covalently binding to the cysteine residue of the KRAS G12C mutation, several molecules inhibiting this mutation have been developed and investigated for their clinical applications preclinically and clinically. The first agent was ARS-1620, which selectively induced tumor regression in patient-derived tumor xenografts with KRAS G12C mutation in vivo.[30] Although ARS-1620 had little potency to continuously inhibit KRAS G12C because of the small size of the switch II pocket in KRAS G12C protein, it has been served as an important tool to investigate the potential therapeutic efficacy of the KRAS G12C inhibitor.[31] The successful clinical development of KRAS G12C inhibition was achieved by sotorasib (AMG 510) and adagrasib (MRTX849), with results of the recent phase 1 and 2 trials mainly for patients with NSCLC and colorectal cancer.[32–35] Sotorasib was the first agent that showed promising clinical efficacy in patients with advanced solid tumors with KRAS G12C through the phase 1 clinical trial. In this study, 129 patients were enrolled, among which 59 were with NSCLC, 42 with colorectal cancer, and 28 with other tumors. Approximately 32% of patients with NSCLC had an objective response and 88% had disease control. On the other hand, among patients with colorectal cancer, only 7% had a confirmed response, and 74% achieved disease control.[32] This disconcordance of response rate (RR) suggests different mechanisms of primary resistance to sotorasib among cancer types. Responses were also seen in other types of tumors, such as pancreatic and endometrial cancers, and malignant melanoma. The efficacy and safety of sotorasib for lung cancers were confirmed through the subsequent phase 2 trial. This study enrolled 126 patients with NSCLC among which most (81.0%) were previously treated with systemic chemotherapy or immune checkpoint inhibitors (ICIs), and demonstrated 37.1% of objective RR with 11.1 months of the median duration of response, leading to the U.S. Food and Drug Administration approval of sotorasib for patients with NSCLC harboring KRAS G12C mutations.[33] Adagrasib, another KRAS G12C inhibitor, initially demonstrated suppression of the downstream MAPK pathway and tumor regression across multiple tumor types in cell lines and PDX models.[36] Subsequently, preliminary results from the KRYSTAL-1 trial showed promising efficacy of adagrasib mainly in patients with NSCLC (RR, 45%; disease control rate [DCR], 96%) and colorectal cancer (RR, 22%; DCR, 87%).[34,35] For heavily pretreated KRAS G12C-mutated advanced pancreatic cancer, encouraging clinical activity was demonstrated by sotorasib (RR, 21% [n = 8 of 38]; DCR, 84% [32 of 38]), and adagrasib (RR, 50% [5/10]; DCR, 100% [10/10]), respectively.[37,38] Sotorasib monotherapy also demonstrated meaningful clinical activity in other gastrointestinal tumors, such as biliary, appendiceal, and gastro-esophageal junction cancer (RR, 35% [6/17]; DCR; 100% [17/17]).[38]
Although these KRAS G12C inhibitors demonstrated meaningful clinical efficacy for KRAS G12C-mutated cancer, most patients with a response eventually became refractory after initiation of these treatments.[33] The mechanisms of primary and acquired resistance to these KRAS G12C inhibitors have been gradually explored. Skoulidis et al[39], reported sotorasib had similar RR among patients with KRAS G12C NSCLC with STK11 comutation (RR with STK11 comutation, 40.0%; without STK11 comutation, 39.1%). Considering STK11 mutation is associated with poor clinical outcome in general, sotorasib for KRAS G12C/STK11 mutations may be an ideal option. In contrast, comutation with KEAP1 was found to have lower RR (RR with KEAP1 comutation, 20.0%; without KEAP1 comutation, 44.0%).[39] Similarly, the KRYSTAL-1 trial also showed favorable RR in patients with NSCLC harboring STK11 mutation but lower RR in those with KEAP1 mutation.[34] These results suggest certain co-genomic alterations are associated with better or worse outcomes for patients treated with KRAS G12C mutation. Several trials combining KRAS G12C inhibitors with other therapy, such as chemotherapy, molecular-targeted therapy including EGFR inhibitors and cyclin-dependent kinase (CKD) 4/6 inhibitors, and ICIs, are ongoing to overcome the resistance and enhance the therapeutic efficacy. Several possible mechanisms of resistance to inhibition of KRAS G12C have been proposed recently in preclinical and clinical studies. Activation of bypass MAPK downstream pathway, epithelial-to-mesenchymal transition, activated proliferative signaling in cancer cells, and diminished antitumor immunity, were suggested as mechanisms of resistance to KRAS G12C inhibition, and a better understanding of resistance mechanisms is needed to explore promising treatment options to overcome resistance to inhibition of KRAS G12C.[40]
MECHANISM OF RESISTANCE TO ICIs IN KRAS-MUTANT LUNG CANCER
Recent advances in immunotherapy, including ICIs and adoptive cell therapies (ACT) such as chimeric antigen receptor therapy and T-cell receptor (TCR) therapy, have revolutionized the paradigm of cancer treatments, and along with the rapid progress of these treatments for a variety of types of cancer, the mechanisms of resistance and strategies to overcome resistance to immunotherapy have also been elucidated. The cytotoxic T-lymphocyte–associated protein 4 (CTLA-4) inhibitor and the programmed cell death 1 (PD-1) and PD-L1 inhibitors were successfully developed and incorporated into the clinical setting, and the mechanisms of resistance have been revealed through both preclinical and clinical studies using these drugs. Resistance mechanisms are primarily categorized into innate and extrinsic resistance; and several mechanisms were reported, such as lack of neoantigen expression, activated or altered cell signaling pathways, an increase in immunosuppressive cells including regulatory T cells and myeloid-derived suppressor cells, activation in epithelial-mesenchymal transition, angiogenesis, and gut microbiome changes.[41–44] Among cancers harboring KRAS mutations, NSCLC was the main tumor type that the strategy with the use of ICI became successful.[45–48] Therefore, mechanisms of resistance to ICIs in KRAS-mutant cancer have been gradually reported through analyses of patients with NSCLC treated with ICI-containing regimens. Several studies revealed that patients with NSCLC or gastrointestinal tumors harboring KRAS mutation respond better to ICIs than those with the wild type or other oncogenic driver mutations, including EGFR and ALK mutations. Indeed, mutations in the RAS-MAPK pathway and TP53 were reported as potential positive predictors of the efficacy of the checkpoint blockade strategy.[49,50] However, most of this proportion shows primary or acquired resistance to checkpoint blockade, and therefore, the risk stratification and identification of promising therapeutic options are needed.[49,51,52] A study that evaluated KRAS-mutant lung adenocarcinoma treated with PD-1 axis inhibitors revealed that STK11/LKB1 alterations are one of the major mechanisms of primary resistance when patients are categorized into three subgroups: STK11/KLB1 comutations, TP53 comutations, and KRAS mutation alone.[53–55] STK11/LKB1 deficiency was also reported to be associated with the accumulation of neutrophils with T-cell suppression, T-cell exhaustion around the tumor microenvironment, and reduced PD-L1 expressions on the surface of tumor cells.[56] Loss of LKB1 also results in suppression of stimulator of interferon genes (STING) expression that usually has a pivotal role in antigen presentation mainly by regulating dendric cells, and in priming of CD8+ T cells.[57] However, recent data showed STK11/LKB1 alterations are not specific as a negative predictor for response to ICI, rather could be a poor prognostic biomarker across different therapies.[58] Overactivation of the RAS/RAF/MAPK pathway contributes to resistance to ICIs through its inhibitory effect on T-cell recruitment and function by producing the vascular endothelial growth factor and other immunosuppressive cytokines, and through the reduction of major histocompatibility complex (MHC) class I expression on tumor cells.[59–61] Oncogenic RAS signaling also promotes tumor immunoresistance and regulates cell-intrinsic PD-L1 expression by stabilizing PD-L1 messenger RNA (mRNA).[62] The PD-L1 expression through activation of the RAS pathway also mediates immune escape in cell models and human tissues of NSCLC.[63] Therefore, inhibition of the RAS signaling pathway might be beneficial to reverse the immunosuppressive tumor microenvironment, leading to improvement of susceptibility to ICIs in KRAS-mutant cancer (Fig. 1).
RATIONALES FOR THE COMBINATION THERAPY OF KRAS G12C INHIBITION WITH ICI
The combination strategies using ICI and molecular-targeted therapy achieved successful development in certain cancer types such as renal cell carcinoma, but ICI plus agents targeting the MAPK pathway have mixed clinical outcomes so far.[64–67] Generally, NSCLC with KRAS mutation responds to ICI monotherapy or ICI with chemotherapy well compared with other mutation types, but most of them become refractory afterward.[68,69] Positive predictive factors in KRAS mutation are high TMB and PD-L1 expression, which may lead to a better response to the ICI treatment. On the other hand, KRAS-mutant cancer also creates immunosuppressive conditions around the tumor microenvironment, as discussed previously, resulting in a poorer response to immunotherapy. To overcome the therapeutic limitations in this context, several rationales for combining checkpoint blockade with KRAS inhibition to augment the efficacy of therapy for this population were proposed through analysis of preclinical models and clinical trials. One important study using sotorasib (AMG 510) revealed that sotorasib not only led to the regression of KRAS G12C tumors but also created a proinflammatory tumor microenvironment resulting in a durable response in mouse models.[70] This finding supports that KRAS G12C inhibitor can reverse the immunosuppressive environment, making cancer cells susceptive to ICIs or other types of immunotherapies. Another pivotal study reported that adagrasib (MRTX 849) increased MHC class I protein expression and decreased immunosuppressive factors. In a mouse model with KRAS G12C mutation, adagrasib increased M1-type tumor-associated macrophages, dendritic cells, and infiltrative T cells, and decreased myeloid-derived suppressor cells. Of note, the combination of adagrasib with ICI was effective even after the tumor cells showed progression with either ICI therapy or adagrasib monotherapy.[71] These findings suggest that KRAS inhibition can potentially make tumor cells more susceptible to ICI therapy by producing an inflammatory tumor microenvironment. Currently, several clinical trials are ongoing to evaluate the combination strategy using KRAS G12C inhibitors with ICIs and are awaiting clinical outcomes (Table 2).
Table 2.
Ongoing clinical trials evaluating the combination of KRAS inhibitors with immune checkpoint inhibitors
|
KRAS
G12C Inhibitor |
Immune Checkpoint Inhibitor
|
Phase
|
Cancer Type
|
Study Description
|
ClinicalTrials.gov Identifer
|
| Sotorasib (AMG 510) | Pembrolizumab (PD-1 inhibitor) | I/II | Advanced solid tumors with KRAS G12C mutation | Sotorasib activity in subjects with advanced solid tumors with KRAS p.G12C mutation (CodeBreak 101) | NCT04185883 |
| Sotorasib (AMG 510) | Anti-PD-1/PD-L1 inhibitors | I/II | Advanced solid tumors with KRAS G12C mutation | A Phase 1/2 study evaluating the safety, tolerability, PK, and efficacy of AMG 510 in subjects with solid tumors with a specific KRAS Mutation (CodeBreaK 100) | NCT03600883 |
| Adagrasib (MRTX849) | Pembrolizumab (PD-1 inhibitor) | II | NSCLC with KRAS G12C mutation | Phase 2 trial of MRTX849 plus pembrolizumab for NSCLC with KRAS G12C mutation KRYSTAL-7 | NCT04613596 |
| Adagrasib (MRTX849) | Pembrolizumab (PD-1 inhibitor) | I/II | Advanced malignancy with KRAS G12C mutation | Phase 1/2 study of MRTX849 in patients with cancer having a KRAS G12C mutation KRYSTAL-1 | NCT03785249 |
| TNO155 (SHP2 inhibitor) | Spartalizumab (PD-1 inhibitor) | Ib | Selected malignancy including KRAS G12C-mutant NSCLC | Phase Ib study of TNO155 in combination with spartalizumab or ribociclib in selected malignancies | NCT04000529 |
KRAS: Kirsten rat sarcoma viral oncogene; NSCLC: non-small-cell lung cancer; PD-1: programmed cell death 1.
To date, the strategies combining ICIs and tyrosine kinase inhibitors (TKIs) targeting the RAS/RAF/MAPK pathway were mainly evaluated in trials for patients with EGFR-mutant NSCLC and BRAF-mutant malignant melanoma. A few studies that evaluated the safety and efficacy of inhibition of BRAF and MEK in addition to ICI revealed that this combination strategy produced a tendency of longer survival in patients with malignant melanoma but led to a significant increase in the incidence of grade 3 or more adverse events.[72–74] Among patients with NSCLC harboring EGFR mutation, EGFR TKIs with immune checkpoint blockade were evaluated in the first or second and beyond line settings. However, the combination of EGFR TKIs and ICIs led to a higher incidence of hepatotoxicity, interstitial lung disease, or pneumonitis with little survival benefit.[75–78] These results suggested difficulty in the development of the combination treatment using RAS/RAF/MAPK pathway inhibitors with immune checkpoint blockade. Tactics targeting KRAS G12C with ICIs could be a breakthrough to overcome these barriers, and ongoing trials are awaiting further safety and efficacy analysis.
Another potential strategy to enhance the efficacy of KRAS inhibitors is the use of an Src homology-2 domain-containing protein tyrosine phosphatase-2 (SHP2) inhibitor. SHP2 is a protein that mediates RAS activation downstream of receptor tyrosine kinase (RTK) and controls downstream signaling of PD-1 of T cells. SHP2 is required for the progression of KRAS-mutant NSCLC and thus, inhibition of SHP2 would be an ideal strategy to restore the sensitivity of KRAS-mutant NSCLC to MEK inhibition.[79,80] Indeed, the combination of SHP2 inhibitors and ARS-1620 (KRAS G12C inhibitor) was shown to be associated with a decrease in GTP-bound KRAS G12C activation, suppression of RTK-mediated MAPK reactivation, AKT and ERK pathways, and an increase in T-cell infiltration.[79–83] Several early-phase trials have just started recently to evaluate the efficacy and safety of SHP2 inhibitors in combination with ICIs such as spartalizumab (PD-1 inhibitor), cyclin-dependent kinase (CDK) 4/6 inhibitors, EGFR inhibitors, or ERK inhibitors (ClinicalTrials.gov Identifiers NCT04000529, NCT04330664, NCT04699188, NCT04670679, NCT04916236, and NCT03114319).
ADAPTIVE T-CELL THERAPY FOR KRAS-MUTANT CANCER
Along with the combination strategy using ICI and KRAS G12C inhibitors, several other strategies such as ACT and vaccine therapy are under investigation through early-phase clinical trials. ACT, including tumor-infiltrating lymphocyte (TIL) therapy, engineered TCR therapy, chimeric antigen receptor T-cell therapy, and natural killer cell therapy, has been recently developed to target small molecules such as tumor-specific neoantigens and clonally expressed molecules on tumor cells, leading to approval for the treatment of malignant lymphoma and multiple myeloma. In KRAS-mutant cancer, successful development of ACT was first identified in a metastatic colorectal cancer case harboring KRAS G12D mutation with an expression of HLA-C*08:02 treated with TIL.[84] HLA-A*11:01 in KRAS G12V and G12D mutations were also found as potential therapeutic targets, and two trials using a murine TCR recognizing these molecules were started but suspended (ClinicalTrials.gov Identifiers NCT03190941, and NCT03745326).[85] Other studies also showed potential targetable molecules such as KRAS G12V/HLA-A*0201 complex and KRAS-G12D neoantigens restricted by HLA-C*08:02 by engineering TCR-mimic antibody-drug conjugates and TIL, respectively.[86–88] These findings paved the way to target other KRAS mutations rather than G12C. A tactic using mRNA vaccine encoding KRAS mutations was also found to induce CD-8 T-cell responses to KRAS tumor antigens in a preclinical study, and a phase I trial evaluating mRNA-5671 (V941) vaccine as monotherapy or in combination with pembrolizumab for patients with NSCLC and colorectal cancer harboring KRAS mutations is ongoing (ClinicalTrials.gov Identifier NCT03948763) (Table 3).
Table 3.
Ongoing clinical trials evaluating adoptive cell therapy and vaccine therapy for KRAS-mutant malignancy
|
Agents
|
Other Agents
|
Phase
|
Cancer Type
|
Study Description
|
ClinicalTrials.gov Identifier
|
| G12V-specific TCR transduced T-cell therapy | • Cyclophosphamide and fludarabine before infusion • Anti-PD-1 inhibitor if needed |
I/II | Advanced pancreatic cancer with KRAS G12V mutation and HLA-A*11:01 allele | Mutant KRAS G12V-specific TCR transduced T Cell therapy for advanced pancreatic cancer | NCT04146298 |
| Anti-KRAS G12V murine TCR | • Cyclophosphamide and fludarabine before infusion • Aldesleukin (high-dose interleukin-2) |
I/II | Advanced cancer harboring KRAS G12V mutation | Administering peripheral blood lymphocytes transduced with a murine t-cell receptor recognizing the G12V variant of mutated RAS in HLA-A*11:01 patients | NCT03190941 |
| Anti-KRAS G12D murine TCR | • Cyclophosphamide and fludarabine before infusion • Aldesleukin (high-dose interleukin-2) |
I/II | Advanced cancer harboring KRAS G12D mutation | Administering peripheral blood lymphocytes transduced with a murine t-cell receptor recognizing the G12D variant of mutated RAS in HLA-A*11:01 patients | NCT03745326 |
| mRNA-5671 vaccine (V941) | • Monotherapy or with pembrolizumab (PD-1 inhibitor) | I | NSCLC, Pancreatic cancer, Colorectal Cancer with KRAS (G12D, G12V, G13D, or G12C) mutation | A study of mRNA-5671/V941 as monotherapy and in combination with pembrolizumab (V941–001) | NCT03948763 |
HLA: human leukocyte antigen; KRAS: Kirsten rat sarcoma viral oncogene homologue; NSCLC: non–small cell lung cancer; PD-1: programmed cell death 1; PD-L1: programmed cell death-ligand 1; PK: pharmacokinetics; TCR: T-cell receptor
CONCLUSION
The successful clinical development of KRAS G12C inhibitor broadened treatment options for NSCLC and will possibly expand the therapeutic possibilities for other cancer types harboring KRAS G12C mutation. At the same time, the fact that many patients face primary or acquired resistance to this targeted therapy requires more effective strategies to augment therapeutic efficacy. This comprehensive review focusing on KRAS inhibitor and its association with the tumor immune microenvironment summarized potential strategies using the combination of KRAS inhibitor with ICIs, and other immunotherapies to overcome resistance to KRAS G12C inhibitor. In addition, there is an unmet need for patients with other KRAS mutations such as G12D and G12V, and further studies are necessary to bring treatment options for this population. Clinical trials using immunotherapy along with KRAS inhibition have just begun, and therefore, more investigations to evaluate clinical outcomes, reveal prognostic factors, and discover further mechanisms of resistance to these treatments, are necessary to achieve a longer duration of response and potential cure for patients with KRAS-mutant cancer.
Funding Statement
Source of Support: None.
Footnotes
Conflict of Interest: Shumei Kato serves as a consultant for Medpace, Foundation Medicine, NeoGenomics, and CureMatch; serves on the advisory board for Pfizer; receives speaker's fees from Roche and Bayer and research funding from ACT Genomics, Sysmex, Konica Minolta, OmniSeq, and Personalis. David S. Hong receives research or grant funding from AbbVie, Adaptimmune, Adlai Nortye, Amgen, AstraZeneca, Bayer, Bristol Myers Squibb, Daiichi-Sankyo, Eisai, Eli Lilly, EMD Serono, Erasca, Fate Therapeutics, Genentech, Genmab, GlaxoSmithKline, Ignyta, Infinity, Kite, Kyowa, LOXO, Merck, MedImmune, Millenium, Mirati, miRNA, Molecular Templates, Mologen, Navier, NCI-CTEP, Novartis, Numab, Pfizer, Seattle Genetics, Takeda, Turning Point Therapeutics, Verstatem, and VM Oncology; travel support from AACR, Amgen, ASCO, AstraZeneca, Bayer, Celgene, Eli Lilly, Genentech, Genmab, GlaxoSmithKline, Janssen, LOXO, miRNA, Pfizer, Philips, SITC, and Takeda Consulting; serves in a speaker or advisory role for Alpha Insights, Acuta, Amgen, Axiom, Adaptimmune, Baxter, Bayer, Boxer Capital, COG, Ecor1, Genentech, GLG, Group H, Guidepoint, HCW Precision, Infinity, Janssen, Merrimack, Medscape, Numab, Pfizer, Prime Oncology, Seattle Genetics, ST Cube, Takeda, Tavistock, Trieza Therapeutics, and WebMD; and has other ownership interests in Molecular Match (advisor), OncoResponse (founder), and Presagia Inc (advisor). Yu Fujiwara has no disclosures.
References
- 1.Hobbs GA, Der CJ, Rossman KL. RAS isoforms and mutations in cancer at a glance. J Cell Sci . 2016;129:1287–1292. doi: 10.1242/jcs.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res . 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasan N, Boyer JL, Herbst RS. A. RAS renaissance: emerging targeted therapies for KRAS-mutated non-small cell lung cancer. Clin Cancer Res . 2014;20:3921–3930. doi: 10.1158/1078-0432.CCR-13-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer . 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 5.Larkin J, Ascierto PA, Dréno B, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med . 2014;371:1867–1876. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 6.Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med . 2017;376:629–640. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato S, Porter R, Okamura R, et al. Functional measurement of mitogen-activated protein kinase pathway activation predicts responsiveness of RAS-mutant cancers to MEK inhibitors. Eur J Cancer . 2021;149:184–192. doi: 10.1016/j.ejca.2021.01.055. [DOI] [PubMed] [Google Scholar]
- 8.Kato S, McFall T, Takahashi K, et al. KRAS-mutated, estrogen receptor-positive low-grade serous ovarian cancer: unraveling an exceptional response mystery. Oncologist . 2021;26:e530–e536. doi: 10.1002/onco.13702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore AR, Rosenberg SC, McCormick F, Malek S. RAStargeted therapies. Nat Rev Drug Discov . 2021. Published online May 14. [DOI] [PubMed]
- 10.Kato S, Okamura R, Sicklick JK, et al. Prognostic implications of RAS alterations in diverse malignancies and impact of targeted therapies. Int J Cancer . 2020;146:3450–3460. doi: 10.1002/ijc.32813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature . 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet . 2016;48:607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tate JG, Bamford S, Jubb HC, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res . 2018;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goody RS, Frech M, Wittinghofer A. Affinity of guanine nucleotide binding proteins for their ligands: facts and artefacts. Trends Biochem Sci . 1991;16:327–328. doi: 10.1016/0968-0004(91)90134-h. [DOI] [PubMed] [Google Scholar]
- 15.Ryan MB, Corcoran RB. Therapeutic strategies to target RAS-mutant cancers. Nat Rev Clin Oncol . 2018;15:709–720. doi: 10.1038/s41571-018-0105-0. [DOI] [PubMed] [Google Scholar]
- 16.Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cell Biol . 2011;13:39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adjei AA, Mauer A, Bruzek L, et al. Phase II study of the farnesyl transferase inhibitor R115777 in patients with advanced non-small-cell lung cancer. J Clin Oncol . 2003;21:1760–1766. doi: 10.1200/JCO.2003.09.075. [DOI] [PubMed] [Google Scholar]
- 18.Heymach JV, Johnson DH, Khuri FR, et al. Phase II study of the farnesyl transferase inhibitor R115777 in patients with sensitive relapse small-cell lung cancer. Ann Oncol . 2004;15:1187–1193. doi: 10.1093/annonc/mdh315. [DOI] [PubMed] [Google Scholar]
- 19.Gilardi M, Wang Z, Proietto M, et al. Tipifarnib as a precision therapy for HRASmutant head and neck squamous cell carcinomas. Mol Cancer Ther . 2020;19:1784. doi: 10.1158/1535-7163.MCT-19-0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho AL, Brana I, Haddad R, et al. Tipifarnib in head and neck squamous cell carcinoma with HRAS mutations. J Clin Oncol . 2021;39:1856–1864. doi: 10.1200/JCO.20.02903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ostrem JM, Peters U, Sos ML, et al. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature . 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arbour KC, Jordan E, Kim HR, et al. Effects of co-occurring genomic alterations on outcomes in patients with KRAS-mutant non-small cell lung cancer. Clin Cancer Res . 2018;24:334–340. doi: 10.1158/1078-0432.CCR-17-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arbour KC, Rizvi H, Plodkowski AJ, et al. Treatment outcomes and clinical characteristics of patients with KRAS-G12C-mutant non-small cell lung cancer. Clin Cancer Res . 2021;27:2209–2215. doi: 10.1158/1078-0432.CCR-20-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dogan S, Shen R, Ang DC, et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res . 2012;18:6169–6177. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones GD, Caso R, Tan KS, et al. KRAS (G12C) mutation is associated with increased risk of recurrence in surgically resected lung adenocarcinoma. Clin Cancer Res . 2021;27:2604–2612. doi: 10.1158/1078-0432.CCR-20-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao G, Liao W, Ma Q, et al. KRAS G12D mutation predicts lower TMB and drives immune suppression in lung adenocarcinoma. Lung Cancer . 2020;149:41–45. doi: 10.1016/j.lungcan.2020.09.004. [DOI] [PubMed] [Google Scholar]
- 27.Gandara DR, Paul SM, Kowanetz M, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med . 2018;24:1441–1448. doi: 10.1038/s41591-018-0134-3. [DOI] [PubMed] [Google Scholar]
- 28.Patelli G, Tosi F, Amatu A, et al. Strategies to tackle RAS-mutated metastatic colorectal cancer. ESMO Open . 2021;6:100156. doi: 10.1016/j.esmoop.2021.100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henry JT, Coker O, Chowdhury S, et al. Comprehensive clinical and molecular characterization of KRAS (G12C)mutant colorectal cancer. JCO Precis Oncol . 2021;5 doi: 10.1200/PO.20.00256. PO.20.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janes MR, Zhang J, Li LS, et al. Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell . 2018;172:578–589.e17. doi: 10.1016/j.cell.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Nagasaka M, Li Y, Sukari A, et al. KRAS G12C Game of Thrones, which direct KRAS inhibitor will claim the iron throne? Cancer Treat Rev . 2020;84:101974. doi: 10.1016/j.ctrv.2020.101974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong DS, Fakih MG, Strickler JH, et al. KRAS(G12C) inhibition with sotorasib in advanced solid tumors. N Engl J Med . 2020;383:1207–1217. doi: 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skoulidis F, Li BT, Dy GK, et al. Sotorasib for lung cancers with KRAS p.G12C mutation. N Engl J Med . 2021;384:2371–2381. doi: 10.1056/NEJMoa2103695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jänne PA, Rybkin II, Spira AI, et al. KRYSTAL-1: activity and safety of adagrasib (MRTX849) in advanced/ metastatic non-small-cell lung cancer (NSCLC) harboring KRAS G12C mutation. Eur J Cancer . 2020;138:S1–S2. [Google Scholar]
- 35.Johnson ML, Ou SHI, Barve M, et al. KRYSTAL-1: activity and safety of adagrasib (MRTX849) in patients with colorectal cancer (CRC) and other solid tumors harboring a KRAS G12C mutation. Eur J Cancer . 2020;138:S2. [Google Scholar]
- 36.Hallin J, Engstrom LD, Hargis L, et al. The KRAS(G12C) inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov . 2020;10:54–71. doi: 10.1158/2159-8290.CD-19-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strickler JH, Satake H, Hollebecque A, et al. First data for sotorasib in patients with pancreatic cancer with KRAS p.G12C mutation: a phase I/II study evaluating efficacy and safety. J Clin Oncol . 2022;40:360490. [Google Scholar]
- 38.Bekaii-Saab TS, Spira AI, Yaeger R, et al. KRYSTAL-1: updated activity and safety of adagrasib (MRTX849) in patients (Pts) with unresectable or metastatic pancreatic cancer (PDAC) and other gastrointestinal (GI) tumors harboring a KRASG12C mutation. J Clin Oncol . 2022;40:519. [Google Scholar]
- 39.Skoulidis F, Li BT, Govindan R, et al. Overall survival and exploratory subgroup analyses from the phase 2 CodeBreaK 100 trial evaluating sotorasib in pretreated KRAS p.G12C mutated non-small cell lung cancer. J Clin Oncol . 2021;39:9003. [Google Scholar]
- 40.Akhave NS, Biter AB, Hong DS. Mechanisms of resistance to KRAS(G12C)-targeted therapy. Cancer Discov . 2021;11:1345–1352. doi: 10.1158/2159-8290.CD-20-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pitt JM, Vétizou M, Daillère R, et al. Resistance mechanisms to immune-checkpoint blockade in cancer: tumor-intrinsic and -extrinsic factors. Immunity . 2016;44:1255–1269. doi: 10.1016/j.immuni.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 42.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell . 2017;168:707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fares CM, Van Allen EM, Drake CG, et al. Mechanisms of resistance to immune checkpoint blockade: why does checkpoint inhibitor immunotherapy not work for all patients? Am Soc Clin Oncol Educ Book . 2019;39:147–164. doi: 10.1200/EDBK_240837. [DOI] [PubMed] [Google Scholar]
- 44.Fujiwara Y, Mittra A, Naqash AR, Takebe N. A review of mechanisms of resistance to immune checkpoint inhibitors and potential strategies for therapy. Cancer Drug Resist . 2020;3:252–275. doi: 10.20517/cdr.2020.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hellmann MD, Paz-Ares L. Bernabe Caro R, et al., editors. Nivolumab plus ipilimumab in advanced non-small-cell lung cancer. N Engl J Med . 2019;381:2020–2031. doi: 10.1056/NEJMoa1910231. [DOI] [PubMed] [Google Scholar]
- 46.Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med . 2015;373:1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gandhi L, Rodríguez-Abreu D, Gadgeel S, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med . 2018;378:2078–2092. doi: 10.1056/NEJMoa1801005. [DOI] [PubMed] [Google Scholar]
- 48.Socinski MA, Jotte RM, Cappuzzo F, et al. Atezolizumab for first-line treatment of metastatic nonsquamous NSCLC. N Engl J Med . 2018;378:2288–2301. doi: 10.1056/NEJMoa1716948. [DOI] [PubMed] [Google Scholar]
- 49.Wang JY, Xiu J, Baca Y, et al. Distinct genomic landscapes of gastroesophageal adenocarcinoma depending on PD-L1 expression identify mutations in RAS-MAPK pathway and TP53 as potential predictors of immunotherapy efficacy. Ann Oncol . 2021;32:906–916. doi: 10.1016/j.annonc.2021.03.203. [DOI] [PubMed] [Google Scholar]
- 50.Dong ZY, Zhong WZ, Zhang XC, et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res . 2017;23:3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 51.Mazieres J, Drilon A, Lusque A, et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann Oncol . 2019;30:1321–1328. doi: 10.1093/annonc/mdz167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gainor JF, Shaw AT, Sequist LV, et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer: a retrospective analysis. Clin Cancer Res . 2016;22:4585–4593. doi: 10.1158/1078-0432.CCR-15-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skoulidis F, Byers LA, Diao L, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov . 2015;5:860–877. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov . 2018;8:822–835. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pore N, Wu S, Standifer N, et al. Resistance to durvalumab and durvalumab plus tremelimumab is associated with functional STK11 mutations in non-small-cell lung cancer patients and is reversed by STAT3 knockdown. Cancer Discov . 2021;11:2828–2845. doi: 10.1158/2159-8290.CD-20-1543. [DOI] [PubMed] [Google Scholar]
- 56.Koyama S, Akbay EA, Li YY, et al. STK11/LKB1 deficiency promotes neutrophil recruitment and proinflammatory cytokine production to suppress T-cell activity in the lung tumor microenvironment. Cancer Res . 2016;76:999–1008. doi: 10.1158/0008-5472.CAN-15-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kitajima S, Ivanova E, Guo S, et al. Suppression of STING associated with LKB1 loss in KRAS-driven lung cancer. Cancer Discov . 2019;9:34–45. doi: 10.1158/2159-8290.CD-18-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papillon-Cavanagh S, Doshi P, Dobrin R, et al. STK11 and KEAP1 mutations as prognostic biomarkers in an observational real-world lung adenocarcinoma cohort. ESMO Open . 2020;5:e000706. doi: 10.1136/esmoopen-2020-000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liao W, Overman MJ, Boutin AT, et al. KRAS-IRF2 axis drives immune suppression and immune therapy resistance in colorectal cancer. Cancer Cell . 2019;35:559–572.e7. doi: 10.1016/j.ccell.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okada F, Rak JW, Croix BS, et al. Impact of oncogenes in tumor angiogenesis: mutant K-ras up-regulation of vascular endothelial growth factor/vascular permeability factor is necessary, but not sufficient for tumorigenicity of human colorectal carcinoma cells. Proc Natl Acad Sci U S A . 1998;95:3609–3614. doi: 10.1073/pnas.95.7.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Loi S, Dushyanthen S, Beavis PA, et al. RAS/MAPK activation is associated with reduced tumor-infiltrating lymphocytes in triple-negative breast cancer: therapeutic cooperation between MEK and PD-1/PD-L1 immune checkpoint inhibitors. Clin Cancer Res . 2016;22:1499–1509. doi: 10.1158/1078-0432.CCR-15-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coelho MA. de Carné Trécesson S, Rana S, et al., editors. Oncogenic RAS signaling promotes tumor immunoresistance by stabilizing PD-L1 mRNA. Immunity . 2017;47:1083–1099.e6. doi: 10.1016/j.immuni.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen N, Fang W, Lin Z, et al. KRAS mutation-induced upregulation of PD-L1 mediates immune escape in human lung adenocarcinoma. Cancer Immunol Immunother . 2017;66:1175–1187. doi: 10.1007/s00262-017-2005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Motzer R, Alekseev B, Rha SY, et al. Lenvatinib plus pembrolizumab or everolimus for advanced renal cell carcinoma. N Engl J Med . 2021;384:1289–1300. doi: 10.1056/NEJMoa2035716. [DOI] [PubMed] [Google Scholar]
- 65.Choueiri TK, Powles T, Burotto M, et al. Nivolumab plus cabozantinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med . 2021;384:829–841. doi: 10.1056/NEJMoa2026982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rini BI, Plimack ER, Stus V, et al. Pembrolizumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med . 2019;380:1116–1127. doi: 10.1056/NEJMoa1816714. [DOI] [PubMed] [Google Scholar]
- 67.Motzer RJ, Penkov K, Haanen J, et al. Avelumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med . 2019;380:1103–1115. doi: 10.1056/NEJMoa1816047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jeanson A, Tomasini P, Souquet-Bressand M, et al. Efficacy of immune checkpoint inhibitors in KRAS-mutant non-small cell lung cancer (NSCLC) J Thorac Oncol . 2019;14:1095–1101. doi: 10.1016/j.jtho.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 69.Passiglia F, Cappuzzo F, Alabiso O, et al. Efficacy of nivolumab in pre-treated non-small-cell lung cancer patients harbouring KRAS mutations. Br J Cancer . 2019;120:57–62. doi: 10.1038/s41416-018-0234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Canon J, Rex K, Saiki AY, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature . 2019;575:217–223. doi: 10.1038/s41586-019-1694-1. [DOI] [PubMed] [Google Scholar]
- 71.Briere DM, Li S, Calinisan A, et al. The KRAS(G12C) inhibitor MRTX849 reconditions the tumor immune microenvironment and sensitizes tumors to checkpoint inhibitor therapy. Mol Cancer Ther . 2021;20:975–985. doi: 10.1158/1535-7163.MCT-20-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ferrucci PF, Di Giacomo AM, Del Vecchio M, et al. KEYNOTE-022 part 3: a randomized, double-blind, phase 2 study of pembrolizumab, dabrafenib, and trametinib in BRAF-mutant melanoma. J Immunother Cancer . 2020;8:e001806. doi: 10.1136/jitc-2020-001806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nathan P, Dummer R, Long GV, et al. LBA43 Spartalizumab plus dabrafenib and trametinib (Sparta-DabTram) in patients (pts) with previously untreated BRAF V600–mutant unresectable or metastatic melanoma: results from the randomized part 3 of the phase III COMBI-i trial. Ann Oncol . 2020;31:S1172. [Google Scholar]
- 74.Gutzmer R, Stroyakovskiy D, Gogas H, et al. Atezolizumab, vemurafenib, and cobimetinib as first-line treatment for unresectable advanced BRAF(V600) mutation-positive melanoma (IMspire150): primary analysis of the randomised, double-blind, placebo-controlled, phase 3 trial. Lancet . 2020;395:1835–1844. doi: 10.1016/S0140-6736(20)30934-X. [DOI] [PubMed] [Google Scholar]
- 75.Creelan BC, Yeh TC, Kim SW, et al. A Phase 1 study of gefitinib combined with durvalumab in EGFR TKI-naive patients with EGFR mutation-positive locally advanced/metastatic non-small-cell lung cancer. Br J Cancer . 2021;124:383–390. doi: 10.1038/s41416-020-01099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang JC, Gadgeel SM, Sequist LV, et al. Pembrolizumab in combination with erlotinib or gefitinib as first-line therapy for advanced NSCLC with sensitizing EGFR mutation. J Thorac Oncol . 2019;14:553–559. doi: 10.1016/j.jtho.2018.11.028. [DOI] [PubMed] [Google Scholar]
- 77.Oxnard GR, Yang JC, Yu H, et al. TATTON: a multi-arm, phase Ib trial of osimertinib combined with selumetinib, savolitinib, or durvalumab in EGFR-mutant lung cancer. Ann Oncol . 2020;31:507–516. doi: 10.1016/j.annonc.2020.01.013. [DOI] [PubMed] [Google Scholar]
- 78.Adderley H, Blackhall FH, Lindsay CR. Toxicity with small molecule and immunotherapy combinations in non-small cell lung cancer. Cancer Immunol Immunother . 2021;70:589–595. doi: 10.1007/s00262-020-02714-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu H, Liu C, Velazquez R, et al. SHP2 inhibition overcomes RTK-mediated pathway reactivation in KRAS-mutant tumors treated with MEK inhibitors. Mol Cancer Ther . 2019;18:1323–1334. doi: 10.1158/1535-7163.MCT-18-0852. [DOI] [PubMed] [Google Scholar]
- 80.Mainardi S, Mulero-Sánchez A, Prahallad A, et al. SHP2 is required for growth of KRAS-mutant non-small-cell lung cancer in vivo. Nat Med . 2018;24:961–967. doi: 10.1038/s41591-018-0023-9. [DOI] [PubMed] [Google Scholar]
- 81.Ryan MB. Fece de la Cruz F, Phat S, et al., editors. Vertical pathway inhibition overcomes adaptive feedback resistance to KRAS(G12C) inhibition. Clin Cancer Res . 2020;26:1633–1643. doi: 10.1158/1078-0432.CCR-19-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fedele C, Li S, Teng KW, et al. SHP2 inhibition diminishes KRASG12C cycling and promotes tumor microenvironment remodeling. J Exp Med . 2021;218:e20201414. doi: 10.1084/jem.20201414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Adachi Y, Ito K, Hayashi Y, et al. Epithelial-to-mesenchymal transition is a cause of both intrinsic and acquired resistance to KRAS G12C inhibitor in KRAS G12C-mutant non-small cell lung cancer. Clin Cancer Res . 2020;26:5962–5973. doi: 10.1158/1078-0432.CCR-20-2077. [DOI] [PubMed] [Google Scholar]
- 84.Tran E, Robbins PF, Lu YC, et al. T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med . 2016;375:2255–2262. doi: 10.1056/NEJMoa1609279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang QJ, Yu Z, Griffith K, et al. Identification of T-cell receptors targeting KRAS-mutated human tumors. Cancer Immunol Res . 2016;4:204–214. doi: 10.1158/2326-6066.CIR-15-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shen Y, Wei X, Jin S, et al. TCR-mimic antibody-drug conjugates targeting intracellular tumor-specific mutant antigen KRAS G12V mutation. Asian J Pharm Sci . 2020;15:777–785. doi: 10.1016/j.ajps.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sim MJW, Lu J, Spencer M, et al. High-affinity oligoclonal TCRs define effective adoptive T cell therapy targeting mutant KRAS-G12D. Proc Natl Acad Sci U S A . 2020;117:12826–12835. doi: 10.1073/pnas.1921964117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dillard P, Casey N, Pollmann S, et al. Targeting KRAS mutations with HLA class II-restricted TCRs for the treatment of solid tumors. Oncoimmunology . 2021;10:1936757. doi: 10.1080/2162402X.2021.1936757. [DOI] [PMC free article] [PubMed] [Google Scholar]