Depletion of mitochondrial iron with the drug ironomycin induced a dramatic loss in mitochondrial respiration and noncanonical BAX/BAK-dependent cell death that synergized with venetoclax in mouse and patient-derived models of leukemia.
Abstract
Cancer cell metabolism is increasingly recognized as providing an exciting therapeutic opportunity. However, a drug that directly couples targeting of a metabolic dependency with the induction of cell death in cancer cells has largely remained elusive. Here we report that the drug-like small-molecule ironomycin reduces the mitochondrial iron load, resulting in the potent disruption of mitochondrial metabolism. Ironomycin promotes the recruitment and activation of BAX/BAK, but the resulting mitochondrial outer membrane permeabilization (MOMP) does not lead to potent activation of the apoptotic caspases, nor is the ensuing cell death prevented by inhibiting the previously established pathways of programmed cell death. Consistent with the fact that ironomycin and BH3 mimetics induce MOMP through independent nonredundant pathways, we find that ironomycin exhibits marked in vitro and in vivo synergy with venetoclax and overcomes venetoclax resistance in primary patient samples.
Significance:
Ironomycin couples targeting of cellular metabolism with cell death by reducing mitochondrial iron, resulting in the alteration of mitochondrial metabolism and the activation of BAX/BAK. Ironomycin induces MOMP through a different mechanism to BH3 mimetics, and consequently combination therapy has marked synergy in cancers such as acute myeloid leukemia.
This article is highlighted in the In This Issue feature, p. 587
Introduction
Mitochondria serve as a central hub orchestrating essential metabolic pathways required to sustain eukaryotic life while concurrently acting as the point of convergence for various stimuli that ultimately initiate cell death (1, 2). This apparent dichotomy, coupling metabolism with cell death, has been proposed to be inexorably linked, and indeed several protagonists have been shown to play an integral role in both arenas. As an example, cytochrome c, an iron–sulfur cluster-containing protein sequestered within the mitochondrial intermembrane space (IMS), has an essential role in the respiratory chain, functioning as an electron shuttle. However, in response to lethal stimuli that lead to mitochondrial outer membrane permeabilization (MOMP), cytochrome c initiates the intrinsic apoptosis pathway (3).
Over the past few decades, advances in medicinal chemistry have seen remarkable progress in targeting mitochondrial proteins to improve the clinical outcome of patients with a range of pathologies including cardiovascular disease and cancer (1). Related to this, in the malignant context, there has been a growing awareness of the importance of cancer metabolism as a critical driver of the malignant phenotype (4). These are best highlighted by the discovery and characterization of oncogenic driver mutations in mitochondrial proteins such as succinate dehydrogenase, fumarate hydratase, and isocitrate dehydrogenase 2 (IDH2). These discoveries have successfully led to the development of novel FDA-approved therapies such as enasidenib for acute myeloid leukemia (AML; ref. 5). The intrinsic apoptotic pathway has also been the subject of intense drug-discovery efforts resulting in the generation of several BH3 mimetics, including venetoclax, which have revolutionized the treatment of several aggressive and often incurable cancers (6–8). Although there has been enormous progress in independently targeting the metabolic and cell death functions of the mitochondria, thus far the identification of small molecules that concurrently link these functional roles has been more elusive.
Results
Ironomycin Induces Cell AML Death through a Noncanonical Pathway
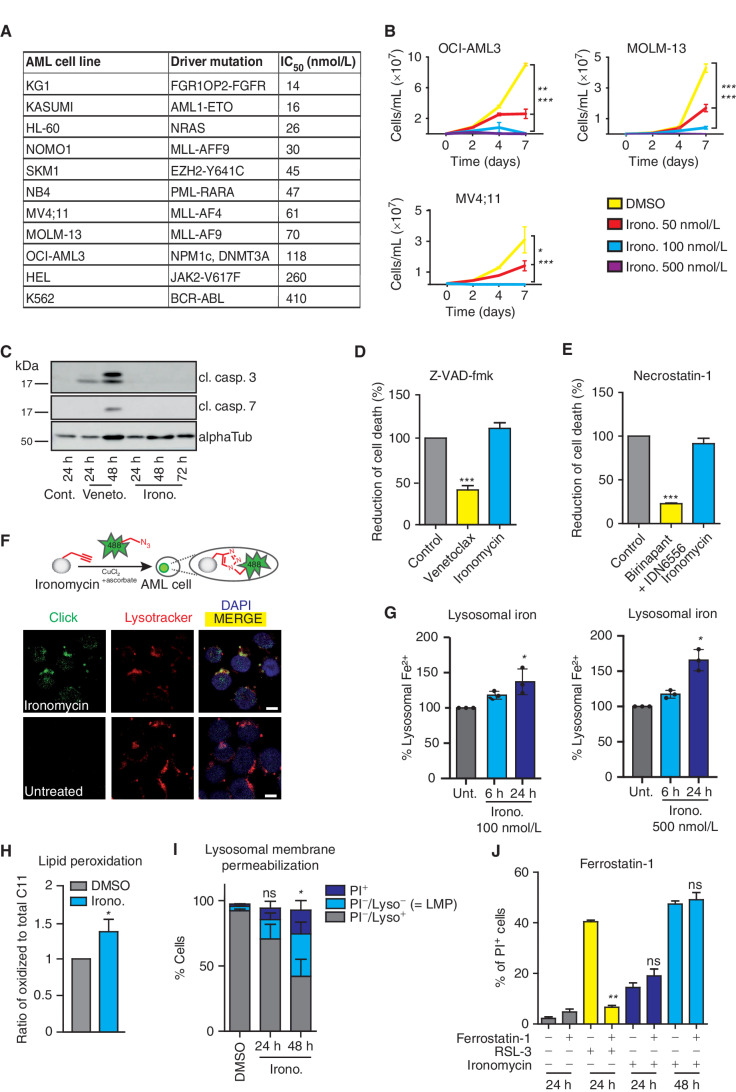
In light of the urgent need to uncover novel therapeutic options that induce cell death pathways in AML, we sought to study the potential role of ironomycin in this disease. Ironomycin (AM5) is a recently described small molecule that is a potent inducer of nonapoptotic cell death. Ironomycin is a synthetic derivative of salinomycin that is more potent and exerts its activity independently of sodium transport. Both ironomycin and salinomycin have been shown to mediate their activity through lysosomal iron targeting (9). Furthermore, we recently extended these findings, illustrating that ironomycin is particularly effective against cancer cells undergoing epithelial–mesenchymal transition (EMT) as these cells exhibit a higher demand on cellular iron and are characterized by an increased α-ketoglutarate–dependent epigenetic plasticity (10). To evaluate the effects of ironomycin in AML, we initially evaluated a diverse panel of AML cell lines spanning a variety of well-characterized AML mutations. These data clearly showed a potent and dose-dependent effect on proliferation, cell-cycle progression, and survival of all AML cell lines (Fig. 1A and B; Supplementary Fig. S1A and S1B). In contrast to venetoclax, the cell death induced by ironomycin did not result in potent caspase activation (Fig. 1C), and the caspase inhibitor Z-VAD-fmk did not prevent cell death (Fig. 1D; Supplementary Fig. S1C). As chemical inhibition of the cellular inhibitors of apoptosis proteins (cIAP1/2) and caspase-8 or ligation of the extrinsic apoptosis death receptors due to cellular stress can also induce an alternative form of nonapoptotic cell death called necroptosis, we investigated whether ironomycin induces necroptosis. Although the chemical induction of necroptosis by clinically used compounds that specifically activate this pathway (i.e., the combination treatment of SMAC-mimetic Birinapant and pan-caspase inhibitor IDN-6556) can be inhibited by the RIPK1 kinase inhibitor necrostatin-1, the activity of ironomycin remained unaffected upon cotreatment with this drug (Fig. 1E).
Figure 1.
Ironomycin induces potent cell death in AML through a noncanonical cell death pathway. A, Half-maximal inhibitory concentration (IC50) of AML cell lines with various genetic background after 72 hours of treatment with ironomycin using resazurin assay (n = 3 biological replicates). B, Proliferation curves of MV4;11, MOLM-13, and OCI-AML3 cell lines treated with ironomycin (n = 3 biological replicates, *, P < 0.05; **, P < 0.01; ***, P < 0.001). C, Protein expression of cleaved caspase-3 (cl. casp. 3) and cleaved caspase-7 shown by immunoblot. MV4;11 cells were treated with DMSO, 50 nmol/L venetoclax, or 500 nmol/L ironomycin. D, Effect on cell death of the pan-caspase inhibitor Z-VAD-fmk. MOLM-13 cells were pretreated with 50 μmol/L Z-VAD-fmk for 30 minutes and treated with 500 nmol/L ironomycin or 50 nmol/L venetoclax for 24 hours. Cell death was assessed by propidium iodide (PI) staining (n = 3 biological replicates; means ± SD; ***, P < 0.001). E, Effect on cell death of the necroptosis inhibitor necrostatin-1. MOLM-13 cells were pretreated with 10 μmol/L necrostatin-1 for 30 minutes and treated with 500 nmol/L ironomycin for 24 hours or 100 nmol/L birinapant plus 5 μmol/L IDN-6556 for 16 hours. Cell death was assessed by PI staining (n = 3 biological replicates; means ± SD; ***, P < 0.001). F, Visualization of the lysosomal localization of ironomycin using click chemistry in AML cells. Top, schematic illustration of the chemical labeling of ironomycin in cells. Bottom, fluorescence microscopy images of labeled ironomycin (Alexa Fluor 488, green), lysosome (lysotracker, red), and nucleus (DAPI, blue) in MOLM-13 cells after 2 hours of 10 μmol/L ironomycin treatment. Scale bar, 10 μm. G, Quantification of lysosomal Fe2+ using a lysosomal turn-on FACS probe in MOLM-13 cells after ironomycin (see Supplemental Data). Fe2+ specifically reduces Rhonox-M to a rhodamine B derivative, which fluoresces (n = 3 biological replicates; *, P < 0.05). H, FACS analysis of LMP using lysotracker (Lyso) and PI in the MOLM-13 cell line. We treated cells with DMSO or 500 nmol/L ironomycin. LMP is associated with a loss of lysotracker staining (n = 3 biological replicates; mean ± SD; *, P < 0.05). I, Analysis of lipid peroxidation by flow-cytometry staining of lipid ROS with the C11 BODIPY 581/591 (BODIPY C11) probe. MOLM-13 cells were treated with DMSO or 500 nmol/L ironomycin for 48 hours. The ratio of oxidized to total C11 median fluorescent intensity (MFI) is shown (n = 4 biological replicates; means ± SD; *, P < 0.05). J, Effect on cell viability of the ferroptosis inhibitor ferrostatin-1 used in combination with ironomycin. MOLM-13 cell line was pretreated with 20 μmol/L ferrostatin-1 for 30 minutes and treated with 500 nmol/L ironomycin or RSL-3 (30 nmol/L). Cell death was assessed by PI staining (n = 3 biological replicates; means ± SD; **, P < 0.01).
We and others have used click chemistry to identify the subcellular localization of small molecules (11–13), and, consistent with previous observations, we found that ironomycin accumulates in the lysosomes of AML cells, leading to retention of iron in this organelle (Fig. 1F and G; Supplementary Fig. S1D). This leads to lipid peroxidation (Fig. 1H; Supplementary Fig. S1E) and lysosomal membrane permeabilization (LMP) that is best observed following 48 hours of drug exposure (Fig. 1I; Supplementary Fig. S1F). Although the lipid peroxidation and cell death induced by ironomycin exhibits features of ferroptosis (9, 10), it is notable that in AML cells, established inhibitors of canonical ferroptosis, including ferrostatin-1 and liproxstatin-1 (14, 15), block the effects of the GPX4 inhibitor RSL-3 but fail to prevent or forestall the activity of ironomycin (Fig. 1J; Supplementary Fig. S1G). Consistent with these findings, neither Z-VAD-fmk, necrostatin-1, nor ferrostatin-1 rescued the cell-cycle arrest induced by ironomycin (Supplementary Fig. S1H). Taken together, our data demonstrate that ironomycin potently induces a block in proliferation and subsequent cell death in AML cells; however, the pathway by which this occurs is not indicative of the most established pathways of programmed cell death.
Genome-Wide CRISPR Screen Identifies Cellular Metabolism and Mitochondrial Homeostasis as Key Regulators of Ironomycin Activity
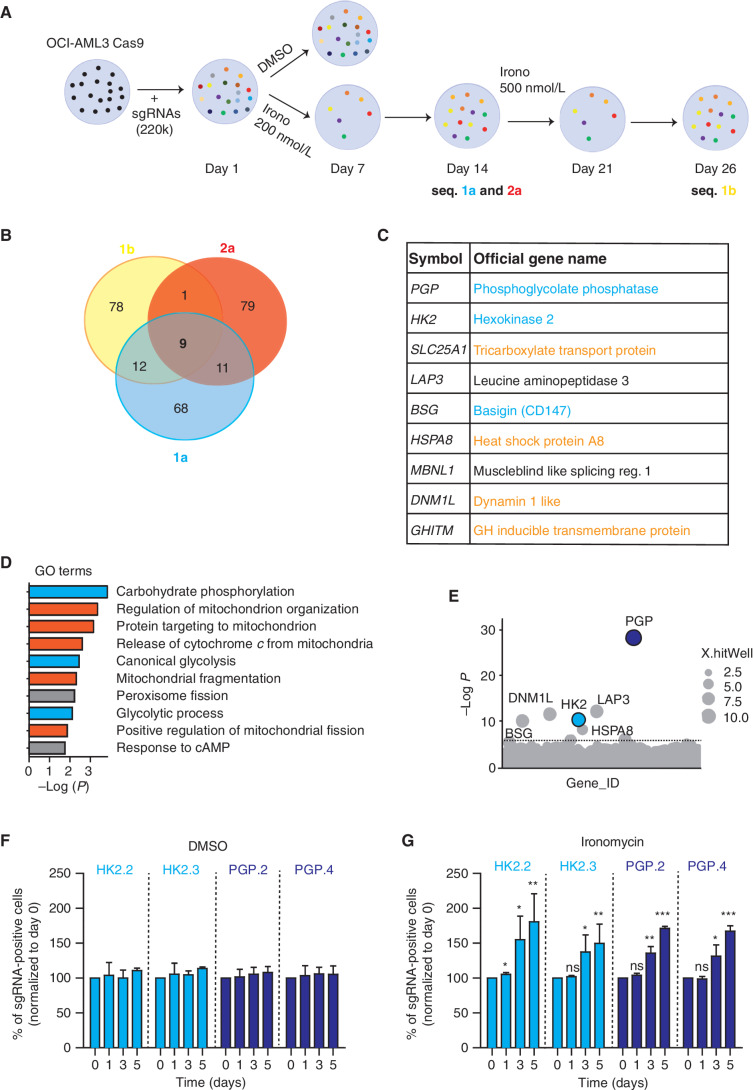
As all cancers have marked genetic and nongenetic intratumor heterogeneity (16), we set out to understand whether ironomycin treatment resulted in selection of a specific subclonal population. To address this, we labeled AML cells with six different fluorochromes, mixed these in equal portion, and treated the cells until cell death was induced. We then washed out the drug and monitored recovery for therapeutic pressure. These data showed that there was no clear selective process in either induction of cell death or recovery following ironomycin treatment (Supplementary Fig. S2A–S2C). Therefore, to gain greater insight into the molecular mechanism of ironomycin in AML cells, we performed a genome-wide positive-selection resistance screen under selection pressure at approximately the IC90 of ironomycin (Fig. 2A). The screen was done in biological replicates in OCI-AML3, as this cell line contains mutations in DNMT3A and NPM1, which are the two most common genes mutated in human AML (17). As expected, ironomycin treatment at 200 nmol/L resulted in near-uniform cell death with only a rare population of cells able to survive to day 7 and subsequently expand to day 14 (Supplementary Fig. S2D). At this point, we collected a sample for sequencing and increased the dose of ironomycin to 500 nmol/L and then took a further sample at day 26 following treatment. This stringent approach enabled us to identify the major genetic determinants able to impede ironomycin-induced cell death. We found nine genes that were consistently enriched in biologically replicate samples whose knockout (KO) conferred reduced sensitivity to ironomycin (Fig. 2B). Interestingly, these data clearly implicated several key components of cellular metabolism, including phosphoglycolate phosphatase (PGP), a central phosphatase involved in glycolysis and pentose phosphate pathway (PPP) regulation (18, 19), and Hexokinase 2 (HK2), the first enzyme of glycolysis. In addition, we also identified Basigin (BSG), a multifunctional transmembrane glycoprotein linked to glucose and lactate cellular transport (20), and several other genes involved in the tricarboxylic acid cycle (TCA) and mitochondrial homeostasis (Fig. 2C). In line with the known activity of several of these targets, gene set enrichment analysis of the major hits found in our screens resulted in Gene Ontology (GO) terms associated with carbohydrate and mitochondrial metabolism (Fig. 2D).
Figure 2.
Genome-wide CRISPR screen identifies cellular metabolism and mitochondrial homeostasis as key regulators of ironomycin activity. A, Schematic outline of the resistance genome-wide CRISPR/Cas9 loss-of-function screen in OCI-AML3 (n = 3 screens from two independent biological replicates). B, Venn diagram showing the common sgRNAs enriched in the ironomycin-resistant cells in the sequencing of the three screens (1a, 2a, and 1b). C, Table displaying the nine common genes from the three screens performed in the OCI-AML3 cell line (1a, 2a, and 1b). Genes related to glycolysis pathway are highlighted in blue and genes related to mitochondria homeostasis are highlighted in orange. D, Enrichment analysis showing GO terms significantly enriched in the top 100 genes from the OCI-AML3 screen. GO terms related to glycolysis are highlighted in blue, and GO terms related to mitochondria homeostasis are highlighted in orange. E, Bubble plots showing the top 1,000 enriched genes identified in the CRISPR screen (replicate 2a). The two hits selected for validation are colored in blue. Dotted line indicates Bonferroni-corrected significance threshold. F and G, Validation of the CRISPR screen in the MV4;11 cell line by competition assays using MV4;11 cells transfected with two independent sgRNAs targeting PGP (PGP.2 and PGP.4) or HK2 (HK2.2 and HK2.3) and a control nontargeted (NT) sgRNA. A 1:1 ratio of mCherry-positive sgRNA cells and mCherry-negative WT MV4;11 cells was treated with DMSO (F) or 500 nmol/L ironomycin (G), and the proportion of mCherry-positive sgRNAs cells was assessed by FACS (n = 2 biological replicates for each KO cell line; means ± SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
As PGP and HK2 were among the strongest hits in our screen (Fig. 2E) and their central role in glycolysis and PPP is clearly established, we chose to focus on understanding the molecular mechanisms by which loss of these key metabolic enzymes was able to confer insensitivity to ironomycin. The results from our screen were independently validated within the FLT3-mutated MV4;11 cell line transfected with single-guide RNAs (sgRNA) targeting PGP or HK2 (Fig. 2F and G; Supplementary Fig. S2E). Although PGP or HK2 KO did not affect cell proliferation by itself (Fig. 2F), these cells have a relative growth and survival advantage only in the presence of ironomycin (Fig. 2G). We also established that ironomycin does not alter HK2 or PGP levels (Supplementary Fig. S2F), suggesting that their effects in mediating resistance cannot be explained by ironomycin altering the expression or stability of these metabolic enzymes. Taken together, these results indicate that the metabolic state of the cancer cell is a major factor that dictates sensitivity to ironomycin.
Metabolic Reprogramming to Reduce Glycolytic Flux Protects Cells against Ironomycin
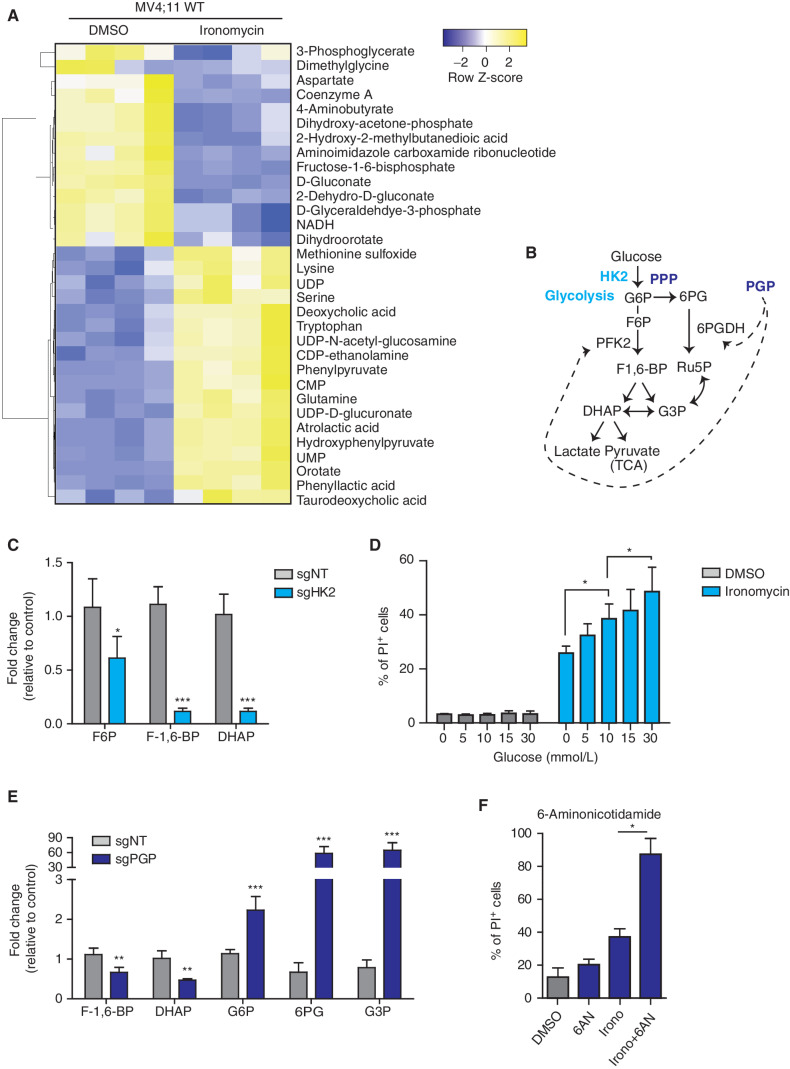
To further investigate the interplay between cellular metabolism and the cell death induced by ironomycin, we performed metabolomics analyses to define the broad metabolic changes seen in AML cells treated with ironomycin in the presence and absence of HK2 and PGP (Fig. 3A; Supplementary Fig. S3A and S3B). The metabolomics analyses clearly highlighted that ironomycin treatment significantly decreased key components of the TCA cycle and consequently the reducing agent nicotinamide adenine dinucleotide (NADH), which mediates the transfer of electrons to fuel ATP generation. In contrast, ironomycin increased the intracellular concentration of amino acids including serine, glutamine, and tryptophan (Fig. 3A). We next compared the metabolite profile of a HK2 KO cell line with a nontargeting (NT) sgRNA control cell line. As anticipated, glycolytic intermediates downstream of HK2, in particular fructose-6-phosphate (F6P), fructose-1,6-bisphosphate (F-1,6-BP), and dihydroxyacetone phosphate (DHAP), were decreased in the HK2 KO cell line (Fig. 3B and C; Supplementary Fig. S3A and Supplementary Table S1). To determine if compromised glycolytic metabolism, in the context of HK2 KO, contributes to ironomycin resistance, we modulated glucose levels in cell culture media and determined cellular sensitivity to ironomycin. First, we observed that glucose level alone did not affect the viability and proliferation of the cells during the experimental timeframe (Supplementary Fig. S3C). However, when ironomycin was added to the cell media, we noted that increased glucose availability was associated with a higher efficacy of ironomycin in killing AML cells (Fig. 3D).
Figure 3.
Metabolic remodeling to reduce glycolytic flux and mitochondrial respiration protects against ironomycin. A, Heat map showing the differential abundance of metabolites in the MV4;11 WT cell line using mass spectrometry. We treated the cells with 500 nmol/L ironomycin or DMSO for 24 hours (n = 4 biological replicates). We selected the metabolites with a log2 fold change >1 and a t test P < 0.05. B, Schematic representation of glycolysis and the branched PPP and function of the two metabolic enzymes hexokinase 2 (HK2) and phosphoglycolate phosphatase (PGP). DHAP, dihydroxyacetone phosphate; F6P, fructose-6-phosphate; F-1,6-BP, fructose-1,6-bisphophate; G3P, glyceraldehyde-3-phosphate; Ru5P, ribulose-5-phosphate; 6PG, 6-phospho-D-glycerate; TCA, tricarboxylic acid cycle. C, Bar graph showing the changes in metabolites expression in the sgHK2 cell line using mass spectrometry (n = 4 biological replicates; means ± SD; *, P < 0.05; ***, P < 0.001). D, Proportion of cell death of ironomycin-treated MV4;11 cells cultured in various glucose concentrations. We performed flow-cytometry analysis using PI. Cells were treated with 500 nmol/L ironomycin for 48 hours in RPMI medium (n = 3 biological replicates; means ± SD; *, P < 0.05). E, Bar graph showing the changes in metabolite expression in the sgPGP cell line. Metabolites downstream phosphofructokinase 2 (PFK2) are decreased such as F-1,6-BP and DHAP. Metabolites upstream PFK2 are increased such as G6P, 6PG, and G3P (n = 3 biological replicates; means ± SD; **, P < 0.01; ***, P < 0.001). F, Proportion of cell death of ironomycin-treated cells in combination with the PPP inhibitor 6AN. We performed flow-cytometry analysis of PI fluorescence in MOLM-13. Cells were pretreated with 10 μmol/L 6AN for 30 minutes and treated with 500 nmol/L ironomycin for 48 hours (n = 3 biological replicates; means ± SD; *, P < 0.05).
We next sought to understand the shared and distinct metabolic features imparted by the loss of PGP that confers protection against ironomycin. PGP is a metabolite repair enzyme that is conserved from yeast to mammals (18). This evolutionarily conserved enzyme acts to eliminate inhibitory intermediate metabolites, which would otherwise accumulate and disable the glycolytic pathway. It has been suggested that in the absence of PGP, 2-phospho-L-lactate, generated as a by-product of pyruvate kinase, can accumulate and inhibit the activity of phosphofructokinase 2 (PFK2; ref. 18). In line with these findings, we found that the PGP KO cells showed a decrease in metabolites downstream of PFK2 (F-1,6-BP, DHAP) and a marked increase in glucose-6-phosphate (G6P), a metabolite upstream of PFK2. The metabolites upstream of PFK2 can be redirected toward the PPP, and consistent with this, we observed that 6-phospho-D-gluconate (6PDG) and glyceraldehyde-3-phosphate (G3P) were also upregulated (Fig. 3E; Supplementary Fig S3B and Supplementary Table S2). To assess if the PPP pathway may be important in providing protection against ironomycin, we used 6-aminonicotidamine (6AN), a well-established inhibitor of the PPP that targets the NADP-dependent enzyme 6-phosphogluconate dehydrogenase (6PGDH). Although a low dose of 6AN by itself was not toxic to AML cells, it markedly potentiated the efficacy of ironomycin in inducing cell death (Fig. 3F).
When considered together, these metabolic changes raised the possibility that ironomycin efficiency takes advantage of the fact that AML cells are highly dependent on glucose for mitochondrial respiration (21–24). Consequently, reduced glycolytic flux along with a diversion to alternative metabolic pathways for energy production is a key adaptive feature that enables tolerance to ironomycin. To test this scenario, we pretreated AML cells with either metformin or phenformin, two biguanides that exhibit pleiotropic effects, including a switch toward anaerobic glycolysis (25, 26). Consistent with the notion that reduced mitochondrial respiration protects against ironomycin, we found that pretreatment with both biguanides curtailed ironomycin-induced cell death in a dose-dependent manner (Supplementary Fig. S3D and S3E).
Ironomycin Induces Mitochondrial Stress through Iron Depletion
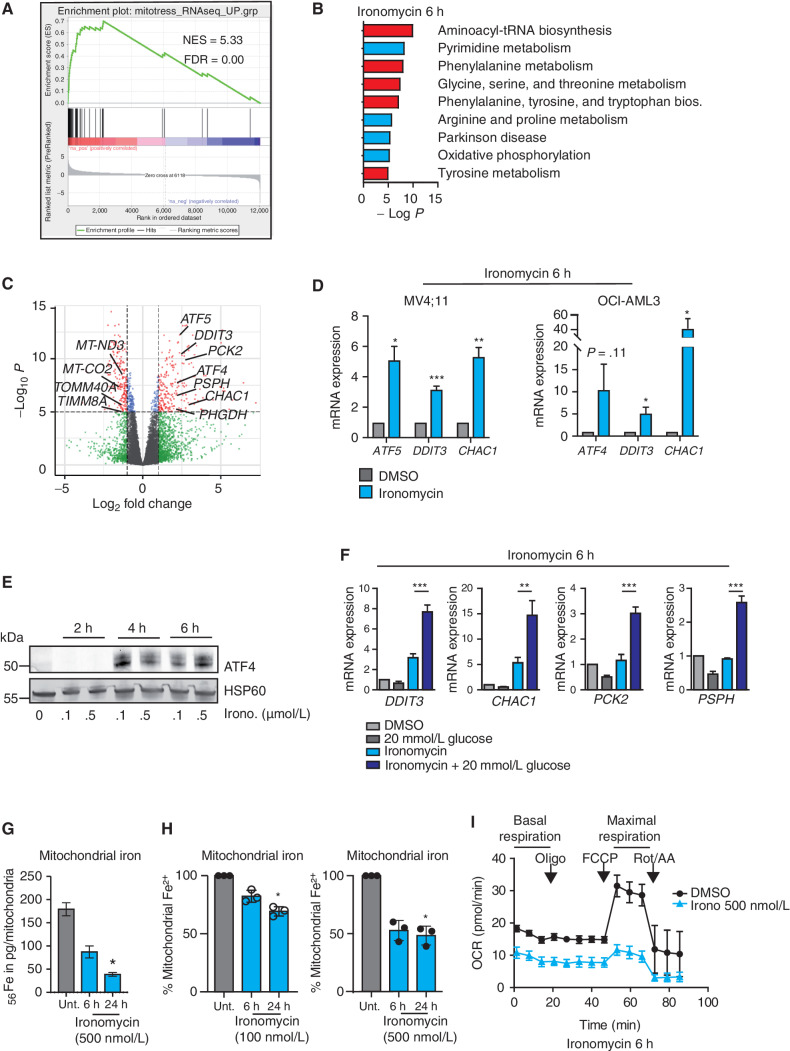
To obtain insights into the molecular activity of ironomycin in AML cells at early time points, we sought to survey the global transcriptional changes induced by ironomycin in both wild-type (WT) cells and PGP and HK2 KO cells. When cells were treated with ironomycin for 6 hours, we observed rapid and specific changes in gene expression that were sustained at 24 hours (Supplementary Fig. S4A). Gene set enrichment analyses showed that these changes were highly concordant with previous transcriptional changes seen in the context of mitochondrial stress (Fig. 4A; Supplementary Fig. S4A). Moreover, consistent with our metabolomic data (Fig. 3; Supplementary Fig. S3), the KEGG pathway terms derived from the RNA-sequencing (RNA-seq) analyses illustrated a decrease in expression of genes associated with oxidative phosphorylation (OxPhos) and an increase in expression of genes associated with alternative metabolic pathways, including amino acid metabolism (Fig. 4B). Notably, the expression of numerous genes related to electronic transport chain (ETC) or genes coding for inner and outer mitochondrial membranes (IMM and OMM) was rapidly altered by ironomycin treatment, such that the mitoCarta 2.0 signature that catalogs the mammalian mitochondrial genes (27, 28) was found to be mainly downregulated (Fig. 4C; Supplementary Fig. S4A). In contrast, the genes rapidly upregulated following ironomycin treatment included activating transcription factor 4 (ATF4) and its paralog activating transcription factor 5 (ATF5) as well as other genes involved in the mitochondrial unfolded protein response (mtUPR), a so-called mitostress signature that is a direct consequence of mitochondrial dysfunction (29, 30). Using orthogonal methods, we validated the results of our RNA-seq data (Fig. 4D and E) and further confirmed that upregulation of the “mitostress” genes was even stronger in high-glucose culture conditions, corroborating our previous data showing that ironomycin takes advantage of glucose-dependent mitochondrial respiration to induce the “mitostress” response (Fig. 4F). Taken together, these transcriptional changes reinforce the results of our CRISPR screen (Fig. 2C) and metabolomic data (Fig. 3; Supplementary Fig. S3) to further demonstrate that ironomycin induces mitochondrial dysfunction.
Figure 4.
Ironomycin induces mitochondrial stress through iron deprivation. A–C, RNA-seq analysis of MV4;11 cells treated with 6 hours of 500 nmol/L ironomycin or vehicle (DMSO, n = 3 biological replicates). A, Gene set enrichment analysis using the “mitostress” signature (30). B, KEGG pathways analysis (pathways upregulated are in red and pathways downregulated are in blue). C, Volcano plots showing the adjusted significance P value (−log10P) versus the fold change (log2). Genes that demonstrate a significant change in expression (P < 0.01) and a significant 2-fold downregulation (left) or 2-fold upregulation (right) are represented in red. Genes selected from the “mitostress” and mitoCarta 2.0 signatures are displayed. D, Validation of RNA-seq analyses by RT qPCR measuring mRNA expression of selected genes from the “mitostress” signature in AML cell lines 6 hours after exposure to 500 nmol/L ironomycin or DMSO. We used β2 m as a housekeeping gene (n = 3 technical replicates; means ± SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001). E, ATF4 protein expression by immunoblot after 500 nmol/L ironomycin treatment. F, Effect of high glucose culture condition (20 mmol/L) on RNA expression of selected genes from the “mitostress” signature upon ironomycin treatment in MV4;11 cells. We used β2 m as a housekeeping gene (n = 3 technical replicates; means ± SD; **, P < 0.01; ***, P < 0.001. G, Quantification of mitochondrial fe2+ by inductively couple plasma-mass spectrometry (ICP-MS) in MOLM-13 cells (n = 3 biological replicates; means ± SD; *, P < 0.05). H, Quantification of mitochondrial iron using a mitochondrial specific probe (see Supplemental Data) by flow cytometry on MOLM-13 cells treated with ironomycin (n = 3 biological replicates; means ± SD; *, P < 0.05). I, Seahorse assay measuring mitochondrial basal and maximal respiration in MV4;11 WT cells. We treated the cells for 6 hours with 500 nmol/L ironomycin (n = 3 biological replicates).
Mitochondria are major hubs of iron accumulation and use. In particular, mitochondrial iron is a major component of the ETC, responsible for mitochondrial respiration (31). Given the ability of ironomycin to sequester iron in lysosomes (9, 10), the distribution hub of iron (Fig. 1F), we next asked whether cell death induced by ironomycin could be mechanistically linked to an iron imbalance in mitochondria. To assess mitochondrial iron levels, we used inductively coupled plasma-mass spectrometry (ICP-MS) on isolated mitochondria after treatment with ironomycin. These data showed a rapid and dose-dependent decrease in mitochondrial iron, which was independently validated using a probe that detects mitochondrial Fe2+ (Fig. 4G and H; Supplementary Fig. S4B). These data sharply contrast with the dose-dependent increase in lysosomal iron sequestration. It is increasingly clear that lysosomal dysfunction is inexorably linked with impaired mitochondrial turnover (32). Given that the ETC is dramatically affected by diminished iron (33), these data raised the prospect that ironomycin impairs OxPhos by mitochondrial iron reduction. In line with this contention, the Seahorse functional assay, which measures mitochondrial respiration, showed a rapid and dramatic decrease in basal and maximal respiration rate with ironomycin (Fig. 4I).
As impaired energy production leads to increased generation of reactive oxygen species (ROS; ref. 32), we next evaluated whether mitochondrial ROS were increased upon ironomycin treatment. These data demonstrated a dose-dependent increase in mitochondrial ROS with ironomycin treatment (Supplementary Fig. S4C); however, in contrast to the ROS-induced cell death from arsenic trioxide (34), the ROS scavenger n-acetylcysteine (NAC) could not rescue cell death from ironomycin (Supplementary Fig. S4D). More-over, as impaired mitochondrial depolarization, a feature of dysfunctional mitochondria, preceded LMP by 24 hours (Supplementary Fig. S4E and S4F), the cell death mechanism was unlikely to be due to LMP in our model.
Ironomycin Induces a Mitochondrial Dysfunction and a BAX/BAK-Dependent Cell Death
All our unbiased orthogonal data derived from whole-genome CRISPR screens, quantitative global metabolomics, and comprehensive transcriptional profiling pointed to mitochondria as being a major organelle disrupted by ironomycin. Therefore, we next examined the ultrastructural appearance of this organelle after ironomycin treatment using transmission electron microscopy (TEM). As a frame of reference, we also examined the ultrastructural appearance of mitochondria following treatment with the BH3 mimetic venetoclax, which induces intrinsic apoptosis. These data clearly demonstrated that morphology was markedly different between ironomycin and venetoclax (Fig. 5A; Supplementary Fig. S5A). Although venetoclax resulted in nuclear condensation, fragmentation, and disruption of the mitochondrial membrane, these classic features of apoptosis were not prominent in ironomycin-treated cells. Instead, ironomycin resulted in marked heterogeneity in mitochondrial size and morphology and was noted to dramatically affect the structural integrity of mitochondria, resulting in abnormal cristae, matrix density changes, mitochondrial membrane blebbing, and features of mitophagy (Fig. 5A). These data raised the possibility that ironomycin may promote MOMP, resulting in cell death without potent caspase activation. The BCL2 effector proteins BCL2-associated X protein (BAX) and BCL2 antagonist or killer (BAK) are the primary executioners of MOMP. Crucially, both BAX and BAK are required for MOMP, and as our CRISPR screen did not allow for simultaneous deletion of more than one gene product, it was unclear if these proteins were involved in ironomycin-induced cell death.
Figure 5.
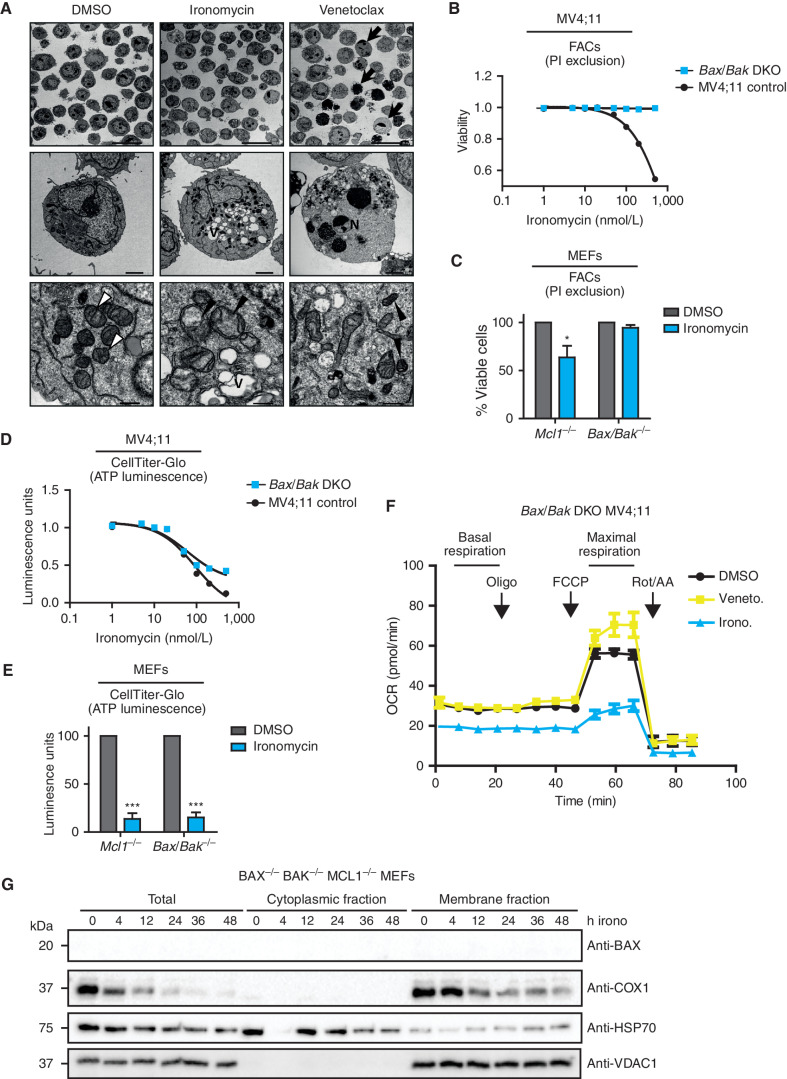
Ironomycin-induced cell death is BAX/BAK dependent. A,TEM images of MV4;11 cells treated with vehicle (DMSO), 500 nmol/L ironomycin or 50 nmol/L venetoclax for 36 hours. Arrows, examples of apoptotic cells; V, vacuolization; N, chromatin condensation and nuclear fragmentation; white arrowheads, standard mitochondrial morphology; black arrowheads, changed mitochondrial morphology (cristae reduction and dilation, fragmentation, or dark condensed matrix). Scale bars, 20 μm, top; 2 μm, middle; 500 nm, bottom. B, Cell death assessed by PI exclusion using FACS in MV4;11 WT and BAX/BAK DKO cell lines after 48 hours of ironomycin (n = 3 biological replicates). C, Cell death assessed by PI exclusion using FACS in MEF cells treated for 48 hours with 2 μmol/L ironomycin. We compared a Bax−/−Bak−/− with a control Mcl1−/− MEF cell line (n = 3 biological replicates; means ± SEM; *, P < 0.05). D, ATP luminescence measured by CellTiter-Glo in MV4;11 WT and BAX/BAK DKO cell lines after 48 hours of ironomycin (n = 3 biological replicates). E, ATP luminescence measured by CellTiter-Glo in MEF cells treated for 48 hours with 2 μmol/L ironomycin. We compared a Bax−/−Bak−/− with a control Mcl1−/− MEF cell line (n = 3 biological replicates; means ± SEM; ***, P < 0.001). F, Seahorse assay measuring mitochondrial basal and maximal respiration in BAX/BAK DKO MV4;11 cells. We treated the cells for 6 hours with 500 nmol/L ironomycin or 50 nmol/L venetoclax (n = 3 biological replicates). G, Fractionation experiment showing BAX and COX1 protein expressions by immunoblot in total cell, cytosolic fraction, and mitochondrial membrane fraction. The nonheme protein VDAC1 was used as a loading marker of membrane fraction and HSP70 as a loading marker of cytoplasmic fraction. We used the Bax−/−Bak−/−Mcl1−/− MEF cell line and treated cells with 500 nmol/L ironomycin.
To address this important point, we deleted both BAX and BAK in AML cells and Bax/Bak mouse embryonic fibroblast (MEF) cells and treated them with ironomycin. Remarkably, we found that these cells are completely protected from the cell death induced by ironomycin (Fig. 5B and C). In contrast, ATP production is equally affected by ironomycin in cells that are either replete or deficient for BAX/BAK (Fig. 5D and E). These effects are specific to ironomycin as venetoclax did not show a discordance between ATP production and cell death in BAX/BAK-deficient cells (Supplementary Fig S5B–S5E). Notably, this result disentangles the impact of ironomycin on cell metabolism and cell death and, moreover, suggests that the metabolic effect of ironomycin occurs prior to BAX/BAK activation. To confirm these data using an orthogonal approach, we performed Seahorse functional assays on BAX/BAK double knockout (DKO) cells. Here again, we observed that ironomycin induced a strong decrease in both basal and maximal mitochondrial respiration (Fig. 5F). In contrast, when cells are treated with venetoclax, cellular respiration and cell death are inexorably coupled and dependent on BAX and BAK activation (Fig. 5F; Supplementary Fig. S5F).
At an ultrastructural level, although BAX/BAK-deficient MV4;11 cells displayed thickened mitochondrial cristae compared with WT cells (Supplementary Fig. S5G), the cellular and mitochondrial morphology of the baseline BAX/BAK-deficient cells is not further altered when treated with venetoclax. In contrast, the BAX/BAK-deficient cells treated with ironomycin still show additional features of mitochondrial dysfunction, including cristae reduction and dilation, condensed matrix, and dilated electron-lucent spots (Supplementary Fig. S5H). Finally, to further illustrate the impact of ironomycin on the functional integrity of mitochondria, we subfractionated BAX/BAK-deficient MEF cells to assess the stability of heme-containing mitochondrial components. Consistent with ironomycin-induced mitochondrial iron deprivation, we observed a marked and rapid decrease in cytochrome C oxidase 1 (COX1), a heme-dependent mitochondrial protein (35). In contrast, the level of non–heme-containing proteins such as VDAC1 remains unaltered (Fig. 5G).
Ironomycin BAX/BAK Activation Is Independent of BH3-Only Proteins
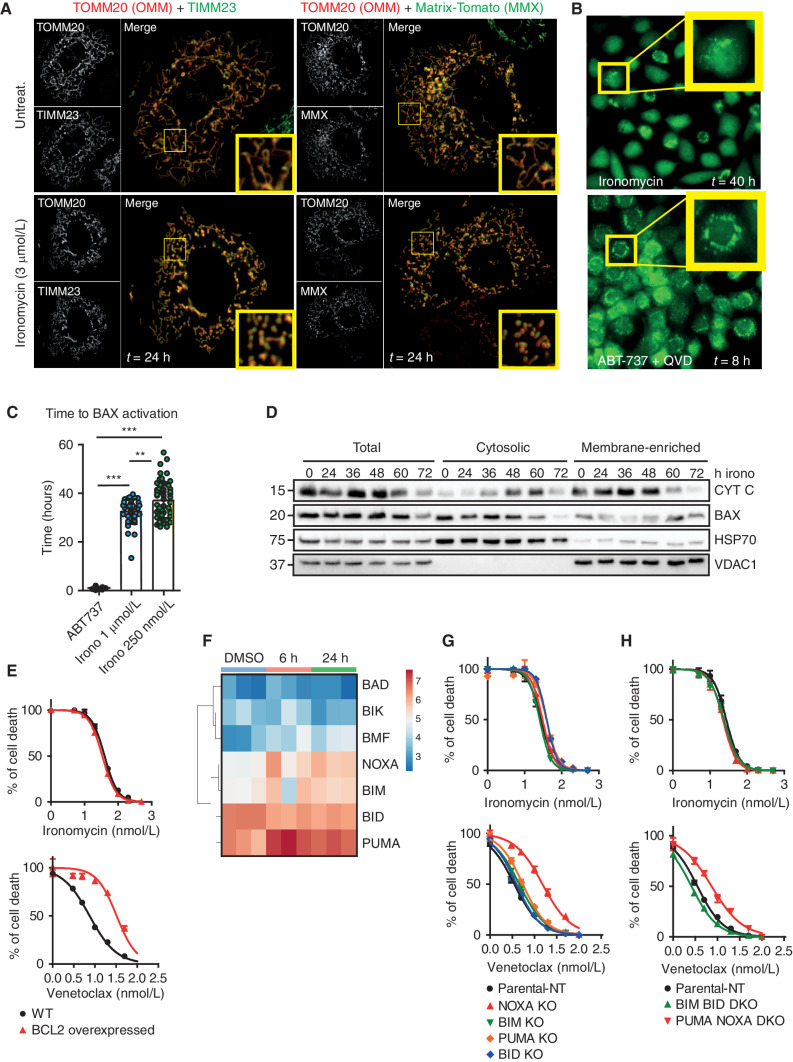
To better visualize the activation of BAX/BAK with ironomycin, we performed imaging in MEF containing fluorescent reporter proteins for BAX and proteins of the OMM (TOMM20), IMM (TIMM23) and mitochondrial matrix (MMX). These data show that within the first 24 hours of treatment with ironomycin, mitochondria become swollen and display fragmented mitochondrial networks (Fig. 6A). Interestingly, there is prominent heterogeneity in the fragmented mitochondrial network after ironomycin treatment, and this again contrasts sharply with the classic features of apoptosis as seen after treatment with BH3 mimetics (Supplementary Fig. S6A). Therefore, to get a better understanding of the temporal sequence of BAX activation with BH3 mimetics and ironomycin, we used a BAX reporter MEF cell line treated with either ABT-737 or ironomycin. These data clearly demonstrate that although BAX activation and recruitment to the mitochondrial membrane occurs with both compounds (Fig. 6B), the temporal nature of BAX activation is clearly delayed with ironomycin, occurring approximately 36 hours after the beginning of treatment (Fig. 6C). Using subcellular fractionation after ironomycin treatment, we clearly observe that the relocalization of BAX from the cytosol to the mitochondrial membrane is associated with a release of cytochrome c, consistent with ironomycin-induced MOMP (Fig. 6D).
Figure 6.
Ironomycin cell death is distinct from canonical apoptosis induced by BH3 mimetics. A, Confocal images of MCL1−/− MEFs expressing tagged-OMM component TOMM20-Halo (JF646, red) with either IMM component TIMM23 (left; mNeonGreen, green) or MMX (right; tdTomato, green) after 24 hours untreated (top) or 3 μmol/L ironomycin (bottom). Insets highlight representative mitochondrial network morphology. B, Snapshots from long-term widefield imaging assay of MCL1−/−Bax−/−Bak−/− MEFs reexpressing mNeonGreen-BAX and TOMM20-Halo, treated with 1 μmol/L ABT-737 + 20 μmol/L QVD-OPH or 500 nmol/L ironomycin, with insets highlighting BAX recruitment to mitochondria prior to cell death (n = 3 independent imaging experiments). C, Scatter plot displaying the time until the appearance of BAX foci, quantified manually, from long-term widefield imaging assay. Each data point represents a single cell (n = 3 biological replicates with >10 cells counted per experiment; means ± SD; **, P < 0.01; ***, P < 0.001). D, Fractionation experiment in MV4;11 cells treated with 500 nmol/L ironomycin showing cytochrome c (CYT C) and BAX protein expressions by immunoblot in total cell, cytosolic fraction, and mitochondrial membrane fraction. VDAC1 is used as a loading marker of mitochondrial membrane fraction and HSP70 as a loading marker of cytoplasmic fraction (n = 3; one representative experiment is shown). E, Cell viability assessed by FACS analysis of PI staining. We treated MV4;11 WT and MV4;11 cells with BCL2 overexpressed for 48 hours with ironomycin (top curve) and venetoclax (bottom curve, n = 3 biological replicates). F, Heat map showing the mRNA expression of BH3-only proteins in MV4;11 cells after 6 and 24 hours of ironomycin. G and H, Cell viability using PI FACS staining. We treated MV4;11 WT and MV4;11 cells with single knockout for the BH3-only proteins NOXA, PUMA, BIM, and BID (G) or double knockouts for BIM/BID and NOXA/PUMA (H) for 72 hours with ironomycin (top curves) and venetoclax (bottom curves; n = 3 biological replicates).
It is generally accepted that BAX/BAK activation requires BCL2 family of proteins (36, 37). Disruption of the binding and sequestration of BAX/BAK by the prosurvival BCL2 proteins, most commonly through the activity of the proapoptotic BH3 proteins, are critical for the oligomerization of BAX/BAK leading to MOMP. However, recent data have raised the possibility that the OMM may activate BAX/BAK even in the absence of the BCL2 family of proteins (37–39). To understand if overexpression of BCL2 can alleviate or forestall the cell death induced by ironomycin, we overexpressed BCL2 and monitored cell death induced by venetoclax or ironomycin. These data show that although the cell death induced by venetoclax is significantly curtailed, BCL2 overexpression had no effect on the cell death induced by ironomycin (Fig. 6E). We next set out to assess whether BH3-only proapoptotic proteins (BIM, BID, BAD, PUMA, NOXA, BIK, BMF, and HRK) were involved in ironomycin-mediated cell death. Monitoring the mRNA expression levels of these proteins in AML cells, we found that only four of the BH3 family members (BIM, BID, PUMA, and NOXA) are expressed at baseline, BIM and NOXA being further increased in expression within 6 hours of treatment (Fig. 6F). Notably, the increased expression at 6 hours is not further increased at 24 hours, and this increase is not temporally associated with MOMP. Thus, we produced isogenic KO cells for BIM, BID, PUMA, and NOXA as well as DKO cells for BIM/BID and NOXA/PUMA (Supplementary Fig. S6B). We found that neither individual KO nor DKO alters the sensitivity of AML cells to ironomycin (Fig 6G and H). In contrast, both single NOXA KO and NOXA/PUMA DKO were associated with venetoclax resistance (Fig. 6G and H), which is consistent with previous findings (40). Furthermore, the potent caspase cleavage that occurs following BH3 mimetic–induced MOMP clearly plays a major role in the BH3 mimetic–induced apoptotic pathway, as evidenced by the marked delay in cell death and delay between BAX activation and death in the presence of the pan-caspase inhibitor QVD-OPH (Supplementary Movie S1, Supplementary Fig. S6C). In contrast, the activity of caspases contributes less to the kinetics of cell death mediated by ironomycin after BAX activation (Supplementary Movie S2, Supplementary Fig. S6D). Taken together, these data further reinforce the fact that although BH3 mimetics and ironomycin induce cell death in a BAX/BAK-dependent manner, the mechanisms by which MOMP is induced and its downstream consequences are distinct.
Ironomycin Shows Marked Synergy with BH3 Mimetics and Overcomes Resistance to Venetoclax
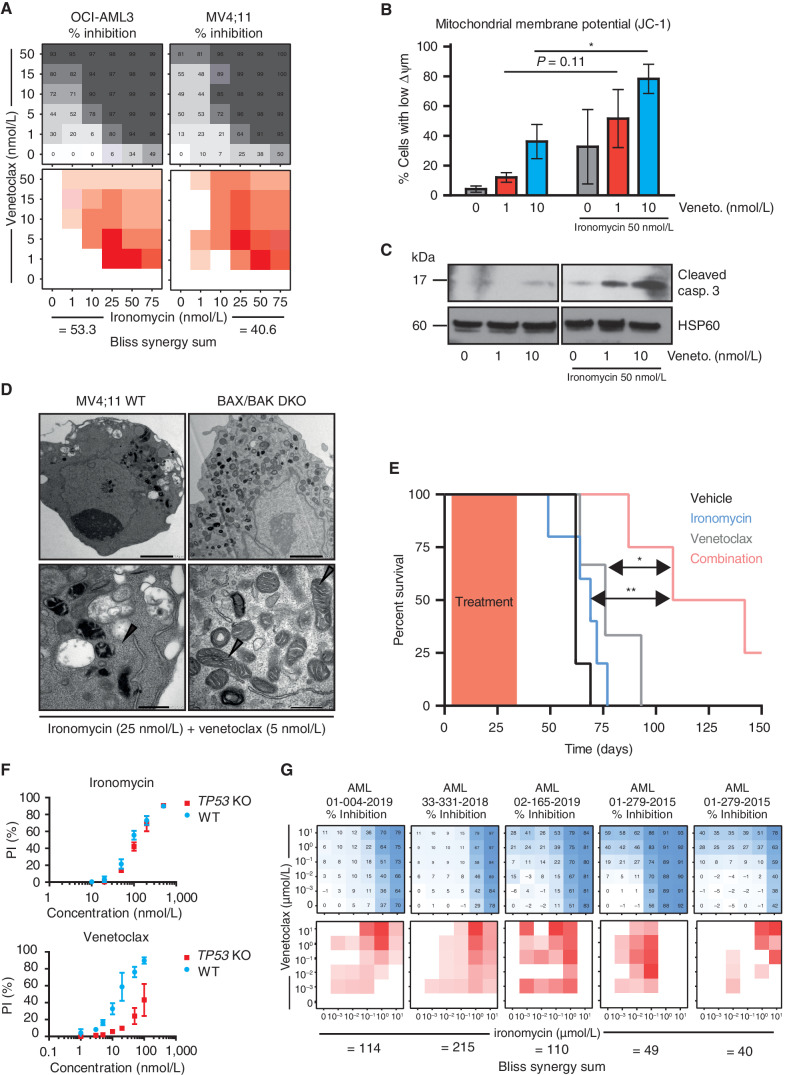
The striking activity of ironomycin in AML cells and the clear distinction in the mechanism of cell death to BH3 mimetics raised the possibility that ironomycin can dramatically enhance the activity of BH3 mimetics. Venetoclax has been paradigm-shifting through its clinical impact across a range of hematologic malignancies, including AML, where it has now been approved by the FDA (6). To assess the effects of ironomycin in combination with venetoclax, we first assessed the activity of the compounds in AML cell lines. These data showed dramatic synergy at low nanomolar concentrations of both drugs (Fig. 7A). Remarkably, in the presence of low doses of ironomycin, even 1 nmol/L of venetoclax is sufficient to result in a marked depolarization of mitochondrial membrane potential and the potent activation of caspases (Fig. 7B and C). When doses of both compounds, which have negligible effects in isolation, are combined, there are dramatic cellular and mitochondrial morphologic changes at an ultrastructural level. Notably, this striking synergy is entirely dependent on the activation of BAX/BAK, because in the absence of these essential mediators of MOMP, the cellular changes seen with combination therapy are completely averted (Fig. 7D).
Figure 7.
Ironomycin shows marked synergy with BH3 mimetics and overcomes resistance to venetoclax. A, Heat maps showing the percentage of inhibition assessed by FACS (PI) using ironomycin and venetoclax as single agent and in combination in two AML cell lines (top) and Bliss calculation measuring synergy between the two drugs (bottom; n = 3 biological replicates). B, Analysis of mitochondrial membrane potential (Δ Ψm) using JC-1 staining assessed by FACS after 24 hours of low-dose venetoclax with or without low-dose ironomycin treatments. Loss of JC-1 staining is associated with a loss of Δ Ψm (n = 3 biological replicates, mean ± SEM; *, P < 0.05). C, Immunoblot showing cleaved caspase-3 after 24 hours of low-dose venetoclax with or without low-dose ironomycin treatments in OCI-AML3 cells. D,TEM images of mitochondria in MV4;11 WT (left) and BAX/BAK DKO cells (right) treated with low doses of ironomycin and venetoclax for 36 hours. Gray arrowheads, mitochondria in BAX/BAK DKO cells; black arrowheads, disrupted mitochondrial integrity. Scale bars, 2 μm, top; 500 nm, bottom. E, Kaplan–Meier analyses showing survival of NSG mice transplanted with MV4;11 cells (n = 5 mice per cohort) treated with ironomycin, venetoclax, and combination of the two drugs (*, P < 0.05; **, P < 0.01). F, Cell viability assessed by FACS analysis of PI staining. We treated MV4;11 WT and MV4;11 cells with TP53 KO for 72 hours with ironomycin (top) and venetoclax (bottom). G, Heat maps showing the effect on cell viability assessed by flow cytometry (PI) in response to escalating doses of ironomycin and venetoclax for 5 days as single agent and in combination in five patients known to be clinically resistant to venetoclax (top) and Bliss calculation measuring synergy between the two drugs (bottom; n = 1 representative experiment).
Although both venetoclax and ironomycin have been shown to exhibit efficacy as single agents in preclinical models of cancer (9, 41), the cornerstone of clinical cancer management is to combine agents with nonoverlapping mechanisms of action to enable the delivery of lower doses of the individual drugs, which in turn minimizes side effects while maximizing efficacy. To test the preclinical validity of combining ironomycin with venetoclax in vivo, we transplanted MV4;11 cells into NOD-SCID IL2Rγ-null (NSG) mice, a model that results in an aggressive and lethal malignancy. Following transplantation, mice were treated with a low dose of venetoclax and ironomycin alone or in combination. As expected, single-agent low-dose therapy did not result in any survival advantage; however, the combination of venetoclax and ironomycin demonstrated significant efficacy (Fig. 7E). Importantly, this highly effective combination therapy was also very well tolerated (Supplementary Fig. S7A–S7D), suggesting no overt toxicity to normal hematopoiesis.
Venetoclax has unquestionably changed the natural history for many patients with AML, with the vast majority of patients deriving a meaningful response to treatment. For subjects with primary or adaptive resistance to venetoclax-based therapy, median survival from treatment failure is only 2.4 months, highlighting the urgent and unmet need for more effective salvage options (6, 42–44). In this regard, it is now well established that patients with AML with TP53 mutation show much poorer outcomes with venetoclax-based combination therapies (6, 42, 45), and our data confirm previous preclinical studies that showed venetoclax resistance in TP53-deficient AML cells (46, 47). Importantly, ironomycin has equal efficacy in AML cells that are either replete or deficient for TP53 (Fig. 7F). These data prompted us to determine if ironomycin had activity in other settings that mediate venetoclax resistance. To address this, we assessed five primary AML samples derived from patients with venetoclax-resistant or venetoclax-refractory disease and varied genetic alterations (Supplementary Fig. S7E). The combination of venetoclax and ironomycin enhanced the magnitude of cell death, and Bliss synergy sum confirmed strong synergy between the two drugs (Fig. 7G). These results demonstrate that ironomycin can be leveraged to resensitize AML cells to venetoclax and substitute for cytotoxic drugs as a more effective therapeutic combination in the salvage setting.
Discussion
Oncogenic reprogramming of cellular metabolism is an established hallmark of cancer (4). In fact, increased glucose consumption to fuel ATP production via the mitochondrial ETC has been a recognized feature of cancer cells for nearly a century (48). AML is an excellent example of a malignancy that has both cell-autonomous and non–cell-autonomous mechanisms to increase glucose availability for the malignant population of cells (24). There is also convincing evidence highlighting the importance of mitochondrial metabolism and particularly a dependency on OxPhos in AML (49, 50). Indeed, the success of combination therapy with azacitidine and venetoclax over venetoclax monotherapy has, at least in part, been attributed to the greater inhibition of OxPhos in AML cells (51). Consistent with these data is supporting evidence showing that perturbation of mitochondrial structure results in an ATF4-mediated mitochondrial stress response, which dramatically sensitizes AML cells to venetoclax (40). Together, these orthogonal lines of evidence clearly converge on mitochondrial function as a critical component necessary for the maintenance of AML and also highlight key opportunities for novel therapeutic intervention.
Central to mitochondrial activity is the availability of iron, particularly as the mitochondrial biosynthetic pathway for the iron–sulfur cluster proteins is essential for the survival of eukaryotic cells (52). These proteins play an essential role in a range of cellular processes, including cellular respiration (52); therefore, alteration in iron availability within the mitochondria is likely to have a profound effect on the functional integrity of mitochondria and consequently cellular metabolism. The cellular labile iron pool involves an intricate equilibrium established between iron trafficking and storage between the cytosol, lysosomes, and mitochondria (53). Critically, it is now apparent that lysosomes function as the major rheostat for cellular iron availability, as impaired lysosomal acidification results in a profound reduction in mitochondrial iron, resulting in a marked impairment of mitochondrial function (54). Consistent with these data, here we provide a pharmacologic approach to leverage this established pathway for therapeutic gain in cancer. We demonstrated that ironomycin uniquely localizes to the lysosome, where it sequesters iron. This in turn results in a reduction of mitochondrial iron and the potent disruption of mitochondrial metabolism. Using a complementary unbiased systems biology approach involving genome-wide CRISPR screening, global transcriptomic and metabolomic analyses, our data clearly converge on mitochondrial metabolism as being central to the activity of ironomycin. Importantly, the targeting of this metabolic dependency in AML is directly coupled to the activation of BAX/BAK and MOMP leading to cell death.
The iron-dependent cell death induced by ironomycin is associated both with prominent lipid peroxidation and on BAX/BAK activation, yet it has few other shared features with the canonical ferroptotic and apoptotic pathways. Until recently, it was generally believed that the activation of BAX/BAK and MOMP was solely governed by the functional interplay between the BCL2 family members (37). However, growing evidence supports the fact that BAX/BAK activation is not solely reliant on the BH3 family members (38), and there is a wealth of accumulating evidence implicating the mitochondrial outer membrane and various lipids in the activation of BAX/BAK (37, 39, 55). Our ultrastructural data and live-cell imaging assays clearly highlight that ironomycin has conspicuous effects on mitochondrial morphology and unquestionably compromises the integrity of the mitochondrial outer membrane. However, the precise stimulus by which ironomycin activates BAX/BAK remains to be established and will be the subject of future work. Nevertheless, here we have provided several lines of evidence to demonstrate that BH3 mimetics and ironomycin activate BAX/BAK through independent nonoverlapping pathways. These findings have potential future clinical application as we find marked synergy with these drugs. Notably, combination with low doses of venetoclax and ironomycin is well tolerated in animals without any discernible systemic toxicity or nadir in peripheral blood counts. Yet at these doses, the combination therapy results in marked anticancer activity, resulting in an impressive survival advantage. Equally important is the fact that ironomycin overcomes resistance to venetoclax in cells of patients who had failed clinical treatment with this agent. Together, our data provide substantial preclinical evidence supporting the fact that reducing mitochondrial iron represents a promising novel strategy to negate the metabolic dependency on mitochondrial metabolism in cancers such as AML and induce MOMP-mediated cell death in a synergistic manner to BH3 mimetics.
Methods
Cell Culture
Human cell lines (MOLM-13, OCI-AML3, and MV4;11) were maintained in RPMI 1640 + glutamine supplemented with 10% FCS, 100 IU/mL penicillin, and 100 μg/mL streptomycin under standard culture conditions (5% CO2, 37°C). All cell lines were regularly tested and verified to be Mycoplasma negative by PCR analysis by in-house genotyping. Human cell lines were authenticated by STR profiling through the Australian Genome Research Facility. Human AML cell lines were obtained from ATCC.
Data and Materials Availability
All RNA-seq data sets generated throughout the course of this study have been uploaded to the NCBI Bioproject under accession number GSE186088.
Click Chemistry
Cells were treated with lysotracker deep red (Thermo Fisher L12492; 50 nmol/L) directly in the culture medium for 1 hour, washed with culture medium once (400G, 4′), and resuspended in culture medium and then treated for 1 hour with ironomycin 10 μmol/L. After two PBS washes, cytospin was performed using 2 × 105 cells (200G, 10′), fixed with 4% PFA PBS in pH 7.4 for 10 minutes at room temperature, and washed three times with PBS. Cells were fixed with PBS containing 0.25% Tween-20 for 10 minutes and washed in PBS-0.1% Tween three times for 5 minutes. The click reaction cocktail was prepared from Click-iT EdU Imaging kits (C10337, Life Technologies) according to the manufacturer's protocol, with slight modifications.
CRISPR Screen
OCI-AML3 cells were transduced with a lentiviral vector encoding Cas9 and selected with blasticidin. For the screen, 108 Cas9 cells were infected with the pooled lentiviral genome-wide sgRNA library at a multiplicity of infection of 0.3. The percentage of cells infected was determined by flow cytometry–based evaluation of mCherry-positive (sgRNA-expressing) cells 72 hours following transduction. Infected cells were selected with 2 mg/mL puromycin for 72 hours, starting 48 hours after transduction. Screen was performed in duplicate with either DMSO or ironomycin 200 nmol/L for 7 days and cultured for an additional 7 days in drug-free medium. Living cells [propidium iodide (PI) negative] were enriched by one round of FACS at day 15 following transduction with the sgRNA library, and a proportion of cells (named 1a in Fig. 2A) was re-treated with 500 nmol/L for 3 days following by another round of living cell FACS (named 1b in Fig. 2A). Genomic DNA was extracted (Puregene Core Kit A, Qiagen) from both the sorted cells and an unselected pool of mutagenized cells grown for the same amount of time. sgRNA sequences were amplified by two rounds of PCR, with the second-round primers containing adaptors for Illumina sequencing. Samples were sequenced with single-end 75-bp reads on an Illumina NextSeq. The sequence reads were trimmed to remove the constant portion of the sgRNA sequences with cutadapt and then mapped to the reference sgRNA library with bowtie2. After filtering to remove multialigning reads, the read counts were computed for each sgRNA. The RSA algorithm was used to rank the genes for which targeting sgRNAs were significantly enriched in the sorted populations compared with the control unsorted populations grown in parallel.
Metabolomics
MV4;11 cells were maintained in full-growth medium, and fresh medium was added at the time cells were treated with ironomycin. Following a 24-hour treatment, 3 × 106 MV4;11 cells were harvested by centrifugation, washed with normal saline, and cell pellets were snap-frozen. For metabolite extraction, cell pellets were resuspended in 500 μL ice-cold MeOH:H2O (80:20) containing internal standards (13C-AMP, 13C-UMP, 13C Sorbitol, and 13C Valine) and vortexed. Samples were incubated on ice for 5 minutes, vortexed, and debris was pelleted by centrifugation at 16,000 × g for 10 minutes. The resulting supernatants were injected and analyzed by hydrophilic interaction liquid chromatography and high-resolution mass spectrometry (Agilent 6545 LC/Q-TOF). Quality control checks were performed using QTOF MassHunter Quant software (Agilent), and metabolite peak calling was conducted using MAVEN analysis software (56).
Long-Term Widefield Imaging Assay
MCL1 −/− Bax −/− Bak −/− MEFs expressing mNeonGreen-tagged BAX and TOMM20-Halo were generated previously (57). Cells were plated in 96-well optical-bottom plates (Nunc, Thermo Fisher 165305) the previous day, before media were replaced with Leibovitz's L-15 medium (GIBCO, Thermo Fisher 21083027) supplemented with 10% FBS, 25 mmol/L HEPES (GIBCO, Thermo Fisher 15630080), penicillin/streptomycin (Sigma P0781, 10 μL/mL), 100 nmol/L JaneliaFluor-646 (JF646) Dye and PI (0.5 μg/mL), and the drug treatments as indicated. Cells were imaged on a Leica DMi8 widefield microscope (within an environment-controlled chamber) using a 63×/1.3 NA glycerol immersion objective, with images captured at 12-minute intervals for 72 hours. Quantification of images was done in a blinded setup, via manual counting using ImageJ, with 10 to 20 cells counted per field of view.
AML Xenograft Model
All mouse studies were conducted with approval from the Alfred Medical Research and Education Precinct Animal Ethics Committee. For in vivo studies, venetoclax was dissolved in 5% DMSO, 50% PEG 300, 5% Tween 80, and ddH2O 60%. Ironomycin was dissolved in PBS + 2.5% DMSO. NSG mice (8–10 weeks old) were obtained from the Animal Resources Centre. MV4;11 cells (1.5 × 105) were transplanted into nonirradiated mice. Three days after transplantation, mice were treated with vehicle, or with ironomycin 1 mg/kg by IP (5 days/week for 4 weeks), venetoclax 75 mg/kg by oral gavage (5 days/week for 4 weeks), or the combination. A toxicity study was performed on nonirradiated NSG mice over 4 weeks with drug treatment as described above. Animals were euthanized at the completion of treatment. Body weight was assessed daily over 4 weeks, and effects on the hematologic system were analyzed via full blood examination of peripheral blood (Cell-Dyn Hematology Analyzer, Abbott) collected weekly.
Primary AML Samples
Samples were collected from patients with AML treated at the Alfred Hospital after obtaining written informed consent. Studies were conducted in accordance with recognized ethical guidelines through the Human Research Ethics Committee of the Alfred Hospital and the Declaration of Helsinki for all subjects. Primary AML cells were cultured in StemSpan SFEM (STEMCELL Technologies) supplemented with 10 ng/mL IL3, 10 ng/mL IL6, 50 ng/mL Stem Cell Factor (SCF), and 50 ng/mL FLT3L (all PeproTech) and 35 nmol/L UM171 and 500 nmol/L stemreginin (STEMCELL Technologies). For primary patient synergy analyses, 50,000 AML cells were plated in each well of a 96-well plate, and cultured with escalating doses of ironomycin and venetoclax for 5 days in medium conditions as described above. After 5 days, viability was assessed via PI staining and flow cytometry (BD FACSCanto). Synergy was determined using the Bliss synergy model.
Supplementary Methods are available in the Supplementary Data file.
Supplementary Material
Acknowledgments
This study was supported by NHMRC Investigator Grant (#1196749, to M.A. Dawson), Cancer Council Victoria Dunlop Fellowship (to M.A. Dawson), Howard Hughes Medical Institute international research scholarship (#55008729, to M.A. Dawson), NHMRC Investigator Grant (#1196755, to S.J. Dawson), and CSL Centenary fellowship (to S.J. Dawson) The R. Rodriguez research group is funded by the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement no. 647973), the Fondation Charles Defforey-Institut de France, Ligue Contre le Cancer (Equipe Labellisée), and Région Ile-de-France. A.G. Cox was supported by an NHMRC Investigator Grant (GNT1176732) and an ARC Discovery Project Grant (DP200102693). K.K. Brown was supported by an NHMRC Project Grant (GNT1146642), a VCA Mid-Career Research Fellowship (MCRF17020), and a Susan G. Komen Career Catalyst Research Grant (CCR18548354). We thank members of the Dawson laboratory, particularly Susan Jackson for technical assistance with experiments. We thank the ICP-MS platform at the Institut de Physique du Globe de Paris. This research used National Collaborative Research Infrastructure Strategy (NCRIS)-enabled Metabolomics Australia infrastructure at the University of Melbourne, funded through BioPlatforms Australia. We thank members of the Y. Collette laboratory and the TrGET Platform, particularly C. Montersino for technical assistance with experiments. We acknowledge S. Anthony and T.L. Saunders for experimental assistance and G. Brumatti for providing Birinapant and IDN-6556. S. Garciaz thanks Nuovo-Soldati for Cancer Research and Fondation de France for the received grants.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
T. Cañeque reports a patent for PCT-EP2021062729 pending. S. Dawson reports personal fees from AstraZeneca and Inivata and grants from Cancer Therapeutics CRC outside the submitted work. A.H. Wei reports grants, personal fees, and other support from Celgene/Bristol Myers Squibb, Novartis, Servier, Astellas, Genentech, AbbVie, Amgen, Syndax, and AstraZeneca, personal fees and other support from Janssen, MacroGenics, Pfizer, and Gilead, and grants from Astex outside the submitted work, as well as patent for venetoclax with royalties paid. R. Rodriguez reports a patent for the use of the lead compound ironomycin for the treatment of AML when used in combination with standard of care pending. M.A. Dawson reports personal fees from GlaxoSmithKline, Storm Therapeutics, Cambridge Epigenetix, CTX CRC, and Celgene and grants from Pfizer outside the submitted work, as well as a patent for 20305750.0 pending. No disclosures were reported by the other authors.
Authors’ Contributions
S. Garciaz: Conceptualization, investigation, writing–original draft, writing–review and editing. A.A. Guirguis: Conceptualization and investigation. S. Muller: Conceptualization and investigation. F.C. Brown: Conceptualization and investigation. Y. Chan: Conceptualization, data curation, and investigation. A. Motazedian: Investigation. C.L. Rowe: Investigation. J.A. Kuzich: Investigation. K. Chan: Investigation. K. Tran: Investigation. L. Smith: Investigation. L. MacPherson: Investigation. B. Liddicoat: Investigation. E.Y.N. Lam: Investigation. T. Cañeque: Investigation. M.L. Burr: Investigation. V. Litalien: Investigation. G. Pomilio: Investigation. M. Poplineau: Investigation. E. Duprez: Resources. S. Dawson: Supervision and funding acquisition. G. Ramm: Resources and validation. A.G. Cox: Conceptualization and investigation. K.K. Brown: Conceptualization and investigation. D.C. Huang: Resources. A.H. Wei: Resources. K. McArthur: Conceptualization, investigation, writing–review and editing. R. Rodriguez: Conceptualization, resources, supervision, funding acquisition, writing–original draft, writing–review and editing. M.A. Dawson: Conceptualization, resources, supervision, writing–original draft, writing–review and editing.
References
- 1. Murphy MP, Hartley RC. Mitochondria as a therapeutic target for common pathologies. Nat Rev Drug Discov 2018;17:865–86. [DOI] [PubMed] [Google Scholar]
- 2. Green DR, Galluzzi L, Kroemer G. Cell biology. Metabolic control of cell death. Science 2014;345:1250256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bhola PD, Letai A. Mitochondria-judges and executioners of cell death sentences. Mol Cell 2016;61:695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab 2016;23:27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stein EM, DiNardo CD, Pollyea DA, Fathi AT, Roboz GJ, Altman JKet al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 2017;130:722–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. DiNardo CD, Jonas BA, Pullarkat V, Thirman MJ, Garcia JS, Wei AHet al. Azacitidine and venetoclax in previously untreated acute myeloid leukemia. N Engl J Med 2020;383:617–29. [DOI] [PubMed] [Google Scholar]
- 7. Roberts AW, Davids MS, Pagel JM, Kahl BS, Puvvada SD, Gerecitano JFet al. Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. N Engl J Med 2016;374:311–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tam CS, Anderson MA, Pott C, Agarwal R, Handunnetti S, Hicks RJet al. Ibrutinib plus venetoclax for the treatment of mantle-cell lymphoma. N Engl J Med 2018;378:1211–23. [DOI] [PubMed] [Google Scholar]
- 9. Mai TT, Hamai A, Hienzsch A, Caneque T, Muller S, Wicinski Jet al. Salinomycin kills cancer stem cells by sequestering iron in lysosomes. Nat Chem 2017;9:1025–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Muller S, Sindikubwabo F, Caneque T, Lafon A, Versini A, Lombard Bet al. CD44 regulates epigenetic plasticity by mediating iron endocytosis. Nat Chem 2020;12:929–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Caneque T, Gomes F, Mai TT, Maestri G, Malacria M, Rodriguez R. Synthesis of marmycin A and investigation into its cellular activity. Nat Chem 2015;7:744–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cañeque T, Müller S, Rodriguez R. Visualizing biologically active small molecules in cells using click chemistry. Nat Rev Chem 2018;2:202–15. [Google Scholar]
- 13. Tyler DS, Vappiani J, Caneque T, Lam EYN, Ward A, Gilan Oet al. Click chemistry enables preclinical evaluation of targeted epigenetic therapies. Science 2017;356:1397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CEet al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012;149:1060–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol 2021;22:266–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marine JC, Dawson SJ, Dawson MA. Non-genetic mechanisms of therapeutic resistance in cancer. Nat Rev Cancer 2020;20:743–56. [DOI] [PubMed] [Google Scholar]
- 17. Cancer Genome Atlas Research Network, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJet al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Collard F, Baldin F, Gerin I, Bolsee J, Noel G, Graff Jet al. A conserved phosphatase destroys toxic glycolytic side products in mammals and yeast. Nat Chem Biol 2016;12:601–7. [DOI] [PubMed] [Google Scholar]
- 19. Mugabo Y, Zhao S, Seifried A, Gezzar S, Al-Mass A, Zhang Det al. Identification of a mammalian glycerol-3-phosphate phosphatase: role in metabolism and signaling in pancreatic beta-cells and hepatocytes. Proc Natl Acad Sci U S A 2016;113:E430–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muramatsu T. Basigin (CD147), a multifunctional transmembrane glycoprotein with various binding partners. J Biochem 2016;159:481–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang YH, Israelsen WJ, Lee D, Yu VWC, Jeanson NT, Clish CBet al. Cell-state-specific metabolic dependency in hematopoiesis and leukemogenesis. Cell 2014;158:1309–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ju HQ, Zhan G, Huang A, Sun Y, Wen S, Yang Jet al. ITD mutation in FLT3 tyrosine kinase promotes Warburg effect and renders therapeutic sensitivity to glycolytic inhibition. Leukemia 2017;31:2143–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rashkovan M, Ferrando A. Metabolic dependencies and vulnerabilities in leukemia. Genes Dev 2019;33:1460–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ye H, Adane B, Khan N, Alexeev E, Nusbacher N, Minhajuddin Met al. Subversion of systemic glucose metabolism as a mechanism to support the growth of leukemia cells. Cancer Cell 2018;34:659–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. El-Mir MY, Nogueira V, Fontaine E, Averet N, Rigoulet M, Leverve X. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J Biol Chem 2000;275:223–8. [DOI] [PubMed] [Google Scholar]
- 26. Janzer A, German NJ, Gonzalez-Herrera KN, Asara JM, Haigis MC, Struhl K. Metformin and phenformin deplete tricarboxylic acid cycle and glycolytic intermediates during cell transformation and NTPs in cancer stem cells. Proc Natl Acad Sci U S A 2014;111:10574–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SEet al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 2008;134:112–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res 2016;44:D1251–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bao XR, Ong SE, Goldberger O, Peng J, Sharma R, Thompson DAet al. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. Elife 2016;5:e10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Quiros PM, Prado MA, Zamboni N, D'Amico D, Williams RW, Finley Det al. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J Cell Biol 2017;216:2027–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Paul BT, Manz DH, Torti FM, Torti SV. Mitochondria and Iron: current questions. Expert Rev Hematol 2017;10:65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Park JT, Lee YS, Cho KA, Park SC. Adjustment of the lysosomal-mitochondrial axis for control of cellular senescence. Ageing Res Rev 2018;47:176–82. [DOI] [PubMed] [Google Scholar]
- 33. Rouault TA, Tong WH. Iron-sulphur cluster biogenesis and mitochondrial iron homeostasis. Nat Rev Mol Cell Biol 2005;6:345–51. [DOI] [PubMed] [Google Scholar]
- 34. Chou WC, Jie C, Kenedy AA, Jones RJ, Trush MA, Dang CV. Role of NADPH oxidase in arsenic-induced reactive oxygen species formation and cytotoxicity in myeloid leukemia cells. Proc Natl Acad Sci U S A 2004;101:4578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim HJ, Khalimonchuk O, Smith PM, Winge DR. Structure, function, and assembly of heme centers in mitochondrial respiratory complexes. Biochim Biophys Acta 2012;1823:1604–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bock FJ, Tait SWG. Mitochondria as multifaceted regulators of cell death. Nat Rev Mol Cell Biol 2020;21:85–100. [DOI] [PubMed] [Google Scholar]
- 37. Luo X, O'Neill KL, Huang K. The third model of Bax/Bak activation: a Bcl-2 family feud finally resolved? F1000Res 2020;9:F1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O'Neill KL, Huang K, Zhang J, Chen Y, Luo X. Inactivation of prosurvival Bcl-2 proteins activates Bax/Bak through the outer mitochondrial membrane. Genes Dev 2016;30:973–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chipuk JE, McStay GP, Bharti A, Kuwana T, Clarke CJ, Siskind LJet al. Sphingolipid metabolism cooperates with BAK and BAX to promote the mitochondrial pathway of apoptosis. Cell 2012;148:988–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen X, Glytsou C, Zhou H, Narang S, Reyna DE, Lopez Aet al. Targeting mitochondrial structure sensitizes acute myeloid leukemia to venetoclax treatment. Cancer Discov 2019;9:890–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen Jet al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med 2013;19:202–8. [DOI] [PubMed] [Google Scholar]
- 42. Chua CC, Roberts AW, Reynolds J, Fong CY, Ting SB, Salmon JMet al. Chemotherapy and venetoclax in elderly acute myeloid leukemia trial (CAVEAT): a phase Ib dose-escalation study of venetoclax combined with modified intensive chemotherapy. J Clin Oncol 2020;38:3506–17. [DOI] [PubMed] [Google Scholar]
- 43. Wei AH, Montesinos P, Ivanov V, DiNardo CD, Novak J, Laribi Ket al. Venetoclax plus LDAC for newly diagnosed AML ineligible for intensive chemotherapy: a phase 3 randomized placebo-controlled trial. Blood 2020;135:2137–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Maiti A, Rausch CR, Cortes JE, Pemmaraju N, Daver NG, Ravandi Fet al. Outcomes of relapsed or refractory acute myeloid leukemia after frontline hypomethylating agent and venetoclax regimens. Haematologica 2021;106:894–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim K, Maiti A, Loghavi S, Pourebrahim R, Kadia TM, Rausch CRet al. Outcomes of TP53-mutant acute myeloid leukemia with decitabine and venetoclax. Cancer 2021;127:3772–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nechiporuk T, Kurtz SE, Nikolova O, Liu T, Jones CL, D'Alessandro Aet al. The TP53 apoptotic network is a primary mediator of resistance to BCL2 inhibition in AML cells. Cancer Discov 2019;9:910–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thijssen R, Diepstraten ST, Moujalled D, Chew E, Flensburg C, Shi MXet al. Intact TP-53 function is essential for sustaining durable responses to BH3-mimetic drugs in leukemias. Blood 2021;137:2721–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol 1927;8:519–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stuani L, Sabatier M, Saland E, Cognet G, Poupin N, Bosc Cet al. Mitochondrial metabolism supports resistance to IDH mutant inhibitors in acute myeloid leukemia. J Exp Med 2021;218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gallipoli P, Giotopoulos G, Tzelepis K, Costa ASH, Vohra S, Medina-Perez Pet al. Glutaminolysis is a metabolic dependency in FLT3(ITD) acute myeloid leukemia unmasked by FLT3 tyrosine kinase inhibition. Blood 2018;131:1639–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pollyea DA, Stevens BM, Jones CL, Winters A, Pei S, Minhajuddin Met al. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat Med 2018;24:1859–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Braymer JJ, Lill R. Iron-sulfur cluster biogenesis and trafficking in mitochondria. J Biol Chem 2017;292:12754–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lv H, Shang P. The significance, trafficking and determination of labile iron in cytosol, mitochondria and lysosomes. Metallomics 2018;10:899–916. [DOI] [PubMed] [Google Scholar]
- 54. Yambire KF, Rostosky C, Watanabe T, Pacheu-Grau D, Torres-Odio S, Sanchez-Guerrero Aet al. Impaired lysosomal acidification triggers iron deficiency and inflammation in vivo. Elife 2019;8:e51031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter Ret al. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 2002;111:331–42. [DOI] [PubMed] [Google Scholar]
- 56. Agrawal S, Kumar S, Sehgal R, George S, Gupta R, Poddar Set al. El-MAVEN: a fast, robust, and user-friendly mass spectrometry data processing engine for metabolomics. Methods Mol Biol 2019;1978:301–21. [DOI] [PubMed] [Google Scholar]
- 57. McArthur K, Whitehead LW, Heddleston JM, Li L, Padman BS, Oorschot Vet al. BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science 2018;359:eaao6047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.