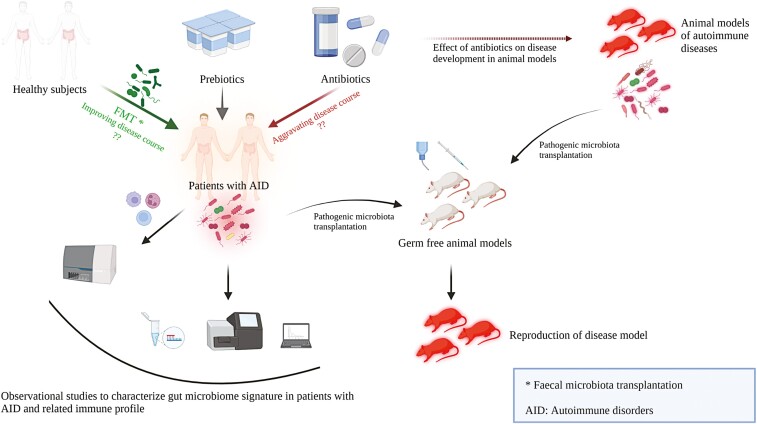
Summary
Autoimmune diseases have long been known to share a common pathogenesis involving a dysregulated immune system with a failure to recognize self from non-self-antigens. This immune dysregulation is now increasingly understood to be induced by environmental triggers in genetically predisposed individuals. Although several external environmental triggers have been defined in different autoimmune diseases, much attention is being paid to the role of the internal micro-environment occupied by the microbiome, which was once termed “the forgotten organ.” In this regard, the gut microbiome, serving as an intermediary between some of those external environmental effectors and the immune system, helps programming of the immune system to be tolerant to innocent external and self-antigens. However, in the presence of perturbed gut microbiota (dysbiosis), the immune system could be erroneously directed in favor of pro-inflammatory pathways to instigate different autoimmune processes.
An accumulating body of evidence, including both experimental and human studies (observational and interventional), points to the role of the gut microbiome in different autoimmune diseases. Such evidence could provide a rationale for gut microbiome manipulation with therapeutic and even preventative intent in patients with established or predisposed to autoimmune diseases, respectively.
Perturbations of the gut microbiome have been delineated in some immune mediated diseases, IBD in particular. However, such patterns of disturbance (microbiome signatures) and related pathogenetic roles of the gut microbiome are context dependent and cannot be generalized in the same exact way to other autoimmune disorders, and the contribution of the gut microbiome to different disease phenotypes has to be precisely defined.
In this review, we revise the evidence for a role of the gut microbiome in various autoimmune diseases and possible mechanisms mediating such a role.
Keywords: Gut microbiome, dysbiosis, immune dysregulation, auto-immune diseases
Several studies point to a role of gut microbiome in the development and progress of different autoimmune diseases. In this review, we revise the evidence and potential mechanism for such a role. This evidence could provide a strong rationale for instigating future clinical trials of gut microbiome manipulation as a therapeutic intervention in patients with established autoimmune diseases.
Graphical Abstract
Graphical Abstract.
Introduction
Autoimmune disorders (AID) have been recognized to be triggered by environmental factors in genetically susceptible individuals with evidence of epigenetic dysregulation being involved in the pathogenesis [1, 2]. Several environmental agents (e.g. heavy metals, smoking, and pesticides) have been identified to have a role in the pathogenesis of autoimmune disorders [3]. The gut microbiome, in particular, has recently been implicated in contributing to autoimmune disorders via the pro-inflammatory and immune deregulatory effects that imbalance (dysbiosis) of the microbiome can induce [4].
While a perturbation of immune homeostasis leading to a predominance of effector Th1, Th17 lymphocytes and plasma cells is an essential pre-requisite for the development of autoimmune disease states, antigen presenting cells APCs (dendritic cells and macrophages) usually instigate the process by sampling different antigens and activating the pro-inflammatory milieu. This is critical for the interplay between gut microbiota and the immune system where these APCs can transport luminal microbiota-derived antigens and toxins and present them to effector T and B lymphocytes, thereby activating them [5].
Although in health, this interaction promotes immune system homeostasis by suppressing pro-inflammatory pathways and increasing immune-modulatory T-Reg cell differentiation and expansion [6, 7], changes in gut microbiota profiles in individuals with a genetic predisposition can lead to pathogenic autoimmune pro-inflammatory responses. An example of this is Th17 activation, with pathologic sequelae, can be induced by certain gut microbiota spp. including Enterococcus gallinarum, Bifidobacterium adolescentis, and Prevotella copri [8–10].
Interestingly, the effects of a specific gut bacterial species on the immune system are context-dependent. For instance, Akkermansia muciniphila mono-colonization in gnotobiotic C57BL/6 mouse models restricts activation of follicular T helper cells (Tfh) to Peyer’s patches with no appreciable activation of other immune cells whereas inoculation of the same species in specific pathogen free (SPF) mice leads to activation of all phenotypes of CD4+ Th cells with extension to the lamina propria as well [11].
The impact of gut microbiota on the immune system extends beyond the effect on T cell repertoire to include over-activation of antibody producing plasma cells, which contributes to autoimmunity [12]
In this article, we review the evidence pointing to gut microbial dysbiosis driving auto immune disease in humans.
Several lines of evidence point to a role for gut microbiota imbalance in the initiation and progression of AID. These include:
(1) Gut microbiota signatures in different AIDs—human cross sectional studies
Different studies have characterized microbiome alterations in different autoimmune diseases, with some of these changes exhibiting diagnostic potential as demonstrated by machine learning models designed to effectively predict the disease state. However, two major challenges exist before adopting any of these models. The first is the inconsistency between these studies regarding the microbiota taxa included in different models. This can be driven by differences in patient cohorts studied, geographical location, and varying methodologies used. The other challenge is the uncertainty regarding association or causation [13]. However, in this regard, the application of some statistical methods such as Mendelian randomization, which integrates genetic variants gleaned from Genome Wide Association Studies (GWAS) and associated with microbiota exposures, to conclude causal associations between gut microbiota and different autoimmune diseases can provide some valuable insight [14, 15]. Furthermore, many of the studies address microbiota dysbiosis in patients with well-established disease, thus introducing the confounding effect of therapies. In addition, with the exception of rheumatoid arthritis, which has been relatively thoroughly studied, few human cross-sectional studies in other diseases have addressed tissue immunology changes in relation to the reported microbiome dysbiosis.
The supplementary table attached demonstrates some of the gut microbiome alterations observed in different AID in humans “Supplementary Table S1”.
Still, the link between gut dysbiosis and consequent aberrant immune system activation has been much better delineated in animal models.
(2) Experimental studies which link gut microbiota changes to the development of AID. In experimental models of liver disease, it was shown that translocation of Enterococcus gallinarum from gut to liver tissues induced autoimmune antibodies and pro-inflammatory consequences in mono-colonized susceptible mice, an effect which was prevented by antibiotic administration or vaccination against E. gallinarum [8]. Another murine model of primary biliary cholangitis (PBC), dnTGFβRII mice, was shown to have gut dysbiosis with higher Lachnospiraceae, Bacteroidiaceae with lower relative abundances of the Ruminococcaceae, Rikenellaceae, S24-7 families compared to wild type mice. Interestingly, dnTGFβRII mice with TLR2 knockout had a significant exaggeration of cholangitis due to bacterial translocation resulting from the impaired gut barrier induced by TLR 2 deficiency. The inflammatory process in both types of these transgenic mice was alleviated with antibiotic administration [16].
In a mouse model of rheumatoid arthritis (RA) K/BxN, specific pathogen-free mice developed arthritis after about a month, whereas the phenotype was greatly abrogated when the mice were kept in germ-free conditions [17]. It was shown that mono-colonization with segmented filamentous bacteria (SFB) promoted inflammation and induced RA-like disease in these transgenic mice via Tfh effector cell activation [17–19]. This again points to the critical interaction between gut dysbiosis and genetic predisposition for the development of autoimmune diseases given that, under normal circumstances, SFB maintain a homeostatic reciprocal relationship with Th17 that contains the gut microbiota and limits Th17 over-expansion in the gut mucosa [20].
In another mouse model of arthritis (collagen-induced arthritis), it was shown that antibiotic treatment (Baytril; enrofloxacin) caused partial depletion of gut microbiota with consequent priming of a pro-inflammatory response (IL-17A, INF-γ) that aggravated arthritis severity. Pretreatment with antibiotics prior to the development of arthritis in this model similarly resulted in an exaggerated immune response [21].
In the context of systemic lupus, Lactobacillus reuteri, showing heterogenous immune impacts, was able to exaggerate the pro-inflammatory pathway in interferon/plasmacytoid dendritic cells (INF/pDC) (even before translocation) in both transgenic lupus-prone-mice and the TLR7 externally-activated lupus mouse model. Interestingly, starch feeding, which suppressed L. reuteri and enriched Clostridiales, prevented the development of lupus and abated the pro-inflammatory pathways [22].
In a study of the relationship between gut microbiota and lupus nephritis activity in MRL/Ipr mice (mouse model of lupus nephritis), it was shown that Lactobacillales were depleted and that administration of a mixture of five strains of lactobacillus, in addition to changing gut microbiota composition toward more Lactobacillales, Clostridiales and Desulfovibrionales taxa, improved kidney function, decreased IL-6, and increased IL-10 levels in gut mucosa. Systemic effects of lactobacilli administration included an increase in serum IL-10 and a shift of Treg/Th17 ratio towards more Treg domination (anti-inflammatory). These effects were observed only in castrated male and female mice suggesting a hormone-dependent effect of probiotics [23]. Similarly, therapeutic manipulations of gut microbiota through diet alterations can have a significant impact on disease progression. This was evident by the slower development and improved course of nephritic syndrome in a mouse model of lupus nephritis (SNF-1) when given acidic water over 34 weeks compared to mice on neutral water, which had a much higher rate of severe nephritis. These effects were mediated by alterations of the immune system driven by significant gut microbiota changes affecting particularly SFB [24].
In the model described above (MRL/Ipr), somewhat paradoxically, administration of an antibiotic cocktail (ampicillin, metronidazole, neomycin, and vancomycin) from 9th week to the 16th week of age after the onset of disease resulted in an attenuated disease course as shown by improvement of kidney function, reduction of proteinuria, and reduction of IgG anti- (ds) DNA levels. The alteration of gut microbiota composition with a decrease of Lachnospiraceae and enrichment of the (pre-treatment depleted), lactobacilli was accompanied by a favorable change in the immune profile, which led to better clinical outcomes in the antibiotic treated arm. The antibiotic cocktail administration was associated with a reduction in Th17, Innate Lymphoid Cells-3 (ICL-3) and double negative cells in the spleen and kidney, which led to a drop in the pro-inflammatory IL-17 and IL-6 cytokines, and up-regulation of anti-inflammatory IL-10 [25]. In contrast to the previous study, antibiotic administration to MRL/Ipr mice before the onset of lupus, changed gut microbiota adversely increasing pathobionts Klebsiella and Proteus while depleting beneficial taxa Lactobacillus, Bifidobacterium, and Bacteroides which exaggerated disease severity. This effect of antibiotics was reversed by fecal microbiota transplantation (FMT), which mitigated disease severity by restoring the gut microbiota composition depleted by antibiotics [26]. These contrasting studies of the effects of antibiotics on the development of lupus highlight the importance of the timing of dysbiosis in relation to the maturity of the mice [25, 26].
Similarly, the role of the gut microbiome in the predisposition to multiple sclerosis (MS) has been suggested by the rapid development of autoimmune encephalitis in the SJL/J mouse model of relapsing remitting MS in specific pathogen free (SPF) mice compared to their germ free counterparts that remained disease free for their lifetime. This could be explained by the critical role of the gut microbiome in the development of a competent immune system capable of initiating an autoimmune response in genetically predisposed mice [27].
In addition, gut microbiota from patients with multiple sclerosis patients decreased anti-inflammatory IL10+ Treg cell expansion and differentiation, inducing pro-inflammatory responses with exaggerated symptoms in a mouse model of autoimmune encephalitis compared to gut microbiota from healthy controls [28]. Furthermore, in mono-colonized mice, Acinetobacter calcoaceticus induced differentiation and expansion of INF+ Th1 response and inhibited FoxP3+ Treg differentiation, in contrast to the immune-modulatory effect of Parabacteroides distasonis (depleted in MS patients), which enhanced IL10 producing CD4+ CD25+ IL10+ T cells and IL-10+ FoxP3+ Tregs [28] [28]. Similarly, fecal microbiota transplantation from MS discordant twin pairs into autoimmune encephalitis GF mouse models resulted in the development of encephalitis along with reduced IL-10 expression in a significant portion of these animals compared to mice transplanted with a healthy human microbiome [29].
Consistent with the above, administration of Lactobacillus reuteri DSM 17938 over 20 days in a mouse model of autoimmune encephalitis significantly and favorably altered gut microbiota composition in these mice to decrease relative abundances of taxa associated with encephalitis (Rikenellaceae, Anaeroplasma, and Clostridium). Also, this probiotic significantly decreased Th1/Th17 populations with their related cytokine levels in these mice models, which significantly inhibited the development of encephalitis [30].
Adoptive T-cell transfer studies also provide an insight into the immune-pathogenesis of AID, highlighting the role of gut microbiota in the pathogenesis of inflammatory bowel disease (IBD). For instance, CD4+ T lymphocytes, harvested from mice treated with antibiotics in early life, hastened the development of IBD in Rag-1 deficient mice. A genome wide analysis of these CD4+ T cells revealed disturbed expression of genes related to cell cycle regulation, cellular stress, and metabolism. Interestingly, cohousing antibiotic-treated mice with control mice decreased the elevated corticosterone levels in antibiotic-treated mice towards normal levels and abated the rapid onset of CD4+ induced experimental colitis observed previously [31]. Although not formally addressed, it seems likely that the gut microbiota stands at the intersection between antibiotics and the immunological effects observed in this study.
Translocation of gut microbiota through a leaky gut epithelial barrier with subsequent activation of Islet reactive T cells has been postulated to result in type 1 diabetes mellitus (T1 DM) in a mouse model of T1 DM, non-obese diabetic (NOD mice). In this experimental study, an impaired gut barrier with down-regulated tight junction proteins and altered mucous layer structure was shown to drive a pro-inflammatory state in the gut mucosa and the differentiation of islet reactive T cells to pro-inflammatory Th1/Th17 phenotypes, which later led to T1 DM. Interestingly, depletion of gut microbiota in BDC2.5XNOD mice by a mixture of antibiotics before induction of significant gut barrier disruption (via DSS colitis) prevented the development of T1 DM in these mice, suggesting the importance of gut microbiota translocation for activation of the pro-inflammatory state [32].
Experimental models of AIDs also provide an invaluable insight on the role of highly selective targeting of gut microbiota by certain antibiotics as a way to alter the course of the disease. For instance, early life exposure to low dose of penicillin, but not enrofloxacin or metronidazole, played a protective role against dextran sulfate induced DSS colitis in mice and dampened the pro-inflammatory Th17 pathways due to selective depletion of SFB [33]. In another study, a cocktail of antibiotics (ampicillin, metronidazole, vancomycin, and neomycin) administered in combination or singularly in B10.RIII mouse model of autoimmune uveitis resulted in improvement of uveitis clinical scores and up-regulation of the immune-modulatory Treg cells in intestinal mucosa and peripheral lymphoid tissues along with a reduction in the pool of effector T lymphocytes. Interestingly, these beneficial effects were seen only when antibiotics were administered orally inducing major gut microbiota shifts and not via the parenteral route where the effect on gut microbiota was minimal [34].
(3) Human intervention studies—probiotics and AID
Probiotics can be classified, based on the immune reactions they induce, into immunostimulatory or immunomodulatory. Immunostimulatory probiotics which induce differentiation and expansion of Th1 and Th17 can help mount an immune response against infections and malignant cells. On the other hand, immunomodulatory probiotics help expansion of Treg cells, leading to suppression and containment of inflammatory responses thereby limiting autoimmune diseases [35]
In a large prospective longitudinal cohort study (TEDDY cohort) involving 8676 children from six centers in the USA and Europe, it was found that early probiotic administration (in the first 27 days of life) was associated with a lower incidence of Islet cell autoimmunity. This effect was most evident in genetically predisposed HLA DR3/4 children but not among other genotypes [36].
In a randomized controlled trial (RCT) studying the therapeutic benefits of 8-week probiotic supplementation in patients with RA (60 patients), it was shown that a probiotic (combination of Lactobacillus casei, acidophilus, and bifidum) supplementation improved the Disease Activity Score of 28 Joints (DAS-28), improved metabolic biomarkers, and reduced high-sensitivity CRP levels significantly in the treated cohort compared to the control group [37]. In another RCT on female patients with RA, L. casei 01 supplementation as a probiotic yielded similar outcomes, with clinical and laboratory improvements in the probiotic arm. In addition, a significant improvement in the cytokine profile was observed with probiotic supplementation [38]. A systematic review and meta-analysis of nine RCTs examining the use of probiotics in RA, showed that IL-6 was significantly lower in patients receiving probiotics compared to control patients, although overall no significant difference in clinical outcomes was detected among the amalgamated groups [39].
In an in vitro study of the effects of gut microbiota from SLE patients on the immune system, co-culturing gut-microbiota-conditioned dendritic cells (DC) with naive CD4+ lymphocytes significantly promoted Th17 differentiation and expansion from CD4+ lymphocytes compared to gut microbiota from healthy controls. Interestingly, enrichment of SLE gut microbiota with either the probiotic Bifidobacterium bifidum or a combination of Ruminococcus obeum and Blautia coccoides abated this effect and decreased the Th17/Th1 ratio [40].
Probiotic administration over 2 months was associated with improvement of reflux, bloating, and distension symptoms in a cohort of otherwise stable 10 systemic sclerosis patients. The administered probiotics included either Lactobacillus GG or Bifidobacterium infantis [41].
In an interesting placebo-controlled RCT of Bifidobacterium longum BB536 administration, over 8 weeks, in patients with mild to moderate active ulcerative colitis (UC), there were significant improvements in different endoscopic activity scores in the probiotic treated group compared to the control arm, although differences between the two groups in clinical remission rates were insignificant. Probiotics were administered as supplementary therapy to their regular medication [42]. On the other hand, in the case of Crohn’s disease, an early systematic review and meta-analysis of eight RCTs on the use of probiotics for maintenance of remission indicated that probiotics failed to maintain clinical or endoscopic remission and did not offer an advantage in preventing relapse [43]. Although these results were reproducible in patients with CD, in another systematic review and meta-analysis of RCTs in patients with IBD, probiotics were able to augment the induction of remission when added to conventional therapies in patients with UC and also to maintain that remission in 21 RCTs in UC. In addition, probiotics also showed an advantage in maintaining antibiotic-induced remission in five RCTs in patients who have inflamed pouches after undergoing ileo-anal pouch anastomosis (IPAA) following subtotal colectomy to treat UC [44].
Consistent with the above, a systematic review and meta-analysis of 22 trials using probiotics in IBD showed that VSL#3 (a combination of 8 microbial strains) was beneficial in inducing remission in patients with active UC. Probiotics were also effective in preventing UC relapses to an extent similar to 5-ASA. However, these benefits of probiotics could not be extended to patients with Crohn’s disease [45]. Another systematic review and meta-analysis of 27 trials of probiotics in IBD yielded similar results, with the conclusion that probiotics, particularly VSL#3, had a significant benefit in patients with UC with varying degrees of colonic involvement. In addition, a combination of lactobacillus with a prebiotic conferred a benefit in UC patients. In Crohn’s disease, the use of a combination of VSL#3, Saccharomyces boulardii, and lactobacillus showed a trend towards benefit, although this was statistically insignificant [46]. In this systematic analysis, significant benefits of probiotics in Crohn’s disease were evident only in patients using steroids and following surgery.
In MS, probiotic administration (a combination of lactobacillales casei, fermentum, acidophilus and Bifidobacterium bifidum) for 12 weeks in a double-blinded placebo-controlled RCT (involving 60 patients), significantly improved clinical scores (Expanded Disability Status Score EDSS, Beck Depression Inventory and Depression Anxiety Scale) along with improvement of different biochemical markers of metabolism and inflammation compared to placebo (such as CRP, HOMA insulin resistance score, and HDL-cholesterol levels) [47]. Almost similar results were reproducible in another placebo-controlled RCT in patients with MS (48 patients), where probiotic administration (Lactobacillales reuteri, casei, plantarum, fermentum, Bifidobacterium infantis, and Bifidobacterium lactis) for 4 months improved the previously mentioned clinical and biochemical markers compared to placebo. In addition, probiotics significantly improved the immune profile in treated patients, decreased IL-6 and increased IL-10 significantly [48].
(4) Human intervention studies—antibiotics and AID
One of the earliest clues to the role of antibiotics in AID was the protective effect of prophylactic antibiotics against rheumatic fever after group A β-hemolytic streptococci infection [49].
From a gut microbiota perspective, the dysbiotic impact induced by antibiotics can be long lasting as shown by Jernberg et al. who showed that one week of clindamycin treatment disturbed human gut microbiota and that this disruption could be detected up to 2 years post treatment [50]. However, the gut microbiota changes induced by antibiotics provide an opportunity for microbiota manipulation with therapeutic intent [51, 52].
The impact of antibiotics on different AID provides a clue to the role of gut microbiota in these disorders given the significant dysbiotic effects of antibiotics. In an early study, the use of specific antimicrobials (phenoxymethylpenicillin and quinolones) by mothers before pregnancy was correlated with the later development of type I DM in their children. Also, frequent use of antibiotics by children (≥7 times) was associated with a higher risk of development of DM compared to those children with a lower frequency of antibiotic intake [53]. In contradistinction to the previous results, antibiotic use in early life or during pregnancy was not associated with the development of type I DM in genetically predisposed children in both the TEDDY cohort group (>15 000 children) and the Norwegian Mother and Child Cohort Study (114 215 children) [54].
Although the American College of Rheumatology included minocycline as one of the Disease Modifying Anti-Rheumatoid Drugs (DMARDs) that can be used to control RA activity [55], this cannot be generalized to other antibiotics where a large case-control study (involving 8482 RA patients and 22 661 control subjects) concluded that antibiotic prescription was associated with a higher risk of RA development in the following years [56]. This association showed a pattern of dose–response relationship where exposure to one antibiotic resulted in an odds ratio (OR) of 1.51 compared to an OR of 2.94 with exposure to >10 antibiotic prescriptions [57]. The previous results were reproducible in another nested case-control study on juvenile idiopathic arthritis patients where bacterial antibiotic, and not antiviral or antifungal, were associated with a significant risk of developing JIA (OR 2.1) with a dose response relationship detected where OR increased to three with more than five antibiotic courses. This effect was independent of the type or frequency of infection [58].
Among the long list of medications implicated in the development of drug-induced lupus, antibiotics and anti-tuberculosis drugs are well-recognized [59]. The mechanism underpinning this has to be explored further in the light of the gut dysbiosis induced by antibiotics as shown in the experimental studies of MRL/Ipr mouse models described above [24, 25]. Also, the xenobiotic metabolism offered by gut microbiota may offer another means by which gut microbiota interact with SLE [60–62].
In a large retrospective study following 1 072 426 subjects over the period from 1994 to 2009, antibiotic exposure in childhood was associated with the development of IBD in a dose-response manner, where exposure to >2 courses of antibiotics had a higher hazard ratio compared to exposure to ≤2 courses of antibiotics. The risk increased by 6% for each antibiotic exposure [63].
In IBD, antibiotics have been used with therapeutic intent in Crohn’s disease and in patients who develop chronic inflammation (pouchitis) following the creation of ileal-pouch anal anastomoses (IPAA) following subtotal colectomy for UC [64, 65]. In Crohn’s disease, antibiotics have been used for the induction of remission with relatively favorable outcomes. Crohn’s disease trials included utilization of metronidazole + ciprofloxacin for 12 weeks [66], ciprofloxacin alone for 6 weeks [67], rifaximin for 16 weeks [68], doxycycline, ciprofloxacin and hydroxychloroquine for 4 weeks followed by doxycycline and hydroxychloroquine for 24 weeks [69].
Collectively, various and sometimes contradictory outcomes of antibiotic use in different AID states provide an insight into the involvement of gut microbiota in the pathogenesis of these diseases given the consistent dysbiotic effect of broad spectrum antibiotics.
The following section assesses what has been learned about the pathogenesis of AID from FMT studies:
(5) Fecal microbiota transplant studies
Fecal microbiota transplantation appears to provide a method to transfer the health or disease phenotypes from the donor into the recipient through the transfer of a healthy or perturbed microbiome niche. This is evident from both experimental studies, where infusion of microbiota from patients with autoimmune diseases instigated similar disease processes into GF or genetically susceptible animal models, and clinical trials of fecal microbiota transplantation from healthy donors into patients with different autoimmune disease processes.
In this context, transfer of microbiota from patients with IBD into gnotobiotic mice induced a Th-17 pro-inflammatory response with down-regulation of the immunomodulatory T-cells in these mice models, with subsequent development of colitis. These changes were reversed upon transfer of gut microbiota from a healthy donor, which corrected dysbiosis and abolished the pro-inflammatory response [70].
Furthermore, FMT from a healthy donor into a rat model of DSS induced colitis improved gut microbiota perturbation in these animal models, restoring diversity and recovering some depleted gut microbiota taxa. This was associated with significant attenuation of colitis. Interestingly, removal of the noxious DSS accelerated the beneficial effects of FMT, suggesting a synergistic effect of anti-inflammatories (represented by stopping DSS) and FMT as a therapeutic approach in colitis [71].
Similarly, transplantation of GF mice with microbiota from MS patients predisposes to the development of more severe autoimmune encephalitis (when induced experimentally 6 weeks after FMT) compared to mice receiving microbiota from healthy donors. This exaggerated response was associated with a limited differentiation of T-Reg cells in mice receiving a diseased microbiome [28].
Although the transfer of fecal microbiota from RA patients into the GF arthritis (SKG) mouse model, did not induce spontaneous severe, it promoted expansion of the Th-17 cell lineage in the large intestine of these mice compared to their counterparts implanted with microbiota from healthy controls. Moreover, after zymosan injection, SKG mice transplanted with gut microbiota from RA patients developed severe arthritis and systemic inflammation as evidenced by higher RF titers, Th-17 response, and enlarged peripheral lymph nodes. Prevotella was the main promoter of such a response [72].
Again, FMT from SLE patients into GF C57/B6J mice led to a pro-inflammatory response as characterized by significantly higher levels of SLE related autoantibodies (Anti-dsDNA, ANA), serum cytokine levels (IL-6, IL-8, TNF-α, and INF-γ), significant expansion of different B-lymphocyte subsets in the intestinal lamina propria, and significant expansion of peripheral Th-17 and CD4+ CXCR3+ cells coupled with a significant reduction of immunomodulatory T-Reg cells in these mice compared to those receiving gut microbiota from healthy controls. Interestingly, FMT from SLE patients altered histidine metabolism in transplanted mice with consequent systemic effects [73].
Although, “yellow soup,” the first reported form of FMT used in treating human diarrhea, dates back more than 1700 years in China, the first formal use of fecal transplant with therapeutic intent in the western world was in 1958 for the treatment of pseudomembranous colitis [74]. Further to the successful use of FMT in the treatment of C. Difficile colitis [75], there has been a surge of trials of FMT with therapeutic intent in patients with IBD in the last decade. A systematic review and meta-analysis estimated the clinical remission and response rates after healthy donor FMT for UC at 28% and 49%, compared to placebo rates of 9% and 28% (OR 3.68, 2.48), respectively [76]. Similar odds ratios for clinical remission and response were reproducible in a later meta-analysis of FMT in UC [77]. This is very promising given the inexpensive and less side effects associated with such a treatment modality.
The immune dysregulation underpinning IBD and the successful use of FMT in these diseases instigated an expanding number of trials of FMT in other AIDs, with at least five trials of FMT in multiple sclerosis, two in RA, two in ankylosing spondylitis, one in Sjogren syndrome and three in type 1 DM currently underway in different phases according to ClinicalTrials.gov as of April 2022.
Recently, autologous fecal microbiota transplantation was shown to preserve residual beta cell function in patients newly diagnosed with type 1 DM. This was associated with a change in gut microbiome structure, metabolomics, gene expression, and more importantly T-cell immunology [78].
The animal experimental and human cross sectional and interventional studies described above demonstrate accumulating evidence for a pathogenetic link between disturbances of the microbiome and the development of AID. For this link to work in practice, gut microbiota components need to transfer across the mucosal/epithelial barrier. The following sections describe what is known about the microbiota and intestinal permeability, and the potential role of bacterial metabolism and molecular mimicry in driving pathogenesis in AID.
Gut permeability and microbial translocation studies
The gut epithelial barrier has been recognized to be impaired in AIDs [79]. This is suggested by a significant body of evidence as follows:
It has been suggested that type 1 DM originates in the gut following gut microbiota alterations by environmental factors (including diet) and impaired gut epithelial barriers that enhance bacterial translocation to prime the immune system against β cells [80]. In an early study of T1 DM patients, gut permeability studies (urinary lactulose/mannitol ratio) and electron microscopy studies of duodenal mucosa suggested significant impairment of gut permeability in those non-celiac patients compared to healthy controls [81]. This increased gut permeability, as measured by increased urinary excretion of the radioactive tracer Ci 51Cr of EDTA, is correlated at least in part to poor glucose control and the development of diabetic neuropathy [82].
In another study of intestinal permeability in diabetic patients, up-regulation of the tight junctional protein zonulin, as indicated by significantly higher serum zonulin levels, was detected in diabetic patients compared to age and sex matched controls. Higher zonulin levels were correlated with higher intestinal permeability (as measured by lactulose/mannitol urinary ratio following the administration of sugar permeability probes) and disturbed expression of other tight junctional proteins such as claudin 1, 2 and myosin IXB. Interestingly, higher serum zonulin level were shown to precede the development of DM by 3.5 ± 0.9 years [83]. This was confirmed in an experimental study on DM prone BB rat models, which showed higher levels of intestinal intraluminal zonulin (35 fold) compared to DM resistant BB rats. Antagonists of zonulin receptors in these DM prone rats significantly decreased the incidence of DM by 70% [84].
Interestingly, enhanced gut permeability could be traced to the preclinical phases of the disease where patients with preclinical islet cell autoimmunity performed similar to those with recent onset and long term diabetes mellitus in terms of higher urinary lactulose/mannitol excretion compared to healthy controls [85]. This again points to a proposed origin of T1 DM in the gut.
In a longitudinal study of 21 neonates over 24 months, it was shown that gut microbiota changes could be linked to gut inflammatory (calprotectin) and permeability (zonulin) biomarker changes during early life. Fecal calprotectin, which decreased progressively after 6 months of life, correlated negatively with alpha diversity and relative abundances of Clostridiales and Ruminococcaceae which increased with age. On the other hand, zonulin expression (as a surrogate marker of permeability) was associated with 19 OTUs, including Ruminococcus torques, bacillales, and clostridiales [86].
In another study, children with recently diagnosed T1 DM or those who have more than two islet autoantibodies (IA) had dysbiotic gut phenotypes with lower relative abundances of Prevotella and Butryicimonas compared to healthy controls. Children with diabetes and those who progressed from expressing autoantibodies to the development of DM had higher intestinal permeability (as measured by serum lactulose/rhamnose ratio) and lower alpha diversity compared to healthy controls and those with antibodies who did not develop diabetes [87].
Patients with Hashimoto’s thyroiditis were shown, in a case control study, to have increased intestinal permeability as demonstrated by the significantly higher zonulin levels in those patients compared to patients with congenital hypothyroidism. Interestingly, serum zonulin levels correlated with levothyroxine dose in Hashimoto’s thyroiditis patients, suggesting a correlation between intestinal permeability and disease severity [88].
An increase in the peripheral pool of B lymphocytes in multiple sclerosis and Crohn’s disease implies excessive antigen exposure with postulated impairment of the gut epithelial barrier, increasing the non-self-antigen exposure load in these patients with subsequent activation of autoimmunity [89].
In an in vitro experiment, it was shown that plasma from patients with active Crohn’s disease, which is rich in pro-inflammatory cytokines (TNF-α) induced enhanced paracellular permeability when incubated with 3D Caco-2 cysts with decreased expression of occludin and ZO-1 tight junction proteins. This effect was mitigated when adalimumab (an anti-TNF monoclonal antibody) or a janus kinase (JNK) inhibitor was added [90]. This suggests that the inflammatory milieu associated with active Crohn’s disease increases gut permeability.
In a cohort study of 50 patients with Crohn’s disease in remission, it was shown that disturbed intestinal permeability as assessed by urinary lactulose/rhamnose excretion was associated with a significantly higher risk of relapse [91]. Abnormal intestinal permeability has been shown to cluster in families of patients with Crohn’s disease affecting, healthy relatives with no distinct or typical familial pattern, where even spouses had higher a prevalence of abnormal intestinal permeability, implying an environmental rather than pure genetic risk [92].
In a prospective study, low dose naltrexone, known for its favorable effects on wound healing, was administered to a group of patients with refractory IBD (47 patients) over 12 weeks. Interestingly, the intervention resulted in clinical improvement and remission in 74.5% and 25.5% of patients, respectively. This was associated with improved intestinal barrier and decreased endoplasmic reticulum stress in the gut mucosa [93].
Interferon gamma (INFγ), a proinflammatory cytokine which is overexpressed in IBD [94], was shown to impair the vascular endothelial barrier in gut mucosa via down-regulation and impairment of VE-Cadherin adherens protein, aggravating permeability and course of IBD. In this regard, imatinib has been shown in experimental colitis models to restore the VE-cadherin junction and alleviate colitis [95].
There is, however, a considerable “chicken or egg” debate regarding the role of intestinal permeability in IBD; whether it is an instigating event that leads to the development of IBD or a consequence that is aggravated by the disease, with substantial argument can be made for either theory [96, 97],
In this regard, it has been postulated that pathogenic microbiota species can enhance gut permeability, providing access for environmental antigens, toxins, and bacterial products to the blood stream where they can activate a pathogenic immune response. Thus, excessive “gram -ve” bacterial LPS translocation (as evidenced by higher soluble CD14 levels) can instigate the evolution and progress of SLE [98]. Also, experimental studies have shown that toll-like receptor 4 (TLR4) activation is another critical mechanism through which gut microbiota can aggravate disease course [99]. TLR2, a receptor of Pathogen Associated and Damage Associated Molecular Patterns (PAMPs and DAMPs), has been shown to be overexpressed by CD4+, CD8+ T lymphocytes, and CD14+ monocytes in patients with SLE. Activation of TLR2 is known to prime a pro-inflammatory milieu by increasing IL-6, IL-17 a, f, and TNF-α. This may provide a clue to the role of the gut microbiota and epithelial barrier dysfunction in the development and exaggeration of SLE [100].
Gut dysbiosis has been clearly demonstrated in patients with SLE [101, 102]. As maintenance of a healthy gut epithelial barrier is dependent, at least in part, on the interactions between a healthy gut microbiota and a homeostatic gut immune system, this could provide a clue to impaired gut barrier in patients with SLE. Patients with SLE are prone to developing protein losing enteropathy, which can be the first manifestation in many cases, and this can provide another pointer to a disrupted gut epithelial barrier given the normal mucosa and absence of lymphangiectasia in SLE patients, compared to other protein-losing enteropathies [103]
Interestingly, retinoic acid (RA) has been shown to augment the intestinal epithelial barrier through regulation of tight junction protein expression and by enriching Lactobacillus spp. which are known to enhance the gut barrier function. It also restores the Th17/Treg balance (which is disturbed in SLE) and inhibits pro-inflammatory cytokine pathways, namely, IL-1 Receptor-Associated Kinase 1 (IRAK-1) and Interferon Regulatory Factor (IRF-7) with a therapeutic potential which could be considered in different AID [104].
Circulating microbiome profiling showed a trend to lower microbiome diversity in both SLE patients and healthy controls, which was statistically significant in first degree relatives (FDR) of patients with SLE compared to healthy controls. Differential microbiome patterns existed among different groups in this study, with Lactobacillus iners and Thermoanaerobacterium saccharolyticum enriched in FDR compared to their control group. Paenibacillus genus (Firmicutes) was, in contrast, enriched in control groups compared to SLE patients or FDR [105].
Contribution of gut microbiota to the development of AID; involved mechanisms
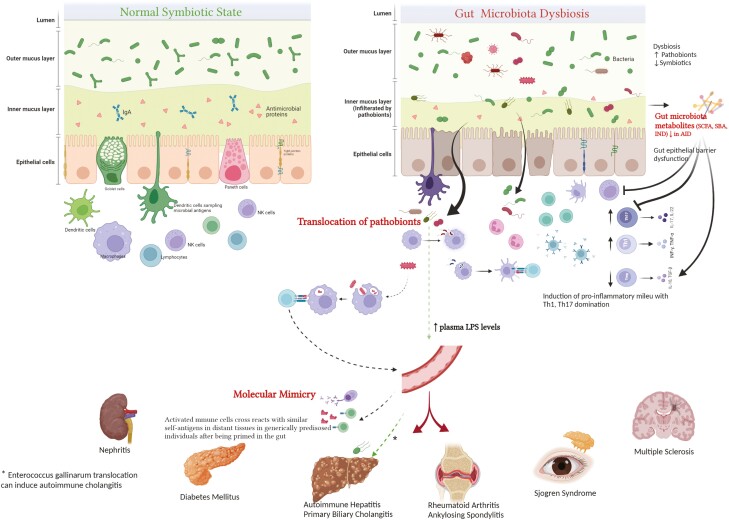
Several mechanisms have been proposed to explain how gut microbiota contribute to the development of autoimmune diseases (Fig. 1). These include:
Figure 1:
gut microbiome and autoimmune diseases (potential mechanisms). Perturbed gut microbiota can contribute to the development of autoimmune diseases through several proposed , including translocation of pathobionts and their proinflammatory products as lipopolysaccharides (LPS), molecular mimicry, which entails similarity between bacterial antigens and self-antigens in genetically predisposed individuals, and disordered metabolome (which normally helps contain inflammatory pathways in healthy state). Normally, different microbiota metabolites (SCFA in particular) can induce expansion of the immunoregulatory T-helper cells (T-Reg), innate lymphoid cells (ILC-3) and their immunoregulatory cytokines (IL-10, 22) while blocking proinflammatory pathways mediated by Th-17. Patients with autoimmune diseases (AID) usually exhibit gut epithelial barrier dysfunction with increased permeability facilitating bacterial translocation.
(1) Evidence for translocation of gut bacteria
Bacterial translocation in AID is evidenced by the detection of bacteria or their products in the circulation or tissues of those patients. Translocation of pathobionts or their products may lead to activation of a pro-inflammatory immune response with a contribution to AID [7].
In a recent case control study, SLE patients and their first-degree relatives (excluding siblings) had significantly higher plasma lipo-polysaccharide (LPS) levels (a TLR4 ligand) compared to healthy controls. Plasma LPS correlated positively with serum anti-dsDNA IgG levels in first-degree relatives (FDR) and healthy controls but not in SLE patients [105]. Also, E. Gallinarum was implicated in the pathogenesis of autoimmune liver diseases through detection of this bacterial DNA in the liver tissues of patients with autoimmune hepatitis. Co-culturing this bacterium with hepatocytes promoted a pro-inflammatory autoimmune response [8].
Furthermore, Zegarra-Ruiz et al. were able to demonstrate the translocation of Lactobacillus Reuteri, which exaggerated pro-inflammatory INF/plasmacytoid DC pathways, to internal organs in spontaneous transgenic and inducible lupus prone mouse models [22].
The oral periodontal pathobiont Porphyromonas gingivalis has been implicated in RA through the generation of citrullinated peptides which promote ACPA formation [106]. This is consistent with an interesting study on 470 individuals from five countries where profiling of oral and gut microbiome showed that transfer to and colonization of large intestine by oral microbiota strains occurs commonly in healthy subjects and that many RA and CRC associated gut pathobionts are, in fact, originating from the oral cavity [107].
(2) Bacterial metabolites induced activation of the immune system
Gut microbiota products, including their metabolites, can have direct effects on the gut mucosal immune system, resulting in either pro-or anti-inflammatory effects. For instance, the SCFA butyrate produced by gut microbiota has been shown to decrease the production of cytokines from invariant NK T cells (iNKT) via histone deacetylase inhibition. This alleviated antibody-induced arthritis (AIA) in wild type mice and not iNKT cell-deficient Jα 18 knockout mice [108].
Metabolomic study of fecal samples from patients with RA and ankylosing spondylitis demonstrated lower fecal SCFA levels [109]. This is consistent with the beneficial immunomodulatory effects of SCFA, which are known to promote the anti-inflammatory response mediated by T-Reg cells and IL-10 [110].
High dietary salt intake (a characteristic of the ‘Western diet’) was shown to promote Th17 expansion along with depletion of Lactobacillus species. with detrimental consequences (hypertension) in both humans and mouse models. Interestingly, Indole-3 Lactic Acid (ILA), a metabolite of the depleted lactobacillus murinus spp. in these mouse models, was shown to significantly reduce Th17 differentiation in vitro [111].
Some strains of Lactobacillus catabolize tryptophan to produce indoles and indole derivatives which are ligands for aryl hydrocarbon receptors (AhR), which when activated and after interaction with the nuclear RORγt receptors, bind to the IL22 locus to up-regulate its expression by ILC-3 (innate lymphoid cells). IL22 belongs to the immunomodulator family IL10 [112]. This highlights the immunoregulatory effects mediated by Lactobacillus spp.
AhR ligands produced by some microbiota members have proven critical in the gut brain communications/axis where, through acting on the central nervous system astrocytes, they help mitigate inflammatory activities in the CNS. This is consistent with their reduced levels in patients with multiple sclerosis [113, 114].
Secondary bile acids, produced by some gut microbiota spp. (Bacteroides fragilis, valgatus, Bifidobacterium, Lactobacillus plantarum, Eubacterium among others), have been shown to control dendritic cell (DC) function via binding to FXR and G Protein Bile Acid Receptor 1 (GPBAR1), thereby inhibiting NF-κB mediated transcription of proinflammatory pathways [115]. Consistent with that, activation of FXR by the FXR agonist obeticholic acid was shown to mitigate intestinal inflammation, promote gut epithelial barrier function and inhibit translocation of pathobionts [116]. This can represent a future prospective towards the prevention of autoimmune processes in susceptible patients.
Enzymatic activities intrinsic to some microbiota species may sometimes be the “first hit” in the autoimmune process. This is exemplified by the peptidylarginine deaminase “PAD” activity of the oral pathobiont, Porphyromonas gingivalis, known to cause periodontitis. This species has been linked to the development of RA along with higher titers of anti-citrullinated peptide antibodies through citrullination of self-peptides by PAD followed by the initiation of the autoimmune strike [117].
(3) Molecular mimicry
Another potential mechanism by which translocating components of the microbiota can effect immune modulation is molecular mimicry. The gut microbial ecosystem is a very rich one, with a massive repertoire of different antigens encoded by more than nine million genes [56].
Molecular mimicry entails similarity between bacterial antigens and self-antigens, which drives cross activation of effector immune cells to attack self-antigens [7]. However, for molecular mimicry to induce an autoimmune response, the contribution of host genetics is critical for HLA-DR related antigen recognition [118]. This supports the concept of microbiota genetic interplay for the development of autoimmune diseases, which is particularly evident with gut microbiota perturbations in early life [119].
Likewise, molecular mimicry entails gut microbiome immune system interactions to generate cross-reactive lymphocytes which can mount autoimmune assaults in remote tissue organs that can involve even immune privileged sites such as the retina [120]. This usually involves different cells of the immune system, both innate and adaptive immune cells, involving and not limited to Th-1, Th17, ILCs, NKT, T-Reg, and B-lymphocytes [121].
Roseburia intestinalis of the gut microbiome has been linked to the evolution of anti-glycoprotein I (Anti-GPI) and autoreactive CD4+ memory T cells in antiphospholipid syndrome (APS). This has been explained through molecular mimicry between Roseburia intestinalis expressed mimotopes and β2 glycoprotein I antigen. Interestingly, anti- RI mimotope antibodies were significantly higher, and correlated positively with anti-β2GPI IgG titers, in patients with APS. Also, colonization of mouse models with R. intestinalis promoted the development of anti-β2GPI IgG with autoimmune consequences [122].
Some microbiota strains have been implicated in the development of collagen-induced arthritis via molecular mimicry of their peptides and collagen type 2. These included Candida albicans [123] and Streptococcus sanguis [124]. In RA, Prevotella copri has been postulated to present antigens, mimicking synovial membrane self-antigens, which promote auto-reactive T lymphocytes to induce autoimmunity. In this context, the filamin A and N-acetylglucosamine-6-sulfatase are two outstanding autoantigens in RA that have been shown to be similar to Prevotella derived epitopes [125].
Another prominent and old example of the importance of molecular mimicry in the development of autoimmune diseases is the cross-reactivity induced by similarity between klebsiella pneumoniae antigens and self HLA-B27 to initiate the development of ankylosing spondylitis [126].
Gut mycobiota and virobiota: the unknown knowns
Although the role of certain viruses such as cytomegalovirus has been well characterized in the development and exacerbation of some AIDs, including UC [127], SLE [128] and MS [129], the role of other integrated viral communities of the gut microbiome has acquired much attention recently. Gut virome alterations have recently been demonstrated in dedicated studies in IBD patients, and a role in the disease has been proposed through interactions with and alteration of gut microbiome compositions by different phages [130, 131]. Lysis of certain gut bacterial species by phages and subsequent release of their components has been implicated in the promotion of an inflammatory response in IBD [132]. The virome in other autoimmune diseases such as RA has recently been explored with compositional variations demonstrated in oral more than gut viromes. Interestingly, an integrated model of gut virome and bacteriome was more accurate in predicting IBD patients than either one of them [130].
Another player in the gut microbiome is the mycobiota, which comprises different fungal communities. Similarly, altered gut mycobiota composition with unbalanced fungal/bacterial ratios has been demonstrated in patients with CD [133, 134]. Still, the altered structure and underpinning role of gut mycobiota in different AIDs have to be further explored in more studies.
Conclusions and the way forward
Data regarding the pivotal role of the gut microbiome in the pathogenesis of chronic autoimmune diseases is accumulating rapidly.
In this review we highlight existing evidence from cross sectional associative studies in human cohorts and animal models, data from studies in which the microbiome has been manipulated, including recent exciting data from studies involving fecal microbiota transplantation. We touched on potential mechanisms by which bacterial components of the gut microbiome could result in systemic inflammation by interacting with intestinal epithelium and some early data regarding the virome in this regard.
Of course, definite mechanisms remain tantalizingly elusive, and much is yet to be learnt regarding the non-bacterial contribution of the complex ecosystem in the gut in this regard. Furthermore, given the lack of studies in patients with incipient disease and longitudinal follow up fundamental questions regarding association as opposed to causality remain uncertain.
Future human studies, perhaps employing FMT as a ‘discovery’ as well as treatment modality with careful mechanistic analyses will undoubtedly shed light on this in the near future.
Supplementary Material
Acknowledgement
WAS dedicates this work to the soul of his father.
Glossary
Abbreviations
- ACPA
anti-citrullinated protein/peptide antibodies
- AhR
aryl hydrocarbon receptors
- AID
autoimmune disorders
- APC
antigen presenting cells
- DAMP
damage associated molecular patterns
- DSS
dextran sulfate sodium
- FMT
fecal microbiota transplantation
- FXR
farnesoid X receptors
- GF
germ free
- GPBAR-1
G Protein Bile Acid Receptor 1
- GWAS
genome wide association studies
- iNKT
invariant NK T cells
- IPAA
ileal pouch anal anastomosis
- IRAK1
IL-1 receptor-associated kinase 1
- IRF-7
interferon regulatory factor 7
- JIA
juvenile idiopathic arthritis
- JNK
janus kinase
- LPS
lipopolysaccharides
- OTU
operational taxonomic unit
- PAD
peptidylarginine deaminase
- PAMP
pathogen associated molecular patterns
- SCFA
short chain fatty acids
- SFB
segmented filamentous bacteria
- SPF
specific pathogen free
- Tfh
follicular T helper
- VE cadherin
vascular endothelial cadherin
Contributor Information
Walaa Abdelaty Shaheen, University of Birmingham Microbiome Treatment Center, Birmingham, UK; Institute of Cancer and Genomic Sciences, University of Birmingham, Birmingham, UK; Gastroenterology Department, Menoufia University, Shebin Elkom, Egypt.
Mohammed Nabil Quraishi, University of Birmingham Microbiome Treatment Center, Birmingham, UK; Institute of Cancer and Genomic Sciences, University of Birmingham, Birmingham, UK; University Hospitals of Birmingham NHS Foundation Trust, Birmingham, UK.
Tariq H Iqbal, University of Birmingham Microbiome Treatment Center, Birmingham, UK; University Hospitals of Birmingham NHS Foundation Trust, Birmingham, UK; Institute of Microbiology and Infection, University of Birmingham, Birmingham, UK.
Funding
The authors received no financial support for the research, authorship and/or Publication of this article.
Conflict of interest
None declared.
Author contributions
WAS performed a literature review and wrote the review. TI and MNQ provided critical feedback on the review. WAS prepared the figures and tables with input from all authors. All authors read, edited, and approved the final draft.
Data availability
All data are incorporated into the article and its online supplementary material. The data underlying this article are available in the article and in its online supplementary material.
References
- 1. Araki Y, Mimura T.. Chapter 12 - Epigenetic basis of autoimmune disorders in humans. In: Tollefsbol TO (ed), Epigenetics in Human Disease, Vol. 6, 2nd edn. Academic Press, 2018, 353–85. [Google Scholar]
- 2. Zhao C-N, Xu Z, Wu G-C, Mao Y-M, Liu L-N, Qian W, et al. Emerging role of air pollution in autoimmune diseases. Autoimmun Rev 2019, 18, 607–14. [DOI] [PubMed] [Google Scholar]
- 3. Khan MF, Wang G.. Environmental agents, oxidative stress and autoimmunity. Curr Opin Toxicol 2018, 7, 22–7. doi: 10.1016/j.cotox.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dehner C, Fine R, Kriegel MA.. The microbiome in systemic autoimmune disease: mechanistic insights from recent studies. Curr Opin Rheumatol 2019, 31, 201–7. doi: 10.1097/BOR.0000000000000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Farache J, Koren I, Milo I, Gurevich I, Kim K-W, Zigmond E, et al. Luminal bacteria recruit CD103+ dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity 2013, 38, 581–95. doi: 10.1016/j.immuni.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mowat AM. To respond or not to respond—a personal perspective of intestinal tolerance. Nat Rev Immunol 2018, 18, 405–15. doi: 10.1038/s41577-018-0002-x. [DOI] [PubMed] [Google Scholar]
- 7. Zhang X, Chen B-d, Zhao L-d, Li H.. The gut microbiota: emerging evidence in autoimmune diseases. Trends Mol Med 2020, 26, 862–73. doi: 10.1016/j.molmed.2020.04.001. [DOI] [PubMed] [Google Scholar]
- 8. Manfredo Vieira S, Hiltensperger M, Kumar V, Zegarra-Ruiz D, Dehner C, Khan N, et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 2018, 359, 1156–61. doi: 10.1126/science.aar7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Asquith MJ, Stauffer P, Davin S, Mitchell C, Lin P, Rosenbaum JT.. Perturbed mucosal immunity and dysbiosis accompany clinical disease in a rat model of spondyloarthritis. Arthritis Rheumatol 2016, 68, 2151–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tan TG, Sefik E, Geva-Zatorsky N, Kua L, Naskar D, Teng F, et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci USA 2016, 113, E8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ansaldo E, Slayden LC, Ching KL, Koch MA, Wolf NK, Plichta DR, et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 2019, 364, 1179–1184. doi: 10.1126/science.aaw7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ma Y, Xu X, Li M, Cai J, Wei Q, Niu H.. Gut microbiota promote the inflammatory response in the pathogenesis of systemic lupus erythematosus. Mol Med 2019, 25, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Walter J, Armet AM, Finlay BB, Shanahan F.. Establishing or exaggerating causality for the gut microbiome: lessons from human microbiota-associated rodents. Cell 2020, 180, 221–32. doi: 10.1016/j.cell.2019.12.025. [DOI] [PubMed] [Google Scholar]
- 14. Xu Q, Ni J-J, Han B-X, Yan S-S, Wei X-T, Feng G-J, et al. Causal relationship between gut microbiota and autoimmune diseases: a two-sample Mendelian randomization study. Front Immunol 2022, 12, 746998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee YH. Causal association of gut microbiome on the risk of rheumatoid arthritis: a Mendelian randomisation study. Ann Rheum Dis 2022, 81, e3. doi: 10.1136/annrheumdis-2019-216747. [DOI] [PubMed] [Google Scholar]
- 16. Ma H-D, Zhao Z-B, Ma W-T, Liu Q-Z, Gao C-Y, Li L, et al. Gut microbiota translocation promotes autoimmune cholangitis. J Autoimmun 2018, 95, 47–57. doi: 10.1016/j.jaut.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 2010, 32, 815–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Block KE, Zheng Z, Dent AL, Kee BL, Huang H.. Gut microbiota regulates K/BxN autoimmune arthritis through follicular helper T but not Th17 cells. J Immunol 2016, 196, 1550–7. doi: 10.4049/jimmunol.1501904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Teng F, Klinger CN, Felix KM, Bradley CP, Wu E, Tran NL, et al. Gut microbiota drive autoimmune arthritis by promoting differentiation and migration of Peyer’s patch T follicular helper cells. Immunity 2016, 44, 875–88. doi: 10.1016/j.immuni.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kumar P, Monin L, Castillo P, Elsegeiny W, Horne W, Eddens T, et al. Intestinal interleukin-17 receptor signaling mediates reciprocal control of the gut microbiota and autoimmune inflammation. Immunity 2016, 44, 659–71. doi: 10.1016/j.immuni.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dorożyńska I, Majewska-Szczepanik M, Marcińska K, Szczepanik M.. Partial depletion of natural gut flora by antibiotic aggravates collagen induced arthritis (CIA) in mice. Pharmacol Rep 2014, 66, 250–5. doi: 10.1016/j.pharep.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 22. Zegarra-Ruiz DF, El Beidaq A, Iñiguez AJ, Lubrano Di Ricco M, Manfredo Vieira S, Ruff WE, et al. A diet-sensitive commensal lactobacillus strain mediates TLR7-dependent systemic autoimmunity. Cell Host Microbe 2019, 25, 113–127.e6. doi: 10.1016/j.chom.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mu Q, Zhang H, Liao X, Lin K, Liu H, Edwards MR, et al. Control of lupus nephritis by changes of gut microbiota. Microbiome 2017, 5, 73. doi: 10.1186/s40168-017-0300-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johnson BM, Gaudreau M-C, Al-Gadban MM, Gudi R, Vasu C.. Impact of dietary deviation on disease progression and gut microbiome composition in lupus-prone SNF1 mice. Clin Exp Immunol 2015, 181, 323–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mu Q, Tavella VJ, Kirby JL, Cecere TE, Chung M, Lee J, et al. Antibiotics ameliorate lupus-like symptoms in mice. Sci Rep 2017, 7, 13675. doi: 10.1038/s41598-017-14223-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang Y, Liu Q, Yu Y, Wang M, Wen C, He Z.. Early and short-term interventions in the gut microbiota affects lupus severity, progression, and treatment in MRL/lpr mice. Front Microbiol 2020, 11, 628. doi: 10.3389/fmicb.2020.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 2011, 479, 538–41. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 28. Cekanaviciute E, Yoo BB, Runia TF, Debelius JW, Singh S, Nelson CA, et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci USA 2017, 114, 10713–8. doi: 10.1073/pnas.1711235114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Berer K, Gerdes LA, Cekanaviciute E, Jia X, Xiao L, Xia Z, et al. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc Natl Acad Sci USA 2017, 114, 10719–24. doi: 10.1073/pnas.1711233114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. He B, Hoang TK, Tian X, Taylor CM, Blanchard E, Luo M, et al. Lactobacillus reuteri reduces the severity of experimental autoimmune encephalomyelitis in mice by modulating gut microbiota. Front Immunol 2018, 10, 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Scheer S, Medina TS, Murison A, Taves MD, Antignano F, Chenery A, et al. Early-life antibiotic treatment enhances the pathogenicity of CD4+ T cells during intestinal inflammation. J Leukoc Biol 2017, 101, 893–900. doi: 10.1189/jlb.3MA0716-334RR. [DOI] [PubMed] [Google Scholar]
- 32. Sorini C, Cosorich I, Lo Conte M, De Giorgi L, Facciotti F, Lucianò R, et al. Loss of gut barrier integrity triggers activation of islet-reactive T cells and autoimmune diabetes. Proc Natl Acad Sci USA 2019, 116, 15140–9. doi: 10.1073/pnas.1814558116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jin S, Zhao D, Cai C, Song D, Shen J, Xu A, et al. Low-dose penicillin exposure in early life decreases Th17 and the susceptibility to DSS colitis in mice through gut microbiota modification. Sci Rep 2017, 7, 43662. doi: 10.1038/srep43662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakamura YK, Metea C, Karstens L, Asquith M, Gruner H, Moscibrocki C, et al. Gut microbial alterations associated with protection from autoimmune uveitis. Invest Ophthalmol Vis Sci 2016, 57, 3747–58. doi: 10.1167/iovs.16-19733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Esmaeili S-A, Mahmoudi M, Momtazi AA, Sahebkar A, Doulabi H, Rastin M.. Tolerogenic probiotics: potential immunoregulators in Systemic Lupus Erythematosus. J Cell Physiol 2017, 232, 1994–2007. doi: 10.1002/jcp.25748. [DOI] [PubMed] [Google Scholar]
- 36. Uusitalo U, Liu X, Yang J, Aronsson CA, Hummel S, Butterworth M, et al. Association of early exposure of probiotics and islet autoimmunity in the TEDDY study. JAMA Pediatr 2016, 170, 20–8. doi: 10.1001/jamapediatrics.2015.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zamani B, Golkar HR, Farshbaf S, Emadi-Baygi M, Tajabadi-Ebrahimi M, Jafari P, et al. Clinical and metabolic response to probiotic supplementation in patients with rheumatoid arthritis: a randomized, double-blind, placebo-controlled trial. Int J Rheum Dis 2016, 19, 869–79. doi: 10.1111/1756-185X.12888. [DOI] [PubMed] [Google Scholar]
- 38. Alipour B, Homayouni-Rad A, Vaghef-Mehrabany E, Sharif SK, Vaghef-Mehrabany L, Asghari-Jafarabadi M, et al. Effects of Lactobacillus casei supplementation on disease activity and inflammatory cytokines in rheumatoid arthritis patients: a randomized double-blind clinical trial. Int J Rheum Dis 2014, 17, 519–27. doi: 10.1111/1756-185X.12333. [DOI] [PubMed] [Google Scholar]
- 39. Mohammed AT, Khattab M, Ahmed AM, Turk T, Sakr N, A MK, et al. The therapeutic effect of probiotics on rheumatoid arthritis: a systematic review and meta-analysis of randomized control trials. Clin Rheumatol 2017, 36, 2697–707. [DOI] [PubMed] [Google Scholar]
- 40. López P, de Paz B, Rodríguez-Carrio J, Hevia A, Sánchez B, Margolles A, et al. Th17 responses and natural IgM antibodies are related to gut microbiota composition in systemic lupus erythematosus patients. Sci Rep 2016, 6, 24072. doi: 10.1038/srep24072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Frech TM, Khanna D, Maranian P, Frech EJ, Sawitzke AD, Murtaugh MA.. Probiotics for the treatment of systemic sclerosis-associated gastrointestinal bloating/distention. Clin Exp Rheumatol 2011, 29, S22–5. [PubMed] [Google Scholar]
- 42. Tamaki H, Nakase H, Inoue S, Kawanami C, Itani T, Ohana M, et al. Efficacy of probiotic treatment with Bifidobacterium longum 536 for induction of remission in active ulcerative colitis: a randomized, double-blinded, placebo-controlled multicenter trial. Dig Endosc 2016, 28, 67–74. doi: 10.1111/den.12553. [DOI] [PubMed] [Google Scholar]
- 43. Rahimi R, Nikfar S, Rahimi F, Elahi B, Derakhshani S, Vafaie M, et al. A meta-analysis on the efficacy of probiotics for maintenance of remission and prevention of clinical and endoscopic relapse in Crohn’s disease. Dig Dis Sci 2008, 53, 2524–31. doi: 10.1007/s10620-007-0171-0. [DOI] [PubMed] [Google Scholar]
- 44. Ghouri YA, Richards DM, Rahimi EF, Krill JT, Jelinek KA, DuPont AW.. Systematic review of randomized controlled trials of probiotics, prebiotics, and synbiotics in inflammatory bowel disease. Clin Exp Gastroenterol 2014, 7, 473–87. doi: 10.2147/CEG.S27530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Derwa Y, Gracie DJ, Hamlin PJ, Ford AC.. Systematic review with meta-analysis: the efficacy of probiotics in inflammatory bowel disease. Aliment Pharmacol Ther 2017, 46, 389–400. doi: 10.1111/apt.14203. [DOI] [PubMed] [Google Scholar]
- 46. Ganji-Arjenaki M, Rafieian-Kopaei M.. Probiotics are a good choice in remission of inflammatory bowel diseases: a meta analysis and systematic review. J Cell Physiol 2018, 233, 2091–103. doi: 10.1002/jcp.25911. [DOI] [PubMed] [Google Scholar]
- 47. Kouchaki E, Tamtaji OR, Salami M, Bahmani F, Daneshvar Kakhaki R, Akbari E, et al. Clinical and metabolic response to probiotic supplementation in patients with multiple sclerosis: a randomized, double-blind, placebo-controlled trial. Clin Nutr 2017, 36, 1245–9. doi: 10.1016/j.clnu.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 48. Salami M, Kouchaki E, Asemi Z, Tamtaji OR.. How probiotic bacteria influence the motor and mental behaviors as well as immunological and oxidative biomarkers in multiple sclerosis? A double blind clinical trial. J Funct Foods 2019, 52, 8–13. doi: 10.1016/j.jff.2018.10.023. [DOI] [Google Scholar]
- 49. Cunningham MW. Autoimmunity and molecular mimicry in the pathogenesis of post-streptococcal heart disease. Front Biosci 2003, 8, s533–43. doi: 10.2741/1067. PMID: 12700052. [DOI] [PubMed] [Google Scholar]
- 50. Jernberg C, Löfmark S, Edlund C, Jansson JK.. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J 2007, 1, 56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 51. Ferrer M, Méndez-García C, Rojo D, Barbas C, Moya A.. Antibiotic use and microbiome function. Biochem Pharmacol 2017, 134, 114–26. doi: 10.1016/j.bcp.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 52. Rosman YL, Shoenfeld Y.. Antibiotic therapy in autoimmune disorders. Clin Pract 2014, 11, 91–103. [Google Scholar]
- 53. Kilkkinen A, Virtanen SM, Klaukka T, Kenward MG, Salkinoja-Salonen M, Gissler M, et al. Use of antimicrobials and risk of type 1 diabetes in a population-based mother–child cohort. Diabetologia 2006, 49, 66–70. doi: 10.1007/s00125-005-0078-2. [DOI] [PubMed] [Google Scholar]
- 54. Kemppainen KM, Vehik K, Lynch KF, Larsson HE, Canepa RJ, Simell V, et al. Association between early-life antibiotic use and the risk of islet or celiac disease autoimmunity. JAMA Pediatr 2017, 171, 1217–25. doi: 10.1001/jamapediatrics.2017.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Singh JA, Furst DE, Bharat A, Curtis JR, Kavanaugh AF, Kremer JM, et al. 2012 update of the 2008 American College of Rheumatology recommendations for the use of disease-modifying antirheumatic drugs and biologic agents in the treatment of rheumatoid arthritis. Arthritis Care Res 2012, 64, 625–39. doi: 10.1002/acr.21641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol 2014, 32, 834–41. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 57. Armstrong D, Dregan A, Ashworth M, White P, McGee C, de Lusignan S.. Influence of prior antibiotic use on risk of rheumatoid arthritis: case control study in general practice. Rheumatology 2019, 59, 1281–7. [DOI] [PubMed] [Google Scholar]
- 58. Horton DB, Scott FI, Haynes K, Putt ME, Rose CD, Lewis JD, et al. Antibiotic exposure and juvenile idiopathic arthritis: a case-control study. Pediatrics 2015, 136, e333–43. doi: 10.1542/peds.2015-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Solhjoo M BP, Goyal A, et al. Drug-Induced Lupus Erythematosus. [Updated 2021 Jan 27]. Treasure Island (FL): StatPearls Publishing. 2021; https://www.ncbi.nlm.nih.gov/books/NBK441889/. [PubMed] [Google Scholar]
- 60. Brown J, Robusto B, Morel L.. Intestinal dysbiosis and tryptophan metabolism in autoimmunity. Front Immunol 2020, 11, 1741. doi: 10.3389/fimmu.2020.01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tapia G, Størdal K, Mårild K, Kahrs CR, Skrivarhaug T, Njølstad PR, et al. Antibiotics, acetaminophen and infections during prenatal and early life in relation to type 1 diabetes. Int J Epidemiol 2018, 47, 1538–48. doi: 10.1093/ije/dyy092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Guthrie L, Kelly L.. Bringing microbiome-drug interaction research into the clinic. EBioMedicine 2019, 44, 708–15. doi: 10.1016/j.ebiom.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kronman MP, Zaoutis TE, Haynes K, Feng R, Coffin SE.. Antibiotic exposure and IBD development among children: a population-based cohort study. Pediatrics 2012, 130, e794–803. doi: 10.1542/peds.2011-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Segal JP, Ding NS, Worley G, Mclaughlin S, Preston S, Faiz OD, et al. Systematic review with meta-analysis: the management of chronic refractory pouchitis with an evidence-based treatment algorithm. Aliment Pharmacol Ther 2017, 45, 581–92. doi: 10.1111/apt.13905. [DOI] [PubMed] [Google Scholar]
- 65. Segal JP, Poo SX, McLaughlin SD, Faiz OD, Clark SK, Hart AL.. Long-term follow-up of the use of maintenance antibiotic therapy for chronic antibiotic-dependent pouchitis. Frontline Gastroenterol 2018, 9, 154–8. doi: 10.1136/flgastro-2017-100913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Prantera C, Zannoni F, Scribano ML, Berto E, Andreoli A, Kohn A, et al. An antibiotic regimen for the treatment of active Crohn’s disease: a randomized, controlled clinical trial of metronidazole plus ciprofloxacin. Am J Gastroenterol 1996, 91, 328–32. [PubMed] [Google Scholar]
- 67. Colombel JF, Lémann M, Cassagnou M, Bouhnik Y, Duclos B, Dupas JL, et al. A controlled trial comparing ciprofloxacin with mesalazine for the treatment of active Crohn’s disease. Groupe d’Etudes Thérapeutiques des Affections Inflammatoires Digestives (GETAID). Am J Gastroenterol 1999, 94, 674–8. doi: 10.1111/j.1572-0241.1999.935_q.x. [DOI] [PubMed] [Google Scholar]
- 68. Shafran I, Johnson LK.. An open-label evaluation of rifaximin in the treatment of active Crohn’s disease. Curr Med Res Opin 2005, 21, 1165–9. doi: 10.1185/030079905x53252. [DOI] [PubMed] [Google Scholar]
- 69. Rhodes JM, Subramanian S, Flanagan PK, Horgan GW, Martin K, Mansfield J, et al. Randomized trial of ciprofloxacin doxycycline and hydroxychloroquine versus budesonide in active Crohn’s disease. Dig Dis Sci 2021, 66, 2700–11. doi: 10.1007/s10620-020-06477-y. [DOI] [PubMed] [Google Scholar]
- 70. Britton GJ, Contijoch EJ, Spindler MP, Aggarwala V, Dogan B, Bongers G, et al. Defined microbiota transplant restores Th17/RORγt+ regulatory T cell balance in mice colonized with inflammatory bowel disease microbiotas. Proc Natl Acad Sci USA 2020, 117, 21536–45. doi: 10.1073/pnas.1922189117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lleal M, Sarrabayrouse G, Willamil J, Santiago A, Pozuelo M, Manichanh C.. A single faecal microbiota transplantation modulates the microbiome and improves clinical manifestations in a rat model of colitis. EBioMedicine 2019, 48, 630–41. doi: 10.1016/j.ebiom.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Maeda Y, Kurakawa T, Umemoto E, Motooka D, Ito Y, Gotoh K, et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol 2016, 68, 2646–61. [DOI] [PubMed] [Google Scholar]
- 73. Ma Y, Guo R, Sun Y, Li X, He L, Li Z, et al. Lupus gut microbiota transplants cause autoimmunity and inflammation. bioRxiv 2021:2021.08.15.456375. [DOI] [PubMed] [Google Scholar]
- 74. Stripling J, Rodriguez M.. Current evidence in delivery and therapeutic uses of fecal microbiota transplantation in human diseases-clostridium difficile disease and beyond. Am J Med Sci 2018, 356, 424–32. doi: 10.1016/j.amjms.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 75. Quraishi MN, Widlak M, Bhala N, Moore D, Price M, Sharma N, et al. Systematic review with meta-analysis: the efficacy of faecal microbiota transplantation for the treatment of recurrent and refractory Clostridium difficile infection. Aliment Pharmacol Ther 2017, 46, 479–93. doi: 10.1111/apt.14201. [DOI] [PubMed] [Google Scholar]
- 76. Costello SP, Soo W, Bryant RV, Jairath V, Hart AL, Andrews JM.. Systematic review with meta-analysis: faecal microbiota transplantation for the induction of remission for active ulcerative colitis. Aliment Pharmacol Ther 2017, 46, 213–24. doi: 10.1111/apt.14173. [DOI] [PubMed] [Google Scholar]
- 77. Dang X, Xu M, Liu D, Zhou D, Yang W.. Assessing the efficacy and safety of fecal microbiota transplantation and probiotic VSL#3 for active ulcerative colitis: a systematic review and meta-analysis. PLoS One 2020, 15, e0228846. doi: 10.1371/journal.pone.0228846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. de Groot P, Nikolic T, Pellegrini S, Sordi V, Imangaliyev S, Rampanelli E, et al. Faecal microbiota transplantation halts progression of human new-onset type 1 diabetes in a randomised controlled trial. Gut 2021, 70, 92–105. doi: 10.1136/gutjnl-2020-322630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Fasano A. Leaky gut and autoimmune diseases. Clin Rev Allergy Immunol 2012, 42, 71–8. doi: 10.1007/s12016-011-8291-x. [DOI] [PubMed] [Google Scholar]
- 80. Vaarala O. Is the origin of type 1 diabetes in the gut?. Immunol Cell Biol 2012, 90, 271–6. doi: 10.1038/icb.2011.115. [DOI] [PubMed] [Google Scholar]
- 81. Secondulfo M, Iafusco D, Carratù R, deMagistris L, Sapone A, Generoso M, et al. Ultrastructural mucosal alterations and increased intestinal permeability in non-celiac, type I diabetic patients. Dig Liver Dis 2004, 36, 35–45. doi: 10.1016/j.dld.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 82. Damci T, Nuhoglu I, Devranoglu G, Osar Z, Demir M, Ilkova H.. Increased intestinal permeability as a cause of fluctuating postprandial blood glucose levels in Type 1 diabetic patients. Eur J Clin Invest 2003, 33, 397–401. doi: 10.1046/j.1365-2362.2003.01161.x. [DOI] [PubMed] [Google Scholar]
- 83. Sapone A, de Magistris L, Pietzak M, Clemente MG, Tripathi A, Cucca F, et al. Zonulin upregulation is associated with increased gut permeability in subjects with type 1 diabetes and their relatives. Diabetes 2006, 55, 1443–9. doi: 10.2337/db05-1593. [DOI] [PubMed] [Google Scholar]
- 84. Watts T, Berti I, Sapone A, Gerarduzzi T, Not T, Zielke R, et al. Role of the intestinal tight junction modulator zonulin in the pathogenesis of type I diabetes in BB diabetic-prone rats. Proc Natl Acad Sci USA 2005, 102, 2916–21. doi: 10.1073/pnas.0500178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bosi E, Molteni L, Radaelli MG, Folini L, Fermo I, Bazzigaluppi E, et al. Increased intestinal permeability precedes clinical onset of type 1 diabetes. Diabetologia 2006, 49, 2824–7. doi: 10.1007/s00125-006-0465-3. [DOI] [PubMed] [Google Scholar]
- 86. Kaczmarczyk M, Löber U, Adamek K, Węgrzyn D, Skonieczna-Żydecka K, Malinowski D, et al. The gut microbiota is associated with the small intestinal paracellular permeability and the development of the immune system in healthy children during the first two years of life. J Transl Med 2021, 19, 177. doi: 10.1186/s12967-021-02839-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Harbison JE, Roth-Schulze AJ, Giles LC, Tran CD, Ngui KM, Penno MA, et al. Gut microbiome dysbiosis and increased intestinal permeability in children with islet autoimmunity and type 1 diabetes: a prospective cohort study. Pediatr Diab 2019, 20, 574–83. doi: 10.1111/pedi.12865. [DOI] [PubMed] [Google Scholar]
- 88. Küçükemre Aydın B, Yıldız M, Akgün A, Topal N, Adal E, Önal H.. Children with Hashimoto’s thyroiditis have increased intestinal permeability: results of a pilot study. J Clin Res Pediatr Endocrinol 2020, 12, 303–7. doi: 10.4274/jcrpe.galenos.2020.2019.0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Fasano A, Shea-Donohue T.. Mechanisms of disease: the role of intestinal barrier function in the pathogenesis of gastrointestinal autoimmune diseases. Nat Clin Pract Gastroenterol Hepatol 2005, 2, 416–22. doi: 10.1038/ncpgasthep0259. [DOI] [PubMed] [Google Scholar]
- 90. Xu P, Elamin E, Elizalde M, Bours P, Pierik MJ, Masclee AAM, et al. Modulation of intestinal epithelial permeability by plasma from patients with Crohn’s disease in a three-dimensional cell culture model. Sci Rep 2019, 9, 2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Arnott ID, Kingstone K, Ghosh S.. Abnormal intestinal permeability predicts relapse in inactive Crohn disease. Scand J Gastroenterol 2000, 35, 1163–9. doi: 10.1080/003655200750056637. [DOI] [PubMed] [Google Scholar]
- 92. Peeters M, Geypens B, Claus D, Nevens H, Ghoos Y, Verbeke G, et al. Clustering of increased small intestinal permeability in families with Crohn’s disease. Gastroenterology 1997, 113, 802–7. doi: 10.1016/s0016-5085(97)70174-4. [DOI] [PubMed] [Google Scholar]
- 93. Lie MRKL, van der Giessen J, Fuhler GM, de Lima A, Peppelenbosch MP, van der Ent C, et al. Low dose naltrexone for induction of remission in inflammatory bowel disease patients. J Transl Med 2018, 16, 55. doi: 10.1186/s12967-018-1427-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol 2014, 14, 329–42. doi: 10.1038/nri3661. [DOI] [PubMed] [Google Scholar]
- 95. Langer V, Vivi E, Regensburger D, Winkler TH, Waldner MJ, Rath T, et al. IFN-γ drives inflammatory bowel disease pathogenesis through VE-cadherin-directed vascular barrier disruption. J Clin Invest 2019, 129, 4691–707. doi: 10.1172/JCI124884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Paray BA, Albeshr MF, Jan AT, Rather IA.. Leaky gut and autoimmunity: an intricate balance in individuals health and the diseased state. Int J Mol Sci 2020, 21, 9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lee SH. Intestinal permeability regulation by tight junction: implication on inflammatory bowel diseases. Intest Res 2015, 13, 11–8. doi: 10.5217/ir.2015.13.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Nockher WA, Wigand R, Schoeppe W, Scherberich JE.. Elevated levels of soluble CD14 in serum of patients with systemic lupus erythematosus. Clin Exp Immunol 1994, 96, 15–9. doi: 10.1111/j.1365-2249.1994.tb06222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Mu Q, Kirby J, Reilly CM, Luo XM.. Leaky gut as a danger signal for autoimmune diseases. Front Immunol 2017, 8, 598. doi: 10.3389/fimmu.2017.00598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Liu Y, Liao J, Zhao M, Wu H, Yung S, Chan TM, et al. Increased expression of TLR2 in CD4(+) T cells from SLE patients enhances immune reactivity and promotes IL-17 expression through histone modifications. Eur J Immunol 2015, 45, 2683–93. doi: 10.1002/eji.201445219. [DOI] [PubMed] [Google Scholar]
- 101. He Z, Shao T, Li H, Xie Z, Wen C.. Alterations of the gut microbiome in Chinese patients with systemic lupus erythematosus. Gut Pathog 2016, 8, 64. doi: 10.1186/s13099-016-0146-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hevia A, Milani C, López P, Cuervo A, Arboleya S, Duranti S, et al. Intestinal dysbiosis associated with systemic lupus erythematosus. mBio 2014, 5, e01548–14. doi: 10.1128/mBio.01548-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Perednia DA, Curosh NA.. Lupus-associated protein-losing enteropathy. Arch Intern Med 1990, 150, 1806–10. [PubMed] [Google Scholar]
- 104. Abdelhamid L, Luo XM.. Retinoic acid, leaky gut, and autoimmune diseases. Nutrients 2018, 10, 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ogunrinde E, Zhou Z, Luo Z, Alekseyenko A, Li Q-Z, Macedo D, et al. A link between plasma microbial translocation, microbiome, and autoantibody development in first-degree relatives of systemic lupus erythematosus patients. Arthritis Rheumatol 2019, 71, 1858–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kriebel K, Hieke C, Müller-Hilke B, Nakata M, Kreikemeyer B.. Oral biofilms from symbiotic to pathogenic interactions and associated disease –connection of periodontitis and rheumatic arthritis by peptidylarginine deiminase. Front Microbiol 2018, 9, 53. doi: 10.3389/fmicb.2018.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Schmidt TS, Hayward MR, Coelho LP, Li SS, Costea PI, Voigt AY, et al. Extensive transmission of microbes along the gastrointestinal tract. Elife 2019, 8, e42693. doi: 10.7554/eLife.42693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Lee S, Koh J, Chang Y, Kim HY, Chung DH.. Invariant NKT cells functionally link microbiota-induced butyrate production and joint inflammation. J Immunol 2019, 203, 3199. doi: 10.4049/jimmunol.1801314. [DOI] [PubMed] [Google Scholar]
- 109. Shao T-j, He Z-x, Xie Z-j, Li H-c, Wang M-j, Wen C-p.. Characterization of ankylosing spondylitis and rheumatoid arthritis using 1 H NMR-based metabolomics of human fecal extracts. Metabolomics 2016, 12, 70. [Google Scholar]
- 110. Gill PA, van Zelm MC, Muir JG, Gibson PR.. Review article: short chain fatty acids as potential therapeutic agents in human gastrointestinal and inflammatory disorders. Aliment Pharmacol Ther 2018, 48, 15–34. doi: 10.1111/apt.14689. [DOI] [PubMed] [Google Scholar]
- 111. Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, et al. Salt-responsive gut commensal modulates T(H)17 axis and disease. Nature 2017, 551, 585–9. doi: 10.1038/nature24628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Wang G, Huang S, Wang Y, Cai S, Yu H, Liu H, et al. Bridging intestinal immunity and gut microbiota by metabolites. Cell Mol Life Sci 2019, 76, 3917–37. doi: 10.1007/s00018-019-03190-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Rothhammer V, Mascanfroni ID, Bunse L, Takenaka MC, Kenison JE, Mayo L, et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med 2016, 22, 586–97. doi: 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Jangi S, Gandhi R, Cox LM, Li N, von Glehn F, Yan R, et al. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun 2016, 7, 12015. doi: 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S.. The bile acid receptor FXR is a modulator of intestinal innate immunity. J Immunol 2009, 183, 6251–61. doi: 10.4049/jimmunol.0803978. [DOI] [PubMed] [Google Scholar]
- 116. Úbeda M, Lario M, Muñoz L, Borrero M-J, Rodríguez-Serrano M, Sánchez-Díaz A-M, et al. Obeticholic acid reduces bacterial translocation and inhibits intestinal inflammation in cirrhotic rats. J Hepatol 2016, 64, 1049–57. doi: 10.1016/j.jhep.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 117. Montgomery AB, Kopec J, Shrestha L, Thezenas M-L, Burgess-Brown NA, Fischer R, et al. Crystal structure of Porphyromonas gingivalis peptidylarginine deiminase: implications for autoimmunity in rheumatoid arthritis. Ann Rheum Dis 2016, 75, 1255–61. doi: 10.1136/annrheumdis-2015-207656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Zhao Z, Ren J, Dai C, Kannapell CC, Wang H, Gaskin F, et al. Nature of T cell epitopes in lupus antigens and HLA-DR determines autoantibody initiation and diversification. Ann Rheum Dis 2019, 78, 380–90. doi: 10.1136/annrheumdis-2018-214125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Örtqvist AK, Lundholm C, Halfvarson J, Ludvigsson JF, Almqvist C.. Fetal and early life antibiotics exposure and very early onset inflammatory bowel disease: a population-based study. Gut 2019, 68, 218–25. doi: 10.1136/gutjnl-2017-314352. [DOI] [PubMed] [Google Scholar]
- 120. Horai R, Zárate-Bladés CR, Dillenburg-Pilla P, Chen J, Kielczewski JL, Silver PB, et al. Microbiota-dependent activation of an autoreactive T cell receptor provokes autoimmunity in an immunologically privileged site. Immunity 2015, 43, 343–53. doi: 10.1016/j.immuni.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Li B, Selmi C, Tang R, Gershwin ME, Ma X.. The microbiome and autoimmunity: a paradigm from the gut–liver axis. Cell Mol Immunol 2018, 15, 595–609. doi: 10.1038/cmi.2018.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Ruff WE, Dehner C, Kim WJ, Pagovich O, Aguiar CL, Yu AT, et al. Pathogenic autoreactive T and B cells cross-react with mimotopes expressed by a common human gut commensal to trigger autoimmunity. Cell Host Microbe 2019, 26, 100–113.e8. doi: 10.1016/j.chom.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Yordanov M, Tchorbanov A, Ivanovska N.. Candida albicans cell-wall fraction exacerbates collagen-induced arthritis in mice. Scand J Immunol 2005, 61, 301–8. doi: 10.1111/j.1365-3083.2005.01575.x. [DOI] [PubMed] [Google Scholar]
- 124. Costalonga M, Hodges JS, Herzberg MC.; Streptococcus sangui modulates type II collagen-induced arthritis in DBA/1J mice. J Immunol 2002, 169, 2189–95. doi: 10.4049/jimmunol.169.4.2189. [DOI] [PubMed] [Google Scholar]
- 125. Pianta A, Arvikar SL, Strle K, Drouin EE, Wang Q, Costello CE, et al. Two rheumatoid arthritis-specific autoantigens correlate microbial immunity with autoimmune responses in joints. J Clin Invest 2017, 127, 2946–56. doi: 10.1172/JCI93450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ebringer A. Molecular mimicry. In: Delves PJ (ed), Encyclopedia of Immunology, 2nd edn. Oxford: Elsevier, 1998, 1736–42. [Google Scholar]
- 127. Al-Zafiri R, Gologan A, Galiatsatos P, Szilagyi A.. Cytomegalovirus complicating inflammatory bowel disease: a 10-year experience in a community-based, university-affiliated hospital. Gastroenterol Hepatol (N Y) 2012, 8, 230–9. [PMC free article] [PubMed] [Google Scholar]
- 128. Choo HMC, Cher WQ, Kwan YH, Fong WWS.. Risk factors for cytomegalovirus disease in systemic lupus erythematosus (SLE): a systematic review. Adv Rheumatol 2019, 59, 12. [DOI] [PubMed] [Google Scholar]
- 129. Langer-Gould A, Wu J, Lucas R, Smith J, Gonzales E, Amezcua L, et al. Epstein-Barr virus, cytomegalovirus, and multiple sclerosis susceptibility: a multiethnic study. Neurology 2017, 89, 1330–7. doi: 10.1212/WNL.0000000000004412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Clooney AG, Sutton TDS, Shkoporov AN, Holohan RK, Daly KM, O’Regan O, et al. Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe 2019, 26, 764–778.e5. doi: 10.1016/j.chom.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 131. Zuo T, Lu XJ, Zhang Y, Cheung CP, Lam S, Zhang F, et al. Gut mucosal virome alterations in ulcerative colitis. Gut 2019, 68, 1169–79. doi: 10.1136/gutjnl-2018-318131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. De Paepe M, Leclerc M, Tinsley CR, Petit MA.. Bacteriophages: an underestimated role in human and animal health?. Front Cell Infect Microbiol 2014, 4, 39. doi: 10.3389/fcimb.2014.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Ott SJ, Kuhbacher T, Musfeldt M, Rosenstiel P, Hellmig S, Rehman A, et al. Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand J Gastroenterol 2008, 43, 831–41. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- 134. Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, et al. Fungal microbiota dysbiosis in IBD. Gut 2017, 66, 1039–48. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are incorporated into the article and its online supplementary material. The data underlying this article are available in the article and in its online supplementary material.