Abstract
Complementation experiments, Tn5 mutagenesis, and DNA sequencing were used to identify a locus (lag-1) that participates in acetylation of Legionella pneumophila serogroup 1 lipopolysaccharide. Nuclear magnetic resonance analyses of lipopolysaccharides from mutant and complemented strains suggest that lag-1 is responsible for O acetylation of serogroup 1 O polysaccharide.
Legionella pneumophila, the causative agent of Legionnaires’ disease, is a gram-negative bacterium that can enter and grow within a variety of eukaryotic cells, including human monocytes and alveolar macrophages (7) and free-living amoebae (4). Barker et al. (1) and Lüneberg et al. (12) showed that structural and serological changes can occur in L. pneumophila lipopolysaccharide (LPS) during intracellular growth in both amoebae and monocyte-like human cell lines. Further, recent evidence suggests that serological changes in serogroup 1 LPS can result in a net reduction in L. pneumophila virulence (12).
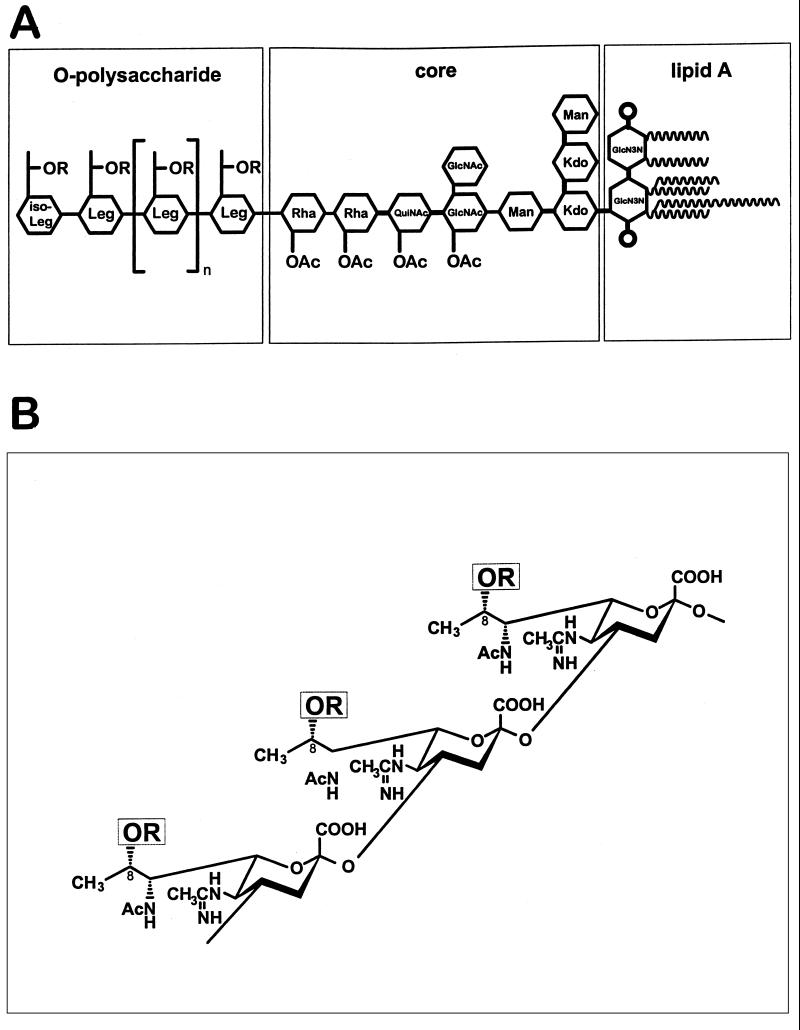
To assess the contribution of LPS to the interaction between L. pneumophila and eukaryotic cells, we isolated a mutant of strain Philadelphia 1, designated CS332, that produced LPS which failed to bind serogroup 1 LPS monoclonal antibodies (MAbs) MAB2 and 33G2 (15, 17). Also, the mutant LPS, in contrast with wild-type LPS, bound the serogroup 1 LPS MAb 144C2 (17). Nuclear magnetic resonance (NMR) spectroscopic analysis performed in the present study revealed that CS332 LPS was missing the O-acetyl group at position 8 of its O-repeat unit (Fig. 1A), which was determined to be 5-acetamidino-7-acetamido-8-O-acetyl-3,5,7,9-tetradeoxy-l-glycero-d-galacto-nonulosonic acid, an N- and O-acylated derivative of legionaminic acid (Fig. 1B) (8, 9, 11, 18, 24).
FIG. 1.
Structure of L. pneumophila serogroup 1 LPS adopted from Knirel et al. (8–11), Zähringer et al. (24), and Moll et al. (18). (A) Schematic representation of the whole LPS structure. (B) Structure of OPS. Sugar abbreviations: GlcN3N, 2,3-diamino-2,3-dideoxy-d-glucose; Kdo, 3-deoxy-d-manno-octulosonic acid; Leg and iso-Leg, derivatives of legionaminic acid and its C4 epimer, respectively; Rha, l-rhamnose; QuiNAc, 2-acetamido-2,6-dideoxy-d-glucose (N-acetylquinovosamine); R = Ac in strains Philadelphia 1 and CS338 or R = H in strain CS332.
In the current study, we used strain CS332 in complementation experiments to identify and characterize a locus, lag-1, that is involved with acetylation of the O polysaccharide (OPS) of L. pneumophila serogroup 1 LPS.
Complementation of strain CS332.
A cosmid library containing genomic DNA from L. pneumophila Philadelphia 1 was constructed in pAM2 (Table 1) as described by Marra et al. (13). The library was mobilized en masse from Escherichia coli DH5α into L. pneumophila CS332 via triparental matings (14), and nine transconjugants were identified that bound serogroup 1 LPS MAb, MAB2, by a colony immunoblot assay (16). Plasmid DNA isolated from the MAB2-positive transconjugants exhibited identical restriction fragment patterns following digestion with several restriction enzymes. A 4.5-kb SphI fragment (pLPS16, Table 1) from one of the recombinant cosmids restored binding of MAB2 and 33G2 and eliminated binding of 144C2 following electroporation (13) into strain CS332.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strain | ||
| Legionella pneumo phila | ||
| AM511 | Philadelphia 1 r− m− Smr | 13 |
| CS332 | Philadelphia 1 LPS mutant Smr | This study |
| CS333 | AM511 harboring pLAW300 | This study |
| CS334 | CS332 harboring pLPS16 | This study |
| CS338 | CS332 harboring pLPS17 | This study |
| CS339 | CS332 harboring pLPS20 | This study |
| Escherichia coli | ||
| DH5α | Nalr r− m+ | C. Collins |
| Plasmid | ||
| pAM2 | IncP cosmid Gmr Tcr mob+ | 13 |
| pRK2073 | ColE1 IncP tra+ | D. Figurski |
| pUC19 | cloning vector ColE1 Apr | New England Biolabs |
| pGEM-7Zf(−) | cloning vector, Apr | Promega |
| pLAW300 | pBlueScript II SK +/− with 939-bp HindIII fragment from Tn903 ColE1 Kmr | H. Shuman |
| pLOI193 | IncQ cloning vector, Cmr Tcr | C. Eddy |
| pLPS16 | 4.5-kb SphI fragment from pLPS15 cloned into pLOI193 | This study |
| pLPS16.1 | 4.5-kb SphI fragment from pLPS15 cloned into pUC19 | This study |
| pLPS17 | 2.3-kb EcoRI fragment from pLPS16.1 cloned into pLAW300 | This study |
| pLPS20 | 1.2-kb EcoRI fragment from pLPS16.1 cloned into pLAW300 | This study |
Apr, ampicillin resistant; Cmr, chloramphenicol resistant; Gmr, gentamicin resistant; Kmr, kanamycin resistant; Nalr, naladixic acid resistant; Smr, streptomycin resistant; Spr, spectinomycin resistant; Tcr, tetracycline resistant; r−, restriction minus; m+, modification plus; m−, modification minus; mob+, mobilization plus; tra+, transfer plus.
There were no obvious differences in the electrophoretic profiles of LPSs isolated from either the mutant or complemented strains (Fig. 2A). However, LPS from strain CS332 that harbored pLPS16 was able to bind MAB2 (Fig. 2B) and 33G2 in Western blotting experiments (25).
FIG. 2.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (A) and Western blot (B) analyses of LPSs from Philadelphia 1 (wild-type) and lag-1 mutant strains. (A) LPS was isolated as described previously (19), resolved on a sodium dodecyl sulfate–14% polyacrylamide gel, and visualized by silver staining. Equal amounts of LPS (∼1.0 μg) were added to each lane. Lanes: 1, S. enterica serovar Typhimurium; 2, Philadelphia 1; 3, CS332 (lag-1 negative); 4, CS334 (CS332/pLPS16 [lag-1 positive]). (B) Equal amounts of LPS from strains Philadelphia 1, CS332, and CS338 (CS332/pLPS17 [lag-1 positive]) were resolved on sodium dodecyl sulfate–14% polyacrylamide gels, transferred to nitrocellulose paper, and probed with MAB2 as described by Mintz and Zou (15). Lanes: 1, Philadelphia 1; 2, CS332; 3, CS338.
Localization and nucleotide sequence of lag-1.
Tn5 mutagenesis (2) of pLPS16.1 (Table 1) showed that the gene(s) responsible for complementation of CS332 was localized to an ∼1.0-kb region of a 2.3-kb EcoRI fragment contained within the cloned 4.5-kb SphI fragment (Fig. 3). This finding was consistent with results of subcloning experiments which showed that the same 2.3-kb EcoRI fragment (contained in pLPS17, Table 1) complemented CS332 to wild-type LPS MAb binding pattern.
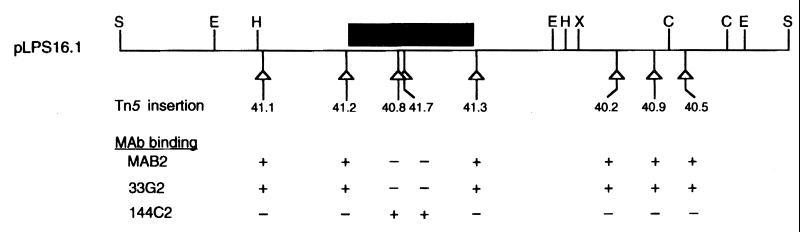
FIG. 3.
Localization of lag-1 by Tn5 mutagenesis. Tn5 insertions were introduced into pLPS16.1 and mapped according to the methods of de Bruijn and Lupinski (2). Each triangle represents the position of a Tn5 insertion in pLPS16.1. Plasmids containing each of the Tn5 insertions were electroporated into strain CS332, and transformants were tested for the ability to bind MAB2, 33G2, and 144C2. (−), insertions eliminating binding of MAB2 and 33G2; (+), insertions having no effect on the binding of MAB2 and 33G2. Wild-type LPS has the following MAb binding pattern: MAB2 (+), 33G2 (+), 144C2 (−). Restriction sites: C, ClaI; E, EcoRI; H, HindIII; S, SphI; X, XhoI. The black box defines the physical location of the lag-1 locus as determined by DNA sequencing experiments.
As shown in Fig. 3, Tn5 insertion no. 40.8 in pLPS16.1 disrupted the gene(s) responsible for complementation of strain CS332. To identify the gene(s) contained within this region, we cloned the two BamHI-EcoRI fragments created by insertion no. 40.8 (Tn5 contains a single BamHI site) into pGEM-7Zf(−) (Promega Corp., Madison, Wis.) and determined the nucleotide sequence of both strands of the 2.3-kb EcoRI fragment from pLPS16.1. The cloned fragment contained a single open reading frame (ORF) spanning 1,074 nucleotides. The ORF, designated lag-1 (lipopolysaccharide-associated gene), encoded a 357-amino-acid protein with a pI of 10.5 and a predicted molecular mass of 40,956 Da. Of interest, we recently determined that strain CS332 contains a deletion mutation in the lag-1 gene (23).
Distribution of the lag-1 locus.
Southern blot experiments (14) with a lag-1 probe and genomic DNA from L. pneumophila serogroups 1 to 5 were performed to determine the distribution of lag-1 in L. pneumophila. Results of these experiments showed that lag-1 could be detected as a single DNA fragment of various sizes in four of eight serogroup 1 LPS isolates (25). LPS from the four serogroup 1 strains containing lag-1 DNA sequences was able to bind MAB2, whereas three of four of these LPSs bound 33G2. In contrast, LPS from the four lag-1-negative serogroup 1 isolates failed to bind MAB2 and 33G2. lag-1 DNA sequences were not detected in chromosomal DNA from LPS serogroups 2 to 5.
lag-1 encodes a polypeptide involved with O acetylation of serogroup 1 OPS.
The predicted amino acid sequence of lag-1 showed strong similarity (54% identity) with a protein called Oac, an O-acetyltransferase encoded by the Shigella flexneri bacteriophage SF6 (22). The Lag-1 polypeptide was slightly larger than the Oac protein (357- versus 333-amino-acid residues). Nevertheless, similarity with Oac extended throughout the entire Lag-1 polypeptide.
The lack of the 8-O-acetyl group from legionaminic acid in LPS produced by the lag-1 mutant CS332 was consistent with the idea that lag-1 encoded an O-acetyltransferase. To test this, we used 1H- and 13C-NMR to characterize the structures of the OPS of LPSs produced by strains CS333 (wild type), CS338 (CS332 plus pLPS17 [lag-1 positive]), and CS339 (strain CS332 plus pLPS20 [lag-1 negative]).
OPSs were isolated from the above-mentioned LPSs and prepared for NMR analysis as described previously (11). 1H- and 13C-NMR spectra of OPSs were recorded at 90.6 MHz with a Bruker AM-360 spectrometer at pH 4 and 45°C (internal standard acetone, delta H 2.225, delta C 31.45) by using standard Bruker software (XWINNMR 1.3).
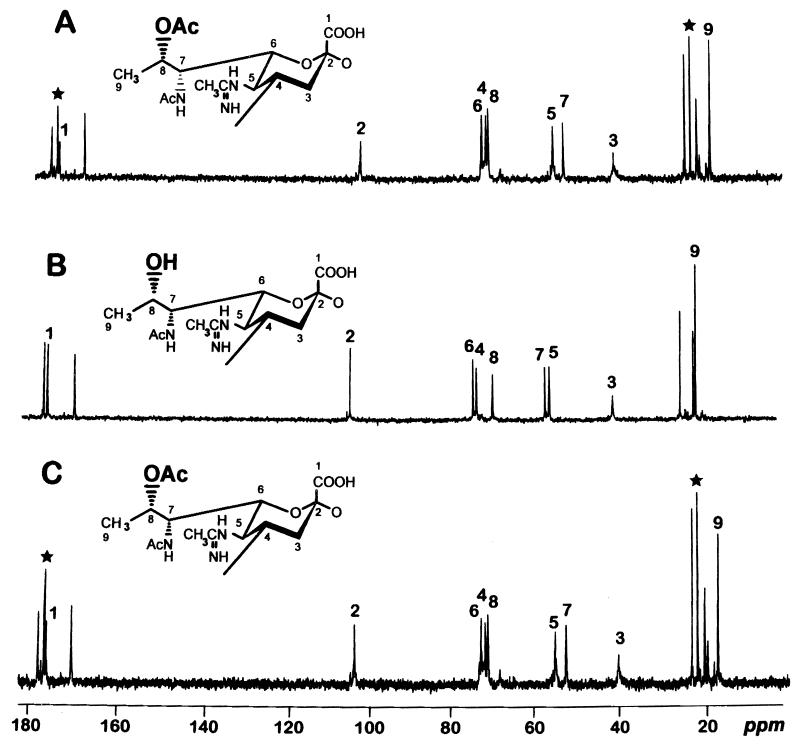
NMR spectroscopy clearly indicated that 8-O-acetylated legionaminic acid was present in the OPSs from lag-1-positive strains CS333 and CS338. In contrast, OPS from the lag-1-negative strain CS339 lacked the O-acetyl group at position 8 of legionaminic acid. Accordingly, signals for an O-acetyl group at δ 22.0 (Me) and 174.4 (CO) were present in the 13C-NMR spectra of OPSs from wild type (CS333) and the complemented strain (CS338) but absent from the mutant (CS339 [Fig. 4]).
FIG. 4.
13C-NMR spectra and corresponding structures of OPSs from LPSs produced by wild-type, mutant, and complemented strains. (A) CS338 (CS332/pLPS17). (B) CS339 (CS332/pLPS20). (C) CS333 (AM511/pLAW300). Signals for the 8-O-acetyl group (Me at δ 22.0 and CO at δ 174.4) are marked by stars. Numbers refer to carbons C1 to C9 of legionaminic acid.
Concluding remarks.
Helbig et al. (5, 6) showed that chemical removal of O-acetyl groups from Philadelphia 1 LPS resulted in loss of reactivity with MAB2 and permitted binding of several MAbs that failed to bind to unmodified LPS. Consistent with the findings of Helbig et al., LPS from strain CS332, which contained 8-O-deacetylated legionaminic acid, failed to bind MAB2 but was able to bind MAb 144C2 (which cannot bind wild-type LPS). Complementation of the lag-1 mutation in strain CS332 resulted in production of OPS that contained 8-O-acetylated legionaminic acid (Fig. 4) which, in turn, restored binding of MAB2 but eliminated binding of MAb 144C2. Our results, along with those of Helbig et al., demonstrate that recognition of serogroup 1 LPS by MAB2 requires an O-acetyl group at position 8 of legionaminic acid and that the 8-O-acetyl group of legionaminic acid can block access to other antigenic determinants contained in serogroup 1 LPS.
The limited distribution of the lag-1 locus among serogroup 1 isolates and its absence in non-serogroup 1 strains suggest that the lag-1 gene product is not essential for biosynthesis of L. pneumophila LPS. Nevertheless, the MAB2 epitope is thought to be associated with the virulence of serogroup 1 LPS strains that produce it (3, 21). However, the contribution of the MAB2 epitope to virulence is unclear, because Mintz and Zou (15) showed that there was no difference in the intracellular growth of a lag-1-positive strain and its isogenic lag-1 mutant in monocyte-like U937 cells and free-living amoebae.
Slauch et al. (20) determined that the Lag-1 polypeptide is a member of a family of proteins that participate in the acylation of exported carbohydrate moieties. This family of proteins includes Oac from phage SF6 (acetylation of Shigella LPS O antigen), OfaA from Salmonella typhimurium (acetylation of LPS O antigen), GumF from Xanthomonas campestris pv. campestris (acetylation of LPS O antigen and xanthan polysaccharide), and several proteins from Rhizobium spp. and Streptomyces spp. that are involved with the acetylation of exopolysaccharides and the acylation of macrolide antibiotics, respectively. Although many regions of these trans-acylases are similar, the most striking homologies can be found in the regions corresponding to amino acids 45 to 89 and 143 to 161 in Lag-1 (20). TMpred analyses of Lag-1 indicated many prominent hydrophobic regions and 10 transmembrane helical domains, suggesting that the Lag-1 polypeptide, like other members of this family of proteins, is an integral membrane protein.
The ability of complemented lag-1 strains to produce OPS containing 8-O-acetylated legionaminic acid, along with the strong similarity between the predicted amino acid sequences of Lag-1 and the Shigella O-acetyltransferase Oac, suggests that lag-1 encodes an O-acetyltransferase. Overexpression of enzymatically active Lag-1 protein and creation of an in vitro assay to measure acetyltransferase activity will be necessary to confirm or refute this hypothesis.
Nucleotide sequence accession number.
The nucleotide sequence of lag-1 has been deposited in Genbank under accession no. U32118.
Acknowledgments
We thank Barry Fields, Dick Miller, Marcus Horwitz, and Joe Plouffe for supplying clinical and environmental isolates of L. pneumophila. We are indebted to Sarah D’Orazio for help with manipulation of lag-1 DNA sequence data. The computing skills of Daniel Gonzalez are gratefully acknowledged.
REFERENCES
- 1.Barker J P, Lamber A, Brown M R W. The influence of intra-amoebic and other growth conditions on the surface properties of Legionella pneumophila. Infect Immun. 1993;61:3503–3510. doi: 10.1128/iai.61.8.3503-3510.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Bruijn F J, Lupinski J R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids. Gene. 1984;27:131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]
- 3.Dournon E, Bibb W F, Rjagopalan P, Desplaces N, McKinney R M. Monoclonal antibody reactivity as a virulence marker for Legionella pneumophila serogroup 1 strains. J Infect Dis. 1988;157:496–501. doi: 10.1093/infdis/157.3.496. [DOI] [PubMed] [Google Scholar]
- 4.Fields B S, Barbaree J M, Shotts E B, Feeley J C, Morrill W, Sanden G S, Dykstra M J. Comparison of the guinea pig and protozoan models for determining virulence of Legionella species. Infect Immun. 1986;53:553–559. doi: 10.1128/iai.53.3.553-559.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helbig J H, Lück P C, Knirel Y A, Witzleb W, Zähringer U. Molecular characterization of a virulence-associated epitope on the lipopolysaccharide of Legionella pneumophila serogroup 1. Epidemiol Infect. 1995;115:71–78. doi: 10.1017/s0950268800058131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helbig J H, Kurtz J B, Pastoris M C, Pelaz C, Lück P C. Antigenic lipopolysaccharide components of Legionella pneumophila recognized by monoclonal antibodies: possibilities and limitations for division of the species into serogroups. J Clin Microbiol. 1997;35:2841–2845. doi: 10.1128/jcm.35.11.2841-2845.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horwitz M A, Silverstein S C. Legionnaires’ disease bacterium (Legionella pneumophila) multiplies intracellularly in human monocytes. J Clin Investig. 1980;66:441–450. doi: 10.1172/JCI109874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knirel Y A, Rietschel E T, Marre R, Zähringer U. The structure of the O-specific chain of Legionella pneumophila serogroup 1 lipopolysaccharide. Eur J Biochem. 1994;221:239–245. doi: 10.1111/j.1432-1033.1994.tb18734.x. [DOI] [PubMed] [Google Scholar]
- 9.Knirel Y A, Helbig J H, Zähringer U. Structure of a decasaccharide isolated by mild acid degradation and dephosphorylation of the lipopolysaccharide of Pseudomonas fluorescens strain ATCC 49271. Carbohydr Res. 1996;283:129–139. doi: 10.1016/0008-6215(95)00401-7. [DOI] [PubMed] [Google Scholar]
- 10.Knirel Y A, Moll H, Zähringer U. Structural study on a highly O-acetylated core of Legionella pneumophila serogroup 1 lipopolysaccharide. Carbohydr Res. 1996;293:223–234. doi: 10.1016/0008-6215(96)00194-2. [DOI] [PubMed] [Google Scholar]
- 11.Knirel Y A, Moll H, Helbig J H, Zähringer U. Chemical characterization of a new 5,7-diamino-3,5,7,9-tetradeoxynonulosonic acid released by mild acid hydrolysis of the Legionella pneumophila serogroup 1 lipopolysaccharide. Carbohydr Res. 1997;304:77–79. doi: 10.1016/s0008-6215(97)00211-5. [DOI] [PubMed] [Google Scholar]
- 12.Lüneberg E, Zähringer U, Knirel Y A, Steinman D, Hartmann M, Steinmetz I, Rohde M, Köhl J, Frosch M. Phase-variable expression of lipopolysaccharide contributes to the virulence of Legionella pneumophila. J Exp Med. 1998;188:49–60. doi: 10.1084/jem.188.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marra A, Blander S J, Horwitz M A, Shuman H A. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc Natl Acad Sci USA. 1992;89:9607–9611. doi: 10.1073/pnas.89.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mintz C S, Shuman H A. Transposition of bacteriophage Mu in the Legionnaires’ disease bacterium. Proc Natl Acad Sci USA. 1987;84:4645–4649. doi: 10.1073/pnas.84.13.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mintz C S, Zou C H. Isolation and characterization of a lipopolysaccharide mutant of Legionella pneumophila. FEMS Microbiol Lett. 1992;93:249–254. doi: 10.1016/0378-1097(92)90470-9. [DOI] [PubMed] [Google Scholar]
- 16.Mintz C S, Fields B S, Zou C H. Isolation and characterization of a conjugative plasmid from Legionella pneumophila. J Gen Microbiol. 1992;138:1379–1386. doi: 10.1099/00221287-138-7-1379. [DOI] [PubMed] [Google Scholar]
- 17.Mintz C S, Zou C H. Abstracts of the 93rd General Meeting of the American Society for Microbiology 1993, Washington, D.C. 1993. Molecular cloning of a locus involved with biosynthesis and expression of Legionella pneumophila lipopolysaccharide, abstr. B-353; p. 89. [Google Scholar]
- 18.Moll H, Knirel Y A, Helbig J H, Zähringer U. Identification of α-d-Man-(1→8)-Kdo disaccharide in the inner core region and the structure of the complete core region of Legionella pneumophila serogroup 1 lipopolysaccharide. Carbohydr Res. 1997;304:91–95. doi: 10.1016/s0008-6215(97)00210-3. [DOI] [PubMed] [Google Scholar]
- 19.Otten S, Iyer S, Johnson W, Montgomery R. Serospecific antigens of Legionella pneumophila. J Bacteriol. 1986;167:893–904. doi: 10.1128/jb.167.3.893-904.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slauch J M, Lee A A, Mahan M J, Mekalanos J J. Molecular characterization of the oafA locus responsible for acetylation of Salmonella typhimurium O-antigen: OafA is a member of a family of integral membrane trans-acylases. J Bacteriol. 1996;178:5904–5909. doi: 10.1128/jb.178.20.5904-5909.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stout J E, Joly J, Para M, Plouffe J, Ciesielsky C, Blaser M J, Yu V L. Comparison of molecular methods for subtyping patients and epidemiologically linked environmental isolates of Legionella pneumophila serogroup 1. J Infect Dis. 1988;157:486–495. doi: 10.1093/infdis/157.3.486. [DOI] [PubMed] [Google Scholar]
- 22.Verma N K, Brandt J M, Verma D J, Lindberg A A. Molecular characterization of the O-acetyl transferase gene of converting bacteriophage SF6 that adds group antigen 6 to Shigella flexneri. Mol Microbiol. 1991;5:71–75. doi: 10.1111/j.1365-2958.1991.tb01827.x. [DOI] [PubMed] [Google Scholar]
- 23.Zaehringer, U., J. H. Helbig, and C. Lueck. Unpublished data.
- 24.Zähringer U, Knirel Y A, Lindner B, Helbig J H, Sonsesson A, Marre R, Rietschel E T. The lipopolysaccharide of Legionella pneumophila serogroup 1 (strain Philadelphia 1): chemical structure and biological significance. In: Levin J, Alving C R, Munford R S, Redl H, editors. Bacterial endotoxins: lipopolysaccharides from genes to therapy. New York, N.Y: Wiley-Liss; 1995. pp. 113–139. [PubMed] [Google Scholar]
- 25.Zou, C. H., and C. S. Mintz. Unpublished data.