Abstract
In recent years, our understanding of the microbial world within us has been revolutionized by the use of culture-independent techniques. The use of multi-omic approaches can now not only comprehensively characterize the microbial environment but also evaluate its functional aspects and its relationship with the host immune response. Advances in bioinformatics have enabled high throughput and in-depth analyses of transcripts, proteins and metabolites and enormously expanded our understanding of the role of the human microbiome in different conditions. Such investigations of the lower airways have specific challenges but as the field develops, new approaches will be facilitated. In this review, we focus on how integrative multi-omics can advance our understanding of the microbial environment and its effects on the host immune tone in the lungs.
Keywords: Lung Microbiome, Host Transcriptome, Metabolome
Introduction
We live in a microbial world. We always have, and our mucosal immune system has co-evolved with it, availing nutrients and critically important molecules that are products of microbial metabolism as well as recognizing a multitude of cues from these microbes that inhabit us. These cues are critical to our own development. Alterations in the microbial composition and/or microbial function in our mucosae, commonly referred to as dysbiosis, leads to compromised mucosal function, altered immune responses, and local and/or distal damage. We have known these facts for several decades now, and they can be convincingly defended on the basis of the knowledge gained from exploring the microbial world using culture-based methods. But more recently, our understanding of the microbial world within us has been revolutionized by the use of culture-independent techniques that have allowed us to characterize both the presence of microbes and their function based on the identification of microbial products (mainly DNA or RNA). Equipped with such armamentarium, we have largely expanded our understanding of the role different microbes play in areas where microbial colonization is well-documented, such as the gut and upper airways.
But the story is different in the lower airways. While the presence of respiratory pathogens is well-documented in the lower airways of patients suffering from various respiratory diseases, the lungs were considered to be sterile for the longest time. More recently, the use of culture-independent techniques has allowed us to harness a complex and dynamic microbial community structure in the lower airways, where pathogens do not exist in isolation but are rather immersed in complex polymicrobial communities. Furthermore, the use of multi-omic approaches can also meticulously characterize the complexity of microbial-host interactions at mucosal sites.
In this review, we will focus on how this approach has advanced our understanding of the microbial environment and its effects on the host immune tone in the lungs. Most notably, we acknowledge that there are myriad ways in which such multi-omic approaches can be utilized. Rather than stipulating a definitive set of guidelines about “best” practices, this review will attempt to provide “conceptual” guidance in a rapidly evolving field that allows plenty of room for optimization and that should be approached with an open technical and analytical mind.
Glossary of lung “omic” nomenclature
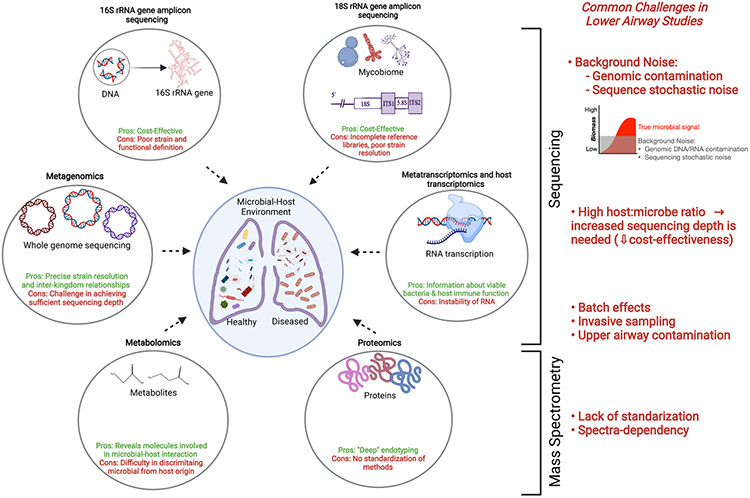
Here we will try to summarize the meaning and specific aspects of different omic approaches as they pertain to the study of the lung environment. Figure 1 summarizes the challenges encountered with different omic technologies that are currently in use for lower airway microbiome studies. We make an illustration of both sequencing-based bioinformatic tools (16S ribosomal RNA (rRNA) gene amplicon sequencing, 18S rRNA gene amplicon sequencing, metagenomics, metatranscriptomics and host transcriptomics) and mass spectrometry-based tools (metabolomics and proteomics); highlighting technology-specific pros and cons. On the right in the figure, we show several key challenges that are commonly shared in varying degrees, between the different omic approaches when studying the lower airways.
Figure 1: Pros and cons of multi -omic approaches applied to lower airway studies.
This figure summarizes the challenges encountered with different omic technologies that are currently in use for lower airway microbiome studies. Here we make an illustration of both sequencing-based bioinformatic tools (16S ribosomal RNA (rRNA) gene amplicon sequencing, 18S rRNA gene amplicon sequencing, metagenomics, metatranscriptomics and host transcriptomics) and mass spectrometry-based tools (metabolomics and proteomics); highlighting technology-specific pros and cons. On the right in the figure, we show several key challenges that are commonly shared in varying degrees between the different omic approaches when studying the lower airways.
“Lung microbial burden:
There are a few methods where targeted measurement of a conserved region of a microbial gene can be utilized for measurement of total microbial burden. Examples include qPCR or digital droplet PCR targeting the 16S rRNA gene for total bacterial load and similar PCR methods for the 18S rRNA gene for total fungal load. These methods offer quantitative data and have been very important in identifying how an increase in bacterial load can be associated with increased lower airway inflammation and an overall poorer prognosis in lung diseases such as asthma, pulmonary fibrosis and critically ill patients with SARS-CoV-2 infection1-6.
Lung microbiota:
This commonly refers to the taxonomic composition of the microbial communities in the lung, frequently explored using short read sequencing that targets variable regions of the 16S rRNA gene. The use of this methodology has led to the recognition that microbial products are found in the lower airways of healthy individuals, challenging a preconception of lung sterility that has dominated the scientific literature for a long time. Early studies involving healthy or diseased individuals used 16S rRNA gene sequencing approaches to taxonomically profile the lungs and identified that there was frequent presence of DNA belonging to oral commensals 7-11. However, these earlier investigations uncovered the challenging reality of study samples with a low microbial biomass: the constant intrusion of background noise in the data. This is predominantly driven by the presence of microbial DNA reads originating from two main potential sources: a) DNA contamination from kits and reagents (true contamination);12 and b) sequencing noise 13. The first one can be distinguished by its inverse relationship between the relative taxonomic abundance of contaminant and microbial DNA load while the second one can be characterized by its stochastic nature (randomness across technical replicates). In both cases, the lower the microbial biomass of the sample, the greater the chances of background noise, a situation commonly present when studying the lungs. Moreover, the premise of background contamination applies in varying degrees to all microbial omic assessments of the lower airways and should be considered in every analytical design. Overall, targeted microbial gene sequencing can be considered a cost-effective approach to taxonomically characterize the lung microbiome. However, it lacks strain resolution and microbial function can only be inferred using analytical pipelines that assume the presence of certain genes based on taxonomic composition (e.g., Phylogenetic Investigation of Communities by Reconstruction of Unobserved States, or “PICRUSt”) 14. Here, it is important to mention that a few studies have compared the results of inferred microbial genomic potential with measured microbial genomics (done by shotgun sequencing) but good correlation between the two datasets could not be demonstrated, raising concerns for the assumptions inherent in this approach15.
Lung mycobiome:
This term refers to the targeted sequencing of fungi and can be done by sequencing amplicons of the 18S rRNA gene or internal transcribed spacer (ITS). Similar to observations made with 16S rRNA, fungi are frequently present in the lower airways, and although overall community composition of the lung mycobiome is distinct from the upper airways, micro-aspiration seems to be the main source of fungi in the lower airways 16. Challenges in the study of the mycobiome include difficulties with DNA isolation and other limitations inherent to different targeted sequences and the less refined reference libraries 17.
Lung metagenome:
This refers to the assessment of the composition of all microbial genes present in the lung environment, usually done through whole genome sequencing (WGS). This method allows us to characterize microbes with a much more precise strain resolution than targeted sequencing and also describes the genomic potential, including potential microbial metabolic functions and antibiotic resistant genes (resistome) 18. There is a paucity of studies that have used this approach to evaluate the lung microbiome due to significant challenges such as insufficient capture of microbial annotated reads. Since there is no amplification of microbial signals prior to sequencing, these methods are also challenged by a lack of sequencing depth required to capture a reasonable number of microbial reads to fully profile the lower airway microbiome. Thus, it is not uncommon to see lung microbiome studies using these techniques aiming to sequence at high depth (e.g., 100 million reads per sample) with the hope of obtaining information from a small percentage of the data that can be assigned to microbial origin. For example, studying the lung microbiome of patients infected with SARS-CoV-2 with metagenomics and metatranscriptomics yielded 1.2% and 0.6% of the total reads assigned to microbes (bacteria, fungi or viruses), respectively 1. One major advantage of these approaches is that they are not restricted to one kingdom, thus allowing for the exploration of inter-kingdom relationships. For example, studies of the lung metagenome have simultaneously explored the presence of bacteria, fungi and viruses in patients with sarcoidosis 19 and SARS-CoV-2 infection 1. Ultimately, such multi-kingdom assessments are often challenging due to discrepancies in reference databases for several microbial components, particularly those of viruses and fungi.
Lung metatranscriptome:
This can be done through RNA sequencing that provides the taxonomic and microbial genomic function of microbes with actual active transcription. Because cell-free RNA is rapidly degraded, this approach can reveal information about viable bacteria in the lower airways. Indeed, studies comparing microbial profiling using metatranscriptome, metagenome and 16S rRNA gene sequencing in the lungs have suggested that the former provides a more accurate representation of viable bacteria in the lower airways by identifying those that actively produce bacterial products such as short chain fatty acids 15. Very few studies have utilized this approach to study the lower airways probably because of similar challenges as those encountered in metagenome studies, and further exacerbated by the instability of RNA.
Lung metabolome and proteome:
This refers to the study of metabolites and proteins and is commonly performed through nuclear magnetic resonance or mass spectrometry (MRS and MS, respectively). Both metabolomic and proteomic approaches can differ vastly and have ‘less targeted’ and ‘more targeted’ approaches that dictate the range of molecules detected. Both technologies have immense untapped value and can add invaluable insight into disease pathogenesis, biomarker discovery, “deep” endotyping and response to therapy in various heterogeneous lung disease states20. Several investigations using lower airway samples have described distinct metabolic signatures in multiple pulmonary conditions such as pulmonary infections 21, HIV 22, acute respiratory distress syndrome 23 and chronic obstructive lung disease 24. While these methods frequently reveal distinct features across different disease states, data interpretation is often difficult due to a lack of standardized measurements in the lower airway samples and the inability to distinguish the relative contributions of microbial and host metabolism to the measured value of the metabolite or protein in question.
A detailed review of such methods pertaining to the lung environment was recently published by the American Thoracic Society 25. Briefly, strategies to improve standardization include plate-based platforms and the use of internal standards and quality control pools to overcome batch effects 26,27. Major challenges include, but are not limited to, data processing and visualization, as no one platform can capture the entire metabolome25. As metabolomic datasets become more and more complicated, tools for biological pathway/network mapping and enrichment analysis of large datasets are being developed; ConceptMetab, Metscape and metabolite set enrichment analysis are examples, to name a few28,29,30.
Lung host transcriptomics:
This is the study of mammalian RNA in the lower airways performed either through targeted approaches (e.g., microarrays, nanostring) or untargeted approaches (e.g., RNA sequencing). A common challenge for studying the lung transcriptome relates to difficulties in obtaining high-quality RNA, especially in samples that are contaminated with saliva due to high levels of salivary RNAse 31,32. Another caveat that warrants special consideration is the cellular composition of the sample obtained given significant topographical variations (e.g., airway brushings vs. bronchoalveolar lavage fluid (BALF)) and innate sample variations (e.g., neutrophilic-predominant BALF vs. macrophage-predominant BALF).
Novel insights about microbial-host interface from lung multi-omic studies
Here we will discuss studies that exemplify how multi-omic approaches can be used to harness the microbial-host interface in health and disease. We will mainly focus on studies that have used lower airway samples. It is important to mention that most lower airway microbiome investigations have focused on studying samples from patients with different disease processes. Due to the invasive nature of sampling, there has been a paucity of investigations that have been conducted on healthy volunteers3,8,33, and this is a shortcoming that may limit our understanding of abnormal microbial host interactions.
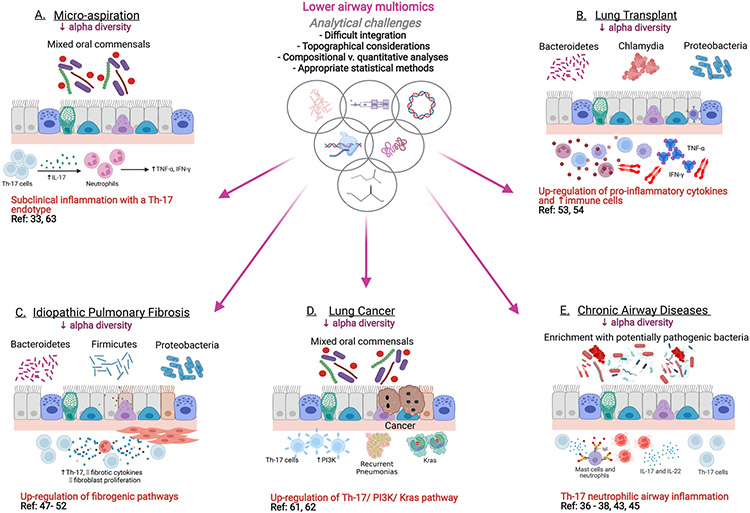
Figure 2 illustrates how various multi-omic studies have helped to unravel the mechanisms underlying pathological conditions such as micro-aspiration, lung transplantation, idiopathic pulmonary fibrosis (IPF), lung cancer and chronic airway diseases. Key discoveries that have been made using multi-omics on lower airway samples are described here.
Figure 2: Approach to multi-omic analyses in lower airways.
Here we exemplify how various multi-omic technologies have helped to unravel the mechanisms underlying pathological conditions such as micro-aspiration, lung transplantation, idiopathic pulmonary fibrosis (IPF), lung cancer and chronic airway diseases. The dysbiotic signals are on the top (luminal side) of the mucosa and the underlying host immune endotype are depicted below the mucosa. References for the studies combining multi-omic datasets in each of these conditions are also displayed.
Multi-omic analyses to uncover lung microbial functions
While the preconception of sterility of the lower airways has been debunked since the identification of microbial signals through culture-independent methods, there is still much debate about whether these microbial signals are representative of true live bacteria or remnants of cleared microbes. Using a combination of lower airway samples to perform 16S rRNA gene sequencing, WGS and RNA metatranscriptome sequencing paired with short chain fatty acid (SCFA) metabolomics, investigators have found that there is a subset of individuals that have viable and metabolically active microbes that produce microbial products with significant immunomodulatory properties such as SCFAs 15. Further, evaluation of the metabolome in BALF from HIV patients revealed specific metabolites such as glycerophospholipid, fatty acids and lineolate that were associated with certain lower airway microbiota and may confer greater risk for pneumonia 22. These studies exemplify how we can start recognizing the presence of viable and metabolically active microbes in the lower airways, at least as a proof-of-concept.
Multi-omic analyses to uncover biomarkers in lung disease
The lack of reliable biomarkers in chronic lung diseases acutely highlights the need for large data integration. Multi-omic-derived biomarkers can also help risk-stratify patients via the integration of genomic, proteomic, transcriptomic and microbiomic datasets 34. Combining multiple omic datasets may generate biomarkers with a high accuracy and positive predictive value. Across several lung diseases, prior studies applying multi-omics have investigated whether diagnostic and prognostic biomarkers can be derived from the lower airways. For example, in patients with lower respiratory tract infections, Langelier et al., 35 utilized a combined metagenome, metatranscriptome, and host transcriptome approach to identify distinct omic specific features in lower airway samples associated with respiratory tract infections.
Role of multi-omic studies at the interface of microbial-host immunity in the characterization of chronic lung diseases
Lower dysbiosis is involved in several chronic airway diseases such as asthma, COPD and cystic fibrosis. When specific “potentially pathogenic bacteria” colonize the airways, the total burden and composition of the microbiota changes and this can trigger the activation of inflammatory pathways such as the Th17- neutrophilic pathway 36. The Th17 pathway and lower airway dysbiosis have a dysregulatory effect on the development of asthma, the severity of asthma and the corticosteroid responsiveness of asthma. This association is particularly strong in the case of neutrophilic asthma, which is characteristically more resistant to steroids 36. Studies comparing the composition of the airway microbiome between steroid-resistant and steroid-sensitive patients have identified differences at the genus level, with gram-negative lipopolysaccharide- producing bacteria with high-endotoxic activity enriched in the BAL of those with steroid-resistant asthma 37. Combining 16S rRNA gene sequencing and microarray data, investigators have found that in severe asthma, enrichment with Proteobacteria in airway brushings is correlated with the up-regulation of Th17-associated genes, whereas Actinobacteria correlated with the bronchial epithelial gene expression of FK506 binding protein, a marker of steroid responsiveness 38. In another study comparing atopic asthma subjects with non-atopic controls, the diagnosis of asthma was associated with differences in the predicted bacterial functions involving amino acid and short-chain fatty acid metabolism, suggesting that alterations in levels of bacterial metabolites with immunomodulatory effects such as SCFAs can affect the clinical phenotype 6. Among patients with cystic fibrosis, there is often an abundance of SCFA-producing anaerobic bacterial species present in the lower airways 39. These SCFAs promote the release of various cytokines and chemokines and their concentration positively correlates with sputum neutrophil counts 39.
The dynamics of the lung microbiome in COPD is constantly in flux, particularly during periods of exacerbation when dramatic changes have been noted in both bacterial strain and species 40. Excessive and persistent inflammation is a major driver of pathogenesis in COPD and a decline in the richness and diversity of the respiratory microbiome is known to be associated with increased immune cell infiltration 41. In one of the earlier studies, Sethi et al.,42 demonstrated that COPD subjects colonized with potentially pathogenic bacteria in the distal airways had greater neutrophil counts and increased concentration of chemokines and matrix metalloproteinases. Using 16S rRNA gene sequencing and metatranscriptome sequencing of BALF samples, three distinct microbial compositions were shown to significantly correlate with lymphocyte proportion, human Th17 immune response and COPD exacerbation frequency 43. In another study using advanced stage emphysematous lung tissue samples, a decline in microbial diversity was found to be associated with emphysematous destruction, remodeling and CD4+ T cell infiltration and specific OTU’s were associated with neutrophils, eosinophils and B cell infiltration 44. In a secondary analysis of BALF microbiome and metabolomic data from patients with early stage COPD (GOLD 0-2), lower lung function and severity of symptoms were positively associated with enrichment with oral commensals, Streptococcus, Neisseria and Veillonella, and with several metabolites, including glycosphingolipids, glycerophospholipids, polyamines and xanthines 45. These observations suggest that the microbiome and its active functional elements play a significant role in immune homeostasis in chronic airway diseases. It is surmisable that a lower airway dysbiotic signature stimulates airway cells and triggers Th-17 responses, higher chemokine levels and recruitment of neutrophils into the airway lumen. A better understanding of the cross-talk between the microbiome and host immune compartments is a relatively untapped territory and future discoveries could have huge implications for optimizing existing treatments in Asthma and COPD.
Studies have also clearly linked dysbiosis with fibrotic lung diseases such as IPF 46. In a seminal study from the UK, Molyneaux at al 2 evaluated BALF and peripheral blood gene expression in IPF patients and matched healthy controls. Over-expression of one gene module was associated with death and disease progression as well as increased BALF bacterial burden. This particular module was also enriched with genes associated with host defense, over-expression of which was associated with worse survival. In the COMET-IPF study, evaluation of BALF from 60 IPF patients showed that relative inhibition of 11 gene signaling pathways was associated with reduced progression free survival time; 8 pathways involved pathogen infection and 3 involved innate immune response receptors 47. Greater relative abundance of a Streptococcal OTU correlated negatively with poor progression free survival. The composition of the lower airway microbiome in patient with IPF has also been associated with higher levels of pro-fibrotic cytokines 48 and pro-inflammatory cytokines such as IL-17 and TNF-α49. Such cytokines can both damage airway epithelial cells and trigger the expression of α- smooth muscle actin gene. A higher bacterial burden has been observed in IPF patients with poorer prognosis but it is especially higher among patients with a minor allele at the promoter of the mucin gene 50,51. In a study using BALF from 68 patients with IPF and a germ-free murine model of pulmonary fibrosis, the loss of microbial diversity correlated with IPF symptoms and spirometric measurements, high serum surfactant protein-D and lactate dehydrogenase levels and elevated pro-inflammatory cytokine levels. These findings portray the mechanistic impact of lower airway dysbiosis on the pathogenesis of this disease 52. Taken together, these data support a possible link between the lung microbiome, an aberrant host response and disease progression in IPF, although further elucidation of the exact directionality of these associations need more experimental investigations.”
Multiomic approaches have also identified lower airway microbial signals of possible importance in lung transplantation. In a study that combined microbiome and host transcriptome from lower airway samples following lung transplantation, investigators showed that the host transcriptome in the allograft presented distinct remodeling profiles, characterized by the expression of genes differentially involved in matrix synthesis (anabolic) or matrix degradation (catabolic) 53. While catabolic remodeling aligned with a microbiota dominated by Staphylococcus, Pseudomonas and Haemophilus, anabolic remodeling was linked to relative abundance of Prevotella, Streptococcus and Veillonella. A study that combined amplicon sequencing and bacterial culturing and gene expression assessment to characterize 234 longitudinal BALF samples from 64 lung transplant recipients established links between viral loads, host gene expression, lung function and clinical stability 54. What the investigators called a ‘balanced’ pneumotype, characterized by a diverse bacterial community, was associated with an immune tolerant host gene signature. The other three pneumotypes, characterized by depletion of taxa from the “balanced” pneumotypes and/or dominated by potential pathogens, were linked to increased immune activity, lower respiratory function and increased risk of infection and rejection. These studies suggest a complex host-microbe interplay affecting inflammatory and remodeling processes in the transplanted lung”.
Multi-omic approaches may reveal a microbial-dependent mechanism of the anti-inflammatory effects of macrolides in lung diseases
Multiple studies have suggested that macrolides have direct anti-inflammatory effects in various airway diseases such as cystic fibrosis, asthma and COPD. Additionally, whenever the effect of macrolides on microbial communities has been evaluated, investigators have found that even among patients with mild COPD without overt infection with a respiratory pathogen, there are substantial macrolide-driven effects on the microbial composition. In a placebo-controlled trial evaluating the microbiome and metabolome in the lower airways, macrolides led to increased levels of microbially-derived anti-inflammatory metabolites, probably due to an antibiotic-driven pressure on the lower airway microbes 55. This increase in anti-inflammatory metabolites also impacted the inflammatory tone of lower airway macrophages. Chronic azithromycin therapy decreased the frequency of exacerbations in COPD, potentially mediated by a reduction in dysbiosis via selective pressure on the lung microbiota 56. In another study, patients with asthma who displayed improved bronchial reactivity after 6 weeks of macrolide treatment had higher baseline bacterial diversity, thus implicating the role of resident microbiota in modulating the outcome of therapeutic interventions 57. Therefore, it is possible that multi-omics can uncover mechanisms by which existing therapies rely upon the lower airway microbiome for providing therapeutic effects.
Integrative microbiomics can help identify exacerbation risk clusters in bronchiectasis
Studies have shown minimal change in dominant taxa during exacerbation of bronchiectasis, thereby questioning the simplistic model in which a single kingdom overgrowth is held culpable 58. Using an integrated multi-biome analysis of the bronchiectasis airway profiles from patients, Mac Aogain et al., 59 showed that patients at highest risk of exacerbations have a multi-biome dominated by antagonistic interactions between microbial kingdoms in which microbes compete rather than cooperate with one another. The study used a subject-to-subject similarity matrix with predictors that depended on the number of subjects rather than molecular features 59, and therefore had superior cluster precision and accuracy in the highly heterogeneous omic datasets of the bronchiectasis cohorts 60. Although lung function and disease severity were similar between the clusters, the frequent exacerbators had lower alpha diversity. This multi-kingdom interactome provided new and previously unrecognized targets for antimicrobial therapy that could be used as an adjunct to or in combination with more established antibiotic-based regimens. The interactome approach may also be used to monitor the outcomes of therapy and to understand the effects of host directed therapy.
Multi-omic approaches have uncovered mechanisms of microbial host interactions in lung cancer
Multiple investigations have looked at whether lung cancer is associated with either differences in lung microbial communities or host immune defects. The use of multi-omics has allowed for the co-evaluation of these two features by facilitating comparisons between patients with malignant and benign neoplasms and between lung cancer patients at different stages. For example, one study demonstrated an association between lung cancer diagnosis and enrichment of the lower airways with oral commensals. Concurrently, there was an upregulation of host transcripts related to inflammatory pathways (or oncogenesis) such as PI3K, Kras and the Th17 cascade 61. As proof of concept, exposure of Kras-mutated epithelial cell lines to similar dysbiotic signals led to an upregulation of similar transcriptomic processes. In the case of studies that evaluated patients with lung cancer, patients with lower airway microbiota enriched with oral commensals were more likely to present with advanced-stage lung cancer and had a poorer prognosis 62. Parallel evaluation of the host transcriptome identified that this dysbiotic signature was associated with an upregulation of the Th17 inflammatory pathway and checkpoint inhibition 62. Extension of these observations with a mouse preclinical lung cancer model showed that lung dysbiosis plays a significant role on tumor progression through a Th17 pathway 62. These two studies exemplify how the application of multi-omic “agnostic” approaches can be used to dissect important associations that can then be explored mechanistically through ex vivo or in vivo experimental models.
How can big data can be integrated to uncover microbial-host interactions in the lung?
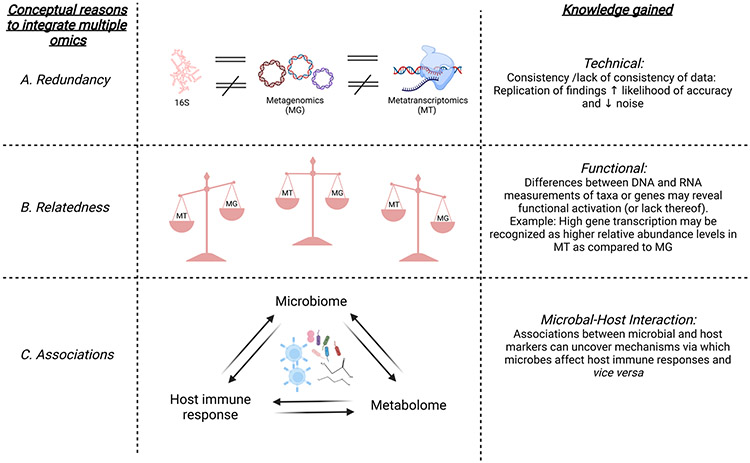
The previous section highlighted some inherent challenges with the various omic approaches available to study the lower airway environment. All omic-centered approaches share some technical challenges such as risk of contamination, topographical differences, and having to measure a large number of variables for a relatively small number of samples. Despite these challenges, omic approaches have blossomed over the last several years and have consistently demonstrated that the lung environment is highly variable between individuals. The next big leap in the -omics multiverse would require an integrated model that can investigate the organization and behavior of all the different layers of the system. The integration of disparate datasets such as microbiomics, genomics, proteomics, metabolomics, metatranscriptomics, and metagenomics, even though computationally challenging, makes this possible. The overwhelming question is therefore: how do we unleash the power of combining -omic approaches to shed light on our understanding of the interaction of microbes with the host in the lung? Whenever using multiple omics (over and above the specific challenges outlined above), investigators need to be cognizant of some common pitfalls that will be encountered during the design of such studies. The research field is still in its infancy and lacks methodological standardization across studies. Here, we discuss some recurrent concerns and potential solutions for frequently encountered issues when combining different omic approaches. In Figure 3, we illustrate our conceptualization of how the comparison of data from different omics technologies can be used to understand the lung microbiome. First, we consider three main conceptual reasons for the integration of multiple omic methods; summarized by what we have called “redundancy”, “relatedness” and “associations”. For example, under “redundancy”, integration of omics helps to assess consistency of data whilst the lack of consistent signals can help identify platform-specific background contaminants. Under “relatedness”, combining different omic methods can uncover differences between the genomic potential vs. transcription of genes, thus revealing functional activation (or lack thereof). As an example, high gene transcription may be recognized as higher relative abundance levels for that gene in metatranscriptomics rather than in metagenomics. The “integration” of different omics can help find meaningful networks via which microbes and host interact; this can help uncover novel associations and mechanisms. Finally, it is important to remember that the confirmation of causality requires experimental studies rather than correlative investigations. Having said that, evaluation of the strength and significance of correlative data might be the first step to identify the most promising signals. In order to use multi-omics to generate new knowledge there are several challenges that should be considered and are discussed below.
Figure 3: Inter-relatedness of various -omic approaches.
In this figure, we illustrate three different approaches to gain knowledge when combining multiple omic data: “redundancy”, “relatedness” and “associations”. A. Integration of omics helps to address “redundancy” by identifying consistency of data. In particular, replication of findings increases the likelihood of accuracy and decreases that of noise. B. The “relatedness” of the different omic methods has the potential to uncover differences related to function. As an example, high rates of gene transcription may be recognized as higher relative abundance in metatranscriptomics rather than in metagenomics for that specific gene, indicating possible functional activation. C. Finally, the integration of different omics can help discover the molecular mechanisms via which microbes affect host immune responses and vice versa.
a). Should different -omic approaches be used in topographically distinct samples?
Topographical variation in microbes and host signals are well-documented. Differences in sample processing across various omic approaches require specific considerations such as how to process and aliquot fresh samples, how to maximize the yield of human cells, and how to account for the potential impact of different preserve media on microbial signatures. Data derived from culture-independent methods have established that micro-aspiration leads to episodic seeding of oral commensals in the lower airways, but topographical differences exist even within each individual. When investigating the lower airways of healthy individuals with BALs, enrichment of the lower airway microbiota with oral commensals such as Prevotella, Streptococcus, Fusobacterium, Rothia and Veillonella is associated with subclinical inflammation and a lower airway inflammatory tone with a Th17 endotype 33. However, topographical microbial differences could affect such assessments and warrants careful planning. Also, it is unclear if the inflammatory signal is due to viable and metabolically active bacteria, non-viable bacteria, or byproducts of bacterial metabolism 63. Differences in topographical environmental conditions, such as oxygen tension, pH and mucus, may affect microbial viability and metabolic activity and is therefore an important variable to consider.
b). Is the data generated within each omic method quantitative, semiquantitative or compositional?
Quite commonly, the nature of the data across different omic datasets varies. For example, the sequencing data for microbial composition (16S rRNA gene, WGS, or metatranscriptome sequencing) is semiquantitative and compositional (no absolute concentrations but rather relative abundance), as well as sparse (frequent null values for many of the taxa assessed). In contrast, metabolomic data is not compositional and can be quantitative when internally labeled standards are added. Integrating these different kinds of datasets into a single analytical pipeline may lead to false assumptions about the behavior of the data and needs to be carefully considered.
c). Are the statistical methods used for analyses similar across different omics?
Some of the multi-omic methods rely on integrating only statistically significant data from the different omic datasets. However, statistical significance may lack uniformity of criteria across various omic methods. In other words, because each omics output can be considered unique in terms of, for example sparsity, data distribution, compositionality, depth, number of features, lack of updated databases; these omics datasets are often not interrogated using the exact same approaches. Thus, even though only statistically significant data is pooled from different omics, the end result of the merged dataset is dependent on how each data was processed prior to analysis and data integration. Wherever possible, multi-omics research warrants special considerations about how to deploy consistent analytical approaches across different omic datasets prior to integration of the data.
d). How do we choose the most appropriate method required for an integrative analysis that combines two or more omic datasets?
The array of multi-omic analytical methods is growing at a pace that is difficult to keep track of. Overall, these methods are all based on correlative analyses between different components of each omic dataset, and although they are all individually valuable, they can be even more useful if integrated appropriately and applied to the dynamic/temporal aspects of disease pathogenesis. Different analytical approaches are built into each multiomic, such as dimensionality reduction methods, correlation-based approaches, regression-based approaches and network-based approaches. In-depth knowledge of each these analytical approaches is needed in order to apply them correctly. Practical examples of the use of such multi-omic methods in lung studies include SparCC 33, Mummichog22, ComPLS61, Mixomics45,64, similarity network fusion 59,60,65, and MMVEC 62. Importantly, differences between these methods rely on how each individual omic dataset is treated and what statistical manipulation and visualization strategies are used. Whenever integrating multi-omic datasets, it is important to not just obtain expert informatic support but also to consider the real difficulties in establishing a statistically robust analytical approach. One such challenge comes from recognizing that some of what is seen as a statistically significant association may be driven by “outliers” 66. Another major challenge is the variation in scale, complexity, and correlation structure between datasets that are inherently different 65. It is therefore paramount that investigators do not simply focus on statistical strength but also consider applying different methods for the integration of disparate datasets and testing the robustness of signals across them. Bayesian data interpretation based on prior knowledge and post-hoc experimental investigations seeking to corroborate and extend associations previously identified are key to selecting “the right” analytical approach.
Future directions
Current multi-omic approaches on cross-sectional data are based on statistical associations and are unable to consider the plausibility of associations. In other words, the statistical significance and strength of the associations between ‘markers/endophenotypes’ within two different -omic datasets often entail the simultaneous consideration of multiple associations. Given the high dimensionality of such datasets, very few associations pass beyond a certain level of statistical stringency.
However, we do have prior knowledge about potential associations between different components of these -omic datasets from epidemiological or immunological studies. For example, we know which microbes and microbial genes contribute to the production of PAMPS. We also know about the microbial genes associated with the production/degradation of metabolites that have immunoregulatory properties and the signals they may elicit or perpetuate in the host immune response. Thus, there is a need for a “Bayesian” integrative analytical approach for multi-omic datasets. Multi-omic approaches may uncover statistically significant associations but directionality and causality need to be explored with other experimental approaches.
As the field develops, we will be better able to characterize the endophenotypes of various chronic lung conditions and this will inevitably lead to new avenues for their diagnosis, management and prevention. However, firstly, multi-omics need to move from cross-sectional descriptive profiling to longitudinal studies during disease progression and treatment. Secondly, we need a better understanding of the effects of different treatments on the microbiome. While probiotics, prebiotics and mixed bacterial transfers (such as fecal material transfer) have enormous promise as new therapies that can modify the gut microbiome, these have not been studied in any significant depth in the lung. In particular, the combination of metabolomics with other transcriptomics can specifically help us identify the interaction between the microbial metabolites and the host immune system and therefore offer the opportunity for a more personalized approach to diagnosis and treatment. Finally, multi-omics can help generate hypotheses for more focused mechanistic studies to help understand disease pathophysiology at a molecular level.
Conclusions
Advances in techniques and bioinformatics have enabled high throughput and in-depth analyses of transcripts, proteins and metabolites and enormously expanded our understanding of the human microbiome and the role it plays in our well-being. Chronic lung disease is a consequence of multiple layers of regulation and dysregulation, many of which are driven or affected by microbes present in the lungs. Large omic studies are able to expeditiously identify the stages that are important in any given disease process. The advent of multi-omics has made possible the generation of data sets that can integrate a broad range of molecular, cellular, pathophysiological and clinical states and thereby shed light on the hitherto unknown relationships between the lung microbiome, host endotype and phenotype. As our approaches evolve it is likely that multi-omics will enter a new phase of clinical application where targeting the lung microbiome will be possible and personalized.
Acknowledgements:
R37 CA244775 (LNS, NCI/NIH), Stony Wold Herbert Inc. Foundation (SS, JGN), ATS COPD Foundation Bronchiectasis Initiative Grant (SS).
Footnotes
Conflict of Interest Statement: No conflicts
References
- 1.Sulaiman I et al. Microbial signatures in the lower airways of mechanically ventilated COVID-19 patients associated with poor clinical outcome. Nature microbiology, doi: 10.1038/s41564-021-00961-5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molyneaux PL et al. Host-Microbial Interactions in Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med 195, 1640–1650, doi: 10.1164/rccm.201607-1408OC (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segal LN et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 1, 19, doi: 10.1186/2049-2618-1-19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Combs MP et al. Lung microbiota predict chronic rejection in healthy lung transplant recipients: a prospective cohort study. The Lancet. Respiratory medicine 9, 601–612, doi: 10.1016/s2213-2600(20)30405-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickson RP et al. Lung Microbiota Predict Clinical Outcomes in Critically Ill Patients. American journal of respiratory and critical care medicine 201, 555–563, doi: 10.1164/rccm.201907-1487OC (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durack J et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. The Journal of allergy and clinical immunology 140, 63–75, doi: 10.1016/j.jaci.2016.08.055 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck JM et al. Multicenter Comparison of Lung and Oral Microbiomes of HIV-infected and HIV-uninfected Individuals. Am J Respir Crit Care Med 192, 1335–1344, doi: 10.1164/rccm.201501-0128OC (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charlson ES et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. American journal of respiratory and critical care medicine 184, 957–963, doi: 10.1164/rccm.201104-0655OC (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bassis CM et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio 6, e00037, doi: 10.1128/mBio.00037-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickson RP et al. Spatial Variation in the Healthy Human Lung Microbiome and the Adapted Island Model of Lung Biogeography. Ann Am Thorac Soc 12, 821–830, doi: 10.1513/AnnalsATS.201501-029OC (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molyneaux PL et al. Outgrowth of the Bacterial Airway Microbiome following Rhinovirus Exacerbation of Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med, doi: 10.1164/rccm.201302-0341OC (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salter SJ et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12, 87, doi: 10.1186/s12915-014-0087-z (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erb-Downward JR et al. Critical Relevance of Stochastic Effects on Low-Bacterial-Biomass 16S rRNA Gene Analysis. mBio 11, doi: 10.1128/mBio.00258-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langille MG et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature biotechnology 31, 814–821, doi: 10.1038/nbt.2676 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sulaiman I et al. Functional lower airways genomic profiling of the microbiome to capture active microbial metabolism. Eur Respir J, doi: 10.1183/13993003.03434-2020 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui L et al. Topographic diversity of the respiratory tract mycobiome and alteration in HIV and lung disease. American journal of respiratory and critical care medicine 191, 932–942, doi: 10.1164/rccm.201409-1583OC (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tipton L, Ghedin E & Morris A The lung mycobiome in the next-generation sequencing era. Virulence 8, 334–341, doi: 10.1080/21505594.2016.1235671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mac Aogain M et al. Metagenomics Reveals a Core Macrolide Resistome Related to Microbiota in Chronic Respiratory Disease. Am J Respir Crit Care Med 202, 433–447, doi: 10.1164/rccm.201911-2202OC (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke EL et al. Microbial Lineages in Sarcoidosis. A Metagenomic Analysis Tailored for Low-Microbial Content Samples. Am J Respir Crit Care Med 197, 225–234, doi: 10.1164/rccm.201705-0891OC (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stringer KA, McKay RT, Karnovsky A, Quémerais B & Lacy P Metabolomics and Its Application to Acute Lung Diseases. Front Immunol 7, 44, doi: 10.3389/fimmu.2016.00044 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui L et al. Metabolomics Investigation Reveals Metabolite Mediators Associated with Acute Lung Injury and Repair in a Murine Model of Influenza Pneumonia. Scientific reports 6, 26076, doi: 10.1038/srep26076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cribbs SK et al. Correlation of the lung microbiota with metabolic profiles in bronchoalveolar lavage fluid in HIV infection. Microbiome 4, 3, doi: 10.1186/s40168-016-0147-4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogers AJ et al. Profiling of ARDS pulmonary edema fluid identifies a metabolically distinct subset. Am J Physiol Lung Cell Mol Physiol 312, L703–L709, doi: 10.1152/ajplung.00438.2016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halper-Stromberg E et al. Bronchoalveolar Lavage Fluid from COPD Patients Reveals More Compounds Associated with Disease than Matched Plasma. Metabolites 9, doi: 10.3390/metabo9080157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowler RP et al. New Strategies and Challenges in Lung Proteomics and Metabolomics. An Official American Thoracic Society Workshop Report. Ann Am Thorac Soc 14, 1721–1743, doi: 10.1513/AnnalsATS.201710-770WS (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carr SA et al. Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol Cell Proteomics 13, 907–917, doi: 10.1074/mcp.M113.036095 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant RP & Hoofnagle AN From lost in translation to paradise found: enabling protein biomarker method transfer by mass spectrometry. Clin Chem 60, 941–944, doi: 10.1373/clinchem.2014.224840 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cavalcante RG et al. ConceptMetab: exploring relationships among metabolite sets to identify links among biomedical concepts. Bioinformatics 32, 1536–1543, doi: 10.1093/bioinformatics/btw016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karnovsky A et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics 28, 373–380, doi: 10.1093/bioinformatics/btr661 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia J & Wishart DS MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res 38, W71–77, doi: 10.1093/nar/gkq329 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan R et al. An optimised saliva collection method to produce high-yield, high-quality RNA for translational research. PLoS One 15, e0229791, doi: 10.1371/journal.pone.0229791 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie Y et al. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer 67, 170–176, doi: 10.1016/j.lungcan.2009.04.004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segal LN et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat Microbiol 1, 16031, doi: 10.1038/nmicrobiol.2016.31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sidhaye VK, Nishida K & Martinez FJ Precision medicine in COPD: where are we and where do we need to go? European respiratory review : an official journal of the European Respiratory Society 27, doi: 10.1183/16000617.0022-2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langelier C et al. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc Natl Acad Sci U S A 115, E12353–E12362, doi: 10.1073/pnas.1809700115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paudel KR et al. Role of Lung Microbiome in Innate Immune Response Associated With Chronic Lung Diseases. Front Med (Lausanne) 7, 554, doi: 10.3389/fmed.2020.00554 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goleva E et al. The effects of airway microbiome on corticosteroid responsiveness in asthma. American journal of respiratory and critical care medicine 188, 1193–1201, doi: 10.1164/rccm.201304-0775OC (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang YJ et al. The airway microbiome in patients with severe asthma: Associations with disease features and severity. The Journal of allergy and clinical immunology 136, 874–884, doi: 10.1016/j.jaci.2015.05.044 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mirković B et al. The Role of Short-Chain Fatty Acids, Produced by Anaerobic Bacteria, in the Cystic Fibrosis Airway. American journal of respiratory and critical care medicine 192, 1314–1324, doi: 10.1164/rccm.201505-0943OC (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcha DS et al. Changes in prevalence and load of airway bacteria using quantitative PCR in stable and exacerbated COPD. Thorax 67, 1075–1080, doi: 10.1136/thoraxjnl-2012-201924 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Sze MA et al. The lung tissue microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 185, 1073–1080, doi: 10.1164/rccm.201111-2075OC (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sethi S, Maloney J, Grove L, Wrona C & Berenson CS Airway inflammation and bronchial bacterial colonization in chronic obstructive pulmonary disease. American journal of respiratory and critical care medicine 173, 991–998, doi: 10.1164/rccm.200509-1525OC (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren L et al. Transcriptionally Active Lung Microbiome and Its Association with Bacterial Biomass and Host Inflammatory Status. mSystems 3, doi: 10.1128/mSystems.00199-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sze MA et al. Host Response to the Lung Microbiome in Chronic Obstructive Pulmonary Disease. American journal of respiratory and critical care medicine 192, 438–445, doi: 10.1164/rccm.201502-0223OC (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madapoosi SS et al. Lung Microbiota and Metabolites Collectively Associate with Clinical Outcomes in Milder Stage COPD. Am J Respir Crit Care Med, doi: 10.1164/rccm.202110-2241OC (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molyneaux PL & Maher TM Respiratory microbiome in IPF: cause, effect, or biomarker? The Lancet. Respiratory medicine 2, 511–513, doi: 10.1016/s2213-2600(14)70088-8 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Huang Y et al. Microbes Are Associated with Host Innate Immune Response in Idiopathic Pulmonary Fibrosis. American journal of respiratory and critical care medicine 196, 208–219, doi: 10.1164/rccm.201607-1525OC (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J et al. Lung microbiome and host immune tone in subjects with idiopathic pulmonary fibrosis treated with inhaled interferon-γ. ERJ Open Res 3, doi: 10.1183/23120541.00008-2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang D et al. Dysregulated Lung Commensal Bacteria Drive Interleukin-17B Production to Promote Pulmonary Fibrosis through Their Outer Membrane Vesicles. Immunity 50, 692–706.e697, doi: 10.1016/j.immuni.2019.02.001 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Seibold MA et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. The New England journal of medicine 364, 1503–1512, doi: 10.1056/NEJMoa1013660 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Helling BA et al. Regulation of MUC5B Expression in Idiopathic Pulmonary Fibrosis. American journal of respiratory cell and molecular biology 57, 91–99, doi: 10.1165/rcmb.2017-0046OC (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Dwyer DN et al. Lung Microbiota Contribute to Pulmonary Inflammation and Disease Progression in Pulmonary Fibrosis. American journal of respiratory and critical care medicine 199, 1127–1138, doi: 10.1164/rccm.201809-1650OC (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mouraux S et al. Airway microbiota signals anabolic and catabolic remodeling in the transplanted lung. The Journal of allergy and clinical immunology 141, 718–729.e717, doi: 10.1016/j.jaci.2017.06.022 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das S et al. A prevalent and culturable microbiota links ecological balance to clinical stability of the human lung after transplantation. Nature communications 12, 2126, doi: 10.1038/s41467-021-22344-4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Segal LN et al. Randomised, double-blind, placebo-controlled trial with azithromycin selects for anti-inflammatory microbial metabolites in the emphysematous lung. Thorax 72, 13–22, doi: 10.1136/thoraxjnl-2016-208599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dickson RP & Morris A Macrolides, inflammation and the lung microbiome: untangling the web of causality. Thorax 72, 10–12, doi: 10.1136/thoraxjnl-2016-209180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang YJ et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. The Journal of allergy and clinical immunology 127, 372–381.e371–373, doi: 10.1016/j.jaci.2010.10.048 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cox MJ et al. Longitudinal assessment of sputum microbiome by sequencing of the 16S rRNA gene in non-cystic fibrosis bronchiectasis patients. PloS one 12, e0170622, doi: 10.1371/journal.pone.0170622 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mac Aogáin M et al. Integrative microbiomics in bronchiectasis exacerbations. Nature medicine 27, 688–699, doi: 10.1038/s41591-021-01289-7 (2021). [DOI] [PubMed] [Google Scholar]
- 60.Narayana JK, Mac Aogáin M, Ali N, Tsaneva-Atanasova K & Chotirmall SH Similarity network fusion for the integration of multi-omics and microbiomes in respiratory disease. Eur Respir J 58, doi: 10.1183/13993003.01016-2021 (2021). [DOI] [PubMed] [Google Scholar]
- 61.Tsay JJ et al. Airway Microbiota Is Associated with Upregulation of the PI3K Pathway in Lung Cancer. American journal of respiratory and critical care medicine 198, 1188–1198, doi: 10.1164/rccm.201710-2118OC (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsay JJ et al. Lower airway dysbiosis affects lung cancer progression. Cancer Discov, doi: 10.1158/2159-8290.CD-20-0263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu BG & Segal LN The Lung Microbiome and Its Role in Pneumonia. Clinics in chest medicine 39, 677–689, doi: 10.1016/j.ccm.2018.07.003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rohart F, Gautier B, Singh A & Le Cao KA mixOmics: An R package for 'omics feature selection and multiple data integration. PLoS computational biology 13, e1005752, doi: 10.1371/journal.pcbi.1005752 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hurgobin B, de Jong E & Bosco A Insights into respiratory disease through bioinformatics. Respirology 23, 1117–1126, doi: 10.1111/resp.13401 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Bu K et al. Identifying correlations driven by influential observations in large datasets. Brief Bioinform, doi: 10.1093/bib/bbab482 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]