Summary
Axon regeneration holds great promise for neural repair of CNS axonopathies, including glaucoma. Pten deletion in retinal ganglion cell (RGC) promotes potent optic nerve regeneration, but only a small population of Pten-null RGCs are actually regenerating RGCs (regRGCs); most surviving RGCs (surRGCs) remain non-regenerative. Here we developed a strategy to specifically label and purify regRGCs and surRGCs respectively from the same Pten deletion mice after optic nerve crush, in which they differ only in their regeneration capability. Smart-Seq2 single cell transcriptome analysis revealed novel regeneration-associated genes that significantly promote axon regeneration. The most potent of these, Anxa2, acts synergistically with its ligand tPA in Pten deletion-induced axon regeneration. Anxa2, its downstream effector ILK, and Mpp1 dramatically protect RGC somata and axons and preserve visual function in a clinically relevant model of glaucoma, demonstrating the exciting potential of this innovative strategy to identify novel effective neural repair candidates.
eTOC blurb:
Combined retrograde regenerating axon tracing and Smart-Seq2 single cell sequencing approaches revealed multiple pro-regeneration genes downstream of Pten deletion. Two of these genes, Anxa2 and Mpp1, also presented striking neuroprotection and visual function preservation in mouse glaucoma model. Additionally, we established the role of tPA-Anxa2-ILK axis in neural repair.
Introduction
Axonopathy is a common early feature of central nervous system (CNS) neurodegenerative diseases, including glaucoma, which is characterized by optic nerve degeneration followed by progressive retinal ganglion cell (RGC) death (Calkins, 2021; Howell et al., 2007; Nickells et al., 2012; Quigley et al., 1995), and is the leading cause of irreversible blindness (Steinmetz et al., 2021; Tham et al., 2014). Glaucomatous neurodegeneration may be initiated by mechanical damage of the optic nerve head due to elevated intraocular pressure (IOP) (Stowell et al., 2017). The axons of adult RGCs do not regenerate spontaneously after degeneration. Therefore, new neural repair therapies are needed, especially because the only currently available treatments act by reducing IOP and fail to completely prevent the progression of glaucomatous neurodegeneration (Garway-Heath et al., 2015; Heijl et al., 2002; Wormald et al., 2020).
Deletion of Pten (phosphatase and tensin homolog) in RGCs remains by far the most potent single gene manipulation strategy to promote optic nerve regeneration after ON crush (ONC) injury (Park et al., 2008; Williams et al., 2020; Zhang et al., 2018). Similar axon regeneration phenotypes after Pten deletion have been reported for mouse cortical motor neurons (Jin et al., 2015; Liu et al., 2010), drosophila sensory neurons (Song et al., 2012), and C. elegans motor neurons (Byrne et al., 2014). However, deregulated Pten activities have been implicated in diverse disorders, including metabolic diseases, tumor formation, cognitive impairment and even senescence (Zoncu et al., 2011). How to manipulate Pten-associated signaling molecules to maximize their axon regeneration activities while minimizing their deleterious side effects is extremely important for developing a clinically useful neural repair strategy. The prerequisite is to understand the specific downstream effectors of Pten in axon regeneration. Our previous studies elucidating the downstream signaling pathways of Pten in axon regeneration demonstrated that AKT coordinates positive signals from PI3K-PDK1 and negative signals from mTORC2 in regulating mTORC1 activation and GSK3β phosphorylation for optic nerve regeneration (Guo et al., 2016; Miao et al., 2016; Yang et al., 2014). However, Pten deletion may also function through an AKT-independent pathway (Huang et al., 2019). The complicated crosstalk and feedback loops among Pten/PI3K/AKT/mTORC1/2 (Zhang et al., 2018) make it very difficult to pinpoint the key regeneration mediators that converge from these pathways. Another challenge is that the majority of Pten knockout (KO) RGCs cannot regenerate their axons even if they survive the injury, bulk RNA-seq of Pten KO RGCs without differentiation of regenerating RGCs from surviving but not regenerating RGCs is not very fruitful. Therefore, we developed a strategy to precisely label and purify regenerating and non-regenerating RGCs separately. Because both sets of RGCs undergo the same genetic manipulation and injury and differ only in axon regeneration capability, comparison of gene expression in these two populations is the most informative way to find genes that are truly associated with axon regeneration. Single cell RNA sequencing (scRNA-seq) is a fast-growing powerful tool for characterizing cell types and states based on signature gene expression patterns (Tanay and Regev, 2017) and has been successfully applied to classify RGC subtypes and identify injury-associated genes (Clark et al., 2019; Hoang et al., 2020; Tran et al., 2019; Yan et al., 2020). We chose a low-throughput strategy, sorting cells with plate-based Smart-Seq2 (Picelli et al., 2013; Picelli et al., 2014) over droplet-based scRNA-seq (Clark et al., 2019; Macosko et al., 2015; Peng et al., 2019; Shekhar et al., 2016) because 1) Droplet-based scRNA-seq, such as 10x Genomics, requires many more cells than we can obtain; 2) Deeper sequencing capability with higher sensitivity and accuracy with Smart-seq2 enables us to detect more genes, especially subtle differences among regenerating and non-regenerating RGCs.
We previously explored RGC and optic nerve neuroprotection in chronic optic neuropathy models (Hu et al., 2012; Huang et al., 2017; Liu et al., 2021; Yang et al., 2016) but only used traumatic ONC model to study axon regeneration (Huang et al., 2019; Miao et al., 2016; Yang et al., 2014). Glaucoma differs from ONC because axon damage is incomplete, and therefore it is difficult to differentiate regenerating axons from surviving axons. However, the pro-regeneration genes identified from regenerating RGCs in the ONC model may be promising candidates for glaucoma neural repair, as showed by a recent study (Lu et al., 2020). Therefore, we first developed a tracing scheme to differentially label and isolate surviving RGCs with or without axon regeneration in Pten KO mice after ONC injury. We then compared the single cell transcriptomes of these RGCs by Smart-Seq2 to systematically cluster genes associated with axon regeneration and identified six novel pro-regeneration genes with various regeneration capacity. Finally, we demonstrated that overexpression of Anxa2 (Annexin A2) in RGCs achieved dramatic neuroprotection and visual function preservation in an experimental model of glaucoma. This proof-of-concept study demonstrates the feasibility and efficacy of this strategy in identifying the targetable downstream effectors of Pten. This strategy can also readily elucidate the downstream effectors of other single or combinatory regeneration approaches, and thus identify effective and safe neural repair targets for glaucoma and other CNS neurodegenerative diseases.
Results
Strategies for labeling, isolating, and single cell transcriptome profiling of regenerating RGCs (regRGCs) and surviving RGCs without axon regeneration (surRGCs)
To specifically label RGCs that send regenerating axons into optic nerve distal to the crush site, we developed an intraorbital optic nerve injection method to label RGC cell bodies by retrograde tracing of dye applied to regenerating axons. This method avoids the need to remove brain tissue, in contrast to a previously described approach (Zhang et al., 2019c). We exposed the intraorbital portion of optic nerve about 2 mm distal to the eyeball by piercing the conjunctiva and tunneling beneath soft tissues and muscles from the lateral side of the mouse eye while avoiding injury to the retro-orbital sinus. Because the ONC site is usually ~ 0.5 mm from the eyeball, we labeled regenerating axons by injecting neuronal tracer 1–1.5 mm distal to the crush site (Fig. 1A). We used dextran after testing several other tracers because of the superiority of its transport speed and retrograde labeling efficiency of RGCs after optic nerve injection (Fig. S1A). In the naïve animal, intraorbital optic nerve injection of dextran labeled about 90% RBPMS+ RGCs (Fig. S1B) but not neighboring AP2α+ amacrine cells (Fig. 1B). The entire retina was labeled uniformly, evidenced by homogenous RGC labeling detected by in vivo SLO retinal fundus imaging and wholemount retina histology (Fig. 1C,F,I). We further confirmed that this method did not label any RGCs in wildtype (WT) mice 14 days post ONC (14dpc) in which there are no regenerating axons (Fig. 1D,G,J). In dramatic contrast, intraorbital optic nerve injection of dextran labeled a distinct population of RGCs in Pten KO mice in which regenerating axons reach at least 1 mm beyond the crush site at 14dpc (Fig. 1E,H,K). These observations substantiate the reliability and efficiency with which this retrograde tracing method labels regenerating RGCs in vivo. We performed the injection at about 1 mm distal to the crush site (~ 1.5 mm from the eyeball) because we found that much fewer regRGCs were labeled by an injection 0.5 mm more distal (~ 2.0 mm from the eyeball), probably because far fewer regenerating axons elongated that distance at 14dpc in Pten KO mice (Fig. S1C). To pre-label the total population of RGCs before ONC to detect surviving RGCs (surRGCs) after crush, we used retrobeads as a neuronal tracer after superior colliculus (SC) injection because of the superior stability of the signal, which persists in RGCs for at least 4 weeks (Fig. S1D), and because they are relatively efficient and labeled about 70% RBPMS+ RGCs (Fig. S1E).
Figure 1. Retrograde tracing of regenerating RGCs by intraorbital optic nerve dye injection.
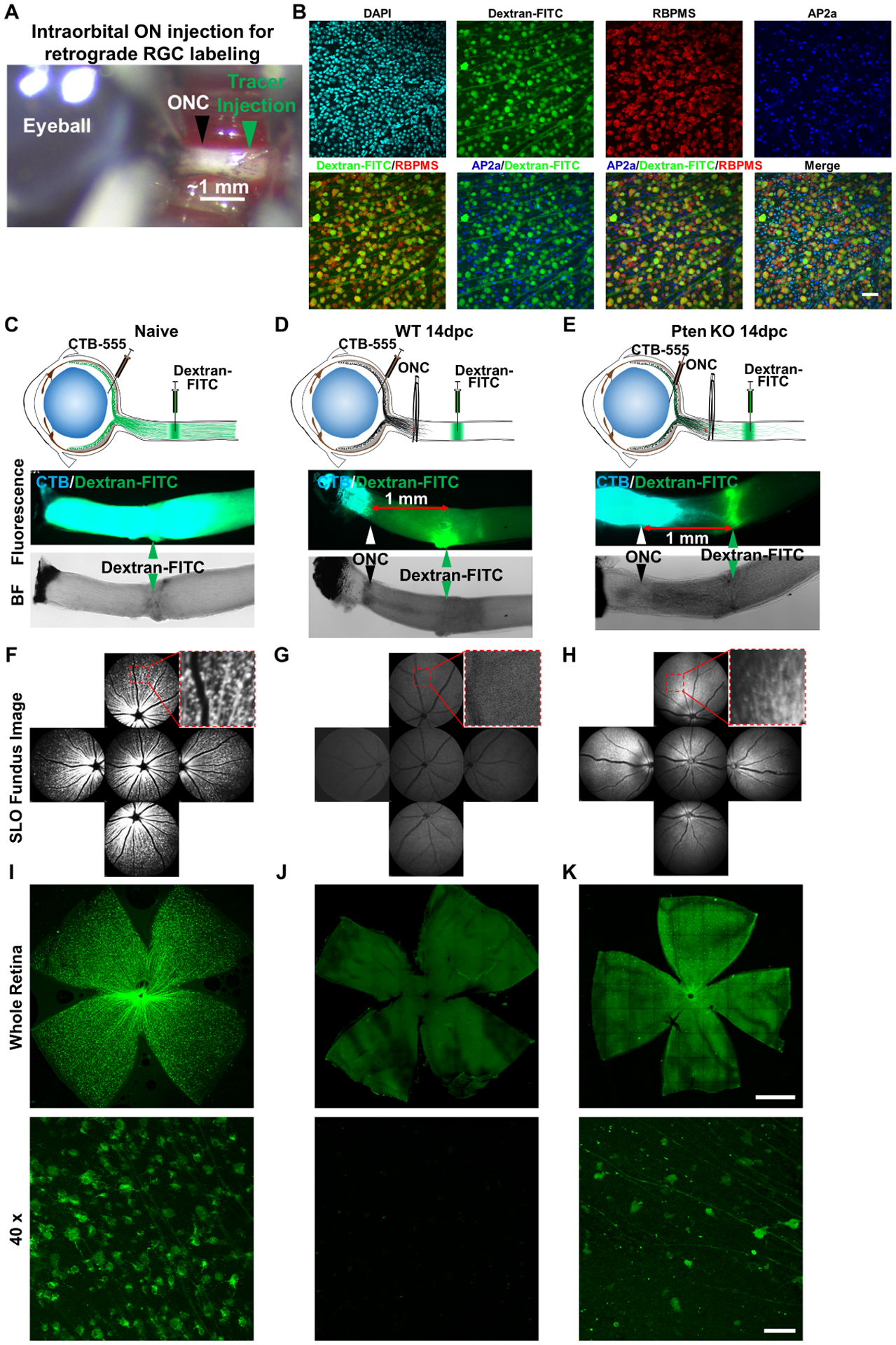
(A) The intraorbital portion of optic nerve (~ 2mm) was exposed by pushing through the ocular muscles and soft tissues under the conjunctiva, without injuring the retro-orbital sinus. The ONC site is about 0.5 mm from the eyeball, leaving 1–1.5mm distal portion for dye injection. (B) Confocal images of retinal wholemounts of naïve mice one day after dextran-FITC intraorbital optic nerve injection, showing co-localization of the dextran and pan-RGC marker (RBPMS)-labeled RGCs, which are distinct from AP2α+ amacrine cells. Scale bar, 50μm. (C, D, E) Cartoon illustration of intravitreal injection of anterograde tracer dye CTB-555 and intraorbital optic nerve injection of retrograde tracer dye dextran-FITC in naïve, ONC injured WT and Pten KO mice at 14dpc; and corresponding fluorescent and bright field (BF) images of optic nerves showing the labeled axons and ONC sites. (F, G, H) SLO retinal fundus images of live animals showing the dextran-FITC labeled RGCs in naïve, ONC injured WT and Pten KO mice at 14dpc. (I, J, K) Retinal wholemounts of naïve, ONC injured WT, and Pten KO mice at 14dpc showing dextran-FITC labeled RGCs. Scale bar, upper panel: 500μm; lower panel: 50μm.
See also Figure S1
We used these retrograde tracing methods to finalize the time line of the labeling and the isolation procedures (Fig. 2A): 1) We induced Pten deletion in RGCs in Pten floxed mice by intravitreal injection of AAV-Cre driven by a mouse γ-synuclein promoter (mSncg), a RGC-specific promoter that we recently identified (Wang et al., 2020a), 14 days before ONC; 2) We injected retrobeads-488 into the SC 2 days before ONC to label all RGCs with green fluorescence; 3) We performed intraorbital optic nerve injection with dextran-Red to label regRGCs with red fluorescence 13 days post ONC (1 day before tissue collection at 14dpc). This strategy detected around 900–1000 regenerating RGCs (regRGCs)/retina and roughly 10,000 surviving RGCs (surRGCs)/retina in Pten KO mice at 14dpc (Fig. 2B,C), similar to published regenerating axon counting (Park et al., 2008; Yang et al., 2014). 4) We used FACS sorting to isolate separately dextran-Red positive regRGCs and retrobeads-Green positive but red negative surRGCs (Fig. 2D). These cells were individually distributed into each well of a 96-well plate and later transferred to a 384-well plate to generate a single cell Smart-Seq2 cDNA library according to an established protocol (Picelli, 2019; Picelli et al., 2014). 384 cells with distinct sequence barcodes were sequenced together in one HiSeq lane to acquire roughly 1 million sequencing reads/cell and repeated once with an independent group of mice (Fig. 2E). Based on the quality control thresholds of more than 900 genes/cell, 0.5 million reads/cell, and fewer than 15% mitochondria genes (Fig. 2F), we acquired 340 regRGCs and 290 surRGCs that had at least one of the 7 pan-RGC markers (RBPMS, Thy1, Slc17a6/vGLUT2, Pou4f1-3/Brn3a-c) and very low or no Pten expression (Fig. 2G).
Figure 2. Regenerating RGCs (regRGCs) and surviving but not regenerating RGCs (surRGCs): labeling, isolation, and Smart-Seq2 scRNA-seq.
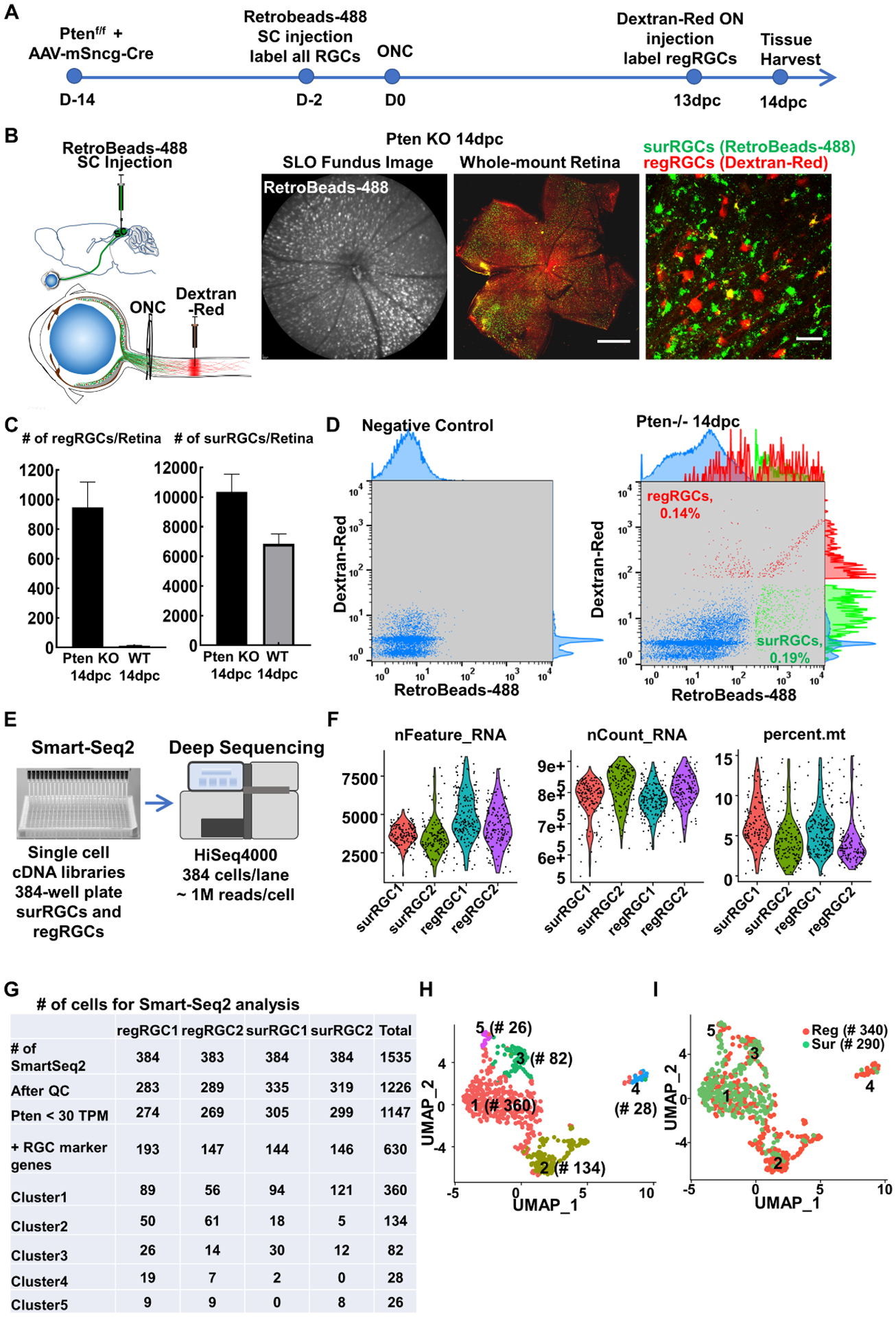
(A) Timeline of experimental design for Pten deletion in RGCs 2 weeks before ONC, SC retrograde labeling of naïve RGCs, optic nerve retrograde tracing regRGCs, and tissue collection at 14dpc. (B) Cartoon illustration of SC injection of retrobeads-488 and intraorbital optic nerve injection of retrograde tracer dye dextran-Red; and SLO retinal fundus image of live animal showing RGCs labeled with retrobeads and retinal wholemounts showing surRGCs (green only) and regRGCs (red or yellow), in Pten KO mice at 14dpc. Scale bar, 500μm in the whole retina, 50μm in the zoom-in. (C) Quantification of the numbers of regRGCs/retina and surRGCs/retina labeled by our tracing strategy in WT and Pten KO mice at 14dpc. n = 6. Data are presented as means ± s.e.m. (D) FACS gating strategy for surRGCs (green only, no red) and regRGCs (red alone or red/green double) purification. (E) Smart-Seq2 preparation for scRNA-seq. (F) Quality control of scRNA-seq data showing number of genes detected/cell (nFeature), total reads/cell (nCount), and percentage of mitochondria genes (percent). (G) Numbers of cells collected for Smart-Seq2 analysis. (H) UMAP (Uniform Manifold Approximation and Projection) visualization of the transcriptional heterogeneity of 630 RGCs (340 regRGCs and 290 surRGCs) isolated from Pten KO mice at 14dpc. Cells are colored into 5 clusters. (I) The 5 RGC clusters are superimposed with regeneration identify: surRGCs are in green and regRGCs are in red.
See also Figures S2, S3
We pooled the Smart-Seq2 transcriptomes of a total of 630 regRGCs and surRGCs and clustered them into 5 molecular clusters (Fig. 2H) based on the distinct gene sets associated with each cluster (Fig. S2A, Table S1). Superimposition of regeneration identities demonstrated that cluster 2 and cluster 4 were enriched with regRGCs (Fig. 2I), suggesting that regRGCs and surRGCs tend to segregate into distinct molecular clusters. Further gene ontology (GO) enrichment analysis showed distinct biological pathways associated with each cluster (Fig. S2B): 1) regRGC-enriched cluster 2 and cluster 4 were associated with lipid transport, cell migration, cell adhesion, and wound healing, or cell mitosis and cell cycle, respectively; 2) Cluster 1 were associated with immune/inflammation responses; cluster 3 genes were associated with visual function and development; and cluster 5 were associated with neuron death, and synapse structure and function. Using the extensive mouse RGC atlas generated by droplet-based scRNA-seq (Tran et al., 2019) as a reference, we confirmed that the majority of Pten KO surRGCs and regRGCs were Spp1+ α-RGCs (Fig. S3A), consistent with a previous report that α-RGC is the general RGC subtype for survival and regeneration after Pten KO in the ONC model (Duan et al., 2015). Other RGC subtypes’ marker genes were also detected in our samples, including Cartpt-RGCs (Cart), F-RGCs (Foxp2), ipRGCs (Opn4/Eomes), N-RGCs (Neurod2/Penk/Satb2), and T-RGCs (Tbr1); some of these (Spp1, Cart, Foxp2, and Opn4) were confirmed by immunostaining (Fig. S3B,C).
Regeneration-associated genes revealed by comparison of Smart-Seq2 transcriptomes of regRGCs and surRGCs
To identify genes that are associated with axon regeneration, we combined all the regRGCs as one group and all the surRGCs as another group, and directly compared the Smart-Seq2 transcriptomes of regRGCs and surRGCs because the only difference between these two populations is whether or not their axons regenerate. This pseudo-bulk comparison revealed 168 upregulated genes (Table S2) and 116 downregulated genes in regRGCs as differentially expressed genes (DEGs) (Table S3). Some of the upregulated DEGs are known to be involved in axon regeneration and cell survival, such as Spp1, Lgals3, Mmp12, and Stmn1 (Abreu et al., 2017; Duan et al., 2015; Grenningloh et al., 2004; Ohkawa et al., 2007; Tran et al., 2019). GO analysis of upregulated DEGs in regRGCs showed enrichment of biological pathways related to regulation of lipid localization, cell adhesion, migration, and metabolic processes (Fig. 3A,C). Based on this association, we marked some of the genes (most significant and enriched in these GO pathways) in the volcano plot (Fig. 3E), and showed their expression in regRGCs and surRGCs, and cluster enrichment, including Anxa2, Iqgap1, Gpnmb, Lgals1, Lgals3, Plin2, Spp1, Mpp1, Acaa2, and Ahnak2 (Fig. 3F, Fig. S4A,B). Based on the online scRNA-seq database of adult mouse RGCs (Tran et al., 2019), these top regeneration-associated upregulated genes are also expressed in many RGC subtypes (Fig. S4C). We further confirmed that Anxa2, Spp1 and Mpp1 were highly expressed in regRGCs by immunostaining (Fig. S5A,B). The downregulated genes were associated with immune responses (Fig. 3B,D–F), such as H2-K1, H2-D1, B2m, and C1q family; we did not pursue them further in this study.
Figure 3. Comparison of Smart-Seq2 transcriptomes of regRGCs and surRGCs reveals biological pathways and DEGs associated with RGC regeneration and survival.
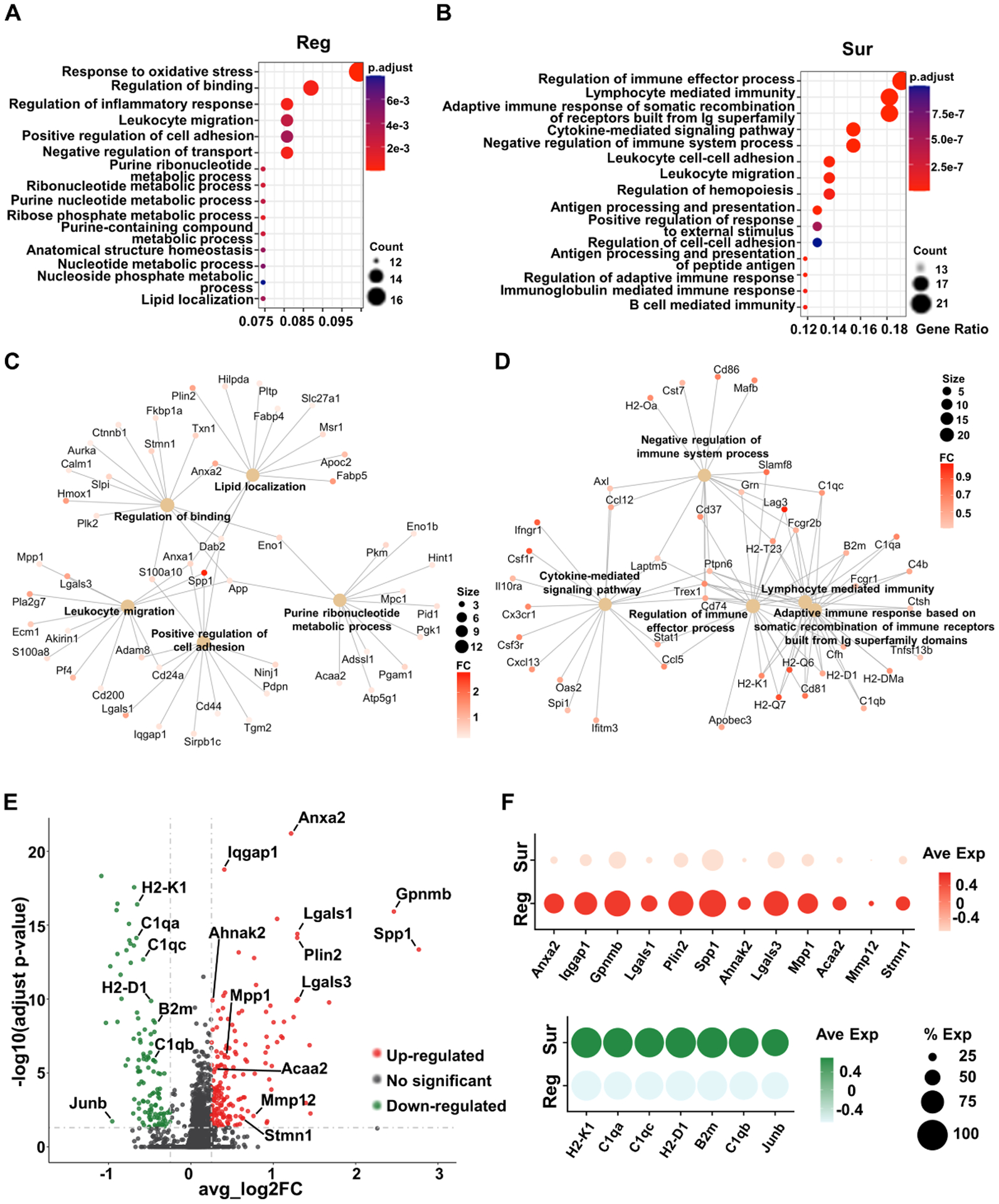
(A, B) Top 15 enriched GO-pathways associated with regRGCs (A) or surRGCs (B). The size of each circle represents the numbers of genes enriched in each pathway, and the color represents the adjusted p value. (C, D) Cnetplot showing the interaction of 5 enriched pathways and their associated genes in regRGCs (C) and surRGCs (D). The size of each circle associated with each pathway represents the numbers of genes enriched in each pathway, and the color of each gene represents the fold change (FC). (E) VolcanoPlot of DEGs of regRGCs. Adjusted p value < 0.05, avg_log2FC > 0.25. The red genes are upregulated, and the green genes are downregulated in regRGCs. The grey genes are not significantly changed. (F) Dotplot showing expression of indicated regeneration-associated genes in regRGCs and surRGCs. The size of each circle represents the percentage of RGCs expressing the gene, and the color represents the expression level.
See also Figures S4, S5
Six of seven tested DEGs promote significant axon regeneration after ONC injury and Anxa2 is the most potent pro-regeneration gene
We successfully generated AAVs to express seven regRGCs-upregulated DEGs driven by the mSncg promoter in RGCs specifically and tested them in axon regeneration after ONC injury (Fig. S5C,D). Because the coding sequences of Iqgap1 and Ahnak2 are too large to fit into AAV vectors, we did not test them. Six out of seven DEGs, Anxa2, Plin2, Mpp1, Acaa2, Spp1, and Lgals1 but not Gpnmb, promoted significant axon regeneration, but to a differing extent (Fig. 4A,B). Anxa2 was the most potent pro-regeneration gene and the only one to increase RGC survival (Fig. S5E,F).
Figure 4. Anxa2 is the most potent of the 6 regeneration-associated genes that promote significant axon regeneration after ONC injury.
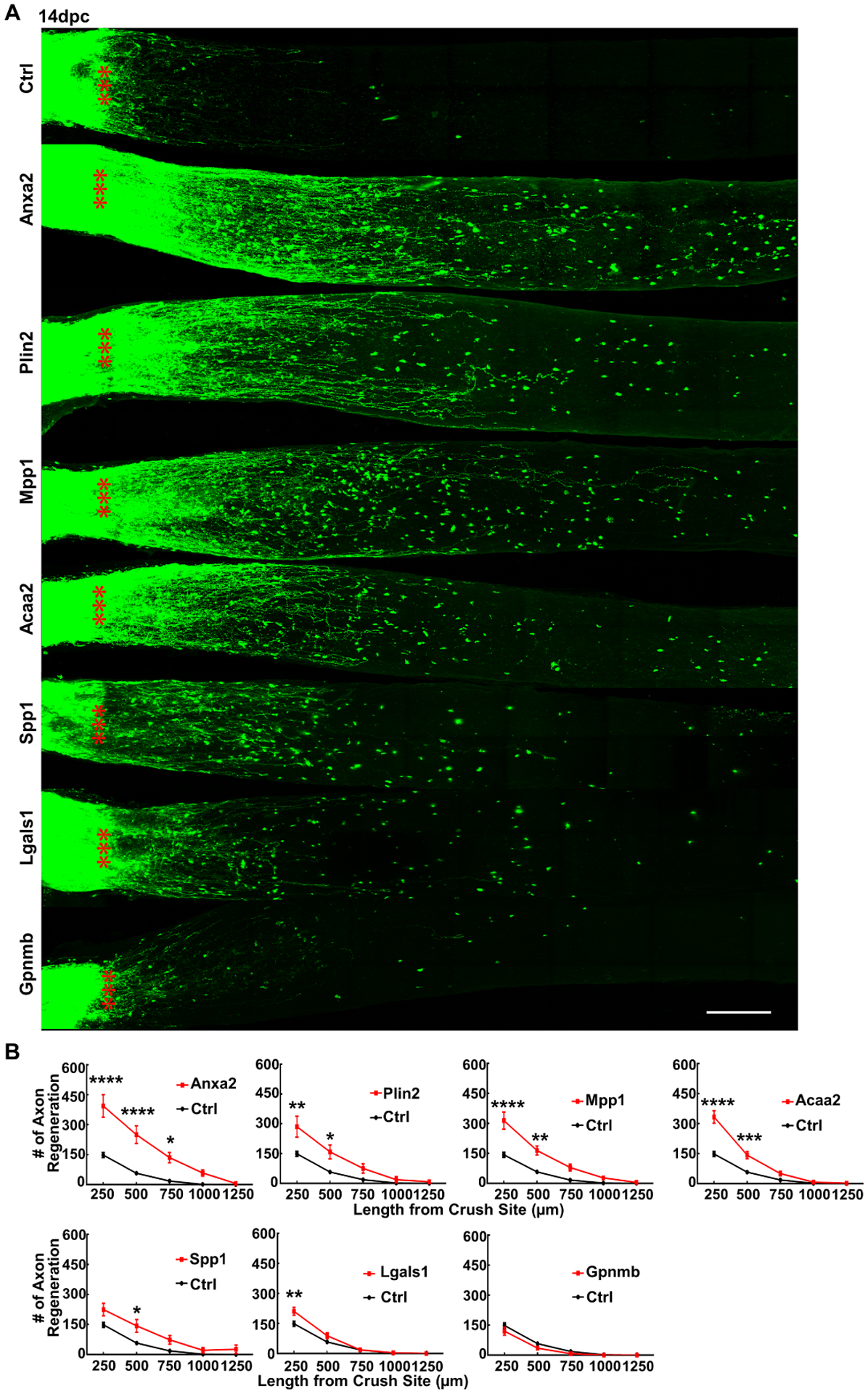
(A) Confocal images of optic nerve wholemounts after optical clearance showing maximum intensity projection of regenerating fibers labeled with CTB-Alexa 555 at 14dpc. Scale bar, 100 μm. *: crush site. (B) Quantification of regenerating fibers at different distances distal to the lesion site. Data are presented as means ± s.e.m, n = 8 in all groups except the Acaa2 group (n = 10). *: p<0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001, two-way ANOVA with Sidak’s multiple comparisons test.
See also Figure S5
Anxa2 and Mpp1 overexpression significantly promote survival of RGC soma and axon, and preserve visual functions in the SOHU glaucoma model.
Next, we asked whether the pro-regeneration genes that we identified through the traumatic ON injury model would enhance neural repair effect of RGC/optic nerve and visual function in the more common, chronic optic neuropathy, glaucoma. We took advantage of our recently developed clinically relevant silicone oil-induced ocular hypertension (SOHU) mouse glaucoma models (Fang et al., 2021; Zhang et al., 2019a; Zhang et al., 2019b) to test whether overexpression of Anxa2, Mpp1, or another two weaker pro-regeneration genes Plin2 and Lgals1, would benefit glaucomatous neurodegeneration. We injected AAV-mSncg-transgene into the vitreous to infect RGCs 14 days before inducing SOHU glaucoma to allow the transgene expression. We then performed intracameral injection of SO into one eye to mimic the secondary glaucoma found in patients when SO induces elevated IOP by pupillary blocking and injected PBS into the contralateral eye as an internal control (Fig. S6A,B). We and others use OCT to measure the thickness of the retinal ganglion cell complex (GCC) in living mice, including retinal nerve fiber layer (RNFL), ganglion cell layer (GCL) and inner plexiform layer (IPL), to serve as an indicator of RGC/ON degeneration in diverse optic neuropathy mouse models (Fang et al., 2021; Guo et al., 2010; Li et al., 2020; Liu et al., 2021; Nakano et al., 2011; Zhang et al., 2019b). In vivo OCT imaging showed significant thinning of the GCC in SOHU eyes compared to contralateral control eyes in control group animals injected with control AAVs, at 3-week post SO injection (3wpi) (Fig. 5A). GCC thinning was concurrent with significant IOP elevation (Fig. S6C). Thus we were able to replicate the SOHU glaucoma model, with stable IOP elevation and severe glaucomatous neurodegeneration (Fang et al., 2021; Zhang et al., 2019b), which faithfully simulates human pupillary blocking glaucoma caused by surgical use of SO (Ichhpujani et al., 2009; Kornmann and Gedde, 2016). Only Anxa2 and Mpp1, but not Plin2 or Lgals1, significantly increased GCC thickness in SOHU eyes (Fig. 5A). Histological analysis of post mortem retina wholemounts consistently showed that treatment with Anxa2 and Mpp1 strikingly increased RGC survival throughout the whole retina (Fig. 5B,C). In contrast, neither Plin2 nor Lgals1 showed a significant RGC protection effect. Quantification of surviving axons in semi-thin sections of optic nerves further confirmed that overexpression of Anxa2 and Mpp1, but not of Plin2 or Lgals1, achieved dramatic axon protection in glaucoma (Fig. 5D,E).
Figure 5. RGC-specific Anxa2 and Mpp1 overexpression significantly promote survival of both RGC somata and axons, and preserves visual functions in SOHU glaucoma model.
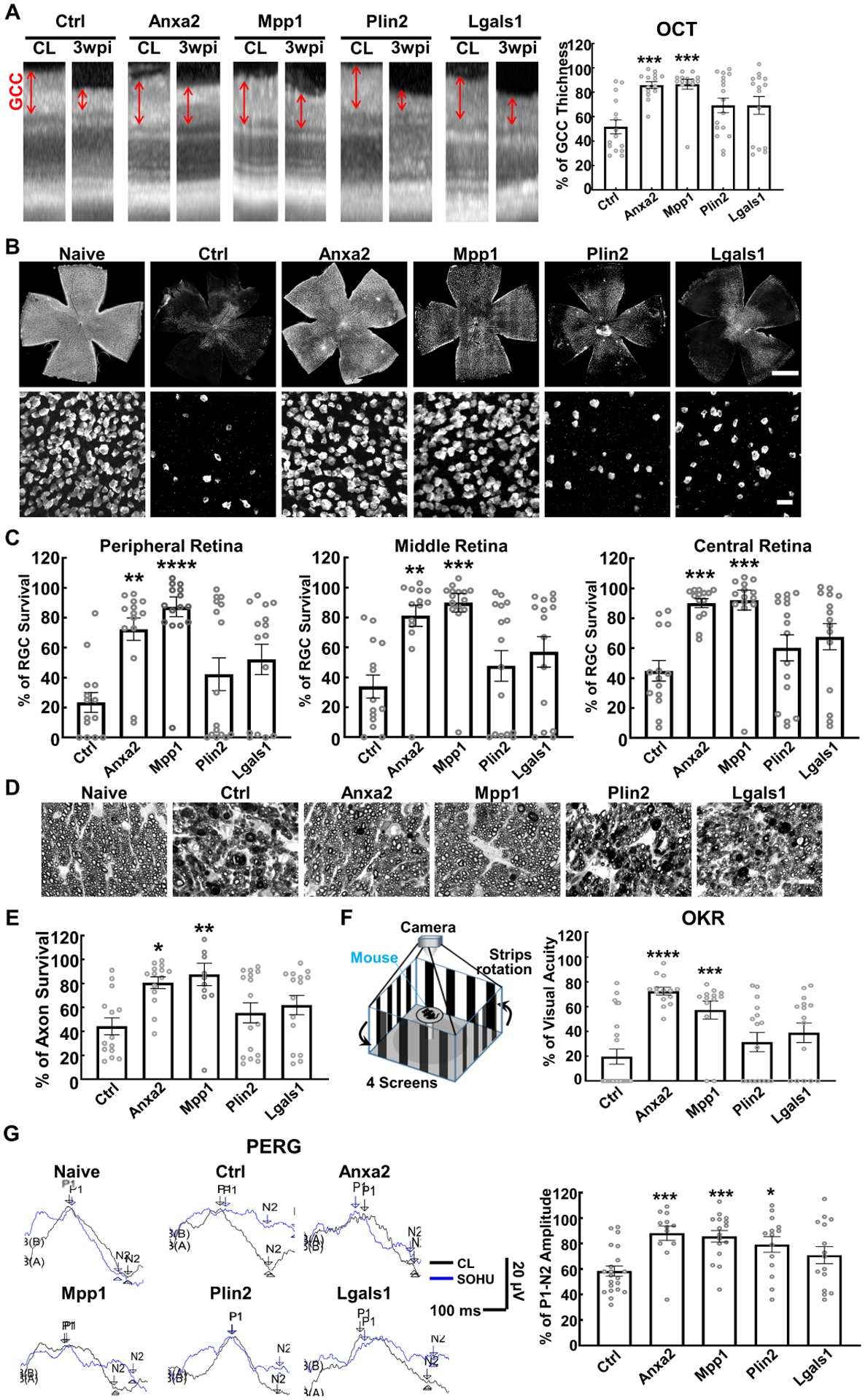
(A) Representative OCT images of mouse retinas from SOHU glaucomatous eyes and contralateral control (CL) eyes at 3 weeks post SO injection (3wpi). GCC: ganglion cell complex, including RNFL, GCL and IPL layers; indicated as double end arrows. Quantification of GCC thickness measured by OCT at 3wpi, represented as percentage of GCC thickness in the SOHU eyes compared to the sham CL eyes. (B) Upper panel, representative confocal images of the whole flat-mounted retinas showing surviving RBPMS-positive RGCs at 3wpi, Scale bar, 500 μm. Lower panel, representative confocal images of peripheral flat-mounted retinas showing surviving RBPMS-labeled RGCs at 3wpi, Scale bar, 50 μm. (C) Quantification of surviving RGC somata in peripheral, middle, and central retinas at 3wpi, represented as percentage of glaucomatous eyes compared to the sham CL eyes. (D) Light microscope images of semi-thin transverse sections of ON with PPD staining at 3wpi. Scale bar, 10 μm. (E) Quantification of surviving axons at 3wpi, represented as percentage of glaucomatous eyes compared to the sham CL eyes. (F) Visual acuity measured by OKR at 3wpi, represented as percentage of glaucomatous eyes compared to the sham CL eyes. (G) Left: representative wave forms of PERG at baseline and 3wpi, blue traces represent glaucomatous eyes, black traces represent contralateral control (CL) eyes. Right: quantification of P1-N2 amplitude of PERG at 3wpi, represented as a percentage of glaucomatous eyes compared to the sham CL eyes. All the quantification, data are presented as means ± s.e.m, n = 13–16 in each group, *: p<0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001, one-way ANOVA with Tukey’s multiple comparisons test.
See also Figure S6A–C
In addition to morphological protection, preservation of visual function is of utmost importance for neural repair. We next investigated whether RGC-specific expression of pro-regeneration genes preserved visual function in glaucomatous mice. Optokinetic tracking response (OKR) is a natural reflex that objectively assesses mouse visual acuity (Douglas et al., 2005; Douglas et al., 2006; Prusky et al., 2004). The mouse eye will only track a grating stimulus that is moving from the temporal to nasal visual field, which allows both eyes to be measured independently. Consistent with our post-mortem histological and in vivo morphological results, Anxa2 and Mpp1 significantly preserved visual acuity of the glaucomatous eyes (Fig. 5F). The pattern electroretinogram (PERG), a sensitive electrophysiological assay of general RGC function (Porciatti, 2015), is obtained in response to a visual stimulus consisting of contrast reversal patterned gratings at constant mean luminance. Because our PERG system can measure both eyes at the same time, there is an internal control to serve as a reference and normalization to minimize variations (Chou et al., 2014). We previously employed PERG successfully to examine the changes of RGC function in the SOHU glaucoma model (Fang et al., 2021; Zhang et al., 2019b). The peak-to-trough (P1-N2) amplitude ratio of the SOHU eyes to contralateral (CL) eyes increased significantly after Anxa2, Mpp1 and Plin2 overexpression in RGCs (Fig. 5G). Taken together, these results show convincingly that Anxa2 and Mpp1 overexpression in RGCs achieve significant neural repair in the SOHU glaucoma model, demonstrated by striking neuroprotection of RGCs and optic nerves, and significant preservation of visual functions.
Anxa2 and its cell surface ligand tissue plasminogen activator (tPA) potentiated the effects of Pten deletion on axon regeneration and neuronal survival
Anxa2 functions as a cell surface co-receptor for tPA and is therefore involved in plasmin-related cellular processes, such as fibrinolysis, extracellular matrix (ECM) degradation, cell migration and cancer cell invasion (Bharadwaj et al., 2013; Bharadwaj et al., 2021a; Bharadwaj et al., 2021b; Gerke et al., 2005). Surprisingly, tPA alone induced significant axon regeneration in the ONC model (Fig.6A,B), suggesting that tPA acts on Anxa2 to promote axon regeneration. Adding tPA further increased Pten KO-induced axon regeneration (Fig. 6A,B). Similarly, RGC-specific Anxa2 overexpression and Pten deletion promoted more axon regeneration than Pten KO alone at 14dpc (Fig. 6A,B), further suggesting that increasing tPA-Anxa2 signaling and inhibiting Pten would act synergistically to enhance axon regeneration. Consistently, triple treatment consisting of Anxa2 overexpression, Pten KO, and tPA administration induced the most potent axon regeneration (Fig. 6A,B), some regenerating axons grow through optic chiasm into optic tract, and some to cross to the contralateral side and grow into contralateral optic nerve at 8-week post crush (8wpc) (Fig. S7A,B). To test a clinically relevant application of tPA-Anxa2 in axon regeneration, we studied delayed treatment (Fig. S7C): Because AAV-mediated gene expression requires at least one week after intravitreal injection, we injected AAV-Anxa2 on the same day as ONC, with or without tPA injection twice a week thereafter. Delayed Anxa2 expression also induced significant axon regeneration, which addition of tPA further increased.
Figure 6. Anxa2 and tPA potentiated Pten KO effects on axon regeneration and RGC survival after ONC.
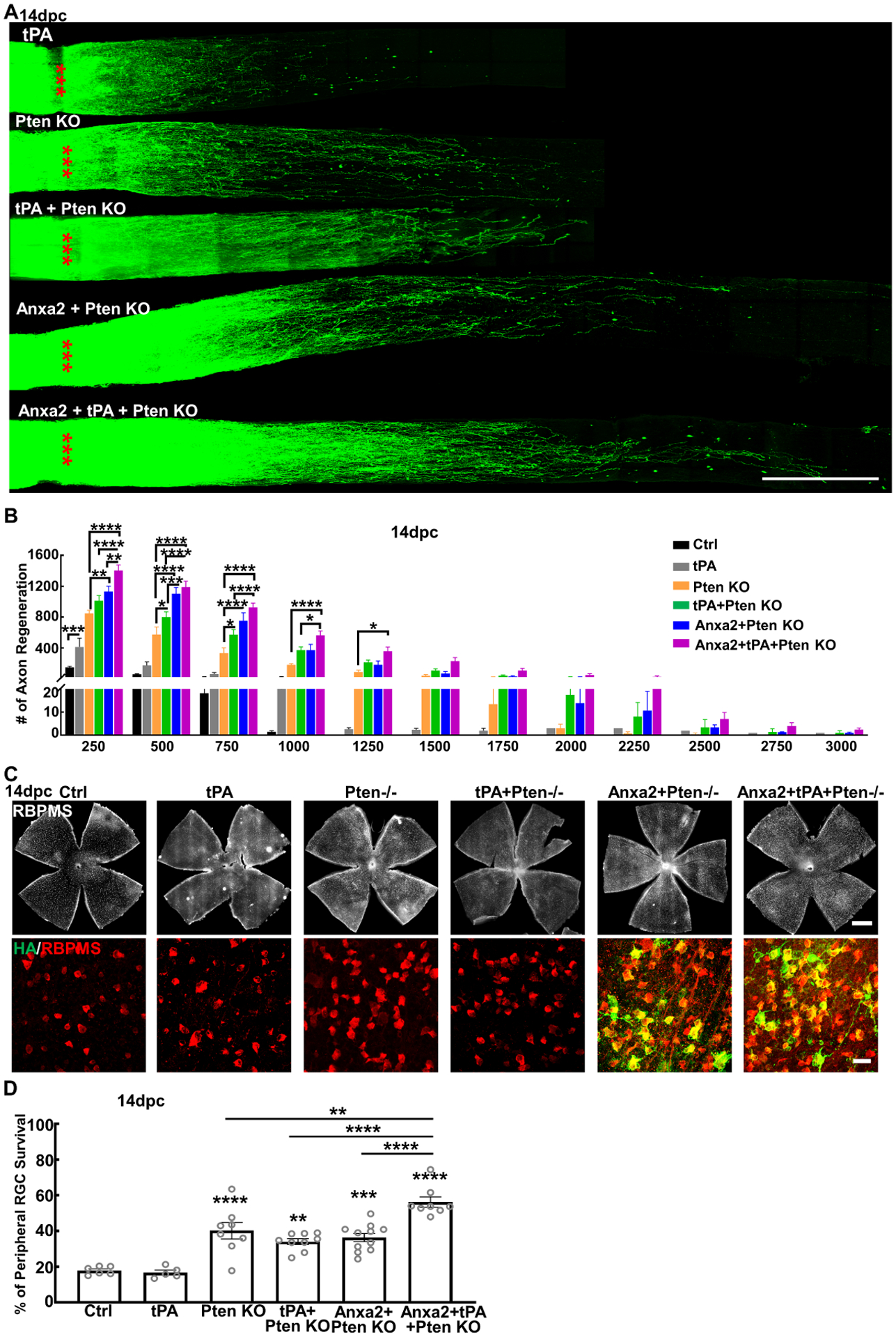
(A) Confocal images of optic nerve wholemounts after optical clearance showing maximum intensity projection of regenerating fibers labeled with CTB-Alexa 555 at 14dpc. Scale bar, 500 μm. *: crush site. (B) Quantification of regenerating fibers at different distances distal to the lesion site. Data are presented as means ± s.e.m, n = 8. *: p<0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001, two-way ANOVA with Tukey’s multiple comparisons test. (C) Confocal images of retinal wholemounts showing RBPMS+ RGCs and HA labeling of Anxa2 at 14dpc. Scale bar, upper panel: 500μm; lower panel: 50μm. (D) Quantification of surviving RGCs somata in peripheral retina at 14dpc, represented as percentage of crushed eyes compared to the sham CL eyes. Data are presented as means ± s.e.m, n = 6–11 in each group. **: p<0.01, ***: p<0.001, ****: p<0.0001, one-way ANOVA with Tukey’s multiple comparisons test.
See also Figure S7
tPA by itself did not increase RGC survival after ONC, but it increased Anxa2-induced RGC survival (Fig. S7C). Neither tPA nor Anxa2 further increased RGC survival in Pten KO mice (Fig. 6C,D). But surprisingly, combining tPA and Anxa2 significantly increased Pten KO-induced RGC survival (Fig. 6C,D), indicating the boosting effect of tPA plus Anxa2. Although current understanding is that neuron survival is not invariably linked proportionately with axon regeneration (Benowitz et al., 2015), we cannot exclude the possibility that tPA/Anxa2 increased Pten KO-induced axon regeneration through increasing RGC survival.
The Anxa2-ILK axis in axon regeneration and neuroprotection.
Anxa2 is known to interact with cell surface integrins to activate integrin linked kinase (ILK) (Lin et al., 2012; Zhang et al., 2020), which is a vital signaling protein that mediates integrin regulation of cell adhesion, proliferation, migration, angiogenesis, and actin-cytoskeleton dynamics (Almasabi et al., 2021). ILK activity is also inhibited by Pten (Morimoto et al., 2000; Persad et al., 2000) and is important for neuronal polarity (Guo et al., 2007). Thus, Anxa2, acting as the downstream effector of Pten inhibition, may promote axon regeneration by activating the integrin-ILK pathway. Consistently, overexpression of wildtype and constitutive active (CA) mutant of ILK (Guo et al., 2007; Pan et al., 2014) in RGCs, also produced significant axon regeneration (Fig. 7A,B). More importantly, blocking ILK with a kinase dead (KD) mutant of ILK significantly decreased Anxa2-induced axon regeneration (Fig. 7A,B), suggesting the involvement of Anxa2-integrin-ILK axis in axon regeneration. ILK did not increase RGC survival after ONC (Fig. S6D,E).
Figure 7. ILK acts downstream of Anxa2 in axon regeneration and glaucoma neuroprotection.
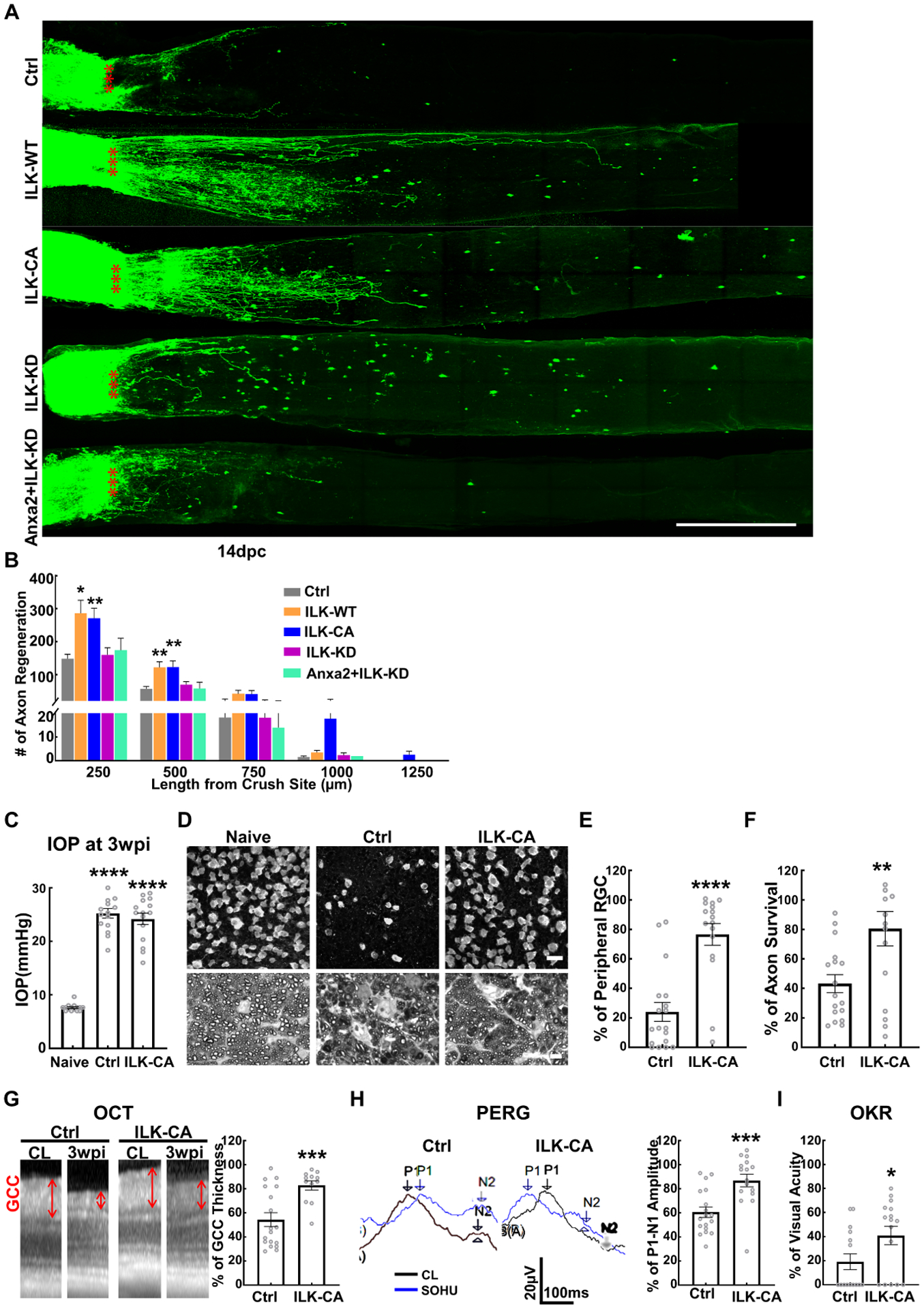
(A) Confocal images of ON wholemounts after optical clearance showing maximum intensity projection of regenerating fibers labeled with CTB-Alexa 555 at 14dpc. Scale bar, 500 μm. *: crush site. (B) Quantification of regenerating fibers at different distances distal to the lesion site. Data are presented as means ± s.e.m, n = 8–10. *: p<0.05, **: p<0.01, two-way ANOVA with Fisher’s LSD test. (C) IOP of naïve and SOHU eyes of control (Ctrl) and AAV-ILK-CA groups at 3wpi. Data are presented as means ± s.e.m, n = 15 in each group. ****: p<0.0001, Student’s t-test. (D) Representative confocal images of peripheral flat-mounted retinas showing surviving RBPMS-labeled RGCs and representative images of semi-thin transverse sections of ON with PPD staining at 3wpi. Upper panel, scale bar, 50 μm; lower panel, scale bar, 10 μm. (E) Quantification of peripheral surviving RGC somata at 3wpi, represented as percentage of glaucomatous eyes compared to the sham CL eyes. Data are presented as means ± s.e.m, n = 16 in each group, ****: p<0.0001, Student’s t-test. (F) Quantification of surviving axons at 3wpi, represented as percentage of glaucomatous eyes compared to the sham CL eyes. Data are presented as means ± s.e.m, n = 16 in each group, **: p<0.01, Student’s t-test. (G) Representative OCT images of mouse retinas from SOHU glaucomatous eyes and contralateral control eyes at 3wpi and quantification of GCC thickness at 3wpi, represented as percentage of GCC thickness in the SOHU eyes compared to the sham CL eyes. Data are presented as means ± s.e.m, n = 16 in each group, ***: p<0.001, Student’s t-test. (H) Left: representative wave forms of PERG at baseline and 3wpi, blue traces represent glaucomatous eyes, black traces represent contralateral control (CL) eyes. Right: quantification of P1-N2 amplitude of PERG at 3wpi, represented as a percentage of glaucomatous eyes compared to the sham CL eyes. Data are presented as means ± s.e.m, n = 16 in each group, ***: p<0.001, Student’s t-test. (I) Visual acuity measured by OKR at 3wpi, represented as percentage of glaucomatous eyes compared to the sham CL eyes. Data are presented as means ± s.e.m, n = 16 in each group, *: p<0.05, Student’s t-test.
See also Figure S6D,E
Since Anxa2 significantly prevented glaucomatous neurodegeneration, we next tested whether ILK activation also would be neuroprotective in the SOHU glaucoma model (Fig. 7C). Like Anxa2, ILK-CA overexpression in RGCs also produced striking protection of RGC somata and axons in glaucoma (Fig. 7D–F). In vivo OCT measurement of retina thickness and visual functional assays (PERG and OKR) also consistently confirmed the significant neuroprotection of ILK-CA in the SOHU glaucoma model (Fig. 7G–I).
Discussion
We developed a novel intraorbital optic nerve retrograde tracing technique, which enabled us for the first time to purify regenerating and non-regenerating RGCs from the same animals with the same genetic background/modification/injuries. These RGCs differ only in their capacity for axon regeneration. Profiling and comparing the transcriptomes of regRGCs and surRGCs at single cell level allowed us to unbiasedly determine genes associated with axon regeneration. Further characterization of the effects of some of these genes on axon regeneration in the ONC model and neuroprotection in the glaucoma model demonstrated the power and efficiency of this comprehensive but very focused strategy. We obtained the following findings: 1) The unique biological pathways associated with distinct clusters of RGCs are readily determined at the single cell molecular level; 2) Direct comparison of regRGCs and surRGCs directly identifies a list of regeneration-associated genes, many of which we showed to be pro-regeneration molecules; 3) The potent pro-regeneration molecules, Anxa2 and Mpp1, both achieve striking neuroprotection in the glaucoma model. This strategy can be used to identify downstream effectors or regulatory networks of other single or combinatory genes that promote significant axon regeneration (Yang et al., 2020b), which will rapidly and significantly advance the axon regeneration field. The common effectors the multiple signaling pathways involved in axon regeneration and neuronal survival will be promising targets for developing effective and safe neural repair therapies for neurodegenerative diseases. Additionally, the common regeneration-associated genes that are not present in non-regenerating RGCs provide potential regeneration markers that are also desperately needed in the field.
Membrane structure/actin binding/signaling molecules are critical axon regeneration
Anxa2 is the most significant regRGCs-enriched gene and promotes the most potent axon regeneration among the genes we tested. It is a Ca2+-dependent phospholipid and F-actin-binding membrane protein with diverse roles in cell migration, cytoskeletal-membrane dynamics, exocytosis, endocytosis, cell polarity, and lipid raft signaling (Gerke et al., 2005). Anxa2 is critical for Ca2+-dependent exocytosis of hormones in neuroendocrine cells (Gabel et al., 2019; Umbrecht-Jenck et al., 2010), indicating its roles in regulating neuronal activity, growth, and plasticity through secretion of neurotransmitters and neuropeptides (Gabel et al., 2015). The most studied function of Anxa2 is related to the co-receptor complex that it forms with S100A10 on the cell surface. This complex attracts tPA locally to generate plasmin and therefore to promote plasmin-mediated ECM degradation (Bharadwaj et al., 2013; Bharadwaj et al., 2021a; Bharadwaj et al., 2021b; Gerke et al., 2005). Interestingly, LRP1 is another tPA receptor that is also involved in axon regeneration (Landowski et al., 2016; Yoon et al., 2013). Our findings that Anxa2 promotes axon regeneration and that tPA further increases Anxa2/Pten modulation-induced axon regeneration, raise the possibility that the Anxa2-tPA complex on the surface of plasma membrane might serve as a proteolytic center that generates plasmin and therefore clears ECM nearby to make room for axons to grow. This action could facilitate regenerating axon elongation, a mechanism that has been confirmed for Anxa2-tPA-mediated cell adhesion, migration, and cancer cell invasion. Anxa2-tPA also regulates integrin-ILK pathway (Bharadwaj et al., 2013; Hu et al., 2007; Lin et al., 2012), which is important for Anxa2-induced axon regeneration. Thus, the Anxa2-tPA-integrin-ILK axis that connects extracellular signaling with intracellular signaling is a potential molecular mechanism of axon regeneration. In addition, Anxa2/S100A10 also interacts with Ahnak to regulate Ca2+-dependent exocytosis, L-type voltage-gated calcium channels, synaptic transmission, and membrane repair. Interestingly, both S100A10 and Ahnak are also in our regRGC-enriched gene list, further supporting the importance of the Anxa2/S100A10/Ahnak/tPA axis in axon regeneration. Anxa2 is also involved in inflammation (Dallacasagrande and Hajjar, 2020). Addition of Anxa2 rescues neuroinflammation and neurological deficits after traumatic brain injury, which deletion exacerbates (Cheng et al., 2021; Liu et al., 2019). Since inflammation also plays a role in optic nerve regeneration (Peterson et al., 2021; Xie et al., 2022; Xie et al., 2021), it is possible that Anxa2 promotes axon regeneration by modulating neuroinflammation.
Three other pro-regeneration genes identified by this study, Mpp1 (Membrane Palmitoylated Protein 1), Spp1 (Secreted Phosphoprotein 1, osteopontin) and Lgals1 (Galectin-1), share many similarities with Anxa2. Mpp1 is also a plasma membrane protein that form diverse protein complexes and is involved in cell structure, polarity, cell adhesion and migration, synaptogenesis, raft formation, and signal transduction (Chytla et al., 2020). Mpp1 is required for insulin-stimulated activation of H-Ras (Podkalicka et al., 2018), a growth factor-initiated signaling pathway that is known to promote pro-axon regeneration (O’Donovan et al., 2014; Zhang et al., 2019c). Spp1 interacts with multiple integrins and is involved in cytokine secretion, cellular differentiation, adhesion, migration, and would healing (Lamort et al., 2019). Galectin 1 (Lgals1), which has known roles in axon growth, axon guidance, and axon regeneration (Higuero et al., 2017; Quinta et al., 2014; Quinta et al., 2016), belongs to a family of glycan-binding proteins that recognize distinct sets of glycosylated proteins or lipids at the cellular surface or ECM (Nio-Kobayashi and Itabashi, 2021). Lgals1 also interacts with β1 integrin to regulate neural progenitor cells (Sakaguchi et al., 2010). Thus, similar to Anxa2, Mpp1, Spp1, and Lgals1 may also promote axon regeneration by activating the integrin-ILK pathway. More broadly, we hypothesize that cell membrane structure molecules that are involved in cell adhesion/migration/invasion/exocytosis are important for axon regeneration, possibly through mediating interaction of intracellular and extracellular signaling and reorganizing local cytoskeleton/ECM. Additional findings that cell surface molecules neural cadherin and thrombospondin 1 (Bray et al., 2019; Ribeiro et al., 2020), complement proteins in the optic nerve locally (Peterson et al., 2021), cytoskeleton stabilization and motor molecules (Hellal et al., 2011; Nawabi et al., 2015; Wang et al., 2020b), promote axon regeneration further support this theme. Another pro-regeneration gene we found, Plin2, is the coat protein of lipid droplets and important for cellular lipid homeostasis (Brasaemle, 2007). Consistently, Lipin 1, an enzyme that inhibits membrane phospholipid, also inhibits axon regeneration (Yang et al., 2020a). Detail profile and characterization of molecules in the growth cones of regenerating axons may shed more light on the molecular mechanisms of local membrane molecules in axon regrowth (Chauhan et al., 2020).
From axon regeneration to neural repair
The striking neuroprotection produced by Anxa2, ILK, and Mpp1 in the glaucoma model is a surprising finding, because they only marginally increase RGC survival after ONC injury. However, although ONC is extensively used as a convenient model of optic neuropathy and often as a surrogate glaucoma model, RGC/optic nerve may respond very differently to the same neural repair treatments depending on whether they are injured by traumatic injury or IOP elevation. One previous study also found that pro-regeneration genetic modulation significantly promotes neural repair in glaucoma model (Lu et al., 2020). The wide range of cellular functions and signaling cascades mediated by Anxa2, ILK, and Mpp1 cited above may contribute to the significant neuroprotection and visual function preservation in the SOHU glaucoma model, but further investigations will be necessary to understand the precise mechanisms. The pro-regeneration activity of these genes may also be critical for the neuroprotection in glaucoma, although it is difficult to appreciate how much axon regeneration is indeed induced by these genes in the glaucoma model in which axon damage is incomplete. The weaker pro-regeneration genes Plin2 and Lgals1 barely increase neuronal survival and visual function in the glaucoma model, which is additional evidence that regeneration capability may be linked to neuroprotection potential, or that the pro-regeneration machinery is also critical for glaucomatous RGC survival. The present results represent an early, but very encouraging and compelling confirmation of this search strategy: identifying pro-regeneration genes associated with regRGCs in the ONC model as promising candidates for neural repair in glaucoma.
In summary, we present a powerful and efficient tracing methodology to distinctly label and isolate regRGCs and surRGCs from the same animals after the same genetic modulation, injury and treatment, which allows us to definitively compare single cell transcriptomes to identify truly regeneration-associated genes. Four out of seven pro-regeneration genes we identified through this study, Anxa2, Mpp1, Spp1, and Lgals1, share many similarities, such as involvement in plasma membrane-binding, local membrane-cytoskeleton dynamic/lipid rafts formation and integrin signaling, and exocytosis. Based on these results, together with the synergistic effect of tPA with Anxa2 and Pten, we hypothesize that plasma membrane-cytoskeleton-ECM remodeling and signaling cascades play critical roles in axon regeneration and regrowth. The differentiation strategy that we employed detects without bias distinct RGC clusters that are preferentially accumulated with regRGCs or surRGCs and have signature genes and biological pathway profiles. Finally, and extremely importantly, we found that Anxa2 is a very promising neuroprotectant due to its dramatic protection of glaucomatous RGCs/optic nerves and visual function deficits, which validates the strategy of searching for neural repair candidates among the potent pro-regeneration molecules. This whole strategy is likely to be applicable to other neurodegenerative diseases or trauma associated with long axons, such as motor neuron degenerative diseases and spinal cord injury.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for protocols, reagents and resources should be directed to and will be fulfilled by the lead contact, Yang Hu (huyang@stanford.edu)
Materials availability
Plasmids generated in this study can be requested to the lead contact.
Data and code availability
All custom scripts have been made available at https://github.com/HuLab-Code/. Any additional information is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice.
C57BL/6J WT and B6.129S4-Ptentm1Hwu/J mutation (Ptenf/f) male and female mice were purchased from Jackson Laboratories (Bar Harbor, Maine). All mice were housed in standard cages on a 12-hour light-dark cycle. All experimental procedures were performed in compliance with animal protocols approved by the IACUC at Stanford University School of Medicine.
METHOD DETAILS
Constructs and AAV production.
The coding regions of Anxa2, Plin2, Mpp1, Acaa2, Spp1, Lgals1, Gpnmb, ILK were amplified from different mouse tissue cDNA (Table S4) with Q5 high-fidelity DNA Polymerase (NEB, M0491L) and cloned into the pAM-AAV-mSncg-3HA-WPRE or pAM-AAV-CS265-3HA-WPRE backbone with EcoRI, MluI, XhoI, or HindIII sites. The pAM-AAV-mSncg-Cre-WPRE plasmid was published previously1. The constitutive active (Ilk-CA, S343D) and kinase dead (Ilk-KD, S343A) mutants were generated by Phusion site-directed mutagenesis kit (Thermo Fisher, F541) with primers (Table S4). The maxi-precipitation of the constructs was performed by following the manual of Endo-Free Plasmid Maxi kit (Omega Bio-tek, D6926-03/101319-342). The detailed procedure of the AAV production has been described previously(Wang et al., 2020a). Briefly, AAV plasmids containing the target genes were co-transfected with pAAV2 (pACG2)-RC triple mutant (Y444, 500, 730F) and the pHelper plasmid (StrateGene) into HEK293T cells by the PolyJet (SignaGen Laboratories, SL100688) transfection reagent. After transfection for 72 hours, the cells were lysed and purified by two rounds of cesium chloride density gradient centrifugation. The AAV titers of target genes were determined by real-time PCR and diluted to 1.5 × 1012 vector genome (vg)/ml for intravitreal injection, 2 μl/eye.
Intravitreal injection.
Mice were anesthetized by xylazine and ketamine based on their body weight (0.01mg xylazine/g + 0.08mg ketamine/g). For AAV intravitreal injection, a pulled and polished microcapillary tube was inserted into the peripheral retina of around 4-week-old mice just behind the ora serrata. Approximately 2 μl of the vitreous was removed to allow injection of 2 μl AAV into the vitreous chamber. The mice were housed for an additional 2 weeks after AAV injection to achieve stable target genes expression. For anterograde tracing of regenerating axons, 2μl of 2μg/μl cholera toxin subunit B-Alexa 555 (CTB555, Invitrogen) was used for intravitreal injection. For the combinatory treatment of Anxa2 overexpression, tissue plasminogen activator (tPA, Sigma, 612200-m) and Pten KO, an AAV mixture with equal amount of Anxa2 and Cre was intravitreally injected into the Pten floxed mouse eyes 14 days before ONC, and 2μl of 30U/μl tPA was intravitreally injected twice/week after ONC.
Optic nerve crush (ONC).
ONC was performed 2 weeks following AAV injection when mice were about 7–8 weeks of age. The optic nerve was exposed intraorbitally at the 12 o’clock position while care was taken not to damage the underlying retro-orbital sinus, and crushed with a jeweler’s forceps (Dumont #5; Fine Science Tools, Foster City, California) for 5 seconds approximately 0.5 mm behind the eyeball. Eye ointment containing neomycin (Akorn, Somerset, New Jersey) was applied to protect the cornea after surgery.
Retrograde labeling of regenerating RGCs by intraorbital optic nerve injection.
Pten KO mice were anesthetized by xylazine and ketamine based on their body weight (0.01mg xylazine/g+0.08mg ketamine/g). The optic nerve was exposed intraorbitally ~1.5–2 mm distal to the eyeball by lateral canthotomy through the conjunctiva and beneath soft tissues and muscles at the 9 o’clock position without injuring the retro-orbital sinus. The optic nerve was immobilized by placing a piece of tissue paper between the optic nerve trunk and surrounding soft tissue and the dura pierced by a 33g needle at the injection site, ~1 mm distal to ONC site. A glass micropipette connected to a 50μL microsyringe (80900, Hamilton) attached to a Micro4 controller was used to deliver ~60nL dextran into the optic nerve through the pre-made hole, at a speed of 100nL/min. Dye leaking at the injection site was removed by the tissue paper. After the injection, ointment containing neomycin was applied to protect the cornea, and mice were placed on heating pad for recovery. Mice were housed for 24 hours before tissue collection.
Retrograde labeling naive RGCs by superior colliculus (SC) injection.
The detailed procedure of the micro-injection into the SC has been described previously (Hu et al., 2012). In brief, the adult mice were anesthetized by xylazine and ketamine based on their body weight (0.01mg xylazine/g+0.08mg ketamine/g) and fixed on a mouse adaptor attached to a digital stereotaxic instrument (68025, RWD Life Science). The bregma was set as the origin of anterior to posterior (AP), medial to lateral (ML) and dorsal to ventral (DV), and the same ML and DV of the lambda was aligned to the bregma. The horizontal plane of the mouse skull was calibrated by adjusting the left hemisphere point (AP:−2.00, ML:2.50) to the same DV as the contralateral point (AP:−2.00, ML:−2.50). The SC coordinates for 4 sites and 3 depths were located and drilled: AP:−3.55, ML:0.6, DV−1.25/−1.60/−2.00, AP:−3.55, ML:−0.6, DV−1.25/−1.60/−2.00, AP:−3.92, ML:0.8, DV−1.25/−1.50/−1.75 and AP:−3.92, ML:−0.8, DV−1.25/−1.50/−1.75. A pulled-glass micropipette fused to a 10 μL syringe (80314, Hamilton) filled with mineral oil was controlled by micro syringe pump (Micro4™, World Precision Instruments, LLC) at the speed of 250 nL/min for 1 minute per site. About 4 μL Dextran-FITC, Retrobeads-488, FluoroGold, or Fast Blue were injected into both SCs.
Retina cell dissociation and FACS purification.
We followed the procedure described in a previous publication (Tran et al., 2019). Briefly, mouse retinas were dissected out in AMES solution (saturated with 95% O2/5% CO2) and then digested in papain (Worthington, LS003126) with L-cysteine (Sigma, C1276–10g) for 10–15 minutes at 37°. The single cell suspension was achieved by manual trituration in ovomucoid solution (Worthington, LS003087) on ice. Cells were spun down at 80 g for 15 minutes at 4° and then resuspended in cold AMES with 4% BSA (Sigma) to a concentration of 1 × 107 cells/ml. Prior to FACS, the cells were filtered through a 40 μm cell strainer (Falcon, 352340). FACS purification with a BD Influx System cell sorter was gated as: Alexa Fluor 488 positive but Texas Red negative RGCs were collected as surRGCs; Texas Red positive RGCs were collected as regRGCs. Individual cells were directly sorted into individual wells of a 96 well-plate with 4ul pre-filled cell lysis buffer, containing 1 U/μl of Recombinant RNase inhibitor (Clontech), 0.1% Triton X-100 (Thermo), 2.5mM dNTP (Thermo), and 2.5 μM oligodT30VN (5’AAGCAGTGGTATCAACGCAGAGTACT30VN-3’, IDT). Cells were immediately spun down after sorting and frozen at −80°C.
Smart-seq2.
Two 384-well plates containing surRGCs or regRGCs were used to generate Smart-Seq2 libraries following the published protocol (Picelli, 2019; Picelli et al., 2013; Picelli et al., 2014) at Stanford Genomics core facility and Chan Zuckerberg Biohub at Stanford. Briefly, Smartscribe (Clontech) was used for reverse transcription of the mRNAs, and then amplified by 23 PCR cycles using a KAPA Hifi HotStart Kit (Roche). Amplified cDNAs were purified by beads cleanup using a Biomek FX automated platform (Beckman), and aliquots run on a Fragment Analyzer (Agilent) for quantitation. Barcoded sequencing libraries were made using the NexteraXT DNA Library Preparation Kit (Illumina), and the PCR was performed as follows: 72°C 3 min, 95 °C 30 s, then 10 cycles of (95°C 10 s, 55°C 30s, 72°C 1min), 72°C 5min. Libraries were cleaned up by 0.8X AMPure XP beads (Beckman Coulter), then diluted to a final concentration of 2nM for sequencing. 384 cells were sequenced in one lane of the Illumina HiSeq 4000 sequencer (Illumina) with 2x 150 bp paired-end configuration.
Data analysis.
Raw data were trimmed by trim-galore (version 0.6.7) to remove adaptor sequences and aligned with Hisat2 (version 2.2.1) to the mouse reference genome (mm10). Transcripts per million reads (TPMs) per gene was calculated by StringTie (version 2.1.7), and genes with TPM > 0 were defined as detected genes. All the downstream analysis was performed by R, using Seurat (version 4.1.0) (Hao et al., 2021) with modifications. Cells with Rbpms, Slc17a6, Sncg, Thy1, Pou4f1, Pou4f2 and Pou4f3, and low Pten expression (TPM < 30) were used for downstream analysis. We filtered out the low-quality cell libraries, only keeping those with (1) percentage of mitochondrial genes, < 15%; (2) number of unique genes, > 900; (3) total reads mapped > 530,000. In total, 630 RGCs (340 regRGCs and 290 surRGCs) were selected for further analysis. We applied the Leiden algorithm (resolution = 0.2) to perform unsupervised clustering of the RGCs using the TPM matrix as input, and obtained five distinct clusters. Differentially expressed genes (DEGs) in each cluster were identified using FindAllMarkers function with default parameters. The top 10 signature genes in each cluster were visualized using heatmap. Gene Ontology (GO) analysis on the DEGs was performed using R package clusterProfiler (Yu et al., 2012). We used Cnetplot to depict the linkage of genes and biological process as a network, showing genes in those most significant enriched terms.
For pseudo-bulk comparison of regRGCs and surRGCs, we used the FindMarkers function, which is implemented within the Seurat, to get DEGs between regRGCs and surRGCs. The DE test was performed based on the Wilcoxon rank-sum test with default parameters. The GO enrichment analysis and Cnetplot were also performed on the DEGs. All raw datasets are shared online via the Gene Expression Omnibus (GEO) database (GSE206626).
Brain-optic nerve clearance and light sheet microscopy imaging.
The attached ON and whole brain were carefully dissected with fine forceps and scissors, and embedded in 1.5% agarose gel block. The tissue embedded gel block was cleared with modified iDISCO method (Renier et al., 2014): PBS for 4 hours; a series of 20%, 40%, 60%, 80%, and 100% methanol in 1xPBS for 1day at each concentration; dichloromethane (DCM)/methanol (2:1) for 1day; 100% DCM for 1day and dibenzyl ether (DBE) for 1day. The ventral side of the tissue gel block was faced up and fixed on a spike holder, then placed into the imaging chamber immersed in the DBE buffer. The ultramicroscope II generated 6 bi-directional 3.89μm thin light sheets to illuminate the tissue gel block from both sides while imaging the excited plane with a 2x objective microscope perpendicular to the sample using a 0.63x zoom for whole tissue and a 6.3x zoom for regenerating axons. Tissue was imaged with the diode 561nm laser, emission filter 620/60nm and sheet numerical aperture (NA) 0.149 through a 2μm step-size of the Z-stack. The multiple optical sliced images of the whole tissue were collected and further maximum projections were processed by Fiji/Image J.
SOHU glaucoma model and IOP measurement.
The detailed procedure has been published before (Fang et al., 2021; Zhang et al., 2019a; Zhang et al., 2019b). In brief, 9-week-old mice were anesthetized by an intraperitoneal injection of Avertin (0.3mg/g) before a tunnel was made by a 32G needle through the layers of the cornea on the superotemporal side close to the limbus to reach the anterior chamber without injuring lens or iris. Then, ~ 2μl silicone oil (1,000 mPa.s, Silikon, Alcon Laboratories, Fort Worth, Texas) was injected slowly into the anterior chamber using a homemade sterile glass micropipette, until the oil droplet expanded to cover most areas of the iris (diameter ~ 1.8–2.2mm). After the injection, veterinary antibiotic ointment (BNP Ophthalmic Ointment, Vetropolycin, Dechra, Overland Park, Kansas) was applied to the surface of the injected eye. The contralateral control eyes received 2μl normal saline to the anterior chamber. Throughout the procedure, artificial tears (Systane Ultra Lubricant Eye Drops, Alcon Laboratories, Fort Worth, Texas) were applied to keep the cornea moist. The IOP of both eyes was measured before SO injection and at 3wpi by the TonoLab tonometer (Colonial Medical Supply, Espoo, Finland) according to product instructions. Briefly, in the morning, mice were anesthetized with a sustained flow of isoflurane (3% isoflurane at 2 L/minute mixed with oxygen) delivered to the nose by a special rodent nose cone (Xenotec, Inc., Rolla, Missouri), which left the eyes exposed for IOP measurement. 1% Tropicamide sterile ophthalmic solution (Akorn, Somerset, New Jersey) was applied three times at 3-minute intervals to fully dilate the pupils (about 10 minutes) before taking measurements. The average of six measurements by the TonoLab was considered as one machine-generated reading and three machine-generated readings were obtained from each eye; the mean was calculated to determine the IOP. During this procedure, artificial tears were applied to keep the cornea moist.
Spectral-domain optical coherence tomography (SD-OCT) imaging and scanning laser ophthalmoscopy (SLO) fundus imaging.
Fundus OCT imaging was performed under OCT mode by switching to a 30° licensed lens (Heidelberg Engineering) as in the previously described procedure (Li et al., 2020; Zhang et al., 2019b). Briefly, the mouse retina was scanned with the ring scan mode centered by the optic nerve head at 100 frames average under high-resolution mode (each B-scan consisted of 1536 A scans). The ganglion cell complex (GCC) includes retinal nerve fiber layer (RNFL), ganglion cell layer (GCL), and inner plexiform layer (IPL). The average thickness of GCC around the optic nerve head was measured manually with the Heidelberg software. The mean of the GCC thickness in the injured retina was compared to that in the contralateral control retina to yield a percentage of GCC thickness value. The investigators who measured the thickness of GCC were blinded to the treatment of the samples. For SLO retinal fundus imaging, the fundus labeled with green fluorescent dye was imaged under FA mode by switching to a 55° non-contact lens and a customized +10D contact lens (3.0 mm diameter, 1.6 mm BC, PMMA clear, Advanced Vision Technologies) (Heidelberg Engineering) as previously described(Li et al., 2020; Zhang et al., 2019b). The mouse retina was imaged under the high-resolution mode (1536 × 1536 pixels) and 100 frames average with 488 nm excitation laser.
Pattern electroretinogram (PERG) recording.
PERG recording of both eyes was performed at the same time with the Miami PERG system (Intelligent Hearing Systems, Miami, FL), as described in our previous publication (Li et al., 2020; Zhang et al., 2019b). In brief, a feedback-controlled heating pad (TCAT-2LV, Physitemp Instruments Inc., Clifton, New Jersey) maintained animal core temperature at 37°C. A small lubricant eye drop (Systane) was applied before recording to prevent corneal opacities. The reference electrode was placed subcutaneously on the back of the head between the two ears, the ground electrode was placed at the root of the tail and the active steel needle electrode was placed subcutaneously on the snout for the simultaneous acquisition of left and right eye responses. Two 14 cm × 14 cm LED-based stimulators were placed in front so that the center of each screen was 10 cm from each eye. The pattern remained at a contrast of 85% and a luminance of 800 cd/m2, and consisted of four cycles of black-gray elements, with a spatial frequency of 0.052 c/d. Upon stimulation, the independent PERG signals were recorded from the snout and simultaneously by asynchronous binocular acquisition. With each trace recording up to 1020 ms, two consecutive recordings of 200 traces were averaged to achieve one readout. The first positive peak in the waveform was designated as P1 and the second negative peak as N2. P1 was typically around 100 ms. The mean amplitude of the P1-N2 amplitude in the injured eye was compared to that in the contralateral control eye to yield a percentage of amplitude change. The investigators who measured the amplitudes were blinded to the treatment of the samples.
OKR measurement.
The spatial vision of both eyes was measured using the opto-kinetic response (OKR) as described in our previous publication (Fang et al., 2021; Zhang et al., 2019b). In brief, living mice were placed unrestrained on a platform in the center of four 17-inch LCD computer monitors (Dell, Phoenix, AZ); their movement was captured by a video camera above the platform. A rotating cylinder with vertical sine wave grating was computed and projected to the four monitors by OptoMotry software (CerebralMechanics Inc., Lethbridge, Alberta, Canada). The sine wave grating provides a virtual-reality environment to measure the spatial acuity of left eye when rotated clockwise and right eye when rotated counterclockwise. When the mouse calmed down and stopped moving, the gray of the monitor immediately switched to a low spatial frequency (0.1 cycle/degree) for five seconds, in which the mouse was assessed by judging whether the head turned to track the grating. The short time frame of assessment ensured that the mice did not adapt to the stimulus, which would lead to false readouts. The mice were judged to be capable of tracking the grating. The spatial frequency was increased repeatedly until a maximum frequency was identified and recorded. The % of vision acuity was yielded by comparing the maximum frequency of the experimental eye to that of the contralateral eye. The mice were tested in the morning and the investigator who judged the OKR was blinded to the treatment of the mice.
QUANTIFICATION AND STATISTICAL ANALYSIS
Optic nerve clearance and axon regeneration quantification.
The CTB anterograde labeled optic nerve was trimmed and cleared by a modified iDISCO method (Renier et al., 2014): PBS for 30mins; a series of 20%, 40%, 60%, 80%, and 100% methanol in 1xPBS for 30mins at each concentration; dichloromethane (DCM)/methanol (2:1) for 30mins; 100% DCM for 30mins and dibenzyl ether (DBE) for 30mins. The cleared optic nerve was mounted on slides between two 22×22mm cover slips supported with DBE, covered with a 22×22mm cover slip, and sealed with clear nail polish. The mounted whole nerve was imaged with a 25x oil immersion objective lens, using the airy scan mode (6μm per stack), Z stack and tile scan. The number of CTB labeled axons was quantified as described previously (Leon et al., 2000; Park et al., 2008; Yang et al., 2014). Briefly, we counted the fibers that crossed perpendicular lines drawn on the optic nerve optical sections distal to the crush site at 250, 500, 1000, 1500, and then every 250 μm till no fibers were visible. 3 Z-stacks at depths of 60, 120 and 180μm were sampled to acquire the mean axon density of the optic nerve, (axon number)/(R*t). The width of the stack (R) was measured at the point (d) at which the counts were taken and used together with the thickness of the optical section (t = 6 μm) to calculate the number of axons/μm2 area of the stack. The mean axon density of the 3 stacks was used to calculate the total axon number, ∑ad= πr2 * mean axon density, r is the radius of the optic nerve. All CTB signals that were in the range of intensity that was set from lowest intensity to the maximum intensity after background subtraction were counted as individual fibers. The investigators who counted the cells or axons were blinded to the treatment of the samples.
Immunohistochemistry of retinal wholemounts and cryo-sections and RGC counts.
After transcardiac perfusion with 4% PFA in PBS, the eyes were dissected out, post-fixed with 4% PFA for 2 hours, at room temperature, and cryoprotected in 30% sucrose overnight. Retinas were then dissected out and washed extensively in PBS before blocking in staining buffer (10% normal donkey serum and 2% Triton X-100 in PBS) for 30 minutes. For cross sections of retina, the eyeballs were dehydrated sequentially with 15% and 30% sucrose solution overnight before embedding in Tissue-PlusTM OCT compound on dry ice. Serial cross sections (25 μm) were cut with a Leica cryostat and collected on Superfrost Plus Slides and stored at −80°C until processed. RBPMS guinea pig antibody (ProSci, California), HA antibody and AP2α mouse antibody were used at 1:4000, 1:500, and 1:100 to label RGCs and amacrine cells, respectively. Floating retinas were incubated with primary antibodies overnight at 4°C and washed 3 times for 30 minutes each with PBS. Secondary antibodies (Alexa Fluor 647-goat anti-guinea pig, Cy3-goat anti-rat and Cy2-goat anti-mouse) were then applied (1:200; Jackson ImmunoResearch, West Grove, Pennsylvania) and incubated for 1 hour at room temperature. Primary antibody of Anxa2 (1:200), Mpp1 (1:200), Spp1 (1:500), Opn4 (1:200), Cart (1:1000), Foxp2 (1:1000) and Kv4.2 (1:200) and secondary antibody (1:200 of Alexa488 donkey anti-rabbit, Alexa647-donkey anti-goat, Alexa647-donkey anti-rabbit or Alexa647-donkey anti-mouse) were used to label mouse RGCs or subtypes in retinal wholemounts or cryo-sections. Retinas were again washed 3 times for 30 minutes each with PBS before a cover slip was attached with Fluoromount-G (Southernbiotech, Birmingham, Alabama). Images of immunostained wholemounts were acquired with a Zeiss M2 epifluorescence microscope and Zeiss confocal microscope (LSM 880) with 20x and 40x oil lens and serial filters (BP410–510 for DAPI or Fast Blue, BP520–550 for Alexa Fluor 488, Dextran-FITC or Cy2, BP565–650 for Cy3 or Dextran Texas red and BP650–750 for Alexa Fluor 647). For RGC counting, 6–9 fields of 332μm×332μm area were sampled on average from peripheral or middle and central regions of each whole retina for imaging and stitching by a 20x lens and a Keyence fluorescence microscope (Itasca, BZ-X800), and RBPMS+ RGCs counted by Fiji/Image J (NIH). The percentage of RGC survival was calculated as the ratio of surviving RGC numbers in injured eyes compared to contralateral uninjured eyes. The investigators who counted the cells were blinded to the treatment of the samples.
Optic nerve semi-thin sections and quantification of surviving axons.
The detailed procedure has been described previously. Briefly, transverse semi-thin (1 μm) sections of optic nerve were cut on an ultramicrotome (EM UC7, Leica, Wetzlar, Germany) from tissue collected 2 mm distal to the eye (about 1.5 mm distal to the crush site) and stained with 1% para-phenylenediamine (PPD) in methanol: isopropanol (1:1). Whole optic nerves were imaged and stitched through a 100x lens of a Keyence fluorescence microscopy. Eight areas of 21.4 μm × 29.1 μm from the entire optic nerve were cropped on average and counted manually with Fiji/ImageJ. After counting all the images taken from a single nerve, the mean of the surviving axon number was calculated for each optic nerve, and compared to that in the contralateral control optic nerve to yield a percentage of axon survival value. The investigators who counted the axons were masked to the treatment of the samples.
Statistical analyses.
GraphPad Prism 7 was used to generate graphs and for statistical analyses. Data are presented as means ± s.e.m. Student’s t-test was used for two groups comparison, One-way ANOVA with post hoc test and Two-way ANOVA were used for multiple comparisons.
Supplementary Material
Table S1 Cluster Specific Genes, Related to Figure 2.
Table S2 Upregulated DEGs in regRGCs, Related to Figure 3.
Table S3 Downregulated DEGs in regRGCs, Related to Figure 3.
Table S4 Primers for PCT of mouse tissue cDNA, Related to STAR Methods section.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | Jackson Laboratories | Cat#JAX000664; RRID: IMSR JAX:000664 |
| Mouse: B6.129S4-Ptentm1Hwu/J | Jackson Laboratories | Cat#JAX006440; RRID:IMSR JAX:006440 |
| Antibodies | ||
| Guinea pig polyclonal anti- RBPMS | Custom-made by ProSci | N/A |
| Mouse monoclonal anti- AP2α | Developmental Studies Hybridoma Bank | Cat#3B5; RRID:AB_2202275 |
| Goat polyclonal anti-Spp1 | R&D | Cat#AF808; RRID:AB_2194992 |
| Rabbit polyclonal anti-Opsin4 | Thermo Fisher Scientific | Cat#PA1780; RRID:AB_2267547 |
| Rabbit polyclonal anti-Mpp1 | Thermo Fisher Scientific | Cat#PA5115589; RRID_AB_2900224 |
| Rabbit monoclonal Anxa2 antibody | Abcam | Cat# ab178677; RRID_AB_2883091 |
| Rabbit polyclonal anti-Cart | Phoenix pharma | Cat#H-00362; RRID AB 2313614 |
| Rabbit polyclonal anti-Foxp2 | Abcam | Cat#ab16046; RRID:AB_2107107 |
| Mouse monoclonal anti-Kv4.2 | NeuroMab | Cat#75361; RRID:AB_2315874 |
| Rat monoclonal anti-3HA | Roche | Cat#11867423001; RRID:AB 390918 |
| Cy2 Goat anti-mouse IgG | Jackson ImmunoResearch | Cat#115225-166; RRID:AB_2338746 |
| Cy3 Goat anti-rat IgG | Jackson ImmunoResearch | Cat#112165-167;RRID:AB_2338251 |
| Alexa488 Donkey anti-goat IgG | Jackson ImmunoResearch | Cat#705545-003;RRID:AB_2340428 |
| DyLight™ 405 Donkey anti-Guinea Pig IgG | Jackson ImmunoResearch | Cat#706475-148;RRID:AB_2340470 |
| Alexa647 Donkey anti-rabbit IgG | Jackson ImmunoResearch | Cat#711605-152;RRID:AB_2492288 |
| Alexa Fluor 647 Goat anti-Guinea Pig IgG | Jackson ImmunoResearch | Cat#106605-003;RRID:AB_2337446 |
| Alexa488 Donkey anti-rabbit IgG | Jackson ImmunoResearch | Cat#711545-152;RRID:AB_2313584 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cholera Toxin Subunit B (Recombinant), Alexa Fluor™ 555 Conjugate | ThermoFisher scientific | Cat#C22843 |
| Dextran-FITC, 3000 MW, anionic, lysine fixable | ThermoFisher Scientific | Cat#D3306 |
| Dextran, Texas Red, 3000 MW, lysine Fixable | ThermoFisher Scientific | Cat#D3328 |
| FluoroGold | ThermoFisher Scientific | Cat#NC1363013 |
| DiI Cell-Labeling Solution | ThermoFisher Scientific | Cat#V22885 |
| FastBlue | PolySciences | Cat#177401 |
| RetroBeads Green (488) Solution | Lumafluor | N/A |
| The tissue plasminogen activator (human) | Sigma-Aldrich | Cat#612200-M |
| Silicone oil | Alcon Laboratories | Cat#1,000 mPa.s,Silikon |
| AMES’ Medium | Sigma-Aldrich | Cat#A142010x1L |
| Papain | Worthington-Fisher | Cat#LS003126 |
| Ovomucoid | Worthington-Fisher | Cat#LS003087 |
| L-cysteine | Sigma-Aldrich | Cat#C127610g |
| Methanol | Fisher Scientific | Cat#A4524 |
| Dichloromethane | Sigma-Aldrich | Cat#348561L |
| Dibenzyl ether | Sigma-Aldrich | Cat#336301L |
| Software and Algorithms | ||
| GraphPad Prism7 | GraphPad Software | N/A |
| Fiji/ImageJ | NIH | N/A |
| R for statistical computing version 4.0.2 | N/A | https://cran.r-project.org |
| StringTie v2.1.7 | Pertea et al., 2016(Pertea et al., 2016) | https://ccb.jhu.edu/software/stringtie/ |
| Hisat2 v2.2.1 | Kim et al., 2019(Kim et al., 2019) | https://daehwankimlab.github.io/hisat2/ |
| Code for Smart-Seq2 DEG | Hu Lab | https://doi.org/10.5281/zenodo.6687970 |
| Other | ||
| LSM 880 confocal microscopy | Zeiss | LSM 880 |
| Keyence fluorescence microscopy | Keyence | BZ-X800 |
| Zeiss M2 fluorescence microscopy | Zeiss | M2 |
| LaVision, Light Sheet Ultramicroscope II | Miltenyi Biotec | N/A |
| Leica CM3050S Cryostat | Leica | CM3050S |
| BD Influx System Cell Sorter | BD Biosciences | N/A |
| Illumina HiSeq 4000 sequencer | Illumina | N/A |
| Heidelberg SLO & OCT | Heidelberg | N/A |
| Pattern electroretinogram (PERG) | Intelligent Hearing Systems | N/A |
| Opto-kinetic response (OKR) | CerebralMechanics | OptoMotry HD |
Highlights.
Powerful single cell regRGCs-seq to profile Pten KO-regeneration-associated genes.
In vivo demonstration of multiple pro-axon regeneration genes.
Striking neuroprotection in glaucoma by pro-regeneration genes Anxa2 and Mpp1.
The tPA-Anxa2-ILK pathway in axon regeneration and neuroprotection.
Acknowledgements
We thank Sopheak Sim, Shelly Huynh and Drs. Stephen Quake and Maurizio Morri for their great technical support with FACS and Smart-Seq2. We thank Drs. Marc Tessier-Lavigne, Aaron Gitler, Jeffrey Goldberg, Alan Tessler and Hu lab members for critical discussion and reading the manuscript. Y.H. is supported by NIH grants EY024932, EY023295, EY028106, EY032518, and grants from Glaucoma Research Foundation (CFC3), BrightFocus Foundation, Chan Zuckerberg Initiative Neurodegeneration Collaborative Pairs Pilot Projects, Stanford SPARK program and Stanford Center for Optic Disc Drusen. L.C. Is supported by National Science Foundation (NSF 1953686), National Institutes of Health (1R35HG011316, 1R01GM141627), and Donald and Delia Baxter Foundation. The light sheet microscope is sponsored by NIH1S10D025091. We are grateful for an unrestricted grant from Research to Prevent Blindness and NEI P30 EY026877 to the Department of Ophthalmology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
YH is a consultant for Janssen BioPharma, Inc.
A patent application has been submitted by Stanford Office of Technology Licensing for candidate neural repair targets identified in this manuscript.
The other authors have declared that no conflict of interest exists.
REFERENCES
- Abreu CA, De Lima SV, Mendonca HR, Goulart CO, and Martinez AM (2017). Absence of galectin-3 promotes neuroprotection in retinal ganglion cells after optic nerve injury. Histol Histopathol 32, 253–262. 10.14670/HH-11-788. [DOI] [PubMed] [Google Scholar]
- Almasabi S, Ahmed AU, Boyd R, and Williams BRG (2021). A Potential Role for Integrin-Linked Kinase in Colorectal Cancer Growth and Progression via Regulating Senescence and Immunity. Frontiers in genetics 12, 638558. 10.3389/fgene.2021.638558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benowitz LI, He Z, and Goldberg JL (2015). Reaching the brain: Advances in optic nerve regeneration. Exp Neurol. 10.1016/j.expneurol.2015.12.015. [DOI] [PubMed] [Google Scholar]
- Bharadwaj A, Bydoun M, Holloway R, and Waisman D (2013). Annexin A2 heterotetramer: structure and function. Int J Mol Sci 14, 6259–6305. 10.3390/ijms14036259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj A, Kempster E, and Waisman DM (2021a). The Annexin A2/S100A10 Complex: The Mutualistic Symbiosis of Two Distinct Proteins. Biomolecules 11. 10.3390/biom11121849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj AG, Kempster E, and Waisman DM (2021b). The ANXA2/S100A10 Complex-Regulation of the Oncogenic Plasminogen Receptor. Biomolecules 11. 10.3390/biom11121772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasaemle DL (2007). Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res 48, 2547–2559. 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- Bray ER, Yungher BJ, Levay K, Ribeiro M, Dvoryanchikov G, Ayupe AC, Thakor K, Marks V, Randolph M, Danzi MC, et al. (2019). Thrombospondin-1 Mediates Axon Regeneration in Retinal Ganglion Cells. Neuron 103, 642–657 e647. 10.1016/j.neuron.2019.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne AB, Walradt T, Gardner KE, Hubbert A, Reinke V, and Hammarlund M (2014). Insulin/IGF1 Signaling Inhibits Age-Dependent Axon Regeneration. Neuron 81, 561–573. 10.1016/j.neuron.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkins DJ (2021). Adaptive responses to neurodegenerative stress in glaucoma. Prog Retin Eye Res, 100953. 10.1016/j.preteyeres.2021.100953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan MZ, Arcuri J, Park KK, Zafar MK, Fatmi R, Hackam AS, Yin Y, Benowitz L, Goldberg JL, Samarah M, and Bhattacharya SK (2020). Multi-Omic Analyses of Growth Cones at Different Developmental Stages Provides Insight into Pathways in Adult Neuroregeneration. iScience 23, 100836. 10.1016/j.isci.2020.100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Wang X, Jiang Y, Li Y, Liao Z, Li W, Yu Z, Whalen MJ, Lok J, Dumont AS, et al. (2021). Recombinant Annexin A2 Administration Improves Neurological Outcomes After Traumatic Brain Injury in Mice. Front Pharmacol 12, 708469. 10.3389/fphar.2021.708469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TH, Bohorquez J, Toft-Nielsen J, Ozdamar O, and Porciatti V (2014). Robust mouse pattern electroretinograms derived simultaneously from each eye using a common snout electrode. Invest Ophthalmol Vis Sci 55, 2469–2475. 10.1167/iovs.14-13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chytla A, Gajdzik-Nowak W, Olszewska P, Biernatowska A, Sikorski AF, and Czogalla A (2020). Not Just Another Scaffolding Protein Family: The Multifaceted MPPs. Molecules 25. 10.3390/molecules25214954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BS, Stein-O’Brien GL, Shiau F, Cannon GH, Davis-Marcisak E, Sherman T, Santiago CP, Hoang TV, Rajaii F, James-Esposito RE, et al. (2019). Single-Cell RNA-Seq Analysis of Retinal Development Identifies NFI Factors as Regulating Mitotic Exit and Late-Born Cell Specification. Neuron 102, 1111–1126 e1115. 10.1016/j.neuron.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallacasagrande V, and Hajjar KA (2020). Annexin A2 in Inflammation and Host Defense. Cells 9. 10.3390/cells9061499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas RM, Alam NM, Silver BD, McGill TJ, Tschetter WW, and Prusky GT (2005). Independent visual threshold measurements in the two eyes of freely moving rats and mice using a virtual-reality optokinetic system. Vis Neurosci 22, 677–684. 10.1017/S0952523805225166. [DOI] [PubMed] [Google Scholar]
- Douglas RM, Neve A, Quittenbaum JP, Alam NM, and Prusky GT (2006). Perception of visual motion coherence by rats and mice. Vision research 46, 2842–2847. 10.1016/j.visres.2006.02.025. [DOI] [PubMed] [Google Scholar]
- Duan X, Qiao M, Bei F, Kim IJ, He Z, and Sanes JR (2015). Subtype-specific regeneration of retinal ganglion cells following axotomy: effects of osteopontin and mTOR signaling. Neuron 85, 1244–1256. 10.1016/j.neuron.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Zhang J, Zhuang P, Liu P, Li L, Huang H, Webber HC, Xu Y, Liu L, Dalal R, et al. (2021). Chronic mild and acute severe glaucomatous neurodegeneration derived from silicone oil-induced ocular hypertension. Scientific reports 11, 9052. 10.1038/s41598-021-88690-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel M, Delavoie F, Demais V, Royer C, Bailly Y, Vitale N, Bader MF, and Chasserot-Golaz S (2015). Annexin A2-dependent actin bundling promotes secretory granule docking to the plasma membrane and exocytosis. J Cell Biol 210, 785–800. 10.1083/jcb.201412030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel M, Delavoie F, Royer C, Tahouly T, Gasman S, Bader MF, Vitale N, and Chasserot-Golaz S (2019). Phosphorylation cycling of Annexin A2 Tyr23 is critical for calcium-regulated exocytosis in neuroendocrine cells. Biochimica et biophysica acta. Molecular cell research 1866, 1207–1217. 10.1016/j.bbamcr.2018.12.013. [DOI] [PubMed] [Google Scholar]
- Garway-Heath DF, Crabb DP, Bunce C, Lascaratos G, Amalfitano F, Anand N, Azuara-Blanco A, Bourne RR, Broadway DC, Cunliffe IA, et al. (2015). Latanoprost for open-angle glaucoma (UKGTS): a randomised, multicentre, placebo-controlled trial. Lancet 385, 1295–1304. 10.1016/S0140-6736(14)62111-5. [DOI] [PubMed] [Google Scholar]
- Gerke V, Creutz CE, and Moss SE (2005). Annexins: linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol 6, 449–461. 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- Grenningloh G, Soehrman S, Bondallaz P, Ruchti E, and Cadas H (2004). Role of the microtubule destabilizing proteins SCG10 and stathmin in neuronal growth. J Neurobiol 58, 60–69. 10.1002/neu.10279. [DOI] [PubMed] [Google Scholar]
- Guo L, Normando EM, Nizari S, Lara D, and Cordeiro MF (2010). Tracking longitudinal retinal changes in experimental ocular hypertension using the cSLO and spectral domain-OCT. Invest Ophthalmol Vis Sci 51, 6504–6513. 10.1167/iovs.10-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Jiang H, Gray V, Dedhar S, and Rao Y (2007). Role of the integrin-linked kinase (ILK) in determining neuronal polarity. Developmental biology 306, 457–468. 10.1016/j.ydbio.2007.03.019. [DOI] [PubMed] [Google Scholar]
- Guo X, Snider WD, and Chen B (2016). GSK3beta regulates AKT-induced central nervous system axon regeneration via an eIF2Bepsilon-dependent, mTORC1-independent pathway. eLife 5, e11903. 10.7554/eLife.11903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 e3529. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijl A, Leske MC, Bengtsson B, Hyman L, Bengtsson B, Hussein M, and Early Manifest Glaucoma Trial, G. (2002). Reduction of intraocular pressure and glaucoma progression: results from the Early Manifest Glaucoma Trial. Arch Ophthalmol 120, 1268–1279. 10.1001/archopht.120.10.1268. [DOI] [PubMed] [Google Scholar]
- Hellal F, Hurtado A, Ruschel J, Flynn KC, Laskowski CJ, Umlauf M, Kapitein LC, Strikis D, Lemmon V, Bixby J, et al. (2011). Microtubule stabilization reduces scarring and causes axon regeneration after spinal cord injury. Science 331, 928–931. 10.1126/science.1201148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuero AM, Diez-Revuelta N, and Abad-Rodriguez J (2017). The sugar code in neuronal physiology. Histochem Cell Biol 147, 257–267. 10.1007/s00418-016-1519-3. [DOI] [PubMed] [Google Scholar]
- Hoang T, Wang J, Boyd P, Wang F, Santiago C, Jiang L, Yoo S, Lahne M, Todd LJ, Jia M, et al. (2020). Gene regulatory networks controlling vertebrate retinal regeneration. Science 370. 10.1126/science.abb8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell GR, Libby RT, Jakobs TC, Smith RS, Phalan FC, Barter JW, Barbay JM, Marchant JK, Mahesh N, Porciatti V, et al. (2007). Axons of retinal ganglion cells are insulted in the optic nerve early in DBA/2J glaucoma. J Cell Biol 179, 1523–1537. 10.1083/jcb.200706181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Wu C, Mars WM, and Liu Y (2007). Tissue-type plasminogen activator promotes murine myofibroblast activation through LDL receptor-related protein 1-mediated integrin signaling. J Clin Invest 117, 3821–3832. 10.1172/JCI32301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Park KK, Yang L, Wei X, Yang Q, Cho KS, Thielen P, Lee AH, Cartoni R, Glimcher LH, et al. (2012). Differential effects of unfolded protein response pathways on axon injury-induced death of retinal ganglion cells. Neuron 73, 445–452. 10.1016/j.neuron.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Miao L, Liang F, Liu X, Xu L, Teng X, Wang Q, Ridder WH 3rd, Shindler KS, Sun Y, and Hu Y (2017). Neuroprotection by eIF2alpha-CHOP inhibition and XBP-1 activation in EAE/optic neuritiss. Cell death & disease 8, e2936. 10.1038/cddis.2017.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Miao L, Yang L, Liang F, Wang Q, Zhuang P, Sun Y, and Hu Y (2019). AKT-dependent and -independent pathways mediate PTEN deletion-induced CNS axon regeneration. Cell death & disease 10, 203. 10.1038/s41419-018-1289-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichhpujani P, Jindal A, and Jay Katz L (2009). Silicone oil induced glaucoma: a review. Graefe’s archive for clinical and experimental ophthalmology = Albrecht von Graefes Archiv fur klinische und experimentelle Ophthalmologie 247, 1585–1593. 10.1007/s00417-009-1155-x. [DOI] [PubMed] [Google Scholar]
- Jin D, Liu Y, Sun F, Wang X, Liu X, and He Z (2015). Restoration of skilled locomotion by sprouting corticospinal axons induced by co-deletion of PTEN and SOCS3. Nature communications 6, 8074. 10.1038/ncomms9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, and Salzberg SL (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature biotechnology 37, 907–915. 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann HL, and Gedde SJ (2016). Glaucoma management after vitreoretinal surgeries. Curr Opin Ophthalmol 27, 125–131. 10.1097/ICU.0000000000000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamort AS, Giopanou I, Psallidas I, and Stathopoulos GT (2019). Osteopontin as a Link between Inflammation and Cancer: The Thorax in the Spotlight. Cells 8. 10.3390/cells8080815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landowski LM, Pavez M, Brown LS, Gasperini R, Taylor BV, West AK, and Foa L (2016). Low-density Lipoprotein Receptor-related Proteins in a Novel Mechanism of Axon Guidance and Peripheral Nerve Regeneration. J Biol Chem 291, 1092–1102. 10.1074/jbc.M115.668996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon S, Yin Y, Nguyen J, Irwin N, and Benowitz LI (2000). Lens injury stimulates axon regeneration in the mature rat optic nerve. J Neurosci 20, 4615–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Huang H, Fang F, Liu L, Sun Y, and Hu Y (2020). Longitudinal Morphological and Functional Assessment of RGC Neurodegeneration After Optic Nerve Crush in Mouse. Frontiers in cellular neuroscience 14, 109. 10.3389/fncel.2020.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Wu C, and Hu K (2012). Tissue Plasminogen Activator Activates NF-κB through a Pathway Involving Annexin A2/CD11b and Integrin-Linked Kinase. Journal of the American Society of Nephrology 23, 1329–1338. 10.1681/asn.2011111123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Lu Y, Lee JK, Samara R, Willenberg R, Sears-Kraxberger I, Tedeschi A, Park KK, Jin D, Cai B, et al. (2010). PTEN deletion enhances the regenerative ability of adult corticospinal neurons. Nat Neurosci 13, 1075–1081. 10.1038/nn.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Jiang Y, Chung JY, Li Y, Yu Z, Kim JW, Lok JM, Whalen MJ, and Wang X (2019). Annexin A2 Deficiency Exacerbates Neuroinflammation and Long-Term Neurological Deficits after Traumatic Brain Injury in Mice. Int J Mol Sci 20. 10.3390/ijms20246125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Huang H, Fang F, Liu L, Li L, Feng X, Chen W, Dalal R, Sun Y, and Hu Y (2021). Neuronal NMNAT2 Overexpression Does Not Achieve Significant Neuroprotection in Experimental Autoimmune Encephalomyelitis/Optic Neuritis. Frontiers in cellular neuroscience 15. 10.3389/fncel.2021.754651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Brommer B, Tian X, Krishnan A, Meer M, Wang C, Vera DL, Zeng Q, Yu D, Bonkowski MS, et al. (2020). Reprogramming to recover youthful epigenetic information and restore vision. Nature 588, 124–129. 10.1038/s41586-020-2975-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214. 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L, Yang L, Huang H, Liang F, Ling C, and Hu Y (2016). mTORC1 is necessary but mTORC2 and GSK3beta are inhibitory for AKT3-induced axon regeneration in the central nervous system. eLife 5, e14908. 10.7554/eLife.14908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto AM, Tomlinson MG, Nakatani K, Bolen JB, Roth RA, and Herbst R (2000). The MMAC1 tumor suppressor phosphatase inhibits phospholipase C and integrin-linked kinase activity. Oncogene 19, 200–209. 10.1038/sj.onc.1203288. [DOI] [PubMed] [Google Scholar]
- Nakano N, Ikeda HO, Hangai M, Muraoka Y, Toda Y, Kakizuka A, and Yoshimura N (2011). Longitudinal and simultaneous imaging of retinal ganglion cells and inner retinal layers in a mouse model of glaucoma induced by N-methyl-D-aspartate. Invest Ophthalmol Vis Sci 52, 8754–8762. 10.1167/iovs.10-6654. [DOI] [PubMed] [Google Scholar]
- Nawabi H, Belin S, Cartoni R, Williams PR, Wang C, Latremoliere A, Wang X, Zhu J, Taub DG, Fu X, et al. (2015). Doublecortin-Like Kinases Promote Neuronal Survival and Induce Growth Cone Reformation via Distinct Mechanisms. Neuron 88, 704–719. 10.1016/j.neuron.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickells RW, Howell GR, Soto I, and John SW (2012). Under pressure: cellular and molecular responses during glaucoma, a common neurodegeneration with axonopathy. Annu Rev Neurosci 35, 153–179. 10.1146/annurev.neuro.051508.135728. [DOI] [PubMed] [Google Scholar]
- Nio-Kobayashi J, and Itabashi T (2021). Galectins and Their Ligand Glycoconjugates in the Central Nervous System Under Physiological and Pathological Conditions. Front Neuroanat 15, 767330. 10.3389/fnana.2021.767330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan KJ, Ma K, Guo H, Wang C, Sun F, Han SB, Kim H, Wong JK, Charron J, Zou H, et al. (2014). B-RAF kinase drives developmental axon growth and promotes axon regeneration in the injured mature CNS. J Exp Med 211, 801–814. 10.1084/jem.20131780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa N, Fujitani K, Tokunaga E, Furuya S, and Inokuchi K (2007). The microtubule destabilizer stathmin mediates the development of dendritic arbors in neuronal cells. Journal of cell science 120, 1447–1456. 10.1242/jcs.001461. [DOI] [PubMed] [Google Scholar]
- Pan L, North HA, Sahni V, Jeong SJ, McGuire TL, Berns EJ, Stupp SI, and Kessler JA (2014). beta1-Integrin and integrin linked kinase regulate astrocytic differentiation of neural stem cells. PLoS One 9, e104335. 10.1371/journal.pone.0104335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, Xu B, Connolly L, Kramvis I, Sahin M, and He Z (2008). Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science 322, 963–966. 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YR, Shekhar K, Yan W, Herrmann D, Sappington A, Bryman GS, van Zyl T, Do MTH, Regev A, and Sanes JR (2019). Molecular Classification and Comparative Taxonomics of Foveal and Peripheral Cells in Primate Retina. Cell 176, 1222–1237 e1222. 10.1016/j.cell.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persad S, Attwell S, Gray V, Delcommenne M, Troussard A, Sanghera J, and Dedhar S (2000). Inhibition of integrin-linked kinase (ILK) suppresses activation of protein kinase B/Akt and induces cell cycle arrest and apoptosis of PTEN-mutant prostate cancer cells. Proc Natl Acad Sci U S A 97, 3207–3212. 10.1073/pnas.060579697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, and Salzberg SL (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nature protocols 11, 1650–1667. 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SL, Li Y, Sun CJ, Wong KA, Leung KS, de Lima S, Hanovice NJ, Yuki K, Stevens B, and Benowitz LI (2021). Retinal Ganglion Cell Axon Regeneration Requires Complement and Myeloid Cell Activity within the Optic Nerve. J Neurosci 41, 8508–8531. 10.1523/JNEUROSCI.0555-21.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S (2019). Full-Length Single-Cell RNA Sequencing with Smart-seq2. Methods Mol Biol 1979, 25–44. 10.1007/978-1-4939-9240-9_3. [DOI] [PubMed] [Google Scholar]
- Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, and Sandberg R (2013). Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nature methods 10, 1096–1098. 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, and Sandberg R (2014). Full-length RNA-seq from single cells using Smart-seq2. Nature protocols 9, 171–181. 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Podkalicka J, Biernatowska A, Olszewska P, Tabaczar S, and Sikorski AF (2018). The microdomain-organizing protein MPP1 is required for insulin-stimulated activation of H-Ras. Oncotarget 9, 18410–18421. 10.18632/oncotarget.24847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porciatti V (2015). Electrophysiological assessment of retinal ganglion cell function. Experimental eye research 141, 164170. 10.1016/j.exer.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusky GT, Alam NM, Beekman S, and Douglas RM (2004). Rapid quantification of adult and developing mouse spatial vision using a virtual optomotor system. Invest Ophthalmol Vis Sci 45, 4611–4616. 10.1167/iovs.04-0541. [DOI] [PubMed] [Google Scholar]
- Quigley HA, Nickells RW, Kerrigan LA, Pease ME, Thibault DJ, and Zack DJ (1995). Retinal ganglion cell death in experimental glaucoma and after axotomy occurs by apoptosis. Invest Ophthalmol Vis Sci 36, 774–786. [PubMed] [Google Scholar]
- Quinta HR, Pasquini JM, Rabinovich GA, and Pasquini LA (2014). Glycan-dependent binding of galectin-1 to neuropilin-1 promotes axonal regeneration after spinal cord injury. Cell Death Differ 21, 941–955. 10.1038/cdd.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinta HR, Wilson C, Blidner AG, Gonzalez-Billault C, Pasquini LA, Rabinovich GA, and Pasquini JM (2016). Ligand-mediated Galectin-1 endocytosis prevents intraneural H2O2 production promoting F-actin dynamics reactivation and axonal re-growth. Exp Neurol 283, 165–178. 10.1016/j.expneurol.2016.06.009. [DOI] [PubMed] [Google Scholar]
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, and Tessier-Lavigne M (2014). iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910. 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Ribeiro M, Levay K, Yon B, Ayupe AC, Salgueiro Y, and Park KK (2020). Neural Cadherin Plays Distinct Roles for Neuronal Survival and Axon Growth under Different Regenerative Conditions. eNeuro 7. 10.1523/ENEURO.0325-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi M, Imaizumi Y, Shingo T, Tada H, Hayama K, Yamada O, Morishita T, Kadoya T, Uchiyama N, Shimazaki T, et al. (2010). Regulation of adult neural progenitor cells by Galectin-1/beta1 Integrin interaction. J Neurochem 113, 1516–1524. 10.1111/j.1471-4159.2010.06712.x. [DOI] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. (2016). Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell 166, 1308–1323 e1330. 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Ori-McKenney KM, Zheng Y, Han C, Jan LY, and Jan YN (2012). Regeneration of Drosophila sensory neuron axons and dendrites is regulated by the Akt pathway involving Pten and microRNA bantam. Genes Dev 26, 1612–1625. 10.1101/gad.193243.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz JD, Bourne RRA, Briant PS, Flaxman SR, Taylor HRB, Jonas JB, Abdoli AA, Abrha WA, Abualhasan A, Abu-Gharbieh EG, et al. (2021). Causes of blindness and vision impairment in 2020 and trends over 30 years, and prevalence of avoidable blindness in relation to VISION 2020: the Right to Sight: an analysis for the Global Burden of Disease Study. The Lancet Global Health 9, e144–e160. 10.1016/s2214-109x(20)30489-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell C, Burgoyne CF, Tamm ER, Ethier CR, Lasker, I.I.o.A., and Glaucomatous Neurodegeneration, P. (2017). Biomechanical aspects of axonal damage in glaucoma: A brief review. Experimental eye research 157, 13–19. 10.1016/j.exer.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A, and Regev A (2017). Scaling single-cell genomics from phenomenology to mechanism. Nature 541, 331–338. 10.1038/nature21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham YC, Li X, Wong TY, Quigley HA, Aung T, and Cheng CY (2014). Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology 121, 2081–2090. 10.1016/j.ophtha.2014.05.013. [DOI] [PubMed] [Google Scholar]
- Tran NM, Shekhar K, Whitney IE, Jacobi A, Benhar I, Hong G, Yan W, Adiconis X, Arnold ME, Lee JM, et al. (2019). Single-Cell Profiles of Retinal Ganglion Cells Differing in Resilience to Injury Reveal Neuroprotective Genes. Neuron. 10.1016/j.neuron.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbrecht-Jenck E, Demais V, Calco V, Bailly Y, Bader MF, and Chasserot-Golaz S (2010). S100A10-mediated translocation of annexin-A2 to SNARE proteins in adrenergic chromaffin cells undergoing exocytosis. Traffic 11, 958–971. 10.1111/j.1600-0854.2010.01065.x. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhuang P, Huang H, Li L, Liu L, Webber HC, Dalal R, Siew L, Fligor CM, Chang KC, et al. (2020a). Mouse gamma-Synuclein Promoter-Mediated Gene Expression and Editing in Mammalian Retinal Ganglion Cells. J Neurosci 40, 3896–3914. 10.1523/JNEUROSCI.0102-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XW, Yang SG, Zhang C, Hu MW, Qian J, Ma JJ, Zhang Y, Yang BB, Weng YL, Ming GL, et al. (2020b). Knocking Out Non-muscle Myosin II in Retinal Ganglion Cells Promotes Long-Distance Optic Nerve Regeneration. Cell reports 31, 107537. 10.1016/j.celrep.2020.107537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PR, Benowitz LI, Goldberg JL, and He Z (2020). Axon Regeneration in the Mammalian Optic Nerve. Annual review of vision science 6, 195–213. 10.1146/annurev-vision-022720-094953. [DOI] [PubMed] [Google Scholar]
- Wormald R, Virgili G, and Azuara-Blanco A (2020). Systematic reviews and randomised controlled trials on open angle glaucoma. Eye (Lond) 34, 161–167. 10.1038/s41433-019-0687-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Cen LP, Li Y, Gilbert HY, Strelko O, Berlinicke C, Stavarache MA, Ma M, Wang Y, Cui Q, et al. (2022). Monocyte-derived SDF1 supports optic nerve regeneration and alters retinal ganglion cells’ response to Pten deletion. Proc Natl Acad Sci U S A 119, e2113751119. 10.1073/pnas.2113751119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Yin Y, and Benowitz L (2021). Chemokine CCL5 promotes robust optic nerve regeneration and mediates many of the effects of CNTF gene therapy. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2017282118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Peng YR, van Zyl T, Regev A, Shekhar K, Juric D, and Sanes JR (2020). Cell Atlas of The Human Fovea and Peripheral Retina. Scientific reports 10, 9802. 10.1038/s41598-020-66092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Wang X, Wang J, Wang X, Chen W, Lu N, Siniossoglou S, Yao Z, and Liu K (2020a). Rewiring Neuronal Glycerolipid Metabolism Determines the Extent of Axon Regeneration. Neuron 105, 276–292 e275. 10.1016/j.neuron.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Li S, Miao L, Huang H, Liang F, Teng X, Xu L, Wang Q, Xiao W, Ridder WH 3rd, et al. (2016). Rescue of Glaucomatous Neurodegeneration by Differentially Modulating Neuronal Endoplasmic Reticulum Stress Molecules. J Neurosci 36, 5891–5903. 10.1523/JNEUROSCI.3709-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Miao L, Liang F, Huang H, Teng X, Li S, Nuriddinov J, Selzer ME, and Hu Y (2014). The mTORC1 effectors S6K1 and 4E-BP play different roles in CNS axon regeneration. Nature communications 5, 5416. 10.1038/ncomms6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SG, Li CP, Peng XQ, Teng ZQ, Liu CM, and Zhou FQ (2020b). Strategies to Promote Long-Distance Optic Nerve Regeneration. Frontiers in cellular neuroscience 14, 119. 10.3389/fncel.2020.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon C, Van Niekerk EA, Henry K, Ishikawa T, Orita S, Tuszynski MH, and Campana WM (2013). Low-density lipoprotein receptor-related protein 1 (LRP1)-dependent cell signaling promotes axonal regeneration. J Biol Chem 288, 26557–26568. 10.1074/jbc.M113.478552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, and He QY (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhou T, Chen Z, Yan M, Li B, Lv H, Wang C, Xiang S, Shi L, Zhu Y, and Ai D (2020). Coupling of Integrin alpha5 to Annexin A2 by Flow Drives Endothelial Activation. Circulation research 127, 1074–1090. 10.1161/CIRCRESAHA.120.316857. [DOI] [PubMed] [Google Scholar]
- Zhang J, Fang F, Li L, Huang H, Webber HC, Sun Y, Mahajan VB, and Hu Y (2019a). A Reversible Silicon Oil-Induced Ocular Hypertension Model in Mice. Journal of visualized experiments : JoVE 153. 10.3791/60409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li L, Huang H, Fang F, Webber HC, Zhuang P, Liu L, Dalal R, Tang PH, Mahajan VB, et al. (2019b). Silicone oil-induced ocular hypertension and glaucomatous neurodegeneration in mouse. eLife 8. 10.7554/eLife.45881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yang D, Huang H, Sun Y, and Hu Y (2018). Coordination of Necessary and Permissive Signals by PTEN Inhibition for CNS Axon Regeneration. Front Neurosci 12, 558. 10.3389/fnins.2018.00558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Williams PR, Jacobi A, Wang C, Goel A, Hirano AA, Brecha NC, Kerschensteiner D, and He Z (2019c). Elevating Growth Factor Responsiveness and Axon Regeneration by Modulating Presynaptic Inputs. Neuron 103, 39–51 e35. 10.1016/j.neuron.2019.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, and Sabatini DM (2011). mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12, 21–35. 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Cluster Specific Genes, Related to Figure 2.
Table S2 Upregulated DEGs in regRGCs, Related to Figure 3.
Table S3 Downregulated DEGs in regRGCs, Related to Figure 3.
Table S4 Primers for PCT of mouse tissue cDNA, Related to STAR Methods section.
Data Availability Statement
All custom scripts have been made available at https://github.com/HuLab-Code/. Any additional information is available from the lead contact upon request.


