Summary
Accurate modeling of the heart electrophysiology to predict arrhythmia susceptibility remains a challenge. Current electrophysiological analyses are hypothesis-driven models drawing conclusions from changes in a small subset of electrophysiological parameters because of the difficulty of handling and understanding large datasets. Thus, we develop a framework to train machine learning classifiers to distinguish between healthy and arrhythmic cardiomyocytes using their calcium cycling properties. By training machine learning classifiers on a generated dataset containing a total of 3,003 healthy derived cardiomyocytes and their various arrhythmic states, the multi-class models achieved >90% accuracy in predicting arrhythmia presence and type. We also demonstrate that a binary classifier trained to distinguish cardiotoxic arrhythmia from healthy electrophysiology could determine the key biological changes associated with that specific arrhythmia. Therefore, machine learning algorithms can be used to characterize underlying arrhythmic patterns in samples to improve in vitro preclinical models and complement current in vivo systems.
Keywords: pluripotent stem cell-derived cardiomyocytes, disease modeling, arrhythmias, machine learning
Highlights
-
•
Calcium reporter enabled analysis of hPSC-derived CM electrophysiology
-
•
Calcium electrophysiology can be decomposed to train machine learning classifiers
-
•
Multi-class classifier distinguishes different forms of arrhythmias
-
•
Binary-class classifiers identify key electrophysiological changes a priori
Current electrophysiological models rely on probing for parameters that are already known to be correlated with arrhythmias. However, not all arrhythmias manifest via the same expected electrophysiological change, and these conditions may blindside electrophysiological screens. Pang and colleagues devised a machine learning platform using calcium electrophysiology and show that it reliably determines changes in electrophysiological parameters affected by arrhythmias.
Introduction
Prediction and prevention of arrhythmic events remains a global health concern. Despite improvements in risk assessment and management of cardiovascular events, manifestations of sudden cardiac death still account for 15%–20% of all deaths (Hayashi et al., 2015). There is an ongoing search for improved modeling platforms to identify aberrant electrophysiology and arrhythmia risk in healthy individuals exhibiting no heart conditions (Hayashi et al., 2015). In addition, improvements in the methodology to screen and identify cardiotoxicity in drugs is needed, as almost 10% of drugs within the past four decades were recalled from the clinical market because of cardiotoxicity not being detected in preclinical studies (Varga et al., 2015). The emergence of somatic cell reprogramming is paving the way to personalized medicine and disease-specific therapeutics (Oh et al., 2012). Assessment of cardiovascular health, sudden cardiac death risk, and cardiotoxicity screens can be done using human pluripotent stem cell-derived cardiomyocytes (hPSC-CMs) as opposed to limited primary cardiomyocyte (CM) tissues (Sallam et al., 2015). As such, adapting hPSC-CM systems to model electrophysiology will complement current clinical assessments in profiling risks among individuals in the population.
Current electrophysiological modeling systems are inherently mechanistic models whereby the causality between input and output parameters is studied in a hypothesis-driven manner (Baker et al., 2018). Genetic variations, cardiovascular structural defects, and chemical/drug exposure serve as the input parameters assessed for potential electrophysiological changes (Ch’en et al., 1998; Clancy and Rudy, 1999; Yap and Camm, 2003). Researchers analyze expected output parameters such as qualitative changes in action potential (AP) waveform, quantitative lengthening of AP duration, and repolarization duration, which are predictive of arrhythmia because of their association with QT interval prolongation and torsades de pointes (TdP) incidence to characterize electrophysiological changes (Fermini et al., 2015; Harris et al., 2013; Ma et al., 2011; Yap and Camm, 2003). However, there are limitations to these mechanistic models despite them being the fundamental workhorse of biological research. Preclinical assessment of drug cardiotoxicity, primarily via assessment of hERG channels for extended AP and repolarization durations, has been shown to be insufficient in capturing all forms of arrhythmogenesis (Fermini et al., 2015). Therefore, unbiased, a priori processing of electrophysiological data would help serve as a hypothesis-generating platform to identify unknown electrophysiological changes.
With the ease of generating large amounts of data, there has been a paradigm shift toward machine learning (ML) models (Baker et al., 2018; Kusumoto and Yuasa, 2019). Researchers carrying out hypothesis-driven research are limited to probing empirically accepted subsets of input parameters, but ML models can take advantage of unbiased artificial intelligence to identify underlying patterns and generating correlations from broadly descriptive input parameters. We propose that a ML platform using electrophysiological data from in vitro cultures can be a complementary screening method to current clinical assessments to profile arrhythmic risk in individuals. In addition, the unbiased ML models can identify the key electrophysiological alterations due to arrhythmic treatments for further mechanistic analysis.
Three established methods exist to generate electrophysiological data suitable for ML model training: patch clamping, multi-electrode arrays (MEAs), and fluorescence reporter tracking of Ca2+ transients (Fermini et al., 2015; Jiang et al., 2018; Ma et al., 2011). Differences in the form of electrophysiological data generated have led us to prefer Ca2+ transient analysis over the other two approaches. Although patch clamping is widely considered as the gold standard for ion channel research, its relatively low throughput and requirement for highly trained personnel limits its ability to generate the large, high-quality datasets. Automated patch-clamp systems could alleviate both limitations and enable higher throughput data generation (Ma et al., 2011). However, these high-end systems are not commonplace and may not be readily available for use by researchers. The MEA system is also suitable to build a high-throughput analysis platform on, with multiple electrodes probing many cells concurrently. However, we experienced that individual electrodes would often be in contact with multiple CMs, resulting in confounding data when asynchronously contracting arrhythmic CMs were analyzed. This limitation can be controlled for when probing for single CMs using Ca2+ transient analysis. Last, mature CMs used for both MEA and patch-clamping systems must be dissociated and replated onto suitable plates or slides for analysis. This replating process is not physiological and would likely cause unintended restructuring of cytoskeletal elements and the development of arrhythmic properties. Fluorescent analysis of Ca2+ transients does not require replating and thus is not subject to this confounding factor.
Conversely, both patch clamping and MEA are more versatile in generating electrophysiological data compared with the limited nature of Ca2+ transient analysis, probing only for perturbations of Ca2+ handling. It is plausible that electrophysiological data for the various ion channel types and field potential could generate better models. However, as the cycling of intracellular Ca2+ ions is the primary regulator enabling the rhythmic cardiac cycles, probing the Ca2+ cycling capabilities alone could suffice as a proxy for electrophysiological health (Bers, 2001). Tracking of the Ca2+ transients in CMs can be done non-invasively through fluorescence imaging systems readily available in biological labs.
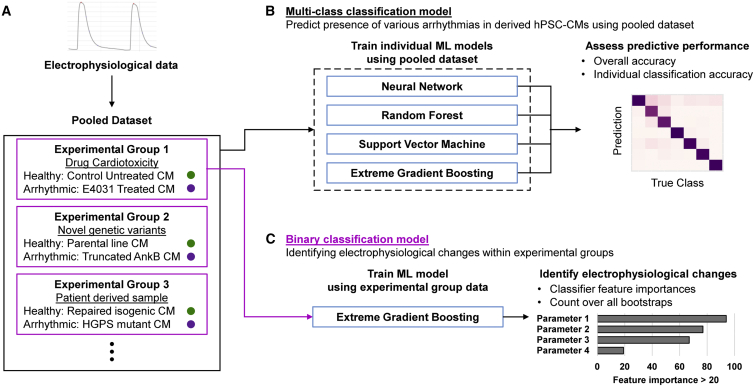
In this study, hPSC-CMs expressing the Ca2+ reporter GCaMP6s were derived using an established protocol to generate electrophysiology data (Lian et al., 2013). We developed a Ca2+ reporter image analysis pipeline that tracks the Ca2+ cycling of CMs at the single-cell level, followed by decomposition of the recorded Ca2+ transient waveforms into 16 Ca2+ handling parameters to be used as inputs to train ML classifiers (Figure 1A). Three experimental groups containing healthy CMs and CMs of arrhythmic subtypes were pooled into a cohort (n = 3,003), and their Ca2+ handling parameters were used as inputs to train four multi-class classifiers to distinguish between healthy CMs and the various arrhythmic subtypes (Figure 1B; Table 1). To identify key features of arrhythmogenesis for specific arrhythmic subtypes, we trained binary classifiers using data within experimental groups, between the healthy CMs and arrhythmic CM subtypes, to determine if the expected biological parameters were used to determine arrhythmogenesis (Figure 1C). The algorithms used to construct this are made accessible to enable other users to similarly dissect and analyze their datasets.
Figure 1.
Framework to train machine learning algorithms to predict arrhythmogenesis and identify electrophysiological changes
(A) Single-cell CM Ca2+ cycling is tracked and processed to obtain the electrophysiological data contained within experimental groups containing specific arrhythmic forms.
(B) For a multi-class arrhythmia prediction model, the pooled dataset is split into multiple train-test splits to train 4 different ML models over 100 bootstraps to identify the best consistently performing ML classifier.
(C) Binary classification models are trained using individual experiment groups containing a healthy CM class and the arrhythmic CM subclass of interest. Assessing the feature importance metric of the trained classifiers can deduce the biological implications of the arrhythmic subclass studied to enable targeted therapy.
Table 1.
Classifications of healthy and arrhythmic CMs from the three independent experimental groups generated
| Experimental groups | Sample name/treatment | Classification | Sample size |
|---|---|---|---|
| Drug-induced arrhythmia | BJ-CM/DMSO control | Healthy | 178 |
| BJ-CM/10 μM E4031 | 10 μM E4031: Arrhythmic | 173 | |
| BJ-CM/40 μM E4031 | 40 μM E4031: Arrhythmic | 169 | |
| Ankyrin-B syndrome | H7-CM | Healthy | 596 |
| H7-CM/ANK frameshift 1 | Mut A1: Arrhythmic | 409 | |
| H7-CM/ANK frameshift 2 | Mut A2: Arrhythmic | 642 | |
| H7-CM/ANK frameshift 3 | Mut A3: Arrhythmic | 274 | |
| Progeria syndrome | HGPS-CM | Mut HGPS: Arrhythmic | 120 |
| Isogenic HGPS-CM/LMNA mutation repaired | Healthy | 211 | |
| BJ-CM | Healthy | 231 |
Results
Ca2+ transients of CMs can be tracked at the single-cell level for arrhythmia risk assessment by stable expression of Ca2+ reporter GCaMP6s
Although Ca2+ transients can be tracked with Ca2+ fluorophores, treatment with the fluorophores is toxic and must be done as an endpoint experiment. Thus, we opted to stably express the genetically encoded Ca2+ indicator GCaMP6s, which allows tracking of Ca2+ cycling at single-cell resolution in CMs over multiple time points. Thus, GCaMP6s was inserted into and expressed in the hPSC lines (H7 ESCs, BJ induced pluripotent stem cells [iPSCs], Hutchinson-Gilford progeria syndrome [HGPS] iPSCs) used in this study using CRISPR (Figures S1A and S1B) (Jiang et al., 2018). Fluorescence was used as a preliminary screen for hPSC knockin, with successful GCaMP6s insertions resulting in a faint detectable green fluorescence in hPSCs (Figure S1C). PCR genotyping was used to screen for homozygous insertions (Figure S1D). Several studies have highlighted incidences of deleterious, unwanted on-target insertions and deletions from CRISPR gene editing that escape conventional PCR and Sanger sequence analysis (Shin et al., 2017). Therefore, the lines generated for this work were validated to be free of unwanted mutations by first probing for fragments of the donor plasmid (Figures S1E and S1F). In addition, quantitative genotyping PCR (qgPCR) was also carried out to confirm that the lines were indeed homozygous and not false positives (Figure S1G) (Simkin et al., 2022).
After clonal isolation of hPSCGCaMP6s+/+ lines, pluripotency maintenance was confirmed by qRT-PCR for pluripotency genes (Figure S2A). As there have been reports on the cell toxicity of GCaMP, karyotyping was also carried out for hPSCGCaMP6s+/+ lines (Figure S2B) (Tallini et al., 2006). To ensure that the CMV-driven GCaMP6s does not undergo gene silencing over long-term cultures, the hPSCGCaMP6s+/+ lines were probed for GCaMP6s gene expression levels (Brooks et al., 2004). No significant changes in gene expression levels were found between early (passage <10) and late (passage ∼30) hPSCGCaMP6s+/+ lines (Figure S2C). In addition, quantification of the fluorescent intensity of individual hPSC colonies in Ca2+ containing media also showed no significant changes between early and late passages (Figure S2D). In this work, to prevent any confounding effects of CMV promoter silencing, hPSCGCaMP6s+/+ lines were differentiated within 20 passages of clonal isolation.
Directed differentiation toward the CM lineage was achieved using an established protocol (Figure S2E) (Lian et al., 2013). Metabolic selection and maturation was carried out using a previously established protocol, via treatment with glucose-free, fatty acid supplemented media, resulting in a >90% pure population of CMs (Figure S2F) (Correia et al., 2017). Fluorescent videos confirmed visual tracking of CM contraction cycles (Figure 2A; Videos S1 and S2). Individual CMs were identified as regions of interest (ROIs), and their fluorescent intensity representing their Ca2+ cycling was quantified over the duration of the recordings (Figure 2B). All CM Ca2+ fluorescent intensity over time data generated in this study were recorded in this manner for downstream analysis.
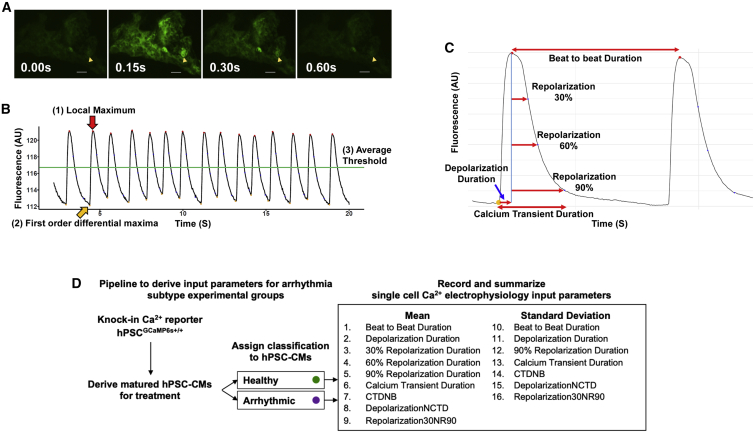
Figure 2.
Functional validation of GCaMP6s Ca2+ reporter by breaking down of Ca2+ cycling readout into biological parameters
(A) Single frame of a video tracking BJ-CM Ca2+ cycling. The arrow marks a single CM ROI tracked. Scale bar, 50 μm.
(B) Fluorescence over time plot of a single CM. Identification of fluorescence peaks such as (1) was automated using the three criteria described.
(C) Annotation of individual Ca2+ transient parameters. Each parameter is computed for every detected Ca2+ transient peak and summarized across the entire Ca2+ cycling readout from a single CM video recording.
(D) Flowchart highlighting the process to generate the electrophysiological data representing the healthy and arrhythmic Ca2+ waveforms in a singular experimental group. Each CM is represented by either healthy or an arrhythmic subtype classification and 16 Ca2+ handling parameters.
Scale bar, 50μm
Scale bar, 250μm
Input parameters can be derived by decomposing Ca2+ fluorescent intensity over time data from cardiomyocytes at single-cell resolution
After video analysis of each individual CM as an ROI, the fluorescent intensity over time data generated was processed to summarize the Ca2+ cycles into parameters that describe the CM electrophysiology. The Ca2+ signal of each CM was processed frame by frame to identify fluorescent peaks corresponding to systole. Peaks are algorithmically identified on the basis of three criteria: (1) the absolute fluorescent intensity at the peak is a local maximum; (2) there is a time point prior to the peak with a first-order differential above a threshold, corresponding to the start of a single depolarization; and (3) the absolute fluorescent intensity at that peak is above a specified threshold, to prevent minor fluctuations from fluorescent minima from being detected as peaks (Figure 2C). These Ca2+ peaks detected represent individual Ca2+ transients.
The following parameters regarding each individual Ca2+ transient were computed using information of the depolarization initiation point corresponding to the initiation of systole, the Ca2+ peak corresponding to the maximum contraction point of systole, and the corresponding fluorescent minima corresponding to diastole (Figure 2C; Table S1). These parameters are analogous to that of AP duration and field potentiation duration changes in patch clamping or multi-electrode array models, respectively, used in preclinical safety assays (Dempsey et al., 2016; Harris et al., 2013):
-
1.
Beat-to-beat duration, the time between one Ca2+ transient peak and the next.
-
2.
Depolarization duration, the time between the first-order derivative maximum and the fluorescent peak.
-
3–5.
Repolarization durations 30%, 60%, and 90%, the times between the fluorescent peak and 30%, 60%, and 90% reduction of fluorescence intensity.
-
6.
Calcium transient duration (CTD), the time between the first-order derivative maximum and the 90% repolarization time point.
In addition to these 6 absolute variables, the following normalizations of some of these absolute variables into compound variables were performed to further describe each Ca2+ waveform (Table S2):
-
7.
CTD normalized to beat-to-beat duration (CTDNB).
-
8.
Depolarization duration normalized to CTD (DepolarizationNCTD).
-
9.
Repolarization 30% normalized to repolarization 90% (Repolarization30NR90).
These 9 parameters were computed across all Ca2+ transients in each single-cell recording. The mean of these 9 parameters together with the SDs of 7 of these parameters were computed to obtain 16 parameters in total to summarize the Ca2+ handling properties of each individual CM. The SDs of repolarization duration 30% and repolarization duration 60% were excluded, as these parameters are highly similar to the SD of the included repolarization duration 90%. Theorized molecular explanations pertaining to the means and SDs of these defined parameters are described at length in Tables S3 and S4. Figure 2D details the process of obtaining electrophysiological parameters to be used as ML inputs for a single experimental group containing both healthy and arrhythmic subtype CMs.
Generation of three experimental groups containing healthy and arrhythmic CMs covering various arrhythmic forms
Studying a particular arrhythmic subtype requires a comparison between healthy CMs and their arrhythmic equivalent CMs. Building a ML risk prediction platform by pooling independent experimental groups in a modular manner will facilitate studying other forms of arrhythmia from external datasets. Thus, three independent experimental groups composed of healthy CMs and their equivalent arrhythmic CMs were generated for this proof-of-concept work. These three arrhythmogenic sources were all expected to confer a bradycardic effect on the CMs, albeit with a different underlying mechanism. Ca2+ handling parameters of CMs from each experimental group were obtained by decomposing their Ca2+ transients into the 16 parameters described above and used as ML inputs (Data S1).
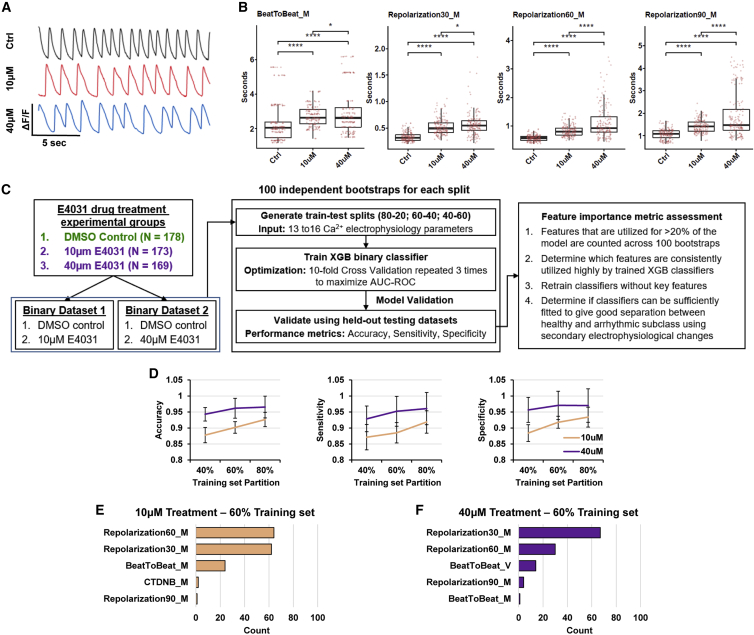
The first experimental group explores effects of drug-induced arrhythmogenicity using E4031, a known cardiotoxic agent that blocks the IKr repolarization current previously used to model long-QT syndrome (Harris et al., 2013). Previously studied as a possible anti-arrhythmic agent, E4031 exerts a slowdown of the cardiac contraction cycle and can be used to prevent tachycardias (Adaniya and Hiraoka, 1990). Healthy BJ iPSC-derived CMs (BJ-CMs) were treated with two concentrations of E4031 (10 μM E4031, n = 173; 40 μM E4031, n = 169) for 2 h and compared with DMSO control (Ctrl; n = 178) treatment (Data S2). Two concentrations of E4031 were used to demonstrate dose effects of the drug-induced arrhythmogenesis.
The second experimental group explores the arrhythmogenic effect of mutation at the ANK2 gene locus encoding for Ankyrin-B (AnkB), generated in vitro via CRISPR-directed mutagenesis. AnkB plays a crucial role in localizing and stabilizing a range of membrane ion channels necessary for Ca2+ cycling in human CMs and is implicated in Ankyrin-B syndrome, which presents with a spectrum of rhythmic irregularities including alternating bradycardia and tachycardia (Cunha and Mohler, 2006; Le Scouarnec et al., 2008). The Cas9 was targeted to exon 40 coding for the death domain, which has mutations linked with abnormal cardiac function (Ichikawa et al., 2016) (Figure S3A). Three biallelic frameshift mutants with truncated AnkB expression were generated, henceforth termed Mut A1, Mut A2, and Mut A3 (Figures S3B–S3D; Table S5). On the basis of flow cytometry, the Mut A1 and Mut A2 lines achieved similar differentiation efficiency compared with the parental H7 embryonic stem cell (ESC) line, while the A3 line exhibited poorer differentiation efficiency (Figure S3E). Western blot confirmed that AnkB expression is lost in A1 and A3 CMs but expressed at a reduced level for A2 CMs (Figure S3F), and all three mutant lines exhibited significant reduction in cardiomyocyte marker TNNT2 expression (Figure S3G). The Ca2+ handling parameters of these three AnkB mutant lines (Mut A1, n = 409; Mut A2, n = 642; Mut A3, n = 274) were compared with the parental H7 ESC-derived CMs (H7-CMs; n = 596) (Data S3).
The third experimental group explores electrophysiological changes present in patients afflicted with congenital Hutchinson-Gilford progeria syndrome. HGPS most commonly occurs because of a single base pair substitution (C1824T, p.G608G) within exon 11 of the lamin A/C gene, LMNA, creating a cryptic splice site leading to the accumulation of a truncated protein termed Progerin. HGPS results in premature death occurring primarily in the second decade of life because of cardiovascular complications (Hennekam, 2006). As HGPS is an ultrarare condition, extensive clinical research is lacking, but cardiac abnormalities including bradycardia and PQ interval/QRS complex prolongation likely contributing to premature death have been demonstrated in progeroid mice (Rivera-Torres et al., 2016). Previously, an iPSC line together with its corrected isogenic control was reprogrammed from a HGPS patient fibroblast carrying the heterozygous point mutation (Zhang et al., 2011) (Figure S4A). Western blot of derived CMs validated the absence of Progerin in the isogenic line (Figure S4B). The Ca2+ handling parameters of both the mutant (Mut HGPS; n = 120) and isogenic (Iso HGPS; n = 211) iPSC-CMs were assessed and compared with a healthy BJ CM control (BJ-CMs; n = 231) cultured together with the HGPS lines (Data S4). Several electrophysiological parameters of the Iso HGPS line improved and were comparable with the healthy BJ-CM control, particularly the mean and variation of both beat-to-beat duration and the CTDNB ratio, a parameter indicative of the refractory period of the CMs (Figure S4C).
Healthy and arrhythmic CMs are not linearly distinguishable on the basis of their electrophysiological parameters
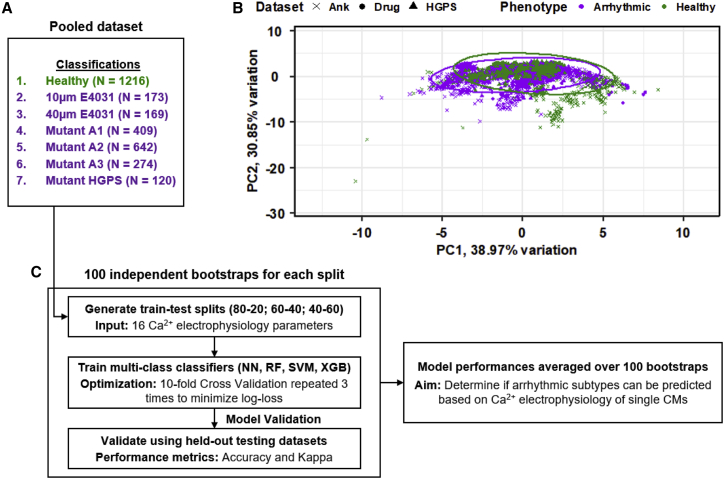
To highlight the basis for applying a ML approach to distinguish between healthy and arrhythmic CMs, all three experimental groups were pooled together into a combined dataset representing healthy cells of different origins, as well as CMs experiencing various forms of arrhythmias (Figure 3A). In the pooled dataset, all the healthy CM lines (Ctrl BJ-CMs, H7-CMs, Iso HGPS, and BJ-CMs) were classified as “Healthy,” while the arrhythmic CMs maintain their arrhythmic subtype classification (Table 1). An unsupervised linear transformation of the electrophysiological parameters defining the CMs using principal-component analysis (PCA) was unable to clearly separate the healthy CMs from arrhythmic classes from each dataset (Figure 3B). Scree plot analysis of the PCA shows that 90% of the variation present in the parameters can be explained using 5 principal components (PCs), but a pairwise relationship plot revealed no obvious distinction between healthy CMs from arrhythmic CMs within these 5 PCs (Figure S5). Therefore, as CMs in a healthy state or an arrhythmic state could not be linearly separated on the basis of their electrophysiological parameters, a more complex, computational approach is warranted.
Figure 3.
Healthy and arrhythmic CMs could not be separated on a linear principal-component analysis
(A) Pooled dataset containing the healthy classification and the six different arrhythmic subclasses from the three experimental groups used in this study.
(B) First two PCs depicting the healthy and arrhythmic CMs and their associated datasets, accounting for 69.8% of variation present. Colors correspond to the arrhythmic/healthy classification. Symbols correspond to the associated datasets.
(C) Overall workflow to identify the most consistent ML classifiers trained on the multi-class pooled dataset and the optimal train-test split to prevent overfitting.
Machine learning classifiers can accurately classify various arrhythmogenic CMs by training on the multi-class pooled dataset
To improve on the separation of healthy CMs from the arrhythmic CMs, four ML algorithms were adapted as multi-class classifiers: neural network (NN), random forest (RF), support vector machine radial kernel (SVM), and extreme gradient boosting (XGB). These four classifiers were trained in parallel using 16 different parameters previously defined (Tables S3 and S4).
The pooled dataset depicted in Table 1, with the 7 classifications (Healthy, 10 μM E4031, 40 μM E4031, Mut A1, Mut A2, Mut A3, and Mut HGPS) were partitioned into train-test splits. Three different train-test ratios were tested (80:20, 60:40, and 40:60), and 100 independent bootstraps were analyzed for each ratio. The performance of the optimized model was assessed using the held-out testing dataset within each bootstrap by computing the accuracy and kappa metrics. Two classification methods were tested, an individualized prediction strategy and a majority voting prediction strategy. For the individual prediction strategy, if the predicted classification of a single CM electrophysiology matches that of its actual classification, the prediction is considered accurate. For the majority voting prediction strategy, all CMs recorded within the same ROI were considered to be from the same individual, and the most common predicted classification across the CMs was determined as the majority. The majority prediction is then used in place of all other predictions of the CMs within the ROI. Figure 3C summarizes the flow of the data analysis, from generation of each independent bootstrap split to the overall analysis of ML classifier performances.
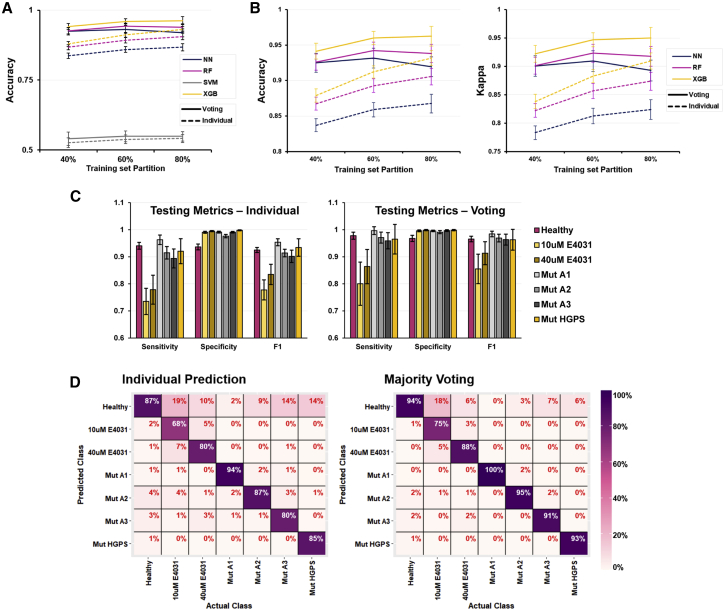
Comparison between the overall performance of the four ML models revealed that the classic SVM algorithm significantly underperformed compared with the other algorithms, achieving only 50%–55% accuracy in classifying unseen test data regardless of train-test splits or prediction strategy (Figure 4A). The main reason behind this underperformance is likely that the base SVM algorithm is a pairwise binary classifier, and its ability to do multi-class classification is an extension of doing multiple pairwise comparisons. Across the remaining three algorithms, the majority voting strategy has a clear performance advantage compared with the individual prediction strategy across all train-test splits (Figure 4B). However, at the 80:20 train-test split, the XGB classifier performed only marginally better than the 60:40 train-test split, while the NN and RF classifiers both performed slightly poorer, a sign of overfitting. This overfitting does not occur even with an 80:20 train-test split for all three classifiers in the individual prediction strategy (Figure 4B). Overall, these results suggest that the XGB classifier is more robust compared with the other ML classifiers in predicting the presence of arrhythmogenesis and is also more resistant to overfitting data as well.
Figure 4.
ML algorithms used to build the multi-class prediction model performed well despite no distinct separation detected from principal-component analysis
(A) Accuracy of all four trained multi-class classification models on the held-out testing datasets. Dashed lines, individual prediction strategy; solid lines, majority voting strategy.
(B) Accuracy and kappa metrices focusing on the three better performing multi-class classification models on the held-out testing datasets.
(C) Sensitivity, specificity, and F1 scores of the XGB multi-class classification models assessed using the held-out testing datasets. Left: individual prediction strategy. Right: majority voting strategy.
(D) Confusion matrixes of the XGB models assessed using the held-out testing datasets. The horizontal axis is the true classification of the CM test data, while the vertical axis is the XGB model predicted classification. Prediction errors fall within the column of the true classifications. Left: individual prediction strategy. Right: majority voting strategy. Mean ± SD is plotted for n = 100 independent bootstraps for all figures.
Assessing the individual classification accuracy of the 60:40 split XGB model, the majority voting strategy outperforms the individual prediction strategy in sensitivity (Figure 4C). Consequently, the F1 score, a performance metric accounting for both sensitivity and specificity of the model, is also higher for the majority voting strategy. Notably, the trained models are less sensitive to the presence of E4031 drug treatment. The heatmaps depicting the confusion matrices across all 100 bootstraps highlight the fact that most prediction errors within the drug treatment classes are mislabeled into the “Healthy” class (Figure 4D). A plausible explanation for this phenomenon is that not all CMs within the treated datasets were equivalently affected by E4031, resulting in some CMs exhibiting electrophysiology close to their healthy CM equivalents. In agreement with this hypothesis, there is an observable dose dependence effect, as the XGB classifiers were more sensitive to the presence of the higher 40 μM of arrhythmogenic E4031 drug treatment (88%) as opposed to the lower 10 μM treatment (75%). Therefore, the XGB classifier can predict arrhythmic risk within these seven subclasses accurately and could potentially differentiate between extents of arrhythmogenesis by assessing its ability to train and perform between classes.
Binary machine learning classifiers distinguish between healthy and arrhythmogenesis using biologically appropriate parameters
To further explore the hypothesis that ML classifiers can classify CMs into various arrhythmic risk categories while considering various extents of arrhythmogenesis, we assessed if these models were grounded in biological basis to make their predictions. Biologically appropriate ML models can be used for patient-specific therapy, providing a hypothesis-generating platform to identify key underlying electrophysiological changes attributed to arrhythmogenic conditions as opposed to classical arrhythmia models whereby only changes in TdP-inducing parameters are considered. The biological appropriateness of the trained models can be determined by probing the features they consistently use highly using the feature importance metric. However, there is little relevance in assessing the feature importance metric in the multi-class prediction model, as each arrhythmic class has differing underlying causes and alterations in the electrophysiology. Even in the selected bradycardic arrhythmias used in this study, apart from an expected increase in beat-to-beat duration, the underlying alterations in electrophysiology might be different. Therefore, training binary classifiers to compare healthy and individual arrhythmic subclasses is necessary to determine if ML models are using features in a biologically appropriate manner.
Within the datasets studied, the E4031 drug-induced arrhythmogenesis experimental group is the most appropriate to assess biological relevance of ML models, as the induced arrhythmogenesis has a known phenotype. It has been reported that the primary arrhythmogenic effect of E4031 is the lengthened repolarization duration due to the blockage of the IKr repolarization current (Harris et al., 2013). Visualizations of the Ca2+ transients show alterations in the waveforms in the 10 μM drug-treated CMs compared with the healthy control CMs, with a more severe effect observed in the higher 40 μM treatment group (Figure 5A). Quantification of the Ca2+ transient parameters confirms the lengthened repolarization durations, as well as a significant reduction in contraction speed attributed to the bradycardic effect of E4031 (Figure 5B). Therefore, both concentrations of E4031 exert the expected biological phenotype known to cause arrhythmogenesis in CMs, and the higher dosage does result in a more severe phenotype.
Figure 5.
Biological changes due to drug-induced arrhythmogenesis are captured by machine learning classifiers
(A) Representative Ca2+ cycling plots depicting qualitative changes in Ca2+ transients.
(B) Boxplots depicting quantitative comparison across treatment groups for beat-to-beat duration (BeatToBeat_M) and 30%, 60%, and 90% repolarization duration parameters (Repolarization30_M, Repolarization60_M, and Repolarization90_M, respectively). Each point represents a single CM. ∗p < 0.05 and ∗∗∗∗p < 0.0001; two-tailed Student’s t test. Ctrl,10 μM, 40 μM plotted with n = 178, 173, 169 CMs respectively.
(C) Overall workflow to validate that XGB classifiers use biologically appropriate features to determine arrhythmogenesis. XGB classifiers were trained on the two drug treatment binary datasets on three train-test splits, and their feature importance was determined.
(D) Performance metrices of the XGB binary classifiers assessed using the held-out testing datasets. Left: accuracy. middle: sensitivity. Right: specificity. Mean ± SD is plotted for n = 100 independent bootstraps for all figures.
(E and F) Total count of all parameters that scored >20 on the feature importance metrics across all XGB model bootstraps of the (E) 10 μM E4031 treatment dataset and the (F) 40 μM E4031 treatment dataset at a 60:40 train-test split.
To validate if the XGB classifiers would use the extended repolarization duration as features to predict E4031-induced arrhythmogenesis, both drug-treated datasets were used to generate binary XGB classifiers (Figure 5C). Similar to the multi-class prediction model, each binary dataset was split evenly across the classes into three train-test splits (80:20, 60:40, and 40:60) for 100 independent bootstraps. Across the train-test splits, the trained XGB classifier consistently performed better in classifying the higher dosage treatment (Figure 5D). Accuracy peaked at 60:40 train-test splits for 40 μM treatment (96.1%), while still marginally improving from 90.2% to 92.7% when increasing to 80:20 for 10 μM treatment. These results agree with the prior multi-class analysis, with 60:40 train-test split being optimized without overfitting and highlighting that the XGB classifiers are sensitive to dose effects and can more accurately predict the presence of higher 40 μM arrhythmogenic drug treatment than the lower 10 μM treatment.
Furthermore, we assessed the feature importance metric across each of the 100 independent models trained to classify the drug-treated CMs. Classifiers trained on the 60:40 train-test splits to distinguish the 10 μM treatment group used the 30% and 60% repolarization duration parameters highly, with >60 bootstraps using these parameters at >20% of the model (Figure 5E). A similar conclusion can be drawn from the classifiers trained to predict the 40 μM treatment group, with 67 and 30 bootstraps using repolarization 30% duration and 60% duration at >20% of the model respectively (Figure 5F). Changing the train-test ratios resulted in classifiers still consistently using either repolarization 30% and 60% duration parameters, suggesting a strong correlation between the extended repolarization duration and electrophysiological changes of the E4031 arrhythmic subclass (Figure S6). As the basic Ca2+ transient analysis did confirm the expected lengthening of Ca2+ repolarization duration due to E4031 treatment, the trained ML classifiers are biologically appropriate by using repolarization duration as the main feature distinguishing the drug-treated group from the healthy CMs. In addition, the prolongation of repolarization of E4031 consequently slows the cardiac contraction rhythm and increases beat-to-beat duration (Adaniya and Hiraoka, 1990). These findings agree with the lengthened beat-to-beat duration feature also consistently used by models across the various train-test splits to distinguish between healthy and E4031-treated CMs, strongly correlating the expected bradycardic phenotype to the electrophysiological changes of the E4031-treated arrhythmic subclass.
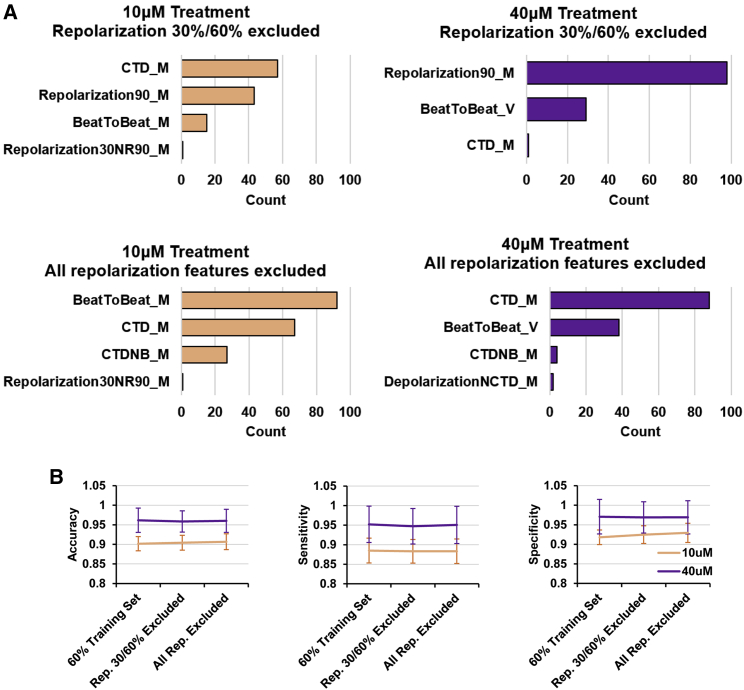
Next, we assessed whether the repolarization duration features could be replaced by the other electrophysiological features input into the model. XGB models were trained at the same 60:40 train-test splits without the repolarization 30% and 60% duration parameters. As expected, the top features used to generate the models became the extension of CTD and repolarization 90% duration (Figure 6A, top). CTD is made up of the depolarization duration and the repolarization 90% duration, and thus it would be the next likely feature that separates healthy CMs and E4031-treated CMs, alongside the repolarization 90% duration feature itself. Removal of the repolarization 90% duration then resulted in models using the increased CTD and beat-to-beat duration, which, as previously mentioned, are the expected secondary electrophysiological changes of the E4031-treated arrhythmic subclass (Figure 6A, bottom). Interestingly, there is no loss in model performance when these repolarization features are removed, suggesting that the secondary electrophysiological changes can sufficiently capture the E4031 treatment arrhythmic phenotype (Figure 6B). Therefore, these results confirm that the trained XGB classifiers are using the key biological changes to distinguish between healthy and the E4031-treated arrhythmic subclass.
Figure 6.
Secondary electrophysiological changes sufficiently recapitulate E4031 treatment arrhythmogenesis
(A) Total count of all parameters that scored >20 on the feature importance metrics across XGB models trained with reduced input variables. Top: repolarization 30% and 60% durations excluded. Bottom: all repolarization duration features excluded.
(B) Performance metrices of the XGB binary classifiers from the 60:40 train-test split compared with models trained with reduced input variables. Left: accuracy. Middle: sensitivity. Right: specificity. Mean ± SD is plotted (n = 100 independent bootstraps).
Discussion
In the field of arrhythmia modeling, there has been increasing interest in ML, with a few recent publications using trained ML models to study CM electrophysiology (Golgooni et al., 2019; Hwang et al., 2020; Juhola et al., 2015). These previous works highlight how single ML classifiers can be trained by the classifications of experienced researchers to accurately determine abnormal FPD or Ca2+ transients. In this work, we further showcase the utility of artificial intelligence in arrhythmia modeling, laying the groundwork by demonstrating that ML classifiers can be trained using single-cell, decomposed Ca2+ transients of CMs derived from both healthy and arrhythmic origins to predict the presence of the specific arrhythmias without any expert intervention. In addition, we show that the binary ML classifiers do use biologically appropriate parameters to make predictions and thus can be used in an unbiased manner to identify key electrophysiological changes due to unknown novel treatments.
The current scope of the developed ML classifiers are limited to the three forms of arrhythmia with bradycardia implications, but with the high accuracy rate (>90%) in identifying arrhythmic CMs using the majority voting strategy, there is confidence to adapt and pool in additional experimental groups for larger scale application. Currently, there is no available database storing electrophysiological data from CMs. Thus, the main limitation of the arrhythmia prediction platform would be to scale up the generation of experimental groups, to include the CM electrophysiological data from patients associated with genetic risk factors, or exposed to other known transient risk factors (Myerburg et al., 1997). Simultaneously, additional experimental groups containing electrophysiological data from other sources of healthy CMs will also be required to overcome population level variability. As Ca2+ tracking of CMs is regularly used as a form of electrophysiological modeling across labs, future collaborative work and post hoc repurposing of experimental data will help build up a more extensive ML arrhythmia prediction platform.
Our developed video-processing pipeline is hassle free, automating away most of the tedium required to generate the large datasets required to train ML classifiers. Apart from manual fluorescence recordings and identification of single cells, all downstream processes are automated to generate the datasets. With the ability to generate large numbers of biological replicates of single CMs using hPSC technology, the whole process is of medium throughput. The process of repurposing other CM Ca2+ fluorescent videos from other research groups can also be streamlined.
With the current limitations on the reprogramming efficiency, patient-specific risk profiling and therapy via reprogramming and deriving CMs will be relatively slow. The improvements to efficiency and next step for personalized medicine will likely occur when direct reprogramming of patient fibroblasts into CMs becomes reliable (Chen et al., 2017). Further improvements will then allow the rapid generation of patient CMs for the ML algorithm to make majority voting predictions for patient risk profiling. Therefore, this ML framework can directly be used to complement conventional, routine patient health check-ups and cardiovascular risk assessments to quantify arrhythmic risks in patients. Further automation for large-scale industrial drug screening can also complement current in vitro electrophysiological analysis and in vivo animal models. Scalability can be obtained by automation of CM derivation, fluorescent recording, and single-cell identification to drive high-throughput data generation. As such, CMs can be exposed to varying concentrations of drugs to conduct cardiotoxicity screens at scale, capturing more facets of possible arrhythmogenesis to improve the efficiency of preclinical studies. As the binary classification model has been shown to use biologically relevant parameters to separate healthy and arrhythmic CMs, it has the potential to be used as an unbiased tool to carry out hypothesis-generating research. Therefore, this study lays the groundwork and justification for future machine learning applications on electrophysiological data from CMs.
Experimental procedures
Fluorescent video capture of single CM Ca2+ cycles
Ca2+ cycles of CMs were imaged at 20× magnification using a Nikon ECLIPSE Ti-S fluorescent microscope and recorded using an Andor Zyla 4.2 sCMOS camera at 30 frames/s for 30 s. Ca2+ transients are detected as fluorescent emissions using a fluorescein isothiocyanate (FITC) filter cube. Video data were analyzed using Nikon’s NIS-Elements AR to isolate single CMs as ROIs and obtain fluorescent intensity over time data for individual CMs per frame.
Video processing of fluorescence to compute Ca2+ handling parameters
Fluorescent intensity values were processed using our self-compiled code in R (RStudio version 1.2.1335/Base R version 3.6.3) to identify fluorescent peaks corresponding to single cardiac contractions. Individual Ca2+ transients were decomposed into a list of output parameters summarizing the shape of the transients mathematically, described in Table S1. Mathematical description of normalized parameters is provided in Table S2. Each sample was defined by the mean and SD of each parameter. For parameters describing absolute values (beat-to-beat duration, depolarization duration, repolarization duration, CTD), the relative SD (RSD) was calculated instead (RSD = SD/mean). All samples, classifications, and output parameters are assembled into datasets and are piped into the ML training algorithm.
Training ML classifiers to predict arrhythmogenesis and determine feature importance
The sample datasets are processed using self-compiled code in R using the caret library for supervised learning (Kuhn, 2015). ML algorithms trained were neural network (library nnet), random forest (library rf), extreme gradient boosting (library xgbTree) and support vector machine, radial kernel (library e1071) (Chang and Lin, 2011; Chen and Guestrin, 2016; Liaw and Wiener, 2002; Venables and Ripley, 2002). Each dataset described in the report is defined by a M × 17 matrix, with M corresponding to the number of individual CMs measured. Each CM is defined by 16 columns of parameters, as previously described, and assigned either “healthy” or the specific arrhythmic subtype classification.
Model training was done using the CaretList function with a tuneLength parameter of 18, except for XGB, which used a tuneLength parameter of 6. Multi-class classifiers were optimized using 10-fold cross-validation repeated three times for minimum logarithmic loss. Class predictions are made using the highest probability choice, and for the majority voting prediction strategy, all predictions were set to be the most common prediction across all CMs within each ROI. Performance of the classifiers was determined using the held-out datasets using the accuracy and kappa metrics. For the training of the XGB binary classifiers, optimization was done using 10-fold cross-validation repeated three times for maximum area under the operator curves. Performance of the classifiers were determined using the held-out datasets using the accuracy, sensitivity and specificity metrics. Feature importance of the built models was derived using the varImp function.
Data and code availability
All data are available in the main text or the supplemental information. Code used for fluorescence processing and ML is available at https://github.com/Jemksp/Pang-Stem-Cell-Reports-2022.
Author contributions
J.K.S.P. designed the research, cloned the plasmids, generated the GCaMP6s knockin lines, carried out the experiments, and ran the machine learning data analysis. J.K.S.P. and S.C. maintained the cells, generated the ANK2 mutant clones, and performed the single-cell Ca2+ video imaging. J.Z., P.S., and C.S. provided the HGPS lines and their expertise on HGPS. H.Y. gave guidance on the machine learning data analysis. W.-K.C., S.Y.N., and B.-S.S. supervised the project. J.K.S.P. and B.-S.S. wrote the manuscript.
Conflicts of interests
The authors declare no competing interests.
Acknowledgments
This work is supported by the Agency for Science, Technology and Research (Singapore) and by a grant to B.-S.S. (HBMS IAF-PP grant H19H6a0026). J.K.S.P. is supported by the National University of Singapore graduate scholarship.
Published: July 14, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.06.005.
Supplemental information
The first worksheet contains the pooled dataset, and every worksheet thereafter corresponds to the boxplot bulk analysis depicted in Data S2 to S4 respectively. Each row corresponds to one individual CM. Each column corresponds to either a user-input identification status or the mean or variation of the decomposed Ca2+ transient input parameters
References
- Adaniya H., Hiraoka M. Effects of a novel class III antiarrhythmic agent, E-4031, on reentrant tachycardias in rabbit right atrium. J. Cardiovasc. Pharmacol. 1990;15:976–982. doi: 10.1097/00005344-199006000-00016. [DOI] [PubMed] [Google Scholar]
- Baker R.E., Peña J.M., Jayamohan J., Jérusalem A. Mechanistic models versus machine learning, a fight worth fighting for the biological community? Biol. Lett. 2018;14:20170660. doi: 10.1098/rsbl.2017.0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bers D. Springer Science & Business Media; 2001. Excitation-contraction Coupling and Cardiac Contractile Force. [Google Scholar]
- Brooks A.R., Harkins R.N., Wang P., Qian H.S., Liu P., Rubanyi G.M. Transcriptional silencing is associated with extensive methylation of the CMV promoter following adenoviral gene delivery to muscle. J. Gene Med. 2004;6:395–404. doi: 10.1002/jgm.516. [DOI] [PubMed] [Google Scholar]
- Ch'en F.F.T., Vaughan-Jones R.D., Clarke K., Noble D. Modelling myocardial ischaemia and reperfusion. Prog. Biophys. Mol. Biol. 1998;69:515–538. doi: 10.1016/S0079-6107(98)00023-6. [DOI] [PubMed] [Google Scholar]
- Chang, C.-C., and Lin, C.-J. (2011). LIBSVM: a library for support vector machines. ACM transactions on intelligent systems and technology (TIST) 2, 1-27, 10.1145/1961189.1961199.
- Chen T., Guestrin C. Xgboost: a scalable tree boosting system. arXiv. 2016:785–794. doi: 10.1145/2939672.2939785. Preprint at. [DOI] [Google Scholar]
- Chen Y., Yang Z., Zhao Z.-A., Shen Z. Direct reprogramming of fibroblasts into cardiomyocytes. Stem Cell Res. Ther. 2017;8:118. doi: 10.1186/s13287-017-0569-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy C.E., Rudy Y. Linking a genetic defect to its cellular phenotype in a cardiac arrhythmia. Nature. 1999;400:566–569. doi: 10.1038/23034. [DOI] [PubMed] [Google Scholar]
- Correia C., Koshkin A., Duarte P., Hu D., Teixeira A., Domian I., Serra M., Alves P.M. Distinct carbon sources affect structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Sci. Rep. 2017;7:8590. doi: 10.1038/s41598-017-08713-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha S.R., Mohler P.J. Cardiac ankyrins: essential components for development and maintenance of excitable membrane domains in heart. Cardiovasc. Res. 2006;71:22–29. doi: 10.1016/j.cardiores.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Dempsey G.T., Chaudhary K.W., Atwater N., Nguyen C., Brown B.S., McNeish J.D., Cohen A.E., Kralj J.M. Cardiotoxicity screening with simultaneous optogenetic pacing, voltage imaging and calcium imaging. J. Pharmacol. Toxicol. Methods. 2016;81:240–250. doi: 10.1016/j.vascn.2016.05.003. [DOI] [PubMed] [Google Scholar]
- Fermini B., Hancox J.C., Abi-Gerges N., Bridgland-Taylor M., Chaudhary K.W., Colatsky T., Correll K., Crumb W., Damiano B., Erdemli G., et al. A new perspective in the field of cardiac safety testing through the comprehensive in vitro proarrhythmia assay paradigm. J. Biomol. Screen. 2015;21:1–11. doi: 10.1177/1087057115594589. [DOI] [PubMed] [Google Scholar]
- Golgooni Z., Mirsadeghi S., Soleymani Baghshah M., Ataee P., Baharvand H., Pahlavan S., Rabiee H.R. Deep learning-based proarrhythmia analysis using field potentials recorded from human pluripotent stem cells derived cardiomyocytes. IEEE J. Transl. Eng. Health Med. 2019;7:1900203. doi: 10.1109/JTEHM.2019.2907945. [DOI] [Google Scholar]
- Harris K., Aylott M., Cui Y., Louttit J.B., McMahon N.C., Sridhar A. Comparison of electrophysiological data from human-induced pluripotent stem cell–derived cardiomyocytes to functional preclinical safety assays. Toxicol. Sci. 2013;134:412–426. doi: 10.1093/toxsci/kft113. [DOI] [PubMed] [Google Scholar]
- Hayashi M., Shimizu W., Albert C.M. The spectrum of epidemiology underlying sudden cardiac death. Circ. Res. 2015;116:1887–1906. doi: 10.1161/CIRCRESAHA.116.304521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennekam R.C. Hutchinson–Gilford progeria syndrome: review of the phenotype. Am. J. Med. Genet. 2006;140A:2603–2624. doi: 10.1002/ajmg.a.31346. [DOI] [PubMed] [Google Scholar]
- Hwang H., Liu R., Maxwell J.T., Yang J., Xu C. Machine learning identifies abnormal Ca2+transients in human induced pluripotent stem cell-derived cardiomyocytes. Sci. Rep. 2020;10:16977. doi: 10.1038/s41598-020-73801-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa M., Aiba T., Ohno S., Shigemizu D., Ozawa J., Sonoda K., Fukuyama M., Itoh H., Miyamoto Y., Tsunoda T., et al. Phenotypic variability of ANK2 mutations in patients with inherited primary arrhythmia syndromes. Circ. J. 2016;80:2435–2442. doi: 10.1253/circj.cj-16-0486. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Zhou Y., Bao X., Chen C., Randolph L.N., Du J., Lian X.L. An ultrasensitive calcium reporter system via CRISPR-Cas9-mediated genome editing in human pluripotent stem cells. iScience. 2018;9:27–35. doi: 10.1016/j.isci.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhola M., Penttinen K., Joutsijoki H., Varpa K., Saarikoski J., Rasku J., Siirtola H., Iltanen K., Laurikkala J., Hyyrö H., et al. Signal analysis and classification methods for the calcium transient data of stem cell-derived cardiomyocytes. Comput. Biol. Med. 2015;61:1–7. doi: 10.1016/j.compbiomed.2015.03.016. [DOI] [PubMed] [Google Scholar]
- Kuhn, M. (2015). Caret: classification and regression training. Astrophysics Source Code Library, ascl: 1505.1003.
- Kusumoto D., Yuasa S. The application of convolutional neural network to stem cell biology. Inflamm. Regen. 2019;39:14. doi: 10.1186/s41232-019-0103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Scouarnec S., Bhasin N., Vieyres C., Hund T.J., Cunha S.R., Koval O., Marionneau C., Chen B., Wu Y., Demolombe S., et al. Dysfunction in ankyrin-B-dependent ion channel and transporter targeting causes human sinus node disease. Proc. Natl. Acad. Sci. USA. 2008;105:15617–15622. doi: 10.1073/pnas.0805500105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Zhang J., Azarin S.M., Zhu K., Hazeltine L.B., Bao X., Hsiao C., Kamp T.J., Palecek S.P. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat. Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw A., Wiener M. Classification and regression by randomForest. R. News. 2002;2:18–22. [Google Scholar]
- Ma J., Guo L., Fiene S.J., Anson B.D., Thomson J.A., Kamp T.J., Kolaja K.L., Swanson B.J., January C.T. High purity human-induced pluripotent stem cell-derived cardiomyocytes: electrophysiological properties of action potentials and ionic currents. Am. J. Physiol. Heart Circ. Physiol. 2011;301:H2006–H2017. doi: 10.1152/ajpheart.00694.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myerburg R.J., Interian A., Jr., Mitrani R.M., Kessler K.M., Castellanos A. Frequency of sudden cardiac death and profiles of risk. Am. J. Cardiol. 1997;80:10F–19F. doi: 10.1016/s0002-9149(97)00477-3. [DOI] [PubMed] [Google Scholar]
- Oh Y., Wei H., Ma D., Sun X., Liew R. Clinical applications of patient-specific induced pluripotent stem cells in cardiovascular medicine. Heart. 2012;98:443–449. doi: 10.1136/heartjnl-2011-301317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Torres J., Calvo C.J., Llach A., Guzmán-Martínez G., Caballero R., González-Gómez C., Jiménez-Borreguero L.J., Guadix J.A., Osorio F.G., López-Otín C., et al. Cardiac electrical defects in progeroid mice and Hutchinson–Gilford progeria syndrome patients with nuclear lamina alterations. Proc. Natl. Acad. Sci. USA. 2016;113:E7250–E7259. doi: 10.1073/pnas.1603754113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallam K., Li Y., Sager P.T., Houser S.R., Wu J.C. Finding the rhythm of sudden cardiac death: new opportunities using induced pluripotent stem cell-derived cardiomyocytes. Circ. Res. 2015;116:1989–2004. doi: 10.1161/CIRCRESAHA.116.304494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H.Y., Wang C., Lee H.K., Yoo K.H., Zeng X., Kuhns T., Yang C.M., Mohr T., Liu C., Hennighausen L. CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat. Commun. 2017;8:15464. doi: 10.1038/ncomms15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin D., Papakis V., Bustos B.I., Ambrosi C.M., Ryan S.J., Baru V., Williams L.A., Dempsey G.T., McManus O.B., Landers J.E., et al. Homozygous might be hemizygous: CRISPR/Cas9 editing in iPSCs results in detrimental on-target defects that escape standard quality controls. Stem Cell Rep. 2022;17:993–1008. doi: 10.1016/j.stemcr.2022.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallini Y.N., Ohkura M., Choi B.-R., Ji G., Imoto K., Doran R., Lee J., Plan P., Wilson J., Xin H.-B., et al. Imaging cellular signals in the heart in vivo: cardiac expression of the high-signal Ca2+ indicator GCaMP2. Proc. Natl. Acad. Sci. USA. 2006;103:4753–4758. doi: 10.1073/pnas.0509378103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z.V., Ferdinandy P., Liaudet L., Pacher P. Drug-induced mitochondrial dysfunction and cardiotoxicity. Am. J. Physiol. Heart Circ. Physiol. 2015;309:H1453–H1467. doi: 10.1152/ajpheart.00554.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables W., Ripley B. In: Fourth S., editor. Springer; New York: 2002. Modern Applied Statistics. [Google Scholar]
- Yap Y.G., Camm A.J. Drug induced QT prolongation and torsades de pointes. Heart. 2003;89:1363–1372. doi: 10.1136/heart.89.11.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Lian Q., Zhu G., Zhou F., Sui L., Tan C., Mutalif R.A., Navasankari R., Zhang Y., Tse H.-F., et al. A human iPSC model of Hutchinson Gilford progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell. 2011;8:31–45. doi: 10.1016/j.stem.2010.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scale bar, 50μm
Scale bar, 250μm
The first worksheet contains the pooled dataset, and every worksheet thereafter corresponds to the boxplot bulk analysis depicted in Data S2 to S4 respectively. Each row corresponds to one individual CM. Each column corresponds to either a user-input identification status or the mean or variation of the decomposed Ca2+ transient input parameters
Data Availability Statement
All data are available in the main text or the supplemental information. Code used for fluorescence processing and ML is available at https://github.com/Jemksp/Pang-Stem-Cell-Reports-2022.