Summary
Fibrolamellar carcinoma (FLC) is a rare, often lethal, liver cancer affecting adolescents and young adults, for which there are no approved therapeutics. The development of therapeutics is hampered by a lack of in vitro models. Organoids have shown utility as a model system for studying many diseases. In this study, tumor tissue and the adjacent non-tumor liver were obtained at the time of surgery. The tissue was dissociated and grown as organoids. We developed 21 patient-derived organoid lines: 12 from metastases, three from the liver tumor and six from adjacent non-tumor liver. These patient-derived FLC organoids recapitulate the histologic morphology, immunohistochemistry, and transcriptome of the patient tumor. Patient-derived FLC organoids were used in a preliminary high-throughput drug screen to show proof of concept for the identification of therapeutics. This model system has the potential to improve our understanding of this rare cancer and holds significant promise for drug testing and development.
Keywords: pediatric cancer, liver cancer, fibrolamellar
Highlights
-
•
Organoids were generated from patient fibrolamellar tumors, a lethal liver cancer
-
•
Tissue source was patient metastases, primary tumors, and adjacent normal tissue
-
•
Organoids recapitulate patient tumor’s histopathology and transcriptome
-
•
Organoids report drug vulnerabilities in high-throughput screen
In this article, Narayan et al. show that organoids generated directly from patient resections recapitulate the transcriptome, pathology, and immunohistochemistry of metastases and primary tumors of the lethal cancer fibrolamellar carcinoma (FLC) as well as adjacent non-transformed tissue. The FLC organoids were implanted as xenografts in mouse and used in a high-throughput screen for drug vulnerabilities.
Introduction
Fibrolamellar carcinoma (FLC) is a rare liver cancer occurring most frequently in adolescents and young adults without prior liver disease or cirrhosis. Because patients with FLC are young and often present with nonspecific symptoms, such as nausea, vague abdominal discomfort, and weight loss (Craig et al., 1980), the majority of patients have advanced disease at diagnosis (Yamashita et al., 2016). Lymphadenopathy is present in 70% of patients at diagnosis, and about 50% eventually develop distant metastases (Kakar et al., 2005). The reported overall 5-year survival ranges from 30% to 45% (Eggert et al., 2013b; El-Serag and Davila, 2004; Kakar et al., 2005; Katzenstein et al., 2003; Njei et al., 2014), and the only established curative therapy is surgery. However, as the rate of advanced disease is high in these patients, surgery is often only palliative (Yamashita et al., 2016).
Grossly, FLC tumors are well circumscribed, tan to yellow in color, and sometimes have an irregular central scar (Graham and Torbenson, 2017). Histologically, the tumor cells are large, with abundant eosinophilic cytoplasm and prominent nucleoli. Dense bands of lamellar fibrosis run through the tumor, creating linear trabeculae of tumor cells (Graham and Torbenson, 2017). Various immunohistochemical stains have been used to assist in the diagnosis of FLC. One of these, cytokeratin 7 (CK7), is very sensitive and has been detected in almost all cases of FLC (Abdul-Al et al., 2010; Graham and Torbenson, 2017; Klein et al., 2005; Van Eyken et al., 1990; Ward et al., 2010). Similarly, CD68 (KP-1 clone) has also been detected in almost all FLC tumors; CD68 stains lysosomes in the cytoplasm of cells, which in FLC are abundant (Graham and Torbenson, 2017; Ross et al., 2011). Taken individually, staining for either CK7 or CD68 is not specific for FLC, but when used in conjunction, along with compatible morphology, co-expression of CK7 and CD68 is diagnostic of FLC (Graham and Torbenson, 2017; Limaiem et al., 2015; Ross et al., 2011).
With an incidence of 1 in 5 million (Eggert et al., 2013a; El-Serag and Davila, 2004; Lalazar and Simon, 2018), FLC comprises approximately 1% of hepatocellular carcinomas (HCCs) in all patients (Graham and Torbenson, 2017; Lin and Yang, 2018) and 25% of HCCs in patients younger than 20 years of age (Edmondson, 1956; Torbenson, 2012). FLC is considered a subtype of HCC (Fritz, 2013), although it has many distinguishing characteristics: distinct demographics, risk factors for HCC, such as viral infection or cirrhosis not seen in FLC, and HCC, and FLC having different tumor markers (Lin and Yang, 2018). HCC and FLC differ in their genomic alterations, in their transcriptome (Darcy et al., 2015; Farber et al., 2018; Simon et al., 2015), and in their proteome (Simon et al., 2015). α-Fetoprotein is elevated in the serum of a subset of patients with HCC but not with FLC. TP53 and β-catenin (CTNNB1) mutations are frequently found in HCC (Kannangai et al., 2005; Terris et al., 1999). In contrast, FLC is characterized by a disruption of the ecology of signaling of protein kinase A (PKA). Patients harbor an ∼400-kb deletion on chromosome 19, producing a fusion between exon 1 of DNAJB1, a heat shock protein, and exons 2–10 of PRKACA, the catalytic subunit of PKA (Honeyman et al., 2014). This fusion, DNAJB1-PRKACA, has been detected in primary tumors and metastases but not in non-tumor “normal” liver of patients with FLC (Graham et al., 2015; Honeyman et al., 2014; Malouf et al., 2015). The DNAJB1-PRKACA fusion kinase maintains the catalytic activity of the wild-type PKA (Honeyman et al., 2014). Other than the fusion chimera, no recurrent mutations have been found in patients with FLC.
Using CRISPR-Cas9 technology to construct the DNAJB1-PRKACA fusion in the livers of mice results in tumors with the histology and transcriptome of FLC (Engelholm et al., 2017; Kastenhuber et al., 2017). Expression of the DNAJB1-PRKACA chimera alone, in trans, was sufficient for pathogenesis, demonstrating that the tumorigenesis is not the result of the deletion (Kastenhuber et al., 2017). Thus, the DNAJB1-PRKACA fusion is believed to be the driver of FLC. Recently, this fusion was also found in biliary and pancreatic neoplasms, suggesting involvement; it can produce tumors outside of the liver (Singhi et al., 2019; Vyas et al., 2019). Other aberrations in PKA have been shown to produce FLC-like histology, although these are less common than the DNAJB1-PRKACA fusion; three patients with deletions in the main regulatory subunit of PKA, PRKAR1A, which causes the Carney complex, were found to have liver tumors with FLC-like histopathology (Graham et al., 2018).
Since many clinicians consider FLC to be a subtype of HCC (Fritz, 2013), it is often treated with the same therapeutics. These therapeutics offer minimal survival advantage in patients with anatomically unresectable disease and/or metastases (Lim et al., 2014; Weeda et al., 2013);, thus the need is to identify new therapeutics to increase survival in patients with advanced FLC. This necessitates a model system that adequately recapitulates the disease. Organoids have emerged as a model for a variety of diseases and are being used for studying disease pathogenesis as well as drug screening. Colorectal cancer organoids have been used to study drug sensitivity and to predict both chemotherapy and chemoradiation response in patients (Ooft et al., 2019; Sasaki and Clevers, 2018; Yao et al., 2020). Organoid technology has also been used to study pathogenesis and/or to test drugs for breast, ovarian, endometrial, lung, liver, pancreatic, kidney, bladder, prostate, neuroendocrine, and brain cancers (Broutier et al., 2017; Calandrini et al., 2020; Driehuis et al., 2019; Drost et al., 2016; Gao et al., 2014; Goldhammer et al., 2019; Jacob et al., 2020; Lee et al., 2018; Li et al., 2020; Liu et al., 2020; Na et al., 2020; Pasch et al., 2019; Puca et al., 2018).
Using technologies from previous work on human liver organoids (Broutier et al., 2017; Hu et al., 2018; Huch et al., 2015), we demonstrate that patient-derived FLC organoids recapitulate the human tumors from which they were derived. We have established 21 patient-derived organoids from patients with FLC, six normal organoid lines, three primary tumor organoid lines, and 12 metastatic organoid lines. These patient-derived FLC organoids recapitulate the histologic morphology, immunohistochemistry, and transcriptome of their corresponding patient tumors. Additionally, we demonstrate the potential of this model system for novel therapeutic discovery via high-throughput drug screening.
Results
Establishment of patient-derived organoid lines
Patient-derived organoids were made from freshly resected human tissue. Tissue was donated by patients with FLC undergoing surgical resection as part of treatment for their cancer and included primary liver tumors, adjacent non-tumor (or “normal”) liver tissue, and metastases. The tissue we acquired was considered “excess tissue,” not needed for clinical care. Thus, we received adjacent non-tumor (“normal”) tissue only when a primary liver tumor, liver recurrence, or liver metastasis was resected. As a result, some FLC organoid lines did not have a corresponding “normal.” Fresh tissue was placed in medium with antibiotics on ice and transported to the laboratory, cut into small pieces, and subjected to enzymatic dissociation into single cells or small groups of cells. The cells were mixed with a basement membrane extracellular matrix and plated as domes on tissue-culture-treated plates.
When the project started, we did not know if the FLC organoids would grow better in cholangiocyte or hepatocyte medium or, if they grew in both, which would be more physiologically relevant. Thus, we tried both with the goal of comparing them for both histopathology and transcriptome. Hepatocyte or cholangiocyte medium was added to the wells and incubated at 37°C (see experimental procedures). When the cells proliferated, they formed 3D structures within the extracellular matrix dome.
Tissue was received from nine patients (range in age 14–36 years, average age 22.6 years old, five male, four female ; Table 1). Two patients were treatment naive prior to surgery, and seven seven patients received at least one chemotherapeutic agent. From these nine patients, a total of 21 distinct organoid lines were derived, including six normal liver, three primary FLC, and 12 metastatic FLC. Seven of the metastatic FLC organoid lines were derived from lymph node metastases from various anatomic locations, (Table 1). One organoid line was established from each of the following locations: liver, lung, abdominal wall, omentum, and ascites.
Table 1.
Patient-derived fibrolamellar hepatocellular carcinoma organoids
| Patient # | ID | -Sex | Normal/primary/metastasis | Previous treatment |
|---|---|---|---|---|
| FLC6 | M1 | F | M-ant. Inf. Pericardial LN | Naive |
| M2 | M-ant. Sup. Pericardial LN | Naive | ||
| FLC7 | N | M | N-liver | Naive |
| P | P-liver | Naive | ||
| M1 | M-r periportal lymph node | Naive | ||
| M2 | M-l periportal lymph node | Naive | ||
| FLC8 | M | F | M-abdominal wall | Experienced |
| FLC9 | N | M | N-liver | Experienced |
| M1 | M-ascites | Experienced | ||
| M2 | M-omental nodule | Experienced | ||
| FLC11 | N | M | N-liver | Experienced |
| M1 | M-lymph node | Experienced | ||
| FLC12 | N | M | N-liver | Experienced |
| M1 | M-liver | Experienced | ||
| M2 | M-lymph node | Experienced | ||
| M3 | M-retroperitoneal lymph node | Experienced | ||
| FLC13 | N | F | N-liver | Experienced |
| P | P-liver | Experienced | ||
| FLC14 | N | M | N-liver | Experienced |
| P | P-liver | Experienced | ||
| FLC15 | M1 | F | M-lung nodule | Experienced |
Twenty-one patient-derived organoid lines cultured from nine patients with fibrolamellar hepatocellular carcinoma including six non-tumor liver (“normal” or N) organoid lines, three primary liver tumor (P) organoid lines, and 12 metastatic (M, M1–3) organoid lines. Patient sex, tumor location, and treatment prior to tumor resection are listed. Experienced, at least one chemotherapeutic agent; naïve, no treatment prior to resection.
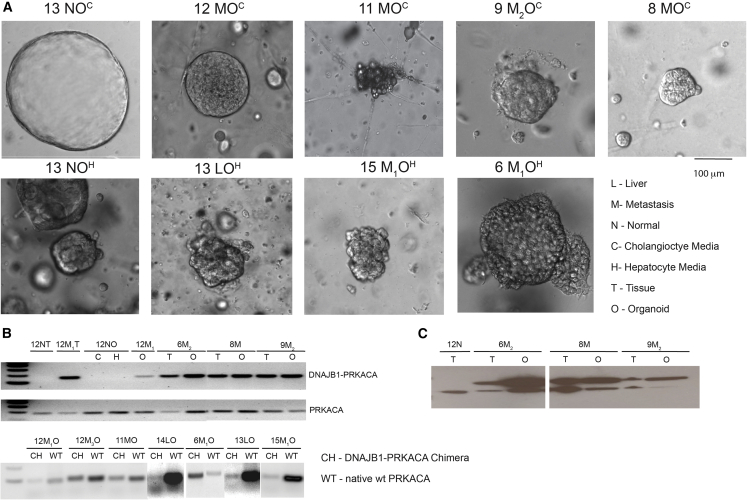
Organoids from normal liver are grown in either cholangiocyte medium or hepatocyte medium (see experimental procedures); these were termed cholangiocyte organoids and hepatocyte organoids. Cholangiocyte organoids are formed from EPCAM-positive ductal cells, whereas EPCAM-negative cells form hepatocyte organoids when placed in medium with the necessary growth factors (Broutier et al., 2017; Hu et al., 2018; Huch et al., 2015). Each patient-derived normal tissue sample was developed into two separate lines: one normal cholangiocyte organoid line and one normal hepatocyte organoid line. On live brightfield imaging, normal cholangiocyte organoids form spherical cystic structures (Figure 1A, top left), which differ significantly from the grape-like cell clusters of the normal hepatocyte organoids (Figure 1A, bottom left).
Figure 1.
Brightfield imaging, RNA transcription, and protein expression of DNAJB1-PRKACA in patient-derived FLC organoids
(A) Brightfield imaging of normal cholangiocyte and hepatocyte organoids compared with FLC primary liver tumor and metastatic organoids. Scale bar, 100 μM in all images.
(B) The DNAJB1-PRKACA fusion transcript was detected in tumor tissue and tumor organoids but not in normal tissue or normal organoids. The wild-type PRKACA transcript was detected in all samples. Top pair of gels: DNAJB1-PRKACA amplicon = 160 bp, wild-type PRKACA amplicon = 184 bp. Bottom: DNAJB1-PRKACA amplicon = 104 bp, wild-type amplicon = 95 bp.
(C) The DNAJB1-PRKACA fusion protein was detected in tumor tissue and tumor organoids but not in normal tissue from patients with FLC. DNAJB1-PRKACA protein = 46 kDa, wild-type PRKACA protein = 41 kDa. N, normal; M, M1, M2, metastasis #; P, primary tumor; O, organoid; T, tissue; C, cholangiocyte; H, hepatocyte.
The organoids derived from the FLC tumor tissue, whether they were derived in cholangiocyte medium or in hepatocyte medium, looked similar to the normal hepatocyte organoids on live brightfield imaging. They formed solid grape-like clusters of cells (Figure 1A), with an occasional cystic component.
Patient-derived FLC organoids recapitulate their original patient FLC tumors
Detection of the DNAJB1-PRKACA fusion
FLC tumors contain the DNAJB1-PRKACA fusion transcript and protein, which has been described as the driver mutation of this type of carcinoma (Engelholm et al., 2017; Honeyman et al., 2014; Lalazar and Simon, 2018). Upon receiving each fresh tissue donation, we verify the presence of the DNAJB1-PRKACA fusion transcript by PCR. To date, all of the FLC tumor samples we have obtained (n = 120) have contained the DNAJB1-PRKACA fusion transcript, while the adjacent normal liver tissue (when available) did not (see Figure 1B). Because this transcript is easily identifiable by PCR and is present in all of the tumor tissue samples, we used this method to confirm the identity of the growing organoids as patient-derived FLC organoids. The DNAJB1-PRKACA fusion transcript was detected in FLC tumor tissue and patient-derived FLC organoids but not in adjacent normal tissue or in patient-derived normal organoids (Figure 1B). The wild-type PRKACA transcript was detected in all samples.
In some of the patient-derived FLC organoid lines, the detected level of the wild-type PRKACA transcript was significantly greater than that of the DNAJB1-PRKACA fusion transcript (13P, 14P, and 15M1; note that when we received multiple metastases from the same patient, they are indicated by the subscript). These samples may be contaminated by normal liver cells (13P and 14P) or normal lung cells (15M1), as we have found that organoids derived from primary liver tumors more frequently are overgrown by normal cells. In the case of 15M1, the patient tissue was a ∼5-mm metastasis within the lung parenchyma, which contained some normal lung, as seen on hematoxylin and eosin (H&E) staining (not shown).
To verify the presence of the DNAJB1-PRKACA fusion protein, we probed with an antibody raised against an epitope in the carboxyl terminus of PRKACA. This antibody identifies both the wild-type PRKACA and the DNAJB1-PRKACA fusion protein (there is currently no antibody that specifically identifies the fusion protein). Again, the DNAJB1-PRKACA fusion was detected in FLC tumor tissue and patient-derived FLC organoids but not in adjacent normal tissue (Figure 1C).
Histology and immunohistochemistry
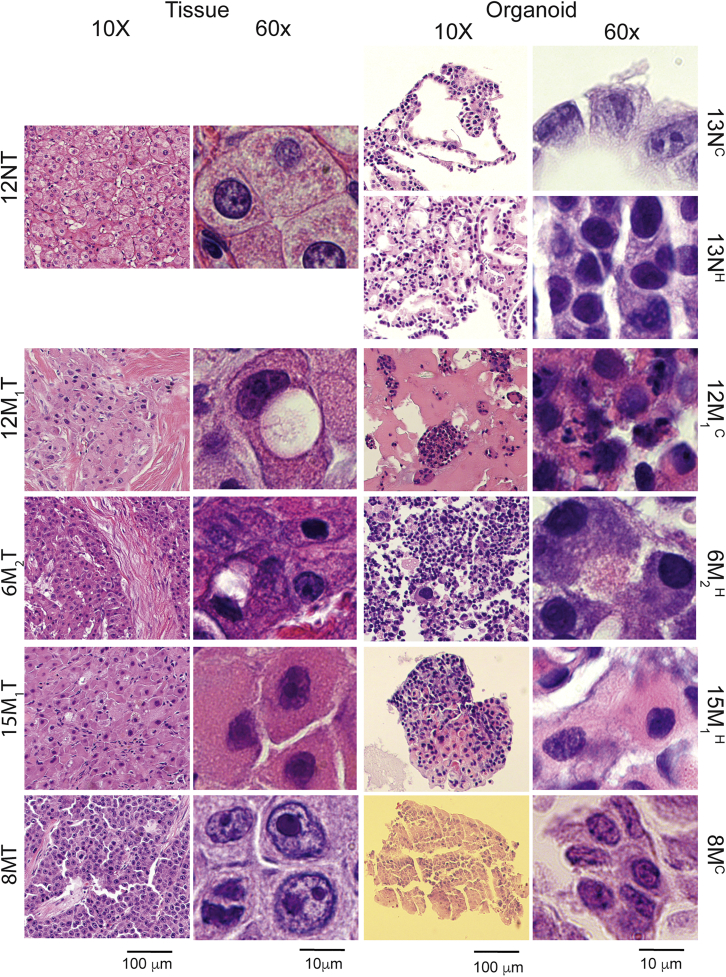
Histologically, FLC cells differ from normal liver cells in several ways. On H&E-stained slides at low magnification, thick bands of fibrous connective tissue (mostly collagen) can be seen running between groups of tumor cells; hence, the name fibrolamellar carcinoma. Compared with normal hepatocytes, FLC cells tend to be larger and polygonal with granular eosinophilic cytoplasm and prominent nucleoli. They may also contain steatosis, pale bodies (cytoplasmic inclusion bodies), and calcifications (Torbenson, 2012). To determine whether the patient-derived FLC organoids histologically resemble their original patient tumors, the organoids were fixed in formalin and embedded in paraffin, and the cut slides were stained with H&E (see experimental procedures). The morphology of the normal tissue, patient-derived normal organoids, patient tumor tissue, and corresponding patient-derived FLC organoids were compared (Figure 2). As expected from their brightfield images, the patient-derived normal cholangiocyte organoids appear cystic with a single layer of cells forming the structure. The patient-derived normal hepatocyte organoids appear in solid groups, forming columns of hepatocytes seen at high magnification. At both low and high magnification, the patient-derived FLC organoids resemble the tumors from which they were derived.
Figure 2.
Histology of FLC patient tumor tissue and patient-derived FLC organoids
H&E-stained slides of normal cholangiocyte and hepatocyte organoids derived from normal tissue compared with FLC tumor organoids derived from their corresponding patient FLC tumor tissue. Slides were imaged at 10× and 60× magnification with scale bars of 100 and 10 μm, respectively. N, normal; M, M1, M2, metastasis #; C, cholangiocyte medium; H, hepatocyte medium; 13NC, passage two cholangiocyte medium; 13NH, passage six hepatocyte medium; 12M1, cholangiocyte medium; 6M2H, hepatocyte medium; 15M1H, hepatocyte medium.
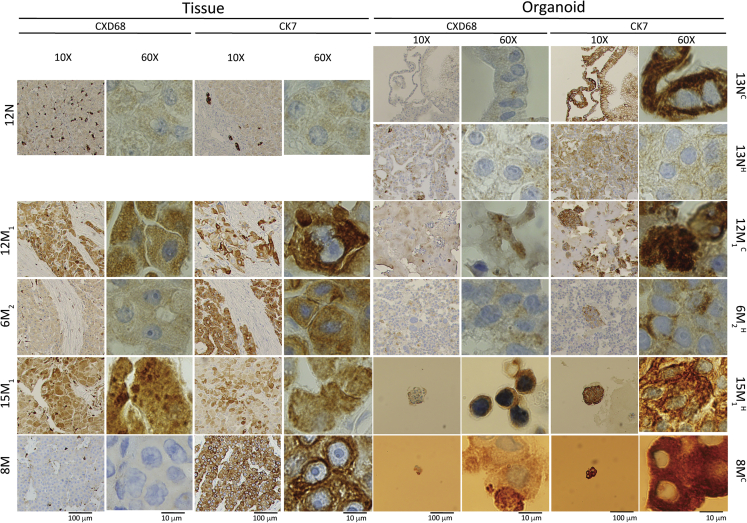
In addition to studying the morphology of the patient-derived FLC organoids, we characterized them by immunohistochemistry. Staining for CK7 and CD68 in combination with the appropriate morphology is specific for FLC (Graham and Torbenson, 2017; Torbenson, 2012). The normal liver tissue stained negative for both CK7 and CD68, except for the biliary structures and Kupffer cells seen at low magnification, respectively (Figure 3). As expected, the patient-derived normal cholangiocyte organoids were negative for CD68 and positive for CK7, and the patient-derived normal hepatocyte organoids were negative for both CD68 and CK7. Most of the patient-derived FLC organoids were positive for both CD68 and CK7, as were the patient tumors from which they were derived (examples shown for 12M1, 15M1, and 6M2). One patient tumor (8M) was positive for CK7 but negative for CD68. This has been reported before in a minority of patients in the literature (Graham and Torbenson, 2017). However, the corresponding patient-derived FLC organoid appeared to be positive for both CD68 and CK7.
Figure 3.
Immunohistochemistry of patient-derived FLC organoids
CD68- and CK7-stained slides of normal cholangiocyte and hepatocyte organoids derived from normal tissue compared with FLC tumor organoids derived from their corresponding patient FLC tumor tissue. Slides were imaged at 10× and 60× magnification with scale bars of 100 and 10 μm, respectively. N, normal; M, M1, M2, metastasis #. 13NC, passage two cholangiocyte medium; 13NH, passage six hepatocyte medium; 12M1C, cholangiocyte medium; 6M2, hepatocyte medium; 15M1, hepatocyte medium.
Transcriptome
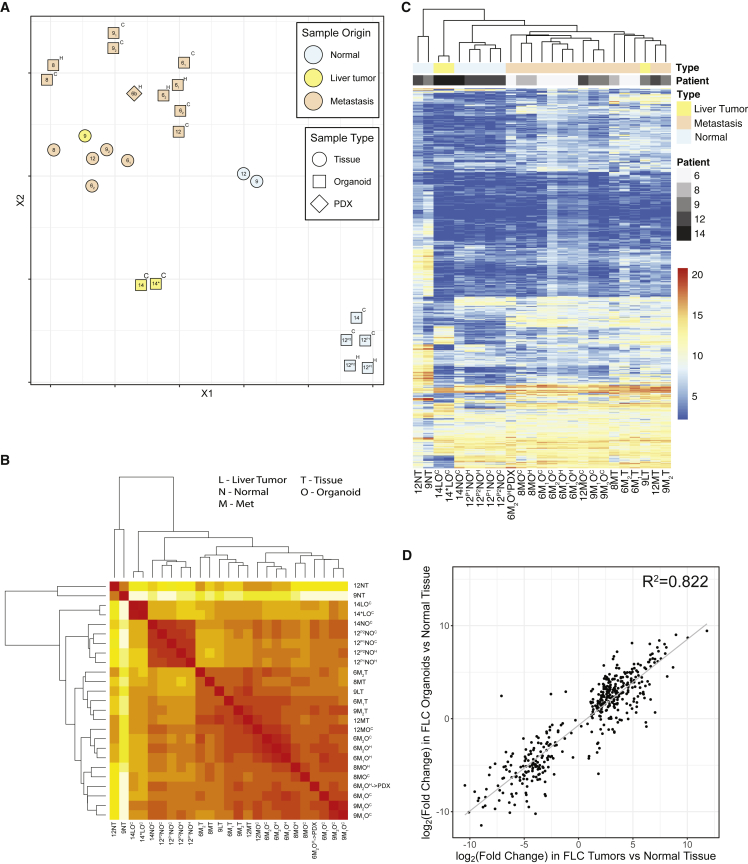
As previously described in the literature (Simon et al., 2015), FLC has a consistent gene profile compared with the normal human genome, with some genes always upregulated and other genes always downregulated. We generated a “transcriptomic profile” of 509 genes that we have found are regularly altered in expression in FLC relative to the adjacent non-tumor tissue (Table S2 in Lalazar et al., 2021). This subset of transcripts was selected based on transcriptome analysis that we did on 91 human tissue samples from 28 patients. To further characterize the patient-derived FLC organoids, we compared their transcriptome by whole RNA sequencing, with focus on this FLC-relevant transcriptomic profile, between normal tissue samples, FLC tumor tissue samples, patient-derived normal organoids, and patient-derived FLC organoids (Figure 4). In the t-distributed stochastic neighbor embedding (t-SNE) plot, the patient FLC tumor tissue clustered together and away from normal liver tissue (Figure 4A). The patient-derived FLC organoids clustered together and close to, but not superimposed on, the patient FLC tumors from which they were derived. When comparing the transcriptome of the FLC tumors versus the FLC organoids, considering only the genes with a base mean above 50 and FDR < 0.05, we found 270 upregulated and 43 downregulated genes out of the 55,148 genes detected. This tight clustering lacked a major transcriptional difference between the organoids in the cholangiocyte or hepatocyte media.
Figure 4.
Transcriptome of patient-derived FLC organoids
(A) Principal-component analysis comparing normal cholangiocyte and hepatocyte organoids derived from normal tissue compared with FLC tumor organoids (in cholangiocyte and hepatocyte media) derived from their corresponding patient FLC tumor tissue.
(B) Heatmap organized by closest similarity between FLC patient normal tissue, FLC patient tumor tissue, patient-derived normal organoids, and FLC organoids in cholangiocyte or hepatocyte media.
(C) Heatmap with known FLC-dysregulated genes comparing FLC patient normal tissue, FLC patient tumor tissue, patient-derived normal organoids, and FLC organoids in cholangiocyte or hepatocyte media. Both heatmaps show unsupervised clustering. N, normal; M, metastasis; L, primary liver tumor; O, organoid; T, tissue; H, grown in hepatocyte medium; C, grown in cholangiocyte medium. When multiple independent metastases were resected and analyzed, we used a subscript. Sample 14∗ was isolated in an alternative medium for liver tumors, which replaced the Noggin, Rspo-1, and Wnt3a with 3 nM dexamethasone (Broutier et al., 2017).
(D) We have generated a “fibrolamellar signature” representing 509 transcripts (Lalazar et al., 2021). The change in expression of these genes in the FLC-derived organoids relative to organoids derived from non-transformed liver tissue was plotted as a function of the expression in FLC tumor relative to the adjacent non-transformed tissue. The Pearson correlation coefficient is > 0.8.
Normal tissue and patient-derived normal organoids clustered away from FLC tumor tissue and the derived FLC organoids. This is seen on the clustering observed in a t-SNE plot or when examined in a heatmap with unsupervised clustering (Figure 4B). Similarly, when we examined the changes in expression of specific transcripts in the FLC gene profile, there was a distinct difference between the normal samples and the FLC samples (Figure 4C). Again, in unsupervised clustering, the patient-derived FLC organoids clustered with the patient tissue from which they were derived.
To contrast the transcriptomic profile in the organoids with the patient tissue, in Figure 4D we compared the expression of these genes in the FLC-derived organoids (y axis) and the FLC tumors (x axis) relative to the adjacent non-transformed tissue. For the 509 genes of the consistent “fibrolamellar signature” that we have previously defined (Lalazar et al., 2021), we obtained a Pearson correlation coefficient of 0.82 (Figure 4D).
Patient-derived FLC organoids proliferate in mice
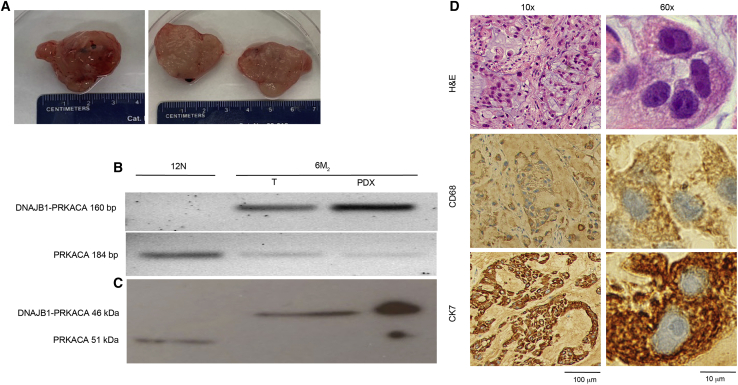
As a final validation that the patient-derived FLC organoids recapitulate the tumors from which they were derived, we injected 6M2 organoids (in extracellular matrix) into the subcutaneous space of three NSG mice (see experimental procedures). Subcutaneous tumors grew in all three mice over a period of 3 months, with an average size of 2 × 2 cm at the time of death (Figure 5A). Grossly, the mouse tumors formed from 6M2 organoids appeared similar to patient FLC tumors, with a yellowish hue. The DNAJB1-PRKACA fusion transcript and protein were detected by PCR and western blot, respectively, in the mouse tumor and in the patient FLC tumor tissue from which the 6M2 organoids were derived (Figures 5B and 5C). Histologically, the mouse tumor displayed typical FLC features, such as prominent nucleoli, granular eosinophilic cytoplasm, and the occasional lamellar band (Figure 5D). When stained with CD68 and CK7, the mouse tumor formed from 6M2 organoids stained positive for both, consistent with the results from the patient tumor tissue from which the 6M2 organoids were derived (Figure 5D).
Figure 5.
Patient-derived FLC organoids form tumors when injected into mice
(A) Gross pathology of 6 M2 tumor after resection from mouse. On the left is the intact tumor and on the right a cross-section.
(B) The DNAJB1-PRKACA fusion transcript (amplicon = 160 bp) was detected in original FLC patient tumor tissue (6M2 T) and tumor formed by injecting patient-derived organoids into mice (6 M2 O→M), but not in tissue from the adjacent non-transformed tissue (normal) from FLC patient (12 N T). The wild-type PRKACA transcript (amplicon = 184 bp) was detected in all samples.
(C) The DNAJB1-PRKACA fusion protein (46 kDa) was detected in original FLC patient tumor tissue (6M2 T) and tumor formed by injecting patient-derived organoids into mice (6M2 PDX), but not in normal tissue from FLC patient (12N T). The wild-type PRKACA protein (41 kDa) was detected in all samples.
(D) H&E-, CD68-, and CK7-stained slides of the tumor formed by injecting patient-derived organoids into mice (6M2 PDX). Scale bars, 100 μm at 10× magnification and 10 μm at 60× magnification. These organoids were grown in hepatocyte medium.
Preliminary drug screening in patient-derived FLC organoids
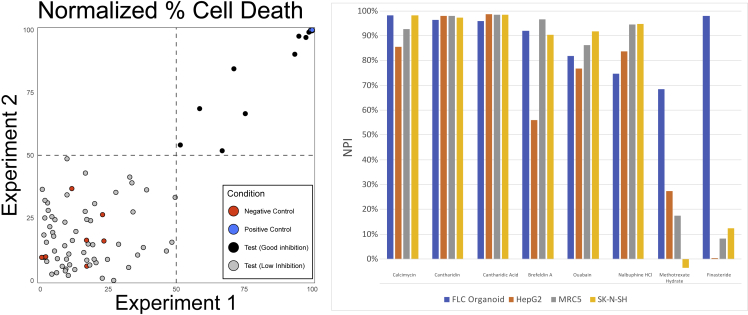
With the successful establishment of this new model system for FLC comes the ability to screen drugs and assay for new therapeutics for FLC, an unmet need. We performed a small pilot screen for toxicity of drugs that could be repurposed for use in FLC patients. Patient-derived FLC organoids (6M2) were plated in 384-well plates, and each well was subjected to a different drug from our drug-repurposing library (about 650 drugs were tested in singlet); this experiment was repeated on two different days (see experimental procedures). The normalized percent inhibition from each experiment was plotted (Figure 6). Chaetocin, the positive control (in blue), showed a consistent normalized percent inhibition of 100%, and DMSO, the negative control (in red), clustered around 0%. Most of the repurposing small-molecule compounds tested (in gray), performed no better than DMSO; there were, however, eight compounds (black) that had a greater than 50% survival inhibition on both experimental days. The compounds screened here were previously screened against a normal lung cell line (MRC5) and two cancer cell lines (HepG2 and SK-N-SH) in monolayer to evaluate for toxicity. Of the eight compound hits in our study, six of them showed toxicity against the normal lung cell line and two other cancer cell lines. However, two of the eight compound hits, finasteride and methotrexate, did not show toxicity in the cell lines. Finasteride is an inhibitor of 5-alpha-reductase type 2, which results in the decreased conversion of testosterone to dihydrotestosterone (DHT) and is primarily used as a treatment for benign prostatic hyperplasia. Finasteride can also inhibit 5-beta-reductase, which is involved in bile acid synthesis. Although it is unclear which mechanism causes toxicity in FLC organoids, it is an interesting drug to consider from this preliminary screen. Methotrexate competitively inhibits dihydrofolate reductase, thereby inhibiting DNA and RNA synthesis, and is currently used in the treatment of several cancers. These hits need to be validated in a dose-response curve experiment using patient-derived normal organoids for comparison. This preliminary drug screen shows the potential of this new organoid system for FLC in discovering new and necessary therapeutics for patients.
Figure 6.
Preliminary drug screening of patient-derived FLC organoids
(A) Correlation plots of screening ∼650 compounds against 6 M2 organoids performed twice (day 1 and day 2), each in singlet (R2 = 0.31; x axis, normalized %survival inhibition on day 1; y axis, normalized %survival inhibition on day 2. Blue dot, positive control (20 μM chaetocin); red dots, negative control (DMSO); gray dots, compounds tested. Compounds in the top right quandrant killed at least 50% 6 M2 organoids in both experiments.
(B) Normalized percent inhibition of some of the most efficacious compounds against FLC organoids, HepG2 cells (from a 15-year-old Caucasian male who had a well-differentiated hepatocellular carcinoma), MRC5 (fibroblast cell line from lung), and SK-N-SH (neuroblastoma).
Discussion
A model system that adequately recapitulates a human disease is necessary for studying the pathogenesis as well as for the development of new therapeutics and/or repurposing of drugs that are currently in clinical use for other diseases. Organoids have emerged as such a model system for a variety of diseases, both benign and malignant. Organoid technology has been used to study cancers, including colorectal, breast, ovarian, endometrial, lung, liver, pancreatic, kidney, bladder, prostate, neuroendocrine, and brain cancers (Broutier et al., 2017; Calandrini et al., 2020; Driehuis et al., 2019; Drost et al., 2016; Gao et al., 2014; Goldhammer et al., 2019; Jacob et al., 2020; Lee et al., 2018; Li et al., 2020; Liu et al., 2020; Na et al., 2020; Pasch et al., 2019; Puca et al., 2018). Many of these organoid lines have been used to identify possible new therapeutics via low- or high-throughput drug screening.
We demonstrate here the proof of concept that patient FLC tissue can be dissociated, cultured, and grown as patient-derived tumor organoids that model the biology of their original human cancers. We have successfully established 21 patient-derived organoid lines from patients with FLC, six normal organoid lines, three primary tumor organoid lines, and 12 metastatic organoid lines. These patient-derived FLC organoids recapitulate the histologic morphology, immunohistochemistry, and transcriptome of their corresponding patient tumors. The driver mutation in FLC, DNAJB1-PRKACA, was detected in all of the patient-derived FLC organoids and not in the patient-derived normal organoids. Histologically, the patient-derived FLC organoids showed morphology consistent with their corresponding patient tumors, such as large cells with granular eosinophilic cytoplasm, and prominent nucleoli.
Whereas many of the stereotypical FLC histology seen in the patient tumor tissue were also in the organoids, lamellar bands were seen only in the original tumor tissue. The lack of lamellar bands could be due to a missing cell type, such as stellate cells, which are responsible for much liver fibrosis. Alternatively, it may be that the regular harvesting and reseeding of the organoids may break up nascent collagen fibers, which might be present only in very long-term cultures. The immunohistochemical staining with CK7 and CD68 was also consistent in patient-derived FLC organoids.
The transcriptomes of the tumor organoids clustered with each other and with the corresponding tumor tissue but away from both normal tissue and normal organoids. In contrast, the organoids from the adjacent non-transformed tissue did not cluster closely with their parental tissue. We observed this previously on other organoids we have developed from liver (Saltsman et al., 2020). We suspect it may be the result of the culture inducing the organoids into a more stem-like state. We do not see this shift in the tumor organoids relative to the parental tumor tissue, possibly because their cell cycle is already dysregulated. As a final validation, when mice were subcutaneously injected with patient-derived FLC organoids, the tumors that grew were consistent with FLC.
This ability of organoids to recapitulate the FLC tumors from which they are derived is a valuable resource for developing new therapeutics through high-throughput drug screening. We have shown that patient-derived FLC organoids can be used in a high-throughput drug screen to identify potential therapeutics. Eight compounds from a portion of the repurposing small-molecule compound library at the High Throughput and Spectroscopy Center at The Rockefeller University were positive hits, which will be further validated through dose-response curves and in vivo. One of the crucial challenges we encountered for FLC organoid high-throughput screening was assay reproducibility. The assay performance can potentially be improved further to include other extracellular plate coatings, longer treatment times, or an imaging-compatible format. However, this pilot study is a proof of concept. Future efforts will focus on improving the generation of organoids compatible with high-throughput drug screening.
Although patient-derived tumor organoids adequately model their parent tumors, they lack an immune system and stromal components. However, these can be included in the model (Cattaneo et al., 2020). Patient-derived xenografts (PDXs) also recapitulate their parent tumors and have the additional benefits of the stromal components as well as being an in vivo system, which makes them an important model. However, PDXs are expensive and time-consuming and can have a low engraftment rate; additionally, they cannot be used in a high-throughput drug screen without surgically removing the tumors and dissociating to single cells, thus removing the 3D architecture from the tumor. Whereas a PDX may take months to grow large enough for an experiment, patient-derived tumor organoids can be grown faster and from fewer tumor cells, making them more accessible for high-throughput drug screening while maintaining the 3D tumoral architecture. Furthermore, patient-derived normal organoid lines can be used as normal controls, whereas normal PDXs do not exist, and appropriate control cells must be found. Positive hits from a high-throughput drug screen using patient-derived organoids can then be validated in a standard low-throughput PDX drug test in mice. Thus, patient-derived tumor organoids and PDXs can be used as complementary systems in the development of new therapeutics.
Conclusions
The patient-derived FLC organoids described here satisfy the criteria of a dependable in vitro model of fibrolamellar carcinoma. They recapitulate the histologic architecture and the transcriptomic profile of their parent FLC tumors and are accessible for high-throughput drug screening. Thus, this organoid model system can be used in the discovery of new therapeutics and/or for predicting patient-specific drug responses.
Experimental procedures
Human tissue samples
With Institutional Review Board approval (Rockefeller IRB# SSI-0797, SSI-0798, and SSI-0855 and MSKCC IRB Protocol #13-010), patients with FLC consented to tissue donation at the time of scheduled tumor resection. A diagnosis of FLC was made by a pathologist at the institution performing the surgical resection and reviewed by a single pathologist with expertise in FLC (M.S.T.). The diagnosis was independently verified by detection of the DNAJB1-PRKACA fusion transcript by RT-PCR and fusion protein by western blotting. Tissue obtained for research purposes included only excess primary and metastatic tissue not necessary for the patient’s diagnosis or treatment. Non-tumor “normal” liver tissue was collected only during a liver tumor resection.
Organoid culture
One-half of the harvested cells were grown in human liver organoid isolation medium to form cholangiocytes, and organoids were prepared as previously described in the literature (Broutier et al., 2017; Hu et al., 2018; Huch et al., 2015) (Advanced DMEM-F12 supplemented with 2% penicillin-streptomycin, 1% Glutamax, 1:50 B27 supplement [without vitamin A], 1:100 N2 supplement, FBS, 1.25 mM N-acetyl-L-cysteine, 10% [v/v] Rspo-1 conditioned medium, 30% [v/v] Wnt3a-conditioned medium, 10 mM nicotinamide, 10 nM recombinant human [Leu15]-gastrin I, 50 ng/mL recombinant human EGF, 100 ng/mL recombinant human FGF10, 25 ng/mL recombinant human HGF, 10 μM forskolin, 5 μM A8301, 25 ng/mL Noggin and 10 μM Y27632). Isolation medium was changed every 48–72 h and switched to human liver organoid expansion medium after 1–2 weeks in culture (Advanced DMEM-F12 supplemented with 2% penicillin-streptomycin, 1% Glutamax, 10 mM HEPES, 1:50 B27 supplement [without vitamin A], 1:100 N2 supplement, 1.25 mM N-acetyl-L-cysteine, 10% [v/v] Rspo-1 conditioned medium, 10 mM nicotinamide, 10 nM recombinant human [Leu15]-gastrin I, 50 ng/mL recombinant human EGF, 100 ng/mL recombinant human FGF10, 25 ng/mL recombinant human HGF, 10 μM forskolin, 5 μM A8301).
One-half of cells was cultured in human hepatocyte medium to form hepatocyte organoids (Advanced DMEM-F12 supplemented with 2% penicillin-streptomycin, 1% Glutamax, 10 mM HEPES, 1:50 B27 supplement [without vitamin A], 1.25 mM N-acetyl-L-cysteine, 15% [v/v] Rspo-1 conditioned medium, 10 mM nicotinamide, 10 nM recombinant human [Leu15]-gastrin I, 50 ng/mL recombinant human EGF, 100 ng/mL recombinant human FGF10, 100 ng/mL recombinant human FGF7 [KGF], 20 ng/mL recombinant human TGFa, 25 ng/mL recombinant human HGF, 2 μM A8301, 3 μM CHIR99021 [Sigma], and 10 μM Y27632). Hepatocyte medium was changed every 48–72 h. Tumor cells were also cultured in both cholangiocyte and hepatocyte media. RSpondin-1 10% (v/v), Wnt3a 30% (v/v), and Noggin 10% (v/v) conditioned media were made in-house (Broutier et al., 2017; Hu et al., 2018; Huch et al., 2015).
Cultures were passaged when organoids became confluent. Organoids were mechanically disrupted by trituration and enzymatic dissociated with TrypLE Express (GIBCO, Grand Island, NY) for 5 min at 37°C. The dissociated organoids were washed in Advanced DMEM-/F12 + 2% penicillin-streptomycin and centrifuged at 300 × g for 5 min at 4°C. Media and excess BME-2 were aspirated before organoids were resuspended in fresh BME-2.
Organoid harvesting
Organoids were harvested 24 h following media changes. Media were aspirated from each well and organoids were washed in 1× PBS. Organoids were mechanically disrupted and incubated in organoid-harvesting solution (Trevigen, Gaithersburg, MD) at 4°C for 45 min. Organoids were centrifuged at 300 × g for 5 min at 4°C, organoid-harvesting solution was aspirated, and organoids were washed in 1× PBS.
Injecting organoids into mice
Patient-derived tumor organoids that were cultured in hepatocyte media were mechanically disrupted by trituration and enzymatic dissociated with TrypLE Express (GIBCO) for 5 min at 37°C. The dissociated organoids were washed in Advanced DMEM/F12 + 2% penicillin/streptomycin and centrifuged at 300 × g for 5 min at 4°C. The organoid pellet was resuspended in 1:1 BME-2 and culture media.
NOD-scid-gamma (NSG) mice (Jackson Laboratories Bar Harbor, ME; NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, strain 005557) were bred at The Rockefeller University animal facility specific-pathogen-free (SPF) immune-core. Mice were kept on a 12-h:12-h light-dark cycle, fed an amoxicillin diet, and had ad libitum access to food and water. With IACUC approval (#17037-H), mice were anesthetized using isoflurane and given buprenorphine for analgesia. The organoids in 1:1 BME-2/medium were injected into the subcutaneous space over the flank (∼1 million organoids per injection) in three mice. The mice were monitored at least biweekly for health and tumor growth. Tumor size was evaluated by palpation. At 3 months following injection, the mice were euthanized, and tumor samples were taken for evaluation.
High-throughput drug screening
Organoids (6M2) were screened as previously published (Francies et al., 2019a; 2019b; Saltsman et al., 2020). Organoids were mechanically disrupted, counted, and resuspended in hepatocyte medium + 2% BME-2. The wells of 384-well plates were coated with 10 μL of 7.5 mg/mL BME-2. The plates were briefly centrifuged, and the BME-2 was allowed to polymerize at 37°C for 30 min. The organoid solution (30 μL; 500 organoids per well) was dispensed into each well. Plates were briefly centrifuged and incubated at 37°C for 4 days prior to drug screen. Following the incubation, cell viability was evaluated using a RealTime-Glo MT cell viability assay (Promega). Luminescence was read (BioTek Neo) and analyzed using Gen5 software (BioTek).
Six hundred fifty compounds from a >5,000 compound drug repurposing library were dispensed onto the organoids in 384-well plates. The plates were incubated at 37°C for 72 h. Each compound was screened in singlets at a final concentration of 10 μM, and the experiment was performed in duplicate. Each plate had negative-control wells (DMSO) and positive control wells treated (20 μM chaetocin, which inhibits lysine methyltransferases and inhibits thioredoxin reductase-1, causing oxidative stress) (Selleck Chemicals S8068). After 72 h, CellTiter-Glo 3D reagent (Promega) was added to each well, and the plates were read for luminescence (BioTek Synergy Neo) and analyzed.
Results were normalized to the number of viable cells per well, using the RealTime-Glo MT cell viability assay reading. To determine the percentage of cell survival inhibition, also called normalized percentage of inhibition (NPI), the CellTiter-Glo 3D reading was normalized against the positive- and negative-control wells in each plate. NPI was calculated using this formula: NPI = [(normalized compound well – mean of the negative control wells)/(mean of the positive control wells – mean of the negative control well)] × 100. Data from all screening studies are archived and were analyzed using the CDD Vault from Collaborative Drug Discovery (Burlingame, CA. http://www.collaborativedrug.com). Graphpad Prism (v.8) and Vortex (Dotmatics) were used for data analysis.
Author contributions
N.J.C.N: conceptualization, methodology, data acquisition, first draft, editing; D.R.: data analysis, methodology, first draft, editing; G.L.: conceptualization, editing; L.R.-E.: methodology, data acquisition; D.N.: methodology, data acquisition; S.L.: methodology, data acquisition; B.S.: methodology, data acquisition; R.W.: methodology, data acquisition; W.J.H.: conceptualization, methodology, data acquisition; J.A.S. III: conceptualization, methodology, data acquisition; H.G.: methodology; M.S.T.: data acquisition, editing; H.C.: methodology;, supervision, editing; M.P.L: supervision, editing; S.M.S.: conceptualization, methodology, data analysis, supervision, funding, first draft, editing.
Conflicts of interest
The authors declare no no competing interests.
Acknowledgments
We are grateful for the support of The Rally Foundation for Childhood Cancer Research grants and the Truth 365 519023 and 401943 (W.J.H., J.A.S., S.M.S.), the support of the NIH/NCI grant 1P50CA210964 and 1U54CA243126 (S.M.S.), NIH/NCI P30CA008748 (N.J.C., M.P.L.), NIH/NCATS grants Clinical and Translation Science Awards UL1TR001866 (G.L., S.M.S.) and UL1TR000043 (W.J.H. and SMS). We acknowledge the use of the Integrated Genomics Operation Core, funded by the NCI Cancer Center Support Grant (CCSG, P30 CA08748), Cycle for Survival, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology. We would like to acknowledge Irina Linkov of the MSKCC Pathology Core Facility for help. We are also grateful for the support and contributions of the patients and caregivers.
Published: July 7, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.06.003.
Supplemental information
Data and code availability
The transcriptome data are available under accession number dbGAP phs002439.v1.
References
- Abdul-Al H.M., Wang G., Makhlouf H.R., Goodman Z.D. Fibrolamellar hepatocellular carcinoma: an immunohistochemical comparison with conventional hepatocellular carcinoma. Int. J. Surg. Pathol. 2010;18:313–318. doi: 10.1177/1066896910364229. [DOI] [PubMed] [Google Scholar]
- Broutier L., Mastrogiovanni G., Verstegen M.M., Francies H.E., Gavarro L.M., Bradshaw C.R., Allen G.E., Arnes-Benito R., Sidorova O., Gaspersz M.P., et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat. Med. 2017;23:1424–1435. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calandrini C., Schutgens F., Oka R., Margaritis T., Candelli T., Mathijsen L., Ammerlaan C., van Ineveld R.L., Derakhshan S., de Haan S., et al. An organoid biobank for childhood kidney cancers that captures disease and tissue heterogeneity. Nat. Commun. 2020;11:1310. doi: 10.1038/s41467-020-15155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo C.M., Dijkstra K.K., Fanchi L.F., Kelderman S., Kaing S., van Rooij N., van den Brink S., Schumacher T.N., Voest E.E. Tumor organoid-T-cell coculture systems. Nat. Protoc. 2020;15:15–39. doi: 10.1038/s41596-019-0232-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig J.R., Peters R.L., Edmondson H.A., Omata M. Fibrolamellar carcinoma of the liver: a tumor of adolescents and young adults with distinctive clinico-pathologic features. Cancer. 1980;46:372–379. doi: 10.1002/1097-0142(19800715)46:2<372::AID-CNCR2820460227>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Darcy D.G., Chiaroni-Clarke R., Murphy J.M., Honeyman J.N., Bhanot U., LaQuaglia M.P., Simon S.M. The genomic landscape of fibrolamellar hepatocellular carcinoma: whole genome sequencing of ten patients. Oncotarget. 2015;6:755–770. doi: 10.18632/oncotarget.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driehuis E., van Hoeck A., Moore K., Kolders S., Francies H.E., Gulersonmez M.C., Stigter E.C.A., Burgering B., Geurts V., Gracanin A., et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc. Natl. Acad. Sci. USA. 2019;116:26580–26590. doi: 10.1073/pnas.1911273116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J., Karthaus W.R., Gao D., Driehuis E., Sawyers C.L., Chen Y., Clevers H. Organoid culture systems for prostate epithelial and cancer tissue. Nat. Protoc. 2016;11:347–358. doi: 10.1038/nprot.2016.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson H. Differential diagnosis of tumors and tumor-like lesions of liver in infancy and childhood. A. M. A. J. Dis. Child. 1956;91:168–186. doi: 10.1001/archpedi.1956.02060020170015. [DOI] [PubMed] [Google Scholar]
- Eggert T., McGlynn K.A., Duffy A., Manns M.P., Greten T.F., Altekruse S.F. Epidemiology of fibrolamellar hepatocellular carcinoma in the USA, 2000-10. Gut. 2013;62:1667–1668. doi: 10.1136/gutjnl-2013-305164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert T., McGlynn K.A., Duffy A., Manns M.P., Greten T.F., Altekruse S.F. Fibrolamellar hepatocellular carcinoma in the USA, 2000-2010: a detailed report on frequency, treatment and outcome based on the Surveillance, Epidemiology, and End Results database. United European Gastroenterol. J. 2013;1:351–357. doi: 10.1177/2050640613501507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Serag H.B., Davila J.A. Is fibrolamellar carcinoma different from hepatocellular carcinoma? A US population-based study. Hepatology. 2004;39:798–803. doi: 10.1002/hep.20096. [DOI] [PubMed] [Google Scholar]
- Engelholm L.H., Riaz A., Serra D., Dagnaes-Hansen F., Johansen J.V., Santoni-Rugiu E., Hansen S.H., Niola F., Frodin M. CRISPR/Cas9 engineering of adult mouse liver demonstrates that the Dnajb1-Prkaca gene fusion is sufficient to induce tumors resembling fibrolamellar hepatocellular carcinoma. Gastroenterology. 2017;153:1662–1673. doi: 10.1053/j.gastro.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber B.A., Lalazar G., Simon E.P., Hammond W.J., Requena D., Bhanot U.K., La Quaglia M.P., Simon S.M. Non coding RNA analysis in fibrolamellar hepatocellular carcinoma. Oncotarget. 2018;9:10211–10227. doi: 10.18632/oncotarget.23325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francies H.E., Barthorpe A., McLaren-Douglas A., Barendt W.J., Garnett M.J. Drug sensitivity assays of human cancer organoid cultures. Methods Mol. Biol. 2019;1576:339–351. doi: 10.1007/7651_2016_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francies H.E., Barthorpe A., McLaren-Douglas A., Barendt W.J., Garnett M.J. Erratum to: drug sensitivity assays of human cancer organoid cultures. Methods Mol. Biol. 2019;1576:353. doi: 10.1007/7651_2018_138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz A.G. Third edition. World Health Organization; 2013. International Classification of Diseases for Oncology: ICD-O. First revision. [Google Scholar]
- Gao D., Vela I., Sboner A., Iaquinta P.J., Karthaus W.R., Gopalan A., Dowling C., Wanjala J.N., Undvall E.A., Arora V.K., et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176–187. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldhammer N., Kim J., Timmermans-Wielenga V., Petersen O.W. Characterization of organoid cultured human breast cancer. Breast Cancer Res. 2019;21:141. doi: 10.1186/s13058-019-1233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.P., Jin L., Knutson D.L., Kloft-Nelson S.M., Greipp P.T., Waldburger N., Roessler S., Longerich T., Roberts L.R., Oliveira A.M., et al. DNAJB1-PRKACA is specific for fibrolamellar carcinoma. Mod. Pathol. 2015;28:822–829. doi: 10.1038/modpathol.2015.4. [DOI] [PubMed] [Google Scholar]
- Graham R.P., Lackner C., Terracciano L., Gonzalez-Cantu Y., Maleszewski J.J., Greipp P.T., Simon S.M., Torbenson M.S. Fibrolamellar carcinoma in the Carney complex: PRKAR1A loss instead of the classic DNAJB1-PRKACA fusion. Hepatology. 2018;68:1441–1447. doi: 10.1002/hep.29719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.P., Torbenson M.S. Fibrolamellar carcinoma: a histologically unique tumor with unique molecular findings. Semin. Diagn. Pathol. 2017;34:146–152. doi: 10.1053/j.semdp.2016.12.010. [DOI] [PubMed] [Google Scholar]
- Honeyman J.N., Simon E.P., Robine N., Chiaroni-Clarke R., Darcy D.G., Lim I.I., Gleason C.E., Murphy J.M., Rosenberg B.R., Teegan L., et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science. 2014;343:1010–1014. doi: 10.1126/science.1249484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Gehart H., Artegiani B., C L.O.-I., Dekkers F., Basak O., van Es J., Chuva de Sousa Lopes S.M., Begthel H., Korving J., et al. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell. 2018;175:1591–1606.e1519. doi: 10.1016/j.cell.2018.11.013. [DOI] [PubMed] [Google Scholar]
- Huch M., Gehart H., van Boxtel R., Hamer K., Blokzijl F., Verstegen M.M., Ellis E., van Wenum M., Fuchs S.A., de Ligt J., et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F., Salinas R.D., Zhang D.Y., Nguyen P.T.T., Schnoll J.G., Wong S.Z.H., Thokala R., Sheikh S., Saxena D., Prokop S., et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell. 2020;180:188–204.e122. doi: 10.1016/j.cell.2019.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakar S., Burgart L.J., Batts K.P., Garcia J., Jain D., Ferrell L.D. Clinicopathologic features and survival in fibrolamellar carcinoma: comparison with conventional hepatocellular carcinoma with and without cirrhosis. Mod. Pathol. 2005;18:1417–1423. doi: 10.1038/modpathol.3800449. [DOI] [PubMed] [Google Scholar]
- Kannangai R., Wang J., Liu Q.Z., Sahin F., Torbenson M. Survivin overexpression in hepatocellular carcinoma is associated with p53 dysregulation. Int. J. Gastrointest. Cancer. 2005;35:53–60. doi: 10.1385/IJGC:35:1:053. [DOI] [PubMed] [Google Scholar]
- Kastenhuber E.R., Lalazar G., Houlihan S.L., Tschaharganeh D.F., Baslan T., Chen C.C., Requena D., Tian S., Bosbach B., Wilkinson J.E., et al. DNAJB1-PRKACA fusion kinase interacts with beta-catenin and the liver regenerative response to drive fibrolamellar hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA. 2017;114:13076–13084. doi: 10.1073/pnas.1716483114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzenstein H.M., Krailo M.D., Malogolowkin M.H., Ortega J.A., Qu W., Douglass E.C., Feusner J.H., Reynolds M., Quinn J.J., Newman K., et al. Fibrolamellar hepatocellular carcinoma in children and adolescents. Cancer. 2003;97:2006–2012. doi: 10.1002/cncr.11292. [DOI] [PubMed] [Google Scholar]
- Klein W.M., Molmenti E.P., Colombani P.M., Grover D.S., Schwarz K.B., Boitnott J., Torbenson M.S. Primary liver carcinoma arising in people younger than 30 years. Am. J. Clin. Pathol. 2005;124:512–518. doi: 10.1309/TT0R7KAL32228E99. [DOI] [PubMed] [Google Scholar]
- Lalazar G., Simon S.M. Fibrolamellar carcinoma: recent advances and unresolved questions on the molecular mechanisms. Semin. Liver Dis. 2018;38:51–59. doi: 10.1055/s-0037-1621710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalazar G., Requena D., Ramos-Espiritu L., Ng D., Bhola P.D., de Jong Y.P., Wang R., Narayan N.J.C., Shebl B., Levin S., et al. Identification of novel therapeutic targets for fibrolamellar carcinoma using patient-derived xenografts and Direct-from-patient screening. Cancer Discov. 2021;11:2544–2563. doi: 10.1158/2159-8290.CD-20-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Hu W., Matulay J.T., Silva M.V., Owczarek T.B., Kim K., Chua C.W., Barlow L.J., Kandoth C., Williams A.B., et al. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell. 2018;173:515–528.e517. doi: 10.1016/j.cell.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Pan B., Song X., Li N., Zhao D., Li M., Zhao Z. Breast cancer organoids from a patient with giant papillary carcinoma as a high-fidelity model. Cancer Cell Int. 2020;20:86. doi: 10.1186/s12935-020-01171-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim I., Farber B.A., LaQuaglia M.P. Advances in fibrolamellar hepatocellular carcinoma: a Review. Eur. J. Pediatr. Surg. 2014;24:461–466. doi: 10.1055/s-0034-1396420. [DOI] [PubMed] [Google Scholar]
- Limaiem F., Bouraoui S., Sboui M., Bouslama S., Lahmar A., Mzabi S. Fibrolamellar carcinoma versus scirrhous hepatocellular carcinoma : diagnostic usefulness of CD68. Acta Gastroenterol. Belg. 2015;78:393–398. [PubMed] [Google Scholar]
- Lin C.C., Yang H.M. Fibrolamellar carcinoma: a concise Review. Arch. Pathol. Lab. Med. 2018;142:1141–1145. doi: 10.5858/arpa.2017-0083-RS. [DOI] [PubMed] [Google Scholar]
- Liu H.D., Xia B.R., Jin M.Z., Lou G. Organoid of ovarian cancer: genomic analysis and drug screening. Clin. Transl. Oncol. 2020;22:1240–1251. doi: 10.1007/s12094-019-02276-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malouf G.G., Tahara T., Paradis V., Fabre M., Guettier C., Yamazaki J., Long H., Lu Y., Raynal N.J., Jelinek J., et al. Methylome sequencing for fibrolamellar hepatocellular carcinoma depicts distinctive features. Epigenetics. 2015;10:872–881. doi: 10.1080/15592294.2015.1076955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na J.C., Kim J.H., Kim S.Y., Gu Y.R., Jun D.Y., Lee H.H., Yoon Y.E., Choi K.H., Hong S.J., Han W.K. Establishment of patient-derived three-dimensional organoid culture in renal cell carcinoma. Investig. Clin. Urol. 2020;61:216–223. doi: 10.4111/icu.2020.61.2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njei B., Konjeti V.R., Ditah I. Prognosis of patients with fibrolamellar hepatocellular carcinoma versus conventional hepatocellular carcinoma: a systematic Review and meta-analysis. Gastrointest. Cancer Res. 2014;7:49–54. [PMC free article] [PubMed] [Google Scholar]
- Ooft S.N., Weeber F., Dijkstra K.K., McLean C.M., Kaing S., van Werkhoven E., Schipper L., Hoes L., Vis D.J., van de Haar J., et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aay2574. [DOI] [PubMed] [Google Scholar]
- Pasch C.A., Favreau P.F., Yueh A.E., Babiarz C.P., Gillette A.A., Sharick J.T., Karim M.R., Nickel K.P., DeZeeuw A.K., Sprackling C.M., et al. Patient-derived cancer organoid cultures to predict sensitivity to chemotherapy and radiation. Clin. Cancer Res. 2019;25:5376–5387. doi: 10.1158/1078-0432.CCR-18-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puca L., Bareja R., Prandi D., Shaw R., Benelli M., Karthaus W.R., Hess J., Sigouros M., Donoghue A., Kossai M., et al. Patient derived organoids to model rare prostate cancer phenotypes. Nat. Commun. 2018;9:2404. doi: 10.1038/s41467-018-04495-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross H.M., Daniel H.D.J., Vivekanandan P., Kannangai R., Yeh M.M., Wu T.-T., Makhlouf H.R., Torbenson M. Fibrolamellar carcinomas are positive for CD68. Mod. Pathol. 2011;24:390–395. doi: 10.1038/modpathol.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltsman J.A., Hammond W.J., Narayan N.J.C., Requena D., Gehart H., Lalazar G., LaQuaglia M.P., Clevers H., Simon S.M. A human organoid model of aggressive hepatoblastoma for disease modeling and drug testing. Cancers (Basel) 2020;12 doi: 10.3390/cancers12092668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki N., Clevers H. Studying cellular heterogeneity and drug sensitivity in colorectal cancer using organoid technology. Curr. Opin. Genet. Dev. 2018;52:117–122. doi: 10.1016/j.gde.2018.09.001. [DOI] [PubMed] [Google Scholar]
- Simon E.P., Freije C.A., Farber B.A., Lalazar G., Darcy D.G., Honeyman J.N., Chiaroni-Clarke R., Dill B.D., Molina H., Bhanot U.K., et al. Transcriptomic characterization of fibrolamellar hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA. 2015;112:E5916–E5925. doi: 10.1073/pnas.1424894112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhi A.D., Wood L.D., Parks E., Torbenson M.S., Felsenstein M., Hruban R.H., Nikiforova M.N., Wald A.I., Kaya C., Nikiforov Y.E., et al. Recurrent rearrangements in PRKACA and PRKACB in intraductal oncocytic papillary neoplasms of the Pancreas and bile duct. Gastroenterology. 2019;158:573–582.e2. doi: 10.1053/j.gastro.2019.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terris B., Pineau P., Bregeaud L., Valla D., Belghiti J., Tiollais P., Degott C., Dejean A. Close correlation between beta-catenin gene alterations and nuclear accumulation of the protein in human hepatocellular carcinomas. Oncogene. 1999;18:6583–6588. doi: 10.1038/sj.onc.1203051. [DOI] [PubMed] [Google Scholar]
- Torbenson M. Fibrolamellar carcinoma: 2012 update. Scientifica. 2012;2012:743790. doi: 10.6064/2012/743790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eyken P., Sciot R., Brock P., Casteels-Van Daele M., Ramaekers F.C., Desmet V.J. Abundant expression of cytokeratin 7 in fibrolamellar carcinoma of the liver. Histopathology. 1990;17:101–107. doi: 10.1111/j.1365-2559.1990.tb00679.x. [DOI] [PubMed] [Google Scholar]
- Vyas M., Hechtman J.F., Zhang Y., Benayed R., Yavas A., Askan G., Shia J., Klimstra D.S., Basturk O. DNAJB1-PRKACA fusions occur in oncocytic pancreatic and biliary neoplasms and are not specific for fibrolamellar hepatocellular carcinoma. Mod. Pathol. 2019;33:648–656. doi: 10.1038/s41379-019-0398-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S.C., Huang J., Tickoo S.K., Thung S.N., Ladanyi M., Klimstra D.S. Fibrolamellar carcinoma of the liver exhibits immunohistochemical evidence of both hepatocyte and bile duct differentiation. Mod. Pathol. 2010;23:1180–1190. doi: 10.1038/modpathol.2010.105. [DOI] [PubMed] [Google Scholar]
- Weeda V.B., Murawski M., McCabe A.J., Maibach R., Brugieres L., Roebuck D., Fabre M., Zimmermann A., Otte J.B., Sullivan M., et al. Fibrolamellar variant of hepatocellular carcinoma does not have a better survival than conventional hepatocellular carcinoma – results and treatment recommendations from the Childhood Liver Tumour Strategy Group (SIOPEL) experience. Eur. J. Cancer. 2013;49:2698–2704. doi: 10.1016/j.ejca.2013.04.012. [DOI] [PubMed] [Google Scholar]
- Yamashita S., Vauthey J.N., Kaseb A.O., Aloia T.A., Conrad C., Hassan M.M., Passot G., Raghav K.P., Shama M.A., Chun Y.S. Prognosis of fibrolamellar carcinoma compared to non-cirrhotic conventional hepatocellular carcinoma. J. Gastrointest. Surg. 2016;20:1725–1731. doi: 10.1007/s11605-016-3216-x. [DOI] [PubMed] [Google Scholar]
- Yao Y., Xu X., Yang L., Zhu J., Wan J., Shen L., Xia F., Fu G., Deng Y., Pan M., et al. Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell. 2020;26:17–26.e16. doi: 10.1016/j.stem.2019.10.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptome data are available under accession number dbGAP phs002439.v1.