Abstract
Uncontrolled epithelial cell proliferation in the prostate transition zone and the hyper-accumulation of mesenchymal-like cells derived from the epithelial-mesenchymal transition (EMT) of prostatic epithelium are two key processes in benign prostatic hyperplasia (BPH). m6A RNA modification affects multiple cellular processes, including cell proliferation, apoptosis, and differentiation. In this study, the aberrant up-regulation of methylase METTL3 in BPH samples suggests its potential role in BPH development. Elevated m6A modification in the prostate of the BPH rat was partially reduced by METTL3 knockdown. METTL3 knockdown also partially reduced the prostatic epithelial thickness and prostate weight, significantly improved the histological features of the prostate, inhibited epithelial proliferation and EMT, and promoted apoptosis. In vitro, METTL3 knockdown decreased TGF-β-stimulated BPH-1 cell proliferation, m6A modification, and EMT, whereas promoted cell apoptosis. METTL3 increased the m6A modification of PTEN and inhibited its expression through the reading protein YTHDF2. PTEN knockdown aggravated the molecular, cellular, and pathological alterations in the prostate of BPH rats and amplified TGF-β-induced changes in BPH-1 cells. More importantly, PTEN knockdown partially abolished the improving effects of METTL3 knockdown both in vivo and in vitro. In conclusion, the level of m6A modification is elevated in BPH; the METTL3/YTHDF2/PTEN axis disturbs the balance between epithelial proliferation and apoptosis, promotes EMT, and accelerates BPH development in an m6A modification-related manner.
Subject terms: Cell biology, Molecular biology
Introduction
Benign prostatic hyperplasia (BPH) is a frequent urinary tract disorder in the elderly male that causes lower urinary tract symptoms [1, 2]. While the specific etiology of BPH is largely unknown, several cellular processes have been reported to be involved in BPH initiation and development. BPH disrupts growth regulation and homeostatic control of proliferative and apoptotic behavior in the periurethral and transition zones, resulting in uncontrolled proliferation and migration of prostatic epithelium and enlargement of the cell proliferating compartment [3, 4]. Furthermore, increased mesenchymal-like cells derived from the prostatic epithelium via epithelial-mesenchymal transition (EMT) also contribute to BPH development [5–7]. Agents or factors that regulate the balance of epithelial proliferation, apoptosis, and EMT might be interesting targets for BPH treatment regimens.
N6-methyladenosine (m6A) has been identified as the most frequent and abundant post-transcriptional modification, notably in eukaryotic messenger RNA (mRNA) [8, 9]. In BPH, hypermethylation has been demonstrated. By performing a comprehensive molecular investigation of BPH, including genomic, transcriptomic, and epigenetic profiling, Liu et al. [10] discovered that global hypermethylation is the dominant process in BPH at the epigenetic level. Integrating transcription and methylation signatures identifies two BPH subgroups with distinct clinical characteristics and signaling pathways, which have been validated in two independent cohorts. For instance, in BPH samples, promoter methylation of 14-3-3, a potential tumor suppressor gene involved in cell cycle control and death following DNA damage, is homogeneous [11]. Changes in CpG island methylation at MDR1 occur with a frequency of 71%, which may promote growth [12]. Reportedly, m6A modification affects multiple cell behaviors, including proliferation, apoptosis, migration, and EMT [13, 14], which all play a critical role in BPH development. Previous studies have shown that m6A regulators such as methyltransferase-like 3/14 (METTL3/14) and YTH domain family members, namely, YTHDC2, YTHDF1, and YTHDF2 played crucial functions in the progress of m6A modifications, such as mRNA decay [15], pre-ribosomal RNA processing [16], and so on. These mechanisms were associated with diverse physiological and pathological processes, such as inducing pluripotent stem cells [17], regulating inflammatory response [18], adipogenesis [19]. Among then, YTHDF2, as a member of m6A “readers”, played a significant role in multiple diseases, such as cardiac hypertrophy [20] and intervertebral disc degeneration [21]. However, the specific effects of m6A modification on BPH and potential mechanisms are still unclear.
This study established the rat BPH model by testosterone injection, and m6A modification levels, m6A loci, and genes with expression differences and altered m6A status were analyzed to identify key methylases and target genes. The in vivo and in vitro effects of identified methylase, the predicted binding between methylase and target gene, and the dynamic effects of the methylase and target gene on the BPH model and TGF-β-stimulated BPH-1 cells were investigated. Collectively, the present study aims to demonstrate the specific effects and the underlying mechanism of critical methylases and target genes in BPH.
Results
Methylases altered in rat BPH model and correlated with BPH etiology
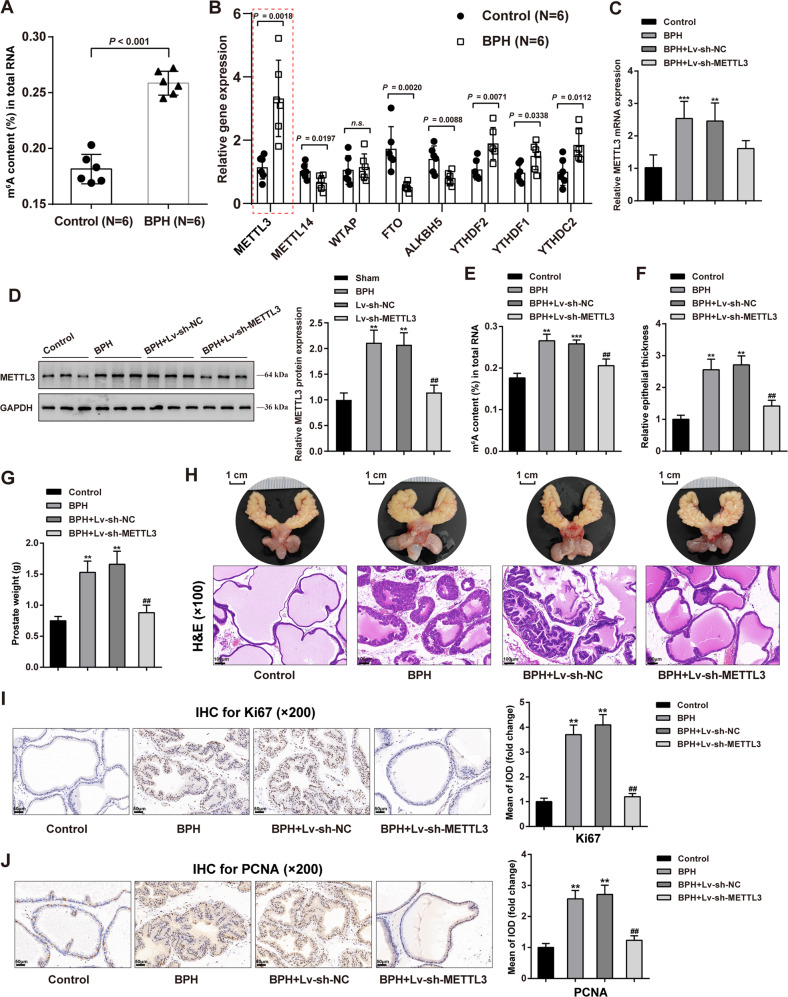
For identifying critical methylases correlated with BPH progression, the m6A status of RNA from normal or hyperplasic prostate was determined. Figure 1A shows that m6A -modified RNA levels were remarkably higher within hyperplasic prostate samples than in normal rat prostate. The expression levels of methylation-associated enzymes, including methylation enzymes: METTL3 and METTL14; m6A methyltransferase complex WTAP; demethylation enzymes: FTO, ALKBH5; and reading proteins: YTHDF2, YTHDF1, YTHDC2 were evaluated. Figure 1B shows that methylase METTL3 expression level was remarkably upregulated within BPH samples (P = 0.0018). Considering the role of METTL3 in BPH development remains unclear, METTL3 was selected for further investigations.
Fig. 1. Methylases altered in BPH and methylase METTL3 regulates prostatic hyperplasia in model rats.
Prostatic hyperplasia model was induced in rats by subcutaneous injection of testosterone as described in the M&M section. A m6A status of RNA from normal or hyperplasic rat prostate were determined using an m6A RNA Methylation Assay Kit. B The expression of methylation-related enzymes was examined in normal and hyperplasic prostate tissues using qRT-PCR, including methylation enzymes: METTL3, METTL14, WTAP; demethylation enzymes: FTO, ALKBH5; reading proteins: YTHDF2, YTHDF1, YTHDC2. C, D METTL3 knockdown was achieved in rat models by transducing lentivirus containing short hairpin targeting METTL3; METTL3 expression was confirmed in rats in each group using qRT-PCR (C) and Immunoblotting (D). E Methylated N6-methyladenosine (m6A) in RNA was evaluated using an m6A RNA Methylation Assay Kit. F, G Prostatic epithelial thickness (F) and prostate weight (G) were evaluated at the end of the modeling and treatment. H A representative prostate with the urinary bladder from each group were shown (upper) and the histopathological characteristics of rats’ prostate from each group were evaluated using H&E staining (down). Scale bar = 1 cm or 100 μm. I, J The contents of Ki67 and PCNA in rats’ prostate tissues were determined using Immunohistochemical (IHC) staining. Scale bar = 50 μm. **P < 0.01, ***P < 0.001, compared with the control group; #P < 0.05, ##P < 0.01, compared with the BPH + Lv-sh-NC group.
Methylase METTL3 regulates prostatic hyperplasia in model rats
For investigating the specific effects of METTL3 on BPH, the prostatic hyperplasia model was induced in rats by subcutaneous injection of testosterone as described, and METTL3 knockdown was achieved in rat models by injecting lentivirus containing sh-METTL3. METTL3 knockdown was confirmed in rats in each group using qRT-PCR and Immunoblotting (Fig. 1C, D). The levels of m6A-modified RNA were evaluated in four groups. Figure 1E shows that the m6A modification level was dramatically increased in the BPH and BPH + Lv-sh-NC groups than in normal control (**P < 0.01, ***P < 0.001), whereas remarkably decreased within the BPH + Lv-sh-METTL3 group than the BPH + Lv-sh-NC group (##P < 0.01). These findings indicate that m6A modification increases in BPH rats, but METTL3 knockdown partially reduces m6A modification in BPH rats.
Regarding related symptoms and pathological characteristics, the prostatic epithelial thickness and prostate weight increased in BPH rats compared with the control group (**P < 0.01), whereas partially decreased by METTL3 knockdown (##P < 0.01; Fig. 1F, G). A representative prostate from each group was shown and the histopathological characteristics of rats’ prostates from each group were evaluated using H&E staining. Figure 1H shows that compared with those of the control group, the thickness of the epithelial layer surrounding glands and the stromal components in the rats’ prostate from the BPH and BPH + Lv-sh-NC groups were significantly enhanced, indicating the presence of BPH. In BPH + Lv-sh-METTL3 group, the prostate’s histological characteristics showed to be significantly enhanced, indicating the underlying anti-BPH therapeutic effect of sh-METTL3 on BPH rats.
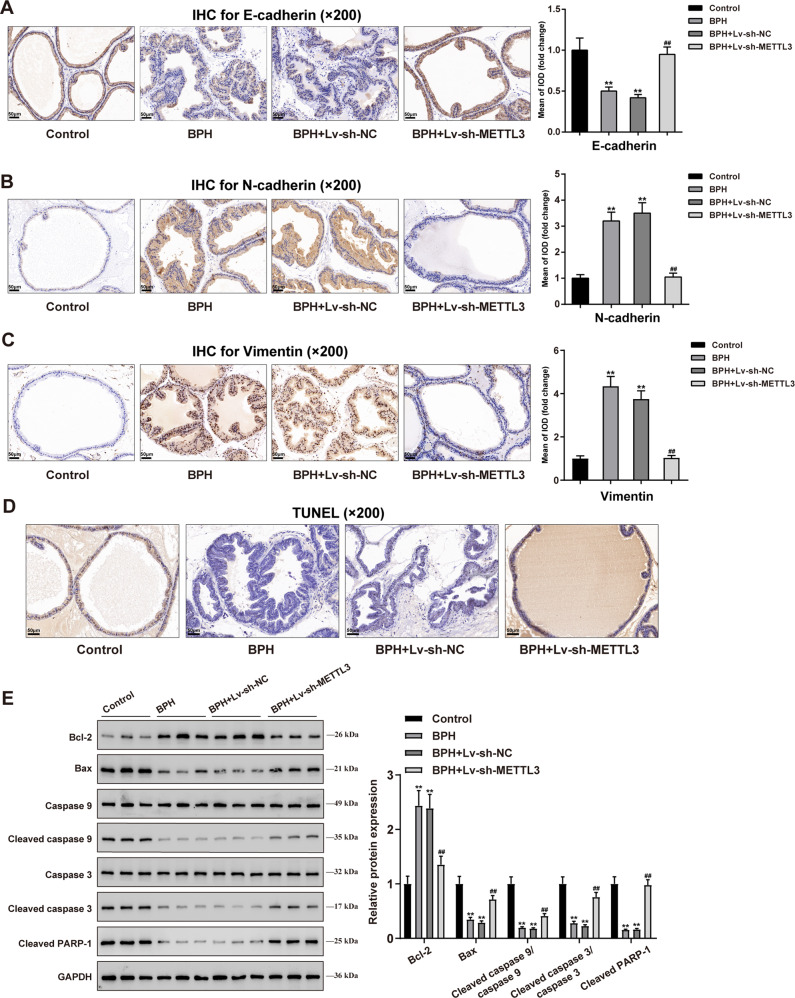
Furthermore, IHC staining was employed to determine the contents of Ki67 and PCNA in rats’ prostate tissue samples. Figure 1I, J shows that Ki67 and PCNA levels were dramatically elevated in BPH rats (**P < 0.01) and remarkably decreased by METTL3 knockdown (##P < 0.01), further suggesting that METTL3 knockdown inhibits cell proliferation in hyperplasic prostate. Moreover, the contents of EMT markers, including E-cadherin, N-cadherin, and vimentin were examined in rats’ prostate tissues. In BPH rats’ prostate, E-cadherin was reduced (**P < 0.01), but N-cadherin and vimentin were elevated (**P < 0.01); after knocking down METTL3, E-cadherin was partially increased, but N-cadherin and vimentin were decreased (##P < 0.01; Fig. 2A–C), suggesting that METTL3 knockdown attenuates the EMT in BPH rats. Moreover, TUNEL assay results indicated cell apoptosis was significantly inhibited in BPH rats but promoted after METTL3 knockdown (Fig. 2D). And the apoptosis-related proteins (Bcl-2, Bax, Caspase 9, cleaved caspase 9, Caspase 3, cleaved caspase 3, and cleaved PARP-1) levels in prostate tissues of each group were detected by Immunoblotting. In BPH rats, Bcl-2 protein level was significantly increased, while Bax, cleaved caspase 9, cleaved caspase 3 and cleaved PARP-1 protein levels were inhibited. But METTL3 knockdown could reverse these results (Fig. 2E). Considering the changes in Ki67 and PCNA and apoptosis-related proteins, METTL3 might disturb the balance between epithelial proliferation and apoptosis.
Fig. 2. METTL3 knockdown inhibits epithelial-mesenchymal transition (EMT) protein expression and promotes apoptosis in rats’ hyperplasic prostate.
A–C The contents of E-cadherin (A), N-cadherin (B), and Vimentin (C) in rats’ prostate tissues were determined using IHC staining. D Cell apoptosis in rats’ prostate tissues was determined using a TUNEL assay. E The apoptosis-related proteins (Bcl-2, Bax, Caspase 9, cleaved caspase 9, Caspase 3, cleaved caspase 3, and cleaved PARP-1) levels in prostate tissues in each group were detected by Immunoblotting. All scale bar = 100 μm. **P < 0.01, compared with the control group; ##P < 0.01, compared with the BPH + Lv-sh-NC group.
Effects of METTL3 knockdown on TGF-β-treated BPH-1 cell proliferation, apoptosis, and EMT
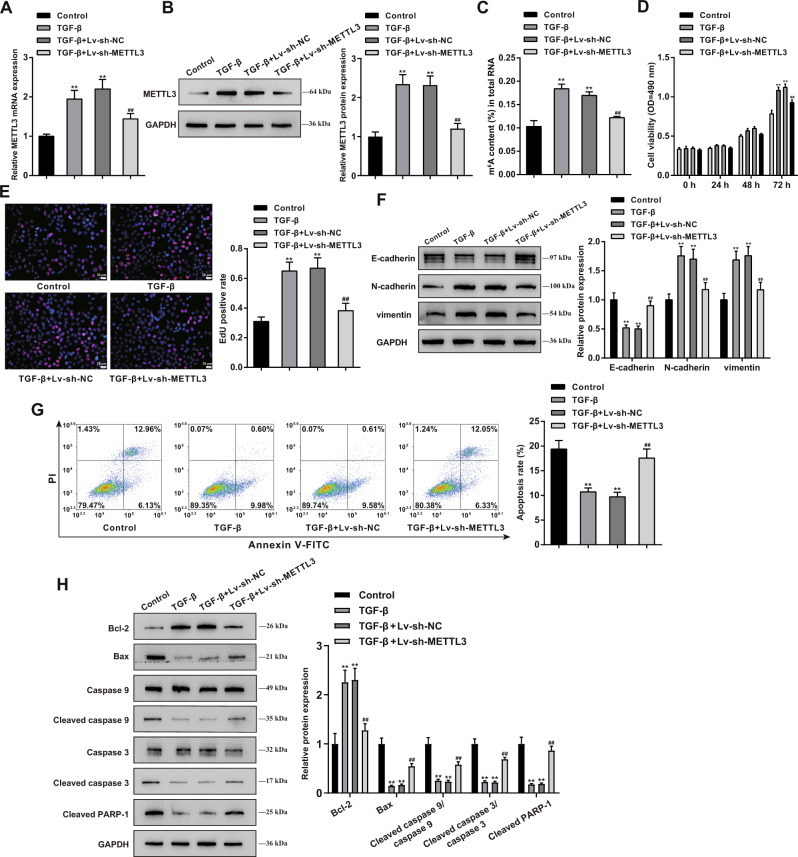
Regarding the cellular effects of METTL3, BPH-1 cells were stimulated with TGF-β for simulating BPH, and METTL3 knockdown was achieved in TGF-β-treated BPH-1 cells by transducing Lv-sh-METTL3. METTL3 knockdown in BPH-1 cells was confirmed using qRT-PCR and Immunoblotting (Fig. 3A, B). Cellular m6A status in RNA was examined. Figure 3C shows that TGF-β stimulation resulted in an obvious elevation within m6A modification (**P < 0.01), which showed to be partially decreased via METTL3 knockdown (##P < 0.01). TGF-β treatment also considerably enhanced cell viability (**P < 0.01; Fig. 3D) and DNA synthesis (**P < 0.01; Fig. 3E), which were partially suppressed by METTL3 knockdown (##P < 0.01; Fig. 3D, E). As for cell EMT, E-cadherin protein contents were reduced, but N-cadherin and vimentin protein contents were elevated in TGF-β-stimulated BPH-1 cells (**P < 0.01); after knocking down METTL3, E-cadherin was elevated, but N-cadherin and vimentin were reduced (##P < 0.01; Fig. 3F). Regarding cell apoptosis, TGF-β stimulation significantly inhibited cell apoptosis (**P < 0.01), whereas METTL3 knockdown partially promoted cell apoptosis (##P < 0.01; Fig. 3G). Moreover, TGF-β treatment notably facilitated Bcl-2 protein level and restrained Bax, cleaved caspase 9, cleaved caspase 3, and cleaved PARP-1 protein levels (**P < 0.01); while METTL3 knockdown exerted the opposite effect with TGF-β treatment (##P < 0.01; Fig. 3H). In addition, the effects of METTL3 knockdown on the cellular survival and apoptosis of the BPH-1 cells under cytotoxic conditions were also determined. As shown in Fig. S1, BPH-1 cells were transduced with sh-METTL3 to achieve METTL3 knockdown and then were stimulated with H2O2. The results indicated that H2O2 stimulation markedly inhibited cell viability (Fig. S1A) and enhanced cell apoptosis (Fig. S1B), and METTL3 knockdown further reinforced the effect of H2O2 treatment on BPH-1 cells.
Fig. 3. Effects of METTL3 knockdown on proliferation, apoptosis, and EMT of TGF-β-treated BPH-1 cells.
METTL3 knockdown was achieved in TGF-β-treated BPH-1 cells by transducing Lv-sh-METTL3 and then examined for A, B METTL3 expression using qRT-PCR (A) and Immunoblotting (B); C m6A status in RNA using an m6A RNA Methylation Assay Kit; D cell viability using MTT assay; E DNA synthesis using EdU assay; Scale bar = 20 μm; F the protein levels of E-cadherin, N-cadherin, vimentin using Immunoblotting; G cell apoptosis using flow cytometry; H the protein levels of Bcl-2, Bax, Caspase 9, cleaved caspase 9, Caspase 3, cleaved caspase 3, and cleaved PARP-1 using Immunoblotting. **P < 0.01, compared with the control group; ##P < 0.01, compared with the TGF-β + Lv-sh-NC group.
PTEN is highly m6A-modified and downregulated in BPH
Methylases exert their functions through modified RNAs. For identifying modified RNA correlated with METTL3 functions, three pairs of prostate tissues were collected from normal control rats’ prostates and BPH rats’ prostates for methylation sequencing analysis. From the map showing m6A modification distribution in the chromosome region, it can be seen that the vast majority of m6A modification loci in the six samples are in the CDS coding region and the 3′-UTR region (Fig. S2A), suggesting the importance of the m6A locus in BPH. The proportion chart also saw a similar trend: most m6A modification loci were in CDS (56.69%), followed by 3′-UTR (38.15%), and only a very small proportion were in 5′-UTR (5.16%) (Fig. S2B). By comparing the m6A peak differences between the control and BPH prostate samples, 35 m6A loci belonging to 30 genes were negatively regulated (logFC < 0.25, P < 0.05), 12 of which were located in CDS and 22 in 3′-UTR; 257 m6A loci belonging to 211 genes were upregulated during BPH (logFC > 1.25, P < 0.05), 135 loci in the CDS and 68 loci in the 3′-UTR (Fig. S2C). These 241 altered genes were then applied for functional enrichment annotation. Fig. S3A shows that these 241 genes were enriched in glandular development, differentiation of myeloid leukocytes, skeletal muscle development, and negative regulation of epithelial cell migration, which were correlated with BPH development. Analysis of protein interaction networks and MCODE modules revealed that core genes regulating BPH are associated with cellular stress response, mRNA metabolic processes, and the regulation of protein kinase activity (Fig. S3B).
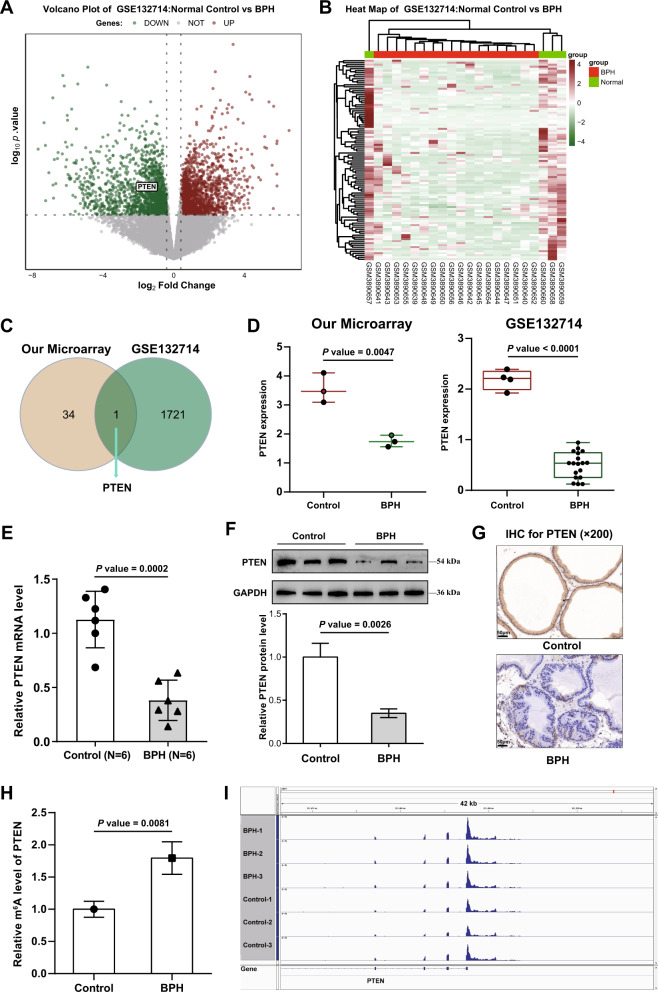
Then, the differential genes were cross-screened in combination with the expression profile between control and BPH samples, and genes with changes in m6A modification and mRNA expression were analyzed. The expression profile between control and BPH samples was analyzed using the R language LIMMA package based on GSE132714. Figure 4A, B shows that 1722 downregulated genes (logFC < −0.4, P < 0.05) and 1677 upregulated genes (logFC > 0.4, P < 0.05) were identified, among which PTEN was with increased m6A modification status and downregulated expression level (Fig. 4C). According to our methylation sequencing analysis and GSE132714, PTEN mRNA expression showed to be remarkably reduced within BPH samples than the normal controls (Fig. 4D). Consistently, PTEN mRNA and protein contents showed remarkably reduced within BPH samples than in the normal control samples, as inferred from qRT-PCR, Immunoblotting, and IHC staining results (Fig. 4E–G). Importantly, the PTEN m6A modification level was dramatically elevated in BPH samples than in the normal control (Fig. 4H). Moreover, most m6A modification loci in PTEN were in the 3′-UTR (Fig. 4I), suggesting that elevated m6A modification in PTEN might play a role in BPH.
Fig. 4. PTEN is highly m6A-modified and downregulated in BPH.
A, B The expression profile between control and BPH samples was analyzed using the R language LIMMA package based on GSE132714. C Genes with altered m6A modification status and altered expression levels were compared, and PTEN was selected. D PTEN expression in normal control and BPH samples according to our microarray and GSE132714. E the mRNA level of PTEN in normal control and BPH samples were determined using RT-PCR. F, G PTEN protein levels in normal control and BPH samples were determined using Immunoblotting and IHC staining. H The m6A modification status of PTEN in normal control and BPH samples was determined using an m6A RNA Methylation Assay Kit. I The m6A modification status in PTEN 3′UTR was analyzed using IGV software.
METTL3 mediates m6A modification of PTEN and regulates its expression through reading protein YTHDF2
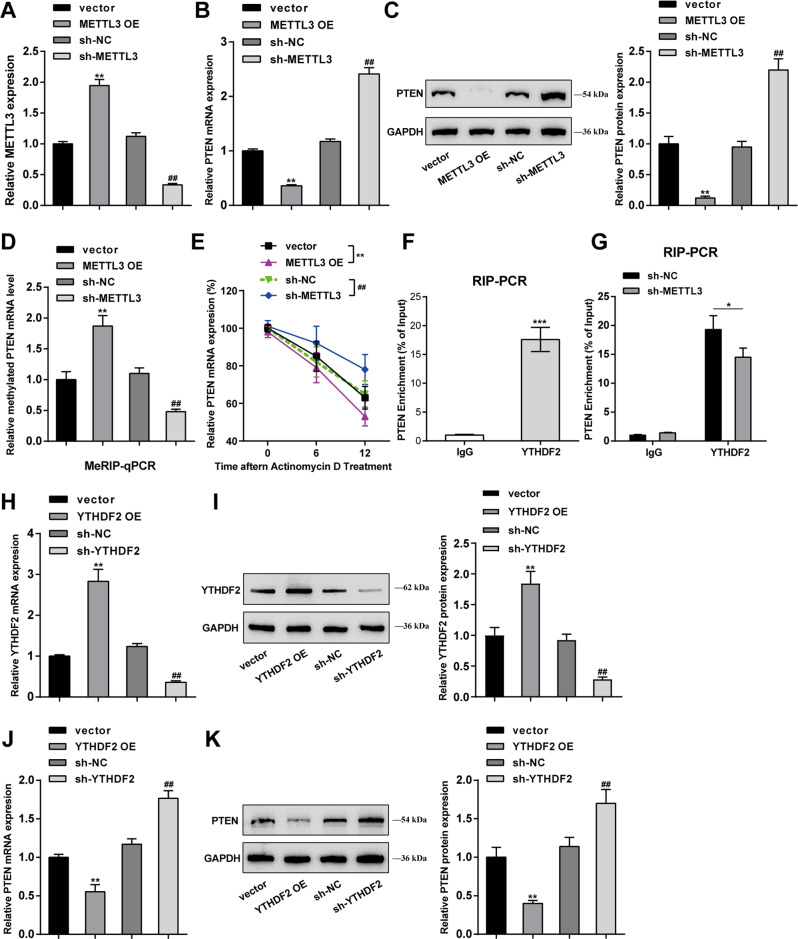
Prior to analyzing the specific effects of PTEN upon BPH, the effects and mechanism of METTL3 in regulating PTEN were first explored. OE-METTL3/sh-METTL3 was transfected to achieve METTL3 overexpression/knockdown in BPH-1 cells, as confirmed by qRT-PCR (Fig. 5A). Within BPH-1 cells, METTL3 overexpression dramatically downregulated PTEN mRNA expression and decreased PTEN protein levels, whereas METTL3 knockdown exerted opposing effects on PTEN (**P < 0.01, compared with the vector group; ##P < 0.01, compared with the sh-NC group; Fig. 5B, C). As for directly investigating the m6A modification status of PTEN by METTL3, the Me-RIP assay was performed, and METTL3 promoted the m6A modification level of PTEN mRNA (**P < 0.01, compared with the vector group; ##P < 0.01, compared with the sh-NC group; Fig. 5D). Furthermore, the actinomycin D RNA synthesis assay showed that PTEN mRNA levels were highly unstable in METTL3-overexpressing cells, while METTL3 knockdown showed the opposite effect (**P < 0.01, compared with the vector group; ##P < 0.01, compared with the sh-NC group; Fig. 5E). Moreover, METTL3-mediated PTEN downregulation has shown to affect the important cancer-associated processes [22, 23], the METTL3 and PTEN expression and their correlation in prostate cancer patients were analyzed based on bioinformatic analysis (Fig. S4). METTL3 expression showed to be dramatically upregulated within prostatic carcinoma samples in comparison with normal control, according to The Cancer Genome Atlas Prostate Adenocarcinoma (TCGA-PRAD) dataset (Fig. S4A) and Gene Expression Omnibus (GEO) dataset (GSE200879) (Fig. S4D). While PTEN expression was observably downregulated in prostatic carcinoma samples when compared to normal control, according to the TCGA-PRAD dataset (Fig. S4B) and GSE200879 dataset (Fig. S4E). Then, Pearson’s correlation analysis based on TCGA-PRAD, and GSE200879 datasets was conducted to analyze the correlation between METTL3 and PTEN expression; according to these two datasets, METTL3 and PTEN were negatively correlated (Fig. S4C, F).
Fig. 5. METTL3 mediates m6A modification of PTEN and regulates its expression through reading protein YTHDF2.
A METTL3 overexpression or knockdown was achieved in BPH-1 cells by transfecting OE-METTL3 or sh-METTL3 and confirmed using qRT-PCR. BPH-1 cells were transduced with OE-METTL3 or sh-METTL3 and examined for PTEN mRNA expression using qRT-PCR (B); PTEN protein levels using Immunoblotting (C). The m6A modification status of PTEN using Me-RIP assay (D); PTEN mRNA synthesis using actinomycin D assay (E). **P < 0.01, compared with the vector group; ##P < 0.01, compared with the sh-NC group. F The binding between YTHDF2 and PTEN was validated in BPH-1 cells using RIP assay with anti-IgG and anti-YTHDF2. ***P < 0.001, compared with the anti-IgG group. G The binding between YTHDF2 and PTEN was validated in sh-NC or sh-METTL3 transduced BPH-1 cells using RIP assay with anti-IgG and anti-YTHDF2. *P < 0.05, compared with the sh-NC group. (H, I) YTHDF2 overexpression or knockdown was achieved in BPH-1 cells by transfecting OE-YTHDF2 or sh-YTHDF2 and confirmed using qRT-PCR and Immunoblotting, respectively. (J, K) BPH-1 cells were transduced with OE-YTHDF2 or sh-YTHDF2 and examined for PTEN mRNA expression and protein levels using qRT-PCR and Immunoblotting, respectively. **P < 0.01, compared with the vector group; ##P < 0.01, compared with the sh-NC group.
YTHDF2 has been reported that could induce target mRNA degradation by reading m6A modification sites [24]. In addition, YTHDF2 was upregulated in BPH model rats (Fig. 1B). To explore the role of YTHDF2 in BPH progress, the effects of YTHDF2 silencing on TGF-β-treated BPH-1 cell proliferation, apoptosis, and EMT were investigated (Fig. S5). Firstly, YTHDF2 knockdown was achieved in TGF-β-treated BPH-1 cells by transducing sh-YTHDF2 and confirmed using qRT-PCR (Fig. S5A). YTHDF2 knockdown observably suppressed TGF-β-treated BPH-1 cell viability (Fig. S5B) and DNA synthesis (Fig. S5C). As for cell EMT, YTHDF2 knockdown promoted E-cadherin protein contents and inhibited N-cadherin and vimentin protein contents (Fig. S5D). Regarding cell apoptosis, YTHDF2 knockdown significantly facilitated cell apoptosis (Fig. S5E) and apoptosis-related protein levels (Fig. S5F). Therefore, the binding between YTHDF2 and PTEN was validated in BPH-1 cells using RIP assay with anti-IgG and anti-YTHDF2. Figure 5F shows that in immunoprecipitate by anti-YTHDF2, the enrichment levels of PTEN were dramatically higher than those in anti-IgG immunoprecipitate (***P < 0.001). Moreover, in immunoprecipitate by anti-YTHDF2 in sh-METTL3 transduced BPH-1 cells, the enrichment levels of PTEN were significantly lower than those in sh-NC transduced BPH-1 cells (*P < 0.05; Fig. 5G), suggesting that METTL3 modulates PTEN expression through YTHDF2. For validating the speculation, OE-YTHDF2/sh-YTHDF2 was transfected to achieve YTHDF2 overexpression/knockdown in BPH-1 cells, as confirmed by qRT-PCR and Immunoblotting (**P < 0.01, compared with the vector group; ##P < 0.01, compared with the sh-NC group; Fig. 5H, I). In BPH-1 cells, YTHDF2 overexpression also significantly downregulated PTEN mRNA expression and decreased PTEN protein levels, whereas YTHDF2 knockdown exerted opposite effects on PTEN (**P < 0.01, compared with the vector group; ##P < 0.01, compared with the sh-NC group; Fig. 5J, K).
METTL3 regulates PTEN and affects cell proliferation and EMT in prostatic hyperplasia model rats
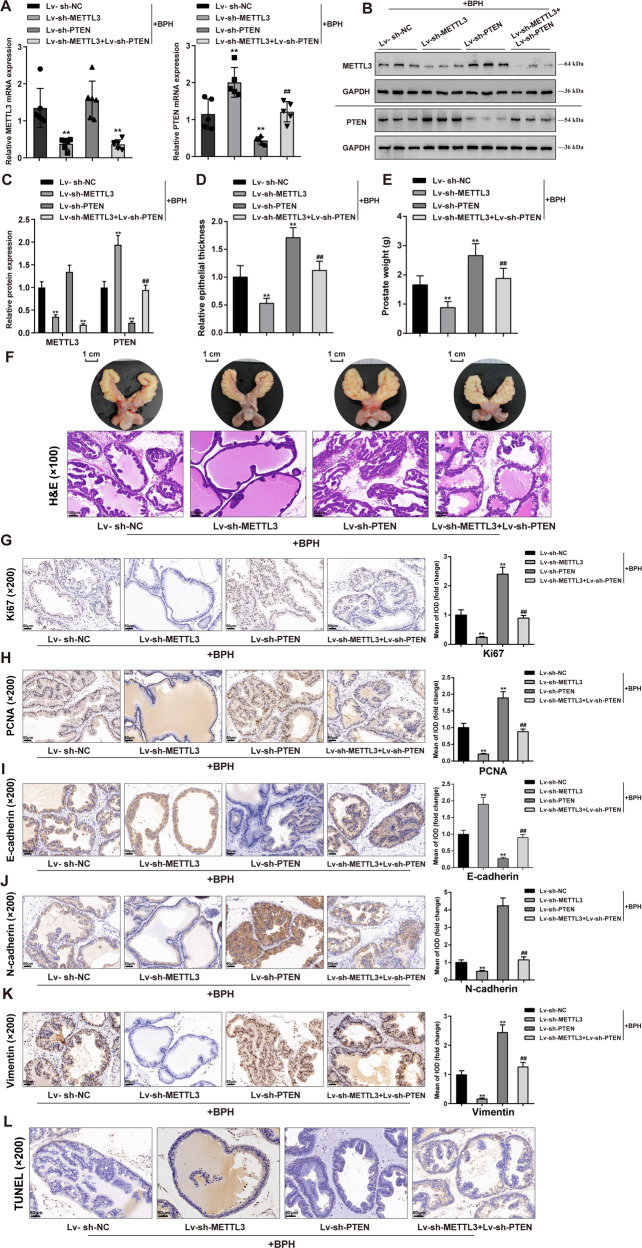
After confirming METTL3 regulating PTEN through YTHDF2, the dynamic effects of METTL3 and PTEN on the BPH model and cell model were investigated. BPH rats were injected with Lv-sh-METTL3 and/or Lv-sh-PTEN and examined for the mRNA and protein expressions of METTL3 and PTEN in rats’ prostate; in BPH rats’ prostate, METTL3 mRNA and protein expressions were downregulated by sh-METTL3; while didn’t affect by sh-PTEN (Fig. 6A–C). And, PTEN mRNA and protein expressions were upregulated after knocking down METTL3 but downregulated after Lv-sh-PTEN injection (**P < 0.01), whereas Lv-sh-PTEN significantly reversed the effects of METTL3 knockdown upon PTEN expression (##P < 0.01; Fig. 6A–C). METTL3 knockdown significantly reduced, whereas PTEN knockdown increased prostatic epithelial thickness and prostate weight (P < 0.01), and the effects of METTL3 knockdown were partially eliminated by PTEN knockdown (##P < 0.01; Fig. 6D, E). A representative prostate from each group was shown (Fig. 6F). H&E staining also indicated the improvement of hyperplasia symptoms by METTL3 knockdown but the exacerbation by PTEN knockdown (P < 0.01). Similarly, the improving effects of METTL3 knockdown were partially abolished by PTEN knockdown (##P < 0.01; Fig. 6F). PTEN knockdown significantly increased the contents of Ki67, PCNA, N-cadherin, and vimentin (P < 0.01), but decreased E-cadherin in rats’ prostate tissues and significantly reversed the effects of METTL3 knockdown upon these proteins (##P < 0.01; Fig. 6G–K). Cell apoptosis investigation saw similar trends: METTL3 knockdown significantly promoted, whereas PTEN knockdown inhibited apoptosis in hyperplasic prostate (P < 0.01), and PTEN knockdown partially attenuated the effects of METTL3 on apoptosis (##P < 0.01; Fig. 6L). Moreover, the dynamic effects of METTL3 and PTEN on the levels of transcription factors orchestrating proliferation, EMT, and apoptosis in BPH model were also investigated (Fig. S6A). METTL3 knockdown significantly inhibited p53, c-Myc, NF-κB, Zeb1, Zeb2, Snail, and Twist1 levels; whereas PTEN knockdown notably promoted p53, c-Myc, NF-κB, Zeb1, Zeb2, Snail, and Twist1 levels, and PTEN knockdown partially attenuated the effects of METTL3 on these transcription factors.
Fig. 6. METTL3 regulates PTEN and affects cell proliferation and EMT in prostatic hyperplasia model rats.
BPH model was established in rats as described above; rats were injected with Lv-sh-METTL3 and/or Lv-sh-PTEN and examined for A–C the mRNA and protein expressions of METTL3 and PTEN in rats’ prostate using qRT-PCR and Immunoblotting; D–E prostatic epithelial thickness and prostate weight at the end of the modeling and treatment; F a representative prostate with the urinary bladder from each group (upper) and the histopathological characteristics of rats’ prostate from each group using H&E staining (down); Scale bar = 1 cm or 100 μm; G–K the contents of Ki67, PCNA, E-cadherin, N-cadherin, and vimentin in rats’ prostate tissues using Immunohistochemical (IHC) staining; L cell apoptosis in rats’ prostate using TUNEL assay; scale bar = 50 μm. **P < 0.01, compared with the Lv-sh-NC group. ##P < 0.01, compared with the Lv-sh-METTL3 group.
METTL3 affects TGF-β-stimulated BPH-1 cell phenotype through PTEN
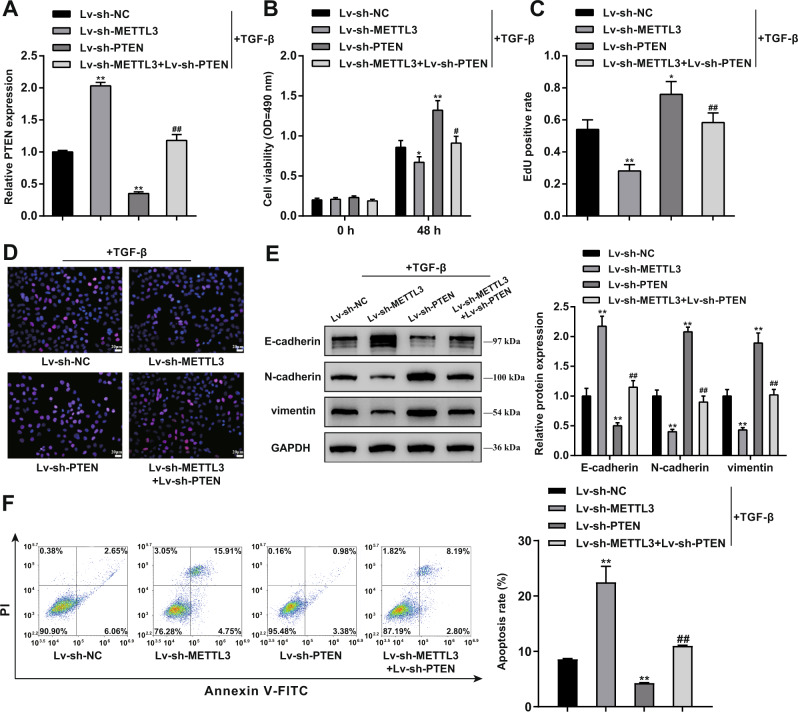
In vitro, BPH-1 cells were co-transduced with Lv-sh-METTL3 and Lv-sh-PTEN and examined for PTEN mRNA expression. Similar to in vivo results, PTEN expression was upregulated after knocking down METTL3 but downregulated after Lv-sh-PTEN transduction (**P < 0.01), whereas Lv-sh-PTEN significantly reversed the effects of METTL3 knockdown upon PTEN expression (Fig. 7A). Regarding cell phenotypes, METTL3 knockdown suppressed, while PTEN knockdown enhanced cell viability and DNA synthesis (**P < 0.01); PTEN knockdown partially attenuated METTL3 knockdown effects on BPH-1 proliferation (Fig. 7B–D). As for EMT markers, METTL3 knockdown upregulated E-cadherin but downregulated N-cadherin and vimentin, whereas PTEN knockdown exerted opposite effects (**P < 0.01); similarly, PTEN knockdown partially abolished METTL3 knockdown effects on EMT markers (Fig. 7E). In contrast, METTL3 knockdown promoted, whereas PTEN knockdown suppressed cell apoptosis (**P < 0.01); PTEN knockdown also partially eliminated the effects of METTL3 knockdown on cell apoptosis (Fig. 7F). Consistent with in vivo pathological alteration, METTL3 knockdown markedly promoted p53 level and inhibited c-Myc, NF-κB, Zeb1, Zeb2, Snail, and Twist1 levels; while PTEN knockdown exerted opposite effects, and PTEN knockdown partially eliminated the effects of METTL3 on these transcription factors (Fig. S6B).
Fig. 7. METTL3 affects the cell phenotypes of TGF-β-stimulated.
BPH-1 cells through PTEN BPH-1 cells were co-transduced with Lv-sh-METTL3 and Lv-sh-PTEN and examined for PTEN mRNA expression using qRT-PCR (A); cell viability using MTT assay (B); DNA synthesis using EdU assay (C, D); Scale bar = 20 μm; the protein levels of E-cadherin, N-cadherin, and vimentin using Immunoblotting (E); cell apoptosis by Flow cytometry (F). **P < 0.01, compared with the Lv-sh-NC group. ##P < 0.01, compared with the Lv-sh-METTL3 group.
Discussion
In this study, methylase METTL3 expression was upregulated in BPH. Elevated m6A modification level was observed in BPH rats’ prostate, which could be partially reduced by METTL3 knockdown. In the prostate of BPH rats, METTL3 knockdown also partially reduced the prostatic epithelial thickness and prostate weight, significantly improved the histological features of the prostate, inhibited epithelial proliferation and EMT, and promoted apoptosis. In vitro, METTL3 knockdown decreased TGF-β-stimulated BPH-1 cell proliferation, m6A modification, and EMT, while promoted cell apoptosis. Furthermore, in BPH rats, METTL3 increases the m6A modification of PTEN and inhibits its expression through the reading protein YTHDF2. PTEN knockdown aggravated the molecular, cellular, and pathological alterations in the prostate of BPH rat and amplified TGF-β-induced changes in BPH-1 cells. Moreover, PTEN knockdown partially abolished the improving effects of METTL3 knockdown both in vivo and in vitro, suggesting METTL3 exerts its functions through PTEN.
Global hypermethylation is the dominant process in BPH at the epigenetic level [10]. For example, methylation of the 5AR2 promoter in some adult prostates is associated with reduced gene expression, which may translate into a slower growth rate of the prostate gland during adulthood [25]. Low or nonexistent expression of 5AR2 in adult human prostate tissues might be due to methylation of the 5AR2 gene promoter. In males with BPH, aging is associated with increased 5AR2 gene promoter methylation and lower protein expression [26]. In males with symptomatic BPH, higher 5-reductase type 2 gene promoter methylation and reduced protein expression are associated with increasing age and BMI [27]. As a result, DNA methylation has been regarded as a tailored medicinal target for the treatment of BPH patients who are resistant to 5-alpha reductase inhibitor medication [28]. However, studies focusing on m6A modification in BPH lack. Herein, the m6A modification levels were significantly elevated in BPH samples, and m6A peaks were enriched around the stop codon, mainly covering CDS and 3′-UTR. Furthermore, signaling and functional enrichment annotation showed that differentially expressed genes with m6A loci were enriched in glandular development and negative regulation of epithelial cell migration, which were correlated with the BPH pathway [29, 30]. During BPH progression, growth factors promote the growth of prostatic epithelial and stromal cells as well as an increase in the expression of molecules related to the EMT, including TGF-β, ZEB-1, and Snail, therefore affecting the onset and progression of BPH [31, 32]. Aberrant proliferation of stromal and epithelial cells causes glandular hypertrophy and bladder outlet obstruction [33]. Considering that EMT and the balance between epithelial proliferation and apoptosis are central events during BPH [34, 35], m6A modification might act on BPH etiology and pathology by affecting these molecular and cellular events.
Among differentially expressed methylases between BPH and normal samples, METTL3 showed to be the most upregulated in BPH. Besides, its role in BPH remains unclear. Notably, METTL3 has been reported to enhance the proliferation and migration but inhibit the apoptosis of several types of cancer cells [36–38]. In this study, in BPH rats, knocking down METTL3 indeed significantly ameliorated the symptoms in model rats, including reduced prostatic epithelial thickness and prostate weight and improved histological features. In the meantime, METTL3 knockdown significantly reduced the m6A modification level, suggesting that METTL3 might act on BPH by affecting m6A modification. Regarding cellular alterations in BPH, as aforementioned, the imbalance between epithelial proliferation and apoptosis is one of the most prominent features of BPH [39]. Loss of apoptosis control increases cell proliferation/cell death ratio, leading to BPH [40, 41]. Consistently, METTL3 knockdown inhibited epithelial proliferation but promoted apoptosis in hyperplastic prostate tissues. In vitro, METTL3 knockdown decreased m6A modification and cell proliferation and promoted cell apoptosis in TGF-β-stimulated BPH-1 cells, suggesting that METTL3 knockdown partially rectifies the imbalance between epithelial proliferation and apoptosis. More importantly, METTL3 knockdown also inhibited EMT in vivo and in vitro. EMT permits a polarized epithelial cell to assume a mesenchymal phenotype, which has increased resistance to apoptosis, an amplified production of ECM components, and the capacity to migrate and invade [42, 43], therefore contributing to BPH [44, 45]. Upon TGF-β stimulation, activation of several transcription factors, including Snail, ZEB-1, or Smad, leads to the expression of mesenchymal genes [46, 47]. Therefore, METTL3 might also regulate EMT to affect BPH development.
As for the underlying molecular mechanism, METTL3, as the core component, catalyzes methylation sites supplemented with other regulatory proteins [48]. The modified m6A sites are recognized and executed by variable readers to produce different functions or events. In this study, PTEN was with increased m6A modification status and downregulated expression level in BPH model rats, therefore being identified as the potential target of METTL3 actions. Previously, the reading protein YTHDF2 has been reported to recognize METTL3-mediated m6A modified PTEN mRNA and enhanced PTEN [49]. In this study, YTHDF2 also mediates the negative regulation of PTEN by METTL3. Mechanically, METTL3 m6A modified PTEN at an m6A loci, which later be recognized by the reading protein YTHDF2, leading to the downregulation of PTEN. Interestingly, PTEN is well-known for its anti-tumor functions in several cancers, inhibiting cell proliferation, enhancing apoptosis, and suppressing the EMT of cancer cells [50, 51], indicating that PTEN might mediate the effects of METTL3 on epithelial cell proliferation, apoptosis, and EMT in BPH in an m6A modification-related manner. As speculated, PTEN knockdown aggravated the molecular, cellular, and pathological alterations in BPH rats’ prostate and amplified TGF-β-induced changes in BPH-1 cells; more importantly, PTEN knockdown partially abolished the improving effects of METTL3 knockdown both in vivo and in vitro, suggesting that METTL3 exerts its functions in BPH through mediating the m6A modification of PTEN.
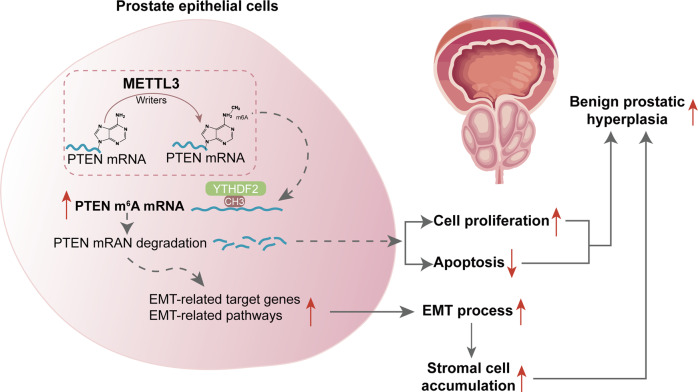
In conclusion, the overall m6A modification level is elevated in BPH. The METTL3/YTHDF2/PTEN axis exerts a crucial effect on promoting BPH development by disturbing the balance between epithelial proliferation and apoptosis and promoting EMT (Fig. 8).
Fig. 8. A schematic diagram indicates the mechanisms of how METTL3 promotes prostatic hyperplasia of BPH.
The METTL3/YTHDF2/PTEN axis disturbs the balance between epithelial proliferation and apoptosis, promotes EMT, and accelerates BPH development in an m6A modification-related manner.
Materials and methods
Experimental animals and BPH model induction
Mature male Sprague Dawley rats weighing 200–250 g (n = 6) were exploited. Rats were maintained in a 23 °C, 50% humidity environment under regular light/dark cycles (12 h light:12 h dark) with full access to food and fluid. All operations were performed as directed by the Medical Sciences Guidelines for Animal Handling at Central South University. The study was carried out according to the Ethics Committee’s Guidelines for laboratory animals of Central South University.
After a 1-week acclimatization, rats were allocated into six groups randomly: normal control, BPH model, BPH + Lv-sh-NC, BPH + Lv-sh-METTL3, BPH + Lv-sh-PTEN, and BPH + Lv-sh-METTL3 + Lv-sh-PTEN. Rats in the BPH groups were administered daily with subcutaneous injections of testosterone (5 mg/kg body weight) for 2 weeks. Rats in the normal control group were given a subcutaneous injection of corn oil of equivalent volume. Gross pathologic assessment and comparison of sonographic pictures were exploited to confirm BPH induction. Rats in the BPH + Lv-sh-NC, BPH + Lv-sh-METTL3, BPH + Lv-sh-PTEN, and BPH + Lv-sh-METTL3 + Lv-sh-PTEN groups received corresponding lentivirus injection 2 weeks following the testosterone injection.
At the end of the experiment, overnight-fasted rats were anesthetized with an intraperitoneal administration of pentobarbital sodium solution (100 mg/kg body weight) and sacrificed for tissue collection. The epithelial thickness and the weight of the prostate were evaluated, each rat’s ventral prostate was preserved in 10% neutral-buffered formalin overnight, and the remaining prostate was snap-frozen in liquid nitrogen for mRNA assays.
m6A RNA methylation modification status in BPH tissues
m6A in RNA was evaluated with an m6A RNA Methylation Assay Kit (Colorimetric) (ab185912, Abcam, Cambridge, MA, USA). Prostate tissues were collected from the normal control and BPH model rats under anesthesia. Total RNA was isolated from target tissues or cells with TRIzol (Invitrogen, CA, USA), and NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA) was employed to determine RNA quality. In general, 200 ng RNA accompanied with m6A standard were coated on assay wells. Next, the capture antibody solution and detection antibody solutions were added to assay wells. The quantification of m6A levels was conducted by detecting absorbance at 450 nm (OD450) in each well and then calculating relative values using the standard curve.
qRT-PCR
Total RNA was extracted from target tissues or cells using Trizol Reagent (Invitrogen). cDNA was obtained using the PrimeScriptTM RT reagent kit (Takara, Dalian, China). qRT-PCR was conducted using an SYBR Premix Ex TaqTM kit (Takara Bio) on an ABI 7900HT Real-Time PCR system (Applied Biosystems Life Technologies, Foster City, CA, USA). All final data were evaluated, and the relative expression levels were calculated using the 2−ΔΔCt method. The primer sequence was listed in Table S1.
Hematoxylin and eosin (H&E) staining
Prostate tissues collected from all groups were H&E stained (Sigma) and evaluated at ×100 magnification using an Olympus microscope (Kyoto, Japan). The existence and extent of epithelial hyperplasia (glandular hyperplasia), stromal edema, inflammation, and degrees were assessed histopathologically in each group by two trial pathologists blinded to the treatment and experimental design.
Immunohistochemical (IHC) staining
Paraffin-embedded prostate tissues were dissected into 4-μm slices. For antigen retrieval, after deparaffinization in xylene, sections were rehydrated through a descending alcohol row for 3 min each time in 100%, 90%, 80%, 75% ethanol, and then microwaved with sodium citrate buffer, followed by blockage with 5% BSA and treatment overnight at 4 °C with anti-Ki67 (27309-1-AP, Proteintech, Wuhan, China), PCNA (10205-2-AP, Proteintech), E-cadherin (BF0219, Affinity, Cincinnati, OH, USA), N-cadherin (AF4039, Affinity), PTEN (22034-1-AP, Proteintech). Next, sections were incubated at ambient temperature for 60 min with a horseradish peroxidase (HRP)-linked secondary antibody (Proteintech), followed by 3,3′-diaminobenzidine development (DAB Substrate Chromogen System; Dako, Denmark) and hematoxylin staining. An inverted microscope was applied to take images after the slices were fixed (Olympus).
TUNEL assay detecting apoptosis in tissue samples
TUNEL labeling was performed on paraffin-embedded slices (5-μm thick) after utilizing a colorimetric TUNEL apoptosis assay kit (Beyotime, China) to identify the apoptosis of prostatic epithelial cells. Positive cells with apoptotic nuclei were shown to be brown. After deparaffinization and dehydration by increasing concentrations of ethanol and xylene, the prostate tissue slices were digested in 20 mg/mL proteinase K solution and kept for 15 min at ambient temperature. Reactions were completed through rinsing and incubating sections for 1 h at 37 °C in the Biotin-dUTP TUNEL reaction mixture at dark. Then, sections were further incubated with streptavidin-HRP and DAB staining reagents. Lastly, an optical microscope was employed to analyze mounted sections upon slides.
Cell lineage, cell culture, and TGF-β stimulation
A human BPH cell line BPH-1 was procured from Merck (SCC256) and cultivated in RPMI 1640 (Gibco, Waltham, MA, USA) containing 20% FBS (Invitrogen, Waltham, MA, USA) at 37 °C in a 5% CO2 atmosphere. BPH-1 cells were stimulated with TGF-β (10 ng/ml) for 24 h following the method described previously [52].
Cell transduction
Silencing of METTL3 was achieved by transducing lentivirus containing short hairpin RNA targeting METTL3 (Lv-sh-METTL3, GenePharma, Shanghai, China). METTL3 overexpression was achieved by transducing plasmid overexpressing METTL3 (METTL3 OE, GenePharma). Silencing of PTEN was achieved by transducing lentivirus containing short hairpin RNA targeting PTEN (Lv-sh-PTEN, GenePharma, Shanghai, China). YTHDF2 silencing or overexpression was achieved by transducing sh-YTHDF2 or YTHDF2 OE vector (GenePharma). Scramble sequence shRNA or empty plasmid was used as negative control (sh-NC/vector). Cell transduction was performed using polybrene (Beyotime) or Lipofectamine 3000 (Invitrogen).
MTT assay detecting cell viability
Cells (2.5 × 103 cells), transduced and/or treated, were sown in each well of 96-well plates. The cells were then treated with 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphynyl-2H-tetrazolium bromide (MTT) dye (5 mg/mL) and kept at 37 °C for 4 h. The supernatants were removed, and the formazan crystals were dissolved by adding 100 μL of dimethyl sulfoxide. A microplate reader with a 490 nm wavelength was used to test the violet solution absorbance.
EdU assay detecting DNA synthesis
Cells, transduced and/or treated, were placed on sterile coverslips in 24-well plates. An EdU Appollo643 kit (RiboBio) was used to measure cell proliferative potential following the product’s protocols, and the nuclei were doubly dyed with EdU and 4’,6-diamidino-2-phenylindole (Beyotime, Shanghai, China). Images were obtained using an inflorescent microscope (Olympus).
Immunoblotting
Total proteins were collected from target cells with a Cell Mitochondria Isolation Kit (Beyotime, Shanghai, China). The samples were incubated at 4 °C for one night with the primary antibodies METTL3 (15073-1-AP, Proteintech), Bcl-2 (ab196495, Abcam), Bax (50599-2-Ig, Proteintech), Caspase 9 (10380-1-AP, Proteintech), Caspase 3 (19677-1-AP, Proteintech), cleaved PARP-1 (ab32064, Abcam), E-cadherin (BF0219, Affinity), N-cadherin (AF4039, Affinity), vimentin (10366-1-AP, Proteintech), PTEN (22034-1-AP, Proteintech), YTHDF2 (ab220163, Abcam), p53 (CSB-PA07889A0Rb, CUSABIO Life Sciences, College Park, USA), c-Myc (10057-1-AP, Proteintech), NF-κb (ab76302, Abcam), Zeb1 (ab180905, Abcam), Zeb2 (ab191364, Abcam), Snail (ab180714, Abcam), Twist1 (ab50887, Abcam); after that, the samples were incubated at room temperature for 1 h with goat anti-rabbit IgG polyclonal antibody (Abcam) or goat anti-mouse IgG polyclonal antibody (Abcam). GAPDH levels were used as an endogenous control for visualization employing the enhanced chemiluminescence (ECL) detection method (Thermo Fisher Scientific).
Flow cytometry for cell apoptosis
An annexin V-FITC/PI apoptosis detection kit was used for apoptosis determination. Cells, transduced and/or treated, were collected and suspended in 500 μl binding buffer and incubated with 5 μl Annexin V-FITC and 5 μl PI for 10 min at room temperature at dark. Then cells were evaluated using Flow cytometry (Novocyte, Agilent, USA).
MeRIP sequencing and RNA sequencing
Total RNA from each sample was isolated using TRIzol reagent (Invitrogen) and then quantified using a NanoDrop ND-1000 instrument. mRNA was isolated first from total RNA samples using the Arraystar Seq-StarTM poly(A) mRNA Isolation Kit and then fragmented into 100-nucleotide-long fragments. The size of fragmented mRNAs was confirmed using agarose electrophoresis. m6A-methylated mRNA fragments were enriched by immunoprecipitation with an anti-m6A antibody (Synaptic Systems, 202003). A portion of the original fragmented mRNAs was kept as input. RNA-seq libraries of m6A antibody-enriched mRNAs and input mRNAs were prepared using the KAPA Stranded mRNA-seq Kit (Illumina) as follows: (1) first-strand cDNA synthesis; (2) second-strand cDNA synthesis; (3) A-tailing; (4) adapter ligation; (5) PCR amplification; and (6) library purification. Quality control of the completed libraries was performed with an Agilent 2100 Bioanalyzer. Libraries were mixed in equal amounts according to the quantification results by qPCR and used for sequencing on the instrument. The DNA fragments in well-mixed libraries were denatured with 0.1 M NaOH to generate single-stranded DNA molecules and loaded onto the reagent cartridge at the appropriate loading concentration. Clustered libraries were then sequenced on the Illumina NovaSeq 6000 following the NovaSeq 6000 S4 Reagent Kit (300 cycles) protocol.
MeRIP-qPCR
Magna MeRIP m6A kit (17–10,499, Millipore) was employed to perform m6A RNA immunoprecipitation (MeRIP) as directed by the manufacturer’s protocol. The IP production was conducted RT-qPCR for PTEN as mentioned above. Primers used for RT-qPCR are presented in Table S1.
Actinomycin D detecting mRNA stabilization
Cells were incubated with 5 μg/ml actinomycin D for 0, 6, and 12 h, and the relative levels of target mRNA were quantified by performing qPCR assays. Target mRNA degradation rate was calculated as directed by a previously reported study [53].
RIP
Cells were washed by cold PBS and lysed using RIP lysis buffer (300 mM NaCl, 0.2% NP-40, 20 mM Tris-HCl PH 7.6, 0.5 mM DTT, protease inhibitor, and RNase inhibitor) at 4 °C via disruptive sonication. Next, the lysis was incubated at 4 °C overnight with 5 μg anti-YTHDF2 Rabbit antibody, or IgG (ProteinTech Group) pre-conjugated protein A/G Magnetic Beads (Millipore) in 500 μl IP buffer (150 mM NaCl, 10 mM Tris-HCl (pH 7.4), 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.5% NP-40) containing RNase inhibitors (Thermo Fisher). The IP complex was treated for 30 min at 55 °C with Proteinase K (Thermo Fisher), and RNA was purified using phenol:chloroform:isoamyl alcohol. Last, RT-qPCR was carried out to determine PTEN mRNA levels in immunoprecipitate by anti-IgG or anti-YTHDF2. Primers used for RT-qPCR are presented in Table S1.
Statistical analysis
GraphPad software (San Diego, CA, USA) was employed to process data from at least three independent experiments. All data were presented as the means ± standard deviation (SD). One-way analysis of variance was employed, followed by Tukey’s multiple comparison test or independent sample t test. The significance level was set at P < 0.05.
Supplementary information
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 81770758) and Natural Science Foundation of Hunan Province (No. 2021JJ31023).
Author contributions
JL and LW performed study concept and design; HY, JH, CL, and YZ performed development of methodology and writing, review and revision of the paper; RX, ZW, and ZL provided acquisition, analysis and interpretation of data, and statistical analysis; JT provided technical and material support. All authors read and approved the final paper.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article.
Competing interests
The authors declare no competing interests.
Ethical approval
The guidelines for the care and use of animals were carried out according to the Ethics Committee’s Guidelines for laboratory animals of Central South University.
Footnotes
Edited by Professor Nickolai Barlev
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jiaren Li, Hanyu Yao, Jin Huang.
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-022-05162-4.
References
- 1.Bianchi-Frias D, Vakar-Lopez F, Coleman IM, Plymate SR, Reed MJ, Nelson PS. The effects of aging on the molecular and cellular composition of the prostate microenvironment. PLoS One. 2010;5:e12501. [DOI] [PMC free article] [PubMed]
- 2.Welliver C, Feinstein L, Ward JB, Fwu CW, Kirkali Z, Bavendam T, et al. Trends in lower urinary tract symptoms associated with benign prostatic hyperplasia, 2004 to 2013: the Urologic Diseases in America Project. J Urol. 2020;203:171–8. doi: 10.1097/JU.0000000000000499. [DOI] [PubMed] [Google Scholar]
- 3.Wendell-Smith C. Terminology of the prostate and related structures. Clin Anat. 2000;13:207–13. doi: 10.1002/(SICI)1098-2353(2000)13:3<207::AID-CA9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 4.McNeal JE. Regional morphology and pathology of the prostate. Am J Clin Pathol. 1968;49:347–57. doi: 10.1093/ajcp/49.3.347. [DOI] [PubMed] [Google Scholar]
- 5.Alonso-Magdalena P, Brossner C, Reiner A, Cheng G, Sugiyama N, Warner M, et al. A role for epithelial-mesenchymal transition in the etiology of benign prostatic hyperplasia. Proc Natl Acad Sci USA. 2009;106:2859–63. doi: 10.1073/pnas.0812666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Popovics P, Schally AV, Salgueiro L, Kovacs K, Rick FG. Antagonists of growth hormone-releasing hormone inhibit proliferation induced by inflammation in prostatic epithelial cells. Proc Natl Acad Sci USA. 2017;114:1359–64.. doi: 10.1073/pnas.1620884114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Zhang Y, Zhao C, Li Y, Hu X, Wu L, et al. The miR-223-3p/MAP1B axis aggravates TGF-beta-induced proliferation and migration of BPH-1 cells. Cell Signal. 2021;84:110004. doi: 10.1016/j.cellsig.2021.110004. [DOI] [PubMed] [Google Scholar]
- 8.Adams JM, Cory S. Modified nucleosides and bizarre 5′-termini in mouse myeloma mRNA. Nature. 1975;255:28–33. doi: 10.1038/255028a0. [DOI] [PubMed] [Google Scholar]
- 9.Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. 1974;71:3971–5. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu D, Shoag JE, Poliak D, Goueli RS, Ravikumar V, Redmond D, et al. Integrative multiplatform molecular profiling of benign prostatic hyperplasia identifies distinct subtypes. Nat Commun. 2020;11:1987. doi: 10.1038/s41467-020-15913-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henrique R, Jeronimo C, Hoque MO, Carvalho AL, Oliveira J, Teixeira MR, et al. Frequent 14-3-3 sigma promoter methylation in benign and malignant prostate lesions. DNA Cell Biol. 2005;24:264–9. doi: 10.1089/dna.2005.24.264. [DOI] [PubMed] [Google Scholar]
- 12.Goessl C, Muller M, Heicappell R, Krause H, Miller K. DNA-based detection of prostate cancer in blood, urine, and ejaculates. Ann N. Y Acad Sci. 2001;945:51–8. doi: 10.1111/j.1749-6632.2001.tb03863.x. [DOI] [PubMed] [Google Scholar]
- 13.Zou D, Dong L, Li C, Yin Z, Rao S, Zhou Q. The m(6)A eraser FTO facilitates proliferation and migration of human cervical cancer cells. Cancer Cell Int. 2019;19:321. doi: 10.1186/s12935-019-1045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taketo K, Konno M, Asai A, Koseki J, Toratani M, Satoh T, et al. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int J Oncol. 2018;52:621–9. doi: 10.3892/ijo.2017.4219. [DOI] [PubMed] [Google Scholar]
- 15.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai X, Gonzalez G, Li L, Li J, You C, Miao W, et al. YTHDF2 Binds to 5-Methylcytosine in RNA and Modulates the Maturation of Ribosomal RNA. Anal Chem. 2020;92:1346–54. doi: 10.1021/acs.analchem.9b04505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu R, Liu Y, Zhao Y, Bi Z, Yao Y, Liu Q, et al. m(6)A methylation controls pluripotency of porcine induced pluripotent stem cells by targeting SOCS3/JAK2/STAT3 pathway in a YTHDF1/YTHDF2-orchestrated manner. Cell Death Dis. 2019;10:171. doi: 10.1038/s41419-019-1417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Q, Wen S, Ye W, Zhao S, Liu X. The potential roles of m(6)A modification in regulating the inflammatory response in microglia. J Neuroinflammation. 2021;18:149. doi: 10.1186/s12974-021-02205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Wu R, Liu Y, Zhao Y, Bi Z, Yao Y, et al. m(6)A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy. 2020;16:1221–35.. doi: 10.1080/15548627.2019.1659617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu H, Wang Z, Chen M, Zhao W, Tao T, Ma L, et al. YTHDF2 alleviates cardiac hypertrophy via regulating Myh7 mRNA decoy. Cell Biosci. 2021;11:132. doi: 10.1186/s13578-021-00649-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, Zhao X, Wang W, Shi H, Pan Q, Lu Z, et al. Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 2018;19:69. doi: 10.1186/s13059-018-1436-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bi X, Lv X, Liu D, Guo H, Yao G, Wang L, et al. METTL3-mediated maturation of miR-126-5p promotes ovarian cancer progression via PTEN-mediated PI3K/Akt/mTOR pathway. Cancer Gene Ther. 2021;28:335–49.. doi: 10.1038/s41417-020-00222-3. [DOI] [PubMed] [Google Scholar]
- 23.Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18:110. doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen M, Wei L, Law CT, Tsang FHC, Shen J, Cheng CLH, et al. RNA N6‐methyladenosine methyltransferase‐like 3 promotes liver cancer progression through YTHDF2‐dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–70.. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 25.Niu Y, Ge R, Hu L, Diaz C, Wang Z, Wu CL, et al. Reduced levels of 5-alpha reductase 2 in adult prostate tissue and implications for BPH therapy. Prostate. 2011;71:1317–24.. doi: 10.1002/pros.21348. [DOI] [PubMed] [Google Scholar]
- 26.Kang PM, Kim YJ, Seo WT, Kang SH, Kim TS, Chun BK, et al. Correlation between 5-alpha reductase type 2 protein expression and methylation of 5-alpha reductase type 2 promotor gene of benign prostatic hyperplasia. World J Urol. 2019;37:709–18.. doi: 10.1007/s00345-018-2422-4. [DOI] [PubMed] [Google Scholar]
- 27.Bechis SK, Otsetov AG, Ge R, Wang Z, Vangel MG, Wu CL, et al. Age and obesity promote methylation and suppression of 5alpha-reductase 2: implications for personalized therapy of benign prostatic hyperplasia. J Urol. 2015;194:1031–7. doi: 10.1016/j.juro.2015.04.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bechis SK, Otsetov AG, Ge R, Olumi AF. Personalized medicine for the management of benign prostatic hyperplasia. J Urol. 2014;192:16–23. doi: 10.1016/j.juro.2014.01.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roehrborn CG. Pathology of benign prostatic hyperplasia. Int J Impot Res. 2008;20:S11–8. doi: 10.1038/ijir.2008.55. [DOI] [PubMed] [Google Scholar]
- 30.Lai KP, Huang CK, Fang LY, Izumi K, Lo CW, Wood R, et al. Targeting stromal androgen receptor suppresses prolactin-driven benign prostatic hyperplasia (BPH) Mol Endocrinol. 2013;27:1617–31. doi: 10.1210/me.2013-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wen S, Chang HC, Tian J, Shang Z, Niu Y, Chang C. Stromal androgen receptor roles in the development of normal prostate, benign prostate hyperplasia, and prostate cancer. Am J Pathol. 2015;185:293–301. doi: 10.1016/j.ajpath.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomson AA, Cunha GR, Marker PC. Prostate development and pathogenesis. Differentiation. 2008;76:559–64. doi: 10.1111/j.1432-0436.2008.00303.x. [DOI] [PubMed] [Google Scholar]
- 33.Gratzke C, Bachmann A, Descazeaud A, Drake MJ, Madersbacher S, Mamoulakis C, et al. EAU guidelines on the assessment of non-neurogenic male lower urinary tract symptoms including benign prostatic obstruction. Eur Urol. 2015;67:1099–109. doi: 10.1016/j.eururo.2014.12.038. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Wang Z, Gan Y, Chen X, Zhang B, Chen Z, et al. Curcumin attenuates prostatic hyperplasia caused by inflammation via up-regulation of bone morphogenetic protein and activin membrane-bound inhibitor. Pharm Biol. 2021;59:1026–35. doi: 10.1080/13880209.2021.1953539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang B, Chen X, Xie C, Chen Z, Liu Y, Ru F, et al. Leptin promotes epithelial-mesenchymal transition in benign prostatic hyperplasia through downregulation of BAMBI. Exp Cell Res. 2020;387:111754. doi: 10.1016/j.yexcr.2019.111754. [DOI] [PubMed] [Google Scholar]
- 36.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62:335–45. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma Z, Li Q, Liu P, Dong W, Zuo Y. METTL3 regulates m6A in endometrioid epithelial ovarian cancer independently of METTl14 and WTAP. Cell Biol Int. 2020;44:2524–31. doi: 10.1002/cbin.11459. [DOI] [PubMed] [Google Scholar]
- 38.Ni XF, Xie QQ, Zhao JM, Xu YJ, Ji M, Hu WW, et al. The hepatic microenvironment promotes lung adenocarcinoma cell proliferation, metastasis, and epithelial-mesenchymal transition via METTL3-mediated N6-methyladenosine modification of YAP1. Aging (Albany NY) 2021;13:4357–69. doi: 10.18632/aging.202397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quiles MT, Arbos MA, Fraga A, de Torres IM, Reventos J, Morote J. Antiproliferative and apoptotic effects of the herbal agent Pygeum africanum on cultured prostate stromal cells from patients with benign prostatic hyperplasia (BPH) Prostate. 2010;70:1044–53. doi: 10.1002/pros.21138. [DOI] [PubMed] [Google Scholar]
- 40.Broster SA, Kyprianou N. Epithelial-mesenchymal transition in prostatic disease. Future Oncol. 2015;11:3197–206. doi: 10.2217/fon.15.253. [DOI] [PubMed] [Google Scholar]
- 41.Kyprianou N, Tu H, Jacobs SC. Apoptotic versus proliferative activities in human benign prostatic hyperplasia. Hum Pathol. 1996;27:668–75. doi: 10.1016/S0046-8177(96)90396-2. [DOI] [PubMed] [Google Scholar]
- 42.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 43.Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol. 2017;11:28–39. doi: 10.1002/1878-0261.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi X, Peng Y, Du X, Liu H, Klocker H, Lin Q, et al. Estradiol promotes epithelial-to-mesenchymal transition in human benign prostatic epithelial cells. Prostate. 2017;77:1424–37. doi: 10.1002/pros.23404. [DOI] [PubMed] [Google Scholar]
- 45.Xu H, Chen Y, Chen Q, Xu H, Wang Y, Yu J, et al. DNMT1 regulates IL-6- and TGF-beta1-induced epithelial mesenchymal transition in prostate epithelial cells. Eur J Histochem. 2017;61:2775. doi: 10.4081/ejh.2017.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–84. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 47.Miyazawa K, Miyazono K. Regulation of TGF-beta family signaling by inhibitory smads. Cold Spring Harb Perspect Biol. 2017;9:a022095. [DOI] [PMC free article] [PubMed]
- 48.Shu B, Zhou YX, Li H, Zhang RZ, He C, Yang X. The METTL3/MALAT1/PTBP1/USP8/TAK1 axis promotes pyroptosis and M1 polarization of macrophages and contributes to liver fibrosis. Cell Death Discov. 2021;7:368. doi: 10.1038/s41420-021-00756-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qin Y, Qiao Y, Li L, Luo E, Wang D, Yao Y, et al. The m(6)A methyltransferase METTL3 promotes hypoxic pulmonary arterial hypertension. Life Sci. 2021;274:119366. doi: 10.1016/j.lfs.2021.119366. [DOI] [PubMed] [Google Scholar]
- 50.Wise HM, Hermida MA, Leslie NR. Prostate cancer, PI3K, PTEN and prognosis. Clin Sci (Lond) 2017;131:197–210. doi: 10.1042/CS20160026. [DOI] [PubMed] [Google Scholar]
- 51.Carnero A, Blanco-Aparicio C, Renner O, Link W, Leal JF. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr Cancer Drug Targets. 2008;8:187–98. doi: 10.2174/156800908784293659. [DOI] [PubMed] [Google Scholar]
- 52.He Y, Ou Z, Chen X, Zu X, Liu L, Li Y, et al. LPS/TLR4 signaling enhances TGF-beta response through downregulating BAMBI during prostatic hyperplasia. Sci Rep. 2016;6:27051. doi: 10.1038/srep27051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–41. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.