Abstract
Combined physical and genetic maps of the genomes of Actinobacillus pleuropneumoniae AP76 (serotype 7 clinical isolate) and of A. pleuropneumoniae ATCC 27088 (serotype 1 reference strain) were constructed by using the restriction endonucleases ApaI, AscI, NotI, and SalI. The chromosome sizes as determined by the addition of estimated fragment sizes were 2.4 Mbp, and both maps had a resolution of approximately 100 kbp. The linkages between the ApaI, AscI, NotI, and SalI fragments and their relative positions were determined by (i) fragment excision and redigestion and (ii) partial digests of defined fragments and Southern blot using end-standing probes. The single SalI site within the chromosome of strain A. pleuropneumoniae AP76 was defined as position 1 of the map; for the map of A. pleuropneumoniae ATCC 27088, the corresponding SalI site was chosen. Putative virulence-associated genes (apx, omlA, sodA, tbpBA, ureC, and a repeat element) and housekeeping genes (glyA, metJ, recA, and rhoAP) were positioned on the physical maps and located on the ApaI and NotI fragments of A. pleuropneumoniae serotype reference strains.
Actinobacillus pleuropneumoniae is the causative agent of porcine pleuropneumonia, a severe and economically important disease occurring worldwide (9). A. pleuropneumoniae isolates can be differentiated into 12 distinct serotypes (29) which are based on polysaccharide antigens of the slime capsule and the lipopolysaccharide of the bacteria (40). The serotypes have been described to be of clonal origin (27). The different serotypes have been also grouped based either on outer membrane protein profiles (32) or on antigenic similarities (28) which are most likely due to shared species-specific antigens such as lipopolysaccharide or membrane proteins (31). More recently, Chevallier and coworkers assessed the phylogenetic relationship among the different serotype reference strains based on restriction enzyme fragment polymorphisms after pulsed-field gel electrophoresis (PFGE) (7).
For A. pleuropneumoniae, several putative virulence-associated factors such as the RTX toxins ApxI, ApxII, and ApxIII (2, 6, 12, 19, 20, 24, 33, 37), the transferrin binding proteins TbpA and TbpB (13, 15, 25, 41), a superoxide dismutase (SodA [23]), a urease (4), an outer membrane lipoprotein (OmlA [5, 14, 17, 18]), and capsular antigens (16, 40) have been characterized. The RTX toxins are distributed in a serotype-specific manner with each serotype containing no more than two of the Apx toxins (12, 20), and the TbpB and OmlA proteins occur in three antigenically distinct isoforms (13, 17).
Despite this detailed information on serotype classification and individual virulence-associated factors no information on the genomic structure of A. pleuropneumoniae is available. As shown for other bacteria (22, 26, 34, 39), such information is required for a detailed understanding of virulence-associated mechanisms. Therefore, we have constructed the first physical and genetic maps of an A. pleuropneumoniae serotype 7 clinical isolate (AP76 [2]) and the A. pleuropneumoniae serotype 1 reference strain (ATCC 27088); these two strains were chosen since A. pleuropneumoniae ATCC 27088 is the strain most frequently investigated, and A. pleuropneumoniae AP76 is the only strain in which spontaneous chromosomal deletions have been shown to occur. In addition, we compared the presence and location of a panel of genes in the other A. pleuropneumoniae serotype reference strains.
MATERIALS AND METHODS
Bacterial strains, plasmids, and probes.
The sources of strains used in this study are given in Table 1; the sources of DNA probes are listed in Table 2. A. pleuropneumoniae strains were cultivated in PPLO (pleuropneumonia-like organism) medium (Difco, Detroit, Mich.) with NAD (10 μg/ml) at 37°C in a 5% CO2 atmosphere. Cultures used for DNA preparation were inoculated with 1/10 volume of an overnight culture, incubated with shaking, and harvested at an optical density at 600 nm of 0.3.
TABLE 1.
A. pleuropneumoniae strains used and their sources
| Strain | Description | Source |
|---|---|---|
| AP76 | A. pleuropneumoniae serotype 7 | Porcine lung isolate provided by Western College of Veterinary Medicine, Saskatoon, Saskatchewan, Canada |
| ATCC 27088 | Reference strain serotype 1 | |
| ATCC 27089 | Reference strain serotype 2 | |
| ATCC 27090 | Reference strain serotype 3 | |
| ATCC 33378 | Reference strain serotype 4 | |
| ATCC 33377 | Reference strain serotype 5A | |
| ATCC 33590 | Reference strain serotype 6 | |
| WF83 | Reference strain serotype 7 | Provided by S. Rosendal, University of Guelph, Guelph, Ontario, Canada |
| 405 | Reference strain serotype 8 | Provided by R. Nielsen, State Veterinary Serum Laboratory, Copenhagen, Denmark |
| CVJ13261 | Reference strain serotype 9 | Provided by R. Nielsen |
| D13039 | Reference strain serotype 10 | Provided by R. Nielsen |
| 56153 | Reference strain serotype 11 | Provided by R. Nielsen |
| 8329 | Reference strain serotype 12 | Provided by R. Nielsen |
TABLE 2.
Genes positioned in the map and origins of probes
| Gene | Encoded | Source | Accession no. | Reference(s) |
|---|---|---|---|---|
| apxICA | Activator C and structural protein A of apxI operon | PCR, 5′-TTGCCTCGCTAGTTGCGGAT-3′ | X68595 | 11 |
| 5′-TCCCAAGTTCGAATGGGCTT-3′ | ||||
| apxIBD | Secretion proteins of apxI operon | PCR, 5′-GTATCGGCGGGATTCCGT-3′ | X68595 | 11 |
| 5′-ATCCGCATCGGCTCCCAA-3′ | ||||
| apxIICA | Activator C and structural protein A of apxII operon | Plasmid pCY76/504, NdeI fragment | M30602 | 2 |
| apxIIICA | Activator C and structural protein A of apxIII operon | PCR, 5′-CCTGGTTCTACAGAAGCGAAAATC-3′ | L12145 | 6 |
| 5′-TTTCGCCCTTAGTTGGATCGA-3′ | ||||
| omlA | Outer membrane lipoprotein | PCR, 5′-TAAGGTTGATATGTCCGCACC-3′ | L06318 | 14 |
| 5′-TAGCACCGATTACGCCTT-3′ | ||||
| tolQ-tbpBA | Colicin transport protein, transferrin binding proteins | Plasmid pTF205/405; BamHI-NsiI fragment | Y17916 | 38, 41 |
| ureC | Large subunit C of bacterial urease | PCR, 5′-GTAAGGATCCATTAACAATCCCACGCAGTCAGTAT-3′ | U89957 | 4 |
| 5′-TCATGTCGACTAGAACAAGAAATAACGCTGTGCAA-3′ | ||||
| sodA | Superoxide dismutase | PCR, 5′-ACGCTTATGATGCGTTAGAGC-3′ | U51441 | 23 |
| 5′-GTCCCAGTTTACCACGTTCCA-3′ | ||||
| glyA | Serine methylase | Plasmid p#4-213-804, HindIII-EcoRI fragment | This work | |
| metJ | Met repressor | Plasmid p#4-213-84; XbaI fragment | This work | |
| recA | Repair and recombination protein | PCR, 5′-GAAAARCAATTYGGTAAAGG-3′ | U32741 | 10 |
| 5′-GCYTTWCCTTGRCCAATTTT-3′ | ||||
| rho | Transcription termination factor | Plasmid pFR100; KpnI-NsiI fragment | Y17915 | 38 |
| Repeat element | Transposon like element | Plasmid pCY76/102; BamHI-BglII fragment | M74588 | 2 |
Sample preparation and PFGE.
Embedded DNA samples were prepared for PFGE essentially as described by Bautsch (3). Briefly, bacteria were harvested by centrifugation, washed, and resuspended in 1/10 culture volume of PET IV (1 M NaCl, 10 mM Tris-HCl [pH 8], 10 mM Na2EDTA). Suspensions were mixed 1:1 with 1.2% agarose at 60°C, and gel plugs were poured into molds. Gel plugs were treated 2 h at 37°C with lysis buffer (1 M NaCl, 10 mM Tris-HCl [pH 8], 0.2 M Na2EDTA, 0.5% N-lauroylsarcosine, 0.2% deoxycholic acid) containing RNase (2 μg/ml) and lysozyme (1 mg/ml). Then plugs were treated overnight at 55°C with proteinase K (1 mg/ml) in ES buffer (0.5 M Na2EDTA, 1% N-lauroylsarcosine). Proteinase K was inactivated by two washes with phenylmethylsulfonyl fluoride (1.5 mM) in TE buffer (10 mM Tris-HCl [pH 8], 1 mM Na2EDTA). Phenylmethylsulfonyl fluoride was eliminated by repeated washing with TE buffer. Restriction enzyme digests were performed after equilibration in buffers supplied by the manufacturer (New England Biolabs, Schwalbach, Germany).
The fragmented DNA was separated on 0.8% agarose (Appligene, Illkirch, France) in 0.5× TBE buffer (45 mM Tris-borate [pH 8], 1 mM Na2EDTA) in a CHEF-DR III pulsed-field electrophoresis system (Bio-Rad Inc., Hercules, Calif.) as recommended by the manufacturer. Running conditions were 24 h at 12°C and 6 V/cm with linear ramped switch times from 5 to 20 s (for the resolution of fragments of 50 to 300 kbp), from 10 to 40 s (100 to 500 kbp), or from 20 to 80 s (500 to 1,000 kbp). DNA was stained with ethidium bromide (0.5 μg/ml) and visualized in an image documentation system (Gel Doc 1000/Multianalyst; Bio-Rad).
Single bands were cut from PFGE gels and restriction cleaved like the original agarose plugs to identify products of double digestion by direct comparison in the same gel. The size of large fragments was determined as the sum of the subfragments by this procedure. Partial enzymatic cleavage was carried out by serial dilution of the appropriate enzyme.
DNA hybridization.
DNA was transferred to a nylon membrane (Positive; Appligene) by capillary blotting (36). Probe DNAs were obtained by PCR or, where available, from recombinant Escherichia coli plasmids (Table 2) by cleavage with appropriate restriction endonucleases.
Small fragments were mapped by being used as probes in Southern blots with partially digested DNA. Fragments were obtained from the gel by adsorption to a silica matrix (Geneclean; Bio 101, Vista, Calif.) and labeled with biotin-dUTP or with [α-32P]dATP by a random priming method (8). Biotin was detected with avidin-alkaline phosphatase conjugate and chemiluminescence substrate (CSPD; Tropix Inc., Bedford, Mass.) according to the manufacturer’s recommendations.
RESULTS
Physical mapping.
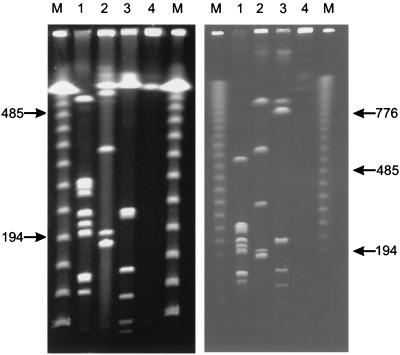
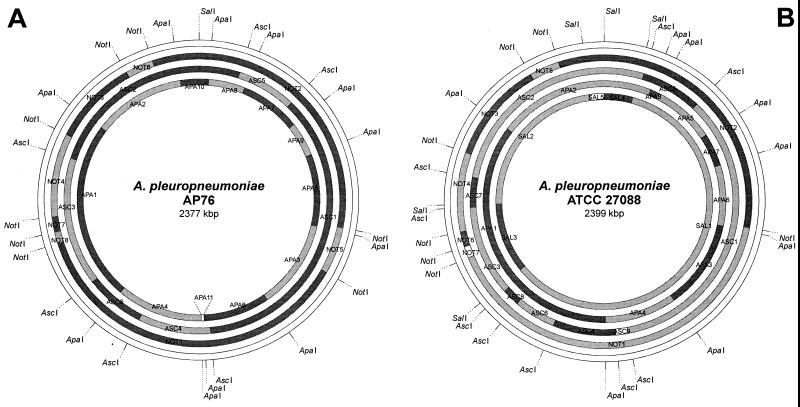
A physical and genetic map was constructed for the porcine lung pathogen A. pleuropneumoniae. The chromosome of A. pleuropneumoniae AP76 serotype 7 was mapped first since it had a single SalI site which was defined as map position 1. The chromosome was found to have a size of approximately 2.4 Mbp containing 11 ApaI, 6 AscI, and 8 NotI fragments with sizes between 9 and 900 kbp (Fig. 1; Table 3). The SalI site was mapped to the fragments APA10, NOT2, and ASC2. The relative position of each fragment was determined by combining results of single and double restriction digests.
FIG. 1.
PFGE separation of A. pleuropneumoniae AP76 DNA, using short (left) and long (right) running conditions. Agarose-embedded DNA was digested with ApaI (lanes 1), AscI (lanes 2), NotI (lanes 3), and SalI (lanes 4). Bacteriophage lambda concatemers were used as molecular size markers (M); the numbers indicate sizes of the corresponding bands in kilobase pairs.
TABLE 3.
Sizes of macrorestriction fragments of the genomes of A. pleuropneumoniae AP76 and A. pleuropneumoniae ATCC 27088
| A. pleuropneumoniae strain |
ApaI
|
AscI
|
NotI
|
SalI
|
||||
|---|---|---|---|---|---|---|---|---|
| Designation | Size (kbp) | Designation | Size (kbp) | Designation | Size (kbp) | Designation | Size (kbp) | |
| AP76 | APA1 | 571 | ASC1 | 886 | NOT1 | 878 | NAa | NA |
| APA2 | 294 | ASC2 | 591 | NOT2 | 794 | |||
| APA3 | 282 | ASC3 | 369 | NOT3 | 225 | |||
| APA4 | 272 | ASC4 | 190 | NOT4 | 219 | |||
| APA5 | 231 | ASC5 | 170 | NOT5 | 117 | |||
| APA6 | 210 | ASC6 | 170 | NOT6 | 73 | |||
| APA7 | 194 | NOT7 | 40 | |||||
| APA8 | 116 | NOT8 | 30 | |||||
| APA9 | 110 | |||||||
| APA10 | 90 | |||||||
| APA11 | 9 | |||||||
| Sum | 2,379 | 2,376 | 2,376 | |||||
| ATCC 27088 | APA1 | 858 | ASC1 | 841 | NOT1 | 970 | SAL1 | 1,436 |
| APA2 | 503 | ASC2 | 644 | NOT2 | 795 | SAL2 | 557 | |
| APA3 | 286 | ASC3 | 266 | NOT3 | 270 | SAL3 | 254 | |
| APA4 | 220 | ASC4 | 182 | NOT4 | 209 | SAL4 | 102 | |
| APA5 | 201 | ASC5 | 175 | NOT5 | 81 | SAL5 | 51 | |
| APA6 | 196 | ASC6 | 121 | NOT6 | 40 | |||
| APA7 | 105 | ASC7 | 87 | NOT7 | 30 | |||
| APA8 | 28 | ASC8 | 51 | |||||
| ASC9 | 36 | |||||||
| Sum | 2,397 | 2,403 | 2,395 | 2,400 | ||||
NA, not applicable (single recognition site only).
By recleaving with NotI, the large ApaI fragment APA1 (Table 3) was found to contain the three NotI fragments NOT4, NOT7, and NOT8 and two additional flanking fragments 68 and 215 kbp in size. The second large ApaI fragment (APA2) was found to contain NOT6 and two flanking fragments of about 157 and 64 kbp. APA3 was cut by NotI into two fragments of 172 and 106 kbp. The largest NotI fragment (NOT1) was cleaved by ApaI into fragments APA4, APA6, the small APA11, and two flanking fragments 172 and 215 kbp in size. The 794-kbp NotI fragment NOT2 contained the small ApaI fragments APA7, APA8, APA9, and APA10 and flanking fragments 64 and 220 kbp in size. Fragment NOT3 was cleaved by ApaI into two fragments of 157 and 68 kbp.
Fragments APA5 and NOT5 were shown to overlap by only 11 kbp. The sizes of the NotI-ApaI subfragments of fragment APA3 were identical to those of the flanking fragments from NOT5 cut with ApaI (106 kbp) and from NOT1 cut with ApaI (172 kbp) and therefore indicated an overlap. The overlap of fragment NOT1 with fragment APA1 ended with the congruence of the flanking fragments of 215 kbp. The region flanking fragments APA1 and APA2 was determined by the cleavage pattern of the linking fragment NOT3 with ApaI into two fragments of 157 and 68 kbp. The overlap from fragment APA2 to fragment NOT2 was determined by the ending linking fragment size of 64 kbp. The overlapping flanking fragment from NOT2 to APA5 was 220 kbp in size and therefore could be confounded with the 215-kbp fragment shared by APA1 and NOT1. This was resolved by Southern hybridization of an omlA-specific probe to the 220-kbp fragment as well as to fragments NOT2 and APA5 (data not shown).
Using these double digests, we could not determine the order of small fragments in three regions. These were the ApaI fragments in NOT1 (APA4, APA6, and APA11) and NOT2 (APA7, APA8, APA9, and APA10) as well as the NotI fragments in APA1 (NOT4, NOT7, and NOT8). These ambiguities were investigated by partial cleavage and hybridization using probes for (i) a 172-kbp NotI-ApaI fragment, (ii) fragment APA11 (sodA), and (iii) fragment APA9 (tolQ). The resulting order of fragments in the map was confirmed by single and double digestion with AscI and Southern hybridization analyses. The resulting physical map had a resolution of approximately 100 kbp (Fig. 2A).
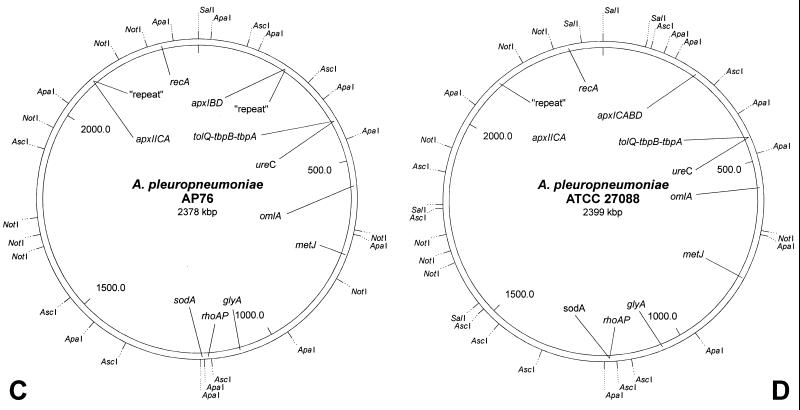
FIG. 2.
Physical (A and B) and genetic (C and D) maps of the genomes of A. pleuropneumoniae AP76 and ATCC 27088 determined by using the restriction endonucleases ApaI, AscI, NotI, and SalI. The single SalI site of the chromosome of A. pleuropneumoniae AP76 and the corresponding site in A. pleuropneumoniae ATCC 27088 have been defined as position 1; sizes of the indicated fragments are listed in Table 2.
The chromosome of A. pleuropneumoniae ATCC 27088 (serotype 1 reference strain) was also found to have a size of 2.4 Mbp containing eight ApaI, nine AscI, seven NotI, and five SalI fragments (Table 3). The initial information about the order of macrorestriction fragments in the A. pleuropneumoniae ATCC 27088 chromosome likewise was obtained by fragment excision and redigestion as well as by partial cleavage as described above. The results of these experiments were confirmed by cross-hybridization particularly using the AscI fragments of A. pleuropneumoniae AP76 as probes for Southern blot analyses. The map was oriented such that the SalI site corresponding to the single SalI site of A. pleuropneumoniae AP76 was position 1. The resulting physical map also had a resolution of approximately 80 kbp (Fig. 2B).
Genetic map.
On the physical maps, several putative virulence-associated genes (apxIA, -IIA, and -IIIA, apxIBD, omlA, sodA, tolQ-tbpBA, ureC, and a repeat element) as well as a variety of housekeeping genes (glyA, metJ, recA, and rho) were located by Southern hybridization (Fig. 2C and D; Fig. 3). It was found that the tolQ-tbpBA region (38) and the urease operon mapped together on the 110-kbp fragment APA9 of strain AP76 and the 105-kbp fragment APA7 of strain ATCC 27088, respectively. The transposon-like repeat element mapped together with the apxIICA genes as expected (2). An additional repeat element mapped together with the apxIBD genes on the 194-kbp ApaI fragment APA7 of strain AP76. In A. pleuropneumoniae ATCC 27088, the complete apxICABD operon was present in a comparable region (on APA5) without an associated repeat sequence. No virulence-associated genes were mapped to approximately one-third of the genome represented by fragments APA1 and APA4 (AP76) or APA1 (ATCC 27088). Therefore, the general genomic arrangement did not differ between the strains A. pleuropneumoniae AP76 (serotype 7) and A. pleuropneumoniae ATCC 27088 (serotype 1) (Fig. 2C and D).
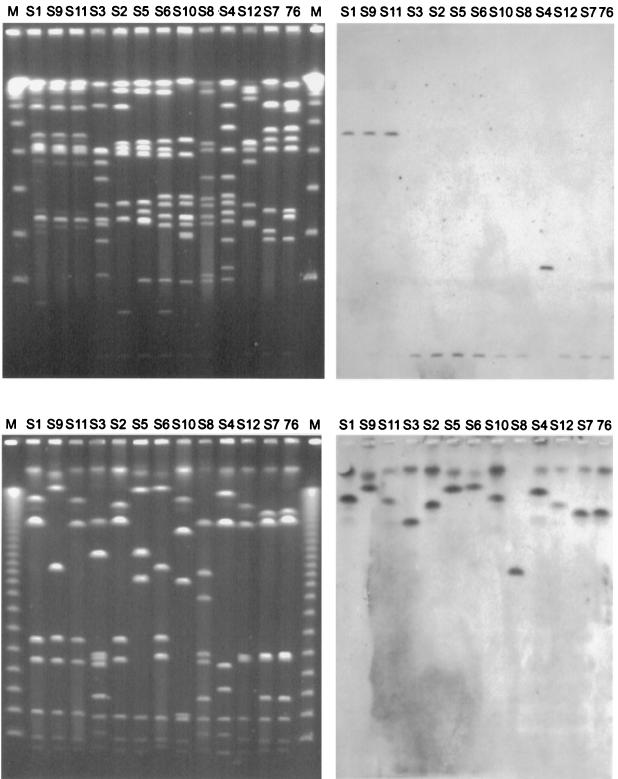
FIG. 3.
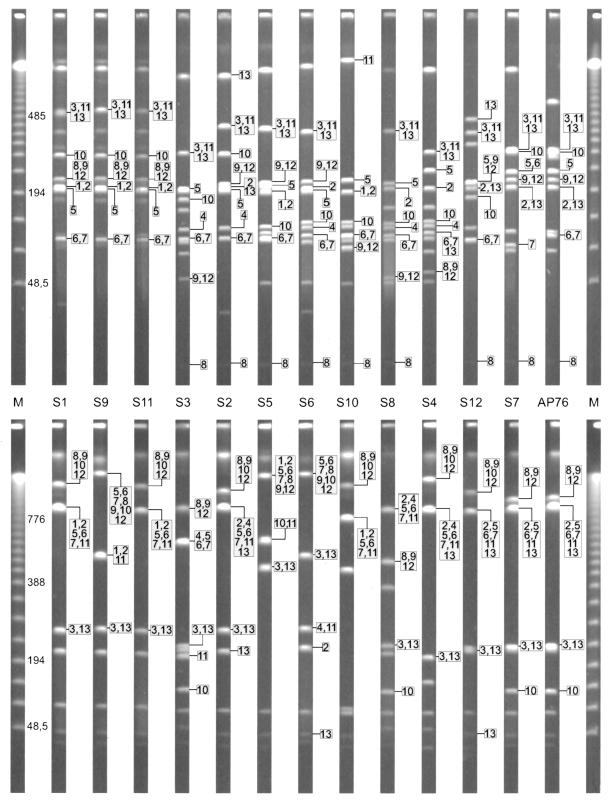
PFGE separation (left) and exemplary Southern blots (right) of the A. pleuropneumoniae serotype reference strains, using the enzymes ApaI (top) and NotI (bottom) and a sodA probe. The top and bottom gels were generated by using short and long running conditions, respectively. The A. pleuropneumoniae serotype reference strains (S1 to S12) are grouped based on their phylogenetic relationship (7). The position of the mapped A. pleuropneumoniae clinical isolate AP76 is indicated by “76.” Lambda concatemers were used as molecular size markers (M).
Strain comparison.
To investigate the genetic homogeneity within the species A. pleuropneumoniae, the hybridization patterns of ApaI- and NotI-restricted genomes of the A. pleuropneumoniae serotype reference strains were compared. The PFGE-separated fragments were hybridized with probes specific for genes positioned on the genetic maps of A. pleuropneumoniae AP76 and ATCC 27088 (Fig. 3 and 4). Using the more frequently cleaving enzyme ApaI, we found that predominantly fragments with comparable sizes were hybridized. In the NotI digest, hybridizing fragments were found to be more variable in size but the linkage of genes appeared to be consistent among the reference strains (Fig. 4). The repeat element was detected in all reference strains in up to three copies. Only the serotype 10 reference strain had no repeat element as assessed by Southern blotting and confirmed by a repeat-specific PCR (data not shown). The repeat element consistently mapped together on the same fragment with the apxIICA toxin genes in the serotype reference strains. The apxIBD genes were also found together on the same fragment with a repeat element in A. pleuropneumoniae serotypes 2, 7, and 12; in the other strains, apxIBD was not linked with a repeat element.
FIG. 4.
Comparative locations of mapped genes on ApaI (top)- and NotI (bottom)-restricted genomes of the 12 A. pleuropneumoniae serotype reference strains. The top gel was run with linear ramped switch times from 5 to 20 s for 13 h and 35 to 70 s for 13 h. The bottom gel was run with switch times from 15 to 30 s for 13 h and 40 to 90 s for 17 h. The A. pleuropneumoniae serotype reference strains (S1 to S12) are grouped based on their phylogenetic relationship (7). Bacteriophage lambda concatemers were used as molecular size markers (M); the unboxed numbers indicate sizes of the corresponding bands in kilobase pairs; the boxed numbers indicate positions of the mapped genes: 1 (apxICA), 2 (apxIBD), 3 (apxIICA), 4 (apxIIICA), 5 (omlA), 6 (ureC), 7 (tolQ-tbpBA), 8 (sodA), 9 (glyA), 10 (metJ), 11 (recA), 12 (rho), and 13 (repeat element).
DISCUSSION
A restriction map with a mean resolution of about 100 kbp has been established for the A. pleuropneumoniae AP76 (serotype 7 clinical isolate) and ATCC 27088 (serotype 1 reference strain). These two strains were chosen since A. pleuropneumoniae ATCC 27088 is the internationally most relevant reference strain and A. pleuropneumoniae AP76 is the only strain in which spontaneous genetic rearrangements have been shown to occur (2). The maps were used to position putative virulence-associated genes as well as some housekeeping genes to their genomic surroundings and to compare their relative locations among the A. pleuropneumoniae serotype reference strains.
The genome size and AscI restriction fragment pattern determined for A. pleuropneumoniae ATCC 27088 corresponded to results presented previously (7). Also, we could confirm the ApaI restriction fragment patterns of the serotype 1, 9, and 11 reference strains (7). Faint bands in the ApaI cleavage pattern of strain ATCC 27088 (7) (Fig. 3 and 4) we considered to be products of incomplete digestion.
In the NotI restriction pattern of A. pleuropneumoniae AP76, no differences from the serotype 7 reference strain A. pleuropneumoniae WF83 were found. Comparing the ApaI restriction patterns, we identified two regions of variation. One variation is most likely due to a loss of the ApaI restriction site in strain AP76 at position 1450 kbp in the map (Fig. 2A), thus joining the two fragments APA1 and APA4 to form a markedly larger fragment (Fig. 4). Another, more complex heterogeneity involves the position of the ureC gene mapping together with the tolQ-tbpBA region (Fig. 2C). In strain WF83, ureC maps to a different fragment, where it is located together with omlA (Fig. 4). Whether this is due to changes of at least two ApaI restriction sites or caused by a small inversion of this part of the chromosome could not be determined from our data. The four variations in fragment sizes in the pattern obtained with AscI are most likely due to the loss and acquisition of singular restriction sites.
The fragment sizes and macrorestriction maps of the two A. pleuropneumoniae strains analyzed showed more differences than similarities (Fig. 2A and B). The arrangements of the genes, however, were found to correspond (Fig. 2C and D). This result confirms the hypothesis of a clonal origin of the A. pleuropneumoniae serotypes proposed by Musser et al. based on multilocus enzyme electrophoresis (27). Thus, only a single difference between the two genetic maps was detected at the position of the genes apxIBD. In A. pleuropneumoniae AP76, a repeat element mapped on the same fragment, whereas in A. pleuropneumoniae ATCC 27088 a complete apxICABD gene cluster was found in this position.
It was found that the apxIICA sequence consistently mapped together with the repeat element as had been described for A. pleuropneumoniae serotype 7 (2). This supports the function of the repeat element as a transposable DNA element and indicates acquisition of the ApxII toxin late in evolution by horizontal gene transfer. In addition, the apxIBD sequence required for toxin transport was found on the same fragment about 200 kbp in size together with another repeat element in serotypes 2, 7, and 12, whereas no association was found in other strains. This is interesting since it might suggest a different evolutionary development for the reference strains carrying the genotype apxIBD apxIICA apxIIICABD without association (serotype 4, 6, and 8) and with possible association (serotype 2) between apxIBD and the repeat element. Thus, the pathogenic A. pleuropneumoniae serotype 2 may have developed from a serotype 3 ancestor with low pathogenicity by acquiring the mobilizable apxIBD. In contrast, serotypes 4, 6, and 8 may have developed from an ancestor with apxICABD by deletion of apxICA as proposed by Frey (12). In this model, A. pleuropneumoniae serotype 10 would be the evolutionarily oldest strain with only apxICABD. The youngest strains in evolutionary terms would be serotypes 7 and 12 carrying only apxIBD and apxIICA, both accompanied by a repeat element.
The results presented here document a high degree of genetic homogeneity and stability within the species A. pleuropneumoniae. Gross rearrangements as described for different serovars of Vibrio cholerae (22, 39) or closely related isolates of Pseudomonas aeruginosa (34) were not observed. Also, there is no indication for the presence of large discrete unstable regions on the A. pleuropneumoniae genome which might be indicative for pathogenicity islands as described for members of the family Enterobacteriaceae (21, 26, 30, 35) and Helicobacter pylori (1). A comparison of our initial genetic map with that of Haemophilus influenzae Rd based on the complete genomic sequence (10) revealed clear differences in the relative order of the genes positioned by us (not shown). This shows that H. influenzae Rd sequences, though useful for identifying homologous A. pleuropneumoniae sequences by PCR or Southern blotting, do not allow a prediction of the relative locations of the corresponding A. pleuropneumoniae genes.
In conclusion, the physical maps presented here will facilitate localization of newly detected genes by Southern hybridization with DNA restriction patterns from single digests with the four restriction endonucleases ApaI, AscI, NotI, and SalI. Particularly in the absence of extensive genomic sequence data, such a completed genetic map will represent a valuable tool for the investigation of A. pleuropneumoniae pathogenicity and for the development of attenuated live vaccines.
ACKNOWLEDGMENTS
This work was supported by grant GE 522/3-1 from the Deutsche Forschungsgemeinschaft, Bonn, Germany, D.V.K. is a fellow of the German Academic Exchange Service, Bonn, Germany.
REFERENCES
- 1.Akoopyants N S, Clifton S W, Kersulyte D, Crabtree J E, Youree B E, Reece C A, Bukanov N O, Drazek E S, Roe B A, Berg D E. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol Microbiol. 1998;28:37–53. doi: 10.1046/j.1365-2958.1998.00770.x. [DOI] [PubMed] [Google Scholar]
- 2.Anderson C, Potter A A, Gerlach G-F. Isolation and molecular characterization of spontaneously occurring cytolysin-negative mutants of Actinobacillus pleuropneumoniae serotype 7. Infect Immun. 1991;59:4110–4116. doi: 10.1128/iai.59.11.4110-4116.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bautsch W. Bacterial genome mapping by two-dimensional pulsed-field gel electrophoresis (2D-PFGE) Mol Biotechnol. 1994;2:29–44. doi: 10.1007/BF02789288. [DOI] [PubMed] [Google Scholar]
- 4.Bosse J T, MacInnes J I. Genetic and biochemical analyses of Actinobacillus pleuropneumoniae urease. Infect Immun. 1997;65:4389–4394. doi: 10.1128/iai.65.11.4389-4394.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bunka S, Christensen C, Potter A A, Willson P J, Gerlach G-F. Cloning and characterization of a protective outer membrane lipoprotein of Actinobacillus pleuropneumoniae serotype 5. Infect Immun. 1995;63:2797–2800. doi: 10.1128/iai.63.7.2797-2800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang Y F, Shi J, Ma D P, Shin S J, Lein D H. Molecular analysis of the Actinobacillus pleuropneumoniae RTX toxin-III gene cluster. DNA Cell Biol. 1993;12:351–362. doi: 10.1089/dna.1993.12.351. [DOI] [PubMed] [Google Scholar]
- 7.Chevallier B, Dugourd D, Tarasiuk K, Harel J, Gottschalk M, Kobisch M, Frey J. Chromosome sizes and phylogenetic relationships between serotypes of Actinobacillus pleuropneumoniae. FEMS Microbiol Lett. 1998;160:209–216. doi: 10.1111/j.1574-6968.1998.tb12913.x. [DOI] [PubMed] [Google Scholar]
- 8.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 9.Fenwick B, Henry S. Porcine pleuropneumonia. J Am Vet Med Assoc. 1994;204:1334–1340. [PubMed] [Google Scholar]
- 10.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrik J M, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 11.Frey J, Haldimann A, Nicolet J, Boffini A, Prentki P. Sequence analysis and transcription of the apxI operon (hemolysin I) from Actinobacillus pleuropneumoniae. Gene. 1994;142:97–102. doi: 10.1016/0378-1119(94)90361-1. [DOI] [PubMed] [Google Scholar]
- 12.Frey J. Virulence in Actinobacillus pleuropneumoniae and RTX toxins. Trends Microbiol. 1995;3:257–261. doi: 10.1016/s0966-842x(00)88939-8. [DOI] [PubMed] [Google Scholar]
- 13.Gerlach G-F, Klashinsky S, Anderson C, Potter A A, Willson P J. Characterization of two genes encoding distinct transferrin-binding proteins in different Actinobacillus pleuropneumoniae isolates. Infect Immun. 1992;60:3253–3261. doi: 10.1128/iai.60.8.3253-3261.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerlach G-F, Anderson C, Klashinsky S, Potter A A, Rossi-Campos A, Willson P J. Molecular characterization of a protective outer membrane lipoprotein (OmlA) from Actinobacillus pleuropneumoniae serotype 1. Infect Immun. 1993;61:565–572. doi: 10.1128/iai.61.2.565-572.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez G C, Yu R H, Rosteck P R, Schryvers A B. Sequence, genetic analysis, and expression of Actinobacillus pleuropneumoniae transferrin receptor genes. Microbiology. 1995;141:2405–2416. doi: 10.1099/13500872-141-10-2405. [DOI] [PubMed] [Google Scholar]
- 16.Inzana T J, Ma J, Workman T, Gogolewsk R P, Anderson P. Virulence properties and protective efficacy of the capsular polymer of Haemophilus (Actinobacillus) pleuropneumoniae serotype 5. Infect Immun. 1988;56:1880–1889. doi: 10.1128/iai.56.8.1880-1889.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito H, Osaki M, Uchida I, Ohya T, Sekizaki T. Demonstration of the third antigenically distinct outer membrane lipoprotein (OmlA) in Actinobacillus pleuropneumoniae serotype 7. FEMS Microbiol Lett. 1998;167:303–308. doi: 10.1111/j.1574-6968.1998.tb13243.x. [DOI] [PubMed] [Google Scholar]
- 18.Ito H, Uchida I, Sekizaki T, Ooishi E, Kawai T, Okabe T, Taneno A, Terakado N. Molecular cloning of an Actinobacillus pleuropneumoniae outer membrane lipoprotein (OmlA) from serotype 5a. Microb Pathog. 1995;18:29–36. [PubMed] [Google Scholar]
- 19.Jansen R, Briaire J, van Geel A B, Kamp E M, Gielkens A L, Smits M A. Genetic map of the Actinobacillus pleuropneumoniae RTX-toxin (Apx) operons: characterization of the ApxIII operons. Infect Immun. 1994;62:4411–4418. doi: 10.1128/iai.62.10.4411-4418.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamp E M, Popma J K, Anakotta J K, Smits M A. Identification of hemolytic and cytotoxic proteins of Actinobacillus pleuropneumoniae by the use of monoclonal antibodies. Infect Immun. 1991;59:3079–3085. doi: 10.1128/iai.59.9.3079-3085.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karaolis, D. K., J. A. Johnson, C. C. Bailey, E. C. Boedeker, J. B. Kaper, and P. R. Reeves. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. USA 95:3134–3139. [DOI] [PMC free article] [PubMed]
- 22.Khetawat G, Bhadra R K, Kar S, Das J. Vibrio cholerae O139 Bengal: combined physical and genetic map and comparative analysis with the genome of V. cholerae O1. J Bacteriol. 1998;180:4516–22. doi: 10.1128/jb.180.17.4516-4522.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langford P R, Loynds B M, Kroll J S. Cloning and molecular characterization of Cu,Zn superoxide dismutase from Actinobacillus pleuropneumoniae. Infect Immun. 1996;64:5035–5041. doi: 10.1128/iai.64.12.5035-5041.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacDonald J, Rycroft A N. Molecular cloning and expression of ptxA, the gene encoding the 120-kilodalton cytotoxin of Actinobacillus pleuropneumoniae serotype 2. Infect Immun. 1992;60:2726–2732. doi: 10.1128/iai.60.7.2726-2732.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medrano A, Querol E, Daban M. Cloning and sequencing of the gene encoding the outer-membrane protein Tbp1 from Actinobacillus pleuropneumoniae. Expression of Tbp1 and Tbp2. Behring Inst Mitt. 1997;98:410–423. [PubMed] [Google Scholar]
- 26.Morschhauser J, Vetter V, Emody L, Hacker J. Adhesin regulatory genes within large, unstable DNA regions of pathogenic Escherichia coli: cross-talk between different adhesin gene clusters. Mol Microbiol. 1994;11:555–566. doi: 10.1111/j.1365-2958.1994.tb00336.x. [DOI] [PubMed] [Google Scholar]
- 27.Musser J M, Rapp V J, Selander R K. Clonal diversity in Haemophilus pleuropneumoniae. Infect Immun. 1987;55:1207–1215. doi: 10.1128/iai.55.5.1207-1215.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicolet J. Taxonomy and serological identification of Actinobacillus pleuropneumoniae. Can Vet J. 1988;29:578–580. [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen R. Serological characterization of Actinobacillus pleuropneumoniae strains and proposal of a new serotype: serotype 12. Acta Vet Scand. 1986;27:452–455. doi: 10.1186/BF03548158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ochman H, Soncini F C, Solomon F, Groisman E A. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perry M B, Altman E, Brisson J-R, Beynon L M, Richards J C. Structural characteristics of the antigenic capsular polysaccharides and lipopolysaccharides involved in the serological classification of Actinobacillus pleuropneumoniae strains. Serodiagn Immun Infect Dis. 1990;4:299–308. [Google Scholar]
- 32.Rapp V J, Mundson R S, Ross R F. Outer membrane protein profiles of Haemophilus pleuropneumoniae. Infect Immun. 1986;52:414–420. doi: 10.1128/iai.52.2.414-420.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reimer D, Frey J, Jansen R, Veit H P, Inzana T J. Molecular investigation of the role of ApxI and ApxII in the virulence of Actinobacillus pleuropneumoniae serotype 5. Microb Pathog. 1995;18:197–209. doi: 10.1016/s0882-4010(95)90049-7. [DOI] [PubMed] [Google Scholar]
- 34.Römling U, Schmidt K D, Tümmler B. Large genome rearrangements discovered by the detailed analysis of 21 Pseudomonas aeruginosa clone C isolates found in environment and disease habitats. J Mol Biol. 1997;271:386–404. doi: 10.1006/jmbi.1997.1186. [DOI] [PubMed] [Google Scholar]
- 35.Schubert S, Rakin A, Karch H, Carniel E, Heesemann J. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect Immun. 1998;66:480–485. doi: 10.1128/iai.66.2.480-485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 37.Tascon R I, Vazquez-Boland J A, Gutierrez-Martin C B, Rodriguiz-Barbosam I, Rodriguez-Ferri E F. The RTX haemolysins ApxI and ApxII are major virulence factors of the swine pathogen Actinobacillus pleuropneumoniae: evidence from mutational analysis. Mol Microbiol. 1994;14:207–216. doi: 10.1111/j.1365-2958.1994.tb01282.x. [DOI] [PubMed] [Google Scholar]
- 38.Thiede S, Goethe R, Gerlach G-F. Proceedings of the 79th Conference of Research Workers in Animal Diseases. 1998. Actinobacillus pleuropneumoniae Rho-factor is an amplifier of transferrin-binding-protein expression; p. 17. [Google Scholar]
- 39.Trucksis M, Michalski J, Deng Y K, Kaper J B. The Vibrio cholerae genome contains two unique circular chromosomes. Proc Natl Acad Sci USA. 1998;95:14464–14469. doi: 10.1073/pnas.95.24.14464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward C K, Lawrence M L, Veit H P, Inzana T J. Cloning and mutagenesis of a serotype-specific DNA region involved in encapsulation and virulence of Actinobacillus pleuropneumoniae serotype 5a: concomitant expression of serotype 5a and 1 capsular polysaccharides in recombinant A. pleuropneumoniae serotype 1. Infect Immun. 1998;66:3326–3336. doi: 10.1128/iai.66.7.3326-3336.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilke M, Franz B, Gerlach G-F. Characterization of a large transferrin-binding protein from Actinobacillus pleuropneumoniae serotype 7. J Vet Med Ser B. 1997;44:73–86. doi: 10.1111/j.1439-0450.1997.tb00953.x. [DOI] [PubMed] [Google Scholar]