Summary
The establishment of effective molecular interventions to improve memory and alleviate memory deficits in disease remains a long-standing challenge despite growing molecular understanding of synaptic plasticity and memory formation. Capitalizing on the fact that long-term potentiation (LTP) requires N-methyl-D-aspartate receptors (NMDARs) and Ca2+/calmodulin-dependent protein kinase II alpha (CaMKIIα), we develop an intrabody that targets NMDARs and show that intrabody-mediated postsynaptic enrichment of CaMKIIα in the hippocampus improves contextual fear memory. This molecular approach suggests a potential demand for effective targeting of postsynaptic molecules to enhance memory and provides insights into studying memory improvement in health and disease.
Subject areas: Biological sciences, Cognitive neuroscience, Neuroscience
Graphical abstract
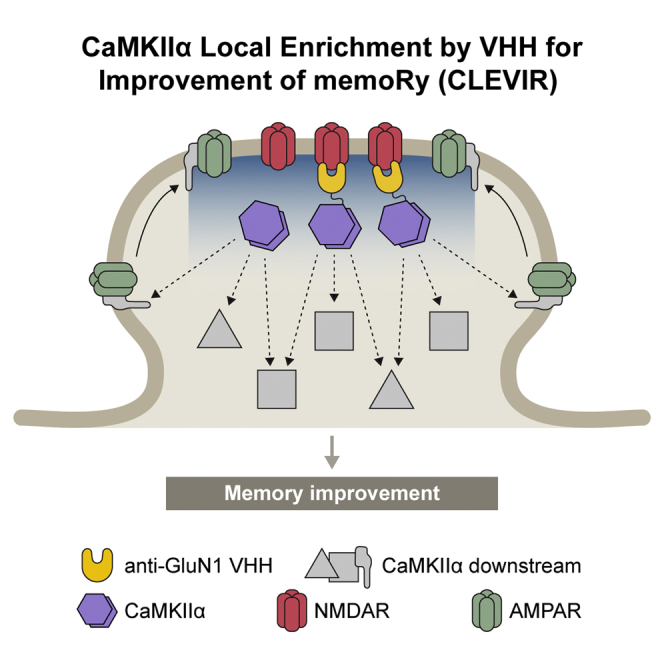
Highlights
-
•
VHHAN1 binds to GluN1, a subunit of NMDA receptors, in cells and in vivo
-
•
Fusion with VHHAN1 orients exogenous CaMKIIα to excitatory postsynaptic regions
-
•
CaMKIIα local enrichment by VHHAN1 in the hippocampus improves contextual memory
-
•
Potential demand for effective modifications of synaptic molecules to modify memory
Biological sciences; Cognitive neuroscience; Neuroscience.
Introduction
Long-term potentiation (LTP) is an activity-dependent strengthening of synaptic transmission that has been considered a cellular mechanism of learning and memory (Huganir and Nicoll, 2013; Nicoll, 2017). In the hippocampus, LTP requires activation of NMDARs, a rise in postsynaptic calcium, and activation of CaMKIIα (Huganir and Nicoll, 2013; Lisman et al., 2002, 2012; Nicoll, 2017; Silva et al., 1992a, 1992b). Knockout and inactive mutant (T286A) knock-in mouse studies have clearly demonstrated the importance of CaMKIIα in LTP and memory (Giese et al., 1998; Irvine et al., 2005; Silva et al., 1992a, 1992b). Recent studies with CaMKIIα phosphorylation substrates (e.g., TARPγ-8 and SynGAP) and a photoactivatable CaMKIIα inhibitor have provided further support regarding the critical links between CaMKIIα and LTP (Araki et al., 2015; Murakoshi et al., 2017; Park et al., 2016).
However, little is known about how CaMKIIα can be utilized for memory improvements. This gap has resulted from limitations in our knowledge and techniques for obtaining an appropriate supply of exogenous CaMKIIα to facilitate memory formation. Virus-mediated hippocampal expression of wild-type CaMKIIα improves spatial memory in water maze tests of rats (∼1.15-fold longer time spent in the target area) (Poulsen et al., 2007). Meanwhile, viral expression of the activated form of CaMKIIα (T286D/T305A/T306A) in the hippocampus impairs place avoidance memory and context discrimination in rodents, similarly to results obtained for a dominant-negative form (K42M) (Rossetti et al., 2017; Ye et al., 2019). We reason that the limited or lack of improvement in memory was due to (1) an inability to respond to the postsynaptic calcium rise in the mutant form and (2) an inadequate postsynaptic supply of CaMKIIα, as overexpressed CaMKIIα diffuses throughout neurons. Thus, there is a need to provide wild-type CaMKIIα with local postsynaptic enrichment proximal to NMDARs, where CaMKIIα is in high demand for LTP induction.
Here, we report an intrabody targeting the cytoplasmic region of a subunit of NMDARs GluN1 (termed VHHAN1). We validated specific targeting of VHHAN1 to GluN1 in heterologous cells and the mouse hippocampus. Employing this intrabody, we supplied the adeno-associated virus (AAV)-mediated expression of CaMKIIα at excitatory postsynaptic regions of the mouse hippocampus, and this approach showed a significant improvement in fear conditioning memory of mice.
Results
Development of an intrabody targeting GluN1
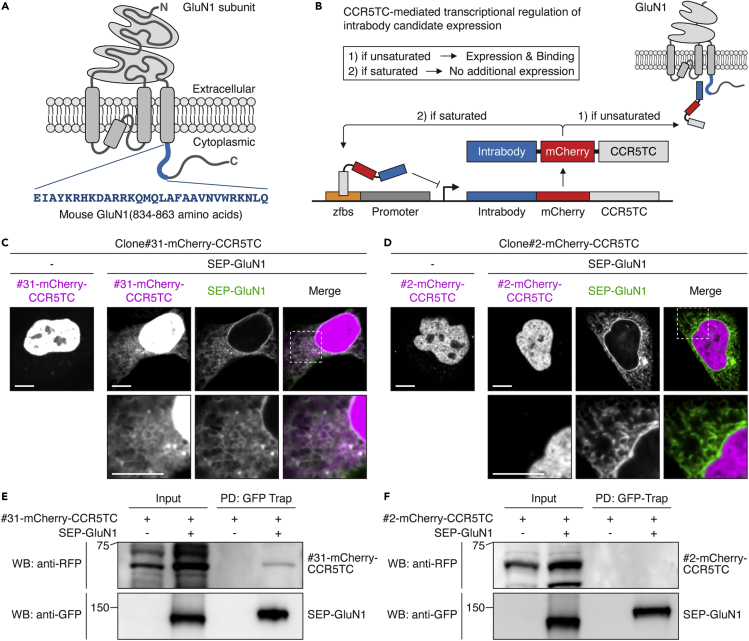
To develop an intrabody targeting endogenous NMDARs in the brain, it is critical to find an appropriate intracellular portion of NMDAR subunits for the intrabody binding. Heterotetrameric NMDARs consist of two subunits of GluN1 and two subunits of either GluN2 (GluN2A through GluN2D) or GluN3 (GluN3A and GluN3B) (Paoletti et al., 2013). The cytoplasmic tail of GluN1 subunits provides an effective platform for intrabody binding, but eight splice variants have been reported (Figure S1A). Therefore, as a target region, we chose a part of the mouse GluN1 cytoplasmic tail conserved throughout all of the splice variants (C0 cassette, 834–863 amino acids of NP_032195.1) (Figure 1A). We purified this fragment (His-AviTag-GluN1C0) from bacteria, immobilized it on streptavidin beads (Figures S1B and S1C), and conducted nanobody screening with a synthetic phage display library (3.0 × 109 nanobodies) (Moutel et al., 2016). We narrowed down the nanobody candidates to 3.1 × 105 by first-phase phage display screening (Figure S1D). We performed further screening via yeast two-hybrid (Y2H) assays to search proteins that bind to the GluN1C0 intracellularly. We conducted two rounds of Y2H screening with the C0 target region and the full-length cytoplasmic tail of GluN1 (834–938 amino acids of NP_032195.1). We obtained 118 positive clones from the Y2H screenings and confirmed three candidate clones after eliminating redundant clones by Sanger sequencing (Figure S1D).
Figure 1.
Development of anti-GluN1 intrabody
(A) Schematic diagram illustrating GluN1 domain structure and the anti-GluN1 intrabody target region (834–863 amino acids).
(B) Illustration of CCR5TC-mediated transcriptional regulation. To avoid overproduction and subsequent random diffusion of intrabodies, intrabody candidates were fused with mCherry and CCR5TC. If the interaction is unsaturated, protein expression continues for further GluN1 binding. If 100% of GluN1 binds to the intrabody, then the newly synthesized intrabodies move to the nucleus and bind to a zinc-finger binding site (zfbs) upstream of a promoter to discontinue protein expression.
(C–F) Colocalization and interaction of Clone#31, but not Clone#2, with GluN1 in heterologous cells. Intrabody candidates (Clone#31 and #2) were fused with mCherry and CCR5TC and expressed in HeLa cells with or without SEP-fused GluN1. In the absence of SEP-GluN1, both Clone#31 and #2 localized in the nucleus due to the nuclear localization signal of CCR5TC (C and D, left panels). When co-expressed with SEP-GluN1 (green), Clone#31 (magenta), but not Clone#2, colocalized with SEP-GluN1, depicting an ER-like structure (C and D, right panels). GFP-Trap pull-down (PD) revealed the specific binding of Clone#31-mCherry-CCR5TC, but not Clone#2, to SEP-GluN1 (E and F). Scale bars, 10 μm.
We next examined colocalization and interaction between GluN1 and the three intrabody candidates in a heterologous system. To circumvent intrabody overproduction that results in diffusion throughout cells, we adopted a previously established system of CCR5TC transcriptional repression (Bensussen et al., 2020; Gross et al., 2013). When an intrabody candidate expresses and binds to less than 100% of GluN1 (if unsaturated), protein expression continues for further GluN1 binding (Figure 1B). When the interaction between GluN1 and the intrabody reaches 100% (if saturated), a newly expressed population of the intrabody candidate moves to the nucleus, binds to a zinc-finger binding site upstream of the promoter, and stops further transcription (Figure 1B). Using this system, we examined the colocalization of super-ecliptic pHluorin (SEP)-fused GluN1 (Kopec et al., 2006) and mCherry- and CCR5TC-fused intrabody candidates in HeLa cells. Spinning disk confocal imaging revealed that a single expression of each intrabody candidate shows its enrichment in the nucleus due to the nuclear localization signal of CCR5TC (Figures 1C, 1D, and S1E, left panels). However, co-expression of SEP-GluN1 in the ER effectively recruited one of the candidates (Clone#31) to show robust colocalization (Figure 1C, right panel). ER retention of GluN1 is expected in the absence of GluN2 subunits (Scott et al., 2001; Standley et al., 2000). In contrast, the other two candidates showed no or minimal colocalization with GluN1 (Figures 1D and S1E, right panels). We also confirmed the interaction between SEP-GluN1 and Clone#31-mCherry-CCR5TC in HeLa cells by a pull-down assay with GFP-Trap beads (Figure 1E). The other intrabody candidates showed no or minimal interaction with GluN1, consistent with the colocalization data (Figures 1F and S1F). These data suggest that one of the intrabody candidates (Clone#31) colocalizes well and binds to GluN1 in heterologous cells; we termed this intrabody VHHAN1 (VHH Anti-GluN1).
VHHAN1 interacts with endogenous GluN1 in vivo
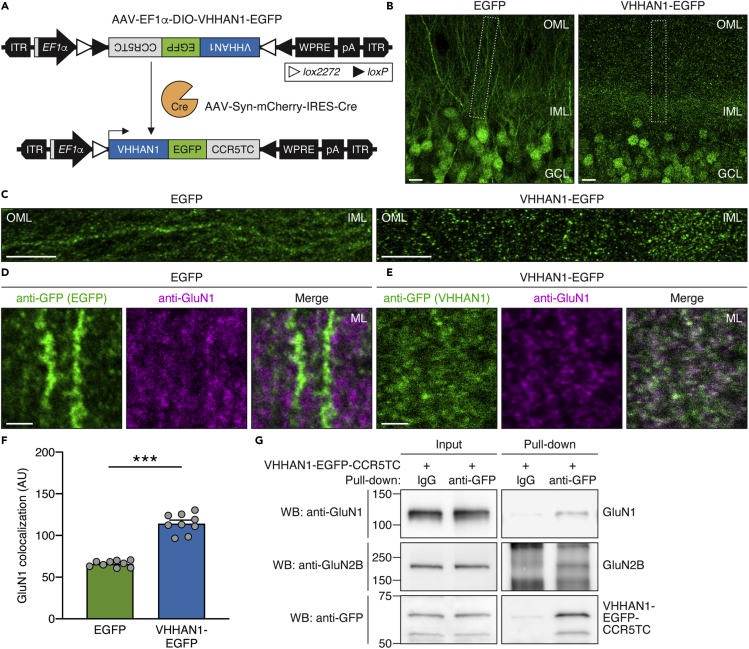
Finally, we validated postsynaptic targeting of VHHAN1 to endogenous GluN1 in vivo. To express VHHAN1 in vivo, we generated an AAV expressing enhanced green fluorescent protein (EGFP)-fused VHHAN1 (Figure 2A). This AAV expresses VHHAN1-EGFP under an elongation factor-1 alpha (EF1α) promoter in a Cre recombinase (Cre)-dependent manner (Figure 2A). To express VHHAN1 in neurons, we co-injected it with an AAV expressing Cre and mCherry under a neuronal promoter (Synapsin) (AAV-Syn-mCherry-IRES-Cre) (Figure 2A). As shown in Figures 2B and 2C, laser scanning confocal microscopy revealed that the mouse hippocampus expressing VHHAN1-EGFP shows regular punctate patterns of VHHAN1-EGFP in the molecular layers, whereas the mouse hippocampus expressing free EGFP shows a diffuse distribution following the soma and neurite projections. The extra GFP signal in the nuclei expressing VHHAN1-EGFP reflects the CCR5TC-mediated transcriptional control (Figures 1B, 2A, and 2B). The VHHAN1-EGFP puncta showed strong colocalization with puncta of the target protein, endogenous GluN1, compared with the limited GluN1 colocalization with the diffuse, free EGFP (Figures 2D and 2E); quantitative analysis revealed a significant difference in this GluN1 colocalization (p < 0.0001) (Figure 2F). We also confirmed VHHAN1 binding to the endogenous NMDAR complex (GluN1 and GluN2B subunits) via co-immunoprecipitation using mouse hippocampal lysates (Figure 2G). Taken together, these results confirm the development of an intrabody VHHAN1 that targets endogenous GluN1 in vivo.
Figure 2.
VHHAN1 associates with endogenous GluN1 in the mouse hippocampus
(A) Illustration of the AAV expressing EGFP- and CCR5TC-fused VHHAN1 (AAV-EF1α-DIO-VHHAN1-EGFP).
(B–F) Comparison of EGFP and VHHAN1-EGFP distribution in the mouse hippocampal dentate gyrus. Anti-GFP immunostaining visualized expression and localization of EGFP (left) and VHHAN1-EGFP (right) (B). OML, outer molecular layer; IML, inner molecular layer; GCL, granule cell layer. Scale bars, 10 μm. The molecular layer regions are shown in (C) as indicated by the dashed boxes in (B). Scale bars, 10 μm. High-magnification confocal images of hippocampal dentate gyrus co-stained with anti-GFP and anti-GluN1 antibodies showed postsynaptic puncta of VHHAN1 (green) that colocalized with endogenous GluN1 puncta (magenta) in the dentate gyrus (D and E). Scale bars, 2 μm. Quantitative analysis of the colocalization of GFP signals with endogenous GluN1 (F). Data are shown as mean ± SEM; AU, arbitrary unit; 9 randomly chosen images from 3 animals; ∗∗∗p < 0.0001; Mann-Whitney test.
(G) The binding of VHHAN1 to the endogenous NMDAR complex was validated by anti-GFP co-immunoprecipitation.
VHHAN1-mediated postsynaptic enrichment of CaMKIIα in the hippocampus
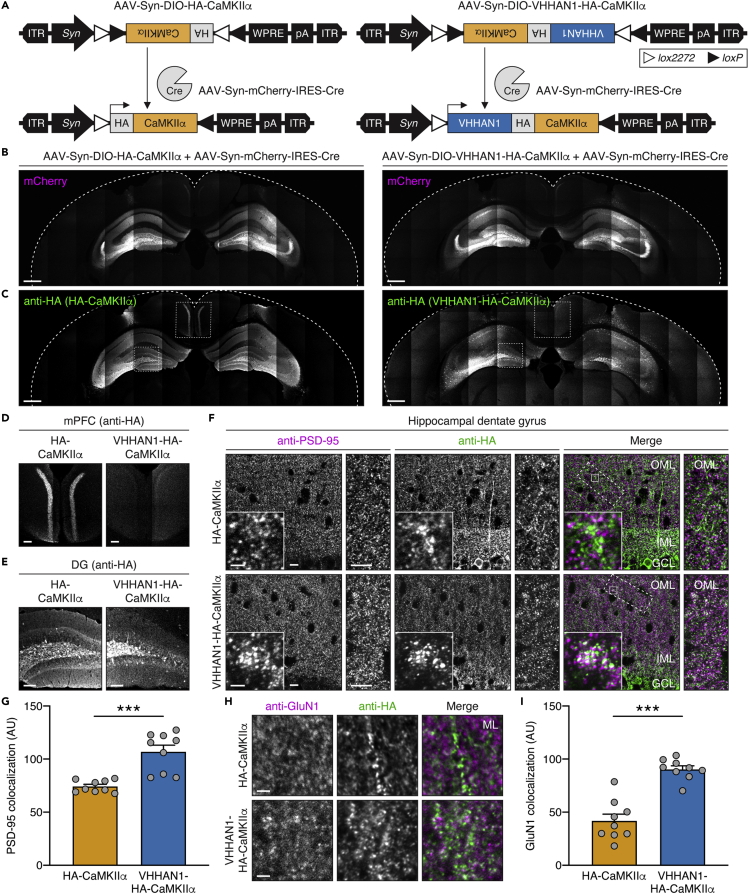
Using this intrabody, we investigated whether wild-type CaMKIIα can be locally enriched at excitatory postsynaptic regions by fusing with VHHAN1. We generated Cre-dependent AAVs expressing either hemagglutinin (HA)-tagged CaMKIIα (AAV-Syn-DIO-HA-CaMKIIα) or both VHHAN1- and HA-tagged CaMKIIα under a Synapsin promoter (AAV-Syn-DIO-VHHAN1-HA-CaMKIIα) (Figure 3A). We selected an N-terminal modification of CaMKIIα with VHHAN1 and an HA-tag to avoid potential disruption of the C-terminal dimerization of CaMKIIα hexamers (Lisman et al., 2012) and because N-terminal protein fusion is known to preserve kinase activity (Park et al., 2016). Co-injections of these AAVs with AAV-Syn-mCherry-IRES-Cre allowed us to achieve the desired CaMKIIα expression and immediately visualize infected brain regions under a fluorescent microscope. As shown in Figure 3B, the coverage of AAV infections in each hippocampus was symmetric within animals and comparable between animals. Immunohistochemistry with anti-HA antibody revealed that HA-CaMKIIα has a broader intracellular distribution than VHHAN1-HA-CaMKIIα in infected neurons (Figure 3C). Specifically, the expression of HA-CaMKIIα showed robust signals following hippocampal axonal projections onto the medial prefrontal cortex (Figure 3D, left panel) and the inner molecular layer of the dentate gyrus (Figure 3E, left panel), whereas VHHAN1-HA-CaMKIIα did not show these patterns (Figures 3D and 3E, right panels), indicating a widespread distribution of HA-CaMKIIα throughout the infected hippocampal neurons (Förster et al., 2006). The mCherry distribution in both conditions was even and comparable throughout the infected hippocampi (Figure 3B), further indicating that the differences in anti-HA immunoreactivity in Figures 3D and 3E result from the distinct subcellular distribution of VHHAN1-HA-CaMKIIα, not the AAV infection coverage. Focusing on the dentate gyrus, laser scanning confocal microscopy revealed regular punctate patterns of VHHAN1-HA-CaMKIIα in the molecular layers compared with a diffuse distribution of HA-CaMKIIα following neurite projections (Figure 3F). The puncta of VHHAN1-HA-CaMKIIα were significantly colocalized with an endogenous excitatory postsynaptic marker, postsynaptic density protein 95 (PSD-95) than HA-CaMKIIα (p < 0.0001) (Figures 3F and 3G). We further evaluated the significant enrichment of VHHAN1-HA-CaMKIIα in the postsynaptic region using co-immunostaining with endogenous GluN1 (Figures 3H and 3I). VHHAN1-HA-CaMKIIα showed a significant increase in GluN1 colocalization (2.2-fold compared with HA-CaMKIIα; p < 0.0001) (Figures 3H and 3I). These data suggest that VHHAN1 orients exogenous wild-type CaMKIIα to excitatory postsynaptic regions in vivo.
Figure 3.
Local enrichment of CaMKIIα at excitatory postsynaptic regions by VHHAN1 in the mouse hippocampus
(A) Illustration of AAVs expressing HA-tagged CaMKIIα (left) and VHHAN1-HA-fused CaMKIIα (right).
(B–E) Each AAV was co-injected with AAV-Syn-mCherry-IRES-Cre into the hippocampus to express HA-CaMKIIα (left) or VHHAN1-HA-CaMKIIα (right) in hippocampal neurons. The symmetric and comparable coverage of AAV infections in each hippocampus was validated by mCherry fluorescence (B). Anti-HA immunostaining visualized expression and localization of HA-CaMKIIα (left) and VHHAN1-HA-CaMKIIα (right) (C). The medial prefrontal cortex (mPFC) and hippocampal dentate gyrus (DG) regions are shown in (D and E) as indicated by the dashed boxes in (C). The lack of a presynaptic distribution of anti-HA signals in the mPFC and the inner molecular layer of the DG from the VHHAN1-HA-CaMKIIα condition (D and E, right panels) indicates the postsynapse-oriented distribution of VHHAN1-HA-CaMKIIα. Scale bars, 500 μm (B and C) and 100 μm (D and E).
(F) High-magnification confocal images of hippocampal DG co-stained with anti-HA and anti-PSD-95 (an excitatory postsynaptic marker) antibodies showed more regular punctate patterns of VHHAN1-HA-CaMKIIα, whereas HA-CaMKIIα displays a more diffuse distribution following neurite projections. Dashed and solid line boxes indicate the magnified regions for inset images. OML, outer molecular layer; IML, inner molecular layer; GCL, granule cell layer. Scale bars, 10 μm for cropped images and 2 μm for square inset images.
(G) Quantitative analysis of the colocalization of HA signals with endogenous PSD-95. Data are shown as mean ± SEM; AU, arbitrary unit; 9 randomly chosen images from 3 animals; ∗∗∗p < 0.0001; Mann-Whitney test.
(H and I) Co-immunostaining of hippocampal DG molecular layers (ML) with anti-HA and anti-GluN1 antibodies showed regular punctate patterns of VHHAN1-HA-CaMKIIα (green) that colocalized with endogenous GluN1 puncta (magenta) more than HA-CaMKIIα (green). Scale bar, 2 μm. Data are shown as mean ± SEM; AU, arbitrary unit; 9 randomly chosen images from 3 to 4 animals; ∗∗∗p < 0.0001; Mann-Whitney test.
CaMKIIα local enrichment by VHHAN1 for improvement of memory
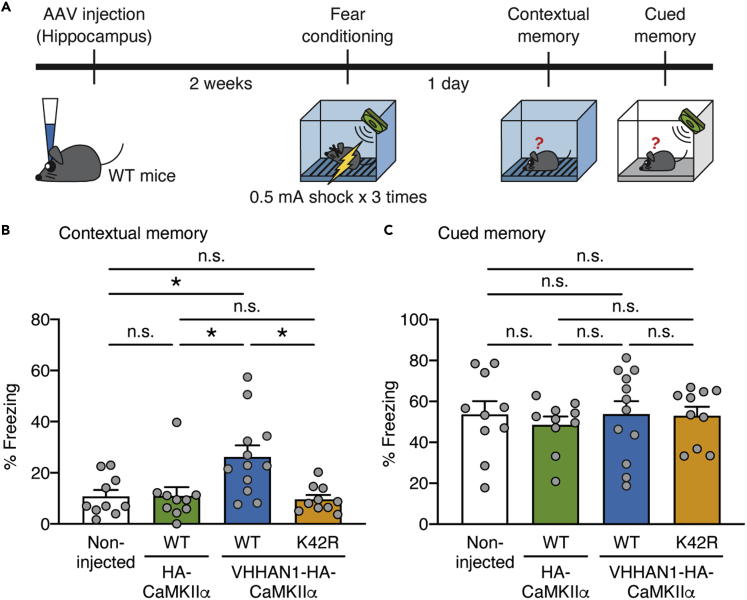
Finally, we examined whether VHHAN1-mediated local postsynaptic enrichment of wild-type CaMKIIα improves memory in mice using a fear conditioning paradigm. We co-injected Cre-dependent AAVs of HA-CaMKIIα, a wild-type form of VHHAN1-HA-CaMKIIα, or a kinase-inactive (K42R) form of VHHAN1-HA-CaMKIIα with AAV-Syn-mCherry-IRES-Cre into both hippocampi of wild-type mice, as confirmed in Figures 3B and 3C. After a two-week period for recovery and protein expression, the mice underwent a fear conditioning paradigm, and the next day, they were subjected to contextual and cued memory tests (Figure 4A). Overexpression of both HA-CaMKIIα and VHHAN1-HA-CaMKIIα had no significant impact on general movement compared with non-injected wild-type mice (Figure S2). Compared with the HA-CaMKIIα and non-injected control groups, the wild-type VHHAN1-HA-CaMKIIα group showed a significant increase in contextual memory (hippocampus-involved) (2.4-fold compared with HA-CaMKIIα and non-injected mice; p = 0.0371 for non-injected versus wild-type VHHAN1-HA-CaMKIIα; p = 0.0260 for HA-CaMKIIα versus wild-type VHHAN1-HA-CaMKIIα; p = 0.0245 for wild-type VHHAN1-HA-CaMKIIα versus VHHAN1-HA-CaMKIIα [K42R]) (Figure 4B) but not cued memory (amygdala-involved) (p > 0.9999) (Figure 4C). However, the VHHAN1-HA-CaMKIIα (K42R) group displayed no significant changes in the contextual and cued fear memory tests compared with the HA-CaMKIIα and non-injected control groups (Figures 4B and 4C). These data suggest that the improved contextual fear memory by VHHAN1-HA-CaMKIIα depends on the CaMKIIα kinase activity (Figure 4B). The symmetric and comparable coverage of AAV infections in each mouse was confirmed by mCherry expression under a fluorescent microscope. Taken together, these data suggest that our molecular approach—which we term CaMKIIα Local Enrichment by VHH for Improvement of memoRy, i.e., CLEVIR—to enrich wild-type CaMKIIα at excitatory postsynaptic regions proximal to NMDARs, where LTP occurs, enhances memory ability in mice.
Figure 4.
CaMKIIα local enrichment by VHHAN1 in the hippocampus improves contextual memory
(A) Schematic diagram depicting the experimental design. AAV-Syn-DIO-HA-CaMKIIα (wild-type, WT), AAV-Syn-DIO-VHHAN1-HA-CaMKIIα (WT), or AAV-Syn-DIO-VHHAN1-HA-CaMKIIα (K42R) was co-injected with AAV-Syn-mCherry-IRES-Cre into the hippocampus of wild-type mice. After two weeks to allow for protein expression, mice underwent a fear conditioning and testing paradigm as described in STAR Methods.
(B and C) Fear memory tests. Mice expressing a wild-type form of VHHAN1-HA-CaMKIIα (WT) in the hippocampus showed a significant increase in contextual memory compared with non-injected, HA-CaMKIIα-expressing, and a kinase-inactive form of VHHAN1-HA-CaMKIIα (K42R)-expressing mice (B). No significant differences were observed in cued fear memory (C). Data are shown as mean ± SEM; n = 10–12 mice per group; ∗p < 0.05; n.s., not significant; Kruskal-Wallis tests with multiple comparisons.
Discussion
Specific visualization and targeting of endogenous synaptic proteins in the brain are important to study synapse biology, but available molecular tools, including intrabodies, remain very limited. Our study reports an intrabody against the GluN1 subunit of NMDARs (i.e., VHHAN1), filling an important gap for studying synapse biology. We demonstrated that this genetically encoded molecule can be readily fused with fluorescent proteins (mCherry and EGFP; Figures 1C, 1E, 2B, 2C, 2E, and 2G) and an enzyme (CaMKIIα; Figure 3). The small size of this intrabody (130 amino acids) is beneficial for delivery via AAVs, which have a length limit for the viral genome being packaged (Wu et al., 2010). We validated the AAV-mediated targeting and visualization of endogenous GluN1 in the mouse hippocampus (Figure 2). We also showed that VHHAN1 drives the AAV-expressed CaMKIIα to orient toward excitatory postsynaptic regions in the mouse hippocampus, whereas CaMKIIα alone diffuses throughout the infected neurons, including axonal projections (Figure 3). Prior work developed two intrabodies against excitatory and inhibitory scaffolding proteins (PSD95.FingR and GPHN.FingR, respectively) (Bensussen et al., 2020; Gross et al., 2013). Considering the differences in protein expression level, subsynaptic localization, and functional roles of GluN1 and PSD-95 (Cheng et al., 2006; Sheng and Kim, 2011; Wegner et al., 2018), it is plausible that each intrabody may be useful for investigating the corresponding subsynaptic compartments in living neurons and animals.
Previous work showed a significant increase in the spatial memory of rats caused by overexpression of wild-type CaMKIIα (Poulsen et al., 2007). However, we observed no significant changes in contextual or cued fear memories of mice with overexpression of HA-CaMKIIα alone (Figures 4B and 4C). Capitalizing on VHHAN1, we supplied an exogenous population of wild-type CaMKIIα biased toward endogenous NMDARs in the hippocampus and demonstrated a more than 2-fold improvement in contextual memory (Figure 4B). The improvement in contextual memory by CaMKIIα local enrichment depended on the CaMKIIα kinase activity (Figure 4B). This finding suggests that there may be a potential demand for postsynaptic enrichment of CaMKIIα to facilitate encoding memory.
Collectively, these molecular tools (VHHAN1 and CLEVIR) are likely to provide a wide range of experimental potential for investigating synapse and memory biology in the field.
Limitations of the study
Although our study focused on the impact of intrabody-mediated local postsynaptic enrichment of CaMKIIα on memory formation, how it modifies synaptic plasticity at the cellular level was not analyzed in the present study. Electrophysiological studies with this molecular approach are needed in future studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-RFP (clone 6G6) | Chromotek | Cat# 6G6; RRID:AB_2631395 |
| Rabbit polyclonal anti-GFP | SYSY | Cat# 132 003; RRID:AB_1834147 |
| Rabbit polyclonal anti-GFP | Abcam | Cat# ab290; RRID:AB_303395 |
| Chicken polyclonal anti-GFP | Aves Labs | Cat# GFP-1020; RRID:AB_10000240 |
| Mouse monoclonal anti-NMDAR1 (clone 54.1) | Millipore | Cat# MAB363; RRID:AB_94946 |
| Rabbit polyclonal anti-NR2B | Millipore | Cat# 06–600; RRID:AB_310193 |
| Rat monoclonal anti-HA (clone 3F10) | Roche | Cat# ROAHAHA; RRID:AB_2687407 |
| Guinea pig polyclonal anti-PSD-95 | Frontier Institute | Cat# PSD95-GP-Af660; RRID:AB_2571539 |
| Normal rabbit IgG | Millipore | Cat# 12–370; RRID:AB_145841 |
| Goat anti-rat IgG, Alexa Fluor 488 | ThermoFisher Scientific | Cat# A-21208; RRID:AB_2535794 |
| Goat anti-rabbit IgG, Alexa Fluor 594 | ThermoFisher Scientific | Cat# A-11037; RRID:AB_2534095 |
| Goat anti-mouse IgG, Alexa Fluor 647 | ThermoFisher Scientific | Cat# A-32728; RRID:AB_2633277 |
| Goat anti-guinea pig IgG, Alexa Fluor 647 | ThermoFisher Scientific | Cat# A-21450; RRID:AB_2735091 |
| Peroxidase AffiniPure goat anti-mouse IgG | Jackson ImmunoResearch | Cat# 115-035-146; RRID:AB_2307392 |
| Peroxidase AffiniPure goat anti-rabbit IgG | Jackson ImmunoResearch | Cat# 111-035-144; RRID:AB_2307391 |
| Bacterial and virus strains | ||
| AAV-EF1α-DIO-VHHAN1-EGFP | This paper | N/A |
| AAV-hSyn-DIO-EGFP | Addgene | Cat# 50457; RRID:Addgene_50457 |
| AAV-Syn-DIO-HA-CaMKIIα (WT) | This paper | N/A |
| AAV-Syn-DIO-VHHAN1-HA-CaMKIIα (WT) | This paper | N/A |
| AAV-Syn-DIO-VHHAN1-HA-CaMKIIα (K42R) | This paper | N/A |
| AAV-Syn-mCherry-IRES-Cre | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Ni-NTA agarose | Qiagen | Cat# 30210 |
| Gateway™ LR Clonase™ II Enzyme mix | Invitrogen | Cat# 11791020 |
| GFP-Trap magnetic agarose | Chromotek | Cat# gtma-20 |
| Protein A Sepharose CL-4B medium | Cytiva | Cat# 17078001 |
| FuGENE HD transfection reagent | Promega | Cat# E2311 |
| Benzonase nuclease | Sigma-Aldrich | Cat# E1014 |
| SuperSignal™ West Pico PLUS Chemiluminescent Substrate | ThermoFisher Scientific | Cat# 34580 |
| Paraformaldehyde | Sigma-Aldrich | Cat# P6148 |
| Pepsin | Agilent | Cat# S300230-2 |
| Normal goat serum blocking solution | Vector Laboratories | Cat# S-1000 |
| DAPI Fluoromount-G | SouthernBiotech | Cat# 0100-20 |
| HiTrap heparin high performance columns | GE Healthcare | Cat# 17-0406-01 |
| Experimental models: Cell lines | ||
| HeLa cells | ATCC | Cat# CCL-2; RRID:CVCL_0030 |
| 293FT cell line | Invitrogen | Cat# R70007; RRID:CVCL_6911 |
| BL21-CodonPlus (DE3)-RIPL competent cells | Agilent | Cat# 230280 |
| Experimental models: Organisms/strains | ||
| C57BL/6J mice | The Jackson Laboratory | Stock# 000664; RRID:IMSR_JAX:000664 |
| Oligonucleotides | ||
| Primers for DNA constructs, see Table S2 | This paper | N/A |
| Recombinant DNA | ||
| pET-28a-AviTag-GluN1(834–863) | This paper | N/A |
| pCAG-VHHAN1(Clone#31)-mCherry-CCR5TC | This paper | N/A |
| pCAG-Clone#2-mCherry-CCR5TC | This paper | N/A |
| pCAG-Clone#69-mCherry-CCR5TC | This paper | N/A |
| pAAV-EF1α-DIO-VHHAN1-EGFP | This paper | N/A |
| pAAV-Syn-DIO-HA-CaMKIIα | This paper | N/A |
| pAAV-Syn-DIO-VHHAN1-HA-CaMKIIα | This paper | N/A |
| pAAV-Syn-mCherry-IRES-Cre | This paper | N/A |
| pCI-SEP-NRI | Kopec et al. (2006) | RRID: Addgene_23999 |
| Software and algorithms | ||
| Prism 9 | GraphPad | N/A |
| Fiji | Schindelin et al. (2012) | http://fiji.sc |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Joongkyu Park (joongkyu.park@wayne.edu).
Materials availability
The plasmids reported in this paper are available from the lead contact upon request.
Experimental model and subject details
Animals
C57BL/6J wild-type mice were obtained from the Jackson Laboratory (Stock #000664). Mice were maintained at the Division of Laboratory Animal Resources facility of Wayne State University and treated under the guidelines of the Institutional Animal Care and Use Committee of Wayne State University. We used both male and female mice for histology and behavioral tests and observed no obvious difference between the groups. Ages of study are 2–3 months of age.
Cell culture
HeLa cells (ATCC #CCL-2) and 293FT cells (Invitrogen #R70007) were obtained directly from ATCC and ThermoFisher Scientific, respectively. The cells were tested by mycoplasma tests on a routine basis.
Method details
Plasmids
To express 6xHis- and AviTag-tagged mouse GluN1 C0 cassette fragment (834–863 amino acids; NP_032195.1) in bacteria, a DNA fragment encoding AviTag-GluN1(834–863) was synthesized by Integrated DNA Technologies (Table S1) and inserted into pET-28a vector (Novagen) using NdeI and XhoI sites.
For mammalian expression of anti-GluN1 intrabody candidates, a CCR5TC transcriptional repression system (a zinc-finger DNA-binding domain and a KRAB(A) transcriptional repressor) was adopted (Gross et al., 2013). Intermediate constructs were generated by inserting each intrabody candidate (Clone#2, #31, and #69) and an mCherry fragment into a linearized pcDNA3 vector using HindIII and XbaI sites. A flexible linker of Gly-Ser-Gly-Ser-Gly was inserted between the intrabody candidates and mCherry. Then, the fragment of intrabody candidate-mCherry was connected with the CCR5TC fragment by overlap extension polymerase chain reaction (PCR) with primers listed in Table S2. The amplified fragments were inserted using KpnI and MluI sites into the pCAG vector backbone containing a zinc-finger binding site upstream of a CAG promoter prepared from pCAG-GPHN.FingR-EGFP-CCR5TC (Addgene #46296) (Gross et al., 2013).
To generate an AAV construct expressing VHHAN1-fused EGFP, we first generated a pENTR construct that contains VHHAN1 and EGFP-CCR5TC fragments. The VHHAN1 fragment was amplified by PCR with the primers listed in Table S2 and digested with HindIII and BamHI. The EGFP-CCR5TC fragment was obtained by a BamHI and XhoI digestion from pCAG-GPHN.FingR-EGFP-CCR5TC (Addgene #46296) (Gross et al., 2013). The DNA fragments of VHHAN1 and EGFP-CCR5TC were inserted into a pENTR vector using HindIII and XhoI sites. This entry clone was recombined by Gateway LR Clonase II enzyme (Invitrogen) with a pAAV-EF1α-DIO-Dest vector, which was generated by replacing a cassette of a ccdB gene and attR1 and attR2 sites with the coding region of AAV-EF1α-DIO-PSD95.FingR-EGFP-CCR5TC (Addgene #126216) (Bensussen et al., 2020).
For cistronic expression of mCherry and Cre under a Synapsin promoter, we first inserted a DNA fragment encoding mCherry-IRES-Cre (synthesized by Integrated DNA Technologies; Table S1) into the pENTR vector using BamHI and EcoRI sites. We then performed Gateway LR recombination of pENTR-mCherry-IRES-Cre and pAAV-Syn-Dest constructs.
For the expression of VHHAN1-HA-CaMKIIα under a Synapsin promoter, an intermediate construct was first generated by Gateway LR recombination of pENTR-VHHAN1 and pAAV-Syn-DIO-Dest constructs, and then a DNA fragment encoding HA-CaMKIIα (synthesized by Integrated DNA Technologies; Table S1) was inserted next to VHHAN1 using BamHI and AscI sites. To express HA-CaMKIIα alone as a control, the DNA fragment encoding VHHAN1 was removed by BamHI and NheI digestion and replaced with an annealed primer linker encoding the Kozak consensus sequence (Table S1). To express a kinase-inactive (K42R) form of VHHAN1-HA-CaMKIIα, the coding region of HA-CaMKIIα (wild-type) was replaced with a DNA fragment encoding HA-CaMKIIα (K42R) (synthesized by Integrated DNA Technologies; Table S1).
pCAG-GPHN.FingR-EGFP-CCR5TC, AAV-EF1α-DIO-PSD95.FingR-EGFP-CCR5TC, pCI-SEP-NRI and pAAV-hSyn-DIO-EGFP were gifts from Don Arnold (Addgene plasmid #46296; http://n2t.net/addgene:46296; RRID: Addgene_46296), Xue Han (Addgene plasmid #126216; http://n2t.net/addgene:126216; RRID: Addgene_126216), Robert Malinow (Addgene plasmid #23999; http://n2t.net/addgene:23999; RRID: Addgene_23999), and Bryan Roth (Addgene plasmid #50457; http://n2t.net/addgene:50457; RRID: Addgene_50457), respectively.
Preparation of anti-GluN1 intrabody target fragment
To design an anti-GluN1 intrabody target fragment, we compared eight splice variants of the mouse GluN1 cytoplasmic tail, which are deposited in the NCBI Reference Sequences database (NP_032195.1, NP_001171127.1, NP_001171128.1, NP_001359487.1, NP_001359488.1, NP_001359489.1, NP_001359490.1, and NP_001359491.1) (Figure S1A). The conserved region (834–863 amino acids; NP_032195.1) was fused with a six-histidine tag (6xHis) and an AviTag (pET-28a-AviTag-GluN1C0) and expressed in BL21-CodonPlus (DE3)-RIPL cells (Agilent). Expression of the His-AviTag-GluN1(834–863) fragment was induced by 1 mM isopropyl b-D-thiogalactopyranoside at 37°C for 3.5 h, and the fragment was purified with Ni-NTA agarose (Qiagen). All eluted samples were then desalted using an Econo-Pac 10DG desalting column (Bio-Rad) and concentrated using an Amicon ultra-centrifugal filter unit (Millipore). The purified fragment was confirmed by resolving on a 17% SDS-PAGE gel and performing Coomassie Brilliant Blue G-250 staining.
Phage display selection and yeast two-hybrid (Y2H) screening
BirA-mediated in vitro biotinylation of the GluN1(834–863) fragment was performed by Hybrigenics Services SAS (www.hybrigenics-services.com). The first-phase screening was conducted using an hs2dAb synthetic library of humanized nanobodies (3.0 × 109 nanobodies) (Moutel et al., 2016). For the Y2H screen, the coding sequences for the C0 cassette (834–863 amino acids; NP_032195.1) and the full-length cytoplasmic tail of mouse GluN1 (834–938 amino acids; NP_032195.1) were subcloned into pB27 as a C-terminal fusion to LexA. Two rounds of Y2H screenings were performed by Hybrigenics Services SAS.
Cell culture and transfection
HeLa and 293FT cells were maintained in DMEM supplemented with 10% (v/v) fetal bovine serum, 100 U/mL penicillin-streptomycin, and 2 mM L-glutamine in a humidified incubator with 37°C and 5% CO2. For confocal imaging and binding assays, transient transfection was performed using the FuGENE HD transfection reagent according to the manufacturer’s instructions (Promega).
Cell imaging
Cells were plated on 35-mm glass-bottom dishes (MatTek Corporation) and imaged under a spinning disk confocal microscope (Zeiss Cell) equipped with a 63x (NA 1.4) objective approximately 18 h after transfection. Before imaging, cells were transferred to a pre-warmed buffer containing 10 mM HEPES (pH 7.4), 137 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, and 1.3 mM MgCl2.
Binding assay and Western blot
Each plasmid encoding an mCherry-CCR5TC-fused anti-GluN1 intrabody candidate (#2, #31, or #69; 2 μg) was co-transfected into HeLa cells (at ∼70% confluency in 35-mm dishes) with or without pCI-SEP-NRI (encoding SEP-GluN1; 1 μg) using FuGENE HD transfection reagent. Cells were lysed in 1 mL of RIPA buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, and 0.2% SDS) containing ∼750 U Benzonase nuclease. After centrifugation (15,000 g for 15 min at 4°C), each lysate was mixed with 15 μL of GFP-Trap magnetic beads overnight at 4°C. The protein complex pulled down by GFP-Trap beads (Chromotek) was washed three times with RIPA buffer and eluted in 30 μL of 1x Laemmli sample buffers by heating at 75°C for 10 min.
To test protein binding in vivo, two hippocampi from the AAV-injected mice were dissected in cold phosphate-buffered saline (PBS) and homogenized 25 times by Dounce A in 40 mM Tris buffer (pH 7.4). The homogenate was lysed by mixing with 1.25x RIPA buffer to make a final 1x RIPA buffer containing ∼750 U Benzonase nuclease. To prevent undesired proteolysis and dephosphorylation, 1x Halt protease and phosphatase inhibitor cocktail (Thermo Scientific) and 5 mM ethylenediaminetetraacetic acid were added into the lysate. After centrifugation (15,000 g for 15 min at 4°C), 0.8 mL of the supernatant was diluted with 3.2 mL of 50 mM Tris (pH 7.4), 150 mM NaCl, and 1% Triton X-100 and mixed with 2 μg of either normal rabbit IgG (Millipore, Cat #12–370) or rabbit polyclonal anti-GFP antibody (Synaptic Systems, Cat #132003) overnight at 4°C. The next day, 30 μL of Protein A Sepharose CL-4B beads (Cytiva) was added for 2 h at 4°C. The protein-immunocomplex pulled down by the beads was washed six times with 1 mL of 50 mM Tris (pH 7.4), 150 mM NaCl, and 1% Triton X-100 and eluted in 30 μL of 1x Laemmli sample buffers by heating at 75°C for 10 min.
The eluted protein complexes were resolved on 7% SDS-PAGE gels and analyzed by Western blot. Briefly, SDS-PAGE gels were transferred to polyvinylidene difluoride membranes, and the membranes were blocked with 3% non-fat dry milk in TBST buffer (20 mM Tris, 150 mM NaCl, and 0.1% Tween 20) for 1 h at room temperature. Membranes were then probed overnight at 4°C with TBST buffer containing 1% non-fat dry milk and primary antibodies, including mouse monoclonal anti-RFP (Chromotek, clone 6G6, Cat #6G6, 1:3,000), rabbit polyclonal anti-GFP (Abcam, Cat #ab290, 1:3,000), mouse monoclonal anti-NMDAR1 (Millipore, clone 54.1, Cat #MAB363, 1:3,000), and rabbit polyclonal anti-NR2B (Millipore, Cat #06–600, 1:3,000). The next day, membranes were washed three times in TBST buffer and incubated for 2 h at room temperature with TBST buffer containing 1% non-fat dry milk and horseradish peroxidase (HRP)-conjugated anti-mouse IgG or anti-rabbit IgG antibodies. The membranes were then washed three times with TBST buffer, and signals were visualized with an enhanced chemiluminescence reagent (Thermo Scientific, SuperSignal™ West Pico PLUS Chemiluminescent Substrate).
AAV production
AAV was generated using the AAV-DJ Helper Free system (Cell Biolabs) as published previously (McClure et al., 2011; Park et al., 2016). Each AAV construct was co-transfected with pAAV-DJ and pHelper into 293FT cells (Invitrogen). Transfected cells were lysed three days after transfection, and AAVs were purified with 1-mL HiTrap heparin high-performance columns (GE Healthcare) as described (McClure et al., 2011). The titer of each AAV was determined by semi-quantitative PCR.
AAV injection
Stereotaxic injections of mouse hippocampi were performed as described previously (Park et al., 2016). The 6- to 9-week-old C57BL/6 wild-type mice were anesthetized with 1.5–3.0% isoflurane and placed in a stereotaxic apparatus (Kopf Instruments). The skull was exposed over the hippocampi based on stereotactic coordinates. Then, 0.7–1.0 μL of AAV was injected into the dorsal hippocampi using a glass pipette (tip diameter 7–10 μm) at a rate of 100 nL/min using a syringe pump (Micro2T; World Precision Instruments). We injected each AAV with the following titer per hippocampus: 1.0 × 106 viral genome (vg) of AAV-EF1α-DIO-VHHAN1-EGFP (or AAV-hSyn-DIO-EGFP) and 4.0 × 107 vg of AAV-Syn-mCherry-IRES-Cre for Figure 2; 3.8 × 107 vg of AAV-Syn-DIO-HA-CaMKIIα, 2.1 × 107 vg of AAV-Syn-DIO-VHHAN1-HA-CaMKIIα (wild-type or K42R), and 2.7 × 107 vg of AAV-Syn-mCherry-IRES-Cre for Figures 3 and 4. The injection site was standardized among animals by using stereotaxic coordinates (ML = ±2.00, AP = −2.20, DV = −1.90, −1.65, and −1.40 for hippocampi) from bregma. At the end of the injections, we waited 10 min before retracting the pipette. One to two weeks after injections, mice were subjected to tissue harvest or behavioral tests.
Immunohistochemistry
Mice were deeply anesthetized and transcardially perfused with 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4). After postfixation for either 3 h (for GluN1) or overnight (for other proteins), 50 μm-thick coronal sections were cut on a vibratome (Leica) at 4°C. To stain postsynaptic proteins, free-floating sections were treated with 1 mg/mL pepsin (Agilent) in 0.2 N HCl at 37°C for 2 min for antigen retrieval and washed in PBS three times. The slices were blocked for 45 min in PBS containing 5% normal goat serum (Vector Laboratories) and 0.3% Triton X-100 and incubated overnight at room temperature with PBS containing 3% normal goat serum, 0.1% Triton X-100, and primary antibodies, including rabbit polyclonal anti-GFP (Synaptic Systems, Cat #132003, 1:1,000), chicken polyclonal anti-GFP (Aves Labs, Cat #GFP-1020, 1:2,000), mouse monoclonal anti-NMDAR1 (Millipore, clone 54.1, Cat #MAB363, 1:1,000), rat monoclonal anti-HA (Roche, clone 3F10, Cat #ROAHAHA, 1:1,000), and guinea pig polyclonal anti-PSD-95 (Frontier Institute, Cat #PSD95-GP-Af660, 1:1,000). Sections were then washed three times with PBS and incubated for 2 h at room temperature with secondary antibodies, including Alexa Fluor 594-conjugated goat anti-rabbit IgG (H+L) highly cross-adsorbed secondary antibody (ThermoFisher, Cat #A11037, 1:1,000), Alexa Fluor 647-conjugated goat anti-mouse IgG (H+L) highly cross-adsorbed secondary antibody (ThermoFisher, Cat #A32728, 1:1,000), Alexa Fluor 488-conjugated donkey anti-rat IgG (H+L) highly cross-adsorbed secondary antibody (ThermoFisher, Cat #A21208, 1:1,000), and Alexa Fluor 647-conjugated goat anti-guinea pig IgG (H+L) highly cross-adsorbed secondary antibody (ThermoFisher, Cat #A21450, 1:1,000). Then, sections were washed three times in PBS and mounted on microscope slides with DAPI Fluoromount-G (SouthernBiotech). Confocal fluorescence images were acquired on a laser scanning confocal microscope (LSM 780; Zeiss) equipped with a 10x (NA 0.3) or 63x (NA 1.4) objective. Images were edited and analyzed using Fiji (http://fiji.sc). For the quantitative analysis of two-channel colocalization, each channel image was filtered with Gaussian blur and adjusted with a threshold. Mask selections were created from one channel (GFP for Figure 2F and HA for Figures 3G and 3I), and the signals from another channel in the selected masks (GluN1 for Figures 2F and 3I and PSD-95 for 3G) were quantified.
Behaviors
All mice were housed on a 12-h light-dark cycle with food and water ad libitum. All behavioral experiments were conducted with 8- to 11-week-old littermates at the time of testing and randomization of individual animals. All behavioral tests and analyses were performed in a blind manner to the AAV genotypes. Mice were tested in four sessions (habituation, training, contextual memory test, and cued memory test) as described previously (Park et al., 2016). For habituation, mice were exposed to a dark plastic chamber with a plastic A-shaped frame insert and a plastic floor cover (Med associates) (as context A). No shocks or cues were delivered. The average motion values in this session were used to monitor potential changes in the general movement of each animal from the surgery for AAV injections and protein overexpression (Figure S2). The lack of a significant change in the general movement suggests that the AAV-injected animals recovered well from the surgery to reach the same activity levels as the non-injected mouse group (Figure S2). In the training sessions, mice were placed in a modified plastic chamber with a house light, removed A-frame insert, and grids for shock delivery (as context B). After 160 s as the baseline, mice underwent three repeats (with 40-s intertrial intervals) of a 20-s tone (85 dB, 2.8 kHz) and a 2-s foot shock (0.5 mA) coincided at the end of each tone. After training, mice were returned to their home cage. The next day, mice were placed again in the same training chamber (context B) to test contextual memory for 6 min. To test cued memory, mice were placed in the context A chamber, and the three rounds of tones (20 s, 85 dB, 2.8 kHz) and intertrial intervals (40 s) were administered as described in the training sessions without foot shocks. Every session was filmed by an infrared video camera (Med associates), and freezing behavior (defined as complete lack of movement between every 1-s frame of videos) was analyzed by Video Freeze (Med associates) software in a blinded manner. No obvious differences in body weight and responses to shocks were observed. Brains from all tested mice were subjected to brain slice sectioning and fluorescent microscopy to confirm the symmetric AAV injections in both hippocampi.
Quantification and statistical analysis
All data are given as mean ± SEM. The sample size was chosen based on the previous studies (Park et al., 2016). Statistical significance between means was calculated using Mann-Whitney tests or Kruskal-Wallis tests with multiple comparisons (Prism 9, GraphPad). Statistical significance is indicated as follows: ∗p < 0.05, ∗∗∗p < 0.0001, and n.s., not significant.
Acknowledgments
We thank Drs. Rodrigo Andrade, Michael J. Bannon, and Sokol V. Todi for providing valuable input. We also thank members of the Park lab for helpful discussions. This work was supported by NIH R21AG068423 (J.P.).
Author contributions
A.C. and J.P. initiated and conceived the study. A.C., J.C., and J.P. designed and performed the experiments. A.C., J.C., and J.P. analyzed the data. J.P. wrote the manuscript. All authors discussed the results and contributed to the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: August 19, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104864.
Supplemental information
Data and code availability
-
•
Data: Data reported in this paper will be shared by the lead contact upon request.
-
•
Code: This paper does not report an original code.
-
•
All other items: Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Araki Y., Zeng M., Zhang M., Huganir R.L. Rapid dispersion of SynGAP from synaptic spines triggers AMPA receptor insertion and spine enlargement during LTP. Neuron. 2015;85:173–189. doi: 10.1016/j.neuron.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensussen S., Shankar S., Ching K.H., Zemel D., Ta T.L., Mount R.A., Shroff S.N., Gritton H.J., Fabris P., Vanbenschoten H., et al. A viral toolbox of genetically encoded fluorescent synaptic tags. iScience. 2020;23:101330. doi: 10.1016/j.isci.2020.101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D., Hoogenraad C.C., Rush J., Ramm E., Schlager M.A., Duong D.M., Xu P., Wijayawardana S.R., Hanfelt J., Nakagawa T., et al. Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol. Cell. Proteomics. 2006;5:1158–1170. doi: 10.1074/mcp.D500009-MCP200. [DOI] [PubMed] [Google Scholar]
- Förster E., Zhao S., Frotscher M. Laminating the hippocampus. Nat. Rev. Neurosci. 2006;7:259–267. doi: 10.1038/nrn1882. [DOI] [PubMed] [Google Scholar]
- Giese K.P., Fedorov N.B., Filipkowski R.K., Silva A.J. Autophosphorylation at Thr286 of the alpha calcium-calmodulin kinase II in LTP and learning. Science. 1998;279:870–873. doi: 10.1126/science.279.5352.870. [DOI] [PubMed] [Google Scholar]
- Gross G.G., Junge J.A., Mora R.J., Kwon H.B., Olson C.A., Takahashi T.T., Liman E.R., Ellis-Davies G.C.R., McGee A.W., Sabatini B.L., et al. Recombinant probes for visualizing endogenous synaptic proteins in living neurons. Neuron. 2013;78:971–985. doi: 10.1016/j.neuron.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huganir R.L., Nicoll R.A. AMPARs and synaptic plasticity: the last 25 years. Neuron. 2013;80:704–717. doi: 10.1016/j.neuron.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine E.E., Vernon J., Giese K.P. AlphaCaMKII autophosphorylation contributes to rapid learning but is not necessary for memory. Nat. Neurosci. 2005;8:411–412. doi: 10.1038/nn1431. [DOI] [PubMed] [Google Scholar]
- Kopec C.D., Li B., Wei W., Boehm J., Malinow R. Glutamate receptor exocytosis and spine enlargement during chemically induced long-term potentiation. J. Neurosci. 2006;26:2000–2009. doi: 10.1523/JNEUROSCI.3918-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J., Schulman H., Cline H. The molecular basis of CaMKII function in synaptic and behavioural memory. Nat. Rev. Neurosci. 2002;3:175–190. doi: 10.1038/nrn753. [DOI] [PubMed] [Google Scholar]
- Lisman J., Yasuda R., Raghavachari S. Mechanisms of CaMKII action in long-term potentiation. Nat. Rev. Neurosci. 2012;13:169–182. doi: 10.1038/nrn3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure C., Cole K.L.H., Wulff P., Klugmann M., Murray A.J. Production and titering of recombinant adeno-associated viral vectors. J. Vis. Exp. 2011:e3348. doi: 10.3791/3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutel S., Bery N., Bernard V., Keller L., Lemesre E., de Marco A., Ligat L., Rain J.C., Favre G., Olichon A., Perez F. NaLi-H1: a universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. Elife. 2016;5:e16228. doi: 10.7554/eLife.16228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi H., Shin M.E., Parra-Bueno P., Szatmari E.M., Shibata A.C.E., Yasuda R. Kinetics of endogenous CaMKII required for synaptic plasticity revealed by optogenetic kinase inhibitor. Neuron. 2017;94:37–47.e5. doi: 10.1016/j.neuron.2017.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoll R.A. A brief history of long-term potentiation. Neuron. 2017;93:281–290. doi: 10.1016/j.neuron.2016.12.015. [DOI] [PubMed] [Google Scholar]
- Paoletti P., Bellone C., Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat. Rev. Neurosci. 2013;14:383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- Park J., Chávez A.E., Mineur Y.S., Morimoto-Tomita M., Lutzu S., Kim K.S., Picciotto M.R., Castillo P.E., Tomita S. CaMKII phosphorylation of TARPγ-8 is a mediator of LTP and learning and memory. Neuron. 2016;92:75–83. doi: 10.1016/j.neuron.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen D.J., Standing D., Bullshields K., Spencer K., Micevych P.E., Babcock A.M. Overexpression of hippocampal Ca2+/calmodulin-dependent protein kinase II improves spatial memory. J. Neurosci. Res. 2007;85:735–739. doi: 10.1002/jnr.21163. [DOI] [PubMed] [Google Scholar]
- Rossetti T., Banerjee S., Kim C., Leubner M., Lamar C., Gupta P., Lee B., Neve R., Lisman J. Memory erasure experiments indicate a critical role of CaMKII in memory storage. Neuron. 2017;96:207–216.e2. doi: 10.1016/j.neuron.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D.B., Blanpied T.A., Swanson G.T., Zhang C., Ehlers M.D. An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J. Neurosci. 2001;21:3063–3072. doi: 10.1523/JNEUROSCI.21-09-03063.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M., Kim E. The postsynaptic organization of synapses. Cold Spring Harb. Perspect. Biol. 2011;3:a005678. doi: 10.1101/cshperspect.a005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A.J., Stevens C.F., Tonegawa S., Wang Y. Deficient hippocampal long-term potentiation in alpha-calcium-calmodulin kinase II mutant mice. Science. 1992;257:201–206. doi: 10.1126/science.1378648. [DOI] [PubMed] [Google Scholar]
- Silva A.J., Paylor R., Wehner J.M., Tonegawa S. Impaired spatial learning in alpha-calcium-calmodulin kinase II mutant mice. Science. 1992;257:206–211. doi: 10.1126/science.1321493. [DOI] [PubMed] [Google Scholar]
- Standley S., Roche K.W., McCallum J., Sans N., Wenthold R.J. PDZ domain suppression of an ER retention signal in NMDA receptor NR1 splice variants. Neuron. 2000;28:887–898. doi: 10.1016/s0896-6273(00)00161-6. [DOI] [PubMed] [Google Scholar]
- Wegner W., Mott A.C., Grant S.G.N., Steffens H., Willig K.I. In vivo STED microscopy visualizes PSD95 sub-structures and morphological changes over several hours in the mouse visual cortex. Sci. Rep. 2018;8:219. doi: 10.1038/s41598-017-18640-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Yang H., Colosi P. Effect of genome size on AAV vector packaging. Mol. Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S., Kim J.I., Kim J., Kaang B.K. Overexpression of activated CaMKII in the CA1 hippocampus impairs context discrimination, but not contextual conditioning. Mol. Brain. 2019;12:32. doi: 10.1186/s13041-019-0454-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data: Data reported in this paper will be shared by the lead contact upon request.
-
•
Code: This paper does not report an original code.
-
•
All other items: Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.