Abstract
Context
Islet autoantibodies (IAbs) are currently the most reliable indicators of islet autoimmunity. However, IAbs do not fully meet the need for the prediction and intervention of type 1 diabetes (T1D). Serological proteins should be great sources for biomarkers.
Objective
This work aimed to identify new proteomic biomarkers with the technology of an expression-based genome-wide association study (eGWAS) in children newly diagnosed with T1D.
Methods
In an attempt to identify additional biomarkers, we performed an eGWAS using microarray data from 169 arrays of the pancreatic islets of T1D rodents (78 T1D cases and 91 controls). We ranked all 16 099 protein-coding genes by the likelihood of differential expression in the pancreatic islets. Our top 20 secreted proteins were screened in 170 children including 100 newly diagnosed T1D, and 50 type 2 diabetes (T2D) and 20 age-matched healthy children. With 6 proteins showing significance, we further conducted a validation study using the second independent set of 400 samples from children including 200 newly diagnosed with T1D, 100 T2D, and 100 age-matched controls.
Results
We identified 2 serum proteins that were significantly changed in T1D vs both control and T2D, and 5 serum proteins were significantly changed both in T1D and T2D vs control. Serum osteopontin (OPN) levels were uniquely higher in T1D (T1D vs controls, P = 1.29E-13 ~ 9.38E-7, T1D vs T2D, P = 2.65E-8 ~ 1.58E-7) with no difference between T2D and healthy control individuals. Serum interleukin 1 receptor antagonist (IL-1RA) levels were lower in T1D compared both with T2D (P = 3.36E-9~0.0236) and healthy participants (P = 1.09E-79 ~ 2.00E-12).
Conclusion
Our results suggest that OPN and IL1-RA could be candidates for useful biomarkers for T1D in children.
Keywords: type 1 diabetes, eGWAS, proteomics, biomarker, osteopontin, IL1-RA
Type 1 diabetes (T1D) is a chronic autoimmune disease with a nonsymptomatic prodromal period characterized by total loss of pancreatic islet insulin-producing β cells in early childhood. Serum islet autoantibodies (IAbs) are currently the most reliable biomarkers of islet autoimmunity, including those known to react to insulin, glutamic acid decarboxylase (GAD65), insulinoma antigen 2 (IA-2), and zinc transporter 8 (ZnT8). However, they also have some limitations. The time between autoantibody emergence and hyperglycemia onset varies from months to more than 10 years (1). Furthermore, currently studied IAbs are not implicated in the disease pathogenesis and are not useful for clinically therapeutic designs. Biomarkers should play essential roles in identifying high-risk populations and, more important, for tailoring and monitoring therapies for the disease (2). Additional serological biomarkers that help to reveal an additional pathogenesis of T1D, enhance disease staging, and provide useful information for therapeutic intervention are urgently required. Multiple studies of serological proteomic biomarkers for T1D have been reported, but the studies were mostly limited to adults or patients with long-term T1D (3-5), which are not representative of T1D in the large cohort of childhood at disease onset. Data in children newly diagnosed with T1D are still absent.
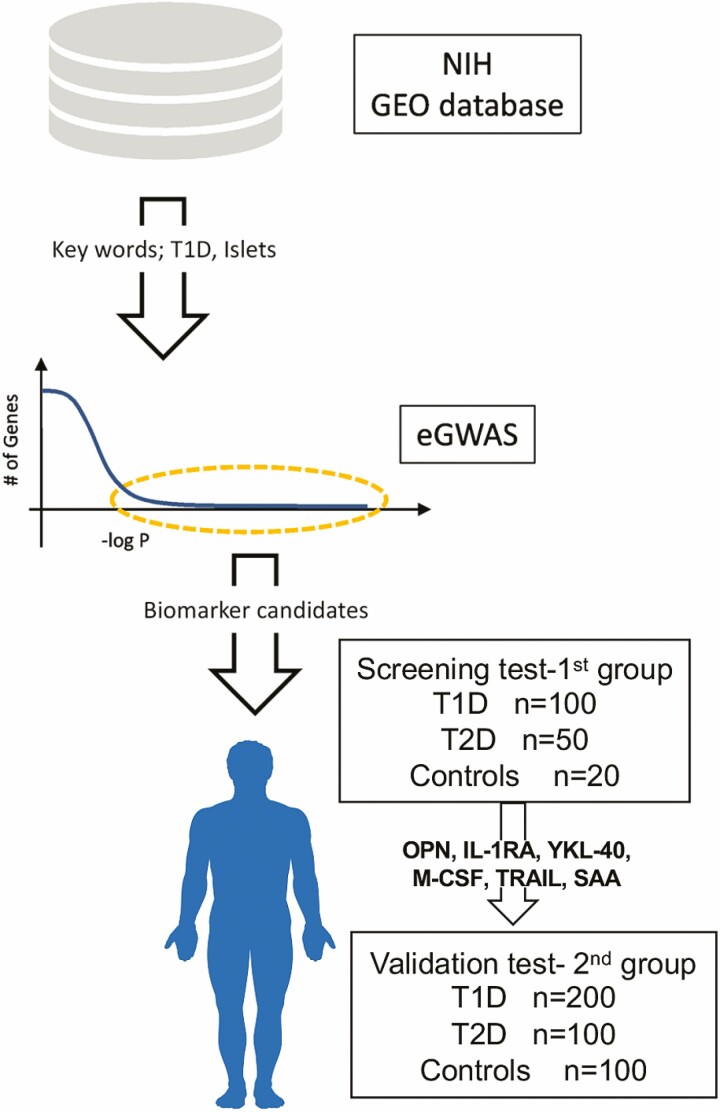
We have previously described a meta-analytic methodology, expression–based genome-wide association study (eGWAS), a computational approach that can be used for calculating the likelihood of differential expression for every gene across a large number of case-control gene-expression microarray experiments collected from a public repository (6, 7). Genes that are most repeatedly implicated across a large set of experimental representations of polygenic diseases serve as data-driven causal disease genes and are candidates for further analysis. With this technology, we have successfully identified multiple candidate molecules in T1D and type 2 diabetes (T2D) (6, 8, 9). In the present study, we attempt to identify potential biomarker candidates by applying our eGWAS methodology to leverage large numbers of raw T1D-related RNA measurements in T1D target tissues from rodent models. Our approach is aided by the increasing amounts of publicly available raw microarray experimental results. We meta-analyzed 21 independent genome-wide gene-expression experiments, totaling 169 cases and control samples, from the pancreatic islets of T1D rodent models. We identified top candidates and successfully validated some serological proteomic biomarkers in newly diagnosed children with T1D and T2D (Fig. 1).
Figure 1.
Study design. Expression-based genome-wide association study (eGWAS) for type 1 diabetes (T1D) was carried out in 169 case-control microarray samples collected from the National Institute of Health (NIH) database (GEO; Gene Expression Omnibus). P values (–log10 [P]) were calculated by comparing dysregulation distribution of genes in pancreatic islets between T1D and control microarrays. Our T1D biomarker candidates extracted from eGWAS were verified by 2 independent groups of human serum samples, the first group for screening protein biomarkers and the second group for validation.
Materials and Methods
Expression-based Genome-wide Association Study
An eGWAS was performed as described (6, 7). All T1D-related genome-wide microarray experiments used for this study were collected from the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/). Studies were found using the following keywords: (“type 1 diabetes” OR “type I diabetes” OR “T1D” OR “insulitis”) AND (“islets”). We identified 169 array samples (78 T1D cases and 91 controls from rodent models) in 21 independent data sets (10).
To estimate differences between samples from diabetic individuals and controls, raw postquantitation microarray data were reanalyzed using a one-tailed t test. We calculated P values for each gene in each of the 21 experiments. We then converted all probe identifiers across the various microarray platforms for mouse and rat to the latest human Entrez Gene identifiers using the AILUN system (11). Gene expression profiles were assigned in our eGWAS database according to the standardized human Entrez Gene ID. There were 16 099 genes in the database in total. We conducted a weighted Z method (12) in our eGWAS database, as previously described (6). The combined P values for each gene were calculated using a weighted Z method; the P values were converted to Z scores. Subsequently, the combined Z scores across all the experiments were calculated for each gene, using a weighted Z method, by weighting each experiment by its sample size (degrees of freedom).
Then, the combined P values for each gene were obtained by converting the weighted Z scores to 2-tailed P values.
Human Participants
Two independent groups of serum samples from newly diagnosed children with diabetes mellitus including T1D and T2D and age-matched healthy controls in each group were used for screening and validating for the top-selected protein biomarkers (see Fig.1 and Table 1). Inclusion criteria were age younger than 18 years at diagnosis, and all serum samples were obtained within 2 weeks of a diagnosis of diabetes. The criteria and classification of diabetes were based on clinical features in line with the American Diabetes Association (13) and International Society of Pediatric and Adolescent Diabetes criteria (14). IAbs (including IAA, GADA, IA-2A, and ZnT8A) were tested with standard radiobinding assays for all the participants at Barbara Davis Center for Diabetes University of Colorado. Participants with T1D had one or more positive IAbs, and participants with T2D or healthy controls were negative for all IAbs. The first group of serum samples used for screening protein biomarkers came from 100 children with newly diagnosed T1D, 50 children with newly diagnosed T2D, and 20 healthy participants. The second group of serum samples used for validating the top-selected protein biomarkers from the first screening included 200 children with newly diagnosed T1D, 100 children with newly diagnosed T2D, and 100 age- and sex-matched healthy participants. Signed written informed consents were obtained from participants and the studies were approved by the institutional review board of the University of Colorado.
Table 1.
Demographic information of study participants
| Group 1 | Group 2 | |||||
|---|---|---|---|---|---|---|
| T1D | T2D | Controls | T1D | T2D | Controls | |
| No. | 100 | 50 | 20 | 200 | 100 | 100 |
| Age, yr | ||||||
| Mean | 10.2 | 14.5 | 12.6 | 10.1 | 11.8 | 10.3 |
| Median | 10.4 | 14.7 | 12.1 | 10.6 | 12.7 | 10.5 |
| Range | 1.0-17.8 | 9.9-17.9 | 0.1-17.5 | 0.4 -7.7 | 2.1-17.9 | 1.9-17.8 |
| Female, % | 55 | 56 | 50 | 46.0 | 42.0 | 56.0 |
Abbreviations: T1D, type 1 diabetes; T2D, type 2 diabetes.
Biomarker Assay
Serum levels of biomarker candidates were measured using a Meso Scale Discovery (MSD) electrochemiluminescence (ECL) assay (Meso Scale Diagnostics; RRID: SCR_020304, https://www.mesoscale.com/en/products/meso-quickplex-sq-120-ai0aa/), as previously described (15-18). This technology is an enzyme-linked immunosorbent assay–based system, measuring individual or multiple analytes within a single well. It includes biotinylated capture antibodies and conjugated Ru Sulfo-NHS detection antibodies to bind with different analytes, and each serum was analyzed in duplicate using customized MSD singleplex and multiplex plates, as per the manufacturer’s recommendations (10). The ECL signals of the MSD 96-well plate were captured by the MSD SECTOR 6000 system (Meso Scale Diagnostics). All samples were coded, and the assay was performed in a blinded fashion. The levels of the biomarker proteins presented in this study were the raw reading data obtained from the MSD system, with unit cps (counts per second).
Statistical Analysis
Statistics were performed using IBM SPSS Statistics software (version 28.0.1.0). The levels of the biomarker proteins were log10-transformed before all statistical analyses to achieve normal distribution. Multiple comparisons between normally distributed levels of biomarkers in different groups were performed using the one-way analysis of variance. Dunnett post hoc comparisons were performed when equivalent variances were assumed, whereas, when the nonequivalent variants were assumed, Games–Howell post hoc comparisons were applied. If the data were not normally distributed, the Kruskal-Wallis H test was used for comparison among groups. This was followed by the Mann-Whitney U 2-independent sample test. The effect of age on serum levels of each biomarker was determined using a linear regression with data stratified by sex and disease status. Logistic regression models with sex and age as covariates were used to assess associations between these biomarkers and T1D. Statistical values were considered significant at P less than .05 after adjusting for multiple testing. All experimental data are represented as mean ± SD unless otherwise noted.
Results
Expression-based Genome-wide Association Study Identifies Type 1 Diabetes Biomarker Candidates
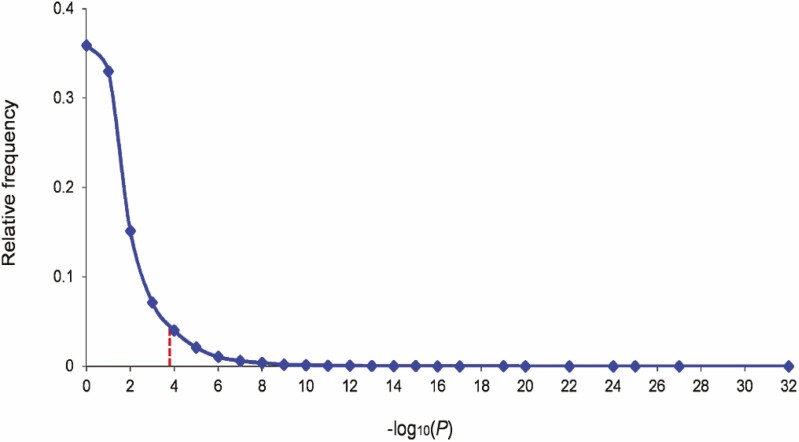
We carried out an eGWAS for T1D using 21 independent microarray experiments, totaling 169 cases and control samples from the pancreatic islets of mouse and rat models (see “Materials and Methods”). Sample data were collected from a public repository (10). For all 16 099 protein coding-genes, we calculated the likelihood that repeated differential expression for every gene was due to chance. P values were calculated using a t test for each gene in each of the 21 experiments. For each gene, we combined the P values from all the experiments. After correction for multiple testing, we considered genes with a Q value (false discovery rate) smaller than 0.001 to be significantly dysregulated in T1D pancreatic islets (Fig. 2). We then filtered the gene list for our biomarker candidates (secreted proteins measurable in the serum samples of T1D patients) using the MSD ECL assay list (https://www.mesoscale.com), yielding 20 genes/proteins (10). These proteins were significantly dysregulated in the pancreatic islets across multiple T1D microarray experiments, and might be changed in the serum of T1D patients.
Figure 2.
Expression-based genome-wide association study (eGWAS) analysis in type 1 diabetes (T1D) pancreatic islets. The relative frequency distribution of –log10 (combined P value). The combined P values for each gene were calculated from 169 T1D case-control microarray samples (78 T1D cases and 91 controls) using a weighted Z method. There were 16 099 genes in total. The –log10 (combined P values) were rounded to the nearest integer, and then the relative frequency distribution was determined. The red line indicates the Q value threshold (Q value < 0.001; P value < 1.6 × 10–4).
Screen for Top 20 Candidate Biomarkers in Children With New-onset Diabetes
Top 20 candidate biomarkers selected from eGWAS were studied for their serum levels using an ECL assay in 170 children including 100 newly diagnosed T1D, 50 newly diagnosed T2D, and 20 age-matched healthy controls. Of 20 proteins measured, 6 protein biomarkers including osteopontin (OPN), interleukin 1 receptor antagonist (IL-1RA), chitinase 3-like 1 (YKL-40), macrophage colony-stimulating factor 1 (M-CSF), tumor necrosis factor superfamily member 10 (TRAIL), and serum amyloid A1 (SAA) were found to have significant differences between children with T1D and healthy controls (P = 4.461E-45 ~ 3.104E-4), and 5 of them were also significantly different between T2D and controls including IL1-RA, YKL-40, M-CSF, TRAIL, and SAA (P = 6.132E-31 ~ .003). The serum levels of 3 protein biomarkers (OPN, IL-1RA, and SAA) in children with T1D not only showed a significant difference compared with healthy controls, but also a significant difference compared with children with T2D (P = 1.577E-7 for OPN, P = 3.362E-9 for IL-1RA, and P = .04 for SAA), whereas the serum levels of OPN showed no difference between children with T2D and healthy controls. The levels of these 6 biomarkers in patients and controls are summarized in Table 2. The rest of the 14 protein markers were not found to have significant differences between patients with diabetes and healthy controls (10).
Table 2.
Serum levels of 6 protein biomarkers from the first group of participants
| Levels of proteins, cps | Controls | T1D | T2D | P (T1D vs controls) | P (T2D vs controls) | P (T1D vs T2D) |
|---|---|---|---|---|---|---|
| OPN | 106 ± 32 (× 102) | 235 ± 135 (× 102) | 134 ± 81 (× 102) | 9.382E-7 | .66 | 1.577E-7 |
| IL-1RA | 3071 ± 2668 | 271 ± 269 | 612 ± 570 | 1.997E-12 | .003 | 3.362E-9 |
| YKL-40 | 1299 ± 639 (× 102) | 308 ± 139 (× 102) | 283 ± 75 (× 102) | 4.439E-29 | 5.677E-18 | .81 |
| M-CSF | 59 ± 30 | 20 ± 9 | 17 ± 8 | 3.104E-4 | 3.872E-4 | .10 |
| TRAIL | 82 ± 38 | 151 ± 44 | 142 ± 36 | 4.461E-45 | 6.132E-31 | .89 |
| SAA | 3372 ± 2436 (× 104) | 803 ± 3726 (× 104) | 1083 ± 3841 (× 104) | 1.133E-25 | 1.846E-18 | .04 |
Abbreviations: cps, counts per second; IL-1RA, interleukin 1 receptor antagonist; M-CSF, macrophage colony-stimulating factor 1; OPN, osteopontin; SAA, serum amyloid A1; T1D, type 1 diabetes; T2D, type 2 diabetes; TRAIL, tumor necrosis factor superfamily member 10; YKL-40, chitinase 3-like 1.
Validation of 6 Candidate Biomarkers in Children With New-onset Diabetes
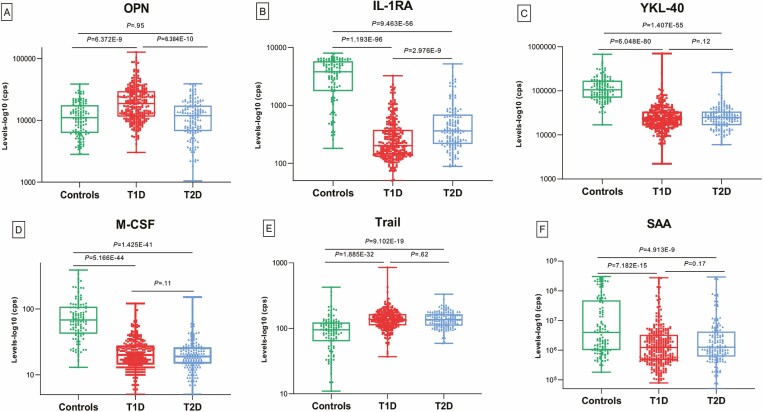
To validate the 6 biomarkers that showed significant differences between patients and healthy controls in screening, we further tested the second independent set of 400 samples from 200 newly diagnosed children with T1D, 100 newly diagnosed children with T2D, and 100 age-matched healthy control children. The outcomes from the validation tests were similar to the results from the screening tests. Serum levels of these 6 biomarkers from the validation tests with 3 subgroups of T1D, T2D, and controls are summarized in Table 3. The results from the first and the second sets were combined and are illustrated in Fig. 3. The combined results (10) (see Fig. 3) show that serum levels of 4 protein biomarkers (YKL-40, M-CSF, TRAIL, and SAA) in children with either T1D or T2D were significantly different from healthy children (T1D vs controls, P = 6.048E-80 ~ 7.182E-15, T2D vs controls, P = 1.407E-55 ~ 4.913E-9), but not significantly different between the T1D group and T2D group. Most interestingly, with consistency, the serum level of OPN was significantly changed only in the T1D group, higher than that both in the control group (P = 6.372E-9) and the T2D group (P = 6.384E-10), but no difference was found between T2D and controls (P = .95). Another protein biomarker, IL-1RA, was significantly lower in children with T1D than both in healthy children (P = 1.193E-96) and children with T2D (P = 2.976E-9), whereas the level of IL-1RA was also lower in children with T2D than in healthy children (P = 9.463E-56).
Table 3.
Serum levels of 6 protein biomarkers from the second group of participants
| Levels of proteins, cps | Controls | T1D | T2D | P (T1D vs controls) | P (T2D vs controls) | P (T1D vs T2D) |
|---|---|---|---|---|---|---|
| OPN | 145 ± 77 (× 102) | 268 ± 193 (× 102) | 162 ± 82 (× 102) | 1.286E-13 | .34 | 2.652E-8 |
| IL-1RA | 4131 ± 2062 | 402 ± 458 | 663 ± 889 | 1.093E-79 | 4.597E-52 | .02 |
| YKL-40 | 1351 ± 1016 (× 102) | 318 ± 548 (× 102) | 321 ± 379 (× 102) | 1.571E-55 | 3.492E-43 | .64 |
| M-CSF | 93 ± 73 | 26 ± 17 | 26 ± 19 | 6.195E-43 | 9.843E-35 | .91 |
| TRAIL | 104 ± 67 | 140 ± 68 | 141 ± 43 | 4.312E-14 | 2.634E-12 | .62 |
| SAA | 3405 ± 7847 (× 104) | 930 ± 3203 (× 104) | 1245 ± 4132 (× 104) | .03 | .08 | .82 |
Abbreviations: cps, counts per second; IL-1RA, interleukin 1 receptor antagonist; M-CSF, macrophage colony-stimulating factor 1; OPN, osteopontin; SAA, serum amyloid A1; T1D, type 1 diabetes; T2D, type 2 diabetes; TRAIL, tumor necrosis factor superfamily member 10; YKL-40, chitinase 3-like 1.
Figure 3.
Serum biomarkers in T1D patients. Serum levels of 6 candidate protein biomarkers: A, OPN; B, IL1-RA; C, YKL-40; D, M-CSF; E, TRAIL; and F, SAA in newly diagnosed children with T1D, T2D, and age-matched healthy controls (combined the results from the first and the second group of participants). The y-axis indicates log-10 transformation of the counts per second (cps) values from the plate counting machine. IL-1RA, interleukin 1 receptor antagonist; M-CSF, macrophage colony-stimulating factor 1; OPN, osteopontin; SAA, serum amyloid A1; T1D, type 1 diabetes; T2D, type 2 diabetes; TRAIL, tumor necrosis factor superfamily member 10; YKL-40, chitinase 3-like 1.
We further analyzed the effect of age and sex on these 6 candidate biomarkers in different groups (10). We observed that age had a significant influence on these protein biomarkers. Some biomarkers had small but significant differences between male and female participants, especially for IL-1RA, YKL-40, TRAIL, and SAA in the second set of T1D patients.
Considering the effect of age and sex, we made logistic regression models including sex and age as covariates to confirm the association of these 6 protein biomarkers and T1D in combined first and second sample data. After adjusting for the confounding variables age and sex, compared with controls, the levels of OPN (P = .001) and TRAIL (P = 2.002E-6) in T1D children significantly increased, and the levels of IL-1RA (P = 9.264E-8), YKL-40 (P = 6.456E-4), and M-CSF (P = .002) decreased significantly, while SAA lost significance (P = .93). Compared with children with T2D, only OPN and IL1-RA had significant differences in children with T1D (P = 1.568E-4 for OPN, P = 3.714E-8 for IL1-RA) (Table 4).
Table 4.
Logistic regression analysis for the association of 6 protein biomarkers with type 1 diabetes in combined first- and second-group data
| Controls vs T1D, OR | P | T2D vs T1D, OR | P | |
|---|---|---|---|---|
| OPN | 0.039 (0.006-0.284) | .001 | 0.201 (0.088-0.462) | 1.567E-4 |
| IL-1RA | 37.314 (9.886-140.840) | 9.264E-8 | 8.022 (3.822-16.838) | 3.713E-8 |
| YKL-40 | 37.820 (4.691-304.888) | 6.457E-4 | 0.443 (0.164-1.199) | .11 |
| M-CSF | 21.325 (2.969-153.157) | .002 | 0.697 (0.100-1.879) | .13 |
| TRAIL | 0.003 (0.000-0.032) | 2.03E-6 | 0.778 (0.175-3.453) | .74 |
| SAA | 0.969 (0.499-1.881) | .93 | 1.308 (0.924-1.850) | .13 |
Age and sex were included as covariates in a logistic regression; OR (odds ratio) and 95% CI are presented.
Abbreviations: IL-1RA, interleukin 1 receptor antagonist; M-CSF, macrophage colony-stimulating factor 1; OPN, osteopontin; SAA, serum amyloid A1T1D, type 1 diabetes; T2D, type 2 diabetes; TRAIL, tumor necrosis factor superfamily member 10; YKL-40, chitinase 3-like 1.
Discussion
T1D is an autoimmune disease resulting from poorly understood cellular and molecular mechanisms in the immune system. It often develops in early childhood and destroys the β cells of the pancreatic islets. T1D susceptibility genes and IAbs have been widely used to identify those at increased risk for the development of T1D. However, existing markers do not fully meet the need for T1D prediction and prevention, and there is especially a lack of tailoring and monitoring rates of progression and therapies for the disease (2). Additional biomarkers are needed to help understanding the cascade of molecular mechanisms in the immune system, which would be useful for more precise predictive models and prevention strategies.
Serum proteins are great sources for biomarkers (2, 19). Since proteins are involved in cellular processes, disease-related proteins detectable in the serum should be great candidates. In this study, we have attempted to identify biomarkers from a pool of gene products expressed in the T1D target tissues. We applied our in silico approach, eGWAS, to leverage large numbers of raw T1D-related RNA measurements in target tissues as a way of identifying new functionally important genes/proteins associated with T1D. We calculated the likelihood of finding repeated differential expression of a gene in disease-related tissues using a large number of case-control genome-wide gene-expression arrays. We performed an eGWAS analysis using data from more than 160 publicly available microarrays from the pancreatic islets of rodent models of T1D. We identified 20 proteins detectable in human blood as our top candidates as biomarkers. Across the microarrays, these proteins were most differentially dysregulated in the pancreatic islets of T1D rodent models and are present and measurable in human blood serum, based on the ECL assay database data.
Multiple prior studies of proteomic biomarkers in T1D have reported some proteins overlapping with those in our present study (4, 5, 20, 21), but the study cohorts were limited in adults or patients with long-term diabetes, which are not representative of the large cohort of childhood T1D at disease onset. We analyzed the serum levels of 20 top biomarker candidates and focused on children newly diagnosed with T1D vs age-matched T2D and healthy controls using 2 independent sets of samples. We identified 6 proteins in children with T1D at disease onset within 2 weeks of diagnosis that were significantly different from age-matched healthy children; 4 of them (YKL-40, M-CSF, TRAIL, and SAA) were also significantly different between T2D and controls, but no differences were observed between T1D and T2D. YKL-40 is a chitin-binding protein involved in acute and chronic inflammatory reactions. YKL-40 was found to be increased in adults with T1D in previous studies (20, 22), while our present study in newly diagnosed children with 2 independent sets of samples clearly showed that YKL-40 was decreased significantly in children with either T1D or T2D, compared with age-matched healthy children. Further studies are needed to confirm these discordant findings in T1D between children and adults and to explore the underlying mechanism. M-CSF is considered a key cytokine in the regulation of microglial inflammatory responses (23), and some studies have reported that M-CSF increased in patients with T2D and diabetic microangiopathy (24). However, in this study, we observed that M-CSF decreased in children with new-onset diabetes. TRAIL is a multifunctional cytokine involved in potent antitumor actions, modulating pathophysiological processes of cardiovascular, metabolic, and autoimmune diseases (25-27). Results from clinical studies on the relationship between circulating TRAIL and diabetes appear to be inconclusive (21, 28). We found the serum level of TRAIL in children with newly diagnosed diabetes increased significantly compared with healthy controls. SAA proteins are members of the acute-phase response protein group that plays a significant role in inflammatory processes, and SAA1 genetic polymorphisms were found to be associated with diabetes, obesity, and cardiovascular disease (29-31). In this study, SAA level decreased in patients with diabetes, but after adjusting for age and sex, SAA had no significant association with T1D. These 4 common markers both in T1D and T2D might be more generally related to hyperglycemia and its related metabolic changes than to diabetes specifically and could be used as indicators of diabetes-related malfunction.
Remarkably, serum levels of 2 proteins (OPN and IL-1RA) in children with T1D were significantly different not only from healthy controls but also from children with T2D, especially for OPN, which uniquely increased in T1D significantly, whereas no difference was found between T2D and healthy controls. OPN is a phosphoprotein with adhesive and cell signaling functions. It plays a vital role in the regulation of immune cell response as it modulates T-cell function by affecting the differentiation of T lymphocytes into Th1 and Th2 cells, regulating the balance between Th1 and Th2, and participating in cell-induced immunological response (32). OPN was demonstrated to induce adipose tissue inflammation and to increase proinflammatory cytokine release into the bloodstream (33). Also, OPN itself acts as a proinflammatory cytokine by chemoattracting monocytes, macrophages, and lymphocytes (34). It also stimulates B lymphocytes to express multiclone antibodies (35). A recent study of OPN null mutant nonobese diabetic mice suggested a protective role of OPN on islet cells (36). An association of alleles at polymorphic sites in the OPN encoding gene in T1D patients has also been reported (37). Previous studies reported some associations of higher serum OPN levels with T1D, but the studies were limited to patients with a long duration of diabetes and most were focused on T1D complications (eg, diabetic kidney disease) (5, 38-42). Serum levels of OPN in children newly diagnosed with T1D have never been studied. This new finding enables us to speculate that OPN might play an important role in the pathogenesis and development of T1D and could be a useful candidate for a serological biomarker for T1D in children.
IL1-RA serum concentrations were significantly lower both in T1D and T2D children compared with healthy controls, and children with T1D were significantly lower than children with T2D. IL1-RA is a member of the interleukin 1 (IL-1) cytokine family and is secreted by immune cells and binds to IL-1 receptors to avoid a cellular response, thereby competitively blocking the proinflammatory effects of IL-1 (43). IL-1 has been shown to promote pancreatic β-cell destruction in the pathogenesis of T1D (44). By binding to β-cell IL-1 receptors, IL-1 signals can promote β-cell dysfunction and death (45-47). IL1-RA (anakinra) was previously examined for therapeutic effect against recent-onset T1D (48). Lower levels of serum IL1-RA were reported in T1D patients with a long duration of hyperglycemia (4). Our present study found significantly lower levels of IL1-RA in circulation in children newly diagnosed with either T1D or T2D, and especially children with T1D had extremely low levels, compared with age-matched healthy children. Low levels of IL1-RA might reflect an enhanced inflammatory pathway to cause the β-cell damage.
Our study has some limitation. Detailed clinical characteristics of participants such as glycemic status and information of body mass index were not available in this study. The metabolic and inflammatory status may affect the level of these serological biomarkers. More detailed clinical information may strengthen the conclusions. Second, the sample size of healthy controls in the first group of participants was limited, which may lead to missing some candidate proteins. The results of our study need to be confirmed in a larger population. The clinical samples used in the present study were limited in children newly diagnosed with T1D. Follow-up studies of these findings on the early stages of preclinical diabetes are anticipated to see how early these biomarkers appear during disease progression and how much predictive value they might add when combined with IAbs to increase the predictive accuracy and the rate of diabetes progression.
In conclusion, using a data-driven candidate gene approach (eGWAS), we discovered that serum levels of 6 proteins were significantly changed in children newly diagnosed with T1D and T2D, while 2 proteins, OPN and IL1-RA, were particularly associated with T1D and could be considered candidates for potential therapeutic targets and additional biomarkers for T1D in children.
Acknowledgment
We thank Ms Yuko Kodama for copyediting and manuscript preparation.
Glossary
Abbreviations
- ECL
electrochemiluminescence assay
- eGWAS
expression-based genome-wide association study
- GAD65
glutamic acid decarboxylase
- GEO
Gene Expression Omnibus
- GWAS
genome-wide association study
- IAb
islet autoantibody
- IA-2
insulinoma antigen 2
- IL-1RA
interleukin 1 receptor antagonist
- M-CSF
macrophage colony-stimulating factor 1
- MSD
Meso Scale Discovery
- NIH
National Institute of Health
- OPN
osteopontin
- OR
odds ratio
- SAA
serum amyloid A1
- T1D
type 1 diabetes
- T2D
type 2 diabetes
- TRAIL
tumor necrosis factor superfamily member 10
- YKL-40
chitinase 3-like 1
- ZnT8
zinc transporter 8
Contributor Information
Xiaofan Jia, Barbara Davis Center for Diabetes, University of Colorado Denver, Aurora, Colorado 80045, USA.
Kyoko Toda, Biomedical Research Center, Kitasato Institute Hospital, Kitasato University, Tokyo 108-8642, Japan.
Ling He, Barbara Davis Center for Diabetes, University of Colorado Denver, Aurora, Colorado 80045, USA.
Dongmei Miao, Barbara Davis Center for Diabetes, University of Colorado Denver, Aurora, Colorado 80045, USA.
Satoru Yamada, Diabetes Center, Kitasato Institute Hospital, Kitasato University, Tokyo 108-8642, Japan.
Liping Yu, Barbara Davis Center for Diabetes, University of Colorado Denver, Aurora, Colorado 80045, USA.
Keiichi Kodama, Health Promotion Team, ORIX Group Health Insurance Society, ORIX Corporation, Tokyo 105-6135, Japan; Department of Endocrinology and Metabolism, Clinical Medicine Research Center, Sanno Medical Center, International University of Health and Welfare, Tokyo 107-8332, Japan.
Financial Support
This work was supported by the Juvenile Diabetes Research Foundation (JDRF grant No. 2-SRA-2019-695-S-B) and the Diabetes Research Center (DRC grant No. P30 DK116073).
Author Contributions
K.K. and L.Y. designed research; X.J., K.T., L.H., D.M., S.Y., L.Y., and K.K. performed research; X.J., K. T., L.H., D.M., S.Y., L.Y., and K.K. analyzed data; all authors participated in data interpretation; K.K. and L.Y. wrote the paper; and all authors provided critical review of the draft and approved the final version. K.K and L.Y. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Disclosures
The authors declared no conflicts of interest.
Data Availability
Some or all data sets generated during and/or analyzed during the present study are not publicly available but are available from the corresponding author on reasonable request.
References
- 1. Knip M, Veijola R, Virtanen SM, Hyöty H, Vaarala O, Akerblom HK. Environmental triggers and determinants of type 1 diabetes. Diabetes. 2005;54(Suppl 2):S125-S136. [DOI] [PubMed] [Google Scholar]
- 2. Carey C, Purohit S, She JX. Advances and challenges in biomarker development for type 1 diabetes prediction and prevention using omic technologies. Expert Opin Med Diagn. 2010;4(5):397-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Purohit S, Sharma A, Zhi W, et al. Proteins of TNF-α and IL6 pathways are elevated in serum of type-1 diabetes patients with microalbuminuria. Front Immunol. 2018;9:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Purohit S, Sharma A, Hopkins D, et al. Large-scale discovery and validation studies demonstrate significant reductions in circulating levels of IL8, IL-1Ra, MCP-1, and MIP-1β in patients with type 1 diabetes. J Clin Endocrinol Metab. 2015;100(9):E1179-E1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barchetta I, Alessandri C, Bertoccini L, et al. Increased circulating osteopontin levels in adult patients with type 1 diabetes mellitus and association with dysmetabolic profile. Eur J Endocrinol. 2016;174(2):187-192. [DOI] [PubMed] [Google Scholar]
- 6. Kodama K, Horikoshi M, Toda K, et al. Expression-based genome-wide association study links the receptor CD44 in adipose tissue with type 2 diabetes. Proc Natl Acad Sci U S A. 2012;109(18):7049-7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kodama K, Zhao Z, Toda K, et al. Expression-based genome-wide association study links vitamin D-binding protein with autoantigenicity in type 1 diabetes. Diabetes. 2016;65(5):1341-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kodama K, Toda K, Morinaga S, Yamada S, Butte AJ. Anti-CD44 antibody treatment lowers hyperglycemia and improves insulin resistance, adipose inflammation, and hepatic steatosis in diet-induced obese mice. Diabetes. 2015;64(3):867-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu LF, Kodama K, Wei K, et al. The receptor CD44 is associated with systemic insulin resistance and proinflammatory macrophages in human adipose tissue. Diabetologia. 2015;58(7):1579-1586. [DOI] [PubMed] [Google Scholar]
- 10. Jia X, Toda K, He L, et al. Supplementary data for “Expression-based Genome-Wide Association Study Links Osteopontin and Interleukin 1 Receptor Antagonist With Newly Diagnosed Type 1 Diabetes in Children.” Figshare 2022. Deposited April 1, 2022https://figshare.com/s/3cad112d96ca5cf18e27 [Google Scholar]
- 11. Chen R, Li L, Butte AJ. AILUN: reannotating gene expression data automatically. Nat Methods. 2007;4(11):879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Whitlock MC. Combining probability from independent tests: the weighted Z-method is superior to Fisher’s approach. J Evol Biol. 2005;18(5):1368-1373. [DOI] [PubMed] [Google Scholar]
- 13. American Diabetes Association. 2. Classification and diagnosis of diabetes: Standards of Medical Care in Diabetes—2019. Diabetes Care. 2019;42(Suppl 1):S13-S28. [DOI] [PubMed] [Google Scholar]
- 14. Couper JJ, Haller MJ, Greenbaum CJ, et al. ISPAD Clinical Practice Consensus Guidelines 2018: stages of type 1 diabetes in children and adolescents. Pediatr Diabetes. 2018;19(Suppl 27):20-27. [DOI] [PubMed] [Google Scholar]
- 15. Choi SH, Kim YH, Hebisch M, et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature. 2014;515(7526):274-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Minter MR, Zhang C, Leone V, et al. Antibiotic-induced perturbations in gut microbial diversity influences neuro-inflammation and amyloidosis in a murine model of Alzheimer’s disease. Sci Rep. 2016;6:30028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Waugh K, Snell-Bergeon J, Michels A, et al. Increased inflammation is associated with islet autoimmunity and type 1 diabetes in the Diabetes Autoimmunity Study in the Young (DAISY). PLoS One. 2017;12(4):e0174840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Purnell JQ, Johnson GS, Wahed AS, et al. Prospective evaluation of insulin and incretin dynamics in obese adults with and without diabetes for 2 years after Roux-en-Y gastric bypass. Diabetologia. 2018;61(5):1142-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Randall SA, McKay MJ, Baker MS, Molloy MP. Evaluation of blood collection tubes using selected reaction monitoring MS: implications for proteomic biomarker studies. Proteomics. 2010;10(10):2050-2056. [DOI] [PubMed] [Google Scholar]
- 20. Rathcke CN, Persson F, Tarnow L, Rossing P, Vestergaard H. YKL-40, a marker of inflammation and endothelial dysfunction, is elevated in patients with type 1 diabetes and increases with levels of albuminuria. Diabetes Care. 2009;32(2):323-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tornese G, Tisato V, Monasta L, Vecchi Brumatti L, Zauli G, Secchiero P. Serum TRAIL levels increase shortly after insulin therapy and metabolic stabilization in children with type 1 diabetes mellitus. Acta Diabetol. 2015;52(5):1003-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Batinic K, Höbaus C, Grujicic M, et al. YKL-40 is elevated in patients with peripheral arterial disease and diabetes or pre-diabetes. Atherosclerosis. 2012;222(2):557-563. [DOI] [PubMed] [Google Scholar]
- 23. Chitu V, Stanley ER. Colony-stimulating factor-1 in immunity and inflammation. Curr Opin Immunol. 2006;18(1):39-48. [DOI] [PubMed] [Google Scholar]
- 24. Wautier MP, Boulanger E, Guillausseau PJ, Massin P, Wautier JL. AGEs, macrophage colony stimulating factor and vascular adhesion molecule blood levels are increased in patients with diabetic microangiopathy. Thromb Haemost. 2004;91(5):879-885. [DOI] [PubMed] [Google Scholar]
- 25. Schaefer U, Voloshanenko O, Willen D, Walczak H. TRAIL: a multifunctional cytokine. Front Biosci. 2007;12:3813-3824. [DOI] [PubMed] [Google Scholar]
- 26. Harith HH, Morris MJ, Kavurma MM. On the TRAIL of obesity and diabetes. Trends Endocrinol Metab. 2013;24(11):578-587. [DOI] [PubMed] [Google Scholar]
- 27. Neve A, Corrado A, Cantatore FP. TNF-related apoptosis-inducing ligand (TRAIL) in rheumatoid arthritis: what’s new? Clin Exp Med. 2014;14(2):115-120. [DOI] [PubMed] [Google Scholar]
- 28. Tornese G, Iafusco D, Monasta L, et al. The levels of circulating TRAIL at the onset of type 1 diabetes are markedly decreased in patients with ketoacidosis and with the highest insulin requirement. Acta Diabetol. 2014;51(2):239-246. [DOI] [PubMed] [Google Scholar]
- 29. Xie X, Ma YT, Yang YN, et al. SAA1 genetic polymorphisms are associated with plasma glucose concentration in non-diabetic subjects. Clin Chem Lab Med. 2013;51(12):2331-2334. [DOI] [PubMed] [Google Scholar]
- 30. Yassine HN, Trenchevska O, He H, et al. Serum amyloid a truncations in type 2 diabetes mellitus. PLoS One. 2015;10(1):e0115320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xie X, Ma YT, Yang YN, et al. Genetic polymorphisms of serum amyloid A1 and coronary artery disease risk. Tissue Antigens. 2015;85(3):168-176. [DOI] [PubMed] [Google Scholar]
- 32. Weber GF, Zawaideh S, Hikita S, Kumar VA, Cantor H, Ashkar S. Phosphorylation-dependent interaction of osteopontin with its receptors regulates macrophage migration and activation. J Leukoc Biol. 2002;72(4):752-761. [PubMed] [Google Scholar]
- 33. Nomiyama T, Perez-Tilve D, Ogawa D, et al. Osteopontin mediates obesity-induced adipose tissue macrophage infiltration and insulin resistance in mice. J Clin Invest. 2007;117(10):2877-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ashkar S, Weber GF, Panoutsakopoulou V, et al. Eta-1 (osteopontin): an early component of type-1 (cell-mediated) immunity. Science. 2000;287(5454):860-864. [DOI] [PubMed] [Google Scholar]
- 35. Wang KX, Denhardt DT. Osteopontin: role in immune regulation and stress responses. Cytokine Growth Factor Rev. 2008;19(5-6):333-345. [DOI] [PubMed] [Google Scholar]
- 36. Melanitou E. Investigation of type 1 diabetes in NOD mice knockout for the osteopontin gene. Gene. 2020;753:144785. [DOI] [PubMed] [Google Scholar]
- 37. Marciano R, D’Annunzio G, Minuto N, et al. Association of alleles at polymorphic sites in the osteopontin encoding gene in young type 1 diabetic patients. Clin Immunol. 2009;131(1):84-91. [DOI] [PubMed] [Google Scholar]
- 38. Gordin D, Forsblom C, Panduru NM, et al. FinnDiane Study Group. Osteopontin is a strong predictor of incipient diabetic nephropathy, cardiovascular disease, and all-cause mortality in patients with type 1 diabetes. Diabetes Care. 2014;37(9):2593-2600. [DOI] [PubMed] [Google Scholar]
- 39. Marcovecchio ML, Colombo M, Dalton RN, et al. AdDIT and the SDRNT1BIO Investigators. Biomarkers associated with early stages of kidney disease in adolescents with type 1 diabetes. Pediatr Diabetes. 2020;21(7):1322-1332. [DOI] [PubMed] [Google Scholar]
- 40. El Dayem SMA, El Bohy AEM, Battah AA, Hamed M, El Aziz SHA. Osteopontin for early detection of microvascular and macrovascular type 1 diabetic complication. Open Access Maced J Med Sci. 2019;7(21):3619-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Talat MA, Sherief LM, El-Saadany HF, Rass AA, Saleh RM, Sakr MM. The role of osteopontin in the pathogenesis and complications of type 1 diabetes mellitus in children. J Clin Res Pediatr Endocrinol. 2016;8(4):399-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. El-Din DSS, Amin AI, Egiza AO. Utility of tissue inhibitor metalloproteinase-1 and osteopontin as prospective biomarkers of early cardiovascular complications in type 2 diabetes. Open Access Maced J Med Sci. 2018;6(2):314-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Perrier S, Darakhshan F, Hajduch E. IL-1 receptor antagonist in metabolic diseases: Dr Jekyll or Mr Hyde? FEBS Lett. 2006;580(27):6289-6294. [DOI] [PubMed] [Google Scholar]
- 44. Sandler S, Eizirik DL, Svensson C, Strandell E, Welsh M, Welsh N. Biochemical and molecular actions of interleukin-1 on pancreatic β-cells. Autoimmunity. 1991;10(3):241-253. [DOI] [PubMed] [Google Scholar]
- 45. Mandrup-Poulsen T, Pickersgill L, Donath MY. Blockade of interleukin 1 in type 1 diabetes mellitus. Nat Rev Endocrinol. 2010;6(3):158-166. [DOI] [PubMed] [Google Scholar]
- 46. Sandler S, Andersson A, Hellerström C. Inhibitory effects of interleukin 1 on insulin secretion, insulin biosynthesis, and oxidative metabolism of isolated rat pancreatic islets. Endocrinology. 1987;121(4):1424-1431. [DOI] [PubMed] [Google Scholar]
- 47. Yamada K, Takane-Gyotoku N, Yuan X, Ichikawa F, Inada C, Nonaka K. Mouse islet cell lysis mediated by interleukin-1-induced Fas. Diabetologia. 1996;39(11):1306-1312. [DOI] [PubMed] [Google Scholar]
- 48. Moran A, Bundy B, Becker DJ, et al. Type 1 Diabetes TrialNet Canakinumab Study Group; AIDA Study Group. Interleukin-1 antagonism in type 1 diabetes of recent onset: two multicentre, randomised, double-blind, placebo-controlled trials. Lancet. 2013;381(9881):1905-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some or all data sets generated during and/or analyzed during the present study are not publicly available but are available from the corresponding author on reasonable request.