Abstract
Temperature has a pleiotropic effect on Yersinia enterocolitica gene expression. Temperature-dependent phenotypes include the switching between two type III protein secretion systems, flagellum biosynthesis (≤30°C) and virulence plasmid-encoded Yop secretion (37°C). The mechanism by which temperature exerts this change in genetic programming is unclear; however, altered gene expression by temperature-dependent changes in DNA topology has been implicated. Here, we present evidence that the Y. enterocolitica virulence plasmid, pYV, undergoes a conformational transition between 30 and 37°C. Using a simplified two-dimensional, single-gel assay, we show that pYV contains multiple regions of intrinsic curvature, including virF, the positive activator of virulence genes. These bends are detectable at 30°C but melt at 37°C, the temperature at which the cells undergo phenotypic switching. We also show that pACYC184, a plasmid used as a reporter of temperature-induced changes in DNA supercoiling, has a single region of intrinsic bending detected by our assay. Topoisomers of pACYC184, with and without this bend, isolated from Y. enterocolitica were resolved by using chloroquine gels. The single bend has a dramatic influence on temperature-dependent DNA supercoiling. These data suggest that the Y. enterocolitica pYV plasmid may undergo a conformational change at the host temperature due to melting of DNA bends followed by compensatory adjustments in superhelical density. Hence, changes in DNA topology may be the temperature-sensing mechanism for virulence gene expression in Y. enterocolitica and other enteric pathogens.
It is now evident that DNA is not merely the repository of genetic information, but that the structure of the molecule itself influences information access. For example, DNA bending (curvature), both intrinsic and induced by protein binding, directly affects transcription. Static or induced bends can determine promoter strength, both positively or negatively, depending on the orientation and distance of the bends relative to the RNA polymerase binding site. Involvement of such ternary structures with regulatory regions appears to be a common trait of most bacterial promoters (reviewed in reference 22).
Intrinsic DNA bending was originally identified with trypanosome kinetoplast DNA. Kinetoplast DNA fragments with phased deoxyribosyladenine (dA) and deoxyribosylthymine (dT) nucleotide tracks display retarded migration in polyacrylamide gels (15). Regions of intrinsic bending identified on the chromosome of Escherichia coli have been predominantly associated with the 5′ regulatory regions of genes (19, 28). The bacterial histone-like protein H-NS (21, 30) preferentially recognizes this topological feature, and this interaction (or binding) can repress gene expression (10, 11, 21). As such, H-NS has been considered a potential generalized repressor for some operons, with gene activation occurring by either competitive displacement of H-NS by an activator protein or displacement of H-NS by changes in DNA supercoiling induced by some environmental signal. Thus, supercoiling, intrinsic or induced bending, and histone-like proteins, individually or synergistically, can modulate gene expression.
Pathogenic bacteria utilize host environmental cues for virulence gene activation (reviewed in references 4 and 16). Temperature is the key environmental parameter for virulence gene induction for the pathogenic Yersinia spp. Shifting Yersinia enterocolitica from ≤30 to 37°C has multiple effects on cell morphology and physiology. Changes after a temperature upshift include the coordinate repression of flagellum synthesis (12, 24) and induction of a set of plasmid-encoded virulence genes (reviewed in reference 7). The virulence plasmid, termed pYV (plasmid of Yersinia virulence) encodes a type III secretion system (ysc and lcr genes) essential for delivery of additional plasmid-borne anti-host factors collectively referred to as Yops (Yersinia outer proteins). Expression of ysc, lcr, and yop genes requires the trans-acting DNA binding protein VirF, a member of the AraC family of positive activators (5). VirF synthesis is induced by elevated temperature (17, 24). However, experiments in which VirF was artificially overexpressed from the tac promoter at 30°C failed to induce yop expression (14). The Yersinia pestis homologue of VirF, LcrF, is constitutively expressed as assayed by lacZ transcriptional fusions, but yop gene transcription is still dependent upon elevated temperature (9). Therefore, Yop expression requires factors in addition to VirF.
Several lines of evidence suggest that DNA topology is involved in Yersinia virulence gene expression. Cornelis et al. (6) showed the role of a putative histone-like protein, YmoA, as having a repressor-like activity on Yop expression. Yop expression becomes VirF independent in ymoA mutants, but expression is still enhanced by elevated temperature. Rohde et al. (24) have shown that temperature-induced modifications in DNA supercoiling are coincident with the temperature phasing of two type III secretion systems (flagellum and Yop synthesis) in Y. enterocolitica. Furthermore, novobiocin (DNA gyrase inhibitor)-resistant mutants include a class that is constitutive for Yop expression, similar to the ymoA mutants mentioned above. Additionally, Yop genes can also be expressed in Y. enterocolitica wild-type cells at a low temperature by inducing supercoiling with subinhibitory levels of novobiocin. Therefore, modulations in supercoiling and the histone-like protein, YmoA, show that temperature activation of Yop expression is partially dependent on changes in DNA topology.
In this report we suggest that a third component of DNA topology may act in concert with temperature-mediated changes in supercoiling and YmoA. First we show that the pYV plasmid contains multiple intrinsic DNA bends. Detection of intrinsic bending is based upon a rapid single-gel assay that may have broad application. Secondly, we show that a single bend in the reporter plasmid pACYC184 dramatically affects supercoiling levels. Third, the intrinsic bends of plasmid pYV are sensitive to melting at 37°C but not at 30°C. A model based on these data taken together is presented to explain temperature induction of virulence genes by the temperature-dependent alteration of plasmid architecture.
MATERIALS AND METHODS
Bacteria, plasmids, and growth conditions.
Y. enterocolitica 8081 pYV+ was grown in brain heart infusion broth or Luria broth (Difco) at either 25 or 37°C. Plasmid pACYC184 (Cmr, Tcr) was electroporated into this strain and maintained on either chloramphenicol (20 μg/ml) or tetracycline (20 μg/ml). Plasmid pACYCΔDraI was constructed by cleaving the pACYC184 plasmid with DraI, purifying the plasmid backbone, and religating to form a circular molecule. The ligation mixture was electroporated into E. coli DH5α, and transformants were selected for Tcr and then screened for chloramphenicol sensitivity. This plasmid was introduced into Y. enterocolitica by electroporation.
Plasmid extraction and radiolabeling.
Plasmid DNA was purified from Y. enterocolitica and E. coli cultures by using the alkaline lysis procedure as described by Sambrook et al. (25). DNA was further extracted with chloroform and purified by CsCl density gradient centrifugation. Protein was not detectable in these preparations by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) performed with Coomassie blue staining. A BamHI library of pYV8081 was constructed with the pBluescript KS+ (Stratagene) cloning vector to facilitate the mapping of specific genes. DNA sequence analysis showed that part of virB (yscN-U operon), virF, and the N-terminal coding sequence for virC (yscA-M operon) are contained within BamHI fragment 5 (4). Additionally, BamHI fragment 6 contains yscV (lcrD) and yopN (lcrE) (data not shown). Radiolabeling of individual fragments of pYV8081 was accomplished by electroelution of the desired fragment from the pBluescript subclone followed by radiolabeling with a random priming kit (U.S. Biochemical) according to the protocol supplied by the manufacturer.
Two-dimensional gel electrophoresis.
A mini-protean II slab gel apparatus (Bio-Rad, Richmond, Calif.) was used to detect DNA intrinsic bends based on a modification of the procedure reported by Mizuno (19). DNA samples to be analyzed (ca. 1 to 2 μg) were digested with the appropriate restriction endonuclease(s), mixed with 1/5 volume bromphenol blue xylene-cyanol marker dye buffer, and loaded in wells on the left-hand side of a 5% acrylamide nondenaturing gel. If multiple samples were analyzed on the same gel, a well was skipped between samples. For the first dimension, the DNA samples were vertically resolved at 60°C at 10 V per cm. At this temperature, DNA bends melt and the fragments migrate at their true molecular weights. The gel spacers were then carefully removed, and the glass-gel sandwich was submerged in a horizontal electrophoresis chamber (2-liter buffer capacity) maintained at 4°C. The samples were electrophoresed in the second horizontal dimension again at 10 V per cm until the xylene-cyanol dye was eluted from the right-hand side of the gel. At 4°C, the intrinsic bends can reform, and at that time such fragments will display retarded migration. The gels were stained with ethidium bromide and photographed. Therefore, in this system, linear molecules form a diagonal line descending from left to right, and nonlinear DNA lags behind this diagonal. For both dimensions Tris-borate-EDTA (TBE) buffer is used (0.045 M Tris-borate, 1 mM EDTA [pH 8.4]).
For analysis of the Y. enterocolitica pYV (70 kb), the plasmid was first digested with three restriction endonucleases to generate fragments within the 1- to 10-kb size range. These fragments were resolved in the first dimension by running both the bromphenol blue-xylene cyanol marker dyes off the gel. After additional running, dye was loaded to the well, and electrophoresis was continued until the xylene cyanol reached the bottom of the gel. For the second dimension, electrophoresis was stopped within 2 to 3 h after the xylene cyanol was run off the gel.
The analysis of individual fragments (<1 kb) for DNA bending was conducted by mixing the fragment with a 1-kb DNA ladder preparation (Gibco/BRL). This ladder preparation forms a straight diagonal and facilitates identification of bent DNA. Sufficient resolution is obtained by running the xylene cyanol marker dye to the bottom of the gel in the first dimension and to the side of the gel in the second dimension. This method also allows multiple samples to be assayed on the same gel for comparative purposes.
Southern transfer.
DNA resolved on two-dimensional gels was transferred to nitrocellulose by the method of Southern (27) with the following modifications. The gels were soaked in denaturant (0.2 N NaOH, 1.5 M NaCl) for 30 min and then neutralized for 30 min (0.5 M Tris-HCl [pH 7.5], 1.5 M NaCl), followed by pH equilibration by soaking in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 30 min before being transferred to nitrocellulose. The 1.1-kb EcoRI fragment (part of the yscN operon and virF) and BamHI fragment 6 (yscVyopN) of pYV8081 were radiolabeled with [α-32P]dATP and used as probes to identify whether these genes reside on fragments with intrinsic curvature.
Chloroquine agarose gel electrophoresis.
Chloroquine gels were used essentially as described by Goldstein and Drlica (8). Y. enterocolitica was transformed with either pACYC184 or the ΔDraI chloramphenocol-sensitive derivative of this plasmid. Cells were grown in Luria-Bertani broth at both 25 and 37°C to mid-exponential growth without antibiotic selection, and plasmid DNA was purified by the alkaline lysis procedure described by Sambrook et al. (25). DNA was extracted with buffered chloroform-isoamyl-phenol to remove protein and precipitated. Further purification was achieved by cesium chloride density ultracentrifugation. Purified plasmid DNA was loaded into the wells of a horizontal 1% agarose gel containing 10 μg of chloroquine/ml and resolved at 4 V per cm for 18 to 24 h in TBE buffer containing 10 μg of chloroquine/ml. Following electrophoresis, the gels were washed 1 to 3 h in distilled water, stained with ethidium bromide, and photographed.
RESULTS
Two-dimensional gel assay development for detection of DNA intrinsic bends.
The method reported by Mizuno (19) for detection of bent DNA fragments utilized a two-dimensional gel system; DNA fragments were first resolved in tube gels at a high temperature (60°C), and the tube gel was then overlaid onto a slab gel and electrophoresed at a low temperature (4°C). Differentiation of bent DNA from linear DNA relied on temperature differences in migration; linear molecules form a straight diagonal with bent DNAs lagging behind this line. Because the same buffer is used for both dimensions, the technique could be simplified by conducting analysis in the same gel, thus eliminating the requirement for tube gels in the first dimension (see Materials and Methods).
To test and standardize this approach, we first analyzed DNA fragments containing the E. coli proU and ompF promoter regions previously reported to contain intrinsic bends (21, 26). For each gene, the respective promoter fragments are retarded in the second dimension of two-dimensional gels compared to linear molecular weight standards (data not shown).
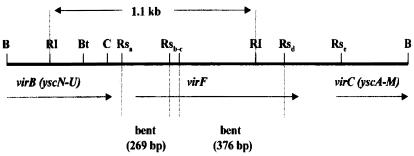
The Y. enterocolitica serotype 08 virF gene is contained within BamHI fragment 5 on the pYV plasmid. We have determined that the basic gene order for this region of pYV (Fig. 1) is the same as reported by Cornelis et al. (4) for pYVe227 (serotype 09). As shown in Fig. 2, the BamHI fragment was digested with RsaI (lane 1) allowing mapping of two bends to the fragments shown in Fig. 1. In lane 2, the 376-bp RsaIc-d fragment was mixed with the 1-kb ladder. Lane 1 also shows additional bent fragments within the 4-kb BamHI fragment 5 localized to virB and the ysc operon. The 269-bp fragment (Fig. 1) includes approximately 70 bp upstream of the virF translation start site.
FIG. 1.
Genetic and physical map of the Y. enterocolitica 08 pYV region encoding virF. The virF gene is contained on a 4-kb BamHI fragment subcloned from pYV. The arrow indicates the direction of virF transcription. When this fragment is digested with RsaI and EcoRI and subjected to two-dimensional gel analysis, two fragments show retarded migration. Restriction sites are denoted as follows: B, BamHI; RI, EcoRI; Rs, RsaI; C, ClaI; and Bt, BstII.
FIG. 2.
The Y. enterocolitica virF gene contains intrinsic bends. (A) The pYV 4.0-kb BamHI fragment 5 contains part of virB (yscN-U operon), virF, and the N-terminal coding region of the yscAM operon. This fragment was digested with RsaI mixed with the 1-kb ladder for reference and subjected to two-dimensional gel analysis (lane 1). Several fragments show retarded migration, including 376-bp and 269-bp fragments which map to virF. The 1.1-kb EcoRI fragment contained within BamHI fragment 5 was subcloned, purified, and then digested with RsaI. The RsaIc-d fragment (Fig. 1) was electroeluted and mixed with the 1-kb ladder (lane 2). (B) Total pYV plasmid DNA was digested with BamHI, EcoRI, and HindIII, and the fragments were resolved on a two-dimensional gel. Multiple DNA fragments show retarded migration. The arrow denotes the DNA fragment hybridizing to the 1.1-kb EcoRI fragment (containing virF; Fig. 1) by Southern blotting.
Analysis of pYV for intrinsic bending.
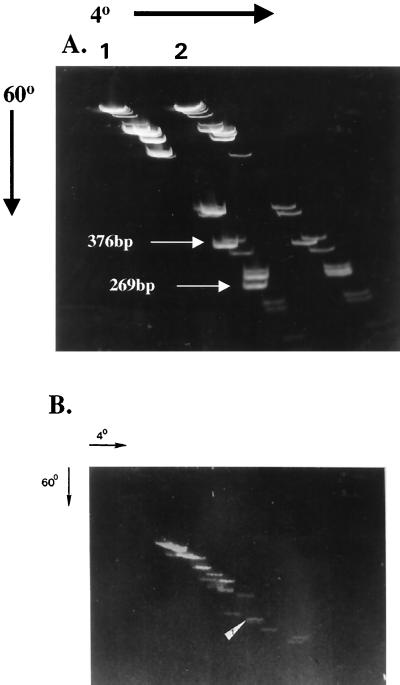
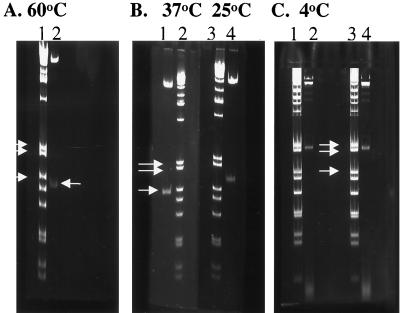
Purified pYV plasmid was digested with BamHI, EcoRI, and HindIII, and the fragments were subjected to two-dimensional gel analysis. As shown in Fig. 2B, this plasmid contains multiple fragments demonstrating pronounced bending based on retarded migration of DNA fragments at low temperature (4°C) in the second dimension. Subsequent transfer of DNA resolved on this gel to nitrocellulose and probed with the EcoRI fragment, which contains part of virB (yscN-U) and virF, determined that the identified fragment (marked by an arrow) was on a nonlinear DNA fragment. Likewise, a two-dimensional gel of restricted pYV DNA (Fig. 3A) was transferred to a membrane and probed with BamHI fragment 6 (containing yscVyopN) determined by DNA sequencing. As shown in Fig. 3B, the yscV and yopN probe hybridizes to fragments that fall to the left of the diagonal line of noncurved DNA. We conclude that both virF and a fragment that contains yopN and yscV genes are contained within fragments that display intrinsic bends.
FIG. 3.
DNA bends associated with pYV. In panel A, purified pYV DNA was digested with HindIII, EcoRI, and BamHI, and the digestion mixture was subjected to two-dimensional gel analysis. A Southern blot was prepared on this gel and probed with radiolabeled pYV BamHI fragment 6, which contains yscV and yopN. These genes are induced by elevated temperature (37°C) and are localized to nonlinear DNA fragments that lag behind the linear fragments of the digest (arrow). The diagonal in panel B was determined by superimposing the X-ray film over a scaled gel photograph.
DNA bending affects supercoiling.
Our previous analysis of temperature-regulated gene expression in Y. enterocolitica determined that phenotypic switching between 25 and 37°C was coincident with thermoinduced modulations of DNA supercoiling (24). Two observations, cited in the conclusion of this report, suggested that other parameters contributed to temperature regulation as follows: (i) other environmental conditions that induce DNA supercoiling in Salmonella typhimurium and E. coli, e.g., increased osmolarity and anaerobiosis, do not mimic the effects seen with temperature. Therefore, supercoiling alone is insufficient to account for the temperature response of Y. enterocolitica. (ii) The mean topoisomer distribution of extracted pACYC184 from Y. enterocolitica cells cultured at 25°C rather than 37°C was greater than the theoretical prediction based on the paper by Goldstein and Drlica (8). That is to say, a change of two linking numbers over this 12°C temperature range is predicted, when in actuality we measured a four to six linking number difference. Therefore, the superhelical density of plasmids in Y. enterocolitica is determined by factors in addition to those that may be predicted.
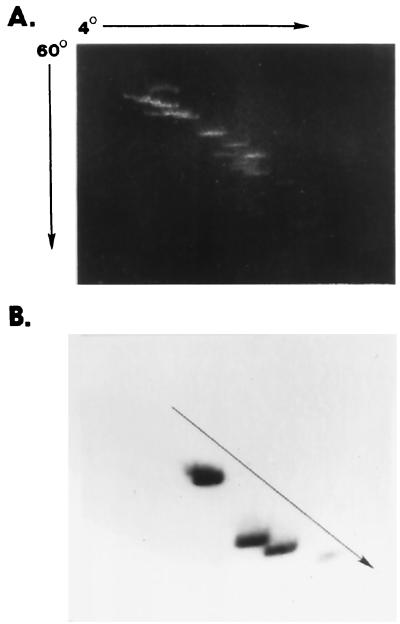
To account for this difference, we examined pACYC184 for regions of intrinsic bends by using the two-dimensional gel assay described. A Sau3A digest showed a single region of bending, which we subsequently mapped to the DraI fragment (bp 3988 to 83, relative to the EcoRI site at position 1). This fragment is internal to the chloramphenicol acetyltransferase (cat) gene of the plasmid. Figure 4A shows a two-dimensional gel of this fragment when pACYC184, digested with DraI, was mixed with a 1-kb molecular size ladder. This fragment shows significant retardation using the two-dimensional gel assay.
FIG. 4.
The pACYC184 DraI fragment is bent, and this fragment affects plasmid supercoiling. (A) Cesium-purified plasmid pACYC184 DNA was digested with DraI to release a 339-bp fragment. The digest was mixed with the 1-kb ladder and analyzed by two-dimensional gel electrophoresis. The DraI fragment, which falls off the linear diagonal generated by the ladder, shows retarded migration. (B) Chloroquine gel electrophoresis was conducted on pACYC184 and pACYCΔDra1 isolated from Y. enterocolitica cells grown at different temperatures. Lanes 1 and 2, pACYC184 isolated from Y. enterocolitica grown at 25 and 37°C, respectively; lanes 3 and 4, plasmid pACYCΔDra1 (339-bp deletion) isolated at the same temperatures. The band across the top in all four lanes is open-circle DNA, showing little difference in migration, whereas the deletion derivative topoisomers migrate much further in the gel.
A DraI deletion of plasmid pACYC184 was constructed and introduced into Y. enterocolitica 8081. Isogenic strains of Y. enterocolitica containing pACYC184 and the DraI deletion construct (pACYCΔDraI) were grown at 25 and 37°C, and the plasmids were purified by cesium chloride density centrifugation and separated on chloroquine gels to resolve topoisomers. Topoisomers can be resolved and visualized by using this assay as a plasmid ladder. Each band differs from adjacent bands by one linking number, with the more negatively supercoiled DNA topoisomers migrating more rapidly in the gel. The results of this experiment are shown in Fig. 4B. The left two lanes show pACYC184 topoisomers resolved at both temperatures. Plasmid pACYC184 topoisomers isolated from Y. enterocolitica are difficult to separate, as the plasmid appears to be in a more relaxed state (less supercoiled) as evidenced by slower migration in the gels. Compared to the same plasmid with the DraI deletion (right two lanes), it can be seen that removal of the bend has a dramatic effect on the overall migration of the plasmid. Part of this effect can be attributed to the removal of 339 bp of DNA (ca. a 7% change in total size). Whereas the difference in migration of supercoiled DNA is significant, the difference in migration of open circles between the two plasmids is negligible. Removal of the bend in this plasmid results in ca. a two linking number increase after a shift from 25 to 37°C as predicted. We conclude that plasmid pACYC184 requires less supercoiling when a bend is present and that this bend accounts for the previous discrepancy in topoisomer distribution noted by Rohde et al. (24).
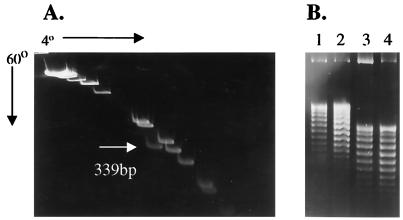
The apparent size of the pACYC184 DraI 339-bp fragment (sequence determination) when separated on a 5% polyacrylamide gel run at 4°C is 490 bp, ca. 1.5 times its actual size (Fig. 5; compare panel A, lane 2 with panel C, lanes 2 and 4). Figure 5 also shows that the DraI fragment shows minimal retardation in gels run at 37°C (compare panel B, lane 1 to panel A, lane 2) but significant retardation in gels run at 25°C (compare lanes 1 and 4 in panel B). These data imply that the bend is maintained at 25°C but melts at 37°C (see next section). This conformational change may therefore account for the significant difference in topoisomer linking number observed over this temperature range.
FIG. 5.
Temperature affects migration of the pACYC184 DraI fragment. Cesium-purified pACYC184 was digested with DraI, and the fragments separated on 5% acrylamide gels run at different temperatures (60, 37, 25, and 4°C) along with a set of molecular size markers. The arrows in panels A, B, and C denote the three reference molecular size markers of 506, 396, and 344 bp. The size of the DraI fragment is 339 bp, based on DNA sequence analysis. This size is also indicated in panel A, lane 2, where it can be seen to migrate just below the 344-bp marker (lane 1) when the gel is run at 60°C. A similar size (ca. 339 bp) is also apparent when this fragment is separated at 37°C (panel B, lane 1) compared to the molecular size standards (panel B, lane 2). Retardation of the DraI fragment is apparent in gels run at 25°C (compare panel B lane 4 to lane 1). In panel C, duplicate digests of the DraI fragment, resolved at 4°C (lanes 2 and 4), migrate with an apparent molecular size of ca. 490 bp relative to the molecular size markers (lanes 1 and 3). Comparison of the migration of the DraI fragment at 60°C (panel A, lane 2) and 37°C (panel B, lane 1) shows that the temperature effect on the retardation of this fragment is minimized around 37°C.
DNA bends melt at 37°C.
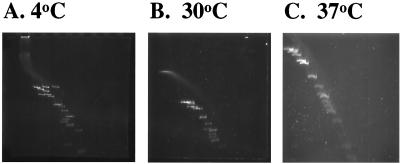
Chan et al. (2, 3) reported that the mean melting temperature in vitro of intrinsically bent synthetic DNA oligomers containing phased dA or dT nucleotide tracts was between 37 and 40°C. Because this is the temperature that induces Y. enterocolitica virulence genes, we utilized the two-dimensional gel assay to determine if pYV bent DNA melts within this temperature range. Figure 6 compares a sample of Y. enterocolitica pYV plasmid digested with three enzymes (BamHI, EcoRI, and HindIII) and resolved at 4, 30, and 37°C, respectively, in the second dimension. As can be seen, the bending is maintained at 30°C but is completely eliminated at 37°C. Similar results were obtained with the DraI fragment of pACYC184 (see Fig. 5); essentially no difference in the molecular weight of this DraI fragment is observed between 37 and 60°C. From these results, we conclude that the presence of intrinsic DNA bends in plasmid DNA can alter the conformation of the plasmid relative to temperature. Furthermore, the most significant change in structure occurs between 30 and 37°C.
FIG. 6.
Plasmid pYV intrinsic DNA bends melt at between 30 and 37°C. This figure illustrates that temperature affects the resolution of pYV DNA fragments in two-dimensional gel analysis. Gels run at 4°C (A) show maximal resolution of bent fragments. Two-dimensional gel analysis conducted at 30°C (B) shows that the bends are still present, but they are completely melted at 37°C (C), as evidenced by a straight diagonal line of fragments.
Plasmids of pathogenic E. coli contain regions of intrinsic bending.
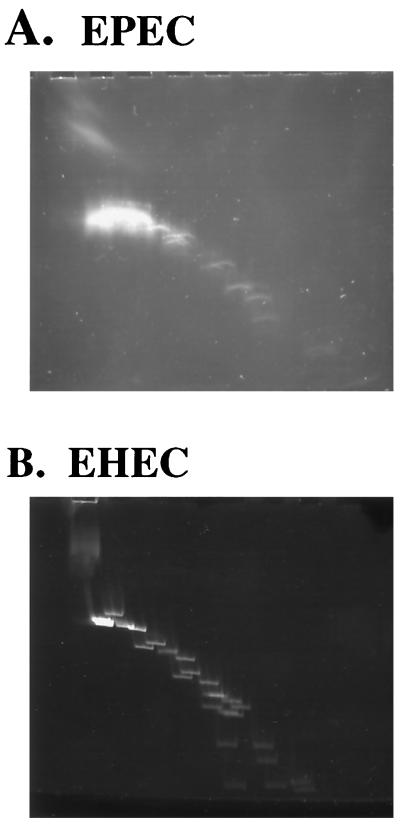
Because temperature regulation of virulence genes is a common theme in facultative pathogens within the family Enterobacteriaceae, we used the two-dimensional gel method to examine the large plasmids of enteropathogenic E. coli (EPEC) and enterohemorrhagic E. coli (EHEC). These plasmids were purified by ultracentrifugation and digested with BamHI, EcoRI, and HindIII, and the fragments separated in two dimensions at 60 and 4°C, respectively. Figure 7 shows that both plasmids are enriched for intrinsic bends.
FIG. 7.
Two-dimensional analysis of E. coli plasmids EAF and pO157. The large plasmids of EPEC and EHEC were cesium purified and digested with EcoRI, BamHI, and HindIII, and the fragments were separated by two-dimensional PAGE. Like the Y. enterocolitica pYV virulence plasmid, both plasmids isolated from pathogenic E. coli strains show multiple fragments with intrinsic bending.
DISCUSSION
One of the remarkable attributes of facultative pathogens is the rapidity with which they respond to the host environment. For many mammalian pathogens, temperature is a key environmental cue in this process (reviewed in reference 16). Over the narrow temperature range of 30 to 37°C, pathogenic Yersinia undergo a significant shift in gene expression involving chromosomal and plasmid loci. Within minutes of exposure to 37°C, plasmid-encoded virulence gene transcripts can be detected, along with the coincident transcriptional arrest of chromosome-encoded flagellar genes. Identifying the cellular thermostat regulating this process may provide key insights into the mechanism of the pathogenesis of Yersinia.
Previous work suggested that temperature-associated changes in DNA structure were involved in phenotypic switching. These studies included correlating the reciprocal repression of flagellar genes and the induction of virulence genes with changes in DNA supercoiling (12, 24) and the involvement of the histone-like protein YmoA (6). The induction of yop genes requires the activator protein VirF, but artificial overexpression of VirF at the nonpermissive temperature (30°C) is insufficient for yop transcription. This observation suggests that the target yop promoters are restricted in some manner at 30°C. On the other hand, Yop promoters in a ymoA virF mutant background are constitutively expressed. Thus, VirF is not an absolute requirement for yop gene expression. Yop genes can also be induced at 30°C in wild-type cells (ymoA+) by modulating DNA supercoiling with subinhibitory concentrations of novobiocin. Virulence gene expression in Yersinia is a complicated process requiring multiple factors. Nonetheless, a common denominator for yop gene expression appears to be DNA topology.
The studies reported here were based upon three observations. First, published pYV DNA sequences of several virulence genes reveal runs of dA or dT nucleotide tracts. Such sequences are known to specify localized DNA intrinsic bending (13). Second, Chan et al. (2, 3) demonstrated that a synthetic 45-bp-long DNA sequence with four segments of phased dA-5 tracts undergoes a temperature-dependent conformational shift. DNA bending of this oligomer is most pronounced at 5°C, but bending is completely eliminated at 40°C, well below the temperature required for strand separation. Because the mean melting temperature for this synthetic bent DNA is a physiological temperature (37°C), these authors posited that this phenomenon might be relevant to DNA topology and function. Finally, our previous work indicated that over the temperature range associated with switching from a low-temperature to a high-temperature phenotype, the degree of supercoiling change associated with the reporter plasmid pACYC184 was greater than expected (24). If temperature-sensitive DNA bending is a factor in temperature regulation, it could account for the observed association between supercoiling and histone-like protein interactions.
To test this hypothesis, we first simplified the two-dimensional gel assay originally reported by Mizuno (19) to assay pYV DNA for bending. Our modifications have two advantages over other methods to detect aberrant migration of DNA fragments due to intrinsic bending. First, only a single gel is required. The method employed by Mizuno (19) requires tube and slab gels. Other methods examining whether a given DNA fragment contains a bend require running two gels at different temperatures for molecular weight comparisons. Second, multiple samples can be analyzed on the same gel. This method could be applied to the rapid mapping of such topological sites on large DNA fragments, particularly if the DNA sequence has been determined.
Using this assay, we have shown that the Y. enterocolitica virulence plasmid has multiple regions of intrinsic bending. Bending is pronounced at 4°C and completely lost at 37°C. Importantly, we show that DNA bending is maintained in the range of 25 to 30°C. Therefore, these static experiments suggest the pYV DNA can undergo a significant conformational change within the temperature range correlating with virulence gene activation.
Does intrinsic DNA bending influence DNA supercoiling in Y. enterocolitica? We have been unsuccessful in resolving pYV topoisomers to monitor changes in temperature-induced supercoiling due to the size of this plasmid. We addressed this question by examining the reporter plasmid pACYC184 for intrinsic bends. As shown, this plasmid has a pronounced bend in the cat gene. Deletion of this bend results in an overall increased supercoiling of the plasmid. This shows that bends can influence plasmid superhelical density in Y. enterocolitica, perhaps substituting for supercoiling requirements in specific regions synergistically with histone-like proteins. The YmoA protein has histone-like characteristics and represses yop gene expression. The YmoA protein is homologous to E. coli Hha; hha complements a ymoA mutant of Y. enterocolitica (18). Furthermore, hha has a repressor-like activity on E. coli hemolysin expression and has been shown to affect DNA supercoiling levels (1). Whether or not YmoA or Hha interact with bent DNA is unknown, but it has been reported that hha hns double mutants of E. coli have a synergistic effect on hemolysin expression (20), suggesting that this may be the case.
Taken together, these results suggest that at temperatures up to 30°C, the conformation of the pYV plasmid is maintained with a specific architecture involving bends, which, we envision, are stabilized by histone-like proteins. This three-dimensional configuration keeps virulence genes in a repressed state. After a shift to 37°C, the intrinsic bends melt and this architecture collapses. Such an event could have a significant effect on superhelical density, requiring compensatory supercoils to reestablish homeostasis. The altered state of the plasmid following such a transition, which occurs within 5 min after a temperature upshift, could promote formation of competent transcriptional complexes, such as the virF promoter and its target yop genes. This model would explain the transcriptional repression of yop genes at 30°C in Y. enterocolitica when VirF is overproduced artificially. Perturbations of plasmid architecture pharmacologically with gyrase inhibitors or genetically (i.e., ymoA or gyrase mutations) may account for the observed loss of temperature regulation under these two conditions.
This model could also account for temperature induction of virulence genes reported for other enteric pathogens. EPEC bundle-forming pili (bfp) expression, encoded by the EAF plasmid, is regulated by temperature (maximal expression occurs between 35 and 37°C). Pili expression requires the positive regulator, BfpT, an AraC homologue like Y. enterocolitica VirF. Puente et al. (23) noted the A and T tracts in the promoter region of this gene and their potential association with intrinsic bending. Two-dimensional gel analysis of the large EAF plasmid shows that this DNA also contains multiple intrinsic bends based on our assay (Fig. 7). Similar results have been obtained with the plasmid pO157H7 of EHEC. Additionally, temperature regulation and phase variation of E. coli pap transcription involves site-specific Dam methylation. At low temperature (25°C), pap expression is maintained in a phase-off configuration due to H-NS protection of Dam methylation sites (29). Based on our hypothesis, because H-NS preferentially binds bent DNA (21, 30), these H-NS-protected sites may become accessible to Dam methylase at high temperature (37°C), due to melting of intrinsic bends and subsequent loss of H-NS recognition. We likewise predict that the virulence plasmid of Shigella, encoding a temperature-inducible type III secretion system, has similar temperature-sensitive intrinsic bends.
The model presented here accounts for temperature activation of gene expression and suggests that the structure of DNA is the thermostat controlling events in early host adaptation. Regions of intrinsic bending within virF and additional temperature-regulated pYV genes are presently being precisely mapped and modified to test this model.
ADDENDUM
A similar model of temperature activation of virulence gene regulation for Shigella and E. coli was recently published by M. Falconi et al. (EMBO J. 17:7033–7043, 1998).
ACKNOWLEDGMENTS
This work was supported by the Idaho State Board of Education and the University of Idaho Agricultural Experiment Station.
We thank Ken Bayles and X. Chen for reviewing the manuscript. Helpful discussion was provided during the course of this work by Phil Youderian, Trish Hartzell, and Greg Bohach. We thank Carolyn Bohach for the EHEC and EPEC strains.
REFERENCES
- 1.Carmona M, Balsalobre C, Munoa F, Mourino M, Jubete Y, De la Cruz F, Juarez A. Escherichia coli hha mutants, DNA supercoiling and expression of the haemolysin genes from the recombinant plasmid pANN202-312. Mol Microbiol. 1993;9:1011–1018. doi: 10.1111/j.1365-2958.1993.tb01230.x. [DOI] [PubMed] [Google Scholar]
- 2.Chan S S, Breslauer K J, Austin R H, Hogan M E. Thermodynamics and premelting conformational changes of phased (dA)5 tracts. Biochemistry. 1993;32:11776–11784. doi: 10.1021/bi00095a005. [DOI] [PubMed] [Google Scholar]
- 3.Chan S S, Breslauer K J, Hogan M E, Kessler D J, Austin R H, Ojemann J, Passner J M, Wiles N C. Physical studies of DNA premelting equilibria in duplexes with and without homo dA.dT tracts: correlations with DNA bending. Biochemistry. 1990;29:6161–6171. doi: 10.1021/bi00478a008. [DOI] [PubMed] [Google Scholar]
- 4.Cornelis G, Laroche Y, Balligand G, Sory M-P. Yersinia enterocolitica, a primary model for bacterial invasiveness. Rev Infect Dis. 1987;9:64–87. doi: 10.1093/clinids/9.1.64. [DOI] [PubMed] [Google Scholar]
- 5.Cornelis G, Sluiters C, DeRouvroit C L, Michiels T. Homology between VirF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J Bacteriol. 1989;171:254–262. doi: 10.1128/jb.171.1.254-262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornelis G R, Sluiters C, Delor I, Geib D, Kaniga K, Lambert de Rouvroit C, Sory M-P, Vanooteghem J C, Michiels T. ymoA, a Yersinia enterocolitica gene modulating the expression of virulence determinants. Mol Microbiol. 1991;5:1023–1034. doi: 10.1111/j.1365-2958.1991.tb01875.x. [DOI] [PubMed] [Google Scholar]
- 7.Cornelis G R, Wolf-Watz H. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 8.Goldstein E, Drlica K. Regulation of bacterial DNA supercoiling: plasmid linking numbers vary with growth temperature. Proc Natl Acad Sci USA. 1984;81:4046–4050. doi: 10.1073/pnas.81.13.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoe N P, Minion F C, Goguen J D. Temperature sensing in Yersinia pestis: regulation of yopE transcription by lcrF. J Bacteriol. 1992;174:4275–4286. doi: 10.1128/jb.174.13.4275-4286.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hulton C S, Seirafi A, Hinton J C, Sidebotham J M, Waddell L, Pavitt G D, Owen-Hughes T, Spassky A, Buc H, Higgins C F. Histone-like protein H1 (H-NS), DNA supercoiling, and gene expression in bacteria. Cell. 1990;63:631–642. doi: 10.1016/0092-8674(90)90458-q. [DOI] [PubMed] [Google Scholar]
- 11.Jordi B J, Dagberg B, de Haan L A, Hamers A M, van der Zeijst B A, Gaastra W, Uhlin B E. The positive regulator CfaD overcomes the repression mediated by histone-like protein H-NS (H1) in the CFA/I fimbrial operon of Escherichia coli. EMBO J. 1992;11:2627–2632. doi: 10.1002/j.1460-2075.1992.tb05328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapatral V, Minnich S A. Co-ordinate, temperature-sensitive regulation of the three Yersinia enterocolitica flagellin genes. Mol Microbiol. 1995;17:49–56. doi: 10.1111/j.1365-2958.1995.mmi_17010049.x. [DOI] [PubMed] [Google Scholar]
- 13.Koo H-S, Wu H M, Crothers D. DNA bending at adenine and thymine tracts. Nature. 1986;320:501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- 14.Lambert de Rouvroit C, Sluiters C, Cornelis G R. Role of the transcriptional activator, VirF, and temperature in the expression of the pYV plasmid genes of Yersinia enterocolitica. Mol Microbiol. 1992;6:395–409. [PubMed] [Google Scholar]
- 15.Marini J C, Levene S D, Crothers D M, Englund P T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci USA. 1982;79:7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maurelli A T. Temperature regulation of virulence genes in pathogenic bacteria: a general strategy for human pathogens? Microb Pathog. 1989;7:1–10. doi: 10.1016/0882-4010(89)90106-x. [DOI] [PubMed] [Google Scholar]
- 17.Michiels T, Vanooteghem J-C, Lambert de Rouvroit C, China B, Gustin A, Boudry P, Cornelis G R. Analysis of virC, an operon involved in the secretion of Yop proteins by Yersinia enterocolitica. J Bacteriol. 1991;173:4994–5009. doi: 10.1128/jb.173.16.4994-5009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mikulskis A V, Cornelis G R. A new class of proteins regulating gene expression in enterobacteria. Mol Microbiol. 1994;11:77–86. doi: 10.1111/j.1365-2958.1994.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 19.Mizuno T. Random cloning of bent DNA from Escherichia coli chromosome and primary characterization of their structures. Nucleic Acids Res. 1987;15:6827–6841. doi: 10.1093/nar/15.17.6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nieto J M, Mourino M, Balsalobre C, Madrid C, Prenafeta A, Munoa F J, Juarez A. Construction of a double hha hns mutant of Escherichia coli: effect on DNA supercoiling and alpha-haemolysin production. FEMS Microbiol Lett. 1997;155:39–44. doi: 10.1111/j.1574-6968.1997.tb12683.x. [DOI] [PubMed] [Google Scholar]
- 21.Owen-Hughes T A, Pavitt G D, Santos D S, Sidebotham J M, Hulton C S J, Hinton J C D, Higgens C F. The chromatin-associated protein H-NS interacts with curved DNA to influence topology and gene expression. Cell. 1992;71:255–265. doi: 10.1016/0092-8674(92)90354-f. [DOI] [PubMed] [Google Scholar]
- 22.Perez-Martin J, de Lorenzo V. Clues and consequences of DNA bending in transcription. Annu Rev Microbiol. 1997;51:593–628. doi: 10.1146/annurev.micro.51.1.593. [DOI] [PubMed] [Google Scholar]
- 23.Puente J L, Bieber D, Ramer S W, Murray W, Schoolnik G K. The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol Microbiol. 1996;20:87–100. doi: 10.1111/j.1365-2958.1996.tb02491.x. [DOI] [PubMed] [Google Scholar]
- 24.Rohde J R, Fox J M, Minnich S A. Thermoregulation in Yersinia enterocolitica is coincident with changes in DNA supercoiling. Mol Microbiol. 1994;12:187–199. doi: 10.1111/j.1365-2958.1994.tb01008.x. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Slauch J M, Silhavy T J. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J Bacteriol. 1991;173:4039–4048. doi: 10.1128/jb.173.13.4039-4048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka K, Muramatsu S, Yamada H, Mizuno T. Systematic characterization of curved DNA segments randomly cloned from Escherichia coli and their functional significance. Mol Gen Genet. 1992;226:367–376. doi: 10.1007/BF00260648. [DOI] [PubMed] [Google Scholar]
- 29.White-Ziegler C A, Hill M L A, Braaten B A, van der Woude M W, Low D A. Thermoregulation of Escherichia coli pap transcription: H-NS is a temperature-dependent DNA methylation blocking factor. Mol Microbiol. 1998;28:1121–1137. doi: 10.1046/j.1365-2958.1998.00872.x. [DOI] [PubMed] [Google Scholar]
- 30.Yamada H, Muramatusu S, Mizuno T. An Escherichia coli protein that preferentially binds to sharply curved DNA. J Biochem. 1990;108:420–425. doi: 10.1093/oxfordjournals.jbchem.a123216. [DOI] [PubMed] [Google Scholar]