Abstract
Genes coding for putative RegA, RegB, and SenC homologues were identified and characterized in the purple nonsulfur photosynthetic bacteria Rhodovulum sulfidophilum and Roseobacter denitrificans, species that demonstrate weak or no oxygen repression of photosystem synthesis. This additional sequence information was then used to perform a comparative analysis with previously sequenced RegA, RegB, and SenC homologues obtained from Rhodobacter capsulatus and Rhodobacter sphaeroides. These are photosynthetic bacteria that exhibit a high level of oxygen repression of photosystem synthesis controlled by the RegA-RegB two-component regulatory system. The response regulator, RegA, exhibits a remarkable 78.7 to 84.2% overall sequence identity, with total conservation within a putative helix-turn-helix DNA-binding motif. The RegB sensor kinase homologues also exhibit a high level of sequence conservation (55.9 to 61.5%) although these additional species give significantly different responses to oxygen. A Rhodovulum sulfidophilum mutant lacking regA or regB was constructed. These mutants produced smaller amounts of photopigments under aerobic and anaerobic conditions, indicating that the RegA-RegB regulon controls photosynthetic gene expression in this bacterium as it does as in Rhodobacter species. Rhodobacter capsulatus regA- or regB-deficient mutants recovered the synthesis of a photosynthetic apparatus that still retained regulation by oxygen tension when complemented with reg genes from Rhodovulum sulfidophilum and Roseobacter denitrificans. These results suggest that differential expression of photosynthetic genes in response to aerobic and anaerobic growth conditions is not the result of altered redox sensing by the sensor kinase protein, RegB.
Many species of purple nonsulfur photosynthetic bacteria regulate the synthesis of their photosynthetic apparatus in response to alterations in oxygen tension and light intensity. Perhaps the best-characterized species with regard to oxygen control are Rhodobacter sphaeroides and Rhodobacter capsulatus, which are known to repress the synthesis of their photosystem almost completely in response to the presence of high levels of oxygen (13). The Rhodobacter response to oxygen contrasts with that of Rhodovulum sulfidophilum and Rhodospirillum centenum, which are known to repress photosystem synthesis only slightly when grown aerobically (17, 69). A group of obligate aerobic bacteria, represented in this study by Roseobacter denitrificans, have been found to synthesize a photosynthetic apparatus aerobically. Interestingly, these “aerobic photosynthetic bacteria” cannot utilize light as the sole energy source for growth. Instead, these organisms rely on respiration (56, 58, 59). Roseobacter denitrificans grows slowly as a result of anaerobic respiration in the presence of alternative electron acceptors such as trimethylamine N-oxide or nitrate (60). However, this bacterium synthesizes much less of the photosynthetic apparatus under these growth conditions than under aerobic conditions (data not shown) (58, 60). The two Rhodobacter species, Rhodovulum sulfidophilum, and Roseobacter denitrificans, belong to the α-3 subgroup of purple bacteria (proteobacteria) (30). Thus, there is a wide range of responses of closely related members of the subgroup to oxygen.
Although the molecular basis for different responses of photosynthetic bacteria to oxygen is unknown, previous studies with Rhodobacter capsulatus and R. sphaeroides have indicated that the molecular mechanism of the oxygen inhibition of photosystem synthesis is controlled, in large part, by regulating the transcription of photosynthesis genes. The regulation of photosynthetic gene expression by oxygen is controlled by many transcriptional factors (1–3, 6, 48, 51). One such factor, RegA, was identified in Rhodobacter capsulatus as a response regulator of a bacterial two-component system that anaerobically activates the light-harvesting and reaction center structural genes located in the puf, puh, and puc operons (42, 55). RegB is a histidine kinase protein that under anaerobic conditions phosphorylates the cognate response regulator RegA, resulting in the activation of puf, puh, and puc operon expression (31). Mutants lacking reg genes are unable to grow under low-intensity light conditions but can grow under high-intensity light conditions (55). In Rhodobacter sphaeroides, genes homologous to regA and regB were found (50) and named prrA and prrB, respectively (19, 20). In contrast to regA-deficient mutant of Rhodobacter capsulatus, the prrA-deficient mutant of R. sphaeroides was unable to grow phototrophically at any light intensity (19). The PrrA-PrrB regulon was shown to regulate puf, puh, and puc operon expression as well as the genes involved in CO2 reduction and synthesis of cytochrome c (19, 52).
It is still not known how RegB senses alterations in oxygen concentration. It was reported that Rhodobacter capsulatus and R. sphaeroides mutants lacking cytochrome oxidase cbb3 exhibit elevated photosynthesis gene expression under both anaerobic and aerobic conditions (9, 71). Cytochrome cbb3 is a dominant terminal cytochrome c oxidase under semiaerobic growth conditions in R. sphaeroides and a sole terminal cytochrome c oxidase in R. capsulatus (22, 64). Although no direct evidence was demonstrated, the electron transfer pathway involving cytochrome c oxidase was suggested to be an important signal for controlling the RegA-RegB phosphorelay cascade (9, 28, 44, 71).
The regulation of photosynthesis gene expression in species other than Rhodobacter has been examined only recently (26, 43). Transcription of the puf operon of Roseobacter denitrificans was observed in atmospheric oxygen tension (43), and that of the puc operon of Rhodovulum sulfidophilum was shown to be weakly repressed by oxygen (26), whereas these transcriptions were markedly suppressed by high-intensity light (26, 43). This is distinctly different from the results obtained with the Rhodobacter species, which show a high degree of repression by oxygen (>30-fold) and weak repression by light (∼2-fold). Thus, the expression of photosynthesis genes in species that can synthesize a photosystem under aerobic growth conditions is somewhat different from that in the Rhodobacter species (17, 26, 29, 43).
In this study, we have cloned and sequenced genes corresponding to regA and regB from the aerobic photosynthetic bacteria Rhodovulum sulfidophilum and Roseobacter denitrificans. Deletion analysis of these genes in Rhodovulum sulfidophilum and complementation analysis of Rhodobacter capsulatus reg mutations were carried out by using reg genes from Rhodovulum sulfidophilum and Roseobacter denitrificans. These results suggest that RegB is not the oxygen-sensing component controlling photosynthesis gene expression in these species.
MATERIALS AND METHODS
Bacteria and growth media.
The bacterial strains and plasmids used in this study are listed in Table 1. Rhodobacter capsulatus was anaerobically grown at 30°C in 30-ml screw-cap bottles filled with RCV medium (67). Rhodovulum sulfidophilum was grown under the same conditions as R. capsulatus in RCV medium supplemented with 2% sodium chloride. Roseobacter denitrificans was aerobically grown at 30°C in a medium containing yeast extract, polypeptone, Casamino Acids, and glycerol (59). Escherichia coli strains were grown at 37°C in a Luria-Bertani medium. Illumination was provided by 60-W tungsten lamps. Aerobic growth of R. capsulatus, Rhodovulum sulfidophilum, and Roseobacter denitrificans was achieved by shaking a 25-ml culture in a 250-ml conical flask at 200 rpm. Antibiotics, when necessary, were added to the E. coli culture to the following final concentrations: ampicillin, 100 μg/ml; kanamycin, 25 μg/ml; tetracycline, 20 μg/ml; spectinomycin, 100 μg/ml; and trimethoprim, 50 μg/ml; they were added to R. capsulatus cultures to the following concentrations: kanamycin, 10 μg/ml; streptomycin, 10 μg/ml; and spectinomycin, 10 μg/ml; and they were added to Rhodovulum sulfidophilum cultures at the following concentrations: kanamycin, 50 μg/ml; tetracycline, 3 μg/ml.
TABLE 1.
Bacteria and plasmids used in this study
| Strain or plasmid | Characteristic(s) | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 λ− | Bethesda Research Laboratories |
| JM109 | endA1 recA1 gyrA96 hsdR17 supE44 λ− Δ(lac-proAB)/F′ [traD36 proAB+ lacZΔM15] | 68 |
| JM109 λ pir | JM109 lysogenized with λ pir bacteriophage | 47 |
| S17-1 | Tpr SmrhsdR pro recA RP4-2-Tc::Mu-Km::Tn7 in chromosome | 61 |
| S17-1 λ pir | S17-1 lysogenized with λ pir bacteriophage | 15 |
| XL1-Blue MR. | recA1 endA1 gyr-96 thi-1 hsdR17 supE44 relA1 lac/F′ [proAB+ lacZΔM15::Tn10 (Tcr)] | 11 |
| C600/pDPT51 | Mobilizing vector | 63 |
| R. capsulatus | ||
| ATCC 11166 | Wild type | 49 |
| TB2 | SB1003 regAΔEcoRI-PflMI::Kmr | 7 |
| SD01 | SB1003 regBΔ640bp N-terminus::Kmr | 18 |
| R. sulfidophilum | ||
| W4 | Wild type | 27 |
| RESA1 | regAΔPinAI-PinAI::Kmr | This study |
| RESB20 | regB::Kmr | This study |
| R. denitrificans | ||
| OCh114 | Wild type | 57 |
| Plasmids | ||
| SuperCos 1 | Apr; cosmid vector | Stratagene |
| pUC118 | Apr; cloning vector | 68 |
| pJP5603 | Kmr; R6K-based suicide vector | 47 |
| pSM3065 | Tcr; pJP5603 derivative | This study |
| pUC7Tc | Apr Tcr; source of the Tcr gene | 33 |
| pUCKM1 | Apr Kmr; source of the Kmr gene | 53 |
| pCB532Ω | Apr Smr Spr; translational fusion of pufQ to lacZ | 4 |
| pJRD215 | Kmr Smr; broad-host-range cosmid vector | 14 |
| pSLA | Apr; 5.0-kb EcoRI fragment (R. sulfidophilum regB-senC-regA) in pUC118 | This study |
| pROA | Apr; 5.5-kb EcoRI fragment (R. denitrificans regB-senC-regA) in pUC118 | This study |
| pSLKm4 | Apr Kmr; 6.3-kb fragment (R. sulfidophilum regAΔPinAI-PinAI::Kmr) in pUC118 | This study |
| pSA10 | Apr Kmr Tcr; 9.4-kb fragment (pSLKm4 digested with KpnI) in pSM3065 | This study |
| pSB07 | Kmr; 0.4-kb SacI-SalI fragment (part of R. sulfidophilum regB) in pJP5603 | This study |
| pMWS3.1 | Kmr Smr; 12-kb fragment (R. capsulatus senC-regA) in pJRD215 | 55 |
| pCSM9e | Kmr Smr; 5.1-kb fragment (R. capsulatus regB-senC) in pJRD215 | 42 |
| pMCS003 | Kmr Smr; 2.8-kb EcoRI-NruI fragment (R. sulfidophilum senC-regA) in pJRD215 | This study |
| pMCS010 | Kmr Smr; 2.6-kb EcoRI-NdeI fragment (R. sulfidophilum regB) in pJRD215 | This study |
| pMCR001 | Kmr Smr; 3.2-kb EcoRI-SacI fragment (R. denitrificans senC-regA) in pJRD215 | This study |
| pMCR010 | Kmr Smr; 2.5-kb HincII fragment (R. denitrificans regB-senC) in pJRD215 | This study |
Preparation of the genomic library.
A genomic library of Rhodovulum sulfidophilum was constructed with the SuperCos 1 cosmid vector kit (Stratagene). The genomic DNA was partially digested by Sau3AI and ligated into the SuperCos 1 cosmid vector at the unique BamHI restriction site. After ligation, the cosmid was packaged into phage particles (Amersham) and then injected into E. coli XL1-Blue MR. Approximately 15,000 clones were pooled and amplified. Cells containing the clones were stored in 15% glycerol solution at −80°C. The genomic library of Roseobacter denitrificans was prepared by the same methods.
Screening and cloning of the regA and regB genes.
Genomic libraries of Rhodovulum sulfidophilum and Roseobacter denitrificans DNA were screened by colony hybridization. A 580-bp EcoRI-EcoRV fragment including the regA gene of Rhodobacter capsulatus (Fig. 1) was used as a probe after labeling with [α-32P]dATP, using a DNA Megalabel labeling kit (TaKaRa). The labeled probe was hybridized to the DNA on the membrane at 60°C. Colonies showing positive signals were isolated. Inserted DNA fragments (more than 40 kb) in cosmid vectors were digested with EcoRI and subcloned into a plasmid pUC118. Clones containing the regA gene were then screened by hybridization with the same probe as described in the cosmid screening and named pSLA (derived from Rhodovulum sulfidophilum DNA) and pROA (derived from Roseobacter denitrificans DNA). Plasmids pSLA and pROA contained 5.0 and 5.5 kb of inserted DNA, respectively (Fig. 1). DNA manipulation and hybridization were carried out by standard methods (37) or as instructed by the enzyme manufacturers.
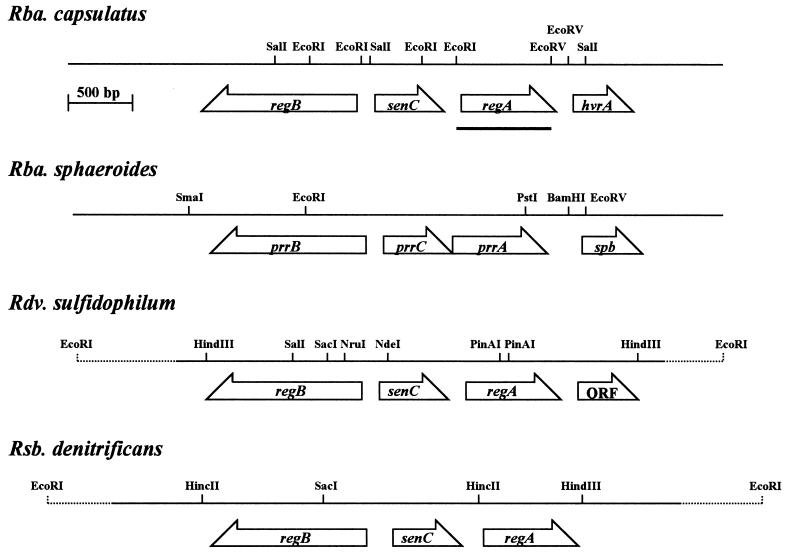
FIG. 1.
Physical and genetic maps of the photosynthetic regulatory gene cluster. ORFs and their directions of transcription are represented by open arrows. The broken lines shown in Rhodovulum (Rdv.) sulfidophilum and Roseobacter (Rsb.) denitrificans indicate the unsequenced regions. The DNA template used in the Southern hybridization is indicated by a thick line shown in Rhodobacter (Rba.) capsulatus. Rhodobacter (Rba.) sphaeroides prrB, prrC, prrA, and spb genes are thought to be equivalent to the homologous regB, senC, regA, and hvrA genes, respectively (see the text).
DNA sequencing.
A deletion kit (TaKaRa) was used to obtain the sequential series of overlapping DNA fragments. Nucleotide sequencing was performed with a 373A DNA sequencer with a Taq dye primer cycle-sequencing kit (Applied Biosystems). Some of the sequencing data were derived from synthetic oligonucleotide primers and a Taq dye terminator cycle-sequencing kit (Applied Biosystems). The data obtained were processed by using a DNASIS (Hitachi) sequence analysis program.
Construction of the Rhodovulum sulfidophilum regA-disrupted strain, RESA1.
For construction of strain RESA1, a suicide vector, pSM3065, was prepared as follows. Suicide vector pJP5603 (47) was digested with BglII and NcoI to isolate a fragment including RP4mob and R6Kori. The BglII-NcoI fragment was blunt ended with T4 DNA polymerase and then ligated with an HincII-HincII fragment containing a tetracycline resistance gene derived from plasmid pUC7Tc (33) to construct suicide vector pSM3065. The kanamycin cassette in the plasmid pUCKM1 (53) was inserted into the PinAI sites of the regA gene of Rhodovulum sulfidophilum in plasmid pSLA to create plasmid pSLKm4. The direction of transcription of the kanamycin resistance gene was the same as that of the regA gene in this construction. Both pSM3065 and pSLKm4 contain unique KpnI sites in the multiple-cloning sites. These two plasmids were digested with KpnI and ligated to construct a plasmid, pSA10. The plasmid was then transferred into Rhodovulum sulfidophilum cells by conjugation with the mobilizing strain S17-1 lysogenized with λpir (47, 61). Kmr Tcs cells were selected as double-crossover candidates, and the chromosomal insertion was confirmed by Southern hybridization. The mutant strain was designated RESA1.
Construction of the Rhodovulum sulfidophilum regB-disrupted strain, RESB20.
The SacI-SalI DNA fragment containing the internal region of regB of Rhodovulum sulfidophilum was cut out from plasmid pSLA and inserted into suicide vector pJP5603 (47) to construct a plasmid, pSB07. The plasmid was then transferred into R. sulfidophilum cells by conjugation with the mobilizing strain S17-1 lysogenized with λpir (47, 61). Cells resistant to kanamycin were selected as single-crossover candidates. The insertion of the plasmid into the chromosome was confirmed by Southern hybridization. The mutant strain was designated RESB20. The single-crossover event in RESB20 took place in the 5′ region of the SacI-SalI regB gene segment (Fig. 1). The direction of transcription of the kanamycin resistance gene was the same as that of the regB gene in the RESB20 chromosome.
Genetic manipulations.
For mobilizing the reporter plasmid pCB532Ω (4) containing a ColE1 origin of replication into Rhodobacter capsulatus, E. coli C600/pDPT51 (63) was used as a mobilizing strain. Plasmid derivatives of pJRD215 (14) were mobilized into R. capsulatus by conjugation with the mobilizing strain S17-1 (61).
Spectral and protein analysis.
Membranes for absorption spectrum measurements were obtained by sonicating cells grown to the mid-logarithmic phase and measuring them with a Shimazu UV 160 spectrophotometer. The bacteriochlorophyll content in the cell suspension was determined with acetone-methanol (7:2) extract as described previously (12). Protein content determination was performed with two assay kits from Bio-Rad (kits 500-0001 and 500-0111) as specified by the manufacturer. The β-galactosidase activity of Rhodobacter capsulatus cells containing a reporter plasmid for gene expression was determined as described by Young et al. (70).
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper are available in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. AB010722 (Rhodovulum sulfidophilum) and AB010723 (Roseobacter denitrificans).
RESULTS
Cloning and sequence analysis of the photosynthesis regulatory gene cluster of Rhodovulum sulfidophilum and Roseobacter denitrificans.
A cosmid DNA library derived from Rhodovulum sulfidophilum and Roseobacter denitrificans chromosomal DNA was probed with a Rhodobacter capsulatus DNA fragment containing regA. DNA fragments in several cosmid clones that gave positive hybridization signals were then subcloned into pUC118. Cloned DNA fragments derived from Rhodovulum sulfidophilum (pSLA) and Roseobacter denitrificans (pROA) were subsequently sequenced (DDBJ, EMBL, and GenBank accession no. AB010722 and AB010723, respectively). Analysis of the primary structures of these nucleotide sequences indicated that these DNA fragments contained at least three open reading frames (ORFs). One ORF showed high sequence identity to the regA gene of Rhodobacter capsulatus (84.2% in Rhodovulum sulfidophilum and 78.7% in Roseobacter denitrificans), while the other two ORFs showed significant sequence identity to regB (56.6% in Rhodovulum sulfidophilum and 55.9% in Roseobacter denitrificans) and to the senC genes of R. capsulatus (51.0% in Rhodovulum sulfidophilum and 47.8% in Roseobacter denitrificans), respectively. The initiation codon for regB of Roseobacter denitrificans is assumed to be GTG based on a comparison of the amino-terminal portion of the sequence with prrB of Rhodobacter sphaeroides and on codon usage after the GTG similar to that observed with the reaction center-core protein genes of Roseobacter denitrificans (36). Although the GTG is an irregular initiation codon, translation initiating from GTG has been found for other proteins (35).
The relative order and directions of the putative regA, regB, and senC were the same in Rhodovulum sulfidophilum and Roseobacter denitrificans as in Rhodobacter capsulatus and R. sphaeroides (Fig. 1) (10, 20, 42). The ORF found downstream of regA in Rhodovulum sulfidophilum has no significant homology to any proteins that have been reported, although Rhodobacter capsulatus and R. sphaeroides have hvrA and spb at that position, respectively, which function as light-responding trans-acting factors for photosynthetic gene expression (10, 40). This result suggests that the counterpart of hvrA or spb has been lost or is located at a distance from the regA-regB gene cluster in Rhodovulum sulfidophilum.
Analysis of RegA sequences.
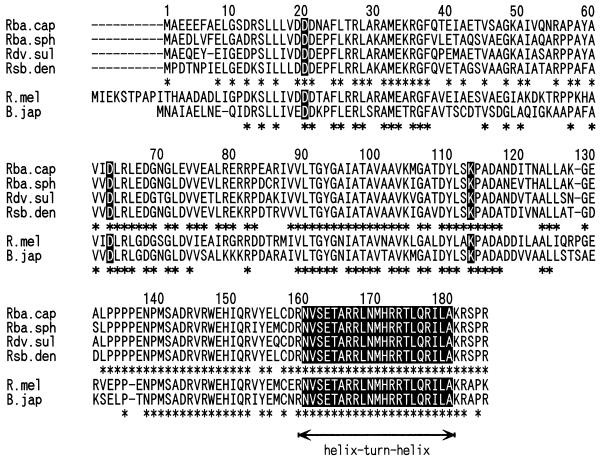
Figure 2 shows the similarity of RegA homologues to those of ActR of Rhizobium meliloti (65) and RegR of Bradyrhizobium japonicum (5). These are response regulator proteins thought to be involved in sensing low pH and controlling nitrogen fixation-associated genes, respectively, to which RegA has a high homology score (5, 65). As demonstrated by the alignment, RegA homologues from Rhodobacter capsulatus, Roseobacter denitrificans, and Rhodobacter sphaeroides exhibit an identical sequence length of 184 amino acid residues whereas RegA from Rhodovulum sulfidophilum lacks only one residue in the amino-terminal region. All of the RegA homologues from photosynthetic bacteria exhibit a high degree of identity throughout their length (78.7 to 84.2%), including the characteristic two Asp residues (positions 20 and 63) and a Lys residue (position 113). These residues are thought to play central roles in phosphorylation, which affects protein activity (19, 24, 55, 65). The RegA homologues also exhibit a conserved series of four prolines (positions 133 to 136), two of which are also conserved in ActR from R. meliloti. This region is immediately followed by a carboxyl-terminal region (positions 137 to 185) exhibiting 93% identity among the RegA homologues. In this region, amino acids 160 to 180 have considerable sequence similarity to known helix-turn-helix DNA-binding motifs (Fig. 2) (5, 18, 38). The putative DNA-binding motif contains an unprecedented 100% sequence identity among RegA homologues in each of these species. Conserved amino acids, which have been suggested to be important in the DNA-binding motifs (16), include Glu at position 163, Ala at position 165, Leu at positions 168, Thr at position 174, and Arg at position 177. As calculated by an amino-acid-versus-position scoring matrix for the evaluation of the helix-turn-helix motif (16), the SD score of the RegA homologues is 5.4, which is well within the range observed for known DNA-binding proteins (2.5 to 7.1) (16).
FIG. 2.
Alignment of amino acid sequences of RegA of Rhodobacter capsulatus (Rba. cap) (55), Rhodovulum sulfidophilum (Rdv. sul), and Roseobacter denitrificans (Rsb. den), PrrA of Rhodobacter sphaeroides (Rba. sph) (19), ActR of Rhizobium meliloti (R. mel) (65), and RegR of Bradyrhizobium japonicum (B. jap) (5). Asterisks indicate identical amino acids. Residues considered functional and a predicted helix-turn-helix DNA-binding motif are boxed.
Analysis of the RegB sequence.
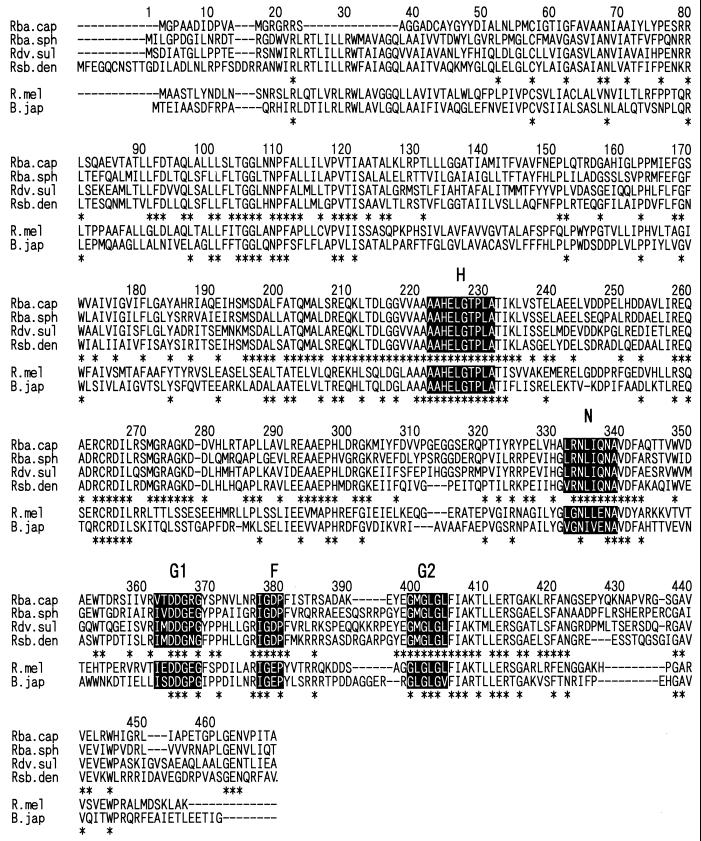
The RegB homologues from all four photosynthetic bacteria are less highly conserved (55.9 to 61.5%) than are the RegA homologues (78.7 to 84.2%). However, sequence conservation is still quite high relative to that observed among other sensor kinases (covered in more detail in Discussion). Figure 3 shows the amino acid sequence alignment of RegB of Rhodobacter capsulatus, Rhodovulum sulfidophilum, and Roseobacter denitrificans and of PrrB of Rhodobacter sphaeroides and ActS and RegS, which are thought to function as sensor kinase proteins responsible for phosphorylating ActR in R. meliloti and RegR in B. japonicum, respectively (5, 65). The region designated the H block, which contains His-224 and has been suggested to be an autophosphorylation site, is highly conserved throughout the RegB homologues and in ActS and RegS (20, 42, 45, 46). Blocks G1, G2, F, and N, known to be conserved in other sensor kinases (45, 46), are also highly conserved. Blocks G1 and G2 are considered the ATP-binding domain because they resemble glycine-rich portions of nucleotide-binding domains (46). Since mutations in block N, G1, or G2 eliminate the autokinase activity of the osmolarity sensor EnvZ in E. coli (32), these blocks are likely to be necessary for the autokinase activity of RegB homologues from the four photosynthetic bacteria as well as of ActS and RegS. Besides the well-characterized G1, G2, F, and N blocks, the cytosolic portion of the protein (C-terminal half) showed extensive sequence similarity among RegB homologues. Some of these homologous portions presumably represent areas involved in docking with RegA, which, as discussed below, is highly conserved.
FIG. 3.
Alignment of amino acid sequences of RegB of Rhodobacter capsulatus (Rba. cap) (42), Rhodovulum sulfidophilum (Rdv. sul), and Roseobacter denitrificans (Rsb. den), PrrB of Rhodobacter sphaeroides (Rba. sph) (20), ActS of Rhizobium meliloti (R. mel) (65), and RegS of Bradyrhizobium japonicum (B. jap) (5). Asterisks indicate identical amino acids. The histidine block (H), an asparagine-rich block (N), glycine-rich domains (G1 and G2), and a variable-length spacer (F) which are roughly conserved in the histidine kinases (45, 46) are boxed.
Analysis of SenC sequences.
Additional genes, i.e., senC in Rhodobacter capsulatus (9) and prrC in R. sphaeroides (20), are also known to be located between regB and regA in these species. As indicated in Fig. 1, senC is also present in a similar position between regB and regA in Rhodovulum sulfidophilum and in Roseobacter denitrificans. The SenC homologues from the four photosynthetic bacteria exhibit 37.4 to 51.0% sequence identity, which is significantly lower than that observed with RegA or RegB homologues. The senC gene product was also previously observed to have high sequence identity to a yeast nucleus-encoded protein, SCO1 (9, 20), which is thought to be an element of the assembly of cytochrome c oxidase in yeast (8, 54). The amino acid sequence alignment of SenC homologues from the four photosynthetic bacteria to SCO1 is shown in Fig. 4. Notable areas of conservation include a very hydrophobic patch among the 38 amino acid residues at the amino-terminal end, as well as a putative iron-binding domain at positions 83 to 89 (9).
FIG. 4.
Alignment of amino acid sequences of SenC of Rhodobacter capsulatus (Rba. cap) (9), Rhodovulum sulfidophilum (Rdv. sul), and Roseobacter denitrificans (Rsb. den), PrrC of Rhodobacter sphaeroides (Rba. sph) (20), and yeast nucleus-encoded protein SCO1 (8, 54). Asterisks indicate identical amino acids. The residues hypothesized to contribute to a predicted iron-binding domain are boxed (9).
Effects of regA and regB disruptions in Rhodovulum sulfidophilum.
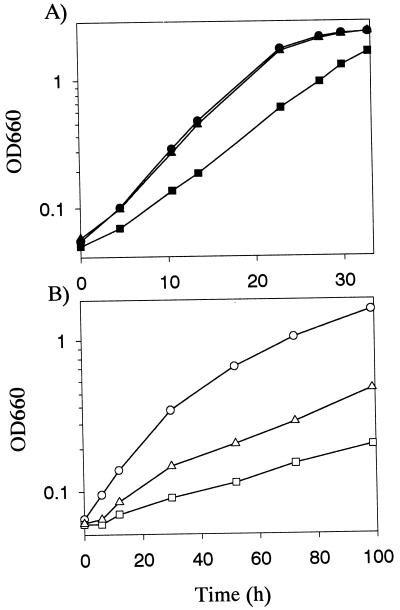
Rhodovulum sulfidophilum regA- and regB-defective mutants were constructed and named RESA1 and RESB20, respectively (see Materials and Methods). The profiles of the photosynthetic growth of these mutants and wild-type Rhodovulum sulfidophilum are shown in Fig. 5. Under high-intensity light conditions (100 W/m2 [Fig. 5A]), RESA1 grew more slowly, with a doubling time of 6.7 h, than wild-type cells, with a doubling time of 4.5 h. The phenotypes were more pronounced when assayed under low-intensity light conditions (3 W/m2 [Fig. 5B]), when the doubling times of RESB20 and RESA1 were three and four times as long, respectively, as that of wild-type cells. These observations are similar to those for Rhodobacter capsulatus mutants in terms of the ability of photosynthetic growth of the regA defective mutant and are different from that of the R. sphaeroides regA (prrA) mutant, which was unable to grow photosynthetically at any light intensity (19, 55).
FIG. 5.
Growth profiles of Rhodovulum sulfidophilum wild-type, regA-disrupted (RESA1), and regB-disrupted (RESB20) strains. Cells were grown under anaerobic high-intensity light conditions (100 W/m2) (solid circles, wild-type cells; solid squares, RESA1 cells; solid triangles, RESB20 cells) (A) and anaerobic low-intensity light conditions (3 W/m2) (open circles, wild-type cells; open squares, RESA1 cells; open triangles, RESB20) (B).
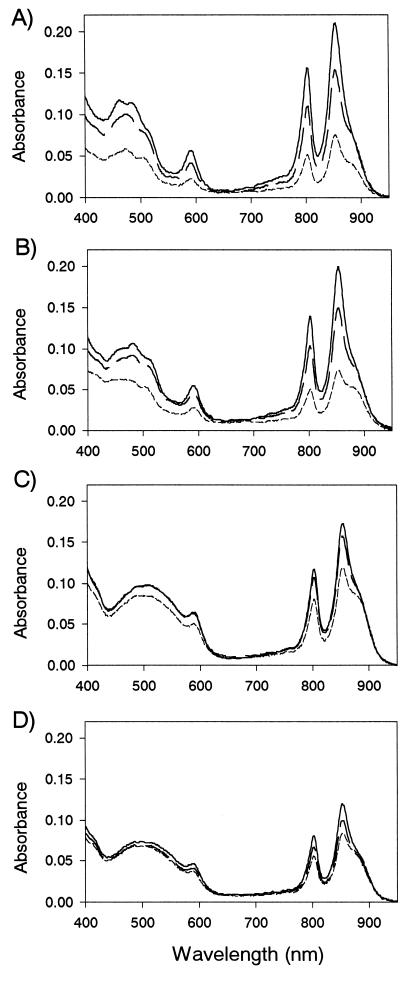
Figure 6 shows the absorption spectra of membranes of Rhodovulum sulfidophilum wild-type, RESA1, and RESB20 cells grown under anaerobic low-intensity light (3 W/m2) (Fig. 6A), anaerobic high-intensity light (100 W/m2) (Fig. 6B), aerobic-dark (Fig. 6C), and aerobic high-intensity light (100 W/m2) (Fig. 6D) conditions. The RESA1 mutant showed reduced synthesis of photopigments compared to that of the wild type under all of these growth conditions. In the RESB20 mutant, the synthesis of photopigments was also reduced to a lesser extent than compared in RESA1. The bacteriochlorophyll (BChl) content per membrane protein of RESA1 and RESB20 cells grown under anaerobic high-intensity light conditions was 40 and 72%, respectively, of that of the wild-type cells. Under aerobic-dark conditions, these mutants synthesized 78 and 91% of the wild-type levels, respectively. These differences between two mutants support the idea that RegA may be phosphorylated via cross talk by a protein kinase other than RegB (23, 42). The effects of the mutations on the expression of light-harvesting complex 2 (LH2), which peaked at 800 and 850 nm, were more pronounced than on the expression of light-harvesting complex 1 (LH1), which appeared as a shoulder at 870 nm (Fig. 6). This observation is similar to that for the Rhodobacter species (19, 55).
FIG. 6.
Absorption spectra of Rhodovulum sulfidophilum wild type (solid lines), RESA1 (short dashed lines) and RESB20 (long dashed lines) grown under anaerobic low-intensity light conditions (3 W/m2) (A), anaerobic high-intensity light conditions (100 W/m2) (B), aerobic-dark conditions (C), and aerobic high-intensity light conditions (100 W/m2) (D). Cells in the mid-logarithmic growth phase were harvested and sonicated, and spectra were measured in membrane preparations. All samples contained 75 μg of protein per ml.
Complementation analysis of reg genes among different species.
We carried out a complementation analysis of Rhodobacter capsulatus reg mutations by using reg genes from Rhodovulum sulfidophilum and Roseobacter denitrificans to examine whether these genes are responsible for the different aerobic and anaerobic expression patterns of photosynthesis genes between Rhodobacter capsulatus, Rhodovulum sulfidophilum, and Roseobacter denitrificans. DNA fragments containing regA or regB from Rhodobacter capsulatus, Rhodovulum sulfidophilum, and Roseobacter denitrificans were cloned into wide-host-range vectors (Table 1) and introduced into the Rhodobacter capsulatus regA or regB disruption mutants TB2 and SD01, respectively. As shown in Table 2, TB2 transconjugants containing the plasmid carrying regA and senC from Rhodobacter capsulatus (pMWS3.1), Rhodovulum sulfidophilum (pMCS003), and Roseobacter denitrificans (pMCR001) all complemented the chromosomal disruption of regA. Similar results were obtained by complementing the regB defect in SD01 with plasmids containing regB and senC from Rhodobacter capsulatus (pCSM9e) and Roseobacter denitrificans (pMCR010) and regB from Rhodovulum sulfidophilum (pMCS010) (Table 2). Furthermore, all of the Rhodobacter capsulatus transconjugants retained the aerobic repression of photosynthesis (puf) gene expression regardless of the species origin of the complementing regA or regB gene (Table 2). The notable difference observed with all of the transconjugants was an increased aerobic level of puf expression in comparison to wild-type cells. This may be due to an increased copy number of regA or regB genes in the cells as a result of being plasmid located, as reported previously (21). Another possible explanation of this difference is that the presence of genes other than regA and regB, such as senC, affects puf operon expression. The increase of puf expression in comparison to wild-type cells was especially prominent in TB2 and SD01 transconjugants containing Rhodovulum sulfidophilum regA-senC and Rhodobacter capsulatus regB-senC, respectively. The puf expression in these transconjugants was, however, still highly repressed by oxygen.
TABLE 2.
Photopigment synthesis and photosynthetic gene expression in reg mutants of Rhodobacter capsulatus complemented with reg genes from various species
| Strain | BChl content (nmol/mg)c
|
Expression of puf operond
|
||
|---|---|---|---|---|
| −O2 | +O2 | −O2 | +O2 | |
| Wild type (R. capsulatus) | 34.5 | 0.5 | 544 ± 30 | 15 ± 5 |
| TB2(pJRD215)a vector only | 13.9 | 0.3 | 40 ± 10 | 9 ± 5 |
| TB2(pMWS3.1) +Rhodobacter regA-senC | 37.2 | 0.7 | 382 ± 50 | 62 ± 20 |
| TB2(pMCS003) +Rhodovulum regA-senC | 51.5 | 0.6 | 1,971 ± 150 | 125 ± 25 |
| TB2(pMCR001) +Roseobacter regA-senC | 42.5 | 0.6 | 315 ± 30 | 60 ± 15 |
| SD01(pJRD215)b vector only | 21.6 | 0.5 | 135 ± 70 | 18 ± 10 |
| SD01(pCSM9e) +Rhodobacter regB-senC | 42.8 | 0.5 | 1,050 ± 145 | 108 ± 40 |
| SD01(pMCS010) +Rhodovulum regB | 45.7 | 0.5 | 855 ± 150 | 60 ± 20 |
| SD01(pMCR010) +Roseobacter regB-senC | 40.1 | 0.4 | 499 ± 125 | 63 ± 25 |
TB2; R. capsulatus (ΔregA).
SD01; R. capsulatus (ΔregB).
BChl content in membranes (nanomoles per milligram of membrane protein). Data are based on the average of three independent assays. Uncertainty limits in this assay are within 5% in all transconjugants and the wild type. −O2, anaerobic photosynthetic growth; +O2, aerobic-dark growth.
Values are β-galactosidase activity (micromoles of o-nitrophenol-β-d-galactoside hydrolyzed per minute per milligram of protein) of strains with the puf::lacZ translational fusion in pCB532Ω (4).
DISCUSSION
The results of this study demonstrate that the regA-senC-regB regulatory gene cluster is present in photosynthetic bacteria that aerobically synthesize the photosynthetic apparatus. As shown in Fig. 2, RegA homologues are highly conserved, with over 78% identity among the four photosynthetic bacteria and over 60% identity to ActR from R. meliloti and RegR from B. japonicum. This is in contrast to the observation that the average identity between two response regulators is 23% based on pairwise alignments of 79 response regulators that have different functions (for example, PhoB and OmpR) (66). The identity score improves only slightly in comparisons of response regulators from different species that have the same function. For example, the identity score is 26% for NtrC homologues, 24% for FixJ homologues, and 37% for OmpR homologues. Thus, the RegA homologues exhibit an unprecedented level of conservation, which suggests that there are significant constraints on the ability of RegA to undergo mutational changes in these species. This could be a function of the diverse roles of RegA (PrrA), which is known to be involved in controlling the expression of the photosynthetic apparatus as well as that of the cytochrome c2 gene and carbon fixation (2, 19, 52).
Comparison of the amino acid sequences of RegA homologues with that of CheY, a well-known response regulator of bacterial chemotaxis (66), shows that at the precise location where CheY ends, the RegA homologues from photosynthetic bacteria contain a stretch of four prolines, presumably providing a flexible region. The stretch of prolines is followed by a highly conserved carboxyl-terminal region (amino acids 137 to 185) exhibiting 93.6% sequence identity among the photosynthetic homologues and 89.8% when including ActR and RegR. An alignment of the conserved carboxyl-terminal region with known DNA-binding sequences (16) reveals a region (NVSETARRLNMHRRTLQRILA) suggestive of a helix-turn-helix DNA-binding motif. This sequence is 100% conserved among the various RegA homologues that have been cloned. Assuming that this region does indeed contain a DNA-binding motif, the remarkable 100% level of sequence identity in this region would indicate that the target DNA sequences are highly conserved in these different species.
Most sensor kinases exhibit significant homology only in the conserved G1, G2, F, N, and H domains located in the cytosolic C-terminal region (Fig. 3). Not surprisingly, the alignment of RegB in Fig. 3 indicates that the six RegB homologues also exhibit extensive homology in these domains. However, further inspection of the alignment indicates that there also exist several additional regions with significant conservation, including extensive sequence conservation (93% identity) in the Q-linker region (positions 196 to 221), which is an area located immediately upstream of the H domain. There is also an additional area of extensive sequence conservation located between the H and N domains (positions 258 to 303). Given that RegA has a very high degree of homology, these additional areas of RegB conservation could represent RegA-docking domains. Indeed, the area between the H and N domains has previously been implicated in the docking of the response regulator CheY to the sensor kinase CheA (62).
There is also a well-conserved (73% identity) region in the amino-terminal membrane-spanning region of RegB (amino acids 91 to 113). This region could be involved in redox signal transduction or in the formation of stable dimers (45, 46). The amino acid sequences of the membrane-spanning region of RegB homologues showed no significant similarities to those of the oxygen-related sensor kinase FixL or the redox-sensitive sensor kinase ArcB (25, 41). The S-boxes that are present in a large family of proteins thought to be involved in sensing the oxygen-redox potential (72) are not present in RegB, whereas they are reported to be present in FixL and ArcB. The heme-binding site located between the membrane-spanning region and the kinase domain in FixL is not located in any of the RegB homologues either.
The occurrence of senC (prrC) between regA and regB in each of the four photosynthetic bacteria suggests an important role for senC in the cascade of the regulation mechanisms of photosynthetic gene expression via the RegA-RegB phosphorelay circuit. Horne et al. (28) suggested that the senC homologue from Rhodobacter sphaeroides senses the redox state of cytochrome c, and this would indirectly influence the PrrB (RegB) activity. Buggy and Bauer (9) demonstrated that strains disrupted in SenC had reduced levels of cytochrome c oxidase as well as of puf, puc, and puh expression. They suggested that SenC may be involved in controlling RegB phosphorylation activity in response to alterations in cytochrome c oxidase activity (9). Clearly, the conservation of senC in each of these photosynthetic species indicates that it plays an important role deserving of further study.
The disruption of regA and regB causes the reduction of photopigment accumulation in Rhodovulum sulfidophilum, indicating that the RegA-RegB regulon plays important roles in photosystem synthesis in this species, as it does as in Rhodobacter species (Fig. 6). Recently, RegA was shown to bind to the promoter regions for the puf (−22 to −80) and puc (−52 to −80) operons in Rhodobacter capsulatus (18). The nucleotide sequence of the Rhodovulum sulfidophilum puc operon was determined and shown to have a similar promoter sequence (−77 to −60) to that of Rhodobacter capsulatus puc operon (26). We sequenced the whole puf operon of Rhodovulum sulfidophilum and found that it contains a similar sequence to the RegA-binding site of the Rhodobacter capsulatus puf operon (39). These findings suggest that the RegA homologue from Rhodovulum sulfidophilum also binds to the puf and puc operon promoters.
Because no consensus sequence was found around the 5′ ends of puf mRNA, the regulation of the transcription of the puf operon in Roseobacter denitrificans has been suggested to be different from that in the Rhodobacter species (43). However, because the transcription initiation sites of the Rhodobacter capsulatus puf operon were far upstream from the stable 5′ end of the puf mRNA (4), the transcription starting point of puf operon in Roseobacter denitrificans may also exist far upstream of the 5′ ends of the stable puf mRNA transcripts. If so, a similar sequence to the RegA-binding site of Rhodobacter capsulatus may be present in its regulatory region. Recently, it was reported that the Roseobacter denitrificans puf operon could be expressed in Rhodobacter capsulatus under the control of its promoter (34). This supports the idea that the puf operon promoter of Roseobacter denitrificans is similar to that of Rhodobacter capsulatus.
Less pigment-protein complex is formed in the RESA1 mutant grown under anaerobic-light conditions than in that grown under aerobic-dark conditions, while the wild type produces more pigment-protein complex under anaerobic-light conditions than under aerobic-dark conditions (Fig. 6A to C). The mutants exhibit a 1.8-fold-higher BChl content per membrane protein under aerobic-dark conditions than under anaerobic high-intensity light conditions. These findings indicate that activation of photosynthetic gene expression caused by the RegA-RegB regulon is more apparent under anaerobic than aerobic conditions. In addition, both the wild type and the RESA1 mutants produce smaller amounts of photopigments under aerobic-light than aerobic-dark conditions (Fig. 6C and D), indicating that the aerobic regulatory system which controls the photosystem construction responding to light intensity seems to be present in Rhodovulum sulfidophilum.
When Rhodobacter capsulatus regA or regB mutants were complemented with regA or regB genes from Rhodovulum sulfidophilum and Roseobacter denitrificans, they produced photosynthetic pigments under anaerobic conditions but not under aerobic conditions, which is the pattern observed with wild-type Rhodobacter capsulatus cells (Table 2). In all transconjugants, expression of the puf operon under aerobic conditions increased compared to that of the wild type, although the BChl contents in the transconjugants under aerobic conditions were almost minimal and did not reflect the increased aerobic puf operon expression. This result indicates that BChl levels continue to be limiting in all transconjugants under aerobic conditions. This may be due to low levels of expression of the genes involved in tetrapyrrole and BChl biosynthesis, which require other regulatory proteins for activation (1). Indeed, all of the transconjugants exhibited a similar 5- to 10-fold-higher level of puf operon expression under anaerobic than aerobic growth conditions (Table 2). Not only does this result indicate functional complementation but also it indicates that RegB from bacteria that aerobically synthesize photopigments also responds to oxygen tension when expressed in Rhodobacter capsulatus. There are several interpretations for this finding. One possibility is that Rhodovulum sulfidophilum and Roseobacter denitrificans cells retain similar reduced states under aerobic and anaerobic growth conditions via a metabolic quirk, such as a high level of respiration, which could effectively scrub out oxygen from these cells. Another possibility is that RegB from Rhodovulum sulfidophilum and Roseobacter denitrificans are incapable of a redox response in their native species but are capable of a redox response in Rhodobacter capsulatus. Perhaps the most intriguing possibility is that RegB is not itself a redox-responding sensor kinase. Instead, its activity may be affected by interacting with another redox-responding protein that is present only in Rhodobacter capsulatus. Alternatively, Rhodobacter capsulatus (but not Rhodovulum sulfidophilum or Roseobacter denitrificans) may have a phosphatase that removes phosphate from RegA in a redox-responsive manner. Clearly, additional in vivo and in vitro studies of RegB activity from these species must be undertaken. Such comparative studies should be useful in clarifying the details of the control mechanisms of anaerobic gene expression in purple bacteria.
ACKNOWLEDGMENTS
We are grateful to Ken-ichiro Takamiya (Tokyo Institute of Technology) for suggestions, Alastair G. McEwan (University of Queensland) for the provision of the E. coli strains used in this study, and Teruya Komano (Tokyo Metropolitan University) for the gift of plasmid pUC7Tc.
This work was supported in part by grants from the Ministry of Education, Science, and Culture of Japan.
REFERENCES
- 1.Bauer C E. Regulation of photosynthesis gene expression. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1221–1234. [Google Scholar]
- 2.Bauer C E, Bird T H. Regulatory circuits controlling photosynthesis gene expression. Cell. 1996;85:5–8. doi: 10.1016/s0092-8674(00)81074-0. [DOI] [PubMed] [Google Scholar]
- 3.Bauer C E, Buggy J, Mosley C. Control of photosystem genes in Rhodobacter capsulatus. Trends Genet. 1993;9:56–60. doi: 10.1016/0168-9525(93)90188-N. [DOI] [PubMed] [Google Scholar]
- 4.Bauer C E, Young D A, Marrs B L. Analysis of the Rhodobacter capsulatus puf operon. J Biol Chem. 1988;263:4820–4827. [PubMed] [Google Scholar]
- 5.Bauer E, Kaspar T, Fischer H, Hennecke H. Expression of the fixR-nifA operon in Bradyrhizobium japonicum depends on a new response regulator, RegR. J Bacteriol. 1998;180:3853–3863. doi: 10.1128/jb.180.15.3853-3863.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beatty J T. Organization of photosynthesis gene transcripts. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1209–1219. [Google Scholar]
- 7.Bird, T., and C. E. Bauer. Unpublished strain construction.
- 8.Buchwald P, Krummeck G, Rodel G. Immunological identification of yeast SCOI protein as a component of the inner mitochondrial membrane. Mol Gen Genet. 1991;229:413–420. doi: 10.1007/BF00267464. [DOI] [PubMed] [Google Scholar]
- 9.Buggy J J, Bauer C E. Cloning and characterization of senC, a gene involved in both aerobic respiration and photosynthesis gene expression in Rhodobacter capsulatus. J Bacteriol. 1995;177:6958–6965. doi: 10.1128/jb.177.23.6958-6965.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buggy J J, Sganga M W, Bauer C E. Characterization of a light-responding trans-activator responsible for differentially controlling reaction center and light-harvesting I gene expression in Rhodobacter capsulatus. J Bacteriol. 1994;176:6936–6943. doi: 10.1128/jb.176.22.6936-6943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bullok W O, Fernandez J M, Short J M. XL1-Blue, a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–379. [Google Scholar]
- 12.Clayton R K. Spectroscopic analysis of bacteriochlorophylls in vitro and in vivo. Photochem Photobiol. 1966;5:669–677. [Google Scholar]
- 13.Cohen-Bazire G, Sistrom W R, Stanier R Y. Kinetic studies of pigment synthesis by non-sulfur purple photosynthetic bacteria. J Cell Comp Physiol. 1957;49:25–68. doi: 10.1002/jcp.1030490104. [DOI] [PubMed] [Google Scholar]
- 14.Davison J, Heusterspreute M, Chevalier N, Ha-Thi V, Brunel F. Vectors with restriction site banks. V. pJRD215, a wide-host-range cosmid vector with multiple cloning sites. Gene. 1987;51:275–280. doi: 10.1016/0378-1119(87)90316-7. [DOI] [PubMed] [Google Scholar]
- 15.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dodd I B, Egan J B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990;18:5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doi M, Shioi Y, Gad’on N, Golecki J R, Drews G. Spectroscopical studies on the light-harvesting pigment-protein complex II from dark-aerobic and light-anaerobic grown cells of Rhodobacter sulfidophilus. Biochim Biophys Acta. 1991;1058:235–241. [Google Scholar]
- 18.Du S, Bird T H, Bauer C E. DNA-binding characteristics of RegA*: a constitutively active anaerobic activator of photosynthesis gene expression in Rhodobacter capsulatus. J Biol Chem. 1998;273:18509–18513. doi: 10.1074/jbc.273.29.18509. [DOI] [PubMed] [Google Scholar]
- 19.Eraso J M, Kaplan S. prrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J Bacteriol. 1994;176:32–43. doi: 10.1128/jb.176.1.32-43.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eraso J M, Kaplan S. Oxygen-insensitive synthesis of the photosynthetic membranes of Rhodobacter sphaeroides: a mutant histidine kinase. J Bacteriol. 1995;177:2695–2706. doi: 10.1128/jb.177.10.2695-2706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eraso J M, Kaplan S. Complex regulatory activities associated with the histidine kinase PrrB in expression of photosynthesis genes in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1996;178:7037–7046. doi: 10.1128/jb.178.24.7037-7046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Horsman J A, Barquera B, Rumbley J, Ma J, Gennis R B. The superfamily of heme-copper respiratory oxidase. J Bacteriol. 1994;176:5587–5600. doi: 10.1128/jb.176.18.5587-5600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomelsky M, Kaplan S. Isolation of regulatory mutants in photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1 and partial complementation of a PrrB mutant by the HupT histidine-kinase. Microbiology. 1995;141:1805–1819. doi: 10.1099/13500872-141-8-1805. [DOI] [PubMed] [Google Scholar]
- 24.Gudrun S L, Binjamin H L, James M M, Ann M S, Jeffry B S. Roles of the highly conserved aspartate and lysine residues in the response regulator of bacterial chemotaxis. J Biol Chem. 1991;266:8348–8354. [PubMed] [Google Scholar]
- 25.Gunsalus R P, Park S J. Aerobic-anaerobic regulation in Escherichia coli: control by the ArcAB and Fnr regulons. Res Microbiol. 1994;145:437–450. doi: 10.1016/0923-2508(94)90092-2. [DOI] [PubMed] [Google Scholar]
- 26.Hagemann G E, Katsiou E K, Forkl H, Steindorf A C J, Tadros M H. Gene cloning and regulation of gene expression of the puc operon from Rhodovulum sulfidophilum. Biochim Biophys Acta. 1997;1351:341–358. doi: 10.1016/s0167-4781(96)00228-x. [DOI] [PubMed] [Google Scholar]
- 27.Hansen T A, Veldkamp H. Rhodopseudomonas sulfidophila, nov. spec., a new species of the purple nonsulfur bacteria. Arch Mikrobiol. 1973;92:45–58. doi: 10.1007/BF00409510. [DOI] [PubMed] [Google Scholar]
- 28.Horne I M, Pemberton J M, McEwan A G. Photosynthesis gene expression in Rhodobacter sphaeroides is regulated by redox changes which are linked to electron transport. Microbiology. 1996;142:2831–2838. [Google Scholar]
- 29.Iba K, Takamiya K. Action spectra for inhibition by light of accumulation of bacteriochlorophyll and carotenoids in marine bacteria. Plant Cell Physiol. 1989;30:471–477. [Google Scholar]
- 30.Imhoff J F. Taxonomy and physiology of phototrophic purple bacteria and green sulfur bacteria. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–15. [Google Scholar]
- 31.Inoue K, Kouadio J K, Mosley C S, Bauer C E. Isolation and in vitro phosphorylation of sensory transduction components controlling anaerobic induction of light harvesting and reaction center gene expression in Rhodobacter capsulatus. Biochemistry. 1995;34:391–396. doi: 10.1021/bi00002a002. [DOI] [PubMed] [Google Scholar]
- 32.Kanamaru K, Aiba H, Mizuno T. Signal transduction and osmoregulation in Escherichia coli. A single amino acid change in the protein kinase, EnvZ, results in loss of its phosphorylation and dephosphorylation abilities with respect to the activator protein, OmpR. J Biol Chem. 1989;264:21633–21637. [PubMed] [Google Scholar]
- 33.Kim S, Funayama N, Komano T. Nucleotide sequence and characterization of the traABCD region of IncI1 plasmid R64. J Bacteriol. 1993;175:5035–5042. doi: 10.1128/jb.175.16.5035-5042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kortluke C, Breese K, Gad’on N, Labahn A, Drews G. Structure of the puf operon of the obligately aerobic, bacteriochlorophyll a-containing bacterium Roseobacter denitrificans OCh114 and its expression in a Rhodobacter capsulatus puf puc deletion mutant. J Bacteriol. 1997;179:5247–5258. doi: 10.1128/jb.179.17.5247-5258.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983;47:1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liebetanz R, Hornberger U, Drews G. Organization of the genes coding for the reaction-centre L and M subunits and B870 antenna polypeptides alpha and beta from the aerobic photosynthetic bacterium Erythrobacter species OCH114. Mol Microbiol. 1991;5:1459–1468. doi: 10.1111/j.1365-2958.1991.tb00792.x. [DOI] [PubMed] [Google Scholar]
- 37.Maniatis T, Fritsch F F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Masuda S, Matsumoto Y, Nagashima K V P, Shimada K, Inoue K, Bauer C E, Matsuura K. Characterization of photosynthetic regulatory genes, regA and regB: Studies among different species. In: Garab G, editor. Photosynthesis: mechanisms and effects. Current research in photosynthesis, in press. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 2881–2884. [Google Scholar]
- 39.Masuda S, Yoshida M, Nagashima K V P, Shimada K, Matsuura K. A new cytochrome subunit bound to the photosynthetic reaction center in the purple bacterium, Rhodovulum sulfidophilum. J Biol Chem. 1999;274:10795–10801. doi: 10.1074/jbc.274.16.10795. [DOI] [PubMed] [Google Scholar]
- 40.Mizoguchi H, Masuda T, Nishimura K, Shimada H, Ohta H, Shioi Y, Takamiya K. Nucleotide sequence and transcriptional analysis of flanking region of the gene (spb) for trans-acting factor that controls light-mediated expression of the puf operon in Rhodobacter sphaeroides. Plant Cell Physiol. 1997;38:558–567. doi: 10.1093/oxfordjournals.pcp.a029205. [DOI] [PubMed] [Google Scholar]
- 41.Monson E K, Ditta G S, Helinski D R. The oxygen sensor protein, FixL, of Rhizobium meliloti: role of histidine residues in heme binding phosphorylation, and signal transduction. J Biol Chem. 1995;270:5243–5250. doi: 10.1074/jbc.270.10.5243. [DOI] [PubMed] [Google Scholar]
- 42.Mosley C S, Suzuki J Y, Bauer C E. Identification and molecular genetic characterization of a sensor kinase responsible for coordinately regulating light harvesting and reaction center gene expression in anaerobiosis. J Bacteriol. 1994;176:7566–7573. doi: 10.1128/jb.176.24.7566-7573.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishimura K, Shimada H, Ohta H, Masuda T, Shioi Y, Takamiya K. Expression of the puf operon in an aerobic photosynthetic bacterium, Roseobacter denitrificans. Plant Cell Physiol. 1996;37:153–159. doi: 10.1093/oxfordjournals.pcp.a028926. [DOI] [PubMed] [Google Scholar]
- 44.O’Gara J P, Eraso J M, Kaplan S. A redox-responsive pathway for aerobic regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1998;180:4044–4050. doi: 10.1128/jb.180.16.4044-4050.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parkinson J S. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 46.Parkinson J S, Kofoid E C. Communication modules in bacterial signalling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 47.Penfold R J, Pemberton J M. An improved suicide vector for construction of chromosomal insertion mutation in bacteria. Gene. 1992;118:145–146. doi: 10.1016/0378-1119(92)90263-o. [DOI] [PubMed] [Google Scholar]
- 48.Penfold R J, Pemberton J M. Sequencing, chromosomal inactivation, and functional expression in Escherichia coli of ppsR, a gene which represses carotenoid and bacteriochlorophyll synthesis in Rhodobacter sphaeroides. J Bacteriol. 1994;176:2869–2876. doi: 10.1128/jb.176.10.2869-2876.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfenning N, Truper H G. Type and neotype strains of the species of phototrophic bacteria maintained in pure culture. Int J Syst Bacteriol. 1971;21:19–24. [Google Scholar]
- 50.Phillips-Jones M K, Hunter C N. Cloning and nucleotide sequence of regA, a putative response regulator gene of Rhodobacter sphaeroides. FEBS Lett. 1994;116:269–276. doi: 10.1111/j.1574-6968.1994.tb06714.x. [DOI] [PubMed] [Google Scholar]
- 51.Ponnampalam S N, Buggy J H, Bauer C E. Characterization of an aerobic repressor that coordinately regulates bacteriochlorophyll, carotenoid, and light harvesting-II expression in Rhodobacter capsulatus. J Bacteriol. 1995;177:2990–2997. doi: 10.1128/jb.177.11.2990-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qian Y, Tabita F R. A global signal transduction system regulates aerobic and anaerobic CO2 fixation in Rhodobacter sphaeroides. J Bacteriol. 1996;178:12–18. doi: 10.1128/jb.178.1.12-18.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saeki K, Suetsugu Y, Tokuda K, Miyatake Y, Young D A, Marrs B L, Matsubara H. Genetic analysis of functional differences among distinct ferredoxins in Rhodobacter capsulatus. J Biol Chem. 1991;266:12889–12895. [PubMed] [Google Scholar]
- 54.Schultze M, Rodel G. Accumulation of the cytochrome c oxidase subunits I and II in yeast requires a mitochondrial membrane-associated protein, encoded by the nuclear scoI gene. Mol Gen Genet. 1989;216:37–43. doi: 10.1007/BF00332228. [DOI] [PubMed] [Google Scholar]
- 55.Sganga M W, Bauer C E. Regulatory factors controlling photosynthetic reaction center and light-harvesting gene expression in Rhodobacter capsulatus. Cell. 1992;68:945–954. doi: 10.1016/0092-8674(92)90037-d. [DOI] [PubMed] [Google Scholar]
- 56.Shiba T. O2 regulation of bacteriochlorophyll synthesis in the aerobic bacterium Erythrobacter. Plant Cell Physiol. 1987;28:1313–1320. [Google Scholar]
- 57.Shiba T, Shimizu U, Taga N. Another aerobic bacterium which contains bacteriochlorophyll a. Bull Jpn Soc Sci Fish. 1979;45:801. [Google Scholar]
- 58.Shimada K. Aerobic anoxygenic phototrophs. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 105–122. [Google Scholar]
- 59.Shioi Y. Growth characteristics and substrate specificity of aerobic photosynthetic bacterium, Erythrobacter sp. (OCH114) Plant Cell Physiol. 1986;27:567–572. [Google Scholar]
- 60.Shioi Y, Doi M. Control of bacteriochlorophyll accumulation by light in an aerobic photosynthetic bacterium, Erythrobacter sp. strain OCh 114. Arch Biochem Biophys. 1988;266:470–477. doi: 10.1016/0003-9861(88)90279-2. [DOI] [PubMed] [Google Scholar]
- 61.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:37–45. [Google Scholar]
- 62.Swanson R V, Schusterm S C, Simon M I. Expression of CheA fragments which defines domains encoding kinase, phosphotransfer, and CheY binding activities. Biochemistry. 1993;32:7623–7629. doi: 10.1021/bi00081a004. [DOI] [PubMed] [Google Scholar]
- 63.Taylor D P, Cohen S N, Clark W G, Marrs B L. Alignment of genetic and restriction maps of the photosynthesis region of the Rhodopseudomonas capsulata chromosome by a conjugation-mediated marker rescue technique. J Bacteriol. 1983;154:580–590. doi: 10.1128/jb.154.2.580-590.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thony-Meyer L, Beck C, Preising O, Hennecke H. The ccoNOQP gene cluster codes for a cb-type cytochrome oxidase that functions in aerobic respiration of Rhodobacter capsulatus. Mol Microbiol. 1994;14:705–716. doi: 10.1111/j.1365-2958.1994.tb01308.x. [DOI] [PubMed] [Google Scholar]
- 65.Tiwari R P, Reeve W G, Dilworth M J, Glenn A R. Acid tolerance in Rhizobium meliloti strain WSM419 involves a two-component sensor-regulator system. Microbiology. 1996;142:1693–1704. doi: 10.1099/13500872-142-7-1693. [DOI] [PubMed] [Google Scholar]
- 66.Volz K. Structural conservation in the CheY superfamily. Biochemistry. 1993;32:11741–11753. doi: 10.1021/bi00095a001. [DOI] [PubMed] [Google Scholar]
- 67.Weaver P F, Wall J D, Gest H. Characterization of Rhodopseudomonas capsulata. Arch Microbiol. 1975;105:207–216. doi: 10.1007/BF00447139. [DOI] [PubMed] [Google Scholar]
- 68.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 69.Yildiz F H, Gest H, Bauer C E. Attenuated effect of oxygen on photopigment synthesis in Rhodospirillum centenum. J Bacteriol. 1991;173:5502–5506. doi: 10.1128/jb.173.17.5502-5506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Young D A, Bauer C E, Williams J C, Marrs B L. Genetic evidence for superoperonal organization of genes for photosynthetic pigments and pigment binding proteins in Rhodobacter capsulatus. Mol Gen Genet. 1989;218:1–12. doi: 10.1007/BF00330558. [DOI] [PubMed] [Google Scholar]
- 71.Zeilstra-Ryalls J, Kaplan S. Control of hemA expression in Rhodobacter sphaeroides 2.4.1: regulation through alterations in the cellular redox state. J Bacteriol. 1996;178:985–993. doi: 10.1128/jb.178.4.985-993.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhulin I B, Taylor B L, Dixon R. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem Sci. 1997;22:331–333. doi: 10.1016/s0968-0004(97)01110-9. [DOI] [PubMed] [Google Scholar]