Abstract
Background
Global publications on Q fever have increased after the 2007 epidemic in the Netherlands. However, the epidemiology of Q fever/coxiellosis in Brazil is still poorly understood. Accordingly, there have been few studies investigating the presence of Coxiella burnetii in dairy products around the world, especially in Brazil, where consumption of fresh cheese made from raw-milk is very high.
Objective
This study was a random survey to assess the prevalence of C. burnetii by PCR in traditional Minas artisanal cheese from the Serro microregion, Brazil, which is manufactured from bovine raw-milk.
Methods
DNA extracted from 53 cheese samples were analyzed by nested PCR with C. burnetii-specific primers and the products confirmed by DNA sequencing.
Results
Out of the 53 cheese samples five (9.43%) were C. burnetii DNA-positive, each coming from one of the respective randomly selected manufacturing agroindustries.
Based on our results, it is estimated that 1.62 tons/day of ready-to-eat cheese made from raw-milk from a total of 16.2 tons produced daily in the study region are contaminated with C. burnetii.
Conclusion
To our knowledge, this is the first report of highly heat-resistant zoonotic pathogen in raw-milk Brazilian artisanal cheese. This food safety hazard has been completely neglected in ready-to-eat raw-milk Brazilian artisanal cheese and could imply potential threats to consumers, since C. burnetii survives in artisanal cheese submitted to long ripening periods. Thus, this work established random and representative baseline prevalence of C. burnetii in this food product in Brazil. Further epidemiological studies, monitoring trends and setting control targets are warranted. Finally, these results point out the importance of including C. burnetii in animal and public health surveillance programs.
Abbreviation: MAC, Minas artisanal cheese
Keywords: Brazil, Coxiella burnetii, Minas artisanal cheese, Zoonosis, Food safety
Introduction
The Minas artisanal cheese (MAC) made from raw-milk is the oldest and most traditional type of cheese in Brazil and represents the main livelihood of several smallholders in this state, which produces about 70,000 tons of cheese per year.1 By using raw-milk in its manufacturing, the MAC can carry particularly zoonotic pathogens such as Coxiella burnetii, which causes Q fever in humans and coxiellosis in animals.2
It is a notifiable disease by the World Organization for Animal Health,2 but in both animals and humans, C. burnetii infections remain poorly understood and their prevalence has been underestimated.3
The most common reservoirs of this pathogen are cattle, sheep, and goats; it is found in urine, feces, milk, and birth fluids of infected animals.
Human infections mainly occur after human inhalation of contaminated aerosols and rarely after the ingestion of unpasteurized milk or cheese.4 C. burnetii is also an important tick-borne pathogen, but their role in human infection is still questionable.2, 3
Global publications on Q fever increased after the 2007 epidemic in the Netherlands.5 Over the past decade, case reports and seroprevalence studies on Q fever have been published in Brazil.6, 7, 8, 9 However, these studies do not yet provide a complete picture of Q fever in this continental country, which does not yet have Q fever/coxiellosis control programs, although case reporting is mandatory for animals since 201310 and for humans since 2014.9 Even so, the cases are under-reported.9, 10
Despite the number of publications, there have been few studies investigating the presence of C. burnetii in dairy products around the world,5 especially in Brazil.10, 11 Therefore, the aim of this study was to estimate the prevalence of C. burnetii by PCR in MAC, which is manufactured using raw-milk.
Methods
Design, location of the study and sample planning
This was a cross-sectional random survey to estimate the prevalence of C. burnetii by PCR in MAC produced by agroindustries of the Serro microregion, Minas Gerais (MG) state, the oldest and most traditional Brazilian cheese. To be included in the survey the agroindustries had to be both MAC manufacturers from a set of single-farm milk and registered in the Minas Gerais Agriculture and Livestock Institute (IMA), the state animal health agency. This study was approved by the Brazilian National Research Ethics Committee (protocol 120466748/2013).
The sample of the target population to estimate the prevalence of C. burnetii was determined by random drawing of 53 (42.4%) MAC samples, each one representing a respective manufacturing agroindustry, from a total of 125 artisanal industries registered in the IMA. This sample size was based on a finite population of 125, 10% margin of error, 95% confidence level and an expected proportion of 50% C. burnetii DNA-positive MAC.
Data and sample collection
MAC producers were interviewed with the use of a structured questionnaire organized by content group, focusing on socio-economic features, animal health, Good Agricultural Practices (GAP) and Good Handling Practices (GHP) for the MAC. The MAC samples were collected with ripening time between three and eight days in their own packaging and kept in isothermal boxes under refrigeration (<4 °C) with recyclable ice during transportation to the laboratory for analyses.
DNA extraction
The cheese samples were fractionated into smaller portions of 100 g, wrapped in sterile bags for solid or liquid samples (INLAB, São Luís, Brazil), sealed, identified, macerated and homogenized. DNA extraction from the cheese samples was performed using the DNeasy Blood & Tissue Kit® (Qiagen, Hilden, Germany).
PCR amplification
Bacterial DNA was detected with C. burnetii-specific primers designed to amplify the IS1111 gene, derived from a transposon-like repeat region of the genome of this bacteria, using the same primers, controls and conditions of a nested PCR standardized by our team.12
DNA sequencing
Properly sized fragments were purified using the Illustra GFX PCR DNA and Gel Band Purification® kit (GE Healthcare, Buckinghamshire, UK). The direct nucleotide sequencing amplicon was performed using the BigDye Terminator v3.1 Cycle Sequencing kit, and purification was performed using the BigDye® X-Terminator Purification kit (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s recommendations. The analyses of the amplicons were performed in an ABI Prism 3730XL with 96 capillaries (Applied Biosystems) and the nucleotide sequences were analyzed using the MEGA7 software (downloaded from www.megasoftware.net). A consensus sequence for each bacterial genome was derived from contiguous sequences assembled by the same software.
Epidemiological analyses
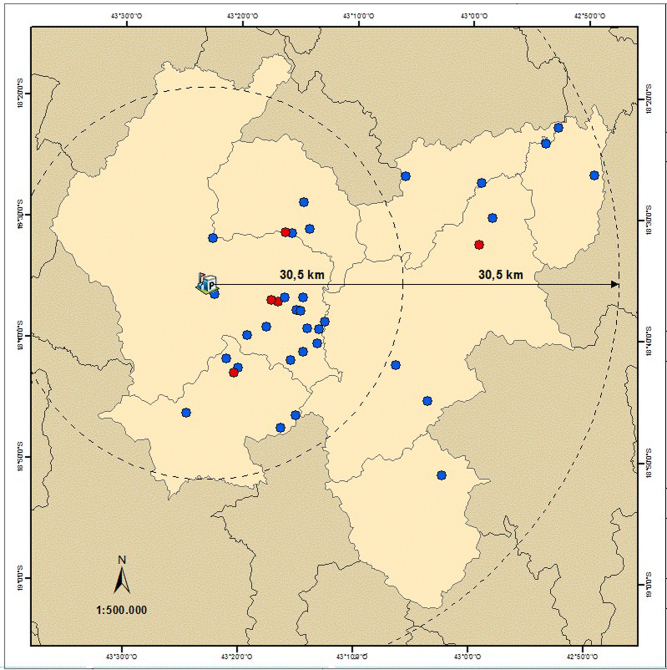
Descriptive epidemiology is presented and the prevalence of C. burnetii DNA-positive MAC is shown with 95% confidence interval. The proportion of agroindustries with C. burnetii DNA-positive MAC between two areas along the Serro microregion was compared by Fisher’s exact test. These areas are the result of the division of a large circle of 61-km radius into two concentric areas. The center of the circle was the municipality of Serro, the main and most producer of the study region.
Results and discussion
Five (9.43%, 95% CI: 3.1–20.7%) C. burnetii DNA-positive MAC were found among 53 samples analyzed. DNA sequences showed 100% identity for the C. burnetii species-specific DNA segment [GenBank CP000890, AE016828, LK937696, CP018005, CP020616] and the accession numbers have been deposited in GenBank (MH920307-183MH920311). This was the first description of this highly infectious and heat-resistant pathogen in ready-to-eat raw-milk MAC in Brazil, although it has been found in cheeses from other countries.13, 14
The distribution of the agroindustries with positive MAC did not present spatial statistical differences (p > 0.05) between the two concentric areas (Fig. 1), presenting evidence that C. burnetii distribution is homogeneous along the region of Serro. Thus, it is estimated that at least 1.62 tons/day (almost 10%) of ready-to-eat raw-milk MAC from a total of 16.2 tons produced daily are contaminated with C. burnetii.
Fig. 1.
Spatial distribution of agroindustries from the region of Serro (light brown) with raw-milk Minas artisanal cheese positive (red) and negative (blue) for Coxiella burnetii DNA, Minas Gerais state, Brazil; elaborated by the Embrapa Dairy Cattle Geoprocessing Department according to the Geographic information system Datum Horizontal SIRGAS 2000 and the Brazilian Institute of Geography and Statistics (IBGE).
Inhalation of contaminated aerosols or dust is the primary source of C. burnetii transmission to humans.4 However, epidemiological evidence strengthens the link between exposure to raw dairy products and this disease. Accordingly, an outbreak affected five consumers of contaminated raw cow’s milk from the same dairy.15 Analysis of the available data, given that many of data required for a quantitative risk assessment are missing even in developed countries, has shown that the risks through consumption of raw dairy products are not negligible.4
A study from 2013 did not find viable C. burnetii in PCR-positive dairy products in France and pointed out that they do not seem to represent a risk for consumers.14 However, in a much worse scenario, another more recent study showed that C. burnetii remained viable after at least eight months of ripening in hard cheese.16
MAC is a minimally processed food. It is made from raw-milk, undergoing no thermal processing and uses non-industrialized native starter culture with an unknown and non-standardized concentration of lactic acid bacteria, most of which are marketed and consumed fresh. Thus, it is believed that the chance of finding viable C. burnetii among fresh MAC is even greater than that verified for hard cheeses by other authors,16 and this requires further studies.
The local regulations determine, for safety reasons, that MAC should be produced from a set of single-farm milk, fermented with a compulsory autochthonous starter culture called “pingo”, which is cheese whey from a previous MAC production that is collected and added in the next production, and ripened for at least 17 days.17 However, according to the producers’ self-reports, approximately half (47.7%) of the studied agroindustries do not use the recommended starter culture for MAC manufacturing and most of them (68.2%) replace it with a banned starter, grated cheese rind, which could potentially introduce C. burnetii spore-like forms.18 Additionally, we observed in situ that MAC was being sold within a median of only four days of ripening, which could potentially increase the risk of human exposure.16
By deduction, if C. burnetii was found in a cheese from a set of single-farm milk in this study, the most likely source is the respective feedstock from positive bovine herd.15
Paradoxically, milk heat treatment is not allowed for MAC manufacturing. Thus, the detection of positive individuals or herds by serological or PCR tests, maintaining closed herds preferably and requiring health certificates when buying animals, immunization, pre-calving separation and destruction of placentas and fetal wraps should be minimum recommendations19 for farmer-owners that produce cheese from raw-milk. As challenges, more than one-third (36.4%) of the herds included in this study are not completely closed and most producers (92.8%) do not require any health certificates to purchase bovine, and this could allow the entry of positive animals, as already reported.20
It is noteworthy that in a recent study, C. burnetii was detected by PCR in 3.57% of the unpasteurized cow’s milk samples in another region of Brazil10 at a concentration within the range count of C. burnetii per mL of milk required to have a 50% probability to start an infection.4
Thus, there is a need for further studies to better understand the epidemiology of this pathogen in Brazil, especially comparing the strains found in cattle and dairy products with those of human cases and assessing the risk of this pathogen in raw-milk cheese for consumers.
Conclusion
We found C. burnetii DNA in ready-to-eat MAC by a random survey. Therefore, this work established representative baseline rates for C. burnetii prevalence in raw-milk MAC in Brazil, to allow monitoring trends, setting control targets and future local risk analysis studies.
Conflicts of interest
The authors declare no conflicts of interest.
Funding
This work was supported by the National Research Council of Brazil (CNPq) (grant numbers 443235/2014-7 and 404762/2016-6), the Mato Grosso do Sul State Agency for Research and Development (FUNDECT) (grant number TO 085/2015), the Minas Gerais State Agency for Research and Development (FAPEMIG) (grant numbers CVZ-APQ-02746-14, CVZ- APQ-03989-17 and CVZ-PPM-00526-16) and EMBRAPA (grant number 02.13.10.007.00.00).
Acknowledgements
We thank the MAC producers who participated in this study and the IMA for doing the fieldwork.
Footnotes
Preliminary results from this study were presented as: (i) a lecture entitled “Coxiella burnetii and other neglected zoonotic hazards in artisanal cheeses made from raw milk in Brazil”, by Marcio Roberto Silva, at the 55th Congress of the Brazilian Society of Tropical Medicine (SBMT) and XXVI Brazilian Congress of Parasitology, 28 to 31 July, 2019, Belo Horizonte, Minas Gerais, Brazil; (ii) as a dissertation entitled “Prevalence and risk factors for Coxiella burnetii in Minas Artisanal Cheese from the micro region of Serro” presented by Letícia Sacafutto de Faria to obtain the Master degree in Science and Technology of Milk by the Federal University of Juiz de Fora (http://www.ufjf.br/mestradoleite/files/2017/06/Disserta%C3%A7%C3%A3o-Final7.pdf).
References
- 1.Emater - Empresa de Assistência Técnica e Extensão Rural do Estado de Minas Gerais Queijo Minas Artesanal: tradição e qualidade que revelam Minas. Rev EMATER MG. 2004;22:8–9. [Google Scholar]
- 2.Porter S.R., Czaplicki G., Mainil J., Guattéo R., Saegerman C. Q fever: current state of knowledge and perspectives of research of a neglected zoonosis. Int J Microbiol. 2011;2011 doi: 10.1155/2011/248418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marrie T.J., Raoult D. Q fever — a review and issues for the next century. Int J Antimicrob Agents. 1997;8:145–161. doi: 10.1016/s0924-8579(96)00369-x. [DOI] [PubMed] [Google Scholar]
- 4.Gale P., Kelly L., Mearns R., Duggan J., Snary E.L. Q fever through 366 consumption of unpasteurized milk and milk products — a risk profile and exposure assessment. J Appl Microbiol. 2015;118:1083–1095. doi: 10.1111/jam.12778. [DOI] [PubMed] [Google Scholar]
- 5.Delsing C.E., Kullberg B.J., Bleeker-Rovers C.P. Q fever in the Netherlands from 2007 to 2010. Neth J Med. 2010;68:382–387. [PubMed] [Google Scholar]
- 6.Epelboin L., Nacher M., Mahamat A., et al. Q fever in French Guiana: tip of the iceberg or epidemiological exception? PLoS Negl Trop Dis. 2016;10 doi: 10.1371/journal.pntd.0004598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa P.S.G., Brigatte M.E., Greco D.B. Questing one Brazilian query: reporting 16 cases of Q fever from Minas Gerais, Brazil. Rev Inst Med Trop. 2006;48:5–9. doi: 10.1590/s0036-46652006000100002. [DOI] [PubMed] [Google Scholar]
- 8.Siciliano R.F., Strabelli T.M., Zeigler R., et al. Infective Endocarditis due to Bartonella spp. and Coxiella burnetii. Ann N Y Acad Sci. 2006;1078:215–222. doi: 10.1196/annals.1374.123. [DOI] [PubMed] [Google Scholar]
- 9.Mares-Guia M.A., Rozental T., Guterres A., et al. Molecular identification of Q fever in patients with a suspected diagnosis of dengue in Brazil in 2013-2014. Am J Trop Med Hyg. 2016;94:1090–1094. doi: 10.4269/ajtmh.15-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mioni M.D.S.R., Ribeiro B.L.D., Peres M.G., et al. Real time quantitative PCR based detection of Coxiella burnetii in unpasteurized cow’s milk sold for human consumption. Zoonoses Public Health. 2019:1–6. doi: 10.1111/zph.12609. [DOI] [PubMed] [Google Scholar]
- 11.Mares-Guia M.A.M.M., Rozental T., Guterres A., et al. Molecular identification of the agent of Q fever – Coxiella burnetii – in domestic animals in State of Rio de Janeiro, Brazil. Rev Soc Bras Med Trop. 2014;47:231–234. doi: 10.1590/0037-8682-0076-2013. [DOI] [PubMed] [Google Scholar]
- 12.Mares-Guia M.A.M.M., Guterres A., Rozental T., Ferreira M.D.S., Lemos E.R.S. Clinical and epidemiological use of nested PCR targeting the repetitive element IS1111 associated with the transposase gene from Coxiella burnetii. Braz J Microbiol. 2018;49:138–143. doi: 10.1016/j.bjm.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galieiro A., Fratini F., Cammà C., et al. Occurrence of Coxiella burnetii in goat and ewe unpasteurized cheeses: screening and genotyping. Int J Food Microbiol. 2016;237:47–54. doi: 10.1016/j.ijfoodmicro.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Eldin C., Angelakis E., Renvoisé A., Raoult D. Coxiella burnetii DNA, but not viable bacteria, in dairy products in France. Am J Trop Med Hyg. 2013;88:765–769. doi: 10.4269/ajtmh.12-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Signs K.A., Stobierski M.G., Gandhi T.N. Q fever cluster among raw milk drinkers in Michigan, 2011. Clin Infect Dis. 2012;55:1387–1389. doi: 10.1093/cid/cis690. [DOI] [PubMed] [Google Scholar]
- 16.Barandika J.F., Alvarez-Alonso R., Jado I., Hurtado A., García-Pérez A.L. Viable Coxiella burnetii in hard cheeses made with unpasteurized milk. Int J Food Microbiol. 2019;303:42–45. doi: 10.1016/j.ijfoodmicro.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 17.Minas Gerais. Secretaria de Estado de Agricultura, Pecuária e Abastecimento. Portaria no 1305, de 30 de abril de 2013. Estabelece diretrizes para a produção do queijo minas artesanal.
- 18.Kersh G.J., Fitzpatrick K.A., Self J.S., et al. Presence and persistence of Coxiella burnetii in the environments of goat farms associated with a Q fever outbreak. Appl Environ Microbiol. 2013;79:1697–1703. doi: 10.1128/AEM.03472-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acha P.N., Szyfres B. Organización Panamericana de la Salud; Washington DC: 2003. Zoonosis y enfermedades transmisibles comunes al hombre y a los animales. [Google Scholar]
- 20.Paul S., Agger J.F., Markussen B., Christoffersen A.B., Agerholm J.S. Factors associated with Coxiella burnetii antibody positivity in Danish dairy cows. Prev Vet Med. 2012;107:57–64. doi: 10.1016/j.prevetmed.2012.05.015. [DOI] [PubMed] [Google Scholar]