Abstract
Escherichia coli formamidopyrimidine (Fpg) DNA glycosylase and MutY DNA glycosylase are base excision repair proteins that work together to protect cells from the mutagenic effects of the commonly oxidized guanine product 7,8-dihydro-8-oxoguanine. The genes encoding these proteins, fpg and mutY, are both cotranscribed as part of complex operons. fpg is the terminal gene in an operon with the gene order radC, rpmB, rpmG, and fpg. This operon has transcription initiation sites upstream of radC, in the radC coding region, and immediately upstream of fpg. There is a strong attenuator in the rpmG-fpg intergenic region and three transcription termination sites downstream of fpg. There is an additional site, in the radC-rpmB intergenic region, that corresponds either to a transcription initiation site or to an RNase E or RNase III cleavage site. mutY is the first gene in an operon with the gene order mutY, yggX, mltC, and nupG. This operon has transcription initiation sites upstream of mutY, in the mutY coding region, and immediately upstream of nupG. There also appear to be attenuators in the yggX-mltC and mltC-nupG intergenic regions. The order of genes in these operons has been conserved or partially conserved only in other closely related gram-negative bacteria, although it is not known whether the genes are cotranscribed in these other organisms.
Oxidative stress has been linked to cancer, aging, and a number of neurodegenerative diseases (for reviews, see references 2 and 12). Cells experience oxidative stress due to reactive oxygen species that are produced during normal oxidative metabolism. Reactive oxygen species cause a variety of damage in cells, including damage to proteins, lipids, and DNA. The last includes single-strand breaks, sites of base loss, and damage to the purine and pyrimidine bases. Oxidative base lesions that block DNA polymerases can be lethal to cells, and lesions that pair with an incorrect base are potentially mutagenic. Cells have evolved an efficient and accurate mechanism, base excision repair, to remove these lesions from DNA. Base excision repair is highly conserved from bacteria to humans.
Escherichia coli formamidopyrimidine (Fpg) DNA glycosylase and MutY DNA glycosylase are base excision repair proteins that work together to protect cells from the mutagenic effects of the commonly oxidized guanine product 7,8-dihydro-8-oxoguanine (8-oxoG) (26). If not repaired prior to DNA replication, 8-oxoG can correctly pair with C or incorrectly pair with A (36). If A is not removed from the 8-oxoG–A pair, a GC-to-TA transversion will be fixed during the following round of replication. Fpg removes 8-oxoG opposite C (9, 39), and after replication, MutY removes any A that was inserted opposite unrepaired 8-oxoG (26, 28), thus giving Fpg another opportunity to remove the 8-oxoG lesion. E. coli fpg and mutY single mutants have elevated rates of GC-to-TA transversions (8, 30). fpg mutY double mutants have GC-to-TA transversion rates 20-fold higher than the sum of the mutation rates for the single mutants (26), showing that Fpg and MutY work together in a synergistic manner. It has recently been shown that endonuclease VIII (nei), identified on the basis of its specificity for oxidized pyrimidines, also removes 8-oxoG. The addition of the nei mutation to the fpg mutY double mutant results in an additional threefold synergy and a further increase in GC-to-TA transversions (3a). MutT is an 8-oxodGTPase that breaks down 8-oxodGTP to 8-oxodGMP in the nucleotide pool, preventing its incorporation into DNA (25). Strains deficient in MutT have elevated rates of AT-to-CG transversions (45). Fpg, MutY, and MutT are often referred to as the GO repair system, an error avoidance pathway devoted to preventing mutations resulting from the presence of 8-oxoG in DNA (GO is a synonym for 8-oxoG) (26–28).
There have been few studies on the regulation of the oxidative DNA glycosylases in E. coli. It has been shown that cells exhibit increased Fpg activity when shifted from anaerobic to aerobic growth conditions and when exposed to the superoxide-generating compound paraquat (20, 21). This response still occurs in mutants defective in SoxR and SoxS, demonstrating that fpg is not part of the SoxRS regulon (21). It has recently been shown that, under anaerobic growth conditions, Fpg activity increases in strains deficient in the global regulators Fur, FNR, and ArcA (21). Possible Fur, FNR, and ArcA binding sites have been identified in the fpg promoter region, suggesting that these proteins may play a negative regulatory role in fpg regulation. In addition, it has been reported that the transcription of fpg is decreased in the presence of thioredoxin and glutathione, and it has been speculated that glutathione-mediated disassembly of iron-sulfur centers in key regulatory proteins may govern the transcriptional regulation of fpg (17). There have been no reported studies on MutY regulation.
Although our major goals are to further delineate the regulatory controls for Fpg and to define any regulatory controls for MutY, we had to first determine whether the fpg and mutY genes in E. coli are transcribed alone or with neighboring genes that have the same orientation. If fpg and mutY are part of operons, the genes with which they are transcribed may play a role in or provide information about their possible regulation. In this paper, we show that fpg is cotranscribed as the terminal gene in a four-gene operon and that mutY is cotranscribed as the first gene in a four-gene operon. Furthermore, both of these are complex operons containing multiple promoters and terminators. The order of genes in the fpg operon is radC, rpmB, rpmG, and fpg. These genes are arranged in a counter-clockwise orientation on the genome. RadC has been suggested to play a role in growth medium-dependent, recA-dependent repair of DNA single-strand breaks after X-irradiation and in postreplication repair after UV irradiation (15). rpmB and rpmG encode the ribosomal proteins (r-proteins) L28 and L33, respectively (22). The order of genes in the mutY operon is mutY, yggX, mltC, and nupG. These genes are arranged in a clockwise orientation on the genome. yggX encodes a protein of unknown function; mltC encodes membrane-bound lytic transglycosylase C, which has been shown to have peptidoglycan hydrolase activity (11); and nupG encodes a high-affinity nucleoside transport protein (42).
MATERIALS AND METHODS
Bacterial strains.
E. coli GC4468 [DE(argF-lac)169 λ− IN(rrnD-rrnE)1 rpsL179(strR)], N3433 [lacZ43(Fs) λ− relA1 spoT1 thi-1], and N3431 [N3433 rne-3071(Ts)] were obtained from the Yale University E. coli Genetic Stock Center. Strain IBPC637 (N3431, rnc-105 nadB51::Tn10) was kindly supplied by Philippe Regnier, Institut de Biologie-Physico-Chimique, Paris, France.
Growth conditions and RNA isolation.
E. coli cultures (1 ml) were grown overnight in Luria-Bertani broth with shaking at 250 rpm. The overnight cultures were diluted 1/100 in fresh Luria-Bertani broth and were grown until they reached the desired optical density at 600 nm (OD600). GC4468 was grown at 37°C in the presence of 10 μg of streptomycin per ml until an OD600 of 0.7 to 0.8 was reached. N3433 and IBPC637 were grown at 30°C until an OD600 of 0.4 to 0.5 was reached, and then they were shifted to 43°C for 30 min. IBPC637 was grown in the presence of 10 μg of tetracycline per ml. Total RNA was isolated with a Qiagen RNeasy kit according to manufacturer’s recommendations. After elution from the RNeasy column, the RNA was treated with DNase, extracted twice with acid-pH phenol, and extracted once with chloroform-isoamyl alcohol. The RNA was precipitated with ammonium acetate and ethanol, washed in 75% ethanol, and resuspended in RNase-free water. The integrity of the 16S and 23S rRNAs was checked on a 1% agarose gel.
Reverse transcription (RT)-PCR.
RNA was reverse transcribed with Gibco BRL SUPERSCRIPT II RNase H-reverse transcriptase. Total RNA (30 μg) and primer (15 pmol) were denatured in water for 10 min at 80°C and then quickly chilled on ice. Gibco BRL 1× first-strand buffer, 10 mM dithiothreitol, and 500 μM each deoxynucleoside triphosphate (dNTP) were added to the annealing mixture and incubated at 47°C for 2 min. SUPERSCRIPT II (1 μl; 200 U) was added to a final volume of 20 μl, and the reaction mixture was incubated for 1 h at 47°C. The RNA was digested with 1 μl of Ambion RNase A-RNase T1 mixture (250 U of RNase A per ml and 10,000 U of RNase T1 per ml). The primer and digested RNA were removed with a Qiagen PCR purification kit. For each primer used in RT, a negative control lacking reverse transcriptase was run. Two microliters of each RT reaction mixture was used as a template for PCR. E. coli genomic DNA was used as a positive control template for PCR. PCR was performed with 50-μl reaction mixtures containing Gibco BRL Taq DNA polymerase and Idaho Technologies 1× buffer with 3 mM MgCl2 and 200 μM each dNTP on an Idaho Technologies Air Thermo-Cycler for 40 cycles with a 10-s denaturing step at 96°C, a 10-s annealing step at 57°C, and a 90-s extension step at 72°C. PCR products were analyzed on a 1% agarose gel with 0.75 μg of ethidium bromide per ml.
RPAs.
RNase protection assays (RPAs) were performed with an Ambion RPA II kit. RNA antisense probes were transcribed with a template containing a T7 phage promoter. The antisense probe template was prepared by PCR with genomic DNA as the template and primer sets with the T7 phage promoter incorporated into the downstream primer. PCR was performed with 50-μl reaction mixtures containing Stratagene Pfu DNA polymerase and Idaho Technologies 1× buffer with 3 mM MgCl2 and 200 μM each dNTP on an Air Thermo-Cycler. PCR products were analyzed on a 1% agarose gel; then, the products were cut out, gel eluted in water, dried under vacuum with centrifugation, and resuspended in 20 μl of water. The template was transcribed with 10 U of Ambion T7 RNA polymerase in a reaction mixture containing Ambion 1× transcription buffer, 2.5 μl of template, 500 μM ATP, 500 μM CTP, 500 μM GTP, 12.5 μM [α-32P]UTP (800 Ci/mmol; 40 mCi/ml), and water in a final reaction volume of 10 μl. The reaction mixture was incubated at 37°C for 1 h and then run on a 5% polyacrylamide gel to purify the probe. E. coli RNA (30 μg) or yeast RNA (30 μg) was hybridized overnight with the labeled RNA probe (10,000 cpm) at 47°C in hybridization buffer. Unhybridized probe was digested with 0.5 U of RNase A and 20 U of RNase T1 in digestion buffer, the RNases were inactivated, and the remaining RNA was precipitated. The pellet was resuspended in formamide gel loading buffer, and the sample was run on a 5% polyacrylamide gel.
Primer extension analysis.
Primers used in the primer extension analysis were 5′ end labeled with Gibco BRL T4 polynucleotide kinase and [γ-32P]ATP (6,000 Ci/mmol; 10 mCi/ml), and the free labeled nucleotide was removed with a Qiagen nucleotide removal kit according to the manufacturer’s recommendations. Total RNA (30 μg) and primer (2 pmol) were annealed in water by heating at 80°C for 10 min and then quickly chilled on ice. Gibco BRL 1× first-strand buffer, 10 mM dithiothreitol, and 500 μM each dNTP were added, and the reaction mixture was heated at 47°C for 2 min. SUPERSCRIPT II (1 μl; 200 U) was added, and the reaction mixture was incubated at 47°C for 1 h. The nucleic acids were precipitated with ammonium acetate and ethanol, washed with 75% ethanol, and resuspended in 5 μl of formamide gel loading buffer. A sequencing ladder was generated from control DNA M13mp18 with a U.S. Biochemicals Sequenase version 2.0 sequencing kit. The sequencing ladder and the 5-μl primer extension reaction mixture were run on a 6% polyacrylamide gel. The gel was dried and exposed to film.
Homology search.
The amino acid sequence for the product of each gene in the fpg and mutY operons was used in a BLAST search at the Unfinished NCBI Microbial Genome BLAST Website (29a). On this web page, the “All” option was chosen to search both the finished and the unfinished genomes. The program used was tblastn. The amino acid sequences for the products of genes in the fpg operon were obtained from GenBank accession no. AE000441, those for mutY and yggX were obtained from accession no. AE000378, and those for mltC and nupG were obtained from accession no. AE000379. The BLAST searches yielded positive results from the following unfinished genomes: Salmonella typhimurium and Yersinia pestis, sequenced at the Sanger Centre through projects funded by Beowulf Genomics (the BLAST servers for these organisms at the Sanger Centre site [35a] were also used; Contig 770 from the S. typhimurium database contains the gene radC and was used to splice together overlapping sequences from this site and GenBank accession no. U23405, which contains S. typhimurium rpmB, rpmG, and fpg); Actinobacillus actinomycetemcomitans, sequenced by the Actinobacillus Genome Sequencing Project supported by a USPHS/NIHS grant from the National Institute of Dental Research; Pseudomonas aeruginosa, sequenced at the University of Washington Genome Center and PathoGenesis Corporation and jointly funded by the Cystic Fibrosis Foundation and PathoGenesis Corporation; and Vibrio cholerae (preliminary sequence data were obtained from the BLAST server at the Institute for Genomic Research [19a]; sequencing of V. cholerae was accomplished with support from NIAID). Sequences from all organisms were aligned with the subject numbers provided by the BLAST searches. In some cases, when genes were located on different contigs, it was possible to align them because of overlapping sequences.
RESULTS
fpg is part of an operon containing four genes.
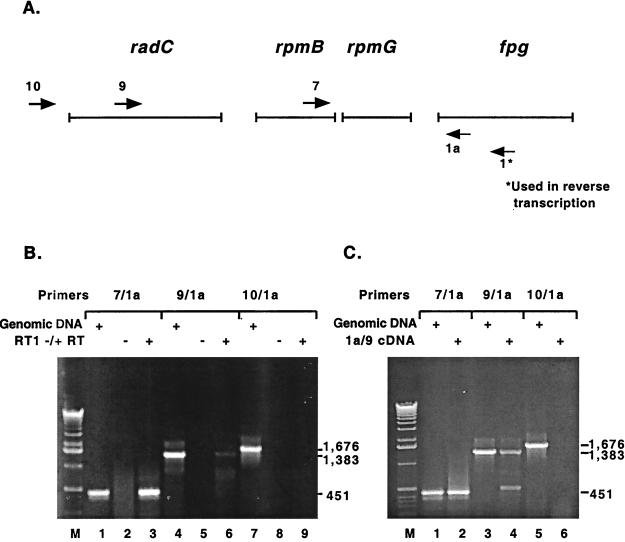
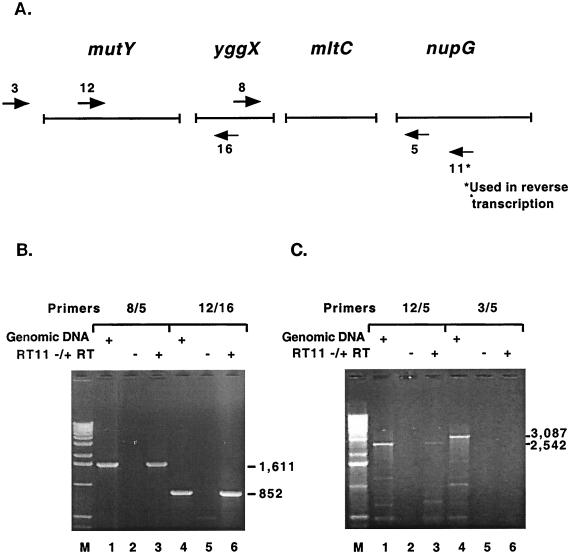
The fpg transcript was initially mapped by RT-PCR to determine if it is transcribed alone or with surrounding genes in the same orientation. The order of the genes that could potentially be transcribed together is radC, rpmB, rpmG, and fpg. rpmB and rpmG have previously been proposed to be transcribed together, and there is a stem-loop structure, including six T residues beginning 15 bases after the termination codon for rpmG; this structure was shown by an S1 nuclease digestion assay to be an RNA polymerase transcription termination site (22). There are 216 bp between the radC stop codon and the rpmB start codon, 20 bp between the rpmB stop codon and the rpmG start codon, and 97 bp between the rpmG stop codon and the fpg start codon. The gene 5′ to radC, dfp, and the gene 3′ to fpg, kdtB, are in opposite orientations, precluding them from being on the same transcript with fpg. E. coli total RNA was reverse transcribed with the primer fpg1, which anneals 357 bp 3′ to the fpg start codon, and PCR was performed to determine the approximate size of the cDNA. Primer fpg1a was the downstream primer and primers fpg7, fpg9, and fpg10 were the upstream primers used in the PCR; Fig. 1A shows approximate annealing locations of PCR primers. Positive PCR controls were run with the three primer sets and genomic DNA as a template (Fig. 1B, lanes 1, 4, and 7; sizes are indicated in the figure). PCR with primer sets fpg7-fpg1a and fpg9-fpg1a and with cDNA as a template yielded a product (Fig. 1B, lanes 3 and 6), indicating that the transcript containing fpg also contained rpmG, rpmB, and radC. PCR with fpg10-fpg1a did not yield a product (Fig. 1B, lane 9), indicating that the upstream transcription initiation site for the transcript was within 148 bp of the proposed start codon for radC. The negative control lanes (the PCR template was an RT reaction lacking reverse transcriptase) were empty, indicating that the RNA used for RT was not contaminated with DNA (Fig. 1B, lanes 2, 5, and 8). To confirm that the PCR product obtained with primer set fpg9-fpg1a was specific for the fpg transcript, the band was cut out, gel eluted, and used as a template in PCRs with the same primer sets. The results were the same: primer sets fpg7-fpg1a and fpg9-fpg1a yielded a product, whereas primer set fpg10-fpg1a did not (Fig. 1C, lanes 2, 4, and 6).
FIG. 1.
Mapping of the fpg transcript by RT-PCR. (A) The arrows numbered 1a, 7, 9, and 10 represent the primers used in PCR. The arrow numbered 1* represents the primer used to reverse transcribe E. coli total RNA. Primer fpg1a anneals 90 bp 3′ to the fpg start codon, fpg7 anneals 336 bp 5′ to the fpg start codon, fpg9 anneals 1,269 bp 5′ to the fpg start codon, and fpg10 anneals 1,561 bp 5′ to the fpg start codon. (B) The primer sets used in PCR are indicated above the lanes. Lanes 1, 4, and 7 are positive controls with E. coli genomic DNA as a PCR template. Lanes 2, 5, and 8 are negative controls with RT reaction mixtures minus reverse transcriptase as a PCR template. Lanes 3, 6, and 9 show PCR results obtained with the cDNA from RT with primer 1* (RT1) as a template. The sizes of the PCR products are indicated on the right. (C) The primer sets used in PCR are indicated above the lanes. Lanes 1, 3, and 5 are positive controls with E. coli genomic DNA as a PCR template. In lanes 2, 4, and 6, the PCR product from panel B, lane 6, was cut out, gel eluted, and used as a PCR template. The sizes of the PCR products are indicated on the right.
The fpg operon contains multiple promoters and terminators.
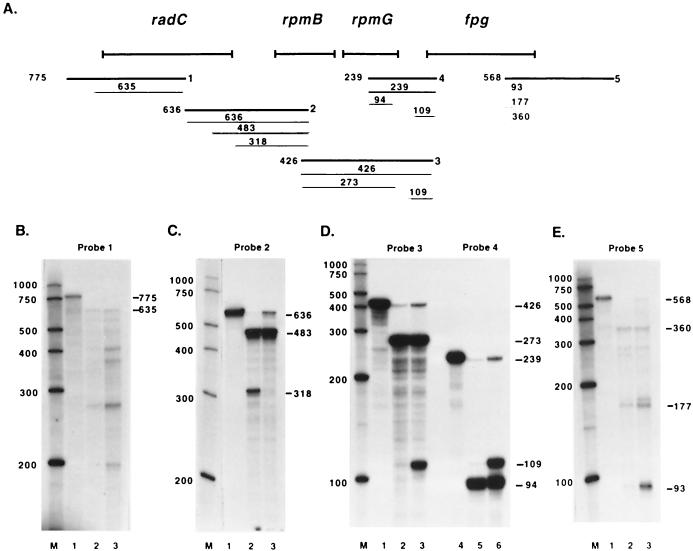
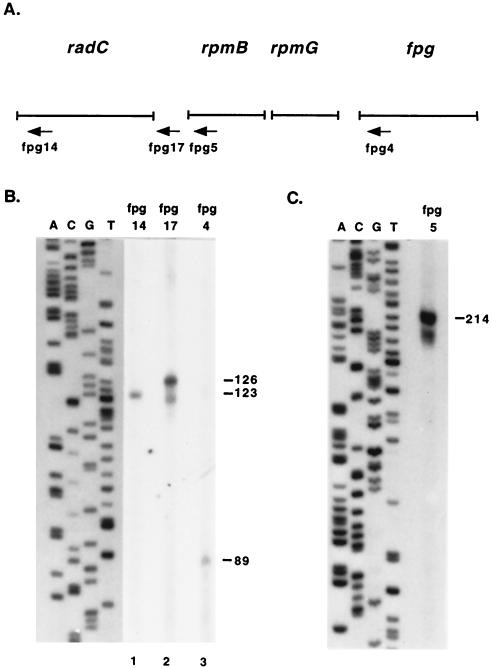
RPAs were performed to confirm the RT-PCR results and to map the locations of promoters, terminators, and transcript processing sites. Several operons containing r-protein genes undergo transcript processing by endonucleases RNase III (rnc) and/or RNase E (rne) (1, 7, 34, 35, 43). For this reason, the fpg RPAs were performed with RNA from wild-type E. coli (N3433) and from rnc rne E. coli (IBPC637). Three reactions were set up for each RNA probe, one with probe plus E. coli RNA and two with probe plus yeast RNA. After overnight hybridization, the E. coli RNA-probe reaction mixture was digested with RNase A and RNase T1. One of the yeast RNA-probe reaction mixtures was also digested with RNase A and RNase T1 to ensure complete digestion of the probe at the RNase concentrations used (data not shown). The other yeast RNA-probe reaction mixture was mock digested; this procedure resulted in a full-length probe when the reaction mixture was run on a gel (only part of the reaction mixture was loaded so the signal would not overpower the other lanes). RNA probes 1, 2, and 3 overlap and cover the length of the transcript from 148 bp 5′ to the proposed start codon for radC to 90 bp 3′ to the fpg start codon (Fig. 2A). The RPA with probe 1 resulted in a 635-nucleotide protected fragment (Fig. 2B, lanes 2 and 3) as predicted with a semilog plot of the distance traveled by the RNA markers. The RPAs with wild-type RNA (Fig. 2B, lane 2) and rnc rne RNA (Fig. 2B, lane 3) looked the same, except for a more pronounced ladder of bands below the 635-nucleotide band in the double mutant. Digestion occurred from the 3′ end of the probe, since the RPA with probe 6 (Fig. 3A) resulted in a full-length product (Fig. 3B, lane 2; this assay was performed with wild-type RNA only). The 635-nucleotide product corresponds to a transcription initiation site about 8 bp upstream from the radC start codon proposed in the GenBank accession no. AE000441 sequence. To more precisely map the site, primer extension analysis was performed with primer fpg14 (Fig. 4A). The primer was extended to a product of 123 nucleotides (Fig. 4B, lane 1), which corresponds to an initiation site −12 bp from the proposed start codon for radC.
FIG. 2.
Mapping of the fpg transcript by RPAs. (A) The thick lines designated 1 to 5 indicate the approximate annealing locations of the probes used in the RPAs. The thin lines indicate the products obtained from RPAs with these probes along with the sizes of the products. The sizes and exact annealing locations of the probes are as follows: probe 1, 775 nucleotides, anneals from 625 bp 3′ to the radC start codon to 148 bp 5′ to the radC start codon; probe 2, 636 nucleotides, anneals from 201 bp 3′ to the rpmB start codon to 219 bp 5′ to the radC stop codon; probe 3, 426 nucleotides, anneals from 90 bp 3′ to the fpg start codon to 51 bp 5′ to the rpmB stop codon; probe 4, 239 nucleotides, anneals from 90 bp 3′ to the fpg stop codon to 52 bp 5′ to the rpmG stop codon; probe 5, 494 nucleotides, anneals from 494 bp 3′ to the fpg stop codon to 74 bp 5′ to the fpg stop codon. (B to E) The probes used in the RPAs are indicated, and lane M contains RNA size markers. Lanes 1 in panels B to E and lane 4 in panel D show the full-length probe (probe hybridized with yeast RNA and mock digested; only part of the reaction mixture was loaded so that the signal would not overpower the other lanes). Lanes 2 in panels B to E and lane 5 in panel D show RPA results obtained wild-type RNA. Lanes 3 in panels B to E and lane 6 in panel D show RPA results obtained with rnc rne RNA. Numbers beside panels are in base pairs.
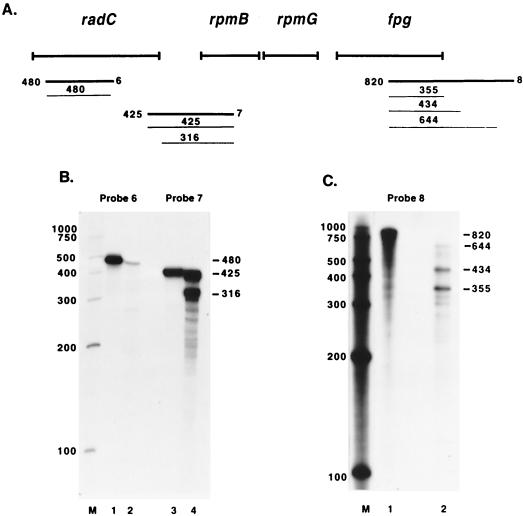
FIG. 3.
Mapping of the fpg transcript by RPAs. (A) The thick lines designated 6 to 8 indicate the approximate annealing locations of the probes used in the RPAs. The thin lines indicate the products obtained from RPAs with these probes along with the sizes of the products. The sizes and exact annealing locations of the probes are as follows: probe 6, 480 nucleotides, anneals from 625 bp 3′ to the radC start codon to 145 bp 3′ to the radC start codon; probe 7, 425 nucleotides, anneals from 201 bp 3′ to the rpmB start codon to 8 bp 5′ to the radC stop codon; probe 8, 820 nucleotides, anneals from 494 bp 3′ to the fpg stop codon to 326 bp 5′ to the fpg stop codon. (B and C) The probes used in the RPAs are indicated, and lane M contains RNA size markers. Lanes 1 in panel B and C and lane 3 in panel B show the full-length probe (probe hybridized with yeast RNA and mock digested; only part of the reaction mixture was loaded so that the signal would not overpower the other lanes). Lanes 2 in panels B and C and lane 4 in panel B show RPA results obtained with wild-type RNA. Numbers beside panels are in base pairs.
FIG. 4.
Mapping of fpg transcription initiation sites by primer extension analysis. (A) Arrows indicate primers used in primer extension analysis. fpg14 anneals 111 bp 3′ to the proposed start codon for radC, fpg17 anneals 22 bp 5′ to the proposed stop codon for radC, fpg5 anneals 76 bp 3′ to the start codon for rpmB, and fpg4 anneals 66 bp 3′ to the start codon for fpg. (B and C) The sequencing ladder was generated with control M13mp18 DNA. (B) Lane 1, primer extension with primer fpg14; lane 2, primer extension with primer fpg17; lane 3, primer extension with primer fpg4. (C) Primer extension with primer fpg5. Numbers beside panels are in base pairs.
The RPA with probe 2 and wild-type RNA resulted in a faint full-length product, a 483-nucleotide product, and a 318-nucleotide product (Fig. 2C, lane 2). The full-length product corresponds to transcript readthrough from radC into rpmB and was more abundant in rnc rne cells (Fig. 2C, lane 3). The 483-nucleotide product was present with equal intensities in both wild-type and double-mutant RNAs, and the 318-nucleotide product was present only in wild-type RNA. To determine if the 483-nucleotide and 318-nucleotide protected products were the result of 5′ or 3′ probe digestion, an RPA was performed with probe 7 (Fig. 3A). If the products were the result of 3′ probe digestion, the 483-nucleotide product would appear as full length and the 318-nucleotide product would remain the same size. This was the case when probe 7 was used (Fig. 3B, lane 4; this assay was performed with wild-type RNA only). The 483-nucleotide product obtained with probe 2 indicates that there is a transcription initiation site about 70 bp 5′ to the radC stop codon. This site was mapped by primer extension analysis with primer fpg17 (Fig. 4A). The primer was extended to a product of 126 nucleotides (Fig. 4B, lane 2), corresponding to a transcription initiation site 104 bp 5′ to the radC stop codon. The 318-nucleotide product seen with wild-type RNA maps to a site about 118 bp 5′ to the rpmB start codon in the radC-rpmB intergenic region. Primer extension analysis was performed with primer fpg5 (Fig. 4A). The primer was extended to a product of 214 nucleotides (Fig. 4C), placing the transcription initiation or cleavage site 138 bp 5′ to the rpmB start codon.
The RPA with probe 3 resulted in a full-length product, a 273-nucleotide product, and a 109-nucleotide product (Fig. 2D, lanes 2 and 3). The full-length product was due to transcript readthrough into fpg and was present in a greater abundance in the double mutant. The 273-nucleotide product was present in equal abundances in wild-type and rnc rne cells; the size of the product, if it resulted from 5′ probe digestion, would correspond to the location of the previously described terminator downstream of rpmG. Probe 4 was used to confirm this result, and the RPA resulted in a full-length product, a 109-nucleotide product, and a 94-nucleotide product of equal intensities in wild-type and double-mutant cells (Fig. 2D, lanes 5 and 6). The 94-nucleotide product corresponds to the proposed terminator, which appears to act as an attenuator, since there is transcript readthrough into fpg. The 109-nucleotide product present with both probes was much more abundant in rnc rne RNA. The size of the product did not shift with two probes, indicating that the product resulted from 3′ probe digestion. This result was confirmed by primer extension analysis with primer fpg4 (Fig. 4A). The primer was extended to an 89-nucleotide product (Fig. 4B, lane 3), placing the transcription initiation site −23 bp from the fpg start codon.
Probe 5 was designed to map the terminator(s) of the operon. The RPA with probe 5 resulted in products of 93, 177, and 360 nucleotides as well as several other, less prominent bands (Fig. 2E, lanes 2 and 3). Since kdtB, the gene 3′ to fpg, begins 39 bp from the fpg stop codon and is in the opposite orientation, probe 5 cannot anneal to kdtB RNA. For this reason, it was assumed that all probe digestion took place from the 5′ end. The patterns of protected products from wild-type cells (Fig. 2E, lane 2) and rnc rne cells (Fig. 2E, lane 3) were similar, except for a faint full-length product and a more abundant 93-nucleotide product in the rnc rne cells. The 93-nucleotide protected product corresponds to a previously predicted region of secondary structure between fpg and kdtB (4) that may act as a bidirectional transcription terminator. This product maps to a site 19 bp 3′ to the fpg stop codon. The 177- and 360-nucleotide products correspond to termination sites 103 bp and 286 bp 3′ to the fpg stop codon, respectively. Both of these termination sites are within the kdtB coding region. The termination sites were confirmed with probe 8 (Fig. 3A); this assay resulted in protected products of 355, 434, and 644 nucleotides and several less prominent protected products (Fig. 3C, lane 2; this assay was performed with wild-type RNA only). These products correspond to termination sites 29, 108, and 318 bp 3′ to the fpg stop codon and, taken together, are within 10 to 30 bp of the sites mapped with probe 5.
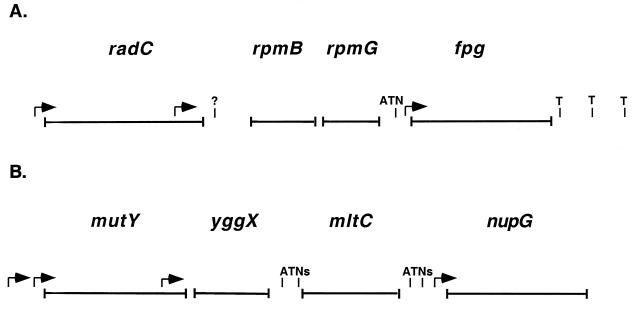
These results indicate that fpg is cotranscribed as the terminal gene in an operon with the gene order radC, rpmB, rpmG, and fpg. There are transcription initiation sites upstream of radC, in the radC coding region, and immediately upstream of fpg (Fig. 5A). In addition, a 5′ transcript end detected in the radC-rpmB intergenic region corresponds to either a promoter or an RNase III or RNase E cleavage site. There is a strong attenuator in the rpmG-fpg intergenic region, and there are three apparent terminators downstream of fpg.
FIG. 5.
Features of the operons. Arrows represent mapped transcription initiation sites. ATN, attenuator—representing mapped termination sites that also allow transcript readthrough. T, mapped termination sites. (A) fpg operon. The question mark represents a mapped 5′ transcript end that corresponds either to a transcription initiation site or to an RNase III or RNase E cleavage site. (B) mutY operon.
mutY is part of an operon containing four genes.
The mutY transcript was initially mapped by RT-PCR to determine if it is transcribed alone or with surrounding genes in the same orientation. The order of genes that could potentially be transcribed together is mutY, yggX, mltC, and nupG. There are 27 bp between the mutY stop codon and the yggX start codon, 61 bp between the yggX stop codon and the mltC start codon, and 153 bp between the mltC stop codon and the nupG start codon. The gene 5′ to mutY, yggH, and the gene 3′ to nupG, speC, are in opposite orientations on the genome, precluding them from being transcribed with mutY. E. coli total RNA was reverse transcribed with primer mutY11, which anneals 465 bp 3′ to the nupG start codon proposed in the GenBank accession no. AE000379 sequence; PCR was performed to determine the approximate size of the cDNA (Fig. 6A). PCR with primers mutY8-mutY5 and mutY12-mutY16 resulted in a product (Fig. 6B, lanes 3 and 6), indicating that mutY, yggX, mltC, and nupG were present on the same transcript. PCR was also performed with primers mutY12-mutY5, which resulted in a product, and mutY3-mutY5, which did not (Fig. 6C, lanes 3 and 6); these results confirm that the four genes were present on the same transcript and that the upstream transcription initiation site for the operon was within 271 bp of the mutY start codon. Positive PCR controls were run with the primers sets and genomic DNA as a template (Fig. 6B, lanes 1 and 4, and Fig. 6C, lanes 1 and 4). The negative control lanes were empty, indicating that the RNA used for RT was not contaminated with DNA (Fig. 6B, lanes 2 and 5, and Fig. 6C, lanes 2 and 5).
FIG. 6.
Mapping of the mutY transcript by RT-PCR. (A) The arrows numbered 5, 16, 3, 12, and 8 represent the primers used in PCR. The arrow numbered 11* represents the primer used to reverse transcribe E. coli total RNA. Primer mutY5 anneals 162 bp 3′ to the nupG start codon, mutY16 anneals 46 bp 3′ to the yggX start codon, mutY3 anneals 271 bp 5′ to the mutY start codon, mutY12 anneals 779 bp 5′ to the mutY stop codon, and mutY8 anneals 151 bp 5′ to the yggX stop codon. (B) The primer sets used in PCR are indicated above the lanes. Lanes 1 and 4 are positive controls with E. coli genomic DNA as a PCR template. Lanes 2 and 5 are negative controls with RT reaction mixtures minus reverse transcriptase as a PCR template. Lanes 3 and 6 show PCR results obtained with the cDNA from RT with primer 11* (RT11) as a template. The sizes of the PCR products are indicated on the right. (C) The primer sets used in PCR are indicated above the lanes. Lanes 1 and 4 are positive controls with E. coli genomic DNA as a PCR template. Lanes 2 and 5 are negative controls with RT reaction mixtures minus reverse transcriptase as a PCR template. Lanes 3 and 6 show PCR results obtained with the cDNA from RT with primer 11* (RT11) as a template. The sizes of the PCR products are indicated on the right.
The mutY operon contains multiple promoters and terminators.
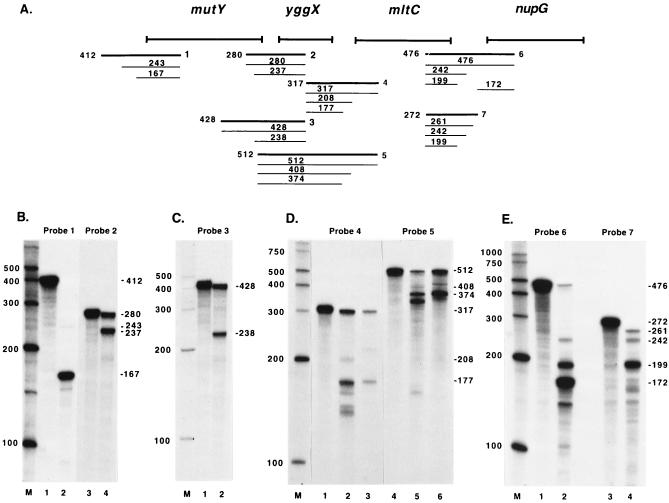
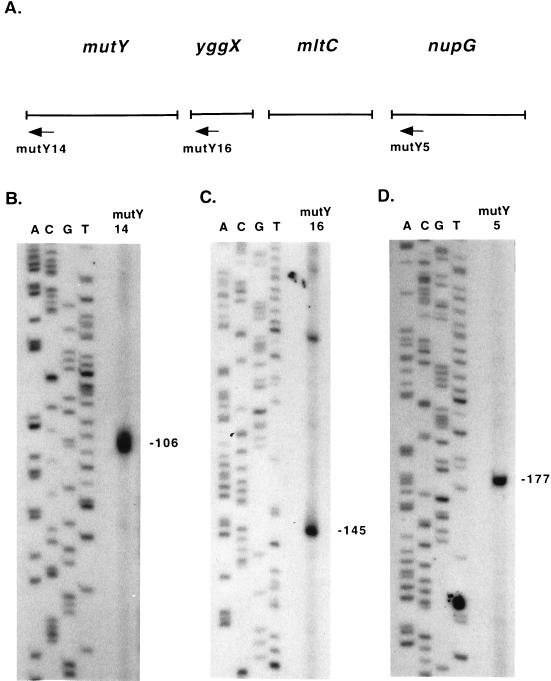
RPAs were performed to confirm the RT-PCR results and to map the locations of promoters and terminators. The assays were performed as described above, except that only RNA from wild-type E. coli (GC4468) was used. Probe 1 (Fig. 7A) was used to map the 5′-most promoter(s) for the operon. The RPA with probe 1 resulted in a faint 243-nucleotide product, a 167-nucleotide product, and a 151-nucleotide product (Fig. 7B, lane 2). The 151-nucleotide product was not consistently seen in other experiments (data not shown) and was not pursued further. The 243-nucleotide product corresponds to a possible minor transcription initiation site about 102 bp 5′ to the mutY start codon, and the 167-nucleotide product corresponds to a possible transcription initiation site about 26 bp 5′ to the mutY start codon. Attempts to map the minor transcription initiation site by primer extension analysis were inconclusive. The site corresponding to the 167-nucleotide product was mapped by primer extension analysis with primer mutY14 (Fig. 8A). The primer was extended to a product of 106 bp (Fig. 8B), corresponding to a transcription initiation site 25 bp 5′ to the mutY start codon.
FIG. 7.
Mapping of the mutY transcript by RPAs. (A) The thick lines designated 1 to 7 indicate the approximate annealing locations of the probes used in the RPAs. The thin lines indicate the products obtained from RPAs with these probes along with the sizes of the products. The sizes and exact annealing locations of the probes are as follows: probe 1, 412 nucleotides, anneals from 141 bp 3′ to the mutY start codon to 271 bp 5′ to the mutY start codon; probe 2, 280 nucleotides, anneals from 140 bp 3′ to the yggX start codon to 113 bp 5′ to the mutY stop codon; probe 3, 428 nucleotides, anneals from 140 bp 3′ to the yggX start codon to 261 bp 5′ to the mutY stop codon; probe 4, 317 nucleotides, anneals from 105 bp 3′ to the mltC start codon to 151 bp 5′ to the yggX stop codon; probe 5, 512 nucleotides, anneals from 105 bp 3′ to the mltC start codon to 43 bp 5′ to the mutY stop codon; probe 6, 476 nucleotides, anneals from 157 bp 3′ to the nupG start codon to 166 bp 5′ to the mltC stop codon; probe 7, 272 nucleotides, anneals from 106 bp 3′ to the mltC stop codon to 166 bp 5′ to the mltC stop codon. (B to E) The probes used in the RPAs are indicated, and lane M contains RNA size markers. These experiments were all done with wild-type RNA. (B) Lanes 1 and 3 show the full-length probe, and lanes 2 and 4 show the RPA results. (C) Lane 1 shows the full-length probe, and lane 2 shows the RPA results. (D) Lanes 1 and 4 show the full-length probe, lanes 2 and 5 show the RPA results, and lanes 3 and 6 show the RPA results after RNase T1 digestion alone instead of RNase A-RNase T1 digestion. (E) Lanes 1 and 3 show the full-length probe, and lanes 2 and 4 show the RPA results. Numbers beside panels are in base pairs.
FIG. 8.
Mapping of mutY transcription initiation sites by primer extension analysis. (A) Arrows indicate primers used in primer extension analysis. mutY14 anneals 81 bp 3′ to the mutY start codon, mutY16 anneals 46 bp 3′ to the yggX start codon, and mutY5 anneals 162 bp 3′ to the nupG start codon. (B to D) The sequencing ladder was generated with control M13mp18 DNA. (B) Primer extension with primer mutY14. (C) Primer extension with primer mutY16. (D) Primer extension with primer mutY5. Numbers beside panels are in base pairs.
The RPA with probe 2 resulted in a full-length product and a 237-nucleotide product (Fig. 7B, lane 4). To determine whether the 237-nucleotide product was the result of 5′ or 3′ probe digestion, an RPA was performed with probe 3. This assay resulted in a full-length product and a 238-nucleotide product (Fig. 7C, lane 2), showing that digestion had occurred from the 3′ end of the probe and that there is a possible transcription initiation site about 70 bp 5′ to the mutY stop codon. This site was mapped by primer extension analysis with primer mutY16 (Fig. 8A). The primer was extended to a 145-nucleotide product (Fig. 8C), which corresponds to a transcription initiation site 72 bp 5′ to the mutY stop codon.
The RPA with probe 4 yielded a full-length product and products of 208, 177, 163, 149, and 143 nucleotides (Fig. 7D, lane 2). The portion of the transcript that this probe anneals to has several regions that are AU rich; this characteristic can result in transient strand separation of the transcript and probe hybrid, leaving it susceptible to cleavage by RNase A, which cleaves 3′ to cytosine and uridine residues. To determine whether any of the protected products were due to RNase A cleavage of separated strands, an assay was performed with probe 4 and RNase T1 digestion only. RNase T1 cleaves 3′ to guanosine residues, a characteristic which should eliminate nonspecific cleavage due to strand separation. After RNase T1 digestion, the 163-, 149-, and 143-nucleotide products disappeared (Fig. 7D, lane 3). The sizes of these products indicate that there were one or two RNase A cleavages of the probe in an AU-rich region surrounding the yggX stop codon. The 177- and 210-nucleotide products seen after RNase A and RNase T1 digestion shifted to 178- and 225-nucleotide products, respectively, after RNase T1 digestion. Less sample was loaded, so the bands look fainter than in Fig. 7D, lane 2.
Probe 5 was used to determine if these products were due to 5′ or 3′ probe digestion. RNase A and RNase T1 digestion resulted in a full-length product, a faint 408-nucleotide product, and 374-, 342-, and 163-nucleotide products (Fig. 7D, lane 5). After RNase T1 digestion alone, the 163- and 342-nucleotide products disappeared (Fig. 7D, lane 6). The sizes of these products indicate that they were due to the same RNase A cleavage events that occurred with probe 4. The 374-nucleotide product shifted to a 375-nucleotide product and the 408-nucleotide product shifted to a 424-nucleotide product after RNase T1 digestion alone. The sizes of the products obtained with probe 5 showed that 5′ digestion had occurred with both probes and indicate that there are two termination sites in the intergenic region between yggX and mltC. The second termination site appears to be minor. The 177-nucleotide product obtained by RNase A and RNase T1 digestion of probe 4 and the 374-nucleotide product obtained by RNase A and RNase T1 digestion of probe 5 map to a C residue 26 bp 3′ to the yggX stop codon and an A residue 28 bp 3′ to the stop codon, respectively. It is not possible to determine with certainty, from these experiments, which residue is the terminating residue, since the size predictions are not exact. A possible stem-loop structure immediately preceding this site may act as an attenuator, although it would appear to be a weak attenuator, since there is substantial transcript readthrough. The minor termination site, corresponding to the 208-nucleotide product obtained by RNase A and RNase T1 digestion of probe 4, and the 408-nucleotide product obtained by RNase A and RNase T1 digestion of probe 5 map to a G residue 3 bp 5′ to the mltC start codon and to the A residue of the mltC start codon, respectively.
The RPA with probe 6 resulted in a full-length product and products of 242, 199, 172, and 144 nucleotides (Fig. 7E, lane 2). The 144-nucleotide product disappeared after RNase T1 digestion (data not shown), indicating that it was due to cleavage of separated hybrid strands. A nupG transcription initiation site was previously mapped by S1 nuclease digestion to an A residue 15 bp 5′ to the predicted start codon (29). 3′ digestion of probe 6 back to this transcription initiation site would result in a product of 172 nucleotides, which is what was seen. To confirm that this product resulted from 3′ probe digestion and to determine whether the other products were due to 5′ or 3′ probe digestion, an RPA was performed with probe 7. This assay resulted in 261-, 242-, and 199-nucleotide products (Fig. 7E, lane 4). The 172-nucleotide product seen with probe 6 was no longer present, confirming that it was due to 3′ probe digestion. The 242- and 199-nucleotide products were present with both probes, indicating that they were due to 5′ probe digestion and that they correspond to termination sites. The 199-nucleotide product maps to a termination site 34 bp 3′ to the mltC stop codon, and the 242-nucleotide product maps to a termination site 76 bp 3′ to the mltC stop codon. Both of these sites are immediately preceded by possible stem-loop structures that may act as terminators. The lack of a full-length product and the presence of a 261-nucleotide product not seen with probe 6 suggest that RNase “nibbling” at the 5′ end of the probe-RNA hybrid may have resulted in the 261-nucleotide product.
To confirm the nupG transcription initiation site, primer extension analysis was performed with primer mutY5 (Fig. 8A). The primer was extended to a product of 177 nucleotides (Fig. 8D), which corresponds to a transcription initiation site at the previously mapped A residue 15 bp 5′ to the nupG start codon. Starting 7 bp downstream of the nupG stop codon, there is a structure with the features of a rho-independent terminator (C+G-rich stem-loop structure followed by a stretch of T residues) (42), but this structure was not mapped in the present study.
These results indicate that mutY is cotranscribed as the first gene in an operon with the gene order mutY, yggX, mltC, and nupG. There are transcription initiation sites upstream of mutY, in the mutY coding region, and immediately upstream of nupG (Fig. 5B). The operon also appears to have attenuators in the yggX-mltC and mltC-nupG intergenic regions.
The genes in the operons are arranged in a similar manner in other organisms.
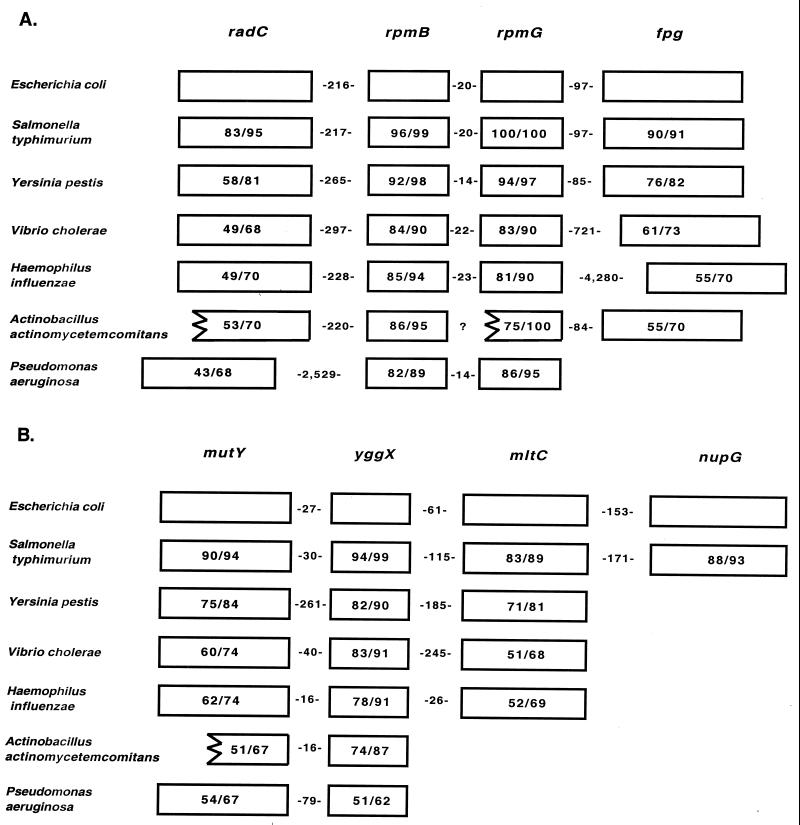
BLAST searches were performed with the finished and unfinished genome databases at the NCBI Microbial Genome BLAST Website and with unfinished genome databases at other sites (see Materials and Methods) by use of the E. coli amino acid sequences for the products of the genes in both operons. The results showed that in S. typhimurium and Y. pestis, the genes of the E. coli fpg operon are arranged in the same order and with similar spacing (Fig. 9A). In V. cholerae and Haemophilus influenzae, radC, rpmB, and rpmG are arranged in the same order and with similar spacing, but fpg is separated from them by 722 and 4,281 bp, respectively. A. actinomycetemcomitans may have the genes arranged in the same order, but it was not possible to confirm this notion. Parts of radC and rpmB were shown to be on the same contig with similar spacing, and the 3′ end of rpmG and all of fpg were shown to be on the same contig with similar spacing. In P. aeruginosa, radC is separated from rpmB and rpmG by 2,530 nucleotides, and the gene for fpg appears to be located elsewhere on the genome.
FIG. 9.
Conservation of gene arrangement. The databases used to compile these charts are listed in Materials and Methods. The numbers in the boxes are percent identity/percent similarity of the proteins relative to the E. coli proteins. The numbers between the boxes represent the numbers of base pairs in the intergenic regions. (A) fpg operon. (B) mutY operon. A question mark indicates unknown.
The genes of the E. coli mutY operon are arranged in the same order and with similar spacing in S. typhimurium (Fig. 9B). In Y. pestis, V. cholerae, and H. influenzae, the first three genes are arranged in the same order and with similar spacing, but these organisms do not appear to have a nupG homolog. In A. actinomycetemcomitans and P. aeruginosa, the first two genes are arranged in the same order and with similar spacing, but neither of these organisms appears to have a mltC or nupG homolog.
DISCUSSION
fpg is transcribed as the terminal gene in the radC-rpmB-rpmG-fpg operon, and mutY is transcribed as the first gene in the mutY-yggX-mltC-nupG operon (Fig. 5). The genes in these operons have no apparent relationship to the function of Fpg or MutY as DNA glycosylases, with the possible exception of radC, the first gene in the fpg operon. A mutation in radC (radC102) sensitizes cells to UV irradiation and X-irradiation, and cells with this mutation are 60% deficient in recombination ability (15). It has been shown that radC102 mutants are deficient in recA-dependent repair of X-ray-induced single-strand breaks and that this deficiency correlates with their X-ray survival deficiency; the mutants also have a small deficiency in repair of X-ray-induced double-strand breaks (16). radC was cloned as a 297-bp open reading frame (ORF) based, in part, on partial restoration of resistance to a radC102 strain carrying a plasmid containing the ORF (14). The present study shows that the mapped size of the radC transcript is more in accordance with an ORF of 675 bp, as assigned in GenBank accession no. U000441, which places the ATG start site 375 bp 5′ to the originally predicted start site.
rpmB and rpmG encode ribosomal proteins L28 and L33, respectively (22), which are 2 of the 52 r-proteins made by E. coli. In E. coli, many of the r-protein genes are arranged in operons, and many of these operons include non–r-protein genes. Three r-protein operons, in particular, are similar to the fpg operon in that they have multiple promoters, undergo transcript processing by RNase III and/or RNase E, and have an attenuator between the r-protein gene(s) and the non–r-protein gene(s). The rplK-rplA-rplJ-rplL-rpoB-rpoC operon encodes r-proteins L11, L1, L10, and L7/12 and RNA polymerase subunits β and β′ (6). This operon has promoters upstream of rplK, in the rplA-rplJ intergenic region, and there may be a minor promoter in the rplL-rpoB intergenic region (6, 23, 44). The attenuator in the rplL-rpoB intergenic region terminates 80% of transcripts, and nonattenuated transcripts are cleaved by RNase III (1, 10). The rpsU-dnaG-rpoD operon encodes r-protein S21, DNA primase, and sigma-70 (7). This operon has two promoters upstream of rpsU and a heat shock promoter in the dnaG coding region (7, 38). The attenuator between rpsU and dnaG terminates 80 to 90% of transcripts, and nonattenuated transcripts are cleaved between dnaG and rpoD by RNase E, after which the dnaG message is rapidly degraded (7, 24, 43). The rpsO-pnp operon encodes r-protein S15 and polynucleotide phosphorylase (37). This operon has promoters upstream of rpsO and upstream of pnp. There is an attenuator between the two genes that terminates 50% of transcripts, and nonattenuated transcripts are processed by both RNase III and RNase E (34, 35, 37). The attenuators in these operons all resemble rho-independent terminators. The obvious difference between these operons and the fpg operon is that there is a non–r-protein gene, radC, preceding the r-protein genes in the fpg operon.
There are at least three promoters in the fpg operon (Fig. 5A). The promoter upstream of radC appears to be a weak one, since very little RNA message originating from it was detected by RPAs in either the wild-type strain or the rnc rne strain (Fig. 2B, lanes 2 and 3). At least some of the transcript originating from this promoter extends through fpg, since it was possible to detect a transcript containing all four genes by RT-PCR. The promoter located in the 3′ end of the radC coding region, upstream of rpmB and rpmG, was expressed at equal strengths in wild-type and rnc rne strains (Fig. 2C, lanes 2 and 3; 483-nucleotide product). There is a possible −10 hexamer (TAAACT) 8 bp upstream of the transcription initiation site and separated by 18 bp from a possible −35 hexamer (TTGTGC). The transcription initiation site upstream of fpg that was identified in this study is 1 bp 5′ to a transcription initiation site mapped in a previous study (5). This initiation site is 2 bp downstream from a proposed −10 hexamer (TTGACT) separated by 16 bp from a proposed −35 hexamer (TTGTTA). The transcript originating from this promoter was present in an approximately 30-fold-larger amount in rnc rne cells than in wild-type cells (Fig. 2D, lanes 2, 3, 5, and 6; 109-nucleotide product), as determined by phosphorimager analysis. At this point it is unclear what is causing this difference. It is possible that a negative regulator that normally represses transcription from the fpg promoter is not active in rnc rne cells. Three repressors of Fpg activity, FNR, Fur, and ArcA, and their possible binding sites in the fpg promoter region have been identified (21), although there are no reports that the transcripts for these proteins require RNase III and/or RNase E processing to be properly expressed. In addition, these regulators appear to repress Fpg activity under anaerobic conditions, so it is unlikely that they would have such a dramatic effect in aerobic growth. It is also possible that RNase III and/or RNase E play a role in the rapid degradation of the fpg transcript in wild-type cells.
The attenuator in the rpmG-fpg intergenic region is located very close to the predicted −35 hexamer for fpg and resembles a rho-independent terminator. One possible secondary structure has a 6-bp G+C-rich stem (with one mismatch) followed by a run of six T residues. The last two T residues in this run are the first two in the predicted TTGTTA −35 hexamer. A second possible secondary structure has up to a 15-bp stem which includes the first stem plus the entire predicted −35 hexamer. This configuration would sequester the −35 hexamer and could result in the poor transcription from the promoter seen in wild-type cells, but it does not help explain the dramatic increase in transcript seen in the double mutants. The sizes of the RPA products corresponding to the attenuated transcripts indicate that the transcripts terminate between 2 and 10 bp before the −35 hexamer (Fig. 2D, lanes 2 and 3 [273-nucleotide product], and lanes 5 and 6 [94-nucleotide product]). The nonattenuated, full-length transcript is present in a 2.6- to 3.6-fold-larger amount in rnc rne cells than in wild-type cells, indicating that RNase III and/or RNase E play a role in processing the readthrough transcript. It has been suggested that the termination frequency at the attenuator between rplL and rpoB in the rplK-rplA-rplJ-rplL-rpoB-rpoC operon can be increased by rho, even though it appears to be a rho-independent terminator, and decreased by NusA (33). It has also been shown in vitro that NusA decreases termination frequency at the attenuator between rpsU and dnaG in the rpsU-dnaG-rpoD operon (31). Studies are under way to determine if conditions that reduce the termination frequency at the attenuator upstream of fpg exist.
An additional site that corresponds to either a promoter or an RNase III or RNase E cleavage site was detected. This 5′ transcript end (Fig. 2C, lane 2; 318-nucleotide product) in the radC-rpmB intergenic region was detected in wild-type cells but not in rnc rne cells and was also detected by primer extension analysis in wild-type cells. This site was previously predicted to be a promoter, and a possible −10 hexamer (TATACT) and −35 hexamer (TTGAGC) were identified (22). If this site is a promoter, it is completely down-regulated in rnc rne cells. Perhaps there is a transcriptional activator which requires the presence of RNase III and/or RNase E for activity. Or this site may be, as it appears, a cleavage site. RNase E cleaves single-stranded RNA, and possible consensus sequences have been proposed. The motif ACAG(A/U)AUUUG was predicted based on sequence similarities between RNase E cleavage sites in 9S RNA and RNA I (40). Other investigators, using site-directed mutagenesis of the phage T4 RNase E cleavage site, proposed (A/G)AUU(A/U) as the consensus sequence (13). The sequence surrounding the possible cleavage site in the fpg operon is GCCACCUUUG. This sequence is 50% homologous to the first predicted RNase E consensus sequence and 60% homologous to the second. RNase III cleaves double-stranded RNA. A preliminary examination of the region surrounding the possible cleavage site with Genetics Computer Group programs Stemloop and Foldrna did not reveal a secondary structure resembling an RNase III cleavage site that would allow the cleaved residue to be in double-stranded form. No 3′ transcript end was detected with probe 2 or 7 that would correspond to a cleavage event (Fig. 2C, lane 2, and Fig. 3B, lane 4). If the RNA is cleaved at this site by RNase E or RNase III, the upstream RNA must be rapidly degraded. This could be part of the reason that radC appears to be poorly transcribed in wild-type cells. There was not an appreciable increase in the radC 635-nucleotide product in rnc rne cells compared to wild-type cells, but the laddering below this band was more intense (Fig. 2B, lanes 2 and 3). It is unclear why this laddering occurred, but it was present with different probe and RNA preparations. A cleavage site between radC and rpmB would explain the increase in the full-length transcript seen in rnc rne cells with probe 2 (Fig. 2C, lanes 2 and 3). If there is a cleavage site, though, less transcript should originate from the promoter in the 3′ end of radC because this would also be processed, and this was not the case (Fig. 2C; 483-nucleotide product). Further experiments are needed to determine whether the 5′ transcript end corresponds to a promoter or a cleavage site. Three termination sites were mapped downstream of the fpg operon. The first one corresponds to a predicted stem-loop structure in the fpg-kdtB intergenic region, and the other two correspond to sites in the kdtB coding region (Fig. 2E and 3C).
The mutY-yggX-mltC-nupG operon consists of genes that have no apparent relationship to each other, although the function of the protein encoded by yggX is unknown. There is a possible minor transcription initiation site 102 bp 5′ to the mutY start codon (Fig. 5B). A second transcription initiation site, 25 bp 5′ to the mutY start codon, corresponds to a previously predicted promoter with a proposed −10 hexamer (TGCAAT) and a proposed −35 hexamer (TTTACA) separated by 17 bp (41). A third transcription initiation site, 72 bp 5′ to the mutY stop codon, is 9 bp downstream from a possible −10 hexamer (TATAAC) and a possible −35 hexamer (TTGATG) separated by 16 bp. The last transcription initiation site in the operon is 15 bp 5′ to the nupG start codon and was previously mapped by an S1 nuclease digestion assay (29). There are termination sites in the yggX-mltC and mltC-nupG intergenic regions. Not all transcripts were terminated at these sites, so there must be secondary structures which act as attenuators. These experiments were performed only with wild-type RNA, so we cannot rule out the possibility that the apparent termination sites correspond to RNase III and/or RNase E cleavage sites.
The arrangement of promoters and terminators in the operon appears to allow different combinations of genes to be transcribed together. From the two 5′-most promoters, mutY can be cotranscribed with yggX, with yggX and mltC, or with all three downstream genes. From the promoter in the 3′ end of mutY, yggX can be transcribed alone, with mltC, or with both downstream genes. nupG can be transcribed from its own independent promoter, and the intensity of the RPA product corresponding to this promoter (Fig. 7E, lane 2; 172-nucleotide product) indicates that it is the most active promoter in the operon. The amount of nonattenuated transcript reading from upstream of nupG into nupG is minor, and it is possible that nupG is predominantly regulated independently of the other genes. It has been shown that the nupG promoter is activated 50-fold by the cyclic AMP-cyclic AMP receptor protein complex and repressed fourfold by CytR and DeoR and that it has binding sites for these proteins (29, 32).
There have been no reports on the regulation of MutY, YggX, or MltC. Since MutY helps repair mispairs resulting from the presence of 8-oxoG in DNA, it is possible that it is up-regulated under conditions that generate the 8-oxoG lesion. MltC is a potentially lytic protein, and it would seem important for cells to control the amount present. The glycan strand of peptidoglycan is made up of N-acetylmuramic acid and N-acetylglucosamine joined together in an alternating sequence by (β1-4) glycosidic bonds. In order for cells to grow and divide, the glycan strand must be cleaved to allow the insertion of new materials. Lytic transglycosylases can cleave the (β1-4) bond between N-acetylmuramic acid and N-acetylglucosamine and transfer the muramyl bond to the carbon-6 hydroxyl group of the same N-acetylmuramic acid, forming (1-6) anhydromuramic acid (19). MltC has been shown to be a membrane-bound lytic glycosylase with the same activity (11). E. coli has three other proteins possessing this activity, one soluble (Slt70) and two membrane bound (MltA and MltB). These proteins have been shown to act as exomuramidases in that they start at the glucosamine end and cleave the glycan strand in a processive manner (for a review, see reference 18). It is not known whether each of these proteins plays a specific role in the cell or whether the redundancy serves as a backup mechanism. Since these proteins can cause cell lysis, it would seem necessary to strictly regulate them, but the soluble lytic transglycosylase can be overexpressed 30-fold without harming cells. It was shown that Slt-dependent autolysis is suppressed by RelA-mediated stringent control, possibly by keeping the enzyme in an inactive form or in an unfavorable topological position (3). It has not been shown whether the enzyme activities of the membrane-bound lytic transglycosylases are controlled in a similar manner or whether any of them is regulated at the transcriptional level. Since mltC does not have an independent promoter, it is possible that the frequency of transcript termination between yggX and mltC plays a role in regulating MltC.
Searches of the genomes of other organisms revealed that the order of genes in both operons is conserved or partially conserved in several closely related gram-negative bacteria, as shown in Fig. 9. Of course, conservation of gene order does not mean that the transcriptional organization has been conserved, and there are no reports on how these genes are transcribed in the organisms identified. It is tempting to speculate that the conserved gene order implies a functional or regulatory significance, but it may just mean that the organisms are so closely related evolutionarily that the genes have not had time to rearrange. We have not seen any obvious homologies to consensus sequence binding sites for LexA, SoxS, OxyR, or KatF in the promoter regions of the fpg or mutY operon. Studies are currently under way to examine the regulation of each of these operons.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant R37 CA33657 awarded by the National Cancer Institute. Christine M. Gifford was supported by Environmental Pathology training grant T32 07122 awarded by the National Institute of Environmental Health Sciences.
REFERENCES
- 1.Barry G, Squires C, Squires C L. Attenuation and processing of RNA from the rplJL-rpoBC transcription unit of Escherichia coli. Proc Natl Acad Sci USA. 1980;77:3331–3335. doi: 10.1073/pnas.77.6.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beckman K B, Ames B N. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 3.Betzner A S, Ferreira L C, Holtje J V, Keck W. Control of the activity of the soluble lytic transglycosylase by the stringent response in Escherichia coli. FEMS Microbiol Lett. 1990;55:161–164. doi: 10.1016/0378-1097(90)90187-u. [DOI] [PubMed] [Google Scholar]
- 3a.Blaisdell, J. O., et al. Submitted for publication.
- 4.Boiteux S, O’Connor T R, Laval J. Formamidopyrimidine-DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 1987;6:3177–3183. doi: 10.1002/j.1460-2075.1987.tb02629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boiteux S, O’Connor T R, Lederer F, Gouyette A, Laval J. Homogeneous Escherichia coli FPG protein. A DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidinic sites. J Biol Chem. 1990;265:3916–3922. [PubMed] [Google Scholar]
- 6.Bruckner R, Matzura H. In vivo synthesis of a polycistronic messenger RNA for the ribosomal proteins L11, L1, L10 and L7/12 in Escherichia coli. Mol Gen Genet. 1981;183:277–282. doi: 10.1007/BF00270629. [DOI] [PubMed] [Google Scholar]
- 7.Burton Z F, Gross C A, Watanabe K K, Burgess R R. The operon that encodes the sigma subunit of RNA polymerase also encodes ribosomal protein S21 and DNA primase in E. coli K12. Cell. 1983;32:335–349. doi: 10.1016/0092-8674(83)90453-1. [DOI] [PubMed] [Google Scholar]
- 8.Cabrera M, Nghiem Y, Miller J H. mutM, a second mutator locus in Escherichia coli that generates G · C→T · A transversions. J Bacteriol. 1988;170:5405–5407. doi: 10.1128/jb.170.11.5405-5407.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung M H, Kasai H, Jones D S, Inoue H, Ishikawa H, Ohtsuka E, Nishimura S. An endonuclease activity of Escherichia coli that specifically removes 8-hydroxyguanine residues from DNA. Mutat Res. 1991;254:1–12. doi: 10.1016/0921-8777(91)90035-n. [DOI] [PubMed] [Google Scholar]
- 10.Dennis P P. Transcription patterns of adjacent segments on the chromosome of Escherichia coli containing genes coding for four 50S ribosomal proteins and the beta and beta′ subunits of RNA polymerase. J Mol Biol. 1977;115:603–625. doi: 10.1016/0022-2836(77)90105-x. [DOI] [PubMed] [Google Scholar]
- 11.Dijkstra A J, Keck W. Identification of new members of the lytic transglycosylase family in Haemophilus influenzae and Escherichia coli. Microb Drug Resist. 1996;2:141–145. doi: 10.1089/mdr.1996.2.141. [DOI] [PubMed] [Google Scholar]
- 12.Dreher D, Junod A F. Role of oxygen free radicals in cancer development. Eur J Cancer. 1996;32A:30–38. doi: 10.1016/0959-8049(95)00531-5. [DOI] [PubMed] [Google Scholar]
- 13.Ehretsmann C P, Carpousis A J, Krisch H M. Specificity of Escherichia coli endoribonuclease RNase E: in vivo and in vitro analysis of mutants in a bacteriophage T4 mRNA processing site. Genes Dev. 1992;6:149–159. doi: 10.1101/gad.6.1.149. [DOI] [PubMed] [Google Scholar]
- 14.Felzenszwalb I, Boiteux S, Laval J. Molecular cloning and DNA sequencing of the radC gene of Escherichia coli K-12. Mutat Res. 1992;273:263–269. doi: 10.1016/0921-8777(92)90088-k. [DOI] [PubMed] [Google Scholar]
- 15.Felzenszwalb I, Sargentini N J, Smith K C. Characterization of a new radiation-sensitive mutant, Escherichia coli K-12 radC102. Radiat Res. 1984;97:615–625. [PubMed] [Google Scholar]
- 16.Felzenszwalb I, Sargentini N J, Smith K C. Escherichia coli radC is deficient in the recA-dependent repair of X-ray-induced DNA strand breaks. Radiat Res. 1986;106:166–170. [PubMed] [Google Scholar]
- 17.Gallardo-Madueno R, Leal J F, Dorado G, Holmgren A, Lopez-Barea J, Pueyo C. In vivo transcription of nrdAB operon and of grxA and fpg genes is triggered in Escherichia coli lacking both thioredoxin and glutaredoxin 1 or thioredoxin and glutathione, respectively. J Biol Chem. 1998;273:18382–18388. doi: 10.1074/jbc.273.29.18382. [DOI] [PubMed] [Google Scholar]
- 18.Holtje J V. From growth to autolysis: the murein hydrolases in Escherichia coli. Arch Microbiol. 1995;164:243–254. doi: 10.1007/BF02529958. [DOI] [PubMed] [Google Scholar]
- 19.Holtje J V, Mirelman D, Sharon N, Schwarz U. Novel type of murein transglycosylase in Escherichia coli. J Bacteriol. 1975;124:1067–1076. doi: 10.1128/jb.124.3.1067-1076.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Institute for Genomic Research. [Online.] http://www.tigr.org. [1 March 1999, last date accessed.]
- 20.Kim H S, Park Y W, Kasai H, Nishimura S, Park C W, Choi K H, Chung M H. Induction of E. coli oh8Gua endonuclease by oxidative stress: its significance in aerobic life. Mutat Res. 1996;363:115–123. doi: 10.1016/0921-8777(96)00006-7. [DOI] [PubMed] [Google Scholar]
- 21.Lee H S, Lee Y S, Kim H S, Choi J Y, Hassan H M, Chung M H. Mechanism of regulation of 8-hydroxyguanine endonuclease by oxidative stress: roles of FNR, ArcA, and Fur. Free Radical Biol Med. 1998;24:1193–1201. doi: 10.1016/s0891-5849(97)00427-9. [DOI] [PubMed] [Google Scholar]
- 22.Lee J S, An G, Friesen J D, Isono K. Cloning and the nucleotide sequence of the genes for Escherichia coli ribosomal proteins L28 (rpmB) and L33 (rpmG) Mol Gen Genet. 1981;184:218–223. doi: 10.1007/BF00272908. [DOI] [PubMed] [Google Scholar]
- 23.Linn T, Scaife J. Identification of a single promoter in E. coli for rplJ, rplL and rpoBC. Nature. 1978;276:33–37. doi: 10.1038/276033a0. [DOI] [PubMed] [Google Scholar]
- 24.Lupski J R, Smiley B L, Godson G N. Regulation of the rpsU-dnaG-rpoD macromolecular synthesis operon and the initiation of DNA replication in Escherichia coli K-12. Mol Gen Genet. 1983;189:48–57. doi: 10.1007/BF00326054. [DOI] [PubMed] [Google Scholar]
- 25.Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 26.Michaels M L, Cruz C, Grollman A P, Miller J H. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci USA. 1992;89:7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michaels M L, Miller J H. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michaels M L, Tchou J, Grollman A P, Miller J H. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry. 1992;31:10964–10968. doi: 10.1021/bi00160a004. [DOI] [PubMed] [Google Scholar]
- 29.Munch-Petersen A, Jensen N. Analysis of the regulatory region of the Escherichia coli nupG gene, encoding a nucleoside-transport protein. Eur J Biochem. 1990;190:547–551. doi: 10.1111/j.1432-1033.1990.tb15608.x. [DOI] [PubMed] [Google Scholar]
- 29a.NCBI Microbial Genome BLAST Website. [Online.] http://www.ncbi.nlm.nih.gov/BLAST/unfinished genome.html. [1 March 1999, last date accessed.]
- 30.Nghiem Y, Cabrera M, Cupples C G, Miller J H. The mutY gene: a mutator locus in Escherichia coli that generates G · C→T · A transversions. Proc Natl Acad Sci USA. 1988;85:2709–2713. doi: 10.1073/pnas.85.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peacock S, Lupski J R, Godson G N, Weissbach H. In vitro stimulation of Escherichia coli RNA polymerase sigma subunit synthesis by NusA protein. Gene. 1985;33:227–234. doi: 10.1016/0378-1119(85)90097-6. [DOI] [PubMed] [Google Scholar]
- 32.Pedersen H, Dall J, Dandanell G, Valentin-Hansen P. Gene-regulatory modules in Escherichia coli: nucleoprotein complexes formed by cAMP-CRP and CytR at the nupG promoter. Mol Microbiol. 1995;17:843–853. doi: 10.1111/j.1365-2958.1995.mmi_17050843.x. [DOI] [PubMed] [Google Scholar]
- 33.Ralling G, Linn T. Evidence that Rho and NusA are involved in termination in the rplL-rpoB intercistronic region. J Bacteriol. 1987;169:2277–2280. doi: 10.1128/jb.169.5.2277-2280.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Regnier P, Hajnsdorf E. Decay of mRNA encoding ribosomal protein S15 of Escherichia coli is initiated by an RNase E-dependent endonucleolytic cleavage that removes the 3′ stabilizing stem and loop structure. J Mol Biol. 1991;217:283–292. doi: 10.1016/0022-2836(91)90542-e. [DOI] [PubMed] [Google Scholar]
- 35.Regnier P, Portier C. Initiation, attenuation and RNase III processing of transcripts from the Escherichia coli operon encoding ribosomal protein S15 and polynucleotide phosphorylase. J Mol Biol. 1986;187:23–32. doi: 10.1016/0022-2836(86)90403-1. [DOI] [PubMed] [Google Scholar]
- 35a.Sanger Centre. [Online.] http://www.sanger.ac.uk/. [1 March 1999, last date accessed.]
- 36.Shibutani S, Takeshita M, Grollman A P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 37.Takata R, Mukai T, Hori K. Attenuation and processing of RNA from the rpsO-pnp transcription unit of Escherichia coli. Nucleic Acids Res. 1985;13:7289–7297. doi: 10.1093/nar/13.20.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor W E, Straus D B, Grossman A D, Burton Z F, Gross C A, Burgess R R. Transcription from a heat-inducible promoter causes heat shock regulation of the sigma subunit of E. coli RNA polymerase. Cell. 1984;38:371–381. doi: 10.1016/0092-8674(84)90492-6. [DOI] [PubMed] [Google Scholar]
- 39.Tchou J, Kasai H, Shibutani S, Chung M H, Laval J, Grollman A P, Nishimura S. 8-Oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc Natl Acad Sci USA. 1991;88:4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomcsanyi T, Apirion D. Processing enzyme ribonuclease E specifically cleaves RNA I. An inhibitor of primer formation in plasmid DNA synthesis. J Mol Biol. 1985;185:713–720. doi: 10.1016/0022-2836(85)90056-7. [DOI] [PubMed] [Google Scholar]
- 41.Tsai-Wu J J, Radicella J P, Lu A L. Nucleotide sequence of the Escherichia coli micA gene required for A/G-specific mismatch repair: identity of micA and mutY. J Bacteriol. 1991;173:1902–1910. doi: 10.1128/jb.173.6.1902-1910.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westh Hansen S E, Jensen N, Munch-Petersen A. Studies on the sequence and structure of the Escherichia coli K-12 nupG gene, encoding a nucleoside-transport system. Eur J Biochem. 1987;168:385–391. doi: 10.1111/j.1432-1033.1987.tb13431.x. [DOI] [PubMed] [Google Scholar]
- 43.Yajnik V, Godson G N. Selective decay of Escherichia coli dnaG messenger RNA is initiated by RNase E. J Biol Chem. 1993;268:13253–13260. [PubMed] [Google Scholar]
- 44.Yamamoto M, Nomura M. Cotranscription of genes for RNA polymerase subunits beta and beta′ with genes for ribosomal proteins in Escherichia coli. Proc Natl Acad Sci USA. 1978;75:3891–3895. doi: 10.1073/pnas.75.8.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanofsky C, Cox E C, Horn V. The unusual mutagenic specificity of an E. coli mutator gene. Proc Natl Acad Sci USA. 1966;55:274–281. doi: 10.1073/pnas.55.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]