Abstract
The hyperthermophilic eubacterium Thermotoga maritima possesses an operon encoding an Hsp70 molecular chaperone protein and a protein with meaningful homology to the small heat shock protein family of chaperones. This represents the first demonstrated co-operon organization for these two important classes of molecular chaperones. We have cloned and initially characterized these proteins as functional chaperones in vitro: the Hsp70 is capable of ATP hydrolysis and substrate binding, and the small heat shock protein can suppress protein aggregation and stably bind a refolding-competent substrate. In addition, the primary sequence of the Hsp70 is used to infer the phylogenetic relationships of T. maritima, one of the deepest-branching eubacteria known.
The 70-kDa heat shock protein (Hsp70) family is an extremely well conserved group of proteins which are present in almost all biota. These proteins serve as essential molecular chaperones which facilitate the correct folding of nonnative polypeptides, the translocation of proteins through membranes, and the formation of multiprotein assemblies (9, 11, 23). All of these functions are tied to the Hsp70 ability to bind short stretches of predominantly hydrophobic amino acids (3, 21, 24) and the modulation of this binding by the hydrolysis of ATP (7, 44, 50). Hsp70 complexed with ATP has faster exchange kinetics for its bound substrate, whereas Hsp70 complexed with ADP has slower substrate exchange kinetics (52).
Most, if not all, of the cellular roles of Hsp70 require it to work in concert with a group of protein cofactors. These cofactors include Hsp40 and the nucleotide exchange factor GrpE (6, 18, 32). The close functional association between these proteins is revealed by their common operon organization in prokaryotes. In all of the over 30 sequenced Hsp70 operons, the Hsp70 gene lies upstream of an Hsp40 gene and is often flanked by a GrpE gene (reviewed in reference 53). In addition to serving as a cofactor to Hsp70, Hsp40 proteins can serve as chaperones in their own right. They have been shown to bind substrate proteins and to suppress their aggregation in an ATP-independent manner (39, 48).
Other heat shock proteins which play a role in the binding and stabilization of substrate proteins include the eukaryotic Hsp90 and the widely distributed small heat shock proteins (sHsp’s) (14, 17). Hsp90s are isolated in stable complexes with Hsp70 and mammalian cellular receptors (51), and sHsp’s are implicated in the sequestration of unfolded proteins during times of cellular stress for future refolding or degradation (17). Of these three classes of chaperones, the ATP-independent Hsp40, sHsp’s, and the partially ATP-independent Hsp90, only sHsp’s have not been closely associated with the primary ATP-utilizing chaperone protein, Hsp70, in vivo.
sHsp’s are ubiquitous proteins which have homology with the mammalian eye lens protein α-crystallin (10, 13). They have been shown to function as molecular chaperones by their ability to suppress the aggregation of chemically denatured or heat-denatured protein substrates in vitro (28, 31) in an ATP-independent manner. More recently, sHsp’s were shown to bind stably to partially unfolded proteins in vitro which could then be productively refolded by ATP-dependent chaperones (16, 37). It is suspected that, in vivo, sHsp’s interact with chaperone proteins which utilize the energy from ATP hydrolysis to catalyze protein refolding (8). These ATP-utilizing chaperones could include Hsp70 or the chaperonin GroEL (eukaryotic Hsp60) (9).
To compare the chaperone properties of a hyperthermophilic Hsp70 system with those of a mesophile, we have cloned the partial Hsp70 operon from the eubacterium Thermotoga maritima. T. maritima was first isolated from geothermally heated sea floor sediments where it grows at temperatures from 50 to 85°C as an obligate anaerobe (29). Because 16S rRNA-based phylogenies place T. maritima as one of the deepest branches of the eubacteria (29), its protein sequences and gene structures are considered primitive and ancestral. Its genes often possess classical operon organization (33), and its well-studied enzymes are extremely thermostable (46). These characteristics are expected to extend to its chaperone proteins. The Hsp70 of T. maritima is also of value for the phylogenetic inferences that may be drawn from its primary sequence (25, 26).
Interestingly, the Hsp70 operon of T. maritima does not contain an Hsp40 or GrpE gene. In this study, we describe the cloning of the T. maritima Hsp70 and its operon partner and putative cochaperone, an sHsp. We demonstrate that these genes encode proteins which are functional molecular chaperones in vitro. The association between these two important groups of molecular chaperones, the Hsp70s and the sHsp’s, should aid in understanding the integration of these different chaperone systems in vivo.
MATERIALS AND METHODS
Hsp70 cloning and purification.
T. maritima (DSM 3109) was grown in a modified medium containing 37.4 g of Difco marine broth per liter, 5 g of Difco yeast extract per liter, 20 ml of seawater per liter, 5 g of glucose per liter, 2 g of NaHCO3 per liter, 0.68 g of cystine per liter, 0.5 g of cysteine per liter, 130 mg of Na2S per liter, and 1 mg of resazurin per liter. Cultures were grown at 80°C for 36 h in sealed bottles under 10 lb/in2 of N2 gas. Cells were harvested, and genomic DNA was extracted as described previously (33), omitting the final CsCl purification step. A degenerate primer (Keystone) (5′-ATGTCCAARATCATCGGWATHGAYCTBGG-3′; R = A or G, W = A or T, H = A, T, or C, Y = C or T, B = G, T, or C) at 2.5 pmol/μl and a nondegenerate primer (5′-GTGACACTATGCTTCTTGGAA-3′) at 1 pmol/μl were used in a PCR amplification mixture containing 500 ng of T. maritima genomic DNA. The amplified Hsp70 gene (1,808 bp total) was TA cloned into the vector pCR2.1 (Invitrogen) as recommended by the manufacturer. DNA sequencing (ABI) gave the complete 5′ sequence, and the entire Hsp70 gene was amplified with a second PCR with nondegenerate primers which created 5′ NdeI and 3′ EagI restriction sites. This PCR product was digested, purified, and ligated into the pET24a expression vector (Novagen). The final construct added a C-terminal hexahistidine extension to the Hsp70 and was used to transform Escherichia coli JM109(DE3). Cells were grown at 37°C, induced at A600 ≈ 0.5 with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside), grown for an additional 3 h, and harvested by centrifugation (JA-10 rotor, 11,000 × g for 20 min at 4°C). Cells were washed once with phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4) and centrifuged as before. Cells were resuspended in lysis buffer (50 mM HEPES [pH 7.5], 200 mM NaCl)–1 mM phenylmethylsulfonyl fluoride, lysed by two passages through an Aminco French press at ∼18,000 lb/in2 at 4°C, and clarified by centrifugation (Ti55.2 rotor, 132,000 × g for 20 min at 4°C). These extracts were heat treated at 70°C for 30 min with stirring, followed by an identical clarification spin, and filtered to 0.2 μm. This solution was loaded onto a nitrilotriacetic acid (Qiagen)-Ni affinity column equilibrated in lysis buffer at 4°C and purified as recommended by the manufacturer. Eluted protein was assayed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and pooled fractions were dialyzed (3 × 1:250) against 50 mM HEPES (pH 7.5)–100 mM KCl–100 mM NaCl and concentrated with Amicon 30 membranes. Reduced Hsp70Tm was prepared by incubating 15 μM Hsp70Tm with 3 mM TCEP [tris-(2-carboxyethyl) phosphine HCl] in 50 mM HEPES (pH 7.5)–100 mM KCl–100 mM NaCl at 40°C for 1 h, followed by buffer exchange on a Sephadex G-25 column (Pharmacia). The final protein concentration was assayed with the bicinchoninic acid reagent (Pierce) as recommended by the manufacturer. All Hsp70Tm samples were aliquoted, quick-frozen in liquid N2, and stored at −70°C.
sHsp cloning and purification.
Downstream sequences of the sHsp were obtained by the use of inverse PCR (42). Briefly, 500 μg of T. maritima genomic DNA was digested to completion with restriction enzymes chosen to yield ∼5-kb fragments. These fragments were ligated with T4 DNA ligase (New England Biolabs) under conditions which favored intramolecular ligation. The resultant circular genomic libraries were used as templates for a PCR amplification outward from known sHsp sequences. A single library (EcoRI digested) yielded a 2.5-kb PCR product which was not present in an identical PCR amplification with unligated genomic DNA as template. This 2.5-kb fragment was TA cloned into the vector pCR2.1 (Invitrogen) as recommended by the manufacturer. DNA sequencing (ABI) gave the complete 3′ sequence of the sHsp gene, as well as additional downstream sequences, and the entire sHsp gene was amplified with a second PCR with nondegenerate primers which created 5′ NdeI and 3′ EagI restriction sites. This PCR product was digested, purified, ligated into the pET24a expression vector (Novagen), and used to transform E. coli JM109(DE3). Cells were grown at 37°C, induced at A600 ≈ 0.5 with 0.5 mM IPTG, grown for an additional 3 h, and harvested by centrifugation (JA-10 rotor, 11,000 × g for 20 min at 4°C). Cells were washed once with phosphate-buffered saline and centrifuged as before. Cells were resuspended in lysis buffer–1 mM phenylmethylsulfonyl fluoride and lysed by two passages through an Aminco French press at ∼18,000 lb/in2 at 4°C. Inclusion bodies were isolated from the lysate by mild centrifugation (JA-20 rotor, 17,600 × g for 20 min at 4°C), washed once with lysis buffer–1% deoxycholate with homogenization, recentrifuged, washed a final time with lysis buffer with homogenization, and recentrifuged. The washed inclusion bodies were then resuspended in 8 M urea–50 mM HEPES (pH 7.5), heated to 70°C for 15 min, and homogenized. This mixture was clarified by centrifugation (Ti55.2 rotor, 132,000 × g for 20 min at 20°C), dialyzed (3 × 1:250) against lysis buffer, filtered to 0.2 μm, and concentrated with Amicon 30 membranes. Soluble protein was precipitated by addition of (NH4)2SO4 to 80% saturation at 4°C. Precipitated protein was collected by centrifugation (JA-20 rotor, 17,600 × g for 20 min at 4°C), and the pellet was resuspended in 6 M guanidine HCl–50 mM HEPES (pH 7.5) and heated to 70°C for 15 min. This solution was cooled to 22°C, filtered to 0.2 μm, and loaded onto a 50- by 2.5-cm Bio-Gel P-60 column (Bio-Rad) preequilibrated in 6 M guanidine HCl–50 mM HEPES (pH 7.5) at 22°C. Fractions corresponding to sHsp by SDS-PAGE were pooled and dialyzed (3 × 1:250) against 8 M urea–50 mM HEPES (pH 7.5), then dialyzed (3 × 1:250) against 50 mM HEPES (pH 7.5)–50 mM NaCl, filtered to 0.2 μm, and concentrated with Amicon 30 membranes. The final protein concentration was assayed with the bicinchoninic acid reagent (Pierce) as recommended by the manufacturer, and sHspTm was aliquoted, quick-frozen in liquid N2, and stored at −70°C.
Hsp70 gel filtration.
Purified Hsp70 (1.5 μM) was injected on a Superose 12 (Pharmacia) gel filtration column preequilibrated with 50 mM HEPES (pH 7.5)–150 mM KCl at 4°C. Incubation with peptide A (150 μM) (20) and/or ATP (1 mM) was at 70°C for 30 min prior to immediate injection.
Hsp70 ATPase.
The assay was performed as described previously (20): Hsp70Tm (1 μM) was combined with 2.5 mM ATP (Sigma) and 50 μM [2,8-3H]ATP (31 Ci/mmol [Amersham]) in ATPase buffer [50 mM HEPES (pH 7.5), 100 mM KCl, 100 mM NaCl, 10 mM (NH4)2SO4, 2 mM MgCl2] and incubated at the stated temperatures. Aliquots were removed at 5-min intervals and analyzed by thin-layer chromatography–scintillation counting, as described previously (20). Mock-purified E. coli extracts were used at a 25-fold-higher relative concentration.
Hsp70 peptide binding.
The peptide with sequence CALLQSR (single-letter amino acid code) was commercially synthesized and high-pressure liquid chromatography purified to ∼70% (Macromolecular Resources). Acrylodan (Molecular Probes) labeling and subsequent purification were as described previously (47). The peptide concentration was assayed by extinction coefficient (ɛ360 = 12,900 M−1 cm−1) (52) and ninhydrin assay. Hsp70Tm (1 μM) binding to labeled peptide (50 nM) was assayed in ATPase buffer with an Aminco 8000 fluorimeter. Excitation was at 370 nm, and emission was monitored at 470 nm for bound peptide and 520 nm for free peptide; all slits were set to 8 nm. Binding curves were fit to double-exponential kinetics with MacCurve Fit software (Kevin Raner Software).
sHsp sedimentation.
One hundred microliters of sHspTm (5.9 μM) was sedimented in a 3.2-ml-total-volume, 5 to 25% sucrose–1× ATPase buffer gradient in an SW Ti55 rotor at 287,000 × g for 5 h at 4°C. One-hundred-forty-microliter fractions were removed after centrifugation, and a 1/10 volume was run on SDS–12.5% PAGE gels. After Coomassie blue staining, sHspTm was scanned, and its density was integrated with NIH Image software. Separate sedimentation reactions with molecular weight standards (Pharmacia) were used to estimate the S value of sHspTm.
sHsp suppression of GFP aggregation.
The green fluorescent protein (GFP) variant S65T–N-terminal His6 (49) was purified on a nitrilotriacetic acid resin column (Qiagen) as described previously (49). GFP (3.6 μM) in 50 mM HEPES (pH 7.5)–50 mM KCl–2 mM MgCl2–10 mM (NH4)2SO4 was combined with sHspTm and incubated at 70°C for 20 min. The tubes were cooled to 22°C and centrifuged for 1 min at 15,000 × g. GFP complexed with sHsp will remain in the supernatant while aggregated GFP will be pelleted under these conditions. The supernatant was decanted and assayed by SDS-PAGE and scanning densitometry with NIH Image software. Control samples with no added GFP were used to calculate unaggregated GFP in experimental samples.
sHsp inhibition of GFP renaturation.
GFP was used as described above. The purified protein was precipitated by addition of (NH4)2SO4 to 70% saturation at 4°C and collected by centrifugation. The resulting pellet was dissolved in sufficient 6 M guanidine HCl–50 mM HEPES (pH 7.5) to yield a 10-mg/ml solution. Renaturation was as described previously (49) with the following changes: 2.5 μl of this solution was pipetted directly into a 1.3-ml stirred-fluorescence cuvette (1:520 dilution) containing 50 mM HEPES (pH 7.5), 50 mM KCl, 2 mM MgCl2, and 10 mM (NH4)2SO4 equilibrated at the specified temperature (final GFP concentration, 675 nM). GFP refolding-fluorescence reacquisition was monitored in an Aminco 8000 fluorimeter. Excitation was at 488 nm, and emission was monitored at 512 nm; all slits were set to 8 nm. Binding curves were fit to double-exponential kinetics with MacCurve Fit software (data not shown).
RESULTS
Hsp70 and sHsp cloning.
We began our analysis with a phagemid clone derived from a T. maritima cDNA library (the generous gift of Jeffrey Miller [33]) which contained 640 bp of the 3′ end of the Hsp70 gene as well as a downstream gene coding for a 16-kDa protein lacking an in-frame termination codon. Because Hsp70s are some of the most conserved proteins known, especially toward the extreme N terminus, it was possible to design a degenerate oligonucleotide primer to the unknown 5′ end of the Hsp70 gene. This degenerate primer and a primer to the known 3′ sequences were used to PCR amplify the entire Hsp70 gene from genomic T. maritima DNA. The Hsp70 protein encoded by this gene will be referred to as Hsp70Tm.
The incomplete downstream gene had sequence similarity to the α-crystallin domain which serves to define sHsp’s (17). In order to obtain the complete downstream gene, and any additional genes in a contiguous operon, the technique of inverse PCR was utilized (43). Outwardly directed oligonucleotide primers to the suspected sHsp gene were used to amplify an additional 2.1 kbp downstream of the Hsp70 gene from a circularized EcoRI-digested T. maritima genomic library. The putative sHsp gene is the final gene in the Hsp70 operon, encodes a protein with meaningful homology to sHsp’s (see below), and will be referred to as sHspTm.
The operon of the T. maritima Hsp70 (Hsp70Tm) and sHsp (sHspTm) spans 2,224 bp and encodes proteins with predicted molecular masses of 65.61 and 17.0 kDa, respectively. This gene arrangement is currently known to exist only in T. maritima. The highly conserved nature of Hsp70 primary sequence and the less-conserved α-crystallin domain which defines the sHsp’s are evident in alignments of the T. maritima Hsp70 (Hsp70Tm) and sHsp (sHspTm) with similar proteins, discussed below.
Protein homology and inferred phylogeny.
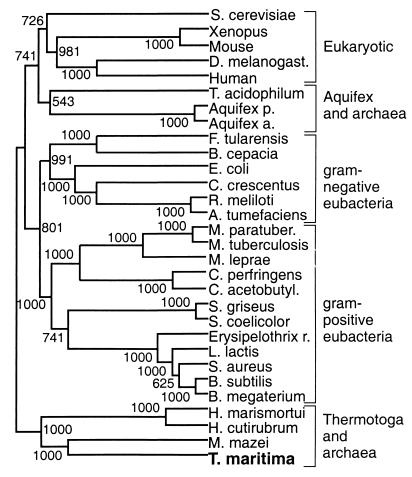
Hsp70s conform to a three-domain structural model. The 44-kDa N-terminal ATPase domain is very highly conserved, with approximately 80% amino acid identity. The adjoining 16-kDa substrate binding domain is slightly less conserved, with approximately 70% amino acid identity. The 10-kDa C-terminal domain has a possible regulatory function and is most divergent, with less than 40% amino acid identity. BLAST alignments (1) of the full-length Hsp70Tm identified the Hsp70 of the archaeon Methanosarcina mazei as being most similar (57% identity and 74% similarity). The highly conserved primary structure and almost universal distribution of Hsp70s have made them of great interest in protein-based phylogenetic studies (25, 26). Integration of the Hsp70Tm in such analysis should help clarify the placement of T. maritima in the tree of life and give insight into the relatedness of primitive bacteria and archaea. An unrooted consensus neighbor-joining tree (55) generated from selected Hsp70 alignments, shown in Fig. 1, reveals T. maritima to consistently branch with three archaebacteria. Gram-positive eubacteria consistently group together and are seen as distinct from the Thermotoga-archaeon clade. This tree (Fig. 1) is in good agreement with other Hsp70-based phylogenies (25, 26), but the newly integrated T. maritima does not branch with gram-positive low-G+C-content eubacteria, while it has been assigned to this group (4, 29). The consistent branching of T. maritima with halobacterial and methanogenic archaea can be seen as evidence of the recent evolutionary divergence of these organisms, and this association is consistent with the assigned deep branching of T. maritima by 16S rRNA-based phylogenies (29). The failure of the archaea to group together can be seen as evidence of the polyphyletic splitting seen in other studies (25), while 16S rRNA studies group the archaea as a coherent domain of life (58).
FIG. 1.
Phylogenetic relationship of T. maritima inferred by using Hsp70 amino acid sequences. An unrooted consensus neighbor-joining tree was generated with 30 Hsp70 sequences over a 532-amino-acid consensus length. Bootstrap scores (out of 1,000) are listed at their respective nodes. The alignment was generated with ClustalW software (55). The organisms include Saccharomyces cerevisiae, Drosophila melanogaster, Thermoplasma acidophilum, Aquifex pyrophilus, Aquifex aeolicus, Francisella tularensis, Burkholderia cepacia, Caulobacter crescentus, Rhizobium meliloti, Agrobacterium tumefaciens, Mycobacterium paratuberculosis, Mycobacterium tuberculosis, Mycobacterium leprae, Clostridium perfringens, Clostridium acetobutylicum, Streptomyces griseus, Streptomyces coelicolor, Erysipelothrix rhusiopathiae, Lactococcus lactis, Staphylococcus aureus, Bacillus subtilis, Bacillus megaterium, Halobacterium marismortui, and Halobacterium cutirubrum, and Methanosarcina mazei.
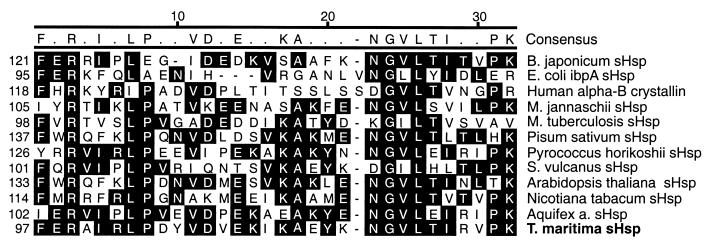
When BLAST alignments (1) of the translated second gene of the T. maritima Hsp70 operon were carried out, the full-length sHspTm protein was shown to have strong sequence similarity to multiple sHsp’s. These sHsp’s have in common the α-crystallin domain, which is comprised of about 40 amino acids toward the C terminus of these relatively small (12 to 30 kDa) proteins (57). The amino acid identity between even closely related species seldom exceeds 30%, although sHsp’s have been found in all organisms studied. sHspTm has the greatest sequence similarity to the recently identified sHsp of Aquifex aeolicus (41% identity and 65% similarity), a primitive eubacterium. The next four most similar proteins in the alignment include the sHsp’s from a proteobacterium, Bradyrhizobium japonicum; higher plants, Nicotiana tabacum and Arabidopsis thaliana; and the archaeon Pyrococcus horikoshii. An alignment of the α-crystallin domain of sHspTm with these and other sHsp’s is shown in Fig. 2.
FIG. 2.
Amino acid sequence alignment of α-crystallin domain from sHspTm and related sHsp’s. Residues with black background are conserved in the majority of sequences aligned. Alignment was generated by the Clustal algorithm within Megalign software (DNAStar). Organisms include Methanococcus jannaschii and Synechococcus volcanus.
Hsp70Tm purification.
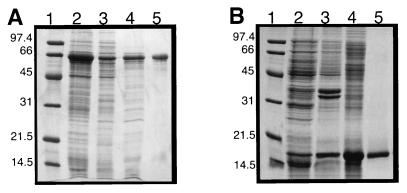
The Hsp70Tm was expressed as a C-terminal hexahistidine-tagged fusion protein; an SDS-PAGE gel used following its purification is shown in Fig. 3A. The E. coli lysates were heated to 70°C to aggregate the majority of endogenous proteins, while Hsp70Tm remained soluble. A single Ni2+ chelate affinity column served to purify Hsp70Tm to approximately 90% (Fig. 3A, lane 5). An additional anion-exchange column did not remove the slightly lower molecular-weight bands seen in the gel; these may represent truncated Hsp70Tm (data not shown). Because the specific ATPase activity of the anion-exchange-purified material was very similar to that of the Ni2+-purified material (data not shown) and Hsp70Tm was prone to aggregate in the low-salt buffer required for the anion exchange, we elected to use the Ni2+-purified Hsp70Tm exclusively in our analysis. Scanning calorimetry of this Hsp70Tm gave a denaturation temperature (Tm) of ∼90°C.
FIG. 3.
Hsp70Tm and sHspTm purification followed by SDS–12.5% PAGE. (A) Hsp70Tm purification. Lanes: 1, molecular weight markers; 2, induced E. coli total whole-cell extract; 3, soluble whole-cell extract; 4, heat-treated and clarified extracts; 5, final Ni2+ affinity-purified Hsp70Tm. (B) sHspTm purification. Lanes: 1, molecular weight markers; 2, induced E. coli total whole-cell extract; 3, total inclusion bodies; 4, renatured inclusion bodies; 5, final denaturing gel filtration-purified sHspTm. Numbers to the left of each gel represent molecular masses (in kilodaltons) of low-range protein markers (Bio-Rad).
Hsp70Tm self-association.
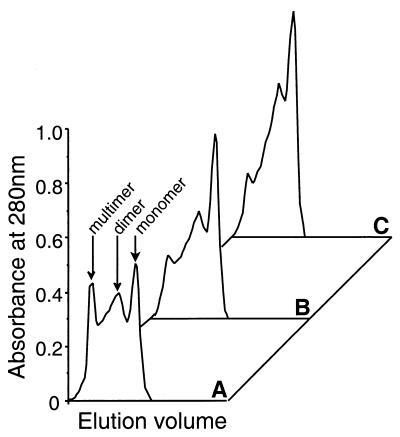
Hsp70 self-association is found in many systems and is related to their ability to bind substrates. Dimers, trimers and higher-order oligomers are detected for many Hsp70s in vitro (2, 54). This self-association can be modulated by nucleotides, substrate peptides, and the cochaperone Hsp40 (22, 56). The addition of peptide-ATP can serve to decrease Hsp70 self-association, while the addition of Hsp40 drives Hsp70 to its slow-substrate-exchanging (ADP-bound) state and can increase Hsp70 self-association. Figure 4 shows the effect of both substrate peptide (150 mM) and ATP (1 mM) on the self-association of Hsp70Tm at 70°C. Size exclusion chromatography revealed a distribution of Hsp70Tm monomers, dimers, and higher-order oligomers (Fig. 4A). Preincubation of Hsp70Tm (1.5 μM) with 150 μM peptide A, a peptide previously demonstrated to bind with high affinity to Hsp70s (20), for 20 min at 70°C prior to injection on the column caused a decrease in the multimeric and dimeric species (Fig. 4B), and similar preincubation with both 150 μM peptide A and 1 mM ATP further decreased Hsp70Tm self-association (Fig. 4C). Incubation with 1 mM ATP alone had a less pronounced effect on the size distribution of Hsp70Tm (data not shown). This lack of a strong disaggregation effect by ATP alone is unusual for an Hsp70 but not unprecedented (35). Preincubation of Hsp70Tm with both peptide and ATP at 22°C did not decrease self-association, and when Hsp70Tm previously incubated with peptide and ATP at 70°C, and thus disaggregated, was further incubated at 22°C for up to 1 h, higher-order oligomers were not reestablished (data not shown). These results suggest that Hsp70Tm does not self-associate at lower temperatures, while at higher temperatures, peptide, and to a lesser extent ATP, serves to decrease the self-association of Hsp70Tm.
FIG. 4.
Effect of peptide and ATP on the self-association of Hsp70Tm. (A) Untreated Hsp70Tm (1.5 μM). (B) Hsp70Tm (1.5 μM) incubated with peptide A (150 μM) at 70°C for 30 min. (C) Hsp70Tm incubated with peptide A (150 μM) and ATP (1 mM) at 70°C for 30 min. Peaks correspond to monomeric, dimeric, and multimeric Hsp70Tm, as labeled in panel A.
Hsp70Tm ATPase.
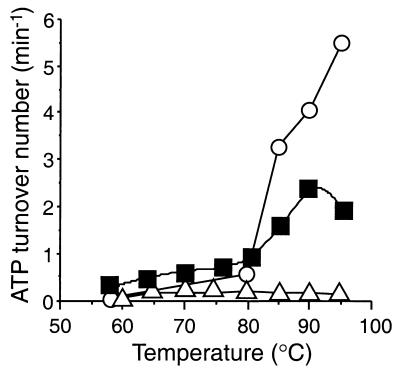
Hsp70s have a weak intrinsic ATPase activity (turnover number on the order from 0.02 to 0.3 min−1) (27, 32). Differences in the binding kinetics of substrate are tied to the nucleotide state of the distal ATPase domain, and peptide binding often stimulates Hsp70 ATPase activity. Hsp70Tm has a low basal ATPase activity over a large portion of the environmental temperature range of T. maritima (Fig. 5). From 58 to 80°C, the ATP hydrolysis rate rises from approximately 2 to 10 pmol min−1 μg−1 (turnover number = 0.13 to 0.65 min−1). At temperatures higher than 80°C, which may be heat shock temperatures for the organism, the ATPase rate of Hsp70Tm increases substantially, peaking at 36 pmol min−1 μg−1 at 90°C (turnover number = 2.35 min−1) before a slight decrease to 33 pmol min−1 μg−1 at 95°C (turnover number = 2.16 min−1). These ATPase rates are high compared to basal ATPase rates of the E. coli Hsp70, DnaK, at 37°C (turnover number, 0.2 min−1), while at 53°C, DnaK had a turnover number of 0.9 min−1 (40). The high ATPase rates of Hsp70Tm at temperatures greater than 80°C are more similar to the peptide-stimulated rates seen for DnaK at 53°C (turnover number of 3.6 to 4.5 min−1) (40).
FIG. 5.
ATPase activity of Hsp70Tm as a function of temperature from 58 to 95°C. Untreated Hsp70Tm (1 μM; closed squares), TCEP (1 μM)-treated and reduced Hsp70Tm; open circles), and a 25-fold-higher relative concentration of mock-purified E. coli extracts (open triangles) were assayed for hydrolysis of ATP (2.7 mM) at the respective temperatures.
While substrate peptides often stimulate the ATPase activity of Hsp70s, addition of three different peptides shown to bind to either Hsp70Tm (see below), DnaK, or other Hsp70s (20) had no effect on the ATPase activity of Hsp70Tm at all assayed temperatures (data not shown). Peptide stimulation of ATPase activity is absent in several Hsp70s (35, 45, 52) or may be highly substrate specific (27). Control reactions with mock-purified E. coli extracts showed a low basal ATPase activity equal to ∼1% of the activity of an equivalent amount of purified Hsp70Tm. Twenty-five-fold-higher concentrations of the mock-purified extracts were required for detection in our standard ATPase assay, as shown in Fig. 5. Thus, Hsp70Tm appears to behave much like other Hsp70s in terms of its ATP hydrolysis but lacks a peptide-stimulated ATPase activity. The relatively high ATPase activity and lack of peptide stimulation are not thought to be due to the presence of contaminating substrates in our Hsp70Tm preparation, in part because of the results of peptide binding experiments discussed in the next section.
The two highest assayed temperatures, 90 and 95°C, may be at or near the Tm of Hsp70Tm; the lower ATPase rate seen at 95°C could indicate thermal inactivation at this temperature. Hsp70Tm which had been reduced with the strong and specific disulfide-reducing agent TCEP showed a greatly increased ATPase activity above 80°C, compared to that of untreated Hsp70Tm. The turnover numbers of 3.2, 4.0, and 5.5 min−1 at 85, 90, and 95°C, respectively, represent an almost threefold increase in ATPase activity over unreduced Hsp70Tm. In addition, there is no evidence of the thermal inactivation observed for the nonreduced Hsp70Tm. Thus, reducing agents have a profound effect on the ATPase activity of Hsp70Tm at temperatures in excess of 80°C and are also shown to affect its peptide binding activity (see below).
Hsp70Tm peptide binding.
The E. coli Hsp70, DnaK, has been shown to tightly bind the fluorescently labeled peptide a-p1 with a Kd of 1.4 μM (52). Binding to DnaK was characterized by a blueshift in the emission wavelength and an increase in the emission maximum of the acrylodan fluorescent probe (52). As seen in Fig. 6, Hsp70Tm binds this labeled peptide appreciably at 80°C. This binding is relatively slow (half-time of binding = 100 s), similar to the ADP-mediated binding seen for DnaK (52). Addition of ATP had no effect on either the rate of binding or the absolute affinity of Hsp70Tm for the labeled peptide (data not shown), while DnaK’s binding rate was shown to increase 540-fold in the presence of ATP (52). The off-rate of peptide bound to Hsp70Tm was also insensitive to ATP (data not shown). Control reactions showed that mock-purified extracts, buffer alone, or bulk protein (RNase A) had no such effect on the labeled peptide emission spectra from 50 to 80°C (data not shown). This demonstrated peptide binding by Hsp70Tm argues that the lack of peptide stimulation of ATPase is not due to the presence of contaminating substrates in the protein preparation. Additionally, preincubation of Hsp70Tm with either unlabeled peptide or denatured RNase A served to block the binding of labeled peptide by Hsp70Tm (data not shown), suggesting that competing substrates are largely absent in the purified Hsp70Tm and, when added, are avidly bound by Hsp70Tm.
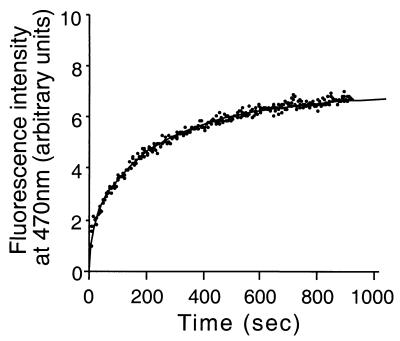
FIG. 6.
Hsp70Tm peptide binding at 80°C. Hsp70Tm (1 μM final concentration) was added to acrylodan (Molecular Probes)-labeled CALLQSR peptide (50 nM final concentration) equilibrated at 80°C in a stirred-fluorescence cuvette. Peptide binding was monitored at 470 nm with excitation at 370 nm. Control reactions with buffer alone, mock-purified extracts, and bulk protein (RNase A) did not enhance the labeled peptide fluorescence at 470 nm by more than 5% of the values obtained with Hsp70Tm (data not shown).
Addition of the reducing agent dithiothreitol or TCEP during Hsp70Tm peptide binding caused a cessation of assayed peptide binding (data not shown), with no decay in the existing signal attributed to previously bound peptide. Hsp70Tm has only one cysteine residue in its N-terminal ATPase domain (C119) that maps by homology to the solvent-exposed nucleotide binding cleft in the crystal structure of Hsc70 (19). The oxidation state of this cysteine may be related to the effect of reducing agents on ATPase activity and peptide binding seen in this study, an effect which has not been reported for any other Hsp70. In spite of the unique sensitivity to reducing agents demonstrated by Hsp70Tm, its appreciable peptide binding activity at 80°C is consistent with its role as a functional chaperone at physiologically relevant temperatures.
sHspTm purification.
To determine if the gene downstream of Hsp70Tm encoded a functional sHsp, we cloned and overexpressed sHspTm in E. coli. sHspTm was found to be sequestered in insoluble inclusion bodies, a phenomenon which has been documented for several other sHsp’s (38). Low-level induction and lowered induction temperatures had no effect on inclusion body formation; however, the inclusion bodies could be readily isolated and solubilized with denaturing agents. The sHsp could then be efficiently renatured. A gel filtration step under denaturing conditions allowed the sHspTm to be separated from most of the higher-molecular-weight contaminating E. coli proteins. The protein was then renatured by dialysis. The final purity of sHspTm was estimated to be greater than 90%, and this purification was followed by SDS-PAGE as shown in Fig. 3B.
sHspTm self-association.
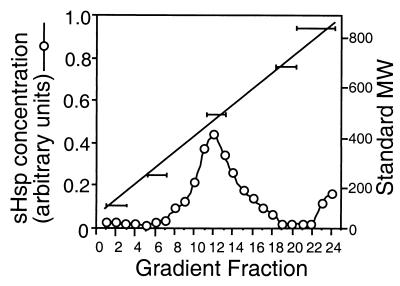
sHspTm has a predicted molecular mass of 17.6 kDa (Fig. 3B). In solution, most sHsp’s exist as higher-order oligomers, a feature which may be essential for proper chaperone activity (36). The sedimentation coefficient of native sHspTm is approximately 15 to 16S (Fig. 7), corresponding to a molecular mass of 400 to 450 kDa (n = 23 to 26 sHspTm subunits). The sHspTm size distribution was not greatly affected by incubation at temperatures up to 80°C for up to 30 min (data not shown). Although not monodispersed, the sedimentation profile is similar to those of other sHsp’s and is consistent with a recently solved sHsp crystal structure (34).
FIG. 7.
Sucrose gradient (5 to 25%) velocity sedimentation of sHspTm. Sedimentation fractions were collected, run on an SDS-PAGE gel, and Coomassie blue stained, and the concentration of sHspTm was calculated by scanning densitometry with NIH Image software (open circles). The right axis corresponds to migration of molecular weight standards performed in parallel with sHspTm sedimentation, and the horizontal bars correspond to fractions with respective molecular weight standards present.
sHspTm inhibition of protein aggregation.
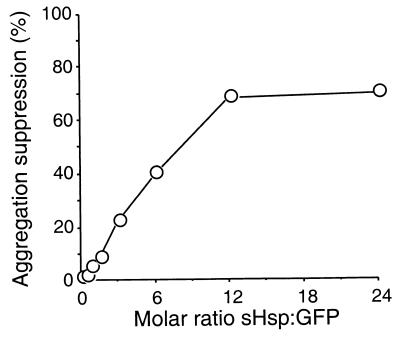
The primary chaperone activity of sHsp’s is the suppression of aggregation in thermally or chemically denatured proteins (31, 34). To determine whether sHspTm behaved as a functional sHsp, we assayed its ability to suppress the thermally induced aggregation of the GFP variant S65T. The Tm of GFP is approximately 73°C (unpublished observation), and incubation at 70°C can cause aggregation. sHspTm was able to suppress this aggregation (Fig. 8). Molar ratios of sHspTm monomers to GFP monomer higher than 3:1 gave some protection from aggregation; higher ratios could suppress aggregation by up to 80%. The highest level of aggregation suppression was achieved at an approximate ratio of 12 sHspTm monomers to 1 GFP monomer (ratio of sHspTm multimer to GFP monomer of approximately 1:2). sHspTm is thus capable of interacting with a thermally unfolding protein and suppressing its aggregation.
FIG. 8.
sHspTm suppresses the heat-induced aggregation of GFP. GFP aggregation suppression (open circles) is shown as a function of sHspTm/GFP molar ratio at 70°C. Ten micrograms of GFP was heated to 70°C for 20 min with and without added sHspTm. After centrifugation to remove aggregated GFP, the amount of GFP remaining in the supernatant was assayed by SDS-PAGE and scanning densitometry with NIH Image software. Comparison to control samples without sHspTm allowed calculation of percent aggregation suppression versus molar ratio of GFP to sHspTm.
sHspTm inhibition of GFP refolding.
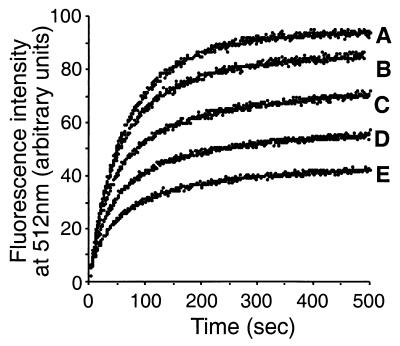
We next examined if sHspTm could bind a refolding-competent protein substrate. GFP denatured with either 8 M urea or 6 M guanidine HCl spontaneously refolds upon dilution into aqueous buffer (49). Refolding may be assayed by the reacquisition of fluorescence at 512 nm, with excitation at 488 nm. Somewhat surprisingly, refolding can occur at temperatures up to 62°C, albeit with a substantial decrease in efficiency (data not shown). Figure 9 shows that at 55°C sHspTm inhibits the refolding of GFP. Increasing amounts of sHspTm increased inhibition of refolding to a maximal level of 80%, while at temperatures below 50°C, sHspTm had no effect on GFP refolding (data not shown). The inhibition of GFP refolding appears to be specific to sHspTm; the addition of bovine serum albumin to the reaction actually gives a twofold increase in the refolding efficiency of GFP at these temperatures. GFP bound to sHspTm was not reactivated by lower temperatures or incubation with Hsp70Tm-ATP (data not shown). Thus, sHspTm appears to stably bind refolding intermediates of GFP and trap them in an intermediate folding state.
FIG. 9.
sHspTm inhibition of GFP refolding at 55°C. (A) No added sHspTm; (B) 2:1 molar ratio of sHspTm to GFP; (C) 8:1 molar ratio of sHspTm to GFP; (D) 16:1 molar ratio of sHspTm to GFP; (E) 32:1 molar ratio of sHspTm to GFP. GFP denatured in 6 M guanidine HCl was diluted 1:520 into a stirred-fluorescence cuvette equilibrated at 55°C to a final concentration of 675 nM, with and without sHspTm. Excitation was at 488 nm, and GFP fluorescence was monitored at 512 nm.
DISCUSSION
We have cloned the partial Hsp70-sHsp operon from the hyperthermophilic eubacterium T. maritima. Its proteins have been expressed, purified, and initially characterized as functional molecular chaperones. To date, this is the only known operon which combines these two ubiquitous and important molecular chaperone genes. All previously described Hsp70 operons have included a downstream Hsp40 gene and often a flanking GrpE nucleotide exchange factor gene (53). These Hsp70 cofactors do appear to exist in T. maritima in a separate, distal operon: a complete operon consisting of HRCA (a heat shock transcriptional repressor), GrpE, and Hsp40 has been constructed from sequence data made available by the ongoing T. maritima genomic sequencing project (30). This unique Hsp70-sHsp operon may be a functional unit of T. maritima’s response to stress conditions. Currently, we have been unable to demonstrate the functional interactions seen for other Hsp70-sHsp systems (16) possibly due to requirements for T. maritima’s Hsp40 and GrpE cofactors or specific protein substrates.
We have shown that the Hsp70 of T. maritima is a functional molecular chaperone. It has self-association behavior, ATPase activity, and peptide binding characteristics similar to those of other studied Hsp70s (3, 40, 52). Hsp70Tm diverges from the majority of characterized Hsp70s in its lack of a strong monomerization response to ATP, its lack of substrate-based stimulation of ATPase activity (35, 45, 52), and its sensitivity to reducing agents. The single cysteine residue (C119) may play a role in disulfide formation in Hsp70Tm; reduction of this bond could be responsible for the up-regulated ATPase activity above 80°C and the observed inhibition of peptide binding. Future mutagenesis experiments will explore this possibility.
The peptide binding of Hsp70Tm is similar to that seen in the Hsp70 of E. coli, DnaK, in its ADP-bound state (52). There is no modulation of peptide exchange kinetics by nucleotide. The only other well-characterized thermostable Hsp70, from Thermus thermophilus, has similar uncoupling of its nucleotide state with substrate binding kinetics. In the case of the Thermus Hsp70, however, the binding is more analogous to that of the ATP-bound form of DnaK, with fast substrate exchange kinetics at temperatures from 25 to 75°C (35). While this uncoupling may be a feature common to thermostable Hsp70s, it is also possible that Hsp70 cofactors, including Hsp40 and GrpE, are necessary to link the nucleotide state of the ATPase domain with the kinetic properties of the substrate binding domain in these Hsp70s. The Thermus Hsp70 has a uniquely intimate association with its Hsp40 cofactor (41); they exist as a stable trimer of trimers complexed with a small (8 kDa) association factor, Daf (42). While there is no evidence of a Daf homologue in T. maritima, or any other organism, it remains possible that Hsp70Tm will require its Hsp40-GrpE cofactors to link its ATPase and substrate binding activities.
We have also shown the Hsp70Tm operon to contain a gene encoding a functional sHsp. sHspTm exists as a large oligomer in solution, an arrangement common in sHsp’s (17, 34) and possibly required for their observed chaperone activity (36). sHspTm is capable of suppressing the thermally induced aggregation of GFP and is also able to bind to refolding-competent GFP at high temperatures. This inhibition of refolding is different from the results of other sHsp studies. Jakob et al. (31) saw an enhancement in refolding yields of both citrate synthase and α-glucosidase in the presence of murine Hsp25 and human Hsp27 at 20°C. Lowering the temperature of GFP-sHspTm complexes, or addition of Hsp70Tm and ATP, did not result in reactivation of GFP fluorescence, or release of GFP by sHspTm (data not shown). Thus, the binding of GFP to sHspTm appears to be quite stable. GFP refolding can be described by double-exponential kinetics (49), and the effect of the sHspTm was to diminish the amplitude of the slow phase of the GFP refolding, while having little effect on the fast-phase amplitude. The net effect is an increase in the overall folding rate; at 55°C the half-time of GFP folding is 150 s without sHspTm and 73 s with maximal sHspTm inhibition. This suggests that slow-phase refolding intermediates are preferentially binding to the sHspTm. The lack of sHspTm inhibition of GFP refolding at temperatures lower than 50°C (data not shown) could be due to either a temperature-dependent activation of sHspTm or a decrease in the amount of slow-phase folding intermediates of GFP which are accessible to sHspTm binding, or a combination of these two effects.
The phylogenetic analysis based on Hsp70 alignments emphasizes the primitive and ancestral nature of T. maritima. Thermotoga consistently branches with halophilic and methanobacterial archaea, while it is seen as distinct from the gram-positive, low-G+C eubacteria which are its phenotypically assigned cohorts (5, 29). The deep branching of Thermotoga seen in 16S rRNA-based phylogenies (29) is supported by this close association between Thermotoga and archaea in the current study. The only deeper-branching eubacterial genus by 16S rRNA analysis, Aquifex, is seen as an out-group in our analysis, distinct from either gram-positive eubacteria or gram-negative proteobacteria and loosely related to the sole thermoacidophilic archaeon in our analysis. The recent genomic sequencing of A. aeolicus has revealed similar difficulties in inferring its phylogeny from protein sequences (12, 15), and this difficulty is not substantially alleviated by the current analysis. Aquifex Hsp70 possesses sequence signatures previously attributed solely to proteobacterial and eukaryotic Hsp70s (25), most notably an insertion of approximately 24 amino acids near its N terminus. This sequence signature, along with Aquifex’s sensitivity to aminoglycoside antibiotics (4), to which Thermotoga is at least partially immune (29), supports the designation of Aquifex as a primitive gram-negative proteobacterium, while Thermotoga is more closely related to primitive gram-positive eubacteria and archaea.
In conclusion, the Hsp70-sHsp operon of the hyperthermophilic eubacterium T. maritima represents a unique genetic association between two major classes of molecular chaperones: the ATP-dependent Hsp70s and the ATP-independent sHsp’s. These genes encode functional chaperone proteins which may interact in vivo. The possibility exists that substrate proteins stably bound to sHspTm can serve as a target for Hsp70Tm mediated refolding and reactivation. The elucidation of this functional interaction may require the presence of Thermotoga’s Hsp40 and GrpE cofactors or require an effective screening procedure for prospective substrate proteins. Future work will address these possibilities.
ACKNOWLEDGMENTS
We thank Eric Bertelsen for critical reading of the manuscript, Walt Baase for assistance with scanning calorimetry, and Yanling Wang for assistance with DNA sequencing.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;17:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blonde-Elguindi S, Cwirla E A, Dower W J, Lipshutz R J, Sprang S R, Sambrook J F, Gething M H. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993;75:12730–12735. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- 3.Blonde-Elguindi S, Fourie A M, Sambrook J F, Gething M H. Peptide-dependent stimulation of the ATPase activity of the molecular chaperone BiP is the result of conversion of oligomers to active monomers. J Biol Chem. 1993;268:12730–12735. [PubMed] [Google Scholar]
- 4.Bocchetta M, Huber R, Cammarano P. Sensitivity of ribosomes of the hyperthermophilic bacterium Aquifex pyrophilus to aminoglycoside antibiotics. J Bacteriol. 1996;178:1762–1765. doi: 10.1128/jb.178.6.1762-1765.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown J R, Masuchi Y, Robb F T, Doolittle W F. Evolutionary relationships of bacterial and archaeal glutamine synthetase genes. J Mol Evol. 1994;38:566–576. doi: 10.1007/BF00175876. [DOI] [PubMed] [Google Scholar]
- 6.Buchberger A, Schroder H, Buttner M, Valencia A, Bukau B. A conserved loop in the ATPase domain of the DnaK chaperone is essential for stable binding of GrpE. Nat Struct Biol. 1994;1:95–101. doi: 10.1038/nsb0294-95. [DOI] [PubMed] [Google Scholar]
- 7.Buchberger A, Valencia A, McMacken R, Sander C, Bukau B. The chaperone function of DnaK requires the coupling of ATPase activity with substrate binding through residue E171. EMBO J. 1994;13:1687–1695. doi: 10.1002/j.1460-2075.1994.tb06433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchner J. Supervising the fold: functional principles of molecular chaperones. FASEB J. 1996;10:10–19. [PubMed] [Google Scholar]
- 9.Bukau B, Horwich A L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 10.Caspers G J, Leunissen J A, de Jong W W. The expanding small heat-shock protein family, and structure predictions of the conserved “alpha-crystallin domain.”. J Mol Evol. 1995;40:238–248. doi: 10.1007/BF00163229. [DOI] [PubMed] [Google Scholar]
- 11.Cyr D M, Neupert W. Roles for Hsp70 in protein translocation across membranes of organelles. EXS. 1996;77:25–40. doi: 10.1007/978-3-0348-9088-5_3. [DOI] [PubMed] [Google Scholar]
- 12.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 13.de Jong W W, Caspers G J, Leunissen J A. Genealogy of the alpha-crystallin—small heat-shock protein superfamily. Int J Biol Macromol. 1998;22:151–162. doi: 10.1016/s0141-8130(98)00013-0. [DOI] [PubMed] [Google Scholar]
- 14.Dittmar K D, Demady D R, Stancato L F, Krishna P, Pratt W B. Folding of the glucocorticoid receptor by the heat shock protein (Hsp) 90-based chaperone machinery. The role of p23 is to stabilize receptor · hsp90 heterocomplexes formed by hsp90 · p60 · Hsp70. J Biol Chem. 1997;272:21213–21220. doi: 10.1074/jbc.272.34.21213. [DOI] [PubMed] [Google Scholar]
- 15.Doolittle R F. Microbial genomes opened up. Nature. 1998;392:339–342. doi: 10.1038/32789. [DOI] [PubMed] [Google Scholar]
- 16.Ehrnsperger M, Graber S, Gaestel M, Buchner J. Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 1997;16:221–229. doi: 10.1093/emboj/16.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrnsperger M, Buchner J, Gaestel M. Structure and function of small heat-shock proteins. In: Fink A L G, Goto Y, editors. Molecular chaperones in the life cycle of proteins. Vol. 1. New York, N.Y: Marcel Dekker, Inc.; 1998. pp. 533–576. [Google Scholar]
- 18.Fink A. The Hsp70 reaction cycle and its role in protein folding. In: Fink A L G, Goto Y, editors. Molecular chaperones in the life cycle of proteins. Vol. 1. New York, N.Y: Marcel Dekker, Inc.; 1998. pp. 123–150. [Google Scholar]
- 19.Flaherty K M, Deluca-Flaherty C, McKay D B. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Science. 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- 20.Flynn G C, Chappell T G, Rothman J E. Peptide binding and release by proteins implicated as catalysts of protein folding. Science. 1989;245:385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- 21.Flynn G C, Pohl J, Flocco M T, Rothman J E. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- 22.Gao B, Eisenberg E, Greene L. Effect of constitutive 70-kDa heat shock protein polymerization on its interaction with protein substrate. J Biol Chem. 1996;271:16792–16797. doi: 10.1074/jbc.271.28.16792. [DOI] [PubMed] [Google Scholar]
- 23.Gething M J, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 24.Gragerov A, Zeng L, Zhao X, Burkholder W, Gottesman M E. Specificity of DnaK-peptide binding. J Mol Biol. 1994;235:848–854. doi: 10.1006/jmbi.1994.1043. [DOI] [PubMed] [Google Scholar]
- 25.Gupta R S, Bustard K, Falah M, Singh D. Sequencing of heat shock protein 70 (DnaK) homologs from Deinococcus proteolyticus and Thermomicrobium roseum and their integration in a protein-based phylogeny of prokaryotes. J Bacteriol. 1997;179:345–357. doi: 10.1128/jb.179.2.345-357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta R S, Golding G B. Evolution of the HSP70 gene and its implications regarding relationships between archaebacteria, eubacteria, and eukaryotes. J Mol Evol. 1993;37:573–582. doi: 10.1007/BF00182743. [DOI] [PubMed] [Google Scholar]
- 27.Ha J H J, Johnson E R, McKay D B, Sousa M C, Takeda S, Wilbanks S M. Structure and properties of the 70-kilodalton heat-shock proteins. In: Fink A L G, editor. Molecular chaperones in the life cycle of proteins. Vol. 1. New York, N.Y: Marcel Dekker, Inc.; 1998. pp. 95–122. [Google Scholar]
- 28.Horwitz J. Alpha-crystallin can function as a molecular chaperone. Proc Natl Acad Sci USA. 1992;89:10449–10453. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huber R, Langworthy T A, König H, Thomm M, Woese C R, Sleytr U B, Stetter K O. Thermotoga maritima sp. nov. represents a new genus of extremely thermophilic eubacteria growing up to 90° C. Arch Microbiol. 1986;144:324–333. [Google Scholar]
- 30.The Institute of Genomic Research. 5 October 1998, posting date. Sequence data. [Online.] http://www.tigr.org. [5 November 1998, last date accessed.]
- 31.Jakob U, Gaestel M, Engel K, Buchner J. Small heat shock proteins are molecular chaperones. J Biol Chem. 1993;268:1517–1520. [PubMed] [Google Scholar]
- 32.Jordan R, McMacken R. Modulation of the ATPase activity of the molecular chaperone DnaK by peptides and the DnaJ and GrpE heat shock proteins. J Biol Chem. 1995;270:4563–4569. doi: 10.1074/jbc.270.9.4563. [DOI] [PubMed] [Google Scholar]
- 33.Kim C W, Markiewicz P, Lee J J, Schierle C F, Miller J H. Studies of the hyperthermophile Thermotoga maritima by random sequencing of cDNA and genomic libraries. J Mol Biol. 1993;231:960–981. doi: 10.1006/jmbi.1993.1345. [DOI] [PubMed] [Google Scholar]
- 34.Kim K K, Kim R, Kim S H. Crystal structure of a small heat-shock protein. Nature. 1998;394:595–599. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- 35.Klostermeier D, Seidel R, Reinstein J. Functional properties of the molecular chaperone DnaK from Thermus thermophilus. J Mol Biol. 1998;279:841–853. doi: 10.1006/jmbi.1998.1816. [DOI] [PubMed] [Google Scholar]
- 36.Kokke B P, Leroux M R, Candido E P, Boelens W C, de Jong W W. Caenorhabditis elegans small heat-shock proteins Hsp12.2 and Hsp12.3 form tetramers and have no chaperone-like activity. FEBS Lett. 1998;433:228–232. doi: 10.1016/s0014-5793(98)00917-x. [DOI] [PubMed] [Google Scholar]
- 37.Lee G J, Roseman A M, Saibil H R, Vierling E. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 1997;16:659–671. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee G J, Vierling E. Expression, purification, and molecular chaperone activity of plant recombinant small heat shock proteins. Methods Enzymol. 1998;290:350–365. doi: 10.1016/s0076-6879(98)90031-3. [DOI] [PubMed] [Google Scholar]
- 39.Lu Z, Cyr D M. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J Biol Chem. 1998;273:5970–5978. doi: 10.1074/jbc.273.10.5970. [DOI] [PubMed] [Google Scholar]
- 40.McCarty J S, Walker G C. DnaK as a thermometer: threonine-199 is site of autophosphorylation and is critical for ATPase activity. Proc Natl Acad Sci USA. 1991;88:9513–9517. doi: 10.1073/pnas.88.21.9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Motohashi K, Taguchi H, Ishii N, Yoshida M. Isolation of the stable hexameric DnaK · DnaJ complex from Thermus thermophilus. J Biol Chem. 1994;269:27074–27079. [PubMed] [Google Scholar]
- 42.Motohashi K, Yohda M, Endo I, Yoshida M. A novel factor required for the assembly of the DnaK and DnaJ chaperones of Thermus thermophilus. J Biol Chem. 1996;271:17343–17348. doi: 10.1074/jbc.271.29.17343. [DOI] [PubMed] [Google Scholar]
- 43.Ochman H, Gerber A S, Hartl D L. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palleros D R, Reid K L, Shi L, Welch W J, Fink A L. ATP-induced protein-Hsp70 complex dissociation requires K+ but not ATP hydrolysis. Nature. 1993;365:664–666. doi: 10.1038/365664a0. [DOI] [PubMed] [Google Scholar]
- 45.Peake P, Basten A, Britton W J. Characterization of the functional properties of the 70-kDa protein of Mycobacterium bovis. J Biol Chem. 1991;266:20828–20832. [PubMed] [Google Scholar]
- 46.Pfeil W, Gesierich U, Kleemann G R, Sterner R. Ferredoxin from the hyperthermophile Thermotoga maritima is stable beyond the boiling point of water. J Mol Biol. 1997;272:591–596. doi: 10.1006/jmbi.1997.1278. [DOI] [PubMed] [Google Scholar]
- 47.Pierpaoli E V, Sandmeier E, Baici A, Schonfeld H J, Gisler S, Christen P. The power stroke of the DnaK/DnaJ/GrpE molecular chaperone system. J Mol Biol. 1997;269:757–768. doi: 10.1006/jmbi.1997.1072. [DOI] [PubMed] [Google Scholar]
- 48.Prip Buus C, Westerman B, Schmitt M, Langer T, Neupert W, Schwarz E. Role of the mitochondrial DnaJ homologue, Mdj1p, in the prevention of heat-induced protein aggregation. FEBS Lett. 1996;380:142–146. doi: 10.1016/0014-5793(96)00049-x. [DOI] [PubMed] [Google Scholar]
- 49.Reid B G, Flynn G C. Chromophore formation in green fluorescent protein. Biochemistry. 1997;36:6786–6791. doi: 10.1021/bi970281w. [DOI] [PubMed] [Google Scholar]
- 50.Russell R, Jordan R, McMacken R. Kinetic characterization of the ATPase cycle of the DnaK molecular chaperone. Biochemistry. 1998;37:596–607. doi: 10.1021/bi972025p. [DOI] [PubMed] [Google Scholar]
- 51.Sanchez E R, Hirst M, Scherrer L C, Tang H Y, Welsh M J, Harmon J M, Simons S S, Jr, Ringold G M, Pratt W B. Hormone-free mouse glucocorticoid receptors overexpressed in Chinese hamster ovary cells are localized to the nucleus and are associated with both Hsp70 and Hsp90. J Biol Chem. 1990;265:20123–20130. [PubMed] [Google Scholar]
- 52.Schmid D, Baici A, Gehrig H, Christen P. Kinetics of molecular chaperone action. Science. 1994;263:971–973. doi: 10.1126/science.8310296. [DOI] [PubMed] [Google Scholar]
- 53.Segal G, Ron E Z. Regulation and organization of the groE and dnaK operons in Eubacteria. FEMS Microbiol Lett. 1996;138:1–10. doi: 10.1111/j.1574-6968.1996.tb08126.x. [DOI] [PubMed] [Google Scholar]
- 54.Shi L, Kataoka M, Fink A L. Conformational characterization of DnaK and its complexes by small-angle X-ray scattering. Biochemistry. 1996;35:3297–3308. doi: 10.1021/bi951984l. [DOI] [PubMed] [Google Scholar]
- 55.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei J, Gaut J R, Hendershot L M. In vitro dissociation of BiP-peptide complexes requires a conformational change in BiP after ATP binding but does not require ATP hydrolysis. J Biol Chem. 1995;270:26677–26682. doi: 10.1074/jbc.270.44.26677. [DOI] [PubMed] [Google Scholar]
- 57.Wistow G. Domain structure and evolution in alpha-crystallins and small heat-shock proteins. FEBS Lett. 1985;181:1–6. doi: 10.1016/0014-5793(85)81102-9. [DOI] [PubMed] [Google Scholar]
- 58.Woese C R. There must be a prokaryote somewhere: microbiology’s search for itself. Microbiol Rev. 1994;58:1–9. doi: 10.1128/mr.58.1.1-9.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]