Abstract
We describe here a new member of the LysR family of transcriptional regulators, AphB, which is required for activation of the Vibrio cholerae ToxR virulence cascade. AphB activates the transcription of the tcpPH operon in response to environmental stimuli, and this process requires cooperation with a second protein, AphA. The expression of neither aphA or aphB is strongly regulated by environmental stimuli, raising the possibility that the activities of the proteins themselves may be influenced under various conditions. Strains of the El Tor biotype of V. cholerae typically exhibit lower expression of ToxR-regulated virulence genes in vitro than classical strains and require specialized culture conditions (AKI medium) to induce high-level expression. We show here that expression of aphB from the tac promoter in El Tor biotype strains dramatically increases virulence gene expression to levels similar to those observed in classical strains under all growth conditions examined. These results suggest that AphB plays a role in the differential regulation of virulence genes between the two disease-causing biotypes.
Cholera is a life-threatening diarrheal disease caused by the gram-negative bacterium Vibrio cholerae. The organism colonizes the upper intestine, and the toxin-coregulated pilus (TCP) is the primary factor involved in this process (36). The severe diarrhea associated with the disease results from the action of the secreted cholera toxin (CT) on intestinal epithelial cells (reviewed in reference 17). The genes required for the biogenesis of TCP are located in an operon on a large pathogenicity island termed the TCP-ACF element, or vibrio pathogenicity island (18, 19). The subunits of CT are encoded by the ctxA and ctxB genes on a separate genetic element which comprises the genome of the lysogenic filamentous bacteriophage CTXφ (38).
Many of the genes involved in the pathogenesis of V. cholerae comprise what is known as the ToxR virulence regulon, since they are coordinately expressed and dependent upon the transcriptional activator ToxR (23, 26). ToxR is a transmembrane DNA binding protein whose activity is enhanced by a second transmembrane protein, ToxS (5, 21, 23). The toxR and toxS genes, which are expressed as an operon, are not associated with either the TCP-ACF or CTX elements but appear to be part of the “ancestral chromosome” and have other important regulatory roles (22). TcpP is a transcriptional activator encoded on the TCP-ACF element which has recently been shown to share significant homology with ToxR and which cooperates with it to initiate gene expression (13, 25). The tcpP gene is coexpressed with a second gene, tcpH, which encodes a protein that enhances the activity of TcpP (2). TcpP and TcpH appear to have a similar membrane topology to ToxR and ToxS.
ToxRS and TcpPH control the expression of the ToxR virulence regulon by their ability to activate the expression of a third transcriptional activator, ToxT, which is also encoded on the TCP-ACF element (7, 13). ToxT is a cytoplasmic protein that is a member of the AraC family of transcriptional activators (15). Once its expression is activated by ToxRS and TcpPH, ToxT then directly activates various genes within the regulon, such as the tcp and ctx operons (3, 7). The toxT gene is located within the tcp operon, and its expression is dependent upon a promoter located immediately upstream of the gene (14) as well as by one located at the beginning of the tcp operon which may function in an autoregulatory capacity (1).
The expression of the tcp and ctx operons are strongly influenced by specific environmental cues such as pH and temperature. Since the expression of tcpPH is also influenced by both of these parameters (2, 32), the mechanisms that regulate the expression of this operon are likely to be of central importance in the control of virulence gene expression by environmental stimuli. AphA is a 20-kDa V. cholerae protein which has recently been shown to be required for expression of the tcpPH operon and for its response to environmental stimuli (32). Since the basal level of tcpPH expression in a ΔaphA mutant still appeared to be influenced by pH and temperature, it was hypothesized that factors in addition to AphA might also play a role in the expression of tcpPH. We describe here a new member of the LysR family of transcriptional regulators, AphB, which is required for transcriptional activation of tcpPH as well as its response to environmental stimuli. AphB functions synergistically with AphA to activate the expression of tcpPH, and it also appears to contribute to the differences in virulence gene expression between the two major disease-causing biotypes, classical and El Tor. Since neither AphA nor AphB is encoded within the TCP-ACF element, these proteins may have other regulatory roles in V. cholerae, and the expression of the tcpPH operon may have evolved to come under their control.
MATERIALS AND METHODS
Bacterial strains and media.
The V. cholerae and Escherichia coli strains and plasmids used in this study are listed in Table 1. Bacteria were maintained at −70°C in Luria-Bertani (LB) medium (20) containing 30% (vol/vol) glycerol. Antibiotics were used at the following concentrations in LB medium or AKI medium (16): ampicillin, 100 μg/ml; kanamycin, 45 μg/ml; tetracycline, 7.5 μg/ml for V. cholerae and 15 μg/ml for E. coli; and streptomycin, 100 μg/ml, except when selecting for loss of integrated plasmids in V. cholerae, where it was used at 1 mg/ml. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was used in LB agar at 40 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Strains | ||
| V. cholerae | ||
| CG842 | O395 (classical Ogawa Smr) ΔlacZ | 4 |
| KSK218 | CG842 ctx-lacZ Smr Cmr | 30 |
| KSK404 | KSK218 aphB::TnphoA | This work |
| GK91 | KSK218 aphB::pGKK17 | This work |
| GK122 | KSK218 ΔaphB1 | This work |
| KSK618 | CG842 tcpP-lacZ | 32 |
| GK121 | KSK618 ΔaphB1 | This work |
| KSK647 | KSK618 ΔaphA1 | 32 |
| KSK805 | KSK618 ΔaphA1 ΔaphB1 | This work |
| KSK666 | CG842 aphA-lacZ | 32 |
| GK130 | CG842 aphB-lacZ | This work |
| C6706str2 | El Tor Inaba Smr | 37 |
| KSK262 | C6706str2 ΔlacZ3 | This work |
| KSK725 | KSK262 tcpP-lacZ | This work |
| GK138 | KSK725 ΔaphB1 | This work |
| GK161 | KSK725 ΔaphA1 | This work |
| GK142 | KSK262 aphB-lacZ | This work |
| E. coli | ||
| MC1061 | Δ (ara-leu)7697 Δ(lac)X74 | Laboratory collection |
| KSK782 | MC1061 λ KSPL1 (tcpP-lacZ) | This work |
| Plasmids | ||
| pKAS64 | pKAS32 ΔrpsL, Apr | 31 |
| pKAS110 | pKAS64 ΔSmaI | This work |
| pGKK17 | pKAS110, 200-bp aphB fragment | This work |
| pGKK18 | pGKK17 chromosomal capture plasmid | This work |
| pGKK25 | pKAS46, ΔaphB1 classical | This work |
| pGKK26 | pGKK25, aphB1-lacZ | This work |
| pGKK28 | pKAS46, ΔaphB1 El Tor | This work |
| pGKK29 | pGKK28, aphB1-lacZ | This work |
| pGKK35 | pKAS46, ΔaphA1 El Tor | This work |
| pKAS48 | pKAS46, ΔlacZ3 | 29 |
| pKAS113 | pKAS46, tcpP-lacZ El Tor | This work |
| pLAFR3 | Tcr expression plasmid | 33 |
| pKAS116 | pLAFR3 aphB (classical), Tcr Gmr | This work |
| pMMB66EH | Apr expression plasmid | 9 |
| pKAS107 | pMMB66EH aphA (classical), Apr | 32 |
| pKAS117 | pMMB66EH aphB (classical), Apr | This work |
| pBAD22 | Apr expression plasmid | 11 |
| pKAS118 | pBAD-TOPO aphB (classical), Apr | This work |
| pKAS119 | pBAD-TOPO aphA (classical), Apr | This work |
| pKAS120 | pBAD-TOPO aphB (El Tor), Apr | This work |
Identification of aphB.
Random insertion of TnphoA into the chromosome of strain KSK218 was as previously described (30, 35, 36). Chromosomal DNA from V. cholerae transposon mutant KSK404 was digested with SphI and ligated into an oriR6K plasmid lacking rpsL (pKAS64). The ligated DNA was subjected to two rounds of PCR amplification, the first using a plasmid-specific primer, ORIR6K (5′-GGTTTAACGGTTGTGGACAAC), and a transposon-specific primer, TNPHOA-1 (30); and the second using ORIR6K with a nested transposon-specific primer, TNPHOA-2 (5′-AGCAGCCCGGTTTTCCAGAAC). The resulting 200-bp fragment, which contained a portion of the aphB open reading frame, was ligated into another oriR6K plasmid lacking rpsL (pKAS110), generating pGKK17. Plasmid pGKK17 was integrated into the aphB gene of KSK218, generating strain GK91. Chromosomal DNA was isolated from GK91, digested with SphI, ligated, and transformed into E. coli. The resulting plasmid, pGKK18, was then used to obtain the complete aphB nucleotide sequence with the ABI PRISM Dye System (Perkin-Elmer).
Construction of in-frame deletions and lacZ fusions.
The in-frame ΔaphB1 mutations in both classical and El Tor biotypes were constructed by PCR amplifying two 200-bp fragments encompassing the regions upstream and downstream of the aphB gene, respectively, from either O395 or C6706str2 (37) by using primer pair CO2-3 (5′-GATCGTCTAGAATGGTTTTCAATAAATCATC) and CO2-4 (5′-GATCGGCGGCCGCATGTCATTGAAGCGAGACGCTC) and primer pair CO2-5 (5′-GATCGGCGGCCGCCTGTATAACCACAAAGATCAC) and CO2-6 (5′-GATCGGAATTCAAGCCATGCAAATGGCGGCC). The resulting fragments were ligated into pKAS46 (29), generating pGKK25 and pGKK28, respectively, and the deletions were introduced into V. cholerae by allelic exchange. To construct the aphB-lacZ fusions, a promoterless E. coli lacZ gene was inserted into the plasmids described above, generating pGKK26 and pGKK29, prior to allelic exchange. The classical ΔaphA1 deletion was previously described (32). The El Tor ΔaphA1 deletion was constructed in a similar manner, except that primer YF-7 (5′-GATCGGAATTCACCATGTCATTACCACACGTTATCC) was used in place of YF-1 and the fragments were ligated into pKAS46, generating pGKK35, prior to allelic exchange.
Construction of chromosomal tcpP-lacZ fusions.
Plasmid pKAS48 (29) was used to construct the ΔlacZ3 deletion in El Tor strain C6706str2 (37) by allelic exchange, generating strain KSK262. The El Tor tcpP-lacZ operon fusion in KSK262 was constructed in a manner similar to that of the classical tcpP-lacZ fusion (32), except that primers TP-BAME (5′-GATCGGGATCCAGTAATGCCGGCTAATTCATG) and TP-SEE (5′-GATCGGTCGACGAATTCCAGCCGTTAGCAGCTTGTAAG) were used in place of TP-BAM and TP-SE for amplification from C6706str2. The resulting fusion in plasmid pKAS113 was introduced into V. cholerae by allelic exchange. The tcpP-lacZ fusion on λ KSPL1 was previously described (32).
Construction and mobilization of expression plasmids.
The expression plasmids constructed in this study are listed in Table 1. The aphB gene was amplified from either the classical (O395) or El Tor (C6706str2) biotypes by using primers CO2-7 (5′-GATCGGAATTCATAAATTAGCGATAGTTGC) and CO2-8 (5′-GATCGAAGCTTGAAAAAGGGCGCGAAGCCC). The aphA gene was amplified from O395 by using primers YF-5 (5′-GATCGGAATTCTAAATGCGTTGATATGCGTGCC) and YF-6 (32). Plasmids derived from pLAFR3 (33), pMMB66EH (9), and pBAD-TOPO (Invitrogen), respectively, were introduced into V. cholerae by mating with E. coli SM10 (28), triparental mating with E. coli MM294 carrying pRK2013 (8), and electroporation.
β-Galactosidase assays.
β-Galactosidase assays (20) were carried out with tcpP-lacZ, aphA-lacZ, or aphB-lacZ fusion strains during mid-logarithmic growth and with ctx-lacZ fusion strains after overnight growth. In AKI medium, cultures were assayed after 4 h without rotation. The bicinchoninic acid procedure (Pierce) was used to determine the total amount of protein in each reaction from the overnight cultures. The data are averaged results from at least two experiments.
Immunoblot analysis.
Cell extracts from overnight cultures were subjected to sodium dodecyl sulfate–12.5% polyacrylamide gel electrophoresis, transferred to nitrocellulose, and probed with anti-TcpA antibody (34) by using the ECL (enhanced chemiluminescence) detection system (Amersham).
Nucleotide sequence accession number.
The accession number for the nucleotide sequence of aphB in GenBank is AF148502.
RESULTS
The aphB gene is required for virulence gene expression.
V. cholerae TnphoA mutant KSK404 was identified as a derivative of the ctx-lacZ fusion strain KSK218, which showed reduced β-galactosidase production under environmental conditions normally optimal for its expression (i.e., LB medium [pH 6.5] at 30°C) and failed to produce TCP. A 200-bp DNA fragment encompassing the region adjacent to the transposon in KSK404 was obtained by ligating restriction-digested chromosomal DNA from the mutant into a plasmid and performing two rounds of PCR with primers specific for the plasmid and for the transposon. The DNA fragment, which contained a portion of the aphB open reading frame, was then inserted into an oriR6K plasmid and used to disrupt the wild-type aphB gene in KSK218. After confirming that the aphB disruption in the resulting strain, GK91, caused a defect in virulence gene expression similar to that of the original transposon mutant, the entire aphB gene was isolated from this strain by using chromosomal capture (30, 31) and sequenced.
To verify that the disruption of aphB was solely responsible for the defect in virulence gene expression in strain GK91, an in-frame deletion of aphB was constructed in KSK218 (ctx-lacZ), strain GK122, and this defect was complemented by inducing a wild-type aphB gene expressed from the tac promoter of plasmid pKAS117. As shown in Fig. 1, the ΔaphB mutation in GK122 significantly reduced the production of β-galactosidase under inducing conditions (LB medium [pH 6.5] at 30°C) and expression of aphB from pKAS117 restored its production to wild-type levels under these conditions. The mutation had only a small effect on the already low levels of β-galactosidase under repressing conditions (LB medium [pH 8.5] at 30 or at 37°C). However, expression of aphB from pKAS117 increased β-galactosidase production at pH 8.5 at 30°C in both the parental strain and the ΔaphB mutant to close to the levels observed under inducing conditions. These results indicate that aphB plays a role in activating ctx expression and that it is also involved in its regulation by environmental stimuli such as pH. Induction of aphB from pKAS117 also increased β-galactosidase production at 37°C, but to a smaller extent than at pH 8.5 at 30°C (Fig. 1).
FIG. 1.
Influence of AphB on the expression of a ctx-lacZ fusion. Cultures were grown in LB medium at pH 6.5 or 8.5 at 30 or 37°C. Those with pKAS117 also contained 1 mM isopropyl-β-d-thiogalactopyranoside. Black bars, KSK218; striped black bars, GK122 (ΔaphB); gray bars, KSK218 with pKAS117 (AphB); striped gray bars, GK122 with pKAS117 (AphB).
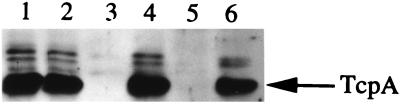
The influence of AphB on the production of TCP, shown in Fig. 2, is similar to the above results observed with ctx. The ΔaphB mutation in strain GK122 prevented the production of the 20.5-kDa major pilin protein TcpA under inducing conditions (Fig. 2, lane 3). Induction of aphB expression from pKAS117 in this mutant restored TcpA production (Fig. 2, lane 4) and permitted the cells to autoagglutinate in culture, a property associated with wild-type levels of TCP. Thus, AphB influences the expression of both the ctx and tcp operons in V. cholerae.
FIG. 2.
AphB influences TcpA production in both classical and El Tor biotypes. Samples were prepared from KSK218 (classical [lane 1]), KSK218 with pKAS117 (AphB [lane 2]), GK122 (classical ΔaphB [lane 3]), GK122 with pKAS117 (AphB [lane 4]), KSK262 (El Tor [lane 5]), and KSK262 with pKAS117 (AphB [lane 6]). Cultures were grown overnight in LB medium (pH 6.5) at 30°C. Those with pKAS117 also contained 1 mM isopropyl-β-d-thiogalactopyranoside. Samples were analyzed by Western blotting with anti-TCP antiserum (34). TcpA is indicated by the arrow to the right.
AphB activates the expression of the tcpPH promoter.
The significant impact of aphB on the expression of the ctx and tcp genes prompted us to investigate whether genes required earlier in the virulence cascade, toxRS, tcpPH, or aphA, were also influenced by AphB. The ΔaphB mutant GK122 did not produce the outer membrane protein OmpT in place of OmpU (data not shown), suggesting that toxR expression was not altered in the strain (22). To assess its effects on the expression of tcpPH and aphA, the ΔaphB mutation was introduced into the classical tcpP-lacZ fusion strain KSK618 and the classical aphA-lacZ fusion strain KSK666 (32). Table 2 shows that the expression of the tcpP-lacZ fusion in the ΔaphB strain, GK121, was significantly reduced under each environmental condition examined relative to the parental strain. Furthermore, the basal level of tcpPH expression in the absence of aphB did not significantly respond to environmental stimuli. When aphB was induced from the tac promoter of pKAS117, the expression of tcpP-lacZ in the ΔaphB mutant GK121 was increased under all environmental conditions examined (Table 2). This increase was most dramatic under the strongest repressive condition, pH 8.5 at 37°C. These results indicate that AphB is required for the activation of the tcpPH operon in V. cholerae and for its response to environmental stimuli. The ΔaphB mutation had no effect on the expression of the aphA-lacZ fusion in V. cholerae (data not shown), indicating that aphB is not influencing tcpPH expression indirectly through AphA.
TABLE 2.
Activation of a classical biotype tcpP-lacZ fusion by AphB and AphA
| Straina | β-Galactosidase activityb at pH and temp:
|
|||
|---|---|---|---|---|
| 6.5, 30°C | 8.5, 30°C | 6.5, 37°C | 8.5, 37°C | |
| KSK618 (tcpP-lacZ) | 3,940 ± 24 | 931 ± 8 | 2,091 ± 31 | 502 ± 11 |
| GK121 (ΔaphB) | 190 ± 22 | 145 ± 3 | 133 ± 8 | 98 ± 15 |
| KSK647 (ΔaphA) | 350 ± 75 | 104 ± 4 | 201 ± 6 | 85 ± 13 |
| KSK805 (ΔaphA ΔaphB) | 117 ± 7 | 97 ± 1 | 94 ± 7 | 74 ± 9 |
| GK121(pKAS117) (AphB) | 6,271 ± 680 | 5,428 ± 968 | 3,415 ± 74 | 2,616 ± 139 |
| KSK647(pKAS117) (AphB) | 3,283 ± 201 | 3,076 ± 408 | 1,669 ± 110 | 1,022 ± 61 |
| GK121(pKAS107) (AphA) | 502 ± 30 | 382 ± 88 | 288 ± 7 | 249 ± 1 |
| KSK647(pKAS107) (AphA) | 5,732 ± 723 | 2,589 ± 142 | 3,660 ± 846 | 1,816 ± 214 |
All plasmids were induced with 1 mM isopropyl-β-d-thiogalactopyranoside.
Units per optical density at 600 nm of culture.
AphB cooperates with AphA to activate tcpPH expression.
AphA has previously been shown to be required for activation of the tcpPH operon (32). The expression of tcpPH in a ΔaphA mutant, strain KSK647 (32), is similar to that of the ΔaphB mutant, except that the basal level of expression is still somewhat responsive to environmental stimuli (Table 2). Thus, loss of either AphA or AphB results in a dramatic decrease in the expression of the tcpPH operon. To determine if increased amounts of either protein could compensate for loss of the other, the aphB expression plasmid pKAS117 was introduced into the ΔaphA mutant KSK647 and the aphA expression plasmid pKAS107 (32) was introduced into the ΔaphB mutant GK121. Interestingly, high levels of AphB in the ΔaphA mutant restored tcpPH expression to close to wild-type levels at pH 6.5 (and to greater than wild-type levels at pH 8.5) (Table 2), whereas high levels of AphA in the ΔaphB mutant increased tcpPH expression somewhat, but did not restore it to wild-type levels. Thus, when present in sufficient amounts, either protein is capable of activating tcpPH transcription in the absence of the other, but AphA still requires AphB to achieve wild-type expression levels.
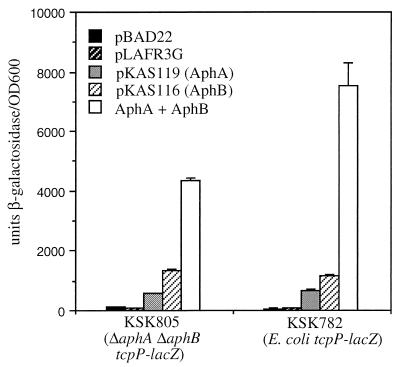
To further address whether AphA and AphB function sequentially or in separate pathways to activate tcpPH expression, the ΔaphA mutation was introduced into the ΔaphB mutant GK121, generating strain KSK805. The finding that the expression of tcpPH is lower in the double mutant under all environmental conditions than in either single mutant (Table 2) suggests that AphA and AphB function cooperatively to activate tcpPH transcription rather than sequentially. This notion is further supported by the results in Fig. 3 which show that in the ΔaphA ΔaphB double mutant, KSK805, and in an E. coli tcpP-lacZ fusion strain, KSK782, the presence of AphA and AphB together from plasmids pKAS119 and pKAS116 results in higher levels of β-galactosidase production than with either protein alone. Thus, it appears that AphA and AphB function synergistically to activate transcription at the tcpPH promoter.
FIG. 3.
Cooperation between AphA and AphB enhances tcpP-lacZ expression. V. cholerae KSK805 (ΔaphA ΔaphB) (left) was grown in LB medium (pH 6.5) at 30°C, and E. coli KSK782 (tcpP-lacZ) (right) was grown in LB medium (pH 7.0) at 37°C. Black bars, pBAD22 plus 0.2% arabinose; striped black bars, pLAFR3G plus 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG); gray bars, pKAS119 (AphA) plus 0.2% arabinose; striped gray bars, pKAS116 (AphB) plus 1 mM IPTG; open bars, pKAS119 plus pKAS116 (AphA plus AphB) plus 0.2% arabinose plus 1 mM IPTG. OD600, optical density at 600 nm.
AphB is a LysR homolog.
The LysR family represents of one of the most common types of prokaryotic transcriptional regulators. These proteins typically interact with small specific signal molecules known as coinducers to activate the expression of divergent or unlinked target genes which function in many diverse processes (for a review, see reference 27). Members of this family show strong homology in their amino-terminal domains, much of which derives from conservation of a helix-turn-helix DNA-binding motif. AphB exhibits significant amino-terminal homology with a large number of these proteins, and an alignment of this region of AphB with several LysR family members is shown in Fig. 4. The aphB gene encodes a protein of 291 amino acids with a predicted molecular mass of 33.3 kDa. Two of the proteins with the strongest overall homology to AphB (27%) are PtxR, a positive regulator of exotoxin A production in Pseudomonas aeruginosa (12); and IrgB from V. cholerae, which positively regulates the expression of irgA in response to iron limitation (10).
FIG. 4.
Alignment of the amino-terminal region of V. cholerae AphB with those of several other members of the LysR family. The helix-turn-helix domain is underlined.
AphB activates tcpPH expression in the El Tor biotype.
The expression of the ToxR virulence regulon in classical biotype strains is maximal in LB medium (pH 6.5) at 30°C. Strains of the El Tor biotype show reduced expression of the regulon under these conditions and require a bicarbonate-containing medium (AKI medium) at 37°C for high-level expression (16). Table 3 shows that the expression of an El Tor tcpP-lacZ fusion, strain KSK725, is significantly reduced in LB medium relative to the classical tcpP-lacZ fusion, strain KSK618 (Table 2), under all of the conditions examined. Although growth of the El Tor strain in AKI medium improved the expression of tcpPH, it was still significantly lower than that of the classical strain in LB medium (pH 6.5) at 30°C. To determine if AphB also activates tcpPH expression in the El Tor biotype, a ΔaphB mutation was introduced into KSK725, generating strain GK138. The ΔaphB mutation in this strain significantly decreased tcpPH expression under AKI conditions (Table 3), but had a smaller effect on the already low levels of expression in LB medium. A similar result was observed with a ΔaphA mutation in this background, strain GK161 (Table 3). Thus, although the response of tcpPH in El Tor strains to environmental stimuli is different from that in classical strains, aphA and aphB play a role in its expression in both biotypes.
TABLE 3.
Activation of an El Tor biotype tcpP-lacZ fusion by AphB and AphA
| Straina | β-Galactosidase activityb at pH and temp:
|
||||
|---|---|---|---|---|---|
| AKI medium | LB medium
|
||||
| 6.5, 30°C | 8.5, 30°C | 6.5, 37°C | 8.5, 37°C | ||
| KSK725 (tcpP-lacZ) | 1,060 ± 27 | 362 ± 8 | 172 ± 0 | 199 ± 8 | 101 ± 2 |
| GK138 (ΔaphB) | 129 ± 8 | 222 ± 6 | 162 ± 8 | 122 ± 8 | 94 ± 1 |
| GK161 (ΔaphA) | 122 ± 3 | 148 ± 1 | 104 ± 1 | 90 ± 3 | 70 ± 1 |
| KSK725(pKAS117) (AphB) | 2,129 ± 37 | 3,785 ± 309 | 1,270 ± 173 | 2,053 ± 168 | 753 ± 197 |
| KSK725(pKAS107) (AphA) | 1,060 ± 26 | 1,689 ± 17 | 730 ± 21 | 621 ± 41 | 284 ± 11 |
All plasmids were induced with 1 mM isopropyl-β-d-thiogalactopyranoside.
Units per optical density at 600 nm of culture.
The mechanisms responsible for the differential expression of tcpPH in classical and El Tor biotype strains are not yet understood. Since expression of either aphA or aphB from the tac promoter significantly increased tcpPH expression under normally nonpermissive expression conditions in the classical biotype, it was of interest to determine whether either of these genes could increase tcpPH expression in the El Tor biotype as well. Table 3 shows that induction of aphB from pKAS117 in the El Tor fusion strain KSK725 increased tcpPH expression in both AKI and LB media. In LB medium, the levels of expression of tcpPH in the presence of pKAS117 were virtually identical to those of the classical tcpP-lacZ fusion strain (Table 2). El Tor strain KSK262 does not produce TCP detectable even by Western blotting when grown in LB medium (pH 6.5) at 30°C (Fig. 2, lane 5). However, induction of aphB expression from pKAS117 in KSK262 increased TCP production in LB medium (pH 6.5) at 30°C to a level similar to that of classical strains (Fig. 2, lane 6) and permitted the cells to autoagglutinate. Expression of aphA from pKAS107 also increased the expression of the El Tor tcpP-lacZ fusion, but to a lesser extent than aphB (Table 3), and did not permit strain KSK262 to produce TCP by Western blotting (data not shown). These findings indicate that the tcpPH promoter can be activated by AphB and, to a lesser extent, AphA, in the El Tor biotype under conditions not normally permissive for its expression.
The AphB protein and its expression appear similar in both biotypes.
The significant effect of inducing aphB expression from pKAS117 on the activation of the El Tor tcpPH promoter in LB medium (pH 6.5) at 30°C suggested that, in this biotype, the AphB protein or its expression might be different from that in the classical biotype. The deduced amino acid sequences of the classical and El Tor AphB proteins, however, were found to be identical. In addition, when either the classical or El Tor aphB gene was induced from an arabinose promoter in plasmid pKAS118 or pKAS120, respectively, both activated an E. coli tcpP-lacZ fusion approximately 30-fold, suggesting that they are equally functional. To assess the expression of aphB in classical and El Tor strains, an aphB-lacZ fusion was constructed in each biotype. Table 4 shows that the levels of expression of aphB in the classical fusion strain GK130 and the El Tor fusion strain GK142 are similar. Since the AphB proteins from the classical and El Tor strains appear to be equally capable of activating tcpPH transcription and the levels of expression of the gene in both biotypes are similar, some other aspect of AphB function may be different in the two biotypes.
TABLE 4.
Comparison of aphB expression in classical and El Tor biotypes
| Strain | β-Galactosidase activitya at pH and temp:
|
||||
|---|---|---|---|---|---|
| AKI medium | LB medium
|
||||
| 6.5, 30°C | 8.5, 30°C | 6.5, 37°C | 8.5, 37°C | ||
| GK130 (classical aphB-lacZ) | 201 ± 6 | 176 ± 3 | 129 ± 1 | 100 ± 3 | 78 ± 1 |
| GK142 (El Tor aphB-lacZ) | 223 ± 2 | 207 ± 2 | 137 ± 1 | 98 ± 6 | 83 ± 7 |
Units per optical density at 600 nm of culture.
It has previously been shown that the expression of aphA is not strongly influenced by either pH or temperature (32). The results in Table 4 indicate that the expression of aphB is also not strongly influenced by these stimuli, nor does it completely reflect the pattern of expression that is observed with tcpPH under the different conditions. For example, expression of the classical aphB-lacZ fusion is not higher at pH 6.5 at 37°C than it is at pH 8.5 at 30°C, and expression of the El Tor aphB-lacZ fusion is not significantly higher in AKI medium than it is in LB medium at pH 6.5 at 30°C. It is also noteworthy that induction of either aphA or aphB from the tac promoter had no effect on the expression of the aphB-lacZ fusion or its response to environmental stimuli (data not shown). Since the expression of tcpPH in response to pH or temperature does not appear to solely depend upon the expression of either aphA or aphB in response to these stimuli, it is possible that the activities of the proteins themselves might be influenced under various conditions.
DISCUSSION
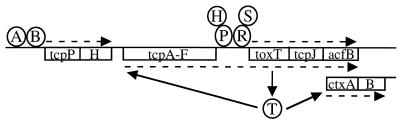
Activation of the ToxR virulence cascade requires multiple factors encoded both within the “ancestral” V. cholerae chromosome and on discrete elements involved in pathogenicity. As shown in Fig. 5, ToxR and ToxS, a chromosomally encoded protein pair, cooperate with the TCP-ACF pathogenicity element-encoded TcpP and TcpH protein pair to positively regulate the expression of the TCP-ACF-encoded regulator, ToxT. ToxT, in turn, activates expression of the ctx and tcp operons as part of a virulence gene regulatory cascade. In this report, we describe another chromosomally encoded protein pair required for the activation of the ToxR virulence cascade. AphB is a new member of the LysR family of transcriptional regulators which cooperates with the recently identified AphA protein (32) to activate the expression of the tcpPH operon.
FIG. 5.
Model of activation of the ToxR virulence cascade. In response to the appropriate environmental conditions, AphA and AphB activate transcription of the tcpPH operon. TcpPH, together with ToxRS, activate transcription of toxT. ToxT, in turn, activates expression of the ctxAB operon as well as expression of the entire tcp operon, including the toxT gene itself. The precise locations of the protein binding sites at the individual promoters have not yet been determined.
V. cholerae strains deficient in either aphA or aphB show reduced expression of the tcpPH operon and as a result do not produce virulence factors such as CT and TCP. That an aphA aphB double mutant shows lower expression of tcpPH than either single mutant suggests that AphA and AphB are not functioning sequentially in the same pathway but that they cooperate to activate tcpPH transcription. When expressed from their natural promoters in V. cholerae, neither protein significantly activates transcription in the absence of the other. When expressed from inducible promoters on plasmids in V. cholerae or E. coli, either protein is capable of activating the transcription of tcpPH in the absence of the other, with AphB showing a stronger effect than AphA and the former even compensating for the latter in V. cholerae. However, the expression of tcpPH is significantly greater with the two proteins together than with either one alone. It is possible that the presence of AphA enhances the ability of AphB to activate transcription.
The ToxR virulence regulon is strongly influenced by environmental cues such as pH and temperature. Although the mechanisms responsible for this regulation are not yet understood, the effect of environmental stimuli on the expression of the regulon may largely be the result of their influence over the expression of tcpPH (24). How pH and temperature control the expression of tcpPH is not yet understood, but AphA and AphB appear to play a role in this process. V. cholerae strains containing plasmids expressing either aphA or aphB show increased tcpPH transcription under both permissive and nonpermissive environmental conditions. Supplying high levels of either of these two proteins in the presence of the other appears to be sufficient to almost completely override environmental regulation by pH and temperature. Since the expression of neither aphA (32) nor aphB is strongly regulated by environmental conditions, it is possible that their activities are influenced by them. Many LysR regulators activate gene expression only in the presence of specific coinducer molecules (27). Interaction of such a molecule with AphB only under certain environmental conditions might render it able to activate tcpPH transcription if AphA is present. High levels of either AphA or AphB might be sufficient to at least partly overcome the need for a coinducer to facilitate transcriptional activation. Alternatively, when present in high levels, AphA or AphB may effectively compete with other proteins that normally function to downregulate tcpPH expression under certain environmental conditions. Additional experiments are necessary in order to distinguish between these possibilities.
It is well established that V. cholerae strains of the El Tor biotype exhibit lower expression of the ToxR virulence regulon in vitro than classical biotype strains. This appears to be the result of reduced expression of toxT and tcpPH in the El Tor biotype relative to the classical biotype (6, 24) (Table 3). Despite the fact that the expression of tcpPH is differentially regulated in classical and El Tor biotypes, aphA and aphB are involved in the activation of tcpPH in both. The observation that expression of aphB from the tac promoter increased tcpPH transcription in the El Tor biotype to classical levels in LB medium and permitted TCP production suggests that AphB might in some respect be different in the two biotypes. However, El Tor biotype strains encode a functional AphB protein and the expression of the gene is similar to that of classical strains. AphA alone does not appear to be responsible for the biotype-specific difference in expression, since induction of the aphA gene in the El Tor biotype did not increase tcpPH transcription to classical levels and it did not permit TCP production. These results raise the possibility that some other aspect of AphB function may be different in the two biotypes, such as the ability of the protein to assume a conformation that allows it to activate transcription at the tcpPH promoter. Experiments to address this issue are currently in progress.
It is not yet known whether AphA and AphB function alone or together in any other regulatory capacity in V. cholerae. It is common for LysR transcriptional regulators to be divergently transcribed from a promoter that is close to or that overlaps a regulated target gene (27). For example, the gene encoding the V. cholerae IrgB protein is divergently transcribed from the gene which it activates, irgA (10). The gene upstream of aphB, which is divergently transcribed, encodes a protein which shows a high degree of homology to response regulators of a number of bacterial two-component systems. Two-component systems frequently regulate gene expression in prokaryotes in response to environmental stimuli. It is tempting to speculate that AphB may also activate the expression of this gene, and experiments to determine this are currently under way.
This study describes a new chromosomal gene, aphB, which encodes a LysR homolog that functions in both biotypes of V. cholerae in concert with a second chromosomally encoded protein, AphA, to activate the expression of a virulence operon within a pathogenicity element. Further understanding of the mechanisms by which AphA and AphB activate gene expression may shed light on a number of questions regarding the pathogenesis of V. cholerae.
ACKNOWLEDGMENTS
We thank Ronald Taylor for many insightful discussions and helpful suggestions.
This work was supported by Public Health Service grant AI-41558 to K.S.
REFERENCES
- 1.Brown R C, Taylor R K. Organization of tcp, acf, and toxT genes within a ToxT-dependent operon. Mol Microbiol. 1995;16:425–439. doi: 10.1111/j.1365-2958.1995.tb02408.x. [DOI] [PubMed] [Google Scholar]
- 2.Carroll P A, Tashima K T, Rogers M B, DiRita V J, Calderwood S B. Phase variation in tcpH modulates expression of the ToxR regulon in Vibrio cholerae. Mol Microbiol. 1997;25:1099–1111. doi: 10.1046/j.1365-2958.1997.5371901.x. [DOI] [PubMed] [Google Scholar]
- 3.Champion G A, Neely M N, Brennan M A, DiRita V J. A branch in the ToxR regulatory cascade of Vibrio cholerae revealed by characterization of toxT mutant strains. Mol Microbiol. 1997;23:323–331. doi: 10.1046/j.1365-2958.1997.2191585.x. [DOI] [PubMed] [Google Scholar]
- 4.Chiang S L, Taylor R K, Koomey M, Mekalanos J J. Single amino acid substitutions in the N-terminus of Vibrio cholerae TcpA affect colonization, autoagglutination, and serum resistance. Mol Microbiol. 1995;17:1133–1142. doi: 10.1111/j.1365-2958.1995.mmi_17061133.x. [DOI] [PubMed] [Google Scholar]
- 5.DiRita V J, Mekalanos J J. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell. 1991;64:29–37. doi: 10.1016/0092-8674(91)90206-e. [DOI] [PubMed] [Google Scholar]
- 6.DiRita V J, Neely M, Taylor R K, Bruss P M. Differential expression of the ToxR regulon in classical and El Tor biotypes of Vibrio cholerae is due to biotype-specific control over toxT expression. Proc Natl Acad Sci USA. 1996;93:7991–7995. doi: 10.1073/pnas.93.15.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiRita V J, Parsot C, Jander G, Mekalanos J J. Regulatory cascade controls virulence in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fürste J P, Pansegrau W, Frank R, Blöcker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 10.Goldberg M B, Boyko S A, Calderwood S B. Positive transcriptional regulation of an iron-regulated virulence gene in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:1125–1129. doi: 10.1073/pnas.88.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamood A N, Colmer J A, Ochsner U A, Vasil M L. Isolation and characterization of a Pseudomonas aeruginosa gene, ptxR, which positively regulates exotoxin A production. Mol Microbiol. 1996;21:97–110. doi: 10.1046/j.1365-2958.1996.6251337.x. [DOI] [PubMed] [Google Scholar]
- 13.Häse C C, Mekalanos J J. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins D E, DiRita V J. Transcriptional control of toxT, a regulatory gene in the ToxR regulon of Vibrio cholerae. Mol Microbiol. 1994;14:17–29. doi: 10.1111/j.1365-2958.1994.tb01263.x. [DOI] [PubMed] [Google Scholar]
- 15.Higgins D E, Nazareno E, DiRita V J. The virulence gene activator ToxT from Vibrio cholerae is a member of the AraC family of transcriptional activators. J Bacteriol. 1992;174:6974–6980. doi: 10.1128/jb.174.21.6974-6980.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwanaga M, Yamamoto K, Higa N, Ichinose Y, Nakasone N, Tanabe M. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol Immunol. 1986;30:1075–1083. doi: 10.1111/j.1348-0421.1986.tb03037.x. [DOI] [PubMed] [Google Scholar]
- 17.Kaper J B, Morris J G, Jr, Levine M M. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karaolis D K R, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovach M E, Shaffer M D, Peterson K M. A putative integrase gene defines the distal end of a large cluster of ToxR-regulated colonization genes in Vibrio cholerae. Microbiology. 1996;142:2165–2174. doi: 10.1099/13500872-142-8-2165. [DOI] [PubMed] [Google Scholar]
- 20.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 21.Miller V L, DiRita V J, Mekalanos J J. Identification of toxS, a regulatory gene whose product enhances ToxR-mediated activation of the cholera toxin promoter. J Bacteriol. 1989;171:1288–1293. doi: 10.1128/jb.171.3.1288-1293.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 24.Murley, Y. M., P. A. Carroll, K. Skorupski, R. K. Taylor, and S. B. Calderwood. Differential transcription of the tcpPH operon confers biotype-specific control of the Vibrio cholerae ToxR virulence regulon. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 25.Ogierman M A, Zabihi S, Mourtzios L, Manning P A. Genetic organization and sequence of the promoter-distal region of the tcp gene cluster of Vibrio cholerae. Gene. 1993;126:51–60. doi: 10.1016/0378-1119(93)90589-u. [DOI] [PubMed] [Google Scholar]
- 26.Peterson K M, Mekalanos J J. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect Immun. 1988;56:2822–2829. doi: 10.1128/iai.56.11.2822-2829.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 28.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis. Biotechnology. 1983;1:784–791. [Google Scholar]
- 29.Skorupski K, Taylor R K. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- 30.Skorupski K, Taylor R K. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin-coregulated pilus in Vibrio cholerae. Proc Natl Acad Sci USA. 1997;94:265–270. doi: 10.1073/pnas.94.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skorupski K, Taylor R K. Sequence and functional analysis of the gene encoding Vibrio cholerae cAMP receptor protein. Gene. 1997;198:297–303. doi: 10.1016/s0378-1119(97)00331-4. [DOI] [PubMed] [Google Scholar]
- 32.Skorupski K, Taylor R K. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol Microbiol. 1999;31:763–771. doi: 10.1046/j.1365-2958.1999.01215.x. [DOI] [PubMed] [Google Scholar]
- 33.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun D, Seyer J M, Kovari I, Sumrada R A, Taylor R K. Localization of protective epitopes within the pilin subunit of the Vibrio cholerae toxin-coregulated pilus. Infect Immun. 1991;59:114–118. doi: 10.1128/iai.59.1.114-118.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor R K, Manoil C, Mekalanos J J. Broad-host-range vectors for delivery of TnphoA: use in genetic analysis of secreted virulence determinants of Vibrio cholerae. J Bacteriol. 1989;171:1870–1878. doi: 10.1128/jb.171.4.1870-1878.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thelin K H, Taylor R K. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect Immun. 1996;64:2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]