Abstract
Aims
Genetic testing is recommended in specific inherited heart diseases but its role remains unclear and it is not currently recommended in unexplained cardiac arrest (UCA). We sought to assess the yield and clinical utility of genetic testing in UCA using whole-exome sequencing (WES).
Methods and results
Survivors of UCA requiring external defibrillation were included from the Cardiac Arrest Survivor with Preserved Ejection fraction Registry. Whole-exome sequencing was performed, followed by assessment of rare variants in previously reported cardiovascular disease genes. A total of 228 UCA survivors (mean age at arrest 39 ± 13 years) were included. The majority were males (66%) and of European ancestry (81%). Following advanced clinical testing at baseline, the likely aetiology of cardiac arrest was determined in 21/228 (9%) cases. Whole-exome sequencing identified a pathogenic or likely pathogenic (P/LP) variant in 23/228 (10%) of UCA survivors overall, increasing the proportion of ‘explained’ cases from 9% only following phenotyping to 18% when combining phenotyping with WES. Notably, 13 (57%) of the 23 P/LP variants identified were located in genes associated with cardiomyopathy, in the absence of a diagnosis of cardiomyopathy at the time of arrest.
Conclusions
Genetic testing identifies a disease-causing variant in 10% of apparent UCA survivors. The majority of disease-causing variants was located in cardiomyopathy-associated genes, highlighting the arrhythmogenic potential of such variants in the absence of an overt cardiomyopathy diagnosis. The present study supports the use of genetic testing including assessment of arrhythmia and cardiomyopathy genes in survivors of UCA.
Keywords: Ventricular fibrillation, Cardiac arrest, Genetic testing, Arrhythmia, Cardiomyopathy, Cardiovascular genetics
Structured Graphical Abstract
Structured Graphical Abstract.
Study flowchart and summary results. ARVC, arrhythmogenic right ventricular cardiomyopathy; BrS, Brugada syndrome; CPVT, catecholaminergic polymorphic ventricular tachycardia; CRDS, RYR2 Ca2+ release deficiency syndrome; DCM, dilated cardiomyopathy; ECG, electrocardiogram; HCM, hypertrophic cardiomyopathy; LQTS, long QT syndrome; LV, left ventricular; MRI, magnetic resonance imaging; MVP, mitral valve prolapse; SQTS, short QT syndrome; UCM, unclassified cardiomyopathy; WES, whole-exome sequencing.
See the editorial comment for this article ‘Explaining the unexplained: applying genetic testing after cardiac arrest and sudden death’, by Elijah R. Behr, https://doi.org/10.1093/eurheartj/ehac172.
Introduction
While most cardiac arrests occur in an older population with coronary artery disease, some occur in young and otherwise healthy individuals.1,2 In the latter group, diagnostic testing with electrocardiography and cardiac imaging may identify the cause of arrest, including heritable electrical or structural disorders [e.g. long QT syndrome (LQTS), cardiomyopathy], and complex disorders (e.g. mitral valve prolapse, coronary spasm).3 When a specific heritable diagnosis is established, genetic testing using targeted panels is recommended,4 mainly to facilitate family screening. In contrast, the role of genetic testing in the absence of a definite diagnosis of inherited arrhythmia and/or cardiomyopathy is questionable, with concerns regarding cost and challenging interpretation of the genetic test results, especially for variants of uncertain significance (VUS). Current guidelines do not support the role of ‘exploratory’ genetic testing in this context.5,6
Despite extensive diagnostic testing, a large proportion of non-coronary cardiac arrest cases remain unexplained. The Cardiac Arrest Survivors with Preserved Ejection fraction Registry (CASPER) was established to improve our understanding of unexplained cardiac arrest (UCA). The registry includes UCA probands as well as their relatives in a longitudinal registry. In addition to undergoing phenotype-driven genetic testing as per local practice, participants can also provide DNA samples for genetic research. Prior retrospective analyses7 using clinician-driven genetic testing support the current recommendations for phenotype-driven testing.5,6 The role of comprehensive genetic testing in an unselected cohort of patients surviving an apparently UCA remains, however, unclear. The current study addresses this knowledge gap in a pragmatic approach. Using systematic whole-exome sequencing (WES) in consecutive consenting UCA survivors from the Canadian multicentre CASPER cohort, we sought to describe the yield and clinical utility of genetic testing in UCA.
Methods
The CASPER study population
CASPER has been described in previous publications3,7 and consists of a national registry and biobank of UCA survivors enrolled from expert cardiogenetics centres throughout Canada (ClinicalTrials.gov ID NCT00292032). Unexplained cardiac arrest survivors are included if they had a cardiac arrest requiring defibrillation that remained of uncertain aetiology following a resting electrocardiogram (ECG), transthoracic echocardiography (TTE), and coronary disease assessment. Exclusion criteria are as follows: obstructive coronary artery disease (any stenosis >50%), reduced left ventricular ejection fraction (LVEF <50%), persistent prolongation of the resting corrected QT (QTc) (>460 ms in males or >480 ms in females), a spontaneous Type 1 Brugada ECG, or a reversible cause of cardiac arrest such as marked hypokalaemia (<2.8 mmol/L) or drug overdose sufficient in gravity without other cause to explain the cardiac arrest.8 All patients included in CASPER have given informed consent to participate in the registry. In addition, all patients were approached for optional participation in a DNA biobank for genetics research. The current study only included CASPER UCA probands that provided DNA samples and consented to such research. DNA was extracted and stored at the Montreal Heart Institute’s Beaulieu-Saucier Pharmacogenomics Center (Montreal, Canada). The protocol was approved by the research ethics board of the Montreal Heart Institute and the University of British Columbia.
Diagnostic assessment
Following cardiac arrest, all patients underwent a cardiovascular evaluation to determine the cause of their cardiac arrest at the discretion of the treating physician according to local practices at the time of arrest. This included standard and high-lead resting ECG, exercise and/or epinephrine testing, procainamide challenge, anatomic coronary assessment, as well as cardiac imaging using TTE and/or cardiac magnetic resonance (CMR).3,7 Some patients were also offered genetic testing as part of their clinical care. All data were collected in the CASPER registry for centralized review. The cause of cardiac arrest was determined by the local investigator and reviewed centrally, using predefined diagnostic criteria (see Supplementary material online, Table S1),8,9 based on published guidelines and consensus statement,4 whenever available. The diagnosis was determined at three stages: (i) following initial advanced phenotypic testing (including CMR, exercise/epinephrine, and procainamide testing), (ii) following initial advanced phenotypic testing and WES, and (iii) at the last available follow-up (see Structured Graphical Abstract). Although WES may have been performed years following the initial cardiac arrest event, the impact of genetic testing on the diagnosis was determined at the time of initial phenotypic testing to address the question of whether genetic testing is useful for diagnostic purposes in the first year following the arrest. Follow-up after the arrest was performed as per local clinical practice and such follow-up data were included in the ongoing CASPER registry. Change in diagnosis was tracked to assess the added value of systematic genetic testing and repeat testing during long-term follow-up in determining the likely cause of cardiac arrest.
Whole-exome sequencing and array genotyping
We performed WES using a Roche capture and Illumina sequencer for all available samples of UCA survivors in CASPER, regardless of whether or not they underwent clinical genetic testing and whether or not a cause of arrest was identified following advanced phenotyping. Specifically, rather than having strict exclusion criteria, we preferred a more inclusive approach where all cases with apparent UCA at presentation would be eligible. This pragmatic approach was selected to improve the generalizability of the findings and allowed us to assess the utility of genetic testing in addition to phenotypic assessment to establish the cause of cardiac arrest. In addition to reporting the yield and utility of genetic testing overall, we also report results separately in cases whose cardiac arrest remains unexplained despite phenotypic testing. Array genotyping was also performed for ancestry mapping. Details on sequencing, genotyping, and bioinformatic analyses are described in the Supplementary material online, Note. Data on gene coverage are provided in Supplementary material online, Table S2.
Virtual panels
Only variants in genes that are previously reported in association with non-syndromic and syndromic heart disease were considered. At the time of analysis planning, the work of the Clinical Genome Resource (ClinGen) cardiovascular clinical domain working group (CDWG) was incomplete. We included the list of genes from the Genomics England crowdsourcing tool PanelApp.10,11 A total of 184 genes were included from two panels signed off by PanelApp consensus: (i) ‘Sudden cardiac death’ (V9.46) and (ii) ‘Cardiomyopathies—including childhood onset’ (V1.5). Genes classified as ‘Green’ (diagnostic-grade) or ‘Amber’ (borderline) were included (see Supplementary material online, Table S3). We recognize that for a majority of those 184 genes, the current evidence implicating them in heart disease is limited. More recently, the ClinGen cardiovascular CDWG completed gene curation for most inherited arrhythmia syndromes and cardiomyopathies.12–17 ClinGen validity grades for those curated genes are shown in Supplementary material online, Table S3. As a secondary analysis, we also report the rate of genetic variants restricted to a subset of 45 genes graded by the ClinGen cardiovascular CDWG as having moderate, strong, or definitive disease evidence in any of the curated arrhythmia syndromes and/or cardiomyopathies, as of August 18th, 2021.
Variant classification
The present analysis focuses on ultrarare variants with presumed large effect sizes (i.e. previously labelled as ‘mutations’). Therefore, we excluded variants that had allele frequencies (AF) higher than the maximal credible population AF to sustain pathogenicity. We calculated the maximal credible AF as previously published,18 setting prevalence to 1 in 10 000 to account for the rarity of UCA and the multiple conditions that could result in UCA.1 We estimated allelic/genetic heterogeneity to be 0.05 to account for the large genetic heterogeneity of cardiac arrest and penetrance to 0.05 since most pathogenic variant carriers would not present with cardiac arrest. Using these rough estimates of prevalence, genetic heterogeneity and penetrance, we calculated the maximal credible AF as 5 × 10−5 for monoallelic (autosomal dominant) genes and 2 × 10−3 for biallelic (autosomal recessive) genes. For X-linked inheritance, we empirically used a 5 × 10−5 maximal credible AF. We excluded variants with a filtering AF [upper bound of the 95% confidence interval (CI)] above the credible AF in any of the five major populations in the Genome Aggregation Database (gnomAD; Non-Finnish European, African, Latino, East Asian, and South Asian). All retained rare variants were classified based on published guidelines.19,20 For genes only involved in autosomal recessive diseases, variant classification was performed only for variants present in homozygous or suspected compound heterozygous states, to reduce the impact of returning incidental findings to the treating physician. Three clinical cardiogenetics experts independently curated all rare variants: two genetic counsellors (S.G. and B.D.) and one cardiologist (R.T.). Discordances were discussed and a consensus was reached.
Comparison of virtual panels
To assess the added yield of increasingly larger genetic panels, we also compared the rate of pathogenic or likely pathogenic (P/LP) variants in the ‘ClinGen’ panel (45 genes) as well as the three predefined PanelApp-derived virtual panels (see Supplementary material online, Table S3): (i) sudden cardiac death panel restricted to ‘Green’ (diagnostic-grade) genes (53 genes); (ii) sudden cardiac death and/or cardiomyopathy (including childhood onset) panels restricted to ‘Green’ (diagnostic-grade) genes (151 genes); and (iii) sudden cardiac death and/or cardiomyopathy (including childhood onset) panels including ‘Green’ (diagnostic-grade) and ‘Amber’ (Borderline) genes (184 genes).
Statistical analyses
The primary objective of the study was to describe the yield and diagnostic utility of genetic testing in UCA. As such, the statistical analyses are mainly descriptive. The rate of genetic variants identified is reported in percentage with the corresponding 95% CIs for the overall cohort, as well as subgroups of cases and/or genes. Categorical variables are reported as N (%) and compared using the Fisher’s exact test. The distribution of continuous variables was assessed using the Shapiro test. Normally distributed continuous variables are reported as mean ± standard deviation and compared using the Student t-test. Non-normally distributed continuous variables are reported as median (quartile 1–quartile 3), and compared using the Wilcoxon rank-sum test. Changes in the rate of P/LP variants over time were assessed using the Cochran-Armitage test for trend.
Results
Clinical characteristics and diagnostic testing
A total of 528 UCA survivors were enrolled in the CASPER registry by February 2018. Of those, 228 also consented to the optional DNA biobanking and genetic sub-study and were included in the present study. The included UCA cases were enrolled in the CASPER registry and biobank between 2004 and 2018, after a median of 0.7 (0.1–2.8) years following their cardiac arrest, and were last seen 7.7 (4.7–11.2) years following the arrest. Table 1 describes the baseline clinical characteristics, diagnostic tests performed, and the suspected diagnoses following advanced phenotypic testing.
Table 1.
Baseline characteristics of the included cardiac arrest cases (N = 228)
| Male sex | 151 (66%) |
| Age at arrest (years) | 39 ± 12 |
| European ancestrya | 162/199 (81%) |
| Sudden death in 1st or 2nd degree relatives | 32/226 (14%) |
| Syncope prior to arrest | 44 (19%) |
| Diagnostic testing performed prior to WES | |
| Cardiac CT and/or coronary angiography | 208 (91%) |
| Cardiac magnetic resonance imaging | 189 (83%) |
| Exercise and/or epinephrine test | 201 (88%) |
| Procainamide test | 153 (67%) |
| Suspected diagnosis following initial diagnostic testing prior to WES b | |
| Unexplained (including IVF, SCVF, and ERS) | 207 (91%) |
| Malignant mitral valve prolapse | 4 |
| Coronary spasm | 4 |
| Catecholaminergic polymorphic VT | 4 |
| Unclassified cardiomyopathy | 4 |
| Brugada syndrome | 2 |
| Long QT syndrome | 2 |
| Arrhythmogenic right ventricular cardiomyopathy | 1 |
Based on genotypic principal component analysis restricted to 199 cases with available array genotyping data.
See Supplementary material online, Table S1 for definitions and Supplementary material online, Table S4 for details and strengths of diagnoses. ERS, early repolarization syndrome; IVF, idiopathic ventricular fibrillation; SCVF, short-coupled ventricular fibrillation; VT, ventricular tachycardia; WES, whole-exome sequencing.
The initial cardiac arrest event occurred at a mean age of 39 ± 12 years. The majority of cases (66%) were males, in line with known sex differences.21 Of the 199 cases with array genotypic data where ancestry could be genetically determined, 81% were of European ancestry (see Supplementary material online, Figure).
The majority of UCA survivors (91%) underwent invasive coronary angiography and/or coronary computed tomography prior to study inclusion. The remaining UCA probands were younger (21 ± 9 years; P < 0.0001), and coronary artery disease/congenital anomalies were excluded based on perfusion imaging, CMR, and/or TTE (in young children). Further clinical assessment to determine the cause of arrest was performed in all UCA survivors at the discretion of the treating physician, including CMR in 189 (83%), exercise and/or epinephrine testing in 201 (88%), and procainamide testing in 153 (67%). Following clinical testing, the likely aetiology of cardiac arrest was determined in 21/228 cases (9%) based on strict diagnostic criteria (Supplementary material online, Table S1), while it remained unexplained in 207 (91%). Specific diagnoses are shown in Table 1, Supplementary material online, Table S4, and the Structured Graphical Abstract.
Whole-exome sequencing
Whole-exome sequencing followed by the analysis of a virtual gene panel was performed in batches from 2014 to 2020 for all 228 cardiac arrest survivors regardless of the suspected diagnosis, to pragmatically assess the yield and benefit of genetic testing in this population. The list of all identified rare variants is shown in Supplementary material online, Table S5. In the overall cohort, a total of 24 P/LP variants in 23/228 patients (10.1%; 95% CI 6.5–14.8%) were identified (Figure 1 and Table 2). One case (#152, discussed below) carried two variants classified as P/LP. The presence of P/LP variants was not significantly associated with patient sex, age at arrest, prior syncopal event, family history of sudden death, or ancestry (European vs. non-European), although the limited number of cases with P/LP variants may have limited statistical power. The proportion of cases with P/LP variants tended to decrease over time (e.g. 8/57, 7/57, 5/57, and 3/57 for the first, second, third, and last quartiles of year of arrest) although the trend was not statistically significant (Cochran-Armitage test for trend Z = −1.7, P = 0.09). A detailed description of cases with P/LP variants is shown in Supplementary material online, Table S6 and data on cascade testing on relatives are shown in Supplementary material online, Table S7.
Figure 1.
Number of rare unique variants identified in each gene. Genes are selected based on two panels signed off by PanelApp consensus: Sudden cardiac death (V 9.46) and cardiomyopathies—including childhood onset (V 1.5). Genes are ordered alphabetically within the panel in which they appear first as well as PanelApp grade (Green or Amber). Green represents diagnostic-grade genes and Amber represents those with borderline clinical actionability. Variants classified as pathogenic/likely pathogenic are shown in dark colour, and variants of uncertain significance are shown in light colour (see legend). Genes where no rare variants are identified are not shown (full list in Supplementary material online, Table S3). Variants that are low quality (see Supplementary material online, Note) are not shown, unless they were validated using Sanger sequencing (a full list of such variants appears in Supplementary material online, Table S5). Genes classified by the ClinGen cardiovascular clinical domain working group as having moderate evidence in cardiac arrhythmia syndromes or cardiomyopathies are marked with as asterisk. CM, cardiomyopathy; P/LP, pathogenic/likely pathogenic variant; SCD, sudden cardiac death; VUS, variant of uncertain significance.
Table 2.
Pathogenic and likely pathogenic variants identified in WES with brief clinical data of carriers
| Case ID | Gene | RefSeq transcript | Nucleic change | Protein change | Sex | Age at arrest | Diagnosis following initial phenotypic testing (strength) | Diagnosis following genetic testing (strength) |
|---|---|---|---|---|---|---|---|---|
| 2 | RYR2 | NM_001035 | c.C13822T | p.Arg4608Trp | F | 25 | IVF (definite) | CRDS (definite) |
| 5 | CACNA1C | NM_000719 | c.G1553A | p.Arg518His | F | 19 | IVF (definite) | LQTS (definite) |
| 6 | LMNA | NM_001257374 | c.C337T | p.Arg113Ter | F | 32 | CPVT (definite) | UCM (probable) |
| 7 | RYR2 | NM_001035 | c.G4938C | p.Glu1646Asp | F | 46 | CPVT (definite) | CPVT (definite) |
| 56 | PTPN11 | NM_002834 | c.A1529G | p.Gln510Arg | F | 22 | IVF (probable) | Rasopathy |
| 70 | SCN5A | NM_198056 | c.T1064G | p.Phe355Cys | M | 42 | BrS (definite) | BrS (definite) |
| 74 | MYH7 | NM_000257 | c.T2207C | p.Ile736Thr | M | 36 | IVF (probable) | UCM (probable) |
| 80 | RYR2 | NM_001035 | c.C364T | p.Arg122Cys | F | 30 | CPVT (definite) | CPVT (definite) |
| 81 | TNNI3K | NM_015978 | c.G2302A | p.Glu768Lys | M | 20 | IVF (definite) | UCM (probable) |
| 88 | PLN | NM_002667 | c.40_42del | p.Arg14del | M | 29 | CPVT (definite) | UCM (probable) |
| 89 | MYBPC3 | NM_000256 | c.2373dupG | p.Trp792fs | M | 59 | IVF (possible) | HCM (definite) |
| 97 | SCN5A | NM_198056 | c.2551_2552insGT | p.Phe851fs | M | 35 | IVF (possible) | BrS (probable) |
| 98 | DSG2 | NM_001943 | c.C941A | p.Ser314Ter | M | 56 | IVF (possible) | ARVC (probable) |
| 101 | MYH7 | NM_000257 | c.G611T | p.Arg204Leu | F | 19 | IVF (definite) | UCM (probable) |
| 120 | SCN5A | NM_198056 | c.2436 + 1G > C | NA | F | 47 | IVF (probable) | BrS (probable) |
| 127 | FLNC | NM_001458 | c.2157delC | p.Ile719fs | M | 43 | IVF (probable) | UCM (probable) |
| 133 | PLN | NM_002667 | c.T116G | p.Leu39Ter | F | 14 | IVF (probable) | UCM (probable) |
| 138 | RBM20 | NM_001134363 | c.1338-1G > T | NA | M | 22 | IVF (definite) | UCM (probable) |
| 145 | FLNC | NM_001458 | c.5584delC | p.Ala1895fs | F | 49 | IVF (possible) | UCM (probable) |
| 152 | RYR2 a | NM_001035 | c.G9352A | p.Gly3118Arg | M | 6 | IVF (possible) | CPVT (definite) |
| 152 | COA6 a | NM_001012985 | c.G63A | p.Trp21Ter | M | 6 | IVF (possible) | CPVT (definite) |
| 164 | MYBPC3 | NM_000256 | c.C1869A | p.Cys623Ter | F | 52 | ERS (definite) | UCM (probable) |
| 191 | SCN5A | NM_198056 | c.C1603T | p.Arg535Ter | F | 22 | IVF (definite) | BrS (probable) |
| 222 | KCNQ1 | NM_000218 | c.A1085G | p.Lys362Arg | M | 57 | UCM (definite) | UCM (definite) |
Additional details are provided in Supplementary material online, Table S6, including extended clinical data, list of prior publications of the listed variants, and ClinVar entry. ARVC, arrhythmogenic right ventricular cardiomyopathy; BrS, Brugada syndrome; CPVT, catecholaminergic polymorphic ventricular tachycardia; ERS, early repolarization syndrome; F, female; HCM, hypertrophic cardiomyopathy; IVF, idiopathic ventricular fibrillation; LQTS, long QT syndrome; M, male; UCM, unclassified cardiomyopathy; CRDS, RYR2 Ca2+ release deficiency syndrome.
Identified in homozygous state in the same case with known parental consanguinity. The presence of the homozygous variant in COA6 associated with mitochondrial cardiomyopathy47 triggered evaluation by a medical geneticist. There was no clinical or biochemical evidence of mitochondrial disease. The variant was later classified as VUS, because (i) it only affects some of the expressed gene isoforms, (ii) the case had no specific clinical features of mitochondrial disease, and (iii) there was an alternative explanation of the cardiac arrest (disease-causing RYR2 variant24).
Subgroup of cases with an identified aetiology prior to whole-exome sequencing
Among the 21 cases with a likely aetiology of cardiac arrest, a P/LP variant was identified in six (28.6%; 95% CI 11.3–52.2%). In 3/6 cases, the genetic finding confirmed the clinical diagnosis: two cases with catecholaminergic polymorphic ventricular tachycardia (CPVT) had RYR2 variants (Cases #7 and #80), and one case with procainamide-induced Brugada syndrome (BrS) had a SCN5A variant (#70). Notably, two cases with a clinical diagnosis of CPVT were found to carry a cardiomyopathy-causing variant, suggesting that CPVT in these cases was in fact a phenocopy of an underlying subclinical cardiomyopathy: Case #6 carried a LMNA variant (p.Arg113Ter), while Case #88 carried a PLN variant (p.Arg14del). Both cases had borderline left ventricular function at baseline TTE (LVEF 55%) but did not undergo CMR to assess for myocardial scar. During long-term follow-up, Case #6 with laminopathy evolved to end-stage dilated cardiomyopathy (DCM) requiring transplant, and Case #88 with PLN-cardiomyopathy also evolved to DCM and later died of electrical storm. Finally, a case (#222) with a baseline diagnosis of definite unclassified cardiomyopathy (UCM) presenting with a left bundle branch block, LVEF 50%, and papillary muscle scarring on CMR was found to carry a likely pathogenic variant in KCNQ1 in heterozygous state (p.Lys362Arg; Clinvar entry #52953), previously identified in multiple patients with autosomal recessive LQTS. The patient had normal QTc at rest and during exercise testing (after correction for QRS prolongation). His sister also carried the variant and had normal QTc at rest and in exercise testing. It was concluded that the heterozygous KCNQ1 variant is likely an incidental finding in this case where the cardiac arrest is due to cardiomyopathy.
Subgroup of cases that remained unexplained following advanced phenotypic testing
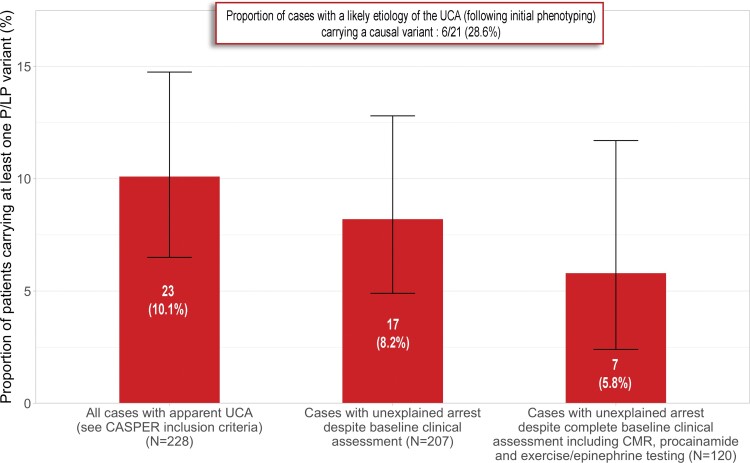
Among the 207 cases in which the cardiac arrest remained of unexplained aetiology despite baseline advanced phenotypic testing, 17 carried P/LP variants (8.2%; 95% CI 4.9–12.9%), including 6 in arrhythmia genes and 11 in cardiomyopathy genes. Of the 207 cases with unexplained arrests, 120 had a complete diagnostic assessment including CMR, stress/epinephrine, and procainamide testing. In this group, P/LP variants were identified in 7/120 (5.8%; 95% CI 2.4–11.7%; Figure 2). All 17 cases with P/LP variants are described briefly below, with more details provided in Supplementary material online, Table S6.
Figure 2.
Proportion of cardiac arrest survivors carrying pathogenic/likely pathogenic variants in the overall study population and clinically relevant subgroups. Proportions are shown as red bars and 95% confidence interval as brackets. Left: proportion of cases carrying a pathogenic/likely pathogenic variant in the overall cohort with apparent unexplained cardiac arrest (23/228). Middle: proportion of cases with pathogenic/likely pathogenic variants among cases where the cardiac arrest remained unexplained following advanced phenotypic testing (17/207). Right: proportion of cases with pathogenic/likely pathogenic variants among those with unexplained arrest despite complete advanced phenotypic testing including cardiac magnetic resonance, stress (or epinephrine), and procainamide testing (7/120).
Six cases with unexplained arrests carried variants in inherited arrhythmia genes: CACNA1C (1), RYR2 (2), and SCN5A (3). Case #5 had borderline QTc prolongation (450–460 ms) and abnormal QTc adaptation during exercise not reaching diagnostic criteria for LQTS and was found to carry a LQTS-causing variant in CACNA1C (p.Arg518His). Two cases had variants in RYR2. Case #2 with a diagnosis of definite idiopathic ventricular fibrillation (IVF) carried a variant in RYR2 (p.Arg4608Trp) causing a functional loss of function,22 compatible with a diagnosis of cardiac ryanodine receptor calcium release deficiency syndrome.23 Case #152 with exertional syncope and cardiac arrest with a non-diagnostic exercise test carried a homozygous variant in RYR2 (p.Gly3118Arg) recently identified in an unrelated family with recessive CPVT.24 Three cases carried loss-of-function SCN5A variants, including two (#120 and #191) that had a negative procainamide challenge, and one (#97) that did not undergo procainamide challenge.
Eleven cases with unexplained arrests carried variants in genes associated with cardiomyopathy, including arrhythmogenic cardiomyopathy [ACM; DSG2, FLNC (2), PLN, RBM20], hypertrophic cardiomyopathy [HCM; MYBPC3 (2), MYH7 (2), PTPN11], and DCM (TNNI3K25). All five cases carrying ACM variants had TTE and four also had CMR, with either normal or non-specific findings (see Supplementary material online, Table S8). Case #98 carrying the DSG2 variant (p.Ser314Ter) had progressive biventricular cardiomyopathy during follow-up, never reaching diagnostic criteria for definite arrhythmogenic right ventricular cardiomyopathy (ARVC). None of the other cases with ACM variants developed overt cardiomyopathy during follow-up (range: 5–21 years of follow-up; age at last follow-up 29–57). Of the five carriers of HCM-causing variants, four cases (#56, #74, #101, #164) had normal left ventricular wall thickness at baseline and at last follow-up (age range 28–58). Case #89 with a pathogenic MYBPC3 variant (p.Trp792fs) had an increased left ventricular wall thickness (14 mm, symmetric) below the HCM diagnostic threshold (15 mm) and initially attributed to systemic hypertension. In this case, HCM was only formally diagnosed after genetic testing. Of interest, Case #56 was found to carry a pathogenic variant in the rasopathy gene PTPN11 (p.Gln510Arg). Medical genetics consultation following genetic testing confirmed the clinical diagnosis of Noonan syndrome with multiple lentigines. Although cardiac arrest in rasopathy patients has been previously reported in the presence of HCM,26,27 this case is the first where cardiac arrest occurred in the absence of overt cardiomyopathy. Finally, Case #81 with a clinical diagnosis of IVF carried a variant in TNNI3K (p.Glu768Lys). This identical variant has been published recently with supportive functional data and strong co-segregation data in three families with variable phenotypes of supraventricular tachyarrhythmias, DCM, and cardiac arrest.25 Transthoracic echocardiography performed at 30 years old, 10 years following cardiac arrest, remains normal. Transthoracic echocardiography and CMR data for all 13 cases with P/LP variants in cardiomyopathy genes (including the two cases with initial diagnoses of CPVT) appear in Supplementary material online, Table S8.
Variants of uncertain significance, ancestry, and virtual panel size
A total of 120 unique VUS with no quality flag (Supplementary material online, Table S5) were identified, with 92/228 patients (40.4%) carrying at least one VUS (69 cases carrying 1 VUS, 17 carrying 2, 5 carrying 3, and 1 case carrying 4 VUS).
Cases of European ancestry had on average 0.53 VUS, compared with 0.46 for non-Europeans (Wilcoxon rank-sum test P = 0.28; difference 2 × 10−5, 95% CI = −9 × 10−6 to 8 × 10−5). Similarly, the proportion of cases with at least one VUS did not differ between those of European ancestry (42%) and non-European ancestry (30%; Fisher exact test P = 0.20).
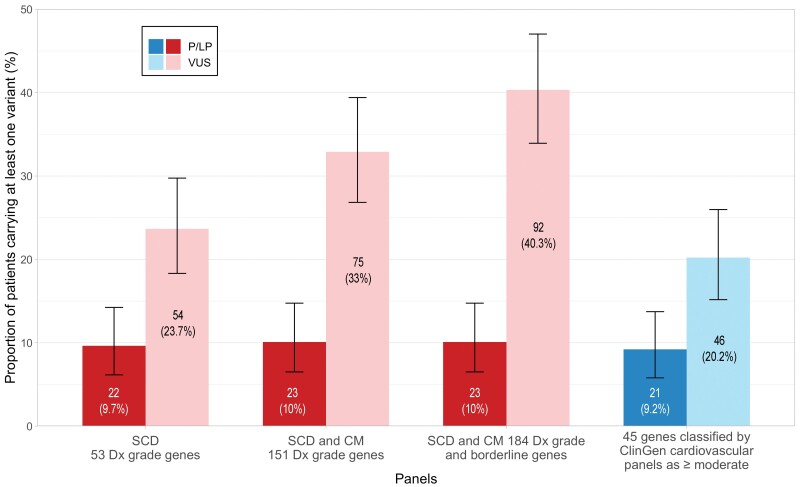
The number of patients carrying at least one rare VUS increased with the number of genes included in the virtual panel, ranging from 24% for a 53-gene panel to 40% for a 184-gene panel (Figure 3). Despite increasing the number of VUS, the larger 184-gene panel only increased the number of patients with at least one P/LP variants from 22 to 23 when compared with the more focused 53-gene panel. This highlights the unfavourable noise-to-signal ratio of large genetic panels in UCA. If only considering the 45 genes currently graded as having moderate, strong, or definitive evidence of disease by the ClinGen cardiovascular CDWG, only 20% of cases would have ≥1 VUS, and all clinically relevant P/LP variants except TNNI3K and PTPN11 would have been detected. TNNI3K has only been curated by ClinGen for isolated DCM.13PTPN11 was curated by the RASopathy CDWG as having definitive evidence in Noonan syndrome with multiple lentigines28 and endorsed by the ClinGen HCM expert panel.12
Figure 3.
Proportion of patients carrying genetic variants by varying virtual gene panels. Proportion of cases carrying at least one variant classified as pathogenic/likely pathogenic and/or at least one variant of uncertain significance for each of the four virtual gene panels based on PanelApp gene panels and restricted to genes classified by ClinGen as having moderate, strong, or definitive evidence in arrhythmias or cardiomyopathies. Note that larger panels increase the rate of patients with variants of uncertain significance with little change in the rate of pathogenic/likely pathogenic variants. Dx, diagnosis/diagnostic; SCD, sudden cardiac death.
Discussion
This study aimed to pragmatically assess the yield and clinical implications of systematic WES-based virtual panel sequencing in patients with apparently UCA. The main findings can be summarized as follows: (i) in a cohort of 228 consecutive unrelated survivors of cardiac arrest from a shockable rhythm without coronary artery disease, significant left ventricular function or a diagnosis of BrS or LQTS on baseline ECG, genetic testing using WES and virtual cardiac panels identifies a P/LP variant in 10% (95% CI 7–15%) of cases; (ii) a significant proportion (∼6%) of cardiac arrest survivors without a clinical diagnosis of cardiomyopathy at baseline carried a P/LP variant in a cardiomyopathy gene, suggesting such variants may result in ventricular arrhythmia in the absence of macroscopic structural heart disease; (iii) increasing gene panel size to include genes with limited evidence in human disease (i.e. PanelApp Amber and/or ClinGen grade < moderate) and genes associated with childhood/syndromic cardiomyopathy increases the rate of VUS nearly two-fold with minimal increase in P/LP detection rate. This increase in VUS to P/LP ratio is unfavourable given the complexity in managing families with VUS with associated diagnostic uncertainty.
The study findings should be interpreted in light of the included patient population. The CASPER cohort includes survivors of apparently UCA, defined as cardiac arrest requiring defibrillation, in the absence of obstructive coronary artery disease, LVEF <50%, persistent resting QTc prolongation, a reversible cause of cardiac arrest, and a spontaneous Type 1 BrS at presentation. Such a population is by definition heterogeneous. Rather than focusing on a specific cardiac arrest aetiology, the CASPER registry and biobank were designed to reflect a real-life clinical presentation where diagnostic uncertainty is common despite extensive phenotyping. The present study aimed to pragmatically scrutinize the role of genetic testing as a diagnostic test in the context of UCA within a multifaceted aetiological assessment framework including CMR, drug challenge, stress testing, etc. In this heterogeneous population, systematic genetic testing identified a P/LP variant in ∼10% of cases. In the subgroup of patients where a likely aetiology was identified following phenotypic testing (N = 21), the proportion of patients carrying a P/LP variant was ∼29%. In contrast, in patients where the cardiac arrest remained unexplained after advanced phenotypic testing (N = 207), the proportion of P/LP variant carriers was ∼8%. In the more selected subgroup of patients where the arrest remained unexplained despite complete baseline advanced phenotypic testing (CMR + stress/epinephrine + procainamide testing; N = 120), the proportion of P/LP variant carriers was ∼6%.
The current study is the first to assess the role of systematic genetic testing in a large cohort (N = 228) of cardiac arrest survivors. In a retrospective analysis of 174 UCA survivors from CASPER that underwent genetic testing at the discretion of the treating physician, we previously showed that the rate of P/LP variants is 17% overall, 25% in phenotype positive cases (e.g. LQTS, BrS, CPVT, ARVC, etc.), and 11% in idiopathic cases.7 Of the 174 UCA probands included in this prior report, 85 with a research DNA sample were also included in the current study. Of interest, 3 of those 85 cases (Cases #2, #74, #101) were found to carry P/LP variants while they had a negative limited gene panel in clinical testing. The higher rate of P/LP variants in our prior retrospective analysis compared with the present study is likely due to selection bias, where genetic testing was more likely to be requested by the treating physician in cases with higher pre-test probability for a genetic disease. Additionally, some variants initially reported as P/LP have since been downgraded to VUS.29 Other groups30–34 have also reported their experience with genetic testing in UCA/IVF. These studies were all retrospective with smaller sample sizes ranging from 24 to 79 subjects, where genetic testing was performed at the discretion of the treating physician. The reported yield of genetic testing was highly variable across studies, ranging from 2 to 48% (weighted average = 19%, based on a total sample size of 213). Such variability reflects heterogeneous definitions of UCA/IVF and indications for genetic testing, as well as variable criteria to assess variant pathogenicity.
As for most diagnostic tests in UCA,9 genetic testing could provide one of the puzzle pieces to establish the cause of cardiac arrest, which would then enable targeted therapy and follow-up, as well as family screening. As such, interpretation of genetic testing results should not only imply establishing the pathogenic potential of rare variants (e.g. using published criteria19) but also integrate the genetic finding with the phenotypic assessment. This is best done within specialized cardiovascular genetics and/or inherited arrhythmia programmes, with expertise in both genetics and clinical evaluation of cardiac arrest.4 In the present study, a total of 24 P/LP variants in 23/228 patients (10.1%) were identified. Of those 24 variants, 3 simply confirmed a clinical diagnoses (RYR2 in CPVT, SCN5A in drug-induced BrS), 15 potentially exposed the cause underlying cardiac arrest, and 2 were thought to be incidental findings (COA6 and KCNQ1) despite being initially classified as P/LP. Importantly, genetic testing does not replace clinical testing to establish the cause of cardiac arrest, but is complementary to proper and extensive phenotyping including sodium-blocker challenge, exercise testing, and CMR with assessment of late gadolinium enhancement.
The high rate of P/LP variants in cardiomyopathy genes is of important clinical and mechanistic interest. Of the 223 survivors of cardiac arrest without a diagnosis of cardiomyopathy at baseline clinical assessment, 13 (6%) were found to carry P/LP variants in genes associated with cardiomyopathy. These include two cases where the initial clinical diagnosis following arrest was CPVT, and 11 cases where the arrest was unexplained despite baseline clinical testing. Prior smaller studies have also observed disease-causing variants in cardiomyopathy genes in patients with documented ventricular arrhythmias without or with non-diagnostic structural anomalies.35–37 Tester et al.37 identified truncating PKP2 variants in 5/18 (28%) cases with gene-elusive clinically diagnosed CPVT, in line with the phenotype of Pkp2 knockout mice showing catecholamine-induced arrhythmia in the absence of structural heart abnormalities.38 In a cohort of 36 idiopathic cardiac arrest survivors, Isbister et al.36 reported that seven (19%) had disease-causing variants in genes associated with ACM and/or HCM, proposing that a ‘concealed cardiomyopathy’ underlies cardiac arrest in these cases. In the absence of a formal nomenclature for such ‘concealed cardiomyopathy’ cases, we used the term ‘probable UCM’ to describe these cases (see Supplementary material online, Table S1). Others have suggested the term ‘unclassified arrhythmogenic cardiomyopathy’. Studies have also identified cardiomyopathy gene variants in victims of sudden unexpected death with normal autopsies, with rates ranging from 2 to 12% in recent studies.39,40 The mechanisms underlying ventricular arrhythmias and sudden death in such cases with cardiomyopathy gene variants without manifest cardiomyopathy remain largely unknown, possibly involving ultrastructural anomalies undetectable by routine clinical imaging41,42 and/or standard autopsy as well as changes in cardiomyocyte electrophysiology and calcium-handling directly mediated by structural proteins43, as was elegantly demonstrated for PKP2.38
The present study has limitations. First, the study is descriptive in nature. The absence of a control population precludes case–control statistical assessment of gene-disease association, differentiating true association from incidental findings, particularly for cardiomyopathy gene variants. Nonetheless, the rate of such variants in this disease cohort and other cohorts36,37,39,40 is most certainly higher than expected in the general population. For instance, 4/228 (1.8%) of UCA probands in this study carried a pathogenic HCM-causing sarcomeric gene variant, while the rate of such variants in the reference population of the UK Biobank is ∼0.25%44 (Fisher test P ∼0.003, odds ratio ∼7.2). Second, the genetic analysis was performed using WES and a virtual panel. Such an analytical approach would not detect deep intronic and intergenic small variants (e.g. the DPP6 haplotype45), large insertions/deletions, as well as exonic variants in other genes. Third, phenotypic evaluation of UCA survivors was performed at the discretion of the treating physician/local investigator based on local practice at the time of arrest, as well as clinical limitations and availability. For instance, performing CMR prior to defibrillator implantation was sometimes not possible in cases where the arrest was initially managed at a remote community hospital. As such, phenotypic testing was incomplete in some cases, which may have resulted in diagnostic misclassification, therefore, increasing the apparent diagnostic utility of genetic testing. In addition, the use of the sodium-blocker procainamide to unmask BrS as opposed to the more potent ajmaline46 may have resulted in under-diagnosis of BrS with an apparent increased diagnostic utility for SCN5A testing. Fourth, the diagnostic utility of genetic testing was determined at the time of arrest. Therefore, its utility may be lesser in cases with UCA that occurred years prior to genetic evaluation, where a specific diagnosis may have been made in the meantime based on repeat phenotypic testing and/or family screening.
In conclusion, systematic genetic testing using WES and a virtual panel identifies a P/LP variant in 10% of apparent UCA survivors, with an added diagnostic value over phenotypic assessment, and may therefore be considered in the diagnostic evaluation of such cases. Genetic variants in cardiomyopathy genes in the absence of a clinical diagnosis of cardiomyopathy are common, supporting the concept of ‘concealed cardiomyopathy’ as a mechanism for cardiac arrest. Variants of uncertain significance are frequent especially with large gene panels and incidental P/LP variants are possible, highlighting the importance of restricting genetic testing to genes with good evidence and within dedicated cardiovascular genetics clinics to maximize the benefit of genetic testing while avoiding misdiagnosis.
Supplementary Material
Acknowledgements
The authors wish to thank the technical expertise of the Beaulieu-Saucier Pharmacogenomics Centre and the Centre d’expertise et de services Génome Québec.
Contributor Information
Steffany Grondin, Cardiovascular Genetics Center, Montreal Heart Institute, Department of Medicine, Université de Montréal, 5000 Belanger, Montreal, QC, Canada H1T 1C8.
Brianna Davies, Center for Cardiovascular Innovation, Division of Cardiology, Department of Medicine, University of British Columbia, Vancouver, BC, Canada.
Julia Cadrin-Tourigny, Cardiovascular Genetics Center, Montreal Heart Institute, Department of Medicine, Université de Montréal, 5000 Belanger, Montreal, QC, Canada H1T 1C8.
Christian Steinberg, Institut universitaire de cardiologie et pneumologie de Québec, Université Laval, Québec City, QC, Canada.
Christopher C Cheung, Center for Cardiovascular Innovation, Division of Cardiology, Department of Medicine, University of British Columbia, Vancouver, BC, Canada.
Paloma Jorda, Cardiovascular Genetics Center, Montreal Heart Institute, Department of Medicine, Université de Montréal, 5000 Belanger, Montreal, QC, Canada H1T 1C8.
Jeffrey S Healey, Population Health Research Institute, McMaster University, and Hamilton Health Sciences, Hamilton, ON, Canada.
Martin S Green, Division of Cardiology, University of Ottawa Heart Institute, Ottawa, ON, Canada.
Shubhayan Sanatani, Division of Pediatric Cardiology, British Columbia Children’s Hospital, Vancouver, BC, Canada.
Wael Alqarawi, Division of Cardiology, University of Ottawa Heart Institute, Ottawa, ON, Canada; Department of Cardiac Sciences, College of Medicine, King Saud University, Riyadh, Saudi Arabia.
Paul Angaran, Cardiac Arrhythmia Service, St Michael’s Hospital, Toronto, ON, Canada.
Laura Arbour, Department of Medical Genetics, University of British Columbia, Vancouver, BC, Canada.
Pavel Antiperovitch, Section of Cardiac Electrophysiology, Division of Cardiology, Department of Medicine, Western University, London, ON, Canada.
Habib Khan, Section of Cardiac Electrophysiology, Division of Cardiology, Department of Medicine, Western University, London, ON, Canada.
Richard Leather, Division of Cardiology, Royal Jubilee Hospital, Victoria, BC, Canada.
Peter G Guerra, Cardiovascular Genetics Center, Montreal Heart Institute, Department of Medicine, Université de Montréal, 5000 Belanger, Montreal, QC, Canada H1T 1C8.
Lena Rivard, Cardiovascular Genetics Center, Montreal Heart Institute, Department of Medicine, Université de Montréal, 5000 Belanger, Montreal, QC, Canada H1T 1C8.
Christopher S Simpson, Department of Medicine, Queen’s University, Kingston, ON, Canada.
Martin Gardner, Queen Elizabeth II Health Sciences Center, Halifax, NS, Canada.
Ciorsti MacIntyre, Queen Elizabeth II Health Sciences Center, Halifax, NS, Canada.
Colette Seifer, St Boniface Hospital, University of Manitoba, Winnipeg, MB, Canada.
Anne Fournier, Ste-Justine Hospital, Université de Montréal, Montreal, QC, Canada.
Jacqueline Joza, Department of Medicine, McGill University Health Center, Montreal, QC, Canada.
Michael H Gollob, Division of Cardiology, University Health Network, Toronto General Hospital, Toronto, ON, Canada.
Guillaume Lettre, Cardiovascular Genetics Center, Montreal Heart Institute, Department of Medicine, Université de Montréal, 5000 Belanger, Montreal, QC, Canada H1T 1C8.
Mario Talajic, Cardiovascular Genetics Center, Montreal Heart Institute, Department of Medicine, Université de Montréal, 5000 Belanger, Montreal, QC, Canada H1T 1C8.
Zachary W Laksman, Center for Cardiovascular Innovation, Division of Cardiology, Department of Medicine, University of British Columbia, Vancouver, BC, Canada.
Jason D Roberts, Population Health Research Institute, McMaster University, and Hamilton Health Sciences, Hamilton, ON, Canada; Section of Cardiac Electrophysiology, Division of Cardiology, Department of Medicine, Western University, London, ON, Canada.
Andrew D Krahn, Center for Cardiovascular Innovation, Division of Cardiology, Department of Medicine, University of British Columbia, Vancouver, BC, Canada.
Rafik Tadros, Cardiovascular Genetics Center, Montreal Heart Institute, Department of Medicine, Université de Montréal, 5000 Belanger, Montreal, QC, Canada H1T 1C8.
Supplementary material
Supplementary material is available at European Heart Journal online.
Funding
The study is supported by the Philippa and Marvin Carsley Chair (J.C.-T., P.G.G. and R.T., Montreal, QC), the Fonds de la Recherche du Québec – Santé (R.T., Principal Investigator, #135055), the Canada Research Chairs program (R.T.), the Heart and Stroke Foundation of Canada (A.D.K, Principal Investigator, G-13-0002775 and G-14-0005732), and Canadian Institutes of Health Research (Hearts in Rhythm Organization, A.D.K., Principal Investigator, RN380020-406814). A.D.K. receives support from the Sauder Family/Heart and Stroke Foundation Chair in Cardiology (Vancouver, BC, Canada), the Paul Brunes Chair in Heart Rhythm Disorders (Vancouver, BC, Canada) and the Paul Albrechtsen Foundation (Winnipeg, MB, Canada).
References
- 1. Hayashi M, Shimizu W, Albert CM. The spectrum of epidemiology underlying sudden cardiac death. Circ Res 2015;116:1887–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Priori SG, Blomström-Lundqvist C, Mazzanti A, Blom N, Borggrefe M, Camm J, et al. 2015 ESC Guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: The Task Force for the Management of Patients with Ventricular Arrhythmias and the Prevention of Sudden Cardiac Death of the European Society of Cardiology (ESC). Endorsed by: Association for European Paediatric and Congenital Cardiology (AEPC). Eur Heart J 2015;36:2793–2867. 10.1093/eurheartj/ehv316. [DOI] [PubMed] [Google Scholar]
- 3. Krahn AD, Healey JS, Chauhan V, Birnie DH, Simpson CS, Champagne J, et al. Systematic assessment of patients with unexplained cardiac arrest: Cardiac Arrest Survivors with Preserved Ejection Fraction Registry (CASPER). Circulation 2009;120:278–285. [DOI] [PubMed] [Google Scholar]
- 4. Priori SG, Wilde AA, Horie M, Cho Y, Behr ER, Berul C, et al. Executive summary: HRS/EHRA/APHRS expert consensus statement on the diagnosis and management of patients with inherited primary arrhythmia syndromes. Europace 2013;15:1389–1406. [DOI] [PubMed] [Google Scholar]
- 5. Ackerman MJ, Priori SG, Willems S, Berul C, Brugada R, Calkins H, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Heart Rhythm 2011;8:1308–1339. [DOI] [PubMed] [Google Scholar]
- 6. Gollob MH, Blier L, Brugada R, Champagne J, Chauhan V, Connors S, et al. Recommendations for the use of genetic testing in the clinical evaluation of inherited cardiac arrhythmias associated with sudden cardiac death: Canadian Cardiovascular Society/Canadian Heart Rhythm Society joint position paper. Can J Cardiol 2011;27:232–245. [DOI] [PubMed] [Google Scholar]
- 7. Mellor G, Laksman ZWM, Tadros R, Roberts JD, Gerull B, Simpson CS, et al. Genetic testing in the evaluation of unexplained cardiac arrest: from the CASPER (Cardiac Arrest Survivors With Preserved Ejection Fraction Registry). Circ Cardiovasc Genet 2017;10:e001686. [DOI] [PubMed] [Google Scholar]
- 8. Davies B, Roberts JD, Tadros R, Green MS, Healey JS, Simpson CS, et al. The Hearts in Rhythm Organization: a Canadian National Cardiogenetics Network. CJC Open 2020;2:652–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alqarawi W, Dewidar O, Tadros R, Roberts JD, Steinberg C, MacIntyre CJ, et al. Defining idiopathic ventricular fibrillation: a systematic review of diagnostic testing yield in apparently unexplained cardiac arrest. Heart Rhythm 2021;18:1178–1185. [DOI] [PubMed] [Google Scholar]
- 10. Martin AR, Williams E, Foulger RE, Leigh S, Daugherty LC, Niblock O, et al. PanelApp crowdsources expert knowledge to establish consensus diagnostic gene panels. Nat Genet 2019;51:1560–1565. [DOI] [PubMed] [Google Scholar]
- 11. Stark Z, Foulger RE, Williams E, Thompson BA, Patel C, Lunke S, et al. Scaling national and international improvement in virtual gene panel curation via a collaborative approach to discordance resolution. Am J Hum Genet 2021;108:1551–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ingles J, Goldstein J, Thaxton C, Caleshu C, Corty EW, Crowley SB, et al. Evaluating the clinical validity of hypertrophic cardiomyopathy genes. Circ Genom Precis Med 2019;12:e002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jordan E, Peterson L, Ai T, Asatryan B, Bronicki L, Brown E, et al. Evidence-based assessment of genes in dilated cardiomyopathy. Circulation 2021;144:7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. James CA, Jongbloed JDH, Hershberger RE, Morales A, Judge DP, Syrris P, et al. International evidence based reappraisal of genes associated with arrhythmogenic right ventricular cardiomyopathy using the clinical genome resource framework. Circ Genom Precis Med 2021;14:e003273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Adler A, Novelli V, Amin AS, Abiusi E, Care M, Nannenberg EA, et al. An international, multicentered, evidence-based reappraisal of genes reported to cause congenital long QT syndrome. Circulation 2020;141:418–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hosseini SM, Kim R, Udupa S, Costain G, Jobling R, Liston E, et al. Reappraisal of reported genes for sudden arrhythmic death: evidence-based evaluation of gene validity for Brugada syndrome. Circulation 2018;138:1195–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Walsh R, Adler A, Amin AS, Abiusi E, Care M, Bikker H, et al. Evaluation of gene validity for CPVT and short QT syndrome in sudden arrhythmic death. Eur Heart J 2022;43:1500–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Whiffin N, Minikel E, Walsh R, O’Donnell-Luria AH, Karczewski K, Ing AY, et al. Using high-resolution variant frequencies to empower clinical genome interpretation. Genet Med 2017;19:1151–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kelly MA, Caleshu C, Morales A, Buchan J, Wolf Z, Harrison SM, et al. Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: recommendations by ClinGen’s Inherited Cardiomyopathy Expert Panel. Genet Med 2018;20:351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tadros R, Ton A-T, Fiset C, Nattel S. Sex differences in cardiac electrophysiology and clinical arrhythmias: epidemiology, therapeutics, and mechanisms. Can J Cardiol 2014;30:783–792. [DOI] [PubMed] [Google Scholar]
- 22. Roston TM, Wei J, Guo W, Li Y, Zhong X, Wang R, et al. Clinical and functional characterization of RyR2 variants implicated in calcium release deficiency syndrome: an international multicenter study. JAMA Cardiol 2022;7:84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sun B, Yao J, Ni M, Wei J, Zhong X, Guo W, et al. Cardiac ryanodine receptor calcium release deficiency syndrome. Sci Transl Med 2021;13:eaba7287. [DOI] [PubMed] [Google Scholar]
- 24. Shauer A, Shor O, Wei J, Elitzur Y, Kucherenko N, Wang R, et al. Novel RyR2 mutation (G3118R) is associated with autosomal recessive ventricular fibrillation and sudden death: clinical, functional, and computational analysis. J Am Heart Assoc 2021;10:e017128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Podliesna S, Delanne J, Miller L, Tester DJ, Uzunyan M, Yano S, et al. Supraventricular tachycardias, conduction disease, and cardiomyopathy in 3 families with the same rare variant in TNNI3K (p.Glu768Lys). Heart Rhythm 2019;16:98–105. [DOI] [PubMed] [Google Scholar]
- 26. Eichhorn C, Voges I, Daubeney PEF. Out-of-hospital cardiac arrest and survival in a patient with Noonan syndrome and multiple lentigines: a case report. J Med Case Rep 2019;13:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aydin A, Yilmazer MS, Gurol T. Sudden death in a patient with Noonan syndrome. Cardiol Young 2011;21:233–234. [DOI] [PubMed] [Google Scholar]
- 28. Grant AR, Cushman BJ, Cave H, Dillon MW, Gelb BD, Gripp KW, et al. Assessing the gene-disease association of 19 genes with the RASopathies using the ClinGen gene curation framework. Hum Mutat 2018;39:1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Davies B, Bartels K, Hathaway J, Xu F, Roberts JD, Tadros R, et al. Variant reinterpretation in survivors of cardiac arrest with preserved ejection fraction (the Cardiac Arrest Survivors With Preserved Ejection Fraction Registry) by clinicians and clinical commercial laboratories. Circ Genom Precis Med 2021;14:e003235. [DOI] [PubMed] [Google Scholar]
- 30. Kumar S, Peters S, Thompson T, Morgan N, Maccicoca I, Trainer A, et al. Familial cardiological and targeted genetic evaluation: low yield in sudden unexplained death and high yield in unexplained cardiac arrest syndromes. Heart Rhythm 2013;10:1653–1660. [DOI] [PubMed] [Google Scholar]
- 31. Giudicessi JR, Ackerman MJ. Role of genetic heart disease in sentinel sudden cardiac arrest survivors across the age spectrum. Int J Cardiol 2018;270:214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jimenez-Jaimez J, Peinado R, Grima EZ, Segura F, Moriña P, Sánchez Muñoz JJ, et al. Diagnostic approach to unexplained cardiac arrest (from the FIVI-Gen Study). Am J Cardiol 2015;116:894–899. [DOI] [PubMed] [Google Scholar]
- 33. Asatryan B, Schaller A, Seiler J, Servatius H, Noti F, Baldinger SH, et al. Usefulness of genetic testing in sudden cardiac arrest survivors with or without previous clinical evidence of heart disease. Am J Cardiol 2019;123:2031–2038. [DOI] [PubMed] [Google Scholar]
- 34. Visser M, van der Heijden JF, van der Smagt JJ, Doevendans PA, Wilde AA, Loh P, et al. Long-term outcome of patients initially diagnosed with idiopathic ventricular fibrillation: a descriptive study. Circ Arrhythm Electrophysiol 2016;9:e004258. [DOI] [PubMed] [Google Scholar]
- 35. Ingles J, Bagnall RD, Yeates L, McGrady M, Berman Y, Whalley D, et al. Concealed arrhythmogenic right ventricular cardiomyopathy in sudden unexplained cardiac death events. Circ Genom Precis Med 2018;11:e002355. [DOI] [PubMed] [Google Scholar]
- 36. Isbister JC, Nowak N, Butters A, Yeates L, Gray B, Sy RW, et al. ‘Concealed cardiomyopathy’ as a cause of previously unexplained sudden cardiac arrest. Int J Cardiol 2021;324:96–101. [DOI] [PubMed] [Google Scholar]
- 37. Tester DJ, Ackerman JP, Giudicessi JR, Ackerman NC, Cerrone M, Delmar M, et al. Plakophilin-2 truncation variants in patients clinically diagnosed with catecholaminergic polymorphic ventricular tachycardia and decedents with exercise-associated autopsy negative sudden unexplained death in the young. JACC Clin Electrophysiol 2019;5:120–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cerrone M, Montnach J, Lin X, Zhao Y-T, Zhang M, Agullo-Pascual E, et al. Plakophilin-2 is required for transcription of genes that control calcium cycling and cardiac rhythm. Nat Commun 2017;8:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guo L, Torii S, Fernandez R, Braumann RE, Fuller DT, Paek K-H, et al. Genetic variants associated with unexplained sudden cardiac death in adult White and African American individuals. JAMA Cardiol 2021;6:1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lahrouchi N, Raju H, Lodder EM, Papatheodorou E, Ware JS, Papadakis M, et al. Utility of post-mortem genetic testing in cases of sudden arrhythmic death syndrome. J Am Coll Cardiol 2017;69:2134–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Groeneveld SA, van der Ree MH, Taha K, de Bruin-Bon RHA, Cramer MJ, Teske AJ, et al. Echocardiographic deformation imaging unmasks global and regional mechanical dysfunction in patients with idiopathic ventricular fibrillation: a multicenter case–control study. Heart Rhythm 2021;18:1666–1672. [DOI] [PubMed] [Google Scholar]
- 42. Haissaguerre M, Hocini M, Cheniti G, Duchateau J, Sacher F, Puyo S, et al. Localized structural alterations underlying a subset of unexplained sudden cardiac death. Circ Arrhythm Electrophysiol 2018;11:e006120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van der Velden J, Stienen GJM. Cardiac disorders and pathophysiology of sarcomeric proteins. Physiol Rev 2019;99:381–426. [DOI] [PubMed] [Google Scholar]
- 44. de Marvao A, McGurk KA, Zheng SL, Thanaj M, Bai W, Duan J, et al. Phenotypic expression and outcomes in individuals with rare genetic variants of hypertrophic cardiomyopathy. J Am Coll Cardiol 2021;78:1097–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alders M, Koopmann TT, Christiaans I, Postema PG, Beekman L, Tanck MWT, et al. Haplotype-sharing analysis implicates chromosome 7q36 harboring DPP6 in familial idiopathic ventricular fibrillation. Am J Hum Genet 2009;84:468–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cheung CC, Mellor G, Deyell MW, Ensam B, Batchvarov V, Papadakis M, et al. Comparison of ajmaline and procainamide provocation tests in the diagnosis of Brugada Syndrome. JACC Clin Electrophysiol 2019;5:504–512. [DOI] [PubMed] [Google Scholar]
- 47. Baertling F, van den Brand MAM, Hertecant JL, Al-Shamsi A, van den Heuvel LP, Distelmaier F, et al. Mutations in COA6 cause cytochrome c oxidase deficiency and neonatal hypertrophic cardiomyopathy. Hum Mutat 2015;36:34–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.