Abstract
Visceral leishmaniasis (VL) is a neglected tropical disease that mainly affects the poor population of the Indian, African, and South American subcontinent. The increasing resistance to antimonial and miltefosine and frequent toxicity of amphotericin B drives an urgent need to develop an anti-leishmanial drug with excellent efficacy and safety profile. In this study, three sequential docking protocols (HTVS, SP, and XP) were performed to screen the secondary metabolites (n = 6519) from the actinomycetes source against five key proteins involved in the metabolic pathway of Leishmania donovani. Those proteins were adenine phosphoribosyltransferase (PDB ID: 1QB7), trypanothione reductase (PDB ID: 2JK6), N-myristoyl transferase (PDB ID: 2WUU), pteridine reductase (PDB ID: 2XOX), and MAP kinase (PDB ID: 4QNY). Although the binding energy of top ligands was predicted using the MM-GBSA module of the Schrödinger suite. SP and XP docking mode resulted in 55 multi-targeted ligands against L donovani. MM-GBSA analysis selected the top 18 ligands with good-binding affinity and the binding-free energy for four proteins, as mentioned earlier, when compared with the miltefosine, paromomycin, and a reference ligand selected for each target. Finally, molecular dynamics simulation, post-MD-binding-free energy (MM-PBSA), and principal component analysis (PCA) proposed three best ligands (Adenosine pentaphosphate, Atetra P, and GDP-4-keto-6-deoxymannose) qualifying the above screening parameters and confirmed as a potential drug candidate to fight against Leishmania donovani parasites.
Keywords: Secondary metabolites, Leishmania donovani, Visceral leishmaniasis infection, Actinomycetes, HTVS, MM-PBSA, PCA
Introduction
Leishmaniasis is a severe infection that affects 0.3 million people living in 97 countries and three territories worldwide (Diseases 2017). There are three forms of leishmaniasis; namely, visceral leishmaniasis (VL), mucocutaneous leishmaniasis (MCL), and cutaneous leishmaniasis (CL). Among these three forms of leishmaniasis, VL or Kala-azar is the most intense and fetal form of leishmaniasis worldwide. If left untreated, it causes inevitable mortality within 2 years after the onset of the disease (Pandey et al. 2015). As per the 2020 WHO report, more than 79% of global cases of VL were reported from only six countries, namely India, Brazil, Ethiopia, Kenya, Eritrea, and Sudan. In India, VL cases abruptly declined by 97% from 1992 to 2020. Death cases have also decreased in the last 20 years, from 1419 deaths in 1992 to 37 in 2020 (Ruiz-Postigo et al. 2020; Gill et al. 2021).
The causative agent of visceral leishmaniasis is the protozoan parasite, mainly Leishmania donovani and L. infantum in the Old World and New World, respectively (Pandey et al. 2016). VL spreads by biting of the hitherto infected female sand-fly vector of the genus Phlebotomine. The Leishmania parasite spends most of its life in the female sand-fly, but it replicates in the mononuclear phagocytes (Killick-Kendrick 1990). Symptoms of VL include swelling of visceral organs like the spleen and liver, followed by pancytopenia, fever, cachexia, and epistaxis.
Only a few anti-leishmanial drugs have been approved to treat VL, but these drugs are associated with severe toxicity, resistance, and high cost, which do not fulfill the objective of anti-leishmanial drug discovery. Pentavalent antimonial was introduced in the early 1940s as a primary treatment option against VL (Prajapati and Pandey 2017). However, it has a long injection schedule of 4 weeks that causes pain. In India's Bihar state, around 1/3rd of VL patients have developed high resistance against this drug, and 7–10% of patients face the life-threatening cardiotoxicity problem. Amphotericin B (first-line parenteral treatment) was introduced as a replacement option against antimonial drugs. It has shown good efficacy, but rigors and chill during infusion and nephrotoxicity were the major hindrances to the success of this drug (Sundar et al. 2004). Miltefosine (alkyl phospholipids) was the first oral drug introduced to treat VL, originally deliberated as an antitumor agent. Nevertheless, it has shown a relapse rate of 7–10 and 20% among the patients at a follow-up period of 6 and 12 months, respectively, in the clinical trials conducted in India and Nepal (Sundar et al. 2012). Paromomycin, an aminoglycoside antibiotic isolated from Streptomyces rimosus, inhibits protein synthesis and is effective against Leishmania infection, and has shown a cure rate of 94.6% (Sundar et al. 2007). The major drawbacks of this drug are the requirement of intramuscular injections for three weeks, serum transaminase monitoring, and unsafe for pregnant women. Therefore, there is a need to develop a new anti-leishmanial drug with good efficacy and safety. This study applied a computational biology approach to finding new anti-leishmanial compounds with good efficacy and negligible toxicity.
Here, we screened the library of actinomycetes secondary metabolites-based novel inhibitors targeting five key leishmania metabolic pathways enzymes. The first enzyme selected was adenine phosphoribosyltransferase (PDB ID-1QB7), which converts the salvage adenine into adenosine-5-monophosphate (Phillips et al. 1999). However, the promastigotes form of L. donovani does not require APRT for adenine salvage (Hwang and Ullman 1997) but APRT has a crucial role in the amastigote form (Looker et al. 1983). Owing to the absence of de novo biosynthetic pathway for purines in Leishmania parasites, they scavenge purines from their host (Hassan and Coombs 1988). Therefore, it can be selected as a novel target for developing the anti-leishmanial drug.
Trypanothione reductase (PDB ID-2JK6) is the second enzyme used for this study, as it maintains the reduced intracellular environment of the Leishmania parasite (Tovar et al. 1998). In humans, instead of TR-trypanothione, glutathione reductase-glutathione is present. Moreover, its presence in the parasite antioxidant system makes it a novel drug target for drug discovery against Leishmania parasite (Pandey et al. 2015). Hence, the specific inhibitors of this enzyme can be used to develop the anti-leishmanial drug.
N-myristoyltransferase (PDB ID-2WUU), the third enzyme included in this study, lapidates the N-terminal specific target proteins through post-translational modification and act as a potential drug target against Leishmania (Tate et al. 2014). It plays a vital role in various cellular processes, including vesicular protein trafficking, signal transduction, and promotes membrane binding. This novel enzyme has the potential to develop a new drug, and it is necessary for cell viability (Brannigan et al. 2010).
The fourth enzyme is Pteridine reductase (PDB ID-2XOX), an NADPH-dependent short-chain reductase, involved in the salvage of pterin and antifolate resistance (Nare et al. 1997). In Leishmania, the PTR1 gene has shown resistance towards methotrexate, but dihydrofolate reductase can inhibit it (Kumar et al. 2008). Hence, it acts as a metabolic by-pass of dihydrofolate reductase thymidylate synthase. Therefore, for the production of anti-leishmanial drugs, dihydrofolate, and pterin can be used as a target. Degradation of PTR1 in Leishmania is an excellent approach for new drug development.
Map kinase (MAPK) (PDB ID-4QNY) is the fifth enzyme that has serine and threonine protein kinases and autophosphorylation activity and is highly conserved in eukaryotes (Soares-Silva et al. 2016). This enzyme is involved in signal transduction cascades and regulates cellular activities such as cell proliferation, differentiation, mortality, stress response, and apoptosis (Soares-Silva et al. 2016; Kaur et al. 2017). Generally, this type of intracellular signaling pathway in the host is targeted by pathogens to inhibit immune responses (Soares-Silva et al. 2016).
Owing to the essential physiological role in Leishmania metabolism, all these proteins were included as a drug target and subjected to the computational study. Actinobacteria is one of the largest taxonomic units of the bacterial domain, and are filamentous, gram-positive bacteria that possess antimicrobial and anti-protozoan activity (Ventura et al. 2007). Actinomycetes are cosmopolitan, but mainly they thrive in the soil. About 45% of the bioactive secondary metabolites (produced by the microorganisms) come from the actinomycetes (Bérdy 2005). Secondary metabolites are not directly involved in the growth or development process, but survive interspecies competition, provide defensive mechanisms against stress, and facilitate reproductive processes prove as a potential source to cure many diseases. Hence, for the development of the new anti-leishmanial drug, we have highlighted the binding interaction of actinomycetes secondary metabolite against five key enzymes of L. donovani, which has already been discussed in the above section.
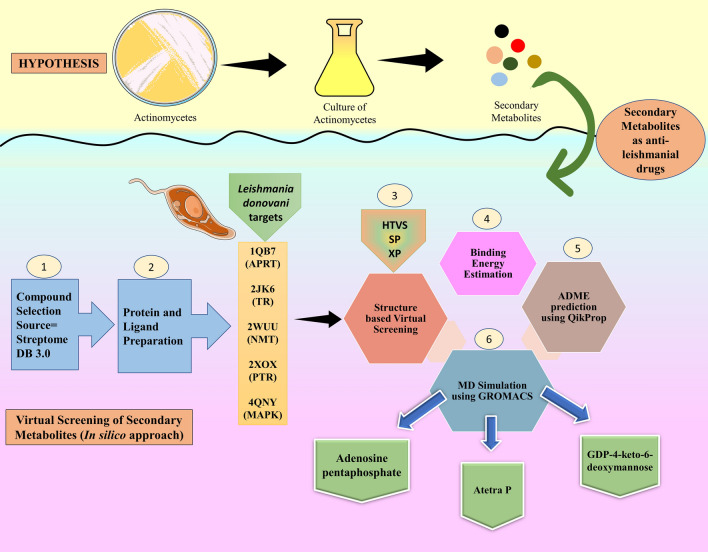
This study starts with the literature search to find actinomycetes secondary metabolites with potent anti-leishmanial activity. We have selected StreptomeDB 3.0, which is an updated compendium of Streptomycetes natural product library (Moumbock et al. 2021). Further, molecular docking and MM-GBSA screening against the aforementioned target proteins were performed to evaluate their binding interaction and energy. Finally, a molecular dynamics simulation study was performed to check the stability of the protein–ligand complex (Fig. 1).
Fig. 1.
Schematic representation of workflow and hypothesis of bioinformatic (in-silico) approach used in virtual screening of actinomycetes derived secondary metabolites. HTVS High-throughput Virtual Screening, SP Special Precision, XP Extra Precision, APRT Adenosine phosphoribosyl transferase, TR Trypanothione reductase, NMT N-Myristoyl Transferase, PTR Pteridine reductase, MAPK MAP Kinase
Materials and methods
This study warrants the search for new anti-leishmanial compounds of actinomycetes origin against five key enzymes of Leishmania donovani metabolic pathways. All secondary metabolites were obtained from StreptomeDB, a library of natural compounds obtained from the Streptomycetes, while proteins were retrieved for the protein data bank. The whole process was performed in a sequential manner, as described below.
Procurement of actinomycetes secondary metabolites library
A literature survey was conducted using the PubMed database (https://www.ncbi.nlm.nih.gov/pubmed/) in search of secondary metabolic compounds. We got a library named StreptomeDB 3.0; this is the third version of the streptomeDB library. This StreptomeDB 3.0 library was published in Nucleic Acid Research on 13 October 2020. This library was initially released in 2012 with 2444 compounds from 1985 organisms (including strains). In the latest release of 2020, there are 6524 secondary metabolites isolated from 3302 organisms/strains (Moumbock et al. 2021). StreptomeDB was initially released in 2012 with 2444 compounds from 1985 organisms, including strains (Lucas et al. 2012).
Miltefosine, Paromomycin was included as a positive control for all the leishmanial proteins. At the same time, specific inhibitors for all selected targets were also included for comparison purposes.
Ligand preparation
The ligands obtained from the database must be prepared before going for the molecular docking studies. A sum of 6524 ligands was obtained from the database are subject to the LigPrep (Chander et al. 2017; Schrödinger Release 2017a, b-4: LigPrep) module of the Schrodinger suite. The motive behind the ligand preparation is to generate a low energy structure with correct chirality in 3D form. Each possible tautomer and ionization states for the input ligand was generated using the Epic, while the Sterioizer module of the Schrodinger suite was used to retain the correct chirality. All the torsional bond of the ligand was set free, and one stereoisomer was prepared for each ligand. Finally, the prepared ligands were optimized by using the OPLS_2005 force field.
ADME prediction for the prepared ligands
ADME stands for the absorption, distribution, metabolism, and elimination from the body. These parameters determine the drugability of the ligand from the pharmaceutical perspective. In detail, these parameters determine the absorption of drugs within our body, followed by distribution to different body parts, the role of the drug in host metabolism, and finally, elimination from the body. Therefore, the ligand output (n = 6524) of LigPrep was used for the ADME prediction using the QikProp module (Pandey and Prajapati 2017; Schrödinger Release 2017a, b-4: QikProp) of the Schrodinger suite. Only ligands passing on the ADME parameters were selected to perform molecular docking experiments.
Protein preparation
The protein selected for the docking study must be prepared to fill missing side chains and loops followed by the hydrogen bond assignment. All five proteins, namely Adenine phosphoribosyltransferase (PDB ID: 1QB7), trypanothione reductase (PDB ID: 2JK6), N-myristoyl transferase (PDB ID: 2WUU), pteridine reductase (PDB ID: 2XOX), and MAP kinase (PDB ID: 4QNY), were subjected to the protein preparation using protein preparation wizard (Madhavi Sastry et al. 2013; Ahluwalia et al. 2017) module of Schrodinger suite. Protein preparation involves the removal of water molecules and co-crystallized ligand, cofactors, and metal ions with missing atoms. After pre-processing, the protein molecules were subjected to the restrained minimization using Optimized Potential for Liquid Simulation (OPLS-2005) force field.
Binding site prediction and grid generation
In order to perform the binding energy prediction of the secondary metabolite against the five target proteins, it was necessary to determine the active site of all five proteins. Binding site residue for four proteins, trypanothione reductase (Pandey et al 2016, 2017a; c), adenine phosphoribosyltransferase (Phillips et al. 1999), N-myristoyl transferase (Brannigan et al. 2010), and pteridine reductase (Barrack et al. 2011), were obtained from work published elsewhere. Although the active site residues for MAP kinase were obtained from the interaction of ligand (Wernimont et al. 2014), which is already present in the PDB file obtained from the protein data bank.
The grid generation module of Glide was used to locate the Grid position in all five proteins. The grid for all the targets was generated using amino acid residues surrounding the active site.
Docking analysis
To screen the compounds with the highest binding energy against target proteins, three sequential docking methods, namely, high-throughput virtual screening (HTVS), standard precision (SP), and extra precision (XP) were used. The main difference between these three modes is the algorithm used to predict the binding affinity. HTVS and SP use the self-same scoring function while XP reduces the thoroughness of the endmost torsional refinement, and sampling also minimizes the number of intermediate conformations throughout the docking. Scoring function XP performs more extensive sampling than SP. XP takes a little longer than SP because it removes the false positive binders. Initially, 6524 secondary metabolites from Streptomycetes and their reference ligands were screened by the HTVS module against all 5 proteins. The top 20% ligands of the HTVS output were selected and subjected to the SP module of docking. Among the top 20% output of SP docking, those ligands with higher binding affinity for all five proteins were screened out and subjected to the XP docking model to remove the false positive ligands. After XP docking, top ligands with good scores were selected as top hits against all five proteins.
Molecular mechanics, the generalized born model, and solvent accessibility
After the docking experiment, top hits were subjected to the MM-GBSA module of the Prime tool to calculate ligand-binding energies and strain energies. The “pv.maegz” file of the protein and ligand complex obtained by XP docking is required by MM-GBSA, which results in ligand ranking based on the predicted binding energies, i.e. MM-GBSA dG binds. The study has been completed by creating a flexible active site of protein with a distance of 5 Å that could be adjusted depending on the input ligand. Prime uses the VSGB 2.0 solvation model, a revolutionary model for high-resolution protein modeling, and the OPLS_2005 force field to determine scores (Naik et al. 2021; Singh et al. 2022). If the calculated binding energy shows a more negative effect, then it indicates a stronger binding affinity. The combined grid file of all five proteins and ligands was imported into the prime MM-GBSA module to determine the relative binding energies of all top selected ligands with the receptor molecule.
Molecular dynamics simulation
Molecular dynamics (MD) simulation method is used to determine the ligand and receptor binding and physical movement of atoms or molecules. To perform the molecular dynamics simulation study, WebGRO for Macromolecular Simulations (https://simlab.uams.edu/) (Balasubramaniam and Williams 2020). GROMACS software package was used to rapidly calculate the bonding and non-bonding interactions. The top three ligands with good docking and MM-GBSA score, and less toxicity were selected to perform the molecular dynamics. For the simulation process GROMOS96, 43a1 force field was used. This force field describes how the molecules/atoms interact with each other within the system. After selecting the force field, the triclinic solvent box and SPC water model were selected (Bekker et al. 1993; Oostenbrink et al. 2004). Using the periodic boundary conditions, the distance between the protein–ligand complex and solvent box was adjusted, and then to neutralize the system, counter ions like Na+ and Cl– were added. Then the energy minimization step was executed with the help of the steepest descent algorithm, the maximum force of 5000 was provided to confirm whether the steric clashes were present or not between the protein–ligand complexes within the solvent system. Next, the equilibration process was performed using the NVT and NPT ensembles. To keep the system at stable environment, temperature and pressure was maintained at 300 K and 1 ATM, respectively. In the final step, molecular dynamics simulations were accomplished for 100 ns. Molecular dynamics simulation was carried out for all the selected complexes, and the entire simulation process was later examined by calculating the structural deviations, RMSD, RMSF, Radius of gyration (Rg), SASA, and hydrogen bonding. RMSD is the measure of average deviation in a backbone atom when superimposed. RMSF denotes the average deviation in amino acid residues during the entire period of dynamics, whereas Rg tells about the compactness of the complex. The output molecular dynamics results were visualized with the help of PyMOL and XMgrace software. Molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) was used to estimate post-MD-binding energy. The MM-PBSA method is commonly used to calculate the binding-free energy of a biomolecular interaction and is often used as a scoring function in computer-aided drug discovery. The MM-PBSA was calculated using the g_mmpbsa tool, which is part of the open-source drug discovery consortium and was developed by Rashmi Kumari (Kumari et al. 2014). G_mmpbsa calculates molecular mechanics energy, polar solvation energy, non-polar solvation energy, and residue-wise energy contribution using the APBS and GROMACS libraries. For one step binding energy calculation, g_mmpbsa requires the molecular dynamics simulation output files (xtc, tpr, and index file) and the input parameter file for polar and non-polar solvation energies (pbsa.mdp). The MmPbSaStat.py (python script) and the output files from one step binding energy calculation (energy_MM.xvg, polar, and apolar.xvg) are required for average binding free energy calculation. Principal component analysis (PCA) is a dimensionality reduction technique and has been used for many decades at least to interpret large datasets with minimal information loss (Jolliffe et al. 2016). PCA was computed using the GROMACS command line and the MD simulation output files, whereas the results were analyzed using XMgrace.
Results
Ligand library construction, preparation, and ADME analysis
This study starts with the literature survey to identify the secondary metabolites of actinomycetes origin or a natural product library from actinomycetes. We selected StreptomeDB 3.0, that contains nearly 6524 compounds from 3302 organisms/strains. Library also contains NMR data, MS data, and ADMET properties of most of the compounds present in the library. These ligands were subjected to the LigPrep module to obtain an energy-minimized structure in 3D form. At the end of ligand preparation, obtained ligands were subjected to the QikProp module for the ADME evaluation. The ADME parameters including this study were molecular weight (range 130–725 Da), the number of Hydrogen bond donors (range 0–6) and acceptor (02–20), octanol–water partition coefficient (– 2 to 6.5), and rule of five (max 4). All these parameters were considered for the ligand evaluation for further study. Miltefosine (PubChem CID: 3599), Paromomycin (PubChem CID:165580) were used as the reference ligands against all the protein targets. Target positive controls specific for each target were summarized in Table 1 along with their PubChem CID and references. Prediction of principle drug characteristic value of potential secondary metabolites from Steptomyces sp are compiled in Table 2, along with PubChem CID, organism from secondary metabolites isolated, molecular weight, SASA, FOSA, FISA, PISA, WPSA, and volume. Table for ADME properties prediction of potential secondary metabolites from Streptomycetes are summarized in Table 3 with complete details.
Table 1.
Target positive controls used in the study for comparative analysis
| S. no. | Protein target | Positive control | PubChem CID | Reference |
|---|---|---|---|---|
| 1 | Trypanothione reductase | Phenothiazine | 7108 | (Chan et al. 1998) |
| 2 | Mitogen-activated protein kinases | PD 169316 | 4712 | (Reddy et al. 2017) |
| 3 | N-Myristoyl transferase | DDD100097 | 58561169 | (Corpas-Lopez et al. 2018) |
| 4 | Pteridine reductase | Thianthrene | 7109 | (Kaur et al. 2012) |
| 5 | Adenine phosphoribosyl transferase | Allopurinol | 135401907 | (Marr and Berens 1977) |
Table 2.
Prediction of principle drug characteristic value of potential secondary metabolites of Streptomyces sp
| S. no. | Potential secondary metabolites | PubChem CID | Organisms | MW | SASA | FOSA | FISA | PISA | WPSA | Volume |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Adenosine pentaphosphate | 46173396 | S aureofaciens | 667.14 | 802.49 | 93.35 | 615.27 | 81.86 | 12.01 | 1475.99 |
| 2 | Arcticoside | 139583920 | S ART5 | 559.48 | 795.57 | 339.14 | 332.95 | 123.48 | 0.00 | 1480.63 |
| 3 | Atetra P | 14003 | S aureofaciens | 587.16 | 699.86 | 61.82 | 528.74 | 95.12 | 14.18 | 1301.73 |
| 4 | Carminomycinol | 43942 | S peucetius SPDHC | 515.52 | 762.60 | 296.88 | 305.06 | 160.66 | 0.00 | 1423.93 |
| 5 | dTDP-6-deoxy-D-allose | 46173788 | S bikiniensis | 548.33 | 760.16 | 377.46 | 359.43 | 18.67 | 4.62 | 1400.20 |
| 6 | GDP-4-keto-6-deoxymannose | 135398621 | S nodosus | 587.33 | 762.49 | 221.64 | 476.06 | 58.72 | 6.07 | 1415.08 |
| 7 | Gobichelin B | 102107335 | S NRRL F-4415 | 689.72 | 1028.47 | 317.55 | 467.15 | 243.77 | 0.00 | 1998.48 |
| 8 | Guanosine pentaphosphate | 135398629 | S griseus;S aureofaciens | 683.14 | 801.31 | 62.69 | 670.36 | 51.93 | 16.34 | 1480.96 |
| 9 | Guanosine tetraphosphate | 135398637 | S griseus;S aureofaciens | 603.16 | 733.93 | 67.25 | 601.64 | 57.41 | 7.63 | 1347.61 |
| 10 | Histomodulin | 10438607, 139585439 | S ruber JCM 3131 | 590.54 | 814.27 | 311.53 | 350.76 | 151.98 | 0.00 | 1563.47 |
| 11 | JBIR-133 | 137332234 | S avermitilis SUKA22 | 284.27 | 529.50 | 66.32 | 241.26 | 221.92 | 0.00 | 886.24 |
| 12 | N-acetylglucosamine | 445675 | S tsukubaensis NRRL 18,488 | 607.36 | 805.20 | 231.41 | 523.26 | 48.72 | 1.81 | 1506.17 |
| 13 | Nikkomycin pseudo-J | 129256, 139588950 | S tendae Tü 901/PF 53 + -3 | 624.56 | 855.59 | 200.30 | 557.05 | 98.25 | 0.00 | 1648.16 |
| 14 | Quercetin glucoside | 44259229 | S sp. KCTC 0041BP | 464.38 | 701.50 | 105.35 | 364.53 | 231.61 | 0.00 | 1258.17 |
| 15 | Sharkquinone | 132487404 | S sp. EGY1 | 336.30 | 543.62 | 88.17 | 219.09 | 236.35 | 0.00 | 948.92 |
| 16 | Solphenazine F | 71524394 | S DL-93 | 518.52 | 766.16 | 314.73 | 217.49 | 233.95 | 0.00 | 1437.66 |
| 17 | N,N',N'-tris(2,3-Dihydroxybenzoyl)-O-L-seryl-O-L-seryl-L-serine | None | S sp. TP-A0874 | 686.58 | 1042.68 | 111.13 | 534.94 | 396.62 | 0.00 | 1918.33 |
| 18 | Naquihexcin A | 132934665 | S sp. HDN-10–293; S sp. KIB3133 | 828.79 | 956.76 | 348.44 | 420.14 | 188.02 | 0.16 | 2012.99 |
Table 3.
Predicted ADME properties of potential secondary metabolites of Streptomyces sp
| S.No | Potential secondary metabolites | Donor HB |
Accpt HB |
QPlog Po/w |
QPlogS | QPlog HERG |
QPlog BB |
QPP Caco |
QPP MDCK |
QPlog Kp |
QPlog Khsa |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Adenosine pentaphosphate | 4.00 | 28.10 | – 3.88 | 1.10 | 5.38 | – 7.33 | 0.00 | 0.00 | – 10.65 | – 4.13 |
| 2 | Arcticoside | 7.00 | 21.75 | – 2.69 | – 1.88 | – 5.52 | – 3.90 | 6.89 | 2.28 | – 5.78 | – 1.70 |
| 3 | Atetra P | 4.00 | 24.10 | – 3.01 | 0.57 | 4.20 | – 5.62 | 0.00 | 0.00 | – 9.30 | – 3.33 |
| 4 | Carminomycinol | 5.00 | 11.80 | 0.42 | – 3.09 | – 6.22 | – 2.80 | 3.16 | 1.09 | – 7.68 | – 0.25 |
| 5 | dTDP-6-deoxy-D-allose | 5.00 | 21.70 | – 2.22 | – 1.54 | – 1.30 | – 3.99 | 0.25 | 0.11 | – 6.83 | – 2.15 |
| 6 | GDP-4-keto-6-deoxymannose | 7.00 | 25.70 | – 4.30 | – 1.06 | – 1.44 | – 5.23 | 0.02 | 0.01 | – 8.74 | – 2.49 |
| 7 | Gobichelin B | 5.25 | 16.60 | – 1.66 | – 1.37 | – 2.03 | – 6.02 | 0.03 | 0.02 | – 9.13 | – 1.65 |
| 8 | Guanosine pentaphosphate | 4.00 | 26.90 | – 3.27 | 0.76 | 7.38 | – 7.87 | 0.00 | 0.00 | – 11.77 | – 4.08 |
| 9 | Guanosine tetraphosphate | 4.00 | 22.90 | – 2.47 | – 0.03 | 5.87 | – 6.65 | 0.00 | 0.00 | – 10.77 | – 3.32 |
| 10 | Histomodulin | 4.00 | 15.20 | 0.30 | – 3.70 | – 5.49 | – 3.87 | 4.67 | 1.50 | – 6.20 | – 0.58 |
| 11 | JBIR-133 | 3.00 | 5.00 | 2.09 | – 3.41 | – 1.52 | – 1.94 | 3.27 | 1.65 | – 4.80 | – 0.48 |
| 12 | N-acetylglucosamine | 7.00 | 25.90 | – 4.97 | – 0.44 | – 0.19 | – 6.00 | 0.01 | 0.00 | – 9.46 | – 2.86 |
| 13 | Nikkomycin pseudo-J | 10.50 | 20.55 | – 6.02 | – 0.63 | 0.22 | – 6.03 | 0.00 | 0.00 | – 11.87 | – 2.35 |
| 14 | Quercetin glucoside | 7.00 | 13.75 | – 1.24 | – 2.89 | – 5.82 | – 3.86 | 3.46 | 1.08 | – 6.37 | – 0.92 |
| 15 | Sharkquinone | 1.00 | 4.00 | 2.33 | – 4.11 | – 4.94 | – 1.66 | 82.84 | 33.50 | – 4.34 | 0.27 |
| 16 | Solphenazine F | 6.00 | 19.00 | – 0.92 | – 2.50 | – 5.82 | – 2.40 | 85.80 | 34.80 | – 3.55 | – 1.27 |
| 17 | N,N',N'-tris(2,3-Dihydroxybenzoyl)-O-L-seryl-O-L-seryl-L-serine | 4.50 | 13.70 | 2.30 | – 6.20 | – 6.21 | – 7.72 | 0.02 | 0.01 | – 7.87 | – 0.20 |
| 18 | Naquihexcin A | 5.00 | 22.75 | 0.04 | – 3.30 | – 3.76 | – 4.65 | 0.26 | 0.08 | – 7.16 | – 1.04 |
Molecular docking study
The output ligands of LigPrep and QikProp, along with reference ligands, were subjected to docking in three consecutive modes (HTVS, SP, and XP), and different numbers of ligands were obtained as output in the case of different proteins. A total 20% of the HTVS output of respective proteins were selected and subjected to the SP mode of docking with the same protein, and their docking score was obtained. In the next step, selected ligands along with three reference ligands were subjected to the XP mode of docking to remove the false positive binder. The output was obtained in excel format, and only good binder ligands, i.e., having more negative docking scores, were selected as the final ligands. The docking score of ligands after XP docking with the Adenine phosphoribosyl transferase, Trypanothione reductase, N-Myristoyl transferase, Pteridine reductase, and Map Kinase, ranging from (– 15.82 to – 0.76), (– 15.86 to – 3.01), (– 11.81 to – 1.59), (– 13.16 to – 3.34), and (– 12.95 to – 4.35) Kcal/mol, respectively. After that, results were narrowed down to search ligands that interact with more than one target protein. We found 55 multi-targeted ligands that interact with more than one protein target. Three ligands target four proteins, 15 ligands target three, and 37 with two protein targets. In Table 4, we have summarized the top 18 ligands targeting three and four leishmanial proteins, along with the docking score of ligands and their positive control. All selected ligands have higher docking scores against five proteins representing the high binding affinity for respective proteins compared to specific reference ligands. The docking score of multi-targeted ligands with the Adenine phosphoribosyl transferase, Trypanothione reductase, N-Myristoyl transferase, Pteridine reductase, and Map Kinase, ranging from (– 15.82 to – 7.68), (– 12.37 to – 7.11), (– 9.32 to – 4.74), (– 13.16 to – 6.79), and (– 12.95 to – 8.73) Kcal/mol, whereas the docking score for specific target positive control were – 3.67, – 4.37, – 3.04, – 3.74, and – 4.40 kcal/mol, respectively. The docking score of another positive control, Miltefosine, was also lower than selected ligands (except for N-Myristoyl transferase, which has the highest docking score of – 11.27 kcal/mol), showing good interaction between protein and ligand. The docking scores of Paromomycin (another positive control) are better than a few ligands, but most of the ligands have a better docking score than Paromomycin (Table 4). The best three ligands (Adenosine pentaphosphate, Atetra P, and GDP-4-keto-6-deoxymannose) interacted with the maximum number of targets (Adenine phosphoribosyl transferase, Map Kinase, Pteridine reductase, and Trypanothione reductase). Ligand interaction diagram and interacting residues for Adenosine pentaphosphate, Atetra P, and GDP-4-keto-6-deoxymannose with Adenine phosphoribosyl transferase, Map Kinase, Pteridine reductase, and Trypanothione reductase were shown in Figs. 2, 3, and 4, respectively.
Table 4.
Estimation of Docking score and binding energies between actinomycetes secondary metabolites and Leishmania donovani proteins
| S. No | Actinomycetes secondary metabolites | Docking & MMGBSA scores against Leishmania donovani target proteins (in Kcal/mol) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N-Myristoyl transferase | Mitogen-activated protein kinases | Adenine phosphoribosyl transferase | Pteridine reductase | Trypanothione reductase | Target proteins | |||||||
| Docking score | MM-GBSA | Docking score | MM-GBSA | Docking score | MM-GBSA | Docking score | MM-GBSA | Docking score | MM-GBSA | |||
| 1 | Arcticoside | – 9.32 | – 53.34 | – 10.98 | – 79.01 | – 8.38 | – 71.38 | 3 | ||||
| 2 | Carminomycinol | – 8.98 | – 76.85 | – 9.72 | – 78.68 | – 10.13 | – 80.74 | 3 | ||||
| 3 | dTDP-6-deoxy-D-allose | – 10.75 | – 75.29 | – 8.50 | – 71.25 | – 10.26 | – 82.02 | 3 | ||||
| 4 | Gobichelin B | – 4.74 | – 52.82 | – 10.44 | – 93.01 | – 7.77 | – 117.58 | 3 | ||||
| 5 | Guanosine pentaphosphate | – 10.92 | – 61.41 | – 12.62 | – 26.40 | – 11.14 | – 82.97 | 3 | ||||
| 6 | Guanosine tetraphosphate | – 10.82 | – 73.03 | – 15.82 | – 62.33 | – 12.13 | – 82.60 | 3 | ||||
| 7 | Histomodulin | – 10.88 | – 67.51 | – 10.03 | – 87.40 | – 8.48 | – 46.70 | 3 | ||||
| 8 | JBIR-133 | – 8.77 | – 46.08 | – 9.83 | – 56.08 | – 6.79 | – 50.21 | 3 | ||||
| 9 | N,N',N'-tris(2,3-Dihydroxybenzoyl)-O-L-seryl-O-L-seryl-L-serine | – 10.53 | – 92.94 | – 9.37 | – 74.27 | – 11.92 | – 105.19 | 3 | ||||
| 10 | N-acetylglucosamine | – 9.35 | – 75.00 | – 11.31 | – 88.38 | – 9.63 | – 109.54 | 3 | ||||
| 11 | Naquihexcin A | – 7.68 | – 75.55 | – 9.51 | – 106.62 | – 9.47 | – 98.87 | 3 | ||||
| 12 | Nikkomycin pseudo-J | – 12.45 | – 80.79 | – 8.22 | – 63.69 | – 8.71 | – 99.39 | 3 | ||||
| 13 | Quercetin glucoside | – 10.86 | – 78.78 | – 8.93 | – 76.02 | – 12.37 | – 93.81 | 3 | ||||
| 14 | Sharkquinone | – 10.24 | – 46.44 | – 7.63 | – 58.80 | – 9.00 | – 63.41 | 3 | ||||
| 15 | Solphenazine F | – 8.73 | – 61.77 | – 10.34 | – 99.81 | – 10.63 | – 104.14 | 3 | ||||
| 16 | Adenosine pentaphosphate | – 12.95 | – 86.04 | – 12.60 | – 77.62 | – 13.16 | – 75.68 | – 11.99 | – 88.82 | 4 | ||
| 17 | Atetra P | – 10.73 | – 59.54 | – 10.55 | – 57.12 | – 11.28 | – 46.80 | – 10.17 | – 85.68 | 4 | ||
| 18 | GDP-4-keto-6-deoxymannose | – 11.32 | – 49.79 | – 9.78 | – 31.85 | – 9.58 | – 43.18 | – 9.50 | – 69.60 | 4 | ||
| 19 | Target Positive Control | – 3.04 | – 80.42 | – 4.40 | – 48.15 | – 3.67 | – 32.83 | – 3.74 | – 40.67 | – 4.37 | – 58.56 | 5 |
| 20 | Miltefosine | – 11.27 | – 123.24 | – 4.35 | – 101.39 | – 0.77 | – 50.06 | – 3.39 | – 53.29 | – 3.01 | – 99.89 | 5 |
| 21 | Paromomycin | – 5.97 | – 71.72 | – 10.92 | – 69.50 | – 7.68 | – 49.24 | – 10.86 | – 76.77 | – 11.07 | – 80.31 | 5 |
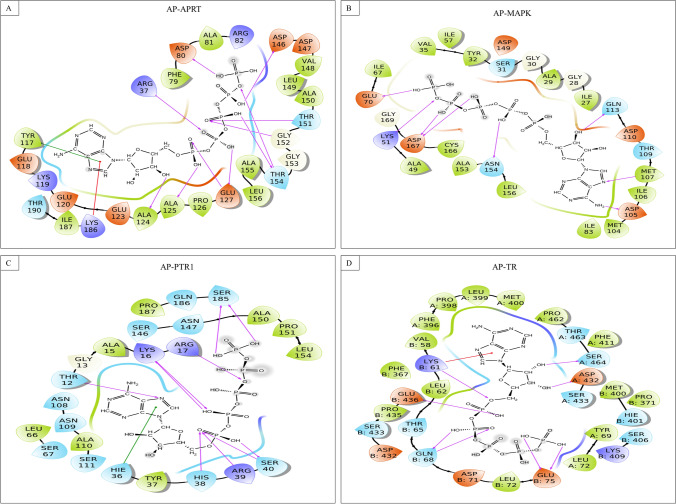
Fig. 2.
Ligand interaction diagram of Adenosine phosphoribosyl transferase with different proteins A Adenosine phosphoribosyl transferase, B MAP Kinase, C Pteridine reductase, and D Trypanothione reductase
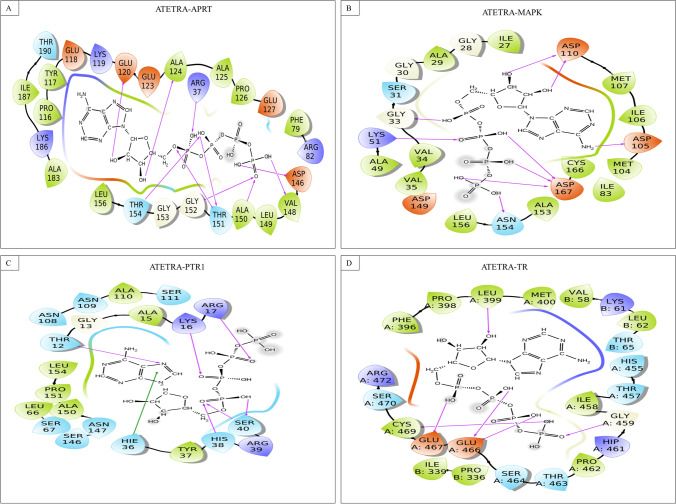
Fig. 3.
Interaction of Atetra P with A Adenosine phosphoribosyl transferase, B MAP Kinase, C Pteridine reductase, and D Trypanothione reductase
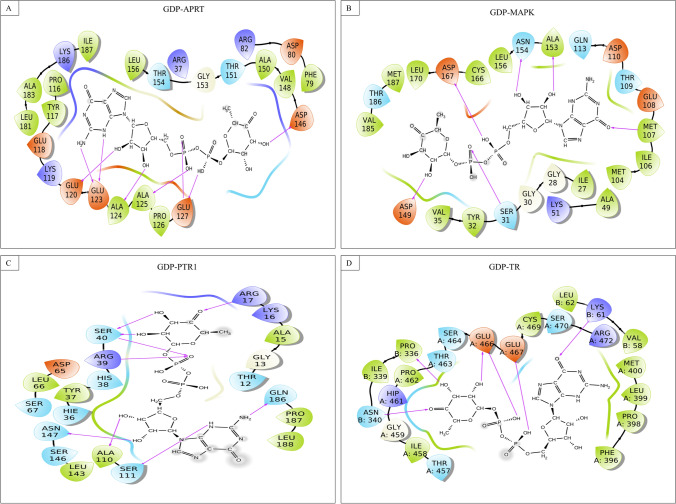
Fig. 4.
GDP-4-keto-6-deoxymannose interacting with A Adenosine phosphoribosyl transferase, B MAP Kinase, C Pteridine reductase, and D Trypanothione reductase
Miltefosine was designated owing to its large success rate as an oral agent in the Indian subcontinent (> 94% cure rate), whereas in Guatemala, miltefosine treatment failed miserably with a cure rate of only 53% (Soto et al. 2004). Miltefosine, an alkylphosphocholine, was initially investigated for cancer and linked to cancer treatment, and accredited for the treatment of leishmaniasis in India (Glantz-Gashai et al. 2017). Li et al., by obstructing the function of molecular chaperones HSP90, miltefosine has been studied as a powerful and extensive HSP90 inhibitor with anticancer activity (Li et al. 2021). As an injectable drug paromomycin has also been approved for the treatment of leishmaniasis in the Indian subcontinent (Jhingran et al. 2009).
Identification of potential multi-targeted inhibitors against Leishmania donovani crucial proteins and their functions
Adenosine pentaphosphate, Atetra P, and GDP-4-keto-6-deoxymannose were the top three ligands, and during molecular docking studies, interacted with these four targets: Adenine phosphoribosyl transferase (APRT), Map Kinase (MAPK), Pteridine reductase (PTR1), and Trypanothione reductase (TR).
Adenosine pentaphosphate
Adenosine pentaphosphate, a structural analog of adenosine 5' diphosphate, may inhibit platelet aggregation and shape change during the human platelet response (Lienhard and Secemski 1973). Some studies claim that this nucleotide polyphosphate is vasoactive in nature, but there is no evidence to support this claim (Lienhard and Secemski 1973; Jiménez et al. 1998).
Adenosine pentaphosphate interacts through a strong hydrogen bond with binding pocket residues of APRT (Arg37, Asp80 & 146, Thr151, Gly152, Thr154, Glu127, Ala124 and 125), MAPK (Lys51, Glu70, Asp105, Met107, Gln113, Asn154, Asp167), PTR1 (Thr12, Lys16, Arg17, His38, Arg39, Ser40 and 185), TR (chain A: Asp432 and Ser464; chain B: Lys61, Gln68, Glu75 and 436); Pi−cation interaction with APRT (Lys186), and TR (chain B: Lys61); whereas Pi−Pi stacking interaction with APRT (Tyr117), and PTR1 (Hie36).
Atetra P
Atetra P is a vasodilator that has been discovered and identified in the human body. They are mostly present in myocardial tissues, which control microcirculations in the heart (Burnstock and Pelleg 2015). Ap4 has also been proposed as a metabolite that facilitates cell signaling in mammals in some studies (Kimura et al. 2018). Additionally, this compound has been shown to significantly reduce platelet aggregation (Agarwal 1996).
Atetra P interacts through strong hydrogen bond with binding pocket residues of APRT (Arg37, Glu120, Ala124, Val148, Val149, Ala150, Thr151, Gly152, and Thr154), MAPK (Gly33, Lys51, Asp105 and 110, Asn154, and Asp167), PTR1 (Thr12, Lys16, Agr17, His38, and Ser40), and TR (chain A: Leu399, Gly459, Glu466 and 467, and Cys469); and Pi−Pi stacking interaction with Hie36 of PTR1.
GDP-4-keto-6-deoxymannose
GDP-4-keto-6-deoxymannose is a natural substance that regulates the formation of GDP-fucose. Fucose-containing glycans serve important roles in mammalian blood transfusion reactions, selectin-mediated leukocyte−endothelial adhesion, host−microbe interactions, and various oncogenic events, such as Notch receptor signaling. Salvage and de novo processes in mammalian cells are known to be involved in GDP synthesis. GMD acts as an enzyme intermediate during the fucose production process (Sullivan et al. 1998). Fucosylation of eukaryotic glycans by humans has been linked to a number of oncogenic signaling pathways, including selectin-mediated leukocyte adhesion to endothelial cells and host−microbe interactions (Nakayama et al. 2003).
GDP-4-keto-6-deoxymannose interacts through a strong hydrogen bond only with the binding pocket residues of APRT (Glu120 and 123, Ala124 and 125, Glu 127, and Asp146), MAPK (Ser31, Met107, Asp149, Ala153, Asn154, and Asp167), PTR1 (Arg17 and 39, Ser40, Ala110, Ser111, Asn147, and Gln186), and TR (chain A: Glu466 and 467; chain B: Lys61, Pro336, and Asn340).
Prime MM-GBSA analysis to determine the binding energy
In order to validate the binding affinity of top ligands received from XP docking against five proteins, binding energy was evaluated using the Prime MM-GBSA module of the Schrödinger suite. The docked posed of proteins and ligands were initially minimized using the Prime tool (integrated tool of Schrödinger software), followed by binding-free energy calculation using the OPLS-2005 force field. The ∆G scores obtained for the ligands ranged from (– 133.46 to – 20.17), (– 141.34 to – 32.46), (– 133.89 to – 13.95), (– 130.81 to – 14.21), and (– 101.38 to – 14.48) Kcal/mol against Adenine phosphoribosyl transferase, Trypanothione reductase, N-Myristoyl transferase, Pteridine reductase, and Map Kinase, respectively. Binding free energy of multi-targeted ligand ranging from (– 99.81 to – 31.85), (– 117.58 to – 63.41), (– 53.34 to – 46.08), and (– 93.01 to – 46.44) Kcal/mol against Adenine phosphoribosyl transferase, Trypanothione reductase, N-Myristoyl transferase, Pteridine reductase, and Map Kinase, respectively. The score obtained for the reference ligands are also comparable. Considering the overall docking score and ∆G score, we found that proteins, namely Trypanothione reductase and MAP Kinase, have good docking and binding free energy scores with three ligands, namely Adenosine pentaphosphate, Atetra P, and GDP-4-keto-6-deoxymannose. Therefore, this ligand complex with Trypanothione reductase and Map Kinase was selected for the molecular dynamics simulation study to determine its complex stability.
Molecular dynamics simulation study
The top hits compounds from the virtual screening and MM-GBSA studies were investigated once again to evaluate the consistency of ligand binding to the active sites of the target proteins, which was confirmed via molecular dynamics. Three compounds (Adenosine pentaphosphate, Atetra P, and GDP-4-keto-6-deoxymannose) in complex with Map Kinase and Trypanothione reductase were subjected to 100 ns dynamics using GROMOS96 43a1 forcefield through WebGRO.
In combination with Map Kinase and Trypanothione reductase, the average RMSD score of Adenosine pentaphosphate was 0.59 and 0.36 nm, Atetra P was 0.61 and 0.35 nm, and the ligand GDP-4-keto-6-deoxymannose was 0.79 and 0.31 nm, respectively. The RMSD of the AP-MAPK complex constantly grows up to 45 ns, but after that it stabilises with little deviations. Similarly, the RMSD of the AP-TR complex consistently increases up to 30 ns, but after that it gets stabilized with minimal deviations (Fig. 5).
Fig. 5.
Graphical representation of Root-mean square deviation of complex A Adenosine pentaphosphate and MAP kinase, B Adenosine pentaphosphate and Trypanothione reductase, C Atetra P and MAP kinase, D Atetra P and Trypanothione reductase, E GDP-4-keto-6-deoxymannose and MAP kinase, F GDP-4-keto-6-deoxymannose and Trypanothione reductase
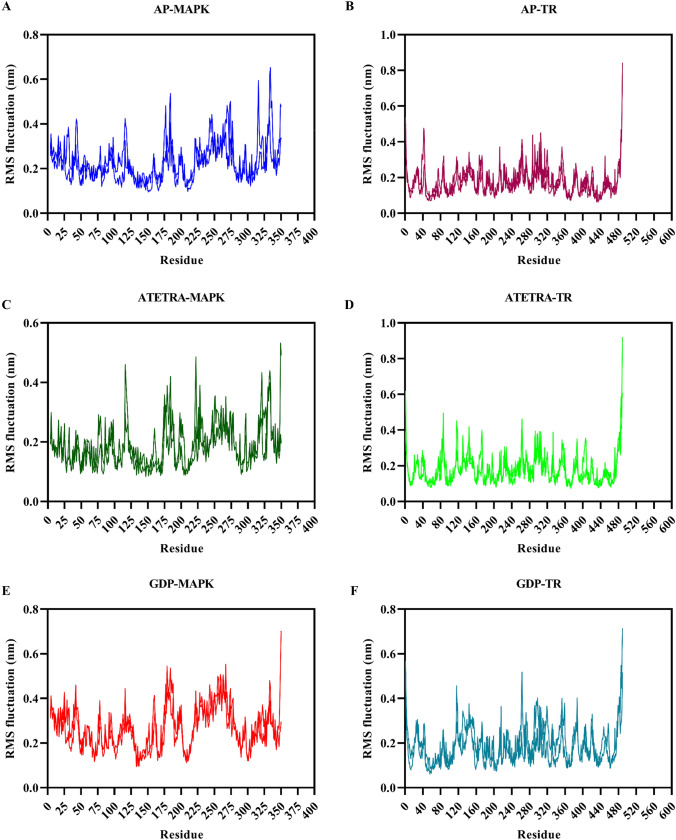
The average RMS fluctuation score of Adenosine pentaphosphate, Atetra P, and the ligand GDP-4-keto-6-deoxymannose in conjunction with Map Kinase was 0.23, 0.19, and 0.27 nm, respectively. The average RMS fluctuation score for Adenosine pentaphosphate was 0.18 nm, Atetra P was 0.18 nm, and the ligand GDP-4-keto-6-deoxymannose was 0.19 nm with Trypanothione reductase, respectively (Fig. 6). All complexes had less variation based on average RMSF scores. The RMS fluctuation of ligands in compound with Trypanothione reductase is lower than that of ligands in complex with MAP Kinase.
Fig. 6.
Graphical representation of Root-mean square fluctuation of complex A Adenosine pentaphosphate and MAP kinase, B Adenosine pentaphosphate and Trypanothione reductase, C Atetra P and MAP kinase, D Atetra P and Trypanothione reductase, E GDP-4-keto-6-deoxymannose and MAP kinase, F GDP-4-keto-6-deoxymannose and Trypanothione reductase
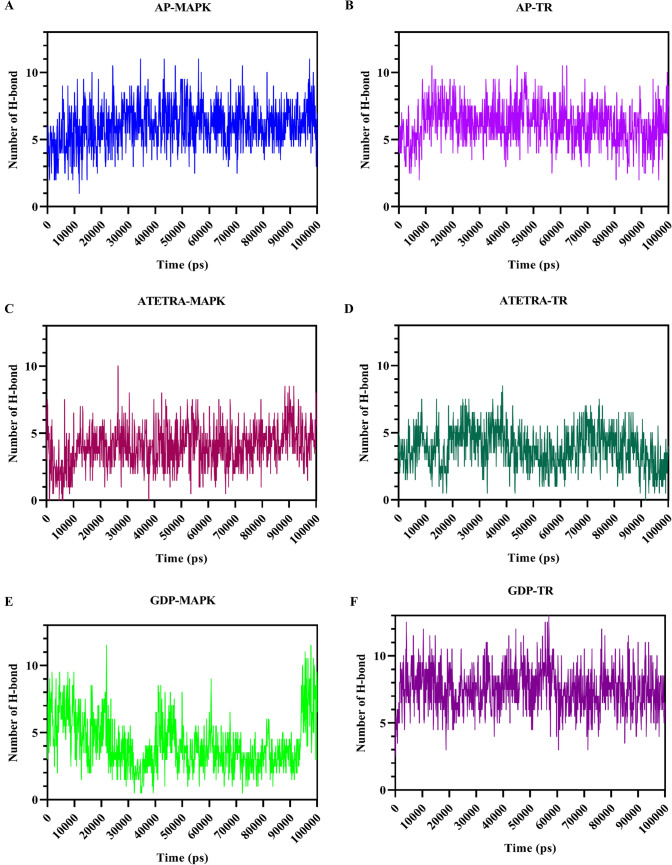
The average number of hydrogen bond formation between protein–ligand complex, during the molecular dynamics period of 100 ns was 3.05 and 6.17 for Adenosine pentaphosphate, 4.00 and 3.87 for Atetra P, and 4.19 and 7.52 for the ligand GDP-4-keto-6-deoxymannose, in complex with Map Kinase and Trypanothione reductase, respectively. In case of MAP Kinase, the number of H-bond was consistent throughout the dynamics, except GDP-MAPK that shows H-bond breaking and formation throughout the period, but in last 50 ns the number of H-bonds were maximum. Similarly, for Trypanothione reductase the number of H-bonds were consistently higher for AP and GDP, whereas ATETRA shows fluctuation in number of H-bond formation and breaking throughout the period (Fig. 7).
Fig. 7.
Illustration of the formation and breaking of hydrogen bonds over the course of a 100-ns molecular dynamics simulation. The following combinations: A Adenosine pentaphosphate and MAP kinase; B Adenosine pentaphosphate and Trypanothione reductase; C Atetra P and MAP kinase; D Atetra P and Trypanothione reductase; E GDP-4-keto-6-deoxymannose and MAP kinase; F GDP-4-keto-6-deoxymannose and Trypanothione reductase. Due to the larger size of Trypanothione reductase compared to MAP kinase, bond formation was higher for this enzyme
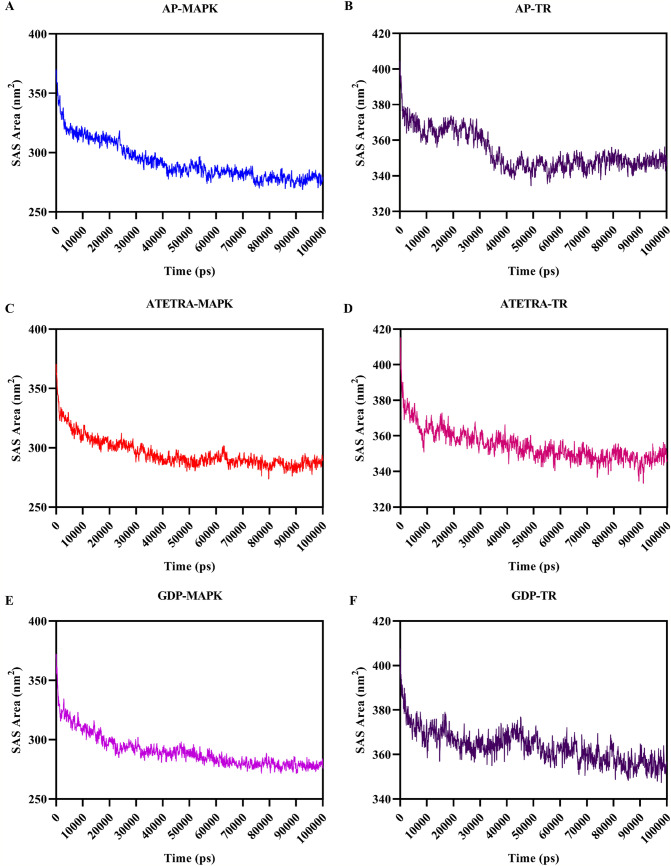
The surface area accessible to the solvent is estimated using the Solvent Accessible Surface Area (SASA) of protein–ligand complexes to determine the interaction of the protein–ligand complex with the solvent molecules. The ligands in complex with MAP Kinase and Trypanothione reductase were estimated with an average SASA score of 292.42 and 353.59 nm2 for Adenosine pentaphosphate, 295.48 and 354.59 nm2 for Atetra P, and 290.14 and 363.52 nm2 for GDP-4-keto-6-deoxymannose, respectively. As the dynamics time goes on, the SAS area of all the complexes decreases till it reaches 100 ns, showing structural uniformity (Fig. 8). The SASA of ligands docked with MAPK is less than 300 nm2, whereas the SASA of ligands in complex with TR is higher than 300 nm2. This may be because of larger size of Leishmania donovani’s TR than the MAPK, which accesses more solvents to bind with the TR.
Fig. 8.
Solvent accessibility surface area during the 100 ns of the molecular dynamics simulation is graphically illustrated. A Adenosine pentaphosphate and MAP kinase, B Adenosine pentaphosphate and Trypanothione reductase, C Atetra P and MAP kinase, D Atetra P and Trypanothione reductase, E GDP-4-keto-6-deoxymannose and MAP kinase, F GDP-4-keto-6-deoxymannose and Trypanothione reductase. SAS area for larger complexes is higher as in case of Trypanothione reductase
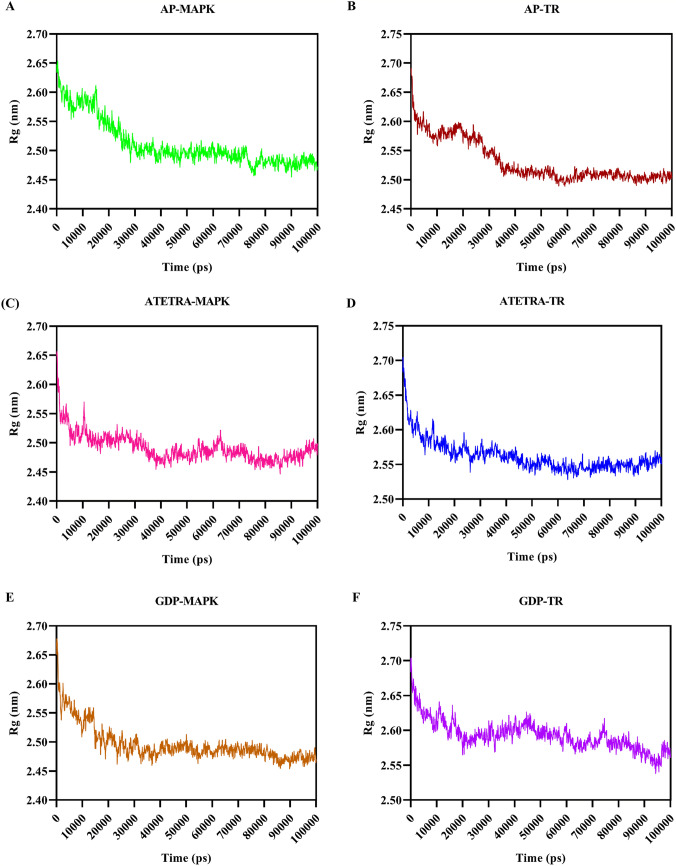
Radius of gyration (Rg) was calculated to estimate the compactness of the structures (protein–ligand complexes) throughout the dynamics period. The average Rg score for Adenosine pentaphosphate was 2.51 and 2.53 nm, for Atetra P 2.49 and 2.56 nm, and for GDP-4-keto-6-deoxymannose was 2.50 and 2.59 nm with MAP Kinase and Trypanothione reductase, respectively (Fig. 9). The estimated Rg score for all the complexes are initially higher, but as the dynamics time goes up the Rg scores getting lower, and the average Rg scores for the structures are nearly same and lies between 2.49 and 2.59 nm this shows the structures getting towards compactness.
Fig. 9.
Graphical representation of Radius of gyration (Rg) of complex A Adenosine pentaphosphate and MAP kinase, B Adenosine pentaphosphate and Trypanothione reductase, C Atetra P and MAP kinase, D Atetra P and Trypanothione reductase, E GDP-4-keto-6-deoxymannose and MAP kinase, F GDP-4-keto-6-deoxymannose and Trypanothione reductase
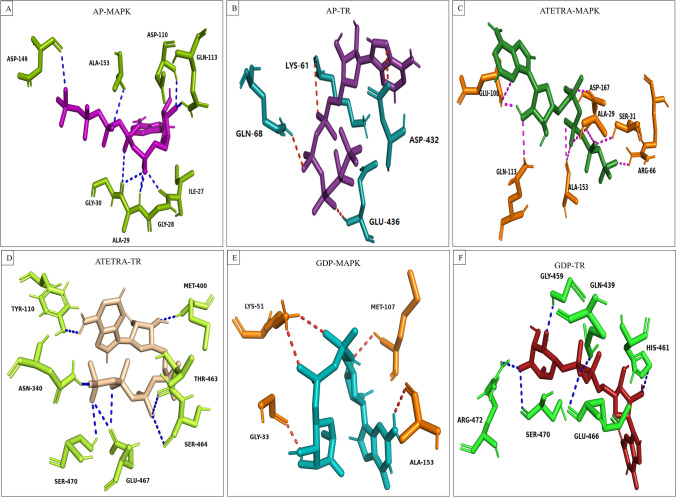
Finally, the final frame (frame number 1001) of the dynamics was retrieved from the allframes.pdb file to examine the protein–ligand hydrogen bond interaction after molecular dynamics. Adenosine pentaphosphate forms the post-MD hydrogen bond with MAPK (Ile27, Gly28, Ala29, Gly30, Asp110, Gln113, Asp149, and Ala153), and TR (Lys61, Gln68, Asp432, and Glu436). Atetra P forms the H-bond with MAPK (Ala29, Ser31, Arg66, Glu108, Gln113, Ala153, and Asp167), TR (Tyr110, Asn340, Met400, Thr463, Ser464, Glu467, and Ser470). Whereas GDP-4-keto-6-deoxymannose interacts with strong hydrogen bond with MAPK (Gly33, Lys51, Met107, and Ala153), and TR (Gln439, Gly459, His461, Glu466, Ser470, and Arg472) (Fig. 10).
Fig. 10.
Following a molecular dynamics simulation, a schematic representation of the hydrogen bond interaction between potential compounds and the therapeutic targets of Leishmania donovani is shown. Diagram was prepared by extracting the 1001 frame using the allframes.pdb file A Adenosine pentaphosphate and MAP kinase, B Adenosine pentaphosphate and Trypanothione reductase, C Atetra P and MAP kinase, D Atetra P and Trypanothione reductase, E GDP-4-keto-6-deoxymannose and MAP kinase, F GDP-4-keto-6-deoxymannose and Trypanothione reductase
All these MD results (RMSD, RMSF, H-bonding, SASA, Rg) shows the specificity our ligands in in-silico studies.
Post-MD binding was calculated for all three potential compounds complexed with each L donovani therapeutic targets using the g_mmpbsa standalone tool. Binding energy is the sum of different energies like van der waal energy, electrostatic energy, polar solvation energy, and SASA energy. Binding energy for Adenosine pentaphosphate with MAPK and TR was estimated – 173.77 and – 192.82 kJ/mol, respectively. For Atetra P in complex with MAPK and TR, binding energy was – 193.57 and – 13.57 kJ/mol; whereas, for GDP-4-keto-6-deoxymannose the binding energy for MAPK and TR was estimated to be – 138.44 and – 194.75 kJ/mol (Table 5).
Table 5.
Binding energy calculation between the therapeutic targets of Leishmania donovani and potential secondary metabolites of the actinomycetes using MM-PBSA in kJ/mol
| S. No. | Potential ligand in complex with Ld therapeutic targets | Binding energy calculation using MM-PBSA in kJ/mol | ||||
|---|---|---|---|---|---|---|
| Van der Waal energy | Electrostatic energy | Polar solvation energy | SASA energy | Binding energy | ||
| 1 | AP-MAPK | – 235.11 | – 65.10 | 142.51 | – 16.06 | – 173.77 |
| 2 | AP-TR | – 306.02 | – 56.07 | 191.13 | – 21.86 | – 192.82 |
| 3 | ATETRA-MAPK | – 308.30 | – 73.67 | 210.79 | – 22.40 | – 193.57 |
| 4 | ATETRA-TR | – 16.46 | – 0.07 | 8.13 | – 5.03 | – 13.435 |
| 5 | GDP-MAPK | – 203.36 | – 43.38 | 123.97 | – 15.670 | – 138.44 |
| 6 | GDP-TR | – 253.77 | – 59.72 | 139.02 | – 20.29 | – 194.75 |
Principal component analysis (PCA) was also calculated for each of the complexes to analyze all the variables in a single/double dimension that helps in understanding the result with minimum data loss for larger set of data. The 2D graph was plotted for projection of eigenvector 1 vs eigenvector 3 for all the complexes (Fig. 11).
Fig. 11.
Principal component analysis (PCA) plotted for all of the complexes, the projection of eigenvector 1 vs. eigenvector 3 was illustrated as a 2D graph. AP-MAPK (black), AP-TR (red), ATETRA-MAPK (green), ATETRA-TR (blue), GDP-MAPK (yellow), and GDP-TR (nude pink)
Discussion
Leishmaniasis is a communicable disease of tropical neglected regions. The principal cause of this disease is poverty or lack of basic human needs (Prajapati et al. 2012). Leishmania parasite has developed resistance towards various drugs (Prajapati and Pandey 2017). However, the treatment currently relies upon highly toxic and non-selective drugs like Antimonial, Miltefosine, Paromomycin, and Amphotericin B. Actinomycetes are lavish producers of secondary metabolites due to their active biological activity, chemical structure, and emergence (Abdelmohsen et al. 2015). In this study, we have identified a library of crucial secondary metabolites from Steptomycetes species of actinomycetes, which show a more significant binding interaction with the target proteins of Leishmania. In order to develop a new anti-Leishmanial drug, we have determined the relative binding affinity of Actinomycetes derived secondary metabolites against the five leishmanial proteins; namely Adenine phosphoribosyl transferase (PDB ID-1QB7), Trypanothione reductase (PDB ID-2JK6), N-Myristoyltransferase (PDB ID- 2WUU), Pteridine reductase (PDB ID-2XOX) and Map Kinase (4QNY). The estimation of binding affinities is accomplished by the computational approach of molecular docking, consisting of three consecutive methods: HTVS, SP, and XP. HTVS and SP method uses the self-same scoring functions, so the final conclusion drawn through XP docking method and 18 ligands were finalized as good binder against all five proteins. The docking score obtained from these 18 ligands against all five proteins is comparable with the other reported works (Pandey et al. 2017b). The highest docking score of – 12.95, – 12.60, – 13.16, and – 11.99 and binding energy of – 86.04, – 77.62, – 75.68, and – 88.82 was obtained for the Adenosine pentaphosphate against MAP Kinase, Adenine phosphoribosyl transferase, Pteridine reductase, and Trypanothione reductase, respectively. Other top two ligand Atetra P and GDP-4-keto-6-deoxymannose also shows good docking and binding energy scores as compared to positive controls (miltefosine, paromomycin, and specific inhibitor for each target).
Following binding affinity and binding energy prediction against five target proteins, molecular dynamics simulation of highest binding affinity ligands (Adenosine pentaphosphate, Atetra P, and GDP-4-keto-6-deoxymannose) was obtained against Map Kinase and Trypanothione reductase. The average RMSD value obtained for Map Kinase (4QNY) and Trypanothione reductase (PDB ID-2JK6) against Adenosine pentaphosphate were 0.46 and 0.40 nm, respectively. Average RMSD for Atetra P with Map Kinase and Trypanothione reductase were 0.54 and 0.37 nm, respectively. GDP-4-keto-6-deoxymannose shows average RMSD of 0.60 and 0.30 nm against protein targets Map Kinase and Trypanothione reductase, respectively. This RMSD value is comparable with the other recently reported literature. From all these aforementioned discussions, we can say that the compound Adenosine pentaphosphate, Atetra P, and GDP-4-keto-6-deoxymannose can be used as a potent inhibitor for the treatment of visceral leishmaniasis in the Indian Subcontinents. Our high-throughput virtual screening study demonstrates that the suggested compounds are superior to currently marketed medications.
Conclusion
This study identifies multi-targeted anti-leishmanial compounds derived from actinomycetes. These compounds have a high affinity for proteins involved in Leishmania donovani metabolic pathways. A high-throughput virtual screening approach was used to screen the actinomycetes secondary metabolite library. The MM-GBSA method was used to calculate free binding energy by integrating contributions from gas-phase energy (MM), electrostatic solvation energy (GB), and non-electrostatic solvation energy (SA). The QikProp application was used to evaluate ADME properties and principal drug-characteristics values to check the compounds' drug-like properties. To determine the ligand−receptor binding and physical movement of atoms or molecules in physiological conditions, the MD simulation method was used. We identified three compounds (Adenosine pentaphosphate, Atetra P, and GDP-4-keto-6-deoxymannose) from the actinomyces natural product library that have the potential to inhibit vital proteins of Leishmania donovani for the treatment of visceral leishmaniasis infection.
Acknowledgements
SS is thankful to Central University of Rajasthan for providing university fellowship. VKP is thankful to the Central University of Rajasthan for lab facilities.
Author contributions
Protocol designed by SS and VKP; Methodology performed by SS and VKP; the manuscript was written by SS, and VKP.
Funding
No funding was received for conducting this study.
Declarations
Conflict of interest
The Authors have declared no competing interest.
References
- Abdelmohsen UR, Grkovic T, Balasubramanian S, Kamel MS, Quinn RJ, Hentschel U. Elicitation of secondary metabolism in actinomycetes. Biotechnol Adv. 2015;33(6, Part 1):798–811. doi: 10.1016/j.biotechadv.2015.06.003. [DOI] [PubMed] [Google Scholar]
- Agarwal KC. Purines and myocardial protection. Boston, MA, US: Springer; 1996. Purines and regulation of platelet activation; pp. 409–418. [Google Scholar]
- Ahluwalia PK, Pandey RK, Sehajpal PK, Prajapati VK. Perturbed microRNA expression by mycobacterium tuberculosis promotes macrophage polarization leading to pro-survival foam cell. Front Immunol. 2017;8:107. doi: 10.3389/fimmu.2017.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramaniam DM, Williams DP (2020) WebGRO for Macromolecular Simulations.
- Barrack KL, Tulloch LB, Burke L-A, Fyfe PK, Hunter WN, Communications C. Structure of recombinant Leishmania donovani pteridine reductase reveals a disordered active site. Acta Crystallogr Sect f: Struct Biol Cryst Commun. 2011;67(1):33–37. doi: 10.1107/S174430911004724X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker H, Berendsen H, Dijkstra E, Achterop S, Vondrumen R, Vanderspoel D, Sijbers A, Keegstra H, Renardus M. 4th International conference on computational physics (PC 92) Singapore: World Scientific Publishing; 1993. Gromacs-a parallel computer for molecular-dynamics simulations; pp. 252–256. [Google Scholar]
- Bérdy J. Bioactive microbial metabolites. J Antibiot. 2005;58(1):1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- Brannigan JA, Smith BA, Yu Z, Brzozowski AM, Hodgkinson MR, Maroof A, Price HP, Meier F, Leatherbarrow RJ, Tate EW, Smith DF, Wilkinson AJ. N-Myristoyltransferase from Leishmania donovani: structural and functional characterisation of a potential drug target for visceral leishmaniasis. J Mol Biol. 2010;396(4):985–999. doi: 10.1016/j.jmb.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnstock G, Pelleg A. Cardiac purinergic signalling in health and disease. Purinergic Signalling. 2015;11(1):1–46. doi: 10.1007/s11302-014-9436-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C, Yin H, Garforth J, McKie JH, Jaouhari R, Speers P, Douglas KT, Rock PJ, Yardley V, Croft SL. Phenothiazine inhibitors of trypanothione reductase as potential antitrypanosomal and antileishmanial drugs. J Med Chem. 1998;41(2):148–156. doi: 10.1021/jm960814j. [DOI] [PubMed] [Google Scholar]
- Chander S, Pandey RK, Penta A, Choudhary BS, Sharma M, Malik R, Prajapati VK, Murugesan S. Molecular docking and molecular dynamics simulation based approach to explore the dual inhibitor against HIV-1 reverse transcriptase and integrase. Comb Chem High Throughput Screen. 2017 doi: 10.2174/1386207320666170615104703. [DOI] [PubMed] [Google Scholar]
- Corpas-Lopez V, Moniz S, Thomas M, Wall RJ, Torrie LS, Zander-Dinse D, Tinti M, Brand S, Stojanovski L, Manthri S. Pharmacological validation of N-myristoyltransferase as a drug target in Leishmania donovani. ACS Infect Dis. 2018;5(1):111–122. doi: 10.1021/acsinfecdis.8b00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diseases (2017) Global leishmaniasis update, 2006–2015: a turning point in leishmaniasis surveillance
- Gill N, Pandey D, Roy N, Dhingra N, Jain S. Kala-azar in India – progress and challenges towards its elimination as a public health problem. Wkly Epidemiol Rec. 2021;96(26):265–280. [Google Scholar]
- Glantz-Gashai Y, Meirson T, Reuveni E, Samson AO. Virtual screening for potential inhibitors of Mcl-1 conformations sampled by normal modes, molecular dynamics, and nuclear magnetic resonance. Drug Des Dev Ther. 2017;11:1803. doi: 10.2147/DDDT.S133127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan HF, Coombs GH. Purine and pyrimidine metabolism in parasitic protozoa. FEMS Microbiol Lett. 1988;54(1):47–83. doi: 10.1016/0378-1097(88)90240-6. [DOI] [PubMed] [Google Scholar]
- Hwang HY, Ullman B. Genetic analysis of purine metabolism in Leishmania donovani. J Biol Chem. 1997;272(31):19488–19496. doi: 10.1074/jbc.272.31.19488. [DOI] [PubMed] [Google Scholar]
- Jhingran A, Chawla B, Saxena S, Barrett MP, Madhubala R. Paromomycin: uptake and resistance in Leishmania donovani. Mol Biochem Parasitol. 2009;164(2):111–117. doi: 10.1016/j.molbiopara.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez AI, Castro E, Delicado EG, Miras-Portugal MT. Potentiation of adenosine 5′-triphosphate calcium responses by diadenosine pentaphosphate in individual rat cerebellar astrocytes. Neurosci Lett. 1998;246(2):109–111. doi: 10.1016/S0304-3940(98)00227-4. [DOI] [PubMed] [Google Scholar]
- Jolliffe IT, Cadima J. Principal component analysis: a review and recent developments. Phil Trans R Soc a: Math, Phy, Eng Sci. 2016;374(2065):20150202. doi: 10.1098/rsta.2015.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J, Dube D, Ramachandran R, Singh P, Singh N (2012) Thianthrene is a novel inhibitor of Leishmaniadonovani pteridine reductase 1 (PTR1). J Mol Biochem 1 (2)
- Kaur P, Garg M, Hombach-Barrigah A, Clos J. MAPK1 of Leishmania donovani interacts and phosphorylates HSP70 and HSP90 subunits of foldosome complex. Sci Rep. 2017;7(1):10202. doi: 10.1038/s41598-017-09725-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killick-Kendrick R. Phlebotomine vectors of the leishmaniases: a review. Med Vet Entomol. 1990;4(1):1–24. doi: 10.1111/j.1365-2915.1990.tb00255.x. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Tanaka C, Oka M. Identification of major enzymes involved in the synthesis of diadenosine tetraphosphate and/or adenosine tetraphosphate in Myxococcus xanthus. Curr Microbiol. 2018;75(7):811–817. doi: 10.1007/s00284-018-1452-x. [DOI] [PubMed] [Google Scholar]
- Kumar P, Kumar A, Verma SS, Dwivedi N, Singh N, Siddiqi MI, Tripathi RP, Dube A, Singh N. Leishmania donovani pteridine reductase 1: biochemical properties and structure-modeling studies. Exp Parasitol. 2008;120(1):73–79. doi: 10.1016/j.exppara.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Kumari R, Kumar R, Lynn A. g_mmpbsa—a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- Li L, Yang M, Li C, Liu Y. Virtual screening based identification of miltefosine and octenidine as inhibitors of heat shock protein 90. Naunyn Schmiedebergs Arch Pharmacol. 2021;394(11):2223–2232. doi: 10.1007/s00210-021-02133-y. [DOI] [PubMed] [Google Scholar]
- Lienhard GE, Secemski II. P1, P5-Di (adenosine-5′) pentaphosphate, a potent multisubstrate inhibitor of adenylate kinase. J Biol Chem. 1973;248(3):1121–1123. doi: 10.1016/S0021-9258(19)44380-9. [DOI] [PubMed] [Google Scholar]
- Looker DL, Berens RL, Marr JJ. Purine metabolism in Leishmania donovani amastigotes and promastigotes. Mol Biochem Parasitol. 1983;9(1):15–28. doi: 10.1016/0166-6851(83)90053-1. [DOI] [PubMed] [Google Scholar]
- Lucas X, Senger C, Erxleben A, Grüning BA, Döring K, Mosch J, Flemming S, Günther S. StreptomeDB: a resource for natural compounds isolated from Streptomyces species. Nucleic Acids Res. 2012;41(D1):D1130–D1136. doi: 10.1093/nar/gks1253%JNucleicAcidsResearch. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavi Sastry G, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27(3):221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- Marr JJ, Berens RL. Antileishmanial effect of allopurinol. II. Relationship of adenine metabolism in Leishmania species to the action of allopurinol. J Infect Dis. 1977;136(6):724–732. doi: 10.1093/infdis/136.6.724. [DOI] [PubMed] [Google Scholar]
- Moumbock AF, Gao M, Qaseem A, Li J, Kirchner PA, Ndingkokhar B, Bekono BD, Simoben CV, Babiaka SB, Malange YI. StreptomeDB 3.0: an updated compendium of streptomycetes natural products. Nucleic Acids Res. 2021;49(D1):D600–D604. doi: 10.1093/nar/gkaa868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik B, Mattaparthi VSK, Gupta N, Ojha R, Das P, Singh S, Prajapati VK, Prusty DJJoBS, Dynamics, Chemical system biology approach to identify multi-targeting FDA inhibitors for treating COVID-19 and associated health complications. J Biomol Struct Dyn. 2021 doi: 10.1080/07391102.2021.1931451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K-i, Maeda Y, Jigami Y. Interaction of GDP-4-keto-6-deoxymannose-3, 5-epimerase-4-reductase with GDP-mannose-4, 6-dehydratase stabilizes the enzyme activity for formation of GDP-fucose from GDP-mannose. Glycobiology. 2003;13(10):673–680. doi: 10.1093/glycob/cwg099. [DOI] [PubMed] [Google Scholar]
- Nare B, Hardy LW, Beverley SM. The roles of pteridine reductase 1 and dihydrofolate reductase-thymidylate synthase in pteridine metabolism in the protozoan parasite Leishmania major. J Biol Chem. 1997;272(21):13883–13891. doi: 10.1074/jbc.272.21.13883. [DOI] [PubMed] [Google Scholar]
- Oostenbrink C, Villa A, Mark AE, Van Gunsteren WF. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J Comput Chem. 2004;25(13):1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- Pandey RK, Prajapati VK. Molecular and immunological toxic effects of nanoparticles. Int J Biol Macromol. 2017 doi: 10.1016/j.ijbiomac.2017.09.110. [DOI] [PubMed] [Google Scholar]
- Pandey RK, Sharma D, Bhatt TK, Sundar S, Prajapati VK. Developing imidazole analogues as potential inhibitor for Leishmania donovani trypanothione reductase: virtual screening, molecular docking, dynamics and ADMET approach. J Biomol Struct Dyn. 2015;33(12):2541–2553. doi: 10.1080/07391102.2015.1085904. [DOI] [PubMed] [Google Scholar]
- Pandey RK, Verma P, Sharma D, Bhatt TK, Sundar S, Prajapati VK. High-throughput virtual screening and quantum mechanics approach to develop imipramine analogues as leads against trypanothione reductase of leishmania. Biomed Pharmacother. 2016;83:141–152. doi: 10.1016/j.biopha.2016.06.010. [DOI] [PubMed] [Google Scholar]
- Pandey RK, Kumbhar BV, Srivastava S, Malik R, Sundar S, Kunwar A, Prajapati VK. Febrifugine analogues as Leishmania donovani trypanothione reductase inhibitors: binding energy analysis assisted by molecular docking, ADMET and molecular dynamics simulation. J Biomol Struct Dyn. 2017;35(1):141–158. doi: 10.1080/07391102.2015.1135298. [DOI] [PubMed] [Google Scholar]
- Pandey RK, Kumbhar BV, Sundar S, Kunwar A, Prajapati VK. Structure-based virtual screening, molecular docking, ADMET and molecular simulations to develop benzoxaborole analogs as potential inhibitor against Leishmania donovani trypanothione reductase. J Recept Signal Transduct Res. 2017;37(1):60–70. doi: 10.3109/10799893.2016.1171344. [DOI] [PubMed] [Google Scholar]
- Pandey RK, Narula A, Naskar M, Srivastava S, Verma P, Malik R, Shah P, Prajapati VK. Exploring dual inhibitory role of febrifugine analogues against Plasmodium utilizing structure-based virtual screening and molecular dynamic simulation. J Biomol Struct Dyn. 2017;35(4):791–804. doi: 10.1080/07391102.2016.1161560. [DOI] [PubMed] [Google Scholar]
- Phillips CL, Ullman B, Brennan RG, Hill CP. Crystal structures of adenine phosphoribosyltransferase from Leishmania donovani. EMBO J. 1999;18(13):3533–3545. doi: 10.1093/emboj/18.13.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapati VK, Pandey RK. Drug design: principles and applications. Singapore: Springer; 2017. Recent advances in the chemotherapy of visceral leishmaniasis; pp. 69–88. [Google Scholar]
- Prajapati VK, Mehrotra S, Gautam S, Rai M, Sundar S. In vitro antileishmanial drug susceptibility of clinical isolates from patients with Indian visceral leishmaniasis–status of newly introduced drugs. Am J Trop Med Hyg. 2012;87(4):655–657. doi: 10.4269/ajtmh.2012.12-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy GS, Mukhopadhyay AG, Dey CS. The p38 MAP kinase inhibitor, PD 169316, inhibits flagellar motility in Leishmania donovani. Biochem Biophys Res Commun. 2017;493(4):1425–1429. doi: 10.1016/j.bbrc.2017.09.135. [DOI] [PubMed] [Google Scholar]
- Ruiz-Postigo JA, Jain S, Mikhailov A, Maia-Elkhoury AN, Valadas S, Warusavithana S, Osman M, Lin Z, Beshah A, Yajima A, Gasimov E. Global leishmaniasis surveillance: 2019–2020, a baseline for the 2030 roadmap. Wkly Epidemiol Rec. 2020;96(35):401–420. [Google Scholar]
- Schrödinger Release 2017a-4 (2017a) LigPrep S, LLC, New York, NY.
- Schrödinger Release 2017b-4 (2017b) QikProp S, LLC, New York, NY
- Singh S, Kumar K, Panda M, Srivastava A, Mishra A, Prajapati VK. High-throughput virtual screening of small-molecule inhibitors targeting immune cell checkpoints to discover new immunotherapeutics for human diseases. Mol Diversity. 2022;28:1–23. doi: 10.1007/s11030-022-10452-2. [DOI] [PubMed] [Google Scholar]
- Soares-Silva M, Diniz FF, Gomes GN, Bahia D. The mitogen-activated protein kinase (MAPK) pathway: role in immune evasion by trypanosomatids. Front Microbiol. 2016;7:183. doi: 10.3389/fmicb.2016.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto J, Arana BA, Toledo J, Rizzo N, Vega JC, Diaz A, Luz M, Gutierrez P, Arboleda M, Berman JD, Junge K, Engel J, Sindermann H. Miltefosine for new world cutaneous leishmaniasis. Clin Infect Dis. 2004;38(9):1266–1272. doi: 10.1086/383321%JClinicalInfectiousDiseases. [DOI] [PubMed] [Google Scholar]
- Sullivan FX, Kumar R, Kriz R, Stahl M, Xu G-Y, Rouse J, Chang X-j, Boodhoo A, Potvin B, Cumming DA. Molecular cloning of human GDP-mannose 4, 6-dehydratase and reconstitution of GDP-fucose biosynthesis in vitro. J Biol Chem. 1998;273(14):8193–8202. doi: 10.1074/jbc.273.14.8193. [DOI] [PubMed] [Google Scholar]
- Sundar S, Mehta H, Suresh AV, Singh SP, Madhukar R, Murray HW. Amphotericin B treatment for Indian visceral leishmaniasis: conventional versus lipid formulations. Clin Infect Dis. 2004;38(3):377–383. doi: 10.1086/380971. [DOI] [PubMed] [Google Scholar]
- Sundar S, Jha TK, Thakur CP, Sinha PK, Bhattacharya SK. Injectable paromomycin for visceral leishmaniasis in India. N Engl J Med. 2007;356(25):2571–2581. doi: 10.1056/NEJMoa066536. [DOI] [PubMed] [Google Scholar]
- Sundar S, Singh A, Rai M, Prajapati VK, Singh AK, Ostyn B, Boelaert M, Dujardin J-C, Chakravarty J. Efficacy of miltefosine in the treatment of visceral leishmaniasis in India after a decade of use. Clin Infect Dis. 2012;55(4):543–550. doi: 10.1093/cid/cis474. [DOI] [PubMed] [Google Scholar]
- Tate EW, Bell AS, Rackham MD, Wright MH. N-Myristoyltransferase as a potential drug target in malaria and leishmaniasis. Parasitology. 2014;141(1):37–49. doi: 10.1017/s0031182013000450. [DOI] [PubMed] [Google Scholar]
- Tovar J, Wilkinson S, Mottram JC, Fairlamb AH. Evidence that trypanothione reductase is an essential enzyme in leishmania by targeted replacement of the tryA gene locus. Mol Microbiol. 1998;29(2):653–660. doi: 10.1046/j.1365-2958.1998.00968.x. [DOI] [PubMed] [Google Scholar]
- Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. Genomics of actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007;71(3):495–548. doi: 10.1128/mmbr.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernimont AK, Loppnau P, Walker JR, Mangos M, El Bakkouri M, Arrowsmith CH, Edwards AM, Bountra C, Hui R, Amani M (2014) Crystal structure of MapK from Leishmaniadonovani, LDBPK_331470. Structural Genomics Consortium (SGC)