Abstract
In an effort to investigate the molecular mechanisms responsible for the drastic morphological changes the mitochondria go through during the life cycle of the aquatic fungus Blastocladiella emersonii, the gene encoding the α subunit of the mitochondrial processing peptidase (α-MPP) was isolated. Nucleotide sequence analysis revealed that the predicted α-MPP polypeptide comprises 474 amino acids with a calculated molecular mass of 51,900 Da, presenting a characteristic mitochondrial signal sequence. Northern blot analysis indicated a single 1.4-kb transcript encoding the B. emersonii α-MPP, whose levels decrease during sporulation, becoming very low in the zoospore, and increase again during germination. Despite these variations in mRNA concentration, B. emersonii α-MPP protein levels do not change significantly during the life cycle of the fungus, as observed in Western blots. Experiments to investigate the submitochondrial localization of B. emersonii α-MPP and β-MPP were also carried out, and the results indicated that both subunits are associated with the mitochondrial inner membrane, possibly as part of the bc1 complex, as described for plants.
Ubiquinol-cytochrome c reductase (bc1 complex) is a component of the eukaryotic respiratory chain, and its function is to catalyze electron transfer from ubiquinol to cytochrome c, which is coupled to the translocation of protons across the mitochondrial inner membrane from the matrix space to the intermembrane space. The mitochondrial bc1 complex was purified from different organisms and contains between 9 and 11 subunits, of which only a few partake directly in electron transport (reviewed in reference 47). The largest subunits devoid of redox centers are the proteins core I and core II, and they have therefore long been assumed to play structural rather than functional roles. Stable mitochondrial bc1 complex cannot be obtained in the absence of the core proteins (27, 35, 48). Structural studies of the bc1 complex from Saccharomyces cerevisiae (10) and Neurospora crassa (22) by electron microscopy of membrane crystals presented strong evidence that the subunits core I and core II are peripherally located in the mitochondrial inner membrane. Recently, Xia et al. (53) determined the crystal structure of the cytochrome bc1 complex from bovine heart mitochondria, on the basis of X-ray diffraction data, and confirmed that more than half of the total molecular mass of the bc1 complex (including the two core proteins) extends 75 Å from the membrane-spanning region into the mitochondrial matrix region.
The proteins core I and core II exhibit sequence similarity and for this reason are considered to have a common phylogenetic origin. On the other hand, the core proteins also show sequence similarity to the two subunits of the general mitochondrial processing peptidase (MPP), which cleaves off the N-terminal targeting signal sequences of nucleus-encoded mitochondrial proteins upon their import into the organelle (6). The MPP is a heterodimeric, metal-dependent endopeptidase, which removes the presequences of mitochondrial precursors in a single step and whose subunit components are designated α-MPP and β-MPP. In S. cerevisiae and mammals, the processing activity is found soluble in the mitochondrial matrix, whereas in plants the MPP is membrane bound and is an integral part of the bc1 complex. The two nonidentical subunits of MPP replace the core I and core II proteins of the complex (5, 7, 17). The filamentous fungus N. crassa represents an intermediate situation, since the β-MPP replaces the core I protein of the bc1 complex and is simultaneously found soluble in the matrix, removing presequences in association with the α-MPP (43). Based on these findings, Braun and Schmitz (5) proposed that the core proteins of the bc1 complex are relics of an ancient processing activity which was originally integrated into the complex.
The chytridiomycete Blastocladiella emersonii is an aquatic fungus notable for the morphogenetic processes which occur during its life cycle. During sporulation, each multinucleated coenocytic vegetative cell gives rise to several small uninucleated motile cells, lacking a cell wall, called zoospores. In the presence of a nutrient-rich substrate, the zoospores germinate, generating new vegetative cells containing a cell wall of chitin (28). The zoospore presents a single giant mitochondrion, which is fragmented into several normal-sized mitochondria during germination in a process which is independent of protein synthesis (8). During sporulation, these multiple individual mitochondria fuse, giving rise to the huge single mitochondrion found in the zoospore (26).
The purpose of this work was to isolate and characterize the gene encoding α-MPP and study its expression at the mRNA and protein levels throughout the life cycle of the fungus and to investigate possible variations during the drastic morphological changes of mitochondria in this organism. Furthermore, considering that B. emersonii is a chytridiomycete, which represents the earliest-diverging lineage between plants and fungi (49, 52), the submitochondrial localization of the α-MPP and β-MPP was investigated to gather further information on the possible evolutionary relationship between the MPP and the core I and core II subunits of the bc1 complex.
MATERIALS AND METHODS
Cloning of the α-MPP gene.
Two degenerate oligonucleotide primers were designed (sense, 5′-TCGAATTCGGBGGBCCBGGBAARGGBATG-3′; antisense, 5′-TCAAGCTTCKVSWYTCSARRTTCAT-3′) based on two highly conserved amino acid sequences of the α-MPP from N. crassa, S. cerevisiae, rat, and potato (7, 18, 21, 23), according to the B. emersonii codon preference, and used to amplify genomic DNA by PCR (32). DNA was amplified with Taq DNA polymerase on a Gene Amp PCR system 2400 (Perkin-Elmer) with the following settings: 35 cycles of 1 min at 95°C, 2 min at 50°C, and 2 min at 72°C, followed by one 6-min extension step at 72°C. A 296-bp fragment was amplified and cloned into pUCBM20 (Boehringer Mannheim) and M13mp19 by using the EcoRI and HindIII restriction sites present in the oligonucleotides (shown in boldface type).
To isolate the complete α-MPP gene, a partial genomic library was constructed in the vector pUCBM21 (Boehringer Mannheim), with B. emersonii DNA fragments obtained from the region of an agarose gel that hybridized with the probe obtained by PCR. The library contained DNA fragments of 4 to 6 kb, obtained from a digestion with SacI, and was analyzed by colony hybridization under high-stringency conditions with a probe consisting of the PCR fragment labeled with 32P by random-primed synthesis (14). The nitrocellulose filters were prehybridized for 2 h at 37°C in 60 mM potassium phosphate (pH 6.2) containing 3× SSC (45 mM sodium citrate, 450 mM NaCl), 10 mM EDTA, 0.2% sodium dodecyl sulfate (SDS), 50% formamide, and 5% nonfat dried milk. Hybridization was performed overnight at 37°C in the same solution after addition of the denatured probe (106 cpm/ml). The filters were sequentially washed at 37°C in 2× SSC–0.1% SDS, 1× SSC–0.1% SDS, 0.5× SSC–0.1% SDS, and 0.1× SSC–0.1% SDS for 30 min each. The filters were air dried and exposed to Kodak X-Omat film with an enhancing screen at −80°C.
DNA sequence analysis.
The 4.7-kb SacI fragment containing the α-MPP gene was subjected to endonuclease restriction analysis, and several restriction fragments were subcloned into M13mp18 and M13mp19 (Bethesda Research Laboratories) for DNA sequence analysis in both strands. The nucleotide sequence was obtained by the dideoxynucleotide chain termination method with the Sequenase DNA-sequencing kit (Amersham). Analysis of sequence data and sequence comparisons were performed with programs devised by Pearson and Lipman (36).
Primer extension mapping of the transcription start site.
An 18-nucleotide (nt) primer, complementary to nt +146 to +163 of the α-MPP coding region, was 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase and hybridized to 50 μg of total B. emersonii RNA isolated from vegetative cells, zoospores, or cells that had germinated for 90 min. The annealing reaction was carried out in 25 μl of 100 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) buffer (pH 7.0)–1 M NaCl–5 mM EDTA at 52°C for 16 h. The nucleic acids were ethanol precipitated and resuspended in 49 μl of 50 mM Tris-HCl buffer (pH 8.3)–5 mM MgCl2–40 mM KCl–2 mM dithiothreitol–0.2 mM (each) dATP, dCTP, dTTP, and dGTP–40 U of RNase inhibitor (Boehringer Mannheim). The annealed primer was extended at 42°C for 90 min with 25 U of avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim). RNA was digested for 30 min at 37°C by the addition of 1 μg of RNase A (Sigma), and the extended products were analyzed by polyacrylamide gel electrophoresis (PAGE) (7 M urea−7.5% polyacrylamide) followed by autoradiography. The fragments were sized by comparison to a dideoxy sequencing ladder of the M13mp18 clone containing the 5′ region of the α-MPP gene, using the same 18-nt oligonucleotide as the primer.
Preparation of antigen and immunization.
The 296-bp PCR-amplified fragment, encoding the middle region (15 kDa) of α-MPP from B. emersonii and containing the restriction sites EcoRI (5′) and HindIII (3′), was cloned in frame into EcoRI-HindIII-digested pET21-a vector (Novagen) under the control of the T7 RNA polymerase. Overnight cultures of BL21DE3 cells transformed with the pET21-a–α-MPP plasmid were diluted 1:200 into 2× TY medium (29) supplemented with 0.4% glycerol and ampicillin (100 μg/ml) and grown at 37°C to an optical density at 600 nm of 0.4. Expression of the fusion protein was induced by addition of isopropyl-1-thio-β-d-galactopyranoside (IPTG) to a final concentration of 0.4 mM, and the incubation was continued up to 3 h. Cells were harvested by centrifugation at 4°C for 10 min at 5,000 × g, and the cell pellets were frozen and stored at −20°C. A bacterial lysate was prepared by thawing and resuspending the cells in 10 mM Tris-HCl (pH 7.0) containing 100 mM NaCl, 1 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride (PMSF) and 50 μM antipain (Sigma). The suspension was sonicated on ice with a Branson sonicator. After centrifugation for 10 min at 5,000 rpm (in a GSA rotor [Sorvall]), the pellet containing most of the α-MPP fusion protein was resuspended in buffer A (50 mM Tris-HCl [pH 8.0], 50 mM NaCl, 5% glycerol, 1 mM PMSF) containing 2% deoxycholate and the suspension was incubated for 10 min at room temperature. After centrifugation at 12,000 × g for 15 min, the pellet obtained was washed twice with buffer A containing 2% deoxycholate. The resulting pellet was then resuspended in buffer A containing 0.3% sodium N-lauroyl sarcosinate (SLS) and left for 40 min at room temperature. The suspension was then centrifuged at 12,000 × g for 10 min at 4°C. The fusion protein was soluble in the supernatant containing 0.3% SLS, and analysis by SDS-PAGE (25) showed that it was 90% pure. One female rabbit was then immunized with approximately 200 μg of the purified α-MPP fusion protein in buffer A containing 0.3% SLS and 0.5 ml of Freund’s complete adjuvant. After 4 weeks, the rabbit received a second injection containing 200 μg of the antigen in Freund’s incomplete adjuvant. Eight days after the second injection, the rabbit was bled from the ear, and the antiserum obtained was tested in Western blots.
Western blot analysis.
Synchronized cells from different stages of Blastocladiella life cycle were isolated as previously described (30). Cell extracts were obtained by the procedure outlined by Silva et al. (44), and proteins were resolved by SDS-PAGE (25) and then transferred to nitrocellulose membranes, as described by Towbin et al. (46). The protein quantification was done both by the Bradford method (4) and by staining the nitrocellulose membrane with Ponceau S, to make sure that equal amounts of protein were present in each lane of the gel. The membranes were analyzed as described previously (2), except for the use of the ECL enhanced-chemiluminescence kit (Amersham).
Carbonate extraction.
Sodium carbonate treatment of membrane fractions was used to solubilize peripherally bound proteins (16). A total of 2 × 108 frozen zoospores were resuspended in 600 μl of cold lysis buffer (100 mM Tris-HCl [pH 7.0] containing 50 mM NaCl, 1 mM β-mercaptoethanol, 1 mM PMSF, and 50 μM antipain), and the suspension was centrifuged for 10 min at 1,000 × g to eliminate unbroken cells. The supernatant was centrifuged at 100,000 × g for 10 min, and the resulting pellet was resuspended in 100 mM Na2CO3 (pH 11.5). The sample was vortexed for 1 min and centrifuged at 100,000 × g for 10 min. All the steps were carried out at 4°C. The final pellet was resuspended in 50 μl of lysis buffer, and the subfractions were analyzed by SDS-PAGE and immunoblotting, as described above.
Northern blot analysis.
Total RNA from synchronized B. emersonii cells, isolated at different stages of the fungal life cycle as described above, was prepared by the method of Maniatis et al. (29) and fractionated by electrophoresis on a 1.5% agarose–2.2 M formaldehyde gel. The resolved RNAs were then blotted to Hybond N+ membranes (Amersham). Before transfer to the membrane, the gel was stained with ethidium bromide to check for the integrity of the RNA. The hybridization probe used was the 296-bp PCR-amplified fragment. As a control, the Northern blot was also hybridized to a 32P-labeled cDNA clone encoding rat glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (15), which was previously shown to be constitutively expressed in B. emersonii (30). The purified fragments were radioactively labeled by random-primed synthesis (14). When the PCR fragment was used as a probe, the filter was hybridized under high-stringency conditions (0.12 M Na2HPO4/NaH2PO4 buffer [pH 7.2]–0.25 M NaCl–7% SDS–1 mM EDTA at 37°C for 16 h). The filter was sequentially washed in 1× SSC–0.1% SDS at 42°C, 0.5× SSC–0.1% SDS at 42°C, and 0.1× SSC–0.1% SDS at 50°C for 30 min each. When rat GAPDH cDNA was used as a probe, the washing procedure was changed to low-stringency conditions, consisting of three sequential washes in 3× SSC–0.1% SDS at 42°C for 30 min each.
Nucleotide sequence accession number.
The nucleotide sequence of the B. emersonii α-MPP gene has been submitted to the GenBank/EMBL data bank under accession no. U90568.
RESULTS
Isolation and sequence analysis of the B. emersonii α-MPP gene.
By using two oligonucleotide primers designed based on highly conserved amino acid segments of the α-MPPs previously described (7, 18, 21, 23) and PCR, a 296-bp B. emersonii genomic fragment was amplified. The PCR product (Fig. 1) was cloned and sequenced, and the deduced amino acid sequence had 57% identity and 72% similarity to the central region of the α-MPP from N. crassa (18). This 296-bp fragment, encoding a portion of the B. emersonii α-MPP, was then 32P labeled by random-primed synthesis and used as a probe to screen a B. emersonii partial genomic library constructed in the pUCBM21 vector, using Blastocladiella DNA fragments isolated from the region of the gel that hybridized in Southern blots to the PCR probe, as described in Materials and Methods. A positive clone presenting a 4.7-kb SacI fragment was isolated (Fig. 1) and subjected to nucleotide sequence analysis. A total of 2,122 bp, encompassing an open reading frame of 1,535 bp, as well as 378 and 209 bp of 5′ and 3′ noncoding regions, respectively, was sequenced. The nucleotide sequence and the deduced amino acid sequence are shown in Fig. 2.
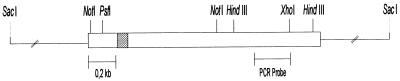
FIG. 1.
Schematic representation of the B. emersonii α-MPP gene. The diagram shows a partial restriction map of the sequenced region (2.2 kb) of the genomic clone SacI-SacI. The open rectangles represent the coding region of the α-MPP gene, the stippled area depicts an intron (112 bp), and the lines represent upstream and downstream regions of the gene. The position of the PCR-amplified fragment, used as a probe to clone the α-MPP gene, is indicated.
FIG. 2.
Nucleotide sequence of the B. emersonii α-MPP gene and the deduced amino acid sequence. Capital letters indicate deoxynucleotides in exons or sequences upstream and downstream of the coding region of the gene; lowercase letters show the deoxynucleotides in the intron. Nucleotide +1 denotes the A of the ATG of the initiator methionine. Residues preceding it are indicated by negative numbers. The deduced amino acid sequence is shown below the nucleotide sequence. The arrows (↓) indicate putative signal sequence cleavage sites. A probable polyadenylation signal is shown in bold.
The predicted B. emersonii α-MPP contains 474 amino acids with a calculated molecular mass of 51.9 kDa, a value comparing very well with the molecular mass of 52 kDa determined by SDS-PAGE. The initiator methionine was chosen based on the presence of an in-frame stop codon (TAA) immediately preceding the ATG. A putative mitochondrial signal sequence, which is rich in basic and hydrophobic amino acids, has been identified (40). Two possible cleavage sites have been observed, one between residues Ser-14 and Thr-15 and another between Leu-20 and Pro-21 (Fig. 2). In both cases an arginine residue (Arg-13 and Arg-19) is present at position −2 relative to the processing site, which is a common theme of such sites (37, 50, 51). The first putative cleavage site seems to be the most probable, since it retains the amino acid Leu-20 in the mature protein, which is a residue conserved in all α-MPPs previously characterized (Fig. 3). B. emersonii α-MPP secondary-structure prediction, using the NNPREDICT program (24, 31), showed a strong tendency for the formation of an α-helix between Ala-5 and Thr-15, followed by an indefinite secondary structure until Glu-73. The presence of an α-helical structure in the amino-terminal region of α-MPP is in agreement with the prediction made for the putative signal peptide from B. emersonii β-MPP (39) and also fulfills the requirements for the MPP to recognize its substrates (40).
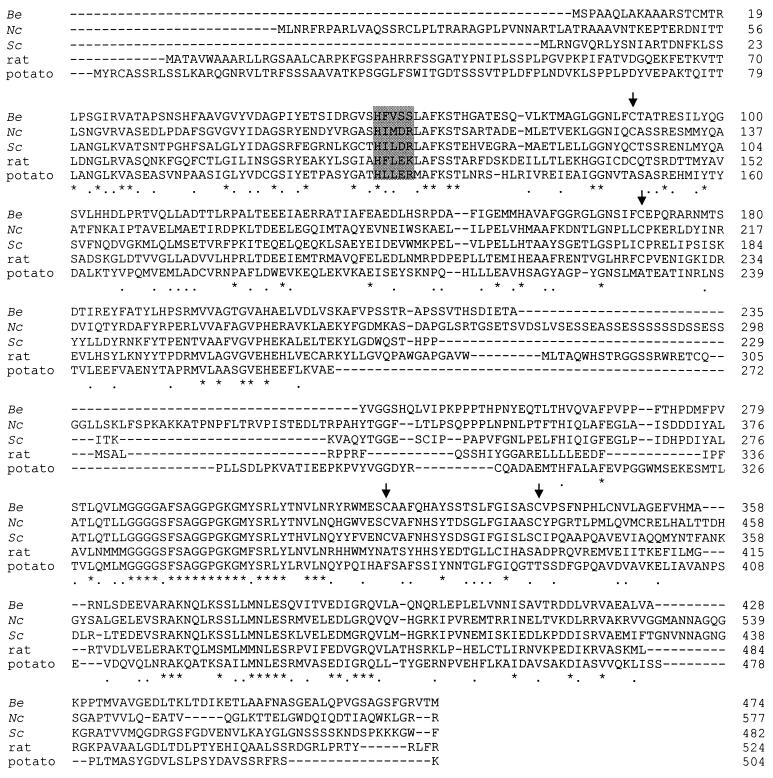
FIG. 3.
Comparison of the α-MPP from different organisms. The amino acid sequence alignment of α-MPP from B. emersonii (Be [this report]), S. cerevisiae (Sc [21]), rat (23), potato (7), and N. crassa (Nc [42]) is shown. Identical amino acids are indicated by asterisks. Conserved residues are indicated by black dots. The four cysteine residues conserved in B. emersonii, S. cerevisiae, and N. crassa are indicated by arrows. The putative metal-binding site motif is delimited by a gray rectangle. Gaps in the alignment are marked by dashes within the sequences. Sequence comparison was performed with the program ClustalV (1).
A single intron, 113 bp in size, is found within the α-MPP coding region, interrupting the highly conserved sequence encoding the amino acid sequence LAFKSTH. The 5′ and 3′ splice sites follow the consensus for B. emersonii introns (9), with a probable branch site (CTGAC) located between nucleotides +271 and +275.
The Blastocladiella α-MPP presents an overall sequence identity of 40.7% to the α-MPP from N. crassa (42), 36.1% to the α-MPP from S. cerevisiae (21, 38), 34.7% to P-55 from rat (34), and 32.6% to subunit III of cytochrome c reductase from Solanum tuberosum (7). It displays the conserved amino acid essential for catalytic activity, the His-58 of the putative metal-binding motif HFLEK (45), and the four cysteine residues (Cys-89, Cys-170, Cys-319, and Cys-338) conserved in all the α-MPPs from fungi (Fig. 3). These cysteine residues could play a functional role in the fungal protein, since they are not conserved in plants and mammals (42, 45). The Blastocladiella putative metal-binding motif presents a serine (Ser-61) in a position where a negatively charged aspartic acid or glutamic acid is usually found (Fig. 3). Nevertheless, Striebel et al. (45) have shown by mutation analysis that replacing the E-112 of rat α-MPP by a glutamine residue, which is its uncharged derivative, improved the performance of MPP beyond wild-type level.
Characterization of the transcription initiation site and analysis of the 5′ noncoding region.
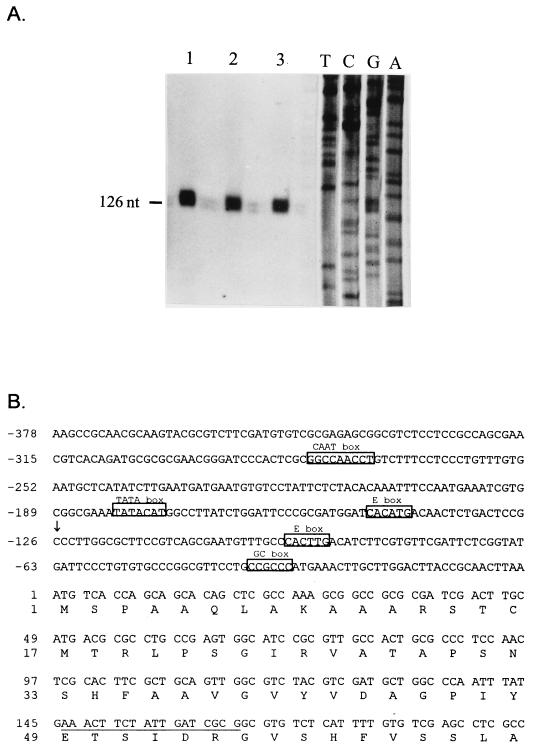
Primer extension experiments were performed to determine the start site of transcription of the Blastocladiella α-MPP gene, and the results are shown in Fig. 4A. A single start site was observed at position −126 from the ATG encoding the initiator methionine. To perform the primer extension assays, an 18-nt primer, complementary to nt +146 to +163 of the coding region (Fig. 4B), was 5′ end labeled with [γ-32P]ATP and hybridized with total RNA isolated from Blastocladiella vegetative cells, zoospores, or germling cells. The hybrids were then extended with avian myeloblastosis virus reverse transcriptase, and the extension products were resolved by PAGE with urea. The fragments were sized by comparison to a dideoxy sequencing ladder of M13mp19 containing the 3.3-kb SacI-XhoI genomic fragment (Fig. 1), with the same oligonucleotide as primer. Detailed examination of the 5′ noncoding region of the Blastocladiella α-MPP gene (Fig. 4B) revealed the presence of some elements usually found in eukaryotic promoters. A putative TATA box was located about 50 nt upstream of the single transcription start site (nt −175 to −181). There was an excellent consensus sequence for a CCAAT box (nt −273 to −281). One copy of the hexanucleotide CCGCCC was also observed (nt −32 to −37). This hexanucleotide is identical to the core sequence recognized by mammalian transcription factor Sp1 (11). Two copies of the consensus core sequence for helix-loop-helix (CANNTG) transcription factor-binding sites (33) were found at positions −90 to −95 and −142 to −147.
FIG. 4.
Transcription start site of the α-MPP gene. (A) Primer extension mapping of the transcription start site. An 18-nt primer, complementary to nt +146 to +163, was 5′ end labeled with [γ-32P]ATP and hybridized to 50 μg of total RNA from B. emersonii vegetative cells (lane 1), zoospores (lane 2), and germling cells (lane 3). The hybrids were then extended with reverse transcriptase, and the extension products were resolved by denaturing gel electrophoresis and autoradiography. The sequencing ladder was generated with the same 18-nt oligonucleotide as a primer and M13mp19 containing the 5′ end of the α-MPP gene (coding strand). (B) Nucleotide sequence of the 5′ region of the α-MPP gene. Nucleotide +1 denotes the A of the ATG encoding the initiator methionine. The underlined region is complementary to the oligonucleotide used for the primer extension experiment. The predicted transcription start point is indicated by an arrow. The putative core sequences representing the binding sites for the TATA-binding protein (TATA box), Sp1 (GC box), CTF/NF1 (CCAAT box), and helix-loop-helix transcription factor (E box) are indicated.
Expression of the α-MPP gene at the mRNA and protein levels.
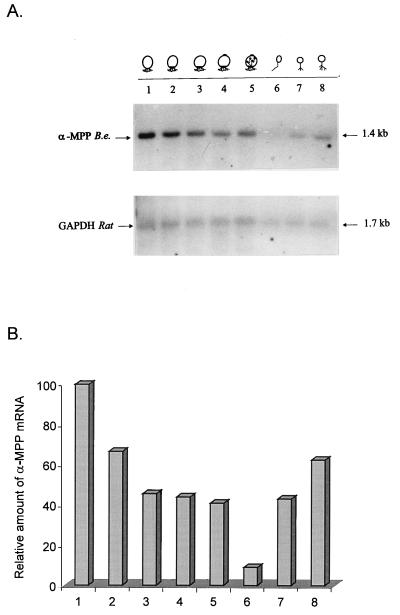
Northern blot analysis was performed to investigate possible changes in the levels of the mRNA encoding the α-MPP during the B. emersonii life cycle. Total RNA isolated from synchronized cells at 0, 60, 90, 120, and 180 min of sporulation, zoospores, or cells at 45 and 90 min of germination was resolved by agarose gel electrophoresis, transferred to Hybond N+ membrane, and probed with the 32P-labeled PCR fragment. A single 1.4-kb transcript encoding the α-MPP, whose levels decreased during sporulation, reaching almost undetectable levels in the zoospore stage, and increased again during germination was observed (Fig. 5). As a control, the same filter was hybridized to a heterologous cDNA probe encoding rat GAPDH, which is constitutively expressed during the life cycle of this fungus (30).
FIG. 5.
α-MPP mRNA levels during B. emersonii development. Total RNA (10 μg/lane) isolated from cells at different stages of the B. emersonii life cycle were subjected to electrophoresis in a formaldehyde-agarose gel, transferred to Hybond N+ membrane, and probed with 32P-labeled PCR-amplified fragment. Lanes: 1 to 5, sporulating cells 0, 60, 90, 120, and 180 min after starvation, respectively; 6, zoospores; 7 and 8, germinating cells 45 and 90 min after inoculation in DM3 medium. As a control, the same blot was hybridized to a 32P-labeled rat GAPDH cDNA. (A) Autoradiograms of the blot. (B) Relative amount of α-MPP mRNA, determined by scanning of the autoradiograms, normalized with respect to the GAPDH signal.
To investigate the levels of the α-MPP protein throughout the B. emersonii life cycle, a specific polyclonal antiserum was obtained from a rabbit immunized with a fusion protein, corresponding to about 15 kDa of the central portion of B. emersonii α-MPP, fused to a histidine tag at the NH2 terminus. Total extracts from synchronized cells, isolated at different times during the B. emersonii developmental cycle, were subjected to Western blot analysis with the α-MPP-specific antiserum. A single 52-kDa band, whose levels did not change significantly during the life cycle of the fungus, was recognized by the antiserum (data not shown). Thus, even though α-MPP mRNA levels vary during B. emersonii life cycle, the amount of the corresponding protein does not change appreciably.
Subcellular localization of α-MPP and β-MPP in B. emersonii.
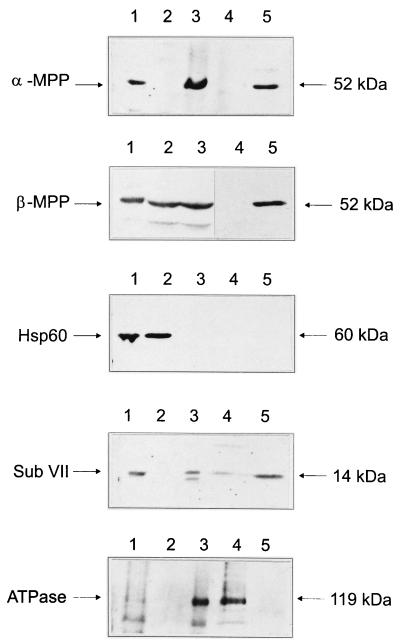
To determine the subcellular localization of B. emersonii α-MPP and β-MPP, total zoospore extracts were fractionated by ultracentrifugation (100,000 × g for 10 min). The soluble fraction was saved, and the particulate fraction was subjected to alkaline treatment with Na2CO3. This procedure, which has been used to investigate the topological organization of the components of S. cerevisiae cytochrome bc1 complex (3), discriminates between peripheral and integral membrane proteins, solubilizing the former ones. In S. cerevisiae, this treatment led to the complete solubilization of the core I and core II proteins, whereas subunits VII and VIII were not extracted from the mitochondrial membrane.
The different fractions obtained from zoospore extracts were analyzed by Western blotting with antisera against B. emersonii α-MPP and β-MPP, anti-Hsp60 antiserum (Sigma), antiserum against subunit VII of the cytochrome bc1 complex of S. cerevisiae, and antiserum against ATPase of B. emersonii. The results, shown in Fig. 6, indicate that α-MPP is completely associated with the insoluble fraction of the extract but can be solubilized by the sodium carbonate treatment. β-MPP was associated partially with the insoluble fraction (60%) and partially with the soluble fraction (40%); it could be totally solubilized by the sodium carbonate treatment. The Hsp60, which in all organisms studied is localized in the mitochondrial matrix, has been detected only in the soluble fraction. This last result indicates that the giant mitochondria of the zoospores were disrupted during cell extract preparation. The B. emersonii P-type ATPase, which is an integral membrane protein, was shown to be totally associated with the particulate fraction and was not solubilized after the alkaline treatment with Na2CO3.
FIG. 6.
Subcellular localization of α-MPP from B. emersonii. Total zoospore extract (lanes 1) was centrifuged at 100,000 × g, resulting in a soluble fraction (lanes 2) and a particulate fraction (lanes 3). The particulate fraction was then subjected to alkaline treatment with 0.1 M Na2CO3, and the suspension was centrifuged at 100,000 × g for 10 min, giving rise to an insoluble fraction (lanes 4) and a soluble fraction (lanes 5). Equal amounts of protein of each fraction (10 μg/lane) were analyzed by SDS-PAGE and Western blotting with anti-B. emersonii α-MPP antiserum, anti-B. emersonii β-MPP antiserum, anti-B. emersonii P-type ATPase antiserum, anti-HSP60 antiserum (Sigma), and antiserum against subunit VII (Sub VII) of S. cerevisiae cytochrome c reductase complex. The blots were developed with the ECL kit (Amersham). The molecular masses (kDa) of the recognized polypeptides are indicated.
The antiserum against the subunit VII of cytochrome bc1 complex cross-reacted with a B. emersonii 14-kDa polypeptide, in agreement with the expected molecular mass for this conserved subunit (20). The putative B. emersonii subunit VII was observed to be completely associated with the insoluble fraction of the zoospore extract and was only partially solubilized after the alkaline treatment (Fig. 6). This result is consistent with the localization of this subunit inside the bc1 complex, whose three-dimensional structure has been determined both by electron microscopy of membrane crystals and X-ray crystallography of the purified complex from N. crassa (22) and bovine heart (53), respectively.
DISCUSSION
The B. emersonii α-MPP gene encodes a precursor protein encompassing 474 amino acids, with a calculated molecular mass of 51,000 Da. The ATG assigned as encoding the initiator methionine is preceded by an in-frame stop codon (TAA). A putative mitochondrial signal sequence has been identified, rich in basic and hydroxylated amino acids and with the potential to form an amphiphilic α-helix. The coding region is interrupted by a single intron, 113 bp in size, located in a conserved α-MPP domain (encoding LAFKSTH) present in all organisms. The conservation of this intron in other α-MPP genes cannot be analyzed since a genomic clone was reported only in S. cerevisiae and no introns were present; in all other cases, just the cDNA was characterized.
Primer extension analysis has indicated a single transcription start site for the α-MPP gene, located at position −126 relative to the ATG. The sequence upstream of the transcription initiation site contains several elements characteristic of eukaryotic promoters such as putative TATA and CCAAT boxes. This observation is in contrast to what was determined for the B. emersonii β-MPP gene, where multiple transcription start sites were found and no sequences resembling a TATA box or a CCAAT box were observed in the 5′ regulatory region of the gene (39). However, possible binding sites for the Sp1 and helix-loop-helix transcription factors were identified in both α-MPP and β-MPP promoter regions.
Northern blot analysis revealed a single 1.4-kb transcript encoding α-MPP, whose levels decrease during sporulation to become almost undetectable at the zoospore stage and increase again during germination. The disappearance of the α-MPP mRNA during the zoospore stage suggests a rapid turnover of this RNA. Transcription and translation are virtually absent in zoospores (28), which might make mRNAs more susceptible to RNases. The presence of AU-rich elements at the 3′ ends of mRNAs has been associated with short half-lives (41), but such sequence elements were not observed in the α-MPP mRNA.
We have previously shown that the β-MPP mRNA, which encodes the β-subunit of the B. emersonii MPP, unlike the α-MPP mRNA, does not vary significantly during the fungal life cycle and is present during all stages of development (39). These results suggest that regulation of expression of α-MPP and β-MPP genes is distinct, which is in agreement with their different 5′ regulatory regions. Nevertheless, α-MPP and β-MPP protein levels do not change significantly during the B. emersonii life cycle, which indicates that the drastic morphological changes in the mitochondria in this fungus do not lead to functional alterations, at least in relation to the processing of mitochondrial presequences.
The analysis of α-MPP and β-MPP secondary structure, using the TMBASE program (19), has indicated that α-MPP presents two domains (amino acids 276 to 294 and amino acids 325 to 347) with a high score for the formation of a transmembrane helix, whereas β-MPP does not present sequences with such characteristics. These structural predictions agree well with the data obtained in the subcellular localization experiments. The fact that B. emersonii α-MPP, like the putative subunit VII of the bc1 complex, is found only in the particulate fraction and that the former is completely solubilized after sodium carbonate treatment whereas the latter is also partially solubilized suggests that α-MPP is associated with the mitochondrial inner membrane like subunit VII but less strongly. On the other hand, 60% of B. emersonii β-MPP has been found in the particulate fraction and was solubilized by the sodium carbonate treatment. These results, in combination, indicate that α-MPP is peripherally associated with the mitochondrial inner membrane whereas the β-MPP is probably anchored to the α-subunit.
The localization of B. emersonii MPP in the inner mitochondrial membrane differs from what was observed in other fungi such as N. crassa or S. cerevisiae. In S. cerevisiae, both protease subunits are found soluble in the matrix, whereas in N. crassa, even though β-MPP is found partially associated with the membrane fraction and partially soluble in the matrix, α-MPP is present only in the mitochondrial matrix.
B. emersonii MPP localization instead resembles the situation observed in plants, where both protease subunits are integrated in the bc1 complex, replacing the core I and core II proteins (12, 13). According to Braun and Schmitz (5), who proposed that the core proteins of the bc1 complex are evolutionary relics of a processing protease, plants exemplify the original situation. Our results suggest that B. emersonii MPP could, as in plants, be a bifunctional protein, playing a structural role in the assembly of the bc1 complex besides its function as the MPP. This hypothesis is strengthened by the fact that Southern blot analysis presented no evidence of other genes similar to the α-MPP and β-MPP genes in B. emersonii (results not shown) and by the position of this fungus at the base of the fungal phylogenetic tree, right where the branching of fungi, animals, and plants occurred (49).
Irrefutable confirmation that the processing activity is integrated into the bc1 complex in B. emersonii will be possible only with the demonstration that the isolated bc1 complex catalyzes the processing of mitochondrial signal sequences; this study is presently under way.
ACKNOWLEDGMENTS
We thank M. V. Marques for stimulating discussions, F. G. Nóbrega for the kind gift of the antiserum against the S. cerevisiae subunit VII of the cytochrome bc1 complex, L. G. Fietto for the antiserum against B. emersonii P-type ATPase, and Elisety de Andrade Silva for manuscript preparation.
This work was supported by grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq-PADCT). C.R.C.R. was a predoctoral fellow of CNPq, and S.L.G. was partially supported by CNPq.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Avedissian M, Gomes S L. Expression of the groESL operon is cell cycle controlled in Caulobacter crescentus. Mol Microbiol. 1996;19:79–89. doi: 10.1046/j.1365-2958.1996.347879.x. [DOI] [PubMed] [Google Scholar]
- 3.Boumans H, Berden J A, Grivell L A. Topological organization of subunits VII and VIII in the ubiquinol-cytochrome c oxidoreductase of Saccharomyces cerevisiae. FEBS Lett. 1996;390:137–141. doi: 10.1016/0014-5793(96)00642-4. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–251. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 5.Braun H P, Schmitz U K. Are the “core” proteins of the mitochondrial bc1 complex evolutionary relics of a processing protease? Trends Biochem Sci. 1995;20:171–175. doi: 10.1016/s0968-0004(00)88999-9. [DOI] [PubMed] [Google Scholar]
- 6.Braun H P, Schmitz U K. Purification and sequencing of cytochrome b from potato reveals methionine cleavage of a mitochondrially encoded protein. FEBS Lett. 1993;316:128–132. doi: 10.1016/0014-5793(93)81200-j. [DOI] [PubMed] [Google Scholar]
- 7.Braun H P, Emmermann M, Kruft V, Schmitz U. The general mitochondrial processing peptidase from potato is an integral part of cytochrome c reductase of the respiratory chain. EMBO J. 1992;11:3219–3227. doi: 10.1002/j.1460-2075.1992.tb05399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bromberg R. Mitochondrial fragmentation during germination in Blastocladiella emersonii. Dev Biol. 1974;36:187–194. doi: 10.1016/0012-1606(74)90201-2. [DOI] [PubMed] [Google Scholar]
- 9.de Oliveira J C F, Borges A C C, Marques M V, Gomes S L. Cloning and characterization of the gene for the catalytic subunit of cAMP-dependent protein kinase in the aquatic fungus Blastocladiella emersonii. Eur J Biochem. 1994;219:555–562. doi: 10.1111/j.1432-1033.1994.tb19971.x. [DOI] [PubMed] [Google Scholar]
- 10.de Vries S, Marres A M. The mitochondrial respiratory chain of yeast. Structure and biosynthesis and the role in cellular metabolism. Biochim Biophys Acta. 1987;895:205–239. doi: 10.1016/s0304-4173(87)80003-4. [DOI] [PubMed] [Google Scholar]
- 11.Dynan W S, Sazer S, Tjian R, Ji T H. Transcription factor Sp1 recognizes a DNA sequence in the mouse dihydrofolate reductase promoter. Nature. 1986;319:246–248. doi: 10.1038/319246a0. [DOI] [PubMed] [Google Scholar]
- 12.Emmermann M, Braun H-P, Arretz M, Schmitz U K. Characterization of the bifunctional cytochrome c reductase-processing peptidase complex from potato mitochondria. J Biol Chem. 1993;268:18936–18942. [PubMed] [Google Scholar]
- 13.Eriksson A C, Sjoling S, Glasser E. Characterization of the bifunctional mitochondrial processing peptidase (MPP)/bc1 complex in Spinacia oleacea. J Bioenerg Biomembr. 1996;28:285–292. doi: 10.1007/BF02110702. [DOI] [PubMed] [Google Scholar]
- 14.Feinberg A P, Volgelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 15.Fort P, Marty L, Piechiczyk M, El Sabrout S, Dani C, Jeanteur P, Blanchard J M. Various rat adult tissues express only major mRNA species from the glyceraldehyde-3-phosphate-dehydrogenase multigenic family. Nucleic Acids Res. 1985;13:1431–1442. doi: 10.1093/nar/13.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujiki Y, Fowler S, Shio H, Hubbard A L, Lazarow P B. Polypeptide and phospholipid composition of the membrane of rat liver peroxisomes: comparison with endoplasmic reticulum and mitochondrial membranes. J Cell Biol. 1982;93:103–110. doi: 10.1083/jcb.93.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glaser E, Eriksson A, Sjoling S. Bifunctional role of the bc1 complex in plants mitochondrial bc1 complex catalyses both electron transport and protein processing. FEBS Lett. 1994;346:83–87. doi: 10.1016/0014-5793(94)00312-2. [DOI] [PubMed] [Google Scholar]
- 18.Hawlitscheck G, Schneider H, Schmidt B, Tropschug M, Hartl F U, Neupert W. Mitochondrial protein import: identification of processing peptidase and of PEP, a processing enhancing protein. Cell. 1988;53:795–806. doi: 10.1016/0092-8674(88)90096-7. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann K, Stoffel W. TMBASE - a database of membrane spanning protein segments. Biol Chem. 1993;374:166. [Google Scholar]
- 20.Japa S, Zhu Q-S, Beattie D S. Subunit VII, the ubiquinone-binding protein, of the cytochrome bc1 complex of yeast mitochondria is involved in electron transport at center o and faces the matrix side of the membrane. J Biol Chem. 1987;262:5441–5444. [PubMed] [Google Scholar]
- 21.Jensen R E, Yaffe M P. Import of proteins into yeast mitochondria: the nuclear MAS2 gene encodes a component of the processing protease that is homologous to the MAS1-encoded subunit. EMBO J. 1988;7:3863–3871. doi: 10.1002/j.1460-2075.1988.tb03272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karlsson B, Hovmoller S, Weiss H, Leonard K. Structural studies of cytochrome reductase—subunit topography determined by electron microscopy of membrane crystals of a subcomplex. J Mol Biol. 1983;165:287–302. doi: 10.1016/s0022-2836(83)80258-7. [DOI] [PubMed] [Google Scholar]
- 23.Kleiber J, Kalousek F, Swaroop M, Rosenberg L E. The general mitochondrial matrix processing protease from rat liver: structural characterization of the catalytic subunit. Proc Natl Acad Sci USA. 1990;87:7978–7982. doi: 10.1073/pnas.87.20.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kneller D G, Cohen F E, Langridge R. Improvements in protein secondary structure prediction by an enhanced neural network. J Mol Biol. 1990;214:171–182. doi: 10.1016/0022-2836(90)90154-E. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;226:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Lessie P E, Lovett J S. Ultrastructural changes during sporangium formation and zoospore differentiation in Blastocladiella emersonii. Am J Bot. 1968;55:220–236. [PubMed] [Google Scholar]
- 27.Linke P, Weiss H. Reconstitution of ubiquinol-cytochrome c reductase from Neurospora mitochondria with regard to subunits I and II. Methods Enzymol. 1984;126:201–211. doi: 10.1016/s0076-6879(86)26022-x. [DOI] [PubMed] [Google Scholar]
- 28.Lovett J S. Growth and differentiation of the water mold Blastocladiella emersonii: cytodifferentiation and the role of ribonucleic acid and protein synthesis. Bacteriol Rev. 1975;39:345–404. doi: 10.1128/br.39.4.345-404.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 30.Marques M V, Borges A C C, de Oliveira J C F, Gomes S L. Coordinate pretranslational control of cAMP-dependent protein kinase subunit expression during development in the water mold Blastocladiella emersonii. Dev Biol. 1992;149:432–439. doi: 10.1016/0012-1606(92)90297-t. [DOI] [PubMed] [Google Scholar]
- 31.McClelland J L, Rumelhart D E. Explorations in parallel distributed processing. Vol. 3. Cambridge, Mass: MIT Press; 1988. pp. 318–362. [Google Scholar]
- 32.Mullis K B, Faloona F A. Specific synthesis of DNA in vitro via a polymerase-catalysed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 33.Murre C, Schonheber P, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 34.Ou W-J, Ito A, Okasaki H, Omura T. Purification and characterization of a processing protease from rat liver mitochondria. EMBO J. 1989;8:2605–2612. doi: 10.1002/j.1460-2075.1989.tb08400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oudshoorn P, Van Steeg H, Swinkeels B W, Schoppink P, Grivell L A. Subunit II of yeast QH2:cytochrome c oxidoreductase. Nucleotide sequence of the gene and features of the protein. Eur J Biochem. 1987;163:97–103. doi: 10.1111/j.1432-1033.1987.tb10741.x. [DOI] [PubMed] [Google Scholar]
- 36.Pearson W R, Lipman D J. Improved tolls for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfanner N, Neupert W. The mitochondrial protein import apparatus. Annu Rev Biochem. 1990;59:331–353. doi: 10.1146/annurev.bi.59.070190.001555. [DOI] [PubMed] [Google Scholar]
- 38.Pollock R A, Hartl F-U, Cheng M Y, Ostermann J, Horwich A, Neupert W. The processing peptidase of yeast mitochondria: the two cooperating components MPP and PEP are structurally related. EMBO J. 1988;7:3493–3500. doi: 10.1002/j.1460-2075.1988.tb03225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rocha C R C, Gomes S L. Isolation, characterization, and expression of the gene encoding the β-subunit of the mitochondrial processing peptidase from Blastocladiella emersonii. J Bacteriol. 1998;180:3967–3972. doi: 10.1128/jb.180.15.3967-3972.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roise D, Schatz G. Mitochondrial presequences. J Biol Chem. 1988;263:4509–4511. [PubMed] [Google Scholar]
- 41.Ross J. Control of messenger RNA stability in higher eukaryotes. Trends Genet. 1996;12:171–175. doi: 10.1016/0168-9525(96)10016-0. [DOI] [PubMed] [Google Scholar]
- 42.Schneider H, Arretz M, Wachter E, Neupert W. Matrix processing peptidase of mitochondria. J Biol Chem. 1990;265:9881–9887. [PubMed] [Google Scholar]
- 43.Schulte U, Arretz M, Sheneider H, Tropschug M, Wachter E, Neupert W, Weiss H. A family of mitochondrial proteins involved in bioenergetics and biogenesis. Nature. 1989;339:147–149. doi: 10.1038/339147a0. [DOI] [PubMed] [Google Scholar]
- 44.Silva A M, Maia J C C, Juliani M H. Developmental changes in translatable RNA species and protein synthesis during sporulation in the aquatic fungus Blastocladiella emersonii. Cell Differ. 1986;18:263–274. doi: 10.1016/0045-6039(86)90058-8. [DOI] [PubMed] [Google Scholar]
- 45.Striebel H-M, Rysavy P, Adamec J, Spizek J, Kalousek F. Mutational analysis of both subunits from rat mitochondrial processing peptidase. Arch Biochem Biophys. 1996;335:211–218. doi: 10.1006/abbi.1996.0500. [DOI] [PubMed] [Google Scholar]
- 46.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedures and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trumpower B L. Cytochrome bc1 complexes of microorganisms. Microbiol Rev. 1990;54:101–129. doi: 10.1128/mr.54.2.101-129.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tzagoloff A, Myers A M. Genetics of mitochondrial biogenesis. Annu Rev Biochem. 1986;55:249–285. doi: 10.1146/annurev.bi.55.070186.001341. [DOI] [PubMed] [Google Scholar]
- 49.van der Auwera G V D, De Wachter R. Large-subunit rRNA sequence of the chytridiomycete Blastocladiella emersonii, and implications for the evolution of zoosporic fungi. J Mol Evol. 1996;43:476–483. doi: 10.1007/BF02337520. [DOI] [PubMed] [Google Scholar]
- 50.von Heijne G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 1986;5:1335–1342. doi: 10.1002/j.1460-2075.1986.tb04364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Heijne G, Steppuhn J, Hermann R G. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- 52.Wainright P O, Hinkle G, Sogin M L, Stickel S K. Monophyletic origins of the metazoa: an evolutionary link with fungi. Science. 1993;260:340–342. doi: 10.1126/science.8469985. [DOI] [PubMed] [Google Scholar]
- 53.Xia D, Yu C A, Kim H, Xia J-Z, Kachurin A M, Zhang L, Yu L, Deisenhofer J. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science. 1997;277:60–66. doi: 10.1126/science.277.5322.60. [DOI] [PubMed] [Google Scholar]