INTRODUCTION
In order to function properly, proteins typically must fold and assume defined native three-dimensional structures. However, disruption of native protein folding may allow abnormal aggregation into self-propagating assemblies (seeds) that can grow by recruiting, and misfolding, additional monomers. Such protein aggregates can be more toxic 1,2 or more infectious 3when relatively small (oligomeric), but continued growth can result in the accumulation of highly ordered amyloid fibrils, and bundles thereof, that comprise the pathological protein deposits that often characterize protein misfolding diseases 4. Protein quality control (proteostasis) mechanisms usually limit the accumulation of abnormally folded and aggregated proteins 5. However, with aging, pathogenic mutations in specific proteins, inoculation of preformed seeds, or perhaps other factors, protein quality control mechanisms can be overwhelmed, allowing protein aggregation to spiral out of control. The accumulation of protein aggregates may, or may not, have devastating consequences for the host 6.
Nervous tissue, with its lack of neuronal turnover, is particularly vulnerable to the effects of pathological protein aggregation. Indeed, the aggregation of specific proteins is known to feature prominently in the pathogenesis of many neurodegenerative diseases including Alzheimer’s, Parkinson’s and prion diseases 1. In genetic forms of these diseases, mutated proteins appear to be more prone to aggregation, leading to the appearance of neurodegenerative disorders in earlier stages of life. Considering the relative inability of neurons to regenerate, accurate diagnosis in the early stages of these diseases should facilitate effective treatment while neuronal damage is not yet too extensive and/or irreversible. In this context, proteopathic seed amplification assays have provided means of detecting minute amounts [attograms to femtograms (ag-fg)] of self-propagating protein aggregates in diverse biological specimens by exploiting their inherent seeded polymerization growth mechanisms 7–15.
The assembly of proteins into amyloid fibrils is akin to “one dimensional crystallization” 16. As with other crystallizations, the de novo, or spontaneous, formation of seeds in a solution of monomers usually takes much longer than the growth of preexisting seeds. In proteopathic seed amplification assays, a biospecimen is incubated in the presence of a vast stoichiometric excess of soluble protein monomers under appropriate conditions. If the specimen contains seeds, the assembly of monomers into amyloid fibrils occurs more rapidly than the de novo assembly that eventually occurs in the absence of seeds. Thus, seeds can be detected by comparing lag phases, or the reaction times required to detect amyloid that is newly formed from the monomeric “substrate” molecules. This use of “substrate” is analogous to the substrate in an enzymatic reaction rather than to a structure onto which something builds. As the substrate molecules are much more abundant than the seeds in a biospecimen, their conversion into amyloid fibrils can, in effect, provide billion-, or even trillion-fold amplifications of the seed 17,18. Such amplification can enable ultrasensitive detection of even a few seed particles in a test sample. Here we highlight many of these types of assays, with an emphasis on their applications to human proteopathies.
PRION SEED AMPLIFICATION ASSAYS
The most highly developed proteopathic seed amplification assays are those for prions, with current assays allowing highly accurate molecular diagnoses of prion diseases in living patients 15,19–21. The ability of infectious prions to induce the conversion of natively folded prion protein (PrPC) into a prion-like protease-resistant form (PrPres) in a cell-free system was first demonstrated in 1994 22. This experimental system revealed prion strain- and sequence- specificities of this prion-seeded conversion reaction 23–27, but did not support the continuous prion propagation that would be required for an amplification assay.
Protein misfolding cyclic amplification
Several years later Soto and coworkers developed a highly sensitive PrPSc (infectious scrapie prion protein) amplification method termed “protein misfolding cyclic amplification” (PMCA) 7. In this assay, attogram amounts of PrPSc present in a tissue sample can be amplified to detectable levels by its incubation with an excess of non-infected brain homogenate, which provides the PrPC monomers needed for the polymerization process. By interleaving incubation and sonication steps, the newly synthesized misfolded aggregate is intermittently broken, providing more seeds, amplifying PrPres and prion infectivity exponentially. The final product is then subjected to a proteinase K (PK)-treatment and proteinase-resistant fragments are revealed by Western blotting with an anti-PrP antibody. PrPSc can be amplified from a 10−12 dilution of infected hamster brain homogenate containing ~26 PrPSc molecules 17. A key feature of PMCA is that it faithfully replicates the PrPSc structure such that the amplified products are fully infectious 28,29. Thus, PMCA has been invaluable as an in vitro experimental system for studying prion propagation. On the other hand, infectious prion amplification can be a disadvantage for routine diagnostic applications when it would be preferable not to generate large amounts of infectivity. Notwithstanding that practical concern, PMCA has clear diagnostic potential as prions have been amplified from blood samples of experimentally infected animals at late 30 and early 31 stages of the disease, as well as in urine samples from animals 32 and humans with variant Creutzfeldt-Jakob disease (vCJD) 33.
To circumvent the need to use brain homogenates as a source of PrPC substrate, Atarashi et al developed PMCA reaction conditions that allowed the use of bacterially expressed recombinant PrPC as the reaction substrate 8. This assay was called rPrP PMCA and allowed for detection of ag-range amounts of PrPSc in 2–3 days compared to the 2–3 weeks required for conventional PMCA assays at that time. A subsequent rPrP PMCA permutation called quaking induced conversion (QuIC) allowed for the substitution of more reproducible shaking for sonication 12.
Amyloid seeding assay
Meanwhile, Colby and colleagues also developed a highly sensitive assay for PrPSc using recombinant PrPC as a substrate called the amyloid seeding assay (ASA) 9. The ASA, which was also shaken rather than sonicated, had the major practical advantages of having a multiwell plate-based assay format and a direct fluorescence readout. The basic principle was that PrPSc seeds in a sample could induce the formation of recombinant PrP amyloid fibrils which were then detected with the amyloid-sensitive dye thioflavin T (ThT). Frequent fluorescence measurements while the reactions progressed in a shaking fluorescence plate reader allowed convenient measurement of the relative lag phases of reactions seeded with prion-infected (e.g. sporadic CJD or sCJD) versus negative control brain homogenates. However, in contrast to most RT-QuIC assays, a phosphotungstate (PTA)-precipitation step was needed in order to purify the seeds prior to its use in the ASA.
RT-QuIC
By combining various aspects of the QuIC and ASA assays and further optimizations, Atarashi and colleagues developed the first real-time QuIC (RT-QuIC) assays which provided easier distinction of prion-infected and uninfected biospecimens such as cerebrospinal fluid (CSF) 10,11, often without the need of doing a pre-clearing of the sample. Many laboratories have since contributed to the development of assays for most known prions of mammals, e.g.34–36 and references therein. The analytical sensitivities of these assays often reach down in the low fg to ag range and typically meet or markedly exceed the sensitivities of animal bioassays for prions. RT-QuIC assays have been adapted to diverse tissues and biological fluids, including brain, skin, olfactory mucosa, blood, CSF, saliva, urine and feces 18,33,37–45. Faster and more sensitive second-generation RT-QuIC assays for human prions allow for nearly 100% accurate antemortem diagnosis of sCJD when either CSF, nasal brushings, or both are tested 21,41,46. Analysis of CSF specimens alone provide 92–96% overall diagnostic sensitivity (the percentage of sCJD cases giving positive assays) and nearly 100% specificity (the percentage of non-prion disease cases giving negative assays) in multiple independent studies by different research groups 21,40,41,46,47. Prior to the availability of these assays, definite diagnoses of CJD required biochemical or immunohistological analysis of brain tissue, which is usually only available postmortem. According to the US Centers for Disease Control and Prevention (CDC), a positive RT-QuIC result in combination with neuropsychiatric symptoms provides a probable diagnosis of CJD, although immunodetection of PrPres is still needed for a definite diagnosis (https://www.cdc.gov/prions/cjd/diagnostic-criteria.html). Similar diagnostic criteria are used by the UK National CJD Research and Surveillance Unit (https://www.cjd.ed.ac.uk/sites/default/files/criteria_0.pdf). Recent demonstrations of the detection of sCJD prions in the skin 43 and multiple components of the eyes 48 of all tested sCJD cases have raised the possibility that these tissues might also be useful diagnostically, as well as being potentially biohazardous sources of prion infectivity. Indeed, studies in prion-infected rodents have shown that prion seeding activity can be detected in skin far in advance of the onset of overt clinical signs of prion disease 38. Detailed protocols for performing prion RT-QuIC assays have been reported elsewhere 20,49
Scrapie cell assay
Klohn and colleagues 50 described a prototypic cellular seeding assay which exploits the ability of prions to infect cells and induce the de novo accumulation of PrPSc. Briefly, neuroblastoma-derived cell lines are incubated with dilutions of prion-infected brain homogenates from rodents, and the proportion of cells accumulating PrPSc (detected by an ELISPOT after three in vitro passages over ~ 2 weeks total) were found to correlate with the prion concentration in the original sample. The standard scrapie cell assay (SSCA) has been automated 51,52 and is as sensitive as the mouse prion bioassay but is faster and less expensive, with the advantage of no use of animals. There have been many revealing applications of the scrapie cell assay, e.g. 52–54, however, so far the assay has been limited to the measurement of rodent-adapted prion strains. Indeed, no cell line capable of supporting replication of human prion strains has been established and made available for research.
SEED AMPLIFICATION ASSAYS FOR SYNUCLEINOPATHIES
αSyn RT-QuIC and PMCA
To provide molecular diagnoses of Parkinson’s disease (PD) and dementia with Lewy bodies (DLB) and multiple system atrophy (MSA), several laboratories have now developed RT-QuIC-like assays for pathological forms of α-synuclein (αSynD) called αSyn RT-QuIC 55–60 or αSyn-PMCA 61. These assays can have unprecedented sensitivity for αSynD down into the low fg range and detect up to 108-fold dilutions of patients’ brain tissue. When applied to CSF specimens collected from living patients, the diagnostic sensitivities for PD and DLB have ranged from 88–96% and specificities from 82–100% 55,57,60–62. Blinded comparisons of the performance of two of these assays on a large set of PD cases and healthy controls revealed a high degree of concordance in diagnostic performance 62. Whereas the earlier assays took 5–13 days, newer permutations take 1–2 d 57 or 3 d 60 with comparable diagnostic performance and the ability to detect αSynD seeds in CSF collected early in the clinical course of PD. Similarly rapid assays have been reported using postmortem submandibular gland tissue from PD and incidental Lewy body disease decedents, giving 100% sensitivity and 94% specificity for synucleinopathy 59. αSynD seeding activity can often be detected in nasal brushings from synucleinopathy patients 63. It is notable that studies have also reported evidence of strain-like differences between types of synucleinopathy-associated seeds 57,58.
Although the multiple initial analyses of αSyn RT-QuIC and related assays have been encouraging, more extensive studies will be required to fully understand their diagnostic and prognostic utilities and the extent to which αSynD seeding activities in accessible biospecimens might vary over time in individual patients. Such information will likely be important in understanding the extent to which the monitoring of αSynD seeding activities might be helpful in setting up and assessing the progress of clinical trials of treatments aimed at reducing αSynD burden in the brain.
HEK cell bioassay for MSA
Woerman and colleagues expressed yellow fluorescent protein-tagged αSyn in human embryonic kidney cells and found that phospshotungstic acid-precipitated extracts of brain homgenates from MSA decedents could be assayed on these cells 4 days after exposure by quantitating cells with fluorescent aggregates 64,65. The assay had an analytical sensitivity of 70 pg/mL, and revealed differences in regional distribution of αSyn seeding activity between MSA patients. Interestingly, this assay was not sensitive to the αSyn seeds of PD cases 64,65. The results provided evidence for the MSA and PD being distinct αSyn prion strains.
Hanabi assay
Recently, an ultrasonication-based assay was described which uses the HANdai Amyloid Burst Inducer (HANABI) system to amplify αSynD 66. Analogous to what is done in classical PMCA, the authors applied ultrasonication to break up αSynD oligomers in order to increase the nuclei from which the polymerization starts. The addition of αSyn pre-formed fibrils in artificial CSF enhanced the reaction speed in a dose-dependent fashion. Although ultrasonication provided faster reactions, it also increased the unseeded polymerization in negative control reactions. The analysis of human CSF from PD and control cases showed overlapping reaction kinetics, complicating the diagnostic sensitivity and specificity of this assay.
SEED AMPLIFICATION ASSAYS FOR TAUOPATHIES
Multiple neurodegenerative diseases involve the pathological accumulation of tau filaments, including AD, chronic traumatic encephalopathy (CTE), progressive supranuclear palsy (PSP), corticobasal degeneration (CBD), frontotemporal dementia and Parkinsonism 17 MAPT (FTDP 17 MAPT), argyrophyllic grain disease (AGD), and Pick disease (PiD). In adult humans, the alternative splicing of the MAPT (microtubule-associated protein tau) gene gives rise to six tau isoforms. These isoforms differ from each other by the presence or absence of inserts near the N-terminus and by the number of microtubule binding domains near the C-terminus [i.e., three (3R) or four (4R) isoforms]. The tau pathologies of different diseases are characterized by the preferential deposition of 3R, 4R or both 3R and 4R tau isoforms. For example, AD and CTE cases accumulate roughly equivalent amounts of both 3R and 4R isoforms because all the isoforms contain the residues comprising the amyloid cores of the paired helical and straight tau filaments of these diseases 67,68. PiD cases accumulate mainly 3R isoforms 69,70,consistent with their compatibility with the distinct amyloid core of PiD filaments 71. In contrast, PSP, CBD, and FTDP 17 MAPT cases preferentially accumulate 4R isoforms, which, in the case of CBD, can again be rationalized by the core structure of the CBD-associated filaments 72.
Tau RT-QuIC assays
Initial studies by Margittai and colleagues showed that in sonicated, multiwell plate-based reactions synthetic fibrils formed with different 3R and 4R tau constructs had distinct seeding activities 73 and also that AD brain homogenates could seed the fibrillization of tau constructs faster than control homogenates 74. Since then, Saijo, Kraus, Metrick, and colleagues have developed ultrasensitive and selective tau RT-QuIC assays for disease-associated 3R, 4R, and 3R+4R tau aggregates, and some subclasses thereof 6,75–78. Tau RT-QuIC performance can be improved, i.e., higher sensitivity can be achieved without compromising specificity, by systematic comparisons of conditions such as salts of the Hofmeister series 77. These tau RT-QuIC assays can detect seeds in 108-1010-fold dilutions of brain tissue, with orders of magnitude of selectivity for the types of tauopathy for which they were optimized. The optimization of each of these assays involved comparisons of various recombinant tau substrate constructs, cofactors and other reaction components and conditions. Use of a 3R tau fragment (K19CFh) as a substrate first allowed the development of the 3R tau RT-QuIC assay which specifically detects PiD seeds in brain tissue and postmortem CSF samples 75. Then, to detect AD and CTE tau filaments, a substrate (τ306) spanning the entire amyloid core of the associated filaments was included in the reaction to allow the amplification of the seeds 6. Seeds of multiple diseases with 4R tauopathy can be detected with the recently developed 4R Tau RT-QuIC 76. This is the first tau RT-QuIC assay to allow detection of tau seeds in antemortem, as well as, postmortem CSF specimens. Finally, we more recently used another tau substrate (K12CFh) to develop a single tau RT-QuIC assay for the detection and discrimination of tau seeds of AD and PiD 78. This assay simplifies the testing for these types of tau seeds and indicates that they differ in conformational templating activity. As with the prion and αSyn RT-QuIC assays, the ability to detect tau seeds in assessible diagnostic specimens will be important for antemortem diagnostic utility of tau RT-QuIC assays. However, at present, further research is necessary to better establish any such utility.
Another valuable feature of the 4R RT-QuIC assay is its ability to differentiate of three classes of 4R tau seeds based on characteristics of the fibrillar reaction products, namely their relative enhancement of ThT fluorescence, and their β-sheet conformations as assessed by Fourier transform infrared (FTIR) spectroscopy: Class A is represented by FTDP-17 with P301L MAPT mutation; Class B includes the FTDP-17 with N279K mutation) and CBD; and Class C includes PSP 76.
In applying tau RT-QuIC assays to biological specimens, it is important to bear in mind that various types of tau aggregates and seeding activities can be found at lower levels (usually by orders of magnitude) in brain tissue, at least, of individuals without primary tauopathies 6,75–78. Thus, the diagnostic specificities of these ultrasensitive assays when applied to biospecimens may depend on quantitative, as well as qualitative, differences in tau seeding activities.
Biosensor cell assays
A highly sensitive and specific biosensor cell seeding assay for tau seeds has also been developed 79,80 by exploiting the ability of tau aggregates to penetrate cells and induce the fibrillization of soluble endogenous tau 81. Briefly, when engineered HEK293T cell lines expressing 3R or 4R tau constructs fused to fluorescent proteins were exposed to exogenous proteopathic tau, these protein aggregates penetrated the cells and seeded the polymerization of intracellular tau 79. The intracellular protein aggregation was detected by Fluorescence Resonance Energy Transfer (FRET), which was measured by fluorescence microscopy or flow cytometry. The exposure of the cells to increasing amounts of recombinant tau fibrils resulted in the gain of FRET signal, in a dose-dependent fashion. Moreover, the incubation of HEK293T cells expressing fluorescent 3R or 4R tau constructs, or both, with specific tauopathy brain lysates (e.g., from AD, CTE, PSP, CBD, AGD) brain lysates triggered the formation of tau inclusions, whereas no tau aggregation was induced by negative controls. This showed that the polymerization was promoted by aggregated tau and not other protein aggregates. Comparison of tau seeds from multiple sources were found to behave like different strains by inducing characteristic intracellular inclusions that could be faithfully propagated through many in vitro passages and then upon inoculation into transgenic mice 82. To compare the sensitivity of this technique to histology (the gold standard postmortem method to diagnose AD 83), a time-course analysis in a tauopathy mouse model was performed. Tau seeding activity preceded histopathological detection by more than four weeks. Although these experimental systems have revealed much about the prion-like propagation and strain-dependent seeding capacity of tau aggregates, the practicality of these cellular models for routine clinical diagnostic purposes in clinical practice may be limited by the need for tissue cultures and either immunostaining or flow cytometry.
AMYLOID-β (Aβ) SEED AGGREGATION ASSAYS
Kinetic aggregation assay
In addition to the aforementioned neurofibrillary tangles of tau, amyloid plaques of the amyloid-β (Aβ) peptide are major lesions found in brains from Alzheimer’s disease patients 84. To detect Aβ aggregates, Kelly and colleagues developed a seeded polymerization-based assay 85. In this “kinetic aggregation assay”, Aβ aggregates from mammalian cell culture media, Caenorhabditis elegans lysates, and AD mouse brain homogenates seeded the fibrillization of monomeric Aβ1–40 peptide, in a cell-free reaction containing ThT to monitor fibril formation over time. The amount of Aβ amyloid fibrils in the sample was proportional to the half time of the growth phase (t50). To prove the specificity of this assay, experiments with αSyn aggregates were also conducted and differences in the kinetic reactions were observed, with reactions seeded with 4.3 μg/mL Aβ1–40 fibrils showing a t50 of ~5h while 14.5 μg/mL α-synuclein fibrils yielded a ~9-h t50. The authors stated that this selectivity is needed to allow the quantification of Aβ amyloid fibrils in tissues which can potentially contain other amyloid seeds; however, human biological specimens from patients with different neurodegenerative disorders were not assessed by this assay in order to prove its specificity.
Aβ-PMCA
Continuing along these lines, Soto and colleagues developed an assay called Aβ-PMCA that detects misfolded Aβ oligomers down to as little as 3 fmol in a multiwell plate-based assay with ThT readout as in RT-QuIC assays 86. Application of Aβ-PMCA to CSF specimens from AD and non-AD cases provided discrimination between the two sets with 90% sensitivity and 92% specificity. As it is well known that cognitively normal or other non-AD cases can have some Aβ deposits in their brain tissue, the specificity of the Aβ-PMCA is presumably dependent upon disease-dependent differences in concentration of Aβ seeds in the CSF. Seed concentration is typically inversely correlated with lag time in seeded protein polymerization reactions, which likely explains the shorter lag phases that were observed with CSF samples from AD cases. This assay promises to provide a useful complement to the AD tau RT-QuIC assay described above in measuring the key pathological protein aggregates of AD.
HUNTINGTON DISEASE SEED AMPLIFICATION ASSAY
The applicability of an amyloid seeding assay of postmortem brain extracts from human Huntington diseases cases, and transgenic mice models thereof, was reported by Gupta and coworkers 87. Misfolded huntingtin partially purified from the diseased brains induced amyloid formation of a largely polyglutamine substrate more rapidly than extracts from negative control brains. Although this assay is inhibited by components of crude brain tissue, hence the need for partial purification of the seeds from brain, it serves as a prototypic test for pathological forms of huntingtin.
CONCLUSIONS
Continuing work by many laboratories is yielding a growing panel of ultrasensitive assays for the various misfolded self-propagating protein aggregates that cause many neurodegenerative diseases. These assays provide important tools for fundamental research into the pathogenesis of these diseases. Moreover, the ability to detect miniscule amounts of such proteopathic aggregates as biomarkers in accessible specimens is allowing more accurate molecular diagnoses of diseases that can otherwise be difficult to discriminate, especially early in pathogenesis while appropriately targeted treatments are more likely to be successful. Importantly, given the frequency with which neurodegenerative disease patients have more than one type of aggregated protein in their CNS, the high sensitivity of these assays allows detection of both primary and secondary proteopathies. Such testing should also facilitate the development of new therapies by allowing clearer identification of cases and controls for clinical trials, as well as the measurement of specific protein seeds as key etiological biomarkers over the course of treatment.
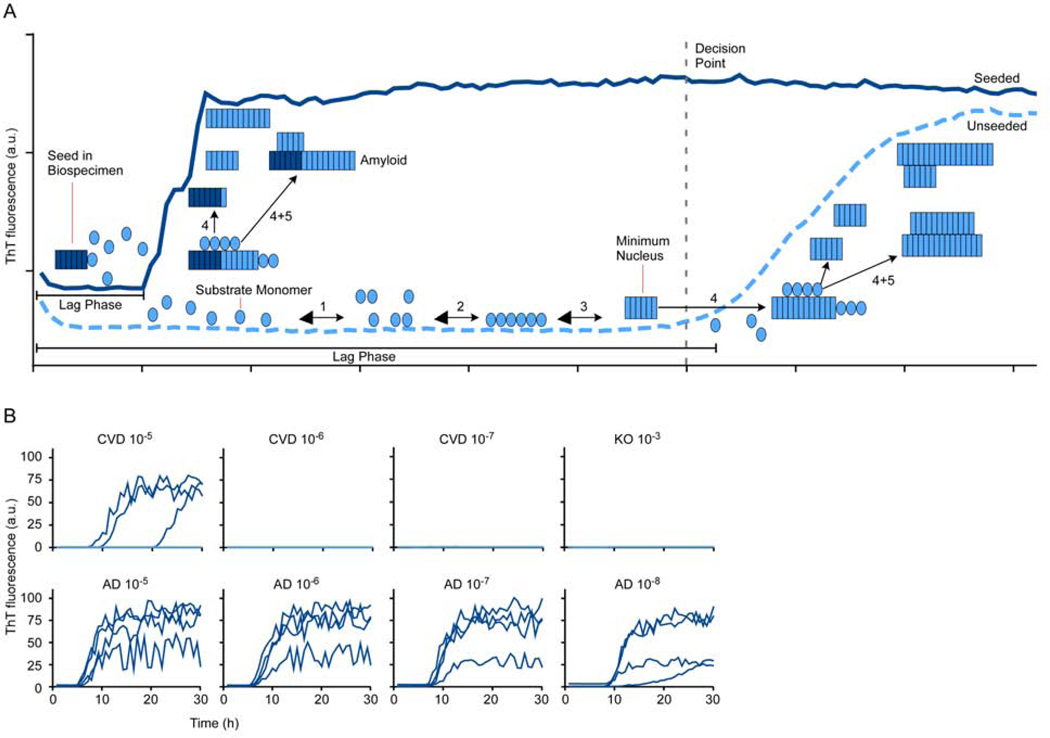
Figure 1. Diagram of seeded polymerization in RT-QuIC reactions.
A. Reactions are set up in multiwell plates by diluting a biospecimen into reaction mixture containing a vast stoichiometric excess of an appropriate protein monomer (substrate) for the type of disease-associated seed being detected. The reaction also contains the amyloid-sensitive fluorescent dye, thioflavin T (ThT). When seeds are present in the biospecimen (left), they immediately start to grow (4) by recruiting new monomers. Elongated fibrils can promote secondary nucleation (5), i.e. the production of new seeding surfaces either by fragmentation or by providing lateral surfaces that can facilitate the ordering and conformational conversion of monomers into additional fibrils. Secondary nucleation contributes to the exponential growth of the new amyloid that enhances ThT fluorescence (dark blue trace). The lag phase in a seeded reaction represents the time it takes for the seeded amyloids to accumulate to levels that are detectable with ThT. Eventually, the reaction plateaus when all of the available monomer is converted to amyloid. In the absence of seeds in a negative control biospecimen (light blue trace), spontaneous nucleation (1–3) may occur, but only after a prolonged lag phase during which the kinetically unfavorable process of forming minimal stable nuclei (in this case a 6-mer) occurs 16. A key to developing an effective assay is finding substrates and assay conditions giving the greatest fold-separation between the lag phases of seeded versus unseeded reactions (e.g. see 77). B. Representative primary AD tau RT-QuIC data comparing seeding by serial 10-fold dilutions of human familial AD (age 44) and cerebrovascular disease (CVD; age 53) brain tissue with reference to a 100 dilution being solid brain tissue. Tau knock-out (KO) mouse brain served as a completely tau-free negative control. Traces from quadruplicate wells for each brain dilution are shown. AD brain could be diluted at least 105-fold and 103-fold further than the KO and CVD brains, respectively, and still seed positive reactions. Although the CVD brain was not recorded as having tau pathology by immunohistochemistry, the typically higher sensitivity of RT-QuIC assays likely allowed detection of tau aggregates that were below the detection limit of immunohistochemistry.
KEY POINTS
Prion-like, self-propagating protein aggregates can cause, and serve as biomarkers, for multiple neurodegenerative diseases.
The seeded polymerization growth mechanism of pathological protein aggregates has been exploited to develop ultrasensitive assays for many of these biomarkers.
Some of these types of assays, e.g. prion and αSyn RT-QuIC assays, can provide highly sensitive and specific diagnoses of prion diseases and α-synucleinopathies when applied to patients’ cerebrospinal fluid or nasal brushings.
Multiple ultrasensitive seed amplification assays have also been developed for many disease-associated types of tau and Aβ aggregates in biospecimens, but these assays are in earlier stages of diagnostic evaluation.
The development of a broad panel of proteopathic seed amplification biomarker assays should facilitate the diagnosis of neurodegenerative diseases as well as the execution of clinical trials of potential therapeutics.
SYNOPSIS
To address the need for etiological biomarkers for neurodegenerative diseases involving protein aggregation, ultrasensitive cellular and cell-free assays have been developed based on the prion-like self-propagating (seeding) capacity of many such aggregates. The most practical and clinically validated of such assays so far are the prion RT-QuIC assays that allow nearly 100% accurate antemortem diagnosis of sporadic Creutzfeldt-Jakob disease using patients’ cerebrospinal fluid or nasal brushings. Analogous assays for synucleinopathies such as Parkinson disease and dementia with Lewy bodies are providing unprecedented diagnostic sensitivity using cerebrospinal fluid. Tau RT-QuIC assays can detect and discriminate different types of tau aggregates associated with Alzheimer disease and other tauopathies. Related assays have also been reported for disease-associated huntingtin and amyloid β aggregates, with the latter being applicable to Alzheimer’s cerebrospinal fluid. The development of a broad panel of seed amplification assays should improve diagnostics as well as the evaluation of potential therapeutics.
ACKNOWLEDGEMENTS
This work was supported by the Intramural Research Program of the NIAID. We thank Anita Mora for graphics assistance.
Footnotes
DISCLOSURE STATEMENT
Byron Caughey is an inventor on patents or patent applications relating to prion, αSyn and tau RT-QuIC assays. Natália do Carmo Ferreira has nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. AnnuRevNeurosci. 2003;26:267–298. [DOI] [PubMed] [Google Scholar]
- 2.Chiti F, Dobson CM. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu Rev Biochem. 2017;109:27–68. [DOI] [PubMed] [Google Scholar]
- 3.Silveira JR, Raymond GJ, Hughson AG, et al. The most infectious prion protein particles. Nature. 2005;437(7056):257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diaz-Villanueva JF, Diaz-Molina R, Garcia-Gonzalez V. Protein Folding and Mechanisms of Proteostasis. Int J Mol Sci. 2015;16(8):17193–17230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. AnnuRevBiochem. 2009;78:959–991. [DOI] [PubMed] [Google Scholar]
- 6.Kraus A, Saijo E, Metrick MAI, et al. Seeding selectivity and ultrasensitive detection of tau aggregate conformers of Alzheimer disease. Acta Neuropathol. 2019;137:585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411(6839):810–813. [DOI] [PubMed] [Google Scholar]
- 8.Atarashi R, Moore RA, Sim VL, et al. Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. NatMethods. 2007;4(8):645–650. [DOI] [PubMed] [Google Scholar]
- 9.Colby DW, Zhang Q, Wang S, et al. Prion detection by an amyloid seeding assay. Proc Natl Acad Sci USA. 2007;104(52):20914–20919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atarashi R, Satoh K, Sano K, et al. Ultrasensitive human prion detection in cerebrospinal fluid by real-time quaking-induced conversion. Nat Med. 2011;17(2):175–178. [DOI] [PubMed] [Google Scholar]
- 11.Wilham JM, Orrú CD, Bessen RA, et al. Rapid End-Point Quantitation of Prion Seeding Activity with Sensitivity Comparable to Bioassays. PLoS Path. 2010;6(12):e1001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atarashi R, Wilham JM, Christensen L, et al. Simplified ultrasensitive prion detection by recombinant PrP conversion with shaking. NatMethods. 2008;5(3):211–212. [DOI] [PubMed] [Google Scholar]
- 13.Barria MA, Gonzalez-Romero D, Soto C. Cyclic amplification of prion protein misfolding. Methods MolBiol. 2012;849:199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caughey B, Orru CD, Groveman BR, et al. Amplified Detection of Prions and Other Amyloids by RT-QuIC in Diagnostics and the Evaluation of Therapeutics and Disinfectants. Prog Mol Biol Transl Sci. 2017;150:375–388. [DOI] [PubMed] [Google Scholar]
- 15.Green AJE, Zanusso G. Prion protein amplification techniques. Handb Clin Neurol. 2018;153:357–370. [DOI] [PubMed] [Google Scholar]
- 16.Jarrett JT, Lansbury PT Jr., Seeding “One-Dimensional Crystallization” of Amyloid: A Pathogenic Mechanism in Alzheimer’s Disease and Scrapie? Cell. 1993;73:1055–1058. [DOI] [PubMed] [Google Scholar]
- 17.Saa P, Castilla J, Soto C. Ultra-efficient replication of infectious prions by automated protein misfolding cyclic amplification. JBiolChem. 2006;281(46):35245–35252. [DOI] [PubMed] [Google Scholar]
- 18.Orru CD, Wilham JM, Raymond LD, et al. Prion disease blood test using immunoprecipitation and improved quaking-induced conversion. mBio. 2011;2(3):e00078–00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zanusso G, Monaco S, Pocchiari M, Caughey B. Advanced tests for early and accurate diagnosis of Creutzfeldt-Jakob disease. Nat Rev Neurol. 2016;12(6):325–333. [DOI] [PubMed] [Google Scholar]
- 20.Schmitz M, Cramm M, Llorens F, et al. The real-time quaking-induced conversion assay for detection of human prion disease and study of other protein misfolding diseases. Nat Protoc. 2016;11(11):2233–2242. [DOI] [PubMed] [Google Scholar]
- 21.Bongianni M, Orrù CD, Groveman BR, et al. Diagnosis of Human Prion Disease Using Real-Time Quaking-Induced Conversion Testing of Olfactory Mucosa and Cerebrospinal Fluid Samples. JAMA Neurology. 2017;74(2):1–8. [DOI] [PubMed] [Google Scholar]
- 22.Kocisko DA, Come JH, Priola SA, et al. Cell-free formation of protease-resistant prion protein. Nature. 1994;370:471–474. [DOI] [PubMed] [Google Scholar]
- 23.Kocisko DA, Priola SA, Raymond GJ, Chesebro B, Lansbury PT Jr., Caughey B. Species specificity in the cell-free conversion of prion protein to protease-resistant forms: a model for the scrapie species barrier. ProcNatlAcadSciUSA. 1995;92:3923–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bessen RA, Kocisko DA, Raymond GJ, Nandan S, Lansbury PT, Jr., Caughey B. Nongenetic propagation of strain-specific phenotypes of scrapie prion protein. Nature. 1995;375:698–700. [DOI] [PubMed] [Google Scholar]
- 25.Bossers A, Belt PBGM, Raymond GJ, Caughey B, DeVries R, Smits MA. Scrapie susceptibility-linked polymorphisms modulate the in vitro conversion of sheep prion protein to protease-resistant forms. Proc Natl Acad Sci USA. 1997;94:4931–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raymond GJ, Hope J, Kocisko DA, et al. Molecular assessment of the transmissibilities of BSE and scrapie to humans. Nature. 1997;388:285–288. [DOI] [PubMed] [Google Scholar]
- 27.Raymond GJ, Bossers A, Raymond LD, et al. Evidence of a molecular barrier limiting susceptibility of humans, cattle and sheep to chronic wasting disease. EMBO J. 2000;19:4425–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castilla J, Saa P, Hetz C, Soto C. In vitro generation of infectious scrapie prions. Cell. 2005;121(2):195–206. [DOI] [PubMed] [Google Scholar]
- 29.Castilla J, Morales R, Saa P, Barria M, Gambetti P, Soto C. Cell-free propagation of prion strains. EMBO J. 2008;27(19):2557–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castilla J, Saa P, Soto C. Detection of prions in blood. Nat Med. 2005;11(9):982985. [DOI] [PubMed] [Google Scholar]
- 31.Soto C, Anderes L, Suardi S, et al. Pre-symptomatic detection of prions by cyclic amplification of protein misfolding. FEBS Lett. 2005;579(3):638–642. [DOI] [PubMed] [Google Scholar]
- 32.Gonzalez-Romero D, Barria MA, Leon P, Morales R, Soto C. Detection of infectious prions in urine. FEBS Lett. 2008;582(21–22):3161–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moda F, Gambetti P, Notari S, et al. Prions in the urine of patients with variant Creutzfeldt-Jakob disease. N Engl J Med. 2014;371(6):530–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orru CD, Groveman BR, Raymond LD, et al. Bank Vole Prion Protein As an Apparently Universal Substrate for RT-QuIC-Based Detection and Discrimination of Prion Strains. PLoS Path. 2015;11(6):e1004983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caughey B, Orru CD, Groveman BR, et al. Detection and diagnosis of prion diseases using RT-QuIC: an update. In: Liberski P, ed. Prions Diseases. Vol 129. Basel: Springer Science+Business Media; 2017:173–181. [Google Scholar]
- 36.Haley NJ, Richt JA, Davenport KA, et al. Design, implementation, and interpretation of amplification studies for prion detection. Prion. 2018;12(2):73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orrú CD, Wilham JM, Hughson AG, et al. Human variant Creutzfeldt-Jakob disease and sheep scrapie PrP(res) detection using seeded conversion of recombinant prion protein. Protein Eng Des Sel. 2009;22(8):515–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z, Manca M, Foutz A, et al. Early preclinical detection of prions in the skin of prion-infected animals. Nat Commun. 2019;10(1):247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGuire LI, Peden AH, Orru CD, et al. RT-QuIC analysis of cerebrospinal fluid in sporadic Creutzfeldt-Jakob disease. Ann Neurol. 2012;72(2):278–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franceschini A, Baiardi S, Hughson AG, et al. High diagnostic value of second generation CSF RT-QuIC across the wide spectrum of CJD prions. Sci Rep. 2017;7(1):10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orru CD, Groveman BR, Hughson AG, Zanusso G, Coulthart MB, Caughey B. Rapid and sensitive RT-QuIC detection of human Creutzfeldt-Jakob disease using cerebrospinal fluid. MBio. 2015;6(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henderson DM, Denkers ND, Hoover CE, Garbino N, Mathiason CK, Hoover EA. Longitudinal Detection of Prion Shedding in Saliva and Urine by Chronic Wasting Disease-Infected Deer by Real-Time Quaking-Induced Conversion. J Virol. 2015;89(18):9338–9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orru CD, Yuan J, Appleby BS, et al. Prion seeding activity and infectivity in skin samples from patients with sporadic Creutzfeldt-Jakob disease. Sci Transl Med. 2017;9(417). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng YC, Hannaoui S, John TR, Dudas S, Czub S, Gilch S. Real-time Quaking-induced Conversion Assay for Detection of CWD Prions in Fecal Material. J Vis Exp. 2017(127). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davenport KA, Mosher BA, Brost BM, et al. Assessment of Chronic Wasting Disease Prion Shedding in Deer Saliva with Occupancy Modeling. J Clin Microbiol. 2018;56(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Groveman BR, Orru CD, Hughson AG, et al. Extended and direct evaluation of RT-QuIC assays for Creutzfeldt-Jakob disease diagnosis. Ann Clin Transl Neurol. 2017;4(2):139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foutz A, Appleby BS, Hamlin C, et al. Diagnostic and prognostic value of human prion detection in cerebrospinal fluid. Ann Neurol. 2017;81(1):79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orru CD, Soldau K, Cordano C, et al. Prion Seeds Distribute throughout the Eyes of Sporadic Creutzfeldt-Jakob Disease Patients. MBio. 2018;9(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saijo E, Groveman BR, Kraus A, et al. Ultrasensitive RT-QuIC Seed Amplification Assays for Disease-Associated Tau, alpha-Synuclein, and Prion Aggregates. Methods Mol Biol. 2019;1873:19–37. [DOI] [PubMed] [Google Scholar]
- 50.Klohn PC, Stoltze L, Flechsig E, Enari M, Weissmann C. A quantitative, highly sensitive cell-based infectivity assay for mouse scrapie prions. ProcNatlAcadSciUSA. 2003;100(20):11666–11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mahal SP, Demczyk CA, Smith EW Jr., Klohn PC, Weissmann C. Assaying prions in cell culture: the standard scrapie cell assay (SSCA) and the scrapie cell assay in end point format (SCEPA). Methods Mol Biol. 2008;459:49–68. [DOI] [PubMed] [Google Scholar]
- 52.Schmidt C, Fizet J, Properzi F, et al. A systematic investigation of production of synthetic prions from recombinant prion protein. Open Biol. 2015;5(12):150165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Merwe J, Aiken J, Westaway D, McKenzie D. The standard scrapie cell assay: development, utility and prospects. Viruses. 2015;7(1):180–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarell CJ, Quarterman E, Yip DC, et al. Soluble Abeta aggregates can inhibit prion propagation. Open Biol. 2017;7(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fairfoul G, McGuire LI, Pal S, et al. Alpha-synuclein RT-QuIC in the CSF of patients with alpha-synucleinopathies. Ann Clin Transl Neurol. 2016;3(10):812–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sano K, Atarashi R, Satoh K, et al. Prion-Like Seeding of Misfolded alpha-Synuclein in the Brains of Dementia with Lewy Body Patients in RT-QUIC. Mol Neurobiol. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Groveman BR, Orru CD, Hughson AG, et al. Rapid and ultra-sensitive quantitation of disease-associated alpha-synuclein seeds in brain and cerebrospinal fluid by alphaSyn RT-QuIC. Acta Neuropathol Commun. 2018;6(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Candelise N, Schmitz M, Llorens F, et al. Seeding variability of different alpha synuclein strains in synucleinopathies. Ann Neurol. 2019;85(5):691–703. [DOI] [PubMed] [Google Scholar]
- 59.Manne S, Kondru N, Jin H, et al. alpha-Synuclein real-time quaking-induced conversion in the submandibular glands of Parkinson’s disease patients. Mov Disord. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bongianni M, Ladogana A, Capaldi S, et al. alpha-Synuclein RT-QuIC assay in cerebrospinal fluid of patients with dementia with Lewy bodies. Ann Clin Transl Neurol. 2019;6(10):2120–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shahnawaz M, Tokuda T, Waragai M, et al. Development of a Biochemical Diagnosis of Parkinson Disease by Detection of alpha-Synuclein Misfolded Aggregates in Cerebrospinal Fluid. JAMA Neurol. 2017;74(2):163–172. [DOI] [PubMed] [Google Scholar]
- 62.Kang UJ, Boehme AK, Fairfoul G, et al. Comparative study of cerebrospinal fluid alpha-synuclein seeding aggregation assays for diagnosis of Parkinson’s disease. Mov Disord. 2019;34(4):536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Luca CMG, Elia AE, Portaleone SM, et al. Efficient RT-QuIC seeding activity for alpha-synuclein in olfactory mucosa samples of patients with Parkinson’s disease and multiple system atrophy. Transl Neurodegener. 2019;8:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Woerman AL, Stohr J, Aoyagi A, et al. Propagation of prions causing synucleinopathies in cultured cells. Proc Natl Acad Sci U S A. 2015;112(35):E4949–4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prusiner SB, Woerman AL, Mordes DA, et al. Evidence for alpha-synuclein prions causing multiple system atrophy in humans with parkinsonism. Proc Natl Acad Sci U S A. 2015;112(38):E5308–5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kakuda K, Ikenaka K, Araki K, et al. Ultrasonication-based rapid amplification of alpha-synuclein aggregates in cerebrospinal fluid. Sci Rep. 2019;9(1):6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fitzpatrick AWP, Falcon B, He S, et al. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature. 2017;547(7662):185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Falcon B, Zivanov J, Zhang W, et al. Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature. 2019;568(7752):420–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arai T, Ikeda K, Akiyama H, et al. Different immunoreactivities of the microtubule-binding region of tau and its molecular basis in brains from patients with Alzheimer’s disease, Pick’s disease, progressive supranuclear palsy and corticobasal degeneration. Acta Neuropathol. 2003;105(5):489–498. [DOI] [PubMed] [Google Scholar]
- 70.Irwin DJ, Brettschneider J, McMillan CT, et al. Deep clinical and neuropathological phenotyping of Pick disease. Ann Neurol. 2016;79(2):272–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Falcon B, Zhang W, Murzin AG, et al. Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature. 2018;561(7721):137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang K, Tarutani A, Newell KL, et al. Novel tau filament fold in corticobasal degeneration, a four-repeat tauopathy. bioRxiv. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dinkel PD, Siddiqua A, Huynh H, Shah M, Margittai M. Variations in filament conformation dictate seeding barrier between three- and four-repeat tau. Biochemistry. 2011;50(20):4330–4336. [DOI] [PubMed] [Google Scholar]
- 74.Meyer V, Dinkel PD, Rickman Hager E, Margittai M. Amplification of Tau fibrils from minute quantities of seeds. Biochemistry. 2014;53(36):5804–5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saijo E, Ghetti B, Zanusso G, et al. Ultrasensitive and selective detection of 3-repeat tau seeding activity in Pick disease brain and cerebrospinal fluid. Acta Neuropathol. 2017;133(5):751–765. [DOI] [PubMed] [Google Scholar]
- 76.Saijo E, Metrick MA 2nd, Koga S, et al. 4-Repeat tau seeds and templating subtypes as brain and CSF biomarkers of frontotemporal lobar degeneration. Acta Neuropathol. 2020;139:63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Metrick MA 2nd, do Carmo Ferreira N, Saijo E, et al. Million-fold sensitivity enhancement in proteopathic seed amplification assays for biospecimens by Hofmeister ion comparisons. Proc Natl Acad Sci U S A. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Metrick MAI, Ferreira NC, Saijo E, et al. A single ultrasensitive assay for detection and discrimination of tau aggregates of Alzheimer and Pick diseases Acta Neuropathologica Communications. 2020;In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Holmes BB, Furman JL, Mahan TE, et al. Proteopathic tau seeding predicts tauopathy in vivo. Proc Natl Acad Sci U S A. 2014;111(41):E4376–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Woerman AL, Aoyagi A, Patel S, et al. Tau prions from Alzheimer’s disease and chronic traumatic encephalopathy patients propagate in cultured cells. Proc Natl Acad Sci U S A. 2016;113(50):E8187-E8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guo JL, Lee VM. Seeding of normal Tau by pathological Tau conformers drives pathogenesis of Alzheimer-like tangles. J Biol Chem. 2011;286(17):15317–15331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kaufman SK, Sanders DW, Thomas TL, et al. Tau Prion Strains Dictate Patterns of Cell Pathology, Progression Rate, and Regional Vulnerability In Vivo. Neuron. 2016;92(4):796–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Montine TJ, Phelps CH, Beach TG, et al. National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease: a practical approach. Acta Neuropathol. 2012;123(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reitz C, Mayeux R. Alzheimer disease: epidemiology, diagnostic criteria, risk factors and biomarkers. Biochem Pharmacol. 2014;88(4):640–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Du D, Murray AN, Cohen E, et al. A kinetic aggregation assay allowing selective and sensitive amyloid-beta quantification in cells and tissues. Biochemistry. 2011;50(10):1607–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Salvadores N, Shahnawaz M, Scarpini E, Tagliavini F, Soto C. Detection of misfolded Abeta oligomers for sensitive biochemical diagnosis of Alzheimer’s disease. Cell Rep. 2014;7(1):261–268. [DOI] [PubMed] [Google Scholar]
- 87.Gupta S, Jie S, Colby DW. Protein misfolding detected early in pathogenesis of transgenic mouse model of Huntington disease using amyloid seeding assay. J Biol Chem. 2012;287(13):9982–9989. [DOI] [PMC free article] [PubMed] [Google Scholar]