Abstract
This study aimed to investigate the correlation between microRNA (miR)-4429 and epidermal growth factor receptor (EGFR), the expression, and clinical significance of miR-4429 in patients with non-small cell lung cancer (NSCLC), and the relationship between miR-4429 and EGFR mutation in NSCLC patients. Blood samples were collected from 122 NSCLC patients and 72 healthy volunteers. miR-4429 expression and EGFR mRNA expression were detected by real-time quantitative polymerase chain reaction. Correlation between miR-4429 and EGFR was evaluated by dual-luciferase reporter assay and the Pearson correlation analysis. The ability of serum miR4429 to discriminate between NSCLC patients and healthy controls, and to discriminate between EGFR wild-type (EGFR-W) and EGFR mutant-type (EGFR-M) patients was assessed using receiver operating characteristic analysis. The relationship between miR-4429 and NSCLC patients’ survival was identified by KaplanMeier survival curves and log-rank test. The prognostic value of miR-4429 in NSCLC patients was evaluated by Cox regression analysis. miR-4429 could directly bind to EGFR. Serum miR-4429, decreased in NSCLC patients, was negatively correlated with serum EGFR mRNA expression in NSCLC patients. In addition, miR-4429 had a high diagnostic value for screening NSCLC patients from healthy controls, and was independently correlated with survival prognosis of NSCLC patients. Moreover, miR4429 was decreased in EGFR-M patients, which had a certain screening ability for EGFRM patients. Our findings indicate that miR-4429 is negatively correlated with EGFR in NSCLC, and may function as a diagnostic and prognostic biomarker for NSCLC patients. Additionally, miR-4429 is associated with EGFR mutation in NSCLC patients.
Keywords: miR-4429, EGFR, non-small cell lung cancer, diagnosis, prognosis, EGFR mutation
INTRODUCTION
Lung cancer is a malignant tumor with high morbidity and mortality [1]. Non-small cell lung cancer (NSCLC) is the major subtype of lung cancer, accounting for 85% of lung cancer cases [2]. Despite current advances in diagnostic and therapeutic approaches, the prognosis of lung cancer remains poor with very low survival rates [3-5]. Therefore, early diagnosis and accurate prognosis are urgent issues in the treatment of NSCLC. Epidermal growth factor receptor (EGFR) is the major and well-studied oncogene of NSCLC [6]. Targeting EGFR is an important method of NSCLC treatment. The EGFR mutation is an important predictor of the effectiveness of targeted drug therapy with tyrosine kinase inhibitors (TKIs). However, drug resistance still occurs in more than 50% of patients [7]. Therefore, probing the molecules related to EGFR is expected to unearth more key molecules in the pathological mechanism of NSCLC, providing new ideas and new targets for the treatment of this disease.
MicroRNAs (miRs) are a type of non-protein coding RNAs that negatively regulate gene expression mainly by inducing targeted messenger RNA (mRNA) degradation or inhibiting targeted mRNA translation [8,9]. miRs have been found to play key roles in the pathogenesis of various diseases [10-12]. Many miRs such as miR-340 [13] and miR-142-3p [14] have been found to be involved in the pathogenesis of NSCLC. In addition, miR-4429 has been found to be downregulated and to inhibit tumor progression in some human malignancies such as ovarian cancer [15], cervical cancer [16], and gastric cancer [17]. In this study, we performed bioinformatics analysis prediction, and the results suggested that the 3’-untranslated region (3’-UTR) of EGFR had the binding sequence of miR-4429. However, the expression profile of miR-4429 in NSCLC, the relationship of miR-4429 with EGFR expression, and EGFR mutation in NSCLC patients are unknown.
Thus, the aim of this study was to explore whether miR-4429 was related to EGFR, measure the expression of miR-4429 in NSCLC, investigate its clinical value in NSCLC, and explore its relationship with EGFR mutation in NSCLC patients. This study is expected to provide a novel biomarker for the diagnosis and survival prognosis of NSCLC patients, and a novel target for the therapy in NSCLC patients.
MATERIALS AND METHODS
Study population and serum collection
Venous blood samples were collected from 122 NSCLC patients enrolled in The Fourth People Hospital of Zibo from 2014 to 2019. Blood samples of 72 healthy volunteers who underwent health physical examination during the same period were also collected as controls. Then, serum separation was performed by centrifugation immediately after blood sample collection to avoid hemolysis, and the serum was immediately stored at −80°C for further use. The inclusion criteria for NSCLC patients were: (1) they had not received any anti-tumor treatment before surgery; (2) they were diagnosed with NSCLC on pathological basis and (3) their clinical medical record data was fully available. Patients were excluded from this study if they: (1) presented with concomitant major organ diseases; (2) had renal, hepatic, or severe cardiac dysfunction; (3) complicated with other malignancies; (4) had incomplete clinical data. The information of clinicopathological characteristics of patients were recorded, including age (was dichotomized according to the age cut-off standard for elderly in China), gender, smoking, tumor size, EGFR mutation, lymph node metastasis, tumor node metastasis (TNM) stage, and histological subtypes. For the EGFR mutation detection, EGFR mutations of exon 18, 19, 20, and 21 were amplified using AmoyDx EGFR Mutations Detection Kit (Amoy Diagnostics, Shanghai, China) according to the principle of the amplification refractory mutation system. Positive or negative results were obtained if the criteria specified by the manufacturer’s protocol were met. All patients underwent a 5-year survival follow-up, and their survival data were recorded for subsequent analysis. This study was approved by the Ethics Committee of The Fourth People Hospital of Zibo (No. 00013269), and all participants provided written informed consent.
Dual-luciferase reporter assay
The binding sequences of miR-4429 to the 3’-UTR region of EGFR were predicted in starBase v2.0 platform (http://starbase.sysu.edu.cn/). The wild-type (WT)-EGFR sequence containing the miR-4429 binding site and mutant-type (MUT)-EGFR sequence was cloned into the pGL3 dual-luciferase reporter vector (Promega, Madison, WI, USA). Then, the WTEGFR and MUTEGFR vectors were co-transfected with miR-4429 mimic or mimic negative control (NC) into HEK293T cells using Lipofectamine 3000 reagent (Invitrogen, CA, USA) following the manufacturer’s instructions. The results were examined after 48 hours.
Total RNA extraction
Total RNA was extracted from the serum samples of NSCLC patients and healthy controls using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and was then purified. The purity and concentration of the obtained RNA were evaluated by NanoDrop 2000 (Thermo Fisher Scientific, Inc.). The obtained RNA could be used for further analysis when the optical density (OD) ratio of 260 nm/280 nm was close to 2.0. The above operation was repeated 3 times. Then, the obtained RNA (1 μg) was reverse transcribed into cDNA using the PrimeScript RT Reagent Kit (TaKaRa, Japan), following the manufacturers’ instructions.
Real-time quantitative polymerase chain reaction (RT-qPCR)
Relative expression of miR-4429 and relative mRNA expression of EGFR was detected by RT-qPCR, which was performed using a SYBR green I Master Mix kit (Invitrogen, Carlsbad, CA, USA) on a 7500 Real-Time PCR System (Applied Biosystems, USA). The cel-miR-39-3p (spike in-control) was used as the internal reference of relative miR-4429 expression, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal reference of relative mRNA expression of EGFR. The primer sequences used for PCR were as follows: miR-4429 forward, 5’-GCCGAGAAAAGCTGGGCTGA-3’ and reverse, 5’-CTCAACTGGTGTCGTGGA-3’; cel-miR-39-3p forward, 5’-UCACCGGGUGUAAAUCAGCUUG-3’ and reverse, 5’-AACGCTTCACGAATTTGCGT-3’; EGFR forward, 5’-TTGCCGCAAAGTGTGTAACG-3’ and reverse, 5’-GTCACCCCTAAATGCCACCG-3’; GAPDH forward, 5’-TGGAAGGACTCATGACCACA-3’ and reverse, 5’-TTCAGCTCAGGGATGACCTT-3’. The PCR was performed 3 times. Relative miR-4429 expression and relative mRNA expression of EGFR were calculated by 2ΔΔCT method [18].
Ethical statement
The experimental procedures were all in accordance with the guideline of the Ethics Committee of The Fourth People Hospital of Zibo and has approved by the Ethics Committee of The Fourth People Hospital of Zibo. This study complies with the Declaration of Helsinki.
A signed written informed consent was obtained from each patient.
Availability of data and materials
The data used to support the findings of this study are available from the corresponding author upon reasonable request.
Statistical analysis
Data analysis results were shown as mean ± standard deviation (SD). All analyses were performed by SPSS 22.0 (IBM Corp.) and GraphPad Prism 7.0 software (GraphPad Software, Inc.). Comparisons between two groups of measurement data were performed using t-tests, and correlation analysis between variables was performed using the Pearson correlation analysis. The Chi-square test was used to analyze the association of miR-4429 with patients’ clinicopathological characteristics. Receiver operating characteristic (ROC) analysis was used to evaluate the clinical significance of serum miR-4429 in distinguishing NSCLC patients from healthy controls, and in distinguishing EGFR WT (EGFR-W) patients from EGFR MUT (EGFRM) patients. Kaplan-Meier curves were used to analyze the relationship of miR-4429 with the survival of NSCLC patients, and the log-rank test was used to analyze the difference between the two curves. Cox regression analysis was used to judge the prognostic value of miR-4429 in NSCLC patients. p < 0.05 indicated a statistically significant difference.
RESULTS
Circulating miR-4429 is negatively correlated with EGFR in NSCLC patients
The results of bioinformatics prediction, shown in Figure 1A, exhibited the binding sites of miR-4429 to EGFR. Subsequently, the luciferase reporter assay results demonstrated that relative luciferase activity of the WT-EGFR group was decreased by miR-4429 mimic in HEK293T cells (p < 0.05), and no significant change was found in the luciferase activity of the MUT-EGFR group, indicating the direct binding between EGFR and miR-4429 (Figure 1B). Moreover, compared to healthy controls, NSCLC patients had significantly lower miR4429 levels (Figure 1C, p < 0.001) and significantly higher EGFR mRNA levels (Figure 1D, p < 0.001). Relative mRNA levels of EGFR were significantly negatively correlated with the relative miR-4429 levels in the serum of NSCLC patients (r = −0.641, p < 0.001; Figure 1E).
FIGURE 1.
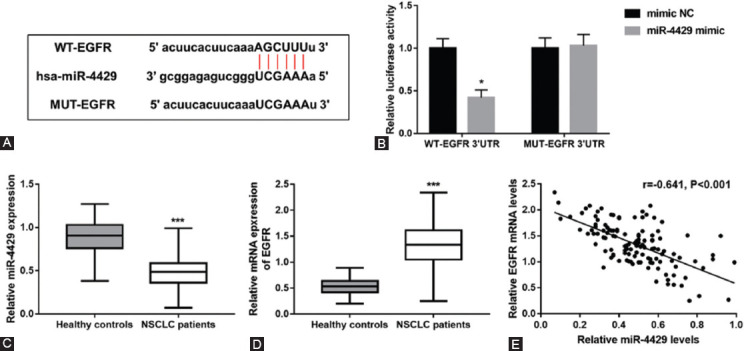
Correlation of miR-4429 with EGFR in NSCLC patients. (A) The binding sites of miR-4429 to EGFR was predicted by starBase v2.0 platform. (B) Relative luciferase activity in WT-EGFR group was decreased by miR-4429 mimic in HEK293T cells. (C) Serum miR-4429 expression in healthy controls and NSCLC patients. (D) Relative mRNA levels of EGFR in healthy controls and NSCLC patients. (E) Relative EGFR mRNA levels were negatively correlated with relative miR-4429 levels in the serum of NSCLC patients (r = -0.641, p < 0.001). *p < 0.05, ***p < 0.001 vs. mimic NC or healthy controls. EGFR, Epidermal growth factor receptor; WT, Wide-type; MUT, Mutant-type; NC, Negative control; NSCLC, Non-small cell lung cancer.
Relationship between miR-4429 and NSCLC patients’ clinicopathological characteristics
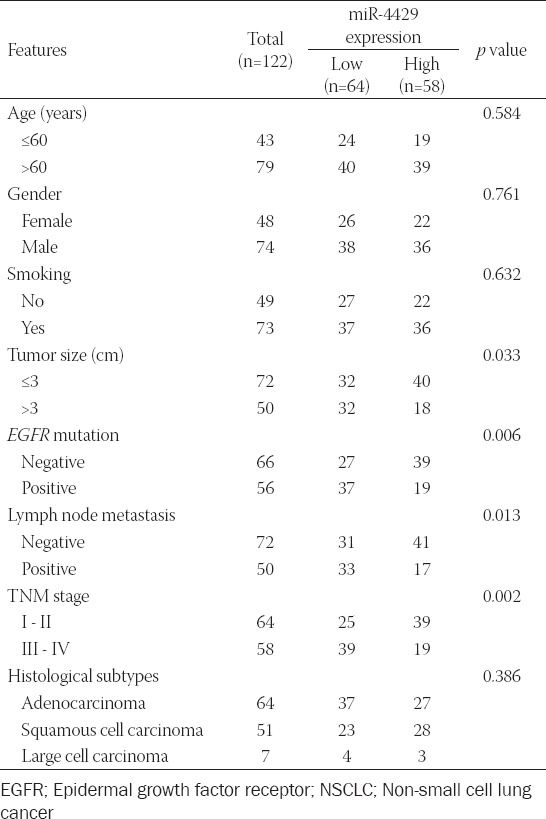
As presented in Table 1, we found that miR-4429 was associated with tumor size (p = 0.033), EGFR mutation (p = 0.006), lymph node metastasis (p = 0.013) and TNM stage (p = 0.002) in NSCLC patients. Nevertheless, no association was found between miR-4429 and other characteristics, including age, gender, smoking, and histological subtypes (all p > 0.05).
TABLE 1.
Relationship between miR-4429 and the clinicopathological characteristics of NSCLC patients
Diagnostic performance of circulating miR-4429 in patients with NSCLC
ROC analysis results, shown in Figure 2, indicated that miR-4429 had high diagnostic potential to screen patients with NSCLC from healthy controls with an area under the ROC curve (AUC) of 0.918. At the optimal cut-off value of 0.700, the sensitivity and specificity were 89.34% and 84.72%, respectively.
FIGURE 2.
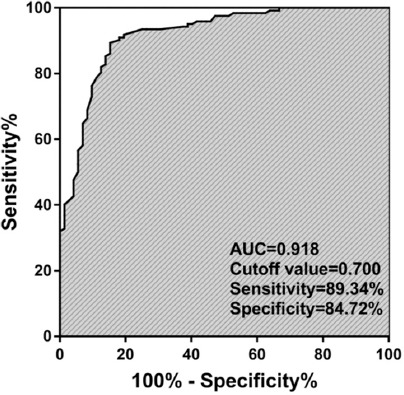
ROC analysis results indicated that serum miR-4429 had a high diagnostic value to screen NSCLC patients from healthy controls with an AUC of 0.918. AUC, Area under the ROC curve; ROC, Receiver operating characteristic; NSCLC, Non-small cell lung cancer.
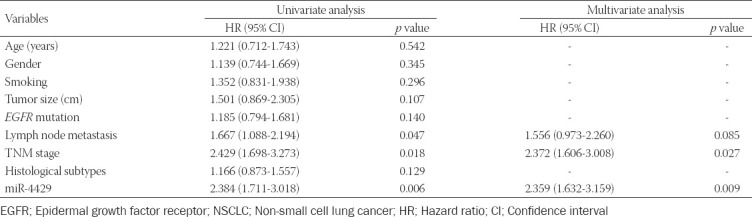
Prognostic value of miR-4429 in predicting the overall survival of NSCLC
As presented in Figure 3, the survival curves indicated that NSCLC patients with low miR4429 levels had poor overall survival (log-rank p = 0.0157). The results of the Cox regression analysis of miR-4429 in predicting the overall survival of NSCLC were shown in Table 2. Univariate Cox regression analysis demonstrated that lymph node metastasis, TNM stage, and miR-4429 were associated with the overall survival of NSCLC patients. Then, the significant variables from univariate analysis results were included in multivariate Cox analysis. Multivariate Cox regression analysis results showed that TNM stage (hazard ratio [HR] = 2.372, 95% confidence interval [CI] = 1.606-3.008, p = 0.027) and miR-4429 (HR = 2.359, 95% CI=1.632-3.159, p = 0.009) were independently associated with the prognosis of NSCLC patients.
FIGURE 3.
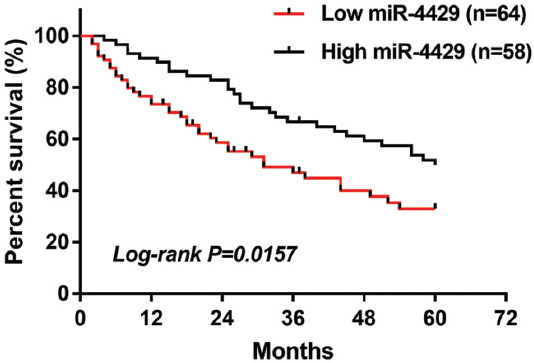
NSCLC patients with low levels of miR-4429 had lower overall survival than patients with high miR-4429 levels (log-rank p = 0.0157). NSCLC, Non-small cell lung cancer.
TABLE 2.
Cox regression analysis for miR-4429 to predict the overall survival of NSCLC
Association of miR-4429 with EGFR mutation in patients with NSCLC
Fifty-six (45.9%) NSCLC patients with EGFR mutation were observed among the 122 NSCLC patients, including 27 (48.2%) cases of exon 19 deletions (19Del), 24 (42.9%) cases of exon 21 mutation (L858R), 3 (5.3%) cases of exon 21 mutation (L861Q) and 2 (3.6%) cases of S768I mutation in exon 21. Serum miR-4429 levels were significantly lower in EGFR-M patients than that in patients with EGFR-W (Figure 4A, p < 0.01). Additionally, no significantly different expression of miR-4429 was observed between patients with different types of EGFR mutations (Figure 4B, p > 0.05). Moreover, ROC analysis results indicated that serum miR4429 had a certain ability to differentiate between EGFR-M patients and EGFR-W patients (AUC = 0.817, Figure 4C).
FIGURE 4.
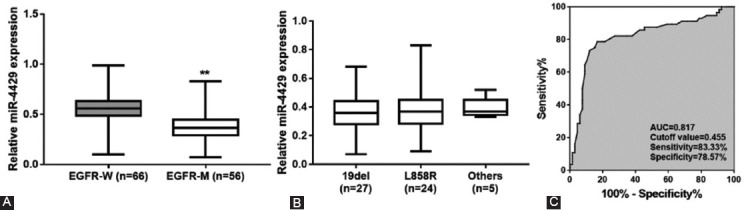
Relationship between miR-4429 and EGFR mutation in NSCLC patients. (A) Serum miR-4429 expression in patients with EGFR-M and patients with EGFR-W. (B) Serum miR4429 expression in patients with different types of EGFR mutations. (C) ROC analysis indicated the ability of serum miR-4429 to discriminate between EGFR-M patients and EGFRW patients. **p < 0.01 vs. EGFR-W; EGFR, Epidermal growth factor receptor; EGFRM, EGFR mutant-type; EGFR-W, EGFR wild-type; AUC, Area under the ROC curve; ROC, Receiver operating characteristic; NSCLC, Non-small cell lung cancer.
DISCUSSION
EGFR is an extensively studied and reported oncogene, thus, searching for molecules related to EGFR is necessary for NSCLC treatment. Currently, many miRs have been demonstrated to affect NSCLC by regulating EGFR. For example, Qi et al. have shown that EGFR is a direct target of miR-146a-5p, and it can reverse the effect of miR146a5p on NSCLC cell lines [19]. Besides, miR-145 has been reported to suppress the proliferation of human lung adenocarcinoma cells by regulating EGFR [20]. A study by Li et al. has revealed that miR-34a suppresses the progression of NSCLC tumors via targeting EGFR [21]. miR-218-5p suppresses NSCLC carcinogenesis by directly inhibiting EGFR [22]. In this study, we found the binding sites between miR-4429 and EGFR through starBase v2.0 platform and further demonstrated their direct binding by luciferase reporter assay. Moreover, downregulated miR-4429 and upregulated EGFR mRNA levels were found in NSCLC patients, and a negative correlation between miR-4429 and EGFR mRNA level was found in the serum of NSCLC patients. The above results indicated that miR-4429 is closely correlated with EGFR in NSCLC. The expression of circulating mRNA and miRNA in serum was measured in this study, including all mRNA and miRNA inside and outside cells. Thus, circulating mRNA and miRNA are not only contained extracellularly, not only from cancer cells, and their secretion includes active and passive secretion. Besides, mRNA digestion consists of total and partial digestion. Furthermore, miR4429 was found to be related to tumor size, EGFR mutation, lymph node metastasis, and TNM stage in NSCLC patients by the Chi-square test. miR-4429 has also been less frequent in other types of cancer, such as ovarian cancer [15], prostate cancer [23], and colorectal cancer [24]. Thus, we conclude that downregulated miR4429 may be involved in the progression of NSCLC.
Circulating miRs used as cancer biomarkers have been widely reported, including NSCLC. For instance, serum miR-203 has been reported to serve as a potential diagnostic and prognostic biomarker for acute myeloid leukemia [25]. A study by Sun et al. has shown that serum miR30a-5p may function as a new biomarker for the diagnosis and prognosis of colorectal cancer [26]. Serum miR-185 [27] and serum miR1246 [28] have been found to be used as diagnostic and prognostic biomarkers for NSCLC.
Given the significant decrease of miR-4429 in NSCLC, this study investigated its clinical significance in NSCLC. First, miR-4429 expression showed a high diagnostic value for screening NSCLC patients (AUC = 0.918), with a sensitivity of 89.34% and a specificity of 84.72%. Kaplan-Meier survival analysis results demonstrated that patients with high miR-4429 expression had significantly longer overall survival. Serum miR-4429 maintained significance as an independent prognostic indicator for the overall survival of NSCLC patients. In addition, miR-4429 has been found to serve as a potential diagnostic biomarker for biliary atresia [29] and is associated with the survival prognosis of other cancers, such as ovarian cancer [15] and clear cell renal cell carcinoma [30]. Thus, serum miR-4429 may function as a diagnostic and prognostic biomarker for NSCLC.
Oncogenic activating mutations of EGFR are associated with the development and progression of lung cancer. Over the past decades, EGFR-TKIs therapy, including gefitinib, erlotinib, and afatinib, has become the standard therapy for NSCLC patients with EGFR activating mutations [31]. Nevertheless, many NSCLC patients develop resistance to targeted drugs in approximately a year [32-34], resulting in poor prognosis and limited therapeutic efficacy. Thus, it is necessary to explore the molecules related to EGFR mutations of NSCLC. Some studies have found that miRs are associated with EGFR mutations of NSCLC [32,35,36]. The present study also found that miR-4429 is associated with EGFR mutation in NSCLC patients. Approximately 30% - 50% Asian and 10% - 15% Caucasian patients with NSCLC have EGFR mutation [37], which provides evidence for such a high EGFR mutation rate (45.9%) in this study. Therefore, miR-4429 may facilitate timely screening of patients with EGFR mutation to facilitate treatment with TKIs. In addition, we considered the reasons why miR-4429 was associated with EGFR mutation. It has been shown that there is a close association between EGFR expression level and EGFR mutation [38], thus, miR-4429 may be linked to the mutation of EGFR by regulating the expression of EGFR. However, how are they specifically related awaits further exploration.
Our study had some limitations. First, the sample size was relatively small, and our findings need further validation in large cohorts. Second, the molecular mechanism underlying the role of miR-4429 in NSCLC needs further exploration. Third, the lack of in vitro experiments and in vivo models to support this study’s conclusion is also a limitation of the study, which should be performed in future studies. Fourth, our study did not screen the entire EGFR gene (just exons 18-21). Fifth, learning if miR-4429 would also be changed in ethylenediaminetetraacetic acid (EDTA)-treated plasma samples can contribute to the potential use of miR-4429 as a biomarker, which will be conducted in the future study. NSCLC patients also harbor other genomic alterations, such as ALK1, ROS1, HER2, BRAF, KRAS, and NTRK mutations. Thus, we will explore the molecules associated with other genomic alterations of NSCLC patients to provide targets for NSCLC.
CONCLUSION
Our study indicates that serum miR-4429 is decreased in NSCLC patients and may serve as a potential biomarker for the diagnosis and prognosis of NSCLC. In addition, there is a correlation between miR-4429 and EGFR mutation in NSCLC patients. Thus, our findings may provide a new effective target for the clinical therapy of NSCLC patients.
Footnotes
Conflicts of interests: The authors declare no conflicts of interest.
Funding: The authors received no specific funding for this work.
REFERENCES
- 1.Amiri A, Pourhanifeh MH, Mirzaei HR, Nahand JS, Moghoofei M, Sahebnasagh R, et al. Exosomes and lung cancer:Roles in pathophysiology, diagnosis and therapeutic applications. Curr Med Chem. 2021;28(2):308–28. doi: 10.2174/0929867327666200204141952. https://doi.org/10.2174/0929867327666200204141952. [DOI] [PubMed] [Google Scholar]
- 2.Inamura K. Lung cancer:Understanding its molecular pathology and the 2015 WHO classification. Front Oncol. 2017;7:193. doi: 10.3389/fonc.2017.00193. https://doi.org/10.3389/fonc.2017.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su Z, Chen M, Ding R, Shui L, Zhao Q, Luo W. Long noncoding RNA HCG11 suppresses the malignant phenotype of nonsmall cell lung cancer cells by targeting a miR875/SATB2 axis. Mol Med Rep. 2021;24(2):552. doi: 10.3892/mmr.2021.12191. https://doi.org/10.3892/mmr.2021.12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salehi M, Movahedpour A, Tayarani A, Shabaninejad Z, Pourhanifeh MH, Mortezapour E, et al. Therapeutic potentials of curcumin in the treatment of non-small-cell lung carcinoma. Phytother Res. 2020;34(10):2557–76. doi: 10.1002/ptr.6704. https://doi.org/10.1002/ptr.6704. [DOI] [PubMed] [Google Scholar]
- 5.Rezaei S, Mahjoubin-Tehran M, Aghaee-Bakhtiari SH, Jalili A, Movahedpour A, Khan H, et al. Autophagy-related MicroRNAs in chronic lung diseases and lung cancer. Crit Rev Oncol Hematol. 2020;153:103063. doi: 10.1016/j.critrevonc.2020.103063. https://doi.org/10.1016/j.critrevonc.2020.103063. [DOI] [PubMed] [Google Scholar]
- 6.Hirsch FR, Bunn PA., Jr EGFR testing in lung cancer is ready for prime time. Lancet Oncol. 2009;10(5):432–3. doi: 10.1016/S1470-2045(09)70110-X. https://doi.org/10.1016/S1470-2045(09)70110-X. [DOI] [PubMed] [Google Scholar]
- 7.Cao S, Li L, Li J, Zhao H. MiR-1299 impedes the progression of non-small-cell lung cancer through EGFR/PI3K/AKT signaling pathway. Onco Targets Ther. 2020;13:7493–502. doi: 10.2147/OTT.S250396. https://doi.org/10.2147/OTT.S250396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iorio MV, Croce CM. MicroRNAs in cancer:Small molecules with a huge impact. J Clin Oncol. 2009;27(34):5848–56. doi: 10.1200/JCO.2009.24.0317. https://doi.org/10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shabaninejad Z, Yousefi F, Movahedpour A, Ghasemi Y, Dokanehiifard S, Rezaei S, et al. Electrochemical-based biosensors for microRNA detection:Nanotechnology comes into view. Anal Biochem. 2019;581:113349. doi: 10.1016/j.ab.2019.113349. https://doi.org/10.1016/j.ab.2019.113349. [DOI] [PubMed] [Google Scholar]
- 10.Khani P, Nasri F, Chamani FK, Saeidi F, Nahand JS, Tabibkhooei A, et al. Genetic and epigenetic contribution to astrocytic gliomas pathogenesis. J Neurochem. 2019;148(2):188–203. doi: 10.1111/jnc.14616. https://doi.org/10.1111/jnc.14616. [DOI] [PubMed] [Google Scholar]
- 11.Mirzaei H, Hamblin MR. Regulation of glycolysis by non-coding RNAs in cancer:Switching on the Warburg effect. Mol Ther Oncolytics. 2020;19:218–39. doi: 10.1016/j.omto.2020.10.003. https://doi.org/10.1016/j.omto.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hashemian SM, Pourhanifeh MH, Fadaei S, Velayati AA, Mirzaei H, Hamblin MR. Non-coding RNAs and exosomes:Their role in the pathogenesis of sepsis. Mol Ther Nucleic Acids. 2020;21:51–74. doi: 10.1016/j.omtn.2020.05.012. https://doi.org/10.1016/j.omtn.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu X, Tian G, Quan J, He P, Liu J. Effects of miR340 overexpression and knockdown on the proliferation and metastasis of NSCLC cell lines. Int J Mol Med. 2019;44(2):643–51. doi: 10.3892/ijmm.2019.4213. https://doi.org/10.3892/ijmm.2019.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Zhou X, Qiao J, Bao A. MiR-142-3p overexpression increases chemo-sensitivity of NSCLC by inhibiting HMGB1-mediated autophagy. Cell Physiol Biochem. 2017;41(4):1370–82. doi: 10.1159/000467896. https://doi.org/10.1159/000467896. [DOI] [PubMed] [Google Scholar]
- 15.Zhu YM, Chen P, Shi L, Zhu T, Chen X. MiR-4429 suppresses the malignant development of ovarian cancer by targeting YOD1. Eur Rev Med Pharmacol Sci. 2020;24(17):8722–30. doi: 10.26355/eurrev_202009_22809. https://doi.org/10.26355/eurrev_202009_22809. [DOI] [PubMed] [Google Scholar]
- 16.Liang L, Zheng YW, Wang YL. miR-4429 regulates the proliferation, migration, invasion, and epithelial-mesenchymal transition of cervical cancer by targeting FOXM1. Cancer Manag Res. 2020;12:5301–12. doi: 10.2147/CMAR.S244167. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.He H, Wu W, Sun Z, Chai L. MiR-4429 prevented gastric cancer progression through targeting METTL3 to inhibit m(6)A-caused stabilization of SEC62. Biochem Biophys Res Commun. 2019;517:581–7. doi: 10.1016/j.bbrc.2019.07.058. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. https://doi.org/10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Qi P, Li Y, Liu X, Jafari FA, Zhang X, Sun Q, et al. Cryptotanshinone suppresses non-small cell lung cancer via microRNA-146a-5p/EGFR axis. Int J Biol Sci. 2019;15(5):1072–9. doi: 10.7150/ijbs.31277. https://doi.org/10.7150/ijbs.31277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho WC, Chow AS, Au JS. MiR-145 inhibits cell proliferation of human lung adenocarcinoma by targeting EGFR and NUDT1. RNA Biol. 2011;8(1):125–31. doi: 10.4161/rna.8.1.14259. https://doi.org/10.4161/rna.8.1.14259. [DOI] [PubMed] [Google Scholar]
- 21.Li YL, Liu XM, Zhang CY, Zhou JB, Shao Y, Liang C, et al. MicroRNA-34a/EGFR axis plays pivotal roles in lung tumorigenesis. Oncogenesis. 2017;6(8):e372. doi: 10.1038/oncsis.2017.50. https://doi.org/10.1038/oncsis.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu K, Ding H, Wang W, Liao Z, Fu Z, Hong Y, et al. Tumor-suppressive miR-218-5p inhibits cancer cell proliferation and migration via EGFR in non-small cell lung cancer. Oncotarget. 2016;7(19):28075–85. doi: 10.18632/oncotarget.8576. https://doi.org/10.18632/oncotarget.ↀ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Xie S, Liu J, Li T, Wang W, Xie Z. MicroRNA-4429 suppresses proliferation of prostate cancer cells by targeting distal-less homeobox 1 and inactivating the Wnt/beta-catenin pathway. BMC Urol. 2021;21(1):40. doi: 10.1186/s12894-021-00810-x. https://doi.org/10.1186/s12894-021-00810-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Liang W, Zhang H, Shui Y, Zhang Z. MicroRNA-4429 restrains colorectal cancer cell invasion and migration via regulating SMAD3-induced epithelial-mesenchymal transition. J Cell Physiol. 2021;236(8):5875–84. doi: 10.1002/jcp.30271. https://doi.org/10.1002/jcp.30271. [DOI] [PubMed] [Google Scholar]
- 25.Zheng Z, Rong G, Li G, Ren F, Ma Y. Diagnostic and prognostic significance of serum miR-203 in patients with acute myeloid leukemia. Int J Clin Exp Pathol. 2019;12(5):1548–56. [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Y, Yang B, Lin M, Yu H, Chen H, Zhang Z. Identification of serum miR-30a-5p as a diagnostic and prognostic biomarker in colorectal cancer. Cancer Biomark. 2019;24(3):299–305. doi: 10.3233/CBM-182129. https://doi.org/10.3233/CBM-182129. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, Han Y, Liu X, Wei S. Serum miR-185 is a diagnostic and prognostic biomarker for non-small cell lung cancer. Technol Cancer Res Treat. 2020;19:1533033820973276. doi: 10.1177/1533033820973276. https://doi.org/10.1177/1533033820973276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang D, Qu D. Early diagnostic and prognostic value of serum exosomal miR-1246 in non-small cell lung cancer. Int J Clin Exp Pathol. 2020;13(7):1601–7. [PMC free article] [PubMed] [Google Scholar]
- 29.Dong R, Shen Z, Zheng C, Chen G, Zheng S. Serum microRNA microarray analysis identifies miR-4429 and miR-4689 are potential diagnostic biomarkers for biliary atresia. Sci Rep. 2016;6:21084. doi: 10.1038/srep21084. https://doi.org/10.1038/srep21084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan H, Hong Y, Yu B, Li L, Zhang X. miR-4429 inhibits tumor progression and epithelial-mesenchymal transition via targeting CDK6 in clear cell renal cell carcinoma. Cancer Biother Radiopharm. 2019;34(5):334–41. doi: 10.1089/cbr.2018.2697. https://doi.org/10.1089/cbr.2018.2697. [DOI] [PubMed] [Google Scholar]
- 31.Lee DH. Treatments for EGFR-mutant non-small cell lung cancer (NSCLC):The road to a success, paved with failures. Pharmacol Ther. 2017;174:1–21. doi: 10.1016/j.pharmthera.2017.02.001. https://doi.org/10.1016/j.pharmthera.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Szpechcinski A, Florczuk M, Duk K, Zdral A, Rudzinski S, Bryl M, et al. The expression of circulating miR-504 in plasma is associated with EGFR mutation status in non-small-cell lung carcinoma patients. Cell Mol Life Sci. 2019;76(18):3641–56. doi: 10.1007/s00018-019-03089-2. https://doi.org/10.1007/s00018-019-03089-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee BS, Park DI, Lee DH, Lee JE, Yeo MK, Park YH, et al. Hippo effector YAP directly regulates the expression of PD-L1 transcripts in EGFR-TKI-resistant lung adenocarcinoma. Biochem Biophys Res Commun. 2017;491(2):493–9. doi: 10.1016/j.bbrc.2017.07.007. https://doi.org/10.1016/j.bbrc.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Yang JJ, Chen HJ, Yan HH, Zhang XC, Zhou Q, Su J, et al. Clinical modes of EGFR tyrosine kinase inhibitor failure and subsequent management in advanced non-small cell lung cancer. Lung Cancer. 2013;79(1):33–9. doi: 10.1016/j.lungcan.2012.09.016. https://doi.org/10.1016/j.lungcan.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 35.Qu L, Li L, Zheng X, Fu H, Tang C, Qin H, et al. Circulating plasma microRNAs as potential markers to identify EGFR mutation status and to monitor epidermal growth factor receptor-tyrosine kinase inhibitor treatment in patients with advanced non-small cell lung cancer. Oncotarget. 2017;8(28):45807–24. doi: 10.18632/oncotarget.17416. https://doi.org/10.18632/oncotarget.17416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ito S, Kamoto Y, Sakai A, Sasai K, Hayashi T, Toyooka S, et al. Unique circulating microRNAs in relation to EGFR mutation status in Japanese smoker male with lung adenocarcinoma. Oncotarget. 2017;8(70):114685–97. doi: 10.18632/oncotarget.21425. https://doi.org/10.18632/oncotarget.21425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan BA, Hughes BG. Targeted therapy for non-small cell lung cancer:Current standards and the promise of the future. Transl Lung Cancer Res. 2015;4(1):36–54. doi: 10.3978/j.issn.2218-6751.2014.05.01. https://doi.org/10.3978/j.issn.2218-6751.2014.05.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang Z, Zhang J, Zeng X, Gao J, Wu S, Liu T. Relationship between EGFR expression, copy number and mutation in lung adenocarcinomas. BMC Cancer. 2010;10:376. doi: 10.1186/1471-2407-10-376. https://doi.org/10.1186/1471-2407-10-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon reasonable request.


