Abstract
Chronic kidney disease (CKD) is a common cause of morbidity and mortality in domestic cats, but the cause is still largely elusive. While some viruses have been associated with this disease, none have been definitively implicated as causative. Recently, Rodent chaphamaparvovirus 1 was recognized as the cause of murine inclusion body nephropathy, a disease reported for over 40 years in laboratory mice. A novel virus belonging to the same genus, Carnivore chaphamapavovirus 2, was recently identified in the feces of cats with diarrhea. The goal of this study was to investigate the possible role of chaphamaparvoviruses including members of Rodent chaphamaparvorivirus 1 and Carnivore chaphamaparvovirus 2 in the development of feline CKD. The presence of these viruses was retrospectively investigated in formalin-fixed paraffin-embedded feline kidney samples using PCR, in situ hybridization, and immunohistochemistry. Cats were divided into three groups: normal (N=24), CKD (N=26), and immunocompromised (N=25). None of the kidney tissues from any of the 75 cats revealed the presence of chaphamaparvovirus DNA, RNA or antigen. We conclude that viruses belonging to the chaphamaparvovirus genus are unlikely to contribute to the occurrence of feline CKD.
Introduction
Chronic kidney disease (CKD) is a common disease of domestic cats, affecting up to 49% of cats older than 15 years, and is a significant cause of morbidity and mortality.3 Many proposed causes have been investigated or implicated, including diet, genetics, and infectious agents, however, to date, no single unifying cause has been confirmed. Recent studies have suggested that feline morbillivirus (FmoPV) or feline immunodeficiency virus (FIV) may play a role in the development of renal disease in cats, however, no direct causal relationship has been established.22,26,32,35 In addition, while feline foamy virus (FFV) was proposed as a possible causal agent of CKD, experimental infections have not shown a clear association between FFV infection and clinically significant renal disease.13,9
After eluding researchers for decades, the cause of inclusion body nephropathy (IBN) in laboratory mice was recently determined to be a novel virus named mouse kidney parvovirus (MKPV).1,16,25 Independent of this discovery in laboratory mice, “murine chapparvovirus” (MuCPV) was concurrently discovered by metagenomic analysis in wild mice trapped in residential buildings in New York City, with an infection prevalence of 13% to 45% depending on the sampling site.33 The MKPV sequences from laboratory mice are 98% identical to those of MuCPV from wild mice, and both viruses were subsequently assigned to a single species, Rodent chaphamaparvovirus 1 as the type species for the new genus Chaphamaparvovirus.21 As of June 2021, the genus Chaphamaparvovirus contains sixteen species identified in samples from a wide range of hosts including rodents, carnivores, primates, marsupials, swine, and birds [ICTV taxonomy database, https://ictv.global/taxonomy/].30 Each of these viruses has been identified in a single animal host, and their host specificity is unknown at this time. Rodent chaphamaparvovirus 1 is the only species for which a clear causal link with clinical signs or lesions has been demonstrated, where a series of experiments that fulfilled Fredrichs and Rellman’s criteria for its role in causing IBN.23 Rodent chaphamaparvovirus 1 has exquisite tropism for kidney, with shedding in the urine and possibly the feces, and experimental horizontal transmission via soiled bedding transfer and co-housing in mice.4,10,14 More recently, a novel chaphamaparvovirus was identified by metagenomic analysis of feces from cats affected by an unexplained outbreak of diarrhea. The virus was named fechavirus and assigned to the species Carnivore chaphamaparvovirus 2. Although there was a strong association between the presence of this virus and diarrhea, a causal relationship has yet to be established.12 At this time, the prevalence or host-specificity of chaphamaparvoviruses in cats and their role in inducing disease is unclear and merits further investigation.
The infection of laboratory mice with a member of the species Rodent chaphamaparvovirus 1 results in persistent viral replication in renal tubular epithelial cells, leading to a spectrum of renal lesions and clinical disease that depends on the host immune status. The histologic features and disease progression of chaphamaparvovirus associated disease in mice is similar to progressive kidney disease in cats.4
Predation and subsequent infection of cats with mouse-borne viral, bacterial or parasitic pathogens has been documented.7,19,24 While the rate of predation indoors is not known, data from outdoor cats along the urban-rural gradient suggests a rate as high as 1 mouse / cat / week, which, adjusted over a 10-year period, would account for 520 predation events.11 Given the large number of mice in New York city, cats could be exposed to chaphamaparvoviruses (as a spillover host) either by ingestion of urine or from urine-contaminated surfaces or food (similar to that shown for mouse-mouse), or by predation of the mouse and ingestion of persistently infected renal tissue.23,34 Given the high PCR prevalence of MuCPV in wild mice living in residential buildings and the strong predator-prey interaction between cats and mice, it is probable that cats are frequently exposed to rodent chaphamaparvoviruses.
We hypothesized that a member of the species Rodent chaphamaparvovirus 1 (RCPV1) may infect cats and contribute to feline CKD. Our second hypothesis was that fechavirus or a currently unknown chaphamaparvovirus with cats as its natural host may contribute to feline CKD. To test the hypotheses, we performed a retrospective study that assessed diseased and non-diseased renal tissues from cats with a normal and compromised immune status, using multiple complementary assays to detect proteins and nucleic acids of RCPV1, fechavirus and potentially other members of the chaphamaparvovirus genus, including viruses that may be currently unknown.
Materials and methods:
Selection of subjects and histologic grading
Samples were obtained from the archives of the Department of Anatomic Pathology at The Animal Medical Center that were submitted from 2008–2018 and had available clinical history and/or medical records and formalin-fixed, paraffin-embedded (FFPE) kidney tissues. Three groups were selected (CKD, immunocompromised, and control) with 25 cats originally attributed to each group, using the inclusion criteria detailed below.
Group 1 (CKD; N=24) inclusion criteria included cats with a histologic diagnosis of chronic renal disease (keywords: chronic tubulointerstitial nephritis and interstitial fibrosis) and azotemia on serum biochemical profile within 3 months prior to postmortem examination. Azotemia was defined as urea and/or creatinine levels above the reference ranges (urea 6–13 mmol/L or 16–37 mg/dL, creatinine 80–221 umol/L or 0.9–2.5 mg/dL). Kidneys were histologically graded using a simplified scheme modified from previously published scales, and is described in Supplemental Table S1.3,17 Group 1 cats required cortical inflammation, cortical fibrosis and tubular degeneration with a grade of at least 3 (Table S1). This group contained kidneys graded as mild (8 cats), moderate (8 cats), and severe (8 cats). Cases with the following diagnoses were excluded: papillary necrosis, renal dysplasia, glomerulonephritis, polycystic kidney disease, obstructive nephropathy, stents or subcutaneous ureteral bypass, nephroliths, bacterial nephritis, amyloidosis and renal neoplasia.
Group 2 (immunocompromised; N=26) required a clinical or histologic diagnosis of diseases associated with alteration of the immune status in cats, with or without renal disease. Briefly, these diseases included viruses or other types of infections in which alterations of immune status or cells within the immune system are proposed to contribute to their pathogenesis. The database was searched for the following keywords: feline immunodeficiency virus (FIV), feline leukemia virus (FeLV), feline infectious peritonitis virus (FIPV), feline panleukopenia virus, feline herpesvirus (FHV-1) and Cryptococcus. FIV and FeLV are retroviruses that are reported to cause immune suppression in infected cats.6 The clinical progression of FIPV depends upon the balance between cellular and humoral immune responses such that cats with reduced or poor cell mediated immunity are more susceptible to infection.28 Active feline panleukopenia virus infection causes lymphoid and bone marrow depletion due to the effects of the virus on rapidly dividing cells.8 FHV-1 is known to establish latency in neurons of the trigeminal ganglia and cause recrudescent disease in animals with altered immune function (e.g. stress, steroid administration).29 A cell mediated adaptive immune response is thought to play a large role in the resolution of cryptococcal infections, thus, immune compromise can contribute to infection by this opportunistic fungal agent.2,15
The severity of the renal lesions from cats in group 2 was also graded according to the modified scheme (Table S1). This group included one cat that was initially assigned to group 1, but upon further evaluation was discovered to be more appropriately categorized in the immunocompromised group given a diagnosis of cryptococcosis.
Group 3 (control; N=25) cats required bloodwork without azotemia within 3 months of postmortem examination, lack of a postmortem gross diagnosis involving the kidneys and a CKD histologic diagnosis of 0 for inflammation, fibrosis and tubular degeneration in the grading scale (Table S1).
DNA extraction, PCR reaction and PCR sensitivity assay
Five 10 μm-thick paraffin scrolls were obtained from each FFPE block and analyzed by PCR to detect the presence of chaphamaparvovirus DNA. Redundant consensus primers (named 961–963) were designed to amplify a 78 bp VP fragment found to be well-conserved in the sequences of viruses of the species Rodent chaphamaparvovirus 1 (GenBank accession MH670588), Rodent chaphamaparvovirus 2 identified in the feces from wild rats in China (GenBank accession KX272741), Chiropteran chaphamaparvovirus 1 identified in kidney of vampire bats (Desmodus rotundus) in Brazil (GenBank accession KX907333.1), and Primate chaphamaparvovirus 1 identified in kidney from a capuchin monkey (Cebus imitator) in Costa Rica (GenBank accession MN265364).10,27,30 Because this sequence is imperfectly conserved in the fechavirus (Carnivore chaphamaparovirus 2), we then designed a pair of redundant primers (named 974–975) optimized to detect fechavirus DNA.
PCR was performed on cat samples from group 1 (N=24) and from group 2 (N=26). Methods for DNA extraction were as previously reported.23
For PCR, 50–100ng of DNA was added to a cocktail comprising Phire Hot Start II DNA Polymerase (Thermo Scientific, Vilnius, Lithuania), 1x Phire Reaction Buffer, 0.2mM dNTP, 0.5μM of forward primer 961 (5’-CARCARYTNGCWATYCAAGG), 0.25μM of reverse primer 962 (5’-ATCRTCTTGTGCTCCTARTG), 0.25μM of reverse primer 963 (5’-ATCRTCTTTGACTCCTARTG) in a total 20μL reaction volume; note that primers 962 and 963 were identical but for the 3 bases underlined. The cycling conditions were: initial denaturation at 98°C for 30s, followed by 10 cycles of “touch-down” PCR with denaturation at 98°C for 5s, annealing from 60°C to 50°C for 5s (−1°C per cycle), extension at 72°C for 15s, followed by 30 cycles of denaturation at 98°C for 5s, annealing at 50°C for 5s and extension at 72°C for 15s, and concluded with a final extension at 72°C for 5min. PCR reactions to specifically detect fechavirus were identical, but replaced primers 961–963 with 0.5μM of forward primer 974 (5’-CARCAACTAGCAATACAAGG) and 0.5μM of reverse primer 975 (5’-ATCATCTTGTGC TCCTAATG). Completed PCR reactions (4μL volume) were analyzed using a Fragment Analyzer 5200 fitted with 33 or 55cm electrophoresis capillaries loaded with matrix capable of resolving dsDNA between 35 and 1500bp (Advanced Analytics, Agilent, Santa Clara, CA, USA). Capillary absorbance traces were analyzed and converted into pseudo-gel images using PROSize 3.0 software (Agilent, Santa Clara, CA, USA). To determine PCR sensitivity, two oligonucleotides (971 and 972) corresponding to nucleotides 2836–2922 of accession MN794869 and to nucleotides 2904–2990 of accession MN396757, respectively (synthesized by Integrated DNA Technologies, Singapore), were serially diluted to provide calibration templates.
Immunohistochemistry and nucleic acid in situ hybridization
For chromogenic immunohistochemistry (IHC) and nucleic acid in situ hybridization (ISH), 14-slot (3×5) tissue microarrays (TMAs) were generated with one slot left blank (for orientation purposes). Two 4 mm-diameter tissue punches were obtained at random from the renal cortices in each FFPE block and inserted into the TMA slots. Punches of renal cortex of a normal pig kidney were included as negative controls. In total, four TMA blocks were generated per group, for a total of 12 TMAs. These blocks were sectioned at 5 μm-thickness and processed for ISH and IHC.
For both methods, appropriate controls were used. Renal tissue from a NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mouse with histologic evidence of IBN and positive for MKPV by PCR, that was housed at Memorial Sloan Kettering Cancer Center (MSKCC) was used as a positive control. The sequence of this virus was previously reported as Genbank accession MH670588. Renal tissue from a histologically normal NSG mouse confirmed negative for MKPV by PCR were included as negative controls.10
Chromogenic IHC for RCPV1 was performed using previously described mouse sera and methods.23 Briefly, slides were incubated with pooled sera derived from three Tac:SW mice co-housed at MSKCC for 3 to 13 weeks with NSG mice shedding MKPV in urine, or with serum from a naïve, non-co-housed Tac:SW, and diluted at 1:1000 dilution. Slides were incubated with the secondary antibody biotin-conjugated horse anti-mouse IgG (Vector Labs, Burlingame CA, USA, Cat #) at 1:500 dilution. Avidin-biotin complete elite was added (part of the Vectastain ABC Elite Kit, Vector Labs, Burlingame, CA, USA), followed by the substrate 3,3’-diaminobenzidine (Sigma, St Louis, MO, USA).23
Chromogenic ISH for RCPV1 was performed as previously described.23 A target probe was designed for a 978-base sequence of MKPV mRNA covering the VP1 and NS1 regions, specifically nucleotides 2444–3421, using RNAscope 2.5 LS Probe (Advanced Cell Diagnostics [ACD], Newark, CA, USA). This probe has been shown to detect multiple variants of MKPV in mice and while it has not been applied to the tissues of wild mice infected by MuCPV, based on the vendor’s proprietary algorithm predicting hybridization when homology is at least 95%, it is expected to detect this virus based on the 98% homology between the MuCPV sequence and the MKPV sequence used to design the probe.10 Feline specific Peptidyl-prolyl cis-trans isomerase B (PPIB; ACD, Newark, CA, USA) was used as a positive control probe and 4-hydroxy-tetrahydrodipicolinate reductase (dapB; ACD, Newark, CA, USA) was used as a negative control probe. Control probes were substituted for the target probe, and adequate hybridization of the controls was confirmed. The slides were examined by two ACVP board-certified pathologists (AOM, TAD).
Results
Results for groups 1, 2 and 3 are summarized in Table 1. Supplemental Table S2 contains all the relevant signalment and clinicopathologic data. Results for group 1 (CKD cats) are summarized in Figures 1–3. The median ages of the cats were 16 years for group 1 (range: 5–19 years), 6 years for group 2 (0.2–18 years), and 6.5 years for group 3 (0.25–13 years).
Table 1.
Summary of IHC, ISH and PCR performed on three groups of cats.
| Group | IHC | ISH | PCR |
|---|---|---|---|
| G1 CKD (24) | 24/24 Neg | 24/24 Neg | 24/24 negative for chaphamaparvovirus DNA |
| G2 Immunocompromised (26) | 26/26 Neg | 26/26 Neg | 26/26 negative for chaphamaparvovirus DNA |
| G3 Control cats (25) | 25/25 Neg | 25/25 Neg | Not Performed |
Group 1: cats with Chronic Kidney Disease (CKD); Group 2: immunocompromised cats; Group 3: control cats with no renal disease or immune compromise.
Figures 1-9.
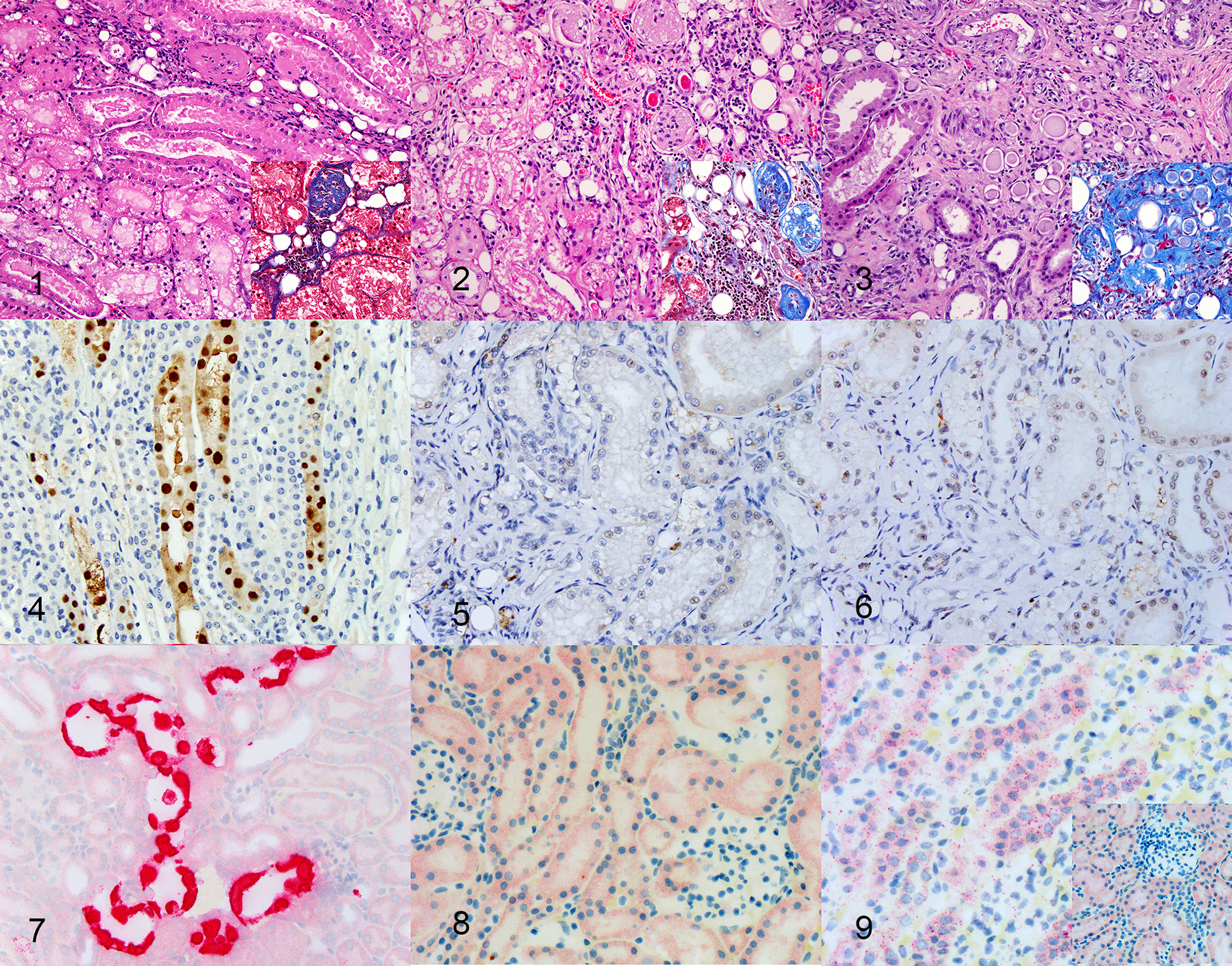
Figures 1–3: Chronic renal disease, kidney, cat. Hematoxylin and eosin (insets: Masson’s trichrome).
Figure 1. Case 5. Small amounts (<25%) of interstitial inflammation and fibrosis within the cortex with focal or scattered tubular degeneration.
Figure 2: Moderate chronic renal disease, kidney, cat, Case 2. Moderate amounts (25–50%) of interstitial inflammation and fibrosis within the cortex with multifocal to coalescing tubular degeneration
Figure 3: Severe chronic renal disease, kidney, cat, Case 16. Severe (>50%) interstitial inflammation and fibrosis with tubular degeneration involving entire nephron units.
Figure 4: Mouse kidney parvovirus (MKPV) infection, kidney, immunodeficient NSG mouse. Strong nuclear and mild cytoplasmic immunoreactivity in tubular epithelial cells. Immunohistochemistry with co-housed mouse serum.
Figures 5, 6: Kidney, cat. Weak, non-specific background chromogen signal, indicating a negative result in a cat with chronic kidney disease (Fig 5) and a normal cat (Fig 6). Immunohistochemistry with co-housed mouse serum.
Figure 7: MKPV infection, kidney, immunodeficient NSG mouse. Strong nuclear and cytoplasmic hybridization in tubular epithelial cells. RNA in situ hybridization with MKPV probe.
Figure 8: Chronic Kidney disease, cat. Weak, non-specific punctate background cytoplasmic chromogen signal indicating a negative result. RNA in situ hybridization with MKPV probe.
Figure 9: Positive control, kidney, cat. Moderate numbers of cytoplasmic intense punctate dots, consistent with hybridization, indicating good RNA sample quality. RNA in situ hybridization with PPIB. Inset: Negative control, kidney, cat. Weak, non-specific punctate background cytoplasmic chromogen signal, similar to MKPV probe (Figure 8). RNA in situ hybridization with DapB.
PCR was negative for all cats from groups 1 and 2. ISH and IHC did not reveal the presence of target nucleic acids or antigen in any of the samples tested (Figs. 4–9). All IHC and ISH positive and negative mouse control tissues showed acceptable positive and negative immunoreactivity and hybridization, respectively. MKPV-infected positive control urine produced a specific product in PCR reactions using broad-specificity primers 961–962/963 or fechavirus-optimized primers 974–975, with sensitivity for as few as 5 templates under ideal conditions (Figs S1–S2). The sections immunolabeled with sera of co-housed and non-co-housed mice had brown chromogen in approximately 2-μm-diameter, round structures within the tubular epithelial cytoplasm, which was interpreted as non-specific chromogen signal (Figs 5–6). Similarly, red chromogen highlighted similar structures when ISH was performed, in addition to diffuse, pinpoint, red chromogen likely representing non-specific signal in the cytoplasm of most samples with no difference between positive (feline PPIB), negative (dapB) and RCPV1 probes (Figs. 8–9). This signal was distinct from the intense punctate hybridization expected for a positive reaction with this assay, as observed in sections hybridized with the probe for feline PPIB (Fig 9). In addition, viral hybridization with RCPV1 is expected to be both cytoplasmic and nuclear (Fig 7).23 The intensity of chromogen signal interpreted as non-specific varied between samples. All samples showed hybridization with feline PPIB probes, confirming adequate preservation of RNA.
Discussion
RCPV1 nucleic acids and protein were not detectable in FFPE renal tissues of cats with CKD, cats without renal disease, or immunocompromised cats. Our PCR results from groups 1 and 2 did not demonstrate the presence of fechavirus or a currently unknown but closely related chaphamaparvovirus, in FFPE renal tissues of cats with CKD or immune compromise. Therefore, these findings do not support the hypotheses that a member of RCPV1 or an unknown chaphamaparvovirus is a prominent cause or cofactor in the pathogenesis of feline CKD.
The pathogenesis of viral nephropathies are often multifactorial and include genetic factors, the renal immunological microenvironment, and cellular responses.22 Previously-implicated viral causes of feline CKD include FmoPV, FFV and FIV.22,26,32,35 One study found that cats younger than 11 years of age with CKD were significantly more likely to be positive for FIV antibodies as compared with cats without CKD.32 Reported renal alterations in FIV-infected cats encompass predominantly glomerular changes including mesangial cell proliferation and glomerulosclerosis, with a smaller percentage showing immune-mediated glomerulonephritis and amyloidosis.22 FmoPV was described with tubulointerstitial nephritis in domestic cats in Hong Kong with detectable antigen observed with IHC in a small number of cases.26,35 None of the cats in our database were tested for FmoPV or FFV, so we were not able to assess renal tropism of these viruses.
Our study is limited by its retrospective nature and usage of FFPE tissues ranging from 2008 to 2018. Antigen, DNA and RNA degradation over time is possible and formalin fixation may decrease sensitivity, although in the case of ISH, appropriate positive controls (Feline PPIB) showed that even the earliest archival tissue had adequate hybridization. In addition, the RNA-scope ISH assay was in part designed to address the issues of nucleic acid quality degradation in FFPE specimens, and it provides a morphologic context that is complementary to PCR.20,31 Similarly, while cost-effective, the use of TMAs to test all animals may have decreased detection sensitivity. Feline CKD has a multifocal tissue distribution, such that sampling two 4 mm-diameter core biopsies per kidney may be insufficient to detect antigen or nucleic acids had they been present.23 To account for this, thick FFPE scrolls from groups 1 and 2 were sampled and tested with primers that targeted a well-conserved region of chaphamaparvoviruses, not limited to RCPV1 (Fig S1). We hypothesize, that a broader limitation to identifying a suspected viral infectious agent with a presumed single-organ tropism as a suspected cause of CKD in situ, is that the massive loss of parenchyma may result in loss of target host cells, which could be the site of viral persistence.18 We addressed this concern by including cats at all stages of CKD spanning a large age distribution in addition to cats with a compromised immune status that could potentially be more susceptible to persistent infection.
In this study, we used three different approaches to detect viral nucleic acids and antigen: ISH, 40-cycle DNA PCR, and chromogenic IHC. This multi-pronged approach to discover DNA viruses specifically, provides higher confidence and validity in a positive or negative result. Although probably less sensitive than 40-cycle PCR (Figs S1–S2), which has a detection limit of <10 RCPV1 genomes, RNA-ISH has higher sensitivity than low cycle-number screening PCR, as some mouse FFPE kidney samples that were negative for RCPV1 by 25-cycle screening PCR were positive by viral nucleic acid ISH.5,10,23 A disadvantage of ISH is that variations in viral nucleotide sequence may result in negative hybridization, given the theoretical requirement of 95% homology between the probe and target. Our previous experience with this probe has indicated that it can detect multiple variants within the Rodent chaphamaparvovirus 1 genus, which share >95% homology, but its unlikely it will detect other related viral species.10 This issue was addressed by PCR from FFPE specimens, using redundant primers designed to recognize several chaphamaparvoviruses previously detected in kidneys, as well as fechavirus, thus covering a broad range of chaphamaparvoviruses. Since DNA is often fragmented or damaged in FFPE samples, PCR sensitivity was maximized by using 40 PCR cycles with a low Tm for the final 30 cycles and by targeting a small amplicon (78bp) with a highly processive polymerase.10,12 By definition, the serum from wild-type Tac:SW mice co-housed with MKPV-infected NSG mice is polyclonal. As such, it was expected to recognize multiple epitopes of this virus, some of which may be conserved among other related chaphamaparvoviruses. However, this presumptive cross-reactivity could not be tested due to the lack of availability of tissues infected by other chaphamaparvoviruses, most of which have been described only by metagenomic analysis without concurrent histopathologic analyses. In addition, the serum would likely recognize any infectious agent to which the Swiss Webster mice have been exposed and that persistently infects any of the cats in our study. While we have clearly demonstrated that this serum can detect MKPV in immunodeficient mice with abundant virus (Fig. 4), the relative sensitivity and specificity of co-housed serum in relation to PCR or ISH is untested.
In our study, we included cats that were affected by CKD (group 1) as well as immunocompromised cats (group 2). In addition, the age range of these 50 veterinary patients was broad (0.2 to 19 years of age). The inclusion of immunocompromised cats was justified by the a high degree of viral persistence and susceptibility to disease development seen in NSG and Rag1 deficient mice infected with RCPV1, which is likely due to their lack of adaptive immunity. We hypothesized that immunocompromised cats may have been at a higher risk for infection by RCPV1, although our results do not support this. Similarly, including a large age-range in both groups, and range of disease severity in the CKD group, decreases the likelihood of false-negative results due to a cleared MKPV infection. However, in NSG mice, MKPV antigen and DNA can still be detected in aged mice with severe CKD lesions, and this has more recently been observed in aged immunocompetent Swiss Webster mice with milder lesions as well (AOM, unpublished data). Finally, these tissue samples were obtained from a defined geographical location (Northeastern USA) and may not reflect global trends in cats with CKD. Replication of these results in cats from a broader geographical origin may be useful.
We conclude that members of RCPV1, fechavirus or another chaphamaparvovirus are not a likely cause or contributing factor to CKD in domestic cats.
Supplementary Material
Supplemental Figures S1-S3: Primer design and 40-cycle PCR sensitivity assay for RCPV1 and related viruses.
Figure S1: Alignment of PCR primers with a VP region conserved in mouse RCPV1 and related chaphamaparvoviruses. Blue boxes indicate the binding regions of primers 961, 962, 963, 974 and 975 - with accession numbers and base positions. A single mismatch between primer 961 and the known cat chaphamaparvoviruses is indicated in red.
Figure S2: Example capillary electrophoretograms of 40-cycle 974–975 PCR products amplified from a dilution series of oligonucleotide 972 templates (5 to 50,000 input templates), 1μL MKPV-infected mouse urine (MKPV+ urine) or 50ng FFPE-extracted cat kidney DNA (cat DNA #1–1; #1–2; #1–3). Specific PCR product is indicated by the grey line; elution of 35 bp (“lower”) and 1,500 bp (“upper”) internal markers is also indicated.
Figure S3: Sensitivity curve (Padé approximant, Prism v8.3.0, GraphPad Software) of normalized peak areas for 40-cycle 974–975 PCR products produced from dilution series of template oligonucleotides 971 or 972. The curves indicate that 40-cycle PCR detected the two known fechavirus variants with similar sensitivity.
Acknowledgments
This work was funded by the Caspary Research Institute of The Animal Medical Center. AOM and SM were supported in part by NCI Cancer Center Support Grant P30 CA008748. TAD was supported by the Caspary Research Institute of the Animal Medical Center. BR, LQ and CJ were supported by Australian National Health and Medical Research Council, the Cancer Institute NSW and the Hillcrest Foundation. The authors would like to thank Amanda Ramkissoon for data collection and performing special stains, John D’Allara for TMA preparation, Iveta Simanska for TMA sectioning, and Maria Jiao for performing IHC and RNA-ISH.
Footnotes
Declaration of conflicting interests: AOM is currently an employee of Regeneron Pharmaceuticals. None of the work herein was performed at or funded by Regeneron. BR is presently an employee at Novartis Institutes for BioMedical Research. Novartis did not fund the study. The authors declare no potential conflicts of interest with respect to the research, authorship and/or publication of this article.
References:
- 1.Baze WB, Steinbach TJ, Fleetwood ML, et al. Karyomegaly and intranuclear inclusions in the renal tubules of sentinel ICR mice (mus musculus). Comp Med. 2006;56: 435–438. [PubMed] [Google Scholar]
- 2.Carroll SF, Guillot L, Qureshi ST. Mammalian model hosts of cryptococcal infection. Comp Med. 2007;57: 9–17. [PubMed] [Google Scholar]
- 3.Chakrabarti S, Syme HM, Brown CA, et al. Histomorphometry of feline chronic kidney disease and correlation with markers of renal dysfunction. Vet Pathol. 2013;50: 147–155. [DOI] [PubMed] [Google Scholar]
- 4.Edmondson EF, Hsieh WT, Kramer JA, et al. Naturally Acquired Mouse Kidney Parvovirus Infection Produces a Persistent Interstitial Nephritis in Immunocompetent Laboratory Mice. Vet Pathol. 2020;57: 915–925. [DOI] [PubMed] [Google Scholar]
- 5.Ge Z, Carrasco SE, Feng Y, et al. Identification of a new strain of mouse kidney parvovirus associated with inclusion body nephropathy in immunocompromised laboratory mice. Emerg Microbes Infect. 2020;9: 1814–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartmann K Clinical aspects of feline retroviruses: a review. Viruses. 2012;4: 2684–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingram WM, Goodrich LM, Robey EA, et al. Mice infected with low-virulence strains of Toxoplasma gondii lose their innate aversion to cat urine, even after extensive parasite clearance. PLoS One. 2013;8: e75246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamm CG, Rezabek GB. Parvovirus infection in domestic companion animals. Vet Clin North Am Small Anim Pract. 2008;38: 837–850, viii-ix. [DOI] [PubMed] [Google Scholar]
- 9.Ledesma-Feliciano C, Troyer RM, Zheng X, et al. Feline Foamy Virus Infection: Characterization of Experimental Infection and Prevalence of Natural Infection in Domestic Cats with and without Chronic Kidney Disease. Viruses. 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Q, Padula MP, Pinello N, et al. Murine and related chapparvoviruses are nephro-tropic and produce novel accessory proteins in infected kidneys. PLoS Pathog. 2020;16: e1008262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lélu M, Langlais M, Poulle ML, et al. Transmission dynamics of Toxoplasma gondii along an urban-rural gradient. Theor Popul Biol. 2010;78: 139–147. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Gordon E, Idle A, et al. Virome of a Feline Outbreak of Diarrhea and Vomiting Includes Bocaviruses and a Novel Chapparvovirus. Viruses. 2020;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin J, Jackson K, Alex CE et al. Investigating the role of feline foamy virus in chronic kidney disease. Proceedings of the annual meeting of the American College of Veterinary Pathologists (ACVP). 2019. [Google Scholar]
- 14.Liu W, Zhang Y, Ma J, et al. Determination of a novel parvovirus pathogen associated with massive mortality in adult tilapia. PLoS Pathog. 2020;16: e1008765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorente-Méndez C, Martínez CM, Corpa JM. Pathology in practice. Systemic cryptococcosis caused by C. neoformans and concomitant severe pulmonary aelurostrongylosis. J Am Vet Med Assoc. 2009;235: 1407–1409. [DOI] [PubMed] [Google Scholar]
- 16.McInnes E, Bennett M, O’Hara M, et al. Intranuclear Inclusions in Renal Tubular Epithelium in Immunodeficient Mice Stain with Antibodies for Bovine Papillomavirus Type 1 L1 Protein. Vet Sci. 2015;2: 84–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLeland SM, Cianciolo RE, Duncan CG, et al. A comparison of biochemical and histopathologic staging in cats with chronic kidney disease. Vet Pathol. 2015;52: 524–534. [DOI] [PubMed] [Google Scholar]
- 18.Nankivell BJ, Renthawa J, Sharma RN, et al. BK Virus Nephropathy: Histological Evolution by Sequential Pathology. Am J Transplant. 2017;17: 2065–2077. [DOI] [PubMed] [Google Scholar]
- 19.Nowotny N, Weissenboeck H, Aberle S, et al. Hantavirus infection in the domestic cat. Jama. 1994;272: 1100–1101. [DOI] [PubMed] [Google Scholar]
- 20.Okello JB, Zurek J, Devault AM, et al. Comparison of methods in the recovery of nucleic acids from archival formalin-fixed paraffin-embedded autopsy tissues. Anal Biochem. 2010;400: 110–117. [DOI] [PubMed] [Google Scholar]
- 21.Pénzes JJ, Söderlund-Venermo M, Canuti M, et al. Reorganizing the family Parvoviridae: a revised taxonomy independent of the canonical approach based on host association. Arch Virol. 2020;165: 2133–2146. [DOI] [PubMed] [Google Scholar]
- 22.Poli A, Tozon N, Guidi G, et al. Renal alterations in feline immunodeficiency virus (FIV)-infected cats: a natural model of lentivirus-induced renal disease changes. Viruses. 2012;4: 1372–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roediger B, Lee Q, Tikoo S, et al. An Atypical Parvovirus Drives Chronic Tubulointerstitial Nephropathy and Kidney Fibrosis. Cell. 2018;175: 530–543.e524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rollag OJ, Skeels MR, Nims LJ, et al. Feline plague in New Mexico: report of five cases. J Am Vet Med Assoc. 1981;179: 1381–1383. [PubMed] [Google Scholar]
- 25.Santagostino SF, Arbona RJR, Nashat MA, et al. Pathology of Aging in NOD scid gamma Female Mice. Vet Pathol. 2017;54: 855–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sieg M, Heenemann K, Rückner A, et al. Discovery of new feline paramyxoviruses in domestic cats with chronic kidney disease. Virus Genes. 2015;51: 294–297. [DOI] [PubMed] [Google Scholar]
- 27.Souza WM, Romeiro MF, Fumagalli MJ, et al. Chapparvoviruses occur in at least three vertebrate classes and have a broad biogeographic distribution. J Gen Virol. 2017;98: 225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tekes G, Thiel HJ. Feline Coronaviruses: Pathogenesis of Feline Infectious Peritonitis. Adv Virus Res. 2016;96: 193–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thiry E, Addie D, Belák S, et al. Feline herpesvirus infection. ABCD guidelines on prevention and management. J Feline Med Surg. 2009;11: 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker PJ, Siddell SG, Lefkowitz EJ, et al. Changes to virus taxonomy and the Statutes ratified by the International Committee on Taxonomy of Viruses (2020). Arch Virol. 2020;165: 2737–2748. [DOI] [PubMed] [Google Scholar]
- 31.Wang F, Flanagan J, Su N, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14: 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White JD, Malik R, Norris JM, et al. Association between naturally occurring chronic kidney disease and feline immunodeficiency virus infection status in cats. J Am Vet Med Assoc. 2010;236: 424–429. [DOI] [PubMed] [Google Scholar]
- 33.Williams SH, Che X, Garcia JA, et al. Viral Diversity of House Mice in New York City. mBio. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams SH, Che X, Paulick A, et al. New York City House Mice (Mus musculus) as Potential Reservoirs for Pathogenic Bacteria and Antimicrobial Resistance Determinants. mBio. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woo PC, Lau SK, Wong BH, et al. Feline morbillivirus, a previously undescribed paramyxovirus associated with tubulointerstitial nephritis in domestic cats. Proc Natl Acad Sci U S A. 2012;109: 5435–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figures S1-S3: Primer design and 40-cycle PCR sensitivity assay for RCPV1 and related viruses.
Figure S1: Alignment of PCR primers with a VP region conserved in mouse RCPV1 and related chaphamaparvoviruses. Blue boxes indicate the binding regions of primers 961, 962, 963, 974 and 975 - with accession numbers and base positions. A single mismatch between primer 961 and the known cat chaphamaparvoviruses is indicated in red.
Figure S2: Example capillary electrophoretograms of 40-cycle 974–975 PCR products amplified from a dilution series of oligonucleotide 972 templates (5 to 50,000 input templates), 1μL MKPV-infected mouse urine (MKPV+ urine) or 50ng FFPE-extracted cat kidney DNA (cat DNA #1–1; #1–2; #1–3). Specific PCR product is indicated by the grey line; elution of 35 bp (“lower”) and 1,500 bp (“upper”) internal markers is also indicated.
Figure S3: Sensitivity curve (Padé approximant, Prism v8.3.0, GraphPad Software) of normalized peak areas for 40-cycle 974–975 PCR products produced from dilution series of template oligonucleotides 971 or 972. The curves indicate that 40-cycle PCR detected the two known fechavirus variants with similar sensitivity.


