Abstract
Alkanes are oxidized in Acinetobacter sp. strain ADP1 by a three-component alkane monooxygenase, composed of alkane hydroxylase, rubredoxin, and rubredoxin reductase. rubA and rubB encode rubredoxin and a NAD(P)H-dependent rubredoxin reductase. We demonstrate here that single base pair substitutions in rubA or rubB lead to defects in alkane degradation, showing that both genes are essential for alkane utilization. Differences in the degradation capacity for hexadecane and dodecane in these mutants are discussed. Two genes, estB and oxyR, are located downstream of rubB, but are not necessary for alkane degradation. estB encodes a functional esterase. oxyR encodes a LysR-type transcriptional regulator, conferring resistance to hydrogen peroxide. rubA, rubB, estB, and oxyR constitute an operon, which is constitutively transcribed from a ς70 promoter, and an estB-oxyR containing message is also transcribed from an internal promoter.
Within gram-negative bacteria, Acinetobacter and Pseudomonas are the most important genera for the degradation of n-alkanes in the environment. Pseudomonas oleovorans, which is able to use medium-chain alkanes ranging from hexane to dodecane as the sole source of carbon and energy (36), contains the alk genes necessary for the conversion of alkanes to acyl coenzyme A separated into two regions on the OCT plasmid. The alkBFGHJKL genes constitute an operon and encode the alkane hydroxylase, two rubredoxins, an aldehyde dehydrogenase, an alcohol dehydrogenase, an acyl coenzyme A synthetase, and an outer membrane protein of unknown function. The second locus contains alkS and alkT, encoding a LuxR-UhpA-like regulator of alk operon transcription and rubredoxin reductase. In the initial degradation step, alkane is converted to the primary alcohol in P. oleovorans by a three-component alkane monooxygenase, composed of alkane hydroxylase, rubredoxin, and rubredoxin reductase. Several alkane oxidation pathways have been described for Acinetobacter spp. An alkane dioxygenase is involved in degradation of long-chain alkanes (C13 to C44) in Acinetobacter sp. strain M-1 (24). The alkane hydroxylase in some Acinetobacter strains able to grow on medium-chain alkanes is a cytochrome P-450 (2), while a rubredoxin- and rubredoxin reductase-dependent alkane hydroxylase is present in Acinetobacter calcoaceticus 69-V growing on long-chain alkanes (C11 to C18) (1).
Acinetobacter sp. strain ADP1 is able to grow on alkanes of 12 or more carbon atoms (31). The genes necessary for alkane degradation are spread on the chromosome in at least three loci: xcpR is a component of the general secretory pathway (29). alkM and the divergent alkR encode the alkane hydroxylase and an activator of the AraC-XylS family, necessary for alkM transcription (30, 31). rubA was mapped on a third locus and encodes a rubredoxin (54 amino acids [aa]) that differs markedly in size from that of P. oleovorans (172 aa) (9). The neighboring gene, rubB, encodes the rubredoxin reductase, as hypothesized on the basis of sequence similarity to NAD(P)H-dependent dehydrogenases (9).
In this paper, we describe sequence analysis of the DNA downstream of rubB, revealing two open reading frames (ORFs), estB and oxyR. We demonstrate that rubA and rubB are necessary for alkane degradation in ADP1 and that they form an operon together with estB and oxyR. estB and oxyR encode a functional esterase and a peroxide response regulator, respectively, which are not necessary for alkane degradation.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Wild-type Acinetobacter sp. strain ADP1 was formerly classified as A. calcoaceticus ADP1 and is synonymously called Acinetobacter sp. strain BD413 (17, 34).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| Acinetobacter | ||
| ADP1 | Wild type | 17, 34 |
| WH362 | rubA(ΔMscI-Tth111I)::lacZ Kmr | 9 |
| WH363 | EMS mutant, alk | 9 |
| WH380 | ORFX(MscI)::lacZ Kmr | This study |
| WH382 | rubB(ΔNarI-NruI)::lacZ Kmr | This study |
| WH384 | estB(ΔNsiI-StyI)::lacZ Kmr | This study |
| WH384ΔoxyR | estB(ΔNsiI-StyI)::lacZ oxyR(ΔAflII-NdeI) Kmr Cmr | This study |
| WH386 | oxyR(ΔAflII-NdeI)::lacZ Kmr | This study |
| WH432 | EMS mutant, alk | 9 |
| WH433 | EMS mutant, alk | 9 |
| WH434 | EMS mutant, alk | 9 |
| E. coli DH5α | recA1 endA1 supE44 gyrA96 thi hsdR17(rK−mK−) relA1 φ80ΔlacZΔM15 Δ(lacZYA-argF)U169 | 13 |
| Plasmids | ||
| pAKA22 | Apr Tcr; 11.6-kbp Sau3A fragment of Acinetobacter DNA containing estB in BclI site of pUN121 | 19 and this study |
| pBluescript II SK+ | Apr | Stratagene, La Jolla, Calif. |
| pKOK6.1 | Apr Kmr; promoterless lacZ | 21 |
| pKT210 | Cmr | 5 |
| pWH891 | Apr; 10,855-bp Acinetobacter DNA | 10 |
| pWH891ΔAflII | Apr; pWH891, deletion of AflII fragment (4,348 bp) containing ORFX, rubA, rubB, and estB | This study |
| pWH891ΔNcoI | AproxyR | This study |
| pWH891SK3 | Apr; HindIII-PvuII fragment (3,079 bp) from pWH891 in EcoRV site of pBluescript II SK+ | This study |
| pWH891SK3ORFX::lacZ | Apr Kmr; ORFX(MscI)::lacZ fusion | This study |
| pWH891SK6 and SK6i | Apr; NruI-EcoRV fragment (2,149 bp) from pWH891 in EcoRV site of pBluescript II SK+ | This study |
| pWH891SK6estB::lacZ | Apr Kmr; estB(ΔNsiI-StyI)::lacZ fusion | This study |
| pWH891SK7i | Apr; HindIII-BamHI fragment (4,727 bp) from pWH891 in EcoRV site of pBluescript II SK+ | This study |
| pWH891SK7ioxyR::Cmr | Apr Cmr; oxyR(ΔAflII-NdeI)::Cmr fusion | This study |
| pWH891SK7ioxyR::lacZ | Apr Kmr; oxyR(ΔAflII-NdeI)::lacZ fusion | This study |
| pWH963ΔMH | Apr; MscI-DraI fragment (1,382 bp) from pWH891 in EcoRV site of pBluescript II SK+ | This study |
| pWH963ΔMHrubB::lacZ | Apr Kmr; rubB(ΔNarI-NruI)::lacZ fusion | This study |
General methods.
Escherichia coli and Acinetobacter were transformed as described previously (13, 28) or electroporated with a Gene Pulser (Bio-Rad Laboratories, Munich, Germany). Total DNA was prepared according to the method of Ausubel et al. (4). Small-scale preparations of plasmids were made by the boiling lysis method (15); large-scale preparations were done with the Nucleobond kit (Macherey-Nagel, Düren, Germany). Total RNA was isolated with the RNeasy Mini kit from Qiagen (Hilden, Germany).
Media and growth conditions.
E. coli was grown at 37°C and Acinetobacter was grown at 28°C. Ampicillin was used at 100 mg/liter for E. coli and 300 mg/liter for Acinetobacter. Kanamycin and chloramphenicol were used for Acinetobacter at 5 and 10 mg/liter, respectively. Growth with alkanes as the carbon source was monitored on plates as described previously (29). Indicator plates for esterase activity contained 1.5% (vol/vol) tributyrin in Difco nutrient broth (NB) (Difco, Detroit, Mich.). Tributyrin was added as a 50% (vol/vol) emulsion in 5% (wt/vol) gum arabic (Sigma, Steinheim, Germany) after sonication with a Branson Sonifier B12 (Braun, Melsungen, Germany).
DNA sequence analysis.
Nucleotide sequences on both strands were determined by the dideoxy chain termination method (33) with Sequenase (U.S. Biochemical Corp., Cleveland, Ohio) and [α-32P]dATP. Successive deletions were done by using the double-stranded nested deletion kit (Pharmacia, Freiburg, Germany). Sequences were analyzed with the GCG software package (6). The GCG program FASTA was used to determine the similarity (percentage of identical amino acids) over the whole protein sequence. Database searches were done by using the Blast 2.0 software offered by the National Center for Biotechnology Information (26a).
Primer extension.
Primer extension reactions were performed as described previously (37). Total RNA (15 μg) was incubated for 5 min at 80°C and hybridized for 5 min with the 5′-end-labeled primer (50 fmol) at 37°C. The reaction mixtures containing 9 U of avian myeloblastosis virus reverse transcriptase (Promega, Madison, Wis.) were incubated for 45 min at 37°C. One-third of the volume was loaded onto a sequencing gel and analyzed with a PhosphorImager (Fujifilm; BAS-1500). The sequence of the rubA-specific primer is 5′-CTTGTGGCCAGCCTTCG-3′.
Southern and Northern hybridization.
Southern and Northern hybridization were performed as described before (11). Total RNA (11 μg per lane) was run on 1% or 2% agarose gels containing 6% formaldehyde and blotted onto a positively charged nylon membrane (Porablot NY Plus; Macherey-Nagel, Düren, Germany) by capillary transfer as described by Sambrook et al. (32). Radioactivity on the membrane was detected with a PhosphorImager. The specific probes were prepared by PCR with [α-32P]dATP amplifying fragments from nucleotides (referring to the sequence under accession no. Z46863) 5778 to 6104 (rubA), 6237 to 7072 (rubB), 7381 to 8289 (estB), and 8323 to 9108 (oxyR).
Chromosomal disruption of ORFX, rubB, estB, and oxyR.
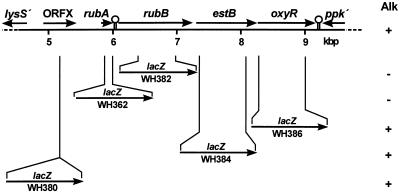
For construction of WH362 (rubA::lacZ) see the article by Geißdörfer et al. (9). DNA fragments harboring the gene to be inactivated were excised from pWH891, filled in with Klenow polymerase, and cloned into the EcoRV site of pBluescript II SK+, resulting in pWH891SK3, pWH963ΔMH, pWH891SK6, and pWH891SK7i (Table 1). The 4.7-kbp lacZ-Kmr cassette was excised with BamHI from pKOK6.1, filled in with Klenow polymerase, and cloned into these plasmids to yield fusions of lacZ to ORFX, rubB, estB, and oxyR on the resulting integration plasmids pWH891SK3ORFX::lacZ, pWH963ΔMHrubB::lacZ, pWH891SK6estB::lacZ, and pWH891SK7ioxyR::lacZ, respectively (Table 1). In the case of rubB, estB, and oxyR, internal fragments of the genes have been deleted during the cloning steps (Fig. 1 and Table 1). The integration plasmids were cut with ApaI and SacII within the vector sequence to prevent integration of the circular plasmids, and the linear DNAs were used to transform Acinetobacter sp. strain ADP1. Transformants were selected on Luria-Bertani (LB) plates with kanamycin, and correct integration of the cassettes was confirmed by Southern hybridization (data not shown). The chromosomal organization of the resulting strains, called WH380 (ORFX::lacZ), WH382 (rubB::lacZ), WH384 (estB::lacZ), and WH386 (oxyR::lacZ), is shown in Fig. 1.
FIG. 1.
Schematic drawing of the relevant DNA from Acinetobacter sp. strain ADP1 cloned on pWH891. Numbers indicate the kilobase pair scale of the sequence in the EMBL database (accession no. Z46863). Inverted repeats are indicated as stem-loop structures. In the lower part, the chromosomal characteristics of the mutants with inserted lacZ-Kmr cassettes (simplified as lacZ) are shown. Alk, phenotype on alkane (+, growth; −, no growth); estB, esterase; oxyR, peroxide response regulator; lysS, lysyl tRNA synthetase; ppk, polyphosphate kinase; rubA, rubredoxin; rubB, rubredoxin reductase.
An additional strain (WH384ΔoxyR), defective in both estB and oxyR, was constructed by transformation of WH384 with pWH891SK7ioxyR::Cmr, linearized with ApaI and PstI, and by selection of transformants on LB plates with chloramphenicol. pWH891SK7ioxyR::Cmr is equivalent to pWH891SK7ioxyR::lacZ, but carries a 3.2-kbp Cmr cassette from pKT210 (PstI fragment, filled in with Klenow polymerase) instead of the lacZ-Kmr cassette. Chromosomal disruptions of estB and oxyR in WH384ΔoxyR were confirmed by Southern hybridization (data not shown). The strains were resistant to kanamycin and chloramphenicol.
Killing zone assay.
For qualitative determination of sensitivity to hydrogen peroxide, the killing zone assay (22) was modified. Two hundred microliters of 1:10,000 dilution of overnight cultures in LB medium were spread on LB plates. After incubation for 2 h, 5 μl of a hydrogen peroxide solution was spotted onto the plates, and the killing zones were measured after incubation for 1 day. Standard deviations obtained from three independent experiments were lower than 15% of the respective means.
Measurement of EstB activity.
E. coli was transformed with plasmid pAKA22 containing estB and grown in nutrient broth with 50 mM K2HPO4 (pH 7.0) and ampicillin to early stationary phase. Cells were washed once in ice-cold 50 mM Tris-HCl (pH 8.0), concentrated twofold by centrifugation, and sonicated on ice at 75 W (duty cycle, 50%) for 3 min with a Branson 250 sonifier. The suspension was immediately used for determination of esterase activity as described previously (19). p-Nitrophenol (pNP) esters with different alkyl chain lengths (20) were used as substrates at a final concentration of 2 mM, and the formation of pNP was measured spectrophotometrically at 410 nm.
Nucleotide sequence accession number.
The nucleotide sequences of estB and oxyR from Acinetobacter sp. strain ADP1 have been deposited in the EMBL database under accession no. Z46863 and X88895.
RESULTS
Sequence analysis of estB and oxyR from Acinetobacter sp. strain ADP1.
The 2,152-bp NruI-EcoRV fragment from pWH891 containing the DNA between rubB (formerly called ORF2 [9]) and ppk (11) was cloned in both orientations into the EcoRV site of pBluescript SK II+, resulting in plasmids pWH891SK6 and pWH891SK6i. Sequence analysis revealed two ORFs, estB and oxyR, in the same orientation as ORFX, rubA, and rubB (Fig. 1). Inverted repeats were found downstream of rubA (AAAAGACCATGT-N7-ACATGGTCTTTT) and oxyR (AAAAAAGGAGTCTTTAAAGACTCCTTTTTT). The estB-oxyR locus was independently cloned on plasmid pAKA22 by transformation of a genomic library of Acinetobacter sp. strain ADP1 into E. coli and screening of the resulting colonies for halo formation on NB plates containing tributyrin. estB encodes a protein of 312 aa with similarity to a putative esterase (LipG [301 aa]) from Mycobacterium tuberculosis (50% identical amino acids [EMBL accession no. Z92772]), poly(3-hydroxyalkanoate) depolymerase (PhaB [283 aa]) (16) from P. oleovorans (30% identical amino acids [SwissProt accession no. P26495), and β-ketoadipate enol-lactone hydrolase (PcaD [260 aa]) from Bradyrhizobium japonicum (28% identical amino acids [EMBL accession no. Y10223]). The EstB sequence contains the GXSXG box (aa 136 to 140), which forms the catalytic triad together with aspartate and histidine residues in serine hydrolases like lipases, esterases, and proteases.
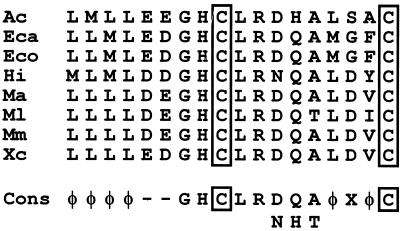
The oxyR-encoded protein (301 aa) is homologous to the functionally characterized OxyR from Xanthomonas campestris (313 aa [35% identical amino acids]) (26), E. coli (305 aa [34% identical aa]) (38), Mycobacterium marinum (311 aa [34% identical amino acids]) (27), Haemophilus influenzae (301 aa [33% identical amino acids]) (23), and Mycobacterium leprae (311 aa [32% identical amino acids]) (7); to the putative OxyR from Erwinia carotovora (302 aa [33% identical amino acids]) (GenBank accession no. U74302) and M. avium (311 aa [31% identical amino acids]) (SwissProt accession no. P52677); and to other members of the LysR family of transcriptional regulators (28 to 24% identical amino acids). OxyR from Acinetobacter shows the highly conserved N-terminal α-helix–turn–α-helix motif (aa 22 to 41) and matches a new signature of 18 aa (OxyR box [Fig. 2]), which specifically identifies OxyR sequences within the databases.
FIG. 2.
Part of a multiple alignment of OxyR sequences and the deduced consensus sequence (Cons) of the OxyR box. Amino acids are shown in the one-letter code. The conserved cysteine residues forming reversible disulfide bonds are boxed. Ac, Acinetobacter sp. strain ADP1; Eca, E. carotovora; Ec, E. coli; Hi, H. influenzae; Ma, M. avium; Ml, M. leprae; Mm, M. marinum; φ, hydrophobic amino acids; −, acidic amino acids; X, any amino acid.
rubA and rubB are necessary for alkane degradation in Acinetobacter sp. strain ADP1.
To examine the function of the ORFs, we inserted a lacZ-Kmr cassette into ORFX, rubB, estB, and oxyR on the chromosome of Acinetobacter sp. strain ADP1 (see Materials and Methods) and tested the resulting strains for growth on minimal medium plates with dodecane or hexadecane as the sole carbon source (Fig. 1). WH380 (ORFX::lacZ), WH384 (estB::lacZ) and WH386 (oxyR::lacZ) are able to use these alkanes as the sole source of carbon, demonstrating that ORFX, estB, and oxyR are not necessary for alkane degradation in Acinetobacter sp. strain ADP1. The alkane-negative phenotypes of WH362 (rubA::lacZ) and WH382 (rubB::lacZ) indicate that rubA and rubB are necessary for alkane degradation.
For further characterization, we used a gap repair strategy (12) to determine the mutations conferring the alkane-negative phenotype in strains WH363, WH432, WH433, and WH434 generated by ethyl methanesulfonate (EMS) mutagenesis of Acinetobacter sp. strain ADP1 (9). The 4.3-kbp AflII fragment containing ORFX, rubA, rubB, and estB was deleted on plasmid pWH891 by restriction with AflII and religation. The resulting plasmid (pWH891ΔAflII) was linearized with AflII and was used to transform Acinetobacter sp. strains ADP1, WH363, WH432, WH433, and WH434 via natural competency. Transformants were selected on LB plates with ampicillin. After passage through E. coli cells, the plasmids were digested with AflII, showing the presence of the respective 4.3-kbp AflII fragments of the transformed strains, and the sequences between ORFX and rubB were determined (nucleotides 4980 to 7295 under accession no. Z46863). The nucleotide sequence of the ADP1 wild type revealed five errors in the published DNA sequence (9): aa 75 (S→L), 77 (D→E), and 199 to 204 (IWRKR→NLEESG) must be corrected in the published RubB alignment (9). The sequences obtained from WH363, WH432, WH433, and WH434 revealed the mutations listed in Table 2. The defects in alkane utilization of WH432 and WH433 are caused by G→D exchanges in RubA and RubB, respectively. Since these mutations have no polar effect, this shows unambiguously that rubA and rubB are necessary for alkane degradation in Acinetobacter sp. strain ADP1. Mutations in rubB present in WH363 and WH434 lead to growth defects with dodecane as the sole carbon source, whereas growth on hexadecane is still possible.
TABLE 2.
Mutations determined for alkane mutants
| Strain | Phenotypea
|
Mutationb
|
||||
|---|---|---|---|---|---|---|
| DD | HD | Plasmid | Gene | Codon | Protein | |
| WH363 | − | + | pWH891rubBL381F | rubB | CTC→TTC | L381F |
| WH434 | − | + | pWH891rubBG204S | rubB | GGT→AGT | G204S |
| WH433 | − | − | pWH891rubBG263D | rubB | GGT→GAT | G263D |
| WH432 | − | − | pWH891rubAG18D | rubA | GGC→GAC | G18D |
The EMS mutants were tested for growth with dodecane (DD) or hexadecane (HD) as the sole carbon source.
The mutated codons and the respective amino acid substitutions are listed.
rubA, rubB, estB, and oxyR constitute an operon that is not regulated by alkanes.
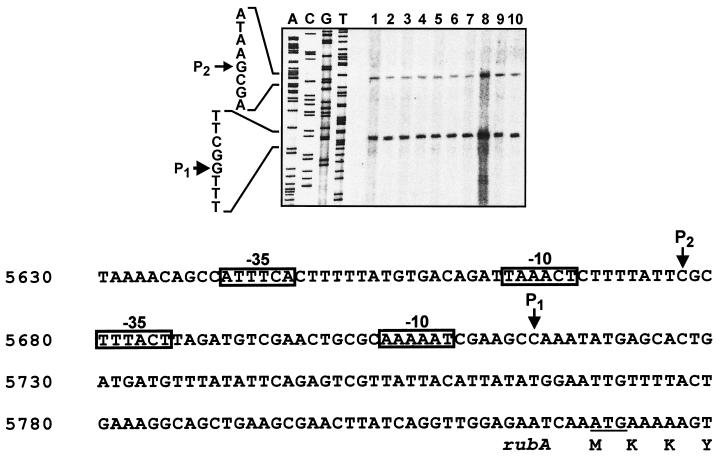
We performed primer extension analysis with a primer hybridizing within the coding region of rubA (Fig. 3). Two major products, P1 and P2 (Fig. 3 [top panel]), indicate transcription from two ς70 promoters (Fig. 3 [bottom panel]). The results obtained with RNAs from Acinetobacter sp. strain ADP1 grown in various media (Fig. 3 [top panel, lanes 1 to 6]) confirm that rubA expression is not induced by alkanes, as was also shown before by β-galactosidase expression from a rubA::lacZ fusion in WH362 (9). No change in rubA transcription is detectable, even when ADP1 was grown on hexadecane as a carbon source (lane 7). However, rubA expression is increased when present in multiple copies (lane 8). Disruption of ORFX or oxyR does not change the efficiency of rubA transcription (lanes 9 and 10).
FIG. 3.
Mapping of the 5′ start site of the rubA mRNA by primer extension. (Top) Autoradiograph. RNA was prepared from cells growing exponentially in LB or minimal medium (MM). Dodecane (DD) and hexadecane (HD) were added at 0.6% (vol/vol). Succinate (Suc) was added at 20 mM. Lanes: 1, ADP1 in LB medium; 2, ADP1 in LB medium plus DD; 3, ADP1 in LB medium plus HD; 4, ADP1 in MM plus Suc; 5, ADP1 in MM plus Suc plus DD; 6, ADP1 in MM plus Suc plus HD; 7, ADP1 in MM plus HD; 8, ADP1 transformed with pWH891 in LB medium; 9, WH380 in LB medium; 10, WH386 in LB medium. Lanes A, C, G, and T show the sequencing products obtained with the same primer. The sequence is shown on the left side. Arrows indicate the start sites of transcription (P1 and P2). (Bottom) Sequence interpretation. The sequence of the coding strand upstream of rubA is shown. The numbers of nucleotide position are given on the left according to Fig. 1. The start codon of rubA is underlined, and the four N-terminal amino acids are shown in the one-letter code. The start sites of transcription (P1 and P2) are indicated by downward arrows, and the sequences with the highest similarity to E. coli ς70 promoter sequences (−10 and −35) are boxed.
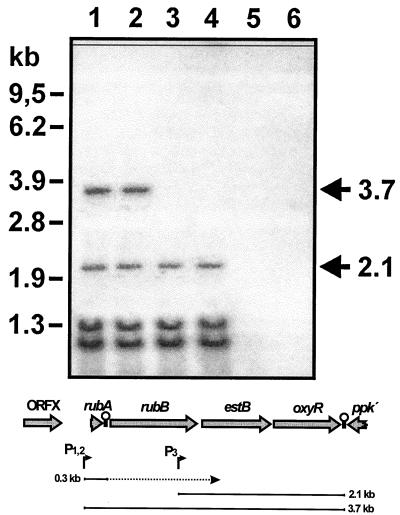
We detected a 3.7-kb RNA in ADP1 and WH380 in Northern blot analyses with an oxyR-specific probe (Fig. 4 [top panel, lanes 1 and 2]). This is in good agreement with an assumed transcript of 3.55 kb extending from the rubA promoter to the putative transcriptional termination sequence downstream of oxyR. This 3.7-kb RNA is not detectable in strains carrying a lacZ cassette integrated into rubA, rubB, estB, or oxyR (lanes 3 to 6). In ADP1 and WH380, an additional 2.1-kb RNA is detectable (lanes 1 and 2). Because this signal is also present in WH362 and WH382 (lanes 3 and 4), where the 3.7-kb RNA is not found, it cannot be explained by degradation of the 3.7-kb RNA, but indicates the presence of an internal promoter for transcription of an estB-oxyR-containing message. Signals corresponding to RNAs of 1.35 and 1.2 kb may be degradation products of the 3.7- or 2.1-kb RNA. Further Northern blot analyses using probes specific for estB and rubB are in agreement with the results obtained with the oxyR-specific probe (data not shown). No RNA is detectable with an ORFX-specific probe (data not shown). An RNA of 310 bases was detected with a rubA-specific probe (data not shown). This RNA ranges from the rubA transcription start site to the inverted repeat downstream of rubA, indicating termination at this stem-loop structure in vivo. The results from Northern blot analyses and primer extensions are summarized in the bottom panel of Fig. 4 and show that rubA, rubB, estB, and oxyR are organized in a constitutively transcribed operon.
FIG. 4.
Detection of oxyR transcripts. (Top) Northern blot hybridized with an oxyR-specific probe. Eleven micrograms of total RNA was run on each lane of a 1% agarose gel. RNA was prepared from the following cells growing exponentially in LB medium: ADP1 (lane 1), WH380 (lane 2), WH362 (lane 3), WH382 (lane 4), WH384 (lane 5), and WH386 (lane 6). On the left, the positions of RNA molecular size marker bands are given in kilobases. On the right, the RNA bands discussed in the text are marked. (Bottom) Interpretation. For explanation of the genomic situation depicted at the top see Fig. 1. In the bottom part, RNA species detected in Northern blot analyses are indicated as bars, with their sizes given in the right and left margins in kilobases. The rubA-specific 0.3-kb RNA was detected in a 2% agarose gel (data not shown). P1,2 and P3, respectively, indicate the promoters P1 and P2 determined by primer extension and promoter P3 proposed on the Northern blot shown in the top panel.
estB encodes a functional esterase.
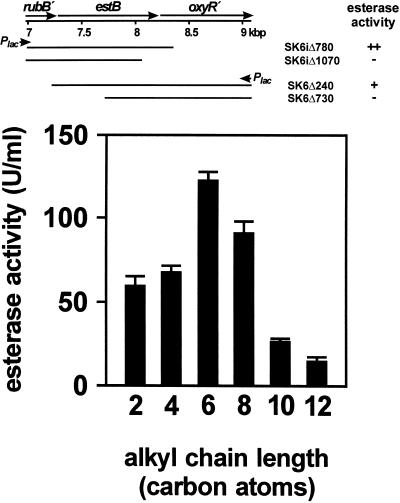
Colonies of E. coli cells, transformed with pWH891SK6 and pWH891SK6i, form clear halos on turbid NB plates with tributyrin after incubation at 37°C for 7 and 5 days, respectively. The different level in esterase activity may be due to the plasmid-borne lac promoter, which is in the same orientation as the estB gene in pWH891SK6i. Transformation of plasmids obtained from nested deletion reactions mapped the DNA relevant for this phenotype into the estB gene (Fig. 5 [top panel]). This shows that estB encodes a functional esterase from Acinetobacter sp. strain ADP1. The substrate specificity of EstB was studied with crude extracts of E. coli, expressing estB from pAKA22, with pNP esters used as substrates. EstB showed activity with pNP esters with acyl chain lengths from 2 to 12 carbon atoms, with a preference around 6 and 8 carbon atoms (Fig. 5 [bottom panel]).
FIG. 5.
EstB activity in E. coli. (Top) Mapping of the DNA conferring esterase activity. For explanation of the genomic situation depicted in the upper part, see Fig. 1. Plasmids with successive deletions were derived from pWH891SK6i and pWHSK6 and analyzed by sequencing. The plasmids were transformed into E. coli, and transformants were analyzed for esterase activity after 7 days at 37°C on indicator plates (−, no halo detectable; + and ++, intensity of halo formation). The fragments present on the deletion plasmids are indicated as bars, with the name of the plasmids on the right indicating the number of base pairs that have been deleted. (Bottom) Acyl chain length specificity of EstB. Activity was measured with crude extracts of E. coli cells expressing estB from pAKA22. No activity was found with pNP esters with chain lengths of 14, 16, and 18 carbons or in the vector controls.
oxyR encodes a peroxide response regulator.
Sequence analysis indicates that oxyR may regulate the peroxide stress response in Acinetobacter sp. strain ADP1. To test this hypothesis, we determined the sensitivity to peroxide of the oxyR mutant WH386 in comparison with that of the wild type by the killing zone assay. For complementation analyses, we constructed plasmid pWH891ΔNcoI, expressing oxyR, by restriction of pWH891 with NcoI and religation, in which the Acinetobacter genes upstream of oxyR (cobQ, sodA, lysS, ORFX, rubA, rubB, and 776 of 936 bp of estB) are deleted and oxyR is placed under control of the tetA promoter on the vector. pWH891ΔNcoI was electroporated into strains WH384ΔoxyR and WH368, and transformants were selected on LB plates with ampicillin. The killing zones for hydrogen peroxide (150 mM) were 30 mm for WH386; 28 mm for WH384ΔoxyR; 22 mm for WH384; 17 mm for WH386/pWH891ΔNcoI; 16 mm for WH384ΔoxyR/pWH891ΔNcoI; 15 mm for WH362, WH382, and WH380; and 14 mm for ADP1. Thus, the sensitivity of mutants with mutations in ORFX, rubA, and rubB to hydrogen peroxide was the same as that of the wild-type, whereas it was clearly increased in the estB mutant and strongly increased in the oxyR mutant. The increased sensitivity to hydrogen peroxide in WH386 is complemented by oxyR; the effect in the estB disruption (WH384) is further increased by additional deletion of oxyR (WH384ΔoxyR) and also is complemented by oxyR. We conclude that absence of estB does not lead to increased sensitivity to hydrogen peroxide and that the effect seen in WH384 results from a polar effect of estB disruption on oxyR expression. The phenotype of oxyR mutants indicates that the OxyR protein regulates the peroxide stress response, as known from other bacteria (8).
DISCUSSION
Previous studies did not prove that rubA and rubB are involved in alkane degradation in Acinetobacter sp. strain ADP1 (9), since the insertion of the lacZ-Kmr cassette into rubA may affect expression of rubB. The single mutations in WH432 and WH433 lead to exchanges of highly conserved glycine to aspartic acid residues in rubredoxin and rubredoxin reductase. This proves the necessity of both proteins for alkane degradation in Acinetobacter sp. strain ADP1. Rubredoxin reductase (AlkT) in P. oleovorans is not essential for alkane degradation, because it can be substituted for by an unknown reductase probably encoded on the chromosome (36). In contrast, ADP1 seems to have only one gene encoding rubredoxin reductase. WH363 and WH434 contain mutations in rubB. A conserved glycine is replaced by a serine in WH363, and a leucine, located in the variable C terminus, is replaced by phenylalanine in WH434. The rubredoxin reductase in both mutants is not completely inactive, because WH363 and WH434 are able to grow on hexadecane. Because rubredoxin reductase serves as an electron transporter for the alkane hydroxylase, it probably does not directly interact with alkanes, and, therefore, a mutation in rubredoxin reductase should not change the utilization spectrum of alkanes. Because the wild type grows faster with hexadecane as the sole carbon source than with dodecane, the latter is a poorer substrate, probably for alkane monooxygenase. Therefore, a rate-limiting rubredoxin reductase activity could reduce alkane turnover below the level necessary for growth on dodecane, whereas growth on hexadecane is still possible. A chain-length-dependent toxicity of alkanes has been postulated for A. calcoaceticus 69-V (1). The reduced turnover could lead to an accumulation of alkanes in the cell, which may be less well tolerated for the more toxic dodecane.
Analysis of the sequence downstream of rubB revealed the genes estB and oxyR, encoding a hydrolase and a transcriptional regulator of the LysR family. We have demonstrated by Northern blot analyses that rubA and rubB constitute an operon together with estB and oxyR (Fig. 4 [top panel]). ORFX is not part of this operon, because insertion of a lacZ-Kmr cassette in ORFX has no polar effect on expression of the rubA-rubB-estB-oxyR operon, as shown by the phenotype of WH380 (ORFX::lacZ) on alkane plates (Fig. 1), primer extension (Fig. 3 [top panel]), and Northern blot analyses (Fig. 4 [top panel]). The presence of a rubA-specific 310-bp RNA indicates that a stem-loop structure downstream of rubA in vivo functions as a transcriptional termination signal or as a stabilizing element preventing degradation of RNA. Primer extension analyses suggest that rubA is transcribed by ς70 RNA polymerase and show that transcription is neither induced by alkane nor subject to repression by succinate or any compound present in LB medium. Taken together, rubA in Acinetobacter sp. strain ADP1 differs from the homologous alkG in P. oleovorans not only in genetic organization and size, but also in regulation, because transcription of alkG is induced by alkane (36). In A. calcoaceticus 69-V, like in P. oleovorans, an alkane-inducible rubredoxin has been found (3). Thus, alkane degradation is regulated differently even within the genus Acinetobacter, assuming the absence of posttranscriptional regulation.
Despite the fact that rubAB, estB, and oxyR are in one operon, there is no indication for a functional relationship. The activity of estB in E. coli, monitored on indicator plates with tributyrin, demonstrates that estB encodes a functional esterase, although tributyrin is probably not the physiological substrate in ADP1. There is no amino-terminal signal peptide in the EstB sequence, indicating that it is a cytoplasmic protein. Tributyrin is hardly internalized by E. coli, which explains that halo formation on indicator plates requires several days, because it depends on cell lysis. Aside from estB, two further genes, lipA and estA, encoding lipolytic enzymes have been cloned from Acinetobacter sp. strain ADP1 (19, 20). An additional esterase, named EstC, with activity for Tween 80 is secreted via the general secretory pathway (29). EstA and LipA differ from EstB in substrate specificity, since they show optimal activity with pNP esters with acyl chains of 4 and 16 carbon atoms, respectively (18, 20). This may indicate that these enzymes have different functions in vivo.
oxyR is separated from its target genes (e.g., katG, ahpCF, and gorA) (35), on the E. coli chromosome, whereas the X. campestris-encoded oxyR is located in an autoregulated ahpF-oxyR-orfX operon (25). The similarity of OxyR from ADP1 to other OxyR proteins (31 to 35% identical amino acids) is only a little higher than that to other members of the LysR family (28% identical amino acids). OxyR proteins, however, are distinguished from all other proteins in the databases by a stretch of amino acids which we called the OxyR box (Fig. 2). This box contains the two cysteine residues, which form reversible disulfide bridges in OxyR from E. coli upon induction by hydrogen peroxide (38). The presence of that sequence in OxyR from ADP1 agrees with its function in regulating the peroxide stress response. oxyR occurs in a unique genetic arrangement in ADP1, together with genes encoding apparently unrelated functions. This resembles the previous observation that genes needed for, e.g., tryptophan biosynthesis are scrambled on the chromosome (14). Thus, the genetic organization in Acinetobacter may be peculiar, because related genes are often apart, and functionally unrelated genes are linked. The consequences of such an arrangement for regulation are clearly seen for the genes encoding alkane degradation, in which only the monooxygenase gene alkM is regulated, whereas rubAB genes are constitutive, unlike the situation in P. oleovorans. It is surprising to conclude that such an arrangement is obviously stable in evolution.
ACKNOWLEDGMENT
This work was supported by the Fonds der chemischen Industrie.
REFERENCES
- 1.Asperger O, Kleber H-P. Metabolism of alkanes by Acinetobacter. In: Towner K J, Bergogne-Berezin E, Fewson C A, editors. The biology of Acinetobacter. New York, N.Y: Plenum Press; 1991. pp. 323–350. [Google Scholar]
- 2.Asperger O, Naumann A, Kleber H-P. Occurrence of cytochrome P-450 in Acinetobacter strains after growth on n-hexadecane. FEMS Microbiol Lett. 1981;11:309–312. [Google Scholar]
- 3.Aurich H, Sorger D, Asperger O. Isolation and characterization of rubredoxin from Acinetobacter calcoaceticus. Acta Biol Med Ger. 1976;35:443–451. [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. I 1994. , II, and III. Greene Publishing Associates, New York, N.Y. [Google Scholar]
- 5.Bagdasarian M, Lurz R, Rückert B, Franklin F C H, Bagdasarian M M, Frey J, Timmis K N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:87–91. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 6.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhandayuthapani S, Mudd M, Deretic V. Interactions of OxyR with the promoter region of the oxyR and ahpC genes from Mycobacterium leprae and Mycobacterium tuberculosis. J Bacteriol. 1997;179:2401–2409. doi: 10.1128/jb.179.7.2401-2409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farr S B, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991;55:561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geißdörfer W, Frosch S C, Haspel G, Ehrt S, Hillen W. Two genes encoding proteins with similarities to rubredoxin and rubredoxin reductase are required for conversion of dodecane to lauric acid in Acinetobacter calcoaceticus ADP1. Microbiology. 1995;141:1425–1432. doi: 10.1099/13500872-141-6-1425. [DOI] [PubMed] [Google Scholar]
- 10.Geißdörfer W, Ratajczak A, Hillen W. Nucleotide sequence of a putative periplasmic Mn superoxide dismutase from Acinetobacter calcoaceticus ADP1. Gene. 1997;186:305–308. doi: 10.1016/s0378-1119(96)00728-7. [DOI] [PubMed] [Google Scholar]
- 11.Geißdörfer W, Ratajczak A, Hillen W. Transcription of ppk from Acinetobacter sp. strain ADP1, encoding a putative polyphosphate kinase, is induced by phosphate starvation. Appl Environ Microbiol. 1998;64:896–901. doi: 10.1128/aem.64.3.896-901.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregg-Jolly L A, Ornston L N. Recovery of DNA from Acinetobacter calcoaceticus chromosome by gap repair. J Bacteriol. 1990;172:6169–6172. doi: 10.1128/jb.172.10.6169-6172.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 14.Haspel G, Kishan V, Hillen W. Organisation, potential regulatory elements and evolution of trp genes in Acinetobacter. In: Towner K J, Bergogne-Berezin E, Fewson C A, editors. The biology of Acinetobacter. New York, N.Y: Plenum Press; 1991. pp. 239–249. [Google Scholar]
- 15.Holmes D S, Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 16.Huisman G W, Wonink E, Meima R, Kazemier B, Terpstra P, Witholt B. Metabolism of poly(3-hydroxyalkanoates) (PHAs) by Pseudomonas oleovorans. Identification and sequences of genes and function of the encoded proteins in the synthesis and degradation of PHA. J Biol Chem. 1991;266:2191–2198. [PubMed] [Google Scholar]
- 17.Juni E, Janik A. Transformation of Acinetobacter calco-aceticus (Bacterium anitratum) J Bacteriol. 1969;98:281–288. doi: 10.1128/jb.98.1.281-288.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kok R G. Lipolytic enzymes in Acinetobacter calcoaceticus. Ph.D. thesis. Amsterdam, The Netherlands: University of Amsterdam; 1995. [Google Scholar]
- 19.Kok R G, Christoffels V M, Vosman B, Hellingwerf K J. Growth-phase-dependent expression of the lipolytic system of Acinetobacter calcoaceticus BD413: cloning of a gene encoding one of the esterases. J Gen Microbiol. 1993;139:2329–2342. doi: 10.1099/00221287-139-10-2329. [DOI] [PubMed] [Google Scholar]
- 20.Kok R G, van Thor J J, Nugteren-Roodzant I M, Brouwer M B, Egmond M R, Nudel C B, Vosman B, Hellingwerf K J. Characterization of the extracellular lipase, LipA, of Acinetobacter calcoaceticus BD413 and sequence analysis of the cloned structural gene. Mol Microbiol. 1995;15:803–818. doi: 10.1111/j.1365-2958.1995.tb02351.x. [DOI] [PubMed] [Google Scholar]
- 21.Kokotek W, Lotz W. Construction of a lacZ-kanamycin-resistance cassette, useful for site-directed mutagenesis and as a promoter probe. Gene. 1989;84:467–471. doi: 10.1016/0378-1119(89)90522-2. [DOI] [PubMed] [Google Scholar]
- 22.Loprasert S, Atichartpongkun S, Whangsuk W, Mongkolsuk S. Isolation and analysis of the Xanthomonas alkyl hydroperoxide reductase gene and the peroxide sensor regulator genes ahpC and ahpF-oxyR-orfX. J Bacteriol. 1997;179:3944–3949. doi: 10.1128/jb.179.12.3944-3949.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maciver I, Hansen E J. Lack of expression of the global regulator OxyR in Haemophilus influenzae has a profound effect on growth phenotype. Infect Immun. 1996;64:4618–4629. doi: 10.1128/iai.64.11.4618-4629.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maeng J H, Sakai Y, Tani Y, Kato N. Isolation and characterization of a novel oxygenase that catalyzes the first step of n-alkane oxidation in Acinetobacter sp. strain M-1. J Bacteriol. 1996;178:3695–3700. doi: 10.1128/jb.178.13.3695-3700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mongkolsuk S, Loprasert S, Whangsuk W, Fuangthong M, Atichartpongkun S. Characterization of transcription organization and analysis of unique expression patterns of an alkyl hydroperoxide reductase C gene (ahpC) and the peroxide regulator operon ahpF-oxyR-orfX from Xanthomonas campestris pv. phaseoli. J Bacteriol. 1997;179:3950–3955. doi: 10.1128/jb.179.12.3950-3955.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mongkolsuk S, Sukchawalit R, Loprasert S, Praituan W, Upaichit A. Construction and physiological analysis of a Xanthomonas mutant to examine the role of the oxyR gene in oxidant-induced protection against peroxide killing. J Bacteriol. 1998;180:3988–3991. doi: 10.1128/jb.180.15.3988-3991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.National Center for Biotechnology Information. 13 April 1999, revision date. [Online.] National Center for Biotechnology Information. http://www.ncbi.nlm.nhi.gov.
- 27.Pagan-Ramos E, Song J, McFalone M, Mudd M H, Deretic V. Oxidative stress response and characterization of the oxyR-ahpC and furA-katG loci in Mycobacterium marinum. J Bacteriol. 1998;180:4856–4864. doi: 10.1128/jb.180.18.4856-4864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmen R, Vosman B, Buijsman P, Breek C K, Hellingwerf K J. Physiological characterization of natural transformation in Acinetobacter calcoaceticus. J Gen Microbiol. 1993;139:295–305. doi: 10.1099/00221287-139-2-295. [DOI] [PubMed] [Google Scholar]
- 29.Parche S, Geißdörfer W, Hillen W. Identification and characterization of xcpR encoding a subunit of the general secretory pathway necessary for dodecane degradation in Acinetobacter calcoaceticus ADP1. J Bacteriol. 1997;179:4631–4634. doi: 10.1128/jb.179.14.4631-4634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ratajczak A, Geißdörfer W, Hillen W. Alkane hydroxylase from Acinetobacter sp. strain ADP1 is encoded by alkM and belongs to a new family of bacterial integral-membrane hydrocarbon hydroxylases. Appl Environ Microbiol. 1998;64:1175–1179. doi: 10.1128/aem.64.4.1175-1179.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ratajczak A, Geißdörfer W, Hillen W. Expression of alkane hydroxylase from Acinetobacter sp. strain ADP1 is induced by a broad range of n-alkanes and requires the transcriptional activator AlkR. J Bacteriol. 1998;180:5822–5827. doi: 10.1128/jb.180.22.5822-5827.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strätz M, Mau M, Timmis K N. System to study horizontal gene exchange among microorganisms without cultivation of recipients. Mol Microbiol. 1996;22:207–215. doi: 10.1046/j.1365-2958.1996.00099.x. [DOI] [PubMed] [Google Scholar]
- 35.Tartaglia L A, Storz G, Ames B N. Identification and molecular analysis of oxyR-regulated promoters important for the bacterial adaptation to oxidative stress. J Mol Biol. 1989;210:709–719. doi: 10.1016/0022-2836(89)90104-6. [DOI] [PubMed] [Google Scholar]
- 36.van Beilen J B, Wubbolts M G, Witholt B. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation. 1994;5:161–174. doi: 10.1007/BF00696457. [DOI] [PubMed] [Google Scholar]
- 37.Williams J G, Mason P J. Hybridization in the analysis of RNA. In: Hames B D, Higgins S J, editors. Nucleic acid hybridization—a practical approach. Oxford, United Kingdom: IRL Press; 1985. pp. 139–160. [Google Scholar]
- 38.Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]