Abstract
The Omicron variant is spreading rapidly throughout several countries. Thus, we comprehensively analyzed Omicron’s mutational landscape and compared mutations with VOC/VOI. We analyzed SNVs throughout the genome, and AA variants (NSP and SP) in VOC/VOI, including Omicron. We generated heat maps to illustrate the AA variants with high mutation prevalence (> 75% frequency) of Omicron, which demonstrated eight mutations with > 90% prevalence in ORF1a and 29 mutations with > 75% prevalence in S-glycoprotein. A scatter plot for Omicron and VOC/VOI’s cluster evaluation was computed. We performed a risk analysis of the antibody-binding risk among four mutations (L452, F490, P681, D614) and observed three mutations (L452R, F490S, D614G) destabilized antibody interactions. Our comparative study evaluated the properties of 28 emerging mutations of the S-glycoprotein of Omicron, and the ΔΔG values. Our results showed K417N with minimum and Q954H with maximum ΔΔG value. Furthermore, six important RBD mutations (G339D, S371L, N440K, G446S, T478K, Q498R) were chosen for comprehensive analysis for stabilizing/destabilizing properties and molecular flexibility. The G339D, S371L, N440K, and T478K were noted as stable mutations with 0.019 kcal/mol, 0.127 kcal/mol, 0.064 kcal/mol, and 1.009 kcal/mol. While, G446S and Q498R mutations showed destabilizing results. Simultaneously, among six RBD mutations, G339D, G446S, and Q498R mutations increased the molecular flexibility of S-glycoprotein. This study depicts the comparative mutational pattern of Omicron and other VOC/VOI, which will help researchers to design and deploy novel vaccines and therapeutic antibodies to fight against VOC/VOI, including Omicron.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-022-00631-2.
Keywords: Omicron, Mutation, SNVs, AA variants, nAb escape
Introduction
A new SARS-CoV-2 variant, Omicron (B.1.1.529), was first identified in South Africa in November 2021 [1]. At that time, scientists noted a sudden rise in COVID-19 cases triggered by Omicron [2–4]. The variant has spread across the USA, Europe, Australia, Canada, and Hong Kong, and has spread to more than 50 countries within a few days [5–7]. Presently, the variant has spread throughout the globe, and several lineages have evolved, such as BA.1, BA.2, BA.3. BA.4, BA.5, in several countries [7–9]. Another recombinant variant, XE variant, was also reported as VOC. The recombinant XE variant contains the genomic part of BA.1 and BA.2 [10]. However, it is not currently designated as VOC by WHO [11]. Due to the Omicron variant and newly evolved lineages, the pandemic has taken a new turn.
With the sudden emergence of a new strain, the virus’s evolutionary trajectory has shifted in a new direction. Generally, when a virus strain like SARS-CoV-2 creates a new line of evolution, it is called an evolutionary trajectory and might be created through positive selection [12]. Jung et al. illustrated in their article that Omicron has not originated from other VOC, and the inference was drawn from phylogenetic analysis. They have also illustrated the amino acid changes that occurred through positive selection pressure [13]. Meanwhile, several lineages have evolved from Omicron [7–9]; thus, Omicron has also created an evolutionary trajectory. Scientists and policymakers are searching for strategies to fight against variants [14, 15]. Through research, they are trying to understand the properties of variants, such as immune escape, antibody escape, and vaccine escape [16, 17]. A recent study by Zhang et al. shows that the Omicron variant exhibits a severe immune escape from the convalescent sera collected from COVID-19 patients. They illustrated that the Omicron variant could escape the neutralized convalescent sera collected in patients infected with the Delta variant or previous strains [18].
The variants have the highest mutations compared to other variants; therefore, it is necessary to understand the impacts of the variants due to their highest mutational load. Quarleri et al. have studied the effect of the high mutational load, and they discussed the mutations in RBD, the S-glycoprotein other than the RBD region, nucleocapsid (N), and deletion in ORF1a [19]. Similarly, Kumar et al. performed a comparative analysis between Omicron and Delta variants of the S-glycoprotein. The relative binding affinity was evaluated using a docking study between the RBD of the S-glycoprotein and the hACE2 receptor in three strains (wild type, Delta, and Omicron variants) to understand the binding affinity with the hACE2 receptor. The docking result showed that the Omicron variant is more likely to interact with hACE2 than the Delta variant. Thus, the Omicron variant has a higher possibility of transmission [20]. It is, therefore, essential to understand all mutations at the molecular level, including their characteristics of mutations. Pascarella et al. assessed the electrostatic potential between the Omicron variant and S-glycoprotein’s other two VOCs (Delta and Delta-plus variants). They evaluated the increased electrostatic potential of the Omicron variant compared to the Delta and Delta-plus variants and indicated that the electrostatic potential favored higher transmissibility leading to augmented infectivity [21]. Recently, the XE variant has been spreading worldwide; this is a recombinant variant of the two sub-variants of Omicron and, therefore, a recombinant variant of BA.1 and BA.2. In addition to the XE variant, two other recombinant variants, XD and XF, have recently emerged [22, 23].
The spread of this recent variant worldwide has created a global concern. Therefore, more understanding and research data are necessary about the molecular determinant factors, such as emerging mutations that provide the Omicron variant’s spreading capacity. Scott et al. suggested capturing data at the molecular level during the spread of this variant. They provided examples, such as collecting S-gene data during the variant from the genome sequence of its different strains. The S-gene data of the Omicron variant may help understand the degree of vaccine escape or the escape from the immune systems of vaccinated individuals [24]. Therefore, it is urgent to unfold all mutations at the molecular level in the Omicron variant, some of which are escape mutations, to help overcome phenomena such as antibody escape and to compare them with the other VOCs and VOIs.
Studies have shown that the Omicron variant has accumulated approximately 50 mutations in different regions of its genome, out of which, 32 mutations have been observed throughout the spike-glycoprotein [20, 25–27]. Preliminary studies have pointed out several overlapping muttons from the other VOCs and VOIs, including Delta, Alpha, Gamma, Beta, Mu, and Lambda. Some overlapping mutations from VOCs or VOIs are N501Y, T478K, N655Y, D614G, N679K, and P681H [28–32]. Therefore, it is crucial to understand the frequency of mutations Omicron variant and compare them with the mutation variants of present VOCs and VOIs.
Notably, some mutations in the spike-glycoprotein are responsible for antibody escape, especially mutations in the RBD region [33, 34]. Therefore, it is essential to map significant mutations from the RBD region that are responsible for escape properties in Omicron and to characterize the molecular and mutational properties of these RBD mutations.
Variants such as Delta (B.1.617.2), Alpha (B.1.1.7), Gamma (P.1), Beta (B.1.351), and Omicron are emerging variants of SARS-CoV-2 that are entitled as variants of concern (VOC) by WHO, CDC (USA), and ECDC (EU) due to some changes compared to the wild type of the Wuhan strain. The differences noted include the increase in virulence or adaptation in clinical disease presentation and an increase in transmissibility [28–30]. Lambda (C.37) and Mu (B.1.621) are entitled variants of interest (VOI). However, WHO categorizes variants as VOCs or VOIs based on their present virulence pattern or pattern change in clinical disease presentation and transmissibility pattern. The status level of VOC or VOI of any variant can be changed. In this study, we analyzed the mutational landscape of Omicron along with the VOCs and VOIs. We considered those variants to be VOCs and VOIs concerning their current status.
In this study, we attempted to comprehend the mutational landscape of the Omicron variant and compare the mutations with those of others. We illustrated single-nucleotide variations (SNVs) throughout the Omicron genome and compared the mutations with other VOCs and VOIs. We also analyzed amino acid (AA) variants in Omicron’s non-structural proteins (NSPs) and other VOCs and VOIs. We also generated heat maps to illustrate the AA variants with a high mutation prevalence (> 75% mutational frequency) in Omicron and other variants. Our study attempted to explain Omicron’s antibody/nAb escape phenomenon and other VOCs and VOIs. A comparative study evaluated the molecular and mutational properties of 28 emerging mutations of the S-glycoprotein of Omicron, and the ΔΔG values were calculated. Furthermore, we chose six important RBD mutations (G339D, S371L, N440K, G446S, T478K, and Q498R) of Omicron and other variants for further comprehensive analysis of the molecular and mutational properties that may be responsible for antibody/nAb escape and partial vaccine escape.
Methods
Data collection of the mutational landscape of the Omicron variant and other VOCs and VOIs
Data associated with the Omicron variant and other VOCs and VOIs were collected from several databases using search engines. We retrieved meaningful information from literature on the SARS-CoV-2 Omicron and other variants from databases such as Web of Science [35], Google Scholar [36], and PubMed [37, 38]. We collected data on the Omicron variants and other VOCs/VOIs from the CDC, ECDC, and WHO [39–42].
We obtained information on the Omicron variants and other VOCs/VOIs from the GISAID database [43, 44] and retrieved data from the RCSBPDB database as PDB files to develop modes for the S-glycoprotein/S-glycoprotein-antibody interactions [45].
Data analysis of the mutational landscape of the Omicron variant and other VOCs and VOIs
Data analysis of the mutation of Omicron variant and other VOCs and VOIs
We applied computational tools, servers, and software to explain the mutational landscape of the Omicron variant and the current VOCs and VOIs. Using PyMOL software, we developed different 3D structural models of the Omicron variant, other VOCs, and VOIs [46, 47]. We used the COVID-19 CG server [48], Nextstrain server [49], Nextstrain (2021)[50], and Outbreak.info [51] for further analysis. For the analysis of SNVs in the Omicron genome, along with other VOCs and VOIs, we used the COVID-19 CG server [48]. For AA variants in the non-structural and structural proteins of Omicron and other VOCs and VOIs, we used the COVID-19 CG server [48]. We used Outbreak.info [51] to provide the heat map-like representation of the high mutation (AA mutation) prevalence (> 75% mutational frequency) of Omicron and other VOCs and VOIs. The Nextstrain server was used to develop a scatter plot for VOCs and VOIs. A 3D model of the S-glycoprotein using PyMOL software and the emerging mutations for antibody interaction were depicted for the Omicron variant, other VOCs, and VOIs [46, 47]. VarEPS was used to analyze emerging mutations related to risk analysis of mutations for antibody-binding, antibody binding sites, and risk analysis of important mutations [52].
The comprehensive prediction of some properties of 28 mutations, including G339D, S371L, N440K, G446S, T478K, and Q498R in the S-glycoprotein of Omicron
We have selected 28 mutations of Omicron’s S-glycoprotein to predict the mutation properties for this study. Researchers widely accept these mutations, and published literature is also available for these mutations from emerging Omicron mutational variants.
We used the DynaMut server to evaluate the 28 mutations in the S-glycoprotein and its characteristic effect [53]. Here, we analyzed several mutation properties (stabilizing/destabilizing events and decreasing molecular flexibility). Properties like fluctuation, interatomic interactions, and deformation of an AA because of a mutation can be represented by the ΔΔG of DDG (Delta-Delta G). The DDG of the AA point mutations of the S-glycoprotein of Omicron were computed to understand its thermodynamic stability. The server illustrated the point mutation into two categories: stabilizing events (ΔΔG value was described as ≥ 0) and destabilizing events (ΔΔG was illustrated as < 0) [53]. Capriotti et al. showed a similar value of the ΔΔG to categorize the point mutations using the stabilizing/destabilizing state [54].
Depiction of plot, graphs, and statistical models
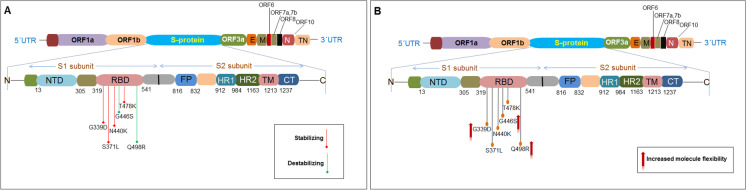
Statistical software (PAST 4.03) was used to generate statistical models [55]. Simultaneously, we used MATLAB to represent the graphs and plots whenever needed [56]. A flow diagram is portrayed to provide an overview of our study to comprehend the mutation mapping of the Omicron variant (Fig. 1A). In addition, the location of the six selected mutations in the S-glycoprotein in our study has been depicted (Fig. 1B).
Fig. 1.
A flow diagram shows the graphical view of our study overall. It also shows the graphic demonstration of the emerging selected mutations from (A) A flow diagram of our study to evaluate the mutational landscape of the Omicron (B.1.1.529) variant. (B) A schematic diagram shows the location of the selected six mutations from the RBD regions of the S-glycoprotein for our study. The study analyzed the molecular and mutational properties of 28 significant mutations, including G339D, S371L, N440K, G446S, T478K, and Q498R
Results
Mutational landscape of Omicron variant and comparison with the mutations of other VOCs and VOIs
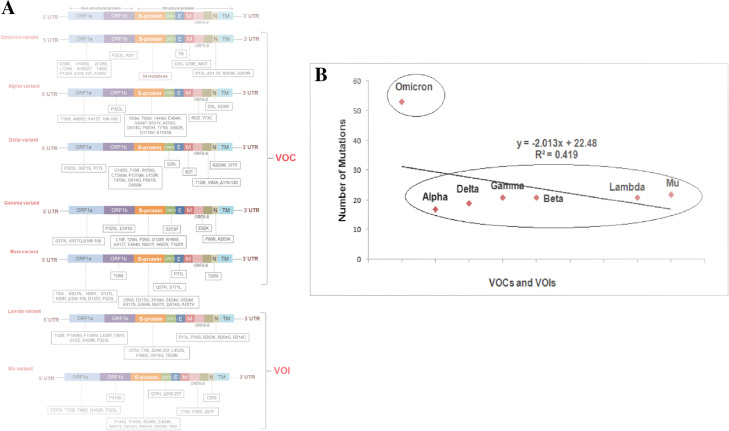
We illustrated the mutational landscape of Omicron and compared the mutations with those of other VOCs. First, a schematic diagram representing all the mutational landscapes throughout the genome and its comparison with VOCs and VOIs was created (Fig. 2A). Our study corroborates previous findings about the immense number of mutations in the Omicron genome compared to other variants. We developed a statistical model using the total number of mutations in the genome. It shows the number of mutations of different VOCs (Alpha, Delta, Gamma, and Beta) and VOIs (Lambda, Mu). In this model, we found all the VOCs and VOIs, including Alpha, Delta, Gamma, Beta, Lambda, and Mu, in a cluster showing the number of mutations other than Omicron. The variant is out of cluster due to more mutations (Fig. 2B). However, from this model, we found that the clustering event formed two groups: the first containing all variants and the second containing only Omicron. Omicron exhibits a high number of mutation patterns compared to other VOCs and VOIs. The mutational landscape of the Omicron variant is presented in Table 1.
Fig. 2.
A schematic diagram representing the mutational landscape throughout the genome of Omicron and its comparison with VOCs and VOIs. (A) The mutational landscape throughout the genome of Omicron and its comparison with VOCs and VOIs. (B) A statistical model illustrates the number of total genome mutations in the VOCs and VOI, including Omicron. This model shows the number of mutations in the genome of VOCs and VOIs Alpha, Delta, Gamma, Beta, Lambda, and Mu formed a cluster regarding the number of mutations. Here, the clustering event formed two groups: The first one contains all variants, and the second one is Omicron. Omicron exhibits a high number of mutation patterns compared to other VOCs and VOIs. For the analysis, we have taken the data from the literature collected from open source databases such as Google Scholar [36] and PubMed [37, 38]. At the same time, we also collected the open source mutation information on CDC’s Omicron variants and other VOCs/VOIs [39–41]; eCDC[41], WHO[42] etc.
Table 1.
Mutational landscape of Omicron variant
| Mutation | Mutation site | Remarks |
|---|---|---|
| A67V | NTD of Spike protein | Ala Val |
| Δ69–70 | Spike protein deletion | Deletion mutations |
| T95I | NTD of Spike protein | Thr Ile |
| G142D | NTD of Spike protein | Gly Asp |
| Δ143–145 | Spike protein deletion | Deletion mutations |
| Δ211 | Spike protein deletion | Deletion mutations |
| L212I | NTD of Spike glycoprotein | Leu Ile |
| ins214EPE | Insertion in Spike glycoprotein | Insertions of Glu, Pro, and Glu amino acids |
| T547K | Spike glycoprotein | Thr Lys |
| D614G | Spike glycoprotein | Asp Gly |
| H655Y | Spike glycoprotein | His Tyr |
| N679K | Spike glycoprotein | Asn Lys |
| P681H | Spike glycoprotein | Pro His |
| N764K | Spike glycoprotein | Asn Lys |
| D796Y | Fusion peptide in Spike protein | Asp Tyr |
| N856K | Spike glycoprotein | Asn Lys |
| Q954H | HR1 in Spike protein | Gln His |
| N969K | HR1 in Spike protein | Asn Lys |
| L981F | HR1 in Spike protein | Leu Phe |
| G339D | RBD in Spike protein | Gly Asp |
| S371L | RBD in Spike protein | Ser Leu |
| S373P | RBD in Spike protein | Ser Pro |
| S375F | RBD in Spike protein | Ser Phe |
| K417N | RBD in Spike protein | Lys Asn |
| N440K | RBD in Spike protein | Asn Lys |
| G446S | RBD in Spike protein | Gly Ser |
| S477N | RBD in Spike protein | Ser Asn |
| T478K | RBD in Spike protein | Thr Lys |
| E484A | RBD in Spike protein | Glu Ala |
| Q493K | RBD in Spike protein | Gln Lys |
| G496S | RBD in Spike protein | Gly Ser |
| Q498R | RBD in Spike protein | Gln Arg |
| N501Y | RBD in Spike protein | Asn Tyr |
| Y505H | RBD in Spike protein | Tyr His |
| K38R | ORF1a | Lys Arg |
| V1069I | ORF1a | Val Ile |
| Δ1265 | Deletion in ORF1a | Deletion mutation |
| L1266I | ORF1a | Leu Ile |
| A1892T | ORF1a | Ala Thr |
| T492I | ORF1a | Thr Ile |
| P132H | ORF1a | Pro His |
| Δ105–107, | Deletions in ORF1a | Deletion mutations |
| A189V | ORF1a | Ala Val |
| P323L | ORF1b | Pro Leu |
| I42V | ORF1b | Ile Val |
| T9I | Envelope protein | Thr Ile |
| D3G | Nucleoprotein | Asp Gly |
| Q19E | Nucleoprotein | Gln Glu |
| A63T | Nucleoprotein | Ala Thr |
| P13L | Nucleocapsid protein | Pro Leu |
| Δ31–33 | Deletions in Nucleocapsid protein | Deletion mutations |
| R203K | Nucleocapsid protein | Arg Lys |
| G204R | Nucleocapsid protein | Gly Arg |
S-glycoprotein substitutions in the Omicron variant and other VOCs and VOIs
We illustrated the mutational landscape of the Omicron S-glycoprotein. A schematic diagram represents all mutational landscapes throughout the genome and its comparison with VOCs and VOIs (Fig. 3A). We constructed a statistical model using the total number of mutations in the S-glycoprotein. The model showed that all VOCs and VOIs formed a cluster other than Omicron (Fig. 3B). Here, the clustering event also formed two groups: the first group contained all variants and the second group contained only Omicron. Similar to the previous model (developed using the number of total genome mutations), Omicron belongs to a different group and shows a different pattern than other variants regarding the number of mutations.
Fig. 3.
A schematic diagram represented the mutational landscape throughout the S-glycoprotein of Omicron and its comparison with VOCs and VOIs. (A) The mutational landscape throughout the S-glycoprotein of Omicron and its comparison with VOCs and VOIs. (B) A statistical model demonstrates the number of mutations in the S-glycoprotein in the VOCs and VOI, including Omicron. This model shows the number of mutations in the S-glycoprotein of VOCs and VOIs Alpha, Delta, Gamma, Beta, Lambda, and Mu formed a cluster regarding the number of mutations. Here, the clustering event also formed two groups: The first one contains all variants, and the second one is Omicron. Like the previous model (developed using the number of total genome mutations), Omicron belongs to a different group and shows a different pattern compared to other variants in terms of the number of mutations. For the analysis, we have taken the data from the literature collected from open source databases such as Google Scholar [36], PubMed[37, 38] etc. At the same time, we also collected the open source mutation information on CDC’s Omicron variants and other VOCs/VOIs [39–41]; eCDC [41], WHO[42] etc.
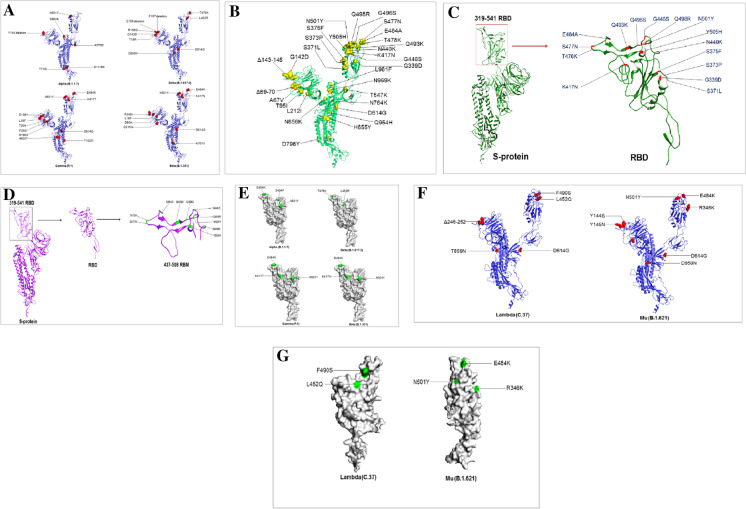
The 3D model deployment of the S-glycoprotein with mutations is significant for the SARS-CoV-2 variant, providing an interactive platform. A 3D model was generated using bioinformatics. The 3D model helps to compare the protein with other variants using 3D space. We developed a 3D model to understand the position of all mutations of S-glycoprotein, the Omicron, and other VOCs and VOIs. Every model was generated to understand every detail of the mutational landscape of the Omicron and other VOCs and VOIs (Alpha, Delta, Gamma, Beta, Lambda, and Mu). We identified mutations of the S-glycoprotein of Omicron in a 3D model (Fig. 4A). Mutations in the RBD of the S-glycoprotein of Omicron were depicted using a 3D model. We found 15 mutations in the RBD region (Fig. 4B). Mutations in the RBM of the S-glycoprotein of Omicron were depicted using a 3D model. We found ten mutations in the RBM region (Fig. 4C). Simultaneously, all mutations of the S-glycoprotein of VOCs (Alpha, Delta, Gamma, and Beta) have been illustrated (Fig. 4D). Mutations of Omicron are a puzzle for researchers, and they have attempted to solve these mutations [54]. Our study is significant in this regard. However, this part of the analysis will assist researchers in understanding the Omicron mutations and those of the Alpha, Delta, Gamma, and Beta variants. The 3D molecular statures of Omicron may help elucidate global takeover events [57]. This structural basis of understanding the Spike protein SARS-CoV-2 will help future researchers further understand this variant’s functional properties. This study informs us of emerging mutations in 3D, and it will assist future researchers in understanding emerging mutations such as E484K, K417T, N501Y, S494P, and L452R for functionality mapping.
Fig. 4.
3D model illustrated the mutational landscape throughout the S-glycoprotein of Omicron and its comparison with VOCs and VOIs. (A) A 3D model that illustrates all the mutations of the S-glycoprotein of Omicron. (B) A 3D model that illustrates all the mutations RBD region of the S-glycoprotein of Omicron. (C) A 3D model describes all the mutations RBM of the S-glycoprotein of Omicron. (D) A 3D model that illustrates all the mutations in the VOCs (Delta (B.1.617.2), Alpha (B.1.1.7), Gamma (P.1), Beta (B.1.351)). (E) A 3D model that illustrates all the RBD mutations in the VOCs (Delta (B.1.617.2), Alpha (B.1.1.7), Gamma (P.1), Beta (B.1.351)). (F) A 3D model that illustrates all the mutations in the VOIs (Lambda (C.37) and Mu (B.1.621). (G) A3D model that illustrates all the RBD mutations in the VOIs (Lambda (C.37) and Mu (B.1.621)). All the 3D models were developed using PyMOL software. For a 3D model generation, we used some PDB files (PDB ID: 6VXX)
All the mutations of the RBD of VOCs (Alpha, Delta, Gamma, and Beta) were identified under the particular position of mutations (Fig. 4E). Similarly, a 3D model was developed to comprehend all mutations of the S-glycoprotein of the VOIs (Lambda and Mu) (Fig. 4F). We illustrated the understanding of all RBD mutations of the S-glycoprotein in the VOIs (Fig. 4G). The mutational landscape for the mutations of the S-glycoprotein has been illustrated for all the VOCs using a table (Table 2). The mutational landscape for the important mutations of the S-glycoprotein has been illustrated for the VOIs in Table 3.
Table 2.
Emerging mutations in S-protein of VOCs of SARS-CoV-2
| Sl No | SARS-CoV-2 VOC | Variants name (WHO label) | SARS-CoV-2 lineages | Mutations in S-protein | |
|---|---|---|---|---|---|
| RBD region | Other than the RBD region | ||||
| 1 | 21 K, GR/484A | Omicron | B.1.1.529 | G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493K, G496S, Q498R, N501Y, Y505H | A67V, Δ69–70, T95I, G142D, Δ143–145, Δ211, L212I, ins214EPE,T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F |
| 2 | 20I/501Y.V1 | Alpha | B.1.1.7 | E484K, S494P, N501Y | 69del, 70del, 144del, A570D, D614G, P681H, T716I, S982A, D1118H, K1191N |
| 3 | 21A, 21I, 21 J/20A/S:478 K | Delta | B.1.617.2 | L452R, T478K | G142D, T19R, R158G, D614G, P681R, D950N, E156del, F157del |
| 4 | 20 J/501Y.V3 | Gamma | P.1 | K417T, E484K, N501Y | L18F, T20N, P26S, D138Y, R190S, H655Y, T1027I |
| 5 | 20H/501Y.V2 | Beta | B.1.351 | K417N, E484K, N501Y | D80A, D215G, 241del, 242del, 243del, D614G, A701V |
Table 3.
Emerging mutations in S-protein of SARS-CoV-2 VOIs
| Sl no | Significant SARS-CoV-2 variants | Variants name (WHO label) | SARS-CoV-2 lineages | Mutations in S-protein | |
|---|---|---|---|---|---|
| RBD region | Other than the RBD region | ||||
| 1 | 21G | Lambda | C.37 | L452Q, F490S | G75V, T76I, Δ246–252, D614G, T859N |
| 2 | 21H | Mu | B.1.621 | R346K, E484K, N501Y | T95I, Y144S, Y145N,D614G, P681H, D950N |
Single-nucleotide variations (SNVs) of 5'UTR, ORF1a, ORF1ab, S, ORF3a, E, M, ORF6, ORF7a, ORF7b, ORF8, N, and 3′UTR regions of Omicron and comparison other VOCs and VOIs
We analyzed SNVs throughout the genome (including 5′UTR, ORF1a, ORF1ab, S, ORF3a, E, M, ORF6, ORF7a, ORF7b, ORF8, N, and 3′UTR regions) of Omicron and compared them with other VOCs and VOIs.
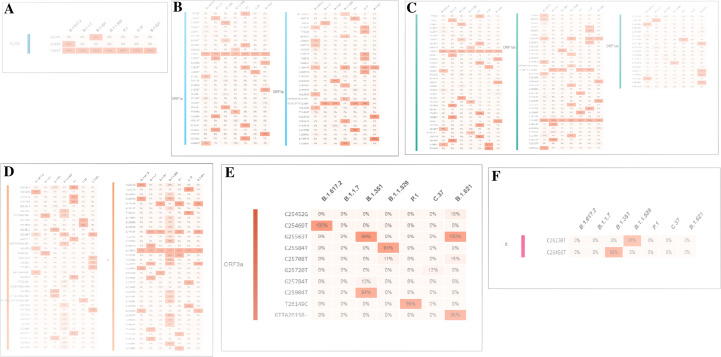
A comparative analysis of SNVs of the 5′UTR region is illustrated in Fig. 5A. We found that SNV with the highest mutational frequency was C241T. For Omicron, the mutational frequency was 94%. However, the mutational frequency was noted in the case of Delta (99%), Alpha (100%), Beta (100%), Gamma (99%), Lambda (100%), and Mu (99%).
Fig. 5.
Single-nucleotide variations (SNVs) of some parts of the genome (5′UTR, ORF1a, ORF1ab, S, ORF3a, E) of Omicron along with other VOCs and VOIs. (A) SNVs of 5′UTR of Omicron along with other VOCs and VOIs. (B) SNVs of ORF1a of Omicron along with other VOCs and VOIs. (C) SNVs of ORF1ab of Omicron along with other VOCs and VOIs. (D) SNVs of S of Omicron along with other VOCs and VOIs. (E) SNVs of ORF3a of Omicron along with other VOCs and VOIs. (F) SNVs of E of Omicron along with other VOCs and VOIs. We have used the COVID-19 CG server for SNVs analysis of these parts of the genome (5′UTR, ORF1a, ORF1ab, S, ORF3a) (E) of Omicron along with other VOCs and VOIs [48]. The server used the data from the GISAID ingestion pipeline, which is open-source data [48]
Similarly, a comparative analysis of SNVs in the ORF1a region is illustrated in (Fig. 5B), showing that SNV with the highest mutational frequency was C3037T. For Omicron, the mutational frequency was 93%. It was 100% for all other VOCs and VOIs.
The comparative analysis of SNVs of the ORF1ab region revealed that the two SNVs with the highest mutational frequencies were C3037T and C14408T (Fig. 5C). For Omicron, the mutational frequencies of these SNVs were 94% and 98%, respectively. However, SNV C3037T was 100% for other VOCs and VOIs. Similarly, the SNV C14408T was 100% for Delta, 100% for Alpha, 90% for Beta, 98% for Gamma, 100% for Lambda, and 99% for Mu.
The comparative analysis of SNVs in the S region is illustrated (Fig. 5D). We found that the SNV with the highest mutational frequency was A23403G. For Omicron, the mutational frequency was 99%. However, it was noted as 100% for all other VOCs and VOIs.
The comparative analysis of SNVs in the ORF3a region is illustrated in Fig. 5E. It was observed that the SNV with the highest mutational frequency of SNV is A23403G. In the case of Omicron, the mutational frequency of the SNV was 99%. It was noted as 100% for all other VOCs and VOIs.
We evaluated the SNVs of the E region (Fig. 5F). We found only one SNV (A26270G) in Omicron, with a mutational frequency of 81%. The SNV was not found for other variants.
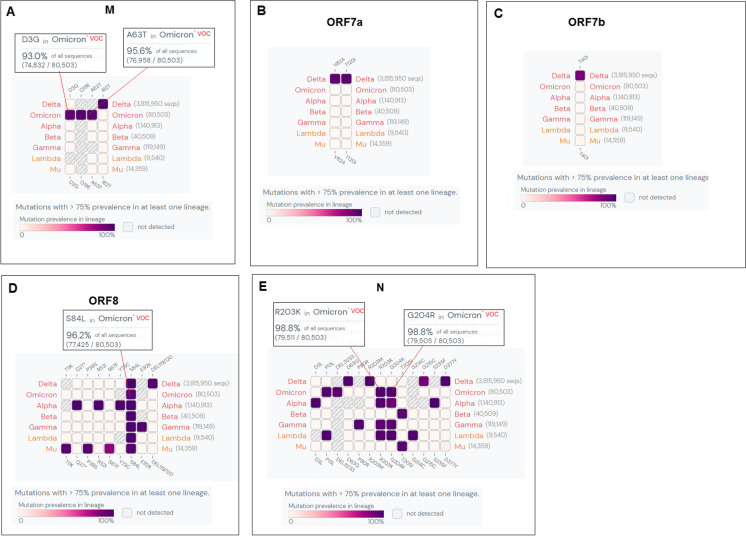
For the M region, we found three SNVs for Omicron, A26530G, C26577G, and G26709A, and the mutational frequencies were 41%, 73%, and 90%, respectively (Fig. 6A). SNV T26767T was also found for Delta with an SNV of 100%.
Fig. 6.
Single-nucleotide variations (SNVs) of some other parts of the genome (M, ORF6, ORF7a, ORF7b, ORF8, 3′UTR) of Omicron along with other VOCs and VOIs. (A) SNVs of M of Omicron along with other VOCs and VOIs. (B) SNVs of ORF6 of Omicron, along with other VOCs and VOIs. (C) SNVs of ORF7a of Omicron, along with other VOCs and VOIs. (D) SNVs of ORF7b of Omicron, along with other VOCs and VOIs. (E) SNVs of ORF8 of Omicron, along with other VOCs and VOIs. (F) SNVs of N of Omicron along with other VOCs and VOIs. (G) SNVs of 3′UTR of Omicron along with other VOCs and VOIs. We have also used the COVID-19 CG server for SNVs analysis of these parts of the genome (M, ORF6, ORF7a, ORF7b, ORF8, 3′UTR) of Omicron along with other VOCs and VOIs [48]. The server used the data from the GISAID ingestion pipeline, which is open-source data [48]
Only one mutational frequency (91%) was found for Omicron for SNV A27259C (Fig. 6B). For the ORF7a region, we found two SNVs. However, the mutational frequencies were insignificant, and these two SNVs could not be considered (Fig. 6C). Here, we found two SNVs for the Delta variant, T27638C, and C27752T, and the mutational frequencies were the same (97%) in both cases. For the ORF7b region, SNV C27807T was found in Omicron, and the mutational frequency was 66% (Fig. 6D). Another SNV C27874T with a mutational frequency of 77% was identified in Delta variant in this region.
In the ORF78 region, we found no significant SNVs for Omicron (Fig. 6E). Here, we identified three SNVs with the highest mutational frequencies for Alpha (C27972T, G28048T, and A28111G), with mutational frequencies of 99% for all SNVs. We also found the two highest mutational frequencies for Mu (C27925A and C28005T), with mutational frequencies of 99% each. For the M region, the two significant SNVs for Omicron (> 75%) were C28311T and G28881A, with mutational frequencies of 80% and 78%, respectively (Fig. 6F).
Lastly, the 3′UTR region analysis showed no significant SNVs for Omicron (Fig. 6G). However, we found one highly significant SNV for the Delta variant, G29742T, with a mutational frequency of 99%. However, we have tried to analyze mutational study events of ORF6. Nevertheless, the used server failed to generate the mutational event.
Recently, Yadav et al. found the Omicron variant in collected samples, and the genome was analyzed using next-generation sequencing. Finally, several SNVs were found in Omicron clinical isolates [58]. Ahmed et al. found the Omicron variant by sequencing a swab sample collected from the nasopharynx of an aircraft traveler. The Omicron variant was also found in subsequent sequencing (ATOPlex and Nanopore) of aircraft wastewater samples. They have attempted to discuss the genome in light of SNVs. However, researchers have not yet fully analyzed SNVs [59]. However, we performed a comprehensive SNV analysis throughout the genome of Omicron and compared it with other VOCs/VOIs.
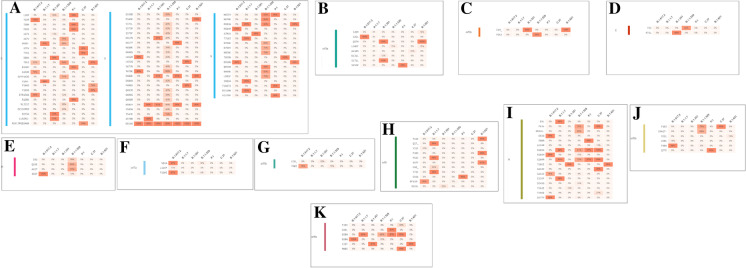
Amino acid (AA) variants in the non-structural proteins of Omicron and comparison with mutations in other VOCs and VOIs
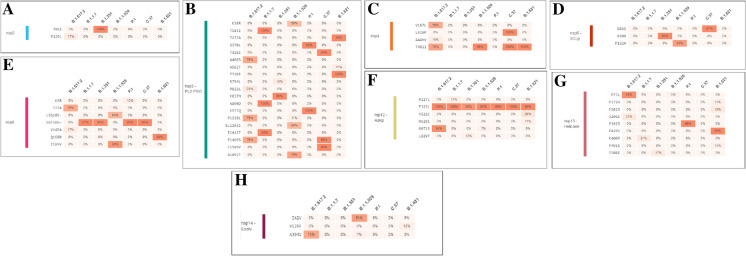
We analyzed AA variants in the non-structural proteins (nsp2, nsp3 (PL2-PRO), nsp4, nsp5 (3CLp), nsp6, nsp12 (RdRp), nsp13 (Helicase), and nsp14) of Omicron and compared the mutations with other VOCs and VOIs. No AA variant was found in the Omicron region of nsp2 (Fig. 7A). We found a T85I AA variant with 100% mutational frequency in the Delta variant. Similarly, in nsp3, the maximum mutational frequency of the Omicron variant was 78% in the A1892T AA variant (Fig. 7B). We found a highly significant mutational frequency of the AA variant in three places in the Alpha variant as well, which was 100% at T183I and A890D and 99% at I1412T. We also found the Mu variant mutational frequency of 100% in T720I AA. In nsp4, one AA variant had a significant mutational frequency (99%) among the Omicron variant in T492I (Fig. 7C). Lambda had two AA variants (L438P and T492I) with 100% mutational frequency. Similarly, Mu has one AA variant (T492I) with 100% mutational frequency. In nsp5, we found one AA variant (P132H) with an 83% mutational frequency in the Omicron variant (Fig. 7D). Additionally, we found one AA variant (K90R) with 99% mutational frequency in the Beta variant and one AA variant (G15S) had 97% mutational frequency in the Lambda variant. Similarly, in nsp6, Omicron had one AA variant (I189V) with an 88% mutational frequency (Fig. 7E). We also found one AA variant (Q160R) with a 99% mutational frequency in the Lambda variant.
Fig. 7.
Amino acid (AA) variants in the non-structural proteins of Omicron along with other VOCs and VOIs. (A) AA variants in nsp2, (B) AA variants in nsp-3 (PL2-PRO), (C) AA variants in nsp4, (D) AA variants in nsp5(3CLp), (E) AA variants in nsp6, (F) AA variants in nsp12 (RdRp), (G) AA variants in nsp13 (Helicase), (H) AA variants in nsp14. In this part of the study, we have also used the COVID-19 CG server to analyze the amino acid (AA) variants of non-structural proteins of Omicron along with other VOCs and VOIs [48]. The server used the data from the GISAID ingestion pipeline, which is open-source data [48]
In nsp12, we found one AA variant (P323L) with a significant mutational frequency for VOCs and VOIs, including Omicron (Fig. 7F). Mutational frequencies were 100% (Delta), 100% (Alpha), 90% (Beta), 97% (Omicron), 100% (Lambda), and 99% (Mu). However, no AA variant was found in Omicron in the nsp13 region (Fig. 7G). Delta had one AA variant (P77L) with a significant mutational frequency of 94%. Similarly, Mu had one AA variant (P419S) with a mutational frequency of 99%. In contrast, one AA variant was found with a mutational frequency of 81% in Omicron in the nsp114 region (Fig. 7H).
3.5. Amino acid (AA) variants of structural proteins and associated regions (S-glycoprotein, ORF3a, ORF3b, E, M, ORF7a, ORF7b, ORF8, N, ORF9b, and ORF9c) of Omicron and comparison with mutations in other VOCs and VOIs
We evaluated AA variants in structural proteins and associated regions (S-glycoprotein, ORF3a, ORF3b, E, M, ORF7a, ORF7b, ORF8, N, ORF9b, and ORF9c) of Omicron and compared the mutations with other VOCs and VOIs. We found several AA variants in the S-glycoprotein with significant mutational frequency in Omicron. The AA variants with mutational frequency are A67V (75%), HV69 (66%), T95I (84%), GVYY142D (71%), T478K (69%), T547K (74%), D614G (99%), H655Y (91%), N679K (91%), P681H (91%), N764K (63%), D796Y (71%), N856K (74%), Q954H (86%), N969K (81%), L981F (81%), G339D (58%), S371L (40%), S373P (42%), S375F (43%), K417N (26%), N440K (34%), G446S (33%), S477N (40%), T478K (69%), E484A (42%), Q493K (43%), G496S (39%), Q498R (43%), N501Y (44%), and Y505H (43%). Interestingly, one AA variant (D614G) was mutated in all VOCs and VOIs, including Omicron (Fig. 8A). Mutation frequencies were 100% (Delta), 100% (Alpha), 100% (Beta), 99% (Omicron), 100% (Lambda), and 100% (Mu).
Fig. 8.
Amino acid (AA) variants in the structural proteins of Omicron along with other VOCs and VOIs. (A) AA variants in S-glycoprotein, (B) AA variants in ORF3a, (C) AA variants in ORF3b, (D) AA variants in E, (E) AA variants in M, (F) AA variants in ORF7a, (G) AA variants in ORF7b, (H) AA variants N, (I) AA variants in ORF8b, (J) AA variants in ORF9b, (K) AA variants in ORF9c. In this part of the study, we have also used the COVID-19 CG server to analyze the amino acid (AA) variants of structural proteins of Omicron along with other VOCs and VOIs [48]. The server used the data from the GISAID ingestion pipeline, which is an open source data [48]
In ORF3a, no AA variant was detected in the Omicron (Fig. 8B). We found S26L (Delta) and Q57H (Mu) AA variants with a 100% mutational frequency. In orf3a, one AA variant (P21S) had a significant mutational frequency (94%) in the Omicron variant (Fig. 8C).
One AA variant (T9I) with 82% mutational frequency was found in Omicron in the M protein (Fig. 8D). Similarly, three AA variants (D3G, Q19E, and A63T) with 39%, 69%, 72% mutational frequencies were found in Omicron in the M protein (Fig. 8E). Simultaneously, one AA variant (I82T) with 100% mutational frequency was found in the M protein of the Delta variant.
In ORF7a, one AA variant (V28A) with meager (10%) mutational frequency in the Omicron variant cannot be considered as an AA variant (Fig. 8F). However, two AA variants (V28A and T120I) were noted, each with 97% mutational frequencies in both cases in the Delta variant.
One AA variant (V28A) with meager (13%) mutational frequency in the Omicron variant in ORF7b cannot be considered an AA variant (Fig. 8G). However, one AA variant (T40I) was noted, with a 76% mutational frequency in the Delta variant.
In ORF8, one AA variant (DF119) with a deficient (7%) mutational frequency in the Omicron variant could not be considered an AA variant (Fig. 8H). However, we found that the same AA variant (DF119) had a significant mutational frequency (86%) in the Delta variant.
Four AA variants (P13L, ERS31, R203K, and G204R) with 78%, 69%, 81%, and 72% mutational frequencies, respectively, were found in Omicron in the N protein (Fig. 8I). Simultaneously, four AA variants (D63G, R203M, G215C, and D377Y) with 99%, 99%, 75%, 99% mutational frequencies respectively were found in the N protein of the Delta variant.
In ORF9b, two AA variants (P10S, ENA27-) with 78% and 69% mutational frequencies were found in Omicron (Fig. 8J). However, we found 99% mutational frequency in one AA variant (T60A) in the Delta variant and one AA variant (Q77E) in the Gamma variant.
Finally, we found one AA variant (G50N) with 69% mutational frequency Omicron in orf9c (Fig. 8K). However, we found 100% mutational frequency in one AA variant (G50W) in the Delta variant. We also found 99% mutational frequency in one AA variant (G50N) in Alpha, two AA variants (G50N, M60I) in Lambda, and one AA variant (L52F) in Mu.
Nikolaidis et al. analyzed the amino acid substitutions (AAS) and found a high rate in the spike of the Omicron [60]. Zhang et al. also evaluated AA mutations in different circulating VOCs, including Omicron. They also illustrated cross-species tropism in cells expressing 18 ACE2 molecules. This study may help to forecast the spread of mutations [61].
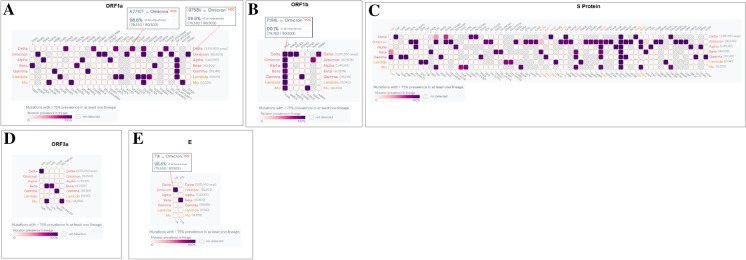
Heat map-like structure of mutation prevalence with > 75% mutational frequency in Omicron and comparison with the mutations with other VOCs and VOIs
Scientists have evaluated and illustrated the SARS-CoV-2 mutations, their occurrence frequencies, and their circumstances [62–64]. This will assist the researchers in understanding the role of this variant in epidemiology.
We analyzed the mutation prevalence of Omicron, compared the mutations with other VOCs and VOIs, and represented them using a heat map. The heat map shows eight mutations with > 90% identity (K856R, S2083I, DEL2084/2084, A2710T, T3255I, P3395H, DEL3674/3676, and I3758V) in ORF1a in Omicron (Fig. 9A). Similarly, the heat map shows one mutation with > 90% (P314L) and one with > 75% (I1566V) mutation prevalence in ORF1b of Omicron (Fig. 9B). Delta had three mutations (P314L, G662S, P1000L) with > 90% and one mutation (A1918V) with > 80% mutation prevalence. Interestingly, the heat map showed 29 mutations with a > 75% mutation prevalence in the S protein (Fig. 9C). No mutations with > 75% mutation prevalence were found in ORF3a in Omicron (Fig. 9D). Simultaneously, we found some mutations (Delta with S26L, Beta with Q57H, and S171L, Gamma with S253P, Mu with Q57H, and Del 256/257) with > 75% mutation prevalence in ORF3a with other VOCs and VOIs. We found one mutation (T9I) with a > 75% mutation prevalence in Omicron in the E protein and compared it with other VOCs and VOIs (Fig. 9E). We found one mutation (P71L) with a > 75% mutation prevalence in Beta. The heat map of the M protein showed that Omicron had three mutations (D3G, Q19E, and A63T) (Fig. 10A). ORF7a, with a > 75% mutation prevalence, was noted in Omicron. However, Delta had two mutation prevalence rates of > 75% (V82A and T120I) (Fig. 10B). ORF7b, with no mutation with > 75% prevalence, was noted in Omicron. However, Delta had a prevalence of > 75%, corresponding to T40I (Fig. 10C). We observed that the prevalence of S84L was > 75% for all ORF8 variants. The only mutation prevalence observed for Omicron with > 75% (Fig. 10D). Finally, we noted that the prevalence of the four Omicron mutations, P13L, DEL31/33, R203K, G204R was > 75% in N (Fig. 10E). We also noted four mutations with a prevalence of > 75% in Delta, Alpha, and Lambda. Characteristic mutations for Omicron and comparison with the mutations of other VOCs and VOIs were noted as deletions or nonsynonymous substitutions, which have taken place in > 75% of sequences within those variants. In this study, we measured the mutations of Omicron with > 75%. Simultaneously, this study assisted in understanding the mutations of VOCs and VOIs with > 75% (Alpha, Beta, Gamma, Delta, Lambda, and Mu). From the heat map, we found that AA variants with high mutation prevalence showed eight mutations with > 90% prevalence in ORF1a and 29 mutations with > 75% prevalence in the S-glycoprotein of Omicron. Our study is significant in this regard.
Fig. 9.
The heat map-like representation shows the high mutation prevalence (> 75% mutational frequency) of Omicron and other VOCs and VOIs in ORF1a, ORF1b, S-glycoprotein, ORF3a, E. (A) Heat map of ORF1a. (B) Heat map of ORF1b. (C) Heat map of S-glycoprotein. (D) Heat map of ORF3a. (E) Heat map of E. Here, we have used outbreak.info [51] to provide the heat map like representation of high mutation (AA mutation) prevalence (> 75% mutational frequency) of Omicron and other VOCs and VOIs of different parts of the genome. The server used GISAID API data and integrated the EpiData pipeline [65] from Johns Hopkins University if needed and finally represented through a graphical interface
Fig. 10.
The heat map-like representation shows the high mutation prevalence (> 75% mutational frequency) of Omicron and other VOCs and VOIs in M, ORF7a, ORF7b, ORF8, N. (A) Heat map of M. (B) Heat map of ORF7a. (C) Heat map of ORF7b. (D) Heat map of ORF8. (E) Heat map of N. Here, we have also used outbreak.info [51] to provide the heat map-like representation of high mutation (AA mutation) prevalence (> 75% mutational frequency) of Omicron and other VOCs and VOIs. The server used GISAID API data and integrated the EpiData pipeline [65] from Johns Hopkins University if needed and finally represented through a graphical interface
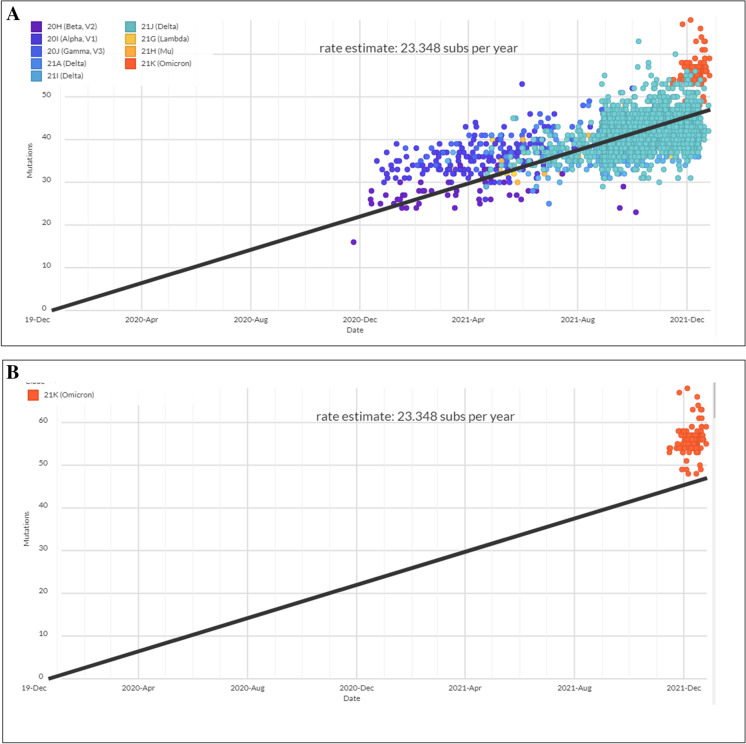
Scatter plot for cluster evaluation of Omicron and other VOCs and VOIs
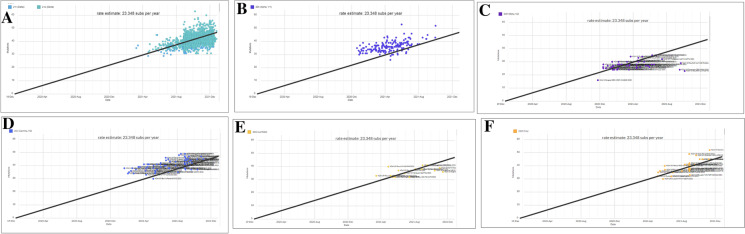
Cluster analysis was performed, and a scatter plot was generated for all current VOCs and VOIs, in conjunction with the Omicron variant. The scatter plot shows the prevalence of all VOCs and VOIs with Omicron and indicates the origin and distribution of all current VOCs and VOIs. A scatter plot with linear regression was developed using the current circulating VOCs and VOIs (Fig. 11A). The model informs us that all the sample values were placed on both sides of the regression line. The figure shows a strong, positive, and linear scatter plot. Simultaneously, the scatter plot of the Omicron variant shows the prevalence of all samples on the upper side of the regression line (Fig. 11B).
Fig. 11.
Scatter plot for cluster evaluation of Omicron and all VOCs and VOIs. (A) Scatter plot for cluster evaluation of all VOCs and VOIs. (B) Scatter plot for cluster evaluation of Omicron. The scatter plot was developed for Omicron and all VOCs and VOIs using the Nextstrain server [49, 50]. The server used the GISAID data
Similarly, the scatter plot of the Delta variant shows the prevalence of all sample points on both sides of the regression line of the plot (Fig. 12A). The scatter plot of the Alpha variant shows the prevalence of all sample points above the regression line. All sample points were in the middle position of the regression line (Fig. 12B). Similarly, the scatter plot of the Beta variant shows the prevalence of all sample points below the regression line. However, a few sample points were placed above the regression line, and all sample points were placed middle in the position of the regression line (Fig. 12C).
Fig. 12.
Scatter plot for cluster evaluation for VOCs and VOIs. (A) Scatter plot for cluster evaluation of Delta. (B) Scatter plot for cluster evaluation of Alpha. (C) Scatter plot for cluster evaluation of Beta. (D) Scatter plot for cluster evaluation of Gamma. (E) Scatter plot for cluster evaluation of Lambda. (F) Scatter plot for cluster evaluation of mu. The scatter plot was developed for VOCs and VOIs using the Nextstrain server [49, 50]. The server used the GISAID data
Similarly, the scatter plot of the Gamma variant shows the prevalence of all sample points in the upper region of the regression line. Most sample points were above the regression line. However, a few sample points are found below the plot’s regression line (Fig. 12D).
Similarly, the scatter plot of the Lambda variant shows the prevalence of all sample points in the upper region of the regression line. Most of the sample points were below the regression line, and however, very few sample points were observed above the regression line (Fig. 12E).
Finally, we developed a scatter plot for the Mu variants. The figure shows the prevalence of all sample points in the upper region of the regression line of the plot. Most of the sample points were below the regression line. However, few sample points were found above the regression line (Fig. 12F). The scatter plots for the Lambda and Mu variants are quite similar. However, it was observed that the Mu variant sample points were densely placed (Fig. 12E, F).
Prevalence of mutations in the neutralizing antibody (NAb) binding site in Omicron and other VOCs and VOIs
Mutations in the nAb binding site or close to the nAb binding site of the S-glycoprotein of the Omicron variant have been noted. We analyzed the mutations in the nAb binding site from an extensive survey of the typical nAb (type-1 to type-4) interaction with the S-glycoprotein and found mutations in the nAb binding site or close to the nAb binding site of the S-glycoprotein. The most emerging mutations in the antibody interaction area were located within the RBD region (Fig. 13A).
Fig. 13.
The figure shows the developed 3D model of S-glycoprotein and the emerging mutations. The major mutations are located within the RBD region, which might involve antibodies interaction area. (A) RBD mutations in Omicron located in antibody interaction region. (B) RBD mutations in VOIs located in antibody interaction region. (C) RBD mutations in VOIs located in the antibody interaction region. The 3D model of S-glycoprotein using PyMOL software and the emerging mutations for antibody interaction was depicted for the Omicron variant, other VOC, and VOI in the 3D model [46, 47]. For a 3D model generation, we used some PDB files (PDB ID: 6VXX)
Similarly, mutations in the nAb binding site or close to the nAb binding site of the S-glycoprotein of the other VOCs are also illustrated (Fig. 13B). We also recorded mutations in the nAb binding site or close to the nAb binding site of the S-glycoprotein of the other VOIs (Fig. 13C). We performed a comparative analysis of mutations in the nAb binding site of all VOCs and VOIs to assess the emerging mutations in the nAb binding site of the S-glycoprotein, and illustrated all crucial mutations in the nAb binding area (Table 4).
Table 4.
Emerging mutations in antibodies interaction area within RBD region of SARS-CoV-2 VOCs and VOIs including Omicron variant
| Sl no | Variants name (WHO label) | SARS-CoV-2 lineages | Mutations in S-protein (RBD region) | Remarks |
|---|---|---|---|---|
| 1 | Omicron | B.1.1.529 | K417N, N440K, G446S, S477N, T478K, E484A, Q493K, G496S, Q498R, N501Y, Y505H |
Comparative analysis from all present VOCs and VOIs noted the following mutations in Ab interaction within the RBD region: K417N, N440K, G446S, S477N, T478K, E484A, Q493K, G496S, Q498R, N501Y, Y505H, S494P, L452R, T478K, K417T, L452Q, F490S, R346K |
| 2 | Alpha | B.1.1.7 | E484K, S494P, N501Y | |
| 3 | Delta | B.1.617.2 | L452R, T478K | |
| 4 | Gamma | P.1 | K417T, E484K, N501Y | |
| 5 | Beta | B.1.351 | K417N, E484K, N501Y | |
| 6 | Lambda | C.37 | L452Q, F490S | |
| 7 | Mu | B.1.621 | R346K, E484K, N501Y |
Zhang et al. illustrated mutations in 11 VOCs and VOIs, including Omicron, and their effect on the nAb binding site. They concluded from their study that mutations near AA 439–448 and AA484 might cause resistance to neutralization [61]. Cui et al. performed an experimental study and developed the Omicron pseudo-virus, attempted to correlate immunogenic areas with high mutation frequencies, and correlated them with properties such as immune escape and infectivity. They identified the ten most critical immunogenic residues. Among them, the three AA substitution residues were Y505H, E484A/K, and Q493R. They have noted some other significant residues, including F456, N487, Y449, Y489, and F486 [66]. Similarly, in another experimental study, Planas et al. illustrated 32 spike mutations, mostly in NTD, which may augment viral fitness and antibody escape [67]. In the present study, we also depicted mutations nAb binding site or close to the nAb binding site of the S-glycoprotein, which may increase viral fitness and nAb escape.
Risk analysis of antibody-binding to significant antibody-binding mutations in the RBD in the S-glycoprotein
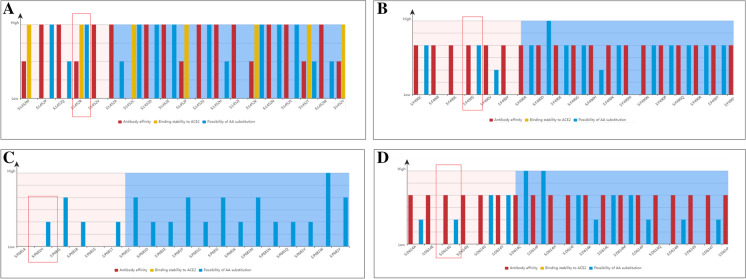
We performed an analysis to understand the risk level of mutations in four positions and found that three AA residues are essential for interactions with the antibody. In this study, we analyzed three positions for mutations (L452, F490, and P681) and a significant mutation (D614G).
The mutation risk of the L452 location was analyzed, and the risk level of 19 different types of mutational variants at a particular site (L452) is illustrated (Fig. 14A). The L452R mutation has a destabilizing effect on the interactions of S-glycoprotein targeting antibodies. Variants of the mutation have also been reported (Fig. S1A). The count for this variant of the L452R mutation is 1657075, and the first variant (L452R) was identified in the USA.
Fig. 14.
Risk analysis of antibody-binding to significant some antibody-binding mutations and its other variants. (A) Risk analysis of antibody-binding to L452 region and L452R mutation. (B) Risk analysis of antibody-binding to F490 region and F490S mutation. (C) Risk analysis of antibody-binding to P681 region and P681H mutation. (D) Risk analysis of antibody-binding to D614 region and D614G mutation. We used the VarEPS server to evaluate the risk analysis of antibody-binding of some antibody-binding mutations [52]. The server used the GISAID data and data from other resources such as CDC China, CDC, USA, WHO
We analyzed the mutation risk at the F490 position, and 19 different types of mutations in different variants at this position (F490) were observed (Fig. 14B). The F490S mutation was found to have a destabilizing effect on antibody interactions. Variants of this mutation have also been reported (Fig. S1B). The count for this variant of the F490S mutation was 13,311, and the first variant (F490S) was identified in Canada.
We analyzed the mutation risk at the P681 position. Similarly, previous mutations and the 19 different types of mutations in different variants (P681) were explained (Fig. 14C). In the P681 mutation, it has been noted that the mutations do not have any destabilizing effect on the antibody interactions. Variants of this mutation have also been reported (Fig. S1C). The count for this variant of the P681H mutation was 130,2751, and the first mutation (P681H) was identified in Nigeria.
Finally, 19 different types of mutations in different variants of D614G are shown (Fig. 14D). The D614G mutation has a destabilizing effect on the antibody interactions. Variants of the mutation have also been reported (Fig. S1D). The count for this variant of the D614G mutation was 4,105,183, and the first variant (D614G) was reported in Germany.
Molecular and mutational properties of some significant RBD mutations in Omicron
Mutations in the RBD region may play a role in antibody, immune, or vaccine escape. In this part of the analysis, we evaluated some RBD mutations’ molecular and mutational properties in the Omicron variants: G339D, S371L, N440K, G446S, T478K, and Q498R. The study illustrated several molecular and mutational parameters such as delta delta G (DDG) or ΔΔG, interatomic interactions in the wild-type and mutant-type residues, and fluctuation and deformation analysis of those mutations. We also analyzed the fluctuation of the wild-type and mutant-type residues, atomic fluctuation in the wild-type and mutant-type of the S-glycoprotein, and deformation energies in the wild-type and mutant-type of the S-glycoprotein in respect of these mutations.
We assessed the molecular and mutational properties of G339D. The molecular and mutational parameters (ΔΔG, interatomic interactions, and fluctuation and deformation analysis) of G339D are illustrated in Fig. 15A. Here, the analysis indicated that the outcome of ΔΔG was 0.019 kcal/mol (stabilizing). The NMA-based predictions showed that ΔΔG ENCoM was − 0.027 kcal/mol (destabilizing). Other structure-based calculations showed that ΔΔG mCSM was − 0.691 kcal/mol (destabilizing), ΔΔG SDM was 0.110 kcal/mol (stabilizing), and ΔΔG DUET was − 0.400 kcal/mol (destabilizing). The ΔΔSVib ENCoM (minute vibrational entropy energy between wild-type and mutant-type) was 0.033 kcal/mol−1 K−1. This model indicates that the mutation increases molecular flexibility, as shown in Fig. 15B. Here, the wild-type and mutant-type residues are colored light green. The model informed the neighboring residues linked to the interaction interface. Figure S2A shows the fluctuation of the wild-type and mutant-type residues. Visual analysis shows atomic fluctuation in the 3D model of the S-glycoprotein, both in the wild-type and mutant-type (Fig. S2B). Simultaneously, the study included a visual analysis of deformation energies to show the amount of local flexibility of the 3D model of both the wild-type and mutant-type S-glycoproteins (Fig. S2C).
Fig. 15.
Mapped molecular and mutational properties of G339D. (A) Prediction of vibrational Entropy Energy of G339D with a visual representation. (B) Prediction of interatomic interactions of wild type to mutant type G339D mutation. In this study, we used the DynaMut server to evaluate the molecular and mutational properties of G339D [53]. For the analysis, we used a model PDB file (PDB ID: 7QO7)
The molecular and mutational properties of S371L were also evaluated. Here, we illustrated the molecular and mutational factors in G339D (Fig. 16A). The analysis indicated that the outcome of ΔΔG was 0.127 kcal/mol (stabilizing). The NMA-based predictions showed that ΔΔG ENCoM was 0.030 kcal/mol (destabilizing). Other structure-based calculations showed that ΔΔG mCSM was − 0.394 kcal/mol (destabilizing), ΔΔG SDM was 1.280 kcal/mol (stabilizing), and ΔΔG DUET was 0.080 kcal/mol (stabilizing). The evaluation of (ΔΔSVib ENCoM) change between wild-type and mutant-type was − 0.037 kcal/mol−1 K−1. The mutational landscape indicated that the mutation decreased molecular flexibility. The molecular contacts between the inter-atoms are illustrated in Fig. 16B. The light green residues indicate the wild-type and mutant-type AA residues in this study. The developed model indicated that adjoining residues were related to the interaction interface. Figure S3A shows the fluctuation of the wild-type and mutant-type residues. Visual analysis shows atomic fluctuation in the 3D model of the S-glycoprotein for both the wild-type and mutant-type (Fig. S3B). We evaluated the visual analysis of deformation energies, showing the quantity of local flexibility of the 3D model of both the wild-type and mutant-type S-glycoprotein (Fig. S3C).
Fig. 16.
Evaluated molecular and mutational properties of S371L. (A) Calculation of vibrational Entropy Energy of S371L with a visual representation. (B) Prediction of interatomic interactions of wild type to mutant type S371L mutation. For this evaluation, the DynaMut server was used to evaluate the molecular and mutational properties of S371L [53]. For the analysis, we used a model PDB file (PDB ID: 7QO7)
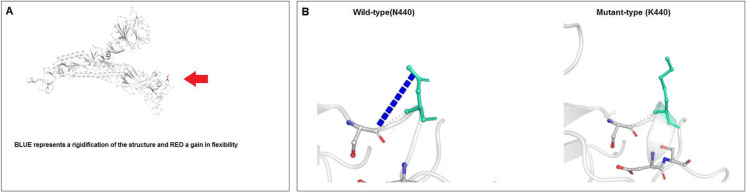
This study analyzed parameters such as ΔΔG, interatomic interactions, and fluctuation and deformation to understand the molecular and mutational properties of the N440K mutation. The results of ΔΔG analysis of N440K mutation are shown in Fig. 17A. Here, the analysis indicated that the outcome of ΔΔG was 0.064 kcal/mol (stabilizing). The NMA-based predictions showed that ΔΔG ENCoM was 0.045 kcal/mol (destabilizing). Other structure-based calculations showed that ΔΔG mCSM was 0.243 kcal/mol (stabilizing), ΔΔG SDM was 0.780 kcal/mol (stabilizing), and ΔΔG DUET was 0.876 kcal/mol (stabilizing). The evaluation of minute vibrational entropy energy between the wild-type and mutant-type (ΔΔSVib ENCoM) indicated as − 0.057 kcal mol−1 K−1. The result of the model predicts that the mutation decreases molecular flexibility. The interactions between the inter-atoms are shown in Fig. 17B. The wild-type and mutant-type residues are marked in light green. The model revealed the adjacent residues involved in the interaction. Figure S4A illustrates the fluctuation of wild-type and mutant-type residues. Visual analysis showed atomic fluctuation in the 3D model of the S-glycoprotein, both in the wild-type and mutant-type (Fig. S4B). Simultaneously, the study evaluated the visual analysis of deformation energies to show the amount of local flexibility of the 3D model of both the wild-type and mutant-type S-glycoproteins (Fig. S4C).
Fig. 17.
Analyzed result of molecular and mutational properties of N440K. (A) Computation of vibrational Entropy Energy of N440K and is represented through a pictorial view. (B) Prediction of interatomic Interactions of wild type to mutant type N440K mutation. In this study, we used the DynaMut server to assess the molecular and mutational properties of N440K [53]. For the analysis, we used a model PDB file (PDB ID: 7QO7)
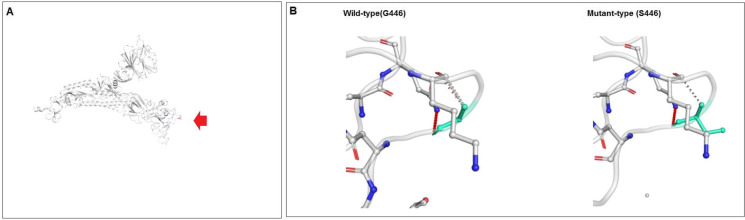
This study analyzed several parameters to evaluate the molecular and mutational features of the G446S mutation. Figure 18A shows the results of ΔΔG analysis of the G446S mutation. In this investigation, the effect of ΔΔG was recorded as − 0.501 kcal/mol (destabilizing). The NMA-based calculation showed that ΔΔG ENCoM was − 0.283 kcal/mol (destabilizing). Additional structure-based computations indicated that ΔΔG mCSM was − 0.593 kcal/mol (destabilizing), ΔΔG SDM was − 0.680 kcal/mol (destabilizing), and ΔΔG DUET was − 0.366 kcal/mol (destabilizing). We evaluated minute vibrational entropy energy change between wild-type and mutant-type, which shows ΔΔSVib ENCoM: − 0.353 kcal/mol−1 K−1. The results of the model indicated a mutational landscape augmentation of molecular flexibility. The inter-atomics interactions are shown in Fig. 18B. The model depicts the wild-type and mutant-type residues as light green and shows the nearby residues involved in the interface. Figure S5A shows the fluctuation of the wild-type and mutant-type residues. Visual analysis showed that the atomic fluctuation in the 3D model indicated both in the wild-type and mutant-type S-glycoprotein (Fig. S5B). We visually analyzed the deformation energies to show the amount of local flexibility through the 3D model of wild-type and mutant-type S-glycoproteins (Fig. S5C).
Fig. 18.
Evaluated outcome of the molecular and mutational properties of G446S. (A) Calculated vibrational Entropy Energy of G446S and its pictorial representation. (B) Illustrated interatomic interactions of wild type to mutant type G446S mutation. For this analysis, the DynaMut server was used to evaluate the molecular and mutational properties of S371L [52]. For the analysis, we used a model PDB file (PDB ID: 7QO7)
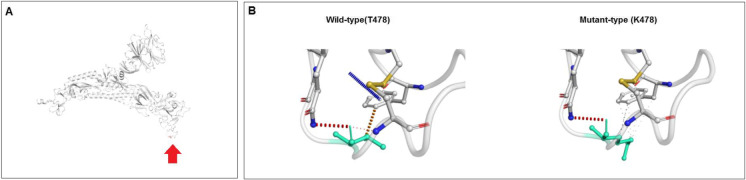
We recorded the results of ΔΔG for the T478K mutation in Fig. 19A. In the computation, the effect of ΔΔG was recoded as 1.009 kcal/mol (stabilizing). The NMA-based estimation showed that ΔΔG ENCoM was 0.181 kcal/mol (destabilizing). Further structure-based analysis indicated three ΔΔG-based parameters ΔΔG mCSM, − 0.695 kcal/mol (destabilizing), ΔΔG SDM (− 0.070 kcal/mol; destabilizing), and ΔΔG DUET (− 0.366 kcal/mol (destabilizing). The ΔΔSVib ENCoM change between wild-type and mutant-type was found to be − 0.227 kcal/mol−1 K−1. The result of the model informed us about mutational flexibility (decreased molecular flexibility). The inter-atom correlations are shown in Fig. 19B. The developed model of the wild-type and mutant-type residue indicated in light green, which informs us that adjunct residues are involved in the interaction interface. The fluctuation of the wild-type and mutant-type residues are shown in Figure S6A. Visual analysis revealed atomic fluctuation of the wild-type and mutant-type of the S-glycoprotein (Fig. S6B). We evaluated the visual analysis of deformation energies to show the amount of local flexibility through a model that provides a pictorial view of both the wild-type and mutant-type S-glycoprotein (Fig. S6C).
Fig. 19.
Analyzed result of the molecular and mutational properties of T478K. (A) Calculated vibrational Entropy Energy of T478K and its visual representation. (B) Illustrated of interatomic interactions of wild type to mutant type T478K mutation. We have used the DynaMut server in this study to explain the molecular and mutational properties of N440K [53]. For the analysis, we used a model PDB file (PDB ID: 7QO7)
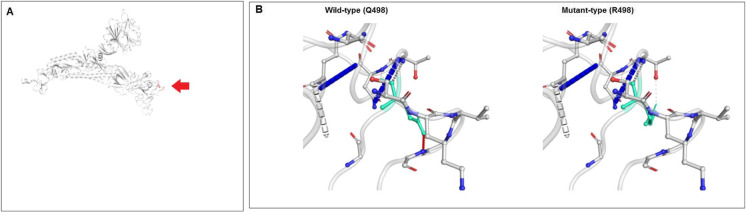
Finally, the mutational features and molecular properties of the Q498R mutation were assessed. We observed the properties of ΔΔG and its effect on the mutant type of the Q498R mutation, as illustrated in Fig. 20A. This analysis recorded the consequence of ΔΔG due to the Q498R mutation as − 0.163 kcal/mol (destabilizing). The computation of NMA-related predictions showed that ΔΔG ENCoM was − 0.289 kcal/mol (destabilizing). Additionally, three structure-based analyses indicated ΔΔG-related features of the mutation. The results were as follows: ΔΔG mCSM was 0.283 kcal/mol (stabilizing), ΔΔG SDM was 0.550 kcal/mol (stabilizing), and ΔΔG DUET was 0.519 kcal/mol (stabilizing). The ΔΔSVib ENCoM, calculated as 0.361 kcal/mol−1 K−1, was noted as increased the molecular flexibility of the S-glycoprotein. We depicted the inter-atom configurations of the mutational landscape of the wild and mutant varieties of Q498R, which is illustrated in Fig. 20B. A model was generated for the wild-type and mutant-type residues, illustrating both residues as light green. The model showed that the residues associated with that particular position were also associated with the interaction interface. Figure S7A shows the fluctuation of the wild-type and mutant-type residues. Furthermore, visual analysis of atomic fluctuation in the mutant residue informed us about the atomic fluctuation of the wild-type and mutant-type spike-glycoproteins (Fig. S7B). Moreover, the experiment illustrated the deformation energies of the visual analysis and provide information on the quantity of local flexibility using a bioinformatics model. The model shows the pictographic view of both the wild-type and mutant-type spike-glycoprotein (Fig. S7C).
Fig. 20.
Representation of the molecular and mutational properties of Q498R. (A) Calculated vibrational Entropy Energy of Q498R and its visual representation. (B) Illustrated interatomic interactions of wild type to mutant type Q498R mutation. We have used the DynaMut server in this study to elucidate the molecular and mutational properties of Q498R [53]. For the analysis, we used a model PDB file (PDB ID: 7QO7)
We have comprehensively analyzed the properties of 28 mutations, and other researchers have tried to analyze other mutations using a similar method [68]. However, our comprehensive study has tried to analyze the 28 mutations along with RBD mutations and concluded how RBD mutations affect receptor binding of Omicron. Finally, we tried to explain how RBD mutations might affect infectivity.
Comparative analysis of delta-delta G (DDG) or ΔΔG of analyzed mutations in the S-glycoprotein
We performed a comprehensive and comparative analysis of ΔΔG for 28 significant mutations, including K417N, S477N, T478K, D796Y, G196D, G446S, E484A, T95I, D614G, H655Y, Q493K, G496S, N501Y, Y505H, N969K, A67V, L981F, N440K, N856K, Q498R, S371L, G339D, L212I, S375F, S373P, N764K, T547K, and Q954H. The ΔΔG values of the 28 analyzed mutations are shown in Table 5. In this study, we performed several cooperative analyses, including ΔΔG ENCoM, ΔΔG, ΔΔG mCSM, ΔΔG DUET, ΔΔG SDM, and ΔΔSVib ENCoM. The NMA-based prediction of ΔΔG ENCoM analysis showed that the K417N mutation had shown a minimum value with the destabilizing result (− 0.677 kcal/mol), and the G196D mutation had a maximum value with a stabilizing result (0.897 kcal/mol). ΔΔG prediction showed that the K417N mutation had a minimum value with a destabilizing impact (− 0.932 kcal/mol) and the Q954H mutation had a maximum value with a stabilizing effect (0.969 kcal/mol). Evaluation of Δ vibrational entropy energy between wild-type and mutant (ΔΔSVib ENCoM) showed that the G196D mutation had a minimum value of − 1.121 kcal/mol−1 K−1. Similarly, the K417N mutation had a maximum value of 0.846 kcal/mol−1 K−1.
Table 5.
Comparative analysis of delta delta G (DDG) or ΔΔG of emerging mutations of S-glycoprotein in Omicron variant
| NMA based prediction | ΔΔG prediction | Other structure-based predictions | Minute vibrational entropy energy change between wild-type and mutant | Remarks | ||||
|---|---|---|---|---|---|---|---|---|
| Sl no | Significant mutations of Omicron variant |
ΔΔG ENCoM (kcal/mol) |
ΔΔG (kcal/mol) |
ΔΔG mCSM (kcal/mol) |
ΔΔG DUET (kcal/mol) |
ΔΔG SDM (kcal/mol) |
ΔΔSVib ENCoM (kcal mol−1 K−1) |
|
| 1 | K417N |
− 0.677, Destabilizing |
− 0.932, Destabilizing |
− 1.138, Destabilizing |
− 1.123, Destabilizing |
− 0.280, Destabilizing |
0.846 | Increase of molecule flexibility increased |
| 2 | S477N |
0.064, Destabilizing |
0.628, Stabilizing |
− 0.215, Destabilizing |
0.235, Stabilizing |
0.780, Stabilizing |
− 0.080 | Decrease of molecule flexibility |
| 3 | T478K |
0.181, Destabilizing |
1.009, Stabilizing |
− 0.695, Destabilizing |
− 0.366, Destabilizing |
− 0.070, Destabilizing |
− 0.227 | Decrease of molecule flexibility |
| 4 | D796Y |
− 0.052, Destabilizing |
0.146, Stabilizing |
0.387, Stabilizing |
0.370, Stabilizing |
− 0.010, Destabilizing |
0.065 | Increase of molecule flexibility |
| 5 | G196D |
0.897, Stabilizing |
0.193, Stabilizing |
− 2.071, Destabilizing |
− 2.248, Destabilizing |
− 2.160, Destabilizing |
− 1.121 | Decrease of molecule flexibility |
| 6 | G446S |
− 0.283, Destabilizing |
− 0.501, Destabilizing |
− 0.593, Destabilizing |
− 0.366, Destabilizing |
− 0.680, Destabilizing |
0.353 | Increase of molecule flexibility |
| 7 | E484A |
− 0.377, Destabilizing |
− 0.456, Destabilizing |
− 0.415, Destabilizing |
− 0.128, Destabilizing |
0.320, Stabilizing |
0.471 | Increase of molecule flexibility |
| 8 | T95I |
0.320, Destabilizing |
1.200, Stabilizing |
− 0.097, Destabilizing |
0.670, Stabilizing |
1.910, Stabilizing |
− 0.400 | Decrease of molecule flexibility |
| 9 | D614G |
− 0.134, Destabilizing |
0.351, Stabilizing |
− 0.514, Destabilizing |
0.171, Stabilizing |
2.510, Stabilizing |
0.168 | Decrease of molecule flexibility |
| 10 | H655Y |
0.158, Destabilizing |
1.728, Stabilizing |
1.302, Stabilizing |
1.516, Stabilizing |
0.400, Stabilizing |
− 0.197 | Decrease of molecule flexibility |
| 11 | Q493K |
0.066, Destabilizing |
0.470, Stabilizing |
0.194, Stabilizing |
0.794, Stabilizing |
0.580, Stabilizing |
− 0.082 | Decrease of molecule flexibility |
| 12 | G496S | − 0.014, Destabilizing | − 0.097, Destabilizing | − 0.763, Destabilizing | − 0.614, Destabilizing | − 1.010, Destabilizing | 0.018 | Decrease of molecule flexibility |
| 13 | N501Y |
− 0.094, Destabilizing |
− 0.203, Destabilizing |
− 0.457, Destabilizing |
− 0.471, Destabilizing |
0.280, Stabilizing |
0.117 | Decrease of molecule flexibility |
| 14 | Y505H |
− 0.305, Destabilizing |
− 0.510, Destabilizing |
− 0.119, Destabilizing |
0.143, Stabilizing |
0.380, Stabilizing |
0.381 | Decrease of molecule flexibility |
| 15 | N969K |
− 0.381, Destabilizing |
0.086, Stabilizing |
0.065, Stabilizing |
0.184, Stabilizing |
− 0.500, Destabilizing |
0.477 | Increase of molecule flexibility |
| 16 | A67V |
0.520, Stabilizing |
0.544, Stabilizing |
− 0.387, Destabilizing |
0.256, Stabilizing |
0.950, Stabilizing |
− 0.650 | Decrease of molecule flexibility |
| 17 | L981F |
0.009, Destabilizing |
0.104, Stabilizing |
− 1.241, Destabilizing |
− 1.289, Destabilizing |
− 0.69, Destabilizing |
0.012 | Increase of molecule flexibility |
| 18 | N440K |
0.045, Destabilizing |
0.064, Stabilizing |
0.243, Stabilizing |
0.876, Stabilizing |
0.780, Stabilizing |
− 0.057 | Decrease of molecule flexibility |
| 19 | N856K |
0.202, Destabilizing |
1.304, Stabilizing |
0.103, Stabilizing |
0.314, Stabilizing |
0.300, Destabilizing |
− 0.252 | Decrease of molecule flexibility |
| 20 | Q498R |
− 0.289, Destabilizing |
− 0.163, Destabilizing |
0.283, Stabilizing |
0.519, Stabilizing |
0.550, Stabilizing |
0.361 | Increase of molecule flexibility |
| 21 | S371L |
0.030, Stabilizing |
0.127, Stabilizing |
− 0.394, Destabilizing |
0.080, Stabilizing |
1.280, Stabilizing |
− 0.037 | Decrease of molecule flexibility |
| 22 | G339D |
− 0.027, Stabilizing |
0.019, Stabilizing |
− 0.69, Destabilizing |
− 0.400, Destabilizing |
0.110, Stabilizing |
0.033 | Increase of molecule flexibility |
| 23 | L212I |
− 0.028, Destabilizing |
0.090, Stabilizing |
− 0.844, Destabilizing |
− 0.334, Destabilizing |
0.490, Stabilizing |
0.035 | Increase of molecule flexibility |
| 24 | S375F |
0.578, Stabilizing |
− 0.125, Destabilizing |
− 0.948, Destabilizing |
− 0.460, Destabilizing |
1.190, Stabilizing |
− 0.723 | Decrease of molecule flexibility |
| 25 | S373P |
− 0.323, Destabilizing |
− 0.797, Destabilizing |
− 0.645, Destabilizing |
− 0.534, Destabilizing |
− 0.030, Destabilizing |
0.404 | Increase of molecule flexibility |
| 26 | N764K |
− 0.344, Destabilizing |
0.189, Stabilizing |
0.089, Stabilizing |
0.475, Stabilizing |
− 0.300, Destabilizing |
0.430 | Increase of molecule flexibility |
| 27 | T547K |
0.072, Destabilizing |
0.462, Stabilizing |
− 0.244, Destabilizing |
0.194, Stabilizing |
− 0.090, Destabilizing |
− 0.090 | Increase of molecule flexibility |
| 28 | Q954H |
0.192, Destabilizing |
0.969, Stabilizing |
− 0.676, Destabilizing |
− 0.491, Destabilizing |
0.060, Stabilizing |
− 0.240 | Decrease of molecule flexibility |
Cui et al. reported a stable Omicron spike that helps to maintain an active conformation, which plays a significant role in receptor recognition [66]. Some mutations were observed with increased flexibility which may also help in receptor interactions and, thus, improve viral fitness. Therefore, our predicted mutations with stabilizing properties in the S-glycoprotein, particularly RBD mutation, play a significant role in receptor recognition, increasing infectivity, and improving viral fitness.
Discussion
The Omicron variant has been spreading rapidly in several countries worldwide. Omicron is an immensely mutated emerging variant that is proving to be a challenge to researchers [25, 26]. This variant has the largest number of mutations among all other VOCs and VOIs of SARS-CoV-2, and researchers have described this variant as a rare collection of mutations [69]. More than 50 mutations have been identified in the genome. Researchers have noted approximately 32 in the spike protein [20, 25–27]. Therefore, it is essential to understand the mutational landscape of the Omicron variant and compare the landscape of mutations with other emerging variants. This study attempted to understand the mutational landscape of the Omicron variant and compared all mutational frequencies of SNVs throughout the genome with those of other emerging variants. We evaluated the AA variants in the Omicron variant and compared them with those in the present VOCs and VOIs. This study found that the D614G AA variant in the S-glycoprotein was present in all VOCs and VOIs at a high frequency (98–100%). Our in silico analysis corroborates the previous finding about the high frequency of D614G mutation in Omicron and other VOI/VOC [31, 32, 63]. Analysis of AA variants of the non-structural protein revealed that P323L in nsp12 was also observed in all VOCs and VOIs with high frequency (97–100%). We illustrated the AA variants using a heat map of mutation prevalence with > 75% mutational frequency of Omicron and compared those mutations with that of other VOCs and VOIs. The heat map helps identify mutations with a high mutational frequency. This study showed eight mutations with > 90% in ORF1a and 29 in S-glycoprotein, with > 75% mutation prevalence in Omicron. Several other studies have attempted to illustrate mutations in different regions of the Omicron. Lupala et al. elucidated the interaction between the RBD region of Omicron and the hACE2 receptor to understand its binding affinity. In this study, they developed a model of a mutated RBD region and an interaction model between the RBD region of Omicron and the hACE2 receptor. Finally, MDS (molecular dynamics simulations) were performed to understand the RBD-hACE2 interaction pattern [70]. In our study, we analyzed 28 mutations and observed that among RBD mutations, G339D, S371L, N440K, and T478K mutations are stabilizing, and G446S and Q498R are destabilizing mutations. At the same time, G339D, G446S, and Q498R mutations have increased molecular flexibility. Cumulative effects of the properties of the mutations (stabilizing, destabilizing, molecular flexibility) of RBD might augment the receptor-binding activity of Omicron, which might help to provide superior infectivity properties of Omicron. Our study has also attempted to understand the complete mutational landscape of Omicron along with the present VOCs and VOIs. It is important to understand the Omicron variant from a mutational point of view.
Emerging mutations in SARS-CoV-2 are a major concern for antibody escape, especially in nAb. Triveri et al. have attempted to describe the escape phenomena of SARS-CoV-2 variants because of mutations in the S-glycoprotein. They considered emerging variants such as the South African (501Y.V2), UK (B.1.1.7), Brazilian (P.1), and D614G variants to understand the correlation between the mutations and escape from immuno-recognition. The study also illustrated the ability of these variants to evade or decrease the recognition capacity of S-glycoprotein targeting antibodies [71]. However, we performed risk analysis for D614G antibody binding. We observed that the mutation in D614 has a high risk related to antibody-binding and can reduce the recognition capability by targeting antibodies to the S-glycoprotein. The outcome of our in silico risk analysis study of D614G corroborates the previous experimental study’s result by Martin et al. [72]. Their study has found that the D614G mutation decreased antibody binding of the S-glycoprotein. Previously, we illustrated that the D614G variant is present in all VOCs and VOIs with high frequency, and the mutant variant may be a positive selection [28, 31, 32]. We are currently attempting to understand the prominent mutations in the S-glycoprotein in SARS-CoV-2 and their role in antibody escape [73, 74].
Several studies have shown that most antibodies bind closer to the NTD or RBD regions [75]. However, some RBD mutations are responsible for antibody escape [33, 34]. We evaluated some mutations, especially mutations in the RBD region in Omicron, and their probable consequence on antibody affinity [76]. In this study, we also attempted to evaluate some molecular and mutational properties of some mutations in the RBD region in Omicron, which may provide deeper insight into the mutations. We evaluated the molecular and mutational properties of some Omicron variants: G339D, S371L, N440K, G446S, T478K, and Q498R. Here, we describe the molecular and mutational properties of these mutations.
Recently, Singh et al. attempted to understand the ΔΔG between the wild-type and mutant proteins in Mycobacterium tuberculosis. We analyzed the ΔΔG value to understand the effects of mutations on molecular flexibility. Several researchers have computed ΔΔG values to understand the mutational properties of mutations in a human pathogen. It aids in the understanding of the properties of point mutations [77]. Othman et al. used the same algorithm to calculate the enthalpy (DDG) and entropy (DDS) to understand the folding energy during the interaction of the SARS-CoV-2 S-glycoprotein and host ACE2 [78]. We also calculated the ΔΔG of some point mutations (G339D, S371L, N440K, G446S, T478K, and Q498R), which are significant in the RBD region of the SARS-CoV-2 Omicron. We found fluctuation in the wild-type and mutant-type residues, atomic fluctuation in the wild-type and mutant-type S-glycoprotein, and deformation energies in the wild-type and mutant-type in the dynamics of the S-glycoprotein with respect to those mutations through the algorithms. Our analysis allowed us to illustrate the harmonic motions of the missense variants in terms of the S-glycoprotein through the analysis of ΔΔG values, which provides insights into accessible conformations of S-glycoprotein and its dynamics; however, we analyzed residue-wise RMS fluctuations, atomic fluctuations, and deformation energies of all mutations (G339D, S371L, N440K, G446S, T478K, and Q498R). The residue-wise RMS fluctuation (Figs. S2A, S3A, S4A, S5A, S6A, and S7A), atomic fluctuation (Figs. S2B, S3B, S4B, S5B, S6B, and S7B), and deformation energies (Figs. S2C, S3C, S4C, S5C, S6C, and S7C) of all these mutations showed a similar result in every category. Presently, scientists are attempting to understand the effects of spike-protein mutations on conformations and dynamics [79–81]. Our study illustrated the essential mutations of the Omicron variant.
The present study chose 28 mutations and six RBD mutations with immense importance in understanding the molecular properties of mutations. These RBD mutations may be associated with some significant properties of Omicron, such as antibody escape and ACE2 interaction. A schematic diagram was used to depict the mutation selection from the RBD region of the S-glycoprotein (Fig. 1B). Finally, we attempted to evaluate all 28 mutations, including the six RBD mutations, to understand mutational properties such as stabilizing or destabilizing events and increased or decreased molecular flexibility.
The study performed a comparative analysis of ΔΔG of the 28 mutations of the S-glycoprotein, including the six RBD mutations, to understand the stabilizing or destabilizing properties and other mutational properties. The comparative outcome is noted in Table 5. ΔΔG analysis and the stabilizing or destabilizing properties were analyzed using the DynaMut server and other mutational properties. We chose six mutations in the RBD of Omicron for a more detailed analysis, G339D, S371L, N440K, G446S, T478K, and Q498R. The research outcome of the six RBD mutations showed that G339D, S371L, N440K, and T478K were stable with 0.019 kcal/mol and 0.127 kcal/mol, 0.064 kcal/mol, and 1.009 kcal/mol, respectively. Other mutations (G446S and Q498R) showed destabilizing results (Fig. 21A). It is noteworthy that, among the six RBD mutations, G339D, G446S, and Q498R mutations increased the molecular flexibility of the S-glycoprotein (Fig. 21B). These RBD mutations may help RBD interact with the ACE2 receptor, thus helping to increase Omicron infectivity. Our study aids future researchers in understanding the mutational landscape of Omicron and in elucidating more about the different properties of Omicron, such as infectivity and antibody escape, using our analyzed properties (stabilizing or destabilizing as well as increased or decreased molecular flexibility of the S-glycoprotein to interact with the mAb/receptor) of those using the 28 emerging mutations along with six RBD mutations. Recently, several researchers have characterized SARS-CoV-2 mutations using the DynaMut server. The DDG of some mutations (L452R, E484Q, and P681R) in the Indian variant (B.1.617) was computed using the DynaMut server [82]. Another study characterized three mutations in an Italian SARS-CoV-2 strain using the DynaMut server [83]. This study analyzed the properties of 28 mutations, including six RBD mutations of the Omicron variant, in a short time. This information may be essential supportive data for future researchers who further unfold the significant properties of Omicron and other VOCs and provide a solution to the pandemic. From this perspective, our study is highly important.
Fig. 21 .
The graphical image shows the emerging mutations of RBD (S-glycoprotein) of the Omicron. It is also shown how it affects flexibility changes and stability alteration due to amino acid point mutation. (A) The schematic diagram illustrates the emerging mutations of RBD (S-glycoprotein) of the Omicron and their effects on stability (stabilizing event or destabilizing) (B) The schematic diagram shows the emerging mutations of RBD (S-glycoprotein) of the Omicron and their impact on flexibility
Limitations
The Omicron variant is a crucial variant termed by the WHO as a VOC because of its infectivity, antibody escape, and partial vaccine escape. Therefore, this variant has created a more challenging pandemic [65, 74, 76, 84]. In this study, we evaluated 28 mutations in Omicron and their properties. The study computed the increased or decreased stability event and the increased or decreased molecular flexibility of the S-glycoprotein because of the mutational change using the DynaMut server in a short time. This may be a limitation of the present study. However, this paper presents a more detailed analysis of these eight mutations. Simultaneously, we computed different data using bioinformatics tools and servers. We cannot calculate some data, such as AA variants of ORF6. This may be another limitation of this study. However, this computational experiment with the mutation of the S-glycoprotein, especially in the RBD region, will help understand the ACE2 binding ability of Omicron and, thus, infectivity. The results generated in a short time frame will also help understand Omicron's antibody escape properties. Our rapidly generated data may be helpful for future research from a pandemic perspective because the Omicron variant is a VOC.
Conclusion
The present study represents the mutational landscape of the Omicron variant along with the emerging VOCs and VOIs, which helps us to understand the pattern of Omicron mutations in the whole genome in terms of SNVs along with the AA variants of structural proteins and non-structural proteins. The study also revealed some molecular properties of the mutations, such as increased or decreased stability events and the increased or decreased molecular flexibility of the S-glycoprotein because of the mutational change.
In this study, we applied computational servers and tools to explain the mutational landscape of the Omicron variant, a newly emerging SARS-CoV-2 variant, and compared the mutations with those in current VOCs and VOIs. In this study, we evaluated all the emerging mutations and categorized them in terms of mutational frequency.
The stability of the S-glycoprotein depends on the different stable mutations, and all the mutations in the region determine the interaction capacity with the host receptor binding and its stability. This study noted that 19 mutations showed stabilizing effects based on the ΔΔG prediction of 28 mutations. Conversely, mutations affect the stability of the S-glycoprotein, which may play a significant role in receptor recognition, increasing infectivity, and improving viral fitness. This study also observed that 12 mutations increased the molecular flexibility and flexibility of the S-glycoprotein. It may play a significant role in receptor binding, increasing infectivity, and improving viral fitness.
Our study also analyzed the mutational frequency of all AA variants in all structural and non-structural proteins of SARS-CoV-2. This study may help future researchers to understand the stability of all the proteins of SARS-CoV-2. Therefore, our study can help future researchers understand the Omicron variant’s fast-spreading properties. Our study attempted to illustrate the molecular and mutational properties of the emerging mutations in the RBD regarding nAb interaction and escape. Furthermore, our study comprehensively mapped RBD mutations in terms of several molecular and mutational parameters, such as ΔΔG, interatomic interactions in the wild-type and mutant-type residues, and fluctuation and deformation analysis. The study will help future researchers to understand the antigenic properties of epitopes of the S-glycoprotein. Finally, this study will help future researchers design and develop novel vaccines and therapeutic antibodies using the SNVs and point mutations with high frequencies in the case of all VOIs and VOCs, including Omicron.
Supplementary Information
Below is the link to the electronic supplementary material.
Diverse AA variants of their antibody-binding, their binding stability with ACE2, and risk of amino acid substitution. (A) AA variants of L452 (B) AA variants of F490 (C) AA variants of L452 (D) AA variants of D614 (PNG 2273 kb)
Calculated atomic fluctuation and deformation energies of the G339D mutation. (A) Residue-wise RMS fluctuation of the G339D mutation. (B) Atomic fluctuation in a virtual approach in wild type to mutant type in G339D mutation. (C) Deformation energies in a virtual manner in wild type to mutant type in G339D mutation (PNG 2596 kb)
Computed atomic fluctuation and deformation energies of S371L mutation. (A) Residue-wise RMS fluctuation of S371L mutation. (B) Atomic fluctuation in a virtual approach in wild type to mutant type in S371L mutation. (C) Deformation energies in a virtual manner in wild type to mutant type in S371L mutation (PNG 2548 kb)
Work out result of the atomic fluctuation and deformation energies of N440K mutation. (A) Residue-wise RMS fluctuation of N440K mutation. (B) Atomic fluctuation in a virtual approach in wild type to mutant type in N440K mutation. (C) Deformation energies in a virtual manner in wild type to mutant type in N440K mutation (PNG 2605 kb)
Work out result of atomic fluctuation and deformation energies of G446S mutation. (A) Residue-wise RMS fluctuation of G446S mutation. (B) Atomic fluctuation in a virtual approach in wild type to mutant type in G446S mutation. (C) Deformation energies in a virtual manner in wild type to mutant type in G446S mutation (PNG 2536 kb)
Computed atomic fluctuation and deformation energies of T478K mutation. (A) Residue-wise RMS fluctuation of T478K mutation. (B) Atomic fluctuation in a virtual approach in wild type to mutant type in T478K mutation. (C) Deformation energies in a virtual manner in wild type to mutant type in T478K mutation (PNG 2505 kb)
Work out a result of atomic fluctuation and deformation energies of Q498R mutation. (A) Residue-wise RMS fluctuation of Q498R mutation. (B) Atomic fluctuation in a virtual approach in wild type to mutant type in Q498R mutation. (C) Deformation energies in a virtual manner in wild type to mutant type in Q498R mutation (PNG 2512 kb)
Abbreviations
- AA
Amino acid
- ACE2
Angiotensin-converting enzyme 2
- GISAID
Global Initiative on Sharing All Influenza Data
- MDS
Molecular dynamics simulation
- nAb
Neutralizing antibody
- NMA
Normal mode analysis
- nsp
Non-structural protein
- NTD
N-terminal domain
- ORF
Open reading frame
- RBD
Receptor binding domain
- RBM
Receptor binding motif
- S-glycoprotein
Spike glycoprotein
- SNV
Single-nucleotide variation
- UTR
Untranslated region
- VOC
Variants of concern
- VOI
Variants of interest
- Δ
Delta
Author contribution
CC conceptualized the study, collected and analyzed the data, and wrote the manuscript. MB did the formal analysis and developed the figures and tables. ARS reviewed and edited the final draft of the manuscript. GA edited the manuscript. KD and CC did the overall supervision. All authors read and approved the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chiranjib Chakraborty and Manojit Bhattacharya contributed equally to this work.
References
- 1.Vaughan A. Omicron emerges. New Sci. 2021;252(3363):7. doi: 10.1016/S0262-4079(21)02140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dyer O. Covid-19: South Africa’s surge in cases deepens alarm over omicron variant. BMJ. 2021;3(375):n3013. doi: 10.1136/bmj.n3013. [DOI] [PubMed] [Google Scholar]
- 3.Karim SSA, Karim QA. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. The Lancet. 2021;398(10317):2126–2128. doi: 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rathinasamy M, Kandhasamy S. An exploratory study on the propagation of SARS-CoV-2 variants: Omicron is the most predominant variant. J Med Virol. 2022;94(6):2414–2421. doi: 10.1002/jmv.27634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abbasi J. Omicron has reached the US—here’s what infectious disease experts know about the variant. JAMA. 2021;326(24):2460–2462. doi: 10.1001/jama.2021.22619. [DOI] [PubMed] [Google Scholar]
- 6.Graham F. Daily briefing: Omicron was already spreading in Europe. Nature. 2021 doi: 10.1038/d41586-021-03610-3. [DOI] [PubMed] [Google Scholar]
- 7.Mohapatra RK, et al. Omicron (B. 11. 529) variant of SARS-CoV-2: concerns, challenges, and recent updates. J Med Virol. 2022;94(6):2336–2342. doi: 10.1002/jmv.27633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohapatra RK, et al. The recently emerged BA. 4 and BA. 5 lineages of Omicron and their global health concerns amid the ongoing wave of COVID-19 pandemic—correspondence. Int J Surg. 2022;103:106698. doi: 10.1016/j.ijsu.2022.106698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan Y, et al. SARS-CoV-2 Omicron variant: recent progress and future perspectives. Signal Transduct Target Ther. 2022;7(1):1–11. doi: 10.1038/s41392-022-00997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakraborty C, et al. Recombinant SARS-CoV-2 variants XD, XE, and XF: the emergence of recombinant variants requires an urgent call for research—correspondence. Int J Surg. 2022;2022(102):106670. doi: 10.1016/j.ijsu.2022.106670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WHO. Tracking SARS-CoV-2 variants. 2022. https://www.who.int/activities/tracking-SARS-CoV-2-variants. Acessed on 13 July 2022.
- 12.Singh J, et al. Evolutionary trajectory of SARS-CoV-2 and emerging variants. Virol J. 2021;18(1):1–21. doi: 10.1186/s12985-021-01633-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung C, et al. Omicron: what makes the latest SARS-CoV-2 variant of concern so concerning? J Virol. 2022;96(6):e02077–e2121. doi: 10.1128/jvi.02077-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desvars-Larrive A. To beat Omicron, Delta and bird flu, Europe must pull together. Nature. 2021;600(7889):386–386. doi: 10.1038/d41586-021-03661-6. [DOI] [PubMed] [Google Scholar]
- 15.McKee M. Public health and politics are inseparable, as Omicron and the UK’s response remind us. BMJ. 2021;375:n3055. doi: 10.1136/bmj.n3055. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, et al. The significant immune escape of pseudotyped SARS-CoV-2 variant Omicron. Emerg Microb Infect. 2022;11(1):1–5.12. doi: 10.1080/22221751.2021.2017757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Callaway E. Omicron likely to weaken COVID vaccine protection. Nature. 2021;600(7889):367–368. doi: 10.1038/d41586-021-03672-3. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, et al. SARS-CoV-2 Omicron strain exhibits potent capabilities for immune evasion and viral entrance. Signal Transduct Target Ther. 2021;6(1):1–3. doi: 10.1038/s41392-021-00852-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quarleri J, Galvan V, Delpino M. Omicron variant of the SARS-CoV-2: a quest to define the consequences of its high mutational load. Geroscience. 2021;44(1):53–56. doi: 10.1007/s11357-021-00500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar S, et al. Omicron and Delta variant of SARS-CoV-2: a comparative computational study of spike protein. J Med Virol. 2022;94(4):1641–1649. doi: 10.1002/jmv.27526. [DOI] [PubMed] [Google Scholar]
- 21.Pascarella S, et al. The electrostatic potential of the omicron variant spike is higher than in delta and delta-plus variants: a hint to higher transmissibility? J Med Virol. 2022;94(4):1277–1280. doi: 10.1002/jmv.27528. [DOI] [PubMed] [Google Scholar]
- 22.Chan JFW, Chu H. Pathogenicity of SARS-CoV-2 Omicron BA. 1.1 in hamsters. eBioMedicine. 2022;80:104035. doi: 10.1016/j.ebiom.2022.104035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohapatra RK, et al. The recombinant variants of SARS-CoV-2: concerns continues amid COVID-19 pandemic. J Med Virol. 2022 doi: 10.1002/jmv.27780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott L, et al. Track Omicron’s spread with molecular data. Science. 2021;374(6574):1454–1455. doi: 10.1126/science.abn4543. [DOI] [PubMed] [Google Scholar]
- 25.Callaway E. Heavily mutated Omicron variant puts scientists on alert. Nature. 2021;600(7887):21–21. doi: 10.1038/d41586-021-03552-w. [DOI] [PubMed] [Google Scholar]
- 26.Song Y, Masaki F. Preparation for the challenge of heavily mutated Omicron variant. Clin Transl Med. 2021;11(12):e679. 10.1101/2021.12.06.471499. [DOI] [PMC free article] [PubMed]
- 27.Miller NL, et al. Insights on the mutational landscape of the SARS-CoV-2 Omicron variant. bioRxiv, 2021. [DOI] [PMC free article] [PubMed]
- 28.Chakraborty C, et al. Evolution, mode of transmission, and mutational landscape of newly emerging SARS-CoV-2 variants. MBio. 2021;12(4):e01140–e1221. doi: 10.1128/mBio.01140-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chakraborty C, Bhattacharya M, Sharma AR. Present variants of concern and variants of interest of severe acute respiratory syndrome coronavirus 2: their significant mutations in S-glycoprotein, infectivity, re-infectivity, immune escape and vaccines activity. Rev Med Virol. 2021;32(2):e2270. 10.1002/rmv.2270
- 30.Chakraborty C, et al. SARS-CoV-2 Brazil variant in Latin America: more serious research urgently needed on public health and vaccine protection. Ann Med Surg. 2021;66:102428. [DOI] [PMC free article] [PubMed]
- 31.Bhattacharya M, et al. D614G mutation and SARS-CoV-2: impact on S-protein structure, function, infectivity, and immunity. Appl Microbiol Biotechnol. 2021;105(24):9035–9045. doi: 10.1007/s00253-021-11676-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakraborty C, et al. D614G mutation eventuates in all VOI and VOC in SARS-CoV-2: is it part of the positive selection pioneered by Darwin? Mol Ther- Nucleic Acids. 2021;26(3):237–241. doi: 10.1016/j.omtn.2021.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L, et al. SARS-CoV-2 variants, RBD mutations, binding affinity, and antibody escape. Int J Mol Sci. 2021;22(22):12114. doi: 10.3390/ijms222212114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greaney AJ, et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat Commun. 2021;12(1):1–14. doi: 10.1038/s41467-021-24435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farooq RK, et al. Bibliometric analysis of coronavirus disease (COVID-19) literature published in Web of Science 2019–2020. J Fam Community Med. 2021;28(1):1. doi: 10.4103/jfcm.JFCM_332_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thoma B, Chan TM. Using Google Scholar to track the scholarly output of research groups. Perspect Med Educ. 2019;8(3):201–205. doi: 10.1007/s40037-019-0515-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuo X, et al. How do we share data in COVID-19 research? A systematic review of COVID-19 datasets in PubMed Central Articles. Brief Bioinform. 2021;22(2):800–811. doi: 10.1093/bib/bbaa331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan J, et al. COVID-19 and literature evidence: should we publish anything and everything? Acta Bio Med: Atenei Parmensis. 2020;91(3):e2020020. doi: 10.23750/abm.v91i3.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.CDC. Omicron variant: what you need to know. 2021. https://www.cdc.gov/coronavirus/2019-ncov/variants/omicron-variant.html. Acessed on 12 Jan 2022.
- 40.CDC. Science brief: Omicron (B.1.1.529) variant. 2021. https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-omicron-variant.html. Acessed on 12 Jan 2022.
- 41.Stockholm E. Implications of the spread of the SARS-CoV-2 B.1.1.529 variant of concern (Omicron) for the EU/EEA—first update. 2 December 2021. 2021.
- 42.WHO. Update on Omicron. 2021. https://www.who.int/news/item/28-11-2021-update-on-omicron. Acessed on 5 Jan 2022.
- 43.Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data—from vision to reality. Eurosurveillance. 2017;22(13):30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elbe S, Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Global Chall. 2017;1(1):33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Velankar S, et al. The protein data bank archive. Methods Mol Biol. 2021;2305:3–21. doi: 10.1007/978-1-0716-1406-8_1. [DOI] [PubMed] [Google Scholar]
- 46.Yuan S, et al. PyMOL and Inkscape bridge the data and the data visualization. Structure. 2016;24(12):2041–2042. doi: 10.1016/j.str.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 47.Rigsby RE, Parker AB. Using the PyMOL application to reinforce visual understanding of protein structure. Biochem Mol Biol Educ. 2016;44(5):433–437. doi: 10.1002/bmb.20966. [DOI] [PubMed] [Google Scholar]
- 48.Chen AT, et al. COVID-19 CG enables SARS-CoV-2 mutation and lineage tracking by locations and dates of interest. Elife. 2021;10:e63409. doi: 10.7554/eLife.63409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mathieu E, et al. A global database of COVID-19 vaccinations. Nat Hum Behav. 2021;5(7):947–953. doi: 10.1038/s41562-021-01122-8. [DOI] [PubMed] [Google Scholar]
- 50.Nextstrain. Nextstrain SARS-CoV-2 resources, on Nextstrain. 2021. https://nextstrain.org/sars-cov-2/. Acessed on 5 Jan 2022.
- 51.Karthik Gangavarapu et al. Outbreak.info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. 2022. 10.1101/2022.01.27.22269965. [DOI] [PMC free article] [PubMed]
- 52.Sun Q, et al. VarEPS: an evaluation and prewarning system of known and virtual variations of SARS-CoV-2 genomes. Nucleic Acids Res. 2021;50(D1):D888–D897. [DOI] [PMC free article] [PubMed]
- 53.Rodrigues CH, Pires DE, Ascher DB. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46(W1):W350–W355. doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Capriotti E, et al. A three-state prediction of single point mutations on protein stability changes. BMC Bioinformatics. 2008;9(2):1–9. doi: 10.1186/1471-2105-9-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hammer Ø, Harper DA, Ryan PD. PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4(1):9. [Google Scholar]
- 56.MathWorks I. MATLAB, High-performance numeric computation and visualization software: user’s guide: for UNIX Workstations. MathWorks; 1992.
- 57.Kwon D. Omicron’s molecular structure could help explain its global takeover. Nature. 2022;602(7897):373–374. doi: 10.1038/d41586-022-00292-3. [DOI] [PubMed] [Google Scholar]
- 58.Yadav PD, et al. Isolation and genomic characterization of SARS-CoV-2 omicron variant obtained from human clinical specimens. Viruses. 2022;14(3):461. [DOI] [PMC free article] [PubMed]
- 59.Ahmed W, et al. Detection of the Omicron (B. 1.1. 529) variant of SARS-CoV-2 in aircraft wastewater. Sci Total Environ. 2022;10(820):153171. [DOI] [PMC free article] [PubMed]
- 60.Nikolaidis M, et al. Comparative Analysis of SARS-CoV-2 Variants of concern, including Omicron, highlights their common and distinctive amino acid substitution patterns, especially at the spike ORF. Viruses. 2022;14(4):707. doi: 10.3390/v14040707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, et al. Cross-species tropism and antigenic landscapes of circulating SARS-CoV-2 variants. Cell Rep. 2022;38(12):110558. doi: 10.1016/j.celrep.2022.110558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mercatelli D, Giorgi FM. Geographic and genomic distribution of SARS-CoV-2 mutations. Front Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Korber B, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khairnar P, et al. Recent highlights on Omicron as a new SARS-COVID-19 variant: evolution, genetic mutation and future perspectives. J Drug Target. 1–33. 10.1080/1061186X.2022.2056187. [DOI] [PMC free article] [PubMed]
- 65.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cui Z, et al. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell. 185(5):860–871. [DOI] [PMC free article] [PubMed]
- 67.Planas D, et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602(7898):671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 68.Jacob JJ, et al. Evolutionary tracking of SARS-CoV-2 genetic variants highlights an intricate balance of stabilizing and destabilizing mutations. MBio. 2021;12(4):e01188–e1221. doi: 10.1128/mBio.01188-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Islam MR, Hossain MJ. Detection of SARS-CoV-2 Omicron (B.1.1.529) variant has created panic among the people across the world: what should we do right now? J Med Virol. 2022;94(5):1768–1769. doi: 10.1002/jmv.27546. [DOI] [PubMed] [Google Scholar]
- 70.Lupala CS, et al. Mutations on RBD of SARS-CoV-2 Omicron variant result in stronger binding to human ACE2 receptor. Biochem Biophys Res Commun. 2021;29(590):34–41. doi: 10.1016/j.bbrc.2021.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Triveri A, et al. SARS-CoV-2 spike protein mutations and escape from antibodies: a computational model of epitope loss in variants of concern. J Chem Inf Model. 2021;61(9):4687–4700. doi: 10.1021/acs.jcim.1c00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martin S, et al. SARS-CoV-2 integral membrane proteins shape the serological responses of patients with COVID-19. Iscience. 2021;24(10):103185. doi: 10.1016/j.isci.2021.103185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chakraborty C, Bhattacharya M, Sharma AR. Emerging mutations in the SARS-CoV-2 variants and their role in antibody escape to small molecule-based therapeutic resistance. Curr Opin Pharmacol. 2022;62:64–73. doi: 10.1016/j.coph.2021.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chakraborty C, et al. A detailed overview of immune escape, antibody escape, partial vaccine escape of SARS-CoV-2 and their emerging variants with escape mutations. Front Immunol. 2022;13:801522–801522. doi: 10.3389/fimmu.2022.801522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu L, et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584(7821):450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 76.Bhattacharya M, et al. Omicron variant (B. 1.1. 529) of SARS-CoV-2: understanding mutations in the genome, S-glycoprotein, and antibody-binding regions. GeroScience. 2022;44(2):619–37. [DOI] [PMC free article] [PubMed]
- 77.Singh P, et al. Computational modeling and bioinformatic analyses of functional mutations in drug target genes in Mycobacterium tuberculosis. Comput Struct Biotechnol J. 2021;19:2423–2446. doi: 10.1016/j.csbj.2021.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Othman H, et al. Interaction of the spike protein RBD from SARS-CoV-2 with ACE2: similarity with SARS-CoV, hot-spot analysis and effect of the receptor polymorphism. Biochem Biophys Res Commun. 2020;527(3):702–708. doi: 10.1016/j.bbrc.2020.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sixto-López Y, et al. Structural insights into SARS-CoV-2 spike protein and its natural mutants found in Mexican population. Sci Rep. 2021;11(1):1–16. doi: 10.1038/s41598-021-84053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Teng S, et al. Systemic effects of missense mutations on SARS-CoV-2 spike glycoprotein stability and receptor-binding affinity. Brief Bioinform. 2021;22(2):1239–1253. doi: 10.1093/bib/bbaa233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Begum F, et al. Analyses of spike protein from first deposited sequences of SARS-CoV2 from West Bengal, India. F1000Research. 2020;9:371. doi: 10.12688/f1000research.23805.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pascarella S, et al. SARS-CoV-2 B. 1.617 Indian variants: are electrostatic potential changes responsible for a higher transmission rate? J Med Virol. 2021;93(12):6551–6556. doi: 10.1002/jmv.27210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Benvenuto D, et al. Analysis of three mutations in Italian strains of SARS-CoV-2: implications for pathogenesis. Chemotherapy. 2021;66(1–2):33–37. doi: 10.1159/000515342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shao W, et al. Challenges and countermeasures brought by Omicron Variant. J Microbiol Immunol Infect. 2022;S1684–1182(22):00054–8. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Diverse AA variants of their antibody-binding, their binding stability with ACE2, and risk of amino acid substitution. (A) AA variants of L452 (B) AA variants of F490 (C) AA variants of L452 (D) AA variants of D614 (PNG 2273 kb)
Calculated atomic fluctuation and deformation energies of the G339D mutation. (A) Residue-wise RMS fluctuation of the G339D mutation. (B) Atomic fluctuation in a virtual approach in wild type to mutant type in G339D mutation. (C) Deformation energies in a virtual manner in wild type to mutant type in G339D mutation (PNG 2596 kb)
Computed atomic fluctuation and deformation energies of S371L mutation. (A) Residue-wise RMS fluctuation of S371L mutation. (B) Atomic fluctuation in a virtual approach in wild type to mutant type in S371L mutation. (C) Deformation energies in a virtual manner in wild type to mutant type in S371L mutation (PNG 2548 kb)
Work out result of the atomic fluctuation and deformation energies of N440K mutation. (A) Residue-wise RMS fluctuation of N440K mutation. (B) Atomic fluctuation in a virtual approach in wild type to mutant type in N440K mutation. (C) Deformation energies in a virtual manner in wild type to mutant type in N440K mutation (PNG 2605 kb)
Work out result of atomic fluctuation and deformation energies of G446S mutation. (A) Residue-wise RMS fluctuation of G446S mutation. (B) Atomic fluctuation in a virtual approach in wild type to mutant type in G446S mutation. (C) Deformation energies in a virtual manner in wild type to mutant type in G446S mutation (PNG 2536 kb)
Computed atomic fluctuation and deformation energies of T478K mutation. (A) Residue-wise RMS fluctuation of T478K mutation. (B) Atomic fluctuation in a virtual approach in wild type to mutant type in T478K mutation. (C) Deformation energies in a virtual manner in wild type to mutant type in T478K mutation (PNG 2505 kb)
Work out a result of atomic fluctuation and deformation energies of Q498R mutation. (A) Residue-wise RMS fluctuation of Q498R mutation. (B) Atomic fluctuation in a virtual approach in wild type to mutant type in Q498R mutation. (C) Deformation energies in a virtual manner in wild type to mutant type in Q498R mutation (PNG 2512 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its supplementary information files).