Abstract
Chest radiographs are widely used in the medical domain and at present, chest X-radiation particularly plays an important role in the diagnosis of medical conditions such as pneumonia and COVID-19 disease. The recent developments of deep learning techniques led to a promising performance in medical image classification and prediction tasks. With the availability of chest X-ray datasets and emerging trends in data engineering techniques, there is a growth in recent related publications. Recently, there have been only a few survey papers that addressed chest X-ray classification using deep learning techniques. However, they lack the analysis of the trends of recent studies. This systematic review paper explores and provides a comprehensive analysis of the related studies that have used deep learning techniques to analyze chest X-ray images. We present the state-of-the-art deep learning based pneumonia and COVID-19 detection solutions, trends in recent studies, publicly available datasets, guidance to follow a deep learning process, challenges and potential future research directions in this domain. The discoveries and the conclusions of the reviewed work have been organized in a way that researchers and developers working in the same domain can use this work to support them in taking decisions on their research.
Keywords: Respiratory diseases, Radiography, Pneumonia, COVID-19, Convolutional Neural networks, Computer-aided diagnostics, Medical image processing, Chest radiography
1. Introduction
Respiratory diseases such as pneumonia is a common lung infection condition and COVID-19 disease has become a life-threatening disease that emerged in later 2019 and has been impacted the entire world. Pneumonia is a fatal lower respiratory infection under the acute diseases category and has been reported to be a major cause of deaths around the world. In 2017, it was accountable for 15% of child deaths that happened during the year [1]. In addition, older people have a high risk of getting pneumonia that leads to critical conditions. However, if diagnosed and treated early, the associated risk can be minimized [2]. Most types of pneumonia infect the large regions of the lungs, while COVID-19 begins with the small areas and uses the lungs’ immune cells to spread. At present, with the recent rapid and massive spread of COVID-19 [3], many researchers around the globe have committed intensive efforts into the research and development of chest X-ray analysis to detect these lung infections conditions.
In general clinical practices, the radiologists look for the white spots called infiltrates in the lungs to identify an infection to diagnose pneumonia using chest radiographs [4]. Moreover, this analysis will help the radiologists to determine whether the patient has any complications related to pneumonia like the pleural effusions which are known as the excess fluid surrounding the lungs [4]. In the COVID-19 diagnosis, characteristics like the airspace opacities of the chest radiographs are analyzed by the radiologists to identify the infection of COVID-19 disease [5].
Chest X-ray image analysis is a cost-effective and non-invasive approach [6], however, the interpretation of chest X-rays requires expert knowledge as distinguishing abnormalities is a challenging task [7]. Thus, computer-aided solutions can be used to identify lung infection conditions by analyzing chest X-ray images as a support tool for an effective and efficient diagnosis process by reducing human error and effort. At present, computational methods play a significant role in decision making across several directions in the field of medical image analysis [8], [9], [10], [11], [12]. The recent advancement in data engineering approaches, particularly deep learning (DL) techniques have shown promising performance in identifying patterns and classifying medical images including chest X-ray images. The state-of-the-art DL techniques are mainly trained on data and learn the features effectively, in comparison to the traditional approaches that require hand-designed features with the knowledge of the domain. With the insight of identifying the lung infection conditions efficiently and effectively, recent studies have addressed chest X-ray image classification using DL techniques.
There are several survey papers published in this field that have summarized and organized the recent research work adding understanding to the status quo in the domain and evaluating the trends, developing a standpoint in the domain [13], [14], [15]. Still, there is a requirement not addressed by these studies to assess and summarize the related work and available resources on lung disease classification with chest X-ray images, to provide accessible evidence to decision-makers. This systematic review addresses these requirements by evaluating and summarizing the findings of the reviewed work. This work explores the trends in recent studies, analyzes the state-of-the-art DL models architectures, provides taxonomies and summaries based on the observations. This systematic review also explores public datasets that have been used in the reviewed work and gives an insight into the use of different datasets. We provide a comprehensive analysis of the related chest X-ray classification studies considering the Pneumonia, COVID-19 and normal conditions. Finally, we suggest guidance to the readers on selecting the suitable models and techniques under different considerations. The work also discusses the open challenges and presents several possible future directions. Developers and researchers can be beneficial from these findings to make decisions on selecting the most appropriate DL techniques, available datasets and possible future directions for research and development.
1.1. Related work
Many research and development based solutions have been addressed chest X-ray image classification using DL techniques to support the diagnosis process of pneumonia and COVID-19 conditions. Table 1 summarizes the available survey papers published during 2020–2021. Although, all the papers have addressed the use of different DL models and related datasets, to the best of our knowledge, only two surveys have presented a comprehensive review in terms of DL [13], [14]. Most of them have not addressed aspects such as loss calculation, optimization, evaluation metrics, the evolution of techniques and model selection guidelines, which we have addressed in this systematic review.
Table 1.
Summary of the related surveys.
Generally, a survey uses published literature of a selected domain to summarize and present an overview knowledge of the domain. On the other hand, a systematic review identifies, evaluates, and summarizes the findings to provide a comprehensive synthesis of the considered studies [20]. Some of the survey papers given in Table 1 have discussed DL approaches for Pneumonia detection [18], [19], while some have focused on COVID-19 detection and have explored the use of DL in medical image processing as well as computational intelligence techniques to detect COVID-19 conditions [16], [17]. However, most of these survey papers have focused only on one disease [16], [21]. On the other hand, a survey conducted by Kieu et al. [13] has discussed four diseases namely COVID-19, pneumonia, lung cancer, and tuberculosis using X-ray, CT scan, histopathology images and sputum smear microscopy. Furthermore, Calli et al. [14] have explored only chest X-ray analysis considering several lung diseases like COVID-19, Pneumonia and Tuberculosis. All referred survey papers have explored different DL algorithms and datasets that are used in the literature. Moreover, the surveys done by Kieu et al. [13] and Tilve et al. [18] have discussed pre-processing techniques and the surveys conducted by Kieu et al. [13] and Calli et al. [14] have addressed data augmentation techniques as well. Only Bhattacharya et al. [16] have discussed evaluation techniques in their survey, while Kieu et al. [13] have presented a taxonomy of the different techniques used in studies. The existing studies have not addressed the aspects such as loss function, optimization, evolution and guidance. Therefore, this study has considered those aspects.
In this systematic review, we mainly discuss the state-of-the-art DL approaches presented in the related literature together with the widely used datasets. Further, this study presents the evolution of trends of the related DL techniques and guide future researchers and developers to make better decisions in achieving promising results. For instance as the main contributions of our study, Table 3 states the recent related development studies that have used multi-models and ensemble models. Table 4 shows the use of different DL architectures in related studies with their accuracy. Here, we discuss the used optimizers, loss functions and give an insight into the percentage studies that have used different DL models. Table 5 explores the widely used publicly available datasets and shows their usage percentage by different studies. Moreover, Table 6, Table 7 and Table 8 summarize the studies on chest X-ray classification with Pneumonia, COVID-19 and both Pneumonia and COVID-19 conditions, respectively. Here, we comprehensively analyze the applied DL technique, loss function, optimization function and the obtained performance values. Also, we give an insight into the percentage of articles that have shown promising results. Fig. 10 provides selection criteria to consider the appropriate DL models for medical image classification from a practical point of view. Finally, we discuss the open challenges and possible future research and development directions.
Table 4.
Summary of related studies with widely used DL techniques.
| Study | VGG | ResNet | InceptionV3 | MobileNetV2 | DenseNet | Caps Net | U-Net | EfficientNet | SqueezeNet | TL | Accuracy% |
|---|---|---|---|---|---|---|---|---|---|---|---|
| [67] | X | – | – | – | – | – | – | – | – | N | 95 |
| [52] | X | – | – | – | – | – | – | – | – | Y | 88.10 |
| [54] | X | – | – | – | – | – | – | – | – | Y | 98.3 |
| [68] | X | – | – | – | – | – | X | – | – | Y | 97.4 |
| [69] | X | – | – | – | – | – | – | – | – | Y | 87 |
| [70] | X | – | – | – | – | – | – | – | – | Y | 96.3 |
| [71] | X | – | – | – | – | – | – | – | – | N | 93.1 |
| [72] | X | – | – | – | – | – | – | – | – | N | 97.36 |
| [73] | – | X | – | – | – | – | – | – | – | N | 97.65 |
| [47] | – | X | – | – | – | – | – | – | – | Y | 96.23 |
| [74] | X | – | – | – | X | – | – | – | – | N | 90, 90 |
| [23] | – | – | – | – | X | – | – | – | – | Y | 76 |
| [75] | – | – | – | – | X | – | – | – | – | Y | 98.45, 98.32 |
| [76] | – | X | – | – | – | – | – | – | – | Y | 98.18 |
| [77] | – | X | – | – | – | – | – | – | – | N | 93.6 |
| [78] | – | X | – | – | – | – | – | – | – | N | 95.33 |
| [79] | – | X | – | – | – | – | – | – | – | Y | 92 |
| [51] | – | – | X | – | – | – | – | – | – | Y | 90.1 |
| [80] | – | – | – | X | – | – | – | – | – | N | 90 |
| [81] | – | – | – | X | – | – | – | – | – | N | 93.4 |
| [10] | – | – | – | X | – | – | – | – | – | Y | 94.72 |
| [82] | – | – | – | X | – | – | – | – | – | Y | 99.1 |
| [83] | – | – | – | X | – | – | – | – | – | N | 98.6 |
| [84] | – | – | – | X | – | – | – | – | – | N | 98.65 |
| [85] | – | – | – | – | X | – | – | – | – | N | 76.80 |
| [86] | – | – | – | – | X | – | – | – | – | Y | 80.02 |
| [87] | – | – | – | – | – | X | – | – | – | N | 84.22 |
| [88] | – | – | – | – | – | X | – | – | – | Y | 98.3 |
| [62] | – | – | – | – | – | – | X | – | – | N | 97.8 |
| [38] | – | – | – | – | – | – | – | X | – | Y | 96.70 |
| [89] | – | – | – | – | – | – | – | X | – | Y | 93.9 |
| [90] | – | – | – | – | – | – | – | X | – | Y | 93.48 |
| [91] | – | – | – | – | – | – | – | – | X | Y | 98.26 |
| [92] | – | – | – | – | – | – | – | – | X | Y | 90.95 |
| [93] | – | X | – | – | – | – | – | – | – | Y | 71.9 |
Table 5.
Summary of chest X-ray datasets.
| Dataset name | Images | Disease | Related studies |
|---|---|---|---|
| Large Dataset of Labeled Optical Coherence Tomography (OCT) [100] | 5856 | Pneumonia | [22], [24], [51], [71], [101], [102], [103] |
| COVID Chestxray Dataset [104] | 646 | COVID-19, Pneumonia | [10], [41], [54], [57], [58], [60], [67], [70], [72], [74], [75], [76], [77], [78], [82], [87], [93], [105], [106], [107], [108] |
| COVID19 Radiography Dataset [109], [110] | 21,165 | COVID-19,Viral Pneumonia, Lung Opacity | [42], [54], [60], [103], [108], [111] |
| Chest X-ray Images (Pneumonia) [112] | 5863 | Virus and Bacterial Pneumonia | [9], [40], [57], [58], [62], [68], [75], [78], [80], [81], [105], [106], [113], [114], [115], [116] |
| Kaggle COVID-19 Patients Lungs X Ray Images 10000 [117] | 100 | COVID-19 | [42] |
| Chest X-ray14 (latest version of chest X-ray8) [118] | 112,120 | Pneumonia Pathology classes | [23], [70], [76], [81], [85], [87], [92], [93], [119] |
| CheXpert [120] | 224,316 | Pneumonia | [23], [57] |
| COVID-19 X rays [121] | 95 | COVID-19 | [10], [59] |
| COVIDx [44] | 13,975 | Bacterial and Viral Pneumonia, COVID-19 | [44], [47], [72], [79], [89] |
| CoronaHack - Chest X-ray-Dataset [122] | 5933 | COVID-19 | [56] |
| Mendeley Augmented COVID-19 X-ray Images Dataset [123] | 1824 | COVID-19 | [67] |
Table 6.
Summary of related studies for chest X-ray classification with pneumonia conditions.
| Study | DL technique | Acc % | Loss function | Optimizer | GPU | Evaluation metric |
|---|---|---|---|---|---|---|
| [40] | CNN | 83.38 | Cross-entropy | Adam | ✓ | Acc |
| [22] | CNN | 96.18 | CCE | Adam | ✓ | Sn, Sp, P, F1-score, Acc, AUROC |
| [23] | DensetNet121 ResNet-50 InceptionV3 |
76 69 61 |
BCE | Adam | ✓ | AUROC |
| [24] | Ensemble model (ResNet18, DenseNet121, InceptionV3, Xception, MobileNetV2) | 98.43 | Cross-entropy | SGD | ✓ | Acc, R, P, F1-Score, AUROC |
| [73] | ResNet-50 | 97.65 | – | – | ✓ | Categorical Accuracy |
| [9] | Ensemble model (AlexNet, DenseNet121, InceptionV3, ResNet18, GoogLeNet) | 96.4 | Cross-entropy | Adam | ✓ | AUROC, R, P, Sp, Acc |
| [80] | MobileNetV2 | 90 | Cross-entropy | – | – | Acc |
| [81] | MobileNetV2 | 93.4 | BCE | Adam | ✓ | AUROC, Acc, Sp, Sn |
| [85] | DenseNet121 | 76.80 | Weighted BCE | Adam | – | F1-score |
| [55] | Ensemble model (ResNet-34 based U-Net, EfficientNet-B4 based U-Net) | 90 | BCE, Dice loss | Ranger optimizer | – | Acc, P, R, F1-score |
| [101] | CNN | 93.73 | – | – | ✓ | Acc |
| [125] | Mask RCNN (ResNet-50+ResNet101) |
– | Multi-task loss | SGD | ✓ | IoU for true positive |
| [119] | CNN | 86 | BCE | – | ✓ | Acc |
| [113] | CNN | 90.68 | BCE | Adam | – | Acc |
| [114] | CNN | 97.34 | Cross-entropy MSE | Gradient Descent | ✓ | Acc |
| [68] | VGG-16 with MLP | 97.4 | – | RMSprop | ✓ | Acc, Sn, Sp, AUROC, F1-score. |
| [126] | CNN | 98.46 | – | – | – | P, R, Acc, F1-Score, AUROC, cross validation |
| [115] | CNN | 92.31 | CCE | Adam | – | Acc, R, F1-score |
| [102] | CNN | 95.30 | CCE | Adam | – | cross validation, Acc, AUROC |
| [128] | CNN MLP |
94.40 92.16 |
Cross-entropy | – | ✓ | 5-fold cross validation, Acc, AUROC |
| [127] | SCN | 80.03 | BCE | Adam | – | Acc, P, R, F1-score |
| [69] | VGG-16 Xception |
87 82 |
CCE | RMSprop | ✓ | Acc, Sp, R, P, F1-score. |
| [86] | DenseNet-169 with SVM |
80.02 | – | – | – | AUROC |
| [51] | InceptionV3 with more layers |
90.1 | CCE | Nadam | ✓ | Acc, P, R, F1-score |
| [62] | CNN with U-Net | 97.8 | – | Adam | – | AUROC, Acc |
Table 7.
Summary of related studies for chest X-ray classification with COVID-19 conditions.
| Study | DL technique | Acc % | Loss function | Optimizer | GPU | Evaluation metric |
|---|---|---|---|---|---|---|
| [41] | CNN | 98 | – | – | – | Acc, P |
| [56] | ResNet-50 +VGG-16 VGG-16 |
99.87 98.93 | CCE | Adam | ✓ | Acc, Sn, Sp |
| [88] | CapsNet | 98.3 | Cross-entropy | Adam | ✓ | Acc, Sn, Sp, AUROC |
| [52] | VGG-16 (nCOVnet) |
88.10 | – | Adam | – | Acc, Sn, Sp, AUROC |
| [70] | VGG19 | 96.3 | BCE | Adam | ✓ | Acc, P, R, F1-score |
| [105] | CNN | 99.5 | BCE | Adam | – | Acc, P, Sn, Sp, F1-score, AUROC |
| [72] | VGG-16 based Faster R-CNN | 97.36 | cross-entropy | Momentum | ✓ | Acc, P, Sn, Sp, F1-score, 10-fold cross validation |
| [75] | DenseNet121 | N - 98.45 C - 98.32 |
cross-entropy | Adamax | ✓ | Acc, P, R, F1-score |
| [71] | DeTraC (VGG19) | 93.1 | cross-entropy | SGD | ✓ | Acc, Sn, Sp, AUROC |
| [82] | MobileNet InceptionResNetV2 VGG-16 VGG19 |
99.1 96.8 93.6 90.8 |
BCE | Adam | ✓ | Acc, P, R, F1-score, AUROC |
| [90] | EfficientNet DenseNet121 Xception NASNet VGG-16 |
93.48 89.96 88.03 85.03 79.01 |
CCE, Weighted BCE | Adam, SGD | ✓ | Acc, P, R, F1-score |
| [93] | ResNet-101 | 71.9 | cross-entropy | – | ✓ | Acc, Sn, Sp, AUROC |
| [83] | MobileNetV2 InceptionV3 Xception ResNet-50 |
98.6 98.1 97.4 82.5 |
– | – | – | Acc, P, Sp, R, F1 score |
| [129] | CNN | 96 | log-loss | SGD | X | Acc, Sn, Sp, MAE, AUROC |
| [77] | ResNet-SVM | 93.6 | BCE | RMSProp | – | Acc, Sn, F1 score, P |
| [74] | VGG19 DenseNet201 |
90 90 |
cross-entropy | – | ✓ | Acc, P, R, F1-score |
| [57] | Ensemble model (ResNet-50, DenseNet201, InceptionV3) | 91.62 | – | Adam | ✓ | 5-fold cross validation, Acc, Sn, F1-score, AUROC |
| [116] | MADE-based CNN | 94.48 | MSE | MADE | ✓ | Acc, Sn, Sp, F1-score, Kappa statistics |
| [78] | ResNet-50 +SVM | 95.33 | – | – | ✓ | Acc, Sn, FPR, F1-score |
| [107] | CNN | 98.5 | – | – | ✓ | Acc, Sn, Sp, AUROC, cross-validation |
| [108] | CNN | 99.49 | – | – | ✓ | Acc, Sn, Sp, 5-fold cross-validation |
Table 8.
Summary of related multi-class classification for chest X-ray with both pneumonia and COVID-19 conditions.
| Study | DL technique | Acc % | Loss function | Optimizer | GPU | Evaluation metric |
|---|---|---|---|---|---|---|
| [42] | CNN | 90.64 | CCE | RMSprop | – | Acc, P, R, F1-Score, 5-fold cross-validation |
| [54] | VGG-16 | 98.3 | CCE | Adam | – | Acc, Sn, Sp |
| [10] | MobileNetV2 | 94.72 | – | Adam | – | Acc, Sn, Sp |
| [47] | ResNet-50 (COVID-ResNet) |
96.23 | – | Adam | – | Acc, Sn, P, F1-score |
| [38] | EfficientNet | 96.70 | – | Adam | ✓ | 10-fold cross validation, Acc, P, R, F1-score |
| [67] | VGG-16 | 95 | CCE | Adam | – | Acc, P, R, F1-score |
| [87] | CapsNet | 84.22 | MSE | Adam | – | Acc, 10-fold cross validation, Sn, Sp, F1-score, P |
| [124] | Darknet-19 (DarkCovidNet) |
87.02 | Cross- entropy |
Adam | – | 5-fold cross validation, Acc, Sn, Sp, P, F1-score |
| [91] | SqueezeNet | 98.3 | – | Bayesian | ✓ | Acc, COR, COM, Sp, F1-score, MCC |
| [44] | COVID-Net | 93.3 | – | Adam | – | Acc, Sn |
| [103] | COV19-ResNet COV19-CNNet | 97.61 94.28 |
– | – | ✓ | Acc, P, R, Sp, F1-score, |
| [76] | ResNet-50 | 98.18 | – | – | ✓ | Acc, P, R, F1-score |
| [59] | VGG-CapsNet | 92 | CCE | SGD | – | Acc, P, R, F1-score, AUROC |
| [89] | EfficientNet B3-X | 93.9 | – | Adam | – | Acc, , + |
| [106] | CoroNet (Xception) | 95 | – | Adam | ✓ | Acc, P, R, Sp, F1-score |
| [37] | CNN SVM DT KNN |
97.14 98.97 96.10 95.76 |
– | Adam, Bayesian | ✓ | 5-fold cross validation, Acc, Sn, Sp, AUROC, F1-score |
| [111] | CNN (CVDNet) | 96.69 | cross-entropy | Adam | – | Acc, P, R, F1-score, 5-fold cross validation |
| [58] | Ensemble (ResNet-50V2, VGG-16, InceptionV3) | 95.49 | CCE | Adam | ✓ | Acc, Sn, Sp, P, AUROC |
| [79] | ResNet-50 | 92 | CCE | Adam | – | Acc, Sn, Sp, F1-score, AUROC |
| [60] | SqueezeNet & MobileNetV2 (Combined features set) | 99.27 | – | – | ✓ | Acc, Sn, Sp, P, F1-score, 5-fold cross validation |
Fig. 10.
Considerations for selecting a technique.
1.2. Scope of the systematic review
This systematic review considers 68 related studies that have applied DL techniques to classify chest X-ray images with pneumonia and COVID-19 conditions. This includes 24 conference papers and 44 journal articles that were published between the years 2017 and 2021. Table 2 states the statistics of the studies considered for this systematic review. Given the timeliness of the solutions, we have selected studies that have focused on normal Pneumonia and COVID-19 Pneumonia detection, so that medical experts or other concerned parties can use this study to better understand the literature. Among many DL techniques, CNNs have proved to be highly effective in the implementation of medical image analysis frameworks. Therefore, we focus on the applicability of CNNs with different approaches such as building CNNs from scratch [22], applying transfer learning with popular models [23] and using ensembles of multiple models [24]. This systematic review provides a comprehensive list of datasets with COVID-19 and Pneumonia images and analyzes the state-of-the-art techniques used in the literature in each phase in the DL process.
Table 2.
Summary of the considered studies.
| Chest X-ray classification using DL techniques |
Conference papers |
Journal articles |
|---|---|---|
| Studies on pneumonia | 18 | 7 |
| Studies on COVID-19 | 3 | 19 |
| Studies on both pneumonia and COVID-19 | 3 | 18 |
Furthermore, this study aims to address the following research questions in the context of classifying chest X-ray images to identify the lung infection conditions due to Pneumonia and COVID-19. Therefore, this work can be utilized by the researchers and developers to obtain a detailed understanding of the evolution of the DL techniques, their features and effectiveness in the area of chest X-ray image classification.
-
RQ1:
What are the trends of recent studies?
-
RQ2:
What are the state-of-the-art DL based models and architectures?
-
RQ3:
What are the potential publicly available datasets?
-
RQ4:
What are the aspects addressed by the related studies?
-
RQ5:
How to select a suitable DL approach?
-
RQ6:
What are the open challenges and possible future research directions?
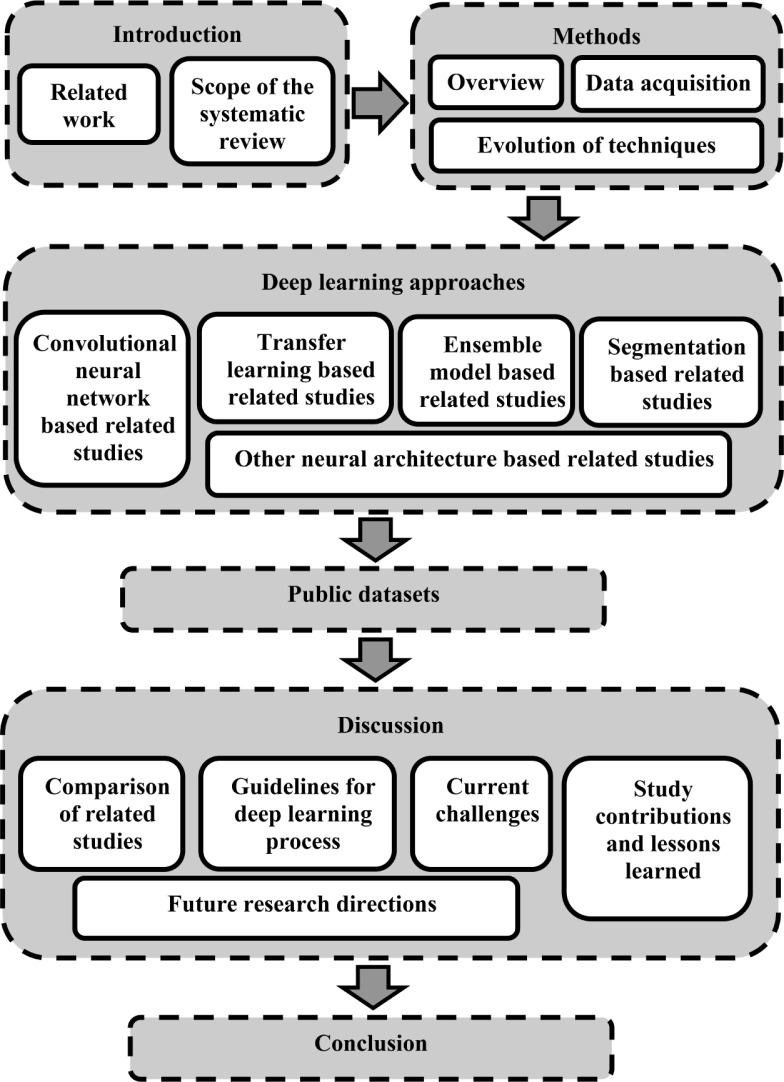
The paper is structured as indicated in Fig. 1 described as follows. Section 1 states the systematic review motivation, main contributions of this research with the scope. Section 2 explains the method followed during the systematic review based on the PRISMA model and the evolution of DL architectures used in the literature. Section 3 describes the DL approaches based on CNNs, transfer learning and other widely used DL architectures. Section 4 presents the publicly available datasets that contain chest X-ray images of pneumonia and COVID-19 conditions. Section 5 elaborates and critically analyzes the existing studies that applied DL for chest X-ray classification in focusing on pneumonia and COVID-19 diseases. In addition, we suggest general criteria to select techniques during the DL process based on different conditions and discusses the open challenges with future potential research directions. Finally, Section 6 concludes the study.
Fig. 1.
High-level structure of the paper.
2. Methods
2.1. Overview
This systematic review explores, summarizes and evaluates the findings in the literature relevant to the domain of lung disease classification with chest X-ray images. This study provides the readers with comprehensive taxonomies and summaries on different techniques, models and datasets used by the reviewed articles. Moreover, it suggests descriptive guidance to select suitable DL approaches using the observations made from the reviewed work as the basis. Further, the authors discuss the open challenges and show specific research directions in the domain that researchers can work on using the knowledge shared through this systematic review. The PRISMA guideline proposed by Tricco et al. [25] was used for conducting the scoping review.
2.2. Data acquisition
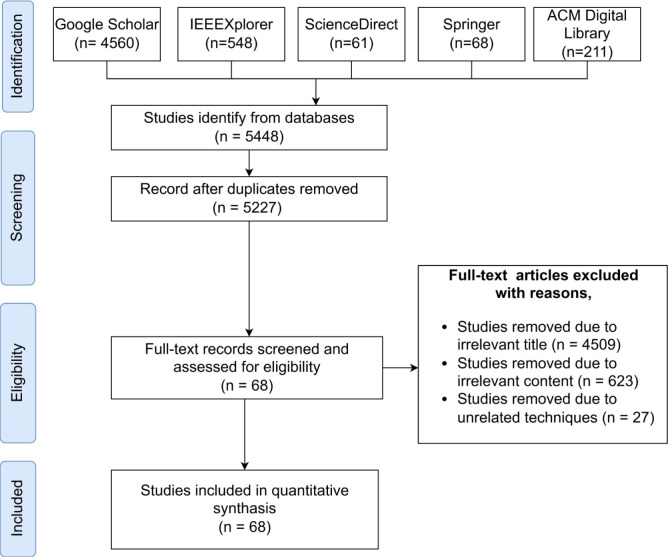
The Preferred Reporting for Systematic Reviews and Meta-Analysis Extension for Scoping Reviews (PRISMA-ScR) guideline proposed by Tricco et al. [25] was used to conduct this systematic review. We have selected the articles, summarized the findings and identified the existing research gaps based on the recommendations presented by this framework. The research scope is defined following the provided checklist of the items that should be reported. The PRISMA-ScR flow diagram is used to explain the search and scoping processes depicted in Fig. 2. The diagram depicts the flow of information through the several stages of the systematic review and concludes the number of articles identified, screened, eligible and eventually included.
Fig. 2.
The PRISMA-ScR protocol algorithm as a numerical flow diagram.
Identification Criteria: Following the PRISMA model, we searched and identified the relevant studies on Pneumonia and COVID-19 detection with chest X-rays using DL techniques. In this systematic review, our search strategy was based on the studies from several databases namely Google Scholar, IEEE Xplore, ACM Digital Library, ScienceDirect and Springer. The main reason behind using these databases was the high chance of finding a large number of relevant studies. Moreover, these databases are widely used for systematic reviews and include most of the reputed sources.
We used the advanced search feature in each of these databases, with keywords like “Pneumonia”, “COVID-19”, “Chest X-ray” and “Deep learning” combined with operations like “AND” and “OR” for the search query. Initially, we used “ AllField:(pneumonia OR covid) AND AllField:(chest X-ray)” as the search strategy. In order to narrow down the search results further, we used additional keywords such as “deep learning” and “detection”. Additionally, the study selection criteria were based on the available filtering options like custom year range, for which we selected from 2017 to 2021, study areas, and articles written in the English language, to narrow down the search scope.
Screening criteria: The screening phase records the number of identified articles and makes the selection process transparent by reporting on decisions made during the systematic review. Mainly we have removed the duplicates. The screening process is mainly done by the second, third and fourth authors. First, we checked the title and abstract of the articles and identified the content covered in the sections and main points discussed in the manuscript. The contradiction opinions were resolved by analyzing those articles by the first author.
Eligibility criteria: The studies obtained through the identification stage were discarded based on irrelevant titles, irrelevant contents and unrelated techniques. Also, we have excluded the research papers that were not written in the English language. The second, third and fourth authors have divided the articles equally among them and performed the data extraction process. The accuracy and consistency were checked by the first author.
Inclusion criteria: In this final stage, the remaining studies after the filtering mechanisms of the eligibility stage were included in this review.
2.3. Evolution of techniques
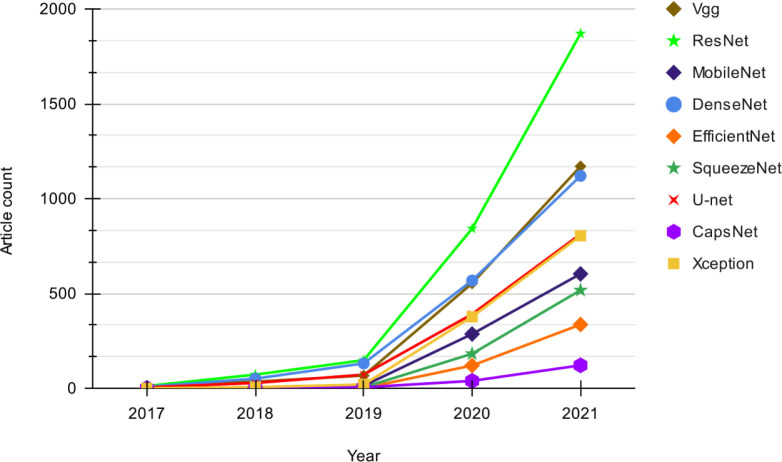
Fig. 3 shows a quantitative view of the use of different DL models in the related literature between the years 2017–2021. We have considered the research papers indexed in Google Scholar for the models in the related studies to give an overview. Our search strategy is based on “chest X-ray” + “<model_name>” + “Pneumonia” OR “covid”. The considered data can have slight flaws due to the associated noise of the search query. For instance, an article can appear for a given DL technique even though the method is discussed in their background literature instead of the methodology. However, we assume the flaws are equally distributed over the search results for all the considered techniques. Thus, the audience can get a comparative view of the usage of the main techniques used in this area.
Fig. 3.
Trend of DL techniques used in chest X-ray based Pneumonia and COVID-19 detection.
As shown in Fig. 3, it is visible that the usage of all the models kept increasing each year irrespective of the type. The noticeable differences in growth in 2019 can be concluded to be due to the emergence of the COVID-19 pandemic. Until the year 2019, both ResNet [26] and DenseNet [27] have shown similar growth, but from 2019 to 2021, ResNet has shown a drastic increase in popularity, exceeding all other models. The moderate growth of ResNet at the beginning could be because it was introduced only in 2016 [26], and due to its effectiveness, ResNet may have gained more popularity later on. Although SqueezeNet was introduced in the same year [28] as ResNet, it has gained less popularity than ResNet. Although VGG [29] and U-Net models have shown somewhat similar growth during 2017–2019, after the year 2019, VGG [29] shows a rapid increase in its growth and manages to gain popularity at the level of DenseNet.
The three models MobileNet [30], Xception [31] and CapsNet [32], which were introduced in 2017, were not used that much at the beginning probably because they were not known widely, but have gained popularity after 2019 with the emergence of studies for COVID-19 detection. Both MobileNet and Xception show remarkable growth compared to CapsNet, where Xception almost reaches the level of U-Net which has been around since 2015 [33]. EfficientNet, which was introduced quite recently in 2019 [34], has gained considerably high popularity within a short period. Most models that were introduced within the considered period were not used in the introduced year. Overall, it can be said that the interest in research in both Pneumonia and COVID-19 detection with all these models have been steadily increased.
3. Deep learning approaches
Deep learning techniques, particularly convolutional neural networks (CNNs) have become popular in the domain of medical image analysis [35]. The ability of DL techniques to learn complex features, store knowledge and reuse for related models has made it possible to implement highly accurate frameworks [36], [37], [38], [39]. This section discusses different DL methods used in the literature to classify chest X-ray images.
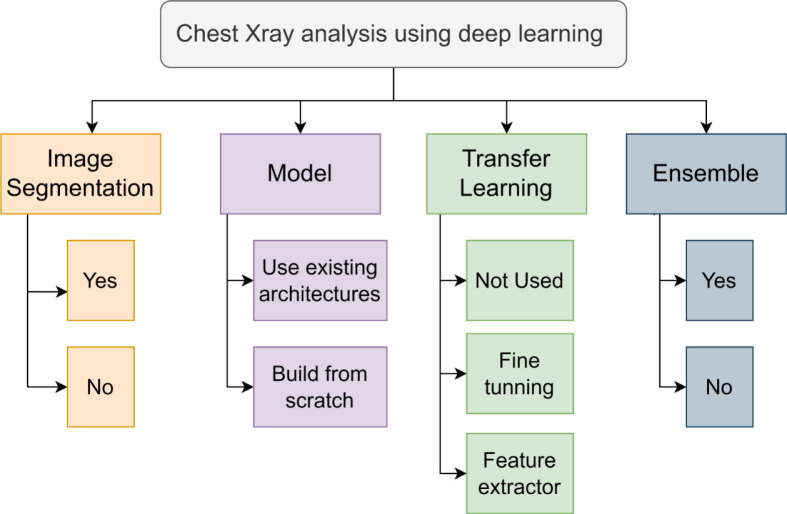
A taxonomy on chest X-ray analysis using DL is depicted in Fig. 4. The work reviewed in this systematic review can be classified based on the employment of image segmentation, usage of an existing DL architecture or built-from-scratch model, application of transfer learning and the presentation of an ensemble model. These key attributes are discussed in the following sections.
Fig. 4.
Taxonomy of chest X-ray classification using deep learning.
3.1. Convolutional Neural Network based related studies
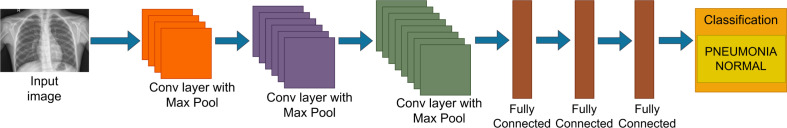
CNN has become a widely used DL technique in medical image classification due to the characteristics such as the ability to learn complex features with a comparatively less number of parameters and the benefits gained by sharing of weights [37]. Several studies in the literature have developed different CNN architectures from scratch for both Pneumonia and COVID-19 classification. For instance, the study by Khoiriyah et al. [40], has presented a CNN architecture comprising of three convolutional layers and three fully connected layers for the detection of Pneumonia in chest radiography as presented in Fig. 5. Using ADAM optimizer they have achieved an accuracy of 83.38% with augmentation. Moreover, a newly developed CNN with two convolutional layers and two fully connected layers with ADAM optimizer was used by Bhatt et al. [22] for chest X-ray based Pneumonia analysis. While using a shallower CNN, they have shown an accuracy of 96.18%, which is much higher than the accuracy obtained by Khoiriyah et al. [40].
Fig. 5.
Representation of the CNN architecture by Khoiriyah et al. [40].
The CNNs developed for chest X-ray classification for diagnosing COVID-19 are also keep emerging. In [41], a CNN built from scratch comprising convolutional layers and max-pooling layers followed by fully connected layers has been developed in a study by Padma and Kumari [41], which has shown an accuracy of 98.3%. Studies have also been done using CNN built from scratch for three-class classification to detect both Pneumonia and COVID-19 to be able to distinguish among them. For instance, Ahmed et al. [42] have presented a CNN model with five convolutional layers, each followed by batch normalization and max-pooling layers, and a dropout where the final layer is fully connected. They have used RMSProp as the optimizer and showed an overall accuracy of 90.64% in detecting COVID-19 and Pneumonia.
Another study done by Kieu et al. [43] has proposed a Multi-CNN model with three CNNs, CNN-128F, CNN-64L, and CNN-64R that were built from scratch to classify chest X-ray images. Each CNN component returns whether the input image is normal or abnormal with a probability value. A fusion rule is applied to these probability values to obtain the final result. Moreover, Wang et al. [44] have presented a tailored architecture named COVID-Net, using depth-wise separable convolution to detect COVID-19 and Pneumonia. They have shown 93.3% accuracy for multi-class classification with the ADAM optimizer. Accordingly, the CNNs have provided state-of-the-art solutions for detecting both Pneumonia and COVID-19 with relatively high accuracy. However, training a CNN from scratch can be computationally expensive and requires more data.
3.2. Transfer learning based related studies
Transfer learning is a popular method used in computer vision where the knowledge acquired from one problem domain is transferred to another similar domain [39]. Transfer learning is used by a variety of studies in DL to create frameworks [45] and models for important analyses while reducing the learning cost. In many studies for chest radiography image classification, pre-trained models are used to achieve better performance. Instead of developing a model from scratch, a model previously trained on another problem is used as the baseline. For instance, Irfan et al. [23] have carried out a deep transfer learning with ResNet-50, InceptionV3 and DenseNet121 models separately for Pneumonia classification using chest X-ray images. They have achieved higher accuracy using transfer learning compared to training from the scratch for each model.
Generally, most of the CNN architectures are pre-trained on the popular dataset ImageNet [46], which is a collection of about 1.2 million training images belonging to 1000 categories. Among many studies, these pre-trained models on ImageNet have been used in studies for Pneumonia classification [24], Pneumonia and COVID-19 detection [10] and the detection of viral Pneumonia, bacterial Pneumonia and COVID-19 [47] from chest radiographs. Moreover, CIFAR [48] and MNIST [49] are some of the other datasets used in pre-training.
Transfer learning can be used in two ways, as a feature extractor and for fine-tuning [39]. When used as a feature extractor, the last fully connected layer is replaced and all the previous layers in the model are frozen to be re-used with the pre-trained weights. Here, the model is used as a fixed feature extractor to take advantage of the learned features from a larger dataset in a similar domain. Only the last classification layer is trained according to the features extracted from previous layers [50]. This method is commonly used when the datasets are small. Several studies have been used transfer learning in the chest X-ray image classification process [9], [10], [51].
Panwar et al. [52] have used VGG-16 as a feature extractor with the ImageNet weights. In the fine-tuning approach, only the initial layers are frozen and the top-level layers are retrained to learn the specific features while generic features are extracted in initial layers and can be applied to other tasks [53]. This method is commonly used when there is a large training dataset available. The study presented by Choudhuri et al. [54] has used a pre-trained VGG-16 model after fine-tuning the top layers to classify chest radiography as Pneumonia or COVID-19. In their study, the CNN built from scratch has shown an accuracy of 96.6%, whereas the VGG-16 based model has achieved a higher accuracy of 98.3%. Although transfer learning has many advantages there are some issues as well. The model needs to be pre-trained on a similar dataset to achieve full performance and a lower learning rate should be chosen to efficiently use pre-trained weights.
3.3. Ensemble model based related studies
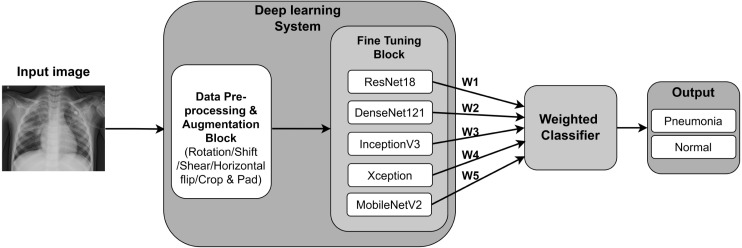
Ensemble classification uses a combination of multiple models to generate accurate results and to improve the robustness as the ensemble reduces the dispersion of the predictions. In this approach, the predictions of each core model are aggregated to generate the final result [36]. The study by Hashmi et al. [24] have used a weighted classifier to calculate the final prediction from an ensemble of the 5 models DenseNet121, ResNet18, Xception, MobileNetV2, and InceptionV3 for the binary classification for Pneumonia. They have used Eq. (1) and Eq. (2), where, denotes the weight corresponding to each model and is the prediction matrix for the two classes. The cross-entropy loss which is also known as the log loss is indicated in Eq. (3). Here, the dataset size is denoted by N and the probability of a given image affected by Pneumonia is denoted by p where y denotes the true label, where y=1 indicate the positive label. As shown in Fig. 6, the output of each model is assigned a weight where the final output is calculated using the weighted classifier. They have used the Stochastic Gradient Descent (SGD) as the optimizer with cross-entropy loss to achieve a final accuracy of 98.43%.
| (1) |
| (2) |
| (3) |
Fig. 6.
Representation of the ensemble model with weighted classifier by Hashmi et al. [24].
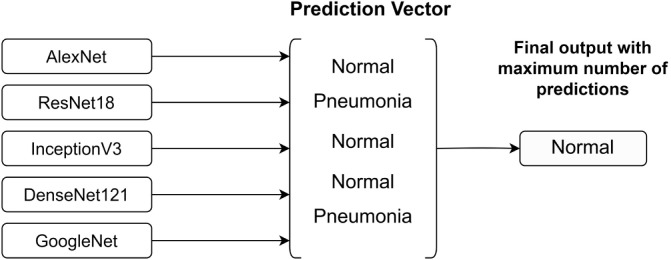
Compared to the weighted classifier method, Chouhan et al. [9] have presented an ensemble model where the final output is taken as the majority vote. They have used a prediction vector from which the maximum number of predictions (normal or Pneumonia) from the five pre-trained models AlexNet, DenseNet121, InceptionV3, ResNet18 and GoogLeNet is taken as the output as depicted in Fig. 7. The final output is calculated only from the majority vote of the outputs of each model without assigning any weights. Compared to the model developed by Hashmi et al. [24], this model has achieved an accuracy of 96.4%. In both studies, DenseNet121 and ResNet18 models have contributed to the ensemble classification model.
Fig. 7.
Prediction Vector used for ensemble model presented by Chouhan et al. [9].
Another study conducted by Pant et al. [55], has used an ensemble model using a U-Net based on ResNet-34 and another U-Net based on EfficientNet-B4. This has achieved an accuracy of 90% with binary cross-entropy loss, Dice loss using the Ranger optimizer. Further, Hilmizen et al. [56] have experimented with DenseNet121, MobileNet, Xception, InceptionV3, ResNet-50 and VGG-16 models separately and concatenating several models. They have tested models concatenating ResNet-50 and VGG-16, then DenseNet121 and MobileNet, and another model which is a concatenation of Xception and InceptionV3. Out of all these concatenations, the concatenation of ResNet-50 and VGG-16 has achieved the highest accuracy of 99.87% using Adam optimizer with categorical cross-entropy.
Table 3 states several related studies that have used a combination of different models. The column TL represents whether the given study is based on transfer learning or not. Here, some of the studies have used an ensemble model using several models [9], [24], [55], [57], [58], while the other studies have concatenated several models to produce the final model [56], [59], [60]. It can be seen that many studies have shown a preference to use ResNet, InceptionV3 and DenseNet to achieve promising results.
Table 3.
Summary of related studies with several models.
| Study | VGG | ResNet | InceptionV3 | MobileNetV2 | DenseNet | CapsNet | U-Net | EfficientNet | SqueezeNet | AlexNet | GoogLeNet | Xception | TL | Acc.% |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [56] | X | X | – | – | – | – | – | – | – | – | – | – | Y | 99.87 |
| [9] | – | X | X | – | X | – | – | – | – | X | X | – | Y | 96.4 |
| [24] | – | X | X | X | X | – | – | – | – | – | – | X | Y | 98.43 |
| [57] | – | X | X | – | X | – | – | – | – | – | – | – | Y | 91.62 |
| [58] | X | X | X | – | – | – | – | – | – | – | – | – | Y | 95.49 |
| [59] | X | – | – | – | – | X | – | – | – | – | – | – | Y | 92 |
| [55] | – | X | – | – | – | – | X | X | – | – | – | – | N | 90 |
| [60] | – | – | – | X | – | – | – | – | X | – | – | – | N | 99.27 |
3.4. Segmentation based related studies
Image segmentation is a method of dividing digital images into multiple fragments [61]. It produces a collection of segments comprising the entire image. In the medical domain, image segmentation has been made the image analysis easier [16]. In a related study, Narayanan et al. [62] have used transfer learning with AlexNet, ResNet, VGG-16 and InceptionV3 models to detect pneumonia conditions using chest X-ray images. Their approach has shown a high performance with the InceptionV3 model with 99% of training accuracy and 98.9% of validation accuracy. Additionally, they have proposed a novel CNN architecture to further analyze the chest X-rays to detect whether the diagnosed Pneumonia condition is due to bacterial or viral effect. They have used image segmentation as a pre-processing step which is conducted using a U-Net architecture. They have trained the U-Net architecture solely with true lung masks provided by Shenzhen Dataset [63], [64]. Their U-Net contains encoding and decoding for three stages. For all the convolutional and pooling operations they have used 3 × 3 and 2 × 2 kernels, while for deconvolution and up-pooling operations they have used bilinear interpolation. They have discovered that the shape of the lung plays an important role in deciding whether bacterial pneumonia or viral pneumonia. The proposed CNN model without lung segmentation has shown 96.7% training accuracy and 96% of validation accuracy, while the same CNN model with lung segmentation has achieved 98.5% training accuracy and 98.3% validation accuracy for detecting bacterial vs viral Pneumonia, showing the performance increase due to the segmentation.
Another study by Lalonde [65] has proposed a convolutional –deconvolutional capsule network architecture for object segmentation, which is an extension of the original capsule network introduced by [32]. They have expanded the capsule network by introducing a new concept, ‘deconvolutional capsules’ and modifying the dynamic routing mechanism in the original network. This novel architecture has improved the results of lung segmentation while reducing the number of parameters required for the task. They have compared the accuracy of their model for lung segmentation with other existing networks using the Dice coefficient and have achieved a relatively high accuracy of 98.47%. Similarly, Bonheur et al. [66], have proposed a semantic segmentation network called ‘Matwo-CapsNet’, which uses the concepts of capsules behind the original Capsule Network. They have introduced several extensions to the original Capsnet [32], which include using matrix encoding instead of vector encoding, combining pose information in each capsule with the matrix encoding, using a dual-routing mechanism instead of dynamic routing and finally extending the SegCaps proposed by Lalonde [65] to multi-label segmentation.
3.5. Other neural architecture based related studies
There are several architectures based on deep neural networks that have been used for image classification. This section discusses the related studies based on these widely used latest architectures. Table 4 shows a summary of related studies with their DL models and obtained accuracy.
VGG: VGG is a classic network architecture for image classification introduced by Simon and Zisserman in 2014 [94]. Among the related studies on Pneumonia diagnosis from chest radiography, several approaches have used VGG-16 model with Adam optimizer [52], [54], [67]. The study by Militante et al. [67], have carried out four class classifications for COVID-19, bacterial Pneumonia, viral Pneumonia, and normal and showed an accuracy of 95% by training the entire model without using transfer learning. In contrast, Choudhuri et al. [54], have used VGG-16 architecture on ImageNet pre-trained weights and fine-tuned the top layers for the classification of three classes for COVID-19, Pneumonia, or normal. Here, the transfer learning with the VGG-16 model has improved the accuracy of the initially proposed CNN from 96.6% to 98.3%. Another study by Ferreira et al. [68], have used VGG-16 with fully-connected layers replaced by customized multilayer perceptron (MLP), whereas Hilmizen et al. [56], have used VGG-16 concatenating with ResNet-50 for detecting COVID-19 conditions using chest X-ray images.
ResNet: ResNet is a modern CNN architecture introduced by He et al. in 2016 [26]. Youssef et al. have used the ResNet-50 architecture in their study for Pneumonia classification [73]. They have added two more convolutional layers with several dropouts and dense layers to the output layer of the ResNet-50 model, achieving an accuracy of 97.65%. A study by Farooq and Hafeez [47], has carried out four class classifications for COVID-19, bacterial Pneumonia, viral Pneumonia and normal classes. They have presented a model called COVID-ResNet based on the ResNet-50 model with the use of pre-trained weights on ImageNet. They have used Adam optimizer achieving 96.23% accuracy. Moreover, Irfan et al. [23], have carried out a deep transfer learning with ResNet-50, Inception V3 and DenseNet121 models separately, to classify chest X-ray images for Pneumonia, where the ResNet-50 model has achieved better accuracy compared to Inception V3.
Inception V3: Inception is a computational and resource-efficient model introduced by Szegedy et al. in 2015 [95] where the evolution of this model resulted in multiple versions such as Inception V1, Inception V2, Inception V3 and Inception-ResNet. However, many studies have been used the Inception V3 model, which was introduced by Szegedy et al. in 2016 together with Inception V2 [96]. A study for binary classification between COVID-19 and normal classes conducted by Hilmizen et al. [56], has used Inception V3 concatenated with the Xception model achieving an accuracy of 98.8%. Several other studies, [9] and [24], have also used Inception V3 for their ensemble models with other pre-trained models for the binary classification of Pneumonia and normal classes, and showed an accuracy of 96.4% and 98.43%, respectively. Moreover, Chouhan et al. [9], have presented a model that concatenated Inception V3 with AlexNet, DenseNet121, ResNet18 and GoogLeNet with Adam optimizer. Further, the study by Hashmi et al. [24] has used DenseNet121, ResNet18, Xception and MobileNetV2 as the other concatenated models while using SGD as their optimizer, signifying its better generalization compared to other adaptive optimizers. In both studies, the cross-entropy loss has been used to calculate the loss.
MobileNet: MobileNet is a lightweight CNN architecture compared to other existing models and was introduced by Howard et al. in 2017 [30]. MobileNet architecture has several versions such as MobileNetV1, MobileNetV2 and MobileNetV3. Many studies have used MobileNetV2 which was introduced by Sandler et al. in 2018 [97]. A study by Tobias et al. [80], for Pneumonia classification with chest X-ray images, has used the MobileNetV2 pre-trained model for its advantageous formulation of bottleneck layers. Hu et al. [81], have used MobileNetV2 architecture for faster and efficient chest radiography classification for binary classes as Pneumonia or normal, same as Tobias et al. [80], resulting in 93.4% accuracy. Another study has used MobileNetV2 based model to classify chest X-rays as Pneumonia, COVID-19 or Normal [84]. They have shown an accuracy of 98.65% and an average recall of 98.15% using a modified architecture by adding three layers namely global average pooling layer, dropout layer and dense layer. A study carried out by Apostolopoulos and Mpesiana [10] has used MobileNetV2 for multi-class classification for COVID-19, Pneumonia or normal by achieving a 94.72% accuracy.
DenseNet: DenseNet is a deep network introduced by Huang et al. in 2017 [27]. Rajpurkar et al. [85] have used the DenseNet121 model for their CNN named ChexNet, which outputs the probability of having Pneumonia in a chest X-ray image and a heat-map that localizes the areas of the image that are most indicative of the disease. This model is extended to detect 14 different thoracic diseases including Pneumonia with promising performance for the detection of 14 classes. Moreover, Chouhan et al. [9] and Hashmi et al. [24], have also used DenseNet121 as one of the models in the ensemble model. A study for Pneumonia classification by Irfan et al. [23], which was carried out a deep transfer learning with ResNet-50, Inception V3 and DenseNet121 models separately, has achieved the highest accuracy from DenseNet121. Varshani et al. [86] have also tried out different classification models like VGG-16, XCeption, ResNet-50, DenseNet121, DenseNet169 with different classifiers and have achieved the high results (AUC of 0.8002) from DenseNet169 with Support Vector Machine (SVM) as the classifier after parameter tuning.
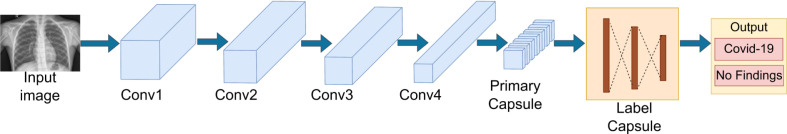
CapsNet: Capsule network (CapsNet) introduced by Sabour et al. in 2017 [32] is an alternative introduced to overcome the limitations associated with CNNs such as the missing of small details in the images due to the max-pooling, that transfer important information between layers [87] and the requirement of large datasets to be trained to achieve high accuracy. A study conducted by Toraman et al. [87], as depicted in Fig. 8, has used CapsNet with four convolutional layers instead of one, unlike in the original CapsNet to have a more effective feature map. These layers are followed by a LabelCaps layer consisting of 16D capsules for both two and three classes. With this model, they have experimented for the two classes COVID, and no findings as well as for the three classes COVID, Pneumonia and no findings achieving the accuracy of 97.04%, 94.57%, respectively. Moreover, Afshar et al. [88] have made use of CapsNet with four convolutional layers and three Capsule layers and achieved 98.3% accuracy with pre-training and 95.7% accuracy without transfer learning approach.
Fig. 8.
Representation of a Capsule Net architecture by Toraman et al. [87].
U-Net: U-Net model, which is introduced by Ronneberger et al. in 2015 [33], is a model that can be applied for biomedical segmentation problems and considered to performs well in various biomedical segmentation applications with only a few annotated images [98]. A study by Pant et al. [55] have used two models, a U-Net model based on ResNet-34 and another U-Net model based on EfficientNet-B4 separately, and ensembled together where the data is simultaneously passed through both models. Ranger optimizer is used with binary cross-entropy loss (BCE) and Dice loss due to its better performance even with imbalanced data. The U-Net model based on EfficientNet-B4 has achieved the highest accuracy, precision, and F1-score whereas the ensemble of the two models has achieved the highest recall. A study conducted by Narayanan et al. [62], has applied a U-Net architecture to implement a lung segmentation algorithm to enhance their model performance. Further, Rahman et al. [99], have presented an architecture adapted from U-Net to generate lung field segmentation. They have evaluated the results of their segmentation using Dice coefficient and Intersection-Over-Union which have yielded the values 94.21% and 91.37%, respectively.
EfficientNet: EfficientNet is a family of models, introduced by Tan and Le in 2019 [34]. Luz et al. [89] have used EfficientNet B0-B5, MobileNet, MobileNetV2, VGG-16, and VGG-19 with transfer learning for multi-class classification of Pneumonia, COVID-19 and normal classes. They were able to obtain higher performance in the EfficientNet B4 model, with an accuracy of 93.9%, a sensitivity of 96.8% and a positive prediction of 100%. Another study for multi-class classification by Marques et al. [38] has used the EfficientNet B4 model achieving 96.70% accuracy. In a study carried out for the binary classification for COVID-19, Nigam et al. [90] have tried out several models and have achieved the highest accuracy of 93.48% for the EfficientNet model.
SqueezeNet: Following the objective of identifying a small CNN with few parameters that can still preserve accuracies achieved by other similar architectures, Iandola et al. introduced SqueezeNet in 2016 [28]. Ucar et al. [91], have used a SqueezeNet based model named as COVIDiagnosis-Net for multi-class classification to detect COVID-19 and Pneumonia using Bayesian optimizer which was able to achieve a higher accuracy value of 98.3%. SqueezeNet is also used by Akpinar et al. [92], for binary classification using Adam optimizer achieving an accuracy of 90.95%.
Considering the studies mentioned in Table 4 it can be seen that VGG, ResNet, MobileNet and DenseNet models have been used by many of the studies. The most frequently used model is VGG. As a percentage, 26.5% of the studies have used the VGG model and 20.6% studies have used ResNet, while MobileNet and DenseNet each has separately been used by 14.7% of the studies considered for this systematic review. These studies have obtained a mean accuracy value of 93.62% with VGG, 92.13% with ResNet, 95.16% with MobileNet, and 84.25% with DenseNet. Moreover, the models U-Net, CapsNet, SqueezeNet, and InceptionV3 seem to be less popular among the works that have been considered in this systematic review, while the rest of the models have been used by a considerable number of studies. The least frequently used model is InceptionV3, which has a usage percentage of 2.9%, while CapsNet, U-Net, and SqueezeNet models are used with a percentage of 5.9%.
Accordingly, as stated in Table 4, it can be seen that many studies have tended to use transfer learning. As a percentage 61.8% of the studies mentioned in the table have used transfer learning in their model, while the rest have not used transfer learning. Another fact to be noticed is that the VGG model is mostly used with transfer learning compared to other studies. As indicated in the reviewed studies, the use of transfer learning can be due to several reasons such as usage of small datasets [54], need to achieve better performance [23] and the need to improve the training time [69]. However, the mean accuracy value of the studies that have used transfer learning is 92.2%, while the mean accuracy of the studies that have not used transfer learning is 92.5%. This analysis indicates that both methods have been able to produce promising results based on the considered dataset, used DL model and the hyperparameter tuning.
It should be noted that some of the studies have experimented with several models indicating the accuracy values obtained from each of the experimented models. When considering such studies, only the model with the highest accuracy value was included in this table. For instance, Hemdan et al. [74] have achieved the same accuracy for both models VGG and DenseNet separately. Moreover, Chauhan et al. [75], have achieved 98.45% accuracy for normal class and 98.32% accuracy for COVID-19 class.
Apart from the discussed learning models, several other models have been used in the reviewed studies with different datasets as shown in Table 5. Among other studies, Ozturk et al. [124], have presented a DarkNet-19 based model named DarkCovidNet with high accuracy of 98.08%. Further, the work presented by Nigam et al. [90] and Khan et al. [106] have used the Xception model in their research and have gained accuracy of 85.03% and 95%, respectively.
4. Public datasets
DL is a way of processing large datasets to extract the important features and provide classifications and predictions. High quality training datasets play a significant role in the field of DL. Generally, to obtain better classification accuracy the dataset should be sufficiently large to train the DL models, unbiased, accessible and taken from a real-world problem domain of interest. Table 5 states widely used publicly available datasets.
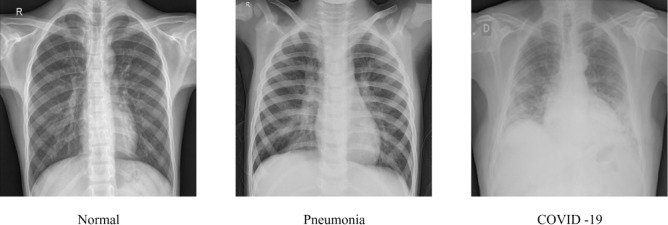
Fig. 9 displays a sample of each class of the chest X-ray images namely normal, Pneumonia and COVID-19 subjects. As summarized in Table 5, many studies that detect Pneumonia have used datasets such as Chest X-ray Images (Pneumonia) [112], Large Dataset of Labeled Optical Coherence Tomography (OCT) [100], and Chest X-ray14 [118]. The Chest X-ray Images (Pneumonia) dataset was collected from Guangzhou Women and Children’s Medical Center, Guangzhou and has been graded by two experts. The Chest X-ray14 dataset has 14 classes of Thorax diseases including Pneumonia.
Fig. 9.
Sample of each class of the chest X-ray.
Among the reviewed studies, 62.7% of the studies were based on only a single dataset, while the remaining 37.3% of studies have used multiple or combined datasets, supporting generalizability. Considering the usage of the available public datasets, the highest number of studies have used the first public COVID-19 image dataset named COVID Chestxray Dataset [104], which is a percentage of 29.5% out of the reviewed papers. The second most used dataset is Chest X-ray Images (Pneumonia) [112], with a percentage of 22.5%. The datasets such as Augmented COVID-19 X-ray Images Dataset [123], COVID-19 Patients Lungs X-ray Images 10000 [117], CoronaHack - Chest X-ray-Dataset [122] have used by approximately 1.4% of the reviewed studies. The reason for the limited use of these datasets could be due to the small number of images. Furthermore, relatively a few studies have used the Mendeley Augmented COVID-19 X-ray Images Dataset [123], as it contains augmented data.
5. Discussion and lessons learned
5.1. Comparison of related studies
This section discusses the chest X-ray classification studies based on DL techniques in three categories of lung infection diseases. Table 6, Table 7 and Table 8 summarize the studies on chest X-ray classification that have considered Pneumonia conditions only, COVID-19 conditions only and both Pneumonia and COVID-19 conditions, respectively. We have stated the applied DL technique, loss function, optimization function and the obtained performance values. The abbreviations used in the table data are as follows: Categorical Cross-entropy (CCE), Siamese Convolutional Network (SCN), Binary Cross-entropy (BCE), Stochastic gradient descent (SGD), Multi-objective Adaptive Differential Evolution (MADE), Forward Positive Rate (FPR), Area Under the Receiver Operating Characteristics curve (AUROC), Intersection over Union (IoU), Mean Squared Error (MSE), Accuracy (Acc), Precision (P), Recall (R), Sensitivity (Sn), Specificity (Sp), Multilayer Perceptron (MLP), Normal (N), COVID-19 (C), COVID-19 sensitivity (), COVID-19 positive prediction (+), fallout = FP/(FN + TP), miss rate = FN/(FN + TP), S-dice = 2TP/(2TP + FP + FN), and Jaccard similarity = TP/(TP + FP + FN).
Table 6 shows that most of the related studies have used existing DL architectures instead of CNN built from scratch. For instance, 44% of the studies indicated in Table 6 have developed their own CNN architecture, while the rest have used existing DL architectures. Some of the studies have built an ensemble model using several existing architectures. Most of these ensemble models have used variations of the ResNet model like ResNet18, ResNet-34, ResNet-50 and ResNet101 [9], [24], [55], [125]. DenseNet121 and InceptionV3 models have also been used in several ensemble models [9], [24]. These ensemble models have reached more than 90% accuracy. For example, the study by Hashmi et al. [24] have achieved a higher accuracy value of 98.43%. Their ensemble model was developed using ResNet18, DenseNet121, InceptionV3, Xception and MobileNetV2 architectures. Therefore, it is observable that the use of an ensemble model can result in high accuracy values in the reviewed studies.
Other studies have used a single existing model or a CNN built from scratch. The highest accuracy of 98.46% is achieved by the study conducted by Mamlook et al. [126], which has used a CNN model and the second-highest accuracy of 98.43% is achieved by the study done by Hashmi et al. [24] using an ensemble model. As a percentage, 36% of the indicated studies have achieved more than 95% accuracy. The models that are used in these studies are ResNet-50 [73], VGG-16 with MLP [68], CNN with U-Net [62], CNNs built from scratch [22], [102], [114], [126] and ensemble models [9], [24]. From this analysis, it can be determined that not only ensemble models but even single models such as ResNet and VGG, as well as newly developed CNNs, can achieve promising results with their datasets.
When training the DL model, the associated optimizer play a significant role to reduce the overall loss and improving the accuracy. Considering the related studies stated in Table 6, 44% of the studies have used Adam optimizer [9], [22], [23], [40], [62], [81], [85], [102], [113], [115], [127]. Adam optimizer is widely used due to the combined benefits of other types of SGD like RMSProp and AdaGrad. That is, Adam optimizer uses both the first and second moments of the gradients when adapting the learning rate. Thus, the Adam optimizer is a method that adaptively calculates the individual learning rates. Performance-wise, the average accuracy achieved by the studies that used Adam optimizer is 88.93%.
Considering the related studies that have achieved more than 95% accuracy, four studies have used Adam optimizer [9], [22], [62], [102], while SGD [24], RMSprop [68] and Gradient Descent [114] has been used by one study each. However, studies have used RMSProp instead of Adam optimizer when the model is VGG-16. The reason could be the failure of the Adam optimizer in model training due to the large number of parameters associated with the VGG network. Moreover, the studies with the VGG-16 model have increased the model accuracy by using multi-layer perception instead of fully connected layers [68], [69]. Accordingly, the DL process uses optimizers to reduce the error in the algorithm. The loss function is used to compute this error by quantifying the actual and predicted output. Considering the loss calculation, many studies have used variants of cross-entropy loss, while some studies have used other types of loss functions like Dice loss and Mean Square Error [9], [114].
Furthermore, different model evaluation techniques have been used in the literature to assess the generalization correctness of models on test data. Model performance are mainly measured using accuracy, precision, recall, sensitivity, specificity and F1 score [22], [24], [51], [55], [69] values, while some have used AUROC [9], [22], [23], [62], [86]. Cross-validation has also been used by several studies [102], [126], [128]. Other methods like IoU and hold-out validation have also been used [43], [125].
Table 7 presents the studies that have been carried out to detect COVID-19 from the chest X-ray images. It can be seen that many studies have used existing architectures rather than developing new CNNs from scratch. As a percentage, only 23.8% of the studies have used CNNs developed from scratch, while the rest of the studies have used existing model architectures. The usage of the models VGG and ResNet, is high among the considered work. For instance, 38% of the studies have used VGG and 28.6% of the studies have used ResNet. Performance-wise, the average of the accuracy values obtained by the studies that have used VGG architecture is 92.7%, while for ResNet it is 89.14%. Compared to the studies conducted for Pneumonia detection in Table 6, there is an increase in accuracy values of the studies conducted for the detection of COVID-19. From the studies indicated in Table 7 it is visible that, 61.9% of the studies have achieved more than 95% accuracy. The models used in these studies are ResNet-50 [56], VGG-16 [56], [72], VGG-19 [70], CapsNet [88], DensNet121 [75], MobileNet [82], MobileNetV2 [83], InceptionV3 [83], Xception [83], InceptionResNetV2 [82] and CNNs developed from scratch [6], [105], [107], [108], [129]. From this analysis, it can be seen that among the studies that have achieved more than 95% accuracy values, only 38.5% of studies have come up with newly developed CNN architectures. GPU is the most widely used hardware accelerator for DL and testing [130]. Most of the studies have used GPU, and the rest of the studies have not mentioned their hardware acceleration details.
The rest of the studies have employed existing model architectures where some of them have introduced new modifications like combinations of architectures. For instance, Hilmizen et al. [56] have used a combination of ResNet-50 and VGG-16, Sethy et al. [78] have applied ResNet-50 with SVM, and Shibly et al. [72] have used a VGG based Faster R-CNN. Another noticeable fact is that some studies have experimented with several models separately and have indicated the achieved accuracy for each of them. Nigam et al. [90] have experimented with five models while Mohammadi et al. [82] and Jabber et al. [83] each have tried with four models. Three out of the four models experimented in the study of Jabber et al. [83] have achieved more than 95% accuracy. Moreover, the study by Mohammadi et al. [82], has shown promising accuracy threshold for two models out of four models. Although the studies that have addressed the classification of chest X-rays with COVID-19 conditions keep emerging, it can be seen that many studies have been able to achieve higher accuracy. Only the study done by Bekhet et al. [129] has not used a GPU, since they mainly focused on building an efficient method that even can run on a normal CPU. Thus, it can be observed that many recent studies have used GPUs for their implementation.
As indicated in Table 7 the reviewed studies have used several optimizers such as Adam, SGD, RMSprop, Momentum, and Adamax to optimize their models. It can be seen that many studies have used Adam optimizer, where the percentage is 38% out of the considered studies. Moreover, the average accuracy of the studies that have used Adam is 95.8%. Moreover, five studies out of the 13 studies that achieved more than 95% accuracy have used Adam optimizer [56], [70], [82], [88], [105], while Momentum [72], Adamax [75] and SGD [129] has been used by one study each. The rest have not mentioned an optimizer in their studies. Accordingly, many studies have used GPU for the implementation. However, some of the studies have not mentioned their hardware acceleration details.
The use of performance metrics for the analysis of chest X-rays for the COVID-19 condition is similar to the metrics used for chest X-ray analysis for pneumonia conditions. For instance, many studies have used cross-entropy loss to calculate the loss function. Many models were evaluated using accuracy, sensitivity and specificity [52], [56], [71], [72], [88], [108]. F1-score is also used in several studies to evaluate their models. As indicated by the reviewed studies, this is due to its ability to compare the results of the two classifiers’ precision and recall, and its eligibility to measure the performance of the models tested with imbalance datasets [70], [72], [82], [90], [105]. Some other studies have evaluated their model using the metric AUC (area under the curve) of the ROC (Receiver Operating Characteristic) due to the better representation of the true positive rate vs. the false positive rate [52], [71], [82], [88], [93].
Accordingly, Table 8 summarizes the chest X-ray classification studies that have considered both pneumonia and COVID-19 conditions. As in previous cases, many studies have used existing CNN architectures and from the studies indicated in the table, the usage of CNNs developed from scratch is only 20%. It can be seen that some studies have tried several techniques separately such as the two models COV19-ResNet, COV19-CNNet used in the study by Keles et al. [103] and the four techniques CNN, SVM, DT, KNN used in the study by Nour et al. [37]. Moreover, some have used a combination of models such as the ensemble model of ResNet50V2, VGG-16, InceptionV3 used in the study by Shorfuzzaman et al. [58] and the model developed with the combined feature set of both SqueezeNet and MobileNetV2 in the study of Togacar et al. [60].
From the studies indicated in the table, 60% of the studies have achieved more than 95% accuracy values. The single models used in these studies are VGG-16 [54], [67], ResNet [47], [76], [103], EfficientNet [38], SqueezeNet [91], Xception [106]. Considering the models of combined feature set of SqueezeNet & MobileNetV2 [60], ensemble model of ResNet50V2, VGG-16, InceptionV3 [58] can be stated. Moreover, CNNs developed from scratch [37], [111], SVM [37], DT [37] and KNN [37] are some of the other approaches used. It can be seen that VGG-16, ResNet, and SqueezeNet are used in several studies which produced promising results of more than 95% of accuracy. Moreover, the study by Nour et al. [37] has applied multiple classification techniques in machine learning such as SVM, Decision Tree, and K-nearest Neighbors along with CNN separately and each of these techniques has been able to produce more than 95% of accuracy.
Similar to the studies with binary classification, most of the studies with multi-class classification have used Adam optimizer. As a percentage, 70% of the studies indicated in Table 8 have used Adam. The rest of the studies have used optimizers like RMSprop, SGD, and Bayesian. An average of 93.9% of accuracy value comes from the accuracy of the studies that have used Adam optimizer. From the 14 studies which produced more than 95% accuracy, eight of the studies have used Adam optimizer [38], [47], [54], [58], [67], [106], [111], while two of the studies have used Bayesian optimizer [37], [91] and the rest have not mentioned an optimizer in their study. From this analysis, it can be concluded that the use of Adam as an optimizer has improved the models to produce promising results.
The Evaluation of the models has mostly been done using accuracy, precision, recall, sensitivity, specificity, and F1 score [47], [67], [76], [103], [106], [111], while some have used AUROC [58], [59], [79]. Cross-validation has also been used by several of the work [87], [111]. Apart from these methods, Ucar and Korkmaz [91], have also used metrics such as correctness, completeness, and Matthew Correlation Coefficient (MCC).
5.2. Guidelines for deep learning process
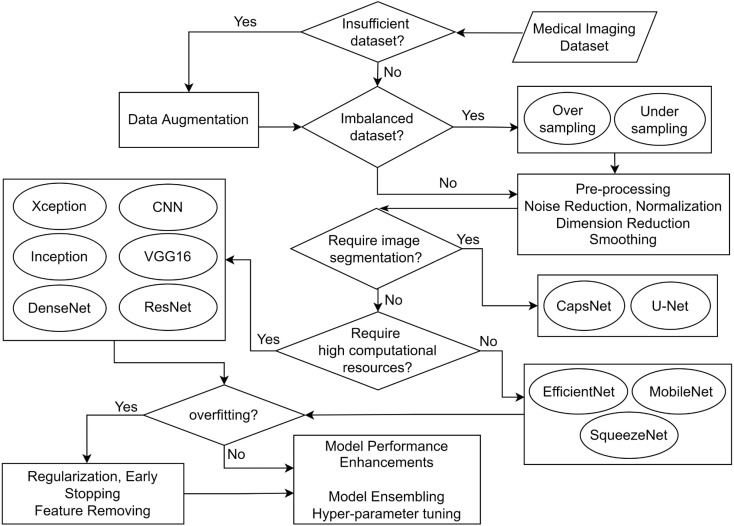
The decision of selection of techniques to be used during a DL process depends on factors such as the type of the problem, dataset and the expected outcome. As shown in Fig. 10, we provide several considerations to review when exploring the appropriate models and techniques for medical image classification from a practical point of view.
When following a DL process for medical image classification, first different pre-processing techniques can be applied to the dataset and prepare it for training. If the dataset consists of an insufficient number of images, different data augmentation techniques can be used to obtain a scaled-up and diverse dataset [24], [67], [85], [113], [125]. Similarly, for imbalanced datasets, the methods like oversampling and undersampling can be used. Consequently, other pre-processing techniques such as noise reduction, normalization, dimension reduction and smoothing can be used to further improve the datasets [54], [55]. The exact combination of these methods would give promising results depending on the type of the dataset and the models being used, and needs to be experimented with to select the most suitable combination of hyper-parameters.
The selection of the most appropriate core DL model is an important task that depends on extensive exploration of the literature. For instance, if the problem requires image segmentation, then the models such as CapsNet or U-net can be used [55], [62]. Another important consideration is the available computational or processing power. The models such as SqueezeNet, MobileNet and EfficientNet can be executed even without high computational power [60], [89], while the models like Xception, Inception, VGG, ResNet and DenseNet require high computational power to be executed [23], [67], [73], [106]. While it is important to consider these factors before selecting a model, the exact requirements specific to the problem need to be given higher priority considering t trade-offs between the accuracy and computational complexity.
Once the model has been selected and trained using the prepared dataset, the model can overfit. To overcome this, techniques like regularization, early stopping and feature removing can be tried out [23], [42], [75], [113]. Finally, model refinements can be applied to improve the performance of the trained model. Hyper-parameter tuning and trying out ensembles of different models are some techniques that can be experimented with [9], [24], [57], [58]. The effectiveness of the aforementioned techniques depends on the data types, application domain, available computational resources and other constraints. These guidelines can be used as an advisory for the practitioners; however, should not be considered as a rigid criterion.
5.3. Current challenges
Several challenges are often encountered when using DL techniques in medical image analysis. One of the challenges in chest X-ray classification is imbalanced datasets, which results in inaccurate outcomes [131]. Class imbalance can also cause model overfitting, as addressed by several studies [55], [125], [132] in binary classification. Similarly, studies on multi-class classifications for Pneumonia and COVID-19 detection have shown overfitting situations [37], [47], [91]. Moreover, the limitation of sufficient datasets and the lack of benchmark dataset makes it challenging to produce more accurate models. This scarcity of publicly available datasets, especially COVID-19 datasets, has become a significant cause for imbalanced datasets for the multi-class classification models.
Handling large size images is another main challenge. Even with powerful GPUs, training with large image sizes can be time-consuming and computationally costly. Most studies have reduced the original image size during training to reduce computational costs and time. For instance, Farooq and Hafeez [47], have used the progressive image re-sizing method to reduce the training time in multi-class classification, whereas Stephan et al. [101] has experimented with five different image sizes to identify the best image size to achieve high accuracy while minimizing the training time and computational cost in binary classification. As stated in Section 2.3 and Section 2.2, the systematic review may have a limitation caused by minor errors associated with the search results.
5.4. Study contributions and lessons learned
We have explored the trends of recent studies (RQ1) by analyzing the used techniques such as transfer learning, use of ensemble models and segmentation and have presented a taxonomy as given in Fig. 4. The employment of different architectural models combined was observed and a summary is provided in Table 3. Moreover, by providing a comparison between the different DL based models used and how parameters such as optimizers, loss functions and evaluation metrics were employed together with the accuracies achieved as stated in Table 4. In this study, we have presented the state-of-the-art DL based models and architectures (RQ2). The publicly available potential datasets have been identified (RQ3) along with the studies that have used them with a comprehensive analysis on the types of diseases and the number of available images as provided in Table 5. As one of the main contributions, We have provided a comprehensive comparison between the related studies (RQ4) by discussing the techniques, models, and metrics used in them along with the accuracy achieved and comparison tables for each of the considered cases. The details were presented in Table 6, Table 7, and Table 8. Furthermore, we have guided the selection of a suitable DL approach as shown in Fig. 10 (RQ5) according to specific requirements using the identified and analyzed approaches. In addition we provided a comprehensive discussion on the facts to be considered when choosing the right techniques. Finally, we have addressed the open challenges and possible future research directions (RQ6) with the analyzed information. The findings of this article will be beneficial for researchers and developers in producing effective and efficient computational solutions for the betterment of the same domain.
5.5. Future research directions
The existing research approaches can be extended in several directions. The accuracy and performance gained from the existing DL architectures can be further improved by implementing an optimization algorithm [133], [134]. Also, both segmentation and classification can be applied for chest X-ray images and develop an ensemble model to produce better results [135], [136].
Additionally, chest X-ray classification with techniques such as deep probabilistic programming, and parallel processing can be considered as promising future research directions [11], [137]. In deep probabilistic programming, an intersection of probabilistic modeling and DL is used for the implementation of new inference algorithms and probabilistic models. These models would enable the computation and modeling of a large number of parameters.
In another point of view, these DL approaches can be extended with explainable models. This will improve the transparency and interpretability of the diagnosis system. Hence the users can understand the system process and enhance the trustworthiness [138], [139]. Local Interpretable Model-Agnostic Explanations (LIME), Gradient-weighted Class Activation Mapping (Grad-CAM) are some techniques that can be used for explainable AI.
Moreover, researchers and developers can focus on developing a system that operates under low resource environments to encourage deployments of such systems in real medical environments. Furthermore, most of the existing models have not been widely tested in real environments. Therefore, these automated chest X-ray classification models can be used in clinical trials for model verification [140].
6. Conclusion
Analysis of chest X-radiation is one of the common methods to identify lung infections. Over the years, many studies have been conducted to identify pneumonia conditions using chest X-ray images. With the growth of data engineering approaches and the spread of COVID-19 disease over the globe, there is a rapid increase in related research and development studies. However, there is a lack of comprehensive systematic reviews available on the current state-of-the-art solutions. This paper has considered the recently published articles on chest X-ray analysis using DL techniques for the diagnosis of pneumonia and COVID-19.
We explored different DL based approaches used in the literature for chest X-ray classification. This study provided taxonomies and compared different techniques and models used by the reviewed articles. In addition, we stated widely used related public datasets used by researchers and developers for the classification of pneumonia and COVID-19. In this review, we extensively analyzed the existing studies and disclosed the current challenges in classifying chest X-ray images. We further suggested possible guidelines for DL based classification and stated future possible directions for research and development. Thereby the future chest X-ray analysis process will thrive with accurate and efficient automated approaches, producing promising results.
CRediT authorship contribution statement
Dulani Meedeniya: Conceptualization, Methodology, Supervision, Writing – review & editing, Project administration. Hashara Kumarasinghe: Data curation, Writing – original draft. Shammi Kolonne: Data curation, Writing – original draft. Chamodi Fernando: Data curation, Writing – original draft. Isabel De la Torre Díez: Methodology, Writing – review & editing. Gonçalo Marques: Methodology, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Biographies

Prof. D. Meedeniya is a Professor in Computer Science and Engineering at the University of Moratuwa, Sri Lanka. She holds a Ph.D. in Computer Science from the University of St Andrews, United Kingdom. She is the director of the Bio-Health Informatics group at her department and engages in many collaborative research. She is a co-author of 100+ publications in indexed journals, peer-reviewed conferences and international book chapters. She serves as a reviewer, program committee and editorial team member in many international conferences and journals. Her main research interests are Software modeling and design, Bio-Health Informatics, Deep Learning and Technology-enhanced learning. She is a Fellow of HEA (UK), MIET, Senior Member IEEE, Member of ACM and a Chartered Engineer registered at EC (UK).

Ms. K.A.S.H. Kumarasinghe is a B.Sc. Eng. graduate from the Department of Computer Science and Engineering, University of Moratuwa, Sri Lanka. Her main research interests are computer vision and deep learning.
Ms. S.L. Kolonne is a B.Sc. Eng. graduate from the Department of Computer Science and Engineering at the University of Moratuwa, Sri Lanka. Her main research interests are computer vision and deep learning.

Ms. K.C.M. Fernando is a B.Sc. Eng. graduate from the Department of Computer Science and Engineering at the University of Moratuwa, Sri Lanka. Her main research interests are computer vision and deep learning.

Prof. Isabel de la Torre Díez is a Professor in the Department of Signal Theory and Communications at the University of Valladolid, Spain. She is the leader of the GTe Research Group (http://sigte.tel.uva.es). She is an author or co-author of more than 210 papers in SCI journals, peer-reviewed conferences proceedings, books and international book chapters. She has coauthored 21 registered innovative software. She has been involved in more than 100 Program committees of international conferences until 2021. She has participated/coordinated in 44 funded European, national and regional research projects. Her research interests include e-health, telemedicine, m-health, sensors, data mining, cloud, Quality of Service (QoS), Quality of Experience (QoE), economical evaluation of e-health services and apps.

Gonçalo Marques holds a Ph.D. in Computer Science Engineering and is member of the Portuguese Engineering Association (Ordem dos Engenheiros). He is currently working as Assistant Professor lecturing courses on programming, multimedia and database systems. Furthermore, he worked as a Software Engineer in the Innovation and Development unit of Groupe PSA automotive industry from 2016 to 2017 and in the IBM group from 2018 to 2019. His current research interests include Internet of Things, Enhanced Living Environments, machine learning, e-health, telemedicine, medical and healthcare systems, indoor air quality monitoring and assessment, and wireless sensor networks. He has more than 80 publications in international journals and conferences, is a frequent reviewer of journals and international conferences and is also involved in several edited books projects.
References
- 1.World Health Organization . 2019. Pneumonia. Online URL https://www.who.int/news-room/fact-sheets/detail/pneumonia. (Accessed 19 July 2021) [Google Scholar]
- 2.Simoes E.A., Cherian T., Chow J., Shahid-Salles S.A., Laxminarayan R., John T.J. Disease Control Priorities in Developing Countries. 2nd. ed. The International Bank for Reconstruction and Development / The World Bank; Washington (DC): 2006. Acute respiratory infections in children; pp. 483–498. [Google Scholar]
- 3.Yang L., Liu S., Liu J., Zhang Z., Wan X., Huang B., Chen Y., Zhang Y. COVID-19: immunopathogenesis and immunotherapeutics. Signal Transduct. Target. Therapy. 2020;5(128):1–8. doi: 10.1038/s41392-020-00243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radiologyinfo.org L. 2021. Pneumonia. Online URL https://www.radiologyinfo.org/en/info/pneumonia. (Accessed 09 March 2022) [Google Scholar]
- 5.Kong W., Agarwal P.P. Chest imaging appearance of COVID-19 infection. Radiol. Cardiothoracic Imaging. 2020;2(1) doi: 10.1148/ryct.2020200028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H., Wang L., Nan Y., Jin F., Wang Q., Pu J. SDFN: Segmentation-based deep fusion network for thoracic disease classification in chest X-ray images. Comput. Med. Imaging Graph. 2019;75:66–73. doi: 10.1016/j.compmedimag.2019.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ginneken B.V., Romeny B.T.H., Viergever M.A. Computer-aided diagnosis in chest radiography: a survey. IEEE Trans. Med. Imaging. 2001;20(12):1228–1241. doi: 10.1109/42.974918. [DOI] [PubMed] [Google Scholar]
- 8.Wijethilake N., Meedeniya D., Chitraranjan C., Perera I., Islam M., Ren H. Glioma survival analysis empowered with data engineering - A survey. IEEE Access. 2021;9:43168–43191. doi: 10.1109/ACCESS.2021.3065965. [DOI] [Google Scholar]
- 9.Chouhan V., Singh S.K., Khamparia A., Gupta D., Tiwari P., Moreira C., Damaševičius R., de Albuquerque V.H.C. A novel transfer learning based approach for pneumonia detection in chest X-ray images. Appl. Sci. 2020;10(2) doi: 10.3390/app10020559. [DOI] [Google Scholar]
- 10.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020;43:635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubasinghe I., Meedeniya D. In: Deep Learning Techniques for Biomedical and Health Informatics. Agarwal B., Balas V., Jain L., Poonia R., Manisha, editors. Elsevier; 2020. Automated neuroscience decision support framework; pp. 305–326. [DOI] [Google Scholar]
- 12.Haputhanthri D., Brihadiswaran G., Gunathilaka S., Meedeniya D., Jayarathna S., Jaime M., Harshaw C. Integration of facial thermography in eeg-based classification of ASD. International Journal of Automation and Computing (IJAC) 2020;17(6):837–854. doi: 10.1007/s11633-020-1231-6. [DOI] [Google Scholar]
- 13.Kieu S.T.H., Bade A., Hijazi M.H.A., Kolivand H. A survey of deep learning for lung disease detection on medical images: State-of-the-art, taxonomy, issues and future directions. J. Imaging. 2020;6(12) doi: 10.3390/jimaging6120131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Çallı E., Sogancioglu E., van Ginneken B., van Leeuwen K.G., Murphy K. Deep learning for chest X-ray analysis: A survey. Med. Image Anal. 2021;72 doi: 10.1016/j.media.2021.102125. [DOI] [PubMed] [Google Scholar]
- 15.Jain A., Mehra N., Khare P. A survey on pneumonia detection methods using computer-aided diagnosis. Int. J. Emerg. Trends Eng. Res. 2021;9(7):890–895. doi: 10.30534/ijeter/2021/09972021. [DOI] [Google Scholar]
- 16.Bhattacharya S., Maddikunta P.K.R., Pham Q.-V., Gadekallu T.R., Chowdhary C.L., Alazab M., Piran M.J., et al. Deep learning and medical image processing for coronavirus (COVID-19) pandemic: A survey. Sustainable Cities Soc. 2021;65 doi: 10.1016/j.scs.2020.102589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tseng V.S., Ying J.J.-C., Wong S.T., Cook D.J., Liu J. Computational intelligence techniques for combating COVID-19: A survey. IEEE Comput. Intell. Mag. 2020;15(4):10–22. doi: 10.1109/MCI.2020.3019873. [DOI] [Google Scholar]
- 18.Tilve A., Nayak S., Vernekar S., Turi D., Shetgaonkar P.R., Aswale S. 2020 International Conference on Emerging Trends in Information Technology and Engineering (Ic-ETITE) IEEE; 2020. Pneumonia detection using deep learning approaches; pp. 1–8. [DOI] [Google Scholar]
- 19.Li Y., Zhang Z., Dai C., Dong Q., Badrigilan S. Accuracy of deep learning for automated detection of pneumonia using chest X-Ray images: A systematic review and meta-analysis. Comput. Biol. Med. 2020;123 doi: 10.1016/j.compbiomed.2020.103898. [DOI] [PubMed] [Google Scholar]
- 20.Aromataris E., Pearson A. The systematic review: An overview. AJN Am. J. Nursing. 2014;114:53–58. doi: 10.1097/01.NAJ.0000444496.24228.2c. [DOI] [PubMed] [Google Scholar]
- 21.Rasheed J., Jamil A., Hameed A.A., Aftab U., Aftab J., Shah S.A., Draheim D. A survey on artificial intelligence approaches in supporting frontline workers and decision makers for the COVID-19 pandemic. Chaos Solitons Fractals. 2020;141 doi: 10.1016/j.chaos.2020.110337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhatt R., Yadav S., Sarvaiya J.N. In: Emerging Technology Trends in Electronics, Communication and Networking. Gupta S., Sarvaiya J.N., editors. Springer Singapore; Singapore: 2020. Convolutional neural network based chest X-Ray image classification for pneumonia diagnosis; pp. 254–266. [DOI] [Google Scholar]
- 23.Irfan A., Adivishnu A.L., Sze-To A., Dehkharghanian T., Rahnamayan S., Tizhoosh H. Classifying pneumonia among chest X-Rays using transfer learning. 2020 42nd Annual International Conference of the IEEE Engineering in Medicine Biology Society; EMBC; IEEE; 2020. pp. 2186–2189. [DOI] [PubMed] [Google Scholar]
- 24.Hashmi M.F., Katiyar S., Keskar A.G., Bokde N.D., Geem Z.W. Efficient pneumonia detection in chest xray images using deep transfer learning. Diagnostics. 2020;10(6) doi: 10.3390/diagnostics10060417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tricco A.C., Lillie E., Zarin W., O’Brien K.K., Colquhoun H., Levac D., Moher D., Peters M.D., Horsley T., Weeks L., et al. PRISMA extension for scoping reviews (PRISMA-ScR): checklist and explanation. Ann. Internal Med. 2018;169(7):467–473. doi: 10.7326/M18-0850. [DOI] [PubMed] [Google Scholar]
- 26.He K., Zhang X., Ren S., Sun J. Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition; CVPR; IEEE; 2016. pp. 770–778. [DOI] [Google Scholar]
- 27.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Densely connected convolutional networks. 2017 IEEE Conference on Computer Vision and Pattern Recognition; CVPR; IEEE; 2017. pp. 2261–2269. [DOI] [Google Scholar]
- 28.Iandola F., Han S., Moskewicz M., Ashraf K., Dally W., Keutzer K. 2016. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and ¡0.5MB model size. [DOI] [Google Scholar]
- 29.Simonyan K., Zisserman A. 2014. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556. [Google Scholar]
- 30.Howard A., Zhu M., Chen B., Kalenichenko D., Wang W., Weyand T., Andreetto M., Adam H. 2017. MobileNets: Efficient convolutional neural networks for mobile vision applications. [Google Scholar]
- 31.F. Chollet, Xception: Deep Learning with Depthwise Separable Convolutions, in: 2017 IEEE Conference on Computer Vision and Pattern Recognition, CVPR, 2017, pp. 1800–1807, 10.1109/CVPR.2017.195. [DOI]
- 32.Sabour S., Frosst N., Hinton G.E. Proceedings of the 31st International Conference on Neural Information Processing Systems. Curran Associates Inc.; Red Hook, NY, USA: 2017. Dynamic routing between capsules; pp. 3859–3869. (NIPS’17). [Google Scholar]
- 33.Ronneberger O., Fischer P., Brox T. In: Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Navab N., Hornegger J., Wells W.M., Frangi A.F., editors. Springer International Publishing; Cham: 2015. U-net: Convolutional networks for biomedical image segmentation; pp. 234–241. [DOI] [Google Scholar]
- 34.Tan M., Le Q. In: Proceedings of the 36th International Conference on Machine Learning. Chaudhuri K., Salakhutdinov R., editors. vol. 97. PMLR; 2019. EfficientNet: Rethinking model scaling for convolutional neural networks; pp. 6105–6114. (Proceedings of Machine Learning Research). [Google Scholar]
- 35.Wong K.K.L., Wang L., Wang D. Recent developments in machine learning for medical imaging applications. Comput. Med. Imaging Graph. 2017;57:1–3. doi: 10.1016/j.compmedimag.2017.04.001. Recent Developments in Machine Learning for Medical Imaging Applications. [DOI] [PubMed] [Google Scholar]
- 36.Chollet F. 2nd ed. Simon and Schuster; 2021. Deep Learning with Python. [Google Scholar]
- 37.Nour M., Cömert Z., Polat K. A novel medical diagnosis model for COVID-19 infection detection based on deep features and Bayesian optimization. Appl. Soft Comput. 2020;97 doi: 10.1016/j.asoc.2020.106580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marques G., Agarwal D., de la Torre Díez I. Automated medical diagnosis of COVID-19 through EfficientNet convolutional neural network. Appl. Soft Comput. 2020;96 doi: 10.1016/j.asoc.2020.106691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Q., Zhang Y., Dai W., Pan S.J. Cambridge University Press; 2020. Transfer Learning. [DOI] [Google Scholar]
- 40.Khoiriyah S.A., Basofi A., Fariza A. Convolutional neural network for automatic pneumonia detection in chest radiography. 2020 International Electronics Symposium; IES; IEEE; 2020. pp. 476–480. [DOI] [Google Scholar]
- 41.Padma T., Kumari C.U. Deep learning based chest X-Ray image as a diagnostic tool for COVID-19. 2020 International Conference on Smart Electronics and Communication; ICOSEC; IEEE; 2020. pp. 589–592. [DOI] [Google Scholar]
- 42.Ahmed F., Bukhari S.A.C., Keshtkar F. A deep learning approach for COVID-19 8 viral pneumonia screening with X-Ray images. Digit. Gov.: Res. Pract. 2021;2(2) doi: 10.1145/3431804. [DOI] [Google Scholar]
- 43.Kieu P.N., Tran H.S., Le T.H., Le T., Nguyen T.T. Applying multi-CNNs model for detecting abnormal problem on chest x-ray images. 2018 10th International Conference on Knowledge and Systems Engineering; KSE; IEEE; 2018. pp. 300–305. [DOI] [Google Scholar]
- 44.Wang L., Lin Z.Q., Wong A. COVID-net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S.W., Chiu Y.-T., Brudnicki P., Bischoff A.M., Jelinek A., Wang J.Z., Bogdanowicz D.R., Laine A.F., Guo J., Lu H.H. 2020. Darwin’s neural network: AI-based strategies for rapid and scalable cell and coronavirus screening. arXiv:2007.11653. [Google Scholar]
- 46.J. Deng, W. Dong, R. Socher, L.-J. Li, K. Li, L. Fei-Fei, ImageNet: A Large-Scale Hierarchical Image Database, in: CVPR09, 2009.
- 47.Farooq M., Hafeez A. 2020. COVID-ResNet: A deep learning framework for screening of COVID19 from radiographs. arXiv arXiv:2003.14395. [Google Scholar]
- 48.Krizhevsky A. University of Toronto; 2009. Learning Multiple Layers of Features from Tiny Images: Tech. rep. [Google Scholar]
- 49.LeCun Y., Cortes C., Burges C. 1998. The MNIST DATABASE of handwritten digits. http://yann.lecun.com/exdb/mnist. [Google Scholar]
- 50.Nogueira K., Penatti O.A., Dos Santos J.A. Towards better exploiting convolutional neural networks for remote sensing scene classification. Pattern Recognit. 2017;61:539–556. [Google Scholar]
- 51.Chhikara P., Singh P., Gupta P., Bhatia T. In: Advances in Bioinformatics, Multimedia, and Electronics Circuits and Signals. Jain L.C., Virvou M., Piuri V., Balas V.E., editors. Springer Singapore; Singapore: 2020. Deep convolutional neural network with transfer learning for detecting pneumonia on chest X-Rays; pp. 155–168. [Google Scholar]
- 52.Panwar H., Gupta P., Siddiqui M.K., Morales-Menendez R., Singh V. Application of deep learning for fast detection of COVID-19 in X-Rays using ncovnet. Chaos Solitons Fractals. 2020;138 doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tajbakhsh N., Shin J.Y., Gurudu S.R., Hurst R.T., Kendall C.B., Gotway M.B., Liang J. Convolutional neural networks for medical image analysis: Full training or fine tuning? IEEE Trans. Med. Imaging. 2016;35(5):1299–1312. doi: 10.1109/TMI.2016.2535302. [DOI] [PubMed] [Google Scholar]
- 54.Choudhuri R., Paul A. Multi class image classification for detection of diseases using chest X ray images. 2021 8th International Conference on Computing for Sustainable Global Development; INDIACom; IEEE; 2021. pp. 769–773. [DOI] [Google Scholar]
- 55.A. Pant, A. Jain, K.C. Nayak, D. Gandhi, B. Prasad, Pneumonia Detection: An Efficient Approach Using Deep Learning, in: 2020 11th International Conference on Computing, Communication and Networking Technologies, ICCCNT, 2020, pp. 1–6, 10.1109/ICCCNT49239.2020.9225543. [DOI]
- 56.Hilmizen N., Bustamam A., Sarwinda D. The multimodal deep learning for diagnosing COVID-19 pneumonia from chest CT-scan and X-Ray images. 2020 3rd International Seminar on Research of Information Technology and Intelligent Systems; ISRITI; IEEE; 2020. pp. 26–31. [DOI] [Google Scholar]
- 57.Das A.K., Ghosh S., Thunder S., Dutta R., Agarwal S., Chakrabarti A. Automatic COVID-19 detection from X-ray images using ensemble learning with convolutional neural network. Pattern Anal. Appl. 2021:1–14. doi: 10.1007/s10044-021-00970-4. [DOI] [Google Scholar]
- 58.Shorfuzzaman M., Masud M., Alhumyani H., Anand D., Singh A. Artificial neural network-based deep learning model for COVID-19 patient detection using X-Ray chest images. J. Healthc. Eng. 2021;2021 doi: 10.1155/2021/5513679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tiwari S., Jain A. Convolutional capsule network for COVID-19 detection using radiography images. Int. J. Imaging Syst. Technol. 2021;31(2):525–539. doi: 10.1002/ima.22566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Toğaçar M., Ergen B., Cömert Z. COVID-19 detection using deep learning models to exploit social mimic optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sultana F., Sufian A., Dutta P. Evolution of image segmentation using deep convolutional neural network: A survey. Knowl.-Based Syst. 2020;201–202 doi: 10.1016/j.knosys.2020.106062. [DOI] [Google Scholar]
- 62.Narayanan B.N., Davuluru V.S.P., Hardie R.C. Medical Imaging 2020: Imaging Informatics for Healthcare, Research, and Applications. vol. 11318. International Society for Optics and Photonics; 2020. Two-stage deep learning architecture for pneumonia detection and its diagnosis in chest radiographs; p. 113180G. [DOI] [Google Scholar]
- 63.Candemir S., Jaeger S., Palaniappan K., Musco J.P., Singh R.K., Xue Z., Karargyris A., Antani S., Thoma G., McDonald C.J. Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Trans. Med. Imaging. 2014;33(2):577–590. doi: 10.1109/TMI.2013.2290491. [DOI] [PubMed] [Google Scholar]
- 64.Jaeger S., Karargyris A., Candemir S., Folio L., Siegelman J., Callaghan F., Xue Z., Palaniappan K., Singh R.K., Antani S., Thoma G., Wang Y.-X., Lu P.-X., McDonald C.J. Automatic tuberculosis screening using chest radiographs. IEEE Trans. Med. Imaging. 2014;33(2):233–245. doi: 10.1109/TMI.2013.2284099. [DOI] [PubMed] [Google Scholar]
- 65.LaLonde R., Bagci U. 2018. Capsules for object segmentation. arXiv arXiv:1804.04241. [Google Scholar]
- 66.Bonheur S., Štern D., Payer C., Pienn M., Olschewski H., Urschler M. In: Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. Shen D., Liu T., Peters T.M., Staib L.H., Essert C., Zhou S., Yap P.-T., Khan A., editors. Springer International Publishing; Cham: 2019. Matwo-CapsNet: A multi-label semantic segmentation capsules network; pp. 664–672. [DOI] [Google Scholar]
- 67.Militante S.V., Dionisio N.V., Sibbaluca B.G. Pneumonia and COVID-19 detection using convolutional neural networks. 2020 Third International Conference on Vocational Education and Electrical Engineering; ICVEE; IEEE; 2020. pp. 1–6. [DOI] [Google Scholar]
- 68.Ferreira J.R., Cardenas D.A.C., Moreno R.A., de Sá Rebelo M.d.F., Krieger J.E., Gutierrez M.A. Multi-view ensemble convolutional neural network to improve classification of pneumonia in low contrast chest X-Ray images. 2020 42nd Annual International Conference of the IEEE Engineering in Medicine Biology Society; EMBC; IEEE; 2020. pp. 1238–1241. [DOI] [PubMed] [Google Scholar]
- 69.Ayan E., Ünver H.M. Diagnosis of pneumonia from chest X-Ray images using deep learning. 2019 Scientific Meeting on Electrical-Electronics Biomedical Engineering and Computer Science; EBBT; IEEE; 2019. pp. 1–5. [DOI] [Google Scholar]
- 70.Vaid S., Kalantar R., Bhandari M. Deep learning COVID-19 detection bias: accuracy through artificial intelligence. Int. Orthopaedics. 2020;44:1539–1542. doi: 10.1007/s00264-020-04609-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abbas A., Abdelsamea M.M., Gaber M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl. Intell. 2021;51(2):854–864. doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shibly K.H., Dey S.K., Islam M.T.-U., Rahman M.M. COVID faster R–CNN: A novel framework to diagnose novel coronavirus disease (COVID-19) in X-Ray images. Inf. Med. Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Youssef T.A., Aissam B., Khalid D., Imane B., Miloud J.E. Classification of chest pneumonia from x-ray images using new architecture based on ResNet. 2020 IEEE 2nd International Conference on Electronics, Control, Optimization and Computer Science; ICECOCS; IEEE; 2020. pp. 1–5. [DOI] [Google Scholar]
- 74.Hemdan E.E.-D., Shouman M.A., Karar M.E. 2020. Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv preprint arXiv:2003.11055. [Google Scholar]
- 75.Chauhan T., Palivela H., Tiwari S. Optimization and fine-tuning of DenseNet model for classification of COVID-19 cases in medical imaging. Int. J. Inf. Manag. Data Insights. 2021;1(2) doi: 10.1016/j.jjimei.2021.100020. [DOI] [Google Scholar]
- 76.Bukhari S.U.K., Bukhari S.S.K., Syed A., Shah S.S.H. 2020. The diagnostic evaluation of convolutional neural network (CNN) for the assessment of chest X-ray of patients infected with COVID-19. medRxiv. [DOI] [Google Scholar]
- 77.Zhou C., Song J., Zhou S., Zhang Z., Xing J. COVID-19 detection based on image regrouping and resnet-SVM using chest X-Ray images. IEEE Access. 2021;9:81902–81912. doi: 10.1109/ACCESS.2021.3086229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sethy P.K., Behera S.K. 2020. Detection of coronavirus disease (covid-19) based on deep features. [Google Scholar]
- 79.Manokaran J., Zabihollahy F., Hamilton-Wright A., Ukwatta E. In: Medical Imaging 2021: Biomedical Applications in Molecular, Structural, and Functional Imaging, vol. 11600. Gimi B.S., Krol A., editors. SPIE; 2021. Deep learning-based detection of COVID-19 from chest x-ray images; pp. 164–170. [DOI] [Google Scholar]
- 80.Tobias R.R.N., De Jesus L.C.M., Mital M.E.G., Lauguico S.C., Guillermo M.A., Sybingco E., Bandala A.A., Dadios E.P. CNN-based deep learning model for chest X-ray health classification using TensorFlow. 2020 RIVF International Conference on Computing and Communication Technologies; RIVF; IEEE; 2020. pp. 1–6. [DOI] [Google Scholar]
- 81.Hu M., Lin H., Fan Z., Gao W., Yang L., Liu C., Song Q. Learning to recognize chest-xray images faster and more efficiently based on multi-kernel depthwise convolution. IEEE Access. 2020;8:37265–37274. doi: 10.1109/ACCESS.2020.2974242. [DOI] [Google Scholar]
- 82.Mohammadi R., Salehi M., Ghaffari H., Rohani A., Reiazi R. Transfer learning-based automatic detection of coronavirus disease 2019 (COVID-19) from chest X-ray images. J. Biomed. Phys. Eng. 2020;10(5):559. doi: 10.31661/jbpe.v0i0.2008-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jabber B., Lingampalli J., Basha C.Z., Krishna A. Detection of Covid-19 patients using chest X-ray images with convolution neural network and mobile net. 2020 3rd International Conference on Intelligent Sustainable Systems; ICISS; IEEE; 2020. pp. 1032–1035. [DOI] [Google Scholar]
- 84.S. Kolonne, H. Kumarasinghe, C. Fernando, D. Meedeniya, MobileNetV2 Based Chest X-Rays Classification, in: Proceedings of International Conference on Decision Aid Sciences and Application, DASA, Bahrain, 2021, pp. 57–61, 10.1109/DASA53625.2021.9682248. [DOI]
- 85.Rajpurkar P., Irvin J., Zhu K., Yang B., Mehta H., Duan T., Ding D.Y., Bagul A., Langlotz C., Shpanskaya K.S., Lungren M.P., Ng A.Y. 2017. CheXNet: Radiologist-level pneumonia detection on chest X-Rays with deep learning. CoRR abs/1711.05225 arXiv:1711.05225. [DOI] [Google Scholar]
- 86.Varshni D., Thakral K., Agarwal L., Nijhawan R., Mittal A. Pneumonia detection using CNN based feature extraction. 2019 IEEE International Conference on Electrical, Computer and Communication Technologies; ICECCT; IEEE; 2019. pp. 1–7. [DOI] [Google Scholar]
- 87.Toraman S., Alakus T.B., Turkoglu I. Convolutional capsnet: A novel artificial neural network approach to detect COVID-19 disease from X-ray images using capsule networks. Chaos Solitons Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Afshar P., Heidarian S., Naderkhani F., Oikonomou A., Plataniotis K.N., Mohammadi A. COVID-CAPS: A capsule network-based framework for identification of COVID-19 cases from X-ray images. Pattern Recognit. Lett. 2020;138:638–643. doi: 10.1016/j.patrec.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Luz E., Silva P., Silva R., Silva L., Guimarães J., Miozzo G., Moreira G., Menotti D. Towards an effective and efficient deep learning model for COVID-19 patterns detection in X-ray images. Res. Biomed. Eng. 2021:1–14. doi: 10.1007/s42600-021-00151-6. [DOI] [Google Scholar]
- 90.Nigam B., Nigam A., Jain R., Dodia S., Arora N., Annappa B. COVID-19: Automatic detection from X-ray images by utilizing deep learning methods. Expert Syst. Appl. 2021;176 doi: 10.1016/j.eswa.2021.114883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ucar F., Korkmaz D. COVIDiagnosis-net: Deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med. Hypotheses. 2020;140 doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Akpinar K.N., Genc S., Karagol S. Chest X-Ray abnormality detection based on SqueezeNet. 2020 International Conference on Electrical, Communication, and Computer Engineering; ICECCE; IEEE; 2020. pp. 1–5. [DOI] [Google Scholar]
- 93.Azemin C., Zulfaezal M., Hassan R., Mohd Tamrin M.I., Md Ali M.A. COVID-19 deep learning prediction model using publicly available radiologist-adjudicated chest X-ray images as training data: preliminary findings. Int. J. Biomed. Imaging. 2020;2020 doi: 10.1155/2020/8828855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.K. Simonyan, A. Zisserman, Very Deep Convolutional Networks for Large-Scale Image Recognition, in: 3rd International Conference on Learning Representations, ICLR, 2015, 10.48550/arXiv.1409.1556. [DOI]
- 95.C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed, D. Anguelov, D. Erhan, V. Vanhoucke, A. Rabinovich, Going Deeper With Convolutions, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, CVPR, 2015.
- 96.C. Szegedy, V. Vanhoucke, S. Ioffe, J. Shlens, Z. Wojna, Rethinking the Inception Architecture for Computer Vision, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, CVPR, 2016.
- 97.M. Sandler, A. Howard, M. Zhu, A. Zhmoginov, L.-C. Chen, MobileNetV2: Inverted Residuals and Linear Bottlenecks, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, CVPR, 2018, pp. 4510–4520, 10.1109/CVPR.2018.00474. [DOI]
- 98.Rubasinghe I., Meedeniya D. Ultrasound nerve segmentation using deep probabilistic programming. J. ICT Res. Appl. 2019;13:241–256. doi: 10.5614/itbj.ict.res.appl.2019.13.3.5. [DOI] [Google Scholar]
- 99.Rahman M.F., Tseng T.-L.B., Pokojovy M., Qian W., Totada B., Xu H. Medical Imaging 2021: Physics of Medical Imaging, vol. 11595. International Society for Optics and Photonics; 2021. An automatic approach to lung region segmentation in chest x-ray images using adapted U-net architecture; p. 115953I. [DOI] [Google Scholar]
- 100.Kermany D., Zhang K., Goldbaum M. 2018. Large dataset of labeled optical coherence tomography (OCT) and chest X-Ray images. [DOI] [Google Scholar]
- 101.Stephen O., Sain M., Maduh U.J., Jeong D.-U. An efficient deep learning approach to pneumonia classification in healthcare. J. Healthcare Eng. 2019;2019:1–7. doi: 10.1155/2019/4180949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Saraiva A.A., Ferreira N.M.F., de Sousa L.L., Costa N.J.C., Sousa J.V.M., Santos D., Valente A., Soares S. Proceedings of the 12th International Joint Conference on Biomedical Engineering Systems and Technologies - BIOIMAGING. SciTePress; 2019. Classification of images of childhood pneumonia using convolutional neural networks; pp. 112–119. [DOI] [Google Scholar]
- 103.Keles A., Keles M.B., Keles A. COV19-CNNet and COV19-ResNet: diagnostic inference engines for early detection of COVID-19. Cogn. Comput. 2021:1–11. doi: 10.1007/s12559-020-09795-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cohen J.P., Morrison P., Dao L., Roth K., Duong T.Q., Ghassemi M. 2020. COVID-19 image data collection: Prospective predictions are the future. arXiv:2006.11988. [Google Scholar]
- 105.Reshi A.A., Rustam F., Mehmood A., Alhossan A., Alrabiah Z., Ahmad A., Alsuwailem H., Choi G.S. An efficient CNN model for COVID-19 disease detection based on X-Ray image classification. Complexity. 2021;2021 [Google Scholar]
- 106.Khan A.I., Shah J.L., Bhat M.M. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Programs Biomed. 2020;196 doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sekeroglu B., Ozsahin I. Detection of COVID-19 from chest X-Ray images using convolutional neural networks. SLAS Technol.: Transl. Life Sci. Innov. 2020;25(6):553–565. doi: 10.1177/2472630320958376. PMID: 32948098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ahsan M., Based M., Haider J., Kowalski M., et al. COVID-19 detection from chest X-ray images using feature fusion and deep learning. Sensors. 2021;21(4):1480. doi: 10.3390/s21041480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chowdhury M.E.H., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., Islam K.R., Khan M.S., Iqbal A., Emadi N.A., Reaz M.B.I., Islam M.T. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. doi: 10.1109/ACCESS.2020.3010287. [DOI] [Google Scholar]
- 110.Rahman T., Khandakar A., Qiblawey Y., Tahir A., Kiranyaz S., Abul Kashem S.B., Islam M.T., Al Maadeed S., Zughaier S.M., Khan M.S., Chowdhury M.E. Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images. Comput. Biol. Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ouchicha C., Ammor O., Meknassi M. CVDNet: A novel deep learning architecture for detection of coronavirus (Covid-19) from chest x-ray images. Chaos Solitons Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mooney P. 2018. Chest X-ray images (Pneumonia) https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia. [Google Scholar]
- 113.Sharma H., Jain J.S., Bansal P., Gupta S. Feature extraction and classification of chest X-Ray images using CNN to detect pneumonia. 2020 10th International Conference on Cloud Computing, Data Science Engineering; Confluence; IEEE; 2020. pp. 227–231. [DOI] [Google Scholar]
- 114.Islam S.R., Maity S.P., Ray A.K., Mandal M. Automatic detection of pneumonia on compressed sensing images using deep learning. 2019 IEEE Canadian Conference of Electrical and Computer Engineering; CCECE; IEEE; 2019. pp. 1–4. [DOI] [Google Scholar]
- 115.Kaushik V.S., Nayyar A., Kataria G., Jain R. Pneumonia detection using convolutional neural networks (CNNs). In: Singh P.K., Pawłowski W., Tanwar S., Kumar N., Rodrigues J.J.P.C., Obaidat M.S., editors. Proceedings of First International Conference on Computing, Communications, and Cyber-Security; IC4S 2019; Singapore: Springer Singapore; 2020. pp. 471–483. [DOI] [Google Scholar]
- 116.Singh D., Kumar V., Yadav V., Kaur M. Deep neural network-based screening model for COVID-19-infected patients using chest X-ray images. Int. J. Pattern Recognit. Artif. Intell. 2021;35(03) doi: 10.1142/S0218001421510046. [DOI] [Google Scholar]
- 117.Sajid N. 2020. COVID-19 patients lungs X ray images 10000. https://www.kaggle.com/nabeelsajid917/covid-19-x-ray-10000-images. [Google Scholar]
- 118.X. Wang, Y. Peng, L. Lu, Z. Lu, M. Bagheri, R.M. Summers, ChestX-ray8: Hospital-Scale Chest X-Ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, CVPR, 2017.
- 119.Kesim E., Dokur Z., Olmez T. X-Ray chest image classification by a small-sized convolutional neural network. 2019 Scientific Meeting on Electrical-Electronics Biomedical Engineering and Computer Science; EBBT; IEEE; 2019. pp. 1–5. [DOI] [Google Scholar]
- 120.J. Irvin, P. Rajpurkar, M. Ko, Y. Yu, S. Ciurea-Ilcus, C. Chute, H. Marklund, B. Haghgoo, R. Ball, K. Shpanskaya, et al., Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison, in: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 33 no. 01, 2019, pp. 590–597.
- 121.Dadario A.M.V. 2020. COVID-19 x rays. https://www.kaggle.com/andrewmvd/convid19-X-rays. [DOI] [Google Scholar]
- 122.Govindaraj P. 2020. CoronaHack - chest X-Ray-dataset. https://www.kaggle.com/praveengovi/coronahack-chest-xraydataset. [Google Scholar]
- 123.Alqudah A.M., Qazan S. 2020. Augmented COVID-19 X-ray images dataset. https://data.mendeley.com/datasets/2fxz4px6d8/4. [DOI] [Google Scholar]
- 124.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jaiswal A.K., Tiwari P., Kumar S., Gupta D., Khanna A., Rodrigues J.J. Identifying pneumonia in chest X-rays: A deep learning approach. Measurement. 2019;145:511–518. doi: 10.1016/j.measurement.2019.05.076. [DOI] [Google Scholar]
- 126.Al Mamlook R.E., Chen S., Bzizi H.F. Investigation of the performance of machine learning classifiers for pneumonia detection in chest X-ray images. 2020 IEEE International Conference on Electro Information Technology; EIT; IEEE; 2020. pp. 098–104. [DOI] [Google Scholar]
- 127.Prayogo K.A., Suryadibrata A., Young J.C. 2020. Classification of pneumonia from X-ray images using siamese convolutional network 18 (3) [DOI] [Google Scholar]
- 128.A. Saraiva, D. Santos, N. Carvalho da Costa, J. Sousa, N. Ferreira, A. Valente, S. Soares, Models of Learning to Classify X-ray Images for the Detection of Pneumonia using Neural Networks, in: Bioimaging, 2019, pp. 76–83, 10.5220/0007346600760083. [DOI]
- 129.Bekhet S., Alkinani M., Reinel Tabares-Soto M.H. An efficient method for Covid-19 detection using light weight convolutional neural network. Comput. Mater. Contin. 2021;69(2):2475–2491. doi: 10.32604/cmc.2021.018514. [DOI] [Google Scholar]
- 130.Mittal S., Vaishay S. A survey of techniques for optimizing deep learning on GPUs. J. Syst. Archit. 2019;99 doi: 10.1016/j.sysarc.2019.101635. [DOI] [Google Scholar]
- 131.Fernández A., García S., Herrera F. Hybrid Artificial Intelligent Systems. Springer Berlin Heidelberg; Berlin, Heidelberg: 2011. Addressing the classification with imbalanced data: Open problems and new challenges on class distribution; pp. 1–10. [DOI] [Google Scholar]
- 132.Li Z., Kamnitsas K., Glocker B. In: Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. Shen D., Liu T., Peters T.M., Staib L.H., Essert C., Zhou S., Yap P.-T., Khan A., editors. Springer International Publishing; Cham: 2019. Overfitting of neural nets under class imbalance: Analysis and improvements for segmentation; pp. 402–410. [DOI] [Google Scholar]
- 133.Adedigba A.P., Adeshina S.A., Aina O.E., Aibinu A.M. Optimal hyperparameter selection of deep learning models for COVID-19 chest X-ray classification. Intell.-Based Med. 2021;5 doi: 10.1016/j.ibmed.2021.100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.C. Fernando, S. Kolonne, H. Kumarasinghe, D. Meedeniya, Chest Radiographs Classification Using Multi-model Deep Learning: A Comparative Study, in: Proceedings of the 2nd International Conference on Advanced Research in Computing, 2022, pp. 165–170, 10.1109/ICARC54489.2022.9753811. [DOI]
- 135.Kumarasinghe H., Kolonne S., Fernando C., Meedeniya D. U-Net based chest X-ray segmentation with ensemble classification for Covid-19 and pneumonia. International Journal of Online and Biomedical Engineering. 2022;18(7):161–174. doi: 10.3991/ijoe.v18i07.30807. [DOI] [Google Scholar]
- 136.Ahmad F., Farooq A., Ghani M.U. Deep ensemble model for classification of novel coronavirus in chest X-ray images. Comput. Intell. Neurosci. 2021;2021 doi: 10.1155/2021/8890226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.D. Tran, M.D. Hoffman, R.A. Saurous, E. Brevdo, K. Murphy, D.M. Blei, Deep probabilistic programming, in: Proceedings of the 5th International Conference on Learning Representations, ICLR, 2017, pp. 1–18, 10.48550/arXiv.1701.03757. [DOI]
- 138.Dasanayaka S., Shantha V., Silva S., Meedeniya D., Ambegoda T. Interpretable machine learning for brain tumour analysis using MRI and whole slide images. Software Impacts. 2022;13:100340. doi: 10.1016/j.simpa.2022.100340. [DOI] [Google Scholar]
- 139.Reyes M., Meier R., Pereira S., Silva C.A., Dahlweid F.-M., Tengg-Kobligk H.v., Summers R.M., Wiest R. On the interpretability of artificial intelligence in radiology: challenges and opportunities. Radiol. Artif. Intell. 2020;2(3) doi: 10.1148/ryai.2020190043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nasser N., Emad-ul Haq Q., Imran M., Ali A., Razzak I., Al-Helali A. A smart healthcare framework for detection and monitoring of COVID-19 using IoT and cloud computing. Neural Comput. Appl. 2021:1–15. doi: 10.1007/s00521-021-06396-7. [DOI] [PMC free article] [PubMed] [Google Scholar]