Abstract
We report here the identification of a new lipoprotein, NlpI, in Escherichia coli K-12. The NlpI structural gene (nlpI) is located between the genes pnp (polynucleotide phosphorylase) and deaD (RNA helicase) at 71 min on the E. coli chromosome. The nlpI gene encodes a putative polypeptide of approximately 34 kDa, and multiple lines of evidence clearly demonstrate that NlpI is indeed a lipoprotein. An nlpI::cm mutation rendered growth of the cells osmotically sensitive, and incubation of the insertion mutant at an elevated temperature resulted in the formation of filaments. The altered phenotype of the mutant was a direct consequence of the mutation in nlpI, since it was complemented by the wild-type nlpI gene alone. Overexpression of the unaltered nlpI gene in wild-type cells resulted in the loss of the rod morphology and the formation of single prolate ellipsoids and pairs of prolate ellipsoids joined by partial constrictions. NlpI may be important for an as-yet-undefined step in the overall process of cell division.
More than 130 different lipoproteins have been identified in a wide variety of both gram-positive and gram-negative bacteria (39). Indeed, more than 16 chromosomally encoded lipoproteins have been identified in Escherichia coli (7, 18, 22, 25, 27, 39, 40). In addition, several lipoproteins that are plasmid encoded have been identified in E. coli and other organisms (11, 35, 39). Although the primary amino acid sequences of the mature forms of these lipoproteins differ considerably, their respective unmodified prolipoprotein forms share certain general structural features. These features include a signal sequence that contains a lipid modification, or “lipobox” sequence, which in turn includes the cysteine residue destined for lipid modification. The consensus lipobox has the sequence Leu(Ala, Val)−4-Leu−3-Ala(Ser)−2-Gly(Ala)−1-Cys+1 (39). The posttranslational lipid modification of these lipoproteins presumably occurs by a common biosynthetic pathway. This pathway involves three posttranslational reactions catalyzed by the enzymes phosphatidylglycerol:prolipoprotein diacylglyceryltransferase, prolipoprotein signal peptidase (signal peptidase II), and phospholipid:apolipoprotein transacylase (N-acyltransferase). These enzymes are encoded by the lgt, lsp, and lnt genes, respectively, and their activities culminate in the synthesis of an N-terminal N-acyl-S-sn-1,2-diacylglyceryl-modified cysteine residue, the signature structural component of bacterial lipoproteins (39).
The isolation and characterization of mutants possessing temperature-sensitive alleles of the lgt, lsp, and lnt genes have revealed that these genes are essential. In addition, the antibiotic globomycin is a specific inhibitor of signal peptidase II, and inhibition of this enzyme by globomycin is lethal (17, 19, 20). These observations have led to the proposal that bacteria possess one or more lipoproteins that are required for normal cell growth and function (39). However, attempts to identify an essential lipoprotein have thus far been unsuccessful.
Previous studies identified a single open reading frame of unknown function located downstream of the pnp (polynucleotide phosphorylase) gene and immediately upstream of the deaD (ATP-dependent RNA helicase) gene at 71 min on the E. coli chromosome (4, 33, 36). The studies reported here demonstrate that this open reading frame encodes a previously unrecognized lipoprotein which we have designated new lipoprotein I (NlpI). Characterization of an nlpI::cm insertion mutant suggests that NlpI may be required for an as-yet-undefined step in the overall process of cell division.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown in Luria-Bertani (LB) broth (30) or in M9 minimal medium (30) containing magnesium sulfate and calcium chloride at final concentrations of 0.1 and 0.01 mM, respectively. Ampicillin and kanamycin were added to culture media when appropriate to give final concentrations of 50 and 25 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids
| Strain, plasmid, or phage | Relevant characteristics | Construction | Source or reference |
|---|---|---|---|
| Strains | |||
| RM4606 | sup0 F+ | R. Maurer | |
| WU620 | As RM4606, except nlpI::cm | MO102(f1R189) × RM4606 | This study |
| WU62 | As RM4606, except nlpI::cm | WU620(P1) × RM4606 | This study |
| WU63 | As RM4606, except nlpI::cm | WU620(P1) × RM4606 | This study |
| MO101 | DH5α carrying plasmid pBU142 | This study | |
| MO102 | JM109 carrying pBIP3-nlpI::cm | This study | |
| DH5α | supE44 ΔlacU169(φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA | Bethesda Research Laboratories | |
| JM109 | F′ traD36 proA+B+ lacIq Δ(lacZ)M15/recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 Δ(lac-proAB) e14− (Mcr−) | New England Biolabs | |
| INVαF′ | F′ endA1 recA1 hsdR17 supE44 thi-1 gyrA96 relA1 φ80lacZΔM15 Δ(lacZYA-argF)U169 | Invitrogen | |
| Plasmids | |||
| pCR2.1 | TA cloning vector; Ampr Kmr | Invitrogen | |
| pBAD24 | Arabinose regulation; Ampr | 10 | |
| pBIP3 | Suicide vector; KmrsacB (sensitivity to sucrose) | R. Maurer | |
| pCU11 | nlpI cloned into pCR2.1 | PCR product (primers U1F1 and U1R) | This study |
| pCU21 | nlpI plus 195 bp immediately upstream of the translational start codon cloned into pCR2.1 | PCR product (primers U1F2 and U1R) | This study |
| pBU142 | nlpI subcloned into pBAD24 | EcoRI-HindIII fragment of pCU11 subcloned into pBAD24 | This study |
| Phages | |||
| f1R189 | Helper phage for transduction (gene II amber mutant that will not replicate in sup+ target strains) | R. Maurer | |
| P1 | Generalized transduction | Laboratory stock |
DNA techniques.
All DNA manipulations were performed by standard techniques (28). Restriction enzymes were purchased from New England Biolabs (Beverly, Mass.). PCR amplifications were carried out with Taq polymerase (Perkin-Elmer, Foster City, Calif., or Takara Shuzo, Shiga, Japan).
Cloning and sequencing of the nlpI gene.
Oligonucleotide primers for PCR amplification of the nlpI gene were designed in accordance with nucleotide sequence data obtained from investigations of the pnp (33) and deaD (36) genes of E. coli K-12. The oligonucleotide primer U1F1 (5′-TCGGGAATTCGAAATGAAGCCT-3′; forward) includes the putative translational start codon (ATG, in bold) of nlpI; the underlined nucleotides indicate alterations of the wild-type sequence which were made in order to create an EcoRI site immediately upstream of the start codon. The oligonucleotide primer U1F2 (5′-CGTATCCGTCTGAGCATTAA-3′; forward) corresponds to nucleotides 2713 to 2732 in the 3′ region of the pnp gene. The oligonucleotide primer U1R (5′-CTGAAGCTTACGTCAGCTATTGC-3′; reverse) is complementary to the 3′ terminus of nlpI as well as to the 16 flanking nucleotides located beyond the translational stop codon; the underlined nucleotides designate changes in the wild-type sequence which were made in order to incorporate a HindIII restriction site. PCR amplifications were carried out with chromosomal DNA obtained from E. coli DH5α and RM4606. The products of PCR amplifications with primer pairs U1F1-U1R and U1F2-U1R were ligated into pCR2.1 to yield pCU11 and pCU21, respectively. The resulting constructs were used to transform E. coli INVαF′. Bidirectional nucleotide sequencing of cloned nlpI was carried out with ABI PRISM dye terminator cycle sequencing kits (Perkin-Elmer), complementary oligonucleotide primer pairs 277F (5′-CGGATATGCCTGAAGTATTCA-3′; forward)-277R (5′-TGAATACTTCAGGCATATCCG-3′; reverse) and 576F (5′-AATCGGATAAGGAACAGTGGG-3′; forward)-576R (5′-CCCACTGTTCCTTATCCGATT-3′; reverse), and M13 forward (−21) and reverse primers.
Construction of nlpI knockout mutants through gene replacement.
Null mutations in the E. coli nlpI gene were generated through allelic exchange with the pBIP3 suicide vector as described by Slater and Maurer (34). Briefly, the 886-bp chloramphenicol resistance determinant of plasmid pACYC184 was obtained by PCR amplification with the forward and reverse primer sequences 5′-CTGAACCGACGACCGGGTCGA-3′ and 5′-TGAGACGTTGATCGGCACGTAAG-3′, respectively. The resulting product was inserted into the middle of the nlpI gene carried by pCU21 by blunt ligation into the unique BstBI site of the nlpI gene. The nlpI::cm insertion was then excised with restriction enzymes ApaI and SpeI and subcloned into the corresponding sites of plasmid pBIP3. The ntp-sacB cassette of plasmid pBIP3 confers resistance to kanamycin and sensitivity to 5% sucrose (34). Thus, the resulting construct was introduced into E. coli JM109 by electroporation, and transformants were selected on LB agar plates containing both chloramphenicol and kanamycin. The pBIP-nlpI::cm derivative was isolated from the transformants, and incorporation of the nlpI::cm construct into the polylinker site of pBIP3 was verified by nucleotide sequencing. Transformants harboring pBIP3-nlpI::cm were infected with helper phage f1R189, and the resulting lysates were used to infect strain RM4606. Chloramphenicol-resistant colonies were plated on salt-free LB agar plates containing 5% sucrose to resolve cointegrates, and colonies resistant to both chloramphenicol and 5% sucrose were screened for sensitivity to kanamycin. Kanamycin-sensitive derivatives were examined by Southern blot analyses with the nlpI and cm genes as probes to identify nlpI::cm insertion mutants. One such mutant, WU620, was used for further studies.
Southern hybridization.
Chromosomal DNA was isolated from wild-type and mutant cells of E. coli with Puregene DNA isolation kits (Gentra Systems, Inc., Minneapolis, Minn.). The isolated DNA was digested with EcoRV at 37°C overnight, and the resulting fragments were separated by electrophoresis in a 0.7% agarose gel (60 V, 5 to 6 h) with TBE buffer (90 mM Tris-borate, 2 mM EDTA [pH 8.0]). DNA fragments were visualized by ethidium bromide staining, and the DNA was subsequently denatured by incubating the gel with gentle shaking in denaturing buffer (0.5 M NaOH, 1.5 M NaCl) for 30 min at room temperature. The gel was next incubated in neutralizing buffer (1 M Tris-HCl [pH 7.4], 1.5 M NaCl), and the denatured DNA fragments were transferred to a Hybond N nylon membrane filter (Amersham Life Science, Inc., Arlington Heights, Ill.) by capillary blotting overnight with 10× SSC buffer (1.5 M NaCl, 0.15 M sodium citrate [pH 7.0]). The filter was air dried, and the DNA was cross-linked to the filter by UV treatment (StratalinkerR; Stratagene, La Jolla, Calif.). Subsequent steps, including the preparation of labeled probes as well as hybridization and detection steps, were carried out with enhanced chemiluminescence direct nucleic acid labeling and detection systems (Amersham Life Science). Probes were prepared from the nlpI coding region (PCR product obtained with primers U1F1 and U1R) and the 886-bp chloramphenicol resistance cassette obtained by PCR amplification as described above.
Expression of nlpI and radiolabeling experiments.
High-level expression of nlpI was made possible by subcloning of the PCR product in pCU11 into the expression vector pBAD24 (10). Subcloning of the PCR product into the multicloning site of this vector was facilitated by use of the EcoRI and HindIII restriction sites that were created in the U1F1 and U1R PCR primers, respectively. The construct resulting from subcloning of this fragment into pBAD24 was designated pBU142. Plasmid pBU142 was introduced into E. coli DH5α by transformation, giving rise to strain MO101.
Radiolabeling of NlpI was carried out with either [9,10-3H]palmitic acid (specific activity, 56.5 Ci/mmol; Dupont NEN, Wilmington, Del.) or [2-3H]glycerol (specific activity, 6.35 Ci/mmol; New England Nuclear Corp., Boston, Mass.). Cells were grown overnight at 37°C in M9 minimal medium containing glucose (0.4%) and supplemented with 5% LB broth (medium A). A portion of the culture (3 ml) was harvested by centrifugation, and the cells were washed with M9 minimal medium containing l-arabinose and LB broth at final concentrations of 0.2 and 5%, respectively (medium B). The cells were then resuspended in 3 ml of medium B to give an A600 of 0.2 to 0.3, and the mixture was incubated with shaking at 37°C for 1 h. Control cultures (3 ml) were grown overnight at 37°C in medium A, washed with medium A, and subsequently resuspended in 3 ml of medium A to give an A600 of 0.2 to 0.3, and the mixture was incubated with shaking at 37°C for 1 h. Ampicillin (50 μg/ml) was included in culture media to ensure the maintenance of plasmids. To each of the cultures was added either [9,10-3H]palmitate (100 μCi) or [2-3H]glycerol (100 μCi), and the cultures were incubated with shaking at 37°C for 5 to 6 h. The cells were harvested by centrifugation, washed with 3 ml of ice-cold 10 mM sodium phosphate buffer (pH 7.0), and resuspended in 3 ml of the same buffer. The labeled cells were subsequently disrupted by sonication, and unbroken cells were removed by centrifugation at 5,000 × g for 5 min at 4°C. The crude cell envelope fraction was isolated by centrifugation of the supernatant solution at 200,000 × g for 1.5 h at 4°C. The cell envelope fraction was washed with 1 ml of cold acetone, and the washed membranes were dried at 60 to 70°C. The dried membranes were solubilized by boiling for 5 min following the addition of 30 to 50 μl of sample buffer (62.5 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 5% 2-mercaptoethanol, 20% glycerol, 0.01% bromophenol blue). The samples were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) either by the procedure of Ito et al. (21) or by the procedure described by Laemmli (23). The resulting gels were soaked in Amplify (Amersham Life Science) with gentle shaking for 20 to 30 min at room temperature and dried, and the location of radioactivity on the gels was determined by fluorography with Kodak X-Omat AR film (Eastman Kodak, New Haven, Conn.).
Inhibition of NlpI posttranslational modification by globomycin.
Strain MO101 was cultured overnight with shaking in medium A at 37°C. The cells contained in 3 ml of the overnight culture were washed with 3 ml of medium B and diluted with medium B to give an A600 of 0.5 to 0.6, and 3-ml portions of the resulting suspension were incubated with shaking at 37°C for 1 h. Globomycin was then added from a stock solution of 10 mg/ml in dimethyl sulfoxide (DMSO) to separate 3-ml cultures to give final concentrations of 0, 50, 100, and 200 μg/ml. The cells were incubated with shaking for an additional 10 min at 37°C, and either [9,10-3H]palmitic acid or [2-3H]glycerol was added to give a final concentration of 100 μCi/ml. The cells were subsequently incubated with the radiolabeled compounds at 37°C for 1 h with vigorous aeration. Radiolabeling was terminated by placing the cultures in an ice bath; this step was followed immediately by the addition of cold trichloroacetic acid to give a final concentration of 5%, and the cells were incubated on ice for 15 min. The cells were harvested by centrifugation, and the pellets were washed with 1 ml of acetone and dried at 60 to 70°C. The dried pellets were resuspended in 25 μl of sample buffer together with 1.25 μl of 5 N NaOH, and radiolabeled NlpI was solubilized by boiling for 5 min. The solubilized samples were analyzed by SDS-PAGE by the procedure of Ito et al. (21) or by SDS-PAGE (12% acrylamide) as described by Laemmli (23). The location of radioactivity on the gels was determined by fluorography as described above.
Purification of radiolabeled NlpI.
SDS-solubilized cell envelope proteins were obtained from strain MO101 following incubation of the cells in medium B in the presence of either [9,10-3H]palmitate or [2-3H]glycerol by the procedures described above. The solubilized proteins were subjected to SDS-PAGE (12% acrylamide) as described by Laemmli (23) and subsequently located on the gels by staining with Coomassie brilliant blue by standard procedures. A thin band containing 32-kDa NlpI was excised from the gel, and the gel slice was placed in a Spectrapor dialysis tube (molecular weight cutoff, 3,500; Spectrum, Los Angeles, Calif.) containing 5 ml of Laemmli running gel buffer (25 mM Tris-glycine [pH 8.4], 1% SDS). Radiolabeled NlpI was electroeluted from the gel slice by electrophoresis for 4 h at a constant voltage (150 V) by use of a horizontal mini-submarine gel apparatus filled with Laemmli running gel buffer. The protein was exhaustively dialyzed against water and lyophilized until further use.
Identification of glyceryl-cysteine.
The occurrence of glyceryl-cysteine in NlpI was demonstrated by previously described procedures (14). Briefly, purified [2-3H]glycerol-labeled NlpI was dissolved in 1 ml of a mixture of H2O2 and 88% formic acid (1:9 [vol/vol]), and the mixture was incubated overnight in a closed screw-cap tube at 4°C. The mixture was transferred to a glass hydrolysis ampoule and lyophilized to remove performic acid. One milliliter of constant-boiling HCl was added to the ampoule, and the ampoule was sealed and incubated in vacuo for 20 h at 100°C. The hydrolysate was lyophilized to remove HCl. The dried sample was dissolved in 20 to 30 μl of electrophoresis buffer (0.47 M formic acid, 1.4 M acetic acid [pH 2.3 to 2.4]) and analyzed by high-voltage paper electrophoresis at 2,000 V for 2 h with cooling by use of Whatman 3MM paper and electrophoresis buffer. Standards of cysteic acid, cysteine, methionine sulfone, and methionine were included in the analyses. The electrophoretic mobilities of radiolabeled components in the sample were determined by cutting the sample lane into 1-cm segments and determining the amount of radioactivity in each segment by standard liquid scintillation counting procedures. The electrophoretic mobilities of standard compounds were determined by spraying the dried electrophoretogram with a ninhydrin solution (0.2% ninhydrin in acetone).
Mild alkali treatment of [9,10-3H]palmitate-labeled NlpI.
A dried sample of purified [9,10-3H]palmitate-labeled NlpI was incubated with 8 μl of 0.5 N NaOH for 2 h at 37°C with occasional mixing. The sample was neutralized by the addition of 10 μl of 0.4 N HCl and subsequently mixed with 9 μl of sample buffer. A second, control sample containing the same amount of radioactivity was incubated with 15 μl of sample buffer. Both samples were analyzed by SDS-PAGE (12% acrylamide) with a minigel apparatus. The gel was dried without staining, and individual lanes were cut into 1-mm segments. The segments were incubated overnight with 0.5 ml of 0.1% SDS at 37°C, and the amount of radioactivity in each segment was determined by standard liquid scintillation counting procedures.
Miscellaneous techniques.
Cellular morphology was examined by light microscopy of crystal violet-stained cells. Nucleoids were stained with 4′,6-diamidino-2-phenylindole (DAPI) and visualized by fluorescence microscopy with a Zeiss type 741 fluorescence microscope interfaced with a Photometrics CE200A camera essentially as described by Hiraga et al. (15). The osmolarity of various media was determined with a 5100B vapor pressure osmometer (Wescor, Inc.).
RESULTS
The nlpI gene encodes a lipoprotein.
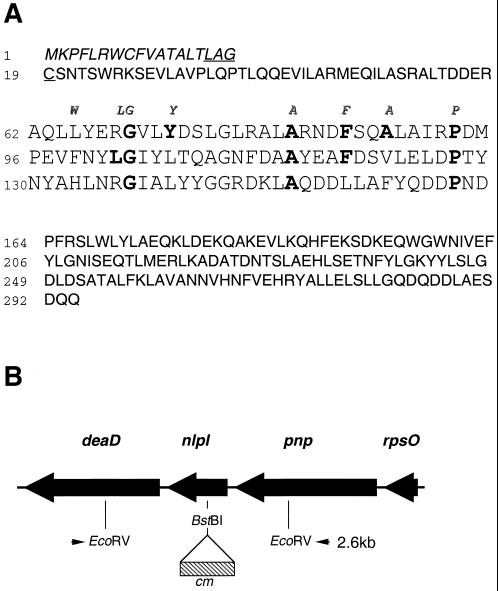
Previous studies reported the occurrence of a gene of unknown function, gene yhbM, located between genes pnp and deaD at 71 min on the E. coli chromosome (4). The predicted polypeptide encoded by yhbM contains 294 amino acid residues and has a calculated molecular mass of 33,619 Da (Fig. 1). Inspection of the amino acid sequence of the predicted polypeptide encoded by yhbM suggested that this gene product is a lipoprotein. Thus, the N terminus possesses the characteristics of a typical bacterial signal sequence; it contains a charged amino acid (lysine) in proximity to the N terminus as well as a short hydrophobic region (residues 3 to 15). In addition, the hydrophobic region is followed by the sequence Leu16-Ala-Gly-Cys19, which conforms to the parameters that define a lipobox, or lipoprotein modification and processing sequence (39).
FIG. 1.
(A) Predicted amino acid sequence of the product of the nlpI gene. The nlpI gene encodes a 34-kDa polypeptide containing 294 amino acids including a putative signal sequence consisting of 18 amino acids (italic type). The amino acids comprising the lipobox sequence, Leu-Ala-Gly-Cys, are underlined. The sequence delineated by amino acid residues 62 to 163 contains three TPRs. The eight consensus amino acids of the TPR motif are indicated in italics above the first TPR. The amino acid residues in NlpI that conform to the consensus TPR motif are indicated in bold type. (B) Genetic map of the region containing the nlpI gene between 71.0 and 71.1 min on the E. coli chromosome. An nlpI::cm insertion mutant was constructed by insertion of an 886-bp chloramphenicol resistance cassette into the BstBI site of nlpI as indicated. Southern blot analyses revealed 2.6- and 3.5-kb fragments in EcoRV digests of chromosomal DNAs obtained from the wild type and the nlpI::cm insertion mutant, respectively.
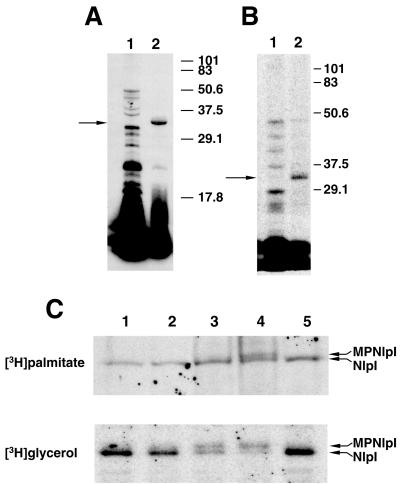
The yhbM gene was amplified by PCR with genomic DNA from E. coli DH5α. It was subsequently subcloned into expression vector pBAD24, placing the expression of yhbM under the control of the araBAD promoter; the resulting construct was designated pBU142. Significant incorporation of radioactivity into a protein with an apparent molecular mass of 32 kDa was observed when strain MO101 (DH5α/pBU142) was incubated with [3H]palmitate under conditions where the expression of yhbM was induced with l-arabinose (Fig. 2A, lane 2). This radiolabeled protein was not detected when strain MO101 was incubated with [3H]palmitate under noninducing conditions (Fig. 2A, lane 1). Similar results were obtained when strain MO101 was incubated with [2-3H]glycerol under inducing and noninducing conditions (Fig. 2B).
FIG. 2.
(A and B) In vivo incorporation of [3H]palmitic acid (A) and [2-3H]glycerol (B) into polypeptides following overexpression of the nlpI gene in E. coli. In vivo labeling and SDS-PAGE were performed as described in Materials and Methods. In each case, strain MO101 was grown overnight in medium A and subsequently incubated in medium A (lane 1) or medium B (lane 2) immediately prior to the addition of radiolabeled palmitic acid or glycerol. The location of 32-kDa NlpI is indicated by an arrow. The electrophoretic mobilities of marker proteins with known molecular masses are indicated in kilodaltons. (C) Effect of globomycin on the maturation of NlpI. Strain MO101 was grown overnight in medium A, and the cells were harvested and washed in medium B. The washed cells were resuspended in medium B containing globomycin at a final concentration of 0 μg/ml (lane 1), 50 μg/ml (lane 2), 100 μg/ml (lane 3), or 200 μg/ml (lane 4). Globomycin was dissolved in DMSO to yield a stock solution of 10 mg/ml; accordingly, 20 μl of DMSO was added to a control culture (lane 5). The cells were subsequently incubated with either [3H]palmitic acid or [2-3H]glycerol, and the incorporation of radiolabel into NlpI was determined by SDS-PAGE and fluorography. Experimental details are provided in Materials and Methods. The upper and lower arrows in each panel indicate the locations of modified pro-NlpI (MPNlpI) and mature NlpI, respectively.
Signal peptidase II is required for the conversion of diacylglyceryl-modified prolipoproteins to apolipoproteins (39). The antibiotic globomycin is a specific inhibitor of signal peptidase II, and this inhibition is characterized by the accumulation of diacylglyceryl-modified prolipoproteins which are slightly larger than their corresponding mature lipoproteins owing to the presence of the signal peptide (17). The incorporation of radioactivity into the 32-kDa protein was clearly inhibited by globomycin following the expression of yhbM in strain MO101 in the presence of either [2-3H]glycerol or [3H]palmitate (Fig. 2C). Partial inhibition was observed when globomycin was added to cultures at a final concentration of 100 μg/ml, whereas significantly greater inhibition occurred when the drug was present at a final concentration of 200 μg/ml. The inhibition of synthesis of the 32-kDa protein was also accompanied by the appearance of a radiolabeled 34-kDa protein. Taken together, the above observations support the conclusion that the yhbM gene encodes a lipoprotein, and we have renamed the gene nlpI, for new lipoprotein I.
Characterization of the lipid modification in NlpI.
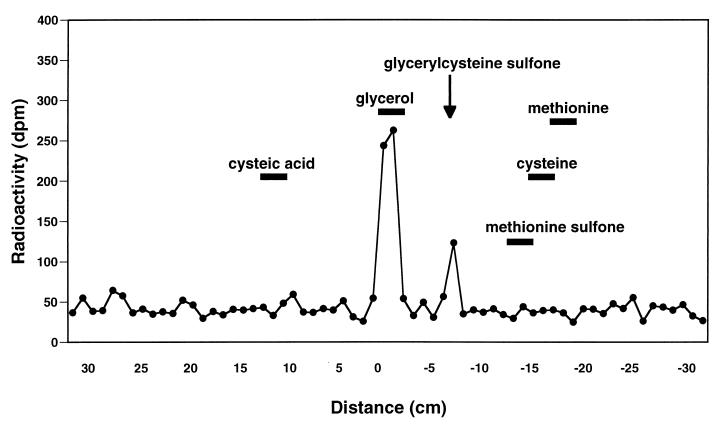
Bacterial lipoproteins are characterized by the occurrence of an N-terminal N-acyl-S-sn-1,2-diacylglyceryl-modified cysteine residue. In E. coli, the N-acyl-linked substituent of Lpp is primarily palmitate; however, other fatty acids have also been found to occur at this position (13). In contrast, the O-acyl-linked substituents of Lpp are similar to those found in bulk phospholipids (13). The thioether linkage in the glyceryl-cysteine moiety is more acid stable than a peptide bond, and performic acid oxidation of bacterial lipoproteins followed by acid hydrolysis characteristically results in the formation of glyceryl-cysteine sulfone (14). Thus, [2-3H]glycerol-labeled NlpI purified by preparative SDS-PAGE was examined for the presence of glyceryl-cysteine by established methods (14). Performic acid oxidation of [2-3H]glycerol-labeled NlpI followed by acid hydrolysis resulted in the release of a radiolabeled compound whose electrophoretic mobility was the same as that reported for glyceryl-cysteine sulfone analyzed by high-voltage paper electrophoresis at pH 2.3 to 2.4 (14) (Fig. 3).
FIG. 3.
Identification of glyceryl-cysteine sulfone in total acid hydrolysates of performic acid-oxidized NlpI. [2-3H]glycerol-labeled NlpI was purified by preparative SDS-PAGE and treated with performic acid at 4°C overnight. Total acid hydrolysates of performic acid-oxidized lipoprotein were then analyzed by high-voltage paper electrophoresis at pH 2.3 to 2.4 along with the indicated standards. The distance of electrophoretic movement (centimeters) from the origin toward the cathode is indicated by the negatively prefixed scale on the abscissa. Additional experimental details are provided in Materials and Methods.
[3H]palmitate-labeled NlpI was also examined for the presence of ester- and amide-linked fatty acyl substituents. Incubation of [3H]palmitate-labeled Lpp with mild alkali results in the hydrolysis of ester-linked fatty acyl moieties, and such treatment typically results in the loss of at least 50% of the radioactivity as free fatty acids. Treatment of [3H]palmitate-radiolabeled NlpI with mild alkali followed by SDS-PAGE resulted in the release of approximately 62% of the radioactivity. The remainder of the radioactivity migrated as a single component with an apparent electrophoretic mobility identical to that of untreated NlpI. These data clearly demonstrate the occurrence of N-acyl-S-diacylglyceryl-modified cysteine in NlpI.
Isolation of an nlpI::cm insertion mutant.
A chloramphenicol resistance cassette was inserted into the E. coli nlpI gene, and the interrupted gene was introduced into the chromosome of E. coli RM4606 by allelic exchange, yielding the insertion mutant WU620. The presence of a single insertionally inactivated nlpI gene in the chromosome was confirmed by Southern blot analyses of WU620 and two transductants, WU62 and WU63; the transductants were obtained by introduction of the mutation in WU620 into the chromosome of the parental strain, RM4606, by P1-mediated transduction. In wild-type cells, the nlpI gene is flanked by EcoRV sites which delineate a 2.6-kb fragment (Fig. 1B). Insertion of the 886-bp Cmr cassette into the BstBI site of the nlpI gene would increase the size of this fragment to approximately 3.5 kb. Southern blot analyses revealed a single DNA fragment of approximately 3.5 kb in an EcoRV digest of the chromosome of each of the transductants and strain WU620 with the chloramphenicol resistance gene as the probe (data not shown). In contrast, this fragment was not detected in an EcoRV digest of the chromosome of the wild-type parental strain, RM4606. Similar results were obtained with labeled nlpI as the probe, except that a single 2.6-kb fragment corresponding to the wild-type nucleotide sequence was detected in an EcoRV digest of the chromosome of strain RM4606.
Phenotype of the nlpI::cm insertion mutant.
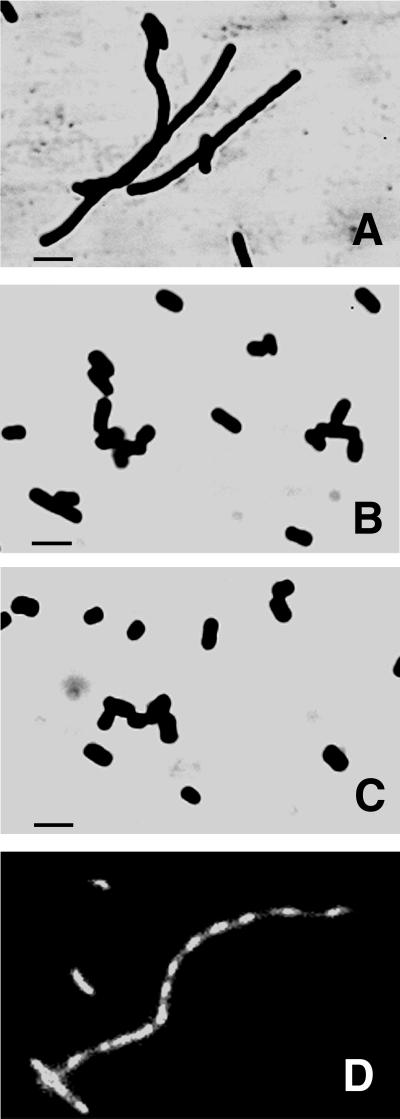
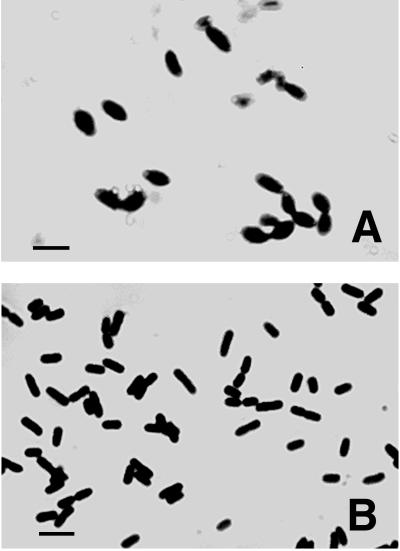
The null mutation in nlpI had no apparent effect on the ability of strain WU62 to grow in standard LB liquid medium or on LB solid medium at 30, 37, and 42°C. However, microscopic examination revealed pronounced filamentation of the mutant cells when broth cultures were incubated at 42°C (Fig. 4A). No obvious morphological differences between mutant and wild-type cells were observed when cells were grown in broth at either 30 or 37°C (Fig. 4B). In addition, the thermosensitive phenotype of the mutant was complemented by plasmid pCU21, which contains the wild-type nlpI gene as well as a 195-bp sequence upstream of the putative translational start codon (Fig. 4C). The filamentation of mutant strain WU62 at an elevated temperature did not appear to affect nucleoid numbers or segregation (Fig. 4D).
FIG. 4.
(A to C) Effect of the nlpI::cm mutation on the morphology of cells at an elevated temperature. Cells were incubated in LB broth at 42°C with shaking until the optical density (A600) reached 0.4 to 0.5. The cells were subsequently stained with crystal violet and visualized by light microscopy. (A) WU62 (nlpI::cm). (B) RM4606 (wild type). (C) WU62 carrying plasmid pCU21. Bars, 2 μm. (D) DNA segregation in strain WU62. Cells were incubated in LB broth at 42°C until the optical density (A600) of the culture reached 0.4 to 0.5, and nucleoids were subsequently visualized by fluorescence microscopy after DAPI staining. Additional details are provided in Materials and Methods.
Although the nlpI::cm insertion did not preclude the growth of strain WU62 in standard LB liquid medium or on LB solid medium containing 0.5% NaCl, the growth of the mutant was severely restricted on low-salt LB medium. The mutant was able to form only small colonies at 30°C on LB agar lacking NaCl, and no growth was observed on this medium at 37 and 42°C (data not shown). The growth of the mutant at an elevated temperature was rescued when NaCl in LB agar was replaced by either KCl or LiCl at the same final concentration (85.86 mM). These observations appear to reflect a sensitivity of the mutant to osmotic conditions, since growth at an elevated temperature on LB medium was also rescued when NaCl in the medium was replaced by 2.5% sucrose. The effect of low salt on the growth of the mutant in LB broth at various temperatures was somewhat more complex. The apparent rate of growth of strain WU62 became progressively lower as the growth temperature was increased from 30 to 42°C, and the growth of the mutant appeared to be almost completely inhibited at 42°C (data not shown). Similarly, cell filamentation became increasingly apparent as the growth temperature was increased; however, filamentation was most pronounced at 42°C.
The phenotype of the nlpI::cm insertion mutant was unaltered following introduction of the recA56 mutant allele into strain WU62. Thus, we conclude that the thermosensitive cell division and osmotic sensitivity of the nlpI::cm insertion mutant are not indirectly due to an induction of the SOS response.
Effect of nlpI overexpression.
The phenotype of the nlpI null mutation prompted an examination of the effect of nlpI overexpression on growth and cellular morphology. Overexpression of nlpI in strain MO101 was clearly evident following growth of the cells at 37°C in medium B (induction with 0.2% arabinose), as indicated by the appearance of the 32-kDa NlpI lipoprotein when cell envelope preparations were analyzed by SDS-PAGE and Coomassie brilliant blue staining (data not shown). The overexpression of nlpI in cells grown in medium B resulted in a loss of the rod morphology, accompanied by the striking appearance of swollen prolate ellipsoids as well as pairs of prolate ellipsoids joined by partial constrictions (Fig. 5A). This morphology was also observed when strain MO101 was grown in LB broth containing 0.2% l-arabinose. The overexpression of nlpI also affected the growth of cells. Accordingly, strain MO101 grew with a decreased generation time in medium B at 37°C, and growth ceased after the apparent cell mass had increased by approximately 1.5 doublings (data not shown). In contrast, the NlpI protein was not detected in cell envelopes obtained from wild-type cells that were grown similarly or from strain MO101 grown at 37°C in medium A (repression with 0.4% glucose) when these preparations were analyzed in the same manner. Furthermore, the growth of strain MO101 was unaffected when cells were incubated at 37°C in medium A, and the morphology of these cells also appeared to be normal (Fig. 5B).
FIG. 5.
Morphology of cells following the overexpression of nlpI. (A) The overexpression of nlpI from the araBAD promoter was induced by the growth of strain MO101 (DH5α/pBU142) at 37°C in medium B with shaking for 4 to 5 h. (B) Control cultures were grown in medium A under the same conditions. The cells were subsequently stained with crystal violet and visualized by light microscopy. Bars, 2 μm.
DISCUSSION
We describe here the identification and characterization of a previously unrecognized lipoprotein in E. coli K-12 which we have designated NlpI. The function of NlpI is not yet known. However, NlpI is the first example of a bacterial lipoprotein whose mutational alteration has a direct effect on cellular morphology. Indeed, the phenotype of the nlpI::cm insertion mutant suggests that this lipoprotein may be important for some step in the overall process of cell division. In this regard, it is interesting that several of the phenotypic characteristics of the nlpI::cm insertion mutant are similar to those expressed by mutants that are conditionally defective in fts (filamenting temperature-sensitive) genes involved in cell division (26). Accordingly, incubation of the nlpI::cm insertion mutant at an elevated temperature resulted in filamentation of the cells and rendered the cells osmotically sensitive. Filamentation of bacterial cells commonly occurs secondarily as a consequence of the SOS response elicited by damage to DNA or as a result of a blockage in DNA replication. However, filamentation of the nlpI::cm insertion mutant at a nonpermissive temperature did not appear to be due to any of these causes, since the synthesis of DNA and the partitioning of chromosomes within filaments were not altered and the same phenotype was observed for recA56 nlpI::cm double mutants. In addition, it is unlikely that the phenotype of the nlpI::cm insertion mutant is due to a polar effect of the insertion in nlpI on downstream genes, since normal cell growth and morphology were restored when the nlpI::cm insertion mutant was complemented by the wild-type nlpI gene alone. Furthermore, the deaD gene is located immediately downstream of nlpI, and it appears to be transcribed independently of the rpsO-pnp genes and the nlpI gene (36).
The overall process of cell division is a complex process that involves the coordinated activities of numerous proteins. Indeed, it is now clear that the actual division event or septum formation requires the coordinated function of several proteins that comprise a macromolecular complex. This complex is localized at the site of circumferential invagination (26), and it has been variously termed a divisome (31), a septalsome (16), and a septator (37). The available data suggest the possibility that NlpI might be a previously unrecognized component of this complex. However, it is important to stress that the identification of NlpI as a component of either the cytoplasmic or the outer membrane remains to be established. Additional studies are also required to ascertain whether or not NlpI is localized in the vicinity of circumferential invagination during cell division. Nevertheless, it is interesting to note that analyses of the deduced amino acid sequence of NlpI have revealed the presence of the tetratricopeptide repeat (TPR) unit (5) (Fig. 1A). The TPR consists of a degenerate 34-amino-acid motif which is tandemly repeated, and this motif is defined by the occurrence of the loosely conserved consensus sequence W…LG…Y…A…F…A…P (6, 9, 24). The TPR motif occurs widely in nature, and TPR-containing proteins participate in a diverse array of related and unrelated biological processes. The TPR motif appears to mediate intermolecular protein-protein interactions, leading to the formation of protein complexes (6), and it has been suggested that the TPR motif may also facilitate intramolecular protein interactions (24).
Altered cell morphology has been observed following the overexpression of several genes known to be involved in cell division and related processes in E. coli. For example, moderate overexpression of ftsZ results in the formation of minicells (2, 26, 38), whereas higher levels of ftsZ expression result in the formation of filaments (26, 38). Additional examples include the formation of spherically shaped cells and chains of cells following the overexpression of dacA (d,d-carboxypeptidase 1A; penicillin-binding protein 5) (29) and cafA (formation of cytoplasmic axial filaments) (32), respectively. The overexpression of nlpI resulted in the loss of rod morphology and the formation of both single prolate ellipsoids and what appear to be pairs of prolate ellipsoids joined by partial constrictions. This result was unexpected, since prolate ellipsoids have only been observed in double mutants conditionally defective in both cell elongation and in an early step of cell division following a shift to nonpermissive conditions. For example, RodA is required for cell elongation and maintenance of the rod shape of E. coli (1), whereas FtsZ is required for the initial step in the assembly of the septal ring structure, the formation of the FtsZ ring (1, 3, 8, 12). Mutants possessing the temperature-sensitive rodA52 allele grow as spherical cells at a nonpermissive temperature (1). However, prolate ellipsoids are formed by double mutants possessing the rodA52 allele and a temperature-sensitive allele of ftsZ when incubated at a nonpermissive temperature (1). This morphology is believed to result from continued peptidoglycan synthesis in the absence of both chain elongation and the formation of the FtsZ ring (1). The formation of prolate ellipsoids following the overexpression of nlpI suggests that increased levels of NlpI might disrupt both of these processes. In addition, the appearance of pairs of prolate ellipsoids joined by partial constrictions may be due to the deleterious effects of elevated NlpI levels on both chain elongation and continued septum formation in cells in which septum formation has already begun prior to the onset of nlpI overexpression.
The basis for the temperature-sensitive phenotype of the nlpI::cm insertion mutant is not yet understood. It is possible that NlpI is required only for normal cell growth and morphology at an elevated temperature. However, this possibility seems unlikely in view of the observation that the nlpI::cm insertion mutation rendered the mutant osmotically sensitive at 30 and 37°C as well as at 42°C. Alternatively, it is possible that the mutant expresses a truncated protein, since the cm insertion is located at the approximate midpoint of the nlpI gene. In this event, the putative truncated protein might be “functional” at the permissive temperature, whereas it is rendered “nonfunctional” at the nonpermissive temperature or in media of reduced osmotic strength. Additional biochemical studies are required to determine whether or not the mutant synthesizes a truncated lipoprotein. In addition, it will also be of interest to determine the phenotype of mutants that either lack the nlpI gene or contain insertions more proximal to the 5′ terminus of the gene. In this regard, the results of numerous investigations have prompted the proposal that E. coli and related organisms possess a minor lipoprotein(s) that is required for growth and viability (39); however, an essential lipoprotein has not yet been identified. Accordingly, attempts to isolate additional nlpI null mutants may provide insights into whether or not NlpI is indeed an essential lipoprotein.
ACKNOWLEDGMENTS
We thank H. Suginaka, M. Sugai, A. Maurelli, Y. Ishi, M. Yamashita, and A. Iwagaki for helpful advice and suggestions. We also thank S. D. Gupta, A. Rahman, K. Barr, and A. DeJesus for technical assistance. Finally, we are grateful to M. Arai for the kind gift of globomycin.
This work was supported by NIGMS grant GM28811 to P.D.R.
REFERENCES
- 1.Begg K, Donachie W D. Cell shape and division in Escherichia coli: experiments with shape and division mutants. J Bacteriol. 1985;163:615–622. doi: 10.1128/jb.163.2.615-622.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Begg K, Nikolaichik Y, Crossland N, Donachie W D. Roles of FtsA and FtsZ in activation of division sites. J Bacteriol. 1998;180:881–884. doi: 10.1128/jb.180.4.881-884.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature (London) 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collade-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Borodovsky M, Rudd K E, Koonin E V. Intrinsic and extrinsic approaches for detecting genes in a bacterial genome. Nucleic Acids Res. 1994;22:4756–4767. doi: 10.1093/nar/22.22.4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das A K, Cohen P T W, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehlert K, Höltje J-V, Templin M F. Cloning and expression of a murein hydrolase lipoprotein from Escherichia coli. Mol Microbiol. 1995;16:761–768. doi: 10.1111/j.1365-2958.1995.tb02437.x. [DOI] [PubMed] [Google Scholar]
- 8.Erickson H P. FtsZ, a prokaryotic homolog of tubulin? Cell. 1995;80:367–370. doi: 10.1016/0092-8674(95)90486-7. [DOI] [PubMed] [Google Scholar]
- 9.Goebl M, Yanagida M. The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem Sci. 1991;16:172–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- 10.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haase J, Kalkum M, Lanka E. TrbK, a small cytoplasmic membrane lipoprotein, functions in entry exclusion of the IncPα plasmid RP4. J Bacteriol. 1996;178:6720–6729. doi: 10.1128/jb.178.23.6720-6729.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hale C A, de Boer P A J. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 13.Hantke K, Braun V. Covalent binding of lipid to protein. Diglyceride and amide-linked fatty acid at the N-terminal end of the murein-lipoprotein of the Escherichia coli outer membrane. Eur J Biochem. 1973;34:284–296. doi: 10.1111/j.1432-1033.1973.tb02757.x. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi S, Wu H C. Identification and characterization of lipid-modified proteins in bacteria. In: Hooper N M, Turner A J, editors. Lipid modification of proteins: a practical approach. Oxford, England: Oxford University Press; 1992. pp. 261–285. [Google Scholar]
- 15.Hiraga S, Niki H, Ogura T, Ichinose C, More H, Ezaki B, Jaffé A. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J Bacteriol. 1989;171:1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holland I B. Genetic analysis of the E. coli division clock. Cell. 1987;48:361–362. doi: 10.1016/0092-8674(87)90183-8. [DOI] [PubMed] [Google Scholar]
- 17.Hussain M, Ichihara S, Mizushima S. Accumulation of glyceride-containing precursor of the outer membrane lipoprotein in the cytoplasmic membrane of Escherichia coli treated with globomycin. J Biol Chem. 1980;255:3707–3712. [PubMed] [Google Scholar]
- 18.Ichihara S, Hussain M, Mizushima S. Characterization of new membrane lipoproteins and their precursors of Escherichia coli. J Biol Chem. 1981;256:3125–3129. [PubMed] [Google Scholar]
- 19.Inukai M, Takeuchi M, Shimizu K, Arai M. Mechanism of action of globomycin. J Antibiot. 1978;31:1203–1205. doi: 10.7164/antibiotics.31.1203. [DOI] [PubMed] [Google Scholar]
- 20.Inukai M, Takeuchi M, Shimizu K, Arai M. Existence of the bound form of prolipoprotein in Escherichia coli B cells treated with globomycin. J Bacteriol. 1979;140:1098–1101. doi: 10.1128/jb.140.3.1098-1101.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito K, Date T, Wickner W. Synthesis, assembly into the cytoplasmic membrane, and proteolytic processing of the precursor of coliphage M13 coat protein. J Biol Chem. 1980;255:2123–2130. [PubMed] [Google Scholar]
- 22.Kraft A R, Templin M F, Höltje J-V. Membrane-bound lytic endotransglycosylase in Escherichia coli. J Bacteriol. 1998;180:3441–3447. doi: 10.1128/jb.180.13.3441-3447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Lamb J R, Tudendreich S, Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- 25.Lommatzsch J, Templin M F, Kraft A R, Vollmer W, Höltje J-V. Outer membrane localization of murein hydrolase: MltA, a third lipoprotein lytic transglycosylase in Escherichia coli. J Bacteriol. 1997;179:5465–5470. doi: 10.1128/jb.179.17.5465-5470.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lutkenhaus J, Mukherjee A. Cell division. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1615–1626. [Google Scholar]
- 27.Ma J, Katsonouri A, Gennis R B. Subunit II of the cytochrome bo3 ubiquinol oxidase from Escherichia coli is a lipoprotein. Biochemistry. 1997;36:11298–11303. doi: 10.1021/bi9709710. [DOI] [PubMed] [Google Scholar]
- 28.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Markiewicz Z, Broome-Smith J K, Schwarz U, Spratt B G. Spherical E. coli due to elevated levels of d-alanine carboxypeptidase. Nature (London) 1982;297:702–704. doi: 10.1038/297702a0. [DOI] [PubMed] [Google Scholar]
- 30.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 31.Nanninga N. Cell division and peptidoglycan assembly in Escherichia coli. Mol Microbiol. 1991;5:791–795. doi: 10.1111/j.1365-2958.1991.tb00751.x. [DOI] [PubMed] [Google Scholar]
- 32.Okada Y, Wachi M, Hirata A, Suzuki K, Nagai K, Matsuhashi M. Cytoplasmic axial filaments in Escherichia coli cells: possible function in the mechanism of chromosome segregation and cell division. J Bacteriol. 1994;176:917–922. doi: 10.1128/jb.176.3.917-922.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Régnier P, Grunberg-Manago M, Portier C. Nucleotide sequence of the pnp gene of Escherichia coli encoding polynucleotide phosphorylase. J Biol Chem. 1987;262:63–68. [PubMed] [Google Scholar]
- 34.Slater S, Maurer R. Simple phagemid-based system for generating allele replacements in Escherichia coli. J Bacteriol. 1993;175:4260–4262. doi: 10.1128/jb.175.13.4260-4262.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Theisen M. Molecular cloning and characterization of nlpH, encoding a novel, surface-exposed, polymorphic, plasmid-encoded 33-kilodalton lipoprotein of Borrelia afzelii. J Bacteriol. 1996;178:6435–6442. doi: 10.1128/jb.178.22.6435-6442.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toone W M, Rudd K E, Friesen J D. deaD, a new Escherichia coli gene encoding a presumed ATP-dependent RNA helicase, can suppress a mutation in rpsB, the gene encoding ribosomal protein S2. J Bacteriol. 1991;173:3291–3302. doi: 10.1128/jb.173.11.3291-3302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vicente M, Kushner S R, Garrido T, Aldea M. The role of the ‘gearbox’ in the transcription of essential genes. Mol Microbiol. 1991;5:2085–2091. doi: 10.1111/j.1365-2958.1991.tb02137.x. [DOI] [PubMed] [Google Scholar]
- 38.Ward J E, Lutkenhaus J. Overproduction of FtsZ induces minicell formation in E. coli. Cell. 1985;42:941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]
- 39.Wu H C. Biosynthesis of lipoproteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1005–1014. [Google Scholar]
- 40.Yu F, Inouye S, Inouye M. Lipoprotein-28, a cytoplasmic membrane lipoprotein from Escherichia coli. Cloning, DNA sequence, and expression of its gene. J Biol Chem. 1986;261:2284–2288. [PubMed] [Google Scholar]